Abstract
We have recently encountered p53 immunohistochemical (IHC) patterns in human papillomavirus (HPV)-associated carcinomas of the gynecologic tract, which were confused with absent (null) or overexpression TP53 mutational staining. We therefore evaluated p53 and p16 IHC in 25 squamous cell carcinomas (SCC) (16 vulva, 4 Bartholin’s gland, and 5 cervix), 20 endocervical adenocarcinomas (EDAC), 14 high-grade squamous intraepithelial lesions (HSIL), 2 adenocarcinoma in situ (AIS), all of which exhibited morphologic features of HPV. Only cases showing diffuse/strong block-like p16 staining were included for further study. All EDACs underwent TP53 sequencing and HPV in situ hybridization (ISH) was performed in selected cases. p53 IHC staining fell into two main patterns. The most common was designated as “markedly reduced (null-like)” (absence or significantly attenuated staining in >70% of cells), which could be confused with true null mutational pattern. This was present in 14/25 (56%) SCCs, 7/14 (50%) HSILs, and 18/20 (90%) EDACs. The second notable pattern was “mid-epithelial (basal sparing)” (distinct absence of staining in basal cells juxtaposed with strong staining in parabasal cells), seen in 10/25 (40%) SCC, 7/14 (50%) HSIL, and none of the EDACs. There was scattered weak to moderate p53 staining (conventional wild type) in 1/25 (4%) SCC and 2/20 (10%) EDAC. No cases showed strong/diffuse overexpression. One EDAC had a TP53 missense mutation and exhibited “markedly reduced (null-like)” staining. HPV ISH revealed an inverse relationship with p53, cells positive for HPV mRNA were negative for p53. Knowledge of these patterns can help pathologists avoid misinterpreting p53 status in the setting of HPVA cancers.
Similar content being viewed by others
Introduction
Carcinomas in the female lower genital tract develop via high-risk human papillomavirus-associated (HPVA) and non-HPVA-associated (NHPVA) etiologic pathways [1,2,3,4,5]. HPV-independent carcinomas follow a comparatively aggressive clinical course [6,7,8,9,10,11], and a subset of these are known to harbor TP53 mutations [10, 12]. In the cervix, morphologic features of HPVA are often sufficiently discriminatory to permit diagnosis based on H&E [13, 14]. By contrast, vulvar squamous cell carcinomas (SCC) and its precursor lesions show morphologic overlap between HPVA and NHPVA tumors in up to 20% of cases [15,16,17,18]. Accurately delineating between HPVA and NHPVA neoplasia in the lower gynecologic tract is important, as it conveys biologic and prognostic information that can inform clinical decision making [6,7,8,9,10,11], and may be predictive of response to radiotherapy [19].
Immunohistochemistry (IHC) can help clarify pathogenesis and improve diagnostic accuracy. IHC for p16INK4A (p16) is a common diagnostic adjunct, used as a surrogate marker of high-risk HPV viral integration in gynecologic neoplasms. While pathologists are familiar with the interpretation and limitations of p16 staining in HPVA carcinomas [13, 20], far less is known about p53 immunostaining in the setting of HPVA disease. Our current knowledge of p53 IHC interpretation has been primarily informed by advances in our understanding of p53 expression in TP53 mutated tubo-ovarian and endometrial carcinomas [21,22,23], with little information available regarding lower genital tract neoplasms.
Accurate interpretation of p53 in vulvar SCC is a growing priority as there appears to be three different prognostic groups based on HPV and p53 stratification: (1) HPV positive, (2) HPV negative and TP53 mutated, and (3) HPV negative and TP53 wild type [12]. It will therefore become important to accurately interpret p53 IHC in vulvar squamous neoplasia. Tessier-Cloutier et al. and Kortekaas et al. have recently proposed and tested a pattern-based framework to assess p53 IHC in vulvar SCC and in situ lesions [24, 25]. This framework describes four p53 IHC patterns in TP53 mutated SCC: (1) diffuse/parabasal overexpression, (2) basal overexpression, (3) absent (null), and (4) cytoplasmic. There were also two p53 staining patterns present in tumors without a TP53 mutation: (1) scattered and (2) Mid-epithelial (absent staining in basal cells and strong staining in parabasal cells). In the original series, only small numbers (n = 6 [4 in Tessier-Cloutier et al. and 2 in Kortekaas et al.]) of p16 positive HPVA SCC were evaluated. Our current study explores a larger series of HPVA tumors and expands on the p53 IHC patterns seen in HPVA neoplasia.
Further justification for this study comes from observations made in our referral practice. We recently reviewed a case initially diagnosed as uterine serous carcinoma on an endometrial biopsy specimen showing a malignant glandular proliferation and “block-like” p16 IHC staining. p53 IHC was initially interpreted as abnormal null staining. The subsequent hysterectomy specimen showed endocervical adenocarcinoma (EDAC) with adjacent adenocarcinoma in situ (AIS) involving the cervix, with classic morphologic features of HPV. HPV in situ hybridization (ISH) was subsequently performed and showed strong signal in >99% of tumor cells. Taken together, these findings highlight the need to expand our understanding of p53 IHC staining in HPVA neoplasia in order to avoid misinterpretation.
Materials and methods
Case selection
For the invasive vulvar and cervical SCC, cases were identified in our institutional archives by searching for excision or hysterectomy specimens with the key words “usual-type vulvar dysplasia (uVIN),” “high-grade squamous intraepithelial lesions (HSIL),” or “p16 positive,” diagnosed between 2015 and 2019. HPVA EDACs from a separate study were also included [26]. Cases treated with neoadjuvant radiation or chemotherapy were excluded. All cases were reviewed by a gynecologic pathologist (LH) and a pathology resident (ET) for consensus diagnosis of HPVA prior to inclusion.
Immunohistochemistry
IHC was performed on 4-μm thick formalin fixed paraffin embedded (FFPE) tissue sections. p53 was performed on whole tissue sections and p16 on whole tissue sections or tissue microarray. IHC staining was performed using methods described previously using the Dako Omnis automated IHC instrument together with the Dako EnVision™ FLEX + detection system (Dako, Carpinteria, CA) [24, 27]. For p53 immunostaining, the D07 mouse anti-p53 monoclonal antibody was used at a dilution of 1:500 (Dako), and for p16, the CINtec E6H4 mouse monoclonal antibody was used at a 1:5 dilution (Roche-Mtm-Laboratories, Heidelberg, Germany). IHC staining was evaluated by a pathology resident (ET) and two gynecologic pathologists (LH and JH). In situ and invasive carcinomas were assessed separately. p16 IHC was considered positive when confluent nuclear and cytoplasmic staining was observed, in accordance with published guidelines [13, 20]. Only cases that demonstrated diffuse/strong “block-like” p16 staining were included for further study. p16 positivity, in addition to the morphologic features, provided confidence these were HPV-associated neoplasms for the purposes of our study. For p53, the percentage of tumor cells showing positive nuclear staining and patterns of staining were documented for all cases.
TP53 sequencing
Our EDACs were selected from another ongoing/unpublished study, where sequencing of TP53 exons 4–9 was already performed with a technique which has been described previously [28, 29]. FFPE tumor tissue samples were extracted using the Qiagen Gene Read DNA FFPE kit per manufacturer’s instructions. DNA was amplified using the Contextual Genomics FIND IT v3.4. assay, a targeted NGS gene panel, followed by sequencing on the Illumina Miseq platform. Results were then analysed through the proprietary Contextual Genomics Analytics pipeline. Acceptable variants had a minimum read depth of 500, an allelic ratio ≥ 5%, a base quality score ≥ 30, and a probability score ≥ 0.90 for single-nucleotide changes or a quality score of ≥1000 for insertion/deletion events.
HPV in situ hybridization
Due to cost restrictions, selected cases which showed varied p53 IHC patterns were sent for HPV ISH. High-risk HPV mRNA expression was tested using Advanced Cell Diagnostics RNA-scope (Newark, CA, USA) and the high-risk HPV probe kit (includes 18 HPV types: 16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 73, and 82), per manufacturer’s instructions.
Results
Cohort description
The cases comprised 25 SCCs (vulva [n = 16], uterine cervix [n = 5], and Bartholin gland [n = 4]), as well as 20 EDACs. Associated in situ neoplasias consisted of vulvar HSIL (n = 10), Bartholin gland HSIL (n = 1), cervical HSIL (n = 3), and EDAC in situ (AIS) (n = 2). All cases were diffusely and strongly p16 positive. One case originally diagnosed as a vulvar SCC with basaloid features was p16 positive, but upon review, it was an infiltrative basal cell carcinoma (subsequently found to be EMA negative and positive for BCL2 and BerEP4 by IHC) and was excluded.
Immunostaining staining patterns
The patterns of p53 staining observed are summarized in Tables 1 and 2 and Figs. 1 and 2, and an expanded results table is available in Supplementary Table 1. The majority of vulvar SCC had more than one pattern of p53 staining.
Null-like patterns
The first notable pattern was “markedly reduced (null-like)”. p53 staining was absent or substantially attenuated in 14/25 (56%) invasive SCCs and 18/20 (90%) EDACs. We found that the majority of tumor cells (at least 70%) showed absence of p53 staining, but with scattered individual tumor cells showing strong staining. In 6/25 (24%) SCC, there were some tumor cells which showed very weak (“blush”) staining. The conspicuous absence or attenuated p53 staining could easily be misinterpreted to represent a TP53 null mutation pattern and thus we designated this pattern of staining as “null-like”.
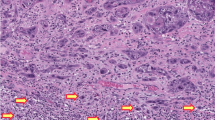
Upon further review, we observed 3 sub-patterns of this markedly reduced (null-like) staining: (a) single cells, (b) clustered, and (c) peripheral (Fig. 3). In the markedly reduced (null-like)—single cells staining pattern, the majority of tumor cells showed complete absence of staining admixed with occasional single tumor cells or lymphocytes with strong positive staining. The markedly reduced (null-like)—clustered pattern was similar, with the majority of tumor cells lacking staining, except for the presence of clusters of tumor cells with strong nuclear staining. In the markedly reduced (null-like)—peripheral pattern, the majority of tumor cells showed complete absence of staining and positive nuclear staining was restricted to tumor cells at the periphery of the infiltrative tumor nests.
a Shows an invasive squamous cell carcinoma (SCC) involving the Bartholin gland (case 6) and “single cells” staining. b Shows a mucinous endocervical carcinoma (EDAC) with scattered “single cells” staining (case 46). c case 3, and d case 20, show scanty “clusters” of cells staining in invasive vulvar SCCs. e show “peripheral” staining in an EDAC (case 37).
Mid-epithelial patterns
The second notable pattern was the mid-epithelial (basal sparing) pattern. This was only seen in the SCCs (9/25, 36%) and not in any of the EDACs.
In the mid-epithelial pattern, there was conspicuous absence of staining or reduced staining of the basal layer (<5% of cells staining) of the invasive squamous nests, accompanied by strong staining in the parabasal cells. We further subdivided the mid-epithelial pattern into (a) classic and (b) central patterns (Fig. 4). In the mid-epithelial—classic pattern, there was absence of staining only in the single basal layer, with strong p53 staining in the parabasal layers. In the mid-epithelial—central pattern, there was absence of staining in the basal layer as well as variable proportions or thicknesses of the parabasal layers. Very often, the only tumor cells showing positive p53 staining were the central portions of the invasive squamous nests. One SCC was initially scored as a mixed pattern of mid-epithelial and aberrant “overexpression” (strong staining in >80% of tumor cells). Upon re-review, this tumor had small tumor cords or single cells at its infiltrative tumor front. In this area, the basal cells were attenuated and the p53 pattern was initially misinterpreted as overexpression. Upon identifying the basal cells, we noted p53 staining was absent in those cells. In adjacent areas of the tumor, the more classic mid-epithelial pattern was appreciated. This case was reclassified as a mid-epithelial pattern only.
a Invasive squamous cell carcinoma (SCC) of cervix (case 19) and b vulvar SCC (case 16) showing “classic” mid-epithelial (basal sparing) staining. c, d Both show the “central” mid-epithelial staining in invasive SCCs of cervix (Cases 21 and 23, respectively) where the basal layer and portion of the parabasal layers show absent staining. e A mixture of central and classic mid-epithelial staining patterns seen in a case of invasive vulvar SCC (case 19). f p53 staining mimic abnormal overexpression in highly infiltrative invasive vulvar SCC, when the basal layer is very attenuated (case 17).
Scattered (wild type) pattern
One (4%) SCC and 2 EDAC (10%) showed scattered (wild type) staining.
In situ neoplasias
The patterns of staining were very similar in HSIL and AIS. In HSIL, we observed markedly reduced (null-like) staining in 7/14 (50.0%) and mid-epithelial staining in 7/14 (50%) cases. In the two AIS, both showed markedly reduced (null-like) staining.
HPV in situ hybridization
HPV RNA ISH was performed on selected cases, the results are demonstrated in Fig. 5. For many of the cases, an inverse association between p53 staining and HPV mRNA ISH signal was observed. Tumor cells which showed absence of p53 IHC staining, appeared to correlate with the cells which were positive by HPV ISH. This inverse pattern was only noted in some areas (not all areas) of the tumors.
Case 46 was an endocervical adenocarcinoma showing a markedly reduced (null-like) p53 staining and b HPV integration in nearly every tumor cell. Case 19 was a keratinizing squamous cell carcinoma of the vulva arising in a background of HSIL. In the invasive component c mid-epithelial classic p53 staining and d HPV viral integration confined to the single basal layer was observed; elsewhere e mid-epithelial central p53 staining and f HPV integration in the basal layer and variable parabasal layers was noted. In the associated HSIL, g mid-epithelial classic p53 staining was observed and h HPV integration was confined to the single basal layer.
TP53 sequencing
Of the 20 EDACs, only 2 had TP53 mutations. One case had a missense mutation (c.530C > T, p.Pro177Leu). This case had markedly reduced (null-like) p53 IHC staining with peripheral accentuation. The peripheral-most rim of tumor cells strongly expressed p53, and the central tumor cells were negative. This odd pattern was seen only focally and subtly in one other EDAC, which did not bear a TP53 mutation.
A different case had a nonsense TP53 mutation (c.991C > T, p.Q331*). It showed scattered (wild type) p53 IHC staining. On re-review, the heterogeneity of staining intensity often seen in a typical p53 wild-type tumor was not as apparent.
Discussion
We assessed p53 IHC staining in HPVA in situ and invasive carcinomas of the vulva and cervix. We categorized p53 staining patterns and found two distinct patterns of IHC staining that could mimic that observed in TP53 mutated cancers. The p53 IHC patterns, markedly reduced (null-like) (mimicker of a TP53 null mutation) and mid-epithelial (mimicker of p53 overexpression/TP53 missense mutation), altogether were observed in 42/45 (93%) of HPV-associated cancers and all HPV-associated in situ lesions (14/14 HSIL and 2/2 AIS) included in this series.
The most common p53 IHC pattern seen in HPV-associated SCC and EDACs was the markedly reduced (null-like) pattern. It was observed in 14/25 (56%) SCC and 18/20 (90%) EDAC. In this pattern, there was absent or significantly reduced p53 staining, often juxtaposed to strong staining in rare tumor cells, that could be mistaken for true p53 null mutation-type staining, to the unaware pathologist. In vitro studies have shown that cells infected with HPV have reduced half-life of wild-type p53 [30, 31]. This occurs through HPV viral oncoprotein E6 forming a complex with E6-associated-protein (E6-AP), which then targets p53 for degradation via the ubiquitin mediated proteolytic pathway [32]. This abrogates the growth-arrest and apoptosis induction roles conferred by p53, resulting in oncogenesis [33]. The absent or attenuated p53 staining seen by IHC is therefore a consequence of HPV mediated p53 degradation. This is also supported by our HPV ISH results, which show an inverse relationship between cells positive for HPV mRNA ISH and negative p53 staining.
To the best of our knowledge, Pilotti et al. were the first to report an inverse relationship between p53 IHC staining and HPV viral integration in gynecologic specimens [34]. In their 1995 paper, 13 HPV-associated VIN and invasive SCCs were regarded as p53 negative, either due to complete absence of p53 staining or due to staining present at levels of <10% [35]. Similarly, Kohlberger et al. noted absence of p53 expression in basaloid/warty type VIN lesions [36]. These studies do not explicitly highlight the possible confusion with null IHC staining, as we do here. Their published images of IHC stained vulvar HSILs, however, do depict staining in keeping with what we have designated as markedly reduced (null-like) staining patterns.
Zielinski et al. evaluated p53 staining using percentage cut-offs in a series of cervical adenocarcinomas [37]. They found that 17/20 (85%) of HPVA and only 1/5 (20%) of NHPVA EDAC had absent (0%) p53 staining i.e. a pronounced tendency toward absence of p53 staining in the HPVA group. In the recent International Endocervical Criteria and Classification (IECC), 5.4% HPVA EDACs showed aberrant p53 staining (overexpression and null expression combined). In that study, IHC was evaluated on tissue microarray, and it is possible that a subset of the cases categorized as null actually could be the markedly reduced (“null-like”) patterns if done on whole sections [14].
The mid-epithelial (basal sparing) pattern was only seen in HPV-associated SCC (10/25 [40%] SCC and 7/14 [50%] HSIL), and not in any of the EDAC. In this pattern, there was notable absence of staining in the basal layer of HSIL or the invasive squamous nests, juxtaposed to strong staining in the parabasal layers. If a pathologist was to miss the absence of staining in the basal layer, this pattern could be misinterpreted at an “overexpression” or TP53 missense mutational pattern. Finding the basal layer in very infiltrative SCC, comprised of single cells or small infiltrative tumor nests, can be a challenge. This mid-epithelial (basal sparing) p53 staining pattern has recently been described by Jeffreys et al. in uVIN/HSIL; basal sparing was observed in many of their cases and is depicted in their Figs. 1b and 3e. Watkins et al. in also noted basal sparing of p53 staining in HSIL with concomitant lichen simplex chronicus (n = 9), depicted in their Fig. 2e. Tessier-Cloutier et al. also noted the mid-epithelial pattern in three HSIL and three HPVA SCC [24].
The pathogenic mechanism accounting for the mid-epithelial (basal sparing) pattern is more difficult to explain. In the squamous lesions of the lower anogenital tract, HPV requires entry into the mitotically active cells of the basal layer, often facilitated through microtrauma of the mucosa in the lower anogenital region [33]. As the basal layer is typically the first site of HPV viral integration, it is therefore intuitive that there is absence of p53 IHC staining in the basal layer. The strong staining in the parabasal layers, may be a compensatory upregulation of wild-type p53 in response to HPV infection, but the precise mechanism remains to be fully explicated.
We had two EDAC that did have an underlying TP53 mutation (one missense and one nonsense mutation, respectively). The case with a TP53 missense mutation would be anticipated to show a strong overexpression pattern, if based in the ovary or endometrium. However, this tumor showed substantial reduction in p53 staining. Although we only have one case to illustrate this, it is probable that mutant p53 protein is also degraded by HPV. Few studies have investigated this possibility. Scheffner et al. did find that a subset of mutant p53 proteins were still able to bind HPV-16 E6 and were targeted for degradation [38]. A larger series of cases is needed to substantiate these findings.
We want to emphasize several messages in this study. The awareness of p53 patterns will avoid confusion with true TP53 IHC mutational patterns in the HPVA tumors. The presence of HPVA related features by histology and interpretation of p53 in conjunction with p16 IHC (as a surrogate for high-risk HPV infection), can help to avoid misinterpretation. We would like to emphasize that the names of the various sub-patterns (“single cells,” “clustered,” and “peripheral” in the markedly reduced category, as well as “classic” and “central” in the mid-epithelial category) are presented only to illustrate the range of patterns that a pathologist may encounter in HPVA carcinomas. The precise terminology is not meant to be used in diagnostic pathology reports for clinicians. In addition, all these patterns fall under the broader umbrella term of “p53 wild type”. In this study, we highlight the range of wild-type p53 patterns that can be seen in HPVA neoplasia. These p53 wild-type patterns look different than the p53 wild-type patterns in non-HPV tumors (that pathologists are much more familiar with).
There are many scenarios where recognition of these p53 patterns in HPVA neoplasia will be helpful. A p53 overexpression pattern (strong and diffuse) should alert the pathologist to consider a non-HPV entity, because a HPVA tumor would be predicted to have reduced p53 staining. For example, the vulvar “basaloid SCC” that was excluded from this study was re-reviewed due to p53 overexpression and this ended up being re-classified as an infiltrative basal cell carcinoma. In certain cases, p53 can serve as an ancillary tool to predict HPV infection, especially because HPV ISH is labor intensive and expensive. In the differential diagnosis of HSIL with superimposed lichen sclerosus versus differentiated vulvar intraepithelial neoplasia (dVIN), a p53 mid-epithelial (basal sparing) pattern will support the former diagnosis without the need for expensive HPV ISH testing. We recently encountered a case of vulvar SCC with basaloid features and HSIL. p16 IHC was performed by a learner and, to our surprise, was negative. p53 IHC was subsequently performed, and showed mid-epithelial (basal sparing) and markedly reduced (null-like) staining, consistent with an HPVA tumor, with the implication that this was an example of an HPVA SCC with loss of heterozygosity (allelic loss of p16) as a secondary event. HPV ISH was subsequently performed and it was positive. Lastly, the markedly reduced (null-like) single cell pattern will be the most difficult to distinguish from true TP53 null-type staining, and this was most apparent in the EDACs. Searching for other HPVA p53 patterns in the same tumor and finding a weak blush of p53 staining in the tumor cells will support a HPVA process, but in some cases, distinction may not be possible without molecular confirmation or HPV ISH.
Finally, this paper highlights the utmost importance of “pattern” versus “positive” in diagnostic pathology. Similar to the situation with p16, positive staining for p53 alone is insufficient to serve as a surrogate marker of TP53 mutation. Consideration of p53 intensity, proportion and pattern all need to be considered. This study shows that p53 staining patterns in HPV-associated carcinomas can mimic those seen in TP53 mutated cancers. Awareness of these patterns can help avoid misclassification of carcinomas in the female lower genital tract and, when present, alert the pathologist to a diagnosis of an HPV-associated carcinoma.
References
Kurman R, Carcangiu M, Herrington C, Young R. WHO classification of tumours of female reproductive organs. 4th ed. Lyon: International Agency for Research on Cancer; 2014.
Toki T, Kurman R, Park J, Kessis T, Daniel R, Shah K. Probable nonpapillomavirus etiology of squamous cell carcinoma of the vulva in older women: a clinicopathologic study using in situ hybridization and polymerase chain reaction. Obstet Gynecol Surv. 1991;46:561–3.
Young R, Clement P. Endocervical adenocarcinoma and its variants: their morphology and differential diagnosis. Histopathology. 2002;41:185–207.
Hoang L, Park K, Soslow R, Murali R. Squamous precursor lesions of the vulva: current classification and diagnostic challenges. Pathology. 2016;48:291–302.
Stolnicu S, Barsan I, Hoang L, Patel P, Terinte C, Pesci A, et al. International Endocervical Adenocarcinoma Criteria and Classification (IECC): a new pathogenetic classification for invasive adenocarcinomas of the endocervix. Am J Surg Pathol. 2018;42:214–26.
Lee LJ, Howitt B, Catalano P, Tanaka C, Murphy R, Cimbak N, et al. Prognostic importance of human papillomavirus (HPV) and p16 positivity in squamous cell carcinoma of the vulva treated with radiotherapy. Gynecol Oncol. 2016;142:293–8.
Hay CM, Lachance JA, Lucas FL, Smith KA, Jones MA. Biomarkers p16, human papillomavirus and p53 predict recurrence and survival in early stage squamous cell carcinoma of the vulva. J Low Genit Trac Dis. 2016;20:252–6.
McAlpine JN, Leung SCY, Cheng A, Miller D, Talhouk A, Gilks CB, et al. Human papillomavirus (HPV)-independent vulvar squamous cell carcinoma has a worse prognosis than HPV-associated disease: a retrospective cohort study. Histopathology. 2017;71:238–46.
Rodriguez-Carunchio L, Soveral I, Steenbergen R, Torne A, Martinez S, Fuste P, et al. HPV-negative carcinoma of the uterine cervix: a distinct type of cervical cancer with poor prognosis. Gynaecol Oncol. 2015;122:119–27.
Karamurzin YS, Kiyokawa T, Parkash V, Jotwani AR, Patel P, Pike MC, et al. Gastric-type endocervical adenocarcinoma: an aggressive tumor with unusual metastatic patterns and poor prognosis. Am J Surg Pathol. 2015;39:1449–57.
Stolnicu S, Hoang L, Chiu D, Hanko-Bauer O, Terinte C, Pesci A, et al. Clinical outcomes of HPV-associated and unassociated endocervical adenocarcinomas categorized by the International Endocervical Adenocarcinoma Criteria and Classification (IECC). Am J Surg Pathol. 2019;43:466–74.
Nooij L, Ter Haar N, Ruano D, Rakislova N, Van Wezel T, Smit V, et al. Genomic characterization of vulvar (pre)cancers identifies distinct molecular subtypes with prognostic significance. Clin Cancer Res. 2017;23:6781–9.
Darragh TM, Colgan TJ, Thomas Cox J, Heller DS, Henry MR, Luff RD, et al. The lower anogenital squamous terminology standardization project for HPV-associated lesions: background and consensus recommendations from the college of American pathologists and the American society for colposcopy and cervical pathology. Int J Gynecol Pathol. 2013;32:76–115.
Stolnicu S, Barsan I, Hoang L, Patel P, Chiriboga L, Terinte C, et al. Diagnostic algorithmic proposal based on comprehensive immunohistochemical evaluation of 297 invasive endocervical adenocarcinomas. Am J Surg Pathol. 2018;42:989–1000.
Ordi J, Alejo M, Fusté V, Lloveras B, Del Pino M, Alonso I, et al. HPV-negative vulvar intraepithelial neoplasia (VIN) with basaloid histologic pattern: an unrecognized variant of simplex (differentiated) VIN. Am J Surg Pathol. 2009;33:1659–65.
Santos M, Landolfi S, Olivella A, Lloveras B, Klaustermeier J, Suárez H, et al. p16 overexpression identifies HPV-positive vulvar squamous cell carcinomas. Am J Surg Pathol. 2006;30:1347–56.
Cheng A, Karnezis A, Jordan S, Singh N, McAlpine J, Gilks B. p16 Immunostaining allows for accurate subclassification of vulvar squamous cell carcinoma into HPV-associated and HPV-independent cases. Int J Gynecol Pathol. 2016;35:385–93.
Dong F, Kojiro S, Borger D, Growdon W, Oliva E. Squamous cell carcinoma of the vulva: a subclassification of 97 Cases by clinicopathologic, immunohistochemical, and molecular features (p16, p53, and EGFR). Am J Surg Pathol. 2015;39:1045–53.
Proctor L, Hoang L, Moore J, Thompson E, Leung S, Natesan D, et al. Association of human papilloma virus status and response to radiotherapy in vulvar squamous cell carcinoma. Int J Gynecol Pathol. 2020;30:100–6.
Singh AN, Gilks CB, Wong RW, Mccluggage WG, Simon C. Interpretation of p16 immunohistochemistry in lower anogenital tract neoplasia; 2018. https://www.thebagp.org/resources/.
Yemelyanova A, Vang R, Kshirsagar M, Lu D, Marks MA, Shih IM, et al. Immunohistochemical staining patterns of p53 can serve as a surrogate marker for TP53 mutations in ovarian carcinoma: an immunohistochemical and nucleotide sequencing analysis. Mod Pathol. 2011;24:1248–53.
Köbel M, Piskorz AM, Lee S, Lui S, LePage C, Marass F, et al. Optimized p53 immunohistochemistry is an accurate predictor of TP53 mutation in ovarian carcinoma. J Pathol Clin Res. 2016;2:247–58.
Köbel M, Ronnett BM, Singh N, Soslow RA, Gilks CB, McCluggage WG. Interpretation of P53 immunohistochemistry in endometrial carcinomas: toward increased reproducibility. Int J Gynecol Pathol. 2019;38:S123–31.
Tessier-Cloutier B, Kortekaas K, Thompson E, Pors J, Chen J, Ho J, et al. Major p53 immunohistochemical patterns in in-situ and invasive squamous cell carcinomas of the vulva and correlation with TP53 mutation status. Mod Pathol. 2019. in press.
Kortekaas K, Solleveld-Westerink N, Tessier-Cloutier B, Rutten T, Van Poelgeest M, Gilks B. et al. Performance of the pattern based interpretation of p53 immunohistochemistry as a surrogate for TP53 mutations in vulvar squamous cell carcinoma (abs 1134). In: USCAP 2020 abstracts: index of abstract authors. Mod Pathol. 2020;33:1925. https://doi.org/10.1038/s41379-020-0486-3.
Ren H, Almadani N, Pors J, Ho J, Chow C, Park KJ, et al. International Endocervical Adenocarcinoma Criteria and Classification (IECC): An Independent Cohort with Clinical and Molecular Relevance (abs 1542). In: USCAP 2020 abstracts: index of abstract authors. Mod Pathol. 2020;33:1935. https://doi.org/10.1038/s41379-020-0486-3.
Aird JJ, Steel MJ, Chow C, Ho J, Wolber R, Gilks CB, et al. Should you repeat mismatch repair testing in tumour recurrences? An evaluation of repeat mismatch repair testing by immunohistochemistry in recurrent tumours of the gastrointestinal and gynaecological tracts. Histopathology. 2020. https://onlinelibrary.wiley.com/doi/abs/10.1111/his.14026. in press.
Pors J, Ho J, Prentice L, Thompson E, Cochrane D, Gibbard E, et al. c-KIT Analysis and Targeted Molecular Sequencing of Mesonephric Carcinomas of the Female Genital Tract. Am J Surg Pathol. 2020. in press. https://journals.lww.com/ajsp/Abstract/publishahead/c_KIT_Analysis_and_Targeted_Molecular_Sequencing.97526.aspx.
Prentice LM, Miller RR, Knaggs J, Mazloomian A, Hernandez RA, Franchini P, et al. Formalin fixation increases deamination mutation signature but should not lead to false positive mutations in clinical practice. PLoS ONE. 2018;13:e0196434.
Hubbert NL, Sedman SA, Schiller JT. Human papillomavirus type 16 E6 increases the degradation rate of p53 in human keratinocytes. J Virol. 1992;66:6237–41.
Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley’ PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–36.
Scheffner M, Huibregtse J, Vierstra R, Howley P. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505.
Longworth MS, Laimins LA. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol Mol Biol Rev. 2004;68:362–72.
Pilotti S, Donghi R, D’Amato L, Giarola M, Longoni A, Della Torre G, et al. Papillomavirus, p53 alteration and primary carcinoma of the vulva. Lett Eur J Cancer. 1993;29A:924–5.
Pilotti S, D’Amato L, Della Torre G, Donghi R, Longoni A, Giarola M, et al. Papillomavirus, p53 alteration, and primary carcinoma of the vulva. Diagn Mol Pathol. 1995;4:239–48.
Kohlberger PD, Kirnbauer R, Bancher D, Gitsch G, Reinthaller A, Leodolter S, et al. Absence of p53 protein overexpression in precancerous lesions of the vulva. Cancer. 1998;82:323–7.
Zielinski GD, Snijders PJF, Rozendaal L, Daalmeijer NF, Risse EKJ, Voorhorst FJ, et al. The presence of high-risk HPV combined with specific p53 and p16INK4a expression patterns points to high-risk HPV as the main causative agent for adenocarcinoma in situ and adenocarcinoma of the cervix. J Pathol. 2003;201:535–43.
Scheffner M, Takahashi T, Huibregtse JM, Minna JD, Howley PM. Interaction of the human papillomavirus type 16 E6 oncoprotein with wild-type and mutant human p53 proteins. J Virol. 1992;66:5100–5.
Acknowledgements
This study has supported by the Carraresi Foundation, Sumiko Kobayashi Marks Memorial Fund, OVCARE and the UBC & VGH Hospital Foundations.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Thompson, E.F., Chen, J., Huvila, J. et al. p53 Immunohistochemical patterns in HPV-related neoplasms of the female lower genital tract can be mistaken for TP53 null or missense mutational patterns. Mod Pathol 33, 1649–1659 (2020). https://doi.org/10.1038/s41379-020-0527-y
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41379-020-0527-y
This article is cited by
-
Clinical and molecular analysis of JCPyV and BKPyV infection and associated risk of urothelial carcinoma development in the upper tract
Virology Journal (2025)
-
Immunoreactivity of LMO7 and other molecular markers as potential prognostic factors in oropharyngeal squamous cell carcinoma
BMC Oral Health (2024)
-
Simultaneous p53 and p16 Immunostaining for Molecular Subclassification of Head and Neck Squamous Cell Carcinomas
Head and Neck Pathology (2024)