Abstract
Talin regulates crucial cellular functions, including cell adhesion and motility, and affects human diseases. Triggered by mechanical forces, talin plays crucial roles in facilitating the formation of focal adhesions and recruiting essential focal adhesion regulatory elements such as vinculin. The structural flexibility allows talin to fine-tune its signaling responses. This study presents our 2.7 Å cryoEM structures of talin, which surprisingly uncovers several auto-inhibitory states. Contrary to previous suggestions, our structures reveal that (1) the first and last three domains are not involved in maintaining talin in its closed state and are mobile, (2) the talin F-actin and membrane binding domain are loosely attached and thus available for binding, and (3) the main force-sensing domain is oriented with its vinculin binding sites ready for release. These structural snapshots offer insights and advancements in understanding the dynamic talin activation mechanism, which is crucial for mediating cell adhesion.
Similar content being viewed by others
Introduction
Talin plays an indispensable role in cell motility and migration by modulating the dynamics of focal adhesions1. Talin facilitates cellular attachment to the extracellular matrix by activating the transmembrane integrin receptors. This binding induces a structural transformation in both integrin polypeptide chains, enhancing integrin in engaging with extracellular matrix ligands2. This activation is critical for cell adhesion and downstream signaling processes that regulate cell growth, survival, and differentiation. Consequently, it is unsurprising that talin is essential for tissue morphogenesis and organogenesis.
Moreover, talin contributes to immune responses by promoting the adhesion of immune cells, such as T cells and neutrophils, to blood vessel walls, thereby enabling their migration to sites of infection or inflammation3. Dysregulation of talin function has been implicated in a spectrum of diseases4,5, including cancer4,5,6,7,8, cardiovascular disorders5,9,10, and autoimmune conditions. A deeper understanding of the involvement of talin in these diseases offers promising avenues for potential therapeutic interventions.
The talin polypeptide chain, consisting of 2541 amino acids, is organized into an amino-terminal head and a carboxy-terminal tail domain connected by approximately eighty residues, which act as a flexible linker11,12,13 (Fig. 1a). The head domain features a distinctive FERM (4.1 protein, ezrin, radixin, moesin) configuration that exhibits the classical FERM subdomains (F1-F3) arranged linearly instead of resembling a cloverleaf, with an additional insertion in F1 and a unique subdomain at the outset, F014 (PDB entries 3ivf15, 6mfs16). SAXS analyses supported the extended conformation of the FERM domain. However, SAXS also indicated additional inter-domain flexibility between the F1 and F2 subdomains. The lack of interdomain contacts between F0–F1 and F2–F3 leads to a slightly twisted arrangement of the F0–F1 and F2–F3 di-domains, suggesting possible alternate conformations in solution (PDB entries 6vgu17, 8fse, 8ftb, and 8t0d). The F3 talin FERM domain interacts with the cytoplasmic tail of β-integrins, thereby enhancing the affinity of integrin for its ligands through an ‘inside-out’ signaling process2,18. The binding of talin to β-integrins depends on favorable membrane electrostatic interactions facilitated by the membrane lipid, phosphatidylinositol 4,5-bisphosphate, which disrupts the talin head-tail interaction and activates talin16,19,20,21.
a The talin polypeptide chain, spanning 2541 amino acids, comprises an amino-terminal head with a FERM domain consisting of the F0-F3 subdomains and a carboxy-terminal ‘rod’ domain containing helix bundles R1-R13 and the carboxy-terminal dimerization α-helix (DD). R8 is inserted between the second and third α-helices of the R7 four-helix bundle. The gray outline highlights the rigid core of talin (encompassing domains F3, R1-R11; labeled in blue font). The decimals (5.5, 3.7, 3.4, and 2.7) refer to the talin5.5, talin3.7, talin3.4, and talin2.7 structures with their ordered domains colored spectrally. Domain boundaries are indicated with residue numbers above or below the domain name. b–g Cartoon drawing of our (b) 2.7 Å, (c) 3.7 Å, (d) 3.4 Å, (e) 3.39 Å, and (f) 5.5 Å talin cryoEM structures, color-coded spectrally and oriented as (g) the previously reported 6.2 Å resolution structure (PDB entry 6r9t)11. h Orthogonal views of our talin5.5 structure.
The carboxy-terminal segment of talin, spanning amino acids 487 to 2541, referred to as the talin rod11, is characterized by sixty-three amphipathic α-helices organized into thirteen bundles (designated as R1 to R13)22, each consisting of four or five α-helices. An extra carboxy-terminal α-helix also serves as the dimerization domain23,24. The talin rod harbors multiple binding sites that influence its activity25,26. The primary interaction between talin and F-actin occurs via the carboxy-terminal dimerization domain and the preceding five-helix bundle (R13). By binding its tail to the actin cytoskeleton and its head to integrins, talin establishes a mechanical link connecting the cytoskeleton to the extracellular matrix (ECM)27,28,29 and orchestrates the organization of the cytoskeleton, which is pivotal for cell shape and movement30.
Detecting mechanical forces by talin enables cells to sense and respond to their mechanical environment, a crucial aspect of tissue development and immune response. Studies employing atomic force microscopy and magnetic tweezer have revealed that applying force to talin enhances its binding affinity for vinculin31. This force-induced mechanism sequentially unfolds numerous helix bundles along the talin rod domain, exposing its vinculin binding sites. R3 demonstrates the lowest stability among the talin domains, exhibiting reversible folding and unfolding on a sub-second timescale when subjected to five pico-newtons of applied force31,32,33. This force magnitude corresponds to a single actomyosin contraction32,33,34 and falls within the range of forces observed on single molecules within focal adhesions. Consequently, the talin R3 domain is poised to initiate focal adhesions by efficiently recruiting vinculin.
Insights from studies in Drosophila suggest that talin may adopt alternative configurations depending on the cell type and developmental stage35,36. This conformational diversity led us to determine the cryoEM structure of murine talin as expressed in mammalian cells (Fig. 1b; Supplementary Fig. 1a), considering that the known 6.2 Å resolution structure was obtained from talin expression in E. coli11. Our collection of high-resolution cryoEM structures has delineated the regions governing the auto-inhibitory states of talin, providing insights into the terminal domains and, notably, the F-actin binding domain R13. This highlights the dynamic nature of sensing mechanical forces, which is more dynamic than previously thought. Given the pivotal roles of the talin domains in coordinating cellular dynamics, our structures contribute significantly to deciphering the complex machinery underlying cell adhesion, motility, and signaling, elucidating the molecular mechanisms of talin as a mechanical bridge between integrins and the actin cytoskeleton.
Results
Talin is a conformationally dynamic protein
The thirteen helical bundle domains (R1–R13; Fig. 1a) provide talin with high flexibility and pose a significant challenge for high-resolution structure determination. A recent attempt resulted in a 6.2 Å resolution cryoEM structure lacking the terminal domains, including residues 1–207 corresponding to the F0-F1 FERM subdomains, and residues 2480–2154 of the dimerization α-helix (PDB entry 6r9t)11. This medium-resolution structure was determined using human talin solubilized in 10% glycerol. We speculated whether murine talin, without glycerol, might offer more stability for high-resolution structure determination. Perhaps unsurprising, given the over 98% sequence identity between murine and human talin, we encountered similar challenges despite extensive optimization efforts, including glutaraldehyde cross-linking and subsequent size exclusion chromatography to remove cross-linked aggregates. Subsequently, we expressed full-length murine talin in mammalian Expi293 cells. We achieved a single chromatography talin peak that exhibited substantial improvement in resolution from the previously published 6.2 Å to eventually our 2.7 Å resolution structure (Fig. 1b; Supplementary Fig. 1a, 2a–d).
Four of our five structures reveal that talin is a compact globular molecule, roughly 15 nm in diameter. It features extensive ion pair interactions between its amino-terminal FERM domain (F2–F3) and its thirteen carboxy-terminal helix-bundle rod sub-domains (R1–R13). The amino-terminal F0-F1 and carboxy-terminal dimerization domain (DD) are disordered. Notably, the integrin-binding site on the talin F3 FERM domain is masked by its interaction with the talin R9 rod domain. These interactions are crucial for maintaining talin in an inhibited state within cells37,38,39.
Additionally, the R4 domain interacts with R1, R5, R9, and R10, which positions R3 to interact with R10 and R11, resulting in an auto-inhibited configuration of R3. Beyond the F3-R9 interaction, the R7-R8-R9 tri-domain conformation associates with R4, R5, R6, and R10 to form the compact quaternary structure of talin. These interactions are consistent with a previously reported 6.2 Å resolution structure (PDB 6r9t)11.
We acquired four structures from two cryoEM samples (Supplementary Figs. 1a–e, 2, 3; Table 1). The initial model was derived after multiple rounds of 2D classifications and segregation through ab initio models and heterogeneous refinements. The core domains (F3 and R1-R11) could be confidently modeled, with side chains clearly resolved. Although residual volume for the F2 and R12 domains persisted in the final 2.7 Å resolution three-dimensional reconstruction, now referred to as talin2.7, the corresponding half maps exhibited features consistent with these two domains. However, the quality of the refined Coulomb potential maps was insufficient to model these F2 and R12 domains, indicating significant particle flexibility in the generated 2.7 Å resolution volume. Our talin2.7 structure delineates an electron potential map well-defined for residues 310–401 (corresponding to the F3 domain) and for residues 485–2131 (corresponding to the R1–R11 domains) (Fig. 1b; Supplementary Figs. 1a, 2d).
To further facilitate accurate modeling of the F2 and R12 domains, the particle stack of one of the ab initio models encompassing these domains underwent heterogeneous refinement to isolate a single class enriched in intact particles containing these domains. Subsequently, the particle stack was subjected to homogeneous refinement followed by non-uniform refinement (Supplementary Fig. 2e). This resulted in the convergence of a talin structure to 3.4 Å resolution, referred to as talin3.4 henceforth, corresponding to an electron potential map that clearly defines residues 208-401 (corresponding to the F2-F3 domains) and residues 485-2291 (corresponding to the R1-R12 domains). Unlike talin2.7, this structure, talin3.4, includes the R12 domain (Fig. 1a, Supplementary Figs. 2e, f). Apart from adding R12, talin3.4 shares essential similarities with talin2.7 and can be superimposed with an r.m.s.d. of 0.468 Å for 9373 atoms. Additionally, talin3.4 accounts for approximately 15% of the initial talin2.7 structure.
During heterogeneous refinement, we observed another class of particles exhibiting additional features near the R12 and F2 domains. Upon further processing (Supplementary Fig. 2f), a third partial model, talin3.39, converged at 3.39 Å resolution with significant additional Coulomb potential adjacent to the R12 domain. The electron potential map in this region allowed for the docking of the R13 domain. However, it did not surpass the noise level sufficiently (with correlation coefficients dropping from an average of 0.9 to about 0.2 for R13) to enable unambiguous modeling of R13 at the residue level.
Through processing our images from a second sample, we identified two more conformations: one at 5.5 Å resolution, referred to as talin5.5, and another at 3.7 Å, referred to as talin3.7 (Supplementary Fig. 3). Unlike talin2.7 and talin3.4, the talin5.5 structure displays the F-actin binding domain (FABD) R13 (Supplementary Figs. 1a, b, d) as previously published (PDB entry 6r9t)11 with a relative R13 tilt (by approximately 6°) towards R12. Further processing of the remaining particles from the heterogeneous refinement yielded a 3.7 Å resolution structure (Supplementary Figs. 1c, 3e), referred to as talin3.7, featuring an intact F2 domain (which is disordered in talin2.7). In contrast to the known 6.2 Å resolution structure, where the Coulomb potential map is mainly missing for R3, our talin structures all exhibit well-defined R3 maps, showcasing the differently oriented force sensing R3 domain, as described below.
Approximately 3% of the total particles selected after 2D classification contributed to the talin5.5 structure. This is two-fold higher than the contribution for the known 6.2 Å resolution structure. Clearly, our amino-terminal GFP tag did not substantially alter the overall conformation of the particles in solution.
All final refined volumes from our 3D reconstructions were analyzed using the 3DFSC server to assess the extent of overfitting or inflated resolution estimates. The resolution estimates were as follows: talin2.7 at 2.79 Å resolution, talin3.4 at 3.54 Å, talin3.39 at 3.58 Å, talin5.5 at 6.05 Å, and talin3.7 at 3.92 Å. These results suggest no significant overfitting from tight masks during the final round of refinement. Additionally, the overall sphericity values for all the 3D reconstructions were greater than 0.84, indicating relatively isotropic views for all samples.
Due to differences in the samples used for grid preparation, the particle stacks from the two datasets were not merged for specific analyses like 3D variability analysis or heterogeneous analysis in CryoDRGN. Moreover, the conformations observed in the 3D reconstructions were not similar enough to justify merging for further studies. Comparing the percentage of particles contributing to the final refined structure for talin confirms these observations.
Talin R3 is the critical regulator of vinculin binding
Despite the crucial role of the F3-R9 domain interaction in maintaining the talin auto-inhibitory conformation, the R3 domain has gained attention due to its potential role as a force sensor40. Atomic force microscopy studies have shown that R3 and R8 are the most unstable among the talin rod domains, and each contains a threonine belt that acts as a tension sensor34,41,42. While the R8 domain is inaccessible due to its unique position within the compact auto-inhibitory conformation, R3 is situated on the exterior, making it accessible and more prominent in sensing and participating in vinculin recruitment upon activation.
However, our structures indicate that R3 is inaccessible to vinculin due to its bent conformation. This observation is supported by biochemical evidence showing that vinculin does not bind talin under low ionic strength conditions11. The three-helix bundle intermediate acts as a tension regulator, a second stable conformation next to the five-helix bundles43. This suggests that upon release of the F3-R9 interactions, R3 could interact with vinculin, revealing an activated state of talin. Within the array of thirteen talin rod domains, R3 is unique due to its ability to unfold under minimal force. It exhibits low mechanical stability and stochastic resonance with folded and unfolded states in rapid equilibrium22,32,42,44. These distinctive characteristics enable R3 to respond to variations in force promptly40. Moreover, the two vinculin-binding sites located in R3 serve as the initial points of engagement with vinculin, thereby initiating adhesion reinforcement45,46,47. Consequently, understanding the function of talin is incomplete without considering the role of R3.
R3 exhibits similarity across all our structures (Fig. 2a; Supplementary Figs. 1a–e). The NMR structure of the isolated R3 domain has been reported (Fig. 2b) (PDB entry 2l7a)22, showing some differences compared to its conformer in the context of the full-length polypeptide chain. Precisely, residues 799-904 of our talin2.7 structure can be superimposed onto the isolated R3 NMR structure with an r.m.s.d. of 1.603 Å for 696 atoms. Apart from minor helical tilt differences, the fourth α-helix of the R3 four-helix bundle extends by an additional turn (residues 905-908) in the NMR structure. These residues form a 90° bent connection to the first α-helix of R4 in our cryoEM talin structures (Fig. 2c, d).
a Superposition of the four α-helices of R3 from our talin2.7, talin3.4, talin3.7, and talin5.5 structures. The R3 four-helix bundle is highlighted and color-coded in orange, yellow, green, and blue, representing α-helices 1 through 4, respectively. VBS, vinculin binding site. b The NMR structure of the isolated R3 domain (PDB entry 2l7a)22 shows the amino terminus (magenta) extending its first α-helix. c In the context of the full-length polypeptide chain, the R3 amino terminus (magenta) is bent to position R3 roughly perpendicular to R2. d Coulomb potential map displaying the connection between R2 and R3 (magenta). e Close-up view of the interactions of R3 (Asn-881, Asp-882, and Gln-887) with R10 (Glu-1959, His-1963, Gln-1969, and Arg-1973) and R11 (Gln-2049). R3 appears responsible for maintaining R11 in its firm state, contrasting with the R12 conformation changes, a notion that is well supported by the local resolution maps.
Regrettably, the published 6.2 Å resolution talin structure (PDB entry 6r9t)11 includes coordinates for the R3 domain, although the Coulomb potential map is mainly absent for R3 (Fig. 1c; Supplementary Fig. 1f). Given that our five talin structures consistently depict R3 rotated to engage in multiple interactions with R10 and R11 (Fig. 2d, e), the orientation and position of R3 in PDB entry 6r9t might warrant reconsideration, as its location is pertinent to its function.
The FERM domain is conformationally dynamic
The talin head domain exhibits a distinctive FERM configuration, featuring an additional insertion in F1 and a unique added subdomain, F014. Several structures of the talin isoform 1 (PDB entries 6r9t11, 3ivf15, and 6vgu17) or isoform 2 (PDB entry 6u4k)48 have suggested that the talin FERM domain consists of one rigid di-domain, F0-F1. At the same time, the F1-F2 and F2-F3 linkers allow for flexibility in the F2 and F3 subdomains. It’s worth noting that crystal contacts could contribute to the observed variations in orientations of F2 and F3 relative to the F0-F1 di-domain.
In our described structures, the F3–R9 inter-domain interaction, which maintains talin in its closed conformation, bears similarity to the crystal structure of F2–F3 in complex with R9 complex (PDB entry 4f7g)49 (Fig. 3a). In this crystal structure (Fig. 3b), temperature factors are notably higher (up to 115 Å2) for the loops connecting α-helices 2 with 3 (residues 1721–1725) and 4 with 5 (residues 1783–1790). These loops are stabilized by their interactions with R7 in the full-length structure that are absent in the crystal structure of the isolated domains. Superposition of the F3 domains of the crystal structure onto our structures reveals a considerable (~10 Å) relative shift of R9 (Fig. 3a, c, d).
a The superposition of the F3 domains of the F2-F3 in complex with R9 crystal structure (PDB entry 4f7g)49 onto our talin2.7 structure reveals the relative domain movement of R9. The individual α-helices of the R9 five-helix bundle are color-coded spectrally, labeled for the crystal structure (PDB entry 4f7g), and underlined for talin2.7. b In the crystal structure (PDB entry 4f7g), temperature factors are below 90 Å2 for F3 and higher (up to ~120 Å2) for R9. c, d Superposition of F3 in our talin2.7 structure (c) and the crystal structure (d) aligns Ser-362 and Ala-360 of F3 but shows a significant relative shift in Leu-1680, Val-1683, and His-1755 of R9.
The F2-R12 contact has been suggested to be a potential secondary head-tail interaction. Notably, the Coulomb potential map of our 2.7 Å resolution cryoEM structure, talin2.7, indicates high disorder for F2 and R12, implying mobile F2-F3 and R11-R12 connections. The R12 domains of our talin3.4 and talin5.5 structures exhibit similarity, with an r.m.s.d. of 0.943 Å for 902 atoms. Moreover, R12 in the earlier 6.2 Å resolution structure (PDB entry 6r9t) also bears resemblance to our talin3.4 (r.m.s.d. of 1.284 Å for 984 atoms) and talin5.5 (r.m.s.d. of 1.238 Å for 987 atoms) structures. Such R12 superpositions position the F2 domains of our talin3.4 and talin5.5 structures similarly. In the 6.2 Å resolution structure (Fig. 4a) and our talin3.4 (Fig. 4b) and talin3.39 (Fig. 4c) structures, F2 residue Lys-272 interacts with R12 residue Glu-2288. However, in our talin5.5 structure, Lys-274 (instead of Lys-272) interacts with Glu-2288 (Fig. 4d). This alternate interaction supports the notion that the F2-R12 interaction is highly dynamic.
a In the previously reported 6.2 Å resolution structure (PDB entry 6r9t)11, the F2-R12 interdomain interactions occur between Lys-272 with Glu-2288 and Lys-274 with Gln-2285. b, c, Our (b) talin3.4 structure and (c) talin2.7 structure display similar interactions as previously reported (see panel a). d In our talin5.5 structure, Lys-274 interacts with Glu-2288 instead of Gln-2285. e Superposition of the F3 and R9 domains that keep talin in its closed conformation shows flexibility in their corresponding F2 and R12 domains. The spheres represent the Cα positions of residues 272 and 2289, respectively.
When comparing the three structures that exhibit the FABD R13, namely talin3.4, talin5.5, and PDB entry 6r9t, the F2 domain is much closer to the R12 domain (by ~4 Å; Fig. 4e) in the 6.2 Å resolution structure compared to our structures. However, caution is required not to overinterpret such differences at this modest resolution.
The previously reported 6.2 Å resolution structure11 showed only one talin conformer that had the phosphatidylinositol 4,5 bisphosphate (PIP2) binding site on the FERM domain covered by the talin R12 rod domain, thereby inhibiting the well-established binding of talin to the plasma membrane16,49,50,51,52,53. However, our structures have shown that R12 is flexible and can transition into disorder, which exposes the PIP2 binding sites (Supplementary Fig. 4).
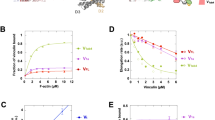
In addition to affecting the conformation of a protein, lipids can affect the location of a protein and its response to mechanical force54,55. To determine the interaction of talin with lipids and their potential contribution to talin’s mechanosensitivity in an intact membrane, we labeled PIP2 and monosialotetrahexosylganglioside (GM1) lipids with fluorescent anti-PIP2 antibody (PIP2−647) and fluorescent cholera toxin B (CTxB-cy3b), respectively, and determined their single molecule localizations (SML) along with eGFP using three-color direct stochastic reconstruction microscopy (dSTORM)54,55. Cross-pair correlation analysis of the SML was used to determine the association of the protein with a particular lipid before and after 3 dynes/cm2 fluid shear. The high resolution is necessary to visualize the short change in distance associated with mechanical force.
Prior to shear, and in agreement with the established talin attachment to the membrane data, eGFP showed a clear association with PIP2 clusters in the membrane of HEK293T cells (Fig. 5a). This can be seen in the high cross-pair correlation at very short distances (of less than 10 nm) and suggests talin localizes to PIP2 containing lipids. Talin also showed a strong pair correlation with GM1 lipids (Fig. 5b), indicating the protein can partition into both lipids.
a, b Three-color dSTORM of talin with a, PIP2, and b, GM1. Single-molecule localizations (SML) were determined using an eGFP label for talin, fluorescent anti-PIP2 antibody, and fluorescent cholera toxin B (GM1 label). Prior to shear, talin pair correlation was high (gray lines) for both lipids, but after 3 dynes/cm2 fluid shear (red lines), pair correlation decreased for GM1 and stayed the same for PIP2. The plot represents the average recordings of independent experiments on cells with n = 7 and n = 10 with shear and no shear, respectively for (a) PIP2, and n = 5 and n = 10 with shear and no shear, respectively for (b) GM1. The error bars represent +/- standard error of mean. Source data are provided as Source Data files. c, d, Representative SML imaging for GM1 (blue) and talin (green), (c) with and (d) without 3 dynes/cm2 fluid shear. The white circles show where GM1 and talin are in proximity. The scale bar is shown in white and represents 2 μm. e Cartoon summarizing the effect of fluid shear on the association of talin with ordered GM1 lipids (blue shading) and PIP2 clusters (purple shading). After shear, GM1 lipids are intact but disrupted (gray bars), causing talin to leave the lipid rafts (black arrow).
Previously, we showed that lipid rafts are mechanosensitive and fluid shear releases palmitoylated proteins55,56. Talin is not palmitoylated but does bind to palmitoylated proteins and integrin, which can be associated with lipid rafts (GM1 containing lipid clusters). Consistent with this binding and mechanosensitivity, applying three dynes/cm2 fluid decreased the talin/GM1 cross-pair correlation (Fig. 5c, d), suggesting the talin is released from GM1 lipids. Interestingly, the pair correlation with PIP2 remained strong during 3 dynes/cm2 shear (Fig. 5e), suggesting PIP2 binding increases relative to GM1 binding during mechanical shear.
The F-actin binding domain, R13, is conformationally dynamic
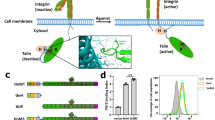
The carboxy-terminal FABD, R13, plays a crucial role in sensing actomyosin-generated forces, thus forming the mechanosensitive talin-vinculin complex necessary for focal adhesion maturation41,57. Our structural analyses reveal not only the flexibility of this FABD but also of its neighboring domains, namely R12 and DD. Notably, the F-actin binding site on R13 remains accessible. Remarkably, both F0-F1 and DD are absent in all structures, while R12 and R13 exhibit two states, either engaged in inter-domain interactions or devoid of such intra-molecular interactions. In its ordered state, R13 is stabilized by R10-R12, while R12 maintains its position through interactions with R11, R13, and F2. Functionally, the FABD R13 is readily released by minor inter-domain movements without force, emphasizing its significance.
Interestingly, upon segregating talin3.4 from talin2.7, we identified a distinct class of particles (Supplementary Fig. 2e) with a feature resembling the R13 domain in a different position. While the residual volume of this talin3.39 structure was adequate to identify the R13 domain at a low (~0.9) contour level, modeling its polypeptide chain without ambiguity proved challenging due to its high mobility. Consequently, further refinement of talin3.39 was not pursued. However, the Coulomb potential map clearly delineates its location (Fig. 6a, b). This alternate suggested position of the R13 domain underscores its significant flexibility, as it appears to dissociate rapidly from the rest of the molecule, possibly facilitated by the sample blotting during grid preparation.
a Coulomb potential map of our partial 3.39 Å resolution talin structure, talin3.39, with its distinct R13 position showing the conventional R13 location with no map coverage. b Although the map in the R13 region is of insufficient quality to assign amino acids unambiguously, the five α-helices can be placed with reasonable certainty. c In contrast, the Coulomb potential map is unambiguous for the remainder of talin3.39 (F2, R1-12). d Our structures show that the talin core (F3, R1-R12; gray) is rigid while the amino- (F0, blue; F1, green, F2, yellow) and carboxy-terminal domains (R12, orange; R13, red) are dynamic. Notably, while the integrin binding site on F3 is inaccessible, the F-actin binding domain, R13, can bind to F-actin. e Multiple states of inactive talin (i, talin5.5; ii, talin3.4; iii, talin3.7; and iv, talin2.7) suggest a loose attachment of the amino- (F0–F2) and carboxy-terminal (R12–R13) domains without affecting the core (F3, R1-R11) that defines the closed state of talin.
Collectively, our structural findings and the previously reported 6.2 Å resolution structure11 demonstrate the flexibility of the three amino-terminal (F0–F2) and carboxy-terminal (R12-R13-DD) domains. Our data suggest that R13 predominantly interacts with R10, R11, and R12, although a minor population of R13 engages in inter-domain interactions with F2 and R12. This contrasts the amino-terminal domains F0 and F1, which remain solvent-exposed and lack a fixed position relative to the rest of the molecule. Thus, while domains F3 and R1–R11 provide rigidity, the flexibility of R12 and R13 underscores their dynamic role in the talin function (Figs. 6d, e; Supplementary Fig. 5).
Discussion
Talin plays a fundamental role in forming multicellular tissues and regulating critical cellular processes such as cell adhesion and motility, which have implications for various human diseases. By recruiting essential focal adhesion regulatory elements, talin is indispensable in facilitating the formation of focal adhesions. Its role in cytoskeletal dynamics that modulate cell adhesion cannot be overstated.
Despite significant progress in understanding talin since its discovery four decades ago58, its activation mechanism at the atomic level remains incompletely understood. Contrary to previous suggestions, the wealth of our structural data reveals several key insights: (1) the first and last three domains are not involved in maintaining talin in its closed state and exhibit mobility akin to beads on a string; (2) among these six domains, only three may or may not engage in inter-domain interactions; (3) the talin F-actin binding domain, R13, displays flexibility, rendering it available for binding; (4) R12 is flexible and can transition into disorder, which exposes the PIP2 binding sites on the talin FERM domain; and (5) the primary force-sensing R3 domain is positioned with its α-helical vinculin binding sites primed for release from the R3 four-helix bundle.
The eagerly awaited human talin structure, recently resolved to 6.2 Å resolution, shed light on correlating its auto-inhibition to its tension sensing and regulating cell adhesion11. While this structure depicted a compact architecture for one talin polypeptide chain, an earlier 25 Å structure of talin obtained from turkey gizzards appeared dimeric59. Since the 6.2 Å resolution structure was derived from human talin expressed in E. coli, we questioned whether talin from mammalian cells exhibits dimerization. Surprisingly, this was not the case as our 2.7 Å talin structure resulting from expressing murine talin in human embryonic kidney cells remains monomeric. Nonetheless, the leap in resolution to near-atomic resolution and the rich structural information gathered from our five structures unveil remarkable mechanistic insights into talin function.
Foremost, our data advance comprehension of the interaction of R3, the sole talin domain that reversibly unfolds within the physiological force range, with vinculin or RIAM32,60. Regrettably, the Coulomb potential map is mainly absent for R3 in the previously published 6.2 Å resolution structure. However, R3 is distinctly resolved in our five structures. Furthermore, whereas the four-helix R3 bundle domain in the 6.2 Å resolution structure is predominantly solvent-exposed, forming a continuous long α-helix with the first α-helix from the subsequent R4 domain, our high-resolution structures reveal a different arrangement. R3 is positioned roughly perpendicular to R4, facilitated by a hinge between the fourth R3 α-helix and the first R4 α-helix. This conformation, stabilized by the tip of the R3 four-helix bundle engaging in hydrogen bonding interactions with residues from R10 and R11, suggests a predisposition for RIAM binding to R3, in line with biochemical and biophysical data22,44, elucidating how R3 promptly responds to force variations40. In contrast, the previously proposed continuation of the fourth R3 α-helix into the first R4 α-helix appears less conducive to such a function.
Furthermore, our five structures consistently delineate the rigid and mobile regions that constitute the auto-inhibited state of talin. While the amino-terminal domains (F0 and F1) and the carboxy-terminal α-helix (DD) were previously recognized as dynamically changing orientations, our high-resolution structures unveil the mobility of additional regions. Specifically, the mobile areas of talin now encompass an additional FERM subdomain, F2, at the amino terminus and two five-helix bundle domains, R12 and R13, at the carboxy terminus. This flexibility endows F0–F2, R12–R13, and DD with the conformational freedom akin to beads on a string. Notably, only three out of these six beads on a string domain may or may not engage in inter-domain interactions, with such interactions exhibiting a low population and suggesting rapid release. These findings carry significant functional implications.
One now recognized loosely attached five-helix bundle domain is particularly significant as the functionally crucial F-actin binding domain, R13. In our talin5.5 structure, the observed interdomain interactions largely resemble those described previously. However, in our talin2.7 structure, the R13 and R12 domains lack inter-domain interactions. Intriguingly, while the known interactions of R13 with R10–R12 are evident in our talin5.5 structure, our talin3.39 structure reveals R13 engaged in entirely different inter-domain interactions involving the amino-terminal F2–F3 FERM subdomains. Consistent with our finding of the flexibility of R13, this domain is absent in our talin2.7 and talin3.4 structures. These discoveries are significant as they offer structural insights into the availability of R13 for binding to F-actin and talin recruitment to nascent focal adhesion sites for stabilizing the adhesion complex.
On the amino terminus, the flexibility of the F2 FERM subdomain is much greater than previously thought, as evidenced by its absence in our talin2.7 structure. Indeed, the flexibility of F2 is corroborated by our local resolution maps and previous crystal structures of the isolated amino-terminal domains15,17,48. A comparison of our three structures with ordered F2 suggests that the flexibility of the F2 domain seems to be independent of the presence (talin3.4 and talin5.5) or absence (talin3.7) of R12. Thus, contrary to previous assumptions, the R12 domain does not significantly influence the release of the F2 FERM subdomain.
Our high-resolution structures collectively reveal that the talin core spans residues 311 through 2134, encompassing domains F3 and R1-R11. Notably, this arrangement renders the integrin binding site on F3 inaccessible while leaving R13 readily available for its binding to F-actin that is not cryptic. Our structures confirm that the known F3-R9 clasps must be severed before the talin F3 domain can bind to the integrin β cytoplasmic tail to activate integrin. However, the F2-R12 closure appears secondary, and its release uncovers the membrane binding site.
The association of talin with both PIP2- and GM1-containing lipid clusters adds a dimension to the binding of talin to the membrane. PIP2 in the cellular membrane is primarily polyunsaturated and resides in the disordered region of the membrane, often separate from saturated lipids like GM1. When talin binds to PIP2, the lipid regulates the proximity or access of talin to potential binding partners and specific lipid environments. This regulation by substrate presentation is distinct from allosteric changes due to binding affinity. The level of PIP2 does not need to change in the membrane. Instead, talin moves to a location with a potentially decreased concentration of PIP2.
The mechanical disruption of ordered lipids affected talin’s location to GM1 lipids but not PIP2. The selective disruption of talin-GM1 interaction suggests that during acute mechanical force, the PIP2 regulation is likely to dominate over the regulation by GM1 lipids. The exact mechanism of forced induce release from GM1 lipids is still poorly understood, but it is thought to involve disruption of ordered lipids. Hence, the flexible regions of talin could have mechanical influence through their binding to the membrane and their effect on location, independent of tension.
Finally, when F2 and R12 are disordered as seen in our talin2.7 structure, no significant differences are observed for the F3–R11 core of talin. This suggests that the release of the minor F2–R12 contacts is insufficient to expose the integrin binding site. Instead, our structures reveal a stepwise domain dissociation at both ends of the polypeptide chain with a core that remains unaffected and that defines the closed state of talin and allows membrane binding.
Methods
Cloning and protein production
Mammalian expression of full-length murine talin was carried out using the plasmid16,48 available from addgene (ID #164838). The full-length murine talin (mtalin1FL) sequence was inserted into a mammalian expression vector (pCMV) containing an amino-terminal (His)10-tag followed by green fluorescence protein (GFP) as an expression marker and a precision protease cleavage site.
Protein production was conducted in suspension cell culture using Expi293 cells (ThermoFisher Scientific Catalog Number A14527) routinely maintained in defined Expi293 media (ThermoFisher Scientific Catalog Number A1435101) without additional supplements. For protein production, the GFP-mtalin1FL plasmid was transfected into 400 ml of Expi293 cells at a concentration of 1 μg/μl using 1 mg/ml polyethyleneimine at a ratio of 1:3. The cells were then incubated at 37 °C with 8% CO2 and shaken at 125 r.p.m. for 72 hours. After incubation, the cells were pelleted at 500 x g for 5 minutes, washed with phosphate-buffered saline, divided into four parts, and stored at −80 °C until purification.
For protein purification, the cell pellet was treated with a hypotonic solution (20 mM Tris-HCl at pH 7.5 containing Pierce protease inhibitor cocktail) and incubated on ice for 30 minutes. Equal volumes of buffer containing 20 mM Tris-HCl at pH 7.5, 50 mM KCl, and 0.2 mM tris(2-carboxyethyl)phosphine (TCEP) were added, followed by sonication for 3 minutes (5 seconds on and 10 seconds off) and centrifugation at 35,000 r.p.m. (100,000 x g) for 30 minutes to recover the supernatant for purification using an AKTATM fast protein liquid chromatography. The clarified lysate was passed through a 1 ml pre-packed Ni-sepharose column pre-equilibrated with 20 mM Tris-HCl at pH 7.5, 20 mM KCl, and 0.2 mM TCEP. After extensive washing, the bound protein was eluted using a combination of stepped (5-column volumes of 25 mM imidazole) and linear gradients of imidazole (25 mM to 100 mM for 20-column volumes and 100 mM to 500 mM for 20-column volumes) to eliminate as many impurities as possible. The peak fractions containing talin, confirmed by SDS-PAGE and GFP fluorescence, were pooled, concentrated to 500 μl, and further subjected to final size exclusion chromatography on a superose 6 10/300 column pre-equilibrated with 20 mM Tris at pH 8, 50 mM KCl, and 0.2 mM TCEP.
Grid preparation
Freshly glow discharged 300 mesh Au-Flat 1.2/1.3 (Protochips) gold grids were employed for automatic plunge freezing using a Leica GP2. A volume of 3.5 μl of freshly purified talin was added to the side of the holey gold alloy film and allowed to incubate for 30 seconds at 4 °C and 95% humidity. One grid was blotted for 4 seconds on the front side, resulting in the talin2.7, talin3.4, and talin3.39 structures. A second grid with talin from a separate elution fraction was blotted for 9 seconds from the back side and utilized in the structures of talin5.5 and talin3.7. Both grids were promptly frozen by plunging in liquid ethane maintained at −183 °C with liquid nitrogen. The frozen grids were stored in liquid nitrogen until screening or data collection.
Data collection and analyses
All data collection was conducted using an in-house Japan Electron Optics Laboratory (JEOL) cryoARM300 transmission electron microscope operating at 300 kV, equipped with an in-column Omega energy filter that is set to a slit width of 20 eV. Electron micrographs were acquired as movies on a Gatan K3 detector using SerialEM61, with an exposure time of two seconds for talin2.7 or three seconds for talin3.5, totaling a dose of 48 e− per Å2 over 40 frames and an equivalent dose per frame of 1.2 e− per Å2. Data were collected in super-resolution and correlated double sampling modes, with a calibrated physical pixel size of 0.72 Å. Multiple data collection sessions were conducted for monomer and dimer samples.
All steps of the data processing workflow were performed using cryoSPARC62 versions 3.4 and 4.2. Patch motion correction with a binning of 2 and patch contrast transfer function (CTF) estimation was performed as implemented in cryoSPARC62. Images with a 5 Å or less CTF fit resolution and a relative ice thickness between a relative value of 1.0 and 1.2, calculated as the beam intensity ratio on the detector in vacuum and the presence of the specimen, were selected via curation for subsequent processing.
Particle picking for talin2.7, talin3.4, and talin3.39 utilized a blob-picker or blob-picker tuner, as implemented in cryoSPARC62, with a particle radius of 200 Å. The particles (~ 3 million) were extracted in a binned box size of 320 pixels (1.152 Å per pixel) for further classifications. After a few rounds of 2D classifications, 646,839 particles were subjected to the generation of initial ab initio models. The ab initio models closely representing the whole protein molecule were further grouped and refined to obtain the final structure or picked individually for conformation heterogeneous refinement to sort out specific conformation and refined further to get the final models. The above processing led to the generation of talin2.7, talin3.4, and tallin3.39.
For talin5.5 and talin3.7, an alternate approach to particle picking was carried out to capture additional views potentially and allow low-contrast particles to be picked using Topaz63, a neural network-based particle picker. After extraction, 281,910 particles were obtained with a final binned box size of 256 pixels ( ~ 1.44 Å per pixel). Particles were used to generate three ab initio models. As determined by features representing the whole protein molecule, the best model was further classified through heterogeneous refinement to segregate particles of different conformations. One model, containing 51,263 particles with a feature corresponding to R13, was further subjected to one round of additional heterogeneous refinement to extract 8318 particles for final refinements to produce the talin5.5 structure. Due to their similarity, the remaining two models were combined (91,849 particles) and subjected to final homogeneous and non-uniform refinements after applying per particle CTF estimates to produce the talin3.7 structure. We also determined the final resolution based on the Gold Standard Fourier Shell Correlation (GSFSC) from the final refinement by cross-verification for overfitting through independent 3DFSC server64 using the respective half maps and refinement masks.
Structure determination
The 6.2 Å resolution talin structure (PDB entry 6r9t)11 was used as the initial coordinates for fitting in Chimera65,66 into our final Coulomb potential maps. Subsequently, flexible fitting and model correction corresponding to the electron potential map were carried out in Coot67. Iterative real-space refinements with simulated annealing were then conducted with Phenix68. Additionally, manual fitting of the atomic coordinates into the Coulomb potential map was performed in Coot67.
Direct stochastic optical reconstruction microscopy (dSTORM)
Human embryonic kidney 293 T (HEK293T, American Type Culture Collection catalog number CRL-32016) cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, CorningTM, #10-013-CV) with 10% fetal bovine serum (FBS, Sigma-Aldrich, #F0926) and 1% penicillin and streptomycin (CorningTM, #30-002-Cl) at 37 °C with 5% CO2 in eight well imaging chambers and transfected with eGFP-talin using Turbofectamine transfection reagent according to the manufacturer’s recommended instructions (40 μl optimum, 2 μl transfection reagent, with DNA added to a well containing 150 μM DMEM).
HEK293T cells expressing eGFP-talin were initially treated and fixed with 4% paraformaldehyde and 0.1% glutaraldehyde for 15 minutes and reduced with 0.1% sodium borohydride for 7 minutes. For sheared experiments, the cells were fixed with 3 dynes/cm2 rotary shear at 37 °C for 20 minutes. For co-labeling the receptors and PIP2 lipids, cells were permeabilized with 0.2% Triton X-100 for 15 minutes. Cells were blocked with 10% bovine serum albumin (BSA) and 0.05% Triton X-100 in PBS for 90 minutes. PIP2 on the membrane was labeled with a PIP2 antibody (Echelon Biosciences, Z-P045). Cells were conjugated with Atto 647 N NHS ester (Sigma-Aldrich, 18373-1MG-F) and CTxB Alexa-555 (Invitrogen, C34776). Following the labeling, cells were washed five times with 1% BSA and 0.05% Triton X-100 in PBS with 15 minutes of shaking in between. The cells were washed with PBS once for 5 minutes and post-fixed with the fixative solution for 15 minutes. Vutara VXL was used for imaging, and Vutara SRX software was used to analyze the imaging data. One thousand frames were collected for each laser channel. Cells were imaged in 10% glucose, 50 mM Tris, and 10 mM NaCl with 1% β-mercaptoethanol, 56 μg/ml glucose oxidase, and 34 μg/ml bovine catalase.
All statistical analyses were performed in GraphPad Prism 10. For the Student’s t-test and nested t-test, significance was calculated using a two-tailed, unpaired parametric test with significance defined as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. All data are expressed as mean ± s.e.m., and each n is a biological replicate. Every experiment is repeated at least twice.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The final coordinates and the Coulomb potential maps generated in this study for the talin2.7 (PDB entry 8vdo; EMD-43152), talin3.4 (8vdp; EMD-43154), talin5.5 (8vdq; EMD-43155), and talin3.7 (8vdr; EMD-43156) structures have been deposited with the Protein Data Bank and Electron Microscopy Data Bank, respectively. The Coulomb potential map for talin3.39 was deposited with EMDB (accession code EMD-44931). The raw movies for both datasets are accessible through accession codes EMPIAR-11881 (talin5.5 and talin3.7 [https://www.ebi.ac.uk/empiar/EMPIAR-11881]) and EMPIAR-11882 (talin2.7, talin3.4, and talin3.39 [https://www.ebi.ac.uk/empiar/EMPIAR-11882]). The expression plasmid is available with Addgene (ID #164838 [https://www.addgene.org/164838/]). Source data are provided with this paper.
References
Cram, E. J., Clark, S. G. & Schwarzbauer, J. E. Talin loss-of-function uncovers roles in cell contractility and migration in C. elegans. J. Cell Sci. 116, 3871–3878 (2003).
Tadokoro, S. et al. Talin binding to integrin β tails: a final common step in integrin activation. Science 302, 103–106 (2003).
Ye, F., Snider, A. K. & Ginsberg, M. H. Talin and kindlin: the one-two punch in integrin activation. Front Med 8, 6–16 (2014).
Haining, A. W., Lieberthal, T. J. & Hernandez, DelRio A.Talin: a mechanosensitive molecule in health and disease. FASEB J. 30, 2073–2085 (2016).
Qi, L., Kolodziej, T., Rajfur, Z. & Huang, C. Roles of talin2 in traction force generation, tumor metastasis and cardiovascular integrity. Curr. Protein Pept. Sci. 19, 1071–1078 (2018).
Desiniotis, A. & Kyprianou, N. Significance of talin in cancer progression and metastasis. Int Rev. Cell Mol. Biol. 289, 117–147 (2011).
Li, L. et al. The role of talin2 in breast cancer tumorigenesis and metastasis. Oncotarget 8, 106876–106887 (2017).
Beaty, B. T. & Condeelis, J. Digging a little deeper: the stages of invadopodium formation and maturation. Eur. J. Cell Biol. 93, 438–444 (2014).
Wu, Q. et al. Talin1 is required for cardiac Z-disk stabilization and endothelial integrity in zebrafish. FASEB J. 29, 4989–5005 (2015).
Zemljic-Harpf, A., Manso, A. M. & Ross, R. S. Vinculin and talin: focus on the myocardium. J. Investig. Med 57, 849–855 (2009).
Dedden, D. et al. The architecture of talin1 reveals an autoinhibition mechanism. Cell 179, 120–131.e113 (2019).
Rees, D. J., Ades, S. E., Singer, S. J. & Hynes, R. O. Sequence and domain structure of talin. Nature 347, 685–689 (1990).
Moser, M., Nieswandt, B., Ussar, S., Pozgajova, M. & Fassler, R. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat. Med 14, 325–330 (2008).
Goult, B. T. et al. Structure of a double ubiquitin-like domain in the talin head: a role in integrin activation. EMBO J. 29, 1069–1080 (2010).
Elliott, P. R. et al. The Structure of the talin head reveals a novel extended conformation of the FERM domain. Structure 18, 1289–1299 (2010).
Chinthalapudi, K., Rangarajan, E. S. & Izard, T. The interaction of talin with the cell membrane is essential for integrin activation and focal adhesion formation. Proc. Natl Acad. Sci. USA 115, 10339–10344 (2018).
Zhang, P. et al. Crystal structure of the FERM-folded talin head reveals the determinants for integrin binding. Proc. Natl Acad. Sci. USA 117, 32402–32412 (2020).
Petrich, B. G. Talin-dependent integrin signalling in vivo. Thromb. Haemost. 101, 1020–1024 (2009).
Martel, V. et al. Conformation, localization, and integrin binding of talin depend on its interaction with phosphoinositides. J. Biol. Chem. 276, 21217–21227 (2001).
Gilmore, A. P. & Burridge, K. Regulation of vinculin binding to talin and actin by phosphatidyl-inositol-4-5-bisphosphate. Nature 381, 531–535 (1996).
Di Paolo, G. et al. Recruitment and regulation of phosphatidylinositol phosphate kinase type 1 γ by the FERM domain of talin. Nature 420, 85–89 (2002).
Goult, B. T. et al. RIAM and vinculin binding to talin are mutually exclusive and regulate adhesion assembly and turnover. J. Biol. Chem. 288, 8238–8249 (2013).
Gingras, A. R. et al. The structure of the C-terminal actin-binding domain of talin. EMBO J. 27, 458–469 (2008).
Gingras, A. R. et al. Structural determinants of integrin binding to the talin rod. J. Biol. Chem. 284, 8866–8876 (2009).
Ye, F., Lagarrigue, F. & Ginsberg, M. H. SnapShot: talin and the modular nature of the integrin adhesome. Cell 156, 1340–1340.e1341 (2014).
Burridge, K. & Mangeat, P. An interaction between vinculin and talin. Nature 308, 744–746 (1984).
Horwitz, A., Duggan, K., Buck, C., Beckerle, M. C. & Burridge, K. Interaction of plasma membrane fibronectin receptor with talin–a transmembrane linkage. Nature 320, 531–533 (1986).
Zhang, X. et al. Talin depletion reveals independence of initial cell spreading from integrin activation and traction. Nat. Cell Biol. 10, 1062–1068 (2008).
Jiang, G., Giannone, G., Critchley, D. R., Fukumoto, E. & Sheetz, M. P. Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature 424, 334–337 (2003).
O’Halloran, T., Beckerle, M. C. & Burridge, K. Identification of talin as a major cytoplasmic protein implicated in platelet activation. Nature 317, 449–451 (1985).
del Rio, A. et al. Stretching single talin rod molecules activates vinculin binding. Science 323, 638–641 (2009).
Yao, M. et al. Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Sci. Rep. 4, 4610 (2014).
Yao, M. et al. Force-dependent conformational switch of alpha-catenin controls vinculin binding. Nat. Commun. 5, 4525 (2014).
Yao, M. et al. The mechanical response of talin. Nat. Commun. 7, 11966 (2016).
Klapholz, B. & Brown, N. H. Talin - the master of integrin adhesions. J. Cell Sci. 130, 2435–2446 (2017).
Klapholz, B. et al. Alternative mechanisms for talin to mediate integrin function. Curr. Biol. 25, 847–857 (2015).
Zhang, H., Chang, Y. C., Huang, Q., Brennan, M. L. & Wu, J. Structural and functional analysis of a talin triple-domain module suggests an alternative talin autoinhibitory configuration. Structure 24, 721–729 (2016).
Haage, A. et al. Talin autoinhibition regulates cell-ECM adhesion dynamics and wound healing in vivo. Cell Rep. 25, 2401–2416.e2405 (2018).
Ellis, S. J. et al. Talin autoinhibition is required for morphogenesis. Curr. Biol. 23, 1825–1833 (2013).
Yan, J., Yao, M., Goult, B. T. & Sheetz, M. P. Talin dependent mechanosensitivity of cell focal adhesions. Cell Mol. Bioeng. 8, 151–159 (2015).
Goult, B. T., Yan, J. & Schwartz, M. A. Talin as a mechanosensitive signaling hub. J. Cell Biol. 217, 3776–3784 (2018).
Haining, A. W., von Essen, M., Attwood, S. J., Hytonen, V. P. & Del Rio Hernandez, A. All subdomains of the talin rod are mechanically vulnerable and may contribute to cellular mechanosensing. ACS Nano 10, 6648–6658 (2016).
Mykuliak, V. V., Haining, A. W. M., von Essen, M., Del Rio Hernandez, A. & Hytonen, V. P. Mechanical unfolding reveals stable 3-helix intermediates in talin and alpha-catenin. PLoS Comput Biol. 14, e1006126 (2018).
Baxter, N. J., Zacharchenko, T., Barsukov, I. L. & Williamson, M. P. Pressure-dependent chemical shifts in the R3 domain of talin show that it is thermodynamically poised for binding to either vinculin or RIAM. Structure 25, 1856–1866.e1852 (2017).
Izard, T. et al. Vinculin activation by talin through helical bundle conversion. Nature 427, 171–175 (2004).
Izard, T. & Vonrhein, C. Structural basis for amplifying vinculin activation by talin. J. Biol. Chem. 279, 27667–27678 (2004).
Bois, P. R., O’Hara, B. P., Nietlispach, D., Kirkpatrick, J. & Izard, T. The vinculin binding sites of talin and α-actinin are sufficient to activate vinculin. J. Biol. Chem. 281, 7228–7236 (2006).
Rangarajan, E. S. et al. A distinct talin2 structure directs isoform specificity in cell adhesion. J. Biol. Chem. 295, 12885–12899 (2020).
Song, X. et al. A novel membrane-dependent on/off switch mechanism of talin FERM domain at sites of cell adhesion. Cell Res. 22, 1533–1545 (2012).
Brown, D. T. & Izard, T. Vinculin-cell membrane interactions. OncoTarget 6, 34043–34044 (2015).
Critchley, D. R. Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu. Rev. biophysics 38, 235–254 (2009).
Saltel, F. et al. New PI(4,5)P2- and membrane proximal integrin-binding motifs in the talin head control beta3-integrin clustering. J. Cell Biol. 187, 715–731 (2009).
Moore, D. T. et al. Affinity of talin-1 for the beta3-integrin cytosolic domain is modulated by its phospholipid bilayer environment. Proc. Natl Acad. Sci. USA 109, 793–798 (2012).
Call, I. M., Bois, J. L. & Hansen, S. B. Super-resolution imaging of potassium channels with genetically encoded EGFP. Preprint at bioRxiv https://doi.org/10.1101/2023.10.13.561998 (2023).
Petersen, E. N. et al. Mechanical activation of TWIK-related potassium channel by nanoscopic movement and rapid second messenger signaling. Elife 12, https://doi.org/10.7554/eLife.89465 (2024).
Petersen, E. N., Chung, H. W., Nayebosadri, A. & Hansen, S. B. Kinetic disruption of lipid rafts is a mechanosensor for phospholipase D. Nat. Commun. 7, 13873 (2016).
Calderwood, D. A., Campbell, I. D. & Critchley, D. R. Talins and kindlins: partners in integrin-mediated adhesion. Nat. Rev. Mol. cell Biol. 14, 503–517 (2013).
Burridge, K. & Connell, L. Talin: a cytoskeletal component concentrated in adhesion plaques and other sites of actin-membrane interaction. Cell Motil. 3, 405–417 (1983).
Goult, B. T. et al. Structural studies on full-length talin1 reveal a compact auto-inhibited dimer: implications for talin activation. J. Struct. Biol. 184, 21–32 (2013).
Tapia-Rojo, R., Alonso-Caballero, A. & Fernandez, J. M. Talin folding as the tuning fork of cellular mechanotransduction. Proc. Natl Acad. Sci. USA 117, 21346–21353 (2020).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Bepler, T., Kelley, K., Noble, A. J. & Berger, B. Topaz-denoise: general deep denoising models for cryoEM and cryoET. Nat. Commun. 11, 5208 (2020).
Tan, Y. Z. et al. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796 (2017).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput Chem. 25, 1605–1612 (2004).
Meng, E. C. et al. UCSF ChimeraX: tools for structure building and analysis. Protein Sci. 32, e4792 (2023).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D: Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. Biol. Crystallogr 66, 213–221 (2010).
Acknowledgements
TI is grateful for the support from grants from the National Institutes of Health (R35 GM139604) and the National Science Foundation (2030119) and start-up funds provided to The Scripps Research Institute by the State of Florida. We are deeply indebted to Drs Ben Goult (University of Liverpool, United Kingdom) and Daniel Lietha (Biological Research Center Margarita Salas, CIB-CSIC, Spain) for countless insightful discussions.
Author information
Authors and Affiliations
Contributions
E.S.R. and T.I. contributed to all stages of the manuscript. S.B.H. designed the dSTORM experiments with mechanical force, analyzed the data, and helped write the manuscript. J.B. performed the talin-GFP expression, fluorescent labeling, and acquisition of the dSTORM data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Rangarajan, E.S., Bois, J.L., Hansen, S.B. et al. High-resolution snapshots of the talin auto-inhibitory states suggest roles in cell adhesion and signaling. Nat Commun 15, 9270 (2024). https://doi.org/10.1038/s41467-024-52581-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-52581-2