Abstract
Invasive micropapillary carcinoma (IMPC) is a special histological subtype of breast cancer, featured with extremely high rates of lymphovascular invasion and lymph node metastasis. Based on a previous series of studies, our team proposed the hypothesis of “clustered metastasis of IMPC tumor cells”. However, the transcriptomics characteristics underlying its metastasis are unknown, especially in spatial transcriptomics (ST). In this paper, we perform ST sequencing on four freshly frozen IMPC samples. We draw the transcriptomic maps of IMPC for the first time and reveal its extensive heterogeneity, associated with metabolic reprogramming. We also find that IMPC subpopulations with abnormal metabolism are arranged in different spatial areas, and higher levels of lipid metabolism are observed in all IMPC hierarchical clusters. Moreover, we find that the stromal regions show varieties of gene expression programs, and this difference depends on their distance from IMPC regions. Furthermore, a total of seven IMPC hierarchical clusters of four samples share a common higher expression level of the SREBF1 gene. Immunohistochemistry results further show that high SREBF1 protein expression is associated with lymph node metastasis and poor survival in IMPC patients. Together, these findings provide a valuable resource for exploring the inter- and intra-tumoral heterogeneity of IMPC and identify a new marker, SREBF1, which may facilitate accurate diagnosis and treatment of this disease.
Similar content being viewed by others
Introduction
Invasive micropapillary carcinoma (IMPC) is a morphologically distinctive form of breast cancer that has a special inverted growth pattern [1]. It has been reported in multiple organs, including the breast [2], lung [3], colon [4], pancreas [5] and bladder [6], etc. Breast IMPC was first identified by Siriaunkgul and Tavassolil in 1993 [2], and is composed of small, hollow or morula-like clusters of tumor cells without fibrovascular cores, surrounded by clear stromal spaces [7]. Compared with invasive ductal carcinoma-not otherwise specified type (IDC-NOS), IMPC is associated with a higher incidence of lymphovascular invasion and lymph node metastasis and a poorer prognosis [8, 9]. The unique clustered growth pattern and aggressive biological behaviors render IMPC a good model for studying tumor invasion and metastasis. Fu et al. [8] further noted that even if an IMPC tumor was small in diameter or consisted of only a few IMPC proportions, tumor cells were more likely to invade and metastasize. Later, a series of studies proposed the hypothesis of “clustered metastasis of IMPC tumor cells” [10,11,12,13,14,15,16,17,18]. However, spatial transcriptomics (ST) characteristics have not yet been reported.
ST technology has been designated as the method of 2020 [19]. Spatially resolved transcriptomics [20] combined with microscopic imaging and mRNA sequencing can provide transcriptome data for every spot from different locations in a tissue section. Spatial visualization exploration was realized by analyzing gene expression levels in corresponding tissue regions. Here, we investigated the gene expression characteristics of IMPC cells at the morphological and transcriptomic scales simultaneously using this popular method for the first time.
Therefore, based on a previous series of studies, we employed a spatial transcriptomics sequencing (ST-seq) approach to assess the transcriptome features of IMPC and dissect IMPC tumor heterogeneity in a relatively homogenous population, thus providing in-depth transcriptional information. Our research identifies a potential biomarker of IMPC and explores key genes associated with the progression of IMPC tumors. Thus, this study provides a further basis for precise diagnosis and treatment of IMPC.
Materials and methods
Freshly frozen IMPC samples
We collected four freshly frozen IMPC samples, all from breast cancer patients undergoing modified radical mastectomy. Another tissue was collected for the permeabilization optimization. All patients were diagnosed as breast IMPC or IDC-NOS with IMPC components using preoperative core needle biopsy and intraoperative cryosection methods. These patients were diagnosed and treated by the Breast Center of Tianjin Medical University Cancer Institute and Hospital.
We collected the five tissues under the supervision of senior pathologists, slowly washed the tissue blocks with 4 °C PBS, and cut out small pieces of about 6.5 × 6.5 × 5 mm3 containing IMPC components. Next, they were embedded in OCT embedding solution (Sakura, #4532, USA), and frozen in isopentane pre-cooled with liquid nitrogen. Cryosections were cut at 4 μm thickness, respectively. Hematoxylin-eosin staining (H&E) (Eosin, Dako CS701, Hematoxylin Dako S3309, bluing buffer CS702) was used to confirm that these tissues contained IMPC once again.
ST barcoded microarray information
The ST library preparation microarray (Visium Gene Expression Slide) used in this study was purchased from 10X Genomics (https://www.10xgenomics.com/). A slide includes four capture areas (the size of each area is 6.5 × 6.5 mm), and a total of four tissue sections can be placed. An optimization slide includes eight capture areas, the same size as the gene expression slide. The diameter of each spot on the ST slide is 55 μm. The distance between the centers of the two spots is 100 μm, and a total of 4992 spots cover an area of 6.5 × 6.5 mm2 (Fig. 1). Each spot with primers that includes: Illumina TruSeq Read 1 (partial read 1 sequencing primer), 16 nt Spatial Barcode (all primers in a specific spot share the same Spatial Barcode), 12 nt unique molecular identifier (UMI), and 30 nt poly(dT) sequence captures poly-adenylated mRNA for cDNA synthesis. In strict accordance with the ST-seq protocol [21], we used a gene expression slide to perform spatial transcriptome sequencing on four cryosections.
Tissue Optimization
Cryosections were cut at 10 μm thickness, and placed on the six capture areas on the slide. The other two areas are used for positive and negative controls. The six cryosections on the slide were subjected to permeabilization at different times to capture mRNA, and the reverse transcription was performed to generate fluorescently labeled cDNA. Cheese the permeabilization time that resulted in maximum fluorescence signal with the lowest signal diffusion. Our result found that the best permeabilization time for this experiment was 18 min.
Cryosection, H&E staining, and brightfield imaging for ST
A −20 °C microtome cryostat (Leica, CM1950) was used to cut tissue cryosections at 10 μm thickness. These sections were mounted onto the gene expression slide. Then the slide was placed on Thermocycler Adaptor with the active surface facing up and incubated 1 min at 37 °C, and fixed for 30 min with methyl alcohol in −20 °C followed by staining with H&E. The brightfield images were taken on a Leica DMI8 whole-slide scanner at 10X resolution. H&E staining cryosections were histologically annotated by three different senior pathologists.
Permeabilization and reverse transcription
Spatial gene expression was processed using Visum Spatial Gene Expression Slide and Reagent Kit (10x Genomics, PN-1000184). Put the slide on the slide cassette to create leakproof wells for adding reagents. For each well, added 70 µl permeabilization enzyme to make it completely cover every tissue for 18 min. Incubated in the Thermocycler Adaptor at 37 °C for 15 min. Finally, aspirated the tissue permeabilization enzyme with a pipette, and 100 µl 0.1X SSC buffer was added to wash for each well. The poly(dT) sequence of each spot will capture the mRNA released from the cells. Then, 75 μl reverse transcription Master Mix was added for cDNA synthesis. RT Master Mix is usually comprised of Nuclease-free Water, RT Reagent, Template Switch Oligo, Reducing Agent B and RT Enzyme D. After incubating at 53 °C for 45 min, reverse transcription was initiated to generate cDNA with spatial barcode information. Removed the Master Mix reagent, added 75 μl 0.08 M KOH to each tissue section, and incubated at room temperature for 5 min. Removed KOH, added 100 µl Qiagen Buffer EB, then added 75 μl Second Strand Mix (including Second Strand Reagent, Second Strand Primer and Second Strand Enzyme) at 65 °C for 15 min. Finally, removed the above-mentioned reaction reagents, and added 100 μl EB Buffer. After removing the EB Buffer, added 35 μl 0.08 M KOH and incubated at room temperature for 10 min. Transferred 35 µl of cDNA from slide to a tube containing 5 μl Tris (1 M, pH 7.0) for amplification and cDNA library construction.
cDNA amplification
Used qPCR to confirm the cycle number, according to the following protocol (Supplementary Table S1). Then added 65 μl cDNA Amplification Mix (including Amp Mix and cDNA Primers) to the cDNA sample and put it in the Thermo Fisher Scientific (#4375786). Performed cDNA amplification according to the protocol in Supplementary Table S2. The number of amplification cycles was determined according to the Cq value of qPCR, as follows: 12.2 corresponds to 12 cycles, 13.5 corresponds to 14 cycles, and 15.7 corresponds to 16 cycles. The amplified cDNA was cleaned up, and finally the results were checked for the size of the amplified fragment and the yield of the amplified product. cDNA amplification was performed on an S1000TM Touch Thermal Cycler (Bio Rad).
Spatial gene expression library construction and sequencing
The spatial gene libraries were constructed using Visum Spatial Library Construction Kit (10X Genomics, PN-1000184). Used chemical methods to break the cDNA into fragments of about 200–300 bp, repaired the ends, added poly(A) tails, and screened the cDNA fragments. Then connected the P7 adapter to the cDNA, introduced the double-ended Index (sample Index) of the sample through PCR amplification, and finally performed fragment screening to obtain the cDNA library. After the library was qualified, 3’ end sequencing was performed using Illumina Novaseq6000 sequencer with a sequencing depth of at least 100,000 reads per spot with pair-end 150 bp (PE150) reading strategy (sequence libraries were generated and sequenced by CapitalBio Technology, Beijing, China).
Reads alignment
The Spaceranger software was obtained from the 10X Genomics website (https://support.10xgenomics.com/spatial-gene-expression/software/downloads/latest). Reads alignment, filtering, barcode counting, and UMI counting were performed with the Spaceranger count module to generate a feature-barcode matrix using the default and recommended parameters. After the library was constructed, the effective length of Read1 was 28 bp, including 16 bp Barcode and 12 bp UMI. The effective length of Read2 was 91 bp. Read2 contained RNA sequences. Space Ranger uses the STAR algorithm (version 2.5.1b) [22] to map Read2 to Genome Reference Consortium Human Build 38.98 (GRCh38.98).
Basic ST data analysis
The Spaceranger software was used to perform effective barcode and UMI counts, and generated a gene-barcode expression matrix for each sample, including barcode-labeled spots and gene expression counts. Through the visualized gene number distribution and UMI number distribution figure, all the spots and expressed genes of the sample were evaluated. Imported these data into Seurat 3.2 (R package) for quality control and downstream analysis. Except for certain situations, default parameters were used in all operations. Removed spots with transcripts less than 1000 and mitochondrial transcripts greater than 20%. Sctransform in Seurat was used to normalize UMI count in each spot.
Hierarchical clustering for each sample
Dimensionality reduction was performed using principal component analysis (PCA) and the first 30 principle components were used to generate clusters by Seurat. Inspired by a graph-based algorithm, we performed Seurat on embedding the spots in the K-nearest neighbor (KNN) [23] graph structure and drew the edges between the spots with similar gene expression patterns. Then tried to decompose the graph into highly interconnected quasi clusters. It first constructed a KNN graph based on Euclidean distance in PCA space. And according to the shared overlap in the Jaccard distance, the edge weight between any two spots was optimized. Two nonlinear dimensionality reduction techniques t-SNE and UMAP were used to visualize the hierarchical clustering results, respectively. The resolution of clustering is 0.8. Next, we used barcodes to generate spatial hierarchical plots. Further, we selected the highly expressed genes with avg_logFC > 0.1 and p_val_adj < 0.05 of every IMPC hierarchical cluster in four samples, and displayed the common highly expressed genes by Venn plot.
Integrated analysis of multiple samples
Integrated analysis of four samples was carried out by Seurat. Seurat could integrate the gene-barcode expression matrix from multiple runs, normalize those counts into the same sequencing depth and get a new integrated matrix. The steps of hierarchical clustering and visualization were detailed in hierarchical clustering for a single sample.
Gene functional enrichment analysis
Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome functional enrichment analyses on the top 50 differentially expressed genes of each hierarchical cluster were performed by ClusterProfiler [24], ReactomePA [25], and DOSE [26], using EnrichProfiler R-packages with Benjamini-Hochberg multiple testing adjustment. The results were visualized using the R package.
Protein–protein interaction analysis
Protein–protein interaction (PPI) was obtained from human protein interaction data of STRING database [27] with combine_score ≥ 400. The interaction of the top 50 genes of every cluster was extracted from the database. The PPI results were visualized by Cytoscape software, respectively.
Transcription factor analysis
The transcription factors were predicted within 2000 bp upstream and 500 bp downstream of the transcription start sites for the top 20 genes of each cluster using JASPAR database [28] and the TFBSTools [29]. The gene and TF network for each cluster was visualized using Cytoscape software, respectively. In the figures, we arranged the genes in descending order of degree, and mapped the first 100 combined transcription factors of the gene.
Gene set enrichment
We considered enrichment in four gene set collections, including GO and KEGG. GSEA was performed by using GSEA software (version 4.0.3) (https://www.gsea-msigdb.org/gsea/index.jsp), which used predefined gene sets from the Molecular Signatures Database (MSigDB v7.1). Gene expression data was calculated by the mean UMI count of gene in one cluster and the rest cluster, respectively. The minimum and maximum criteria for the selection of gene sets from the collection were 0 and 500 genes, respectively. Next, calculated the enrichment score (ES), normalized enrichment score (NES), and false discovery rate (FDR). The FDR value of 25% was selected to determine whether the enriched functional gene set was appropriate.
Immunohistochemistry staining of FFPE tissue
A total of 132 IMPC and 121 IDC-NOS formalin-fixed and paraffin-embedded (FFPE) tumor tissues were continuously sectioned into 4 μm thickness. Primary antibodies against SREBF1(1:200 dilution, ab191857, Abcam), and FASN (1:3000 dilution, ab128870, Abcam) were used to evaluate protein expression and were reviewed by at least three pathologists. SREBF1 and FASN were graded according to the percentage of positive tumor cells (0 = 0%; 1 < 25%; 2 = 25–50%; 3 > 50%) and the intensity of staining in the tumor (0 = no staining; 1 = weak; 2 = moderate; 3 = high); the two scores were multiplied to obtain an overall score. If the product of the two scores was > 4, they were considered positively stained. And primary antibodies against CD45 (ZM-0183, ZSGB-BIO), CD3 (ZA-0503, ZSGB-BIO), CD8(ZA-0508, ZSGB-BIO), and CD20 (TA800394, ZSGB-BIO) and were also used to independently evaluate protein expression by three pathologists. The immunoreaction was graded as follows: (−) = no positive cells, (+) = 1–25% of the cells stained, (++) = 26–50% of the cells stained and (+++) = 51–100% of the cells stained. The brightfield images were taken on a Leica DMI8 whole-slide scanner at 40X resolution.
RT-qPCR
Another ten IMPC and nine IDC-NOS tumor samples were collected, and RNA was extracted and reversed, respectively. RT-qPCR was used to detect the expression of SREBF1 in the IMPC compared with IDC-NOS tumor tissues. SREBF1 primer is
Forward primer: CGGCGCTGCTGACCGACATC
Reverse primer: CCCTGCCCCACTCCCAGCAT.
FASN primer is:
Forward primer: CCATCTACAACATCGACACCA
Reverse primer: CTTCCACACTATGCTCAGGTAG
RT-qPCR was conducted as follows: 95 °C for 30 s, 40 cycles of 95 °C for 5 s and 60 °C for 30 s, then 95 °C for 10 s, 60 °C for 5 s, 95 °C for 5 s. Reaction volume is 10 μl.
TCGA data analysis
The correlation analysis result was obtained from GEPIA2 website (http://gepia2.cancer-pku.cn/#index). The Pearson test was used for the comparison and P value < 0.05 was considered statistically different.
Statistical analysis
All statistical analyses were performed using R (http://www.r-project.org) and SPSS version 25.0 (SPSS Inc., Chicago, IL, USA). The clinicopathological characteristics were compared using the Pearson χ2, Fisher’s exact test and Mann–Whitney U-test. Correlations were analyzed using the Spearman or Pearson rank test. Survival analysis (overall survival and disease-free survival) of 82 IMPC and 80 IDC-NOS patients was performed using Kaplan–Meier analysis. Log-rank test, two-sided. P < 0.05 was considered to indicate statistical significance.
Results
Basic data analysis of transcriptomes
To generate an unbiased hierarchical spatial map of the transcriptome, we collected four freshly frozen samples from IMPC patients (Fig. 1). The clinicopathological information of the four IMPC patients is provided in Supplementary Table S3. The RNA integrity numbers (RIN) of all samples were in the range of 6.67–7.80 and met our requirements (Supplementary Table S4). We acquired transcriptome data for a total of 7242 spots from the four samples. In addition, 10,000 UMIs (median) and 5000 genes (median) per spot were ascertained (Supplementary Table S5). These sequencing data results were consistent with those of other published papers using ST-seq [21, 30, 31]. The numbers of genes and UMIs and the mitochondrial ratio are shown in Supplementary Fig. S1. The spots with a high proportion of mitochondrial genes were filtered out. Next, we demultiplexed the reads and identified their spatial locations in the four tissue samples using location-specific barcodes.
Spatial genes expression visualization
We applied PCA on highly variable expressed genes across all spots of each sample. Two methods, t-SNE [32] and UMAP [33], were used to visualize the hierarchical clustering results. Next, the clusters were classified into regions using specific barcodes to generate ST maps of IMPC (Fig. 2A–F, samples 1 and 4). The maps of samples 2 and 3 are shown in Supplementary Fig. S2A–F. Different hierarchical clusters were assigned different colors, and the cluster identity was interpolated across the morphology to visualize major spatial patterns within each IMPC sample. The tissue regions were pathologically identified by three senior pathologists. The spatial maps were consistent with the pathological annotations, reflecting the feasibility of distinguishing different spatial regions within a section based on gene expression.
A H&E-stained tissue images of sample 1 with marked IMPC (red), IDC-NOS and IMPC-like (black), and stroma (yellow) tissue regions. B Hierarchical clustering of the spatial features in sample 1. Each cluster was assigned a color. The cluster 0 (red) and cluster 2 (green) represent interstitial regions. The cluster 1 (yellow) represents IDC-NOS region. The cluster 3 (wathet) is IMPC-like region. Clusters 4 (blue) and 5 (pink) are the IMPC area. C t-SNE color visualization of hierarchical clustering profile in sample 1. D H&E-stained tissue images of sample 4 with marked IMPC (red), IDC-NOS (black), and stroma (yellow) tissue regions. E Hierarchical clustering of the spatial features in sample 4. Each cluster was assigned a color. The cluster 0 (red) and cluster 3 (cyan) represent IDC-NOS regions. The cluster 1 (brown) and cluster 6 (pink) are stromal regions. The cluster 2 (green), 4 (blue), and 5 (purple) are the IMPC area. F t-SNE color visualization of hierarchical clustering profile in sample 4. G, H Heatmap plots of sample 1 and sample 4. t-SNE, t-distributed statistical neighbor embedding.
IMPC clusters were anatomically separate in spatial maps
In sample 1, we identified six main clusters, including cluster 0 and 2 (stromal region), cluster 1 (IDC-NOS region), cluster 3 (IMPC-like region [34], the same as IMPC in morphology), and clusters 4 and 5 (IMPC region) (Fig. 2A, B). The top 20 differentially expressed genes in every cluster are shown in heatmap plots (Fig. 2G).
A total of seven clusters were identified in sample 4 (Fig. 2E). Heatmap plots of the top 20 highly expressed genes are shown in Fig. 2H. Clusters 2, 4, and 5 were located at the IMPC region. Clusters 0 and 3 were identified in the IDC-NOS region. The heatmap plots of samples 2 and 3 are shown in Supplementary Fig. S2G, H. In addition to cancer regions, sample 2 could also distinguish regions of breast ductal epithelial cells (cluster 6) with high expression levels of PTN, RGS2, CLU, and KRT15 (Supplementary Fig. S2G).
Furthermore, we used the JASPAR databases and TFBSTools to predict transcription factors. Several highly expressed genes in IMPC regions were regulated by ZNF354C, SOX10, and MZF1 (Fig. 3A). These three transcription factors have been reported to be related to tumorigenesis and progression [35,36,37] and may be the key factors in the progression of IMPC.
A Highlight spatial hierarchical plot, GO enrichment (biological process, BP), KEGG enrichment and transcription factor analysis among top 50 gene in each IMPC cluster. GO, Gene Ontology. KEGG, Kyoto Encyclopedia of Genes and Genomes. The 30 terms with the lowest p.adjust values of the enrichment results were selected to draw the enrichment plot. The horizontal axis is gene and the vertical axis is term. The color represents the gene’s logFC value. p.adjust: use BH for multiple hypothesis testing, adjusted P value. p.adjust < 0.05 is a significant difference. Network diagram of highly expressed genes and transcription factors, the blue dots represent differential genes, and the purple dots represent transcription factors. The larger the node, the more nodes interacted to it. The arrows represent the enrichment terms related to metabolism, tumor immune response, and some important signaling pathways. B The t-SNE diagram of the integrated sequencing data. The four samples were completely distinguished. C, D The protein–protein interaction (PPI) plots of top 50 highly expressed genes in cluster 5 of sample 1 and cluster 2 of sample 4. The circles are nodes, and the size represents the size of the degree.
IMPC tumor regions in samples 1 and 4 were composed of multiple hierarchical clusters. Different IMPC clusters exhibited the characteristics of anatomical separation, and were grouped together rather than intermixed (Fig. 2B, E and Supplementary Fig. S2B, E). In contrast, IMPC regions in samples 2 and 3 were composed of only one cluster. Subsequently, we integrated the sequencing data of four samples to generate a merged gene-barcode expression matrix for PCA and visualization (Fig. 3B). The four samples were also clearly separated in the t-SNE plot, further demonstrating extensive heterogeneity between different samples.
This study first delineated the spatial transcriptome of IMPC, visually displaying the gene expression diversity in IMPC regions, and also revealed inter- and intra-tumoral heterogeneity and uncovered the unexplored landscape of the IMPC tumor region, where multiple tumor subclones converge in different spatial regions.
Genetic heterogeneity was related to metabolic reprogramming in IMPC
Two clusters represented two different metabolic states in the IMPC region of sample 1. Sample 1 has six hierarchical clusters, two of which are IMPC clusters (i.e., clusters 4 and 5). The enrichment analyses showed that the top 50 up-regulated genes in cluster 4 of sample 1 were mainly enriched for lipid metabolism, such as unsaturated fatty acid metabolic, and long-chain fatty acid metabolic (Fig. 3A-a). GSEA showed a strong association with fatty acid beta oxidation and cilium assembly, etc (Supplementary Fig. S3A). We observed higher expression of SCP2 in cluster 4 than in the other clusters, which was associated with lipid metabolism (Fig. 2G). Cluster 4 was an enhanced lipid metabolism group.
The functional enrichment results also showed that cluster 5 was associated with lipid metabolism and carbohydrate metabolism, including biosynthesis of unsaturated fatty acids, and glycolysis/gluconeogenesis (Fig. 3A-b). High levels of lipid metabolism-related genes (e.g., APOD, ALOX15B, SCD, LTF, and FASN) and carbohydrate metabolism-related genes (e.g., TFF3 and PGK1) were found in cluster 5 (Fig. 2G). Among the top 50 differentially expressed genes of cluster 5 in sample 1 and cluster 2 in sample 4, a protein interaction relationship was identified among SREBF1, FASN, and SCD (Fig. 3C, D).
In contrast, the enrichment results of cluster 1(IDC-NOS region) showed correlations in the tumor immune response and some other signaling pathways (Supplementary Fig. S4A). Cluster 3 (IMPC-like region) was a special histology area, and the enrichment results of the top 50 up-regulated genes were enriched for the tumor immune response, cell-substrate adhesion and some signaling pathways (Supplementary Fig. S3B), similar to cluster 1.
Only one IMPC cluster represented one metabolic state in sample 2 or sample 3. Sample 2 has seven hierarchical clusters. A total of five clusters were identified in sample 3. However, in sample 2 or sample 3, only one cluster of metabolic enhancement was found in the IMPC region. Notably, the top 50 differentially expressed genes for cluster 4 in sample 2 were enriched for lipid metabolism, amino acid metabolism, cilium, and the tumor immune response (Fig. 3A-c and Supplementary Fig. S3C). Specifically, highly expressed genes of cluster 4 in sample 2 were enriched for microvillus. We observed increased expression of lipid metabolism-related genes PLPP5, XBP1, and ESR1 and amino acid metabolism-related gene GATA3 in cluster 4 of sample 2 (Supplementary Fig. S2G).
The differentially expressed genes for cluster 2 in sample 3 were enriched for lipid metabolism and carbohydrate metabolism (Fig. 3A-d). In sample 3, the IMPC region (cluster 2) was highly expressed in ADAMTS1, a gene related to lipid metabolism, and PFKFB3, ENOSF1, and GPI, which are related to carbohydrate metabolism (Fig. S2H). The enrichment results of highly expressed genes in the IDC-NOS region (cluster 0 of sample 3) were related to signaling pathways and immune responses (Supplementary Fig. S4B).
Three IMPC clusters represented three different metabolic states in sample 4. Sample 4 has seven hierarchical clusters, three of which are IMPC clusters. Furthermore, sample 4 was characterized by obvious heterogeneity within the IMPC region, including clusters 2, 4, and 5. In cluster 2, the functional enrichment analyses showed that the top 50 genes were correlated with lipid metabolism, amino acid metabolism, cilium assembly and microvillus (Fig. 3A-e and Supplementary Fig. S3D). Higher expression levels of lipid metabolism-related genes (SREBF1 and SCD) and amino acid metabolism-related genes (GLUL) were observed in cluster 2 (Fig. 2H).
We performed enrichment analysis on cluster 4. Lipid metabolism was found to be the main term (Fig. 3A-f and Supplementary Fig. S3E), which was further supported by high expression of the lipid metabolism gene LTF (Fig. 2H). Genes for cluster 5 were enriched for lipid metabolism, carbohydrate metabolism and immune response terms (Fig. 3A-g and Supplementary Fig. S3F). We observed higher expression levels of some genes in cluster 5 (Fig. 2H), such as SERPINA3, LTF, PLAAT4 (lipid metabolism), and ALDOA (carbohydrate metabolism).
IDC-NOS regions (cluster 0 and 3) were dominated by enriched terms related to multiple metabolisms, immune response, and other cancer signaling pathways (Supplementary Fig. S4C, D). Our results elucidated that the immune response term was enriched in all IDC-NOS clusters. Next, 50 IMPC and 41 IDC-NOS FFPE tumor tissues were used for immunohistochemical staining of CD45, CD3, CD8, and CD20. We found significantly higher expression levels of CD45, CD3, and CD8 in the IDC-NOS group than in the IMPC group (Mann–Whitney U-test, all P < 0.05), but no difference in CD20 was observed (P > 0.05) (Supplementary Table S6). And T lymphocytes were more numerous in IMPC and IDC-NOS tumors than B lymphocytes (Mann–Whitney U-test, both P < 0.01, Supplementary Table S7). It suggests that IDC-NOS tumors have noticeable numbers of tumor infiltrating lymphocytes representing the immune response.
Polyclonal samples 1 and 4 originated from pN1 and pN2 patients with lymphovascular invasion, respectively. In contrast, patients 2 and 3 had no lymph node metastasis and no lymphovascular invasion (Supplementary Table S3). Extensive intra-tumor heterogeneity correlates with a higher N stage and a higher rate of lymphovascular invasion.
Spatial visualization characteristics of the tumor microenvironment
In sample 1, the stromal region has two clusters, namely, cluster 0 and cluster 2. In the spatial map, cluster 0 was close to the IMPC region, and cluster 2 was farther away from the IMPC region (Fig. 4A). IGKC, IGHG4, IGHG3, and IGHA1 were highly expressed in cluster 0 (Fig. 4B–E). Functional enrichment analysis of cluster 0 showed that it was associated with cellular interaction, complement pathway and immune response (Fig. 4F, G and Supplementary Fig. S5A–H). The complement system plays a major and complex role in the tumor microenvironment. For one thing, the complement system was found to be related to immune surveillance for antitumor [21, 38]. For another thing, some studies have reported that the complement system was distinctly important for promoting tumor growth, which has an immunosuppressive effect in tumor immunity [39, 40]. Our study revealed that the complement pathways was enriched in the tumor reactive stromal region (cluster 0), which played a vital role in the growth of IMPC cells.
A The spatial distribution position of two stromal clusters (cluster 0 and 2). The stroma (green cluster) is far from IMPC regions (blue and pink clusters). Another stroma (red cluster) is near IMPC regions. B–E The expression level of IGKC, IGHG4, IGHG3, and IGHA1 genes on the spatial plots. F, G The enrichment results of cluster 0 highly expressed top 50 genes on GO (BP) and KEGG. H, I The enrichment results of cluster 2 highly expressed top 50 genes on GO (BP), and KEGG. The arrows represent that these terms were linked to immune response, cellular interaction, complement pathway, and oxidative phosphorylation, etc.
In contrast, genes for cluster 2 were enriched for oxidative phosphorylation, extracelluar and intercellular transport terms (Fig. 4H, I and Supplementary Fig. S5I). Although both are stromal clusters, their enrichment terms are totally different. In particular, the terms interstitial support, transduction and tumor immune response were mainly enriched in cluster 0 (close to the IMPC region in spatial position), while cluster 2 (far away from the IMPC area) was not. In addition, only one metabolic enhancement occurred when many interstitial cells were mixed in the IMPC region (e.g., cluster 4). However, when fewer mesenchymal cells were present (such as cluster 5), two types of metabolic enhancement occurred simultaneously. Therefore, the difference in spatial distribution between the stroma and the tumor may affect the immune response of stromal region. The metabolic mode of IMPC also changes when more mesenchymal cells are close to the IMPC region.
High IMPC-specific SREBF1 expression was associated with malignant biological behavior
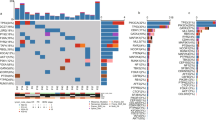
Excitingly, SREBF1 was a common highly expressed gene in IMPC clusters of all samples (“Methods”, Fig. 5A, B) and was the only gene that was highly expressed in all IMPC clusters. Furthermore, the expression level of SREBF1 in IMPC clusters was significantly higher than that in IDC-NOS clusters (Student’s t-test, P < 0.05) (Fig. 5C).
A The Venn plot showed that SREBF1 was a common high expression gene in all IMPC clusters of the 4 samples. B SREBF1 was highly expressed in IMPC clusters of each sample in spatial plots. C Boxplot of SREBF1 expression level in IMPC clusters versus IDC-NOS clusters. Bar represents median, and boxplot represents quartiles; scale in log10(RPM). Student’s t-test for comparison between two groups in each sample. The difference was significant in sample 1 (P < 2e-16), sample 3 (P = 2.1e-08) and sample 4 (P < 2e-16). D The correlation analysis between SREBF1 and FASN using TCGA database. The Pearson correlation coefficient was used, two-sided. P < 0.05 was considered to indicate statistical significance. E SREBF1 was more highly expressed in IMPC tumor tissues using RT-qPCR. Bar represents median, and boxplot represents quartiles. Student’s t-test for comparison. The two groups were significantly different (P = 0.013).
FASN is a target gene of SREBF1 [41], and a positive correlation was found between them (R = 0.400, P = 0.000) using the TCGA database (Fig. 5D). In addition, in IMPC clusters of samples 1 and 4, the expression levels of FASN were significantly higher than those in IDC-NOS (Student’s t-test, P < 0.05) (Supplementary Fig. S6A–G).
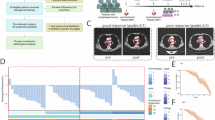
The RT-qPCR results revealed that SREBF1 and FASN showed significantly high expression in ten IMPC and nine IDC-NOS tumor tissues, respectively (Student’s t-test, P = 0.013 and 0.037) (Fig. 5E and Supplementary Fig. S6H). In all, 82 IMPC and 80 IDC-NOS FFPE tissues were used for immunohistochemical staining of SREBF1 and FASN. The expression of SREBF1 and FASN was significantly high in IMPC, and the difference was statistically significant (Pearson χ2 test, P = 0.000) (Fig. 6A and Supplementary Fig. S7A). The expression of SREBF1 and FASN proteins was significantly associated with a higher level of lymph node metastasis in IMPC (SREBF1, R = 0.405, P = 0.000; FASN, R = 0.521, P = 0.000) (Supplementary Fig. S7B). The high expression levels of the SREBF1 and FASN proteins indicated a poor prognosis for overall survival and disease-free survival time (based on the log-rank test, P < 0.05 for both) (Fig. 6B).
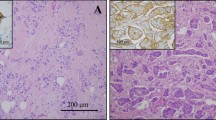
A Images of immunohistochemical staining for SREBF1 and FASN in patients with IMPC (n = 82) and IDC-NOS (n = 80). The upper panel shows high expression of SREBF1 and FASN in patients with IMPC; the lower panel shows low expression of the SREBF1 and FASN proteins in patients with IDC-NOS, x400 magnification. B The association of SREBF1 and FASN protein expression with disease-free survival (DFS) and overall survival (OS). The light panel shows SREBF1, and the right panel shows FASN, with patients with IMPC stratified by high and low protein expression. Log-rank test, two-sided. The P values for DFS of SREBF1 and FASN were 0.022, and 0.009, respectively; the P values for OS of SREBF1 and FASN were 0.044 and 0.015, respectively. P < 0.05 was considered to indicate statistical significance.
Discussion
Breast cancer accounts for 24.5% of the total number of new cancer cases in women worldwide, overtaking lung cancer as the most common cancer in the world [42]. IMPC is classified as a special histologic subtype of breast cancer. The unique growth pattern and histological morphology caused by polarity reversal, facilitate IMPC tumor cell clusters’ detachment, and these clusters break into single cells or smaller clusters, thereby promoting invasion and metastasis [10]. To explore the inter- and intra-tumoral heterogeneity of IMPC and understand why IMPCs have aggressive biological behaviors, such as very high rates of lymph vascular invasion and lymph node metastasis [8, 11], we conducted this study to investigate the gene expression characteristics of IMPC at the spatial transcriptome level.
Single-cell RNA-seq can identify oncogenic drivers, explore the heterogeneity and reconstruction of evolutionary lineages [43,44,45]. Although breakthroughs in the immune treatment and chemoresistance of various malignant tumors, such as breast cancer [46] and hepatocellular carcinoma [47], have been achieved, information on the spatial locations of tumor tissues is lacking. In our study, we applied ST-seq, which quantified an array of transcriptomes across a tissue section, to unravel gene spatial expression characteristics in IMPC.
Some studies have reported that the tumor heterogeneity of breast cancer is correlated with changes in chromosomes and genes [48]. Heterogeneity affects multiple developmental stages of tumor. Nicholas et al. [49] observed that tumor subpopulations may be anatomically separate or intermixed. That is, a “tumor subclone” can be separated in different areas, or scattered and crossed in the same region. Studying the distribution characteristics of tumors subclones enables us to determine the developmental organization of tumor growth and the migratory pattern. In our study, two of the four samples had different clusters in the IMPC region, and “subclones” were clustered in different regions and separated from each other. The patterns of special growth and development characterized by IMPC subpopulations require further research and analysis.
Several studies have found that tumor cells can re-adapt to the environment through metabolic reprogramming to maintain advantages in proliferation and metastasis [50, 51]. All IMPC clusters had abnormal lipid metabolism enhancement. Fatty acids are the main energy substances of the body in the tumor-bearing state, and may support the growth of tumor cells by providing metabolic substrates for energy production [52]. Under the energy stress of tumor cells, the fatty acid β oxidation pathway can also provide energy for tumor cells. This form may be an important metabolic change in tumor development [53]. Abnormal carbohydrate metabolism enhancement is also a characteristic change in IMPC. The Warburg effect [51] holds that glycolysis in tumor cells is abnormally active, and is preferentially used to provide cells with energy. The citric acid cycle (TCA) is the key means for prostate cells to transform into malignant tumor cells [54]. Some researchers have found that the fatty acid-β oxidation pathway is enhanced in prostate cancer and provides ATP and acetyl-CoA, which can also accelerate the oxidative metabolism of citric acid [53]. Metabolic reprogramming is an important feature that distinguishes IMPC and IDC-NOS and may also be a key mechanism leading to the special growth mode and high rates of invasion and metastasis.
Highly expressed genes in the IDC-NOS regions of the four samples were mainly enriched in the immune response of the tumor. The immunohistochemistry results also confirmed the presence of more lymphocyte infiltration in IDC-NOS regions than in IMPC regions. The IMPC region lacks the immune-surveillance antitumor effect of T lymphocyte infiltration [55]. A fierce interaction occurs between immune cells and tumor cells in IDC-NOS regions. Tumor cells interact with the immune microenvironment, and immune cells play an important role in tumor immunity, indirectly or directly inhibiting the growth and invasion of tumor cells. In addition, the highly expressed genes of IDC-NOS were also related to the formation of extracellular matrix and cell-cell interaction. Reports in the literature indicate that the extracellular matrix of breast cancer stroma is remodeled, which plays an important role in the invasion and metastasis of tumor cells [56]. Taken together, the IDC region has a stronger tumor immune response and more lymphocyte infiltration than the IMPC. The lack of an antitumor effect of lymphocyte infiltration may be one of the reasons for the malignant biological behavior of IMPC.
In our study, a new gene expression characterization of the stroma in the proximity of IMPC was elucidated by spatial mapping. The stromal regions show different gene expression characteristics, which are related to the distance between them and IMPC. This study revealed high levels of immunoglobulin-related genes within the stroma close to IMPC, whereas the stroma far away from IMPC was related to oxidative phosphorylation. A high level of immunoglobulin secretion indicates that the immune effect of lymphocytes in the cancer area is active. This type of stroma is regarded as an emerging hallmark of tumor invasion and metastasis [21]. The phenomenon of different interstitial functions caused by different spatial distributions can be clearly observed by spatial atlas gene expression.
Microvilli are abundant on the outer surfaces of the IMPC tumor cell clusters under electron microscopy, and most of the microvilli are gathered in the movement direction of the IMPC cell clusters [10]. The assembly and formation of cilia can significantly promote the invasion and metastasis of pancreatic cancer, lung cancer, lymphoma and other tumors [57].
SREBF1 is a key transcription factor regulating FASN in lipid metabolism [58] and is regulated by the PI3K-mTORC1-AKT signaling pathway [59]. Phosphorylation of mTORC1-dependent phospholipid-1 can promote SREBF1 to enter the nucleus [60]. FASN is a key enzyme for the de novo synthesis of long-chain fatty acids. Under pathological conditions, SREBF1 and FASN have been reported to be a pair of metabolically related oncogenes [58], which have been confirmed to be related to prostate cancer and breast cancer [61,62,63,64]. In recent years, antitumor treatments targeting enzymes or transcription factors in cell metabolism have become a research hot spot. SREBF1/FASN may play an extremely important role in the invasion and metastasis of IMPC. The specific mechanism and whether they can be used as target genes for IMPC precision therapy require further research and verification.
In summary, this study reports ST maps of IMPC for the first time and further improves the research hypothesis of “clustered metastasis of IMPC tumor cells” postulated in our previous research. Furthermore, we revealed that IMPC tumor cells have extensive heterogeneity, closely related to metabolic reprogramming based on lipid metabolism. SREBF1 is associated with the aggressive biological behavior of IMPC. These results provide a powerful basis and targeted molecules for the precise diagnosis and treatment of IMPC.
Data availability
Sequencing data are deposited at the Genome Sequence Archive (GSA), under the accession number HRA001442.
Code availability
R and other custom scripts used to analyze the data are available upon request.
References
Acs G, Esposito NN, Rakosy Z, Laronga C, Zhang PJ. Invasive ductal carcinomas of the breast showing partial reversed cell polarity are associated with lymphatic tumor spread and may represent part of a spectrum of invasive micropapillary carcinoma. Am J Surg Pathol. 2010;34:1637–46.
Siriaunkgul S, Tavassoli FA. Invasive micropapillary carcinoma of the breast. Mod Pathol. 1993;6:660–2.
Kuroda N, Hamaguchi N, Ohara M, Hirouchi T, Miyzaki E, Mizuno K. Intracytoplasmic lumina in invasive micropapillary carcinoma of the lung. Diagn Cytopathol. 2006;34:224–6.
Sakamoto K, Watanabe M, Cruz CDL, Honda H, Ise H, Mitsui K, et al. Primary invasive micropapillary carcinoma of the colon. Histopathology 2005;47:479–84.
Kitagawa H, Nakamura M, Tani T, Tajima H, Nakagawara H, Ohnishi I, et al. A pure invasive micropapillary carcinoma of the pancreatic head: long disease-free survival after pancreatoduodenectomy and adjuvant chemotherapy with gemcitabine. Pancreas 2007;35:190–2.
Amin MB, Ro JY, el-Sharkawy T, Lee KM, Troncoso P, Silva EG, et al. Micropapillary variant of transitional cell carcinoma of the urinary bladder. Histologic pattern resembling ovarian papillary serous carcinoma. Am J Surg Pathol. 1994;18:1224–32.
Yang Y-L, Liu B-B, Zhang X, Fu L. Invasive micropapillary carcinoma of the breast: an update. Arch Pathol Lab Med. 2016;140:799–805.
Chen L, Fan Y, Lang RG, Guo XJ, Sun YL, Cui LF, et al. Breast carcinoma with micropapillary features: clinicopathologic study and long-term follow-up of 100 cases. Int J Surg Pathol. 2008;16:155–63.
Li W, Han Y, Wang C, Guo X, Shen B, Liu F, et al. Precise pathologic diagnosis and individualized treatment improve the outcomes of invasive micropapillary carcinoma of the breast: a 12-year prospective clinical study. Mod Pathol. 2018;31:956–64.
Fu L, Ikuo M, Fu XY, Liu TH, Shinichi T. Relationship between biologic behavior and morphologic features of invasive micropapillary carcinoma of the breast. Zhonghua Bing Li Xue Za Zhi. 2004;33:21–5.
Guo X, Chen L, Lang R, Fan Y, Zhang X, Fu L. Invasive micropapillary carcinoma of the breast: association of pathologic features with lymph node metastasis. Am J Clin Pathol. 2006;126:740–6.
Cui LF, Guo XJ, Wei J, Liu FF, Fu L. Overexpression of TNF‐α and TNFRII in invasive micropapillary carcinoma of the breast: clinicopathological correlations. Histopathology 2010;53:381–8.
Cui LF, Guo XJ, Wei J, Liu FF, Fu L. Significance of interleukin-1beta expression and microvascular density in invasive micropapillary carcinoma of breast. Zhonghua Bing Li Xue Za Zhi. 2008;37:599–603.
Liu F, Lang R, Wei J, Fan Y, Cui L, Gu F, et al. Increased expression of SDF‐1/CXCR4 is associated with lymph node metastasis of invasive micropapillary carcinoma of the breast. Histopathology 2009;54:741–50.
Li W, Liu F, Lei T, Xu X, Liu B, Cui L, et al. The clinicopathological significance of CD44+/CD24−/low and CD24+ tumor cells in invasive micropapillary carcinoma of the breast. Pathol Res Pract. 2010;206:828–34.
Li W, Yang D, Wang S, Guo X, Lang R, Fan Y, et al. Increased expression of CD146 and microvessel density (MVD) in invasive micropapillary carcinoma of the breast: comparative study with invasive ductal carcinoma-not otherwise specified. Pathol Res Pract. 2011;207:739–46.
Li S, Yang C, Zhai L, Zhang W, Yu J, Gu F, et al. Deep sequencing reveals small RNA characterization of invasive micropapillary carcinomas of the breast. Breast Cancer Res Treat. 2012;136:77–87.
Wang X-X, Liu B-B, Wu X, Su D, Zhu Z, Fu L. Loss of leucine zipper putative tumor suppressor 1 (LZTS1) expression contributes to lymph node metastasis of breast invasive micropapillary carcinoma. Pathol Oncol Res. 2015;21:1021–6.
Method of the Year 2020: spatially resolved transcriptomics. Nat Methods. 2021;18:1.
Ståhl PL, Salmen F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016;353:78–82.
Berglund E, Maaskola J, Schultz N, Friedrich S, Lundeberg J. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat Commun 2018;9:2419.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21.
Wagner F, Yan Y, Yanai I. K-nearest neighbor smoothing for high-throughput single-cell RNA-Seq data. Preprint at https://doi.org/10.1101/217737 (2017).
Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–7.
Yu G, He Q-Y. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst. 2016;12:477–9.
Yu G, Wang L-G, Yan G-R, He Q-Y. DOSE: an R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics 2015;31:608–9.
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–52.
Sandelin A, Alkema W, Engström P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–4.
Tan G, Lenhard B. TFBSTools: an R/bioconductor package for transcription factor binding site analysis. Bioinformatics 2016;32:1555–6.
Thrane K, Eriksson H, Maaskola J, Hansson J, Lundeberg J. Spatially resolved transcriptomics enables dissection of genetic heterogeneity in stage III cutaneous malignant melanoma. Cancer Res. 2018;78:5970–9.
Moncada R, Barkley D, Wagner F, Chiodin M, Devlin JC, Baron M, et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat Biotechnol. 2020;38:333–42.
Maaten LJPVD, Hinton GE. Visualizing high-dimensional data using t-SNE. J Mach Learn Res. 2008;9:2579–605.
Yu D, Huber W, Vitek O. Shrinkage estimation of dispersion in negative binomial models for RNA-seq experiments with small sample size. Bioinformatics 2013;29:1275–82.
Acs G, Dumoff KL, Solin LJ, Pasha T, Xu X, Zhang PJ. Extensive retraction artifact correlates with lymphatic invasion and nodal metastasis and predicts poor outcome in early stage breast carcinoma. Am J Surg Pathol. 2007;31:129–40.
Oo JA, Irmer B, Günther S, Warwick T, Pálfi K, Ponce JI, et al. ZNF354C is a transcriptional repressor that inhibits endothelial angiogenic sprouting. Sci Rep. 2020;10:19079.
Han S, Ren Y, He W, Liu H, Zhi Z, Zhu X, et al. ERK-mediated phosphorylation regulates SOX10 sumoylation and targets expression in mutant BRAF melanoma. Nat Commun. 2018;9:28.
Brix DM, Clemmensen K, Kallunki T. Zinc finger transcription factor MZF1-A specific regulator of cancer invasion. Cells 2020;9:223.
Fishelson Z, Donin N, Zell S, Schultz S, Kirschfink M. Obstacles to cancer immunotherapy: expression of membrane complement regulatory proteins (mCRPs) in tumors. Mol Immunol. 2003;40:109–23.
Cho MS, Vasquez HG, Rupaimoole R, Pradeep S, Wu S, Zand B, et al. Autocrine effects of tumor-derived complement. Cell Rep. 2014;6:1085–95.
Afshar-Kharghan V. The role of the complement system in cancer. J Clin Invest. 2017;127:780–9.
Zhao M, Bu Y, Feng J, Zhang H, Chen Y, Yang G, et al. SPIN1 triggers abnormal lipid metabolism and enhances tumor growth in liver cancer. Cancer Lett. 2020;470:54–63.
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49.
Zhang P, Yang M, Zhang Y, Xiao S, Lai X, Tan A, et al. Dissecting the single-cell transcriptome network underlying gastric premalignant lesions and early gastric cancer. Cell Rep. 2019;27:1934–47.
Chung W, Eum HH, Lee H-O, Lee K-M, Lee H-B, Kim K-T, et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat Commun. 2017;8:15081.
Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck. Cancer Cell 2017;171:1611–24.
Kim C, Gao R, Sei E, Brandt R, Hartman J, Hatschek T, et al. Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell 2018;173:879–93.
Zheng C, Zheng L, Yoo J-K, Guo H, Zhang Y, Guo X, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 2017;169:1342–56.
Bertucci F, Ng CKY, Patsouris A, Droin N, Piscuoglio S, Carbuccia N, et al. Genomic characterization of metastatic breast cancers. Nature 2019;569:560–4.
Navin N, Krasnitz A, Rodgers L, Cook K, Meth J, Kendall J, et al. Inferring tumor progression from genomic heterogeneity. Genome Res. 2010;20:68–80.
Currie E, Schulze A, Zechner R, Walther TC, Farese RV. Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153–61.
Warburg O. On the origin of cancer cells. Science 1956;123:309–14.
Kuhajda FP. Fatty acid synthase and cancer: new application of an old pathway. Cancer Res. 2006;66:5977–80.
Liu Y. Fatty acid oxidation is a dominant bioenergetic pathway in prostate cancer. Prostate Cancer Prostatic Dis. 2006;9:230–4.
Costello LC, Franklin RB. The intermediary metabolism of the prostate: a key to understanding the pathogenesis and progression of prostate malignancy. Oncology 2000;59:269–82.
Zitvogel L, Galluzzi L, Kepp O, Smyth MJ, Kroemer G. Type I interferons in anticancer immunity. Nat Rev Immunol. 2015;15:405–14.
Trujillo KA, Heaphy CM, Mai M, Vargas KM, Jones AC, Vo P, et al. Markers of fibrosis and epithelial to mesenchymal transition demonstrate field cancerization in histologically normal tissue adjacent to breast tumors. Int J Cancer. 2011;129:1310–21.
Yasar B, Linton K, Slater C, Byers R. Primary cilia are increased in number and demonstrate structural abnormalities in human cancer. J Clin Pathol. 2017;70:571–4.
Menendez JA, Decker JP, Lupu R. In support of fatty acid synthase (FAS) as a metabolic oncogene: extracellular acidosis acts in an epigenetic fashion activating FAS gene expression in cancer cells. J Cell Biochem. 2005;94:1–4.
Jeon T-I, Osborne TF. SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol Metab. 2012;23:65–72.
Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 2011;146:408–20.
Swinnen JV, Heemers H, Sande TVD, Schrijver ED, Brusselmans K, Heyns W, et al. Androgens, lipogenesis and prostate cancer. J Steroid Biochem Mol Biol. 2004;92:273–9.
Ettinger SL, Sobel R, Whitmore TG, Akbari M, Bradley DR, Gleave ME, et al. Dysregulation of sterol response element-binding proteins and downstream effectors in prostate cancer during progression to androgen independence. Cancer Res. 2004;64:2212–21.
Huang W-C, Li X, Liu J, Lin J, Chung LWK. Activation of androgen receptor, lipogenesis, and oxidative stress converged by SREBP-1 is responsible for regulating growth and progression of prostate cancer cells. Mol Cancer Res. 2012;10:133–42.
Zhang N, Zhang H, Liu Y, Su P, Zhang J, Wang X, et al. SREBP1, targeted by miR-18a-5p, modulates epithelial-mesenchymal transition in breast cancer via forming a co-repressor complex with Snail and HDAC1/2. Cell Death Differ. 2019;26:843–59.
Acknowledgements
We thank all the participants included in this study.
Funding
This study was supported by the grants from the National Natural Science Foundation of China (Nos. 81672637 and 81872164).
Author information
Authors and Affiliations
Contributions
J.K.L., Q.Q.S., F.G., and L.F. designed the study. J.K.L., G.S.G., and L.F. collected and prepared the samples. J.K.L., Q.Q.S., Y.W.H., W.D.L., H.J.L., J.Y.Z., C.N., G.S.G., Y.R.F., R.Y.Z., K.L.W., S.L., F.G., and L.F. collected the data. J.K.L., Q.Q.S., Y.W.H., F.G., and L.F. analyzed and interpreted the data. J.K.L. and L.F. wrote the paper. J.K.L., Q.Q.S., Y.W.H., F.G., and L.F. revised the manuscript. All authors have reviewed the manuscript and approved the final version.
Corresponding authors
Ethics declarations
Ethics statement
The study was approved by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Professor Gennaro Ciliberto
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lv, J., Shi, Q., Han, Y. et al. Spatial transcriptomics reveals gene expression characteristics in invasive micropapillary carcinoma of the breast. Cell Death Dis 12, 1095 (2021). https://doi.org/10.1038/s41419-021-04380-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-021-04380-6
This article is cited by
-
Spatial transcriptomics reveals regionally altered gene expression that drives retinal degeneration
Communications Biology (2025)
-
Granzyme K+CD8+ T cells interact with fibroblasts to promote neutrophilic inflammation in nasal polyps
Nature Communications (2024)
-
Metabolic shifts in lipid utilization and reciprocal interactions within the lung metastatic niche of triple-negative breast cancer revealed by spatial multi-omics
Cell Death & Disease (2024)