Abstract
Richter’s transformation (RT) is an aggressive lymphoma occurring upon progression from chronic lymphocytic leukemia (CLL). Despite advances in deciphering the RT genetic architecture, the mechanisms driving this disease remain unknown. BCOR disruptive mutations were found in CLL and frequently associated with NOTCH1 aberrations, a common feature in CLL and RT. We engineered mice to knock-out Bcor in B and CLL cells of Eμ-TCL1 mice. Bcor loss resulted in alterations of the B cell compartment and favored CLL transformation into an aggressive lymphoma with reduced survival in Eμ-TCL1 mice. RNA-sequencing demonstrated a molecular signature reminiscent of human RT and implied the involvement of the T cell tumour microenvironment in the disease onset. Bcor deficiency was associated with Notch1 activation in splenic CD19 + CD5+ cells to accelerate Eμ-TCL1 mice lymphoproliferation. Notch1 inhibition progressively reduced circulating CD19+ CD5+ and RT cells infiltrating the spleen of diseased mice with concomitant reduction of PD-1 expressing T cells and improved survival. Our data demonstrated an interplay between the tumour suppressor activity of Bcor and Notch1 in RT pathogenesis with potential for tumour targeting. This model represented a new platform to uncover promising alternatives for this incurable tumour.
Similar content being viewed by others
Introduction
Richter Transformation (RT) is the occurrence of an aggressive lymphoma with poor prognosis in CLL patients. The advent of targeted therapies has improved patients’ management in CLL, whereas these advancements do not apply to RT. This also depended on the lack of preclinical models that have limited the understanding of RT pathogenesis and therapeutic developments. In these years, genome-wide sequencing deciphered the genetic architecture of CLL and RT, allowing the identification of recurrent mutations in these conditions [1,2,3]. Within the genetic heterogeneity of CLL, disruptive mutations of the BCL6 co-repressor (BCOR) were found in up to 2% of cases [4] and have been included among the lymphoma driver genes [1]. Moreover, BCOR mutations frequently co-occurred with mutated NOTCH1 [5, 6], a hallmark alteration associated with RT [7]. BCOR is a transcription factor involved in NOTCH1 signaling suppression during embryogenesis and lymphoid development [8]. It has been demonstrated that Bcor acts as a tumor suppressor gene by increasing the transactivating ability of NOTCH to promote leukemogenesis in mice [9]. Furthermore, Bcor deletion enhances nuclear accumulation of cleaved Notch1 in murine splenocytes [10], suggesting a mechanistic connection between BCOR and NOTCH1 cleavage. Here, we determined the in vivo role of Bcor loss in normal and leukemic B cells using Eµ-TCL1 transgenic mice. We generated a new mouse model of RT and highlighted a pathogenic mechanism involving the interplay between Bcor and Notch1 with potential for targeted treatment.
Methods
Mouse strains and transplantation procedure
Experiments were performed according to murine ethical approval (authorization n°971/2020-PR – n°253/2024-PR). For all the experiments, we used homozygous Bcorflox/flox female and hemizygous Bcorflox/Y male mice on a C57BL/6 N genetic background [11]. Bcorflox/flox and Bcorflox/Y mice were intercrossed with CD19-Cre mice (Cre) [12] to generate Bcorflox/flox;Cre+ and Bcorflox/Y;Cre+ mice in order to obtain a complete Bcor loss-of-function. Hereafter, Bcorflox/flox;Cre+ homozygous and Bcorflox/Y;Cre+ hemizygous mice will be referred as Bcor−/− and Bcorflox/flox;Cre- and Bcorflox/Y;Cre− used as controls will be indicated as Bcor+/+ mice. Bcor−/− mice were further intercrossed with homozygous Eµ-TCL1 (TCL1) models [13] to generate Bcor−/−; TCL1 mice. In adoptive transfer experiments, C57BL/6 N recipients were transplanted with 2–2.5 × 107 splenic cells from one original leukemic mouse donor of TCL1 and Bcor−/−; TCL1 strain, by intravenous injection (I.V.). Mice at the 2° round of transplantation were used in this study. Experiments were performed in at least three replicates using at least three original mouse donors for each strain.
Immunophenotyping
Samples isolated from peripheral blood (PB), bone marrow (BM), spleen and liver were used for a complete blood count and immunophenotyping. The latter was conducted by flow cytometry (FC) using a 2-colors panel for the detection of CLL markers, or 3- up to 5-colors panels (supplementary Figs. 1, 2) for the characterization of B- and T-cell subpopulations in spleen and BM.
Immunoblot analysis
Immunoblots of murine samples were performed using the monoclonal antibodies against Notch1, Hes1 and Myc.
PCR-based analyses
-
To verify the occurrence of Cre-mediated excision event of the Bcor locus, we performed a standard PCR on gDNA samples of splenic flow-sorted B cells, as previously described [11].
-
To verify the loss of murine Bcor transcript, for first, and investigate the involvement of Notch1 signaling, total mRNA was extracted from splenic flow-sorted B cells, retrotranscribed, and murine mBcor or mNotch1, mHes1 and mMyc expression levels, respectively, were evaluated by RT-qPCR.
-
To investigate the clonality of neoplastic cells from Bcor−/−; TCL1, before and after serial transplantion, genomic DNA was isolated from splenic flow-sorted B cells and analysed for three murine IGHV families (VHJ558, VH7183, VH52Q), by standard PCR and, if necessary, Sanger sequencing.
Histology and Immunohistochemistry
Hematoxylin and eosin (H&E) staining was performed for morphological analyses on liver and splenic sections prepared from tissues originated by non- and transplanted mice, also after Bepridil treatment. Immunohistochemistry (IHC) was performed on the same samples using the monoclonal antibody against Notch1 adopted for the immunoblot analysis. Images were acquired and interpretated with the support of an expert pathologist.
Transcriptomic profiling
Total RNA was extracted from splenic flow-sorted B cells of transplanted mice (N = 3) and a precise quantity of RNA was characterized at transcriptional level by Standard RNA sequencing (RNA-Seq) with the support of Genewiz-Azenta Life Sciences. Unique Differentially Expressed Genes (DEGs) identified were used for an enrichment analysis conducted by an expert bioinformatician.
Drug administration
Three C57BL/6 N female recipients per treatment arm (group #1: Vehicle; group #2: Bepridil) were I.V. injected with 2–2.5 × 107 splenic cells from three original leukemic mouse female donors of Bcor−/−; TCL1 strain (leukemic burden >50%). Transplanted mice were injected intraperitoneally with DMSO (vehicle) or bepridil (5 mg/kg), respectively, once daily, 5 days/week, over a 1-month period. Only mice at second round of transplantation received the treatment.
Additional methods
Further details on mouse models, experimental conditions, bioinformatic analyses, reagents and instrumentation information are provided in supplementary materials
Results
Bcor deficiency perturbs B cell differentiation but is not sufficient to drive lymphoid malignancies in mice
We crossed homozygous Bcorflox/flox female mice and hemizygous Bcorflox/Y male mice with CD19-Cre mice (Cre+) to obtain a complete deletion of Bcor in B cells. Both Bcorflox/flox;Cre+ homozygous (Bcor−/−) and Bcorflox/Y;Cre+ (Bcor−/−) hemizygous mice were characterized by a complete Bcor loss-of-function. Analysis of splenic B cells confirmed the deletion of Bcor at DNA, mRNA and protein levels (supplementary Fig. 3A). Flow cytometric analyses of Bcor−/− splenocytes revealed a significant 1.35-fold reduction of total B220+ cells compared with Bcor+/+ mice (35.13 × 106 ± 11.56 vs 47.57 × 106 ± 15.34 cells, respectively; supplementary Fig. 3B) along with an altered distribution of B-cell subpopulations, characterized by a 1.6-fold reduction of the follicular (Fo) population (8.76 × 106 ± 5.78 vs 14.03 × 106 ± 10.74 cells; supplementary Fig. 3C) and a 1.45-fold increase of marginal zone (MZ) B cells (1.58 × 106 ± 1.05 vs 1.05 × 106 ± 0.68 cells, respectively; supplementary Fig. 3D). BM mature recirculating B cells were 1.6-fold reduced in Bcor−/− mice than controls (1.69 × 106 ± 0.69 vs 2.69 × 106 ± 1.47 cells, respectively; supplementary Fig. 3E).
During a 18-months follow up, Bcor−/− mice did not present alterations in PB counts (supplementary Fig. 4A), in the number of CD19+ CD5+ cells (79.83 ± 41.9 vs 72.85 ± 41.31 cells/µL of Bcor+/+; supplementary Fig. 4B) or sign of lymphoid disease leading to death (supplementary Fig. 4C), suggesting that Bcor deficiency is insufficient to drive lymphoid malignancies in mice.
Bcor loss sustains transformation of Eμ-TCL1 leukemia toward a high-grade lymphoid malignancy mimicking human RT
To explore the impact of Bcor loss in CLL, we bred Bcor−/− with Eµ-TCL1 (TCL1) mice (supplementary Fig. 5A). As shown in Fig. 1A, we found a higher number of CD19+ CD5+ B cells in Bcor−/−;TCL1 compared to TCL1 mice at different time-points of age in PB (0–6 months: 582 cells/mL ± 477.8 vs 305.7 cells/mL ± 242.2, respectively; 6–12 months: 9315 cells/mL ± 17250 vs 5324 cells/mL ± 6174, respectively; >12 months: 14665 cells/mL ± 15639 vs 12290 cells/mL ± 16826, respectively), spleen (0–6 months: 8.59 cells × 106 ± 3.42 vs 3.75 cells × 106 ± 2.04; 6–12 months: 47.75 cells × 106 ± 21.70 vs 25.75 cells × 106 ± 18.19; >12 months: 79.68 cells × 106 ± 52.38 vs 34.36 cells × 106 ± 31.93) and BM (0–6 months: 2.02 cells × 106 ± 1.16 vs 0.83 cells × 106 ± 0.75; 6–12 months: 10.21 cells × 106 ± 5.64 vs 4.30 cells × 106 ± 2.69; >12 months: 14.38 cells × 106 ± 18.46 vs 4.74 cells × 106 ± 5.24). To have a proof of clonality of the lymphoproliferations highlighted by flow cytometry, we conducted an analysis of IGHV rearrangements in splenic sorted B cells, revealing monoclonality with a perfect match of 100% to the IGHV germline sequences in Bcor−/−; TCL1 mice (supplementary Fig. 5B). Concomitant Bcor loss and TCL1 overexpression enhanced alterations of splenic Fo and BM recirculating B cells of Bcor−/− mice (supplementary Fig. 5C). At necropsy, Bcor−/−; TCL1 mice showed splenomegaly, due to a diffuse infiltration by medium-large size cells with scattered mitotic figures, completely disrupting organs’ architecture (Fig. 1B). This observation was confirmed in 15 analyzed Bcor−/−; TCL1 mice. These cells were characterized by nuclei with dispersed chromatin, well-defined nucleoli and broad cytoplasm. TCL1 mice presented a prevalence of small B lymphocytes with round nuclei with condensed chromatin and scant nucleoplasms. Survival of Bcor−/−;TCL1 mice (median survival 334 days with a range of 30–361 and 28% of survival at day 365) was significantly reduced compared to Bcor−/−(median survival undefined and 100% of survival at day 365), TCL1 (median survival undefined, range 131–355 and 65% of survival at day 365) and Bcor+/+ mice (median survival undefined and 100% of survival at day 365) (Fig. 1C).
A FC1 analysis of CD19+ CD5+ cells in TCL1 vs Bcor−/−;TCL1 mice at different time-points in (i) PB2 at 0–-6 months (N = 17, 14), 6–12 months (N = 10, 10) and >12 months (N = 11, 9); (ii) spleen at 0-6 months (N = 17, 14), 6–12 months (N = 9, 9) and >12 months (N = 11, 12); (iii) BM3 at 0-6 months (N = 17, 14), 6–12 months (N = 9, 9) and >12 months (N = 11, 12); Mean ± SD4. P values are indicated above each graph according to Mann-Whitney U test. (B) Representative images of H&E5 staining of spleen magnification 20× (N = 2 for each genotype; left panels) and 40× (N = 3 for each genotype; right panels): sections from TCL1 (upper panels) and Bcor−/−;TCL1 (bottom panels) mice. UPlanApo 40×/0.85 NA objective, Olympus BX-51 microscope. (C) OS6 curve of Bcor−/−;TCL1 (N = 25) compared to Bcor+/+ (N = 19), Bcor−/−(N = 25) and TCL1 (N = 20) controls. Survival curves are compared using a Long-rank (Mantel-Cox) test. 1Flow cytometry; 2Peripheral Blood; 3Bone Marrow; 4Standard Deviation; 5Hematoxylin and Eosin; 6Overall survival.
Adoptive transfer of Bcor −/− ; TCL1 leukemic splenocytes induces a more rapidly lethal disease in mice
To promote a disease with accelerated kinetics we performed transplantation of splenocytes from diseased Bcor−/−; TCL1 into C57BL/6 mice (supplementary Fig. 6A). In Bcor−/−; TCL1 transplanted mice we found significant leukocytosis (WBC count: 14.71 ± 13.41 vs 4.18 ± 1.58 cells/µL), anemia (HGB values: 9.73 ± 4.09 vs 13.34 ± 0.64 g/dL) and thrombocytopenia (PLTs count: 209.7 × 103 ± 113.9 vs 395.2 × 103/µL ± 152.7) compared to TCL1 transplanted mice (supplementary Fig. 6B). Bcor−/−; TCL1 transplanted mice had significantly enlarged liver and spleen compared to TCL1 transplanted mice (spleen/body weight ratios: 0.036 ± 0.016 vs 0.013 ± 0.005, respectively; Fig. 2A). Lymph nodes were not significantly involved in the neoplastic phenotype of Bcor−/−; TCL1 mice. Histopathology of spleen and liver of transplanted mice was consistent with the non-transplanted counterpart in terms of infiltration by medium-large size cells mimicking RT (Fig. 2B). Bcor−/−; TCL1 transplanted mice showed higher levels of CD19+ CD5+ cells compared to TCL1 transplanted mice in spleen (275.3 × 106 cells ± 172.3 vs 53.13 × 106 cells ± 97.80, respectively), BM (8.68 cells × 106 ± 8.52 vs 1.10 cells × 106 ± 0.99, respectively) and liver (61.14% ± 29.84 vs 4.85% ± 3.69, respectively) (supplementary Fig. 6C). Clonality of splenic CD19+ CD5+ cells was also confirmed in Bcor−/−; TCL1 transplanted mice (supplementary Fig. 6D). Additionally, neoplastic cells of Bcor−/−;TCL1 transplanted mice presented significant increased Ki-67 levels compared to TCL1 mutant in spleen (19.4% ± 6.44 vs 10.7% ± 3.2, respectively) and liver (47.24% ± 27.05 vs 13.15% ± 4.64, respectively) (supplementary Fig. 6E), suggesting the transformation of TCL1 CLLs towards a high-proliferating disease reminiscent of RT. Survival of Bcor−/−;TCL1 transplanted mice was significantly reduced (median survival of 36, range 30–69; 5.56% of survival at day 75) compared to TCL1 transplanted mice (median survival of 74, range 67–74; 20% of survival at day 75) (Fig. 2C). These data indicated that Bcor−/−; TCL1 mice mimic both clinical and phenotypic features of human RT.
(A) Representative images of spleen (left) and scatter dot plot graph showing spleen/body weight ratios (right) of Bcor−/−; TCL1 (N = 10) compared to TCL1 (N = 8) transplanted mice. P value is indicated above the graph according to Mann-Whitney U test. (B) Representative images of H&E1 staining of spleen (left; magnification 40×) and liver (right; magnification 20×) sections from TCL1 (upper panels) and Bcor−/−;TCL1 (bottom panels) transplanted mice (N = 3, respectively). UPlanApo 40×/0.85 NA objective, Olympus B×-51 microscope. (C) OS2 curves of Bcor−/−;TCL1 (N = 18) compared to TCL1 (N = 9) transplanted mice showing a median survival of 36 (range 30–69; 5.56% of survival at day 75) vs 74 (range 67–74; 20% of survival at day 75) days, respectively. Survival curves were compared using a Long-rank (Mantel-Cox) test. 1Hematoxylin and Eosin; 2Overall survival.
Splenic B cells of Bcor −/− ; TCL1 mice exhibit a transcriptional signature reminiscent of human RT
To dissect the molecular mechanisms underlying CLL-to-RT transition, we performed RNA-seq in transplanted splenic B cells identifying 1338 upregulated and 1674 downregulated genes in Bcor−/−; TCL1 compared to TCL1 mice (N = 3 for all genotypes) (supplementary Table 1). Enrichment analysis showed that Bcor deletion cooperates with TCL1 to activate transcriptional programs sustaining cell proliferation, cell cycle progression and contrasting apoptosis. This signature included the upregulation of cyclins (Ccnd2, Ccnb1, Ccnd3) and cyclin dependent kinases (Cdk1, Cdk6), and the downregulation of cyclin dependent kinase inhibitors (Cdkn1a, Cdkn2b) (Fig. 3A). Furthermore, gene set analysis revealed upregulation of oncogenic and metabolic processes in association with downregulation of immunological pathways, like oxidative phosphorylation (OXPHOS) and DNA Damage Repair (DDR) processes (Fig. 3B). These data indicated that Bcor−/−; TCL1 mice closely mimic RT found in humans at the transcriptional level [1, 14, 15].
A Heatmap of selected DEGs1 (FDR2 q < 0.05) showing upregulation of Ccnd2 (log2FoldChange = 1.887; padj < 0.0001), Ccnb1 (log2FC = 1.505; padj < 0.0001), Ccnd3 (log2FC = 1.546; padj < 0.0029), Cdk1 (log2FC = 1.328; padj = 0.0001), Cdk6 (log2FC = 2.324; padj < 0.0001), and downregulation of Cdkn1a (log2FC = −1.885; padj = 0.0003), Cdkn2b (log2FC = −1.621; padj < 0.0001) in splenic B cells sorted from Bcor−/−;TCL1 compared to TCL1 mice (N = 3, both). B Bar graph showing NES3 of deregulated pathways (“Oxidative Phosphorylation” NES = 1.19; “Glycolysis” NES = 1.06; “Generation of precursor metabolites and energy” NES = 1.15; “DNA Damage Repair” NES = 1.13; “mTORC1 Signalling” NES = 1.26; “Aerobic Respiration” NES = 1.41; “G2M_Checkpoint” NES = 1.66; “E2F Targets” NES = 1.92; “MYC_Targets” NES = 2.07; “Plasma Membrane Organization” NES = −1.64; “BCR Signaling Pathway” NES = −1.63; “Antigen Activates BCR Signaling” NES = −1.73; “Adaptive_Immune_System” NES = −1.89 “Mitochondrial_Translation” NES = 1.42) resulted from pathway enrichment analysis in splenic sorted B cells from Bcor−/−;TCL1 compared to TCL1 mice (N = 3, both) after RNA-seq analysis. 1Differential Expressed Genes; 2False Discovery Rate; 3Normalized Enriched Score.
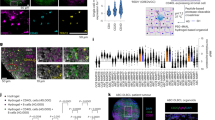
To further validate the role of BCOR deregulations in RT patients, we analysed metadata from three recent studies [1, 14, 15] comparing longitudinal samples from CLL patients who developed RT. At genomic level, whole-genome sequencing and whole-exome sequencing did not identify mutations in the BCOR gene within the bulk tumor populations of RT samples. Conversely, the integration of single-cell DNA sequencing (scDNA-seq), copy number alteration analysis, and RNA-seq data led to the identification of BCOR gene mutations, loss of heterozygosity (LOH) within the BCOR locus, and transcriptomic deregulations of BCOR molecular partners involved in the Polycomb Repressive Complex (PRC) in 6 out of 19 RT patients. Specifically, the DNA sequencing at single-cell level, led to the identification of RT subclones carrying a BCOR mutation, in 1 out of 4 patients analysed (supplementary Fig. 7A). At the structural level, the copy number alteration analysis revealed loss of heterozygosity (LOH) at the locus on the chromosome X, where the BCOR gene is located, in 1 out of 19 RT patients analysed (supplementary Fig. 7A). This structural variation may be relevant to RT oncogenic processes, potentially reducing the tumor suppressor function of BCOR. At transcriptomic level, RNA-seq data from 6 patients with paired CLL and RT samples revealed downregulation of genes encoding several molecular partners of BCOR within the PRC in 6 out of 6 RT patients (supplementary Fig. 7B). The PRC is a multiprotein complex involved in the transcriptional repression of target genes through histone modifications. Proteins within the PRC include BCOR, EZH1, LCOR, CBX4, CBX7, PHC1, PHC3, JARID2 and USP7 (supplementary Fig. 7C). Additionally, only male patients exhibited direct alterations in the BCOR gene, including subclonal mutations or loss of heterozygosity (LOH), while females (n = 3) displayed impaired transcriptomic profiles associated with PRC genes.
Bcor −/− ; TCL1 RT mice exhibit alterations of the T cell compartment
To further explore the biological impact of Bcor loss in B cells of the TCL1 model, we analyzed the tumor microenvironment of Bcor−/−; TCL1 mice. Pathway enrichment analysis performed with Reactome and KEGG databases revealed an impaired immune-microenvironment regulation in transplanted Bcor−/−; TCL1 compared to TCL1 mice. Specifically, Bcor−/−; TCL1 malignant cells showed upregulation of Ccl17 and Ccl22 chemokines that attract regulatory T (Treg) and Th2 cells in tumor microenvironment (TME). We also found overexpression in genes involved in microenvironment-dependent growth of neoplastic B cells, such as Il6ra, Cx3cl1 and Tgfbr1. Conversely, Bcor−/−; TCL1 samples exhibited downregulation of H2-Q2 and Lair1 genes, involved in adaptive immune system and regulating the BCR activation induced by TME interactions (Fig. 4A). These data suggested that RT developed in Bcor−/−; TCL1 mice involved a deregulation of the local immune response. Based on this observation, we analyzed the T-cell compartment within the BM ecosystem of Bcor−/−;TCL1 demonstrating a significant 2-, 2.67- and 1.86- fold increase of CD4+ (1.55 × 106 ± 1.15), CD4 + CD25+ (0.29 × 106 ± 0.08) and CD8+ (0.71 × 106 ± 0.10) cells compared to TCL1 mice (0.39 × 106 ± 0.15 vs 0.11 × 106 ± 0.09 vs 0.35 × 106 ± 0.15, respectively; Fig. 4B). Bcor−/−; TCL1 mice showed a significantly higher expression of PD-1 compared to TCL1 mice in CD4+ (1.26% ± 0.95 vs 0.22% ± 0.11, respectively), CD4 + CD25+ (0.13% ± 0.01 vs 0.04% ± 0.02) and CD8+ cells (2.00% ± 1.14 vs 0.51% ± 0.61; Fig. 4C). Our results indicated that an impaired T cells function was associated with the RT phenotype of Bcor−/−; TCL1 mice with potential pathogenic implications.
A Heatmap of selected DEGs1 (FDR2 < 0.05, p adj3 < 0.05 and log Fold Change threshold of 1) showing upregulation of Ccl17 (log2FC = 5.71; padj < 0.0001), Ccl22 (log2FC = 7.73; padj < 0.0001), Il6ra (log2FC = 1.36; padj = 0.001), Cx3cl1 (log2FC = 6.86; padj < 0.0001), Tgfbr1 (log2FC = 1.04; padj = 0.003) and dowregulation of H2-Q2 (log2FC = −3.50; padj < 0.0001) and Lair1 (log2FC = −5.26; padj < 0.0001) in splenic-sorted B cells from Bcor−/−;TCL1 compared to TCL1 mice (N = 3, both). B FC4 analysis of the BM5 T-cell compartment of TCL1 vs Bcor−/−; TCL1 mice showing altered (i) CD4+ (N = 5), (ii) CD4 + CD25+ (N = 5) and (iii) CD8+ (N = 5) cells frequencies and relative CD4+ (N = 5), CD4 + CD25+ (N = 5) and CD8+ (N = 5) cell number. Mean ± SD6. P values are indicated above each graph according to Mann-Whitney U test. C FC4 analysis of TCL1 vs Bcor−/−; TCL1 mice BM5 samples showing the frequencies of the PD-1 marker gated on CD4+ (N = 5), CD4 + CD25 + (N = 5), and CD8+ (N = 5) cell populations. Mean ± SD6. P values are indicated above each graph according to Mann-Whitney U test. 1Differential Expressed Genes; 2False Discovery Rate; 3Adjusted p-value; 4Flow Cytometry; 5Bone Marrow; 6Standard Deviation.
Neoplastic B cells of Bcor −/− ; TCL1 RT mice exhibit Notch1 signaling activation
NOTCH1 mutation has been described as a driving oncogenic event associated with high-risk CLL and RT onset. NOTCH1 mutation prevents its proteasomal degradation thus enhancing its own activation and transcription of some pro-survival genes [1,2,3,4,5,6,7]. Based on this, we investigated the involvement of Notch1 signaling activation in the disease of Bcor−/−; TCL1 mice. RT-qPCR analyses of splenic B cells revealed a significant increase of the Notch1 mRNA (2.76-fold increase) along with its downstream targets Hes1 (3.36-fold increase) and Myc (2.40-fold increase) (Fig. 5A) while western blot data showed a significant increase of the activated Notch1 (3.11-fold increase) protein levels in Bcor−/−; TCL1 compared to TCL1 mice (Fig. 5B). IHC detection of the Notch1 Intracellular Domain (NICD), the active fragment of the NOTCH1 protein, showed a strong reactivity in nearly 100% RT large B cells infiltrating the spleen of Bcor−/−; TCL1 mice compared to a less intense and more variable positivity in TCL1 sections (Fig. 5C).
A Real-time qPCR analysis of Notch1, Hes1 and Myc mRNA levels in splenic flow sorted B cells from TCL1 and Bcor−/−; TCL1 transplanted mice (N = 5, both). Mean ± SD1. P values are indicated above each graph according to Mann-Whitney U test. B Representative WB2 (left) analysis and relative densitometry graphs (right) of NICD3 in splenic flow sorted B cells from TCL1 and Bcor−/−; TCL1 transplanted mice. Densitometry analyses are normalized to β-Actin and performed using Image Lab software. C Representative IHC4 staining showing NICD3- positive cells on splenic sections from TCL1 and Bcor−/−; TCL1 transplanted mice (N = 3, both). Magnification 40× (UPlanApo 40×/0.85 NA objective, Olympus BX-51 microscope). 1Standard Deviation; 2Western Blot; 3Notch1 Intracellular Domain: 4Immunohystochemestry.
Notch1-inhibition delayed CLL-to-RT transformation by affecting both neoplastic B cells and T cells in Bcor −/− ; TCL1 mice
In order to modulate Notch1 overactivation, we treated RT mice with bepridil that we recently reported as anti-NOTCH1 molecule with anti-leukemic effects in CLL models [16, 17]. In vivo, bepridil treatment progressively reduced PB CD19 + CD5+ cells compared to the untreated mice (day 0: 91.98 cells/mL ± 87.68 vs 163.3 cells/mL ± 172, respectively; day 14: 772.5 cells/mL ± 625 vs 466.9 cells/mL ± 322.7, respectively; day 21: 2220 cells/mL ± 1,021 vs 211.7 cells/mL ± 148.6, respectively; day 28: 3195 cells/mL ± 282 vs 1023 cells/mL ± 440, respectively; Fig. 6A). At sacrifice, we observed a 1.46-fold reduction of spleen size (spleen/body weight ratios of 0.0045 ± 0.0007 of treated mice vs 0.0065 ± 0.0003 of control; Fig. 6B) associated with a reduced infiltration by RT large B cells and a prevalence of small monomorphic cells more similar to the histo-pathology of TCL1 mice (Fig. 6C). IHC analysis of splenic B cells showed a reduction of 80% in NICD levels in treated mice compared to vehicle (Fig. 6D). WB analysis further showed a reduction of NICD (0.29-fold-reduction) leading to the downregulation of its targets Myc (0.54 fold-reduction) and Hes1 (0.38 fold-reduction) at the protein level in treated mice compared to vehicle (Fig. 6E). Furthermore, bepridil treatment negatively modulated the frequency of CD4+ PD-1+ (0.21 × 106 ± 0.01 vs 0.33 × 106 ± 0.06 cells of the control) and CD4+ CD25+ PD-1+ (0.05 × 106 ± 0.012 vs 0.1 × 106 ± 0.013 cells of the control) (Fig. 6F). Our data indicated that the inhibition of Notch1 impacted on the RT phenotype by acting on both neoplastic B cells and TME.
A Curve graph showing the time course differences between vehicle (N = 3) vs bepridil-treated (N = 3) groups of Bcor−/−; TCL1 transplanted mice in number of PB1 CD19+ CD5+ : day 0, 91.98 cells/mL ± 87.68 vs 163.3 cells/mL ± 172, respectively, p = 0.645; day 14, 772.5 cells/mL ± 625 vs 466.9 cells/mL ± 322.7, respectively, p = 0.629; day 21, 2,220 cells/mL ± 1021 vs 211.7 cells/mL ± 148.6, respectively, p = 0.008; day 28, 3195 cells/mL ± 282 vs 1023 cells/mL ± 440, respectively, p = 0.002. Mean ± SD2. P values are calculated for each time-point according to U t-test and indicated above the graph in case of statistical significance. (B) Scatter dot plot graph showing the spleen/body weight ratios of transplanted Bcor−/−; TCL1 mice after treatment with vehicle vs bepridil (0.0065 ± 0.0003 vs 0.0045 ± 0.0007; p = 0.0107; N = 3, both). Mean ± SD2. P value is indicated above the graph according to U t-test. (C) Representative images of H&E3 staining ( ~ 2x digital magnification of the 40X magnification acquisitions) of splenic sections from Bcor−/−; TCL1 transplanted mice treated with vehicle (upper panel, N = 3) or bepridil (bottom panel, N = 3). Images evidence a change in cell-morphology with a prevalence of small monomorphic CLL4 cells after bepridil treatment (bottom panel), reminding the leukemic cells of TCL1 mice (shown in Fig. 1), in place of the large sized cells characterizing the RT5 phenotype of Bcor−/−; TCL1, which are present in the spleen of vehicle mice instead (upper panel). Magnification 40X (UPlanApo 40×/0.85 NA objective, Olympus BX-51 microscope). (D) Representative IHC6 staining on splenic sections from Bcor−/−; TCL1 mice showing the variation in number of NICD7-positive cells between vehicle (upper panel; N = 3) and bepridil-treated (bottom panel; N = 3) groups. Magnification 40X (UPlanApo 40×/0.85 NA objective, Olympus BX-51 microscope). (E) Representative WB8 analysis (top position) and relative densitometry graphs (bottom position) showing the differences in NICD7, MYC and HES1 protein levels in splenic-sorted B cells from a Bcor−/−; TCL1 transplanted mouse after vehicle (N = 3) vs bepridil (N = 3) treatment. Densitometry analyses are normalized to β-Actin and performed using Image Lab software. (F) FC9 analysis of the PD-1 marker in BM10 samples of Bcor−/−;TCL1 transplanted mice treated with bepridil (N = 3) compared to the vehicle (N = 3), gated on CD4+ (46.77% ± 2.67 vs 35.37% ± 0.49, respectively, and absolute number 0.33 × 106 ± 0.06 vs 0.21 × 106 ± 0.01 cells, respectively; left) and CD4 + CD25+ (0.1% ± 0.013 vs 0.05% ± 0.012, respectively, and absolute number 0.07 × 106 ± 0.007 vs 0.04 × 106 ± 0.01 cells, respectively; right) T-cell populations. Mean ± SEM11. P values are indicated above the relative graphs according to U t-test. 1Peripheral Blood; 2Standard Deviation; 3Hematoxylin and Eosin; 4Chronic Lymphocytic Leukemia; 5Richter Transformation; 6Immunohystochemestry; 7Notch1 Intracellular Domain; 8Western Blot; 9Flow Cytometry; 10Bone Marrow; 11Standard Error of the Mean.
Discussion
BCOR disruptive mutations have been detected in several hematological malignancies, including B cell lymphomas and CLL [6, 8]. In these conditions, BCOR mutations often cooperate with other genetic alterations to drive the progression of the disease. Based on the tumor-suppressor role of BCOR [9, 18], we investigated the impact of Bcor loss in the context of CLL evolution through the generation of an in vivo pre-clinical model. Our data indicated that Bcor deficiency triggered the CLL phenotype of Eµ-TCL1 mice toward a more rapidly lethal malignancy characterized by an increased number of proliferating large-sized cells, reminiscent of RT in humans. In a recent study, Nadeu et al. [1] listed BCOR as a driver gene in CLL and B-cell lymphomas, suggesting a potential role for BCOR deregulations in RT onset. Moreover, a comprehensive genome-wide sequencing analysis defined a high frequency of Bcor mutations in murine B-cell lymphomas, demonstrating that genetic disruption of Bcor accelerates the MYC-driven lymphomagenesis in the Eµ-Myc mouse model [18], further supporting the tumor suppressor role of Bcor in RT mice. These observations provide further evidence of the reliability of our Bcor−/−; TCL1 model in recapitulating a RT disease.
Transcriptomic analysis of splenic B cells from Bcor−/−; TCL1 mice showed alterations of key regulators of cell cycle and apoptosis that are considered molecular hallmarks of human RT together with TP53 and NOTCH1 mutations, as well as MYC amplification [1, 7, 14, 15]. This data further corroborates the reliability of our RT model, even at the transcriptional level. In addition, the transcriptional landscape identified in Bcor−/−; TCL1 mice included deregulated metabolic processes and immunological pathways such as OXPHOS upregulation and BCR signalling downregulation. These results were consistent with recent studies showing that RT cells have a notable shift in the transcriptional program that converges into the activation of the OXPHOS pathway and downregulation of BCR signaling, the latter potentially compensated by activating Toll-like, MYC and MAPK pathways [1, 15]. Metabolic alterations have been identified in CLL as a result of disturbed calcium homeostasis [19], which could affect OXPHOS. This is due to the pivotal role of calcium in regulating mitochondrial ATP production.
Neoplastic B cells of Bcor−/−; TCL1 mice also showed enrichment for genes involved in the DDR pathway, described to be a dominant mechanism driving RT transformation. Previous RNA profiling showed that DDR pathway genes are differentially regulated in human RT compared with CLL, and cells harbouring deregulations in the DDR pathways demonstrate high clonal expansion probability [14]. In line with these observations, our data indicated that Bcor−/−; TCL1 mice closely mimic RT found in humans at the transcriptional and phenotypic level. Moreover, the integration of multiomics analysis performed with paired CLL and RT human samples (1) revealed that RT patients presented alterations of BCOR and its molecular partners involved in the PRC. The PRC has been shown to play a key role in normal hematopoiesis, and its dysregulation was closely associated with the pathogenesis of hematological malignancies. Notably, the alterations in BCOR and PRC genes identified through the metadata align with gender-specific expectations in RT patients. Validation in human samples support the relevance of our RT murine model and unleashes the need for future studies on BCOR and its partners within the PRC in human RT pathogenesis.
It has been demonstrated that CLL progression depends on interactions between neoplastic cells and tumor infiltrating lymphocytes within the TME [20, 21]. Our model showed upregulation of chemokines attracting Treg and Th2 cells in the TME and overexpression in genes involved in microenvironment-dependent growth of neoplastic B cells [22,23,24,25]. Conversely, Bcor−/−; TCL1 samples exhibited downregulation of genes involved in adaptive immune system and regulating the BCR activation induced by TME interactions [26, 27]. Based on this observation, we analyzed the T-cell compartment within the BM ecosystem demonstrating the increase of CD4 + , CD4 + CD25+ and CD8+ cells compared to TCL1 mice. Wierz et al. [28] described the expansion of Tregs with an enhanced immunosuppressive phenotype within the TME of a pre-clinical CLL model obtained by performing adoptive transfer of splenocytes from diseased TCL1 into C57BL/6 recipient mice. This model was characterized by activated T cells displaying exhaustion features, such as the overexpression of PD1, LAG3, TIM3, and CTLA4. Interestingly, Tregs from Bcor−/−; TCL1 mice showed a significantly higher expression of PD-1 compared to TCL1 mice. In this regard, it has been demonstrated that resistance to anti-PD1 drugs in vivo was correlated with the molecular silencing of Bcor, indicating a potential role of BCOR in immune checkpoint regulation [29]. Based on the role of the PD-1/PD-L1 pathway in tumor immune evasion [30], our results indicated an impaired T cells function in contributing to the RT phenotype in the context of Bcor deficiency.
A crosstalk between RT cells and TME has been recently described in Eμ-TCL1 mice with Akt [31] overactivation. In this model, Akt orchestrated the development of RT via the induction of Notch1 signaling in B cells, fuelled by microenvironmental T cells. Splenic B cells from Bcor−/−; TCL1 mice revealed enhanced Notch1 signaling demonstrated by a significant increase of both transcript and protein levels of Notch1 and mRNA of its downstream target Hes1, compared to TCL1 mice. These results support a mechanistic association between Bcor function and NOTCH1 signalling in CLL progression toward RT.
Most BCOR-mutated CLL cases are found in co-occurrence with high-risk prognostic factors, such as IGHV unmutated, trisomy 12 and NOTCH1 aberrations [6, 32]. Furthermore, BCOR has been described as a tumor suppressor gene with the ability to transactivate NOTCH in the development of T-ALL in mice [9]. Thus, we assumed that activated Notch1 could be responsible for disease exacerbation in Bcor−/−; TCL1 mice by reprogramming the immunological niche within the CLL-RT continuum.
To investigate the role of NOTCH1 signalling activation in our model, we inhibited NOTCH1 signalling in RT mice using the calcium-channel modulator bepridil [16, 17]. Bepridil has been shown to inhibit NOTCH1 in CLL cells more specifically than gamma secretase inhibitors (GSI), whose effects also impact NOTCH2 [16]. Splenic B cells of treated mice showed reduced levels of active NOTCH1 and of its targets MYC and HES1, compared to vehicle protein levels. After treatment, these mice exhibited a change in cell-morphology with a prevalence of small monomorphic CLL cells which remind the leukemic cells of TCL1 mice, in place of the medium-large sized cells characterizing our mouse model. The driving oncogenic role of NOTCH1 deregulations have been widely explored in CLL [1,2,3,4,5,6,7, 14,15,16,17, 21, 31,32,33,34,35,36,37], pointing out NOTCH1 as a potential mechanism in RT evolution. Several preclinical studies have explored the effects of NOTCH1 inhibition in CLL models, showing promising results in terms of suppressing CLL cell proliferation and inducing apoptosis [16, 17]. In CLL, NOTCH1 activation have been described as an alternative mechanism underlying acquired resistance to ibrutinib, independent of point mutations in the BTK binding sites [17]. In this context, the antileukemic effects of ibrutinib was associated with NOTCH1 downregulation [17], suggesting that NOTCH1 inhibition could overcome the selective pressure of target therapies in resistant clones that may undergo RT progression. Growing evidence have been also provided about the critical role of NOTCH1 in driving resistances to anti-CD20 antibodies and BCL2 inhibitors, providing a rationale for NOTCH1 targeting in high-risk CLL and RT patients [36]. Additionally, the identification of NOTCH1 aberrations in hematopoietic stem cells of CLL patients highlighted the contribution of NOTCH1 in driving the clonal expansion of cancer cells [38], suggesting that NOTCH1 inhibition could potentially lead to CLL regression or contrasting RT progression. Inhibition of NOTCH1 was demonstrated to be effective also in other NOTCH1 dependent tumors [39] further supporting its oncogenic role.
In addition to direct antineoplastic effects, the inhibition of Notch1 induced by bepridil negatively modulated the frequency of CD4 + PD-1+ and CD4 + CD25 + PD-1+ cells in Bcor−/−; TCL1 mice. These results were consistent with previous findings describing the involvement of NOTCH1 in regulating T-cell responses and Tregs differentiation [40] as well as PD-1 expression [41]. These data suggested that NOTCH1 inhibition could represent an effective therapeutic option acting on both the neoplastic clone and the immune deregulations found in RT.
In conclusion our study provides the first evidence of the tumor suppressor role of Bcor coupled with Notch1 deregulation in a pre-clinical model of high-grade lymphoid malignancy mimicking human RT. Our model represents a robust system to study new pathogenic mechanisms and interrogate novel therapies in a challenging disease with an extremely dismal outcome.
Data availability
For original data, please contact paolo.sportoletti@unipg.it.
References
Nadeu F, Royo R, Massoni-Badosa R, Playa-Albinyana H, Garcia-Torre B, Duran-Ferrer M, et al. Detection of early seeding of Richter transformation in chronic lymphocytic leukemia. Nat Med. 2022;28:1662–71.
Puente XS, Pinyol M, Quesada V, Conde L, Ordóñez GR, Villamor N, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–5.
Sportoletti P, Baldoni S, Del Papa B, Aureli P, Dorillo E, Ruggeri L, et al. A revised NOTCH1 mutation frequency still impacts survival while the allele burden predicts early progression in chronic lymphocytic leukemia. Leukemia. 2014;28:436–9.
Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–26.
Rosati E, Baldoni S, De Falco F, Del Papa B, Dorillo E, Rompietti C, et al. NOTCH1 aberrations in chronic lymphocytic leukemia. Front Oncol. 2018;8:229.
Kim JA, Hwang B, Park SN, Huh S, Im K, Choi S, et al. Genomic profile of chronic lymphocytic leukemia in korea identified by targeted sequencing. PLoS One. 2016;11:e0167641.
Fabbri G, Khiabanian H, Holmes AB, Wang J, Messina M, Mullighan CG, et al. Genetic lesions associated with chronic lymphocytic leukemia transformation to Richter syndrome. J Exp Med. 2013;210:2273–88. https://doi.org/10.1084/jem.20131448.
Sportoletti P, Sorcini D, Falini B. BCOR gene alterations in hematologic diseases. Blood. 2021;138:2455–68.
Tanaka T, Nakajima-Takagi Y, Aoyama K, Tara S, Oshima M, Saraya A, et al. Internal deletion of BCOR reveals a tumor suppressor function for BCOR in T lymphocyte malignancies. J Exp Med. 2017;214:2901–13.
Tara S, Isshiki Y, Nakajima-Takagi Y, Oshima M, Aoyama K, Tanaka T, et al. Bcor insufficiency promotes initiation and progression of myelodysplastic syndrome. Blood. 2018;132:2470–83.
Sportoletti P, Sorcini D, Guzman AG, Reyes JM, Stella A, Marra A, et al. Bcor deficiency perturbs erythro-megakaryopoiesis and cooperates with Dnmt3a loss in acute erythroid leukemia onset in mice. Leukemia. 2021;35:1949–63.
Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25:1317–8.
Bichi R, Shinton SA, Martin ES, Koval A, Calin GA, Cesari R, et al. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc Natl Acad Sci USA. 2002;99:6955–60.
Parry EM, Leshchiner I, Guièze R, Johnson C, Tausch E, Parikh SA, et al. Evolutionary history of transformation from chronic lymphocytic leukemia to Richter syndrome. Nat Med. 2023;29:158–69.
Klintman J, Appleby N, Stamatopoulos B, Ridout K, Eyre TA, Robbe P, et al. Genomic and transcriptomic correlates of Richter transformation in chronic lymphocytic leukemia. Blood. 2021;137:2800–16.
Baldoni S, Del Papa B, Dorillo E, Aureli P, De Falco F, Rompietti C, et al. Bepridil exhibits anti-leukemic activity associated with NOTCH1 pathway inhibition in chronic lymphocytic leukemia. Int J Cancer. 2018;143:958–70.
Del Papa B, Baldoni S, Dorillo E, De Falco F, Rompietti C, Cecchini D, et al. Decreased NOTCH1 Activation correlates with response to ibrutinib in chronic lymphocytic leukemia. Clin Cancer Res. 2019;25:7540–53.
Lefebure M, Tothill RW, Kruse E, Hawkins ED, Shortt J, Matthews GM, et al. Genomic characterisation of Eμ-Myc mouse lymphomas identifies Bcor as a Myc co-operative tumour-suppressor gene. Nat Commun. 2017;8:14581.
Adamo FM, Silva Barcelos EC, De Falco F, Dorillo E, Rompietti C, Sorcini D, et al. Therapeutic targeting potential of novel silver nanoparticles coated with anti-CD20 antibody against chronic lymphocytic leukemia. Cancers. 2023;15:3618.
Svanberg R, Janum S, Patten PEM, Ramsay AG, Niemann CU. Targeting the tumor microenvironment in chronic lymphocytic leukemia. Haematologica. 2021;106:2312–24.
De Falco F, Sabatini R, Del Papa B, Falzetti F, Di Ianni M, Sportoletti P, et al. Notch signaling sustains the expression of Mcl-1 and the activity of eIF4E to promote cell survival in CLL. Oncotarget. 2015;6:16559–72.
Takegawa S, Jin Z, Nakayama T, Oyama T, Hieshima K, Nagakubo D, et al. Expression of CCL17 and CCL22 by latent membrane protein 1-positive tumor cells in age-related Epstein-Barr virus-associated B-cell lymphoproliferative disorder. Cancer Sci. 2008;99:296–302.
Hashwah H, Bertram K, Stirm K, Stelling A, Wu CT, Kasser S, et al. The IL-6 signaling complex is a critical driver, negative prognostic factor, and therapeutic target in diffuse large B-cell lymphoma. EMBO Mol Med. 2019;11:e10576.
Ferretti E, Pistoia V, Corcione A. Role of fractalkine/CX3CL1 and its receptor in the pathogenesis of inflammatory and malignant diseases with emphasis on B cell malignancies. Mediat Inflamm. 2014;2014:480941.
Timmins MA, Ringshausen I. Transforming growth factor-beta orchestrates tumour and bystander cells in B-cell non-Hodgkin lymphoma. Cancers. 2022;14:1772.
Fangazio M, Ladewig E, Gomez K, Garcia-Ibanez L, Kumar R, Teruya-Feldstein J, et al. Genetic mechanisms of HLA-I loss and immune escape in diffuse large B cell lymphoma. Proc Natl Acad Sci USA. 2021;118:e2104504118.
Perbellini O, Falisi E, Giaretta I, Boscaro E, Novella E, Facco M, et al. Clinical significance of LAIR1 (CD305) as assessed by flow cytometry in a prospective series of patients with chronic lymphocytic leukemia. Haematologica. 2014;99:881–7.
Wierz M, Pierson S, Guyonnet L, Viry E, Lequeux A, Oudin A, et al. Dual PD1/LAG3 immune checkpoint blockade limits tumor development in a murine model of chronic lymphocytic leukemia. Blood. 2018;131:1617–21.
Wang G, Chow RD, Zhu L, Bai Z, Ye L, Zhang F, et al. CRISPR-GEMM pooled mutagenic screening identifies KMT2D as a major modulator of immune checkpoint blockade. Cancer Discov. 2020;10:1912–33.
McClanahan F, Riches JC, Miller S, Day WP, Kotsiou E, Neuberg D, et al. Mechanisms of PD-L1/PD-1-mediated CD8 T-cell dysfunction in the context of aging-related immune defects in the Eµ-TCL1 CLL mouse model. Blood. 2015;126:212–21.
Kohlhaas V, Blakemore SJ, Al-Maarri M, Nickel N, Pal M, Roth A, et al. Active Akt signaling triggers CLL toward Richter transformation via overactivation of Notch1. Blood. 2021;137:646–60.
Puente XS, Beà S, Valdés-Mas R, Villamor N, Gutiérrez-Abril J, Martín-Subero JI, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526:519–24.
Baldoni S, Del Papa B, De Falco F, Dorillo E, Sorrentino C, Rompietti C, et al. NOTCH1 activation negatively impacts on chronic lymphocytic leukemia outcome and is not correlated to the NOTCH1 and IGHV mutational status. Front Oncol. 2021;11:668573.
Silva Barcelos EC, Rompietti C, Adamo FM, Dorillo E, De Falco F, Del Papa B, et al. NOTCH1-mutated chronic lymphocytic leukemia displays high endoplasmic reticulum stress response with druggable potential. Front Oncol. 2023;13:1218989.
De Falco F, Rompietti C, Sorcini D, Esposito A, Scialdone A, Baldoni S, et al. GSK3β is a critical, druggable component of the network regulating the active NOTCH1 protein and cell viability in CLL. Cell Death Dis. 2022;13:755.
De Falco F, Sabatini R, Falzetti F, Di Ianni M, Sportoletti P, Baldoni S, et al. Constitutive phosphorylation of the active Notch1 intracellular domain in chronic lymphocytic leukemia cells with NOTCH1 mutation. Leukemia. 2015;29:994–8.
Herling, Abedpour CD, Weiss N, Schmitt J, Jachimowicz A, Merkel RD, et al. Clonal dynamics towards the development of venetoclax resistance in chronic lymphocytic leukemia. Nat Commun. 2018;9:727.
Di Ianni M, Baldoni S, Del Papa B, Aureli P, Dorillo E, De Falco F, et al. NOTCH1 Is aberrantly activated in chronic lymphocytic leukemia hematopoietic stem cells. Front Oncol. 2018;8:105.
Marchesini M, Gherli A, Montanaro A, Patrizi L, Sorrentino C, Pagliaro L, et al. Blockade of oncogenic NOTCH1 with the SERCA inhibitor CAD204520 in T cell acute lymphoblastic leukemia. Cell Chem Biol. 2020;27:678–97.
Di Ianni M, Del Papa B, Baldoni S, Di Tommaso A, Fabi B, Rosati E, et al. NOTCH and graft-versus-host disease. Front Immunol. 2018;9:1825.
Mathieu M, Cotta-Grand N, Daudelin JF, Thébault P, Labrecque N. Notch signaling regulates PD-1 expression during CD8(+) T-cell activation. Immunol Cell Biol. 2013;91:82–8.
Acknowledgements
Eμ-TCL1 transgenic mice were a generous gift from Carlo M. Croce (Comprehensive Cancer Center, Ohio State University, Columbus, OH). FMA acknowledge the PhD program at University of Perugia and Post Graduate School in Clinical Pathology and Clinical Biochemistry at University of Salerno.
Funding
The research leading to these results has received funding from AIRC under IG 2018-ID. 21352 and IG 2024 - ID. 30768 project – P.I. Sportoletti Paolo; FIRC- AIRC (3-years fellowship “Filomena Todini” ID.23928 – P.I. Chiara Rompietti; 3-years fellowship “Leonino Fontana and Maria Lionello” ID.26617 – P.I. Francesco Maria Adamo); European Union – FSE-REACT-EU, PON Research and Innovation 2014–2020 DM1062/2021 (P.I. Filomena De Falco).
Author information
Authors and Affiliations
Contributions
PS, CR and FMA conceived the study. PS, CR and FMA wrote the manuscript. CR, DS, FMA and FDF designed the experiments and analyzed the data. CR, FMA, DS, FDF, ED, GM, AS, CG, RA, AE, ECSB and AS performed the experiments. CR, DS, FMA, CG and RA performed cytometric analyses. CR, ED and AS performed molecular analyses. CR, FMA and ECSB executed bioinformatic analyses. CR and GM carried out histological analyses. CR, FMA, AS and DS realized murine manipulation procedures. LS, PG and JB provided animal models. PS, CR, FMA, ER and FDF contributed to funding acquisition. PS and BF performed critical review of the manuscript. All authors approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All methods were performed in accordance with the relevant guidelines and regulations. The Institutional Animal Care and Use Committee approved all the procedures. Experiments were performed with the approval of the Italian Ministry of Health (authorization n°971/2020-PR and n°253/2024-PR).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rompietti, C., Adamo, F.M., Sorcini, D. et al. Bcor loss promotes Richter transformation of chronic lymphocytic leukemia associated with Notch1 activation in mice. Leukemia 39, 1157–1168 (2025). https://doi.org/10.1038/s41375-025-02557-y
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41375-025-02557-y