Abstract
IgA antibodies play an important role in mucosal immunity. However, there is still no effective way to consistently boost mucosal IgA responses, and the factors influencing these responses are not fully understood. We observed that colonization with the murine intestinal symbiotic protozoan Tritrichomonas musculis (T.mu) boosted antigen-specific mucosal IgA responses in wild-type C57BL/6 mice. This enhancement was attributed to the accumulation of free arachidonic acid (ARA) in the intestinal lumen, which served as a signal to stimulate the production of antigen-specific mucosal IgA. When ARA was prevented from undergoing its downstream metabolic transformation using the 5-lipoxygenase inhibitor zileuton or by blocking its downstream biological signaling through genetic deletion of the Leukotriene B4 receptor 1 (Blt1), the T.mu-mediated enhancement of antigen-specific mucosal IgA production was suppressed. Moreover, both T.mu transfer and dietary supplementation of ARA augmented the efficacy of an oral vaccine against Salmonella infection, with this effect being dependent on Blt1. Our findings elucidate a tripartite circuit linking nutrients from the diet or intestinal microbiota, host lipid metabolism, and the mucosal humoral immune response.
Similar content being viewed by others
Introduction
Immunoglobulin A (IgA) is the predominant class of immunoglobulins produced and secreted in the mucosa of the gastrointestinal (GI) and respiratory tract. IgA plays a crucial role in maintaining host homeostasis and defending mucosal surfaces against pathogens1,2,3,4,5. Recent evidence suggests that both dietary factors and microbial signals in the intestine can influence the magnitude of the IgA effector responses, both in baseline conditions and in response to enteric pathogens6,7,8. The gut microbiota is a very complex community, which includes a diverse collection of viruses, bacteria, fungi, and protists. While much attention has been directed towards understanding the interaction between IgA and the bacterial microbiome, which represents the majority of the human intestinal ecosystem, it is important to note that various commensal protists also play a significant role in shaping microbial community structures and mucosal immune homeostasis9,10,11. This underscores the necessity for a deeper exploration of the functions of the eukaryotic protozoal microbiota in the gut.
Tritrichomonas musculis (T.mu) is a flagellated anaerobic parabasalid that naturally colonizes mice and is considered a commensal in the murine gut. Related parabasalids have also been found in humans12. Similar to certain keystone bacteria such as segmented filamentous bacteria and Helicobacter species, eukaryotic T.mu and its closely related Tritrichomonas spp. have the ability to modulate the ‘immunologic tone’ of the host12. For instance, this commensal protozoan can increase susceptibility to T-cell-driven colitis and contribute to mucosal defenses against bacterial infections by inducing the release of interleukin (Il)-18 from gut epithelial cells and promoting dendritic cell-driven T helper (Th)1 and Th17 immunity12,13. The colonization of Tritrichomonas in the gut is facilitated by dietary polysaccharides and influenced by commensal bacteria14,15,16. Once established in the gut, it actively produces and releases succinate, which in turn stimulates the release of IL-25 from intestinal tuft cells. IL-25 derived from Tuft cells then activates lamina propria innate lymphoid cell type 2 (ILC2) to secrete IL-13, leading to the expansion of goblet and tuft cells, which is part of the ‘weep and sweep’ response to parasitic helminths14,17,18. Notably, mice harboring T.mu in the gut exhibit elevated levels of IgA in both the bloodstream and the intestinal tissues12. However, the precise relationship between IgA and this commensal protozoan remains unclear.
In the present study, we have demonstrated that the eukaryotic commensal T.mu has the capacity to enhance host antigen-specific mucosal IgA responses. This enhancement appears to be related to T.mu’s ability to modulate the intestinal microenvironment by altering the intestinal metabolome, particularly the levels of luminal ARA. Both dietary supplementation of ARA and colonization with T.mu were found to amplify antigen-specific mucosal IgA responses, contingent upon an optimal host metabolism and subsequent activation of the Blt1-dependent pathway. Our findings offer insight into a tripartite interaction involving nutrients from the diet or intestinal eukaryotic microbiota, host lipid metabolism, and the humoral immune response. This underscores the intricate interplay between dietary factors, microbial influences, and host immune function in the context of mucosal immunity.
Results
T.mu colonization promotes host IgA response
We previously demonstrated the presence of T.mu in the GI tract of our in-house bred C57BL/6 (B6) mouse colony (Supplementary Fig. S1A)15,19. Compared to T.mu-free B6 mice purchased from Vital River Laboratories (VRL, Beijing, China), our in-house animals exhibited significantly elevated levels of both serum and cecal luminal total IgA (p-value less than 0.01; Supplementary Fig. S1B, C). Conversely, the total serum IgG and IgM levels remained similar in both types of mice (Supplementary Fig. S1D, E). This prompted an investigation into the relationship between T.mu and IgA.
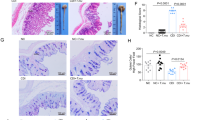
To directly assess whether intestinal colonization by the commensal protozoan facilitates IgA induction, we orally gavaged ex vivo purified T.mu (1 × 106 T.mu protozoa/mouse) (Supplementary Fig. S2) into the GI tract of VRL B6 mice. One week post T.mu transfer, the frequency of T.mu colonization in the recipients matched that in our in-house T.mu-positive mice (Fig. 1A). T.mu colonization increased the relative levels of both serum and cecal content total IgA but had no significant impact on total IgG and IgM levels in the recipient mice (Fig. 1B–G). Control mice displayed mean total IgA levels of approximately 50 µg/mL for serum and 70 µg/g for cecal content, while T.mu-colonized mice showed levels around 300 µg/mL for serum and 450 µg/g for cecal content (Supplementary Fig. S3A, B). Furthermore, immunofluorescence staining for IgA revealed a significant increase in IgA-positive cells in the Peyer’s Patches (PPs) and the lamina propria (LP) of the small intestine in T.mu-colonized mice (Fig. 1H, I). Flow cytometry analysis further confirmed and extended these results, demonstrating increased fractions of IgA+, but not of IgG1+, B cells among CD45+B220+ cells and IgA+ plasma cells among CD45+CD138+ cells not only in the PPs and LP of the small intestine but also in the bone marrow (BM) of T.mu-colonized mice (Fig. 1J–P, and Supplementary Fig. S4). The total number of IgA+ plasma cells in the LP doubled in T.mu-colonized mice (Fig. 1P), with a trend of increase of IgA+ plasma cell numbers in the BM (p = 0.07) (Fig. 1P).
T.mu isolated from our in-house B6 mice was used to inoculate VRL B6 mice via oral gavage (1 × 106 T.mu protozoa/mouse; referred to as the T.mu group). The control group was orally gavaged with an equal volume of PBS (referred to as the Ctrl group). A The number of T.mu protozoa in the cecal content 7 days post T.mu transfer was quantified using a hemocytometer (n = 5 mice/group). For comparison, the number of T.mu present in our in-house reared animals is also shown. B, C The relative total IgA levels present in serum (B) and cecal content (C) 7 days post T.mu transfer (n = 16 mice per group). D–G The relative levels of total IgG (D, E) and IgM (F, G) in the serum and cecal content of the indicated animals (n = 5 mice/group). H Immunofluorescent staining of IgA (green) and DAPI (blue) in the Peyer’s Patches (PPs) of control and T.mu-colonized mice (n = 5 mice/group). Scale bar: 200 μm; inset = 50 µm. I Immunofluorescent staining of IgA (green) and DAPI (blue) in the distal part of the small intestine of the control and T.mu-colonized mice (n = 5 mice/group). DAPI stains the nuclei in the animal cells. Scale bar: 100 μm. J–P Flow cytometry analysis of the IgA+B220+ B cell fractions among total CD45+B220+ B cells in the PPs (J, N) of the small intestine and bone marrow (BM) of the femur bone (K, N), and the IgA+CD138+ plasma cells among total CD45+CD138+ plasma cells in the lamina propria of the small intestine (LPL) (L, O, P) and BM (M, O, P) from the control and T.mu-colonized mice (n = 5 mice/group) in (J, K, N). n = 6 mice/group in (L, M, O, P). All data are shown as mean ± SEM. One-way ANOVA with Tukey’s post hoc test (A) or two-sided Student’s t-test (B–G, N–P) was performed. AU arbitrary unit. Source data are provided as a Source Data file.
To investigate whether T.mu colonization drives the IgA response to a specific antigen, mice were orally administered either vehicle or T.mu followed by exposure to the model antigen chicken ovalbumin (OVA) mixed with adjuvants via various routes (i.e., intraperitoneal, oral, subcutaneous, or intramuscular route) (Fig. 2A). Ten to fourteen days post intraperitoneal immunization with OVA mixed with either complete Freund’s adjuvant (CFA) or Aluminum (Alum) adjuvant, T.mu-colonized mice consistently produced significantly more (p < 0.01) serum and mucosal anti-OVA IgA antibodies than control mice, while relative serum anti-OVA IgG and IgM levels remained similar (Fig. 2B–E). In addition, ELISPOT assays revealed that following intraperitoneal immunization with OVA plus CFA, the intestinal LP of T.mu-colonized mice harbored more OVA-specific IgA-producing cells compared to control mice (Fig. 2F). A single oral immunization with OVA (100 μg) mixed with a relatively weaker mucosal adjuvant cholera toxin B subunit (CTB)20 did not induce detectable serum IgA against OVA (Fig. 2G); however, intestine luminal OVA-specific IgA levels were about four-fold higher in T.mu-colonized mice than in controls (Fig. 2H). Intramuscular immunization with OVA plus Alum enhanced serum anti-OVA IgA but failed to induce mucosal anti-OVA IgA in T.mu-colonized mice (Fig. 2G, H). Similarly, subcutaneous immunization with OVA plus CFA did not induce strong IgA responses in the serum or intestinal lumen of T.mu-colonized mice (Fig. 2G, H). Additionally, T.mu itself exhibited weak natural adjuvant properties for IgA production, as evidenced by higher anti-OVA IgA levels in the serum and cecal contents of T.mu-colonized mice that received intraperitoneal immunization with OVA alone without any added artificial adjuvant (Supplementary Fig. S5). Based on these findings, intraperitoneal immunization with CFA as adjuvant was chosen for the subsequent tests.
A Schematic of the experimental setup. B, C The relative levels of serum (B) and cecal content (C) anti-OVA Igs after intraperitoneal immunization of the control and T.mu-colonized mice with OVA plus CFA for 14 days (n = 5 mice/group for IgG/IgM, n = 10 mice for IgA in Ctrl group and n = 9 mice in T.mu group). D, E The relative levels of anti-OVA Igs in serum (D) and cecal content (E) after intraperitoneal immunization of the control and T.mu-colonized mice with OVA plus Alum for 10 days (n = 6 mice/group). F The numbers of anti-OVA IgA+ plasma cells were determined by ELISPOT assay after intraperitoneal immunization of control and T.mu-colonized mice with OVA plus CFA for 14 days (n = 5 mice/group). G, H The concentrations of serum (G) and cecal content (H) anti-OVA IgA after the indicated route (i.p., intraperitoneal; i.g., intragastric; s.c., subcutaneous; i.m., intramuscular) of immunization of the control and T.mu-colonized mice with OVA plus CFA, Alum, or CTB (n = 12 mice/group for CFA i.p., n = 6 mice/group for Alum i.p. and i.m., n = 5 mice/group for CTB i.g. and CFA s.c.). I–L The relative levels of serum (I–K) and cecal content (L) anti-NP Igs after intraperitoneal immunization of the control and T.mu-colonized mice with the TD antigen NP-KLH plus CFA for 14 days (n = 6 mice/group) in (I–K). n = 10 mice for Ctrl and 9 for T.mu group in (L). M–P The relative levels of serum (M–O) and cecal content (P) anti-TNP Igs after intraperitoneal immunization of the control and T.mu-colonized mice with the TI antigen TNP-LPS (n = 6 mice/group for IgG/IgM, n = 6 mice/group for unimmunized IgA and n = 12 mice/group for immunized IgA). All data are shown as mean ± SEM. Two-way ANOVA with Sidak’s multiple comparisons test (B, D, F, I–K, and M–O) or two-sided Student’s t-test (C, E, G, H, L, and P) were performed. AU arbitrary unit. Source data are provided as a Source Data file.
Both thymus-dependent (TD) and thymus-independent (TI) antigens can elicit the production of isotype-switched antibodies such as IgA21,22,23. To determine the antigen type regulated by T.mu for IgA production, T.mu-free or -colonized B6 mice were immunized with either the model TD antigen nitrophenyl-keyhole limpet hemocyanin (NP-KLH) or the model TI antigen 2,4,6-trinitrophenyl-lipopolysaccharide (TNP-LPS). Post intraperitoneal immunization with NP-KLH, T.mu-colonized mice produced more NP-specific IgA, not IgG and IgM, in both serum and cecal content (Fig. 2I–L). Similarly, post intraperitoneal immunization with TNP-LPS, T.mu-colonized mice produced more TNP-specific IgA, not IgG and IgM, in both serum and cecal content (Fig. 2M–P). Taken together, these observations suggest that T.mu colonization enhances mucosal and systemic IgA responses induced by both TD and TI antigens.
The role of T.mu and the gut bacterial microbiota in the T.mu-colonization-induced increase of antigen-specific IgA response
We have previously demonstrated that T.mu colonization alters the host gut bacterial microbiota15. To investigate whether the gut bacterial microbiota is involved in the T.mu-colonization-induced increase of antigen-specific IgA response, we colonized WT B6 mice with T.mu, and initiated treatment with either vancomycin to deplete Gram-positive bacteria or nalidixic acid/colistin to deplete Gram-negative bacteria 1 week later24,25, and intraperitoneally immunized the mice with OVA plus CFA four days after antibiotic treatment commencement (Fig. 3A). The antibiotics treatment did not influence T.mu colonization (Supplementary Figs. S6 and S7A). However, nalidixic acid/colistin treatment led to a reduction in systemic and mucosal anti-OVA (Fig. 3B, C) and total (Fig. 3D, E) IgA levels. Nevertheless, T.mu-colonized mice treated with nalidixic acid/colistin still produced more serum and mucosal anti-OVA IgA antibodies than T.mu-free control mice treated with nalidixic acid/colistin (Fig. 3B–E). Conversely, serum and mucosal anti-OVA and total IgA levels did not differ between vancomycin-treated and untreated T.mu-bearing mice (Supplementary Fig. S7B–E). These findings collectively suggest that while T.mu and nalidixic acid/colistin-sensitive bacteria work together to promote the IgA-high phenotype, T.mu alone is adequate to enhance the antigen-specific IgA response.
A Schematic of the experimental setup for T.mu colonization, antibiotics treatment, and antigen immunization. NA nalidixic acid, CS colistin sulfate, Vanco vancomycin, MDZ metronidazole. B–E The relative levels of serum anti-OVA IgA (B), cecal content anti-OVA IgA (C), serum total IgA (D), and cecal content total IgA (E) of control (Ctrl) and T.mu-colonized mice with or without NA+CS treatments were determined 14 days after intraperitoneal immunization (n = 6 mice/group). F–I The relative levels of serum anti-OVA IgA (F), cecal content anti-OVA IgA (G), serum total IgA (H), and cecal content total IgA (I) of the indicated mice treated or nontreated with MDZ were determined 14 days after immunization (n = 6 mice/group). All data are shown as mean ± SEM. One-way ANOVA with Tukey’s post hoc test (B–I) was performed. AU arbitrary unit. Source data are provided as a Source Data file.
To explore the impact of nalidixic acid/colistin treatment on the gut microbiota, we conducted bacterial 16S rRNA gene sequencing of cecal samples obtained from mice treated with or without antibiotics, with or without T.mu inoculation. Linear discriminant analysis effect size (LEfSe) was utilized to identify variations in bacterial composition. As demonstrated in Supplementary Fig. S8A, the administration of nalidixic acid/colistin had minimal effects on the overall landscape of the bacterial microbiota, as indicated by principal co-ordinates analysis (PCoA) conducted on operational taxonomic units (OUT) abundances in the samples. In the normal T.mu-free control group (CNC), the dominant differential bacterial families included Lactobacillaceae, Streptococcaceae, Helicobacteraceae, and Peptostreptococcaceae (Supplementary Fig. S8B), whereas in the T.mu-colonized control group (TNC), the dominant families were Muribaculaceae, Prevotellaceae, and Sutterellaceae (Supplementary Fig. S8B). Following antibiotic treatment, the dominant bacterial families shifted in the T.mu-free mice (CNACS group) to Lachnospiraceae, Comamonadaceae, Rikenellaceae, Peptococcaceae, and Butyricicoccaceae (Supplementary Fig. S8B), and in T.mu-colonized mice (TNACS group) to Ruminococcaceae, Anaerofustaceae, Eggerthellaceae, and Atopobiaceae (Supplementary Fig. S8B). Notably, the relative abundance of Lactobacillaceae (Gram-positive) and Akkmansiaceae (Gram-negative) decreased in the CNACS group compared to the CNC group (Supplementary Fig. S8C), while the relative abundance of Muribaculum (Gram-negative) and Tyzzerella (Gram-negative) decreased in the TNACS group compared to the TNC group (Supplementary Fig. S8D). It is worth noting that Muribaculum belongs to the family Muribaculaceae, which is known to possess fermentation pathways capable of producing succinate, acetate, and propionate26. These metabolites have been reported to have the ability to promote IgA production, particularly under specific experimental conditions such as antibiotic treatment or a diet with specially formulated dietary fiber27,28,29,30.
To further confirm the involvement of T.mu in promoting antigen-specific IgA responses, we initially inoculated WT B6 mice with T.mu on day 11. After allowing 1 week for T.mu to establish stable colonization, which concurrently induced increased total serum and cecal IgA (as described in Fig. 1), we commenced treatment with 3 g/L metronidazole in drinking water on day 4 for two weeks to deplete T.mu. On day 0, we performed intraperitoneal immunization with OVA plus CFA (Fig. 3A). The 2-week metronidazole treatment effectively eradicated T.mu (Supplementary Fig. S9), leading to the inability of mice to enhance anti-OVA IgA responses (Fig. 3F, G). This suggests a significant correlation between the presence of T.mu and an augmented antigen-specific mucosal IgA response. Interestingly, the total IgA levels, which had already risen prior to T.mu depletion, did not show a significant decrease after the 2-week metronidazole treatment (Fig. 3H, I). This observation is likely attributable to the relatively long half-life of total IgA.
Furthermore, besides depleting T.mu, the administration of metronidazole had a significant impact on the overall composition of the bacterial microbiota (Supplementary Fig. S10A, B). Specifically, there was a decrease in the relative abundance of Muribaculaceae and Ruminococcaceae (short-chain fatty acids producer31), while the relative abundance of Enterobacteriaceae and Enterococcaceae increased following metronidazole treatment (Supplementary Fig. S10C). Hence, the potential coordinated contributions of the T.mu-shaped bacterial microbiota to an enhanced antigen-specific mucosal IgA response cannot be disregarded.
T.mu colonization induces a favorable intestinal niche for IgA production
To elucidate how exposure to the unique but natural commensal protozoan T.mu enhances IgA levels in conventional mice, we conducted a comparative transcriptome analysis of small intestinal tissues from T.mu-free and -colonized B6 mice using bulk RNA sequencing. In B6 mice supplemented with T.mu in their GI tracts, a substantial change in gene expression was observed on day 7 post T.mu inoculation, with 617 genes upregulated and 324 genes down-regulated more than two-fold (q-value less than 0.05) compared to the controls (Fig. 4A). Pathway analysis highlighted that significant up-regulation of various Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways related to immune responses, particularly the pathway involved in the intestinal immune network for IgA production (Fig. 4B). Expression of numerous IgA-inducing genes was upregulated, including those known to contribute to B cell proliferation, differentiation, class-switching, intestinal migration, and plasma cell survival. These genes encompassed several members of the TNF-receptor superfamily (Tnfrsf17, also known as Bcma; Tnfrsf13b, also known as Taci; Tnfrsf13c, also known as Baffr), Cd40/Cd40 ligand, Inducible T Cell Co-stimulator (Icos), interleukin (Il)-10, integrin α4β7, and Ccr9/Ccr10 (Fig. 4C). Subsequent Ki67 and IgA co-immunostaining confirmed a significant increase in proliferative Ki67+IgA+ cells in the intestinal lamina propria of T.mu-colonized mice (Supplementary Fig. S11). Collectively, these data suggest that T.mu colonization may create a conducive intestinal environment for IgA production.
A Volcano plots depicting the differentially expressed genes (DEGs) in the small intestine of the T.mu-colonized mice compared to the control mice. Red = log2 fold change > 1, green = log2 fold change < −1. B KEGG pathway enrichment analysis based on all genes that were upregulated in the small intestine of T.mu-colonized mice. Top 10 KEGG pathways are shown based on enrichment score. C Heatmap depicting all the upregulated genes involved in regulating IgA production (fold change > 2). D KEGG pathway enrichment analysis based on all genes with down-regulated expression in the small intestine of T.mu-colonized mice. Top 10 KEGG pathways are shown based on enrichment score. Genes involved in linoleic acid metabolism were also listed (right panel) (n = 5 mice/group for A–D). E Untargeted metabolomic analysis of the cecal content of control and T.mu-colonized mice. Heatmap depicting all the upregulated metabolites in T.mu-colonized mice compared to control mice. Arachidonic acid is highlighted in bold red (n = 10 mice/group). F, G Heat-inactivated T.mu (2 × 108 /mouse) was inoculated into VRL B6 mice once every other day via oral gavage. On day 7, mice were immunized with OVA plus CFA, intraperitoneally. The relative levels of serum (F) and cecal content (G) anti-OVA IgA were determined 14 days after immunization (n = 6 mice/group). All data are shown as mean ± SEM. One-way ANOVA with Tukey’s post hoc test (F, G) was performed. AU arbitrary unit. Source data are provided as a Source Data file.
Further pathway analysis revealed that the majority of KEGG pathways that were significantly down-regulated in the intestine after T.mu colonization were associated with lipid metabolism, including pathways such as steroid hormone synthesis, retinol metabolism, and linoleic acid metabolism. These pathways encompassed genes encoding many cytochrome P450 (Cyp450) enzymes known to be involved in the metabolism of steroids, fatty acids, vitamins, and other chemicals (Fig. 4D). The data suggest that T.mu colonization may induce remodeling of intestinal lipid metabolism.
In contrast to the down-regulated lipid metabolic genes in the T.mu-colonized host intestinal tissue, metabolomic profiling demonstrated increased levels of various lipid types in the luminal fecal content (Fig. 4E). Lipidomic analysis of ex vivo purified T.mu revealed that T.mu contained about 7.9 µg LA and 1.6 µg ARA per 107 cells (Supplementary Fig. S12A). By comparison, the calculated concentration of ARA in human platelets is ~0.3 µg/107 cells32. Notably, T.mu also contained trans-fatty acids (e.g., trans-C18:1∆11, 5.7 µg per 107 cells) (Supplementary Fig. S12A). Considering that one gram of cecal content of T.mu-colonized conventional B6 mice contained about 4*108 T.mu cells, T.mu cells alone accounted for about 316 µg LA/g cecal content (~58% of total amount of LA found in the cecal content of T.mu- colonized mice) and 64 µg ARA/g cecal content (~51% of total amount of ARA found in the cecal content of T.mu-colonized mice) (Supplementary Fig. S12B). Furthermore, the animal chow diet used in our study contained about 5.8 mg/g LA and 24 µg/g ARA. Thus, T.mu has relatively more abundant ARA than the diet (Supplementary Fig. S12C). Moreover, T.mu colonization led to a two- to four-fold increase of fecal ARA levels in conventional WT mice (Supplementary Fig. S12B). The data collectively suggest that T.mu colonization induces distinct fecal metabolomes, especially providing a relatively rich resource of ARA.
We hypothesized that the increased lipid metabolites induced by T.mu colonization might suppress the expression of certain host lipid metabolic genes by negative feedback inhibition, thereby remodeling the host’s GI tract lipid metabolic flow to create a favorable niche for IgA production. To evaluate the importance of live T.mu for its IgA-boosting effect, we orally administered heat-killed T.mu to VRL B6 mice, which did not result in increased IgA production (Fig. 4F, G).
Intestinal luminal arachidonic acid enhances antigen-specific IgA production
To evaluate whether the metabolomic alterations induced by T.mu in the intestinal lumen impact the IgA response, we examined several metabolites that were found to be increased after T.mu colonization (Supplementary Fig. S13A). Among these candidates, we initially investigated succinate, given Tritrichomonas’ capacity for active succinate production and secretion, and previous evidence showing that 150 mM succinate in drinking water can stimulate mucosal type 2 immunity14,17,18. However, supplementation of 2.5% (i.e., 150 mM) succinate in drinking water for 7 days, followed by intraperitoneal immunization with OVA plus CFA, did not lead to heightened anti-OVA IgA production (Supplementary Fig. S13B, C).
Tritrichomonas is capable of generating acetate14, and acetate has been shown to enhance gut IgA production through G protein-coupled receptor 43 (GPR43)-dependent mechanisms under antibiotic treatment conditions29. Nevertheless, supplementation of 300 mM acetate in drinking water for WT mice without any antibiotic treatment did not elevate the antigen-specific IgA response post-immunization (Supplementary Fig. S13D, E).
Processes such as B cell activation, proliferation, differentiation, and antibody secretion are intricately governed by glucose metabolism33. Previously, our group demonstrated that T.mu disturbs host glucose homeostasis by promoting free choline generation19. In light of this, we administered 2% or 5% choline in drinking water on day 0, followed by intraperitoneal immunization with OVA plus CFA one week later. However, choline supplementation did not lead to increased anti-OVA IgA production (Supplementary Fig. S13F, G).
Considering the significant accumulation of ARA in our T.mu-cultured media and in the GI tracts of T.mu-colonized animals, we investigated whether the increased ARA levels following T.mu colonization contribute to the enhanced IgA response. To examine this, we divided VRL B6 mice into two groups: one group was fed a normal control diet, while the other group received an ARA-supplemented diet. Subsequently, the mice were immunized with OVA. The mice fed the ARA-supplemented diet produced significantly higher levels of serum and mucosal OVA-specific IgA compared to the control diet-fed mice (Fig. 5A, B). However, the levels of serum OVA-specific IgG and IgM were similar between the two groups (Supplementary Fig. S14). These findings suggest that the heightened luminal ARA levels observed in T.mu-colonized conditions likely contribute to T.mu’s role in promoting the antigen-specific IgA response. Nevertheless, unlike T.mu, dietary supplementation of ARA did not increase the levels of serum and cecal content total IgA (Fig. 5C, D), suggesting that the regulation of total IgA pools involves a different mechanism independent of ARA. Importantly, when VRL B6 mice were supplemented with ARA via intraperitoneal injection, there was no observed increase in the antigen-specific IgA response (Fig. 5E). This can be attributed to the fact that the intraperitoneal injection regimen used in our study only elevated serum ARA levels but did not affect ARA levels in intestinal tissue (Supplementary Fig. S15A, B), suggesting that locally increased free ARA pools in the gut are necessary to promote the antigen-specific IgA response. Consistently, our nontargeted metabolomic analysis indicated that T.mu-colonized mice did not exhibit increased serum ARA levels (Supplementary Fig. S15C).
A–D Groups of VRL B6 mice were fed either with a control diet (Ctrl group) or an ARA-supplemented diet (ARA group) [0.4% (v/w)] for 7 days and then intraperitoneally immunized once with OVA mixed with CFA on day 7. The relative levels of serum and cecal content anti-OVA (A and B, n = 10 mice in the Ctrl group and 9 mice in the ARA group) and total (C and D, n = 5 mice/group) IgA were determined on day 21. E Groups of VRL B6 mice were treated daily with vehicle or ARA (20 mg /day) intraperitoneally. On day 7, mice were immunized once with OVA plus CFA. The relative levels of serum anti-OVA IgA were determined on day 21 (n = 4 mice/group). F Schematic of the three major arachidonic acid metabolic pathways. G Schematic of the experimental setup for T.mu colonization or dietary ARA supplementation, selective inhibitors administration, and antigen challenge. H–K The effect of zileuton on the relative levels of serum (H, J) and cecal content (I, K) anti-OVA IgA 14 days post antigen challenge (n = 5 mice/group for Ctrl and Ctrl+Zileuton groups in (H) and (I), n = 9 mice for T.mu group and n = 10 mice for T.mu+Zileuton groups in (H) and (I). n = 6 mice/group in J and K). L, M The effect of aspirin on the relative levels of serum (L) and cecal content (M) anti-OVA IgA 14 days post antigen challenge (n = 5 mice/group). N, O The effect of MS-PPOH on the relative levels of serum (N) and cecal content (O) anti-OVA IgA 14 days post antigen challenge (n = 6 mice/group). All data are shown as mean ± SEM. Two-sided Student’s t-test (A–E) or One-way ANOVA with Tukey’s post hoc test (H–O) were performed. AU arbitrary unit. Source data are provided as a Source Data file.
Our metabolomic analysis revealed that T.mu colonization can elevate luminal levels of docosahexaenoic acid (DHA) (Supplementary Fig. S12), a ω-3 polyunsaturated fatty acid (PUFA) that has the potential to compete with ARA in terms of incorporation into cell membrane phospholipids and as a substrate for cyclooxygenases (Cox) pathway34. However, even with the supplementation of 0.4% DHA in the diet, there was no observed increase in the antigen-specific IgA response (Supplementary Fig. S13H, I).
ARA can undergo metabolism by host cyclooxygenases (Coxs), lipoxygenases (Loxs), and cytochrome P450 (Cyp450) enzymes, leading to the production of various bioactive mediators (Fig. 5F)35. To investigate the role of these ARA metabolic pathways in T.mu’s ability to enhance the IgA response, we utilized small molecule inhibitors to block these pathways and examined their effects on T.mu-mediated IgA response enhancement (Fig. 5G). When we inhibited the Lox pathway using the selective 5-lipoxygenase (5-Alox) inhibitor zileuton, we observed suppression of both dietary ARA- and T.mu-induced enhancement of OVA-specific IgA production (Fig. 5H–K). However, this inhibition did not affect T.mu colonization efficiency (Supplementary Fig. S16A). In contrast, inhibiting either the Cox pathway with the Cox inhibitor aspirin or the Cyp450 pathway with the selective Cyp450 epoxygenase inhibitor N-(methylsulfonyl)-2-(2-propynyloxy)-benzenehexanamide (MS-PPOH), which prevents the conversion of ARA to epoxyeicosatrienoic acid, had no effect on T.mu’s ability to boost antigen-specific IgA production (Fig. 5L–O, Supplementary Fig. S16B, C). These findings suggest that ARA may enhance antigen-specific IgA production through its transformation into leukotrienes via the ARA lipoxygenase pathway. In line with this, both dietary ARA supplementation and T.mu colonization led to increased levels of the ARA lipoxygenase pathway intermediate leukotriene B4 (LTB4) in intestinal tissue (Supplementary Fig. S17).
T.mu colonization enhances mucosal antigen-specific IgA production in a Blt1-dependent manner
In mammalian cells, free ARA can undergo conversion into LTA4 by 5-Alox. Subsequently, LTA4 can be rapidly converted either into LTB4 by LTA4 hydrolase (Lta4h), or into LTC4 through a reaction catalyzed by LTC4 synthase (Ltc4s), or into lipoxin A4 (LXA4) by the enzymes 12- and 15-Alox. LTC4 further converts into LTD4 and LTE4, collectively constituting cysteinyl leukotrienes (cysLTs). These bioactive mediators, including LXA4, cysLTs, and LTB4, bind to their respective receptors, triggering various biological effects (Fig. 6A)36. When we restricted the conversion of ARA to LXA4 using the selective 12-Alox inhibitor ML355, it did not affect the T.mu-mediated enhancement of OVA-specific IgA production (Fig. 6B, C, and Supplementary Fig. S18A). However, it increased the OVA-specific IgA production in control T.mu-free mice, emphasizing the importance of intestinal ARA metabolic flow in controlling antigen-specific IgA response and the potential lipid metabolism difference between the control and T.mu-colonized mice (Fig. 6B, C). Furthermore, blocking cysLTs signaling by using Montelukast, a selective antagonist of cysteinyl leukotriene receptor 1 (cysLtr1), also failed to influence T.mu-mediated enhancement of OVA-specific IgA production (Fig. 6D, E, and Supplementary Fig. S18B).
A Schematic of arachidonic acid lipoxygenase (Lox) pathway. B–E Groups of mice were administered once with either PBS (Ctrl) or T.mu via the oral route on day 0. From day 6 and thereafter, the mice were treated with vehicle, ML-355 (B, C), or Montelukast (D, E) once daily by oral gavage. On day 7, the mice were intraperitoneally challenged with OVA plus CFA. The relative levels of serum (B, D) and cecal content (C, E) anti-OVA IgA were determined on day 21 (n = 6 mice/group) in (B) and (C). n = 12 mice/group for Ctrl and T.mu groups in (D) and (E), and n = 6 mice/group for Ctrl+Montelukast and T.mu+Montelukast groups in (D) and (E). F, G Groups of Blt1−/− and the littermate Blt1+/− mice were orally administered once with either PBS or T.mu on day 0. On day 7, the mice were intraperitoneally challenged with OVA plus CFA. The relative levels of serum (F) and cecal content (G) anti-OVA IgA were determined on day 21 (n = 7 mice/group for Blt1+/− and Blt1+/−-T.mu groups, n = 5 mice/group for Blt1−/− and Blt1−/−-T.mu groups). Data are shown as mean ± SEM. One-way ANOVA with Tukey’s post hoc test (B–G) was performed. AU arbitrary unit. Source data are provided as a Source Data file.
We then investigated the significance of LTB4 signaling in the T.mu-induced IgA response. Among the ARA lipoxygenase pathway genes we examined, we observed a slight increase in the expression of the LTB4-producing gene Lta4h in the small intestine of T.mu-colonized mice (Supplementary Fig. S18C). LTB4 acts on leukocytes via the G protein-coupled receptor Blt1 (also known as Ltb4r1)37. To assess the role of Blt1, we transferred T.mu to Blt1−/− mice and their littermate controls (Blt1+/−), followed by challenge with OVA plus the adjuvant CFA. Deletion of host Blt1 did not influence the frequency of T.mu colonization (Supplementary Fig. S18D) or the increase in T.mu-induced serum anti-OVA IgA levels (Fig. 6F). However, it completely abolished the T.mu-mediated enhancement of intestine luminal anti-OVA IgA production (Fig. 6G). Moreover, mucosal IgA levels were reduced in Blt1-deficiency mice, even in the absence of T.mu colonization (Fig. 6G), indicating the importance of LTB4 signaling for mucosal IgA responses even under T.mu-free condition. Consistently, dietary supplementation of ARA to T.mu-free Blt1−/− mice showed a trend toward increased serum anti-OVA IgA (p = 0.0714) but failed to increase mucosal anti-OVA IgA levels (Supplementary Fig. S18E, F). However, it’s important to note that since the microbiomes of Blt−/− and Blt+/− were not directly compared, a possible indirect effect of Blt through changes in the microbiome can’t be ruled out at this stage.
T.mu colonization enhanced the effectiveness of an oral vaccine against a mucosal pathogen
Based on the above findings, we propose that the T.mu-mediated boosting effect on IgA production might enhance the effectiveness of oral vaccines against enteropathogens. Therefore, to test this hypothesis, we utilized a vaccination model using heat-killed Salmonella enterica serotype Typhimurium (S. Typhimurium). WT B6 mice were colonized with T.mu on day 7 and subsequently orally vaccinated with two oral doses of heat-killed S. Typhimurium on day 0 and day 14, respectively. On day 21, the mice were challenged with live S. Typhimurium (Fig. 7A). Eight days after the second dose of oral vaccination, the relative level of S. Typhimurium-specific IgA antibodies showed only a slight increase in the T.mu-free control mice (Fig. 7B–D). Furthermore, the overall survival of the T.mu-free mice after infection was not improved compared to unvaccinated controls (Supplementary Fig. S19A, B). These results indicate that the heat-inactivated S. Typhimurium vaccine was ineffective in protecting T.mu-free mice from a lethal dose of infection. In contrast, T.mu-colonized mice exhibited over a 10-fold increase in serum and intestine luminal relative titers of anti-S. Typhimurium IgA after vaccination (Fig. 7B–D). Moreover, in comparison to T.mu-free mice, T.mu-colonized mice that had been vaccinated showed reduced S. Typhimurium titers in the mesenteric lymph node and significantly improved survival compared to T.mu-free mice (Fig. 7E–G). The results suggest the composition of the gut microbiome, specifically the presence of T.mu, influences the intestinal mucosal IgA response and the efficacy of the mucosal vaccination.
A Schematic of the experimental setup for (A–K). B–D The relative levels of serum (B) and cecal content (C) IgA or serum IgG (D) 3 weeks after the introduction of the heat-inactivated S. Typhimurium vaccine (n = 6 mice/group). E Survival curve (n = 11 mice/group). F, G S. Typhimurium colony-forming units (CFU) in the cecal content (F) and mLN (G) 24 h post-infection (n = 6 mice/group). H, I The mice were treated intraperitoneally with vehicle or zileuton once daily from day 1. H The relative levels of serum IgA were determined 3 weeks after the first dose of vaccine. I Survival curve (n = 6 mice/group). J, K Groups of mice (n = 12 mice/group) were fed with either a control diet or an ARA-supplemented diet from day 7 and treated with heat-inactivated S. Typhimurium vaccine orally on day 0 and day 14, followed by live S. Typhimurium challenge. J The relative levels of serum IgA were determined 3 weeks after the first dose of vaccine. K Survival curve. L–N Blt1−/− mice and the littermate controls (Blt1+/− mice) were colonized with T.mu, 1 week later were vaccinated with heat-inactivated S. Typhimurium, and two more weeks later injected the second dose of vaccine. 7 days after the second vaccination, mice were challenged with live S. Typhimurium. L The colonic transcription of Il18 two days after infection was determined by qRT-PCR (n = 6 mice/group). M The fecal anti-S. Typhimurium IgA titers 8 days after the second vaccination (n = 8 mice/group). N Survival curve (n = 8 mice/group). All data are shown as mean ± SEM. Log-rank (Mantel-Cox) test (E, I, K, and N), Two-way ANOVA with Sidak’s multiple comparisons test (B and D), or One-way ANOVA with Tukey’s post hoc test (H and M), or two-sided Student’s t-test (C, F, G, J and L) were performed. AU arbitrary unit. Source data are provided as a Source Data file.
Previous research indicates that T.mu-induced Il-18 confers protection from Salmonella-induced intestinal inflammation and tissue damage within 2 days of infection12. However, our observation revealed that in the absence of prior vaccination with heat-killed S. Typhimurium, all control WT mice succumbed to Salmonella infection within 7 days, irrespective of T.mu presence (Supplementary Fig. S19B). This suggests that without vaccination, T.mu colonization alone, or T.mu-related innate immunity, including Il-18 production, is insufficient to ensure host survival.
To further demonstrate that T.mu-enhanced IgA response is responsible for conferring protection against Salmonella infection, we administered the proteasome inhibitor bortezomib, which can deplete plasma cells38 to suppress IgA production. Remarkably, bortezomib treatment completely inhibited T.mu-boosted anti-Salmonella mucosal IgA production and concurrently abrogated the protective effect of the vaccine in T.mu-colonized mice (Supplementary Fig. S20A–E). In addition, the treatment also inhibited vaccine-induced basal anti-Salmonella IgA production and accelerated the mortality rate in T.mu-free mice (Supplementary Fig. S20B). Therefore, T.mu-enhanced IgA response is highly correlated with its boosting effect on vaccine efficacy. However, it should be mentioned that bortezomib affects not only plasma cells but also many other important immune cell types39, and whether the other immune cell types play a role in T.mu-enhanced vaccine efficacy requires further investigation. Notably, the T.mu-promoted Il-18 expression was not inhibited by bortezomib treatment (Supplementary Fig. S20E), further indicating that Il-18 alone is insufficient to ensure host survival.
Furthermore, to confirm the involvement of ARA metabolism in the improved efficacy of mucosal vaccines promoted by T.mu, we administrated zileuton i.p. once daily for the entire 2-week period between the first and the second vaccination doses. Zileuton treatment inhibited the T.mu-mediated enhancement of S. Typhimurium-specific IgA production, and concurrently, the vaccine’s protective effect disappeared (Fig. 7H, I). Therefore, the effects of T.mu on the intestinal mucosal IgA response and the effectiveness of the mucosal vaccination rely on ARA metabolism.
Moreover, dietary supplementation of ARA to T.mu-free VRL WT B6 mice not only promoted anti-S. Typhimurium IgA antibody production after oral vaccination with heat-inactivated S. Typhimurium but also improved survival for T.mu-free WT mice after live S. Typhimurium challenge (Fig. 7J, K).
In parallel, we tested whether ARA downstream Blt1 signaling dictates the improved efficacy of mucosal vaccines promoted by T.mu. Despite the intact T.mu-mediated induction of colonic tissue Il18 expression in Blt1−/− mice (Fig. 7L), the T.mu-mediated enhancement of vaccine-induced IgA production and protective effect against Salmonella infection was diminished in Blt1−/− mice (Fig. 7M, N). This suggests that Blt1-dependent signals, rather than Il-18-related events, contribute to the enhanced vaccine protection against mucosal bacterial infections mediated by T.mu. It is known that Blt1−/−neutrophils lack the ability to swarm and cluster at a localized injury site40. However, Blt1−/− mice, with or without T.mu colonization, did not exhibit accelerated mortality compared to control mice after live Salmonella infection in the absence of prior vaccination with heat-inactivated salmonella (Supplementary Fig. S21), suggesting that the lack of improvement in oral vaccine efficacy in Blt1−/− mice by T.mu is unlikely due to neutrophil deficiencies in these mice. Moreover, daily passive immunization through oral administration for 4 days following Salmonella infection with cecal lavage fluid obtained from vaccinated T.mu-colonized WT mice resulted in prolonged survival of unvaccinated Blt1−/− mice without inducing Il-18 expression (Supplementary Fig. S22A, B), indicating that T.mu-mediated enhancement of humoral effectors, which are lacking in Blt1−/− mice, confers protection against Salmonella infection. Indeed, adoptive transfer of pan B cells from vaccinated T.mu-colonized WT mice into naïve Blt1−/− mice followed by live Salmonella challenge extended the survival of the unvaccinated Blt1−/− recipient mice post-infection (Supplementary Fig. S23).
These findings collectively suggest that T.mu or dietary ARA enables more effective mucosal IgA responses upon vaccination and imparts protective advantages against live enteropathogen infection through a Blt1-dependent mechanism.
Discussion
The mucosa surface serves as the primary barrier against pathogen entry into the host, and there is a pressing need for strategies to enhance mucosal IgA alongside systemic immunity to combat emerging mucosal infectious diseases41. Both diet and the microbiota play crucial roles in regulating IgA production in the gut6,42. Understanding the mechanisms through which they influence the IgA response is vital for the development of IgA-boosting adjuvants. In this study, we present an example of how the presence of a murine eukaryotic commensal protist in the gut can influence host mucosal IgA responses to foreign antigens and mucosal vaccine efficacy. Our data suggest that colonization of the gut by the T.mu protozoan increases luminal ARA levels, which can act as a signal to enhance antigen-specific mucosal IgA responses. However, the boosting effect of ARA on mucosal antigen-specific IgA requires optimal downstream metabolism. Inhibiting ARA’s downstream transformation by 5-lipoxygenase using zileuton or blocking its downstream biological signaling through genetic deletion of Blt1 suppressed T.mu-mediated enhancement of mucosal antigen-specific IgA production. The exact mechanism through which BLT1 signaling enhances antigen-specific intestinal IgA responses remains to be elucidated. BLT1 is expressed in various cell types, including myeloid cells, Tregs, activated effector T cells, ILC2, and even gut epithelial cells43,44,45,46,47. Notably, mouse B cells acquire BLT1 expression during their differentiation into IgA+ B cells and plasma cells in PPs and the small intestinal LP48. Future investigations are needed to determine which cell types expressing BLT1 are essential for promoting mucosal IgA production. Additionally, the possibility of an indirect effect of BLT1 through changes in the microbiome cannot be ruled out and warrants further exploration.
We demonstrated that T.mu colonization leads to increases in both total and antigen-specific IgA levels. However, dietary supplementation with ARA can only increase antigen-specific IgA. Thus, T.mu may employ yet-to-be-discovered mechanisms to regulate overall host IgA pools. Our transcriptome analysis indicated that T.mu colonization upregulated the expression of genes such as Cd40/Cd40l and Taci, known to be involved in IgA production. Specifically, Taci signaling is associated with T-independent IgA production, while Cd40l/Cd40 is linked to T-dependent IgA production49,50,51. Furthermore, gut microbiota-derived extracellular vesicles may promote cytokines that facilitate IgA production28,52. A recent study demonstrated that T.mu colonization induces serum total IgA in an Icos- and T-cell-dependent manner53. However, the precise mechanisms by which T.mu promotes both total and antigen-specific IgA antibody responses remain to be elucidated in future research.
The purpose of increased IgA production induced by T.mu remains unclear. We postulate that the T.mu-mediated increase in total IgA may benefit its commensalism within the host intestinal lumen by altering both the protist itself and the trans-kingdom cross-talk between commensal bacteria, protozoa, and the host. It is worth noting that T.mu colonization can stimulate luminal IgA production that binds to itself and can directly influence the composition of commensal bacteria15. Additionally, a recent study indicated that T.mu metabolism and the expression of putative virulence factors are influenced by host B cells16. Therefore, the anti-protozoal IgA induced by intestinal luminal protists may function similarly to the antifungal IgA induced by commensal fungi, which regulates fungal commensalism by coating fungal morphotypes associated with virulence54,55.
ARA, a ω-6 very long-chain polyunsaturated fatty acid (VLCPUFA), holds significant nutritional importance and cannot be synthesized de novo in mammals56. It is primarily obtained from dietary sources or synthesized via desaturation and chain elongation of the plant-rich essential fatty acid, LA. However, various microorganisms, including bacteria, microalgae, primitive fungi, and protists, possess the capability to synthesize a range of VLCPUFAs, including ARA. Interestingly, serum lipid levels of ARA have been found to be closely associated with specific fecal flora in patients with liver cirrhosis57. Our findings suggest that the eukaryotic commensal protozoa T.mu harbors a diverse array of lipids, among which ARA is notable. Conventional mice colonized with T.mu showed increased levels of cecal ARA (Supplementary Fig. S12B) and intestinal tissue LTB4 (Supplementary Fig. S17A). Notably, when considering all cecal T.mu cells together, they accounted for approximately 51% of the total cecal luminal ARA in T.mu-colonized mice (Supplementary Fig. S12B). The mechanisms underlying the abundant presence of ARA in T.mu cells remain elusive. Due to the great challenge of culturing T.mu axenically, to date we are not able to figure out whether the protist can synthesize ARA de novo or not. Alternatively, T.mu may absorb and store ARA from its environment, releasing it as needed. Phospholipids, a component of mucus58, could serve as a potential ARA resource in the GI tract. Previous evidence suggests that T.mu possesses the capability to break down phospholipids19. As commensal protists persist and proliferate in the gut, their potential role in storing and recycling VLCPUFAs for the host should not be underestimated. It’s worth noting that the human pathogen T. vaginalis has been reported to release LTB4 and CysLTs to activate host innate immune cells during infection59.
ARA has traditionally been viewed as an inflammation-inducing molecule; however, emerging data indicate that it plays a crucial role in maintaining host immune homeostasis. Both dietary and microbiota-derived free ARA are fundamental for mammalian health. ARA has been found to suppress pyrin inflammasome activation by activating the serine-threonine kinases PKNs60. Additionally, ARA-derived prostaglandin E2 inhibits NLRP3 inflammasome activation through the EP4 receptor and intracellular cAMP in human macrophages61. Furthermore, ARA-derived resolvins, a newly identified family of mediators with anti-inflammatory, pro-resolving, and protective properties, aid in attenuating inflammation and promoting wound and lesion healing62,63. Other ARA-derived metabolites, such as prostaglandin D2, leukotriene D4 and E4, stimulate ILC2 expansion and effector functions, while lipoxin A4 inhibits these processes64. This suggests that the metabolic flow of ARA, rather than ARA itself, acts as a crucial regulator of type 2 immune responses. Therefore, it is plausible that variations in host metabolic activities, both in steady states and under diseased conditions, may impact the magnitude of mucosal IgA responses. Whether such variations in host metabolic activities also influence mucosal vaccine efficacy is an intriguing question that warrants investigation in future studies.
Our findings indicate that dietary supplementation with ARA can effectively enhance antigen-specific IgA responses. Interestingly, previous research has revealed that the diurnal rhythmicity of intestinal IgA secretion aligns with the timing of food consumption65. Moreover, the absorption and conversion of dietary cholesterol to 7α,25-dihydroxycholesterol in gut epithelial cells play a crucial role in restraining intestinal plasma cells from secreting IgA6. These findings, coupled with our own, suggest that nutrient availability from both diet and the microbiota can exert either positive or negative regulatory effects on IgA responses. This underscores the intricate interplay between intestinal luminal dietary cues, gut microbiota composition, and host metabolism and immunity.
Although our study provides valuable insights, it also has certain limitations. We primarily utilized intraperitoneal and oral vaccination models in mice, and further investigation is needed to assess the translational potential of our findings in human mucosal vaccinations, particularly by manipulating dietary or microbial nutrients and host metabolic activities. Additionally, future studies are warranted to explore whether the ARA 5-ALOX pathway and Blt1 signaling play a role in promoting antigen-specific mucosal IgA responses in humans. These avenues of research will be crucial for advancing our understanding of strategies to restore mucosal homeostasis, particularly in the context of intestinal infection and inflammation.
Methods
Animal studies
All animal work was conducted with the approval of the Animal Care Committee of Xuzhou Medical University. The work was performed at the Laboratory Animal Centre, Xuzhou Medical University and adhered to the mandatory laboratory health and safety procedures according to IACUC Issue No. 202207S068. Mice were housed in a temperature-controlled room (22 ± 2 °C) with 40–70% humidity, and subjected to a 12-h light/dark cycle, with free access to water and food. Both male and female C57BL/6 mice (6 to 8 weeks old) were purchased from Vital River Laboratory Animal Technology Co. Ltd. (Beijing, China). The Blt1-deficient Blt1−/− male and female mice were developed from the C57BL/6 background and purchased from the Shanghai Model Organisms Center (Shanghai, China).
Mice were fed on a normal chow diet (1010002, Jiangsu Xietong Pharmaceutical Bio-engineering Co., Ltd, Nanjing, China) unless specified otherwise. The ARA-supplemented diet was prepared by adding 0.4% ARA (HY-109590, MCE, Shanghai, China; A875622, Macklin, Shanghai, China) or DHA (v/w) (D875665, Macklin, Shanghai, China) into the normal chow diet, and 0.2% vitamin C (w/w) (A800296, Macklin, Shanghai, China) was also added to prevent the oxidation of ARA. We also added 0.2% vitamin C (w/w) into the control chow diet.
For in vivo succinate, acetate, or choline treatment, mice were fed in drinking water with either 150 mM sodium succinate (S818135, Macklin, Shanghai, China), 300 mM sodium acetate (adjust pH to 7) (A601611, Sangon Biotech, Shanghai, China), or 2 to 5% choline (C7527, Merck KGaA, Darmstadt, Germany).
For in vivo suppression of 5-Alox, 10 mg/kg/day zileuton (HY-14164, MCE, Shanghai, China) dissolved in 10% Dimethyl Sulfoxide (DMSO) plus 90% corn oil was injected intraperitoneally 1 day before OVA (A003049, Sangon Biotech, Shanghai, China) immunization. For in vivo inhibition of Coxs or Cyp450s, either 5 mg/kg/day aspirin (HY-14654, MCE, Shanghai, China) or 20 mg/kg/day MS-PPOH (206052-02-0, Cayman Chemical, Ann Arbor, USA), respectively, dissolved in 10% DMSO plus 90% corn oil, and administrated intraperitoneally 1 day before OVA immunization. For in vivo inhibition of 12-ALox or blocking of cysLtr1signaling, 15 mg/kg/12 h ML-355 (HY-12341, MCE, Shanghai, China; T4347, TOPSCIENCE, Shanghai, China) dissolved in 10% DMSO plus 90% corn oil or 1 mg/kg/day Montelukast sodium (HY-13315A, TOPSCIENCE, Shanghai, China) in PBS, respectively, was administered via oral gavage 1 day before OVA immunization.
To deplete T.mu from mice, 3 g/L metronidazole (A600633, Sangon Biotech, Shanghai, China) was added to the drinking water.
To deplete Gram-negative bacteria from mice, 200 mg/L nalidixic acid (A610363, Sangon Biotech, Shanghai, China) was added to the drinking water accompanied by daily colistin sulfate (HY-A0089, MCE, Shanghai, China; T1245, TOPSCIENCE, Shanghai, China) gavage (600 μg/mouse). To deplete Gram-positive bacteria, 0.5 g/L vancomycin (A600983, Sangon Biotech, Shanghai, China) was added to the drinking water.
To deplete plasma cells in mice, bortezomib (HY-10227, MCE, Shanghai, China) was administered intraperitoneally to the specified mice. The bortezomib solution was freshly prepared before injection and given at a dose of 1 mg·kg−1 in a vehicle comprising 2% DMSO and 30% polyethylene glycol 300 (P9570, Solarbio, Beijing, China), with injections administered every 3 days, beginning 1 day before T.mu inoculation.
T.mu isolation and in vivo transfer
The cecal contents of our in-house T.mu-colonized mice were harvested with sterile PBS and filtered three times through a 100 µm cell strainer. The filtrate was centrifuged at 200×g for 5 min at 4 °C, and the pellet was washed twice with PBS. T.mu was further purified by a 40%/80% Percoll gradient. To further purify the protist, T.mu harvested from the Percoll gradient was suspended in BHI broth (CM1135, Oxoid, Basingstoke, UK) containing a cocktail of broad-spectrum antibiotics [100 mg/mL streptomycin (A100382, Sangon Biotech, Shanghai, China), 100 U/mL penicillin (A613460, Sangon Biotech, Shanghai, China), 50 mg/mL vancomycin (A600983, Sangon Biotech, Shanghai, China), 10 mg/mL ciprofloxacin (A600310, Sangon Biotech, Shanghai, China), 20 mg/mL gentamicin (A506614, Sangon Biotech, Shanghai, China), and 0.5 mg/mL amphotericin B (171375, Sangon Biotech, Shanghai, China)]. The culture was incubated in an anaerobic workstation (Don Whitley Scientific, West Yorkshire, UK) at 37 °C for 2 days.
For the transfer of T.mu, each mouse was inoculated with 1 × 106 T.mu via oral gavage.
For the preparation of heat-killed T.mu, purified T.mu was incubated in a 65 °C water bath for at least 60 min.
Quantification of T.mu in the cecal content
Cecal content of the control and infected mice was collected and suspended in PBS. The suspension was transferred to a hemocytometer, and the number of protists present in the cecal content was counted under a microscope.
Wright-Giemsa staining
The cecal content was diluted in 10 volumes of PBS and filtered through a 100 µm cell strainer. The filtrate was spread on a microscope slide and air-dried. Samples were then stained using the Wright-Giemsa staining kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer’s instructions.
Immunization
For systemic immunization, 2 mg/mL OVA (A003049, Sangon Biotech, Shanghai, China) in PBS was mixed with Complete Freund’s adjuvant (F5881-6X10ML, Merck KGaA, Darmstadt, Germany) or Imject Alum adjuvant (A6435, Merck KGaA, Darmstadt, Germany), and the homogenates (100 μL/mouse) were injected intraperitoneally, subcutaneously, or intramuscularly. For mucosal immunization, mice were immunized with 100 μg OVA plus 10 μg CTB (SAE0069, Merck KGaA, Darmstadt, Germany; abs80001, Absin, Shanghai, China) in PBS by oral gavage. For NP-KLH immunization, 100 μg/mouse NP-KLH (N-5060, LGC Biosearch Technologies, California, USA) was administrated via intraperitoneal injection with CFA as adjuvant. For the TI antigen TNP-LPS immunization, 50 μg per mouse TNP-LPS (T-5065, LGC Biosearch Technologies, California, USA) was injected intraperitoneally.
Salmonella infection
Salmonella enterica serovar Typhimurium SL1344 strain was kindly provided by Dr. Huan Yang (Xuzhou Medical University). S. Typhimurium was initially cultured on Luria-Bertani (LB) plates with 50 μg/mL streptomycin. A single colony was then picked and cultured in liquid LB media. After two rounds of culture in LB broth, the bacteria were used for the infection of animals.
For the preparation of the oral vaccine, S. Typhimurium was heat-inactivated for 120 min at 65 °C.
For S. Typhimurium oral vaccination, each mouse received 1 × 108 heat-inactivated S. Typhimurium twice via oral gavage on day 7 and day 21 post T.mu transfer or ARA diet feeding. Live S. Typhimurium (1 × 107/mouse) was then orally administrated 1 week after the secondary vaccination. To assess mice survival, a lethal dose of 1 × 108 live S. Typhimurium was used. For in vivo inhibition of 5-Alox, zileuton was administrated 1 day before the first vaccination and followed by daily intraperitoneal injections.
For passive immunization, cecal contents from vaccinated T.mu-free (Ctrl) or T.mu-colonized (T.mu) WT mice were suspended in 4 times the volume of sterilized PBS. After vortexing, the homogenates were centrifuged at 12,000×g for 5 min at 4 °C. The resulting supernatants were collected and administered to unvaccinated Blt−/− mice (200 µL/mouse) via oral gavage. One hour later, the mice were challenged with live Salmonella. The cecal lavage fluids were administered to the mice once daily for a total of 4 days.
For B cell adoptive transfer experiment, pan B cells from LPL of vaccinated T.mu-free (Ctrl) or T.mu-colonized (T.mu) WT mice were isolated using the MojoSort™ Mouse Pan B Cell Isolation Kit (480052, BioLegend, San Diego, USA) according to the manufacturer’s instructions. The isolated B cells were resuspended in sterile PBS followed by retro-orbital injection into naïve Blt1−/− mice (~1 × 106 B cells/mouse). Two days later, the recipient mice were challenged orally with a lethal dose of live Salmonella, the survival rate was then monitored daily.
ELISA
For total IgG /IgM /IgA tests, goat anti-mouse Ig (1010-01, SouthernBiotech, Birmingham, USA) was coated on high-binding plates (CLS3922, Corning, New York, USA; V1096HL, VICMED, Xuzhou, China). Anti-IgG/IgM/IgA ELISA was performed using the anti-mouse IgG-HRP antibody (1030-05, SouthernBiotech, Birmingham, USA), anti-mouse IgM-HRP antibody (1020-05, SouthernBiotech, Birmingham, USA), anti-mouse IgA-HRP antibody (ab97235, Abcam, Cambridge, UK), and TMB Chromogen Solution (E661007, Sangon Biotech, Shanghai, China) was selected to assess the Igs levels according to the manufacturer’s protocol.
For antigen-specific IgG/IgM/IgA tests, 2 μg/well OVA/NP-BSA (N-5050, LGC Biosearch Technologies, California, USA)/TNP-BSA (T-5050, LGC Biosearch Technologies, California, USA) was coated on high-binding plates for OVA, NP, or TNP-specific Ig tests, respectively.
For IgA absolute value quantification, the mouse IgA standard (88-50450-22, Invitrogen, MA, USA) was used. For relative qualification, one arbitrary unit (1 AU) of IgA was defined as: 1 AU = (the average OD450 absorbance value of control group) − (background OD450 absorbance value).
For S. Typhimurium-specific IgG/IgA tests, total soluble S. Typhimurium antigen was prepared as the following. After washing with PBS twice, the bacteria were resuspended in coating buffer (C1050, Solarbio, Beijing, China) supplemented with protease inhibitor cocktail (P1010, Beyotime, Nantong, China). The bacterial cells were shattered via ultrasonication on ice followed by centrifugation at 12,000×g for 10 min at 4 °C. Then the supernatant was collected, and the protein concentration was determined by the BCA method. Finally, the S. Typhimurium protein was diluted to 20 μg/mL with coating buffer, and 100 μL/well protein solution was added to high-binding plates and incubated overnight at 4 °C.
For cecal contents or feces processing, the collected samples were weighed and resuspended in 10× volume of PBS with protease inhibitor cocktail (P1010, Beyotime, Nantong, China). After vortex and ultrasonic processing, the homogenates were centrifuged at 12,000×g for 5 min at 4 °C. The supernatants were harvested for Igs tests.
For total Igs: serum and cecal/fecal samples were diluted 10,000-fold for total IgA test, 100,000-fold for total serum IgM, and 1,000,000-fold serum total IgG. For anti-OVA Igs: serum was diluted 20–50 times for anti-OVA IgA test, 500 times for anti-OVA IgM, and 1000 times for anti-OVA IgG, and cecal/fecal samples were diluted 10–50 times for anti-OVA IgA test. For anti-S. Typhimurium Igs: serum was diluted 20 times for anti-S. Typhimurium IgA test, and 100 times for anti-S. Typhimurium IgG test and fecal samples were diluted 10 times for anti-S. Typhimurium IgA test.
For LTB4 quantification, the weighed ileocecal segment of the small intestine was suspended in 2 times volumes of PBS supplemented with protease inhibitor cocktail (P1010, Beyotime, Nantong, China) followed by homogenate. Then the samples were centrifuged at 12,000×g for 5 min, at 4 °C. Finally, the supernatants were harvested for ELISA tests with a mouse LTB4 ELISA kit (TW8165, Shanghai Tongwei Industrial Co., Ltd, Shanghai, China) according to the manufacturer’s instructions.
Immunofluorescence
The 4% formaldehyde-fixed small intestine tissues obtained from the control and T.mu-colonized mice were embedded in paraffin, followed by sectioning, de-paraffining and rehydration. After antigen retrieval with 10 mM sodium citrate (pH 6.0), the samples were blocked with 3% Bovine Serum Albumin (A8020, BSA, Solarbio, Beijing, China). Antibodies against IgA (PAA546MU01, Cloud-Clone Corp., Houston, USA), Ki67 (GB121141, Servicebio, Wuhan, China), Alexa Fluor 488-linked secondary antibody (A0423, Beyotime, Nantong, China), and Cy3-linked secondary antibody (A0521, Beyotime, Nantong, China) were used to detect IgA and Ki67. 4’,6-diamidino-2-phenylindole (C1005, DAPI, Beyotime, Nantong, China) was used for nuclear staining. High-resolution images were scanned by the digital slide scanner Pannoramic SCAN (3DHISTECH Ltd, Budapest, Hungary).
Bacterial microbiome analysis
Total DNA was extracted from cecal content samples by using the DNeasy PowerSoil Pro Kit (47014, QIAGEN, Dusseldorf, Germany). The concentration and purity were monitored using a NanoDrop2000 (Thermo Fisher Scientific, USA). The 16S bacterial rRNA genes were amplified by polymerase chain reaction (PCR) using the primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). The PCR reaction conditions were as follows: 3 min at 95 °C for initialization, 30 cycles of 30 s denaturation at 95 °C, 30 s annealing at 52 °C, and 42 s extension at 72 °C, followed by a final elongation at 72 °C for 10 min. The length and concentration of the PCR products were monitored by agarose gel electrophoresis. PCR products were mixed in equidensity ratios according to the GeneTools Analysis Software (Version 4.03.05.0, SynGene), and the mixture was purified using the AxyPrep DNA Gel Extraction Kit (AP-GX-250, Axygen, San Francisco, USA). Sequencing libraries were generated using the NEXTFLEX Rapid DNA-Seq Kit (5144-08, Bioo Scientific, Austin, USA) according to the manufacturer’s protocol. The library was sequenced on a Miseq PE300/NovaSeq PE250 platform (Shanghai Majorbio Technology Co., Ltd. Shanghai, China). Illumina paired-end reads were merged into single reads by using Flash 1.2.11 software (https://ccb.jhu.edu/software/FLASH). Fastp software (https://github.com/OpenGene/fastp, version 0.20.0) was used to quality control of original sequencing sequence. UPARSE was the method used to cluster the sequencing results into operational taxonomic units (OTUs) using an identity threshold of 97%. Taxonomic annotation of OTUs was according to the Ribosomal Database Project (RDP) classifier ratio against the Silva138.1/16s_bacteria gene database with a 70% confidence threshold. All bioinformatics analyses were conducted by Shanghai Majorbio Technology using USEARCH version 11 (https://www.drive5.com/usearch/). Further analysis was carried out using the Majorbio Cloud platform (https://cloud.majorbio.com). Qiime (http://qiime.org/install/index.html) was used to calculate the distance of beta diversity based on Bray-Curtis distance metrics. The PCoA (Principal Co-ordinates Analysis) graph was drawn with R-3.3.1 (vegan). The linear discriminant analysis (LDA) effect size (LEfSe) (http://huttenhower.sph.harvard.edu/LEfSe) was performed to identify the significantly abundant taxa of bacteria among the different groups [LDA score (log10) > 3, P < 0.05]. Clean reads were deposited in the NCBI Sequence Read Archive under the accession number PRJNA1109335 (SRR28981040-87).
RNA-seq analysis
The fresh ileocecal segment of small intestines with Peyer’s patches (PPs) removed or PPs alone were put into 1 mL TRIzol reagent (15596018, Life Technologies, California, USA) and kept for 5 min at room temperature, then the samples were quick-frozen in liquid nitrogen. Total RNA was extracted using TRIzol reagent according to the manufacturer’s instructions. After removing genomic DNA by gDNA digester (H2901291, YEASEN, Shanghai, China), the RNA quality and quantity were determined using a Nanodrop spectrophotometer. RNA sequencing was performed by Beijing Genome Institute (BGI; Shenzhen, China) using the BGISEQ platform. Before RNA sequencing, the final quality of RNA was assessed by Beijing Genome Institute including concentration, 28S/18S, RIN or RQN through Agilent 2100, Fragment Analyzer and Qsep-400. KEGG pathway analysis was performed on Dr. Tom Data Visualisation Solution (https://biosys.bgi.com/). Clean reads were deposited in the NCBI Sequence Read Archive under the accession number PRJNA977570 (SRR24760665-84).
Flow cytometry analysis
PPs and bone marrow were collected freshly from the control and T.mu-colonized mice and ice-cold PBS was added, followed by mashing and filtering through 70-µm cell strainers. The cell suspensions were centrifuged at 400×g for 5 min at 4 °C, and the pellets were resuspended in Ammonium-Chloride-Potassium (ACK) cell lysing buffer to remove erythrocytes. After lysis, the residual cells were resuspended in FACS buffer (PBS supplemented with 1% BSA), followed by further filtration through 70-µm cell strainers. For live/dead cell staining, the filtered cells were washed three times with FACS buffer, followed by staining with Zombie Aqua (423101, BioLegend, San Diego, USA) according to the manufacturer’s instructions. After washing three times with FACS buffer, the cells were incubated with unlabeled CD16/32 antibody (14-0161-82, eBioscience, California, USA) for 15 min. The cells were then washed and stained for cell surface molecules, followed by cell fixation and permeabilization, and finally, stained for intracellular IgA. Unstained sample and single fluorescent antibody-labeled samples were used to calibrate detection parameter. The following fluorophore-conjugated antibodies (dilution times 1: 100) were used: anti-CD45-PE-Cy7 (103113, Clone: 30-F11, Biolegend, San Diego, USA), anti-CD45-APC (561018, Clone: 30-F11, BD Pharmingen, New Jersey, USA), anti-B220-APC (103212, Clone: RA3-6B2, Biolegend, San Diego, USA), anti-IgA-FITC (11-4204-81, Clone: mA-6E1, Invitrogen, California, USA), anti-IgG1-Percp-Cy5.5 (406612, Clone: RMG1-1, Biolegend, San Diego, USA), anti-CD138-PE (142503, Clone: 281-2, Biolegend, San Diego, USA). The hemocytometer was used for total cell count. BD FACSAria III flow cytometer (BD Biosciences) and BD FACSCanton II (BD Biosciences) were used for flow cytometry analysis. Data analysis was performed with FlowJo_V10.
For lamina propria IgA+ plasma cell analysis, LPL was isolated according to a reported method described by Kim et al.66. Briefly, the distal 15 cm of the small intestine was collected, after excising PPs, the epithelial cells as well as intraepithelial lymphocytes were wiped off by 5 mM EDTA and 1 mM DTT. Then the residual fragments were digested by 50 μg/mL Dnase I (11284932001, Merck KGaA, Darmstadt, Germany) and 1.5 mg/mL Collagenase IV (abs47048003, Absin, Shanghai, China). After filtration, the filtrates were layered by Percoll solution, and LPL was harvested at the interface of 40/80% Percoll solution for the following flow cytometry analysis after washed.
ELISPOT
After 14 days of OVA immunization, LPLs from the small intestine were freshly isolated and resuspended in RPMI-1640 medium (SH30809, Hyclone, Logan, USA) containing 10% FBS (16000-044, Gibco, New York, USA). For anti-OVA IgA ELISPOT assays, MultiScreen plates (MSIPS4510, Merk KGaA, Darmstadt, Germany) were activated with 35% ethanol and coated with OVA (2 μg/well) overnight at 4 °C. The plates were then washed three times with PBS and blocked with 5% BSA for 120 min at room temperature. After another three washes with RPMI-1640 medium containing 10% FBS, the gradient-diluted LPLs were added to the MultiScreen plates and cultured overnight at 37 °C in a 5% CO2 incubator. The plates were washed three times with PBST (PBS containing 0.1% Tween-20), followed by the addition of anti-mouse IgA-HRP antibody, which was incubated for 60 min. After six washes with PBST, the anti-OVA IgA spots were visualized using an ECL Chemiluminescent Substrate kit (BL523B, Biosharp, Beijing, China) or a DAB Peroxidase Substrate kit (CR2402044, Servicebio, Wuhan, China).
Untargeted metabolomics analysis
Untargeted metabolomics analysis was performed by Shanghai Applied Protein Technology (Shanghai, China). For the serum samples (n = 10 mice/group; one group from T.mu-free mice, and the other group from T.mu-colonized mice), each sample was diluted with 4 times the volume of pre-chilled methanol/acetonitrile solution (1:1, v/v) and mixed by vortexing. After 10 min incubation at −20 °C, the mixtures were centrifuged at 14,000×g, 4 °C for 15 min, and the supernatants were collected, followed by vacuum drying. Finally, the samples were reconstituted in acetonitrile aqueous solution (acetonitrile: water = 1:1, v/v). For the cecal content samples (n = 10 mice/group; one group from T.mu-free mice, and the other group from T.mu-colonized mice), 60 mg cecal content was homogenized in 200 μL ultrapure water, after which 800 μL of methanol/acetonitrile (1:1, v/v) was added, followed by sonication. After sonicating on ice for 30 min, the samples were incubated at −20 °C for a further 60 min. The samples were centrifuged at 14,000×g, 4 °C for 15 min to precipitate proteins, and supernatants were freeze-dried for the subsequent mass spectrometry analysis. The sample was separated using an Agilent 1290 Infinity LC ultra-high performance liquid chromatography system (UHPLC) with a HILIC column; column temperature was set at 25 °C; flow rate was 0.3 mL/min; injection volume was 2 μL; the mobile phase composition was A: water + 25 mM ammonium acetate + 25 mM ammonia, B: acetonitrile. The gradient elution program was as follows: 0–1 min, 95% B; 1–14 min, B linearly decreased from 95% to 65%; 14–16 min, B linearly decreased from 65% to 40%; 16–18 min, B was held at 40%; 18–18.1 min, B linearly increased from 40% to 95%; 18.1–23 min, B was maintained at 95%. Throughout the analysis, samples were kept in an automatic sampler at 4 °C. To avoid the impact of fluctuations in instrument detection signals, samples were continuously analyzed in a random order. QC samples were inserted into the sample queue to monitor and evaluate the stability of the system and the reliability of the experimental data.
The separated samples were analyzed on a Triple TOF 5600 mass spectrometer (AB SCIEX, Boston, USA) with electrospray ionization (ESI)-positive ion and -negative ion modes used for detection. The ESI source conditions after HILIC chromatography separation are as follows: Ion Source Gas1 (Gas1): 60, Ion Source Gas2 (Gas2): 60, Curtain gas (CUR): 30, source temperature: 600 °C, Ion Spray Voltage Floating (ISVF) ± 5500 V (positive and negative modes); TOF MS scan m/z range: 60–1000 Da, product ion scan m/z range: 25–1000 Da, TOF MS scan accumulation time 0.20 s/spectrum, product ion scan accumulation time 0.05 s/spectrum; the second-stage mass spectrometry is obtained using information-dependent acquisition (IDA) and is conducted in high sensitivity mode, Declustering potential (DP): ±60 V (positive and negative modes), Collision Energy: 35 ± 15 eV, IDA settings are as follows: Exclude isotopes within 4 Da, Candidate ions to monitor per cycle: 6.
The raw data was converted to mzXML format using ProteoWizard (https://proteowizard.sourceforge.io/), and then peak alignment, retention time correction, and peak area extraction were performed using the XCMS program (https://bioconductor.org/packages/3.20/bioc/html/xcms.html). Metabolite structure identification was conducted through precise mass matching (within <25 ppm) and secondary spectrum matching, utilizing a self-built laboratory database. For the data extracted by XCMS, ion peaks with a total group sum greater than 2/3 were removed. Pattern recognition was performed using SIMCA-P 14.1 (Umetrics, Umeå, Sweden) after preprocessing the data with Pareto scaling. Univariate statistical analyses included Student’s t-test and fold change analysis.
Targeted free fatty acids metabolome analysis
The targeted free fatty acids metabolome analysis was performed by Shanghai Biotree Biomedical Technology Co., Ltd. (Shanghai, China). Briefly, the weighed cecal content, cecum tissue, serum, animal chow diet powder, or enumerated T.mu cells (n = 1 sample per group; one group accumulated from 6 T.mu-free WT mice, and the other group accumulated from 6 T.mu-colonized WT mice) were transferred into new 2 mL EP tubes followed extraction with 500 μL extracting solution [Isopropanol: n-Hexane = 2:3 (V:V)], including 0.2 mg/L internal standard. After homogenized in a ball mill for 4 min at 40 Hz and a 5 min ultrasound treatment, the samples were centrifuged 16,200×g for 15 min at 4 °C. Then the supernatants were transferred into new 2 mL EP tubes followed with nitrogen blow dry. The dried samples were resuspended in 500 μL of methanol: trimethylsilyl diazomethane solution (1:2), after standing 30 min at room temperature, another nitrogen blow dry was performed. Then, the samples were resuspended in 160 μL of n-hexane, followed by a centrifugation of 16,200×g for 1 min. Finally, the supernatants were transferred into new vials for GC-MS analysis. GC-MS analysis was performed using an Agilent 7890B gas chromatograph system coupled with an Agilent 5977B mass spectrometer. The system employed a DB-FastFAME capillary column (90 m × 0.25 mm × 0.25 µm, Agilent Technologies). A 1 µL aliquot of the derivatized sample was injected in split mode (5:1), using helium as the carrier gas. The gas flow rate was set to 3 mL/min, with the pressure maintained at a constant 46 psi. The initial column temperature was held at 75 °C for 1 min, after which it was increased to 200 °C at a rate of 50 °C/min and held for 15 min. The temperature was then raised to 210 °C at a rate of 2 °C/min and maintained for 1 min, followed by a final increase to 230 °C at a rate of 10 °C/min, which was held for 16.5 min. The temperatures for the injector, transfer line, quadrupole, and ion source were set at 240 °C, 240 °C, 230 °C, and 150 °C, respectively. Ionization was conducted in electron impact mode at 70 eV. Mass spectrometry data were acquired in Scan/SIM mode over an m/z range of 33–400 after a solvent delay of 7 min. Fatty acid methyl esters (FAMEs) were identified by comparing them to commercial FAME standards (ANPEL, Shanghai, China, Cat# CDAA-252). The absolute concentrations of FAMEs were calculated using calibration curves based on the internal standard method.
Quantitation of ARA
Serum, small intestine tissue, and feces were suspended in methanol followed by shaking, homogenizing, and sonicating, then the suspensions were centrifuged at 12,000×g for 15 min, and supernatants were collected for quantification of ARA by UPLC-MS/MS (Waters Acquity I class UPLC, Waters XEVO TQD) with methanol plus 0.1% formic acid as mobile phase.
Quantitative real-time PCR (qPCR)
DNA was extracted by the TIANamp Stool DNA Kit (DP328, Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions. Total RNA was extracted from tissues homogenized in Trizol (15596018, Thermo Fisher Scientific, Waltham, USA); 1 μg of purified RNA was used to generate cDNA with a High-Capacity cDNA Reverse Transcription Kit (RR047A, Takara, Dalian, China). The qPCR reaction was performed using the AceQ qPCR SYBR Green Master Mix kit (Q111, Vazyme, Nanjing, China). The relative mRNA expression levels were determined by the 2−ΔΔCt method with β-Actin as the internal reference control, while the relative total bacterial abundance was determined by 16S rDNA with equal amounts of templates. Primer sequences are listed as follow: mβ-Actin (F: 5′ CGTTGACATCCGTAAAGACC 3′, R: 5′ AACAGTCCGCCTAGAAGCA 3′), mIl18 (F: 5′ GTTCTCTGTGGTTCCATGCTTTC 3′, R: 5′ GTATTACTGCGGTTGTACAGTGAAGTC 3′), Bacterial 16S rRNA gene (F: 5′ CGGTGAATACGTTCCCGG 3′, R: 5′ TACGGCTACCTTGTTACGACTT 3′).
Statistical analysis
The GraphPad Prism 9.0 software was used to perform statistical analysis. All graphs were shown as means ± SEM. For all the in vivo experiments, each symbol represents an individual mouse. Student’s t-test (two-tailed), One-way ANOVA with Tukey’s post hoc test, Two-way ANOVA with Sidak’s multiple comparisons test, or Log-rank (Mantel-Cox) test were used to determine significance. Statistical significance is indicated by *p < 0.05, **p < 0.01, ***p < 0.001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The transcriptome data generated in this study have been deposited in the NCBI SRA database under accession code: SRR24760665-84 (https://www.ncbi.nlm.nih.gov/search/all/?term=PRJNA977570). The 16S bacterial microbiome analysis data generated in this study have been deposited in the NCBI SRA database under accession code: SRR28981040-87 (https://www.ncbi.nlm.nih.gov/search/all/?term=PRJNA1109335). The untargeted metabolome data were generated in 2018; however, no raw data were provided to us by the mass spectrometry service company at that time. The processed untargeted metabolome data have been deposited in the online Figshare data repository: https://doi.org/10.6084/m9.figshare.23632281. The processed free fatty acids metabolome data generated in this study have been included as source data. Additionally, the raw data have been deposited in the online Figshare data repository: https://doi.org/10.6084/m9.figshare.23632281, as well as in the EMBL-EBI MetaboLights database under the identifier MTBLS9299 and the Metabolomics Workbench database under Study ID ST003425 (https://doi.org/10.21228/M8ZV6G). All remaining data generated or analyzed in this study are provided in the Source Data file. Source data are provided with this paper.
References
Sterlin, D. et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. 13, eabd2223 (2021).
Wang, Z. et al. Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci. Transl. Med. 13, eabf1555 (2021).
Liew, F. Y., Russell, S. M., Appleyard, G., Brand, C. M. & Beale, J. Cross-protection in mice infected with influenza A virus by the respiratory route is correlated with local IgA antibody rather than serum antibody or cytotoxic T cell reactivity. Eur. J. Immunol. 14, 350–356 (1984).
Asahi-Ozaki, Y. et al. Secretory IgA antibodies provide cross-protection against infection with different strains of influenza B virus. J. Med. Virol. 74, 328–335 (2004).
Okuya, K. et al. Potential role of nonneutralizing IgA antibodies in cross-protective immunity against influenza A viruses of multiple hemagglutinin subtypes. J. Virol. 94, e00408–20 (2020).
Ceglia, S. et al. An epithelial cell-derived metabolite tunes immunoglobulin A secretion by gut-resident plasma cells. Nat. Immunol. 24, 531–544 (2023).
Goguyer-Deschaumes, R., Waeckel, L., Killian, M., Rochereau, N. & Paul, S. Metabolites and secretory immunoglobulins: messengers and effectors of the host-microbiota intestinal equilibrium. Trends Immunol. 43, 63–77 (2022).
Li, Y. et al. Identification of trypsin-degrading commensals in the large intestine. Nature 609, 582–589 (2022).
Lukes, J., Stensvold, C. R., Jirku-Pomajbikova, K. & Wegener Parfrey, L. Are human intestinal eukaryotes beneficial or commensals? PLoS Pathog. 11, e1005039 (2015).
Nieves-Ramirez, M. E. et al. Asymptomatic intestinal colonization with protist blastocystis is strongly associated with distinct microbiome ecological patterns. mSystems 3, e00007–e00018 (2018).
Scanlan, P. D. et al. The microbial eukaryote Blastocystis is a prevalent and diverse member of the healthy human gut microbiota. FEMS Microbiol. Ecol. 90, 326–330 (2014).
Chudnovskiy, A. et al. Host-protozoan interactions protect from mucosal infections through activation of the inflammasome. Cell 167, 444–456.e414 (2016).
Escalante, N. K. et al. The common mouse protozoa Tritrichomonas muris alters mucosal T cell homeostasis and colitis susceptibility. J. Exp. Med. 213, 2841–2850 (2016).
Schneider, C. et al. A metabolite-triggered tuft cell-ILC2 circuit drives small intestinal remodeling. Cell 174, 271–284.e214 (2018).
Wei, Y. et al. Commensal bacteria impact a protozoan’s integration into the murine gut microbiota in a dietary nutrient-dependent manner. Appl. Environ. Microbiol. 86, e00303–e00320 (2020).
Popovic, A. et al. The commensal protist Tritrichomonas musculus exhibits a dynamic life cycle that induces extensive remodeling of the gut microbiota. ISME J. 18, wrae023 (2024).
Howitt, M. R. et al. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 351, 1329–1333 (2016).
Nadjsombati, M. S. et al. Detection of succinate by intestinal tuft cells triggers a type 2 innate immune circuit. Immunity 49, 33–41.e37 (2018).
Kou, Y. et al. A murine commensal protozoan influences host glucose homeostasis by facilitating free choline generation. Appl. Environ. Microbiol. 88, e0241321 (2022).
Vajdy, M. & Lycke, N. Y. Cholera toxin adjuvant promotes long-term immunological memory in the gut mucosa to unrelated immunogens after oral immunization. Immunology 75, 488–492 (1992).
Bunker, J. J. et al. Natural polyreactive IgA antibodies coat the intestinal microbiota. Science 358, eaan6619 (2017).
Xu, Z., Zan, H., Pone, E. J., Mai, T. & Casali, P. Immunoglobulin class-switch DNA recombination: induction, targeting and beyond. Nat. Rev. Immunol. 12, 517–531 (2012).
Cerutti, A., Puga, I. & Cols, M. Innate control of B cell responses. Trends Immunol. 32, 202–211 (2011).
Loho, T. & Dharmayanti, A. Colistin: an antibiotic and its role in multiresistant Gram-negative infections. Acta Med. Indones. 47, 157–168 (2015).
Bruniera, F. R. et al. The use of vancomycin with its therapeutic and adverse effects: a review. Eur. Rev. Med. Pharm. Sci. 19, 694–700 (2015).
Ormerod, K. L. et al. Genomic characterization of the uncultured Bacteroidales family S24-7 inhabiting the guts of homeothermic animals. Microbiome 4, 36 (2016).
Takeuchi, T. et al. Acetate differentially regulates IgA reactivity to commensal bacteria. Nature 595, 560 (2021).
Tan, J. et al. Dietary protein increases T-cell-independent sIgA production through changes in gut microbiota-derived extracellular vesicles. Nat. Commun. 13, 4336 (2022).
Wu, W. et al. Microbiota metabolite short-chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal Immunol. 10, 946–956 (2017).
Kim, M., Qie, Y. Q., Park, J. & Kim, C. H. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe 20, 202–214 (2016).
Louis, P. & Flint, H. J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 19, 29–41 (2017).
Brash, A. R. Arachidonic acid as a bioactive molecule. J. Clin. Invest. 107, 1339–1345 (2001).
Jellusova, J. Metabolic control of B cell immune responses. Curr. Opin. Immunol. 63, 21–28 (2020).
Lands, W. E. et al. Maintenance of lower proportions of (n − 6) eicosanoid precursors in phospholipids of human plasma in response to added dietary (n − 3) fatty acids. Biochim. Biophys. Acta 1180, 147–162 (1992).
Wang, B. et al. Metabolism pathways of arachidonic acids: mechanisms and potential therapeutic targets. Signal Transduct. Target. Ther. 6, 94 (2021).
Haeggstrom, J. Z. & Funk, C. D. Lipoxygenase and leukotriene pathways: biochemistry, biology, and roles in disease. Chem. Rev. 111, 5866–5898 (2011).
Tager, A. M. et al. Leukotriene B4 receptor BLT1 mediates early effector T cell recruitment. Nat. Immunol. 4, 982–990 (2003).
Neubert, K. et al. The proteasome inhibitor bortezomib depletes plasma cells and protects mice with lupus-like disease from nephritis. Nat. Med. 14, 748–755 (2008).
Pellom, S. T., Dudimah, D. F., Thounaojam, M. C., Sayers, T. J. & Shanker, A. Modulatory effects of bortezomib on host immune cell functions. Immunotherapy 7, 1011–1022 (2015).
Lammermann, T. et al. Neutrophil swarms require LTB4 and integrins at sites of cell death in vivo. Nature 498, 371–375 (2013).
Boyaka, P. N. Inducing mucosal IgA: a challenge for vaccine adjuvants and delivery systems. J. Immunol. 199, 9–16 (2017).
Hara, S. et al. Dietary antigens induce germinal center responses in Peyer’s patches and antigen-specific IgA production. Front. Immunol. 10, 2432 (2019).
von Moltke, J. et al. Leukotrienes provide an NFAT-dependent signal that synergizes with IL-33 to activate ILC2s. J. Exp. Med. 214, 27–37 (2017).
Hayashi, S. et al. Intestinal epithelial BLT1 promotes mucosal repair. JCI Insight 7, e162392 (2022).
Tager, A. M. & Luster, A. D. BLT1 and BLT2: the leukotriene B4 receptors. Prostaglandins Leukotr. Ess. Fatty Acids 69, 123–134 (2003).
Saeki, K. & Yokomizo, T. Identification, signaling, and functions of LTB4 receptors. Semin. Immunol. 33, 30–36 (2017).
Wang, L. M. et al. BLT1-dependent alveolar recruitment of CD4+CD25+ Foxp3+ regulatory T cells is important for resolution of acute lung injury. Am. J. Resp. Crit. Care 186, 989–998 (2012).
Nagatake, T. et al. BLT1 mediates commensal bacteria-dependent innate immune signals to enhance antigen-specific intestinal IgA responses. Mucosal Immunol. 12, 1082–1091 (2019).
Grasset, E. K. et al. Gut T cell-independent IgA responses to commensal bacteria require engagement of the TACI receptor on B cells. Sci. Immunol. 5, eaat7117 (2020).
He, B. et al. Intestinal bacteria trigger T cell-independent immunoglobulin A2 class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity 26, 812–826 (2007).
Reboldi, A. et al. IgA production requires B cell interaction with subepithelial dendritic cells in Peyer’s patches. Science 352, aaf4822 (2016).
Macia, L., Nanan, R., Hosseini-Beheshti, E. & Grau, G. E. Host- and microbiota-derived extracellular vesicles, immune function, and disease development. Int. J. Mol. Sci. 21, 107 (2019).
Cao, E. Y. et al. The protozoan commensal Tritrichomonas musculis is a natural adjuvant for mucosal IgA. Preprint at bioRxiv https://doi.org/10.1101/2022.10.08.511442 (2022).
Doron, I. et al. Mycobiota-induced IgA antibodies regulate fungal commensalism in the gut and are dysregulated in Crohn’s disease. Nat. Microbiol. 6, 1493–1504 (2021).
Ost, K. S. et al. Adaptive immunity induces mutualism between commensal eukaryotes. Nature 596, 114–118 (2021).
Qiu, X., Xie, X. & Meesapyodsuk, D. Molecular mechanisms for biosynthesis and assembly of nutritionally important very long chain polyunsaturated fatty acids in microorganisms. Prog. Lipid Res. 79, 101047 (2020).
Usami, M., Miyoshi, M., & Yamashita, H. Gut microbiota and host metabolism in liver cirrhosis. World J. Gastroenterol. 21, 11597–11608 (2015).
Stremmel, W. et al. Retarded release phosphatidylcholine benefits patients with chronic active ulcerative colitis. Gut 54, 966–971 (2005).
Lee, Y. A., Nam, Y. H., Min, A. & Shin, M. H. Trichomonas vaginalis-secreted cysteinyl leukotrienes promote migration, degranulation and MCP-1 production in mast cells. Parasite Immunol. 42, e12789 (2020).
Park, Y. H., Wood, G., Kastner, D. L. & Chae, J. J. Pyrin inflammasome activation and RhoA signaling in the autoinflammatory diseases FMF and HIDS. Nat. Immunol. 17, 914 (2016).
Sokolowska, M. et al. Prostaglandin E-2 inhibits NLRP3 inflammasome activation through EP4 receptor and intracellular cyclic AMP in human macrophages. J. Immunol. 194, 5472–5487 (2015).
Barnig, C. et al. Lipoxin A4 regulates natural killer cell and type 2 innate lymphoid cell activation in asthma. Sci. Transl. Med. 5, 174ra126 (2013).
Reis, M. B. et al. Lipoxin A4 encapsulated in PLGA microparticles accelerates wound healing of skin ulcers. PLoS ONE 12, e0182381 (2017).
Tallima, H. & El Ridi, R. Arachidonic acid: physiological roles and potential health benefits—a review. J. Adv. Res. 11, 33–41 (2018).
Penny, H. A. et al. Rhythmicity of intestinal IgA responses confers oscillatory commensal microbiota mutualism. Sci. Immunol. 7, eabk2541 (2022).
Kim, E., Tran, M., Sun, Y. & Huh, J. R. Isolation and analyses of lamina propria lymphocytes from mouse intestines. STAR Protoc. 3, 101366 (2022).
Acknowledgements
The authors would like to express their gratitude to EditSprings (https://www.editsprings.cn) for the expert linguistic services provided. This work was supported by the National Natural Science Foundation of China (Grant numbers 81770853 to Y.Wang and 81700774 to Y.K.), Natural Science Foundation of Jiangsu Province (BK20231172 to Y.K.), Special funds of Xuzhou for Promoting Scientific and Technological Innovation (KC23057 to Y.K.), Open Competition Grant of Xuzhou Medical University (JBGS202202 to the team) and the Starting Foundation for Talents of Xuzhou Medical University (Grant numbers D2016029 to Y.K. and D2016018 to Y.Wang).
Author information
Authors and Affiliations
Contributions
Y.K., H.W. and Y.Wang were responsible for the study design; Y.K. and Y.Wang were responsible for the supervision, and manuscript preparation; Y.K., S.Z., J.C., Y.S., Z.Z., H.H., Y.M., Y.X., L.L., J.Z. and W.C. were responsible for the animal experiments; Y.K., Z.L. and Y.Wei were responsible for sequencing analysis; S.Z., Y.K. and H.Y. were responsible for bacterial culture. Y.K. and Y.Z. were responsible for mass spectrometry detection and analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Crystal Loving and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kou, Y., Zhang, S., Chen, J. et al. A mouse protozoan boosts antigen-specific mucosal IgA responses in a specific lipid metabolism- and signaling-dependent manner. Nat Commun 15, 7914 (2024). https://doi.org/10.1038/s41467-024-52336-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-52336-z