Abstract
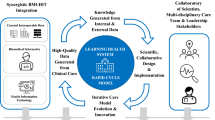
The concept of a Learning Health System (LHS) has been widely discussed in academic literature, yet its practical implementation remains a challenge. This paper describes the institutional journey, leadership structure, data governance policies, and technical innovations that together support a scalable and sustainable Research-Oriented LHS. Additionally, we propose an expanded data vision that aligns with interdisciplinary and translational research needs. Supplementary materials provide technical details for those interested in implementing such a model.
Similar content being viewed by others
Introduction
The Learning Health System (LHS) has been defined as an organizational approach to healthcare delivery in which continuous improvement and innovation are grounded in systematic learning from the populations served1,2. The construction of an LHS within an institution requires not only a conceptual transformation but also a pragmatic and operational shift in the enterprise. In practice, an LHS seeks to optimize structures and processes of care delivery so that the right treatment reaches the right patient at the right time, with feedback loops enabling research to inform practice and practice to inform research2. While advances in electronic health records (EHRs) and data infrastructure provide critical foundations, building a true LHS requires the alignment of governance, culture, and workforce around the principle that data-driven learning must permeate clinical care, operations, and research alike.
Within this broader framework, the concept of a Research-Oriented Learning Health System (RO-LHS) was pioneered as a distinctive model for the LHS. Whereas many implementations of the LHS emphasize quality improvement—often described as problem-driven and focused on immediate operational outcomes—an RO-LHS frames improvement efforts in terms of their potential to generate generalizable knowledge and contribute to the scientific evidence base2. This reorientation transforms quality improvement and research from parallel or siloed activities into interdependent, symbiotic processes. Integration is prioritized over isolation, and generalizability over local optimization, thereby advancing the greater good of health systems that face similar challenges.
Transitioning from a siloed, discipline-specific research paradigm to an integrated RO-LHS thus requires a fundamental reimagining of institutional priorities. It entails embedding research in the fabric of clinical operations, creating governance structures that bridge the demands of care delivery and discovery, and committing resources to infrastructures that support reproducibility, scalability, and dissemination. The Ohio State University Wexner Medical Center (OSUWMC), in collaboration with Nationwide Children’s Hospital (NCH), has pursued this vision through deliberate investments in governance transformation, technical infrastructure, and cultural change.
The OSUWMC and NCH are independent healthcare systems joined as collaborative partners under a National Center for Advancing Translational Sciences (NCATS) Clinical and Translational Science Award (CTSA); that have embraced data democratization and developed and deployed tools that support this journey. Central to that approach has been the explicit effort to knit parallel RO-LHS efforts into a coordinated—but not hierarchical—learning ecosystem, the institutions came together to reimagine leadership, infrastructure, and vision for the technical and information components of the biomedical research mission. By framing research as an enterprise-wide responsibility—rather than as the activity of a specialized few—the RO-LHS model provides a pathway for leveraging the everyday clinical environment as a laboratory for continuous, generalizable learning.
Institutional leadership recognized that a fragmented approach to research infrastructure would limit innovation, and a redesigned leadership model was developed to place greater authority in a single leader, The Chief Research Information Officer (CRIO) was matched in terms of formal role in both the Clinical and Translational Science Institute (CTSI) as the Director of Biomedical Informatics as well as the College of Medicine as the Associate Dean of Research. In the first six months of their appointment, the CRIO consulted stakeholders and developed a strategic vision (Supplement 2) focused on reducing the transaction costs of engaging in research within OSU and across institutions that outlined an overarching direction (see Box 1). At OSU, the university moved the informatics service center out of the department of Biomedical Informatics to the Office of Research in the College of Medicine, with an ongoing 2.5 M annual financial commitment used to resource the newly developed Department of Research Information Technology. Key outcomes of this collaborative governance model included:
-
1.
Purpose-built Research IT divisions at OSU to supply expanded services that researchers can access through formal service agreements, preserving institutional autonomy;
-
2.
Joint data-access framework via an interinstitutional Data Use Agreement, an expanded Honest Broker Protocol and compatible Research Health Information (RHI) and Protected Health Information (PHI) policies to enable compliant data sharing while each organization retains its own privacy oversight; and a
-
3.
Shared investment pipeline that included coordinated—but separately budgeted—funding that supports cross-institutional cloud and HPC resources without shifting financial control.
[Box 2] In this framework, the CRIO operates as a convener and infrastructure catalyst, ensuring interoperable platforms and policies while respecting the independence of both OSUWMC and NCH (Supplement 4). This transition was marked by a deliberate and strategic commitment to respectful governance infrastructure development. Governance reviews to safeguard data took significant legal, governance, and technical efforts. For example, OSUWMC is one of the largest Epic Community Connect providers, in an electronic health record (EHR)-as-a-service model, in the United States where we provide the EHR for 14 non-affiliated hospitals. By agreement, data held by OSUWMC for Community Connect sites are not allowed to be used for research without explicit consent of the organizations. On the other hand, NCH has Neonatal Intensive Care Unit operations at multiple healthcare systems that are market competitors of OSU, and significant concerns were raised about securing that data from OSU clinical operations. Robust governance structures were established to respectfully bridge the gap between research and clinical operations with the intent to use that synergy to systematically advance the clinical mission, in the context of complicated agreements, polices, and applicable law.
The ability of our RO-LHS to function depends on both strong governance and the strategic investment in robust technology platforms – spanning the EHR, cloud-based discovery environments, shared high-performance computing, and advanced analytics. For example, hospital alarms often fail to distinguish between critical and non-critical events, leading to confusion, overrides, and delayed responses. By redesigning alarm sounds and displays around urgency, predictive value, and source, within the research framework, we worked to describe how the work done at OSU can more broadly improve clarity, reduce false alarms, and ensure faster, safer patient care. To that end, OSUWMC established interdisciplinary stakeholder committees to set research priorities, oversee implementation, and ensure alignment with institutional goals. Through these committees, we aim to ensure that resources are managed in a manner that reduces transaction costs and staffed by a dedicated informatics and data management workforce3. By defining clear roles, responsibilities, and communication pathways for these shared services, OSUWMC avoids cost-prohibitive “concierge” models and instead fosters efficient, cross-functional collaboration among clinicians, nurses, researchers, and IT professionals.
A flagship example of this dual governance-and-resource strategy was the transfer of 250 high-performance computing nodes, inclusive of both GPU resources, from OSUWMC’s direct management to the Ohio Supercomputer Center (OSC) – an independent agency of the State of Ohio. [Box 3] This arrangement is supported by a Business Associates Agreement (BAA) where OSUWMC data is held in PHI-compliant storage and OSC manages the high-performance computational cluster on behalf of the Medical Center. By offloading the management of infrastructure to domain experts, this arrangement created one of the few PHI-approved enclaves available on a campus supercomputer—an unusually rigorous compliance posture that broadens secure data handling for clinical and translational studies. Whether streamlining discovery workflows or provisioning shared computational assets in ways that scale sustainably across the enterprise, the continuous refinement of this integrated governance-and-resource framework is an ongoing but essential component to maintaining leadership in translational research and healthcare innovation.
Research Health Information (RHI) vs. Protected Health Information (PHI): A Defining Distinction
Via the leadership and governance transformation, the enterprise introduced a critical distinction between PHI under The Health Insurance Portability and Accountability Act of 1996 (HIPAA)4, and research data related to the health of the participant. Adopted in 2020, an environmental scan revealed peer institutions—such as the University of Delaware and University of Miami—had already implemented similar RHI frameworks5,6,7. RHI is a purpose-built classification that is comprised of identified or coded datasets that meet HIPAA criteria or identified data collected under an approved Institutional Review Board (IRB) protocol and governed by the Common Rule8 where the subject of the data pertains to the health of the respondent. OSUWMC adopted the following core principles for RHI:
-
Regulatory alignment. RHI data must comply with federal research regulations (the Common Rule) and institutional policy.
-
Controlled separation. Data extracted directly from the EHR retains PHI status unless first vetted by an honest broker or transferred into a research-only environment.
-
Broker-mediated release. Any dataset—identified, coded, or limited—released by an honest broker under an approved IRB is classified as RHI.
To operationalize these distinctions, OSUWMC deployed both technical and professional controls. The IT team built segregated storage and data-transfer pipelines that keep PHI within clinical systems, while curated RHI is hosted in secure research enclaves and storage. Meanwhile, the Privacy Office and IRB jointly authored detailed guidance on RHI handling, supplemented by publicly available process documentation, training modules, and decision-support tools (Supplement 5)9,10. Implementing this layered approach required broad policy revisions across the university. However, it clarified accountability:
-
RHI incidents are reported to the Data Incident Response Team under research governance protocols associated with University requirements related to IRB issues.
-
PHI incidents remain under the purview of the WMCOSU Privacy Office, with escalation to the Office for Civil Rights as needed.
By establishing a shared vocabulary and clearly defining roles—technical controls managed by Information Security at OSUWMC, professional controls required by the IRB, and oversight by both privacy and research governance bodies—OSUWMC laid the groundwork for an effective, integrated data sharing approach that fosters interdisciplinary collaboration, accelerates ethical data sharing, and sustains continuous innovation in translational research.
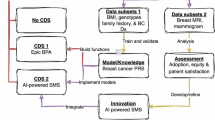
One of the most sensitive challenges in a RO-LHS is ensuring compliant, efficient use of clinical data for research11,12. To address this, OSUWMC established an honest broker protocol that periodically reviews representative request samples to define concrete boundaries for data release. Illustrated in Fig. 1, under this model the Honest Broker protocol (Supplement 1) serves as the IRB of record for:
-
Expedited releases of fully de-identified or limited datasets that pose minimal re-identification risk which are not transferred to third parties outside the University which are handled directly by honest brokers.
-
Intermediate-risk requests—for example, those involving limited PHI elements or small cohort sizes—require Privacy Officer approval based on predefined criteria (dataset scope, data sensitivity, research purpose).
-
High-risk or novel scenarios—such as cross-institutional data sharing, rare disease cohorts, or requests involving genomic or geolocation data—are automatically escalated to the Data Governance Committee (or IRB) for formal review.
These granular decision pathways mean that honest brokers can act autonomously within clearly defined performance envelopes, referring only truly complex or sensitive requests for committee review. By balancing researcher autonomy with responsible, tiered oversight, OSUWMC and its partners can streamline data provisioning—reducing duplicate reviews and unnecessary delays—so investigators remain focused on study objectives and quality improvement. The result is faster, more predictable access to approved data while preserving patient privacy and institutional compliance particularly related to the delivery of concierge data requests. Continuous refinement of these governance protocols ensures we maintain both innovation velocity and the highest standards of data stewardship (Figs. 2–4).
LifeScale:
A Unified Data Integration System
Despite near-universal adoption of EHRs, many academic medical centers continue to face systemic barriers that limit the integration of clinical and research data into real-time systems. Fragmentation across data custodians, incompatible schemas, protracted legal negotiations, and dependence on bespoke informatics extractions constrain the pace of discovery and impair the scalability of translational research. Taken together, innovations and improvements in governance and technology has allowed for novel approaches to bring data and technology closer to real time data transparency across the Academic Medical Center Enterprise. That is, in the face of these structural limitations, OSUWMC and NCH developed LifeScale—a unified, enterprise-grade data integration platform designed to enable scalable, ethical, and efficient data use across operational, research, and educational missions.
At its core, LifeScale consolidates diverse institutional data assets into a governed research enclave, replicating a multiorganizational integration of data from OSUWMC and NCH into a centralized Microsoft Azure data lake or connections to such data via Delta Sharing using Parquet file structures. This architecture supports both native Epic schemas (i.e., Caboodle and components of Clarity) and the Observational Medical Outcomes Partnership (OMOP) Common Data Model as well as curated data on telemetry and alarms, social determinants of health via geolocated curation data13, allowing investigators to pivot between detailed clinical data and standardized observational frameworks without duplicative extract-transform-load (ETL) processes [Fig. 2]. LifeScale is an IRB-approved, honest broker-mediated, data repository where linkage is available across longitudinal multi-institutional pediatric-adult datasets, facilitating mother-baby linkages, transition-of-care studies, and lifespan research that have historically been very challenging to execute at scale (Supplement 6).
Beyond the clinical record, LifeScale systematically integrates structural social determinants of health (SDoH), environmental exposures, registries, and primary research data to support whole-person, context-aware analyses facilitated through honest brokerage by the HIPAA covered entity. The platform’s scope reflects a deliberate strategy to extend analytic capacity beyond biomedical phenotyping, acknowledging the growing evidence that non-medical drivers of health are central to effective LHS interventions. By embedding these diverse data types into a common analytic environment, LifeScale supports multi-level studies that address the complexity of health and healthcare delivery across populations and over time.
A single, Reliant IRB protocol, combined with a jointly negotiated Business Associate/Data Use Agreement between OSUWMC and NCH, streamlines data sharing while ensuring HIPAA compliance and institutional risk management (Supplement 6). The designation of RHI within LifeScale provides investigators with access to coded-limited datasets that maintain regulatory protections while allowing analyses at scale without repeated expert determinations or bespoke de-identification workflows. This governance model effectively reduces transaction costs and regulatory latency, aligning data access processes with the rapid analytic cycles required in modern translational research.
Technically, LifeScale operates as a fully provisioned secure analytic enclave within Azure, supporting Databricks workspaces, John Snow Labs natural language processing pipelines, Datavant privacy-preserving linkage, and leveraging Medicom imaging de-identification services. The system’s real-time, metadata-driven data pipelines enable continuous updates, ensuring that both clinical and research stakeholders operate on the most current data available. Importantly, the platform eliminates the traditional dependency on informatician-dependent data extractions by providing self-service cohort identification and shared analytic workspaces with role-based permissions, multi-factor authentication, and rigorous audit trails. This design facilitates team-based science while preserving strict data governance, supporting both reproducibility and institutional compliance. LifeScale’s development is underpinned by a comprehensive governance framework that enables secure, compliant data access while minimizing operational friction. This framework provides the opportunity to expand to other organizations, serving as a backbone for Data Coordinating Centers that include other research partners.
Artificial intelligence (AI) models in LifeScale are deployed into RO-LHS workflows through governance-driven integration with the health system’s analytic center of excellence and Wexner Medical Center Information Technology (WMCIT). Guided by clinical champions, governance committees, and change management professionals, models are aligned to the “Five Rights” framework for clinical decision support—ensuring the right information reaches the right person, in the right format, through the right channel, and at the right time in the workflow. This partnership ensures that model deployment is technically feasible, clinically relevant, and ethically sound, while embedding continuous monitoring and recalibration into real-world practice. By situating AI models within existing operational and analytic infrastructures, LifeScale enables scalable and sustainable implementation that supports both local improvement and the generation of generalizable knowledge, fulfilling the core aims of the RO-LHS.
The integration of NCH into LifeScale marked a significant expansion of the platform’s capabilities, connecting pediatric and adult records to cover maternal-child linkage and self-self-linkage for transition of care. Through privacy-preserving record linkage technologies—leveraging Datavant tokenization14,15,16,17—the platform enables cross-institutional follow-up for longitudinal studies that capture patient trajectories across complex care pathways. This capability is particularly salient in pediatrics, where continuity of care frequently spans institutional boundaries and requires integrated data for analyses of chronic disease management and developmental outcomes. LifeScale’s cross-institutional integration thus enables comprehensive cohort discovery and multi-site observational studies that were previously infeasible due to structural and legal barriers.
At NCH, adoption has accelerated with investigators across eight pediatric subspecialties (e.g., neonatology, primary care, clinical genetics) are actively using LifeScale for clinical research. NCH-originating data requests increased throughout 2025 before any formal introduction of LifeScale was publicized. Projects now include observational studies and AI development that leverage pediatric–adult linked cohorts. Engagement spans research-focused faculty, clinician-investigators, and operational leaders, demonstrating broad applicability across roles.
The LifeScale initiative reflects a deliberate institutional investment in reusable infrastructure that moves beyond project-specific data provisioning to create durable capacity for continuous discovery, operational learning, and responsible innovation. It is grounded in a broader data vision to include operations, research, and education. The associated data architecture was developed to identify clinical sources of data and establish pipelines to move it into a common platform for operations and discovery. Importantly, LifeScale was intentionally architected as an enterprise platform rather than a research-specific DataMart.
Its development illustrates a replicable model for academic medical centers seeking to operationalize LHS principles through sustained governance transformation, technical innovation, and inter-institutional collaboration. As real-world data continues to expand in both scale and complexity, platforms such as LifeScale will be essential to enabling ethically grounded, scientifically rigorous, and operationally sustainable translational research at scale. By aligning operational, research, and education infrastructures, the platform reduces translational lag between clinical innovation and empirical validation, while simultaneously preparing the institution for emerging AI and multimodal analytics workloads. LifeScale’s governed data architecture provides the controlled environment necessary to support natural language processing, predictive modeling, and advanced machine learning applications while safeguarding patient privacy and institutional integrity.
The vision of aligning and making a version of operations data available to researchers was explicitly focused on reducing the time from research to practice18,19,20. In LifeScale, we removed the traditional role of the informatician in data extraction and instead refocused them towards reusable infrastructure and data models. Initially, differential privacy approaches were explored with vendors, but many could not provide expert determination related to HIPAA for de-identification. Consequently, the use of a coded-limited dataset was initiated—an investment that was impractical using grant project funds–at scale that was viewed as an opportunity to leverage our data in a meaningful way.
Strategic Evolution, Governance Operations, and Institutional Alignment
The development of LifeScale required not only technical innovation but also sustained institutional coordination to establish scalable, compliant, and operationally durable research infrastructure. OSUWMC and NCH approached LifeScale as an enterprise-level investment, deliberately aligning data integration efforts with institutional priorities in clinical care, research, education, and regulatory compliance. This convergence of strategic planning, governance design, and leadership commitment allowed LifeScale to evolve into a robust platform capable of supporting real-world translational research at scale.
Operational governance within LifeScale is grounded in multi-layered controls that ensure responsible data access and sustained regulatory compliance. All users of LifeScale are required to complete mandatory training on data privacy, compliance standards, and analytic best practices prior to receiving access credentials. Access is provisioned through role-based authorization models that restrict users to the minimum necessary data for their research activities, with continuous audit trails and real-time monitoring to detect unauthorized access or inappropriate use. Oversight is provided by a multidisciplinary data governance board composed of informatics leaders, regulatory experts, faculty representatives, legal counsel, and information security officers from both OSU and NCH. This board reviews data access requests, ensures ongoing adherence to institutional policies, and maintains alignment between data use and institutional research priorities.
The legal framework for LifeScale required more than a year of BAA/DUA negotiation between OSUWMC and NCH to align governance and risk tolerance across legal, compliance, informatics, and executive teams. Including the security controls detailed elsewhere, we note that consequential contractual terms concerned shared operational standards and escalation mechanisms. The resulting agreements establish a durable, enterprise-grade foundation for multi-institutional data sharing in support of research.
To meet the security demands of multi-institutional data sharing, LifeScale’s information security architecture was designed to exceed baseline HIPAA requirements by adopting a security framework aligned to the NIST SP 800-53 controls at the Federal Information Security Modernization Act (FISMA) Moderate level. This compliance posture reflects the high-risk profile of integrating identified clinical data across research and operational domains. All data assets are encrypted both at rest and in transit, with strict key management protocols and system-wide vulnerability monitoring. Identity and access management incorporates multi-factor authentication, least-privilege role assignments, and continuous behavioral auditing to detect anomalous activity. In areas where partner institutions had differing security postures, compromise frameworks were jointly developed to harmonize breach notification timelines, incident response protocols, encryption standards, and audit logging procedures while preserving institutional autonomy. This security architecture ensures that LifeScale remains both research-enabled and enterprise-hardened, providing a compliant, scalable foundation for data-intensive translational research.
As LifeScale continues to expand its institutional footprint and analytic capabilities, its governance framework remains adaptable to emerging challenges. Ongoing refinements include expanded audit mechanisms, strengthened analytic transparency, and continuous alignment with evolving federal and institutional regulations. LifeScale’s governance model balances the demands of translational research with the obligations of regulatory compliance, operational security, and public trust, offering a replicable model for scalable enterprise data integration across the LHS landscape.
LifeScale and AI
Building upon the integrated infrastructure, LifeScale is deliberately positioned to serve as a platform for AI and machine learning (ML)-enabled translational research. A persistent challenge in AI-driven healthcare research is securing access to sufficiently large, diverse, and well-curated data sources that support both model development and external validity. LifeScale directly addresses this challenge by creating a centralized, longitudinal, and multi-institutional data environment that supports advanced analytic workloads while maintaining strict adherence to privacy and governance standards.
AI and ML applications require not only scale but diversity in patient representation to avoid model bias and ensure clinical applicability across heterogeneous populations. LifeScale’s integrated and layered data architecture, incorporating structured EHR data, unstructured clinical notes, and social determinants of health, with the capability of also including imaging studies, creates a rich analytic substrate that enables the development of generalizable AI models. The integration of NCH extends this capability across the lifespan, supporting pediatric, adult, and maternal-child linkages that historically, have been rarely available in unified analytic environments.
Recognizing the importance of data interoperability for reproducible machine learning, LifeScale operates both Epic-native data models and the OMOP Common Data Model in parallel. This dual-model approach allows researchers to harmonize analytic workflows across institutions and compare model performance across differing data representations, while simultaneously identifying potential sources of bias or e-iatrogenesis that may arise from model portability. The alignment of OMOP with Epic schemas further enables cross-institutional collaborations that leverage common data frameworks while preserving institution-specific operational data fidelity.
The platform incorporates several specialized AI-enabling technologies. Databricks provides scalable compute infrastructure for training and validating large-scale ML models; John Snow Labs’ natural language processing pipelines allow extraction of clinically relevant features from unstructured physician notes, discharge summaries, and pathology reports and also serves as the platform used to deidentify our clinical notes and patient messaging; and Medicom enables de-identification of high-resolution medical imaging, converting radiologic, pathologic, and cardiologic image archives into research-ready formats without compromising patient confidentiality. Together, these tools extend LifeScale’s analytic capacity beyond structured tabular data into multimodal AI, supporting deep learning models that integrate both structured and unstructured sources for comprehensive phenotyping and predictive modeling.
Importantly, LifeScale’s analytic enclave supports a zero-data-movement model, where data remains within secure governed environments, and analytic pipelines are deployed into proximity with the data. This architecture not only reduces data movement risk but ensures compliance with regulatory frameworks while maintaining analytic agility. Synthetic data generation further expands LifeScale’s training and educational utility, providing realistic yet privacy-preserving datasets that allow for model prototyping, algorithm refinement, and skills development without accessing identified clinical records. The availability of synthetic data has proven valuable for training the next generation of data scientists, clinicians, and biomedical informaticians in advanced analytic methods while preserving patient confidentiality.
As AI models mature within LifeScale, they are directly positioned for translational deployment within our RO-LHS framework. Risk stratification, predictive modeling, and decision support algorithms derived from LifeScale data can be rapidly embedded into operational workflows, enabling earlier identification of at-risk patients, optimizing care delivery, and supporting clinical decision-making at the point of care. The deliberate coupling of AI development to governed enterprise data architecture ensures that innovation remains aligned with institutional governance, ethical standards, and clinical priorities.
Through LifeScale, OSUWMC and NCH have established an enterprise data platform that not only advances traditional clinical research but serves as a durable, scalable foundation for responsible AI innovation in the LHS context. As AI continues to transform healthcare delivery, LifeScale offers a replicable model for institutions seeking to integrate AI research, ethical data governance, and translational clinical impact within a unified infrastructure.
Improving the Clinical Record through Automation:
A PARTNERed-enabled approach
A key component of the LHS involved enhancing the quality of data in the EHR particularly during patient engagement. LifeScale leverages clinical data which means that the quality of data collection in the clinical engagement affects downstream research usage. To explore ways to improve that data collection workflow, we designed an IRB-approved protocol called PARTNER to create a learning laboratory to explore the interfaces between patients and their clinical experience.
The PARTNER protocol (Supplement 7), a research-oriented LHS approach to innovation, is a repository protocol. The protocol is expansive and includes both data and biospecimens in its design and has a goal to engage patients in building a better LHS. At OSU, IT builds follow either an IRB-approved track or a clinical use track; therefore, agreement on building general use technology infrastructure to support research can be a challenge. An IRB-approved protocol that is amended for the purposes of testing infrastructure has been a novel use of research infrastructure to advance technological discovery. Within that framework, we have the capabilities to perform Plan-Do-Study-Act (PDSA) testing by User Experience and Design experts tasked with identifying best practices in the context of research infrastructure design. For instance, we have used PARTNER as the basis of deploying and testing consent prototypes in REDCap for electronic consent with the goal of building template projects that can be used to quickly advance and deploy general use infrastructure as best practice.
We have also leveraged PARTNER as a patient-inclusive learning laboratory to explore workflow opportunities that chain technological systems—Azure Document AI, REDCap, Epic SmartElements, Epic Our Practice Advisories (OPAs), and clinical office scanners—to enable teams to provide patient-reported outcome forms to patients and research participants more efficiently. Leveraging in-house design experts, we created forms that are optimized for digital capture on paper, which are then distributed to research and clinical teams. Patients complete these forms and return them to the front desk, where a label is affixed to the first page before being uploaded via email using a simple hotkey function on the scanner. Microsoft Azure Document AI processes the document by separating it into component parts, identifying the forms involved, reordering them appropriately, and extracting the responses into REDCap. Furthermore, these data are prepared for integration into our Epic environment by triggering a BPA based on a SmartData Element, which is activated when new patient data (identified by the label) is available in the system. The BPA performs a call that transfers the data into Epic Flowsheets and can also present that information in a dashboard [Fig. 3].
This type of practice redesign, that is patient centered, is the hallmark of patient centered clinical decision support (PC CDS) integrates digital tools into clinical care to facilitate shared decision-making that accounts for individual patient circumstances, values, and preferences. PC CDS enables patients, caregivers, and clinicians to jointly evaluate health-related choices by combining evidence-based research, such as patient-centered outcomes research (PCOR) and comparative effectiveness research, with patient-specific data. These data can include patient-reported outcomes, patient-generated health data, social determinants of health, and individual preferences. We use PARTNER to experiment on ways to create effective PC CDS that enables the institution to engage patients and caregivers directly through applications, patient portals, and point-of-care interfaces, creating opportunities for meaningful dialogue and collaborative decision-making between patients and clinicians. By creating a workspace where approaches for supporting these interactions can occur, we can create lower transaction cost routes for PC CDS to enhance the delivery of care that is aligned with each patient’s unique needs and goals, and subsequently improve data capture and reduce costs, thereby improving both the clinical and research work of our organization through higher data quality.
Further, we can deploy forms in diverse languages that are pre-programmed to render in Epic in the appropriate data elements, reduce workloads associated with data transcription, and present data in a manner that is more patient-centric. At OSU, the HT2 clinical trial found that paper-based workflows had a 99.5% completion rate while tablet-mediated approaches had only an 80% completion rate21. The data quality gap created by technology-enabled workflows may lower organizational data collection costs by avoiding costly data entry costs - frequently by overextended clinical staff - but result in lower quality data collection. This is often exacerbated by the additional data loss created by clinicians potentially using the paper forms in a clinical encounter without entering that data into the EHR. By focusing technology on mediating the effort to make paper data digital (a workflow we call paper-to-plastic) we support patient-centered engagement in research and practice.
In this context, the whole system change approach – one where the goal is to move beyond the silos where positive deviance in practice change is regularly practiced, the need for approaches that generalize to the enterprise becomes critical. Although this might not be the experience of every organization, working within existing frameworks is crucial for advancing infrastructure. Earlier efforts to adopt technology-mediated privacy-preserving machine learning (PPML) illustrate an attempt to introduce innovations outside the organization’s usual practices. Despite efforts to familiarize the organization with such technology, the opaque nature of PPML placed it outside the organization’s acceptable risk parameters. After 12 months of attempting to deploy the technology, these efforts were redirected by the CRIO towards more familiar processes involving coded-limited data. Recently, the organization has reconsidered approaches to PPML as it has gained wider acceptance. Often, the research mission paves the way for innovative approaches, and while they may not initially be adopted, revisiting previous presentations can help establish understanding among senior leaders.
Methods
Governance and Oversight
A unified governance model integrates privacy, compliance, and research oversight. The Honest Broker protocol (Supplement 1) defines data extraction and de-identification workflows under the Research Health Information (RHI)framework, which differentiates research data from Protected Health Information (PHI). The LifeScale repository protocol (Supplement 6) enables federated data integration through a jointly executed Business Associate/Data Use Agreement, while the PARTNER protocol (Supplement 7) provides a patient-inclusive environment for testing user experience and workflow innovations. All protocols align with the Common Rule and HIPAA Privacy Rule.
Data Sources and Security
The LifeScale platform integrates Epic-derived EHR data, registries, social determinants of health, and environmental data within an Azure-based, PHI-compliant data lake. De-identification and linkage use Datavant tokenization, with data access is mediated through honest brokers. All storage and transmission are encrypted (AES-256, TLS 1.3), and access is role-based with multi-factor authentication and continuous auditing.
Computational Environment
Analyses are performed within governed analytic enclaves on Azure Databricks and the OSC’s Ascend cluster, both operating under institutional business agreements and aligned to NIST SP 800-53 (FISMA-Moderate) standards.
Ethical Compliance
IRB approvals included LifeScale (#2020H01234), Honest Broker (#2021H00456), and PARTNER (#2022H00089). All methods followed relevant regulations and institutional policies; de-identified or coded-limited data were used where appropriate.
Discussion
Taken together, these opportunities enable the institution to leverage research to advance our collective discourse on what is possible, reduce transaction costs of such deployments, and generalize that knowledge across projects [Fig. 4]. The LHS model isn’t simply about moving from clinical operations to research, but rather to lower the barriers for engagement through learning across the system. If research is part of our LHS, then repurposing infrastructure is a critical component of that approach. The CRIO’s office is responsible for looking at the research experience to ask the value question related to large scale research investments that could benefit from expanded investments across the portfolios. Two examples below – Portal and MPRT – represent such examples that we explicitly invest in as an organization to enable new kinds of research or expand our capabilities to serve as an agent of change locally, within our state, or nationally.
As part of the HEALing Communities Study (HCS), RIT developed technology to create community-level data coordinating center infrastructure22. Named Portal, this technology has been repurposed for use in our data coordinating center infrastructure as part of a broader vision to support such endeavors23. While a research project might produce specific artifacts, investing in sustainability at the institutional level allows the organization to learn from domain-specific projects designed for research. In the case of HCS, the dashboard project was transferred to the State of Ohio to aid their efforts in providing data on the Opioid crisis24. Portal (portal.osu.edu) disseminates public announcements, papers, dashboards, documents, and calendars on a public website, while creating a restricted data compliant secure backend that includes study-level and site-level microsites. This infrastructure is deployed as a REDCap external module with lightweight tools aimed at simplifying engagement. Using this system, research projects can quickly and efficiently establish the necessary infrastructure, moving swiftly from concept to implementation. As an illustration, we established the LifeScale community of practice website in just 2 days (http://lifescale.osu.edu), and the technology currently supports 11 data coordinating centers.
As part of Ohio Medicaid’s Infant Mortality Research Partnership (IMRP), we developed the Medicaid Perinatal Risk Toolkit (MPRT), a cloud-native, electronic health record–integrated system designed to support perinatal care coordination and risk stratification. MPRT leverages Fast Healthcare Interoperability Resources (FHIR)-based data extraction to populate a predictive model, derived from a linked dataset of over 500,000 Ohio pregnancies, which estimates the probability of adverse maternal and infant health outcomes. The toolkit delivers these risk estimates and relevant clinical data to a centralized State of Ohio hub, facilitating timely collaboration with Medicaid Managed Care Organizations. Hosted within Ohio State’s Microsoft Azure environment, MPRT is deployed in hospitals across the state and includes both clinician-facing and patient-facing components to support decision-making at the point of care.
To ensure scalability and resilience, MPRT was architected using standardized data models and continuous integration/continuous deployment (CI/CD) workflows—methods more typical of enterprise-grade systems than academic prototypes. Recognizing that traditional research IT environments often lack the governance structures needed for production-scale deployment, we established an internal Cloud Center of Excellence to formalize infrastructure, security, and compliance processes. This framework not only enabled rapid and responsible adoption of cloud technologies for MPRT but also served as a reusable governance model for other research technologies. By balancing innovation with operational discipline, we demonstrated how research IT can create shared infrastructure that accelerates institutional readiness, facilitates broader clinical integration, and produces public goods in the form of scalable, standards-based technology (Box 4).
Conclusion
This case study offers a roadmap for institutions seeking to build or refine their own RO-LHS. The initiatives described—spanning data governance innovations, AI-ready infrastructure, patient-centered CDS, and cloud-based scalability—are not unique to OSUWMC in their need or importance. Rather, they represent transferable strategies that can be adapted to diverse organizational contexts. From the deployment of LifeScale as a multi-institutional research platform to the operationalization of new information security and compliance models, this work provides tangible examples of how institutions can navigate complexity while maintaining alignment with regulatory frameworks and research imperatives.
Importantly, the transformation at OSUWMC has shown that research IT can function as a catalyst—not a constraint—when supported by clear governance, institutional leadership, and a culture of collaboration. Efforts to integrate patient-generated data, facilitate shared decision-making, and support federated AI model development demonstrate that it is possible to meet the demands of modern translational science while upholding ethical, equitable, and patient-centered values.
Looking ahead, the path to a fully realized RO-LHS will vary by institution, but the principles outlined here—standardization, governance, interoperability, and sustained cross-sector engagement—are broadly applicable. As healthcare systems continue to grapple with fragmented data, operational silos, and evolving regulatory expectations, the OSUWMC/NCH experience offers both practical insights and aspirational guidance. We remain optimistic that as more institutions adopt similar strategies, a national ecosystem of interconnected, research-enabled LHSs will emerge—one capable of accelerating discovery, improving population health, and transforming care delivery for decades to come.
In summary, the development of a RO-LHS at OSUWMC and NCH demonstrates the feasibility and value of aligning research infrastructure, clinical operations, and data governance within a unified strategic framework. By combining robust policy development, agile yet compliant data integration strategies, and a deep commitment to interdisciplinary collaboration, OSUWMC has built a durable foundation that supports continuous learning, accelerates discovery, and improves clinical outcomes.
Data availability
No datasets were generated or analysed during the current study.
References
Embi, P. J., Tachinardi, U., Lussier, Y., Starren, J. & Silverstein, J. Integrating governance of research informatics and health care IT across an enterprise: experiences from the trenches. AMIA Jt Summits Transl. Sci. Proc. 2013, 60–62 (2013).
Moffatt-Bruce, S., Huerta, T., Gaughan, A. & McAlearney, A. S. IDEA4PS: the development of a research-oriented learning healthcare system. Am. J. Med Qual. 33, 420–425 (2018).
Patterson, E. S., Rayo, M. F., Edworthy, J. R. & Moffatt-Bruce, S. D. Applying Human Factors Engineering to Address the Telemetry Alarm Problem in a Large Medical Center. Hum. Factors 64, 126–142 (2022).
Health Insurance Portability and Accountability Act, Pub. L. No. 104-191, 110 Stat. 1936 (1996).
UCLA. (2021). HIPAA Research Guidelines and Information. https://ohrpp.research.ucla.edu/hipaa/#:~:text=Research%20Health%20Information%20(RHI)%20is,HIPAA%20Privacy%20and%20security%20Rules.
University of Delaware. (2025). HIPAA Hybrid Statement. https://www1.udel.edu/research/preparing/hybrid-hipaa-ud.html
Blake, H., Bital, E. (2015). IRB Grand Rounds Unlocking Information: From PHI to RHI. University of Miami Health System. <https://hsro.uresearch.miami.edu/_assets/pdf/20150609_unlockinginformationfromphitorhi-blakebital.pdf>.
Services, U. S. D. o. H. a. H. (Washington, DC, 2018).
Office of Technology and Digital Innovation, The Ohio State University. (n.d.). Research Health Information. https://it.osu.edu/security/research-support/research-health-information
University, T. O. S. RHI Questions and Answers. https://u.osu.edu/controvertedissues/2022/04/26/rhi-q-and-a/ (2025).
Liu, J. et al. Toward a fully de-identified biomedical information warehouse. AMIA Annu Symp. Proc. 2009, 370–374 (2009).
Obeid, J. S. et al. A survey of practices for the use of electronic health records to support research recruitment. J. Clin. Transl. Sci. 1, 246–252 (2017).
Lawrence, J., Rayo, M. & Huerta, T. ALarms, Outcomes Telemetry with Timing (ALOTT): a Bedside-EMR Database (version 1.0.0). PhysioNet https://doi.org/10.13026/sbq5-dy17 (2025).
Graham, R. J. et al. Real-world analysis of healthcare resource utilization by patients with X-linked myotubular myopathy (XLMTM) in the United States. Orphanet J. Rare Dis. 18, 138 (2023).
Afshar, M. et al. Creation of a data commons for substance misuse related health research through privacy-preserving patient record linkage between hospitals and state agencies. JAMIA Open 6, ooad092 (2023).
Kiernan, D. et al. Establishing a framework for privacy-preserving record linkage among electronic health record and administrative claims databases within PCORnet((R)), the National Patient-Centered Clinical Research Network. BMC Res Notes 15, 337 (2022).
Mirel, L. B., Resnick, D. M., Aram, J. & Cox, C. S. A methodological assessment of privacy preserving record linkage using survey and administrative data. Stat. J. IAOS 38, 413–421 (2022).
Green, L. W., Ottoson, J. M., Garcia, C. & Hiatt, R. A. Diffusion theory and knowledge dissemination, utilization, and integration in public health. Annu Rev. Public Health 30, 151–174 (2009).
Balas, E. A. & Boren, S. A. Managing Clinical Knowledge for Health Care Improvement. Yearb Med Inform, 65-70 (2000).
Grant, J., Green, L. & Mason, B. Basic research and health: a reassessment of the scientific basis for the support of biomedical science. Res Eval. 12, 217–224 (2003).
Gregory, M. E., Sova, L. N., Huerta, T. R. & McAlearney, A. S. Implications for electronic surveys in inpatient settings based on patient survey response patterns: cross-sectional study. J. Med Int. Res. 25, e48236 (2023).
Slavova, S. et al. HEALing Communities Study: Data measures for supporting a community-based intervention to reduce opioid overdose deaths. Drug Alcohol Depend. 274, 112738 (2025).
Wu, E. et al. Community dashboards to support data-informed decision-making in the HEALing communities study. Drug Alcohol Depend. 217, 108331 (2020).
Huerta, T. & Fareed, N. State of Ohio Integrated Behavioral Health Dashboard. https://data.ohio.gov/wps/portal/gov/data/view/ohio-ibhd (2022).
Acknowledgements
Several components of the work described in this publication and its associated work were supported, in part, by The Ohio State University Clinical and Translational Science Institute (CTSI) and the National Center for Advancing Translational Sciences of the National Institutes of Health under Grant Number UM1TR004548. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors would like to explicitly acknowledge the role of staff at both The Ohio State University Wexner Medical Center (OSUWMC) and Nationwide Children’s Hospital (NCH) that have made these advancements possible. This work includes appendices created to support this transformational effort, and their contributions have been both great and small to the greater good. Most notably this includes staff at the Department of Research Information Technology at OSUWMC, RISI at NCH, and Information Technology at OSUWMC. The authors also explicitly acknowledge the work of Dr. Rebecca Jackson who, as the founding Principal Investigator of the NCATS CTSA grant at OSU, laid the foundation for this work through both her direct engagement and leadership. Further, we acknowledge the extensive work of OSUWMC IT and NCH staff in executing on this vision. While not authors on this paper, their work has been essential to executing on our collective vision and translating ideas into practice.
Author information
Authors and Affiliations
Contributions
TRH, CB, TB, AAF, ASM, AC, CRB, CO, CW, CAG, GA, HL, JFPB, LL, LP, PM, RVS, SEM, WES, YH, DMB were involved in conception and design of the learning healthcare system (LHS) infrastructure and implementation. TRH, CB, AAF, HL, PM, MM, and XN were involved in involved in the development of new software or code that supports LHS operations. TRH, CB, AAF, ASM, CO, CW, HL, JAJ, MM, PM, SEM, TG, WES, YH, DMB were involved in creating a new data infrastructure to support LHS operations. TRH, CB, AAF, AC, CW, LP, RVS, SEM, TG, YH, DMB were involved in data acquisition. TRH, CB, ASM, DMB were involved in data analysis. TRH, CB, JAJ were involved in project funding. TRH, CB, AAF, ASM, CO, CW, CAG, EAM, GA, JFPB, JAJ, LP, NVK, PZ, QM, SEM, SG, WES, XN, YH, DMB were involved in drafting, substantial review, or revision of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Huerta, T.R., Bartlett, C.W., Alain, G. et al. Operationalizing a research-oriented learning healthcare system across covered entities: cross-institutional strategies and innovations. npj Health Syst. 2, 47 (2025). https://doi.org/10.1038/s44401-025-00051-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s44401-025-00051-2