Abstract
Methionine adenosyltransferase 2 A (MAT2A) and MAT2B are essential for hepatic stellate cells (HSCs) activation. Forkhead box M1 (FOXM1) transgenic mice develop liver inflammation and fibrosis. Here we examine if they crosstalk in male mice. We found FOXM1/MAT2A/2B are upregulated after bile duct ligation (BDL) and carbon tetrachloride (CCl4) treatment in hepatocytes, HSCs and Kupffer cells (KCs). FDI-6, a FOXM1 inhibitor, attenuates the development and reverses the progression of CCl4-induced fibrosis while lowering the expression of FOXM1/MAT2A/2B, which exert reciprocal positive regulation on each other transcriptionally. Knocking down any of them lowers HSCs and KCs activation. Deletion of FOXM1 in hepatocytes, HSCs, and KCs protects from BDL-mediated inflammation and fibrosis comparably. Interestingly, HSCs from Foxm1Hep−/−, hepatocytes from Foxm1HSC−/−, and HSCs and hepatocytes from Foxm1KC−/− have lower FOXM1/MAT2A/2B after BDL. This may be partly due to transfer of extracellular vesicles between different cell types. Altogether, FOXM1/MAT2A/MAT2B axis drives liver inflammation and fibrosis.
Similar content being viewed by others
Introduction
Hepatic fibrosis is characterized by the accumulation of extracellular matrix (ECM) components such as collagens type I (COL1A1) and type III, which may further lead to cirrhosis and liver cancer1. Activation of hepatic stellate cells (HSCs) is a crucial event in liver fibrosis. Forkhead box M1 (FOXM1) is a member of the forkhead box family with winged helix DNA binding domain that works as a crucial transcription factor in proliferation and oncogenesis2,3. Recent studies have reported that FOXM1 could promote multiple organ fibrosis including liver fibrosis4,5,6. Furthermore, Kurahashi et al.3 demonstrated that overexpression of FOXM1 in hepatocytes caused spontaneous liver injury, inflammation, fibrosis, and hepatocellular carcinoma (HCC). Forkhead domain inhibitory-6 (FDI-6), a small molecule that selectively targets the FOXM1 DNA-binding domain, could inhibit the transcription of FOXM1 target genes7. FDI-6 inhibited corneal inflammation and fibrosis after alkali burn in rats8 and attenuated subconjunctival fibrosis in a trabeculectomy rabbit model9. These studies suggest that targeting FOXM1 may be a potential therapeutic strategy in liver inflammation and fibrosis. However, this has not been examined.
Methionine adenosyltransferase (MAT) is an essential enzyme that catalyzes the biosynthesis of S-adenosylmethionine (SAMe)10. MAT2A encodes for the α2 catalytic subunit and MAT2B gene encodes the β regulatory subunit of the MATII isoenzyme10. MAT2A and MAT2B are expressed in extrahepatic tissues and non-parenchymal cells of the liver, such as HSCs and Kupffer cells (KCs)11. MAT2A expression is induced in cultured activated HSCs, and in livers treated with carbon tetrachloride (CCl4) or thioacetamide (TAA)12. MAT2A and MAT2B affect HSCs activation through changes in SAMe levels and extracellular-regulated kinase and phosphoinositide 3-kinase signaling mechanisms10. In addition, MAT2B siRNA treatment reduced collagen level and inhibited HSCs activation in TAA-induced liver fibrosis in mice13. Wang et al.14 also found that knockdown of MAT2A alleviated CCl4 and transforming growth factor-beta 1 (TGF-β1)-induced HSCs activation, whereas overexpression of MAT2A facilitated hepatic fibrosis. Here we investigated whether there is a crosstalk between FOXM1, MAT2A and MAT2B; to better understand the role of FOXM1 in a cell-type specific manner, we established Foxm1Hep−/−, Foxm1HSC−/−, and Foxm1KC−/− mice and examined the role of the FOXM1/MAT2A/MAT2B axis in vitro and vivo.
In this work, we found that FOXM1, MAT2A and MAT2B expression are upregulated in hepatocytes, HSCs, and KCs after bile duct ligation (BDL) in mice. Treatment with FDI-6 significantly attenuates the development of liver injury and fibrosis but more remarkably, reverses liver fibrosis that was already established. We found FOXM1, MATα2 and MAT2β directly interact, and they positively regulate each other at the transcriptional level via FOX elements present in their promoters. Knocking down any of the three inhibits TGF-β1-induced HSCs activation as well as lipopolysaccharide (LPS)-induced macrophage activation. Interestingly, deleting FOXM1 from hepatocytes, HSCs or KCs all protect from liver injury and fibrosis induced by BDL comparably. This may be in part due to exchange of FOXM1, MATα2 and MAT2β in secreted extracellular vesicles (EVs) amongst the different cell types. Taken together, our results suggest targeting any of the components of the FOXM1/MAT2A/MAT2B axis may be a potential therapeutic strategy in liver fibrosis.
Results
FOXM1/MAT2A/MAT2B axis is induced in liver fibrosis
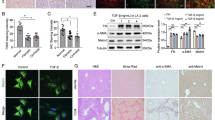
MATα2, MAT2β and FOXM1 are involved in liver fibrosis3,13,14. Here we found the upregulation of FOXM1, MAT2A and MAT2B in hepatic cells and small bile ducts in human and murine fibrotic livers (Supplementary Fig. 1a–e). Hepatocytes express lower levels of Mat2a and Mat2b as compared to HSCs and KCs at baseline, but they are more induced following BDL and CCl4 treatments (Supplementary Fig. 1e). Interestingly, Foxm1 was expressed at a lower level in HSCs than in hepatocytes and KCs but it was induced in all three cell types after BDL and CCl4 treatment (Supplementary Fig. 1e). These proteins are also induced in primary biliary cholangitis (PBC) and primary sclerosing cholangitis (PSC) (Supplementary Fig. 2a–c). Furthermore, FOXM1 expression increased from day 1 to day 14 after bile duct ligation (BDL) while expression of MATα2 and MAT2β increased from day 2 to day 14 after BDL (Fig. 1a), suggesting FOXM1 may be upstream of MATα2 and MAT2β. Primary HSCs isolated from BDL mice at day 7 showed higher cytoplasmic and nuclear FOXM1, MATα2, and MAT2β staining compared with sham control (Fig. 1b). FDI-6 treatment reduced the mRNA and protein levels of FOXM1, MAT2A, and MAT2B in the day-5 culture activated HSCs and LX-2 cell line (Fig. 1c, d, densitometry is shown in Supplementary Fig. 3a, b). FDI-6 also reduced cytoplasmic and nuclear staining of FOXM1, MATα2, and MAT2β in LX-2 cells (Fig. 1e), and in BDL livers (Fig. 1f, densitometry is shown in Supplementary Fig. 3c, d). FDI-6 also lowered the proliferation (Fig. 1g) and migration (Fig. 1h) of LX-2 cells.
a Western blots show the time course of protein expression of FOXM1, MATα2, and MAT2β in liver tissues after BDL (n = 3 independent experiments). b Immunofluorescence (IF) of FOXM1, MATα2, and MAT2β in primary hepatic stellate cells (HSCs) isolated from sham and BDL mice at day 5. The top row shows DAPI staining. The second and third rows show the antibody (AB) staining. The fourth row shows merged images of DAPI and FOXM1, MATα2 or MAT2β, and the fifth row shows high magnification (HM) from the merged image (n = 3 independent experiments). c Expression of mRNA (top) and protein (bottom) of FOXM1, MATα2, MAT2β, α-SMA, and COL1A1 in HSCs isolated from WT mice and cultured for up to 5 days and FDI-6 treatment for 24 h starting at day 4. Data presented as mean ± SEM (n = 3 per group), mRNA levels of Foxm1, Mat2a, Mat2b, Acta2 and Col1a1 in HSCs at day 5 vs. day 1, p = 0.0044, p = 0.0029, p = 0.0056, p = 0.0010 and p = 0.00002, respectively. mRNA levels of Foxm1, Mat2a, Mat2b, Acta2 and Col1a1 in HSCs at day 5 + FDI-6 vs. day 1, p = 0.0431, p = 0.0152, p = 0.0245, p = 0.0083, and p = 0.0082, respectively. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See Supplementary Fig. 3a for densitometric values of the western blots. d Expression of mRNA and protein of FOXM1, MATα2, MAT2β, α-SMA, and COL1A1 after FDI-6 treatment in LX-2 cells. Data presented as mean ± SEM (n = 3 per group), mRNA levels of Foxm1, Mat2a, Mat2b, Acta2 and Col1a1 in LX2 cells with DMSO treatment vs. FDI-6, p = 0.0187, p = 0.0023, p = 0.0122, p = 0.0108 and p = 0.0124, respectively. *p < 0.05, **p < 0.01 vs. DMSO. See Supplementary Fig. 3b for densitometric values of the western blots. e IF of LX-2 cells after treatment with FDI-6. HM, high magnification from the merged images (n = 3 independent experiments). f FOXM1, MATα2, and MAT2β in cytoplasm and nucleus from HSCs isolated from sham and BDL mice with or without FDI-6 treatment (n = 3 independent experiments). Densitometry for cytoplasmic protein levels is summarized in Supplementary Fig. 3c and nuclear protein levels is summarized in Supplementary Fig. 3d. Proliferation (g) and migration (h) of LX-2 cells in vitro after FDI-6 treatment for 24 h. Data presented as mean ± SEM (n = 3 per group). p = 0.00016, p = 0.00002 vs.DMSO. Statistical significance was determined by using two-tailed unpaired Student’s t-test. ***p < 0.001, ****p < 0.0001 vs. DMSO (n = 3). Abbreviations: BDL bile duct ligation, DMSO dimethylsulfoxide. Source data are provided as a Source Data file.
FDI-6 in the development and progression of liver fibrosis
To examine how inhibiting FOXM1 will impact on CCl4-induced liver fibrosis, we divided the mice into two groups (prevention and treatment). In the prevention group, FDI-6 was administered from the start of CCl4 treatment for three weeks. In the treatment group, FDI-6 was started after liver fibrosis was already established (after three weeks of CCl4) and continued with CCl4 for two additional weeks. In these two groups, immunohistochemistry (IHC) showed that FDI-6 treatment reduced the expression of COL1A1, F4/80, α-SMA, FOXM1, MATα2, and MAT2β induced by CCl4 (Fig. 2a). FDI-6 also reduced the levels of alanine transaminase (ALT), aspartate transferase (AST), and hydroxyproline induced by CCl4 (Fig. 2b–d). Furthermore, FDI-6 reduced the mRNA (Fig. 2e) and protein levels (Fig. 2f, densitometry is shown in Supplementary Fig. 3e) of FOXM1, MAT2A, MAT2B, α-SMA, F4/80, and COL1A1. Note that FDI-6 lowered fibrosis markers such as hydroxyproline and COL1A1 to below the levels seen with CCl4 alone for three weeks, which suggests it can reverse already existing fibrosis. CCl4 treatment also increased the interaction between FOXM1 and MATα2 and MAT2β, which was reduced by FDI-6 (Supplementary Fig. 3f, g).
a Liver sections from prevention groups of the corn oil (Oil) + DMSO, Oil + FDI-6, CCl4 + DMSO, and CCl4 + FDI-6 for three weeks, and treatment groups of Oil + DMSO, Oil + FDI-6, CCl4 + DMSO, and CCl4 + FDI-6 treated for two weeks after CCl4 treatment for three weeks. IHC stained with antibodies of COL1A1, F4/80, α–SMA, FOXM1, MATα2, and MAT2β. H&E is shown in the top row. b, c show changes in ALT (n = 6 per group) (b) and AST (n = 6 in prevention group of Oil + DMSO, n = 5 in prevention group of Oil + FDI-6, n = 3 in prevention group of CCl4 + DMSO and treatment group of Oil + FDI-6, n = 4 in prevention group of CCl4 + FDI-6, and treatment groups of Oil + DMSO, CCl4 + DMSO, and CCl4 + FDI-6) (c) levels after FDI-6 administration in the prevention and treatment groups. Data presented as mean ± SEM, **p < 0.01, ****p < 0.0001. d Hydroxyproline content was measured in the livers from prevention and treatment groups with or without FDI-6 administration. Data presented as mean ± SEM, **p < 0.01, ****p < 0.0001 (n = 5 per group). e mRNA levels of Foxm1, Mat2a, and Mat2b in the livers after FDI-6 administration in the prevention and treatment groups. Data presented as mean ± SEM (n = 4), **p < 0.01, ****p < 0.0001 (n = 4 in prevention and treatment groups of Oil + DMSO for Foxm1 mRNA; n = 6 in prevention and treatment groups of Oil + DMSO for Mat2α and Mat2b mRNA; n = 4 in prevention and treatment groups of Oil+ FDI-6 for Foxm1 mRNA; n = 6 in prevention and treatment groups of Oil + FDI-6 for Mat2α and Mat2b mRNA level; n = 4 in prevention group of CCl4 + DMSO for Foxm1 mRNA level; n = 6 in prevention and treatment groups of CCl4 + DMSO for Mat2α and Mat2b mRNA and treatment group of CCl4 + DMSO for Foxm1 mRNA; n = 5 in treatment group of CCl4 + FDI-6 for Foxm1 mRNA level; n = 6 in prevention and treatment groups of CCl4 + FDI-6 for Mat2α and Mat2b mRNA and prevention group of CCl4+FDI-6 for Foxm1 mRNA. f Protein levels of FOXM1, MATα2, MAT2β, α–SMA, F4/80, and COL1A1 from livers after prevention and treatment with FDI-6 (n = 3 independent experiments). Densitometry values for protein levels are summarized in Supplementary Fig. 3e, f. Statistical significance was determined by using two-tailed, unpaired Student’s t-test. Source data are provided as a Source Data file. Abbreviations: ALT alanine transaminase, AST aspartate aminotransferase.
Reciprocal positive regulation between FOXM1/MAT2A/MAT2B
To better understand how they crosstalk, we examined the relationship between FOXM1 and MAT2A/2B in LX-2 cells and primary cholangiocytes isolated from BDL livers. FOX binding sites are present in the human MAT2A (Supplementary Fig. 4a), MAT2B (Supplementary Fig. 4b), and FOXM1 promoter regions (Supplementary Fig. 5). Different fragments of these human promoter constructs were transfected in LX-2 cells and primary cholangiocytes and treatment with FDI-6 (5 μM) or FOXM1 siRNA significantly decreased MAT2A promoter activity with maximal effect seen with promoter fragment −271/+60 in LX-2 cells (Fig. 3a, left), MAT2B promoter activity with maximal effect seen with fragment −250/+3 (Fig. 3b, left), and FOXM1 promoter −1333/+107 fragment (Fig. 3c, left). Similar attenuation was seen in all three promoters in primary cholangiocytes after these treatments (Fig. 3a, right for Mat2a, Fig. 3b, right for Mat2b, Fig. 3c, right for Foxm1), suggesting important elements are within those regions of the MAT2A/MAT2B/FOXM1 promoters. Mutation at each of the FOX binding sites significantly attenuated the ability of FDI-6 and FOXM1 siRNA treatments to lower MAT2A/MAT2B/FOXM1 promoter activities (Fig. 3d–f). These results show that FOXM1 regulates MAT2A, MAT2B, and FOXM1 mainly via FOX binding sites.
MAT2A (a), MAT2B (b) and FOXM1 (c) promoter activities in LX-2 cells and primary cholangiocytes ± FDI-6 or siRNA treatment as described in Methods. Effects of mutating FOX elements in LX-2 cells are shown in (d) MAT2A, (e) MAT2B, and (f) FOXM1. Cells transfected with WT and mutant constructs were treated with siRNA against MAT2A, MAT2B, and FOXM1 and reporter activities were measured. Data presented as mean ± SEM (n = 3 per group). For a, Mat2a promoter activities of D-271/ + 60, D-671/ + 60 and D-1329/ + 60 in LX2 cell, SC + DMSO vs. SC + FDI-6, p = 0.0002, p = 0.0052 and p = 0.0948, respectively; SC + DMASO vs. SC + FOXM1 si, p = 0.0001, p = 0.0022 and p = 0.0003, respectively; Mat2a promoter activities of D-271/ + 60 in cholangiocytes, SC + DMSO vs. SC + FDI-6 or SC+Foxm1si, p = 0.0286 and 0.0149, respectively. For b, Mat2b promoter activities of D-25−/+3, D-713/+3, D-990/+3 and D-1319/+3 in LX2 cell, SC + DMSO vs. SC + FDI-6, p = 0.0051, p = 0.0316, p = 0.016 and p = 0.0034, respectively; SC + DMSO vs. SC + FOXM1 si, p = 0.0106, p = 0.0233, p = 0.0019 and p = 0.0049 respectively; Mat2b promoter activities of D-250/+3 in cholangiocytes, SC + DMSO vs. SC + FDI-6 or SC + Foxm1 si, p = 0.0002 and 0.023, respectively. For c, Foxm1 promoter activities of D-312/+107 and D-1333/+107 in LX2 cell, SC + DMSO vs. SC + FDI-6, p = 0.5799 and p = 0.0020, respectively; SC + DMSO vs. SC + FOXM1 si, p = 0.7684 and p = 0.0047 respectively; Foxm1 promoter activities of D-1333/ + 107 in cholangiocytes, SC + DMSO vs. SC + FDI-6 or SC + Foxm1 si, p = 0.0163 and 0.0131, respectively. For d, MAT2A promoter activities (−270/+60) of WT and MU, SC vs. SC, p = 0.015 and p = 0.28 respectively; FOXM1si vs. SC, p = 0.0032 and p = 0.042 respectively; MAT2Asi vs. SC, p = 0.0033 and p = 0.066 respectively. For e, MAT2B promoter activities (−250/+3) of WT and MU, SC vs. SC, p = 0.0017 and p = 0.11 respectively; FOXM1si vs. SC, p = 0.0018 and p = 0.022 respectively; MAT2Asi vs. SC, p = 0.0015 and p = 0.026 respectively. For f, FOXM1 promoter activities (−1333/+107) of WT and MU, SC vs. SC, p = 0.000017 and p = 0.0078 respectively; FOXM1si vs. SC, p = 0.000014 and p = 0.0063 respectively; MAT2Asi vs. SC, p = 0.00020 and p = 0.019 respectively. g ChIP assay was performed by spanning two FOX regions of the FOXM1 promoter in LX-2 cells using FOXM1, MATα2 and MAT2β antibodies after treatments that varied the expression of FOXM1, MAT2A or MAT2B in the top three rows. Seq-ChIP with anti-MATα2 and MAT2β antibodies after FOXM1 ChIP was performed as described in Methods. Representative results from three experiments are shown. h qPCR analysis of the ChIP assay from (g). For h, ChIP and Seq-ChIP percentage of input DNA, FOXM1 si vs. SC, MAT2A si vs. SC, MAT2B si vs. SC, FOXM1 OV vs. EV, MAT2A OV vs. EV, and MAT2B OV vs. EV, for FOXM1 ChIP, p = 0.00089, p = 0.011, p = 0.0045, p = 0.0000053, p = 0.00024, and p = 0.000039; for MAT2A seq-ChIP p = 0.0015, p = 0.0023, p = 0.0037, p = 0.000037, p = 0.00076, and p = 0.00032; for MAT2B seq-ChIP p = 0.0036, p = 0.018, p = 0.0092, p = 0.0036, p = 0.0000098, p = 0.0013 respectively. Data presented as mean ± SEM, *p < 0.05, **p < 0.01, and ***p < 0.001 vs. SC or EV (n = 3 independent experiments). i EMSA was done using labeled probes containing two FOX binding motifs of the FOXM1 promoter as shown in (f) and 100 ug of nuclear protein from LX-2 cells after treatments that varied FOXM1/MAT2A/MAT2B (n = 3 independent experiments). j Super shifts were done using 100 ng of recombinant proteins of FOXM1, MATα2, MAT2β alone or combined, and antibodies to FOXM1, MATα2 and MAT2β. Probe and IgG only served as negative controls. Results represent three independent experiments. k In vitro pull-down shows direct interaction between MATα2, MAT2β and FOXM1 using recombinant MATα2, MAT2β and FOXM1 proteins (n = 3 independent experiments). l MATα2, MAT2β and FOXM1 interaction in Flox control (WT), Foxm1Hep−/−, with or without BDL was detected by Co-IP and western blotting (n = 3 independent experiments). Statistical significance was determined by using two-tailed, unpaired Student’s t-test. Source data are provided as a Source Data file. Abbreviations: AB antibody, EV empty vector, IP immunoprecipitation, OV overexpression, si siRNA, WT wild type, MU mutants.
Since we found interaction between FOXM1, MATα2, and MAT2β in liver lysates (Supplementary Fig. 3f), we next examine if they interact at the two FOX binding sites of the FOXM1 promoter using chromatin immunoprecipitation (ChIP) and sequential ChIP (Seq-ChIP). We found that FOXM1 but not MATα2 or MAT2β can bind to the FOX region (Fig. 3g). However, in the presence of FOXM1, MATα2 and MAT2β co-occupy this region on Seq-ChIP (Fig. 3g). Furthermore, MAT2A, MAT2B, and FOXM1 overexpression (OV) led to an increase in FOXM1 and MATα2/2β binding and the opposite was true with MAT2A, MAT2B, and FOXM1 knockdown in LX-2 cells (Fig. 3g, h). FDI-6 treatment had the same results as FOXM1, MAT2A and MAT2B knockdown in LX-2 cells and cholangiocytes (Supplementary Fig. 6a, b). To confirm that these proteins were binding to the FOX element, we used electrophoretic mobility shift assay (EMSA), which confirmed that FOXM1, MAT2A and MAT2B siRNAs decreased and their overexpression increased protein binding to the FOX elements (Fig. 3i). Recombinant protein (r) of MATα2 or MAT2β cannot bind to the FOX element by itself. However, they can bind when combined with rFOXM1 as shown by supershift assay (Fig. 3j). Using co-immunoprecipitation (Co-IP) with purified recombinant proteins we found direct interaction between FOXM1 and MATα2 and MAT2β (Fig. 3k), and BDL increased their interaction in the liver (Fig. 3l). These results suggest these three proteins all interact at the FOX elements to activate the transcription of each other. Consistently, primary cholangiocytes isolated from BDL and sham control livers showed BDL increased cytoplasmic and nuclear FOXM1, MATα2 and MAT2β content (Supplementary Fig. 6c). Furthermore, FOXM1, MAT2A, and MAT2B overexpression increased each other’s protein expression and mRNA levels in LX-2 cells (Supplementary Fig. 7a), and the opposite was true with FOXM1, MAT2A, and MAT2B knockdown (Supplementary Fig. 7b). Consistent with the importance of FOXM1/MAT2A/MAT2B in HSCs activation, their siRNAs inhibited while overexpression raised migration of LX-2 cells (Supplementary Fig. 8). Lastly, FOXM1 (Supplementary Fig. 9a), MAT2A (Supplementary Fig. 9b) and MAT2B (Supplementary Fig. 9c) mRNA levels are all elevated in human liver cirrhosis (n = 31) and they positively correlate with each other (Supplementary Fig. 9d–f). Their high expressions in liver cirrhosis correlates with hepatitis B virus (HBV) infection (Supplementary Table 1).
Foxm1 Hep−/− mice have lower BDL-induced injury and fibrosis
Hepatocellular injury is a central mechanism of inflammation and disease progression in chronic liver diseases15. Hepatocytes, activated HSCs, and KCs cooperate in the establishment and resolution of liver fibrosis16. To explore how hepatocytes’ FOXM1 is involved in liver fibrosis, we generated Foxm1Hep−/− mice (Supplementary Fig. 10a). Foxm1Hep−/− mice subjected to BDL have lower CK19 (biliary/progenitor cell marker), α-SMA (a biomarker for myofibroblast differentiation), F4/80 (a marker of macrophages) and Sirius red staining (Fig. 4a, b), hydroxyproline (Fig. 4c), ALT (Fig. 4d), and AST (Fig. 4e) levels, lower F4/80 positive cells (Fig. 4f), CK19 and α-SMA area/total area (Fig. 4g) as compared to Flox controls after BDL. We next measured the expression levels of FOXM1, MATα2 and MAT2β in a cell-type specific manner from Foxm1Hep−/− mice at baseline and after BDL. Foxm1 mRNA and protein are not detected in hepatocytes or cholangiocytes (Fig. 4h–j). Foxm1Hep−/− mice have reduced protein and mRNA levels of Mat2a, and Mat2b in hepatocytes (Fig. 4h, i), cholangiocytes (Fig. 4h–j), and HSCs (Fig. 4h–k) after BDL but unchanged in KCs (Fig. 4h–l) as compared to Flox controls after BDL. See Supplementary Fig. 11a–d for a summary of densitometric changes of Fig. 4h.
a H&E, Sirius red, CK19, α-SMA, and F4/80 staining in Flox control and Foxm1Hep−/− mice after BDL as compared to sham surgery. Liver fibrosis was measured by Sirius red staining (n = 7 per group) (b) and hydroxyproline assay (n = 5 per group) (c), liver injury by ALT (n = 6 per group) (d) and AST (n = 6 per group) (e) levels, macrophage number by F4/80 (n = 6 per group) (f), and ductular proliferation by CK19 (n = 4 per group) and myofibroblast differentiation by α-SMA staining (n = 3 per group) (g). For b, Sirus red area/total area for Flox BDL vs. Sham and Hep−/− BDL vs. Flox BDL, p = 0.00000000011 and p = 0.0000015, respectively. For c, hydroxyproline (ug/g, liver) for Flox BDL vs. Sham and Hep−/− BDL vs. Flox BDL, p = 0.000058 and p = 0.0034, respectively. For d, ALT level (ug/L) for Flox BDL vs. Sham and Hep−/− BDL vs. Flox BDL, p = 0.000000000014 and p = 0.00000000097, respectively. For e, AST level (ug/L) for Flox BDL vs. Sham and Hep−/− BDL vs. Flox BDL, p = 0.0000000000001 and p = 0.00000025, respectively. For f, F4/80 positive number for Flox BDL vs. Sham and Hep−/− BDL vs. Flox BDL, p = 0.0000000020 and p = 0.00080, respectively. For g, CK19/total area for Flox BDL vs. Sham and Hep−/− BDL vs. Flox BDL, p = 0.00000078 and p = 0.00026, respectively. Data are shown as mean ± SEM, **p < 0.01, ***p < 0.001, ****p < 0.0001. h Protein expression of FOXM1, MATα2, and MAT2β in hepatocytes, cholangiocytes, HSCs, and KCs isolated from Flox control and Foxm1Hep−/− mice ± BDL, Densitometry values for protein levels are summarized in Supplementary Fig. 11a–d. mRNA levels of Foxm1, Mat2a, and Mat2b in hepatocytes (n = 6 animals in Flox + Sham, Foxm1hep−/− + Sham, Flox + BDL groups and Foxm1hep−/− + BDL group for foxm1 and mat2b mRNA; n = 4 in Flox + Sham, Foxm1hep−/− + Sham, Flox + BDL groups and n = 6 in foxm1hep−/−+BDL group for mat2a mRNA) (i), cholangiocytes (n = 4 animals in Flox + Sham, foxm1hep−/− + Sham, Flox + BDL groups and foxm1hep−/− + BDL groups for foxm1 mRNA; n = 5 in Flox + Sham and foxm1hep−/− + Sham groups and n = 4 in Flox + BDL and Foxm1hep-−/− + BDL groups for mat2a mRNA; n = 6 in Flox + Sham, Foxm1hep−/− + Sham, Flox + BDL groups and foxm1hep−/− + BDL group for mat2b mRNA) (j), HSCs (n = 6 animals) (k), and KCs (n = 4 animals in Flox + Sham, Flox + BDL groups, foxm1hep−/− + BDL groups and n = 5 in foxm1hep−/− + Sham for foxm1 mRNA; n = 5 in Flox + Sham and foxm1hep−/− + Sham groups, Flox + BDL and Foxm1hep−/− + BDL groups for mat2a mRNA; n = 5 in Flox + Sham, Flox + BDL and Foxm1hep−/− + BDL groups and n = 6 in foxm1hep−/− + Sham group for mat2b mRNA). l isolated from Flox control, Foxm1Hep−/− mice ± BDL. Data are shown as mean fold of Flox control ± SEM, *p < 0.05, ***p < 0.001, ****p < 0.0001. p values obtained via two-tailed unpaired Student’s t tests. For i, mRNA levels in hepatocytes, fold of Flox con of FOXM1, MAT2A, and MAT2B of Flox BDL vs. Sham p = 0.0000045, p = 0.000030, and p = 0.000028 respectively; of Hep−/− BDL vs. Flox BDL p = 0.0000000099, p = 0.00024, p = 0.000099 respectively. For j, mRNA levels in cholangiocytes, fold of Flox con of FOXM1, MAT2A, and MAT2B of Flox BDL vs. Sham p = 0.00023, p = 0.00000026, and p = 0.000000011, respectively; of Hep−/− BDL vs. Flox BDL p = 0.000041, p = 0.013, and p = 0.0000031, respectively. For k, mRNA levels in HSCs, fold of Flox con of FOXM1, MAT2A, and MAT2B of Flox BDL vs. Sham, p = 0.0000000000005, p = 0.000000067, and p = 0.0000000000 respectively; of Hep−/− BDL vs. Flox BDL p = 0.00000000050, p = 0.00025, and p = 0.0000000011 respectively. For l, mRNA levels in KCs, fold of Flox con of FOXM1, MAT2A, and MAT2B of Flox BDL vs. Sham, p = 0.000041, p = 0.000010, and p = 0.00000035 respectively; of Hep−/− BDL vs. Flox BDL p = 0.84, p = 0.16, and p = 0.62 respectively. Statistical significance was determined by using two-tailed unpaired Student’s t-test. Source data are provided as a Source Data file. Abbreviations: ALT alanine transaminase, AST aspartate aminotransferase, BDL bile duct ligation, Cho cholangiocytes, HSCs hepatic stellate cells, Hep hepatocytes, KCs Kupffer cells.
Foxm1 HSC−/− mice have lower BDL-induced injury and fibrosis
Activated HSCs release ECM, which is considered a key event in liver fibrosis. To better understand the role of HSCs FOXM1 in liver fibrosis, we generated Foxm1HSC−/− mice (Supplementary Fig. 10b). IHC results showed Foxm1HSC−/− mice have lower α-SMA and Sirius Red staining (Fig. 5a, b) and hydroxyproline level (Fig. 5c) after BDL, but F4/80 and CK19 staining were similar to Flox controls after BDL (Fig. 5a). ALT (Fig. 5d) and AST (Fig. 5e) levels were lower, F4/80 positive number (Fig. 5f) and CK19 area/total area (Fig. 5g) were similar but α-SMA area/total area was lower than Flox controls after BDL (Fig. 5g). FOXM1 protein (Fig. 5h) and mRNA (Fig. 5k) in HSCs after BDL are undetectable from Foxm1HSC−/−. Foxm1HSC−/− mice have lower FOXM1, MATα2 and MAT2β protein (Fig. 5h) and mRNA (Fig. 5i) levels in hepatocytes as compared to Flox controls after BDL but cholangiocytes (Fig. 5h–j) and KCs (Fig. 5h–l) were similar. See Supplementary Fig. 11e–h for a summary of densitometric changes of Fig. 5h.
a H&E, Sirius red, CK19, α-SMA, and F4/80 staining in Flox control and Foxm1HSC−/− mice after BDL as compared to sham surgery. Liver fibrosis was measured by Sirius red staining (n = 8 animals per group) (b) and hydroxyproline assay (n = 6 animals per group) (c), liver injury by ALT (n = 6 animals in Flox + Sham, Flox + BDL and Foxm1HSC−/− + BDL groups and n = 5 in Foxm1HSC−/− + Sham) (d) and AST (n = 7 in Flox + Sham, Flox + BDL groups and n = 6 in Flox + BDL group and n = 5 in foxm1HSC−/− + BDL) (e) levels, macrophage number by F4/80 (n = 4 animals per group) (f), and ductular proliferation by CK19 staining (n = 6 animals per group) and myofibroblast differentiation by α-SMA staining (n = 3 animals per group) (g). For b, Sirius red area/total area of Flox BDL vs. Sham and HSC−/− BDL vs. Flox BDL, p = 0.0000000015 and p = 0.0000060 respectively. For c, hydroxyproline (ug/g, liver) for Flox BDL vs. Sham and HSC−/− BDL vs. Flox BDL, p = 0.00000012 and p = 0.000094 respectively. For d, ALT level (ug/L) for Flox BDL vs. Sham and HSC−/− BDL vs. Flox BDL, p = 0.000000041 and p = 0.0000013 respectively. For e, AST level (ug/L) for Flox BDL vs. Sham and HSC−/− BDL vs. Flox BDL, p = 0.00000000066 and p = 0.00044 respectively. For f, F4/80 positive number for Flox BDL vs. Sham and HSC−/− BDL vs. Flox BDL, p = 0.0000000071 and p = 0.0073 respectively. For g, Flox BDL vs. Sham and HSC−/− BDL vs. Flox BDL for CK19/total area, p = 0.000000000075 and p = 0.080 respectively; for α-SMA area/total area, p = 0.000026 and p = 0.00011 respectively. Data are shown as mean ± SEM, ***p < 0.001, ****p < 0.0001. h Protein expression of FOXM1, MATα2, and MAT2β in hepatocytes, cholangiocytes, HSCs, and KCs isolated from Flox control and Foxm1HSC−/− mice ± BDL, Densitometry values for protein levels are summarized in Supplementary Fig. 11e, f. mRNA levels of Foxm1, Mat2a, and Mat2b in hepatocytes (n = 6 per group) (i), cholangiocytes (n = 6 per group) (j), HSCs (n = 6 per group) (k), and KCs (n = 6 per group) (l) isolated from Flox control and Foxm1HSC−/− mice ± BDL. Data are shown as mean fold of Flox control ± SEM, ****p < 0.0001, and ns not significant. p values obtained via two-tailed unpaired Student’s t tests. For i, mRNA levels in Hepatocytes, fold of Flox con of FOXM1, MAT2A, MAT2B for Flox BDL vs. Sham, p = 0.0000000000004, p = 0.0000000000001 and p = 0.0000000000072 respectively; for HSC−/− BDL vs Flox BDL, p = 0.000000000022, p = 0.000000000020, and p = 0.000000054 respectively. For j, mRNA levels in Cholangiocytes, fold of Flox con of FOXM1, MAT2A, and MAT2B for Flox BDL vs. Sham p = 0.0000000000000, p = 0.000000028, and p = 0.0000000085 respectively; for HSC−/− BDL vs Flox BDL, p = 0.17, p = 0.42, and p = 0.46 respectively. For k, mRNA levels in HSCs, fold of Flox con of FOXM1, MAT2A, and MAT2B for Flox BDL vs. Sham p = 0.0000052, p = 0.0000000000, and p = 0.0000000000 respectively; for HSC−/− BDL vs Flox BDL, p = 0.00000056, p = 0.00018, and p = 0.00000000002 respectively. For l, mRNA levels in KCs, fold of Flox con of FOXM1, MAT2A, and MAT2B for Flox BDL vs. Sham p = 0.000000000091, p = 0.00036, and p = 0.00000010 respectively; for HSC−/− BDL vs Flox BDL, p = 0.086, p = 0.61, and p = 0.15 respectively. Statistical significance was determined by using two-tailed unpaired Student’s t-test. Source data are provided as a Source Data file. Abbreviations: ALT alanine transaminase, AST aspartate aminotransferase, BDL bile duct ligation, Cho cholangiocytes, HSCs hepatic stellate cells; Hep hepatocytes, KCs Kupffer cells.
Foxm1 KC−/− mice have lower BDL-induced injury and fibrosis
KCs are resident macrophages that are localized within the lumen of the liver sinusoids, which play a vital role in liver inflammation16. In response to hepatocyte injury, KCs become activated and express cytokines and signaling molecules, such as tumor necrosis factor α (TNF-α), interleukin-1β (IL-1β), and interleukin-6 (IL-6)16. To examine the role of macrophage-derived FOXM1, we created Foxm1KC−/− mice (Supplementary Fig. 10c). IHC results showed Foxm1KC−/− mice have lower F4/80, Sirius red, and α-SMA staining after BDL but no change in CK19 staining (Fig. 6a, b, f, g). They have lower levels of hydroxyproline (Fig. 6c), ALT (Fig. 6d) and AST (Fig. 6e). FOXM1 protein (Fig. 6h) and mRNA (Fig. 6l) in KCs after BDL are undetectable in Foxm1KC−/− mice. However, Foxm1KC−/− mice have lower protein (Fig. 6h) and mRNA level of Foxm1, Mat2a, and Mat2b in hepatocytes (Fig. 6i) and HSCs (Fig. 6k) as compared to Flox controls after BDL, except for cholangiocytes (Fig. 6j), which were similar. See Supplementary Fig. 12a–d for a summary of densitometric changes of Fig. 6h.
a H&E, Sirius red, CK19, α-SMA, and F4/80 staining in Flox control and Foxm1KC−/− mice after BDL as compared to sham surgery. Liver fibrosis was measured by Sirius red staining (n = 8 animals per group), b and hydroxyproline assay (n = 5 animals in Flox + Sham group and n = 6 in foxm1KC−/− + Sham, Flox + BDL and foxm1KC−/− + BDL groups) (c), liver injury by ALT (n = 6 animals per group) (d) and AST (n = 6 animals per group) (e) levels, macrophage number by F4/80 (n = 6 animals per group) (f), and ductular proliferation by CK19 (n = 4 animals per group) and myofibroblast differentiation by α-SMA staining (n = 3 animals per group) (g). For b, Sirius red area/total area of Flox BDL vs. Sham and KC−/− BDL vs. Flox BDL, p = 0.00000000049 and p = 0.00014 respectively. For c, hydroxyproline (ug/g, liver) for Flox BDL vs. Sham and KC−/− BDL vs. Flox BDL, p = 0.00030 and p = 0.012 respectively. For d, ALT level (ug/L) for Flox BDL vs. Sham and KC−/− BDL vs. Flox BDL, p = 0.0000000000000 and p = 0.000000000081 respectively. For e, AST level (ug/L) for Flox BDL vs. Sham and KC−/− BDL vs. Flox BDL, p = 0.0000000000001 and p = 0.00000025 respectively. For f, F4/80 positive number for Flox BDL vs. Sham and KC−/− BDL vs. Flox BDL, p = 0.0000000067 and p = 0.0000094 respectively. For g, Flox BDL vs. Sham and KC−/− BDL vs. Flox BDL for CK19/total area, p = 0.00000010 and p = 0.039 respectively; for α-SMA area/total area, p = 0.000026 and p = 0.00033 respectively. Data are shown as mean ± SEM, *p < 0.05, ***p < 0.001, ****p < 0.0001. h Protein levels of FOXM1, MATα2, and MAT2β in hepatocytes, cholangiocytes, HSCs, and KCs isolated from Flox and Foxm1KC−/− mice ± BDL, Densitometry values for protein levels are summarized in Supplementary Fig. 12a–d. mRNA levels of Foxm1, Mat2a, and Mat2b in hepatocytes (n = 6 per group) (i), cholangiocytes (n = 6 per group) (j), HSCs (n = 6 per group) (k), and KCs (n = 6 per group) (l) isolated from Flox control and Foxm1KC−/− mice ± BDL. Data are shown as mean fold of Flox control ± SEM, ***p < 0.001, ****p < 0.0001, ns not significant. For i, mRNA levels in hepatocytes, fold of Flox con of FOXM1, MAT2A, MAT2B for Flox BDL vs. Sham, p = 0.000000012, p = 0.0000019 and p = 0.50 respectively; for KC−/− BDL vs Flox BDL, p = 0.00000086, p = 0.00060, and p = 0.0000000036 respectively. For j, mRNA levels in cholangiocytes, fold of Flox con of FOXM1, MAT2A, and MAT2B for Flox BDL vs. Sham p = 0.0000000000001, p = 0.0000000000005, and p = 0.00000000046 respectively; for KC−/− BDL vs Flox BDL, p = 0.071, p = 0.064, and p = 0.061 respectively. For k, mRNA levels in HSCs, fold of Flox con of FOXM1, MAT2A, and MAT2B for Flox BDL vs. Sham p = 0.0000000086, p = 0.0000000002, and p = 0.000000000 respectively; for KC−/− BDL vs Flox BDL, p = 0.0000032, p = 0.0000095, and p = 0.0000078 respectively. For l, mRNA levels in KCs, fold of Flox con of FOXM1, MAT2A, and MAT2B for Flox BDL vs. Sham p = 0.000000040, p = 0.000000033, and p = 0.00000075 respectively; for KC−/− BDL vs Flox BDL, p = 0.00000000071, p = 0.00000097, and p = 0.0000059, respectively. Statistical significance was determined by using two-tailed unpaired Student’s t-test. Source data are provided as a Source Data file. Abbreviations: ALT alanine transaminase, AST aspartate aminotransferase, BDL bile duct ligation, Cho cholangiocytes, HSCs hepatic stellate cells, Hep hepatocytes, KCs Kupffer cells.
FOXM1-MAT2A-MAT2B interplay drives fibrosis and inflammation
TGF-β1 promotes liver fibrosis by activating HSCs17. SMAD3 is the core transcription factor that mediates TGF-β signaling18. To see if MAT2A and MAT2B are important in FOXM1-driven fibrosis, we treated LX-2 cells with TGF-β1 (20 ng/ml) and found FOXM1, MATα2, MAT2β and p-SMAD3 levels were increased three hours after treatment, along with an increase in fibrosis markers such as α-SMA and COL1A1 (Fig. 7a, see Supplementary Fig. 12e for densitometric changes). Knocking down FOXM1, MAT2A, or MAT2B reduced protein expression of MATα2, MAT2β, SMAD3, p-SMAD3, α-SMA and COL1A1 (Fig. 7b, see Supplementary Fig. 12f for densitometric changes). To determine the effect of the FOXM1/MAT2A/MAT2B axis on TGF-β1-mediated HSC activation, we treated the LX-2 cells with MAT2A siRNA plus TGF-β1 or MAT2B siRNA plus TGF-β1. Knocking down either MAT2A or MAT2B attenuated TGF-β1-mediated induction of FOXM1, MATα2, MAT2β, SMAD3, p-SMAD3, α-SMA and COL1A1. FOXM1 overexpression further augmented TGF-β1-induced protein expression of MATα2, MAT2β, SMAD3, p-SMAD3, α-SMA and COL1A1, but not if MAT2A or MAT2B were silenced (Fig. 7c, see Supplementary Fig. 13a for densitometric changes). These results indicate the FOXM1/MAT2A/MAT2B axis is important in liver fibrosis induced by the TGF-β1 signaling pathway and MAT2A and MAT2B are required for FOXM1 to exert its pro-fibrogenic effect.
a Time courses of protein expression of FOXM1, MATα2, MAT2β, SMAD3, α–SMA, and COL1A1 after TGF-β1 treatment in LX-2 cells. b FOXM1, MATα2, MAT2β, SMAD3, α-SMA and COL1A1 protein levels after siRNA knockdown of FOXM1, MAT2A, or MAT2B in LX-2 cells after 24 h. c Protein levels of FOXM1, MATα2, MAT2β, SMAD3, α-SMA, and COL1A1 after FOXM1 overexpression with or without MAT2A or MAT2B siRNA knockdown and TGF–β1 treatment (20 ng/ml) for 24 h in LX-2 cells. d Effect of LPS on protein expression of FOXM1, MATα2, MAT2β, TNF-α, and IL-6 with or without siRNA knockdown of FOXM1, MAT2A or MAT2B in RAW 264.7 cells for 24 h. e Effects of LPS on protein expression of FOXM1, MATα2, MAT2β, TNF-α, and IL-6 with FOXM1 overexpression and MAT2A or MAT2B siRNA treatment for 24 h in RAW 264.7 cells (left panel) and KCs isolated from Flox control mice (right panel). Densitometry values for protein levels are summarized in Supplementary Fig. 12e, f and Supplementary Fig. 13a–d. n = 3 independent experiments. Source data are provided as a Source Data file.
Macrophages have emerged as the central players in sustaining and amplifying chronic inflammation, which is the hallmark of liver fibrosis19. Lipopolysaccharide (LPS) stimulates RAW 264.7 macrophages to release pro-inflammatory activation, such as TNF-α and IL-620. We next examined the FOXM1/MAT2A/MAT2B axis in inflammation, first using RAW 246.7 cells, and then confirming the results in primary KCs. Interestingly, knocking down FOXM1, MAT2A or MAT2B reduced the protein expression of FOXM1, MATα2, MAT2β, TNF-α, and IL-6 at baseline and attenuated or completely blocked LPS-induced expression of these proteins (Fig. 7d, see Supplementary Fig. 13b for densitometric changes). Importantly, FOXM1 overexpression recapitulated the effects of LPS on TNF-α, and IL-6 but silencing MAT2A or MAT2B significantly attenuated FOXM1-mediated induction (Fig. 7e, see Supplementary Fig. 13c, d for densitometric changes). Silencing Foxm1, Mat2a or Mat2b attenuated LPS-induced TNF-α, and IL-6 release (Supplementary Fig. 13e). These results indicate the FOXM1/MAT2A/MAT2B axis also participates in inflammation.
Liver cells secrete EVs containing FOXM1/MATα2/MAT2β
To understand why HSCs from Foxm1Hep−/−, hepatocytes from Foxm1HSC−/− and HSCs and hepatocytes from Foxm1KC−/− all have lower levels of FOXM1, MATα2 and MAT2β after BDL, we examined the possibility that these proteins are secreted in EVs and transferred to other cells. First, we isolated and cultured hepatocytes, HSCs, and KCs from WT and their respective KOs at day 14 of BDL. The following day EVs were isolated from culture medium and used to treat KO HSCs or hepatocytes with the following combinations: 1. HSCs from Foxm1HSC−/− treated with EVs from hepatocytes of WT and Foxm1Hep−/−, 2. hepatocytes from Foxm1Hep−/− treated with EVs from HSCs of WT and Foxm1HSC−/−, 3. HSCs from Foxm1HSC−/− treated with EVs from KCs of WT and Foxm1KC−/−, and 4. hepatocytes from Foxm1Hep−/− treated with EVs from KCs of WT and Foxm1KC−/−. We found cells treated with EVs from cell-type specific knockouts all resulted in lower expression of FOXM1, MATα2, and MAT2β as compared to cells treated with EVs from WT cells after BDL (Fig. 8a–d, see Supplementary Fig. 14a–d for densitometric changes). We also evaluated whether hepatocytes and HSCs can transfer EVs to KCs in vitro. Contrary to what we observed in vivo, the treatment of KCs with EVs from hepatocytes or HSCs from Flox mice after BDL showed an increase in the expression of FOXM1, MATα2 and MAT2β when compared to EVs from hepatocytes from Foxm1Hep−/− or HSCs from Foxm1HSC−/− after BDL (Fig. 8e, f, see Supplementary Fig. 14e, f for densitometric changes). After BDL, we also found that EVs isolated from hepatocytes of Foxm1Hep−/−, HSCs of Foxm1HSC−/−, and KCs of Foxm1KC−/− have lower protein expression of FOXM1, MATα2, and MAT2β (Supplementary Fig. 14g) as compared to EVs from respective WT cells. Characterization of these EVs suggest they are most likely exosomes based on the size distribution (Supplementary Fig. 14h). These results suggest that FOXM1, MATα2 and MAT2β are released by hepatocytes, HSCs and KCs in EVs and they can influence the expression of these proteins in other liver cell types.
a Following BDL and culture of hepatocytes from flox mice and Foxm1Hep−/−, EVs were extracted from the medium and used to treat HSCs from Foxm1HSC−/−. b Following BDL and culture of HSCs from flox mice and Foxm1HSC−/−, EVs were extracted from the medium and used to treat hepatocytes from Foxm1Hep−/−. c, d Following BDL and culture of KCs from flox mice and Foxm1KC−/−, EVs were extracted from the medium and used to treat (c) HSCs from Foxm1HSC−/− or (d) hepatocytes from Foxm1Hep−/−. e EVs extracted from hepatocytes of Flox and Foxm1Hep−/− after BDL were used to treat KCs from Foxm1KC−/− and (f) EVs extracted from HSCs of Flox and Foxm1HSC−/− after BDL were used to treat KCs from Foxm1KC−/−. All EV treatments were for 24 h after which protein expression of FOXM1, MATα2 and MAT2β were measured in cell lysates by western blotting. Densitometry values for protein levels are summarized in Supplementary Fig. 14a–f, n = 3 independent experiments (g) Summary of key findings showing the FOXM1/MAT2A/MAT2B axis in the different liver cell types driving liver inflammation and fibrosis. Source data are provided as a Source Data file.
Discussion
Liver fibrosis is associated with high morbidity and mortality, and liver transplantation is currently the most effective way to cure liver fibrosis21. Although FOXM1 is involved in multiple diseases22, such as pulmonary fibrosis, diabetes, vascular diseases, and psoriasis, earlier studies regarding FOXM1 mainly focused on its function in oncogenesis2,3,23,24,25,26. In recent studies, Kurahashi et al.3 identified hepatocyte FOXM1 as a crucial driver of chronic liver inflammation as hepatocyte specific FOXM1 transgenic mice developed spontaneous liver inflammation, fibrosis, and HCC. However, the level of expression in a transgenic model is not physiologic so the role of endogenous FOXM1 in liver fibrogenesis is still not confirmed. FDI-6 could inhibit corneal inflammation, and subsequently attenuated subconjunctival fibrosis8,9. However, how FOXM1 causes liver inflammation and fibrosis is uncertain and whether inhibiting FOXM1 can be effective in the treatment of existing liver fibrosis has not been investigated. We and others have shown MAT2A and MAT2B are essential for HSCs activation and liver fibrosis10,12,13,14. In the current work, we examined if there is interplay between FOXM1 and MAT2A/MAT2B. We also investigated the contribution of cell-type-specific FOXM1 expression in liver inflammation and fibrosis.
Consistent with previous reports, the expression of FOXM1, MAT2A and MAT2B is induced in human and murine liver fibrosis. Following BDL, the cytoplasmic and nuclear content of all three proteins increased. MAT2A is best known as the gene that encodes the α2 catalytic subunit of MATII, which catalyzes the biosynthesis of SAMe in non-hepatic cells and non-parenchymal cells of the liver11. We and others have shown that MATα2 is present in the nucleus where it can regulate gene expression via epigenetics and as a transcription factor27,28. MAT2B is best known as the gene encoding for the regulatory subunit β that regulates the activity of MATII by lowering its Km for methionine and Ki for SAMe10. MAT2A and MAT2B are often regulated in parallel, and both are overexpressed in many cancers10. MATα2 and MAT2β also stabilize each other29. Here we found they also regulate each other positively at the transcriptional level and form a positive regulatory loop with FOXM1 so that knocking down any of them can be effective in stopping the progression of liver fibrosis. Consistently, FDI-6 treatment, which is a specific inhibitor of FOXM1, lowered the cytoplasmic and nuclear expression of FOXM1, MATα2 and MAT2β in activated HSCs, LX-2 cells, and cholangiocytes after BDL. Importantly, FDI-6 was effective in preventing as well as stopping the progression and regressing already established liver fibrosis that was induced by three weeks of CCl4 treatment. These results suggest FDI-6 may be a potential therapy in liver fibrosis through inhibiting the FOXM1/MAT2A/2B axis.
FOXM1 activates transcription of many oncogenes, such as cell cycle-related genes, and interacts with proteins to enhance the oncogenesis via its winged helix DNA-binding domain3,30. We also showed that FOXM1 activates NF-κB and forms a complex with NF-κB to bind to the FOX elements of the FOXM1 and MAT1A promoters to activate the former but repress the latter, thereby explaining why FOXM1 and MAT1A exert reciprocal negative regulation against each other2. NF-κB subunit p65 was reported to trans-activate the MAT2A promoter10,14, and we have shown that overexpression of MAT2A increased NF-κB reporter activity28, suggesting NF-κB and MAT2A form a positive regulatory loop. In this work, we found that FOXM1, MAT2A and MAT2B exert reciprocal positive regulation on each other via FOX binding sites present in their promoters. MATα2 and MAT2β are not able to bind to the FOX element alone, but they were able to in the presence of FOXM1. This is because they can directly interact with FOXM1, as demonstrated by using recombinant proteins in Co-IP experiments. Interestingly, FDI-6 not only lowered the expression of the FOXM1, MAT2A and MAT2B, it also lowered their interaction. Human cirrhosis tissues also confirmed direct correlation between FOXM1, MAT2A, and MAT2B mRNA levels and upregulation of all three at the mRNA level. Interestingly, high levels of all three were noted in those with HBV infection. One plausible explanation is that the X protein of HBV was shown to increase MAT2A expression at the transcriptional level31. This could then further increase the expression of FOXM1 and MAT2B.
Hepatocytes damage underlies multiple acute and chronic liver diseases and contribute to disease progression15,32, such as inflammation and liver fibrosis. HSCs activation forms the basis of liver fibrosis. KCs also enhance hepatic fibrosis by promoting activated HSCs survival in an NF-κB-dependent manner16,33. In addition, IL-1 and TNF-α mediate the crosstalk between KCs and HSCs16. To better understand the role of FOXM1 in a cell-type specific manner, we constructed Foxm1Hep−/−, Foxm1HSC−/−, and Foxm1KC−/− mice. We found that all three cell-type-specific knockouts were able to attenuate liver injury as measured by ALT and AST to comparable degrees and lowered liver fibrosis induced by BDL. Foxm1Hep−/− mice also had less F4/80 number and ductular proliferation, which were unchanged in Foxm1HSC−/− mice whereas Foxm1KC−/− mice had less F4/80 number but no change in ductular proliferation. These results seem to suggest lowering F4/80 number or ductular proliferation were not the key determinants of protection in this model.
Unexpectedly we found deletion of Foxm1 in a cell-type-specific manner influenced the expression of FOXM1 in other cell types after BDL. Although we cannot rule out the possibility that there was leakiness in the Cre-lox system, our results suggest another explanation, namely exchange/transfer of FOXM1, MATα2 and MAT2β amongst different cell types via EVs. Consistently, HSCs and hepatocytes from respective knockouts had lower levels of FOXM1, MATα2 and MAT2β when treated with EVs from knockouts as compared to WTs after BDL. Indeed, EVs and their cargo transfer various cellular materials and signals (RNA, DNA, proteins, and lipids) between neighboring or distant cells, playing vital roles in the development of metabolic diseases and cancer34,35,36,37. Transfer of EVs enriched in FOXM1, MATα2 and MAT2β between hepatocytes, HSCs, and KCs can maintain sustained activation of HSCs and KCs. Other studies have shown hepatocytes, HSCs, and KCs interaction via EVs38,39. We cannot exclude the possibility that other components of the EVs are also involved to modulate the changes in the expression of these three proteins.
Also, it is interesting that in vivo after BDL, hepatocyte-specific and HSC-specific Foxm1 deletion attenuated Foxm1/Mat2a/Mat2b induction in each other but they had no influence on their expression in KCs. This is in contrast to in vitro treatment of KCs with EVs from hepatocytes or HSCs after BDL that clearly showed an increase in the expression of FOXM1, MATα2 and MAT2β as compared to EVs from KO hepatocytes or HSCs. One consideration is the spatial relationship of these cells. For instance, hepatocytes and HSCs are in close contact with each other but KCs are in the sinusoids, separated from the other two cell types by sinusoidal endothelial cells. However, KC-specific Foxm1 KO mice had lower expression of Foxm1/Mat2a/Mat2b in hepatocytes and HSCs after BDL. One possibility is that Foxm1 deleted KCs are releasing less proinflammatory cytokines which are known to induce Foxm1 and Mat2a2,10. Another observation is that In vivo the cholangiocytes do not appear to be influenced by the EVs released by the other three cell types. At present we are not sure of the underlying mechanisms, but one speculation is how the different cells receive their blood supply. Bile duct epithelial cells receive their blood supply from the hepatic artery40. It is possible that hepatocytes release EVs mainly into sinusoidal blood under our experimental conditions.
Since FOXM1, MAT2A and MAT2B are positively regulating each other and their encoded proteins all physically interact, we resorted to testing how essential they are in fibrogenesis and inflammation using overexpression and knockdown. TGF-β1 pathway plays an essential role in the pathogenesis of liver fibrosis and it is known to activate NF-κB subunit p65, which contributes to TGF-β1-induced HSCs activation10,14. Macrophage-derived TGF-β1 is the most potent known fibrogenic agonist16. LPS increases liver injury and inflammation via the release of inflammatory cytokines41, such as TNF-α and IL-6. We found FOXM1, MATα2 and MAT2β, α-SMA and COL1A1 were all increased at three hours and since TGF-β1 activates NF-κB, which activates both FOXM12 and MAT2A14, they may be induced simultaneously. However, after BDL, the induction in FOXM1 occurred slightly earlier, suggesting FOXM1 is induced first in vivo. Knocking down any of the three reduced fibrogenic markers and although FOXM1 overexpression can augment TGF-β1 signaling, this requires participation of MAT2A and MAT2B. A similar scenario also occurs with LPS-induced proinflammatory cytokines in macrophages, where knocking down any of the three attenuated/eliminated LPS’ effect and MAT2A and MAT2B are required for FOXM1 to exert its full pro-inflammatory effect. These results support the notion that the FOXM1-MAT2A-MAT2B axis is essential for liver fibrosis and inflammation.
Methods
The study protocol was approved by the Institutional Animal Care and Use Committee of CSMC (No. 8850).
Materials and reagents
See Supplementary Table 2 for a list of the reagents and sources.
Human samples
Microarrays for 16 healthy human liver (HHL), 24 cases of hepatitis and 28 cases of cirrhosis tissues were from US biolab Inc. (Cat# DLV20812A, Rockville, MD, https://usbiolab.com/index.php/tissue-array/product/liver/DLV20812a). Four cases of PSC and three cases of PBC human liver specimens obtained from liver biopsy from 2014–2019, and five healthy liver tissues obtained from surgical resection for patients suffering from intrahepatic ductal stones from 2018-2019 were obtained from the department of Pathology at the Xiangya Hospital Central South University, Changsha, Hunan province, China. The samples were embedded in paraffin and stored in the institutional biobank. PSC, PBC and healthy tissues were evaluated histologically to confirm presence or absence of cancer.
For mRNA assays of FOXM1, MAT2A and MAT2B in healthy human livers (NHL), ten case of healthy control samples were obtained from surgical resection of hepatic hemangioma. None of the healthy control individuals underwent preoperative chemotherapy, and liver histology demonstrated absence of both cirrhosis and malignancy. Liver tissues were immediately frozen in liquid nitrogen. Thirty-one patients with cirrhosis were diagnosed at Xiangya Hospital Central South University from 2016–2021. There was no bias in the selection of patients. The sex of the participants was self-reported, and neither sex nor gender was considered during participant selection. Liver biopsies were evaluated blindly by two expert hepatopathologists. Hepatitis B surface antigen (HBsAg) was measured with a commercial enzyme immunoassay (Shanghai Kehua Bio-engineering Co. Ltd Shanghai). Total RNA of NHL and cirrhosis was extracted from stored frozen liver specimens using QIAzol (Qiagen) reagent according to the manufacturer’s recommendations, and RNA quality and integrity were assessed using the RNA 6000 Nano assay on the Agilent 2100 Bioanalyzer. mRNA levels of FOXM1, MAT2A and MAT2B were measured by TaqMan. The research was conducted under both the Declarations of Helsinki and Istanbul, and the study protocol was approved by the IRB and the Medical Ethical Committee of Xiangya Hospital Central South University. All human materials were obtained with patients’ informed written consent.
Mice
Four-month-old male C57BL/6 littermates were used for this study. Male mice were used because they are more susceptible to liver fibrosis. Mice were housed with ad libitum access to food (diet: CAT#5053, Picolab, San Francisco, CA) and water and kept on a 12 h (8 a.m. to 8 p.m.) light-dark cycle in a temperature (72–770F) and humidity (42%) controlled room at the animal facility of the Cedars-Sinai Medical Center (CSMC). Care of animals was within institutional animal care committee guidelines. Mice were euthanized using ketamine (100 mg/kg, intraperitoneally).
Generation of cell-type-specific Foxm1 knockout mice
Generation of Foxm1 Hep−/− mice
-
1.
Foxm1fl/fl mice (Jackson Laboratory) were cross bred with Alb-Cre mice (Jackson Laboratory) transgenic mice. Foxm1fl/flCre-negative littermates served as WT controls. Cell type-specific knockouts were confirmed by isolation of liver cells and messenger RNA (mRNA) expression. Twelve-week-old mice were used for the BDL experiments. All animal experiments were approved by the Institutional Review Board of the Cedars-Sinai Medical Center. Sequences of primers of forward 5’-TGGCTTCCCAGCAGTACAAATC-3’ and reverse 5’-TGCTTACAAAAGACACACTTGGACG’. The PCR reaction consisted of an initial denaturation at 94 °C for 3 min followed by 35 cycles at 95 °C for 30 s, annealing at 60 °C for one minute and extension at 72 °C for one minute using the 2XTaq red master (Apex, El Cajon, CA, Cat #: 42-138). PCR analysis of Foxm1Hep−/−, Foxm1Hep+/−and Foxm1Hep+/+ depicted in Supplementary Fig. 10a.
Generation of Foxm1 HSC−/− mice
-
2.
Foxm1fl/fl mice were crossed with Lrat-Cre transgenic mice (kindly provided by Dr. Robert Schwabe, Columbia University). Mice were back-crossed at least 10 generations onto the C57BL/6 background. Foxm1fl/flCre-negative littermates served as WT controls. Sequences of primers of forward 5’-CCTTTCTTTGACCCCCTGCAG-3’ and reverse 5’-GACCGGCAAACGGACAGAAG’. The PCR reaction is same as Foxm1Hep−/−. PCR analysis of Foxm1HSC−/−, Foxm1HSC+/−and Foxm1HSC+/+ depicted Supplementary Fig. 10b.
Generation of Foxm1 KC−/− mice
-
3.
Foxm1fl/fl mice were cross bred with Clec4f-Cre (Jackson Laboratory) transgenic mice. Foxm1fl/flCre-negative littermates served as WT controls. Sequences of primers for mutant reverse 5’-ACACCGGCCTTATTCCAAG-3’, wild type reverse 5’-GAAAGACCCAAGGGAAGGAG-3’ and common 5’-CAAGAAGTCCACAGGGTGGT-3’. The PCR reaction is same as Foxm1Hep−/−. PCR analysis of Foxm1KC−/−, Foxm1KC+/−and Foxm1KC+/+ depicted in Supplementary Fig. 10c.
Genomic DNA from mouse ears was isolated and amplified by PCR for analysis of Foxm1Hep−/−, Foxm1HSC−/−, and Foxm1KC−/− genotypes.
Carbon tetrachloride treatment and BDL
Mice were housed in a room maintained at a temperature of 22 °C and relative humidity of 50 ± 10% with alternating 12 h of light and darkness. Mice were acclimatized for one week prior to use and had free access to food and water during the entire experiment. In the CCl4 groups, mice were intraperitoneally (i.p) injected with 0.3% CCl4 (10 mL/kg, dissolved in corn oil). 64 mice at 8-weeks of age were randomly assigned to two groups: a prevention group and a treatment group. Prevention and treatment groups were divided into subgroups of Oil + DMSO, Oil + FDI-6, Oil + CCl4 and CCl4 + FDI-6. CCl4 was given via i.p. injection three times/week for 3 weeks in prevention group and 5 weeks in the treatment group. FDI-6 was dissolved in 10% DMSO and diluted in 90% corn oil (final concentration: 2.5 mg/mL). FDI-6 treatment was given at 25 mg/kg/d via direct i.p. injection three times/week started at the same time as CCl4 in the prevention group versus after 3 weeks of CCl4 in the treatment group and continued with CCl4 for 2 weeks.
For BDL surgery, 32 mice at 3-months of age from Foxm1Hep−/−, Foxm1HSC−/−, Foxm1KC−/− or flox controls were fed chow ad libitum and housed at constant temperature (22 °C) with alternating 12 h of light and darkness. Following i.p. injection of ketamine (80 mg/kg) and xylazine (10 mg/kg), the common bile duct was exposed through a midline abdominal incision, ligated in two places with a silk thread and sectioned between the ligatures. Sham operated mice had anesthesia and laparotomy only. The serum and partial liver tissues were used for assays of ALT, AST, hydroxyproline and IHC. The hepatic cells were isolated for studies described below and used for western blots and mRNA determinations.
For time-curse of FOXM1, MAT2α and MAT2β during BDL, C57BL/6 mice (9–10 weeks) were anesthetized with isoflurane and subjected to midline laparotomy (~1 cm) to expose the common bile duct (CBD). Two knots were made on the CBD using a 5–0 silk suture and then, the CBD was cut between the two knots. The peritoneum was re-aligned, and the underneath muscle layers and skin were closed individually using a 5–0 silk suture. Mice. Briefly, mice were fed ad libitum with diet for 5 days and then divided into six groups of the day 0, 1, 2, 3, 7 and 14 (n = 6 per group, 36 total). Sham operation was on day 0. Sera and liver tissues were collected for various assays and cell isolation. Eight Flox male mice at three months of age were used for cholangiocyte isolation and promoter assays.
All procedure protocols, use, and the care of the animals were reviewed and approved by the Institutional Animal Care and Use Committee at Cedars-Sinai Medical Center and all experiments involving research animals were conducted in accordance with all relevant ethical regulations. All mice were housed under 12-h light/12-h dark cycle at an average temperature of 74 0F and 40% humidity.
Isolation of different liver cell types
Foxm1Hep−/−, Foxm1HSC−/−, Foxm1KC−/−, and their corresponding Flox are subjected to BDL for seven days. Isolations of primary HSCs from three-months old male knockout and wild-type animals were performed by pronase-collagenase perfusion followed by density gradient centrifugation. Briefly, after in situ perfusion of the liver with 1 mg/ml pronase (CAT#: 1145963001, Roche, Indianapolis, IN), followed by 0.35 mg/mL collagenase (CAT#: C9891-1G, Sigma), dispersed cell suspensions were layered on a discontinuous density gradient Optiprep solution (CAT#: 07820, Serumwerk, Bernburg) and washed with 1XHBSS (CAT#: 21-021-CV, CORNING). HSCs were collected from the gradient interface, and cell viability was verified by phase-contrast microscopy as well as trypan blue staining. The viability of all cell cultures used for the studies was >95%. For EVs isolation, the cells were plated at a density of 4,000,000/10 cm dish. The cells were kept in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) and antibiotics in a humidified atmosphere with 5% CO2 and 21% O2 at 37 °C for 24 h and then changed to serum free DMEM media for 24 h. To isolate Kupffer cells, animals were performed by collagenase perfusion followed by low spin at 50 g for 5 min. The supernatants were centrifugated at 800 g for 10 min. The cell pellet was followed by density gradient centrifugation at a speed of 16,000 for 15 min. The cell fraction at middle layer was incubated with anti-F4/80 microbeads (Milenyl Biotec, Auburn, CA) antibody for 15 min in the dark in the refrigerator (2–8 °C). The cells were washed with 1–2 ml of buffer (a solution containing phosphate-buffered saline (PBS), pH 7.2, 0.5% bovine serum albumin (BSA) and 2 mM EDTA) and spun at 300 g for 10 min and aspirated supernatant completely. For EVs isolation, the cells were plated at a density of 5,000,000/10 cm dish. To isolate hepatocytes, animals were performed by collagenase perfusion followed by low spin at 50 g for 5 min. The cell pellet was washed by Hanks’ balanced salt solution (HBSS) twice. The cells were plated at a density of 3,000,000/10 cm dish. Cell culture condition for KCs and hepatocytes was the same as HSCs. Cell pellets were immediately stored at −80 °C for RNA or protein measurements.
Cell lines
LX-2 cells were kindly provided by Dr. Ekihiro Seki and the mouse macrophage cell line RAW 264.7 was purchased from American Type Cell Collection (ATCC, Manassas, VA). Both cell lines were cultured in DMEM containing 10% fetal bovine serum (FBS) and antibiotics (2 mM glutamine, 50 mM penicillin, and 50 mg/ml streptomycin sulfate).
Measurements of liver injury and fibrosis
Liver samples from CCl4 with or without FDI-6 treatments with three kinds of Foxm1 cell specific knockout or Flox mice were homogenized in 6 N HCl, hydrolyzed overnight at 100 °C, and centrifuged at 10,000 × g for 3 min. Supernatants were transferred to a 96-well plate and analyzed following the Hydroxyproline assay kit protocol (MAR008-1KT, Sigma). Hydroxyproline content was measured using a CLARIO Star machine (absorbance at 560 nm) and normalized to liver weight. The ALT and AST levels in serum were determined using ALT kit (CAT# TR71121, ThermoFisher) and AST kit (CAT# TR70121, ThermoFisher), respectively. The collagen proportional area (CPA) was defined as the percentage of the area positive for Sirius red staining. Sirius Red area/total area was quantified according to the value of CPA42.
Real-time quantitative PCR (qPCR)
Total RNA was extracted using TRIzol reagent (ThermoFisher) or Quick-RNA miniPrep kit (Zymo Research, Irvine, CA) from liver tissues, hepatic cells of Foxm1Hep−/−, Foxm1HSC−/−, and Foxm1KC−/−, or LX-2 (human activated HSCs) and RAW 264.7 (murine macrophages) cells. Human and mouse probes for MAT2A, MAT2B and FOXM1, and the PCR Supermix were purchased from ThermoFisher. Hypoxanthine phosphoribosyltransferase 1 (HPRT1) was included as a housekeeping gene. The thermal profile comprised of initial denaturation at 95 °C for 3 min followed by 40 cycles at 95 °C for 3 s and at 60 °C for 30 s. The cycle threshold (Ct value) of the target gene was normalized to that of the HPRT1 gene to obtain the delta Ct (DCt). Relative genes expression was calculated with 2−ΔΔCT method and normalized to HPRT1 expression (ΔΔCt = ΔCt of target genes – ΔCt of HPRT1).
Western blot analysis
Western blot analysis was done with antibodies against FOXM1 (ab207298; CAT#: 13147-1-AP, Proteintech), MATα2 (55309-1-AP), MAT2β (BNP1-82797), α-SMA (ab5831), COL1A1 (ab270993), β-actin (ab8226), F4/80 (ab300421), SMAD3 (ab52903), tumor necrosis factor alpha (TNF-α, ab183218), tubulin (ab18251) and interleukin 6 (IL-6, ab259341). The membranes were incubated with secondary antibody (mouse anti-rabbit IgG, Cell Signaling, #7074, USA) for 40 min at room temperature. Membranes were stripped and re-probed with antibodies against β-actin or histone H3 as housekeeping control or CD9 as EVs control. A chemiluminescence system (Millipore Corporation, Billerica, MA, USA) was used for signal detection. All results were quantified by ImageJ software (versions 1.51 and 1.54j). Statistical significance was defined as p < 0.05.
Morphological and immunohistochemical analysis
All human and mouse liver tissues were embedded in paraffin, sectioned, and stained with hematoxylin & eosin (H&E), Sirius red, and F4/80. Five μM paraffin sections of the liver tissue samples were used for IHC staining. Slides were immunostained with antibodies to FOXM1, MATα2, MAT2β, COL1A1, EMR1 (F4/80), α-SMA, Keratin 19 (CK19, ab52625), and IgG according to an IHC detection kit (ab64264). All sections were counterstained with hematoxylin for 30 s and were observed under Digital Inverted Fluorescence Microscope (ThermoFisher, AMF4300). No staining occurred with normal mouse IgG. For every slide, 10 representative fields were captured randomly and analyzed by Image-Pro Plus v6.0 software.
Promoter analysis
The −1329/+60 PCR product representing the MAT2A 5’-flanking region was cloned in the sense orientation upstream of the luciferase coding sequence of the pGL3-basic vector (Promega)43. To produce 5’-deletion constructs, MAT2A promoter was digested with NheI and Afl II to produce a −939 to /+60 deletion, NheI and EcoRV to produce a −571 to /+60 deletion, and NheI and Apa I (−47 to /+60 deletion. −271/ + 60 bp deletion was produced using forward primer 5’-CATCAAACAAGGAAGAGCAATCC-3’ (−250 to −271 bp relative to transcription start site) and reverse primer 5’-AGCTGCGGACAGCGTTCTACTCG-3’ ( + 39 to +60 bp relative to transcription start site). Mutagenesis of the MAT2A promoter FOX binding sites were performed as follows: 5’- CTATAAA-3’ (−16 to −22) was changed to 5’-CTATCAA-3’ using forward primer 5’- GGCGCTGCTCTATCAATACCGGGCC-3’ (−7 to −30 bp relative to transcription start site); 5’- CATCAAACA-3’ (−264 to −272) was changed to 5’- CATCCAACA -3’ using forward primer 5’- TTTCTCCCACATCCAACAAGGAAGAGC -3’, (−255 to −281). A −1319/ + 3 5′-flanking region of the human MAT2B was cloned into the SmaI site of pGL-3 basic vector43. MAT2B promoter was subcloned with forward primers from −990 to –967, −713 to –688, and −250 to –225, and reverse primer was +3 to −20 to generate deletion constructs −990/+3, −713/+3, and −250/+3 MAT2B-LUC, respectively. All sequences are relative to the ATG start codon. Mutagenesis of the MAT2B promoter FOX binding site was performed as follows: 5’- CAAATA-3’ was changed to 5’-CAACTA-3’ (−140 to −145) using forward primer 5’-AATAAAAAGCACTCAACTAAAATCTCCGAAAC-3’ (−127 to −158) with QuikChange multisite-Directed mutagenesis kit (#200515-5) from Agilent Technologies (Carpinteria, CA). The mutant strand synthesis reaction consisted of an initial denaturation at 95 °C for 30 s followed by 16 cycles at 95 °C for 30 s, annealing at 55 °C for one minute and extension at 68 °C for one minute/kb of plasmid length using the PfuUltra HF DNA polymerase. DpnI digestion of the amplified PCR products and transformation of XL10-gold ultracompetent cells were done in accordance with their suggested protocol (Agilent Technologies). The human 1.4 kb FOXM1 promoter was purchased from GeneCopoeia (Rockville, MD). FOXM1 promoter was subcloned with forward primers from −1304 to –1282 and −123 to –102, and reverse primer was +89 to +107 to generate deletion constructs −1304/+107, and −123/+107 FOXM1-LUC, respectively. Mutagenesis of the FOX binding sites in the FOXM1 promoter were performed as follows: 5’-TGTTTG-3’ (−1022 to −1027) was changed to 5’-GGTTTG-3’ using forward primer 5’- GCAATAATTCAACATTGGTTTGTTTTGGAGAC-3’ (−1012 to −1043 bp relative to transcription start site), and 5’-TGTTTA-3’ (-732 to -737) was changed to 5’-TGTTGA-3’ using forward primer 5’-GCCCACATTTGTTGATTTGATTAAAATGTC-3’, (−717 to −751), with QuikChange multisite-Directed mutagenesis kit (Agilent Technologies, Inc. Santa Clara, CA). Plasmids were transfected into cells using Lipofectamine 2000 reagent (Invitrogen, #11668019, MA). Promoter constructs and pGL3/pLuc-MCS/PEZ/SV40 (1 μg) were then transfected into LX-2 and primary cholangiocytes using JetPRIME transfection reagent (Polyplus Transfection, #101000027, USA). Luciferase assays were performed by the Dual Luciferase Reporter Assay System (Promega, #E190, USA) 24 h after transfection.
Isolation of extracellular vesicles (EVs) and characterization
Since FOXM1, MATα2 and MAT2β expression increased dramatically in hepatocytes, HSCs, and KCs after BDL, we isolated EVs from these cells after BDL and used them to treat other liver cell types. We used commercial kits (Thermo Fisher cat# 4484450, 4478360; SBI cat# EQ806A-1) for EVs isolation. Briefly, serum-free media was collected and centrifuged at 2000 g for 30 min to remove the cell debris. Then we further concentrated the 150 ml supernatant media to 1 ml with MilliporeSigma UFC701008 Centricon Plus-70 Centrifugal Filter (100 kd)44. The concentrated medium was then processed with the SBI kit according to the provided protocol. The concentration and size distribution of EVs were assessed using Nanoparticle Tracking Analysis (NTA) via NanoSight NS300. Validation of EVs was performed through western blot analysis employing EVs markers45.
We used 10 μg of EVs to treat hepatocytes, HSCs, and KCs. HSCs from Foxm1HSC−/− were treated with EVs from hepatocytes of Foxm1Hep−/− and Flox. Hepatocytes from Foxm1Hep−/− were treated with EVs from HSCs of Foxm1HSC−/− and Flox. Hepatocytes from Foxm1Hep−/− and HSCs from Foxm1HSC−/− were treated with EVs from Flox and Foxm1KC−/−. Expression of FOXM1, MATα2, and MAT2β were measured by western blot in HSCs, hepatocytes, and KCs.
ChIP and sequential-ChIP (Seq-ChIP)
LX-2 cells were crosslinked with formaldehyde to bind the proteins to the DNA. The fragmentation of DNA was made by cell sonication (shearing of DNA into small fragments ranging from 50 to 500 bp). Immunoprecipitation (IP) using FOXM1, MATα2, and MAT2β and lgG antibodies enriches the fragments bound by the protein. Then, crosslink reversal frees the DNA fragments for further processing. ChIP and Seq-ChIP were performed using a EpiTect ChIP OneDay kit (Qiagen, Germantown, MD) in accordance with the kit instructions. For the FOX element of the human FOXM1 promoter, DNA immunoprecipitation (IP) was performed using antibodies against MATα2, MAT2β and FOXM1. lgG was used for control. This was followed by a second round of IP using antibodies against MATα2, MAT2β and FOXM1. The purified DNA was detected by PCR analysis. PCR primers for promoter regions containing FOX binding sites were FOXM1 - forward 5’- CATTTGTTTGTTTTGGAGACGGTGTC-3’ (−1006 to −1031) and reverse 5’-GGAAGAGGGGCACAGACATTTTAATC-3’ (−703 to −728) (GenBank® accession no. NM-001243088). All PCR products were electrophoresed on 2% agarose gel. The PCR protocol involved initial denaturation at 94 °C for 3 min and 25 cycles at 94 °C for 30 s, followed by annealing and extension at 65 °C for 90 s using the Advantage GC 2 PCR kit (Clontech, Mountain View, CA).
For ChIP-qPCR assays, chromatin was prepared from LX-2 cells treated with scrambled control, siRNAs of MAT2A, MAT2B and FOXM1, overexpression of MAT2A, MAT2B and FOXM1, or empty vector. Chromatin was enzymatically sheared and immunoprecipitated with anti-MATα2, anti-MAT2β and anti-FOXM1 using Pierce™ Magnetic ChIP Kit (Cat#: 26157, ThermoFisher Scientific) in accordance with the kit’s instructions. Rabbit IgG was used as a mock antibody for negative control. Immunoprecipitated chromatin was subjected to real-time qPCR with the SYBR Premix DimerEraser (RR091A, Takara). Two ChIP-qPCR primer pairs (forward primer: 5’- CATTTGTTTGTTTTGGAGACGGTGTC-3’ (−1006 to −1031) and reverse primer: 5’- ATAAACAAATGTGGGCTGGGCATAG-3’ (−730 to −755)) overlap two FOX binding sites of the human FOXM1 promoter region. Samples were run in triplicates, and data from MATα2, MAT2β and FOXM1 IP, and control IP were presented as enrichment relative to input DNA. ChIP-qPCR was repeated triplicates to confirm the reproducibility of results. The quality of chromatin enzymatically sheared was assessed using agarose gel electrophoresis. The percent input is used to normalize ChIP-qPCR data. 1% of starting chromatin is used as input. ChIP-qPCR data is normalized for both background levels and input chromatin going into the ChIP. Pierce™ Magnetic ChIP Kit (Cat#: 26157).
Electrophoretic mobility shift assay (EMSA)
In 10 cm dishes, LX-2 cells were transfected with scrambled control, siRNA (si) against FOXM1, MAT2A, or MAT2B, empty vector (EV), FOXM1 OV, MAT2A OV, or MAT2B OV for 48 h. LX-2 cells were grown to 70–80% and after removing the medium, cells were washed twice with ice-cold PBS and scraped into a 15 mL conical tube and centrifuged at 1000 rpm for 5 min. The nuclear proteins from LX-2 cells were isolated using the Nuclear-Extraction kit (Abcam, ab113473). Double FOX binding sites (5’-ACATTTGTTTGTTTTGACATTTGTTTATTTGA-3’) were used for gel shift and supper shift probe. EMSA assays were done by following the EMSA Kit (ThermoFisher Scientific, E33075). The shift bands were detected from the gel images with three biological replicates. The shifted bands were compared with that of the non-shifted band in each lane46,47.
MTT and migration assays
For MTT assays, LX-2 Cells were cultured at 1 × 106 cells per ml in a transparent plate and rinsed twice with PBS. 50 µL serum-free medium and 50 µL MTT reagent was added according to manuscript (ab211091, USA). After 3 h of incubation at 37 °C, 150 µL of MTT solvent was added to each well. Plate was wrapped in foil and shaken for 15 min on an orbital shaker. Data was read at OD = 590 nm. For migration assays, LX-2 cells (1 × 105) were seeded into transwell inserts (8-μm pore size, BD Falcon). After 24 h of culture for the migration assays, the wells were stained with crystal violet (Beyotime, C0121). Migrated cells number was quantified using the ImageJ software.
Immunofluorescence staining
Primary HSCs and LX-2 cells were plated on 6-well plates containing coverslips. Cells were fixed with 4% paraformaldehyde for 30 min at room temperature and ice-cold methanol for 15 min. Cells were then blocked by incubation in PBS with 0.05% TWEEN® 20 (PBST, Sigma, P3563) containing 10% goat serum and 3% BSA for 45 min. Samples were incubated with FOXM1, MATα2, and MAT2β antibodies at 250-time dilution (1:250) in PBS for 1 h at 37 °C and incubated with fluorescent secondary antibodies (Invitrogen, Waltham, MA) for 3 h. Negative control was used with IgG antibody (Abcam, ab171870; Rabbit). VECTASHIELD mounting medium (Sigma, #DUO82040) containing DAPI was used to mount slides. EVOS™ FL and EVOS™ FL color Imaging systems (ThermoFisher Scientific, AMF4300) were used to capture images. Photoshop (version 23.5.2) overlay method was used to contrast and overlay images.
FOXM1-MATα2-MAT2β interactions
40 μL of A/G beads (Cat#: SC-2003, Santa Cruz Biotechnology), 500 μL IP buffer (150 mM NaCl, 50 mM Tris-HCl PH7.6, 1 mM EDTA PH7.0, 1 mM EGTA7.4, 25 mM NaF, 1% NP-40), and 2 μg of MATα2, MAT2β or FOXM1 antibody were mixed and rotated at 4 °C for one hour. The beads were washed 6 times with 500 μL IP buffer. For recombinant proteins (FOXM1, MATα2, and MAT2β) interactions, after the final wash with 500 μL of IP buffer above, 1 μg of recombinant protein was added. For the extracted proteins, 500 μL of IP buffer and 500 μg lysates were needed. 500 μL of IP buffer and 0.5 μL of rabbit IgG were mixed as control. All the reaction tubes were rotated at 4 °C overnight. Then the beads were washed 6 times with 500 μL IP buffer. After the last wash buffer was removed and beads were boiled in 2× SDS sample buffer for 10 min at 95 °C, and proteins were separated on 10% SDS-PAGE followed by Immunoblot analysis for MATα2, MAT2β and FOXM1.
Enzyme-linked immunosorbent assay (ELISA)
Raw 264.7 cells were cultivated in high-glucose DMEM containing 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37 °C in a humidified incubator containing 5% CO2. After overnight culture in a 96-well plate (2 × 104 cells/well, 200 μL medium/well), cells were transfected with siRNAs of scrambled, FOXM1, MAT2A and MAT2B for 8 h and treated with LPS (Escherichia coli, Sigma-Aldrich L4130 0111: B4) at 1 μg/ml for an additional 16 h. The culture supernatant from each well was collected at the end of scheduled experiments and used to measure TNF-α (KE10002) and IL-6 (KE10091, proteintech, Rosemont, IL, USA) concentration by ELISA according to the manufacturer’s instructions, and the absorbance was measured at 450 nm and 630 nm, respectively, using a microplate reader.
Statistical analysis
All sample data were presented as mean ± standard error of the mean (SEM). Sample data statistical analysis was used Student t-test and one-way ANOVA, followed by Fisher’s test for multiple comparisons. Chi-square test was used to analyze mRNA levels of FOXM1, MAT2A or MAT2B and clinicopathological features in liver cirrhosis. Pearson correlation was used to measure the strength of the linear relationship between FOXM1 and MAT2A, FOXM1 and MAT2B, MAT2A and MAT2B in human liver cirrhosis tissues. All experiments were repeated at least three times, and p < 0.05 was considered statistical significance. Excel (16.54) and Graphpad Prism 9.0.0 were used for calculations.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the findings described in this manuscript are available in the article and in the Supplementary Information and from the corresponding author upon request. Source data are provided with this paper.
References
Friedman, S. L. Liver fibrosis – from bench to bedside. J. Hepatol. 38, S38–S53 (2003).
Li, Y. et al. Reciprocal regulation between forkhead box M1/NF-κB and methionine adenosyltransferase 1A drives liver cancer. Hepatology 72, 1682–1700 (2020).
Kurahashi, T. et al. Forkhead box M1 transcription factor drives liver inflammation linking to hepatocarcinogenesis in mice. Cell. Mol. Gastroenterol. Hepatol. 9, 425–446 (2020).
Sun, W. et al. ALKBH5 promotes lung fibroblast activation and silica-induced pulmonary fibrosis through miR-320a-3p and FOXM1. Cell. Mol. Biol. Lett. 27, 26 (2022).
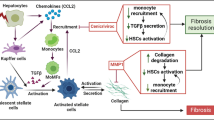
Filliol, A. & Schwabe, R. F. FoxM1 induces CCl2 secretion from hepatocytes triggering hepatic inflammation, injury, fibrosis, and liver cancer. Cell. Mol. Gastroenterol. Hepatol. 9, 555–556 (2020).
Wang, Y., Zhou, Q., Tang, R., Huang, Y. & He, T. FoxM1 inhibition ameliorates renal interstitial fibrosis by decreasing extracellular matrix and epithelial-mesenchymal transition. J. Pharmacol. Sci. 143, 281–289 (2020).
Gormally, M. V. et al. Suppression of the FOXM1 transcriptional programme via novel small molecule inhibition. Nat. Commun. 5, 5165 (2014).
Lan, C. et al. Forkhead domain inhibitor-6 suppresses corneal neovascularization and subsequent fibrosis after alkali burn in rats. Invest. Ophthalmol. Vis. Sci. 63, 14 (2022).
Lan, C. et al. Forkhead domain inhibitory-6 attenuates subconjunctival fibrosis in rabbit model with trabeculectomy. Exp. Eye. Res. 210, 108725 (2021).
Murray, B., Barbier-Torres, L., Fan, W., Mato, J. M. & Lu, S. C. Methionine adenosyltransferases in liver cancer. World J. Gastroenterol. 25, 4300–4319 (2019).
Mato, J. M. & Lu, S. C. Role of S-adenosyl-L-methionine in liver health and injury. Hepatology 45, 1306–1312 (2007).
Ramani, K., Donoyan, S., Tomasi, M. L. & Park, S. Role of methionine adenosyltransferase α2 and β phosphorylation and stabilization in human hepatic stellate cell trans-differentiation. J. Cell. Physiol. 230, 1075–1085 (2015).
Hu, X. & Zhou, Y. Curcumin reduces methionine adenosyltransferase 2B expression by interrupting phosphorylation of p38 MAPK in hepatic stellate cells. Eur. J. Pharmacol. 886, 173424 (2020).
Wang, K. et al. TGF-β1/p65/MAT2A pathway regulates liver fibrogenesis via intracellular SAM. EBioMedicine 42, 458–469 (2019).
Luedde, T., Kaplowitz, N. & Schwabe, R. F. Cell death and cell death responses in liver disease: mechanisms and clinical relevance. Gastroenterology 147, 765–783 (2014).
Koyama, Y. & Brenner, D. A. Liver inflammation and fibrosis. J. Clin. Invest 127, 55–64 (2017).
Dewidar, B., Meyer, C., Dooley, S. & Meindl-Beinker, A. N. TGF-β in hepatic stellate cell activation and liver fibrogenesis-updated 2019. Cells 8, 1419 (2019).
Zehender, A. et al. TGFβ promotes fibrosis by MYST1-dependent epigenetic regulation of autophagy. Nat. Commun. 12, 4404 (2021).
Binatti, E., Gerussi, A., Barisani, D. & Invernizzi, P. The role of macrophages in liver fibrosis: new therapeutic opportunities. Int. J. Mol. Sci. 23, 6649 (2022).
Yamaguchi, M., Hashimoto, K., Jijiwa, M. & Murata, T. The inflammatory macrophages repress the growth of bone metastatic human prostate cancer cells via TNF-α and IL-6 signaling: involvement of cell signaling regulator regucalcin. Cell. Signal. 107, 110663 (2023).
Rho, H., Terry, A. R., Chronis, C. & Hay, N. Hexokinase 2-mediated gene expression via histone lactylation is required for hepatic stellate cell activation and liver fibrosis. Cell. Metab. 35, 1406–1423 (2023).
Zhang, Z., Li, M., Sun, T., Zhang, Z. & Liu, C. FOXM1: functional roles of FOXM1 in non-malignant diseases. Biomolecules 13, 857 (2023).
Hu, G. et al. FOXM1 promotes hepatocellular carcinoma progression by regulating KIF4A expression. J. Exp. Clin. Cancer Res. 38, 188 (2019).
Khan, M. A., Khan, P., Ahmad, A., Fatima, M. & Nasser, M. W. FOXM1: a small fox that makes more tracks for cancer progression and metastasis. Semin. Cancer Biol. 92, 1–15 (2023).
Liang, S. K. et al. FOXM1 is required for small cell lung cancer tumorigenesis and associated with poor clinical prognosis. Oncogene 40, 4847–4858 (2021).
Wang, K. et al. Peptide-based PROTAC degrader of FOXM1 suppresses cancer and decreases GLUT1 and PD-L1 expression. J. Exp. Clin. Cancer Res. 41, 289 (2022).
Tomasi, M. L. et al. Methionine adenosyltransferase α2 sumoylation positively regulate Bcl-2 expression in human colon and liver cancer cells. Oncotarget 6, 37706–37723 (2015).
Liu, T. et al. Mechanisms of MAFG dysregulation in cholestatic liver injury and development of liver cancer. Gastroenterology 155, 557–571 (2018).
Tomasi, M. L. et al. S-adenosylmethionine and methylthioadenosine inhibit cancer metastasis by targeting microRNA 34a/b-methionine adenosyltransferase 2A/2B axis. Oncotarget 8, 78851–78869 (2017).
Gartel, A. L. FOXM1 in cancer: interactions and vulnerabilities. Cancer Res. 77, 3135–3139 (2017).
Liu, Q. et al. The X protein of hepatitis B virus inhibits apoptosis in hepatoma cells through enhancing the methionine adenosyltransferase 2A gene expression and reducing S-adenosylmethionine production. J. Biol. Chem. 286, 17168–17180 (2011).
Wree, A., Mehal, W. Z. & Feldstein, A. E. Targeting cell death and sterile inflammation loop for the treatment of nonalcoholic steatohepatitis. Semin. Liver. Dis. 36, 27–36 (2016).
Pradere, J. P. et al. Hepatic macrophages but not dendritic cells contribute to liver fibrosis by promoting the survival of activated hepatic stellate cells in mice. Hepatology 58, 1461–1473 (2013).
Kalluri, R. & LeBleu, V. S. The biology, function, and biomedical applications of exosomes. Science 367, eaau6977 (2020).
Luo, Z. et al. CRIg(+) Macrophages prevent gut microbial DNA-containing extracellular vesicle-induced tissue inflammation and insulin resistance. Gastroenterology 160, 863–874 (2021).
Isaac, R., Reis, F. C. G., Ying, W. & Olefsky, J. M. Exosomes as mediators of intercellular crosstalk in metabolism. Cell. Metab. 33, 1744–1762 (2021).
Mathieu, M., Martin-Jaular, L., Lavieu, G. & Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell. Biol. 21, 9–17 (2019).
Cho, Y. E., Song, B. J., Akbar, M. & Baek, M. C. Extracellular vesicles as potential biomarkers for alcohol- and drug-induced liver injury and their therapeutic applications. Pharmacol. Ther. 187, 180–194 (2018).
Hernández, A. et al. Chemical hypoxia induces pro-inflammatory signals in fat-laden hepatocytes and contributes to cellular crosstalk with Kupffer cells through extracellular vesicles. Biochim. Biophys. Acta Mol. Basis. Dis. 1866, 165753 (2020).
Tabibian, J. H., Masyuk, A. I., Masyuk, T. V., O’Hara, S. P. & LaRusso, N. F. Physiology of cholangiocytes. Compr. Physiol. 3, 541–565 (2013).
Perea, L. et al. Pentraxin-3 modulates lipopolysaccharide-induced inflammatory response and attenuates liver injury. Hepatology 66, 953–968 (2017).
Tsochatzis, E. et al. Collagen proportionate area is superior to other histological methods for sub-classifying cirrhosis and determining prognosis. J. Hepatol. 60, 948–954 (2014).
Yang, H. et al. Expression pattern, regulation, and functions of methionine adenosyltransferase 2beta splicing variants in hepatoma cells. Gastroenterology 134, 281–291 (2008).
Chhoy, P., Brown, C. W., Amante, J. J. & Mercurio, A. M. Protocol for the separation of extracellular vesicles by ultracentrifugation from in vitro cell culture models. Star. Protoc. 2, 100303 (2021).
Liu, J. et al. Extracellular vesicles-encapsulated let-7i shed from bone mesenchymal stem cells suppress lung cancer via KDM3A/DCLK1/FXYD3 axis. J. Cell. Mol. Med. 25, 1911–1926 (2021).
Yang, B. et al. CSNK2A1-mediated MAX phosphorylation upregulates HMGB1 and IL-6 expression in cholangiocarcinoma progression. Hepatol. Commun. 7, e00144 (2023).
Lu, L. et al. Deregulated 14-3-3ζ and methionine adenosyltransferase α1 interplay promotes liver cancer tumorigenesis in mice and humans. Oncogene 40, 5866–5879 (2021).
Acknowledgements
This work was financially supported by NIH grant P01CA233452 (S.C. Lu, H. Yang, E. Seki), Plan Nacional of I + D SAF2017-88041-R (J.M. Mato), National Natural Science Foundation of China 82070632 (Liu T.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
B.Y., L.L., T.X., H.Y., J.H., W.F., T.T., D.C., J.Z. and H. P. performed the experiments and analyzed the data. B.Y., L.L., T.X. and H.Y. reviewed the literature, drafted the manuscript. L.L. and T.L. collected the clinic sample and data. J. W. performed cell culture, analysis, and interpretation. N. M. and M.L.T. bred the mice. L.B.-T., J.M.M., X.Y., J.C., S.S. and K.R. provided critical reading and editing of the manuscript. V.K. provided the Foxm1 flox mice. E.S. provided the L-rat-Cre mice and critical reading of the manuscript. H.Y. supervised the experiments. S.C.L. obtained funding, conceptualized the experiments, provided overall direction, and edited the manuscript. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Chang Liu, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, B., Lu, L., Xiong, T. et al. The role of forkhead box M1-methionine adenosyltransferase 2 A/2B axis in liver inflammation and fibrosis. Nat Commun 15, 8388 (2024). https://doi.org/10.1038/s41467-024-52527-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-52527-8