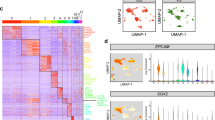
Abstract
Vestibular hair cells are mechanoreceptors critical for detecting head position and motion. In mammals, hair cell loss causes vestibular dysfunction as spontaneous regeneration is nearly absent. Constitutive expression of exogenous ATOH1, a hair cell transcription factor, increases hair cell regeneration, however, these cells fail to fully mature. Here, we profiled mouse utricles at 14 time points, and defined transcriptomes of developing and mature vestibular hair cells. To mimic native hair cells which downregulate endogenous ATOH1 as they mature, we engineered viral vectors carrying the supporting cell promoters GFAP and RLBP1. In utricles damaged ex vivo, both CMV-ATOH1 and GFAP-ATOH1 increased regeneration more effectively than RLBP1-ATOH1, while GFAP-ATOH1 and RLBP1-ATOH1 induced hair cells with more mature transcriptomes. In utricles damaged in vivo, GFAP-ATOH1 induced regeneration of hair cells expressing genes indicative of maturing type II hair cells, and more hair cells with bundles and synapses than untreated organs. Together our results demonstrate the efficacy of spatiotemporal control of ATOH1 overexpression in inner ear hair cell regeneration.
Similar content being viewed by others
Introduction
Loss of vestibular hair cells renders the inner ear balance organs dysfunctional. A variety of insults, including ototoxic drugs, infection, genetic mutations, and ageing can damage vestibular hair cells1,2, causing vestibular deficits that affect more than 35% of the US population aged 40 and above3. Since hair cells are the primary mechanoreceptors and are highly susceptible to damage, hair cell degeneration is the main pathology underlying human vestibulopathy4. At present, we lack effective treatments for vestibulopathy; therefore, there is a pressing need for a biological treatment—therapies that regenerate vestibular hair cells can serve this unmet need.
In non-mammalian species, hair cells are terminally differentiated, whereas supporting cells, accessory epithelial cells that interdigitate with hair cells within the sensory epithelium, have a remarkable ability to regenerate lost hair cells allowing recovery of auditory and vestibular function5,6,7,8,9,10,11. While the capacity to regenerate hair cells is substantially less in the mammalian inner ear, vestibular supporting cells in mammals have retained some latent potential for hair cell regeneration, even at mature ages12,13,14,15. This contrasts sharply with adult cochlear supporting cells, which show little to no regenerative capacity in the absence of external factors16.
While mice can regenerate up to 30% of lost vestibular hair cells after damage, regenerated hair cells appear immature, and it remains unclear if vestibular function can be fully restored12,17,18. Nevertheless, multiple studies have demonstrated that this spontaneous regenerative response can be augmented with exogenous factors, with some reporting modest recovery of function13,19. Thus, the underlying regenerative machinery in the mammalian vestibular system appears to be intact and manipulatable.
One promising therapeutic target is the basic helix-loop-helix transcription factor, ATOH1, which is both necessary and sufficient for inducing hair cell fate during development16. Multiple studies in rodents and human vestibular organs have demonstrated that transgenic or viral overexpression of ATOH1 forces the direct conversion of vestibular supporting cells into new hair cells at adult ages13,19,20,21,22,23,24. However, the reported degrees of differentiation of the regenerated hair cells were mixed, with their transcriptomes undetermined at the single cell level. Nevertheless, improvements in rotarod performance, vestibulo-ocular reflex (VOR) gain, and vestibular evoked potential (VsEP) thresholds have been reported13,19,22,23, suggesting some functional recovery.
During hair cell development, ATOH1 is transiently expressed in progenitor cells and immature hair cells and becomes downregulated or silenced as hair cells differentiate25,26,27. In the cochlea, persistent overexpression of ATOH1 in differentiating hair cells delays development and can be cytotoxic28,29. ATOH1 is also a potent determinant of cell fate in the cerebellum, spinal cord, and skin30,31,32. Thus, the level, temporal control, and cell type-specific distribution of ATOH1 expression are critical for hair cell specification and maturation.
With the aim of developing an optimized AAV-based ATOH1 gene therapy, we used single-cell RNA-Seq to transcriptionally map the innate maturation of vestibular hair cells and explored the impact of defined promoters on ATOH1 transgene expression level, timing, effects in distinct cell types, and the maturity of regenerated vestibular hair cells ex vivo and in vivo. We found that the level of ATOH1 overexpression in supporting cells strongly correlates with new hair cell formation, with high levels of ATOH1 expression needed for efficient cell fate conversion. Furthermore, we provide single-cell transcriptomic data demonstrating that silencing the ATOH1 transgene enables regenerated hair cells to undergo a greater degree of maturation. Using transcriptomes of hair cells in the developing and mature utricle as a benchmark, we found that ATOH1-mediated transdifferentiation of supporting cells induced the production of cells which resemble endogenous type II hair cells. Collectively, our results underscore the importance of spatiotemporal transcriptional control of ATOH1 in hair cell regeneration and maturation and should guide future design of ATOH1-based gene therapies.
Results
The level and duration of ATOH1 transgene expression determine supporting-cell-to-hair-cell conversion in the adult mouse utricle ex vivo
Adenoviral-based overexpression of mouse or human ATOH1 converts supporting cells into new hair cells in cultured utricles from adult mice, rats, and humans14,21,33,34. Early work comparing promoters delivered via adenoviral vectors concluded that lower levels of ATOH1 expression resulted in more regenerated hair cells compared to higher levels34. To begin to determine how spatiotemporal expression of ATOH1 affects hair cell regeneration, we cultured adult mouse utricle, lesioned hair cells with the aminoglycoside gentamicin, and subsequently transduced them with escalating titers of AAV-CMV-ATOH1 (Supplementary Figs. 1 and 2a–f). We observed a dose-dependent conversion of supporting cells to Pou4f3+ hair cells (Supplementary Fig. 2g–i), consistent with previous results20.
Higher titers of AAV increased the total number of cells transduced (Supplementary Fig. 2g–i), and may enhance the efficiency of AAV-CMV-ATOH1 in inducing hair cell conversion. We reasoned that gene regulatory elements (e.g., promoters) leading to different levels of gene expression (termed strengths hereon) and are specific to supporting cells may allow for the titration of the level of ATOH1 transgene expression at equivalent AAV doses. To identify such regulatory elements, candidate sequences were identified from single-cell genomics data35, cloned into AAV constructs driving an H2B-EGFP transgene (Supplementary Fig. 1), and tested in adult utricle explants. The final constructs tested were AAV8-CMV-H2B-EGFP, AAV8-glial fibrillary acidic protein (GFAP)-H2B-EGFP, and AAV8-retinaldehyde binding protein 1 (RLBP1)-H2B-EGFP (4.0 × 1011 vg/ml). We selected the AAV8 serotype as the vector because it has been shown to transduce utricular hair cells and supporting cells at high rates in vivo20,36. We transduced utricle explants from adult mice with AAV8 carrying the regulatory sequences associated with CMV, GFAP, and RLBP1 for 3 days and found different levels of EGFP intensity within both hair cells and supporting cells among these promoters (Supplementary Fig. 3a–f). When ATOH1-GFP under different promoters was delivered, the CMV promoter induced relatively higher ATOH1-GFP intensity in supporting cells than GFAP or RLBP1 (Fig. 1b–d, f–h, k). CMV-ATOH1-GFP fluorescence intensity in supporting cells was 1.5 and 6.6-fold higher than GFAP and RLBP1, respectively, and GFAP was 4.4-fold more intense than RLBP1 at 12 days ex vivo. These differences increased to 2.2, 12.1, and 5.5-fold, respectively, at 20 days ex vivo (Fig. 1k). Furthermore, the CMV promoter drove robust EGFP expression in both hair cells and supporting cells, while with GFAP and RLBP1 expression was mostly restricted to supporting cells, suggesting that these regulatory elements are not active in hair cells as expected (Supplementary Fig. 3b–f).
a Utricles from 4-10-week-old mice were treated with gentamicin (1 mM) followed by AAV8-CMV-ATOH1-H2B-EGFP, AAV8-GFAP-ATOH1-H2B-EGFP or AAV8-RLBP1-ATOH1-H2B-EGFP (4 × 1011 vg/mL), then examined at day 12 and day 20. b–i Utricles treated with CMV-ATOH1, GFAP-ATOH1, RLBP1-ATOH1 or gentamicin control for 12 and 20 days. Insets showing the GFP signals. j, k Mean GFP intensity in hair cells and supporting cells after treatment with CMV, GFAP and RLBP1 promoters. l Quantification shows Pou4f3-positive hair cell counts at day 12 and 20 after treatment with CMV-ATOH1, GFAP-ATOH1, RLBP1-ATOH1 or gentamicin control. Over the time, hair cell number significantly increased with CMV-ATOH1 and GFAP-ATOH1, but not RLBP1-ATOH1 treatment. Data shown as mean ± S.D., compared using two-way ANOVA. p = 0.0063 between CMV-ATOH1 and GFAP-ATOH1 at day 12, and p < 0.0001 for all the others in (j), p = 0.0043 between CMV-ATOH1 and RLBP1-ATOH1 at day 12, p = 0.0001 between GFAP-ATOH1 and RLBP1-ATOH1 at day 20 and p < 0.0001 for all the others in (k). p = 0.0074 between CMV-ATOH1 and RLBP1-ATOH1 at day 12, p = 0.0014 and 0.0191 between day 12 and day 20 with CMV-ATOH1 or GFAP-ATOH1 treatment, respectively. p = 0.006 between RLBP1-ATOH1 and gentamicin control at day 20 and p < 0.0001 for all the others in (l). n = 4, 4, 3 for CMV, GFAP or RLBP1-ATOH1 at 12 days and 5, 5 and 6 at 20 days in (j). n = 4, 4 and 4 for CMV, GFAP or RLBP1-ATOH1 at 12 days and 5, 5 and 6 at 20 days in (k). n = 6, 5, 5 and 5 for CMV, GFAP, RLBP1-ATOH1 or gentamicin control at 12 days and 5, 6, 6 and 4 at 20 days in (l). Scale bars = 100 μm in (b–g). Source data are provided as a Source Data file.
Having identified a set of regulatory elements driving varying degrees of EGFP expression, we next tested the relationship between levels of ATOH1 transgene expression and hair cell regeneration. Gentamicin-damaged utricle explants37,38 were transduced with AAV8-CMV-ATOH1-H2B-EGFP, AAV8-GFAP-ATOH1-H2B-EGFP, or AAV8-RLBP1-ATOH1-H2B-EGFP (4.0 × 1011 vg/mL) and then cultured for an additional 5 or 13 days to allow time for hair cell regeneration to occur (Fig. 1a–i). Significantly more Pou4f3+ hair cells were observed in all three treatment groups compared to gentamicin only control, with the quantity increasing in the order (CMV > GFAP > RLBP1) in line with their corresponding EGFP intensity at 5 and 13 days after AAV transduction (Fig. 1b–i, l).
The number of Pou4f3+ hair cells increased significantly between the two time points for CMV- and GFAP-ATOH1, but not RLBP1-ATOH1 treatment (Fig. 1b–i, l), indicating that more new hair cells were generated with longer culture periods under the control of defined promoters. Furthermore, the number of Pou4f3+ hair cells remained higher for CMV than GFAP and RLBP1 at both 5 and 13 days after AAV transduction. At the end of the culture period, the number of Pou4f3+ hair cells were 157.6 ± 16.6 for CMV-ATOH1, 127.2 ± 39.7 for GFAP-ATOH1, and 60.5 ± 17.1 for RLBP1-ATOH1 per 10,000 µm2 (Fig. 1l). Taken together, these data suggest that CMV is a stronger promoter than both GFAP and RLBP1 and induced more hair cell regeneration in damaged mouse utricles ex vivo.
scRNA-seq reveals transcriptome-wide changes during hair cell development and regeneration
To characterize the transcriptomes of hair cells in the developing and mature utricles, we isolated and analyzed mouse utricles using single-cell RNA-sequencing (scRNA-Seq). Mouse utricles were collected on embryonic day (E)14.5, 15.5, 16.5, 18.5, postnatal day (P) 0, 1, 2, 3, 4, 5, 6, 10, 12 and 16. A total of 260,808 high-quality cells were profiled at these 14 developmental time-points, with 6543-33,910 cells collected at each stage (Supplementary Fig. 4a, c). Based on expression of known markers39,40, we identified 9 distinct cell types including hair cells (Pou4f3), supporting cells (Agr3), vascular cells (Cd34), muscle cells (Kcnj8), glia cells (Ncam1), epithelial cells (Vmo1), dark cells (Kcne1), immune cells (Cfn), and mesenchymal cells (Dcn, Supplementary Fig. 4b). Both putative hair cells and supporting cells were abundantly represented, with 16,212 cells identified as high-quality hair cells or precursors (Supplementary Fig. 4d).
Mammalian vestibular organs contain two types of hair cells, type I and type II, that are found throughout the organ, along with a central striolar region and a peripheral extrastriolar zone41,42. The functional significance of hair cell subtypes and zonal distribution have yet to be fully elucidated, but type I hair cells in the striolar region are associated with fast, phasic responses and type II hair cells with more static, tonic responses43,44. To distinguish the transcriptomes of developing type I from those of type II hair cells, we compiled 18 and 20 known marker genes of each hair cell subtype as benchmarks for hair cell maturity (Supplementary Data 1), and plotted putative hair cells and hair cell precursors at 14 developmental time points according to these two hair cell maturity indices (Supplementary Fig. 4c, d). We found that cells derived from embryonic and neonatal mouse utricle scored qualitatively lower for both type I and II markers relative to those from P12 and P16 utricles (Supplementary Fig. 4d). We next used this trajectory as a reference to assess the maturity of regenerated hair cells.
To assess the transcriptomic profiles of regenerated hair cells, we first treated gentamicin-damaged utricles with the ATOH1 transgene. Utricular explants were transduced with AAV8-CMV-ATOH1, AAV8-GFAP-ATOH1, or AAV8-RLBP1-ATOH1 (4.0 × 1011 vg/mL) and then were maintained in culture for 7, 12, or 20 days before being processed for scRNA-Seq (Fig. 2a). A total of 34,574, 30,428, and 21,891 high-quality cells were collected for each regulatory element, respectively (Supplementary Fig. 5a–d). After merging these datasets and using marker gene expression to enrich supporting cells and regenerating hair cells (Supplementary Fig. 5a–i), the final dataset consisted of 6901 cells. UMAP and heatmap visualization of the merged dataset from cells taken at all timepoints revealed six distinct cell clusters that were separated based on gene expression into distinct phases of supporting cell-to-hair cell conversion (Fig. 2b-b’, c). The six clusters were organized by differentiation status along an unbiased pseudotime gradient (Supplementary Fig. 6b), with maturation of regenerated hair cells progressing over time in culture (Fig. 2g). Within these six clusters we found one cluster of immature and two clusters of mature regenerated hair cells, which we designated as mature hair cell α and β. These different mature hair cell clusters primarily arose from either CMV or GFAP/RLBP1 treatments, respectively, suggesting a divergence of cell fates and possibly different maturation states (Fig. 2b, d–f). Compared to 7 days in vitro (DIV) when only immature hair cells were present, more mature hair cell α were observed with CMV treatment and more mature hair cell β with GFAP/RLBP1 treatments at 12 DIV and 20 DIV (Fig. 2g). While all six clusters were observed after transduction with any of the three regulatory elements, higher proportions of converting supporting cells and mature hair cells were found after treatment with ATOH1 driven by CMV and GFAP than by RLBP1 (Fig. 2g).
a Utricles from 4-10-week-old mice were treated with gentamicin (1mM) followed by CMV-ATOH1, GFAP-ATOH1 or RLBP1-ATOH1 (4 × 1011 vg/mL), and processed for single-cell RNA sequencing at day 7, 12 and 20. b UMAP plot of integrated dataset of 3 different virus treatments from all timepoints showing 6901 single cells following all quality control steps including only hair cells (HC) and supporting cells (SC). Six cell clusters including, converting SC1, converting SC2, immature HC, mature HCα and mature HCβ were identified. b’ Schematic showing supporting cell-to-hair cell conversion. c Heatmap showing differentially expressed genes among the 6 cell clusters. The top differentially expressed genes of each cluster are shown on the right. A full list of these genes is found in the source data file. The heatmap is colored by relative expression from −2 (magenta) to 2 (yellow). d–f UMAP plot colored by CMV-ATOH1 (ruby), GFAP-ATOH1 (blue) or RLBP1-ATOH1 (green) treatment. g The fraction of cells from different treatment groups and different timepoints that were assigned to each cell type. n = 7–8 utricles per treatment, per timepoint. Source data are provided as a Source Data file.
Supporting cells that responded to ATOH1 overexpression (stage 1 and 2 converting SC) initially downregulated known supporting cell genes (Otol1, Otog, Tspan8, and Gsn), and upregulated Notch pathway genes (Hey1, Hes5, Jag1, and Notch1) prior to the onset of expression of early hair cell genes (Pou4f3, Lhx3, Hes6, and Dlk2) (Fig. 3n and Supplementary Fig. 6c, d). A “Notch Activity Score” based on expression of 57 genes associated with the canonical Notch pathway (see Methods, Supplementary Data 2) showed that Notch pathway activity is the highest in the stage 1 and 2 converting supporting cell clusters (Supplementary Fig. 6c, d, p < 0.0001, one-way ANOVA). As Notch signaling suppresses supporting cells from acquiring a hair cell fate45,46,47, these findings suggest that Notch gene expression becomes upregulated in converting supporting cells possibly in response to exogenous ATOH1. At 12 and 20 DIV, there were substantially higher percentages of stage 1 and 2 converting supporting cells (Fig. 2g). Interestingly, many supporting cells did not show any signs of conversion despite displaying levels of ATOH1 transgene expression comparable to converting SCs, with 31.5% of SCs at 20 DIV remaining in the supporting cell cluster (Figs. 2g and 3a). These ATOH1-transgene+, non-converting SCs exhibited lower Notch activity scores than stage 1 and 2 converting supporting cells (Supplementary Fig. 6c, d).
a Violin plot depicting ATOH1-EGFP transgene expression levels in 6 cell clusters. b UMAP plot of ATOH1-EGFP transgene expression level. c, d Schematic showing persistently and transiently upregulated ATOH1-EGFP transgene in supporting cells and regenerated hair cells after CMV-ATOH1 treatment, and GFAP-ATOH1 or RLBP1-ATOH1 treatment, respectively. e Violin plot of endogenous ATOH1 expression level in 6 different cell clusters. f UMAP plot of endogenous ATOH1 expression level. g–j Ridge plots of hair cell maturity, immaturity score, type II hair cell and type I hair cell scores of 6 different cell clusters. k Levels of expression of type I and type II hair cell genes for immature HC (yellow), mature HCα (magenta), mature HCβ (green), and developing and mature hair cells (gray). l Levels of expression of type I and type II hair cell genes for regenerated hair cells after CMV-ATOH1 (ruby), GFAP-ATOH1 (blue) or RLBP1-ATOH1 (green) treatment. Developing and mature hair cells depicted in gray. m Dot plots showing top 18 highly enriched mature hair cell genes from each cell cluster. n Dot plots showing top 10 highly enriched immature hair cell genes from each cell cluster.
Consistent with EGFP intensity corresponding to the strength of each regulatory element (Fig. 1b–d, f–h and Supplementary Fig. 3a–f), the levels of ATOH1 transgene in supporting cells was similarly ordered (average across all cells, CMV = 18.6 normalized read counts; GFAP = 3.3; RLBP1 = 1.2; Supplementary Fig. 6a). Moreover, the percentage of regenerated hair cells (immature hair cells, mature hair cells α and β) increased with regulatory elements associated with higher ATOH1 transgene expression level (CMV = 28.7%; GFAP = 24.9%; RLBP1 = 12.2%; Fig. 2g).
ATOH1 transgene expression under control of a supporting-cell-specific regulatory element becomes silenced in regenerated hair cells
Two of the regulatory elements used to drive ATOH1 expression, GFAP and RLBP1, led to limited EGFP expression in hair cells (Fig. 1j and Supplementary Fig. 3e, f). Based on these results, ATOH1 transgene expression was expected to become downregulated as supporting cells convert into hair cells (Fig. 3a–d). ATOH1 transgene expression was detected in several clusters (supporting cells, stage 1 and 2 converting supporting cells, immature hair cells, and mature hair cells α) for all three regulatory elements utilized, indicating that they are active during the early stages of supporting cell conversion and hair cell specification (Fig. 3a). In contrast, the mature hair cell β cluster segregated into distinct groups visible on the UMAP based on regulatory element and ATOH1 transgene level (Figs. 2d–f and 3b). Hair cells from the CMV group almost exclusively comprised the mature hair cell α cluster (99.8% of cells), whereas the GFAP and RLBP1 groups comprised the mature hair cell β cluster (92.9% of cells; Fig. 2d–f). As predicted, the ATOH1 transgene was expressed at high levels in the mature hair cell α cluster, but it was significantly reduced or absent in mature hair cell β cluster (Fig. 3a, b).
ATOH1 autoregulates its own gene expression by binding to regulatory regions in the 3’ enhancer of the endogenous gene48,49. We distinguished Atoh1 transcripts originating from the endogenous locus from vector transcripts via the WPRE element at the 3’ end of the ATOH1 transgene. Interestingly, expression of endogenous Atoh1 was detected in the immature hair cell, mature hair cell α, and mature hair cell β clusters, but not supporting cell or either converting supporting cell clusters (Fig. 3e, f). This finding suggests that ATOH1 protein produced from the vector transgene induced expression of endogenous Atoh1, and endogenous Atoh1 expression persisted in the mature hair cell β cluster after Atoh1 transgene expression from the GFAP and RLBP1 regulatory elements became silenced.
Silencing of AAV-ATOH1 transgene from supporting cell-specific regulatory elements promotes maturation of regenerated vestibular hair cells ex vivo
Atoh1 is expressed transiently in hair cells during development, and misexpression of Atoh1 in developing hair cells delays maturation50,51. We compared the transcriptomes of the mature hair cell α and β clusters to assess whether silencing of Atoh1 transgene expression driven by supporting cell-specific regulatory elements promotes maturation. As a measure of maturation, “hair cell maturity indices” were created based on the top 200 mature hair cell and top 100 immature hair cell genes that are up- or down-regulated as hair cells differentiate during development (see Methods, Supplementary Data 2) and were used to score cell clusters along the supporting cell-hair cell axis. The mature hair cell β cluster scored higher on the “Mature hair cell” and lower on the “Immature hair cell” indexes compared to mature hair cell α cluster (p < 0.0001, one-way ANOVA, Fig. 3g–j), confirming that the former is more mature at the transcriptome level. The expression of select index genes is shown in Fig. 3m, n, highlighting the differentially expressed genes in the mature hair cell β cluster including those critical for synaptogenesis (Otof, Slc17a8), stereocilia formation (Espn, Fscn2), and mechanotransduction (Clrn1) (Supplementary Fig. 6e, f).
To assess the extent to which regenerated hair cells differentiate into specialized hair cell subtypes, we analyzed them using scores based on transcriptomes of developing and mature type I and type II hair cells and found that the mature hair cell β cluster scored higher on the type II hair cell index (Sox2, S100a1 and Ptn) compared to the mature hair cell α cluster (Fig. 3i and Supplementary Data 2). Cells from both the mature hair cell α and β clusters scored relatively low on type I hair cell index (Adam11, Atp2b2, Map4k4), indicating that regenerated hair cells were not adopting a type I hair cell fate ex vivo (Fig. 3j and Supplementary Data 2). We also assessed the maturity of regenerated hair cells against the criteria that we had previously established for developing and mature hair cells (Supplementary Data 1). Compared to mature hair cells α with CMV treatment, mature hair cells β with GFAP/RLBP1 treatments displayed higher type II gene scores (Fig. 3k, l). These data indicate that the expression of the ATOH1 transgene under control of supporting cell-specific regulatory elements becomes silenced in regenerating hair cells as they differentiate, and this reduction in transgene expression promotes further maturation towards a more mature type II hair cell fate.
AAV8-GFAP-ATOH1 drives supporting cell-specific transgene expression in adult mouse vestibular organs in vivo
So far, the ex vivo findings indicated that ATOH1 under control a supporting cell-specific regulatory element enhances maturation of regenerated hair cells, thereby serving as a promising gene therapy candidate for vestibular hair cell regeneration in vivo. Amongst the three regulatory elements tested ex vivo, GFAP was deemed superior to RLBP1 and CMV because it led to relatively strong and targeted ATOH1 expression in supporting cells and robust regeneration of hair cells that display transcriptomes resembling maturing type II hair cells. To further assess the potential utility of GFAP as a regulatory element for ATOH1 gene therapy, its activity profile was assessed in the adult mouse utricle and cristae in vivo. AAV8-GFAP-H2B-EGFP was administered locally at two doses (1 × 1010 and 3 × 1010 vg/mL) via the posterior semicircular canal of adult mice (Supplementary Fig. 7a). Two weeks later utricles showed robust EGFP in supporting cells, with little to no detectable EGFP expression in hair cells (Supplementary Fig. 7b). No statistically significant difference was observed between the two doses (Supplementary Fig. 7c–g). These results indicate that GFAP drove robust EGFP selectively in supporting cells in vivo.
AAV8-GFAP-ATOH1 induces regeneration of vestibular HCs with stereocilia bundles and presynaptic ribbons in vivo
To assess whether AAV8-GFAP-ATOH1 promotes regeneration of vestibular hair cells in damaged mouse utricles in vivo, we first assessed an in vivo hair cell damage model. 3,3’-iminodipropionitrile (IDPN) is a well-described vestibulotoxin that induces dose-dependent hair cell death after a single systemic administration to adult mice in vivo13,52. In 4-10-week-old mice, IDPN administration resulted in significant loss of hair cells in utricular maculae (67% loss in extrastriola, 84% loss in striola, respectively, p < 0.001) and cristae (61% loss in peripheral zone, 93% loss in central zone, respectively, p < 0.01) within one week (Supplementary Fig. 8a–d, g, h). Hair cell loss was higher in the striolar than extrastriolar regions of utricles and higher in the central zone than peripheral zone of cristae, the lesions across both ears were highly symmetric (Supplementary Fig. 8d, f, h, i). Immunolabeling with antibodies against the type II hair cell marker (Myosin7a and Sox2) demonstrated that IDPN preferentially killed type I hair cells as 96% of the surviving hair cells were Sox2+ (Supplementary Fig. 8e).
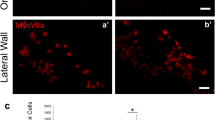
Next, AAV8-GFAP-ATOH1 (1.71 × 1013 vg/mL) was administered unilaterally one or 14 days after IDPN treatment. Vestibular organs were harvested and examined 2-4 weeks after AAV administration (Fig. 4a). Virus injection resulted in robust expression of EGFP in both utricles and cristae, whereas no EGFP was detected in non-injected contralateral ears (Fig. 4b–f”, insets). Utricles and cristae from injected ears also displayed more Myosin7a+ HCs in comparison to contralateral controls (Fig. 4b–f” and Supplementary Fig. 9a–c, e–g). Quantification showed significantly more Myosin7a+ HCs in both the striolar and extrastriolar regions of utricles from injected ears compared to contralateral controls at 2 weeks (70.6 ± 51.5 vs. 26.9 ± 28.0 and 109.7 ± 62.3 vs. 68.6 ± 43.8 cells per 10,000 µm2 in striola and extrastriola, respectively) and 1 month (142.7 ± 83.5 vs. 47.0 ± 36.7 and 176.4 ± 50.8 vs. 93.7 ± 36.2 cells per 10,000 µm2 in striola and extrastriola, respectively) (Fig. 4b, c”, g, h). Similarly, significantly more regenerated hair cells were observed in the peripheral and central regions of cristae 2 weeks (22.0 ± 8.0 vs. 7.6 ± 3.6 and 16.4 ± 3.6 vs. 1.6 ± 1.7 cells per 10,000 µm2 in peripheral and central regions, respectively) and 1 month after AAV administration (26.8 ± 7.1 vs. 12.0 ± 7.6 and 18.0 ± 6.4 vs. 2.8 ± 1.5 cells per 10,000 µm2 in peripheral and central regions, respectively) (Fig. 4e, f and Supplementary Fig. 9h, i), suggesting that AAV8-GFAP-ATOH1 promoted regeneration of vestibular hair cells in vivo. In addition, the numbers of Myosin7a+ HCs increased with time in both the striolar (70.6 ± 51.5 vs. 142.7 ± 83.5 cells per 10,000 µm2 at 2 weeks and 1 month, respectively) and extrastriolar (109.7 ± 62.3 vs. 176.4 ± 50.8 cells 10,000 µm2 at 2 weeks and 1 month, respectively) regions of the utricle (Fig. 4g, h and Supplementary Fig. 9d).
a 4-10-week-old mice were treated with IDPN (4 or 5 mg/g) at day 1, AAV8-GFAP-ATOH1 (1.71 × 1013 vg/mL) were injected at day 2 or 15. Organs were examined at 2 weeks and 1 month after injection. b, c” Treated utricles had more Myosin7a+ (red) hair cells compared to contralateral ear. d, d’ High magnification images of injected and contralateral ears (from b and b”). GFP+ hair cells (arrowhead) were observed after GFAP-ATOH1 injection. e, f” Crista ampullaris from injected ear has more Myosin7a+ (red) hair cells compared to contralateral ear. g, h GFAP-ATOH1 significantly increased Myosin7a+ (red) hair cells compared to contralateral ear. i, j’ Utricles from injected ear had more hair bundles (phalloidin, green) than contralateral ear. i’, j’ High magnification pictures in boxes. k Phalloidin-labeled bundles increased significantly in utricles from injected ear compared to contralateral controls. l–n Representative images of bundles less than 6 µm, between 6 µm and 12 µm, and more than 12 µm. o Measurements of bundle height. p–r Expression of Ctbp2+ puncta (gray) on the basolateral surfaces of utricular hair cells from naive, injected ear and contralateral ear. Insets are the 3D view of hair cells at 1 month. s Quantification of Ctbp2+ puncta in Myosin7a+ (green) hair cells. Number of Ctbp2+ puncta per hair cell increased significantly in contralateral ear compared to naive and injected ear. Data shown as mean ± S.D, compared using two-tailed paired Student’s t tests and ordinary one-way ANOVA. p = 0.0033 and 0.005 in extrastriola and striola, respectively in (g), p < 0.0001 and =0.0003 in extrastriola and striola, respectively in (h), p = 0.0110 in (k), p = 0.002 between naive and injected ear in striola and p < 0.0001 for all the others in (s). n = 8 at 2 weeks and =10 at 1 month in (g) and (h). n = 4 in (k). n = 114 and 79 bundles from injected and contralateral ears in (o). n = 36 cells from 3 naive mice ear, 72 cells from 6 injected mice ear and 72 cells from 6 contralateral mice ear in (s). Scale bars = 100 μm in (b–f”, i, j), 20 μm in (d, d’, p–r), 10 μm in (i’, j’), 2 μm in (l–n), 5 μm in (p–r) inserts. Source data are provided as a Source Data file.
Hair cells require stereociliary bundles for mechanoreception and synapses to relay information centrally via afferent neurons53,54. One month after AAV8-GFAP-ATOH1 administration to IDPN-treated mice, the density of phalloidin-labeled bundles throughout the utricle of injected ears was 1.8-fold higher than contralateral control ears (p < 0.05; Fig. 4i–k). In the injected ears, we found bundles with various heights (<6, 6 to 12, and >12 µm), and that there were more short (<6 µm) and more long (>12 µm) bundles than contralateral control ears (Fig. 4l–o).
Ribbon synapses form along the basolateral surface of vestibular hair cells and are identified by the presynaptic marker Ctbp255. Surprisingly, utricular hair cells in non-injected ears from IDPN-treated mice showed a significant increase in the density of Ctbp2-labeled puncta relative to hair cells from naive, undamaged mice (Fig. 4p–s). This phenomenon has been reported in surviving hair cells before56, but whether it resulted from damage-induced fragmentation of pre-existing synapses, an additive process, or remodeling was unclear. In contrast, utricular hair cells from ears administered AAV8-GFAP-ATOH1 showed comparable Ctpb2 puncta density relative to naive hair cells (Fig. 4s). Among 144 hair cells examined in the virus-treated utricle (n = 6), all of them displayed Ctbp2+ puncta, suggesting that they expressed components needed for synaptic transmission.
AAV8-GFAP-ATOH1 induces regeneration of mature type II HCs in vivo
We further characterized in vivo vestibular hair cell regeneration after AAV8-GFAP-ATOH1 using scRNA-seq. Utricles and cristae were collected from IDPN-damaged mice treated with AAV8-GFAP-ATOH1 (IDPN and GFAP-ATOH1), IDPN-damaged (IDPN), or naive mice (Control; Fig. 5a and Supplementary Fig. 12a) at 2 weeks and 4 weeks post-treatment (4 and 6 weeks after IDPN administration). The organs were pooled by group, single cells were isolated and processed for scRNA-seq. A total of 12,130, 11,111, and 15,146 cells were collected for the control, IDPN, and IDPN and GFAP-ATOH1 treated utricles, respectively, as well as 18,102, 27,072, and 32,927 cells for the cristae (from 6 mice). Marker gene identification in unbiased clusters of the merged groups revealed rich datasets that included all the various cell types that constitute the utricle and crista: hair cells, supporting cells, transitional epithelial cells (TECs), dark cells, melanocytes, mesenchymal cells, glia, endothelial cells, smooth muscle cells, and immune cells (Supplementary Figs. 10e–j and 13e–j). In the GFAP-ATOH1-treated utricle and crista, the ATOH1 transgene was detected in mesenchymal cells, TECs, supporting cells, and supporting cells presumably converting into hair cells (Supplementary Figs. 10f, k and 13f, k). GFAP regulatory element activity in mesenchymal cells was expected as GFAP expression has been reported in this cell population57.
a 4-10-week-old mice were treated with IDPN (5 mg/g) at day 1, AAV8-GFAP-ATOH1 (1.71 × 1013 vg/mL) at day 15. Single-cell RNA sequencing was performed at 2 weeks and 1 month after injection. b UMAP plot of integrated dataset of 3 different treatment groups (control, IDPN, IDPN and GFAP-ATOH1) from both time points. There were 8399 single cells following all quality control steps including only hair cells and supporting cells. Six cell clusters including supporting cells, stage 1, stage 2, stage 3 regenerated hair cells, type II and type I hair cells were identified. c Heatmap showing differentially expressed genes among the 6 cell clusters. The top differentially expressed genes of each cluster are shown on the right. A full list of these genes is found in the source data file. The heatmap is colored by relative expression from −2 (magenta) to 2 (yellow). d–f UMAP plot colored by control (red), IDPN (green) or IDPN or GFAP-ATOH1 (blue) treatment. g The fraction of cells from different treatment groups and different time points that were assigned to each cell type. n = 6 ears per treatment per time point. Source data are provided as a Source Data file.
To examine supporting cells converting into hair cells, transcriptomes of supporting cells, hair cell precursors, and hair cells from all groups were examined and re-clustered (8399 cells from the utricles and 7855 cells from the cristae). Unbiased clustering identified six clusters in both the utricle and cristae (Fig. 5b, c and Supplementary Fig. 12b, c). Marker gene expression revealed three of the clusters were supporting cells, mature type I and II hair cells. Supporting cells and type II hair cells were present in similar proportions for all three treatment groups (Fig. 5g and Supplementary Fig. 12g). In contrast, type I hair cells were scant in the IDPN only and IDPN and GFAP-ATOH1 groups, consistent with the finding that IDPN preferentially ablates type I hair cells (Fig. 5g and Supplementary Figs. 8e and 12g)58.
The other three clusters (stage 1/2/3 regenerated HCs) in the utricle primarily comprised cells from the IDPN and IDPN + GFAP-ATOH1 groups, with a higher proportion in the latter (Fig. 5g). Gene expression analysis indicated that these groups contained cells representing various stages of supporting cell-to-hair cell conversion, thereby revealing the transcriptional map along the supporting cell-to-hair cell axis after in vivo treatment with GFAP-ATOH1. The cells were then ordered along a pseudotime trajectory and were designated stage 1-3 regenerating hair cells, with stage 1 being the earliest stage of conversion and stage 3 being the most mature state (Fig. 5b and Supplementary Fig. 11a). Out of Stage 1-3 regenerated hair cells, 86.2% came from utricles treated with IDPN + GFAP-ATOH1 (Fig. 5g). These clusters were compared to transcriptomes of hair cells and hair cell precursors in the developing and mature utricles (Supplementary Fig. 4d) Compared to undamaged controls, there was an increase in the proportion of stage 1-3 hair cells in the IDPN only group, consistent with a low level of spontaneous regeneration after IDPN damage alone13. Further, there were substantially more regenerating hair cells with ATOH1 overexpression, than after IDPN alone (Fig. 5g). Similar to the ex vivo results, ATOH1 transgene expression levels were highest in supporting cells and stage 1 regenerated hair cells, and then was gradually downregulated in stage 2 and 3 regenerated hair cells (Fig. 6a, b). Thus, like ex vivo, the GFAP regulatory element also became silenced as supporting cells convert into hair cells in vivo (Fig. 6a, b). Interestingly, expression of endogenous Atoh1 was detected in the stage 2 regenerated hair cell, stage 3 regenerated hair cell, and type II hair cell clusters (Fig. 6c, d). As the ATOH1 transgene became downregulated, the transcriptomes of the regenerated hair cells (stage 1-3) scored higher in maturity indices, suggesting a more mature phenotype (Fig. 6e, f, k, l). In addition, based on type I and type II hair cell indices, regenerating hair cells primarily adopted a type II fate as they progressed from stage 1 to 3 (Fig. 6g, h). No significant expression of type I hair cell marker genes or increase in type I index score was detected as regenerating hair cell differentiated, indicating that ATOH1 overexpression mainly induces a type II hair cell fate in vivo (Fig. 6g, h). Relative to transcriptomes of type I and II hair cells in the developing and mature utricles, stage 3 regenerated hair cells and most GFAP-ATOH1-induced hair cells resemble maturing type II hair cells (Fig. 6i, j and Supplementary Fig. 4d).
a Violin plot of ATOH1-EGFP transgene expression level in 6 cell clusters. b UMAP plot of ATOH1-EGFP transgene expression levels. c Violin plot of endogenous ATOH1 expression level in 6 different cell clusters. d UMAP plot of endogenous ATOH1 expression level. e–h Ridge plots of hair cell maturity, immature hair cell, type II hair cell and type I hair cell scores of 6 different cell clusters. i Expression levels of type I and type II hair cell genes for stage 1 regenerated HC (yellow), stage 2 regenerated HC (green), stage 3 regenerated HC (cyan), type I HC (blue), type II HC (magenta), and developing and mature hair cells (gray). j Levels of expression of type I and type II hair cell genes for hair cells in control (red), IDPN only (green) or IDPN and GFAP-ATOH1 (blue). Developing and mature hair cells are depicted in gray. k Dot plots showing top 18 highly enriched mature hair cell genes from each cell cluster. l Dot plots showing top 10 highly enriched immature hair cell genes from each cell cluster.
In parallel, we analyzed the cristae by isolating and examining 7855 supporting cells, hair cells, and putative hair cell precursors from untreated, IDPN, and IDPN and ATOH1-treated organs, revealing six clusters that are comparable to those found in the utricle (Supplementary Figs. 12 and 13). In cristae, ATOH1 transgene expression decreased along the trajectory from supporting cells to hair cells (Supplementary Fig. 14a, b), and hair cell maturity score increased (Supplementary Fig. 14e, f). As in the utricle, endogenous Atoh1 was detected in the stage 2 regenerated hair cell, stage 3 regenerated hair cell, and type II hair cell clusters (Supplementary Fig. 14c, d).
In both the utricle and crista, the expression of Notch pathway genes is highest in supporting cells and stage 1 regenerated HCs and is decreased with increasing maturity (Supplementary Figs. 11b, c and 15b). This agrees with the ex vivo results, where Notch activity decreases as hair cells mature. Furthermore, stage 3 regenerated HCs exhibited genes related to hair bundle and synapse formation that were similar to those found in maturing type II hair cells (Supplementary Figs. 11d, e and 15c, d).
In summary, these data demonstrate that use of the supporting cell-specific promoter GFAP to drive transient ATOH1 overexpression is effective in enhancing maturation of the regenerated type II-like hair cells.
Discussion
Vestibular hair cells are required for detecting head acceleration and rotation. Their loss causes vestibular hypofunction. Because there is minimal spontaneous regeneration in the mature, mammalian vestibular organs, vestibular hypofunction is deemed irreversible. Independent studies have found that ATOH1 overexpression either via transgenic approaches or viral transduction increases hair cell regeneration in the damaged mature mouse utricle13,19,20. However, regenerated hair cells failed to fully mature, in part because these approaches have employed constitutive overexpression of ATOH1. As developing hair cells naturally downregulate ATOH125,26,27 and ATOH1 overexpression has been reported to impede maturation of cochlear hair cells28,29, we have engineered an ATOH1 transgene to be driven by supporting cell promoters to test whether transient overexpression of ATOH1 can induce both hair cell regeneration and subsequent maturation. In damaged utricles ex vivo and in vivo, we found that constitutive and ubiquitous overexpression of ATOH1 using AAV8-CMV-ATOH1 induced more regenerated hair cells than transient overexpression via AAV8-GFAP-ATOH1 and AAV8-RLBP1-ATOH1. In addition to the transient expression of ATOH1 mediated by AAV8-GFAP-ATOH1 and AAV8-RLBP1-ATOH1, supporting cells in this model also downregulated Notch signaling, upregulated endogenous ATOH1, and converted into hair cells that display transcriptome consistent with maturing type II hair cells. Together, our results suggest that transient ATOH1 expression is a promising approach to initiate mature vestibular hair cell regeneration. Our results are also consistent with a similar approach that transiently overexpresses ATOH1 in the cochlea59.
Responsiveness of supporting cells to ATOH1 overexpression
Relative to cochlear supporting cells, vestibular supporting cells in mature mouse are more responsive to singular overexpression of the hair cell transcription factor ATOH114,60. In our experiments with AAV8-GFAP-ATOH1, we observed different stages of converting supporting cells and maturing regenerated hair cells that expressed the ATOH1 transgene, clearly indicating that supporting cells responded to AAV8-GFAP-ATOH1 and acquired a hair cell fate. One interpretation that there are different stages of converting supporting cells and maturing hair cells is that supporting cell-to-hair cell conversion was asynchronized. Alternatively, supporting cell-to-hair cell conversion may occur in parallel but at different rates. Both possibilities are supported by the findings that the hair cell number was higher at the later than earlier time points in vivo.
While it is expected that mesenchymal cells do not convert and become hair cells despite expression of the ATOH1 transgene, some supporting cells also failed to convert despite expression of the ATOH1 transgene. Considering Notch signaling is known to govern lateral inhibition in the mature utricle46,61, one explanation is that there is active Notch signaling in supporting cells as a result of some surviving hair cells, which are present in both the gentamicin and IDPN damage paradigms. One may speculate that regions with higher density of surviving hair cells will contain supporting cells expressing greater levels of Notch target genes, which we have observed in a subset of supporting cells in our single-cell dataset. We postulate that only supporting cells with high levels of exogenous and endogenous ATOH1 can overcome the inhibitory Notch activity and drive transdifferentiation of supporting cells to hair cells, whereas supporting cells with lower ATOH1 levels, would be unable to overcome Notch signaling and as such would remain in a supporting cell state.
Differentiation of regenerated hair cells
The mature mammalian vestibular organs harbor type I and II hair cells which feature distinct cell body and bundle morphology, transcriptomes, basolateral potassium currents, and innervation patterns41,42. While there are reports claiming the relative importance of each hair cell subtype, both likely contribute to the overall organ function. After damage to the mouse utricle, spontaneously regenerated hair cells demonstrate features of immature type I and II hair cells13,17,18. Our study builds on the previous findings39 and provides a comprehensive transcriptomic atlas of sensory and non-sensory cell types of the developing and mature mouse utricle, and a new transcriptomic atlas of the mature mouse cristae. Using transcriptomes of native developing and mature type I and II hair cells and supporting cells of the utricle as benchmarks, we found that AAV8-GFAP-ATOH1 induces robust supporting cell-to-hair cell conversion. Remarkably, regenerated hair cells mature as they downregulate the ATOH1 transgene and transiently upregulate endogenous ATOH1, and express mature hair cell genes including those representing the maturing type II hair cells. These results are consistent with the previous report that ATOH1 overexpression resulted in primarily regenerated type II hair cells13. Moreover, GFAP-ATOH1 treatment resulted in an increase in bundle-bearing hair cells and regenerated hair cells expressing a subset of bundle genes. Lastly, our study shows that regenerated hair cells after AAV8-GFAP-ATOH1 treatment express some synapse genes, as well as having synapse counts comparable to native hair cells. These results suggest that the regenerated hair cells can partially mature and display some features necessary for mechanotransduction and synaptic transmission. Since damaged hair cells have been reported to self-repair56, these results should be further validated with fate-mapping experiments.
It is remarkable that the endogenous Atoh1 levels remain low even after high exogenous Atoh1 transgene level was achieved with CMV-ATOH1 treatment. It is possible that endogenous Atoh1 levels in regenerating hair cells are tightly regulated, similar to those in cochlear hair cells by Hic1 or Huwe162,63, such that exogenous Atoh1 transgene is unable to increase endogenous Atoh1 further. While the enhancers for Atoh1 in the cochlea were recently described64, whether the same enhancers regulate Atoh1 in the regenerating vestibular system is not known. Future work can better characterize these interactions.
Previous studies using VsEP and behavioral assays suggest some functional recovery of the mouse utricle months after ATOH1-induced hair cell regeneration13,19. Our study is significantly shorter in duration and did not assess the function of regenerated hair cells, thus the long-term function of the regenerated utricle after transient ATOH1 overexpression is unknown. As VsEP mainly measures function of striolar type I hair cells65, other vestibular function measurements66 may be necessary to assess organ function of AAV-GFAP-ATOH1 treatment in the future.
In summary, our study describes the effectiveness and limitation of gene therapy using supporting cell promoters to drive transient overexpression of ATOH1 in vestibular hair cell regeneration. Our results provide a high-resolution transcriptomic atlas of the mature mouse vestibular organs and a promising approach to regenerate mature type II hair cells.
Methods
Ethics statement
All protocols were approved by the Animal Care and Use Committee of the Stanford University School of Medicine (#18606, #26925) and Decibel Therapeutics (#2019-009, #2018-004).
Mice
4-10-week-old CD-1 wild type mice (#022, Charles River; #030, Envigo), and C57BL/6J wild type mice (#000664, The Jackson Laboratory) were used. Both male and female were both include in this study. IDPN (3,3’-iminodipropionitrile, Cat #AV54D, TCI America, Portland, OR, USA) was given via a single intraperitoneal (IP) injection (4-5 mg/g, 200 µl) to adult mice for vestibular hair cell ablation. Animal holding rooms are at 12 hours cycle. Lights turn on at 7AM and off at 7PM. Room temperatures are maintained between 68-72 °F. Humidity is within 30-70% range.
Virus preparation
All transgenic elements were packaged in an adeno-associated virus vector containing a promoter (Supplementary Fig. 1). In short, vectors were generated and produced by the Viral Core at Boston Children’s Hospital (BCH) in compliance with the BCH Institutional Biosafety Committee (protocol # IBC-P00000447). AAV vectors were purified by iodixanol gradient ultracentrifuge, followed by ion exchange chromatography. Titers were calculated by quantitative PCR with eGFP primers (F-GACCTTTGGTCGCCCGGCCT, R-GAGTTGGCCACTCCCTCTCTGC). Vectors were aliquoted and stored at −80 °C.
Ex vivo utricle culture
After microdissection from the temporal bone, both the roof and otolithic membrane were removed from utricles prior to culturing in DMEM/F-12 (Cat #10565018, Thermo Fisher Scientific Inc., Waltham, MA, USA) with 5% FBS (Cat #A4766801, Thermo Fisher Scientific Inc.,) and 10 µg/mL ciprofloxacin (Cat #449620050, Thermo Fisher Scientific Inc.,). Gentamicin (Item #17861615, Aspen Veterinary Resources LTD, Loveland, CO, USA) (0.1-1 mM, final concentration) was administered to the cultures on the day of dissection. After 24 hours, gentamicin was washed out and utricles were cultured with AAV for 3 more days. The utricles were then washed repeatedly to remove the AAV and were cultured for an additional 5-13 days before being fixed with 4% paraformaldehyde overnight at 4 °C and processed for immunohistochemistry. AAV carrying mouse Atoh1 was used for all ex vivo experiments.
In vivo drug delivery
Animal preparation: animals were prepared for surgery using sterile techniques. Briefly, hair was removed from the post-auricular region, which was prepped with 70% ethanol and draped. Animals were anesthetized with isoflurane in O2 inhalation (2-3% induction, 2-2.5% maintenance) via induction chamber followed by nose cone. Animal’s body temperature was maintained at 37.5 °C with a closed-loop heating pad and monitored using a rectal probe. Under an operating microscope, an incision was made, the muscles separated, and the posterior semicircular canal (PSC) exposed. A small hole was made on the PSC with a micro tool (PMT, Micro tool 100M2X300S), and the tip of the polyimide tube (Microlumen, code 039-I x12” long (ref. lot 024105-000)) inserted into the PSC toward the crus commune. Before delivery the therapeutic agent, the polyimide tube was be sealed in the PSC with tissue glue (Fisher Scientific #NC0304169). The tightness of the seal was visually confirmed by absence of perilymph leakage. Then the animals were given agents of interest at rate of 0.1 µL/minute for 10 min, giving infused total volumes of 1 µl. To allow the therapeutic agent to spread throughout the inner ear and stabilize, the tube was left in place for 3 min after the injection. The tube was then cut, folded, and left inside. The wound was thoroughly irrigated with saline and closed in layers by suturing or tissue glue. Carprofen (5 mg/kg, subcutaneous [SC]) was infiltrated into the subcutaneous layer to act as a local analgesic just prior to surgery. The total surgery duration was approximately 30-45 min. AAV carrying human ATOH1 was used for all in vivo experiments.
Immunohistochemistry
Utricles were fixed for 40 min in 4% paraformaldehyde (in PBS, pH 7.4; Electron Microscopy Services) at room temperature. Tissues were blocked with 5% donkey or goat serum, 0.1% TritonX-100, 1% bovine serum albumin (BSA), and 0.02% sodium azide (NaN3) in PBS at pH 7.4 for 1 hour at room temperature after washing with PBS, followed by incubation with primary antibodies diluted in the same blocking solution overnight at 4˚C. The next day, after washing with PBS, tissues were incubated with secondary antibodies diluted in 0.1% TritonX-100, 0.1% BSA, and 0.02% NaN3 solution in PBS for 2 hours at room temperature. After PBS washing, tissues were mounted onto slides in antifade Fluorescence Mounting Medium (DAKO or Leica). Antibodies against the following markers were used: Myosin7a (1:1000; Cat #256790, Proteus Bioscience), Pou4f3 (1:200, Cat # sc-81980, Santa Cruz Biotechnology Inc.), Sall2 (1:200; Cat # HPA004162, Sigma Aldrich), Sox2 (1:250; Cat #AF2018, R&D), Ctbp2 (1:200; Cat # 612044, BD Biosciences), GFP (1:200; ThermoFisher; A-11122). Secondary antibodies were conjugated with Alexa Fluor 488 (1:500, Thermo Fisher Scientific), Alexa Fluor 546 (1:500, Thermo Fisher Scientific), Alexa Fluor 647 (1:250, Thermo Fisher Scientific). Fluorescence-conjugated phalloidin (1:1000; Cat #A22287, Thermo Fisher Scientific) and DAPI (1:10000; Cat #D1306, Thermo Fisher Scientific) were also used.
For a subset of animals, they were perfused with 10% formalin, temporal bones were harvested and fixed with 10% formalin overnight. Temporal bones were then decalcified on shaker for 3-5 days at room temperature, placed in 1x PBS for 30 min, and then dehydrated stepwise in ethanol and then xylene. Tissue was embedded in paraffin and processed to yield 4 μm sections. Candidate sections were baked overnight at 65 ˚C. Sections were processed and prepared for staining on a BOND Rxm Stainer (Leica). This process included an epitope retrieval step (25 min; 94 C; EDTA pH 9.0). Blocking in 3% NGS and staining with an anti-GFP antibody (1:200; A-11122, ThermoFisher), an AP-conjugated secondary (Goat anti-rabbit; 31346, ThermoFisher) and detected with Fast Red (Abcam). Sections were then washed in PBS and H2O and mounted (CV Ultra Mounting Medium; Leica) before scanning.
Image acquisition, cell quantification and statistics
Images were captured using a 10x or 63x objective on a Zeiss LSM 700 or 880 confocal microscope (Zen Black, Zeiss). Images were taken from 1 representative area from striolar (central) and extrastriolar (peripheral) regions or the whole sensory epithelium for analysis. Whole utricle images were acquired using tile scanning via Z-stack images tiled across the length and width of the entire utricle at 63x magnification on the Zeiss LSM 880 confocal microscope. Zen Blue was used for image processing. The image was then stitched during post-processing to fuse the individual tiles. Imaris software (v9.8.2, Oxford Instruments, Manchester, UK) was used to generate ortho slicer (ortho-stack view) and 3D images. All cells were quantified from z-stack images of 2500 or 10,000 µm2 or the whole sensory epithelium using Fiji and Imaris softwares. Pou4f3+ hair cells, Sall2+ supporting cells were quantified using a semi-automated protocol developed with Imaris imaging analysis software (Oxford Instruments, United Kingdom). The spot function was implemented using a size threshold of 3.5 µm XY diameter on the Pou4f3 or Sall2 channel. Thresholds were manually adjusted to maximize the accuracy of the spots overlayed to the Pou4f3+ or Sall2+ nuclei. Any spots clearly in error were manually deleted and thus were not quantified. Myosin 7a+ hair cells, phalloidin-labeled bundles and Ctbp2 positive puncta per hair cell were manual counted by using Fiji (v2.0.0, NIH, Bethesda, Maryland, USA). GFP intensity in individual Pou4f3+ hair cells and Sall2+ supporting cells were auto measured by using Imaris software. Multiple observers were blinded to do the manual quantification. For all experiments, n values represent the number of cells or mice examined unless otherwise stated. Statistical analyses were conducted using Microsoft Excel (Microsoft), GraphPad Prism 9.0 software (GraphPad) when necessary. Two-tailed, paired Student’s t-tests, unpaired Student’s t-tests, one-way and two-way ANOVA were used to determine statistical significance. p < 0.05 was considered statistically significant: *p < 0.05, **p < 0.01, ***p < 0.001 Data shown as mean ± S.D or mean ± SEM.
Tissue dissection, processing, and single-cell RNA-seq
For scRNA-Seq, samples were processed fresh on the day of collection. For explant studies, 7-8 utricles were used per treatment group. For in vivo studies, cristae and utricles from 6 treated ears were microdissected in ice-cold DMEM/F12 media and collected in separate tubes, on ice. For transcriptional developmental trajectory studies, vestibular organs from 14 developmental time-points (E14.5, E15.5, E16.5, E18, P0, P1, P2, P3, P4, P5, P6, P10, P12, and P16) were microdissected in ice-cold DMEM/F12 media. For each timepoint, utricles from 5 mice were pooled into one tube for single cell dissociation. For the E14.5 timepoint, the entire vestibular tissue was collected as the utricle and cristae have not separated yet. The pooled tissue was then placed in a pre-warmed dissociation buffer of EBSS solution containing 33 units of papain (Worthington Biochemical), 1.67 mM L-cysteine, 0.83 mM EDTA, 15 mM HEPES, and 10 Kunitz/ml DNase I. The tissues were incubated for 45–60 min at 37 °C, triturating with a 200 μL pipette every 10 min for tissue dissociation. At the end of the 37 °C incubation, the solution was passed through a 25 G needle a few times to ensure a single cell suspension. An equal volume of DMEM/F12 media containing 20% ovomucoid protease inhibitor (Worthington Biochemical) was added, and the dissociated cells were passed through a 40 μm filter (pluriSelect) to remove debris. The cells were pelleted at 300 g for 5 min, washed twice with DMEM/F12 containing 0.04% BSA, and then pelleted and resuspended in the DMEM/F12 with BSA solution. The cells were counted with the Luna-FL automated fluorescence cell counter using an AO/PI viability assay (Logos Biosystems) and immediately loaded on the 10x Chromium controller (10x Genomics) for partitioning single cells into droplets. For each sample, 15,000 cells were loaded in one channel of a Chromium Chip (10x Genomics). 3’ v2 chemistry was used to profile one explant study and 3’ v3 chemistry was used to profile all other samples. cDNA and gene expression libraries were generated according to the manufacturer’s instructions (10x Genomics). cDNA and gene expression library fragment sizes were assessed with a DNA High Sensitivity Bioanalyzer Chip (Agilent). cDNA and gene expression libraries were quantified using the Qubit dsDNA High Sensitivity assay kit (Thermo Fisher Scientific). Gene expression libraries were multiplexed and sequenced on the Novaseq 6000 (Illumina) with the following read structure: Read 1: 28 cycles, Read 2: 72 - 100 cycles, Index Read 1: 8 cycles.
scRNA-seq expression matrices
Cell Ranger mkfastq (10x Genomics) was used to demultiplex the raw sequencing reads, and Cell Ranger count was used to align sequencing reads and generate a counts matrix. Reads were aligned to a custom-built mouse mm10 and transgene RNA reference.
scRNA-seq analysis of explant data
Data were processed with Seurat v4.1.1 (Stuart et. al., 2019) using R 4.0.4. Cells from 3 timepoints (day 7, 12, and 20) with the Atoh1 transgene combined with three promoters (CMV, GFAP, and RLBP1) were merged into one Seurat object. Cells with fewer than 2000 or greater than 6000 genes detected, or greater than 30% of UMIs mapped to mitochondrial genes were filtered out. On the remaining cells, 2000 variable genes were selected using the FindVariableFeatures function with default parameters. Data were scaled using ScaleData with default parameters and principal component analysis (PCA) was performed on the scaled data for the variable genes. Cells were clustered in PCA space using Seurat’s FindNeighbors on the top 50 PCs with an error bound of 0.1, followed by FindClusters with a resolution of 0.5. Cell types were assigned to all cells by evaluating the expression of known marker genes and comparing to previous scRNA-seq datasets, and cells were visualized on a UMAP using the top 50 PCs. Supporting cells and hair cells were retained. Two of the clusters annotated as supporting cells did not express canonical markers (i.e., Slc6a14, Agr3, or Lfng) and so were excluded, as well as one cluster of fully mature hair cells that were assumed to have escaped gentamicin damage and therefore not be regenerated. Using just the remaining cells, variable genes were chosen, data were scaled, and PCA performed using the same method as above. The data were then batch-corrected for the chemistry used (10X kit version) using Harmony v1.067 with default parameters. FindNeighbors (with an error bound of 0.1), FindClusters (with a resolution of 0.8), and UMAP dimensionality reduction were performed on the top 50 Harmony dimensions. Two clusters were removed, one because it showed high expression of glial markers and one because mixed marker gene expression and high numbers of UMIs led to the suspicion that it consisted of multiplets, and the procedure from choosing variable genes through creating a UMAP was repeated, this time with a clustering resolution of 0.4. Upregulated genes in each cluster were calculated using FindAllMarkers with the ROC method, and the genes with the top 10 AUC scores were plotted in a heatmap. In each dataset, the expression of genes related to synapses and bundles were plotted35,68.
scRNA-seq analysis of development and in vivo data
Data were processed with Seurat v4.1.169 using R 4.0.4. Datasets from each timepoint and treatment group were merged and processed together. On the full dataset, 2000 variable genes were selected using the FindVariableFeatures function with the VST method and default parameters. Data were scaled using ScaleData with default parameters and principal component analysis (PCA) was performed on the scaled data for the variable genes. Cells were clustered in PCA space using Seurat’s FindNeighbors on the top 50 PCs with an error bound of 0.1, followed by FindClusters with a resolution of 0.5. Cell types were assigned to each cluster by manual inspection of the expression levels of known marker genes for each cell type. For the in vivo data, supporting cells, hair cells and hATOH1+ SC/HC cluster (Supplementary Fig. 10f) or supporting cells, hair cells and hATOH1+ SC cluster (Supplementary Fig. 13f) were retained. Using just these cells, variable genes were chosen, data were scaled, PCA performed, and clusters assigned using the same method as above. Several small clusters were removed as suspected doublets as they expressed markers of cell types other than supporting cells and hair cells. Finally, the cells from each individual 10X lane and sequencing run were separately scaled and PCA was run, and then they were integrated together using Seurat’s IntegrateData on the top 75 PCs, using default parameters. This integrated dataset was scaled and clustered as before. A final round of very small clusters was removed as suspected doublets, followed by a final round of scaling and clustering as before. A UMAP was calculated on the remaining integrated cells, and clusters were assigned more specific cell types (Stage 1 through 3 regenerating hair cells) based on upregulated genes per cluster, calculated with FindAllMarkers using default parameters.
Maturation score
To estimate the maturity of regenerated cells, we used our dataset of mouse hair cells from embryonic and adult vestibule (Supplementary Fig. 4, see also GEO under accession codes GSE251854). Using this developmental dataset, we calculated the top 100 genes upregulated in immature, 200 genes upregulated in mature hair cells (independent of hair cell subtype), 100 genes upregulated in type I and type II hair cells, and 57 Notch genes (see complete list of genes in Supplementary Data 2). Signature scores were calculated for each cell in the regeneration datasets by isolating genes from those gene lists, taking normalized gene counts, scaling and centering counts per gene in each cell, and then averaging across all genes in the signature list. Signature scores were further added to converting hair cells using genes previously identified as Notch targets.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All relevant data and source data are included in this article and Supplementary Information. The single cell RNA sequencing data generated in this study have been deposited in NCBI Gene Expression Omnibus database under accession code GSE251854. Source data are provided with this paper.
Code availability
The following publicly available algorithms and software were used for data analysis presented in this manuscript. All implemented in R (v4.0.4) where applicable: Seurat (v4.1.1), Harmony (v1.0), and Cellranger (10X genomics, v6). Publicly available vignettes and code were used as well as default settings unless otherwise stated in the text. There were no custom algorithms generated.
References
Tsuji, K. et al. Temporal bone studies of the human peripheral vestibular system. Aminoglycoside ototoxicity. Ann. Otol. Rhinol. Laryngol. Suppl. 181, 20–25 (2000).
Rauch, S. D., Velazquez-Villasenor, L., Dimitri, P. S. & Merchant, S. N. Decreasing hair cell counts in aging humans. Ann. N. Y. Acad. Sci. 942, 220–227 (2001).
Agrawal, Y., Carey, J. P., Della Santina, C. C., Schubert, M. C. & Minor, L. B. Disorders of balance and vestibular function in US adults: data from the National Health and Nutrition Examination Survey, 2001-2004. Arch. Intern. Med. 169, 938–944 (2009).
Sayyid, Z. N., Kim, G. S. & Cheng, A. G. Molecular therapy for genetic and degenerative vestibular disorders. Curr. Opin. Otolaryngol. Head. Neck Surg. 26, 307–311 (2018).
Corwin, J. T. Postembryonic production and aging in inner ear hair cells in sharks. J. Comp. Neurol. 201, 541–553 (1981).
Popper, A. N. & Hoxter, B. Growth of a fish ear: 1. Quantitative analysis of hair cell and ganglion cell proliferation. Hear Res. 15, 133–142 (1984).
Jorgensen, J. M. & Mathiesen, C. The avian inner ear. Continuous production of hair cells in vestibular sensory organs, but not in the auditory papilla. Naturwissenschaften 75, 319–320 (1988).
Corwin, J. T. & Cotanche, D. A. Regeneration of sensory hair cells after acoustic trauma. Science 240, 1772–1774 (1988).
Ryals, B. M. & Rubel, E. W. Hair cell regeneration after acoustic trauma in adult Coturnix quail. Science 240, 1774–1776 (1988).
Dooling, R. J., Ryals, B. M. & Manabe, K. Recovery of hearing and vocal behavior after hair-cell regeneration. Proc. Natl Acad. Sci. USA 94, 14206–14210 (1997).
Smolders, J. W. Functional recovery in the avian ear after hair cell regeneration. Audio. Neurootol. 4, 286–302 (1999).
Golub, J. S. et al. Hair cell replacement in adult mouse utricles after targeted ablation of hair cells with diphtheria toxin. J. Neurosci. 32, 15093–15105 (2012).
Sayyid, Z. N., Wang, T., Chen, L., Jones, S. M. & Cheng, A. G. Atoh1 directs regeneration and functional recovery of the mature mouse vestibular system. Cell Rep. 28, 312–324.e314 (2019).
Jen, H. I. et al. Transcriptomic and epigenetic regulation of hair cell regeneration in the mouse utricle and its potentiation by Atoh1. Elife 8, https://doi.org/10.7554/eLife.44328 (2019).
Kawamoto, K., Izumikawa, M., Beyer, L. A., Atkin, G. M. & Raphael, Y. Spontaneous hair cell regeneration in the mouse utricle following gentamicin ototoxicity. Hear Res. 247, 17–26 (2009).
Atkinson, P. J., Huarcaya Najarro, E., Sayyid, Z. N. & Cheng, A. G. Sensory hair cell development and regeneration: similarities and differences. Development 142, 1561–1571 (2015).
Gonzalez-Garrido, A. et al. The differentiation status of hair cells that regenerate naturally in the vestibular inner ear of the adult mouse. J. Neurosci. 41, 7779–7796 (2021).
Wang, T. et al. Uncoordinated maturation of developing and regenerating postnatal mammalian vestibular hair cells. PLoS Biol. 17, e3000326 (2019).
Schlecker, C. et al. Selective atonal gene delivery improves balance function in a mouse model of vestibular disease. Gene Ther. 18, 884–890 (2011).
Guo, J. Y. et al. AAV8-mediated Atoh1 overexpression induces dose-dependent regeneration of vestibular hair cells in adult mice. Neurosci. Lett. 747, 135679 (2021).
Shou, J., Zheng, J. L. & Gao, W. Q. Robust generation of new hair cells in the mature mammalian inner ear by adenoviral expression of Hath1. Mol. Cell Neurosci. 23, 169–179 (2003).
Staecker, H., Praetorius, M., Baker, K. & Brough, D. E. Vestibular hair cell regeneration and restoration of balance function induced by math1 gene transfer. Otol. Neurotol. 28, 223–231 (2007).
Staecker, H. et al. Optimizing atoh1-induced vestibular hair cell regeneration. Laryngoscope 124, S1–S12 (2014).
Taylor, R. R. et al. Characterizing human vestibular sensory epithelia for experimental studies: new hair bundles on old tissue and implications for therapeutic interventions in ageing. Neurobiol. Aging 36, 2068–2084 (2015).
Yang, H., Xie, X., Deng, M., Chen, X. & Gan, L. Generation and characterization of Atoh1-Cre knock-in mouse line. Genesis 48, 407–413 (2010).
Pan, N. et al. A novel Atoh1 “self-terminating” mouse model reveals the necessity of proper Atoh1 level and duration for hair cell differentiation and viability. PLoS ONE 7, e30358 (2012).
Liu, Z. et al. Age-dependent in vivo conversion of mouse cochlear pillar and Deiters’ cells to immature hair cells by Atoh1 ectopic expression. J. Neurosci. 32, 6600–6610 (2012).
Zheng, J. L. & Gao, W. Q. Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat. Neurosci. 3, 580–586 (2000).
Gubbels, S. P., Woessner, D. W., Mitchell, J. C., Ricci, A. J. & Brigande, J. V. Functional auditory hair cells produced in the mammalian cochlea by in utero gene transfer. Nature 455, 537–541 (2008).
Ben-Arie, N. et al. Math1 is essential for genesis of cerebellar granule neurons. Nature 390, 169–172 (1997).
Miesegaes, G. R. et al. Identification and subclassification of new Atoh1 derived cell populations during mouse spinal cord development. Dev. Biol. 327, 339–351 (2009).
Wright, M. C. et al. Unipotent, Atoh1+ progenitors maintain the Merkel cell population in embryonic and adult mice. J. Cell Biol. 208, 367–379 (2015).
Taylor, R. R. et al. Regenerating hair cells in vestibular sensory epithelia from humans. Elife 7, https://doi.org/10.7554/eLife.34817 (2018).
Praetorius, M. et al. Adenovector-mediated hair cell regeneration is affected by promoter type. Acta Otolaryngol. 130, 215–222 (2010).
Jan, T. A. et al. Spatiotemporal dynamics of inner ear sensory and non-sensory cells revealed by single-cell transcriptomics. Cell Rep. 36, 109358 (2021).
Wang, G. P. et al. Adeno-associated virus-mediated gene transfer targeting normal and traumatized mouse utricle. Gene Ther. 21, 958–966 (2014).
Cunningham, L. L., Cheng, A. G. & Rubel, E. W. Caspase activation in hair cells of the mouse utricle exposed to neomycin. J. Neurosci. 22, 8532–8540 (2002).
Forge, A., Li, L. & Nevill, G. Hair cell recovery in the vestibular sensory epithelia of mature guinea pigs. J. Comp. Neurol. 397, 69–88 (1998).
McInturff, S., Burns, J. C. & Kelley, M. W. Characterization of spatial and temporal development of type I and type II hair cells in the mouse utricle using new cell-type-specific markers. Biol. Open 7, https://doi.org/10.1242/bio.038083 (2018).
Burns, J. C., Kelly, M. C., Hoa, M., Morell, R. J. & Kelley, M. W. Single-cell RNA-Seq resolves cellular complexity in sensory organs from the neonatal inner ear. Nat. Commun. 6, 8557 (2015).
Desai, S. S., Zeh, C. & Lysakowski, A. Comparative morphology of rodent vestibular periphery. I. Saccular and utricular maculae. J. Neurophysiol. 93, 251–266 (2005).
Desai, S. S., Ali, H. & Lysakowski, A. Comparative morphology of rodent vestibular periphery. II. Cristae ampullares. J. Neurophysiol. 93, 267–280 (2005).
Eatock, R. A. & Songer, J. E. Vestibular hair cells and afferents: two channels for head motion signals. Annu. Rev. Neurosci. 34, 501–534 (2011).
Contini, D. et al. Intercellular K(+) accumulation depolarizes type I vestibular hair cells and their associated afferent nerve calyx. Neuroscience 227, 232–246 (2012).
Collado, M. S. et al. The postnatal accumulation of junctional E-cadherin is inversely correlated with the capacity for supporting cells to convert directly into sensory hair cells in mammalian balance organs. J. Neurosci. 31, 11855–11866 (2011).
Lin, V. et al. Inhibition of Notch activity promotes nonmitotic regeneration of hair cells in the adult mouse utricles. J. Neurosci. 31, 15329–15339 (2011).
Wang, G. P. et al. Notch signaling and Atoh1 expression during hair cell regeneration in the mouse utricle. Hear Res. 267, 61–70 (2010).
Helms, A. W., Abney, A. L., Ben-Arie, N., Zoghbi, H. Y. & Johnson, J. E. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development 127, 1185–1196 (2000).
Shi, F., Cheng, Y. F., Wang, X. L. & Edge, A. S. Beta-catenin up-regulates Atoh1 expression in neural progenitor cells by interaction with an Atoh1 3’ enhancer. J. Biol. Chem. 285, 392–400 (2010).
Kelly, M. C., Chang, Q., Pan, A., Lin, X. & Chen, P. Atoh1 directs the formation of sensory mosaics and induces cell proliferation in the postnatal mammalian cochlea in vivo. J. Neurosci. 32, 6699–6710 (2012).
Cai, T. et al. Characterization of the transcriptome of nascent hair cells and identification of direct targets of the Atoh1 transcription factor. J. Neurosci. 35, 5870–5883 (2015).
Soler-Martin, C., Diez-Padrisa, N., Boadas-Vaello, P. & Llorens, J. Behavioral disturbances and hair cell loss in the inner ear following nitrile exposure in mice, guinea pigs, and frogs. Toxicol. Sci. 96, 123–132 (2007).
Pacentine, I., Chatterjee, P. & Barr-Gillespie, P. G. Stereocilia Rootlets: actin-based structures that are essential for structural stability of the hair bundle. Int. J. Mol. Sci. 21, 324 (2020).
Nouvian, R., Beutner, D., Parsons, T. D. & Moser, T. Structure and function of the hair cell ribbon synapse. J. Membr. Biol. 209, 153–165 (2006).
Schug, N. et al. Differential expression of otoferlin in brain, vestibular system, immature and mature cochlea of the rat. Eur. J. Neurosci. 24, 3372–3380 (2006).
Kim, G. S. et al. Repair of surviving hair cells in the damaged mouse utricle. Proc. Natl Acad. Sci. USA 119, e2116973119 (2022).
Rio, C., Dikkes, P., Liberman, M. C. & Corfas, G. Glial fibrillary acidic protein expression and promoter activity in the inner ear of developing and adult mice. J. Comp. Neurol. 442, 156–162 (2002).
Llorens, J. & Dememes, D. Hair cell degeneration resulting from 3,3’-iminodipropionitrile toxicity in the rat vestibular epithelia. Hear Res. 76, 78–86 (1994).
Bi, Z. et al. Development and transdifferentiation into inner hair cells require Tbx2. Natl Sci. Rev. 9, nwac156 (2022).
Atkinson, P. J., Kim, G. S. & Cheng, A. G. Direct cellular reprogramming and inner ear regeneration. Expert Opin. Biol. Ther. 19, 129–139 (2019).
Collado, M. S., Burns, J. C., Hu, Z. & Corwin, J. T. Recent advances in hair cell regeneration research. Curr. Opin. Otolaryngol. Head Neck Surg. 16, 465–471 (2008).
Cheng, Y. F., Tong, M. & Edge, A. S. Destabilization of Atoh1 by E3 ubiquitin ligase Huwe1 and casein kinase 1 is essential for normal sensory hair cell development. J. Biol. Chem. 291, 21096–21109 (2016).
Abdul-Aziz, D., Hathiramani, N., Phung, L., Sykopetrites, V. & Edge, A. S. B. HIC1 represses Atoh1 transcription and hair cell differentiation in the cochlea. Stem Cell Rep. 16, 797–809 (2021).
Luo, Z. et al. Three distinct Atoh1 enhancers cooperate for sound receptor hair cell development. Proc. Natl Acad. Sci. USA 119, e2119850119 (2022).
Ono, K. et al. Retinoic acid degradation shapes zonal development of vestibular organs and sensitivity to transient linear accelerations. Nat. Commun. 11, 63 (2020).
Ratzan, E. M. et al. TMC function, dysfunction, and restoration in mouse vestibular organs. Front. Neurol. 15, 1356614 (2024).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Johnson, S. L., Safieddine, S., Mustapha, M. & Marcotti, W. Hair cell afferent synapses: function and dysfunction. Cold Spring Harb. Perspect. Med. 9, https://doi.org/10.1101/cshperspect.a033175 (2019).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e1821 (2019).
Acknowledgements
We thank the Cheng Lab, P. Atkinson, S. Billings, and T. Jan for insightful comments and A. Palermo, K. Ellis, J. Sellon and D. Gobron for excellent technical support on the manuscript. This work was supported by National Natural Science Foundation of China 82071056 (T.W.), R01DC016919, RO1DC021110, K24DC020986 (to A.G.C.), and generous support by the Yu and Ogawa families and the Bill and Susan Oberndorf Foundation.
Author information
Authors and Affiliations
Contributions
T.W., T.Y., X.W., K.S., L.B., N.P., J.H., B.H., and M.K. performed experiments; T.W., T.Y., J.B., and A.G.C. designed experiments; T.W., T.Y., T.G., G.P., R.M., L.C., A.M., S.N., N.D., A.K., J.B., and A.G.C. analyzed data; T.W., T.Y., G.P., R.M., N.D., A.K., J.B., and A.G.C. prepared the manuscript.
Corresponding authors
Ethics declarations
Competing interests
T.Y., A.K., G.P., R.M., B.H., X.W., L.B., N.P., K.S., S.N., M.K., T.G., N.D., and J.B. were employees of Decibel Therapeutics. G.P., R.M., L.B., N.P., and K.S. are current employees of Regeneron Pharmaceuticals. A.G.C. was a member of Decibel Therapeutics’ scientific advisory board. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Heiko Locher, Adam Carlton and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, T., Yang, T., Kedaigle, A. et al. Precise genetic control of ATOH1 enhances maturation of regenerated hair cells in the mature mouse utricle. Nat Commun 15, 9166 (2024). https://doi.org/10.1038/s41467-024-53153-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-53153-0
This article is cited by
-
Pou4f3 Deficiency Obstructs the Subtype Differentiation of Vestibular Hair Cells
Neuroscience Bulletin (2025)