Abstract
Invasive lobular carcinoma with extracellular mucin (ILCEM) is a rare histologic subtype of breast cancer. Little is known about the pathologic or genomic signatures that distinguish ILCEM from classic invasive lobular carcinoma (ILC) or mucinous carcinoma. We studied 17 breast cancers with lobular morphology and extracellular mucin. Thirteen tumors with sufficient tissue for DNA extraction were analyzed by a next generation sequencing (NGS) assay that interrogates 447 genes for mutations and copy number variations (CNVs). Median patient age was 66 yrs (range: 31–77 yrs). Sixteen patients presented with masses, 7 of which were >2 cm. Seven patients had lymph node metastases. The cases of ILCEM were moderately (n = 13) or poorly differentiated (n = 4), frequently exhibiting variant morphology that has not been previously described or emphasized, including grade 3 nuclei (n = 11), diffuse signet ring cells (n = 10), solid growth (n = 4), tumor necrosis (n = 3) or apocrine features (n = 2). All tumors showed absent or reduced membranous E-cadherin expression. Concurrent lobular carcinoma in situ (LCIS) was seen in 11/17 cases, 1 of which was a striking example of signet ring cell LCIS with extracellular mucin. Receptor profiles were ER+/HER2− (n = 15) and ER+/HER2+ (n = 2). With a median follow-up of 83.5 months (range: 3–171 months) in 12 patients with available information, 8 patients had recurrences resulting in 4 cancer-related deaths. The most common CNVs were 16q loss (n = 11) and 1q gain (n = 9). CDH1 gene-level alterations were detected in all but one case, including frameshift (n = 7), nonsense (n = 2), and donor splice site (n = 1) mutations and indels (n = 2). Recurrent mutations were also seen in PIK3CA (n = 3), POLQ (n = 3), TP53 (n = 3), ERBB3 (n = 3), ERBB2 (n = 2), and RUNX1 (n = 2). Genes with recurrent amplifications included GATA3 (n = 4), FOXA1 (n = 3), CCND1 (n = 2). Our data highlights ILCEM as a distinct variant of ILC that often presents with higher-grade and variant morphologic features and is associated with an aggressive clinical course. NGS data support an overall lobular-type molecular profile and reveal potentially targetable alterations in a subset of cases with recurrence.
Similar content being viewed by others
Introduction
Invasive lobular carcinoma (ILC) is a special histologic type of invasive breast cancer which accounts for 5–15% of breast cancer diagnoses1,2,3,4. As first described by Foote and Stewart5, classic ILC has a distinctive discohesive morphology characterized by uniform cells with round nuclei and inconspicuous nucleoli arranged as single cells and single file arrays. A hallmark of invasive and in situ forms of lobular neoplasia is the loss of expression or dysfunction of E-cadherin, a transmembrane protein of the adherens-type junction responsible for cell-cell cohesion, encoded by the CDH1 gene on the short arm of chromosome 16. Loss of E-cadherin is typically accompanied by abnormal expression of other members of the cadherin-catenin complex, such as loss of membranous β-catenin and/or aberrant cytoplasmic localization of p120 catenin6,7. Most examples of classic ILC are low-to-intermediate nuclear grade, hormone receptor-positive (>95%) and lack human growth factor receptor 2 (HER2; ERBB2) protein overexpression and gene amplification (<5%)8,9. Prognosis of ILC is similar to invasive ductal carcinoma (IDC) when matched for stage and grade10,11. Several morphologic variants of ILC have been described, including those with pleomorphic, histiocytoid or signet ring cell cytologic features as well as those with tubulolobular, solid or alveolar architectural patterns. Recognition of certain variants (e.g., pleomorphic, signet ring cell and solid) is important as they may have altered biomarker profiles and/or a worse prognosis than classic ILC8.
Invasive lobular carcinoma with extracellular mucin (ILCEM) is a newly appreciated variant that was first reported by Rosa et al. in 200912. Although intracellular mucin in the form of a targetoid intracytoplasmic vacuoles and signet ring cells is a well-recognized feature of lobular carcinoma, extracellular mucin was historically accepted as an indication of a “ductal” phenotype13. As such, it is likely that this entity is underrecognized and not infrequently diagnosed as mucinous carcinoma (MC) or invasive ductal carcinoma (IDC) with mucinous features (“mixed” MC). To our knowledge, 21 cases have been reported in the literature to date in a few case reports12,14,15,16,17,18,19,20,21 and small series22,23. Given that little is known about ILCEM and its distinction from classic ILC and pure MC, we performed a comprehensive clinicopathologic and molecular characterization of the largest series reported to date of this unusual variant.
Materials and methods
Clinicopathologic evaluation of ILCEM
Following Institutional Review Board approval, cases diagnosed at Brigham and Women’s Hospital and Brigham and Women’s Faulkner Hospital from 2001 to 2018 were identified via a key word search for “carcinoma” and “lobular” and words containing “mucin” in reports in the pathology information system. Additional cases were contributed from the authors’ affiliated institutions (Massachusetts General Hospital, Beth Israel Deaconess Medical Center, and New York University Langone Medical Center). ILCEM was defined as invasive carcinoma with a lobular growth pattern and the presence of extracellular mucin pools. Cases were included in this study following confirmation of the diagnosis by histologic review (BTH and TRS). Clinicopathologic data was abstracted from the electronic medical records and pathology reports, including patient demographic factors, clinical and radiologic presentation, and follow-up status; tumor histologic features, grade, and size; presence of lymphovascular invasion or an in-situ component; nodal status; pathologic T, N and M stage; and estrogen receptor (ER), progesterone receptor (PR) and HER2 status.
Slides were reviewed to confirm the reported pathologic factors, including the Nottingham combined histologic grade (modified Scarff-Bloom-Richardson grade)24, and to further characterize the histologic features such as the presence of variant morphology and the proportion of the tumor associated with extracellular mucin (% mucinous component). Cytologic features were classified as follows: (1) classic: tumor cells of low-to-intermediate nuclear grade with round-to-oval nuclei with relatively smooth contours, fine chromatin and indistinct nucleoli; (2) pleomorphic: tumor cells with marked nuclear pleomorphism, defined as nuclei greater than 4 times the size of lymphocytes, equivalent to high nuclear grade1; (3) apocrine: tumor cells with abundant granular eosinophilic cytoplasm, eccentric round-to-oval nuclei and prominent nucleoli; and (4) signet ring cell: tumor cells with a single, large intracytoplasmic mucin-filled vacuole and a peripherally displaced and compressed crescent-shaped nucleus. Growth patterns were classified as: (1) conventional (classic): single cells and single file arrays of cells; (2) solid: sheets of cells; (3) alveolar: nests of tumor cells (at least 20 cells in aggregates1) separated by delicate fibrovascular septae; and (4) expansile: mucin-producing lobular carcinoma in situ (LCIS) with massive acinar expansion, attenuation of the peripheral myoepithelial cell layer, and transition to contiguous pools of mucinous lobular carcinoma at points of rupture and extravasation.
Immunohistochemical assessment of ILCEM
Immunohistochemical studies were performed on 4μm-sections of formalin-fixed paraffin-embedded (FFPE) tissue using antibodies against ER (1:50 dilution; SP1 rabbit monoclonal antibody from Thermo Fisher Scientific, Waltham, MA), PR (1:50 dilution; PgR 636 rabbit monoclonal antibody, DAKO, Santa Clara, CA), HER2 (1:40 dilution; SP3 rabbit monoclonal antibody, Thermo Scientific, Rocklin, CA), E-cadherin (1:50 dilution; mouse monoclonal antibody clone NCH-38, DAKO, Santa Clara, CA), p120 catenin (1:500 dilution; mouse monoclonal antibody 98/PP120, BD Biosciences, San Jose, CA), beta-catenin (1:1000 dilution; mouse monoclonal antibody clone 14/beta-catenin, BD Biosciences, San Jose, CA), MUC-2 (1:800 dilution; mouse monoclonal antibody clone Ccp58, Vector Laboratories, Inc, Burlingame, California) and MUC-6 (1:300 dilution; mouse monoclonal antibody, Leica Biosystems, Nussloch, Germany). Antigen retrieval was performed with 1 mM EDTA (pH 8.0) in a pressure cooker. External controls were assessed for appropriate immunoreactivity.
Interpretation of ER, PR and HER2 immunohistochemical stains was performed in accordance with the most recent American Society of Clinical Oncology/College of American Pathologists guidelines for breast cancer25,26,27. ER and PR were scored as “positive” (nuclear reactivity in >10% of tumor cells), “low positive” (nuclear reactivity in ≥1 to 10% of tumor cells) or “negative” (<1%), whereas HER2 was scored as “positive” (intense, complete membranous staining in a contiguous focus representing ≥10% of the tumor), “equivocal” (weak-to-moderate complete membranous staining in ≥10% of the tumor), or “negative” (lesser degrees of staining). A final HER2 status of “positive” or “negative” was assigned to equivocal (2+) cases by integrating available florescence in situ hybridization (FISH) results.
Molecular profiling of ILCEM
Samples were processed in the Center for Advanced Molecular Diagnostics (CAMD), a CLIA-certified laboratory in the Department of Pathology at Brigham and Women’s Hospital. DNA from archival FFPE with at least 20% tumor cellularity was analyzed by an internally developed hybrid-capture next generation sequencing (NGS) assay (OncoPanel platform) that interrogates the full coding sequences of 447 genes for mutations and copy number variations (CNV), as well as 191 selected introns across 60 genes for rearrangements28,29. The complete list of genes is listed in Supplementary Table 1. Samples with a mean target coverage of <50× were excluded from the study. One sample with 15% tumor cellularity was attempted with >400x depth to identify genomic variants.
Targeted sequences were captured using a solution phase Agilent SureSelect hybrid capture kit (Agilent Technologies, Santa Clara, CA, USA) and massively parallel sequencing performed on an Illumina HiSeq 2500 sequencer (Illumina, San Diego, CA, USA). The sequence reads were aligned and processed through a bioinformatics pipeline to identify single-nucleotide variations and small insertions–deletions (indels). Mutation calls were made using Mutect v.1.1.4 with annotations by Oncotator and indels using Indelocator (Broad Institute, Cambridge, MA, USA)30,31. Since testing was not performed on paired germline DNA, variants present at >0.1% in Exome Variant Server, NHLBI GO Exome Sequencing Project, Seattle, WA, USA (URL: http://evs.gs.washington.edu/EVS/) or gnomAD (https://gnomad.broadinstitute.org/about) were filtered and removed from analysis. Any filtered variants that were reported in Catalog of Somatic Mutations in Cancer32 (COSMIC; cancer.sanger.ac.uk) more than twice were rescued and presented for manual review. Single nucleotide variants (SNVs) and indels were manually reviewed for biologically significant mutations including (i) loss of function mutations (splice site disruption, frameshift, nonsense) as well as hotspot missense mutations for tumor suppressor genes, (ii) missense hotspot mutations for oncogenes, (iii) pathogenic gene mutations listed in COSMIC32 (v95, released 24-Nov-21), ClinVar33 (https://www.ncbi.nlm.nih.gov/clinvar), and/or cBioPortal34,35 (v3.7.27; https://www.cbioportal.org/). Novel variants or those of uncertain biologic significance were included in the analysis only if evidence in the literature existed for a role of the altered gene in breast cancer, in particular lobular or mucinous neoplasia. Tumor mutational burden (TMB) was calculated by determining the number of nonsynonymous somatic mutations that occur per megabase of exonic sequence data across all genes on the panel.
Copy number variants (CNVs) and detection of structural variants were identified using an internally developed bioinformatics pipeline and algorithms, RobustCNV and BreaKmer36, respectively. For copy number analysis, Robust CNV was used to calculate the fractional coverage of specified genomic intervals compared with the median fractional coverage obtained in a panel of 152 FFPE non-neoplastic samples. Copy number (CN) was calculated using the formula, CN = (2* (AGCR-1)/P) + 2, where AGCR is the average gene copy ratio and P is the lesion purity. CN is a function of the subjective visual assessment of lesion purity, and as such, represents an estimate. For the purposes of this study, an estimated copy number of greater than or equal to 6 was employed as the threshold for a reportable gain28,29,31, with copy number of 6-9 considered as low-level amplification and copy number greater than and equal to 10 as high-level amplification.
Statistical analyses
Comparison of categorical clinicopathologic variables was done using Chi-squared and Fisher’s exact tests. Wilcoxon signed rank test was used for comparing continuous variables including tumor size, mutational burden, and frequencies of genetic alterations with correction for multiple comparisons when appropriate. All analyses were done using Stata/SE version 15 (StataCorp, College Station, TX). p-values of <0.05 were considered statistically significant.
Results
Clinicopathologic features of ILCEM
Seventeen cases were included in the study. All patients were women and the median age at diagnosis was 66 years (range: 31–77 years). At clinical presentation, most patients (16/17; 94%) were found to have a radiologic mass: 5 screen-detected, 5 palpable, and 6 without documented clinical features. One patient presented with skin changes consistent with inflammatory breast cancer. All cases were unilateral, with nearly half (8/17; 47%) demonstrating multifocal disease on pathologic assessment. Seven cases (7/17; 41%) had tumors greater than 2 cm in size (at least pT2) and seven cases (7/17; 41%) were found to have metastasis to axillary lymph node(s) (pN1 to pN2) (Table 1). Primary tumor sizes ranged from 0.7 cm to 10.0 cm (median: 2.6 cm) (Table 2). None of the patients presented with distant metastases at the time of initial diagnosis. In 2 cases, the patients had a prior history of ipsilateral ILC treated with breast conserving therapy, which suggested that the ILCEM represented an in-breast recurrence. There was no note made to the presence of extracellular mucin, although one of the prior carcinomas was referred to as a “signet ring cell carcinoma”. Two patients had a history of contralateral carcinoma of a different histologic subtype (tubular carcinoma and invasive ductal carcinoma).
Follow-up data were available for 12 cases, with median follow-up of 83.5 months (range: 3–171 months). Recurrences were noted in 8 women (8/12; 67%), including 2 local recurrences in the ipsilateral breast, 4 loco-regional recurrences involving lymph nodes, and 5 cases with distant metastases (Table 1). More than half (4/7) of those with lymph node metastases at initial presentation developed recurrences during follow-up. Four women died of breast cancer during follow-up (Table 1).
Histopathologic features of ILCEM
All cases included in this study demonstrated lobular morphology, classic or variant, throughout the non-mucinous and mucinous components of the tumor, without any evidence of tubule formation, cohesive nests or micropapillae (Figs. 1 and 2). Extracellular mucin was present in all cases (range: 5–95%), involving at least 25% of the tumor in 11 cases (65%). Among 8 cases for which both core needle biopsies (CNB) and subsequent resections were available for review, 5 did not have conspicuous extracellular mucin in the CNB sample. No significant association was identified between extent of extracellular mucin and other histologic or clinical features.
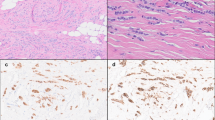
(a, b) Case 9. The ILCEM is present as tumor cells suspended in small pools of mucin associated with a classic component. (c–h) Case 2. The ILCEM is a mixture of solid and classic components. There are solid cellular tumor nodules with scant extracellular mucin (c) and large pools of mucin (c, d), both consisting of predominantly pleomorphic signet ring cells (e). Classic ILC is also present (f). Immunohistochemical stains show loss of E-cadherin expression (g) and aberrant cytoplasmic p120 expression (h) in tumor cells.
(a–d) Case 14. ILCEM in which the mucinous component consists of solid cellular nodules of signet ring cells in extracellular mucin (a, b). The tumor also contains ILC with a conventional growth pattern and pleomorphic LCIS (c, d). (e, f) Case 6. ILCEM is composed of rhabdoid pleomorphic apocrine tumor cells in an unusual discohesive alveolar growth pattern. (g, h) Case 8. Expansile nodules of mucin-producing signet ring cell LCIS transitioning to ILCEM. The signet ring cell LCIS is associated with abundant extracellular mucin and massive acinar expansion, with foci of rupture and contiguous pools of extravasated mucinous lobular carcinoma, and with foci of interspersed irregular pools of frankly invasive mucinous lobular carcinoma (g). A p63 immunohistochemical stain demonstrates the presence of a myoepithelial cell layer at the periphery of LCIS (short arrow) and the loss of this layer at the edge of the contiguous pool of extravasated tumor-containing mucin (long arrow) (h). The signet ring cell LCIS in this case represents the only example of mucin-producing LCIS.
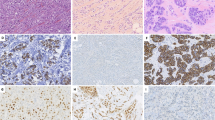
ILCEM had a Nottingham histologic grade of moderately (n = 13) or poorly differentiated (n = 4), with a high nuclear grade (score 3 of 3) in 11 cases (65%). As such, 11 cases were considered pleomorphic ILC, 2 of which exhibited apocrine cytologic features. Diffuse signet ring cell features in ≥20% of the tumor were identified in 10 (59%) cases. All but one case of ILC at least focally displayed a conventional growth pattern, characterized by poorly cohesive tumor cells arranged as dispersed single cells and single file arrays (Fig. 1a, b), while a subset of cases also had solid (4 cases; 24%) (Figs. 1c–h, and 2a–d) and/or alveolar (2 cases; 12%) growth pattern (Fig. 2e, f). The solid component in three cases was present as distinctive cellular mucinous nodules with diffuse signet ring cell features (Figs. 1c–e, and 2a, b). The alveolar component seen in two cases was unusual, exhibiting areas vaguely reminiscent of alveolar rhabdomyosarcoma with “rhabdoid” pleomorphic apocrine neoplastic cells arranged in a hobnail fashion at the periphery of empty to mucin-filled nests separated by fibrous septae (Fig. 2e, f). The single case lacking a conventional growth pattern was a striking example of circumscribed expansile nodules of mucin-producing signet ring cell LCIS transitioning to ILCEM. An immunohistochemical stain for p63 highlighted an attenuated myoepithelial cell layer at the periphery of massively dilated acini (up to 0.3 cm), while it demonstrated the absence of this layer at the periphery of contiguous pools of extravasated mucinous lobular carcinoma as well as interspersed small irregular foci of frankly invasive mucinous lobular carcinoma (Fig. 2g, h). An immunohistochemical stain for smooth muscle myosin heavy chain was negative throughout the lesion. This phenomenon bears resemblance to solid papillary carcinoma with extracellular mucin production transitioning to mucinous carcinoma. This was the only example of LCIS with mucin production. Overall, concurrent LCIS was present in a majority of cases (11/17; 65%) (Fig. 2d, h), 2 of which were pleomorphic, while ductal carcinoma in situ (DCIS) was not identified in any case. Lymphovascular invasion was found in 3 (18%) cases. Three (18%) of the invasive carcinomas displayed areas of necrosis. Of the cases with necrosis, all were Nottingham grade 2, but two were pleomorphic (high nuclear grade) and one had a mitotic score >1. Histopathologic features are detailed in Table 2.
All tumors were ER-positive and the majority (12/17; 71%) were PR-positive. Two (18%) tumors were HER2 positive (3+) by immunohistochemistry. Absent or reduced membranous expression of E-cadherin was seen in all 17 cases across non-mucinous and mucinous components of the invasive carcinoma as well as in the associated carcinoma in situ (Fig. 1g). Twelve of 17 cases had sufficient material for additional immunohistochemical stains, of which most showed absent or reduced membranous expression of beta-catenin (11/12 cases) and/or cytoplasmic expression of p120 (11/12 cases) (Fig. 1h). Intact membranous expression of p120 and beta-catenin was observed in the one case with reduced rather than absent membranous E-cadherin expression; this case did not have material available for sequencing. All 12 cases stained for MUC2, while only 4 stained for MUC6 (data not shown).
Genomic profiles of ILCEM
Of the 17 cases included in the study, 13 (76%) had available FFPE tissue and sufficient DNA extracted for our NGS assay. Most (12/13; 92%) of the samples were derived from primary tumors, with specimens including 11 resections and 1 core biopsy. In one case, the sample was obtained from a core biopsy of breast with recurrent tumor. Biopsy samples were utilized only when access to FFPE tissue from a primary resection was lacking.
Specimens were sequenced to a median depth of 266x. Sequencing revealed 154 nonsynonymous variants, 38 of which were included in this analysis due to predicted or potential biologic significance. The median tumor mutational burden was 6.8 mut/Mb (range: 3.0 to 18.3), with 2 tumors that were TMB-high (≥10mut/Mb). The median SNV frequency detected was 11 (range: 2 to 30). The most common SNVs with predicted or potential biologic significance were seen involving CDH1 (10/13 cases; 77%), followed by PIK3CA (3/13 cases; 31%), POLQ (3/13 cases; 23%), TP53 (3/13 cases; 23%), ERBB3 (3/13 cases; 23%), ERBB2 (2/13 cases; 15%), and RUNX1 (2/13 cases; 15%). Other genes with isolated SNVs of potential significance included AKT1, FANCD2, GATA3, MAP3K1, MUTYH, PTEN, RB1, and SF3B1 (Figs. 3 and 5; Supplementary Table 2). Gene-level alterations of CDH1 were seen in all but one case, including frameshift mutations (n = 7), nonsense mutations (n = 2), donor splice site mutation (n = 1), insertion (n = 1) and deletion(n = 1). CDH1 mutations were detected across the gene coding regions. Biallelic inactivation with concurrent CDH1 mutations and 16q full arm or partial arm loss was seen in 11 of 13 cases (Fig. 4). Of the cases lacking evidence of biallelic inactivation, one harbored 16q loss without a detectable CDH1 gene alteration and the other harbored CDH1 gene alteration without a detectable 16q loss. Tumors from both cases were characterized by signet ring cell features, as well as loss of E-cadherin and beta-catenin membrane expression and aberrant p120 cytoplasmic expression. CDH1 promoter hypermethylation cannot be excluded in these cases, as this was not assessed. A deletion involving GNAS was also identified in one case.
Recurrent gene amplifications independent of chromosome arm changes were identified in association with GATA3 (4/13; 31%), FOXA1 (3/13; 23%), CCND1 (2/13; 15%), VEGFA (2/13; 15%), KAT6A (2/13; 15%) and POLB (2/13; 15%). Isolated gene-level amplifications were also seen with ERBB2, CDK12, RUNX1, AURKA, ZNF217, ESR1, FGFR1, WHSC1L1, amongst others (Fig. 5). ERBB2 amplification was seen in the only HER2 IHC positive (3+) case sequenced. Partial arm or focal amplifications were identified in 3 cases, involving chromosomes 8p12-8p11 (2/13; 15%), 6p21-22 (1/13; 8%), 6q (1/13; 8%), 7q (1/13; 8%), 14q32 (1/13; 8%), 16p11-13 (1/13; 8%), and 20q13 (1/13; 8%).
Arm loss of 16q was present in 11 of 13 (85%) of sequenced cases and constituted the most common chromosomal change in the series. Partial 16q arm loss involving 16q22-q24 was also seen in one additional case. Full arm 1q gain/amplification was seen in 9 of 13 (69%) cases, with all 9 showing concurrent 1q gain and 16q loss. Other recurrent arm-level chromosomal changes included 17p loss (5/13; 38%), 18q loss (4/13; 31%) and 22q loss (5/13; 38%).
ILCEM tumors with lymph node metastases and/or subsequent recurrences were associated with more mutations with biologic significance in breast carcinoma than ILCEM with no recurrence (Fig. 5), although the difference did not reach statistical significance (p = 0.07). There were no significant associations between the number of gene amplifications or chromosome arm level changes, mutational burden, SNVs, and insertions/deletions and the pathologic or clinical features.
Discussion
This is the largest reported series of ILCEM and the first to provide genomic analyses. Consistent with prior reports of this entity12,14,15,16,17,18,19,20,21,22,23 (Table 3), our results show that a majority of cases present in older, post-menopausal women, often as multifocal disease with sizable breast tumors and nodal involvement. All reported tumors have been hormone receptor positive, with a higher rate of HER2 positivity (>10%) than seen in classic ILC and pure MC. Most cases of ILCEM included in this and other reports have demonstrated loss of normal membranous expression of E-cadherin with or without alterations in expression of other members of the cadherin-catenin complex, a hallmark of the lobular phenotype, supporting its classification as a variant of ILC. Although ILCEM has been previously described as having classic or solid lobular morphology, our findings expand upon those descriptions to emphasize a broader spectrum of variant cytological and architectural features. A significant proportion of cases exhibited pleomorphic and/or diffuse signet ring cell features, some of which contained distinctive alveolar areas of apocrine pleomorphic ILC, nodular sheets of signet ring cells, or expansile nodules transitioning from mucin-producing signet ring cell LCIS. Although the presence of signet ring cells has been noted in most other reported cases, the extent has been either reported as limited or not described in detail. Beyond an enhanced appreciation of the tumor morphology, our series provides the most information to date regarding the natural history of ILCEM, which appears to have a relatively high rate of recurrence, especially among those with lymph node involvement at the time of initial diagnosis. This suggests that ILCEM has more aggressive biology than either classic ILC or pure MC, which may be related to the variant solid, pleomorphic and signet ring cell features of ILC known to be associated with worse prognosis8.
Sequencing of the ILCEM cases in our series revealed genetic alterations consistent with a lobular phenotype. Most important, CDH1 alterations were identified in all but one of the sequenced cases of ILCEM, most of which had demonstrable biallelic inactivation of CDH1 due to concurrent CDH1 mutation and whole or partial 16q arm loss. Concurrent 16q loss and 1q gain were frequent, more characteristic of ILC37 than MC38,39. Our findings are consistent with those from the most comprehensive molecular profiling to date of ILC by the Cancer Genome Atlas (TCGA) Research Network37, in which the most commonly mutated gene was CDH1 (63%), almost invariably co-occurring with heterozygous loss of 16q. Besides biallelic inactivation of CDH1 inactivation, the genetic hallmark of ILC, the TCGA study also demonstrated that ILC, predominantly ER-positive (ER+) and luminal A, was enriched for mutations affecting PIK3CA (48%), PTEN (13%), RUNX1 (10%), TBX3 (9%) and FOXA1 (7%) as well as strongly activated PI3K/Akt signaling. While ILC was characterized by FOXA1 mutations correlating with increased protein expression and activity, GATA3 mutations were more common in luminal A IDC [28], suggesting differential modulation of ER activity in these tumor types. In our study, ILCEM harbored PIK3CA (n = 3), RUNX1 (n = 2), AKT1 (n = 2) and PTEN (n = 1) mutations, whereas FOXA1 mutations were not detected; instead, FOXA1 amplifications were identified in 3 cases, all coupled with GATA3 amplifications. Beyond indicating that a subset of ILCEM likely has a luminal phenotype, the significance of this observation regarding ILCEM and its ER modulation remains uncertain. The presence of recurrent PIK3CA mutations is notable as it may offer the opportunity for treatment with recently FDA-approved or investigational PI3K and Akt inhibitors40.
ILCEM cases in our series, especially those with recurrences during follow up, harbored additional clinically or biologically relevant genomic alterations (e.g., ERBB2, ERBB3, TP53, POLQ, CCND1 and FGFR1), some not typically seen in classic ILC, which may drive pathogenesis and contribute to its aggressive clinicopathologic features. In the TCGA study of ILC, TP53 mutations were uncommon (8%) and ERBB2 mutations were rare (2%)37. In contrast, frequent mutation, or amplification of ERBB2 and ERBB3 has been reported in pleomorphic ILC41,42,43, as well as relapsed classic CDH1-mutant ILC44, with implications for HER2-targeted therapy via monoclonal antibodies and tyrosine kinase inhibitors. Interestingly, ERBB2 or ERBB3 alterations were found in 5 ILCEM cases (4 of 12 profiled cases and presumed in another unsequenced case with IHC 3+), most (n = 4) of which displayed pleomorphic features, while all 3 cases of ILCEM with pathogenic TP53 mutations were pleomorphic. Mutations in POLQ, a gene encoding DNA polymerase theta involved in an error-prone DNA repair pathway response to double strand breaks45, were observed in one classic and 2 pleomorphic ILCEM cases, but had not been emphasized in previous profiling studies of ILC. This finding is worth noting due to the correlation of POLQ-encoded protein overexpression with high tumor grade and poorer survival in breast cancer45,46. Of the patients in our series with death due to disease, one had a POLQ-mutant tumor (variant of uncertain significance), and two others had TP53-mutant tumors, with loss of heterozygosity in one of them due to concurrent 17p loss.
ILCEM displayed few amplifications at genomic loci that are known to drive breast tumorigenesis and have been reported at varying frequencies in lobular and other ER + breast carcinomas, including at 8p11-p12 (n = 2, e.g., FGFR1), 11q13 (n = 2, e.g., CCND1), and 20q13 (n = 2, ZNF217). In our series, recurrent 8p11-12 amplifications were detected in 2 cases, involving FGFR1, WHSC1L1, KAT6A, and POLB genes, a finding in breast cancer that has been associated with worse survival47,48. FGFR1, in particular, has been suggested to be an amplicon driver49, and its amplification has been associated with sensitivity to FGFR inhibitors in preclinical models49,50. Two additional ILCEM cases harbored amplifications of CCND1, encoding cyclin D1, which complexes with and activates cyclin-dependent kinases (CDKs) and mediates cell cycle progression. CCND1 amplification is frequently observed in ER + breast cancer51,52, providing rationale for targeted therapy with CDK4/6 inhibitors53,54 as well as predicting tamoxifen resistance55. It is interesting to note that ILCEM harbors concurrent 16q loss and 1q gain, the hallmark of the low-grade breast neoplasia pathway, along with additional genetic alterations typical of high-grade breast carcinoma. Our molecular findings for ILCEM are consistent with those from prior studies of pleomorphic ILC and LCIS which suggest that high-grade variants of lobular carcinoma evolve from classic lobular neoplasia56,57. This represents an exception to the commonly accepted model of breast cancer pathogenesis in which low-grade and high-grade neoplasia are thought to largely develop via independent pathways58.
Of note, amplifications of CCND1 and FGFR1, as well as alterations in ERBB2, have also been reported to be enriched in a subset of pure MCs with an aggressive and metastatic clinical course59, further underscoring the role of these genes in the progression of ER + breast cancer. Otherwise, conventional pure MC, a relatively indolent tumor, appears to be molecularly distinct from IDC and ILC, including ILCEM, given its lack of PIK3CA and TP53 mutations and concurrent 1q gains and 16q losses38,39. GATA3 is the most frequently mutated gene in MC39, a finding more similar to IDC than ILC. Mutations in SF3B1, which encodes a member of the SF3B complex involved in pre-mRNA splicing60, have been recognized in MC39,60,61 and were detected in one of the ILCEM cases. Pathogenic SF3B1 mutations (e.g., hotspot K700E) are thought to result in alternative splicing and to potentially constitute an oncogenic driver and therapeutic target in breast cancer60,62; however, the biologic significance of the missense mutation (V184I) identified in our ILCEM series is not entirely certain (predicted pathogenic in COSMIC only). Although our findings suggest that the molecular underpinnings of ILCEM differ from those of MC, it should be noted that the lobular and mucinous components were not microdissected nor independently evaluated, and therefore, it is possible that the genetic features are biased toward those of the lobular component.
It must be acknowledged that significant overlap exists between the cases of ILCEM in this series and the entity previously referred to as primary signet ring cell carcinoma of the breast63,64,65. In the most recent World Health Organization classification system66, primary signet ring cell carcinoma is no longer considered to represent a distinct entity, as signet ring cell features can be seen in lobular, ductal or mucinous carcinoma, and tumors should be classified according to one of those subtypes. Historically, although published criteria were variable63,64,65, primary signet ring cell carcinoma was most commonly defined as any breast carcinoma in which signet ring cells comprised greater than 20% of the tumor64. As some authors considered it as a variant of lobular carcinoma64,67, ILC with signet ring cell features was well-represented in prior studies. Descriptions of this entity included some cases with extracellular mucin, ranging in extent from “rather minimal collections”64 to “lakes”63. It is important to recognize that many ILCEM cases in our series would fit this diagnosis given that breast signet ring cell carcinoma has been associated with aggressive behavior and poor prognosis in both early and contemporary series63,64,65,68. Furthermore, the genomic profile of ILCEM described herein reflects to some extent that of breast signet ring cell carcinoma as previously recognized.
Entities in the differential diagnosis of ILCEM include metastatic signet ring cell carcinoma, matrix-producing metaplastic carcinoma, and IDC with mucinous features. Metastatic signet ring cell carcinoma of gastric origin should be considered when any ER-negative carcinoma with signet ring cell features is encountered in the breast in the absence of an in-situ component. It can be distinguished from ILCEM by immunohistochemical staining for CK20 and CDX2, and lack of staining for ER, GATA3, mammaglobin, and GCDFP-1569. Matrix-producing metaplastic carcinoma with single cells dispersed in a myxoid matrix may bear a striking resemblance to ILCEM, and complicating the matter, we have encountered cases that exhibit loss of membranous E-cadherin expression and aberrant catenin expression, perhaps consistent with the phenomenon of epithelial-to-mesenchymal transition in metaplastic carcinoma70,71. The diagnosis of matrix-producing metaplastic carcinoma is favored by the recognition of cohesive areas of the tumor and myxoid matrix, as well as by the triple negative immunophenotype, which has not been reported in ILCEM to date.
Finally, it is likely that many cases of ILCEM have been confused for IDC with mucinous features (mixed MC) due to the common belief that extracellular mucin is a “ductal” attribute. Expert opinion is that the distinction of ILC from IDC is first and foremost based on morphology, with the diagnosis of ILC rendered in the presence of discohesive single cells and single file arrays of cells. Routine use of E-cadherin immunohistochemistry is not advised due to intact membranous staining in up to 15% of cases with ILC morphology; however, it should be acknowledged that a negative E-cadherin stain may be helpful in confirming the morphologic impression of ILC, especially in the setting of variant features6. Most cases of ILCEM in this study demonstrated loss of membranous E-cadherin staining, which obviates controversy regarding the nature of the included cases. One exception is a single case included based on morphology, which demonstrated reduced rather than absent E-cadherin staining and intact membranous staining for p120 and beta-catenin; molecular profiling data is unfortunately not available for this case.
ILCEM is thus far an exceedingly rare entity, and although our multi-institutional series represents the largest to date, conclusions are still limited by the small sample size. In comparison to prior reports, this cohort is enriched for pleomorphic and signet ring cell rather than classic features, which may not only expand upon our understanding of ILCEM, but also bias it. If ILCEM with classic morphology were better represented in our series, as in prior reports, it is possible that different conclusions would be drawn regarding the clinicopathologic and genomic profile of this entity. Another limitation is that our molecular characterization of this entity is based on a targeted NGS platform with a customized panel of genes. Nonetheless, this platform allows for the detection of SNVs in over 400 genes, including most of the driver mutations known to play a role in breast cancer pathogenesis and prognosis. Future studies that assess the whole genome or exome as well as transcriptomic and epigenetic profiles would provide a more complete molecular landscape of ILCEM.
In summary, this series adds substantially to the emerging literature on ILCEM highlighting this tumor as a distinct variant of ILC that often presents with pleomorphic (high nuclear grade) and signet ring cell features and more aggressive clinical course than classic ILC or conventional pure MC. Our genomic data provides further evidence in support of a lobular rather than ductal or mucinous phenotype and reveals potentially targetable molecular alterations predominantly found in a subset of cases with recurrence. It is hoped that this study facilitates better recognition of mucin-producing ILC in clinical practice, and in turn, enables future studies to further elucidate the natural history and molecular pathogenesis of this entity.
Data availability
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
References
Shin SJ, D. C., Kristiansen G, Reis-Filho JS, Sasano H. Invasive lobular carcinoma In: WHO Classification of Tumours Editorial Board (ed). Breast Tumours WHO Classification of Tumours 114-118 (International Agency for Research on Cancer: Lyon, France, 2019).
Sastre-Garau, X., Jouve, M., Asselain, B., Vincent-Salomon, A., Beuzeboc, P., Dorval, T. et al. Infiltrating lobular carcinoma of the breast. Clinicopathologic analysis of 975 cases with reference to data on conservative therapy and metastatic patterns. Cancer 77, 113–120 (1996).
Ellis, I. O., Galea, M., Broughton, N., Locker, A., Blamey, R. W. & Elston, C. W. Pathological prognostic factors in breast cancer. II. Histological type. Relationship with survival in a large study with long-term follow-up. Histopathology 20, 479–489 (1992).
Li, C. I., Anderson, B. O., Daling, J. R. & Moe, R. E. Trends in incidence rates of invasive lobular and ductal breast carcinoma. JAMA 289, 1421–1424 (2003).
Foote, F. W. & Stewart, F. W. Lobular carcinoma in situ: A rare form of mammary cancer. Am J Pathol 17, 491–496 493 (1941).
Dabbs, D. J., Schnitt, S. J., Geyer, F. C., Weigelt, B., Baehner, F. L., Decker, T. et al. Lobular neoplasia of the breast revisited with emphasis on the role of E-cadherin immunohistochemistry. Am J Surg Pathol 37, e1–11 (2013).
Canas-Marques, R. & Schnitt, S. J. E-cadherin immunohistochemistry in breast pathology: uses and pitfalls. Histopathology 68, 57–69 (2016).
Rakha, E. A. & Ellis, I. O. Lobular breast carcinoma and its variants. Semin Diagn Pathol 27, 49–61 (2010).
Mamtani, A. & King, T. A. Lobular Breast Cancer: Different Disease, Different Algorithms? Surg Oncol Clin N Am 27, 81–94 (2018).
Li, C. I. & Daling, J. R. Changes in breast cancer incidence rates in the United States by histologic subtype and race/ethnicity, 1995 to 2004. Cancer Epidemiol Biomarkers Prev 16, 2773–2780 (2007).
Arpino, G., Bardou, V. J., Clark, G. M. & Elledge, R. M. Infiltrating lobular carcinoma of the breast: tumor characteristics and clinical outcome. Breast Cancer Res 6, R149–156 (2004).
Rosa, M., Mohammadi, A. & Masood, S. Lobular carcinoma of the breast with extracellular mucin: new variant of mucin-producing carcinomas? Pathol Int 59, 405–409 (2009).
Gad, A. & Azzopardi, J. G. Lobular carcinoma of the breast: a special variant of mucin-secreting carcinoma. J Clin Pathol 28, 711–716 (1975).
Bari, V. B., Bholay, S. U. & Sane, K. C. Invasive lobular carcinoma of the breast with extracellular mucin- a new rare variant. J Clin Diagn Res 9, ED05-06 (2015).
Boukhechba, M., Kadiri, H. & El Khannoussi, B. Invasive Lobular carcinoma of the breast with extracellular mucin: case report of a new variant of lobular carcinoma of the breast. Case Rep Pathol 2018, 5362951 (2018).
Gomez Macias, G. S., Perez Saucedo, J. E., Cardona Huerta, S., Garza Montemayor, M., Villarreal Garza, C. & Garcia Hernandez, I. Invasive lobular carcinoma of the breast with extracellular mucin: A case report. Int J Surg Case Rep 25, 33-36 (2016).
Haltas, H., Bayrak, R., Yenidunya, S., Kosehan, D., Sen, M. & Akin, K. Invasive lobular carcinoma with extracellular mucin as a distinct variant of lobular carcinoma: a case report. Diagn Pathol 7, 91 (2012).
Koufopoulos, N., Antoniadou, F., Kokkali, S., Pigadioti, E. & Khaldi, L. Invasive lobular carcinoma with extracellular mucin production: description of a case and review of the literature. Cureus 11, e5550 (2019).
Yu, J., Bhargava, R. & Dabbs, D. J. Invasive lobular carcinoma with extracellular mucin production and HER-2 overexpression: a case report and further case studies. Diagn Pathol 5, 36 (2010).
Baig, A., Omeroglu-Altinel, G. & Omeroglu, A. Invasive pleomorphic-type lobular carcinoma of the breast presenting as a mucinous carcinoma. Case Rep Pathol 2019, 1839208 (2019).
Burky, M. J., Ray, E. M., Ollila, D. W., O’Connor, S. M., Hertel, J. D. & Calhoun, B. C. Pleomorphic invasive lobular carcinoma of the breast with extracellular mucin and HER2 amplification. Breast Cancer 14, 1178223420976383 (2020).
Cserni, G., Floris, G., Koufopoulos, N., Kovacs, A., Nonni, A., Regitnig, P. et al. Invasive lobular carcinoma with extracellular mucin production-a novel pattern of lobular carcinomas of the breast. Clinico-pathological description of eight cases. Virchows Arch 471, 3–12 (2017).
Singh, K., DiazGomez, B., Wang, Y., Ou, J. & Hansen, K. Invasive lobular carcinoma with extracellular mucin: not all mucinous mammary carcinomas are ductal! Int J Surg Pathol 27, 55–58 (2019).
Elston, C. W. & Ellis, I. O. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 19, 403–410 (1991).
Allison, K. H., Hammond, M. E. H., Dowsett, M., McKernin, S. E., Carey, L. A., Fitzgibbons, P. L. et al. Estrogen and progesterone receptor testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists Guideline Update. Arch Pathol Lab Med 144, 545–563 (2020).
Wolff, A. C., Hammond, M. E. H., Allison, K. H., Harvey, B. E., McShane, L. M. & Dowsett, M. HER2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update Summary. J Oncol Pract 14, 437–441 (2018).
Wolff, A. C., Hammond, M. E., Hicks, D. G., Dowsett, M., McShane, L. M., Allison, K. H. et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med 138, 241–256 (2014).
Garcia, E. P., Minkovsky, A., Jia, Y., Ducar, M. D., Shivdasani, P., Gong, X. et al. Validation of OncoPanel: A Targeted Next-Generation Sequencing Assay for the Detection of Somatic Variants in Cancer. Arch Pathol Lab Med 141, 751–758 (2017).
Sholl, L. M., Do, K., Shivdasani, P., Cerami, E., Dubuc, A. M., Kuo, F. C. et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight 1, e87062 (2016).
Cibulskis, K., Lawrence, M. S., Carter, S. L., Sivachenko, A., Jaffe, D., Sougnez, C. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31, 213–219 (2013).
Ramos, A. H., Lichtenstein, L., Gupta, M., Lawrence, M. S., Pugh, T. J., Saksena, G. et al. Oncotator: cancer variant annotation tool. Hum Mutat 36, E2423–2429 (2015).
Tate, J. G., Bamford, S., Jubb, H. C., Sondka, Z., Beare, D. M., Bindal, N. et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res 47, D941–D947 (2019).
Landrum, M. J., Lee, J. M., Benson, M., Brown, G. R., Chao, C., Chitipiralla, S. et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 46, D1062–D1067 (2018).
Cerami, E., Gao, J., Dogrusoz, U., Gross, B. E., Sumer, S. O., Aksoy, B. A. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404 (2012).
Gao, J., Aksoy, B. A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S. O. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6, pl1 (2013).
Abo, R. P., Ducar, M., Garcia, E. P., Thorner, A. R., Rojas-Rudilla, V., Lin, L. et al. BreaKmer: detection of structural variation in targeted massively parallel sequencing data using kmers. Nucleic Acids Res 43, e19 (2015).
Ciriello, G., Gatza, M. L., Beck, A. H., Wilkerson, M. D., Rhie, S. K., Pastore, A. et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell 163, 506-519 (2015).
Nguyen, B., Sanchez-Vega, F., Fong, C. J., Chatila, W. K., Boroujeni, A. M., Pareja, F. et al. The genomic landscape of carcinomas with mucinous differentiation. Sci Rep 11, 9478 (2021).
Pareja, F., Lee, J. Y., Brown, D. N., Piscuoglio, S., Gularte-Merida, R., Selenica, P. et al. The genomic landscape of mucinous breast cancer. J Natl Cancer Inst 111, 737-741 (2019).
Verret, B., Cortes, J., Bachelot, T., Andre, F. & Arnedos, M. Efficacy of PI3K inhibitors in advanced breast cancer. Ann Oncol 30 Suppl 10, x12-x20 (2019).
Lien, H. C., Chen, Y. L., Juang, Y. L. & Jeng, Y. M. Frequent alterations of HER2 through mutation, amplification, or overexpression in pleomorphic lobular carcinoma of the breast. Breast Cancer Res Treat 150, 447–455 (2015).
Rosa-Rosa, J. M., Caniego-Casas, T., Leskela, S., Cristobal, E., Gonzalez-Martinez, S., Moreno-Moreno, E. et al. High Frequency of ERBB2 Activating Mutations in Invasive Lobular Breast Carcinoma with Pleomorphic Features. Cancers 11 (2019).
Christgen, M., Bartels, S., Radner, M., Raap, M., Rieger, L., Christgen, H. et al. ERBB2 mutation frequency in lobular breast cancer with pleomorphic histology or high-risk characteristics by molecular expression profiling. Genes Chromosomes Cancer 58, 175–185 (2019).
Ross, J. S., Wang, K., Sheehan, C. E., Boguniewicz, A. B., Otto, G., Downing, S. R. et al. Relapsed classic E-cadherin (CDH1)-mutated invasive lobular breast cancer shows a high frequency of HER2 (ERBB2) gene mutations. Clin Cancer Res 19, 2668–2676 (2013).
Higgins, G. S., Harris, A. L., Prevo, R., Helleday, T., McKenna, W. G. & Buffa, F. M. Overexpression of POLQ confers a poor prognosis in early breast cancer patients. Oncotarget 1, 175–184 (2010).
Lemee, F., Bergoglio, V., Fernandez-Vidal, A., Machado-Silva, A., Pillaire, M. J., Bieth, A. et al. DNA polymerase theta up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. Proc Natl Acad Sci USA 107, 13390–13395 (2010).
Gelsi-Boyer, V., Orsetti, B., Cervera, N., Finetti, P., Sircoulomb, F., Rouge, C. et al. Comprehensive profiling of 8p11-12 amplification in breast cancer. Mol Cancer Res 3, 655–667 (2005).
Irish, J. C., Mills, J. N., Turner-Ivey, B., Wilson, R. C., Guest, S. T., Rutkovsky, A. et al. Amplification of WHSC1L1 regulates expression and estrogen-independent activation of ERalpha in SUM-44 breast cancer cells and is associated with ERalpha over-expression in breast cancer. Mol Oncol 10, 850–865 (2016).
Reis-Filho, J. S., Simpson, P. T., Turner, N. C., Lambros, M. B., Jones, C., Mackay, A. et al. FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas. Clin Cancer Res 12, 6652–6662 (2006).
Gozgit, J. M., Wong, M. J., Moran, L., Wardwell, S., Mohemmad, Q. K., Narasimhan, N. I. et al. Ponatinib (AP24534), a multitargeted pan-FGFR inhibitor with activity in multiple FGFR-amplified or mutated cancer models. Mol Cancer Ther 11, 690–699 (2012).
Al-Kuraya, K., Schraml, P., Torhorst, J., Tapia, C., Zaharieva, B., Novotny, H. et al. Prognostic relevance of gene amplifications and coamplifications in breast cancer. Cancer Res 64, 8534–8540 (2004).
Wilkerson, P. M. & Reis-Filho, J. S. The 11q13-q14 amplicon: clinicopathological correlations and potential drivers. Genes Chromosomes Cancer 52, 333–355 (2013).
Finn, R. S., Crown, J. P., Lang, I., Boer, K., Bondarenko, I. M., Kulyk, S. O. et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol 16, 25–35 (2015).
VanArsdale, T., Boshoff, C., Arndt, K. T. & Abraham, R. T. Molecular pathways: targeting the cyclin D-CDK4/6 axis for cancer treatment. Clin Cancer Res 21, 2905–2910 (2015).
Bostner, J., Ahnstrom Waltersson, M., Fornander, T., Skoog, L., Nordenskjold, B. & Stal, O. Amplification of CCND1 and PAK1 as predictors of recurrence and tamoxifen resistance in postmenopausal breast cancer. Oncogene 26, 6997–7005 (2007).
Harrison, B. T., Nakhlis, F., Dillon, D. A., Soong, T. R., Garcia, E. P., Schnitt, S. J. et al. Genomic profiling of pleomorphic and florid lobular carcinoma in situ reveals highly recurrent ERBB2 and ERRB3 alterations. Mod Pathol 33, 1287–1297 (2020).
Simpson, P. T., Reis-Filho, J. S., Lambros, M. B., Jones, C., Steele, D., Mackay, A. et al. Molecular profiling pleomorphic lobular carcinomas of the breast: evidence for a common molecular genetic pathway with classic lobular carcinomas. J Pathol 215, 231–244 (2008).
Simpson, P. T., Reis-Filho, J. S., Gale, T. & Lakhani, S. R. Molecular evolution of breast cancer. J Pathol 205, 248–254 (2005).
Ross, J. S., Gay, L. M., Nozad, S., Wang, K., Ali, S. M., Boguniewicz, A. et al. Clinically advanced and metastatic pure mucinous carcinoma of the breast: a comprehensive genomic profiling study. Breast Cancer Res Treat 155, 405–413 (2016).
Maguire, S. L., Leonidou, A., Wai, P., Marchio, C., Ng, C. K., Sapino, A. et al. SF3B1 mutations constitute a novel therapeutic target in breast cancer. J Pathol 235, 571–580 (2015).
Sun, P., Zhong, Z., Lu, Q., Li, M., Chao, X., Chen, D. et al. Mucinous carcinoma with micropapillary features is morphologically, clinically and genetically distinct from pure mucinous carcinoma of breast. Mod Pathol 33, 1945–1960 (2020).
Fu, X., Tian, M., Gu, J., Cheng, T., Ma, D., Feng, L. et al. SF3B1 mutation is a poor prognostic indicator in luminal B and progesterone receptor-negative breast cancer patients. Oncotarget 8, 115018–115027 (2017).
Hull, M. T., Seo, I. S., Battersby, J. S. & Csicsko, J. F. Signet-ring cell carcinoma of the breast: a clinicopathologic study of 24 cases. Am J Clin Pathol 73, 31–35 (1980).
Steinbrecher, J. S. & Silverberg, S. G. Signet-ring cell carcinoma of the breast. The mucinous variant of infiltrating lobular carcinoma? Cancer 37, 828–840 (1976).
Merino, M. J. & Livolsi, V. A. Signet ring carcinoma of the female breast: a clinicopathologic analysis of 24 cases. Cancer 48, 1830–1837 (1981).
Breast Tumours. 5th edn, (International Agency for Research on Cancer Lyon, France, 2019).
Eltorky, M., Hall, J. C., Osborne, P. T. & el Zeky, F. Signet-ring cell variant of invasive lobular carcinoma of the breast. A clinicopathologic study of 11 cases. Arch Pathol Lab Med 118, 245–248 (1994).
Wang, T., Shen, B., Wang, L. & Liu, F. Primary signet ring cell carcinoma of the breast: A rare entity with unique biological behavior-A clinical study based on pure signet ring cell carcinoma cohort. Pathol Res Pract 216, 152948 (2020).
Buerba-Vieregge, H. H., Fernandez-Ferreira, R., Soberanis-Pina, P. D., De la Pena-Lopez, I. R., Navarro-Garcia, L. M. & Macari-Jorge, A. Breast metastasis of gastric signet ring cell carcinoma: a case report and literature review. Case Rep Oncol 14, 165–172 (2021).
Lien, H. C., Hsiao, Y. H., Lin, Y. S., Yao, Y. T., Juan, H. F., Kuo, W. H. et al. Molecular signatures of metaplastic carcinoma of the breast by large-scale transcriptional profiling: identification of genes potentially related to epithelial-mesenchymal transition. Oncogene 26, 7859–7871 (2007).
Sarrio, D., Rodriguez-Pinilla, S. M., Hardisson, D., Cano, A., Moreno-Bueno, G. & Palacios, J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res 68, 989–997 (2008).
Acknowledgements
We are grateful to the Center of Advanced Molecular Diagnostics of Brigham and Women’s Hospital for their technical support and contributions to this study.
Funding
This study was completed with the support of the Department of Pathology of Brigham and Women’s Hospital. The authors received no specific funding for this work.
Author information
Authors and Affiliations
Contributions
T.R.S. and B.T.H. performed study concept and design; T.L.R.S., T.J.W., G.M.B., L.C.C., S.C.L. and S.J.S. provided material support; T.R.S., D.A.D., S.J.S. and B.T.H. participated in data analysis; T.R.S. and B.T.H. prepared the manuscript, tables, and figures; all authors read and approved of the final paper.
Corresponding author
Ethics declarations
Competing interests
D.A.D. consults for Novartis and is on the Academic Advisory Board of Oncology Analytics, Inc. The other authors declare that they have no conflict of interest.
Ethics approval and consent to participate
This study was performed with the approval of the Institutional Review Board of Partners HealthCare/Mass General Brigham in accordance with the Declaration of Helsinki.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Soong, T.R., Dillon, D.A., Rice-Stitt, T.L. et al. Invasive lobular carcinoma with extracellular mucin (ILCEM): clinicopathologic and molecular characterization of a rare entity. Mod Pathol 35, 1370–1382 (2022). https://doi.org/10.1038/s41379-022-01084-w
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41379-022-01084-w
This article is cited by
-
Unraveling complexity and leveraging opportunities in uncommon breast cancer subtypes
npj Breast Cancer (2025)