Abstract
RNA surveillance systems degrade aberrant RNAs that result from defective transcriptional termination, splicing, and polyadenylation. Defective RNAs in the nucleus are recognized by RNA-binding proteins and MTR4, and are degraded by the RNA exosome complex. Here, we detect aberrant RNAs in MTR4-depleted cells using long-read direct RNA sequencing and 3′ sequencing. MTR4 destabilizes intronic polyadenylated transcripts generated by transcriptional read-through over one or more exons, termed 3′ eXtended Transcripts (3XTs). MTR4 also associates with hnRNPK, which recognizes 3XTs with multiple exons. Moreover, the aberrant protein translated from KCTD13 3XT is a target of the hnRNPK-MTR4-RNA exosome pathway and forms aberrant condensates, which we name KCTD13 3eXtended Transcript-derived protein (KeXT) bodies. Our results suggest that RNA surveillance in human cells inhibits the formation of condensates of a defective polyadenylated transcript-derived protein.
Similar content being viewed by others
Introduction
RNA polymerase II transcribes various types of premature RNA that are then processed into mature RNAs through a maturation process that includes 5′ capping, splicing, cleavage, and polyadenylation at the 3′ end1,2,3,4,5,6. Alternative cleavage and polyadenylation (APA) generates RNA isoforms in a gene-specific and cell-type-specific manner6. Moreover, APA plays an important role in regulating cellular processes under diverse physiological conditions7. Dysregulation of APA has been attributed to many pathological processes, including cancer and hematological, immunological, and neurological diseases5,6. APA can be classified into four types based on the localization of polyadenylation sites: “3′-untranslated region (UTR) APA” generates multiple isoforms that differ in their 3′-UTR lengths and encode the same protein; “alternative terminal exon APA,” in which alternative splicing produces isoforms with different last exons; “internal exon APA,” which involves premature polyadenylation within the coding region; and “intronic APA,” in which APA occurs in introns and tends to produce different proteins7.
Transcripts derived from intronic APA have physiological roles and contribute to proteome diversity by producing proteins that vary in coding potential, stability, translational efficiency, and localization6,8. Polyadenylation is strictly connected with splicing, and intronic APA is likely to compete with splicing9. In addition, cancer-specific intronic APAs, which are generated by mutations in DNA, cleavage and polyadenylation factors, and RNA polymerase II interacting factors, produce aberrant RNAs, resulting in the production of truncated proteins that lack or gain essential domains10,11,12. Although aberrant RNAs with dysregulated intronic APA are thought to be frequently produced, the overall generation and turnover of aberrant RNAs remains unclear.
Premature transcripts in the nucleus are recognized by surveillance systems to prevent accumulation and translation of aberrant pathogenic transcripts. Degradation of aberrant nuclear RNA is dominated by the evolutionarily conserved 3′–5′ exo- and endo-nucleolytic RNA exosome complex, which is important in regulating transcriptome quality13,14,15. The human RNA exosome is composed of a catalytically inactive barrel structure of nine core subunits (known as EXO9) that achieves its activity by association with the exonuclease EXOSC10 and the exo/endo-nuclease DIS314,15,16,17,18. The RNA exosome associates with adaptor proteins to provide target RNA specificity. The RNA helicase MTR4 (SKIV2L2, MTREX), serves as a platform for forming nucleoplasmic adaptor complexes, such as the Trf4p/5p-Air1p/2p-Mtr4p polyadenylation (TRAMP) complex, which consists of the zinc finger protein ZCCHC7 and the poly(A) RNA polymerase PAPD5, the nuclear-exosome-targeting (NEXT) complex, which consists of the zinc finger protein ZCCHC8 and the RNA-binding protein RBM7, and the pA-tail-exosome-targeting (PAXT) complex, which is composed of the zinc-finger protein ZFC3H1 and the nuclear polyA-binding protein PABPN114,19,20,21,22,23,24. In general, the NEXT complex mediates the decay of short RNAs that lack polyA tails, whereas the PAXT complex targets unspliced and polyadenylated RNAs for degradation20,21,23,25. Moreover, the TRAMP complex is involved in polyadenylation, processing, and degradation of pre-rRNA, pre-snoRNA, and tRNA19,26,27. However, the degradation machinery of aberrant polyadenylated RNAs with splicing events regulated by MTR4 has not been clarified.
In the present study, we attempted to identify aberrant RNAs with defects in polyadenylation that were destabilized by MTR4 by combining data from long-read direct RNA sequencing and 3′ sequencing. We found that MTR4 destabilizes aberrant RNAs with several types of APA. We focused on intronic APA isoforms that are transcribed over one or more exons and terminated by polyadenylation sites in introns. We termed the intronic APA isoforms 3′ eXtended Transcripts (3XTs) and divided them into two types, with or without splicing events: multi- or mono-exon 3XTs, respectively. We further show that the RNA exosome cooperating with the PAXT, but not the NEXT complex, degrades both mono- and multi-exon 3XTs. Moreover, we show that hnRNPK interacts with MTR4 and regulates the degradation of multi-exon 3XTs, but not mono-exon 3XTs, by recognizing their primarily intronic regions that extend from the existing exonic regions in the last exon of the 3XTs, termed 3′ eXtended Regions (3XRs). We also demonstrate that the protein translated from KCTD13 3XT, an MTR4-target multi-exon 3XT, forms aberrant condensates, termed KCTD13 3eXtended Transcript-derived protein (KeXT) bodies. Our results indicate that nuclear RNA surveillance has a crucial roles in interfering with the formation of condensates of a defective polyadenylated transcript-derived protein.
Results
MTR4 destabilizes 3XTs
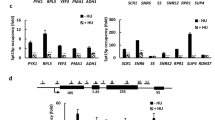
To identify aberrant RNAs with defects in splicing and polyadenylation that are regulated by MTR4, we searched for transcripts that are both increased in abundance and stabilized by MTR4 depletion, termed “MTR4-target transcripts,” by combining the data from long-read direct RNA sequencing, short-read RNA-sequencing, and 3′ sequencing (Fig. 1a and Supplementary Fig. 1a). We applied a bioinformatic method, FLAIR (fast local ancestry estimation), to identify and quantify novel and known transcripts using long- and short-read RNA sequence data (Fig. 1a, Supplementary Fig. 1b, and Supplementary Data 1). We then examined their stabilization by MTR4 depletion using 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB) chase-combined 3′ sequencing data (Fig. 1a). We found 8,043 transcripts were increased in abundance and that 1164 transcripts were stabilized by MTR4 depletion. Cross-referencing these data, we identified 464 MTR4-target transcripts (Fig. 1a and Supplementary Data 2). We next categorized aberrant RNAs with APA into the following six types according to the status of the last exon; 3′ extended, both extended, 5′ extended, known exon, intron-exonized, and intergenic (Supplementary Fig. 1c). Except for the known exon-type transcripts, MTR4 mainly destabilized the intergenic- and 3′ extended-type transcripts (Supplementary Fig. 1d). We next focused on 3′ extended-type transcripts whose exonic region was extended into a primarily intronic region, termed 3′ eXtended Transcripts (3XTs) (Supplementary Data 3). Interestingly, MTR4 destabilized two types of 3XT; one was an intronic APA isoform with one exon and the other was an intronic APA isoform with multiple exons, termed mono- and multi-exon 3XTs, respectively (Fig. 1b and Supplementary Data 3–5). We also defined a 3′ eXtended Region (3XR) as the intronic region extending from the existing exonic region in the last exon of a 3XT (Fig. 1b). Consistent with our results, MTR4 has been shown to prevent the accumulation of prematurely terminated RNAs, similar to mono-exon 3XTs21,23. Although the regulatory mechanisms of some transcripts, such as mono-exon 3XTs and prematurely terminated RNAs, are well studied10,21,23,28,29, those of multi-3XTs have not been determined. We next selected the 3XTs transcribed from ATP23, TP53TG1, and USP45 as MTR4-target mono-exon 3XTs, and those from HECTD2, SPRED2, and KCTD13 as MTR4-target multi-exon 3XTs (Fig. 1c and Supplementary Fig. 1e–j). qRT-PCR analysis confirmed that MTR4 knockdown using siRNA resulted in a marked increase in the levels of mono- and multi-exon 3XTs, but not of their originally annotated mature mRNAs (Fig. 1d and Supplementary Fig. 1k). In addition, we performed RNA immunoprecipitation (RIP) assays with an anti-MTR4 antibody using lysates from HeLa cells. qRT-PCR analysis of the immunoprecipitates revealed that MTR4 associated with endogenous HECTD2 3XT, SPRED2 3XT, and KCTD13 3XT (Supplementary Fig. 1l).
a Schematic illustration of the identification of RNAs destabilized by MTR4. (left) To identify transcript variant repertoire upregulated upon MTR4 depletion, HeLa cells transfected with siCont or siMTR4 were subjected to short- and long-read sequencing, as presented in Supplementary Fig. 1b. Pale-colored boxes are transcripts identified in this study. (right) To identify the transcript variant stabilized by MTR4 depletion, HeLa cells transfected with siCont or siMTR4 were treated with 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB), a transcription inhibitor, for the indicated times and then subjected to 3′-sequencing to estimate stability. By combining these data, MTR4-target transcripts were determined. b Schematic representation of two types of 3′ eXtended Transcript (3XT) and 3′ eXtended Regions (3XRs). The middle exon is the exon other than the first or last exon in annotated transcripts. 3XTs are transcripts that have an extended last exon with (multi-exon 3XT) or without (mono-exon 3XT) splicing events. c A violin plot from NanoBlot results displaying the distribution of sequenced read lengths of a representative 3XT. ATP23 (left, blue) and HECTD2 (right, red) are examples of genes with mono- and multi- exon 3XTs, respectively. Arrowheads indicate the lengths of ATP23 (blue) and HECTD2 (red) 3XT. d qRT-PCR analysis of mono- (blue) and multi-exon (red) 3XT expression in HeLa cells transfected with siRNA targeting MTR4. Results are expressed as the mean ± s.d. (n = 3 biologically independent replicates). Paired two-sided Student’s t-test; *p < 0.05. The exact p-values are ATP23 3XT: p = 0.00916 (siCont vs siMTR4#1), p = 0.00074 (siCont vs siMTR4#2), TP53TG1 3XT: p = 0.00077 (siCont vs siMTR4#1), p = 0.00227 (siCont vs siMTR4#2), USP45 3XT: p = 0.00792 (siCont vs siMTR4#1), p = 0.01991 (siCont vs siMTR4#2), HECTD2 3XT: p = 0.00441 (siCont vs siMTR4#1), p = 0.00667 (siCont vs siMTR4#2), SPRED2 3XT: p = 0.00794 (siCont vs siMTR4#1), p = 0.00122 (siCont vs siMTR4#2), KCTD13 3XT: p = 0.02597 (siCont vs siMTR4#1), p = 0.02677 (siCont vs siMTR4#2). Source data are provided as a Source Data file.
PAXT, but not NEXT and TRAMP, regulates the expression of both mono- and multi-exon 3XTs
MTR4, in association with the cofactor complexes PAXT, NEXT, and TRAMP, facilitates RNA incorporation into the RNA exosome by unwinding complicated RNA structures in the nucleus20,21. We examined whether the RNA exosome, PAXT, NEXT, and TRAMP complexes are involved in regulating the expression of MTR4-target 3XTs in HeLa cells. qRT-PCR analysis showed that knockdown of EXOSC5 and DIS3, which are RNA exosome components, upregulated both MTR4-target mono- and multi-exon 3XTs (Fig. 2a and Supplementary Fig. 2a,b). Moreover, MTR4-target 3XTs were upregulated by knockdown of PABPN1 and ZFC3H1, which are PAXT components, but not of RBM7 and ZCCHC8, which are NEXT components, or of ZCCHC7 and PAPD5, which are TRAMP components (Fig. 2b–d and Supplementary Fig. 2c–g). RNA-seq analysis also revealed that the expression of MTR4-target 3XTs was upregulated in HeLa cells in which PABPN1, but not RBM7, was knocked down21 (Supplementary Fig. 2h). These results indicate that both mono- and multi-exon 3XTs destabilized by MTR4 are recognized by the PAXT complex, but not by the NEXT and TRAMP complexes. They are then degraded by the RNA exosome complex.
a–d (left) qRT-PCR analysis of mono- (blue) and multi-exon (red) 3XT expression in HeLa cells transfected with siRNA targeting EXOSC5 (a), PABPN1 (b), RBM7 (c) or ZCCHC7 (d). qRT-PCR results are expressed as the mean ± s.d. (n = 3 biologically independent replicates). Paired two-sided Student’s t-test; *p < 0.05. (middle) qRT-PCR analysis of EXOSC5, PABPN1 or RBM7 expression in HeLa cells transfected with siRNA targeting EXOSC5 (a), PABPN1 (b), RBM7 (c), or ZCCHC7 (d). qRT-PCR results are expressed as the mean ± s.d. (n = 3 biologically independent replicates). Paired two-sided Student’s t-test; *p < 0.05. (right) Cell lysates from HeLa cells transfected with siRNA targeting the indicated genes were subjected to immunoblotting analysis with anti-EXOSC5 (a), anti-PABPN1 (b), anti-RBM7 (c), anti-ZCCHC7 (d), or anti-GAPDH antibodies. GAPDH was used as a loading control. The exact p-values are a ATP23 3XT: p = 0.00833 (siCont vs siEXOSC5#1), p = 0.00927 (siCont vs siEXOSC5#2), TP53TG1 3XT: p = 0.00048 (siCont vs siEXOSC5#1), p = 0.00067 (siCont vs siEXOSC5#2), USP45 3XT: p = 0.00187 (siCont vs siEXOSC5#1), p = 0.00312 (siCont vs siEXOSC5#2), HECTD2 3XT: p = 0.00187 (siCont vs siEXOSC5#1), p = 0.00312 (siCont vs siEXOSC5#2), SPRED2 3XT: p = 0.00977 (siCont vs siEXOSC5#1), p = 0.02162 (siCont vs siEXOSC5#2), KCTD13 3XT: p = 0.00443 (siCont vs siEXOSC5#1), p = 0.00792 (siCont vs siEXOSC5#2), EXOSC5: p = 0.00010 (siCont vs siEXOSC5#1), p = 0.00007 (siCont vs siEXOSC5#2); b ATP23 3XT: p = 0.00054 (siCont vs siPABPN1#1), p = 0.01888 (siCont vs siPABPN1#2), TP53TG1 3XT: p = 0.02140 (siCont vs siPABPN1#1), p = 0.04520 (siCont vs siPABPN1#2), USP45 3XT: p = 0.00101 (siCont vs siPABPN1#1), p = 0.00695 (siCont vs siPABPN1#2), HECTD2 3XT: p = 0.00956 (siCont vs siPABPN1#1), p = 0.03642 (siCont vs siPABPN1#2), SPRED2 3XT: p = 0.01983 (siCont vs siPABPN1#1), p = 0.01966 (siCont vs siPABPN1#2), KCTD13 3XT: p = 0.01838 (siCont vs siPABPN1#1), p = 0.03199 (siCont vs siPABPN1#2), PABPN1: p = 0.00088 (siCont vs siPABPN1#1), p = 0.00021 (siCont vs siPABPN1#2); c RBM7: p = 0.00033 (siCont vs siRBM7#1), p = 0.00006 (siCont vs siRBM7#2); d ZCCHC7: p = 7.8721E-07 (siCont vs si ZCCHC7#1), p = 0.00009 (siCont vs si ZCCHC7#2). Source data are provided as a Source Data file.
hnRNPK regulates the expression of multi- but not mono-exon 3XTs
To clarify the mechanisms underlying MTR4-mediated destabilization and degradation of multi-exon 3XTs in HeLa cells, we searched for proteins that recognize multi-exon 3XRs in host 3XTs. MEME motif analysis revealed that the CCWSCC motif was enriched in MTR4-target multi-exon 3XRs (Fig. 3a). We next compared this C-rich motif to known motifs of RNA-binding proteins using the Tomtom motif comparison tool30. Tomtom analysis showed that heterogeneous nuclear ribonucleoprotein K (hnRNPK) or poly(RC) binding protein (PCBP2) can associate with the CCWSCC motif (Fig. 3b and Supplementary Data 6). In contrast, RBMX, hnRNPH2, SRSF9, and SRSF4, which are nuclear RNA-binding proteins that regulate splicing and transcription31,32, can associate with MTR4-target mono-exon 3XRs (Supplementary Fig. 3a and Supplementary Data 6). We then examined the effect of knocking down these proteins on the expression of MTR4-target multi-exon 3XTs. We observed that knockdown of hnRNPK, but not PCBP2, caused upregulation of the multi-exon 3XTs (Fig. 3c and Supplementary Fig. 3b–f). We also found that hnRNPK depletion had no effect on the expression of the mono-exon 3XTs, on SNHG9 and SNHG19, which are PAXT targets, or on proRBM39, proINFAR, and proSNRNP70, which are NEXT targets (Supplementary Fig. 3g, h). hnRNPK is a DNA/RNA-binding protein that controls several biological processes and the development of numerous diseases33,34,35. Similar to MTR4, hnRNPK is mainly localized in the nucleus of HeLa cells36 (Supplementary Fig. 3i). hnRNPK contains three K homology (KH) domains, which recognize RNA or single-stranded DNA, a nuclear-localization signal, and a nuclear shuttling domain. hnRNPK is involved in transcription, mRNA splicing, stability, transport, and translation33,35. However, hnRNPK has not been reported to be involved in nuclear RNA degradation.
a The C-rich motif is enriched in MTR4-target multi-exon 3XRs. The table shows the results of MEME motif enrichment analysis. E-value; the expected number of motifs with the given log likelihood ratio (or higher), and with the same width and site count that one would find in a similarly sized set of random sequences. Sites; the number of sites contributing to the construction of the motif. b Venn diagram showing the CCWSCC-matched RNA-binding proteins speculated by Tomtom using different distance metrics. Pearson; Pearson correlation coefficient. Euclidean; Euclidean distance. Sandelin; Sandelin-Wasserman similarity. W; A or T. S; G or C. c qRT-PCR analysis of mono- (blue) and multi-exon (red) 3XT expression in HeLa cells transfected with si-hnRNPK. Results are expressed as the mean ± s.d. (n = 3 biologically independent replicates). Paired two-sided Student’s t-test; *p < 0.05. d Lysates from HeLa cells were subjected to immunoprecipitation with an anti-hnRNPK antibody or rabbit IgG, followed by qRT-PCR analysis to detect the indicated RNAs. SNHG9 and SNHG19 RNA were used as negative controls. Results are expressed as the mean ± s.d. (n = 3 biologically independent replicates). Paired two-sided Student’s t-test; *p < 0.05. e hnRNPK associates with MTR4 in vivo. Lysates from HeLa cells were subjected to immunoprecipitation with an anti-MTR4 antibody, an anti-hnRNPK antibody or rabbit IgG followed by immunoblotting analysis with anti-MTR4, anti-hnRNPK or anti-GAPDH antibodies. GAPDH was used as a negative control. This experiment was repeated independently two times with similar results. The exact p-values are c HECTD2 3XT: p = 0.03689 (siCont vs si-hnRNPK#1), p = 0.04965 (siCont vs si-hnRNPK2), SPRED2 3XT: p = 0.01871 (siCont vs si-hnRNPK#1), p = 0.02218 (siCont vs si-hnRNPK#2), KCTD13 3XT: p = 0.01953 (siCont vs si-hnRNPK#1), p = 0.04751 (siCont vs si-hnRNPK#2); d HECTD2 3XT: p = 0.01229 (rabbit IgG vs hnRNPK IP), SPRED2 3XT: p = 0.00157 (rabbit IgG vs hnRNPK IP), KCTD13 3XT: p = 0.01079 (rabbit IgG vs hnRNPK IP). Source data are provided as a Source Data file.
To clarify the association between hnRNPK and multi-3XTs, we performed RIP assays with an anti-hnRNPK antibody using lysates from HeLa cells. qRT-PCR analysis of the immunoprecipitates revealed that hnRNPK associated with endogenous HECTD2 3XT, SPRED2 3XT, and KCTD13 3XT but not with SNHG9 or SNHG19 RNA (Fig. 3d). In addition, MTR4 knockdown did not alter the association between hnRNPK and multi-exon 3XTs (Supplementary Fig. 3j). We also found that hnRNPK associated with originally annotated mature HECTD2, SPRED2, and KCTD13 mRNAs; however, depletion of hnRNPK or PABPN1 did not cause upregulation of their expression (Supplementary 3k–m). By contrast, hnRNPK knockdown downregulated the association between MTR4 and multi-exon 3XTs (Supplementary Fig. 3n). We next examined whether hnRNPK associates with MTR4 in HeLa cells. Pull-down assays using HeLa cell lysates treated with RNase A/T1 and anti-MTR4 or anti-hnRNPK antibodies revealed that hnRNPK associated with MTR4 in the absence of RNA (Fig. 3e). We also found that hnRNPK associated with the PAXT components, PABPN1 and ZFC3H1 (Supplementary Fig. 3o). Consistent with these results, hnRNPK was listed as an interactor candidate of MTR4, PABPN1 and ZFC3H1 by an affinity capture and mass spectrometry approach20,23,37,38. Taken together, these results indicate that hnRNPK associates with and downregulates multi-exon 3XTs by interacting with MTR4 in the nucleus.
hnRNPK promotes the degradation of multi-exon 3XTs by recognizing their 3XRs
We next examined whether 3XRs derived from multi-exon 3XTs are involved in the hnRNPK-mediated degradation of multi-3XTs. qRT-PCR analysis revealed that the fusion of multi-exon 3XRs downregulated β-globin mRNA in HeLa cells (Fig. 4a). Moreover, the overexpression of both hnRNPK and MTR4 resulted in the downregulation of β-globin mRNA (Supplementary Fig. 4a). In addition, RIP analyzes with an anti-Flag antibody using lysates from HeLa cells transfected with Flag-tagged hnRNPK and with anti-hnRNPK antibody using lysates from HeLa cells revealed that hnRNPK also associated with β-globin-fused multi-exon 3XRs, but not with SNHG19 RNA (Fig. 4b and Supplementary Fig. 4b). Moreover, RIP assays using a series of hnRNPK deletion mutants revealed that the regions spanning amino acid 42 to 104 (KH1 in Fig. 4c) and 387 to 451 (KH3 in Fig. 4c) were required for the association of hnRNPK with β-globin-fused KCTD13 3XR (Fig. 4c, d and Supplementary Fig. 4c, d). Consistent with these results, the structure of hnRNPK predicted by AlphaFold indicated that KH1 (purple) and KH3 (pink) domains are spatially closed and that there is space between them that could accommodate RNAs (Supplementary Fig. 4e). We also found that hnRNPK depletion led to the cytoplasmic accumulation of KCTD13 3XTs in HeLa cells (Supplementary Fig. 4f).
a HeLa cells were transfected with a 3XR-fused rabbit β-globin gene and then subjected to qRT-PCR analysis. Results are expressed as the mean ± s.d. (n = 3 biologically independent replicates). Paired two-sided Student’s t-test; *p < 0.05. b Lysates from HeLa cells transfected with a 3XR-fused rabbit β-globin gene and Flag-tagged hnRNPK were subjected to immunoprecipitation with an anti-Flag antibody followed by qRT-PCR analysis to detect the indicated RNAs. SNHG19 RNA was used as a negative control. Results are expressed as the mean ± s.d. (n = 3 biologically independent replicates). Paired two-sided Student’s t-test; *p < 0.05. c Schematic representation of wild-type (WT) and deletion mutant (delKH1 ~ 3) hnRNPK proteins. KH; K homology. d Lysates from HCT116 cells transfected with a KCTD13 3XR-fused rabbit β-globin gene along with wild-type or mutant Flag-tagged hnRNPK were subjected to immunoprecipitation with an anti-Flag antibody followed by qRT-PCR analysis to detect β-globin mRNA and SNHG19 RNA. SNHG19 RNA was used as a negative control. Results are expressed as the mean ± s.d. (n = 3 biologically independent replicates). Paired two-sided Student’s t-test; *p < 0.05. The exact p-values are a β-globin: p = 0.00002 (Mock vs HECTD2 3XR), p = 0.00147 (Mock vs SPRED2 3XR), p = 0.00036 (Mock vs KCTD13 3XR); b β-globin: p = 0.01727 (Mock vs HECTD2 3XR), p = 0.03705 (Mock vs SPRED2 3XR), p = 0.00146 (Mock vs KCTD13 3XR); d β-globin: p = 0.01415 (WT vs delKH1), p = 0.01123 (WT vs delKH3). Source data are provided as a Source Data file.
To elucidate the direct function of hnRNPK in RNA degradation, we performed a tethering assay using lambdaN-HA-tagged hnRNPK and β-globin RNA fused with 5BoxB. qRT-PCR analysis showed that hnRNPK led to the degradation of β-globin RNA fused with 5BoxB in HeLa cells (Supplementary Fig. 4g). Moreover, MTR4 facilitated the degradation of β-globin RNA fused with 5BoxB by hnRNPK (Supplementary Fig. 4h). These results indicate that hnRNPK recognizes multi-exon 3XRs to incorporate them into the RNA decay machinery in the nucleus.
The protein translated from KCTD13 3XT forms condensates
Recent findings suggest that disease-related aberrant proteins produced by gene mutation or fusion form abnormal biomolecular condensates via aberrant liquid–liquid phase separation (LLPS) and transition (LLPT) in cancer development and neurodegenerative diseases39,40,41,42. We hypothesized that some translation products from multi-exon 3XTs, which are destabilized by the MTR4-hnRNPK complex under normal conditions, could form aberrant biomolecular condensates. There are several computational tools and resources available for predicting LLPS-related molecular characteristics within protein, RNA, and DNA sequences43; therefore, to test the hypothesis, we predicted the key regions of aberrant proteins from MTR4-target multi-exon 3XTs for LLPS using a machine learning algorithm. We first annotated the coding sequences of MTR4-target multi-exon 3XTs using TransDecoder44,45 (Supplementary Data 7). Only transcripts with the longest coding region for each multi-exon 3XT were retained, and only multi-exon 3XTs with complete coding regions were used for the phase separation prediction by dSCOPE, which can reveal phase separation regions (PSRs) in protein sequences. The dSCOPE prediction showed that three proteins of multi-exon 3XTs have potential phase separation regions (Supplementary Data 8). We focused our analysis on KCTD13 3XT-derived protein because it had the highest dSCOPE score among the proteins analyzed (Supplementary Data 8). We found that the peptide encoded by the 3XR of KCTD13 contains a PSR (Supplementary Fig. 5a). Moreover, the KCTD13 3XT protein structure determined by AlphaFold indicated that its PSR is disordered (Fig. 5a).
a The structure of the protein derived from KCTD13 3XT is predicted by AlphaFold and the PSR region (pink) translated from the KCTD13 CDS contains a disorder region. b Detection of KeXT bodies in HeLa cells transfected with siMTR4#2. Representative maximum projection immunofluorescence image of HeLa cells transfected with siMTR4#2 and immunolabeled with KCTD13 3XT protein (orange). Nuclei were stained with Hoechst33258 (blue). Scale bar, 10 µm. This experiment was repeated independently three times with similar results. c Detection of KeXT bodies in HeLa cells transfected with Flag-tagged KCTD13 3XT protein. Immunofluorescence images of HeLa cells transfected with Flag-tagged KCTD13 3XT protein (WT) or Flag-control (Mock) and immunostained with anti-Flag antibody (orange). Nuclei were stained with Hoechst33258 (blue). Scale bar, 10 µm. This experiment was repeated independently three times with similar results. d The 3XR is required for KeXT body assembly. Immunofluorescence images of HeLa cells transfected with PSR-deletion mutant Flag-tagged KCTD13 3XT protein (del-PSR) or Flag-control (Mock) and immunostained with anti-Flag antibody (orange). Nuclei were stained with DAPI (blue). Scale bar, 10 µm. This experiment was repeated independently three times with similar results. e KeXT bodies in HeLa cells transfected with siMTR4#2 in the presence of 1,6-hexandiol or 2,5-hexandiol. Representative maximum projection immunofluorescence image of HeLa cells transfected with siMTR4#2, treated with or without 1,6-hexandiol or 2,5-hexandiol, and immunolabeled with KCTD13 3XT protein (orange). Nuclei were stained with Hoechst33258 (blue). Scale bar, 10 µm. This experiment was repeated independently three times with similar results.
We then examined whether KCTD13 3XT proteins form biomolecular condensates in HeLa cells. We found that endogenous KCTD13 3XT protein formed granule-like structures, which we name KCTD13 3eXtended Transcript-derived protein (KeXT) bodies, primarily in the cytoplasm of MTR4-knockdown HeLa cells (Fig. 5b and Supplementary Fig. 5b). We also found that the Flag-tagged KCTD13 3XT protein formed KeXT bodies, whereas deletion of the PSR led to diffusion of KCTD13 3XT coding sequences into the nucleoplasm, indicating that KeXT bodies are formed through a PSR-dependent mechanism (Fig. 5c, d and Supplementary Fig. 5a). In addition, treatment with 3% 1,6-hexanediol, which disrupts liquid-like assemblies by disturbing multivalent hydrophobic interactions46, resulted in both endogenous and exogenous KeXT body dissociation, whereas treatment with 2,5-hexanediol, an aliphatic alcohol-like 1,6-hexanediol, did not affect the formation of KeXT bodies, indicating that both endogenous and exogenous KeXT bodies are likely phase separated (Fig. 5e and Supplementary Fig. 5c–f).
Taken together, these results indicate that KCTD13 3XT protein may accumulate and form condensates by acquiring the KCTD13 3XR, a read-through region from the last exon that is necessary for KCTD13 3XT degradation.
Discussion
A screen for aberrant polyadenylated transcripts that are destabilized by MTR4 in HeLa cells identified mono- and multi-exon 3XTs, which are among the intronic APA isoforms. We found that the PAXT complex, but not the NEXT or TRAMP complexes, was required for the degradation of MTR4-target 3XTs by the RNA exosome. We also found that hnRNPK interacted with MTR4 and regulated the expression of multi-exon 3XTs via association with their 3XRs. Further analysis showed that 3XRs were necessary and sufficient to recruit hnRNPK onto target RNAs. Moreover, we found that the protein translated from KCTD13 3XT, a multi-exon 3XT that was retained in the nucleus by hnRNPK, formed biomolecular condensates, named KeXT bodies. Our findings therefore indicate that hnRNPK-MTR4 complex-mediated destabilization of KCTD13 3XT may be important for the inhibition of aberrant condensate formation (Fig. 6).
A schematic model showing the mechanism of multi-exon 3XT degradation by the hnRNPK-MTR4-RNA exosome complex. MTR4 interacts with hnRNPK, which recognizes 3XRs of multi-exon 3XTs, and degrades them via the RNA exosome complex in the nucleus of cells under normal conditions. RNA surveillance dysregulation, in which MTR4/hnRNPK is depleted, leads to the production of multi-exon 3XT-derived proteins, resulting in the formation of aberrant biomolecular condensates.
We classified aberrant RNAs with APA into six types according to the status of the last exon (Supplementary Fig. 1c). According to APA classification in the present study, transcripts categorized as “known exon” are consistent with transcript isoforms with 3′-UTR APA or alternative terminal exon APA, the transcripts with “5′-extended” last exon are the same as isoforms with “internal exon APA,” and the transcripts with “3′-extended” or “both extended” are categorized as isoforms with intronic APA7. In addition to the transcripts with known APA features, we also found transcripts with “intron-exonized” or “intergenic” last exon in HeLa cells, some of which were destabilized by MTR4 (Fig. 1b and Supplementary Data 2). In our future studies, we aim to clarify the physiological functions and regulatory mechanisms of these different types of aberrant transcript with APA features.
To elucidate the mechanism by which MTR4 destabilizes multi-exon 3XTs, we searched for multi-exon 3XR-recognition proteins and found that hnRNPK interacted with multi-exon 3XTs via their 3XRs to retain them in the nucleus. Consistent with our results, hnRNPK promotes the nuclear and chromatin retention of RNAs with C-rich motifs, including SIRLOIN element and XIST34,47,48. Although hnRNPK can induce transcription by interacting directly with the RNA polymerase machinery33,35, we found that hnRNPK interacts with MTR4 to degrade multi-exon 3XTs. However, the CCWSCC motif was not enriched in MTR4-target mono-exon 3XRs, and hnRNPK knockdown had no effect on the expression of the mono-exon 3XTs, indicating that other RNA binding proteins, such as RBMX and hnRNPH2, may recognize the mono-exon 3XR to promote degradation by MTR4 and the RNA exosome. From these results, we suggest that hnRNPK retains aberrant RNAs in the nucleus and recruits MTR4 and the RNA exosome to degrade them in the nucleus.
Our combined analysis of long-read direct RNA sequencing and 3′ sequencing revealed a role for MTR4 in regulating the destabilization of aberrant transcripts with various types of APA. Long-read sequencing technologies and/or 3′ end detecting approaches have also been applied to search for distinct RNA isoforms through alternative RNA processing49,50. Alfonso-Gonzalez et al. performed a combination of long-read sequencing techniques, including cDNA and direct RNA sequencing using devices from Oxford Nanopore Technologies, and Iso-seq and FLAM-seq using a Sequel I system from Pacific Biosciences, to reveal alternative polyadenylation sites coupled to alternative transcription start sites50. Tang et al. performed metabolic labeling-coupled polyadenylation site-seq and Nanopore long-read RNA-sequencing using RNA from different cell compartments to show that longer APA isoforms terminated at distal polyadenylation sites and anchored in the chromatin/nuclear matrix serve as precursors for further processing at the proximal polyadenylation sites49. In addition to the long-read sequencing technologies, 3′-tagged single-cell or single-nucleus RNA-seq techniques, as well as various computational pipelines, hold promise in quantifying gene and 3′-UTR isoform expression in various types of cell51,52,53,54. Future advances in technology and bioinformatics will provide more detailed insights into APA-related biological phenomena and disease phenotypes.
Phase separation prediction by dSCOPE revealed that KCTD13 3XT-derived proteins have a potential PSR in the peptide encoded by their 3XR. In addition, we found that KCTD13 3XT protein formed KeXT bodies primarily in the cytoplasm of MTR4-knockdown HeLa cells. However, we currently do not understand the impact of KeXT body formation on cell fate and its molecular functions. Recent studies suggest that gene mutation or fusion can induce aberrant phase separation, leading to either gain or loss of biomolecular condensates in cancer development and neurodegenerative diseases39,40,43. Aberrant phase separation driven by EWS-FLI155 and EML4–ALK56 fusion genes promotes aberrant condensates, oncogenic gene expression programs, and oncogenic RAS/MAPK signaling, whereas the oncogenic fusion gene DnaJB1–PKA abolishes PKA-mediated phase separation57. In addition, amyotrophic lateral sclerosis patient mutations in prion-like domains of FUS cause liquid-to-solid phase transition58 and disease mutations in the prion-like domains of hnRNPA2 and hnRNPA1 promote incorporation of these proteins into stress granules and drive degenerative disease59. Considering prior studies on aberrant condensates, we assume that the formation of KeXT bodies, aberrant condensates degraded by the RNA surveillance system under normal conditions, impede proper cellular function. Interestingly, RNA exosome defects are associated with neurodegenerative diseases60. Mutations in EXOSC3 and EXOSC8 cause a progressive disease characterized by spinal motor neuron dysfunction, pontocerebellar hypoplasia, cerebellar and corpus callosum hypoplasia, and abnormal myelination61,62. hnRNPK is also an essential factor for the development and differentiation of neurons63,64. Our findings, in conjugation with prior studies, indicate an important link between KeXT bodies and pathological developments, including neuronal disease. However, further detailed studies are needed to confirm this.
In conclusion, we have shown that MTR4 plays a critical role in the degradation of aberrant transcripts containing several types of APA features in HeLa cells. We further elucidated that MTR4 cooperates with hnRNPK in a complex to destabilize KCTD13 3XT, a multi-exon 3XT, and thus prevent the formation of KeXT bodies. Our findings provide important insight into the regulation and function of aberrant transcripts that have alternative polyadenylation and splicing defects.
Methods
Cell culture
HeLa cells (631183, Takara) were cultured in Dulbecco’s modified Eagle’s medium (043-30085, Wako) supplemented with 10% fetal bovine serum at 37 °C in a humidified incubator with 5% CO2.
Antibodies and reagents
Anti-MTR4 (A5576, Lot 00202580201, 1:1,000 dilution) antibody was obtained from Abclonal. Anti-hnRNPK (11426-1-AP, Lot 00022887, 1:1,000 dilution) and anti-PAPD5 (55197-1-AP, Lot 09000346, 1:1,000 dilution) antibodies were obtained from Proteintech. Anti-GAPDH (MAB374, clone 6C5, Lot 3725986, 1:1,000 dilution) antibody was obtained from Merck Millipore. Anti- ZFC3H1 (NB100-68267, Lot R33689, 1:1,000 dilution) antibody was obtained from Novus Biologicals. Anti-EXOSC5 (ab168804, Lot GR138334-1, 1:1,000 dilution) and anti-PABPN1 (ab75855, Lot GR32937-10, 1:1,000 dilution) antibodies were obtained from Abcam. Anti-DIS3 (HPA039281, Lot A95805, 1:1,000 dilution), anti-RBM7 (HPA013993, Lot F119176, 1:1,000 dilution), anti-ZCCHC7 (HPA021088, Lot A117087, 1:1,000 dilution), anti-ZCCHC8 (SAB1102086, Lot 080M2750, 1:1,000 dilution), and anti-FLAG (F3165, clone M2, Lot 058K6113, 1:1,000 dilution) antibodies were obtained from Sigma. Anti-UHRF1 (612264, clone 28, Lot 83172, 1:1,000 dilution) antibody was obtained from BD Biosciences. Anti-α-tubulin (CP-06, clone DM1A, Lot D00175772, 1:1,000 dilution) antibody was obtained from CALBIOCHEM. Secondary antibodies were purchased from GE Healthcare (NA934, Lot 9599347, 1:5,000 dilution; NA931, Lot 384924, 1:5,000 dilution) and Cell Signaling (7074S, Lot 29, 1:5,000 dilution). Normal rabbit IgG (PM035, Lot 007) was obtained from MBL. Antisera against human KCTD13 3XT protein were raised in rabbits by immunization with a chemically synthesized peptide of KCTD13 3XR (LGELPKQVLEQKKGDMGYG) by Eurofins Genomics (Hokudo, 1:500 dilution for IF). ECL-plus was purchased from GE Healthcare. VeriBlot for IP Detection Reagent (HRP) (ab131366, Abcam) was used to avoid the detection of heavy and light chains.
qRT-PCR analysis
Total RNA was isolated using a NucleoSpin RNA mini kit (740955, MACHEREY-NAGEL) with the EconoSpin column for RNA (EP-21201, Epoch Life Science) and reverse transcribed using the PrimeScript RT Reagent Kit with gDNA eraser (RR047B, Takara Bio). qRT-PCR analysis of cDNA was performed on a Thermal Cycler Dice Real-Time System (Takara Bio) using TB Green Premix Ex Taq II (RR820L, Takara Bio). Prior to fold-change calculation, the values were normalized to the signal generated from GAPDH mRNA. Primer sequences are listed in Supplementary Data 9.
RNA interference
siRNA duplexes targeting MTR4 were purchased from Hokkaido System Science (HSS). Stealth siRNA duplexes targeting ZCCHC8 and RBM7 were purchased from Invitrogen. siRNA duplexes targeting hnRNPK, PCBP2, EXOSC5, ZFC3H1, PABPN1, ZCCHC7, and PAPD5 were purchased from Ambion. siRNA duplexes targeting DIS3 were purchased from Ambion and GeneDesign. Cells were transfected with RNA duplexes using Lipofectamine RNAiMAX (13778150, Invitrogen). For DRB chase experiments, a final concentration of 100 µM 5,6-dichloro-1-β-D-ribofuranosyl-1H-benzimidazole (DRB, 10010302, Cayman Chemical) in DMSO (047-29353, FUJIFILM) was added to cells 72 h after siRNA transfection. Sequences or IDs of siRNAs are shown in Supplementary Data 10.
Constructs and transfection
Flag-tagged hnRNPK and its deletion mutants were amplified by PCR and cloned into pcDNA3.1(+). Flag-tagged MTR4 was amplified by PCR and cloned into pcDNA3.1(+). Lambda N-HA tagged hnRNPK was cloned into pCAGGS using the In-Fusion HD cloning kit (Takara). The 3XRs of HECTD2, SPRED2 and KCTD13 were amplified by PCR and cloned into pTet-BBB. The flag-tagged CDS region of KCTD13 3XT and its deletion mutants were synthesized and cloned into pcDNA3.1(+) using GeneArt (Thermo Fisher Scientific). Primer sequences are listed in Supplementary Data 11. Plasmids were transfected into cells using Lipofectamine 2000 (11668019, Invitrogen).
Immunoblot analysis
Cells were lysed with Lysis buffer (50 mM HEPES pH 7.5, 150 mM KCl, 0.5% NP-40, 2 mM EDTA, 1 mM NaF) containing Protease Inhibitor Cocktail (P8340, Sigma) for 20 min on ice. Following centrifugation at 13,000 × g, an equal volume of SDS sample buffer (100 mM Tris-HCl pH 6.8, 20% glycerol, 4% SDS, 12% β-mercaptoethanol, 0.1% bromophenol blue) was added to the supernatant. After heating to 95 °C for 5 min, samples were resolved by SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon-P, IPVH00010, Merck Millipore), and analyzed by immunoblotting using HRP-conjugated secondary antibodies. Membranes were blocked with 5% bovine serum albumin (BSA, 011-27055, Wako) in TBST buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1% Tween-20) at room temperature for one hour or with Blocking One (03953-95, Nacalai Tesque) for 5 min at room temperature before probing with antibodies. After each incubation, membranes were washed three times with TBST buffer. Visualization was performed using HRP Substrate (WBKLS0500, Merck Millipore) and the LAS-4000UVmini Luminescent Image Analyzer (FUJIFILM).
Immunoprecipitation
HeLa cells (107 cells) were lysed with Lysis buffer (50 mM HEPES pH 7.5, 150 mM KCl, 0.5% NP-40, 2 mM EDTA, 1 mM NaF) containing Protease Inhibitor Cocktail (P8340, Sigma) for 20 min on ice. Following centrifugation at 13,000 × g for 20 min, the supernatants were treated with RNase A/T1 (1 µg/µL RNase A, 40 U/µl RNase T1, 50 mM Tris HCl, 50 mM NaCl, 50% glycerol) for 15 min on ice. RNase-treated lysates were incubated with anti-hnRNPK (1.375 µg) antibody, anti-MTR4 (2 µg) antibody, or normal rabbit IgG for 2 h at 4 °C with gentle rotation. Twenty microliters of Dynabeads Protein G (10009D, Invitrogen) were added and incubated for 1 h at 4 °C with gentle rotation. The beads were washed three times with Wash buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.05% NP-40) containing Protease Inhibitor Cocktail (P8340, Sigma) and then twice with TBST (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1% Tween-20) containing Protease Inhibitor Cocktail (P8340, Sigma). After washing, immunocomplexes were eluted with 1× SDS sample buffer (50 mM Tris-HCl pH 6.8, 10% glycerol, 2% SDS, 6% β-mercaptoethanol, 0.05% bromophenol blue) containing Protease Inhibitor Cocktail (P8340, Sigma) and immunoblotted with antibodies against target proteins.
RNA Immunoprecipitation
RIP assays was performed as described previously65,66,67 with minor modifications. HeLa cells (1 × 107 cells, for targeting endogenous proteins; 1 × 106 cells, for targeting overexpressed proteins) were lysed with Lysis buffer (50 mM HEPES pH 7.5, 150 mM KCl, 0.5% NP-40, 2 mM EDTA, 1 mM NaF) containing Protease Inhibitor Cocktail (P8340, Sigma) and Recombinant RNase Inhibitor (2313 A, Takara) for 20 min on ice. Following centrifugation at 13,000 × g, the supernatants were incubated with anti-hnRNPK (1.375 µg) or anti-MTR4 (2 µg) antibodies, or normal rabbit IgG for 2 h at 4 °C with gentle rotation. Thirty microliters of Dynabeads Protein G (10009D, Invitrogen) were added and incubated for one hour at 4 °C with gentle rotation. For Flag-IP analysis, the supernatants were incubated with 10 µL of Anti-FLAG M2 Magnetic Beads (M8823, Sigma, Lot 091M6291V) for one hour at 4 °C with gentle rotation. The beads were washed three times with Wash buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.05% NP-40) and then twice with TBST (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1% Tween-20) containing Protease Inhibitor Cocktail (P8340, Sigma) and Recombinant RNase Inhibitor (2313 A, Takara). After washing, RNA was isolated using a NucleoSpin RNA mini kit (740955, MACHEREY-NAGEL), treated with Recombinant DNase I (2270 A, Takara) and Recombinant RNase Inhibitor (2313 A, Takara) for 20 min at 37 °C, and purified using a NucleoSpin RNA clean-up kit (740948, MACHEREY-NAGEL). qRT-PCR was performed as described above. Primer sequences for qRT-PCR are shown in Supplementary Data 9.
Immunofluorescence
HeLa Cells were seeded on coverslips in 12-well plates 24 h before transfection with the Flag-tagged hnRNPK, KCTD13 3XT coding sequence or their mutants. At 24 h after transfection, cells were washed twice with PBS plus 0.1% Tween20 and fixed with cold 100% methanol for 5 min at room temperature. The coverslips were then washed three times with PBS, permeabilized in 0.1% Triton-X100/PBS for 5 min at 4 °C, and washed three times with PBS. After blocking with Blocking One Histo (06349-64, Nacalai Tesque) for 5 min at room temperature, the cells were incubated in a 20-fold dilution of Blocking One Histo containing anti-Flag or anti-KCTD13 3XT protein antibody at 4 °C overnight. After washing three times with PBS plus 0.1% Tween20, a 20-fold dilution of Blocking One Histo with Alexa 488- (ThermoFisher Scientific, A11029 Lot 56649 A, 1: 1000 dilution) or 555-conjugated (ThermoFisher Scientific, A31570, Lot 2387458, 1: 1000 dilution) secondary antibody was added to the cells, and the samples were incubated for 1 h at room temperature. After washing three times with PBS plus 0.1% Tween20, the cells were incubated in DAPI (340-07971, Dojindo)/PBS or Hoechst33258 (H1343, TCI)/PBS for 10 min at room temperature. The coverslips were mounted with ProLong gold (P36934, Thermo Fisher Scientific), and images were acquired with a ZEISS LSM 980 with Airyscan 2. Airyscan processing and Z-axis projection were performed using Zeiss ZEN 3.4 (blue) (version 3.4.91.00000).
For the evaluation of the LLPS properties, cells were treated with 1,6-hexanediol (087-00432, Wako) or 2,5-hexanediol (11904-10 G, Sigma) dissolved in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum at room temperature for 5 min. After these treatments, the cells were fixed with cold 100% methanol (137-01823, Wako) for 5 min at room temperature, followed by the immunofluorescence procedure described above.
Tethering assay
HeLa cells were transfected with the β-globin-5BoxB reporter or the β-globin control, a plasmid expressing EGFP, Flag-tagged MTR4, and plasmids expressing λN-HA-hnRNPK or λN-HA control. At 24 h after transfection, RNA was isolated from the cells using a NucleoSpin RNA mini kit (740955, MACHEREY-NAGEL) with the EconoSpin column for RNA (EP-21201, Epoch Life Science), treated with Recombinant DNase I (2270 A, Takara) and Recombinant RNase Inhibitor (2313 A, Takara) for 20 min at 37 °C, and purified using a NucleoSpin RNA Clean-up Kit (740948, MACHEREY-NAGEL). qRT-PCR was performed as described above. Prior to fold-change calculation, the values were normalized to the signal generated from EGFP mRNA. Primer sequences for qRT-PCR are shown in Supplementary Data 9.
Subcellular fractionation
Cell pellets were resuspended in 1 packed cell volume of Hypotonic buffer (10 mM HEPES pH 7.5, 10 mM KCl, 1.5 mM MgCl2). After incubation on ice for 15 min, cells were disrupted by 10 passages through a 25-gauge needle. Cells were centrifuged for 10 min at 1000 g at 4 °C and the supernatant containing the cytoplasmic fraction was collected by further centrifugation at 15,000 × g for 20 min. The remaining pellets were washed thrice with Hypotonic buffer, resuspended in Hypotonic-NP-40 buffer (10 mM HEPES pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 0.5% NP-40) and centrifuged for 10 min at 6000 × g at 4 °C. The remaining pellets were washed once with Hypotonic buffer, resuspended in Hypertonic buffer (20 mM HEPES pH 7.5, 250 mM KCl, 1.5 mM MgCl2, 0.5% NP-40) and incubated at 4 °C for 30 min with gentle rotation. Cells were centrifuged for 3 min at 6800 × g at 4 °C and the supernatant containing the nucleoplasm fraction was collected by centrifugation at 15,000 × g for 15 min.
Sequence analysis
RNA was isolated using a NucleoSpin RNA mini kit (740955, MACHEREY-NAGEL), treated with Recombinant DNase I (2270 A, Takara) and Recombinant RNase Inhibitor (2313 A, Takara) for 20 min at 37 °C, and purified using a Nucleospin RNA Clean-up Kit (740948, MACHEREY-NAGEL). For RNA sequencing analyzes of MTR4-depleted HeLa cells, libraries were prepared with polyA-selected RNA using a KAPA mRNA Capture Kit (KK8440, KAPA Biosystems) and an MGIEasy RNA Directional Library Prep Set (1000006385, MGI). Libraries were sequenced on the DNBSEQ-T7RS platform with 150-bp paired-end reads by GenomeLead Co., Ltd. Direct RNA sequencing was performed on the PromethION platform (Oxford Nanopore Technologies). 3′-Tag-sequencing (3′-Tag-seq) was performed on the NextSeq system by the DNA Technologies & Expression Analysis Core at the UC Davis Genome Center.
The classification of aberrant RNAs with APA events
All last exons from alternative isoforms were extracted and compared with basic gene annotations (Fig. 1b). The last exons were classified into the following six categories in descending order of priority, according to their relative position to the annotated exons. (a) Intergenic: the last exon is included in the intergenic region. (b) Intron-exonized: the last exon is included in or identical to the intronic region. (c) Known exon: the last exon is identical to an annotated exon. (d) 5′-Extended: the 5′ end extends to an intronic/intergenic region, while the 3′ end is included in an annotated exon. (e) Both extended: the last exon overlaps with annotated exon(s), with its 5′/3′ ends extending to intronic/intergenic regions. (f) 3′-Extended: the opposite of 5′-extended. The above classification was conducted by custom scripts.
Transcript and alternative isoform detection
RNA-seq data (DNBSEQ) were used to mark reliable splicing junctions. The human genome (hg38) and basic gene annotations (GENCODE v32) were downloaded from GENCODE68. The reads were mapped to the genome using STAR with the following parameters: --outFilterMultimapNmax 1 --outFilterMismatchNmax 10 --alignSJoverhangMin 20. Junctions and supporting reads were extracted from the mapped reads of all samples by junctionsfromsam.py in FLAIR (v1.4.0)69. Junctions with fewer than 50 supporting reads were removed.
Long-read direct RNA sequencing data were used to identify alternative isoforms. First, the reads were mapped to the human genome using Minimap2 (v2.17-r954-dirty)70 with the following parameters: -ax splice -uf -k14. For subsequent processing, the mapped reads were converted to sorted, indexed BAM format files using Samtools (v1.11)71 and then further converted to BED format files by bam2Bed12.py in FLAIR. Second, the FLAIR correct subcommand corrected misaligned splicing junctions in these mapped reads using gene annotations and reliable splicing junctions. The corrected reads in all samples were merged, and the redundancies were eliminated. Finally, the FLAIR collapse subcommand built high-confidence alternative isoforms from the genome, gene annotations, the reads from PromethION, and corrected reads with the following parameter: -s 10. The FLAIR quantify subcommand calculated the expression of alternative isoforms in each sample based on the reads from PromethION. A custom script normalized the read counts to CPM (counters per million mapped reads) which were calculated by excluding reads that did not map to annotated transcripts from the total mapped reads in the denominator. Genes that were upregulated by ≥ 1.5-fold due to MTR4 knockdown, using two different siRNAs, were considered differentially expressed. RNA isoforms from long-read direct RNA sequencing data were visualized using NanoBlot72.
RNA-seq data were retrieved from NCBI’s GEO or SRA databases (siPABPN1 and siRBM7, GSE8417221; si-hnRNPK, SRP11175647; siPCBP2, GSE13056473) using SRA Run Selector. After removing low-quality reads and adaptor sequences using fastp v0.23.474, the remaining reads were mapped to the genome using STAR v2.7.10b75. Expression count estimates and transcripts per million (TPM) were generated using RSEM v1.3.376 with the transcript reference generated from the above FLAIR analysis.
3′-Tag-seq analysis
3′-Tag-seq reads were trimmed using Cutadapt (v2.6)77 with the following parameters: -m 30 -a AGATCGGAAGAG. Trimmed reads were mapped to the human genome (hg38) using STAR, and uniquely mapped reads were kept. PCR duplicates were removed with Picard (v2.18.1, http://broadinstitute.github.io/picard/). Given that 3′-Tag-seq data are expected to have read coverage only at the 3′ ends of transcripts, the 100 bp-long 3′ ends from the above alternative isoforms were taken for transcript quantification. Read counting was performed using featureCounts (v2.0.1)78 with the following parameters: -p -M -O --fraction. Then, a custom script normalized the read counts to CPM (counts per million mapped reads) which were calculated by excluding reads that did not map to annotated transcripts from the total mapped reads in the denominator.
Sequence motif analysis and quantitative motif comparison
Motif analysis for MTR4-target multi-exon 3XRs was carried out using MEME v5.4.179 with the following parameters: -mod anr -nmotifs 5 -minw 6 -maxw 7 -objfun classic -markov_order 0. To compare known motifs with the motif derived from MEME analysis, a pair-wise motif comparison was performed using Tomtom motif comparison search tool30 and the known motifs in the human known RBP motifs dataset from the CISBP-RNA database80.
Protein annotation
Coding sequences within the multi-exon 3XTs were predicted and extracted using the TransDecoder tool (version-5.5.0) (https://github.com/TransDecoder/TransDecoder) with the default parameters. BLASTp programs (version-2.12.0 + ) were used for homology-based similarity searches of the transcripts and to predict proteins against the latest human protein sequence dataset from the Ensemble database (https://asia.ensembl.org/info/data/ftp/index.html, Last modified, 2022-12-13) with a maximum e-value of 1e−10. The coding sequences and lengths of multi-exon 3XTs are listed in Supplementary Data 7.
Prediction of phase separation region
dSCOPE81 was used to predict the potential phase separation regions of the predicted proteins from multi-exon 3XTs.
AlphaFold2 structure prediction
We predicted structures for the KCTD13 3XT coding sequence using AlphaFold Colab v2.3.282. The wild-type hnRNPK structure was retrieved from the AlphaFold Protein Structure Database (AlphaFold DB)82,83.
Statistical analysis
Statistical analysis was performed using a paired two-tailed Student’s t-test (Fig. 1d; Fig. 2a–d; Fig. 3c, d; Fig. 4a, b, d; Supplementary Fig. 1k, l; Supplementary Fig. 2a–g; Supplementary Fig. 3b–e, g, h, j–n; Supplementary Fig. 4a, b, f–h; Supplementary Fig. 5b, c, e), an unpaired two-tailed Student’s t-test (Supplementary Fig. 3f), or the Brunner-Munzel test (Supplementary Fig. 2h). A p-value < 0.05 was considered to be statistically significant. All experiments were independently repeated three times, as stated in figure legends. Statistical analysis was performed with R 4.3.1 or Microsoft Excel.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data supporting the findings of this study are available from the corresponding authors upon request. Short-read RNA sequencing, long-read direct RNA sequencing, and 3′Tag-seq data are deposited in the DNA Data Bank of Japan Sequence Read Archive (DRA) database under accession code DRA014762). Source data are provided with this paper.
Code availability
Custom scripts used in this study are available from https://doi.org/10.6084/m9.figshare.2640173884.
References
Richard, P. & Manley, J. L. Transcription termination by nuclear RNA polymerases. Genes Dev. 23, 1247–1269 (2009).
Licatalosi, D. D. & Darnell, R. B. RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet 11, 75–87 (2010).
Manning, K. S. & Cooper, T. A. The roles of RNA processing in translating genotype to phenotype. Nat. Rev. Mol. Cell Biol. 18, 102–114 (2017).
Tian, B. & Manley, J. L. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 18, 18–30 (2017).
Gruber, A. J. & Zavolan, M. Alternative cleavage and polyadenylation in health and disease. Nat. Rev. Genet 20, 599–614 (2019).
Mitschka, S. & Mayr, C. Context-specific regulation and function of mRNA alternative polyadenylation. Nat. Rev. Mol. Cell Biol. 23, 779–796 (2022).
Elkon, R., Ugalde, A. P. & Agami, R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat. Rev. Genet 14, 496–506 (2013).
Tian, B., Pan, Z. & Ju, Y. L. Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Res 17, 156–165 (2007).
Li, W. et al. Systematic profiling of poly(A)+ transcripts modulated by core 3’ end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLoS Genet 11, e1005166 (2015).
Insco, M. L. et al. Oncogenic CDK13 mutations impede nuclear RNA surveillance. Science 380, eabn7625 (2023).
Dubbury, S. J., Boutz, P. L. & Sharp, P. A. CDK12 regulates DNA repair genes by suppressing intronic polyadenylation. Nature 564, 141–145 (2018).
Lee, S. H. et al. Widespread intronic polyadenylation inactivates tumour suppressor genes in leukaemia. Nature 561, 127–131 (2018).
Jensen, T. H., Jacquier, A. & Libri, D. Dealing with pervasive transcription. Mol. Cell 52, 473–484 (2013).
Schmid, M. & Jensen, T. H. Controlling nuclear RNA levels. Nat. Rev. Genet 19, 518–529 (2018).
Kilchert, C., Wittmann, S. & Vasiljeva, L. The regulation and functions of the nuclear RNA exosome complex. Nat. Rev. Mol. Cell Biol. 17, 227–239 (2016).
Vanacova, S. & Stef, R. The exosome and RNA quality control in the nucleus. EMBO Rep. 8, 651–657 (2007).
Januszyk, K. & Lima, C. D. The eukaryotic RNA exosome. Curr. Opin. Struct. Biol. 24, 132–140 (2014).
Schneider, C. & Tollervey, D. Threading the barrel of the RNA exosome. Trends Biochem Sci. 38, 485–493 (2013).
Zinder, J. C. & Lima, C. D. Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev. 31, 88–100 (2017).
Lubas, M. et al. Interaction profiling identifies the human nuclear exosome targeting complex. Mol. Cell 43, 624–637 (2011).
Meola, N. et al. Identification of a nuclear exosome decay pathway for processed transcripts. Mol. Cell 64, 520–533 (2016).
Tanu, T. et al. hnRNPH1-MTR4 complex-mediated regulation of NEAT1v2 stability is critical for IL8 expression. RNA Biol. 18, 537–547 (2021).
Ogami, K. et al. An Mtr4/ZFC3H1 complex facilitates turnover of unstable nuclear RNAs to prevent their cytoplasmic transport and global translational repression. Genes Dev. 31, 1257–1271 (2017).
Kadaba, S. et al. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 18, 1227–1240 (2004).
Lubas, M. et al. The human nuclear exosome targeting complex is loaded onto newly synthesized RNA to direct early ribonucleolysis. Cell Rep. 10, 178–192 (2015).
LaCava, J. et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121, 713–724 (2005).
Wyers, F. et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121, 725–737 (2005).
Kaida, D. et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468, 664–668 (2010).
Berg, M. G. et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell 150, 53–64 (2012).
Gupta, S., Stamatoyannopoulos, J. A., Bailey, T. L. & Noble, W. S. Quantifying similarity between motifs. Genome Biol. 8, R24 (2007).
Elliott, D. J., Dalgliesh, C., Hysenaj, G. & Ehrmann, I. RBMX family proteins connect the fields of nuclear RNA processing, disease and sex chromosome biology. Int J. Biochem Cell Biol. 108, 1–6 (2019).
Brownmiller, T. & Caplen, N. J. The HNRNPF/H RNA binding proteins and disease. Wiley Interdiscip. Rev. RNA 14, e1788 (2023).
Bomsztyk, K., Denisenko, O. & Ostrowski, J. hnRNP K: one protein multiple processes. Bioessays 26, 629–638 (2004).
Wang, Z. et al. The emerging roles of hnRNPK. J. Cell Physiol. 235, 1995–2008 (2020).
Sugimasa, H. et al. Heterogeneous nuclear ribonucleoprotein K upregulates the kinetochore complex component NUF2 and promotes the tumorigenicity of colon cancer cells. Biochem Biophys. Res Commun. 459, 29–35 (2015).
Silla, T., Karadoulama, E., Mąkosa, D., Lubas, M. & Jensen, T. H. The RNA exosome adaptor zfc3h1 functionally competes with nuclear export activity to retain target transcripts. Cell Rep. 23, 2199–2210 (2018).
Rolland, T. et al. A proteome-scale map of the human interactome network. Cell 159, 1212–1226 (2014).
Kwiatek, L., Landry-Voyer, A. M., Latour, M., Yague-Sanz, C. & Bachand, F. PABPN1 prevents the nuclear export of an unspliced RNA with a constitutive transport element and controls human gene expression via intron retention. RNA 29, 644–662 (2023).
Taniue, K. & Akimitsu, N. Aberrant phase separation and cancer. FEBS J. 289, 17–39 (2021).
Taniue, K. & Akimitsu, N. Fusion genes and RNAs in cancer development. Non-coding RNA 7, 1–14 (2021).
Shin, Y. & Brangwynne, C. P. Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017).
Boeynaems, S. et al. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 28, 420–435 (2018).
Chakravarty, A. K. et al. Biomolecular Condensation: a new phase in cancer research. Cancer Discov. 12, 2031–2043 (2022).
Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013).
GitHub - TransDecoder/TransDecoder: TransDecoder source. https://github.com/TransDecoder/TransDecoder.
Lin, Y. et al. Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target lc domain polymers. Cell 167, 789–802.e12 (2016).
Lubelsky, Y. & Ulitsky, I. Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature 555, 107–111 (2018).
Pintacuda, G. et al. hnRNPK Recruits PCGF3/5-PRC1 to the Xist RNA B-Repeat to Establish Polycomb-Mediated Chromosomal Silencing. Mol. Cell 68, 955–969.e10 (2017).
Tang, P. et al. Alternative polyadenylation by sequential activation of distal and proximal PolyA sites. Nat. Struct. Mol. Biol. 29, 21–31 (2022).
Alfonso-Gonzalez, C. et al. Sites of transcription initiation drive mRNA isoform selection. Cell 186, 2438–2455.e22 (2023).
Diag, A., Schilling, M., Klironomos, F., Ayoub, S. & Rajewsky, N. Spatiotemporal m(i)RNA Architecture and 3’ UTR Regulation in the C. elegans Germline. Dev. Cell 47, 785–800.e8 (2018).
Agarwal, V., Lopez-Darwin, S., Kelley, D. R. & Shendure, J. The landscape of alternative polyadenylation in single cells of the developing mouse embryo. Nat. Commun. 12, 5101 (2021).
Patrick, R. et al. Sierra: discovery of differential transcript usage from polyA-captured single-cell RNA-seq data. Genome Biol. 21, 167 (2020).
Lee, S. et al. Diverse cell-specific patterns of alternative polyadenylation in Drosophila. Nat. Commun. 13, 5372 (2022).
Boulay, G. et al. Cancer-specific retargeting of BAF complexes by a prion-like domain. Cell 171, 163–178.e19 (2017).
Tulpule, A. et al. Kinase-mediated RAS signaling via membraneless cytoplasmic protein granules. Cell 184, 2649–2664.e18 (2021).
Zhang, J. Z. et al. Phase separation of a PKA regulatory subunit controls cAMP compartmentation and Oncogenic signaling. Cell 182, 1531–1544.e15 (2020).
Patel, A. et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 162, 1066–1077 (2015).
Kim, H. J. et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495, 467–473 (2013).
Laffleur, B. & Basu, U. Biology of RNA Surveillance in Development and Disease. Trends Cell Biol. 29, 428–445 (2019).
Wan, J. et al. Mutations in the RNA exosome component gene EXOSC3 cause pontocerebellar hypoplasia and spinal motor neuron degeneration. Nat. Genet 44, 704–708 (2012).
Boczonadi, V. et al. EXOSC8 mutations alter mRNA metabolism and cause hypomyelination with spinal muscular atrophy and cerebellar hypoplasia. Nat. Commun. 5, 4287 (2014).
Ciarlo, M. et al. Regulation of neuroendocrine differentiation by AKT/hnRNPK/AR/β-catenin signaling in prostate cancer cells. Int J. cancer 131, 582–590 (2012).
Yano, M., Okano, H. J. & Okano, H. Involvement of Hu and heterogeneous nuclear ribonucleoprotein K in neuronal differentiation through p21 mRNA post-transcriptional regulation. J. Biol. Chem. 280, 12690–12699 (2005).
Taniue, K. et al. Long noncoding RNA UPAT promotes colon tumorigenesis by inhibiting degradation of UHRF1. Proc. Natl Acad. Sci. 113, 1273–1278 (2016).
Taniue, K. et al. ASBEL –TCF3 complex is required for the tumorigenicity of colorectal cancer cells. Proc. Natl Acad. Sci. 113, 12739–12744 (2016).
Taniue, K. et al. LncRNA ZNNT1 induces p53 degradation by interfering with the interaction between p53 and the SART3-USP15 complex. PNAS nexus 2, pgad220 (2023).
Frankish, A. et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 47, D766–D773 (2019).
Tang, A. D. et al. Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. Nat. Commun. 11, 1438 (2020).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Li, H. et al. The sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Demario, S., Xu, K., He, K. & Chanfreau, G. F. Nanoblot: an R-package for visualization of RNA isoforms from long-read RNA-sequencing data. RNA 29, 1099–1107 (2023).
Wang, X. et al. PCBP2 posttranscriptional modifications induce breast cancer progression via upregulation of UFD1 and NT5E. Mol. Cancer Res 19, 86–98 (2021).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma. 12, 323 (2011).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Bailey, T. L. & Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int Conf. Intell. Syst. Mol. Biol. 2, 28–36 (1994).
Ray, D. et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499, 172–177 (2013).
Yu, K. et al. dSCOPE: a software to detect sequences critical for liquid-liquid phase separation. Brief. Bioinform 24, bbac550 (2023).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Varadi, M. et al. Alphafold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res 50, D439–D444 (2022).
Kenzui, T. et al. The MTR4/hnRNPK complex surveils aberrant polyadenylated RNAs with multiple exons. figshare. https://doi.org/10.6084/m9.figshare.26401738 (2024).
Acknowledgements
We thank T. Jensen of Aarhus University for helpful discussion and feedback. This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI [Grant Numbers: JP17KK0163 (N.A.), JP20H00624 (M.H), JP21H00243 (N.A.), JP21H04792 (N.A.), JP21H02758 (K.T.), JP21K19402 (K.T.), JP22KK0285 (K.T), JP22H04925 (M.H), JP22K15093 (C.Z), JP23H00509 (M.H), and JP23H04955 (N.A)] and the Japan Agency for Medical Research and Development [JP21ae0121049 (M.H.)]. N.A. and K.T. were supported by the Takeda Science Foundation and the Uehara Memorial Foundation. N.A. was supported by the Novartis Foundation. K.T. was supported by the MSD Life Science Foundation, the Kobayashi Foundation, the ONO Medical Research Foundation, and the Naito Foundation. We also thank Jeremy Allen, PhD, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.
Author information
Authors and Affiliations
Contributions
K.T., H.H., and N.A. designed research. K.T., A.S., H.H., X.G., and Y.S. performed experiments with help from A.N.O. and R.O-M. K.T., C.Z., and M.H. performed bioinformatic analyzes. M.S., and Y.S. performed sequencing analysis. K.T. and N.A. wrote and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Guillaume Chanfreau, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Taniue, K., Sugawara, A., Zeng, C. et al. The MTR4/hnRNPK complex surveils aberrant polyadenylated RNAs with multiple exons. Nat Commun 15, 8684 (2024). https://doi.org/10.1038/s41467-024-51981-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-51981-8