Abstract
Mutations in CHD8 (chromodomain-helicase-DNA binding protein 8) are highly associated with autism spectrum disorders. It has been well established that CHD8 has a prominent role in the development of neurons. However, there is little knowledge of its specific roles in microglia, and its possible roles in cellular functions after development, i.e. adulthood. In addition, while microglial dysfunction has been characterized in autism, the roles of autism-associated genes in microglial function have not been well characterized. Using conditional knockdown technology in C57BL6 mice models, we determined that adulthood deletion of Chd8 in microglia induces robust changes in behavior, including anxiety, social deficits, and depression-like behavior, in association with changes in microglial activation and robust microglial gene expression changes in the whole brain, including expression of cytokines. Of great interest, many of these changes were seen specifically in male deletion mice, and not female deletion mice. In contrast, adulthood neuron knockdown had more subtle effects on behavior, mainly on depression-like behavior in males. In addition, neuronal knockdown leads to upregulation of genes associated with Hedgehog and Wnt/Beta-catenin pathways in the hippocampus specifically in males. In summary, CHD8 is particularly important for microglial function in adulthood and has cellular effects that are specific to males in C57BL6 mice.
Similar content being viewed by others
Introduction
Autism spectrum disorder is a group of neurodevelopmental disorders that begin in early childhood. Behavior patterns and etiologies of autism are highly heterogeneous [1, 2], and prevalence is four times higher among boys than girls. Clinical diagnosis is characterized by impaired social communication and language development, as well as repetitive and stereotypical behaviors, and prevalence has risen during the last decades to 1 out of 44 eight-year-old children in the USA [3] and about 1% of the worldwide population. CHD8 mutations are among the most common mutations associated with autism and cause approximately 0.5% of all cases. While CHD8 has been found in individuals not diagnosed with autism, multiple studies have shown relatively high levels of penetrance for CHD8 in autism [4,5,6,7,8,9]. Chromodomain helicase DNA-binding protein 8 (CHD8) is located on chromosome 14q11.2 in humans and has two isoforms, a large isoform and a small isoform [10, 11]. Individuals carrying CHD8 mutations have unique phenotypes including atypical face, macrocephaly, tall and slender stature, gastrointestinal disturbance, anxiety, and sleep problems [12,13,14,15,16,17].
CHD8 is a chromatin-binding factor and was found to directly interact with β-catenin and negatively regulate its targeted genes, including WNT signaling [10, 18]. In addition, CHD8 was found to regulate the expression of many genes considered to be autism risk factors and involved in human neurodevelopment. According to ChIP-seq analysis, CHD8 directly binds genomic sites in neurodevelopment-related and autism-risk genes [5, 19]. Interestingly, some studies found that mutations in CHD8 show a male bias effect in mice (in a ratio of (~85:15), but the reason is unclear [7, 20,21,22,23].
Several Chd8 knockdown mouse models have been developed to study the role of CHD8 in brain function and development. However, most studies did not investigate if CHD8 mutations affect behavioral or molecular phenotypes in a sex-specific manner [17, 22, 24,25,26,27,28]. Several models of Chd8 Haploinsufficiency during development displayed increased total brain volume [21, 24, 29, 30], increased absolute volume of the cerebral cortex, hippocampus and amygdala [29, 30] increased cortical and hippocampal connectivity (rsfMRI) [30] and sexually dimorphic neuronal firing and synaptic transmission in the hippocampus [21]. One study found that the function and structure of synapses in the prefrontal cortex were predominantly altered in male Chd8 haploinsufficiency mouse models [31]. In a study of genetic heterogeneity in Chd8 haploinsufficiency mouse models, a significant sex effect on behavioral changes was observed in multiple behaviors [32]. However, nearly all these studies determined phenotypes in models where CHD8 is perturbed from early developmental time points, and did not look into possible effects of CHD8 perturbation specifically in adulthood.
Within the central nervous system, microglia are the primary phagocytotic cells, accounting for 5–20% of all glial cells [33]. They play a critical role in brain development [34] and are involved in neurogenesis, axonal migration, synapse formation, and programmed cell death [35, 36]. As integral components of the immune system, they play a crucial role in regulating neuroinflammatory responses [33, 37, 38]. Microglia were found to be involved in many neurodevelopmental disorders, including autism [34, 39,40,41,42,43,44,45]. Initial human studies found evidence for microglial activation in children diagnosed with autism [46]. Furthermore, animal studies have found that environmental factors, such as maternal immune activation during pregnancy, can induce activation of microglia associated with behavioral and neurodevelopmental changes in offspring [47]. Therefore, microglial dysregulation has been proposed as a biological mechanism linking environmental factors to autism. However, few studies have studied the link between genetic factors involved in autism and microglial regulation.
While downstream effects of CHD8 perturbation in early development have been well studied, it is not well known if CHD8 perturbation specifically in adulthood will induce molecular or behavioral phenotypes. This study aimed to shed light on several aspects of CHD8 regulation in the brain, with a primary focus on its role during adulthood and its specific roles in neurons and microglia in adulthood of C57BL6 mice. In addition, we aim to decipher the sex-specific effects of CHD8 in these cellular subtypes.
Results
Generation and characterization of adulthood-specific Chd8 knockdown mice in microglia and excitatory neurons
To generate adulthood-specific knockdown models of Chd8 in distinct cell populations, Chd8flox C57BL6 mice were crossbred with Cx3cr1Cre-ERT/Chd8flox or CamKIIaCre-ERT /Chd8flox transgenic C57BL6 lines (Fig. 1a). Inducible microglial Chd8 knockdown (imKO mice) were obtained by tamoxifen administration to 8-week-old Chd8flox/Cx3cr1Cre-ERT+ mice. Tamoxifen-treated CHD8flox/Cx3cr1Cre-ERT- mice were used as controls. Four weeks post-treatment, immunohistochemical analysis in the infralimbic region of the Prefrontal Cortex (PFC) confirmed the reduction of Chd8 expression specifically in microglial cells, and not in surrounding IBA-1 negative cells (Fig. 1b, c, Supplementary Fig. 1) in both males and females. In addition, in wild-type mice, CHD8 is found at slightly lower levels in IBA-1+ positive than in IBA-1 negative cells (Supplementary Fig. 1c). Concurrently, adult-onset Chd8 knockdown in excitatory neurons (inKO mice) was achieved by administering tamoxifen to 8-week-old Chd8flox/CamKIIaCre-ERT+ mice and Chd8flox/CamKIIaCre-ERT- controls. Immunohistochemical analysis performed four weeks post-treatment revealed a robust Chd8 depletion distinctively within neurons in the hippocampus (Fig. 1d, e) and in the infralimbic region of the prefrontal cortex (Supplementary Fig. 2).
a Timeline of tamoxifen administration and animal sacrifice of two animal models for immunohistochemistry analysis. b, c Immunohistochemistry of the infralimbic cortex with antibodies targeting microglia (IbaI positive) and CHD8 in male b and female c mice. Nuclei are stained in blue (Hoechst). d, e Immunohistochemistry of hippocampus with antibodies targeting neurons (NeuN), and CHD8 in male d and female e mice. Nuclei are stained in blue (Hoechst).
Sex-specific effect of CHD8 on imKO and inKO mice
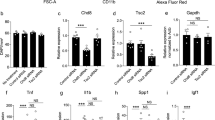
A panel of behavioral tests was performed on the imKO (microglia-specific knockdown) in both male and female cohorts. Male imKO mice exhibited reduced time spent in the center of the open field (Fig. 2a) in the light zone of the dark-light chamber (Fig. 2b) compared to wild-type controls, and in the open arms of the elevated plus maze (Fig. 2c). There were no differences between the genotypes in tests for repetitive behaviors (Fig. 2d, e). However, male imKO mice displayed depression-like behaviors evident by increased immobility in the tail suspension test (Fig. 2f) and forced swimming test compared to wild-type controls (Fig. 2g). There were no differences between genotypes in general motor function in the rotarod test (Fig. 2h) and in distance in open field or beam tests (Supplementary Fig. 3). In the cue fear conditioning test (Fig. 2i), imKO male mice had reduced freezing responses, indicating possible memory and learning impairments. Moreover, alterations in sociability behaviors were evident, as imKO male mice exhibited a lack of preference for the stranger versus an empty cage (Fig. 2j). Therefore, male imKO mice displayed several behavioral phenotypes, including an increase in anxiety and depression-like behavior.
Chd8-imKO males: a Time spent in the center of the open field. (*p = 0.0205, t = 2.554, df = 17, two-way student’s t-test) b Time spent in the light area of the dark-light exploration test. (*p = 0.0473, t = 2.149, df = 16, two-way student’s t-test) c Time spent in the open arm of the elevated plus maze (*p = 0.0242, t = 2.527, df = 14, two-way student’s t-test). d Time self-grooming within a 10 min time period. e Number of marbles buried in 30 min time period. f Time spent immobile in tail suspension test (*p = 0.0195, t = 2.580, df = 17, two-way student’s t-test) g Time spent immobile in forced swim test (**p = 0.0037, t = 3.437, df = 15, two-way student’s t-test). h Motor coordination and balance were assessed using the rotarod. Time spent before falling off rotarod. i Percent freezing in each of three tones in cue fear conditioning test (Two way ANOVA, Genotype effect: p = 0.0055, F(1, 17) = 10.09, Tukey’s post hoc test, *p = 0.0172, **p = 0.0016, *p = 0.0173). j Time spent in interaction zone with stranger mice and empty chamber in three-chamber social interaction test (Two way ANOVA, Stranger effect: p < 0.0001, F(1, 62) = 22.68, Tukey’s post hoc test, ***p < 0.0001, nsp = 0.1589). n = wt 9–11 imKO 7–8. *p < 0.05, **p < 0.01, ***p < 0.001. Chd8-imKO females: k Time spent in the center of the open field (*p = 0.0239, t = 2.479, df = 17, two-way student’s t-test). l Time spent in the light area of the dark-light exploration test. m Time spent in the open arm of the elevated plus maze. n Time self-grooming within a 10 min time period. o Number of marbles buried in 30 min time period. p Time spent immobile in tail suspension test q Time spent immobile in forced swim test. r Motor coordination and balance were assessed using the rotarod. Time spent before falling off rotarod. s Percent freezing in each of three tones in cue fear conditioning test t Time spent in interaction zone with stranger mice and empty chamber in three-chamber social interaction test (Two way ANOVA, Stranger effect: p < 0.0001, F(1, 34) = 24.59, Tukey’s post hoc test *p = 0.0169, **p = 0.0027). Further tests determined no differences between experimental groups. (l- dark-light, m- elevated plus maze, p- tail suspension test, q- forced swimming test). n = wt 9–10 imKO 9–10. *p < 0.05, **p < 0.01.
The same behavioral tests were performed in female imKO mice. Female imKO mice displayed decreased time spent in the center of the open field (Fig. 2k). However, in all other behavioral assessments, including repetitive behavior tests, depression-related tests, fear conditioning, and sociability tests, there were no significant differences between female imKO mice and their wild-type controls (Fig. 2l–t, Supplementary Fig. 3). Therefore, Chd8 deletion in microglia during adulthood induces a strong behavioral phenotype specifically in males.
To study the role of CHD8 within excitatory neurons, we performed the same behavioral tests in inKO mice. InKO male mice displayed reduced exploration in the center of the open field (Fig. 3a), but no differences between phenotypes were displayed in the dark light (Fig. 3b), elevated plus maze (Fig. 3c), and grooming tests (Fig. 3d). However, there was a decrease in the number of buried marbles (Fig. 3e). Moreover, inKO male mice exhibited increased immobility during the forced swimming test (Fig. 3f) and tail suspension test (Fig. 3g), suggesting depression-like behavior. Notably, motor function assessments remained unaltered in rotorod, distance travelled in open field and wire hanging tests (Fig. 3h, Supplementary Fig. 4a,b). However, inKO mice spent significantly more time crossing the beam test, which may be indicative of subtle anxiety or apathy behaviors (Supplementary Fig. 4c,d). No significant differences were observed in the cue fear conditioning test (Fig. 3i) or sociability test (Fig. 3j). These results lead us to conclude that inKO male mice specifically display behaviors of increased depression-like phenotype. The decrease in center exploration in the open field, as well as decreased marble burying, may also suggest a slight anxiety-like phenotype, as seen in Shank3 mouse models [48, 49].
Chd8-inKO male: a Time spent in the center of the open field was decreased in inKO (***p = 0.0003, t = 4.234, df = 24, two-way student’s t-test). b Time spent in the light area of the dark-light exploration test. c Time spent in the open arm of the elevated plus maze. d Time self-grooming within a 10 min time period. e Number of marbles buried in 30 min time period. (**p = 0.0022, t = 3.426, df = 24, two-way student’s t-test). f Time spent immobile in tail suspension test (****p < 0.0001, t = 5.364, df = 28, two-way student’s t-test) g Time spent immobile in forced swim test (*p = 0.0474, t = 2.073, df = 28, two-way student’s t-test). h Motor coordination and balance were assessed using the rotarod. Time spent before falling off rotarod. i Percent freezing in each of three tones in cue fear conditioning test j Time spent in interaction zone with stranger mice and empty chamber in three-chamber social interaction test (Two way ANOVA, Stranger effect: p < 0.0001, F(1, 42) = 70.45, Tukey’s post hoc test ****p < 0.0001). n = wt 11–13 inKO 13–15. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Chd8-inKO Female Mice: k Time spent in the center of the open field. l Time spent in the light area of the dark-light exploration test. m Time spent in the open arm of the elevated plus maze. n Time self-grooming within a 10 min time period. (****p < 0.0001, t = 5.738, df = 23, two-way student’s t-test). o Number of marbles buried in 30 min time period. p Time spent immobile in tail suspension test q Time spent immobile in forced swim test. r Motor coordination and balance were assessed using the rotarod. Time spent before falling off rotarod. s Percent freezing in each of three tones in cue fear conditioning test t Time spent in interaction zone with stranger mice and empty chamber in three-chamber social interaction test (Two way ANOVA, Stranger effect: p < 0.0001, F(1, 48) = 40.56, Tukey’s post hoc test ***p = 0.001, ***p = 0.0006). n = wt10-13 inKO13–15. ***p < 0.001, ****p < 0.0001.
InKO female mice exhibited minor behavioral alterations characterized by a reduction in total distance moved during the open field test (Supplementary Fig. 4e) and heightened grooming behaviors (Fig. 3n). Interestingly, assessments of other anxiety-related behaviors (Fig. 3k–m) and the number of buried marbles (Fig. 3o) in inKO female mice yielded results comparable to the wt group. In contrast to male mice, inKO female mice did not display any depression-like behaviors (Fig. 3p, q). Additionally, motor function assessments (Fig. 3r), cue fear conditioning tests (Fig. 3s), and sociability assessments (Fig. 3t) in female inKO mice were consistent with those of control groups. These results demonstrate a sex-specific imbalance in which CHD8 adulthood knockdown in excitatory neurons induces depression-like behavior exclusively in male mice.
Sex-specific alterations in CHD8-imKO mice: microglial morphology and transcriptomic profile
Microglia morphology was studied in male and female imKO mice. The hippocampus was studied due to its central role in depression-like behavior in mice, as well as the frontal cortex due to the additional change in social behavior. Male imKO mice exhibited a reduction in the number of microglial cells within the hippocampus and PFC (Fig. 4a–d). TUNEL staining determined no increase in apoptosis in male imKO mice, suggesting that the population reduction might not be attributed to apoptosis (Supplementary Fig. 5). Moreover, microglial ramification was significantly decreased, evident as reduced intersection numbers from the soma (Fig. 4e) and lower length of processes (Fig. 4f, l). A decrease in ramification is a hallmark of a pro-inflammatory state of the microglia. Conversely, imKO female mice did not display any significant differences in the microglial population (Fig. 4a, b, g, h) or characteristics (Fig. 4i, j, m). This analysis highlights the influence of CHD8 in microglia particularly in males.
Microglial morphology was assessed in the hippocampus and prefrontal cortex (PFC) of male and female Chd8-imKO mice. a Representative pictures of IBA-1 positive cells in imKO and wt mice in male and female PFC. b Representative pictures of IBA-1 positive cells in imKO and wt mice in male and female hippocampus. c Microglia number (IBA-1+ cells) in hippocampus of male mice (**p = 0.0070, t = 2.868, df = 34, n = wt-17 imKO-19 two-way student’s t test) d IBA-1 positive cells in PFC of male mice (***p = 0.0007, t = 4.128, df = 17, n = wt-11 imKO-8, two-way student’s t test). e Sholl analysis (two-way ANOVA, genotype effect, ****p < 0.0001, F(1, 3212) = 144.1) and f dendrite length of microglial cells in the PFC indicates a reduction in cell complexity (***p = 0.0001, t = 3.990, df = 95, two-way student’s t-test). g IBA-1+ cells in hippocampus of female mice (n = 15 wild type, 15 imKO). h IBA-1+ cells in PFC of female mice (n = 11 wild type, 15 imKO). i Sholl analysis in the PFC of female mice j Measurement of microglial dendrite length in the PFC of female mice. Representative Tracking of microglia processes in k–m WT, male imKO, and female imKO mice using Imaris. **p < 0.01, ***p < 0.001.
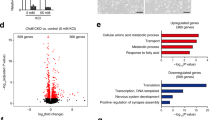
RNA sequencing was employed to study transcriptomic alterations in microglia after CHD8 depletion. Given the limited population of microglial cells in the brain, we performed FACS-mediated isolation of microglia from the entire brain, followed by whole transcriptome analysis (Fig. 5a). CD45+, TMEM119+, and Cd11b+ antibodies were used for identification and sorting of microglia (Fig. 5b, Supplementary Fig. 6). Both genotype and sex had effects on gene transcription (Supplementary Fig. 7). In male mice, there were a total of 1381 genes that were differentially expressed between Chd8 imKO microglia and wild-type microglia (Fig. 5c, Supplementary Table 1). Of those, 773 were upregulated in the imKO microglia and 608 were downregulated. In female mice, only 75 genes were differentially expressed between Chd8 imKO microglia and wild-type microglia (Fig. 5d, Supplementary Table 2). Of those, 18 were upregulated in imKO microglia, and 57 were downregulated. Therefore, CHD8 had a more profound effect on gene expression in the microglia in males compared to females. Marsh et al. previously identified specific microglial genes that are affected by the enzymatic extraction method used in our protocol. Of those 66 genes, we found five genes that were differentially expressed in our male group and five that were differentially expressed in females. Those genes were removed from our list before further analysis [50]. The Gene Ontology (GO) analysis revealed many alterations in males, including the upregulation of chemokines, cytokines, cell cycle-related genes, ATPase activity-dependent DNA processes, and positive regulation of neuron development (Fig. 5e). Cytokine-related genes that were upregulated include CCL2, CCL5, CCL12, CX3CL1, among others. Cell cycle-related genes that were upregulated include multiple genes that downregulate cell cycle progression, including CHEK2 and MTUS1. These findings correlate with the increase in the inflammatory state of the microglia but a decrease in the number of activated microglia. Concurrently, there was a discernible downregulation of ribosomal and structural proteins, which could support the decrease in microglial numbers. In comparison, females displayed comparatively fewer pathway changes, mainly marked by the upregulation of ubiquitin and enzymatic binding pathways, alongside downregulation observed in ribosomal and translational genes (Fig. 5f). These findings underscore a distinct sex-specific response to Chd8 depletion in microglia, with males exhibiting a more substantial transcriptional reprogramming compared to females.
RNA-Seq analysis was performed on microglia from the whole brain tissues obtained from male and female Chd8-imKO mice. a Microglia sorting from the whole brain and b FACS analysis by three antibodies (CD45+, TMEM119+, Cd11b+) that together identify specifically microglia. The presorted CD45+ population is presented. TMEM119 is represented on the Y axis, and CD11b on the X axis. Blue cells are TMEM+CD11b+, red cells are TMEM-CD11b-, and purple cells are TMEM+CD11b-. c, d Volcano plot illustrating differentially expressed genes in the microglia of c male and d female Chd8-inKO mice. Male microglia displayed 1381 differentially expressed genes (Of those, 773 were upregulated in the imKO microglia and 608 were downregulated) and female microglia displayed 75 differentially expressed genes (Of those, 18 were upregulated in the imKO microglia and 57 were downregulated. e, f Gene ontology of differentially expressed genes in e male and f female imKO mice. Principle Component Analysis showing distribution of all samples in this analysis (n = 4–5 per group) found in Supplementary Fig. 7.
Assessment of neuronal morphology and transcriptomic profiling in inKO mice
While fewer behavioral changes were seen in the inKO mice, compared to the imKO mice, further experimentation was performed to understand the possible mechanisms underlying existing dysregulation in the inKO mice. GOLGI staining was used to analyze the neuronal morphology in both male and female inKO mice, evaluating dendrite length and complexity. Hippocampus was studied due to its central role in depression-like behavior, which was the main phenotype in these mice. Surprisingly, our findings revealed no significant alterations in neuron morphology in male or female inKO mice (Fig. 6). This analysis suggests that Chd8 depletion during adulthood does not significantly impact dendritic morphology.
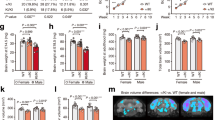
a Dendrite length in hippocampus in male wt and inKO mice (n = 17 wild type, 20 inKO, student’s t-test, not significant). b Sholl analysis of dendritic complexity in male wt and inKO mice (n = 17 wild type, 20 inKO, student’s t test, not significant). c Dendrite length in hippocampus in female wt inKO mice (n = 25 wild type, 19 inKO, two way ANOVA, not significant). d Sholl analysis of dendritic complexity in female wt and inKO mice (n = 25 wild type, 19 inKO, two way ANOVA, not significant). Representative pictures and tracks of e WT and f inKO neurons.
RNA-seq analysis of bulk hippocampus in inKO mice revealed only a few statistically significant changes between wt and inKO mice. There were only 10 differentially expressed genes in inKO males and six differentially expressed genes in inKO females (Fig. 7a, b, Supplementary Tables 3, 4). These differentially expressed genes in both groups were not enriched in any gene ontology categories. Gene Set Enrichment Analysis (GSEA) was further performed on the dataset to discover if any biological pathways were enriched in genes with high fold change, regardless of the statistical significance of individual genes. GSEA discovered the heightened expression of genes associated with the Hedgehog and Wnt/Beta-catenin pathways specifically in male inKO mice (Fig. 7c). Remarkably, these alterations were absent in the hippocampal transcriptome of inKO female mice (Fig. 7d). These findings from RNA-Seq further strengthen sex-specific alterations in gene expression profiles in the hippocampus of Chd8-inKO mice.
RNA-Seq analysis of hippocampal samples from Chd8-inKO male and female mice. Volcano plots highlight minor transcriptional alterations in Chd8-inKO a males and b females. c, d Gene Set Enrichment Analysis (GSEA) detects enrichment for genes within the Hedgehog and Wnt/Beta-catenin pathways that are upregulated specifically in males c but not changed in females d.
Discussion
This study explored the sex-dependent role of CHD8 during adulthood in microglia and excitatory neurons. To achieve this, we generated conditional knockdowns specifically in microglia and excitatory neurons during adulthood. We utilized Chd8flox/Cx3cr1Cre-ERT+ (imKO) C57BL6 mice for microglia and Chd8flox/CamKIIaCre-ERT+ (inKO) C57BL6 mice for excitatory neurons. These newly developed mouse lines allowed us to study the involvement of CHD8 within distinct cell populations during adulthood, considering both male and female subjects.
Deletion of CHD8 had a more pronounced effect in males compared to females in both inKO and imKO mice. This was particularly apparent in the microglia, where male imKO displayed significantly higher levels of dysregulation compared to female imKO mice at the behavioral, cellular, and gene expression levels. Microglial depletion of CHD8 in males manifests in a comprehensive behavioral phenotype, characterized by impaired sociability, anxiety, deficient learning and memory, and depressive behaviors. Conversely, in inKO mice, CHD8 depletion in males primarily leads only to a depressive phenotype. Jung et al. previously observed that CHD8 depletion in Chd8+/N237k mice predominantly impacted male behavior [21]. However, while there were often opposite dysregulations among the sexes at the level of neuronal physiology, including mIPSC frequency, female Chd8+/N237k mice actually displayed more dysregulated genes in the whole brain and hippocampus at P25. Differences in gene expression among our study and earlier studies of Chd8 mutations, including the Chd8+/N237k mice, are likely because our knockdown was during adulthood. Therefore, the lack of significant genes in the neuronal knockdown in our current study suggests that CHD8 has its most important effect on gene expression in neurons during development. It is important to note that Tabbaa et al. found that sex-specific effects in Chd8 mouse models were strain-dependent [32]. In other words, C57BL6 Chd8 mutant female mice showed resilience to some of the behavioral deficits, while female Chd8 mutant mice in many other strains showed similar deficits to those seen in males. Therefore, it is possible that the sex-specific effects of Chd8 deletion seen in our model are specific to the C57BL6 background, and won’t necessarily generalize to other mouse strains.
Only a limited number of studies have explored CHD8’s influence on depression. Cherepanov et al. reported a depressive phenotype exclusively in female Chd8+/∆SL mice, however, they used a haploinsufficiency developmental model [51]. Males exhibited an increased autistic-like phenotype compared to females, including anxiety and fear conditioning dysregulation, in Chd8 haploinsufficiency mouse models [31]. Tabbaa et al. found that the CHD8 sex effect differs between traits and different genetic backgrounds [32]. Shiraishi et al. suggested that the type of Chd8 mutations affects sex dimorphism [52]. Lee et al. found that in the Chd8-S62X mouse model, the CHD8 sex effect is affected by age [7]. However, in Chd8+/N2373K mice, sex dimorphism was observed regardless of age [53]. It is important to note that these studies focused on haploinsufficiency mutations, meaning they investigated the effects of CHD8 absence starting from the developmental stage. In contrast, this study focuses on specific cell types during adulthood. Most other studies have not included or reported depression-related tests in their behavioral tests. Therefore, we cannot rule out the possibility that these phenotypes might also be found in developmental models. Moreover, emerging evidence suggests a role for activated microglia in depression [54]. In our gene expression analysis, we found an increase in several cytokines, including CCL2, in imKO mice. CCL2 has been implicated as a driver of depression-like behavior [55].
The current study reveals that CHD8 depletion in microglia induces both morphological and transcriptional alterations. Our results align with existing studies that have reported gene expression changes linked to immune response activation in the context of autism [42, 56]. Activated microglia exhibit a dynamic morphological transition between pro-inflammatory and anti-inflammatory stages, in which activated microglia display less branching, otherwise known as decreased ramification. Microglia characterized by a short ramification morphology release pro-inflammatory cytokines and chemokines, potentially contributing to neuronal damage [38, 41, 57,58,59]. Chd8 imKO microglia displayed decreased ramification at the morphological level, in parallel to increased transcription of cytokines. In male Chd8-imKO mice, cytokine-related genes that were upregulated included CCL2, CCL5, CCL12, and CX3CL1, all of which are associated with pro-inflammatory functions [60, 61]. Therefore, Chd8 knockdown induced an activation of microglia. In a separate study of CHD8 knockdown in human cerebral organoids, Astorkia et al. demonstrated that CDH8 reduction leads to impaired neuronal-glial communication [62]. The decrease in number of microglia, in comparison to the activation of microglia, is more difficult to interpret. The transcriptional data determined dysregulation of several cell-cycle-related genes, including genes responsible for inhibiting cell-cycle progression. This may be the reason for the fall in microglia number, particularly since no apoptosis was detected in the tissue. The microglia of female Chd8 imKO mice displayed subtle gene expression changes which were enriched for ribosomal-related genes. While these changes did not correlate with major behavioral or morphological phenotypes in the female mice, it is possible that phenotypes could emerge under the effects of inflammatory environmental insults, such as LPS or Poly I:C treatment.
Multiple previous studies have found evidence for sex-specific differences in microglial function. Villa et al. found multiple gene expression alterations specifically in female microglia, compared to male microglia, indicating a neuroprotective phenotype of microglia in females. In addition, transplantation of female microglia in males helped to treat damage caused by acute cerebral ischemia [58]. Another study found that females have increased anti-inflammatory microglia during adulthood [59]. In addition, Xu et al. revealed that exaggerated protein translation in microglia impaired sociability behavior in males, not females, accompanied by microglia morphological changes and a reduction in the number of microglia in PFC in males only. RNA-sequencing found that this was accompanied by upregulation of cytokines and chemokines expression in microglia KO males only [63]. Therefore, male microglia seem to be particularly vulnerable to dysregulation, although it is still not completely clear why. A reason for the sex-specific effects of microglial Chd8 knockdown would be the differential binding of CHD8 on genomic regions between male and female microglia. Future experimentation should look into the possible sex-specific binding of CHD8 in microglia.
This study unveils sex-specific variations in both the morphology and transcriptional profile of microglia, offering pivotal insights into the consequences of CHD8 deficiency on microglial populations. In contrast to the negligible impact observed in excitatory neuron morphology and transcriptomic following CHD8 depletion, microglial depletion resulted in a reduction in cell number in the prefrontal cortex (PFC) and hippocampus. Notably, reduced ramification was observed exclusively in males, underscoring the sex-specific nature of these morphological changes. Moreover, a substantial alteration in gene expression was evident, with the effects being more pronounced in males. Since microglia are known to regulate synaptic structure and activity, it would be interesting to decipher if there is dysregulation of synaptic or neuronal morphology in the Chd8 imKO. The interplay between microglial dysfunction and neuronal morphology and activity can be an avenue of future research in this animal model.
Various Chd8 mouse models have reported changes in neuronal gene expression [21, 22, 25, 29, 30], although the changes in adult animals are often very minimal. The current study revealed elevated gene transcription associated with Hedgehog and Wnt/β-catenin signaling pathways according to the GSEA analysis. Wnt/β-catenin signaling is crucial for diverse processes in the central nervous system, including neuronal differentiation, synaptic maintenance, and axonal remodeling, has been associated with autism [64]. CHD8 is a β-catenin binding protein, and several studies have shown that CHD8 is a negative regulator of the β-catenin pathway [18, 65]. Some previous studies have also found that deletion of Chd8 is associated with upregulated inWnt/β-catenin signaling in neurons [66, 67]. Platt. et. al. also reported an increase in Wnt/β-catenin-associated gene transcription specifically in the nucleus accumbens of mice with a Chd8 mutation [25]. While CHD8 is expressed at high levels in neurons during the embryonic stage, peaking at E16–18, its expression dramatically decreases postnatally [25, 68, 69]. Along with the fact that neurons do not proliferate during adulthood, this may explain the limited impact of CHD8 depletion in adulthood on neuronal morphology and gene transcription. An important limitation is that RNA sequencing in the neuronal model was conducted on bulk hippocampal samples rather than purified neurons. However, since neurons constitute a large proportion of the cells in the hippocampus, they likely contribute significantly to the overall gene expression profile.
This novel study determined how CHD8 affects behavior and gene transcription during adulthood and comparatively in microglia and neurons in a sex-specific manner. Interestingly, CHD8 depletion in microglia causes multiple behavioral phenotypes in adult males. On the other hand, CHD8 depletion in excitatory neurons mainly leads to a depression-like phenotype. Also, a male bias, as seen in humans, was observed in these mouse models. This research gives us new information about how CHD8 can influence depression-like and social behaviors in adults, and its effect on specific cell populations.
Methods
Mouse models
C57BL6 Mice were housed according to Federation of Laboratory Animal Science Associations (FELASA) guidelines. Mice were maintained in a vivarium at 22 C with a reversed light cycle (lights on at 19:00 h, off at 07:00 h). Food and water were provided ad libitum, except during behavioral testing. Testing occurred between (9:00 and 16:00 h). Littermate mice were randomly assigned to experimental groups.
Ethics approval and consent to participate
All experimentation was approved by the Institutional Animal Care and Use Committee (IACUC) under protocol number 46-07-2021 and experimentation was performed in accordance with the relevant guidelines and regulations (FELASA). There was no human experimentation that requires consent to participate.
inKO mouse model
To induce conditional knockdown of Chd8 in excitatory neurons, the CamKiia-Cre-Ert C57BL6 mouse line (kindly provided by the laboratory of Prof. Alon Chen, Weizmann Institute of Science, Israel) was crossed with Chd8lx/flx C57BL6 mice (The Jackson Laboratory, Strain #:031555) to generate the Chd8lx/flxCamk2aCreErt+/− cKO mice. Then, these were further bred with Chd8flx/flx mice to produce both the Chd8flx/flxCamk2aCreErt+/− (cKO) and Chd8flx/flxCamk2aCreErt−/− (wild-type) mice, forming the basis for the experiments. All offspring maintained the Chd8flx/flx genotype, with half carrying the Camk2aCreErt+/− allele. The Camk2aCreErt−/− mice served as control animals throughout the experiments.
imKO mouse model
In parallel, the Cx3cr1-Cre-Ert mouse line (kindly provided by the laboratory of Prof. Stephen Jung, Weizmann Institute of Science, Israel) was used to specifically target Chd8 in microglia. These mice were crossed with Chd8flx/flx mice (The Jackson Laboratory, Strain #:031555) to generate the Chd8flx/flxCx3cr1CreErt+/− (cKO) andChd8flx/flxCx3cr1CreErt−/− (wild-type) mice. All experimental studies involved crossing these two lines, ensuring that all offspring maintained the Chd8flx/flx genotype, with half carrying the Cx3cr1CreErt+/− allele.
DNA extraction and genotyping
Genotyping was carried out using DNA isolated from ear tissue followed by PCR. DNA Extraction: Ear tissue samples from mice were suspended for 30 min at 95 °C in the DNA extraction buffer. The extraction buffer consisted of NaOH (5 M) and EDTA (0.5 M) at pH 7.6–8. Specific primers were utilized for each target gene: Chd8 Primers: Forward Primer: 5′ TGG GGT GCT GGG AAC AGT A 3′. Reverse Primer: 5′ GCT CAC ACG AAT ATA ACC TCA CA 3′. Camk2a-Cre-Ert Primers: Forward Primer: 5′ GGT TCT CCG TTT GCA CTC AGG A 3′. Reverse Primers: Reverse 1: 5′ CTG CAT GCA CGG GAC AGC TCT 3′. Reverse 2: 5′ GCT TGC AGG TAC AGG AGG TAG T 3′. Cx3cr1-Cre-Ert Primers: Common Forward Primer: 5′ ACG CCC AGA CTA ATG GTG AC 3′. Mutant Reverse Primers: 5′ GTT AAT GAC CTG CAG CCA AG 3′. PCR amplification was performed using Master Mix 2xPCRBIO HS Taq Mix Red (PBo45618-032-17).
Tamoxifen treatment for inducing Cre-ERT expression
To activate the Cre-ERT system, all groups were given Tamoxifen treatment at 8 weeks old including both cKO and wt (wild-type) mice. Tamoxifen was administered once daily for four consecutive days at a dosage of 100 mg/kg per day via oral gavage. Four weeks post-treatment immunofluorescence was carried out followed by behavioral tests.
Animal brain fixation
A 100 μl injection of Petal veterinary solution (pentobarbitone sodium 200 mg/ml CTS) diluted with saline (1:10) was administered i.p. to the mice. The abdominal cavity was opened and a needle was placed in the left ventricle. Blood was washed out with PBS administration using a paristaltic pump into the heart, followed by fixation using 4% Paraformaldehyde (PFA) injection. The fixed brains were extracted and incubated overnight in 4% PFA, followed by another overnight incubation in 30% sucrose with 4% PFA. Brain slices of 30μm thickness were obtained using a sliding microtome (HM430) and stored in PBS with sodium azide at 4 °C.
Immunostaining
PBS washes were performed on the brain slices, followed by a 10 min incubation in Sodium citrate buffer (10 mM Sodium citrate, 0.05% Tween 20, pH 6.0) for antigen retrieval. Further washing steps included PBST (0.3% Triton) and PBS. Blocking was carried out using normal horse serum for 1 h at room temperature. Overnight incubation at 4˚C was performed with primary antibodies diluted to (1:200) in PBS. After three washes with PBST and PBS, slices were incubated with secondary antibodies for 1 h. Subsequent washes were performed, and the slices were mounted on glass slides using immu-mount. Throughout the procedure, incubations were conducted in shaking mode. Imaging was conducted using an Upright Microscope ApoTome or Slide Scanner.
Neurons were labeled using the anti-NeuN Antibody (clone A60, MAB377 Sigma-Aldrich, Host-mouse) as the primary antibody and Cy3 (Donkey anti-mouse IgG, Jackson, 715165150) as the secondary antibody. Microglia was marked by anti-IbaI (Goat anti-mouse IgG, Abcom, ab5076) as the primary antibody and Donkey anti-goat Cy3 as a secondary antibody. For CHD8, the anti-CHD8 antibody (ab84527, Rabbit polyclonal to CHD8, Isotype: IgG) served as the primary antibody, while Alexa Fluor® 488 (Goat anti-rabbit, Jackson, 11545144) was used as the secondary antibody. Nuclei were marked with Hoechst stain (Sigma, 1:10,000). Staining in the prefrontal cortex was specifically in the infralimbic region of the prefrontal cortex (bregma 1.75). Staining of the hippocampus was specifically in the dentate gyrus (bregma −2.00).
Slices were scanned using the ZEISS Axio Scan.Z1 slide scanner and cell numbers and staining intensity was measured using ZEN blue image analysis software.
Behavioral tests
Mice, aged between 8–10 weeks, were administered a 100 mg/kg tamoxifen solution per day via gavage. Four weeks post the initial tamoxifen administration, behavioral tests were carried out. A camera was employed to record the mice’s movements. Tracking and analysis of animal behavior were conducted using EthoVision XT 10-Noldus. Animals of the same genotype were randomly assigned to each experimental group. Exact numbers in each experiment is described in each figure. Animals were excluded from analysis only if they left the experimental arena. A separate set of mice were used for forced swim test and tail suspension test in order not to perform too many stressful tests on one set of animals.
Open field test and grooming
The open field test was performed to evaluate anxiety-like behavior in mice within a novel environment. Each mouse was placed in a 50 × 50 × 30 cm open field box, positioned in a corner, under a light intensity of approximately ~120 lux. The first 10 min of the test were analyzed for cumulative duration spent in the center and border zones, along with the total distance moved by the mouse. The subsequent 10 min of the session were dedicated to manually assessing grooming behaviors.
Dark-light test
The dark-light test was conducted as an additional measure to evaluate anxiety-related behaviors in mice. A two-chambered cage setup was utilized for this test, consisting of a dark chamber made from black plastic with a closable top, and a light chamber made from white plastic that remained open at the top. The light intensity in the light chamber was maintained at approximately ~1200 lux. Initially, the mouse was placed in the dark chamber with the top cover closed. Access to the light chamber was provided by opening the entryway, allowing the mouse to freely move between chambers for 10 min. The analysis involved tracking the frequency and cumulative duration of the mouse’s presence within the light arena.
Elevated plus maze test
This test was used to evaluate anxiety-related behaviors in mice, focusing on their exploration of the safe and unsafe zones of the maze. An X-shaped apparatus comprising two open arms (30 cm by 5 cm, white plastic) and two closed arms (15 cm high walls) was utilized for the test. The maze was positioned 55 cm above the floor, maintaining a light intensity of 20 lux. The mouse was initially placed in the central square (5 by 5 cm) of the maze and allowed to navigate among the arms for 5 min. Analysis included the frequency and cumulative duration of visits to the center, open, and closed arms.
Marble burying test
The marble burying test was carried out to evaluate repetitive behaviors exhibited by mice. The cage was lined with a 5 cm layer of sawdust, and 20 marbles of uniform size and color were evenly arranged on the surface. Lighting conditions were maintained at 20 lux throughout the test. The mouse was placed in a corner of the cage and allowed to explore the environment freely for 30 min. Marbles that were buried to a depth ranging between 70–100% beneath the sawdust layer were counted.
Tail suspension test
During the tail suspension test, the mouse was suspended by the last cm of its tail, allowing it to hang at 60 cm above the floor within a chamber (20 × 40 × 60 cm). Video recording captured the mouse behavior during the 6 min test, focusing on total immobility.
Forced swimming test
A dark chamber was filled with water at room temperature to a depth of 15 cm. The mouse was placed in the water for a total duration of 6 min, with the first 2 min dedicated to habituation, and was video recorded. The last 4 min of the test were analyzed, focusing on the observation of mouse immobility.
Rotarod test
Mice were placed on a rotating cylinder apparatus (Med Associates, St. Albins, VT) set at a speed of 40 rpm, with a maximum test duration of 300 s. The time that each mouse remained on the rotating cylinder was measured as a marker of motor function.
Wire hanging test
Mice were given a 2 mm thick horizontal metal wire to grip onto. The test was performed five times for each mouse to ensure accuracy and consistency. The time until the mouse fell from the wire, was recorded during each trial. Based on the five trials, an average latency to fall was calculated to represent its grip motor functions.
Beam test- 6 and 12 mm
Mice were placed on a 1-meter-long horizontal beam, varying in thickness between 6 mm and 12 mm. The time taken by the mouse to reach a designated safety zone from the starting point on the beam was measured. Additionally, the number of falls the mouse experienced while traversing the beam was recorded. Before the test, mice underwent a training phase where they were familiarized with navigating through the beam. Training sessions were carried out over two consecutive days, with mice undergoing two sessions per day to ensure familiarity and consistency.
Cue fear conditioning test
The cue fear conditioning test was utilized to study memory and learning abilities in mice. The initial day was allocated for training and habituation. Mice were introduced into the test chamber (10.5 × 10.5 × 10.5 cm) for 5 min. A 75 dB tone served as the conditioned stimulus for 30 s, followed by a brief, mild foot shock (0.7 mA) lasting 2 s as the unconditioned stimulus. After a 1 min interval, another tone-shock pair was administered, and subsequently, the mouse was returned to its home cage 1 min after the second tone-shock pairing. The following day, mice were placed in a different chamber with novel sensory cues (odor, flooring, and lighting) for cue-dependent memory assessment. After a 2 min habituation period, the tone was presented three times for 30 s each, with 1 min intervals between tones. Freezing behavior during the three-tone presentations was recorded as a measure of cue-dependent memory recall.
Sociability assessment
The three-chambered social test aimed to evaluate sociability behavior in mice through social interaction assessment. The testing apparatus consisted of a three-chambered cage divided by two entrances, each chamber measuring 20 by 40 by 22 cm. A clear plastic cylinder was positioned between the right and left chambers to facilitate controlled social interaction. Two mice of identical genotypes were introduced into the clear plastic cylinder for 10 min each day over 5 days for habituation purposes. The subject mouse was positioned in the center of the middle chamber. Initially, an unfamiliar mouse (stranger) was placed in the right cylinder for a 5 min habituation phase. Following this habituation, gates providing access to the right and left chambers were opened, enabling the tested mouse to freely explore all three chambers for 10 min. Behavioral metrics included measuring the time and distance spent by the tested mouse in each chamber and within a predefined sniffing zone.
Golgi staining for excitatory neuron morphology
GOLGI-Cox stain kit from Bioenno Tech, LLC was employed to analyze the morphology of excitatory neurons following the manufacturer’s protocol. 12-week-old mice brains were extracted and freshly dissected brains were sectioned at 180 µm thickness using a vibratome (Leica VT 1200 s). Excitatory neurons were randomly selected for analysis, and their images were captured using a bright-field microscope. IMARIS 9.1.2 software was utilized to measure dendritic length and conduct Sholl analysis for neuron morphology assessment. Statistical analysis was performed using two-way ANOVA via GraphPad Prism software.
Sample collection and bulk hippocampal RNA sequencing
12-week-old mice were decapitated and brains were harvested and washed with PBS buffer and the hippocampal tissue was dissected. Hippocampus was extracted bilaterally using a 14 gauge needle. RNA extraction was performed utilizing the RNeasy Micro Kit (QIAGEN, 74004) following the manufacturer’s protocol. To prepare libraries, NEBNext® Poly(A) mRNA Magnetic Isolation Module was used to isolate mRNA. This was followed by library preparation using NEBNext® Ultra™ II RNA Library Prep Kit for Illumina. A total of 30 libraries, accounting for 20.84 Gbp, were generated and subjected to initial quality control analysis. Quality assessment was conducted, and the samples were trimmed accordingly. A secondary quality evaluation of the reads was performed. Fastq-Screen software (v0.14.1) was utilized to evaluate contamination by aligning randomly selected reads to various reference genomes. Reads were mapped to the mouse genome (GCF_000001635.27_GRCm39 assembly) using the STAR software (v.27.9a). Uniquely mapped reads were quantified based on their corresponding transcripts using FeatureCounts software (V2.0.3). Assessment of DNA contamination was conducted by analyzing mapped reads based on their genomic location (Exons/Introns). Removal of PCR duplicates was executed using the DupRadar r-package to ensure data integrity. Raw counts obtained were employed to identify differentially expressed genes across the experimental groups (FcKO, FWT, McKO, and MWT). Differential expression analysis was performed using the EdgeR R-package (v3.23.5) with genes filtered based on LogFC and FDR criteria (Adjusted p-value < 0.05). GSEA software was used for gene network analysis. Genes were preranked by logfold2 change and then subject to GSEA analysis.
TUNEL staining
The In-Situ Cell Death Detection Kit (Roche Life Science) was used to identify apoptotic processes within the brain tissue. 30 µm thick floating sections obtained from 4% formaldehyde-fixed brains were utilized for the staining analysis. The staining procedure followed the instructions provided by the manufacturer.
Microglial sorting via FACS
12-week-old mice were anesthetized (pentobarbitone sodium 200 mg/ml), and brains were extracted following PBS perfusion. Brain tissues underwent enzymatic digestion by incubating at 37 °C with 1 ml of digestion buffer for 20 min (pipet after 10 min), followed by passage through a grey mesh to homogenize. After enzymatic digestion, the sample was washed using 10 ml MACS buffer. Gentlely pounding of the mesh with a syringe’s rubber end assisted tissue passage. To halt collagenase action, a total of 20–30 ml MACS buffer was added, followed by centrifugation at 2200 rpm, 4 °C for 5 min. The pellet was resuspended in 1 ml of 40% percol solution and transferred to a tube containing 2 ml of 40% percol, totaling 3 ml in a 15 ml tube. Centrifugation was performed at 900 g, room temperature, no breaks for 15 min. Percol was aspirated, resuspended the pellet in ~100ul MACS buffer, avoiding debris, and transferred through a white mesh into a fresh FACS tube containing 5 ml cold MACS buffer on ice. Subsequent centrifugation was conducted at 1400 rpm, 4 °C for 5 min. The supernatant was discarded, and the pellet was gently vortexed. Fc blocks (10 ul of 1:20 in FACS buffer) were added and incubated for 15 min on ice. Extracellular staining was performed without washing by adding 50 ul of antibodies (CD45-Alexa 488, CD11b-BV 605, Tmem119-Alexa 647 at (1:100) dilution in MACS buffer) and incubating for 15 min on ice in the dark. Washed cells were centrifuged at 1400 rpm, 4 °C for 5 min. The supernatant was discarded, and the pellet was resuspended in 300 ul MACS buffer. The cell suspension was transferred into a new FACS tube and sorted using Aria-Sorter based on specific antibodies (CD45, CD11b, Tmem119) identifying microglia.
Microglial RNA sequencing
Following cell sorting, RNA was extracted from the isolated microglial cells. RNA-seq libraries were prepared at the Crown Genomics Institute of the Nancy and Stephen Grand Israel National Center for Personalized Medicine, Weizmann Institute of Science. A bulk adaptation of the MARS-Seq protocol (Jaitin et al., Science, 2014; Keren-Shaul et al., Nature Protocols, 2019) was used to generate RNA-Seq libraries for expression profiling. Briefly, (30 ng of input) RNA from each sample was barcoded during reverse transcription and pooled. Following Agencourct Ampure XP beads cleanup (Beckman Coulter), the pooled samples underwent second-strand synthesis and were linearly amplified by T7 in vitro transcription. The resulting RNA was fragmented and converted into a sequencing-ready library by tagging the samples with Illumina sequences during ligation, RT, and PCR. Libraries were quantified by Qubit and TapeStation as well as by qPCR for the mouse Actin B gene as previously described (Jaitin et al., Science, 2014; Keren-Shaul et al., Nature Protocols, 2019). Sequencing was done on a Nova-Seq6000 using SP, 100 cycles kit mode, allocating 800 M reads in total (Illumina). Poly-A/T stretches and Illumina adapters were trimmed from the reads using cutadapt]; resulting reads shorter than 30 bp were discarded. Remaining reads were mapped onto 3′ UTR regions (1000 bases) of the M. musculus, GRCm39 genome according to Refseq annotations, using STAR with EndToEnd option and outFilterMismatchNoverLmax was set to 0.05. Deduplication was carried out by flagging all reads that were mapped to the same gene and had the same UMI. Counts for each gene were quantified using htseq-count, using the gtf above. UMI counts were corrected for saturation by considering the expected number of unique elements when sampling without replacement. Differentially expressed genes were identified using DESeq2 with the betaPrior, cooksCutoff and independentFiltering parameters set to False. Raw P values were adjusted for multiple testing using the procedure of Benjamini and Hochberg. The raw data is available online at GSE262286.
Statistics
Statistical analysis of behavior and immunohistochemistry quantification was performed using Graphpad prism 9.3 software. Statistical test used depends on experiment and is mentioned in figure legend. Main tests used were two-tailed unpaired T-test or two way ANOVA. Levene’s test was carried out before each test to verify homogeneity of variance. Data are presented as mean ± standard error of the mean (SEM).
Data availability
The RNA-seq data produced in this study is available at GSE262286. Any other data can be provided by request from the corresponding author.
References
Pina-Camacho L, Villero S, Fraguas D, Boada L, Janssen J, Navas-Sánchez FJ, et al. Autism spectrum disorder: does neuroimaging support the DSM-5 proposal for a symptom dyad? a systematic review of functional magnetic resonance imaging and diffusion tensor imaging studies. J Autism Dev Disord. 2012;42:1326–41. https://doi.org/10.1007/s10803-011-1360-4.
Kereszturi É. Diversity and classification of genetic variations in autism spectrum disorder. Int J Mol Sci. 2023;24:16768 https://doi.org/10.3390/ijms242316768.
Maenner MJ, Shaw KA, Bakian AV, Bilder DA, Durkin MS, Esler A, et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years - autism and developmental disabilities monitoring network, 11 sites, United States, 2018. MMWR Surveill Summ. 2021;70:1–16. https://doi.org/10.15585/mmwr.ss7011a1.
Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–5. https://doi.org/10.1038/nature11011.
Wilkinson B, Grepo N, Thompson BL, Kim J, Wang K, Evgrafov OV, et al. The autism-associated gene chromodomain helicase DNA-binding protein 8 (CHD8) regulates noncoding RNAs and autism-related genes. Transl Psychiatry. 2015;5:e568 https://doi.org/10.1038/tp.2015.62.
Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An J-Y, et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020;180:568–84.e23. https://doi.org/10.1016/j.cell.2019.12.036.
Lee SY, Kweon H, Kang H, Kim E. Age-differential sexual dimorphism in CHD8-S62X-mutant mouse behaviors. Front Mol Neurosci. 2022;15:1022306 https://doi.org/10.3389/fnmol.2022.1022306.
Kweon H, Jung WB, Im GH, Ryoo J, Lee J-H, Do H, et al. Excitatory neuronal CHD8 in the regulation of neocortical development and sensory-motor behaviors. Cell Rep. 2021;34:108780 https://doi.org/10.1016/j.celrep.2021.108780.
Wang T, Hoekzema K, Vecchio D, Wu H, Sulovari A, Coe BP, et al. Large-scale targeted sequencing identifies risk genes for neurodevelopmental disorders. Nat Commun. 2020;11:4932 https://doi.org/10.1038/s41467-020-18723-y.
Thompson BA, Tremblay V, Lin G, Bochar DA. CHD8 is an ATP-dependent chromatin remodeling factor that regulates beta-catenin target genes. Mol Cell Biol. 2008;28:3894–904. https://doi.org/10.1128/MCB.00322-08.
Zahir F, Firth HV, Baross A, Delaney AD, Eydoux P, Gibson WT, et al. Novel deletions of 14q11.2 associated with developmental delay, cognitive impairment and similar minor anomalies in three children. J Med Genet. 2007;44:556–61. https://doi.org/10.1136/jmg.2007.050823.
O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–22. https://doi.org/10.1126/science.1227764.
An Y, Zhang L, Liu W, Jiang Y, Chen X, Lan X, et al. De novo variants in the Helicase-C domain of CHD8 are associated with severe phenotypes including autism, language disability and overgrowth. Hum Genet. 2020;139:499–512. https://doi.org/10.1007/s00439-020-02115-9.
Douzgou S, Liang HW, Metcalfe K, Somarathi S, Tischkowitz M, Mohamed W, et al. The clinical presentation caused by truncating CHD8 variants. Clin Genet. 2019;96:72–84. https://doi.org/10.1111/cge.13554.
Wang T, Guo H, Xiong B, Stessman HAF, Wu H, Coe BP, et al. De novo genic mutations among a Chinese autism spectrum disorder cohort. Nat Commun. 2016;7:13316 https://doi.org/10.1038/ncomms13316.
Stolerman ES, Smith B, Chaubey A, Jones JR. CHD8 intragenic deletion associated with autism spectrum disorder. Eur J Med Genet. 2016;59:189–94. https://doi.org/10.1016/j.ejmg.2016.02.010.
Weissberg O, Elliott E. The mechanisms of CHD8 in neurodevelopment and autism spectrum disorders. Genes. 2021;12:1133 https://doi.org/10.3390/genes12081133.
Nishiyama M, Skoultchi AI, Nakayama KI. Histone H1 recruitment by CHD8 is essential for suppression of the Wnt-β-catenin signaling pathway. Mol Cell Biol. 2012;32:501–12. https://doi.org/10.1128/MCB.06409-11.
Cotney J, Muhle RA, Sanders SJ, Liu L, Willsey AJ, Niu W, et al. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat Commun. 2015;6:6404 https://doi.org/10.1038/ncomms7404.
Cerase A, Avner P. From X-inactivation to neurodevelopment: CHD8-transcription factors (TFs) competitive binding at regulatory regions of CHD8 target genes can contribute to correct neuronal differentiation. Curr Res Neurobiol. 2023;5:100114 https://doi.org/10.1016/j.crneur.2023.100114.
Jung H, Park H, Choi Y, Kang H, Lee E, Kweon H, et al. Sexually dimorphic behavior, neuronal activity, and gene expression in Chd8-mutant mice. Nat Neurosci. 2018;21:1218–28. https://doi.org/10.1038/s41593-018-0208-z.
Jiménez JA, Ptacek TS, Tuttle AH, Schmid RS, Moy SS, Simon JM, et al. Chd8 haploinsufficiency impairs early brain development and protein homeostasis later in life. Mol Autism. 2020;11:74 https://doi.org/10.1186/s13229-020-00369-8.
Ostrowski PJ, Zachariou A, Loveday C, Beleza-Meireles A, Bertoli M, Dean J, et al. The CHD8 overgrowth syndrome: A detailed evaluation of an emerging overgrowth phenotype in 27 patients. Am J Med Genet C Semin Med Genet. 2019;181:557–64. https://doi.org/10.1002/ajmg.c.31749.
Katayama Y, Nishiyama M, Shoji H, Ohkawa Y, Kawamura A, Sato T, et al. CHD8 haploinsufficiency results in autistic-like phenotypes in mice. Nature. 2016;537:675–9. https://doi.org/10.1038/nature19357.
Platt RJ, Zhou Y, Slaymaker IM, Shetty AS, Weisbach NR, Kim J-A, et al. Chd8 mutation leads to autistic-like behaviors and impaired striatal circuits. Cell Rep. 2017;19:335–50. https://doi.org/10.1016/j.celrep.2017.03.052.
Zhao C, Dong C, Frah M, Deng Y, Marie C, Zhang F, et al. Dual requirement of CHD8 for chromatin landscape establishment and histone methyltransferase recruitment to promote CNS myelination and repair. Dev Cell. 2018;45:753–68.e8. https://doi.org/10.1016/j.devcel.2018.05.022.
Hulbert SW, Wang X, Gbadegesin SO, Xu Q, Xu X, Jiang Y-H. A novel chd8 mutant mouse displays altered ultrasonic vocalizations and enhanced motor coordination. Autism Res. 2020;13:1685–97. https://doi.org/10.1002/aur.2353.
Hurley S, Mohan C, Suetterlin P, Ellingford R, Riegman KLH, Ellegood J, et al. Distinct, dosage-sensitive requirements for the autism-associated factor CHD8 during cortical development. Mol Autism. 2021;12:16 https://doi.org/10.1186/s13229-020-00409-3.
Gompers AL, Su-Feher L, Ellegood J, Copping NA, Riyadh MA, Stradleigh TW, et al. Germline Chd8 haploinsufficiency alters brain development in mouse. Nat Neurosci. 2017;20:1062–73. https://doi.org/10.1038/nn.4592.
Suetterlin P, Hurley S, Mohan C, Riegman KLH, Pagani M, Caruso A, et al. Altered neocortical gene expression, brain overgrowth and functional over-connectivity in Chd8 haploinsufficient mice. Cereb Cortex. 2018;28:2192–206. https://doi.org/10.1093/cercor/bhy058.
Ellingford RA, Tojo M, Basson MA, Andreae LC. Male-dominant effects of Chd8 haploinsufficiency on synaptic phenotypes during development in mouse prefrontal cortex. ACS Chem Neurosci. 2024;15:1635–42. https://doi.org/10.1021/acschemneuro.3c00690.
Tabbaa M, Knoll A, Levitt P. Mouse population genetics phenocopies heterogeneity of human Chd8 haploinsufficiency. Neuron. 2023;111:539–56.e5. https://doi.org/10.1016/j.neuron.2023.01.009.
Kierdorf K, Prinz M. Microglia in steady state. J Clin Invest. 2017;127:3201–9. https://doi.org/10.1172/JCI90602.
Lukens JR, Eyo UB. Microglia and neurodevelopmental disorders. Annu Rev Neurosci. 2022;45:425–45. https://doi.org/10.1146/annurev-neuro-110920-023056.
Fan G, Ma J, Ma R, Suo M, Chen Y, Zhang S, et al. Microglia modulate neurodevelopment in autism spectrum disorder and schizophrenia. Int J Mol Sci. 2023;24:17297 https://doi.org/10.3390/ijms242417297.
Schwarz JM, Sholar PW, Bilbo SD. Sex differences in microglial colonization of the developing rat brain. J Neurochem. 2012;120:948–63. https://doi.org/10.1111/j.1471-4159.2011.07630.x.
Carson MJ, Doose JM, Melchior B, Schmid CD, Ploix CC. CNS immune privilege: hiding in plain sight. Immunol Rev. 2006;213:48–65. https://doi.org/10.1111/j.1600-065X.2006.00441.x.
Yang R, Wang H, Wen J, Ma K, Chen D, Chen Z, et al. Regulation of microglial process elongation, a featured characteristic of microglial plasticity. Pharmacol Res. 2019;139:286–97. https://doi.org/10.1016/j.phrs.2018.11.028.
McDougle CJ, Landino SM, Vahabzadeh A, O’Rourke J, Zurcher NR, Finger BC, et al. Toward an immune-mediated subtype of autism spectrum disorder. Brain Res. 2015;1617:72–92. https://doi.org/10.1016/j.brainres.2014.09.048.
Fernández de Cossío L, Guzmán A, van der Veldt S, Luheshi GN. Prenatal infection leads to ASD-like behavior and altered synaptic pruning in the mouse offspring. Brain Behav Immun. 2017;63:88–98. https://doi.org/10.1016/j.bbi.2016.09.028.
Morgan JT, Chana G, Pardo CA, Achim C, Semendeferi K, Buckwalter J, et al. Microglial activation and increased microglial density observed in the dorsolateral prefrontal cortex in autism. Biol Psychiatry. 2010;68:368–76. https://doi.org/10.1016/j.biopsych.2010.05.024.
Gupta S, Ellis SE, Ashar FN, Moes A, Bader JS, Zhan J, et al. Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism. Nat Commun. 2014;5:5748 https://doi.org/10.1038/ncomms6748.
McCarthy MM, Wright CL. Convergence of sex differences and the neuroimmune system in autism spectrum disorder. Biol Psychiatry. 2017;81:402–10. https://doi.org/10.1016/j.biopsych.2016.10.004.
Xiao L, Yan J, Feng D, Ye S, Yang T, Wei H, et al. Critical role of TLR4 on the microglia activation induced by maternal LPS exposure leading to ASD-like behavior of offspring. Front Cell Dev Biol. 2021;9:634837 https://doi.org/10.3389/fcell.2021.634837.
Zhao D, Mokhtari R, Pedrosa E, Birnbaum R, Zheng D, Lachman HM. Transcriptome analysis of microglia in a mouse model of Rett syndrome: differential expression of genes associated with microglia/macrophage activation and cellular stress. Mol Autism. 2017;8:17 https://doi.org/10.1186/s13229-017-0134-z.
Suzuki K, Sugihara G, Ouchi Y, Nakamura K, Futatsubashi M, Takebayashi K, et al. Microglial activation in young adults with autism spectrum disorder. JAMA Psychiatry. 2013;70:49–58. https://doi.org/10.1001/jamapsychiatry.2013.272.
Schaafsma W, Basterra LB, Jacobs S, Brouwer N, Meerlo P, Schaafsma A, et al. Maternal inflammation induces immune activation of fetal microglia and leads to disrupted microglia immune responses, behavior, and learning performance in adulthood. Neurobiol Dis. 2017;106:291–300. https://doi.org/10.1016/j.nbd.2017.07.017.
Dhamne SC, Silverman JL, Super CE, Lammers SHT, Hameed MQ, Modi ME, et al. Replicable in vivo physiological and behavioral phenotypes of the Shank3B null mutant mouse model of autism. Mol Autism. 2017;8:26 https://doi.org/10.1186/s13229-017-0142-z.
Jaramillo TC, Speed HE, Xuan Z, Reimers JM, Escamilla CO, Weaver TP, et al. Novel Shank3 mutant exhibits behaviors with face validity for autism and altered striatal and hippocampal function. Autism Res. 2017;10:42–65. https://doi.org/10.1002/aur.1664.
Marsh SE, Walker AJ, Kamath T, Dissing-Olesen L, Hammond TR, de Soysa TY, et al. Dissection of artifactual and confounding glial signatures by single-cell sequencing of mouse and human brain. Nat Neurosci. 2022;25:306–16. https://doi.org/10.1038/s41593-022-01022-8.
Cherepanov SM, Gerasimenko M, Yuhi T, Furuhara K, Tsuji C, Yokoyama S, et al. Oxytocin ameliorates impaired social behavior in a Chd8 haploinsufficiency mouse model of autism. BMC Neurosci. 2021;22:32 https://doi.org/10.1186/s12868-021-00631-6.
Shiraishi T, Katayama Y, Nishiyama M, Shoji H, Miyakawa T, Mizoo T, et al. The complex etiology of autism spectrum disorder due to missense mutations of CHD8. Mol Psychiatry. 2024;29:2145–60. https://doi.org/10.1038/s41380-024-02491-y.
Lee SY, Kweon H, Kang H, Kim E. Age-differential sexual dimorphisms in CHD8-S62X-mutant mouse synapses and transcriptomes. Front Mol Neurosci. 2023;16:1111388 https://doi.org/10.3389/fnmol.2023.1111388.
Huang C, Wang P, Xu X, Zhang Y, Gong Y, Hu W, et al. The ketone body metabolite β-hydroxybutyrate induces an antidepression-associated ramification of microglia via HDACs inhibition-triggered Akt-small RhoGTPase activation. Glia. 2018;66:256–78. https://doi.org/10.1002/glia.23241.
Curzytek K, Leśkiewicz M. Targeting the CCL2-CCR2 axis in depressive disorders. Pharmacol Rep. 2021;73:1052–62. https://doi.org/10.1007/s43440-021-00280-w.
Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–4. https://doi.org/10.1038/nature10110.
Benedusi V, Della Torre S, Mitro N, Caruso D, Oberto A, Tronel C, et al. Liver ERα regulates AgRP neuronal activity in the arcuate nucleus of female mice. Sci Rep. 2017;7:1194 https://doi.org/10.1038/s41598-017-01393-0.
Villa A, Gelosa P, Castiglioni L, Cimino M, Rizzi N, Pepe G, et al. Sex-specific features of microglia from adult mice. Cell Rep. 2018;23:3501–11. https://doi.org/10.1016/j.celrep.2018.05.048.
McCarthy MM, Pickett LA, VanRyzin JW, Kight KE. Surprising origins of sex differences in the brain. Horm Behav. 2015;76:3–10. https://doi.org/10.1016/j.yhbeh.2015.04.013.
Wood S, Jayaraman V, Huelsmann EJ, Bonish B, Burgad D, Sivaramakrishnan G, et al. Pro-inflammatory chemokine CCL2 (MCP-1) promotes healing in diabetic wounds by restoring the macrophage response. PLoS ONE. 2014;9:e91574 https://doi.org/10.1371/journal.pone.0091574.
Takada YK, Fujita M, Takada Y. Pro-inflammatory chemokines CCL5, CXCL12, and CX3CL1 bind to and activate platelet integrin αIIbβ3 in an allosteric manner. Cells. 2022;11:3059 https://doi.org/10.3390/cells11193059.
Astorkia M, Liu Y, Pedrosa EM, Lachman HM, Zheng D. Molecular and network disruptions in neurodevelopment uncovered by single cell transcriptomics analysis of CHD8 heterozygous cerebral organoids. Heliyon. 2024;10:e34862 https://doi.org/10.1016/j.heliyon.2024.e34862.
Xu Z-X, Kim GH, Tan J-W, Riso AE, Sun Y, Xu EY, et al. Elevated protein synthesis in microglia causes autism-like synaptic and behavioral aberrations. Nat Commun. 2020;11:1797 https://doi.org/10.1038/s41467-020-15530-3.
Caracci MO, Avila ME, Espinoza-Cavieres FA, López HR, Ugarte GD, De Ferrari GV. Wnt/β-catenin-dependent transcription in autism spectrum disorders. Front Mol Neurosci. 2021;14:764756 https://doi.org/10.3389/fnmol.2021.764756.
Kobayashi M, Kishida S, Fukui A, Michiue T, Miyamoto Y, Okamoto T, et al. Nuclear localization of Duplin, a beta-catenin-binding protein, is essential for its inhibitory activity on the Wnt signaling pathway. J Biol Chem. 2002;277:5816–22. https://doi.org/10.1074/jbc.M108433200.
Wang P, Lin M, Pedrosa E, Hrabovsky A, Zhang Z, Guo W, et al. CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in neurodevelopment. Mol Autism. 2015;6:55 https://doi.org/10.1186/s13229-015-0048-6.
Wang P, Mokhtari R, Pedrosa E, Kirschenbaum M, Bayrak C, Zheng D, et al. CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in cerebral organoids derived from iPS cells. Mol Autism. 2017;8:11 https://doi.org/10.1186/s13229-017-0124-1.
Xu Q, Liu Y-Y, Wang X, Tan G-H, Li H-P, Hulbert SW, et al. Autism-associated CHD8 deficiency impairs axon development and migration of cortical neurons. Mol Autism. 2018;9:65 https://doi.org/10.1186/s13229-018-0244-2.
Durak O, Gao F, Kaeser-Woo YJ, Rueda R, Martorell AJ, Nott A, et al. Chd8 mediates cortical neurogenesis via transcriptional regulation of cell cycle and Wnt signaling. Nat Neurosci. 2016;19:1477–88. https://doi.org/10.1038/nn.4400.
Acknowledgements
We thank the Israel National Center for Personalized Medicine in the Weizmann Institute of Science for help with microglial RNA sequencing. This study was funded by grants from the Israel Science Foundation (898/17 and 1159/22).
Author information
Authors and Affiliations
Contributions
OW performed all the behavioral and molecular experimentation and wrote the manuscript. RH performed analysis of cell morphology. CI performed immunohistochemistry analysis. DG analyzed portions of the behavioral experimentation. EE designed the study, supervised the study, and made major contributions to the manuscript. All authors edited and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Weissberg, O., Harari, R., Dogun, C. et al. CHD8 adulthood microglial knockdown in C57BL6 mice induces behavioral, morphological, and transcriptional changes in a sex-dependent manner. Transl Psychiatry 15, 245 (2025). https://doi.org/10.1038/s41398-025-03468-3
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41398-025-03468-3