Abstract
The etiology of primary open angle glaucoma is constituted by both intraocular pressure-dependent and intraocular pressure-independent mechanisms. However, GWASs of traits affecting primary open angle glaucoma through mechanisms independent of intraocular pressure remains limited. Here, we address this gap by subtracting the genetic effects of a GWAS for intraocular pressure from a GWAS for primary open angle glaucoma to reveal the genetic contribution to primary open angle glaucoma via intraocular pressure-independent mechanisms. Seventeen independent genome-wide significant SNPs were associated with the intraocular pressure-independent component of primary open angle glaucoma. Of these, 7 are located outside known normal tension glaucoma loci, 11 are located outside known intraocular pressure loci, and 2 are novel primary open angle glaucoma loci. The intraocular pressure-independent genetic component of primary open angle glaucoma is associated with glaucoma endophenotypes, while the intraocular pressure-dependent component is associated with blood pressure and vascular permeability. A genetic risk score for the intraocular pressure-independent component of primary open angle glaucoma is associated with 26 different retinal micro-vascular features, which contrasts with the genetic risk score for the intraocular pressure-dependent component. Increased understanding of these intraocular pressure-dependent and intraocular pressure-independent components provides insights into the pathogenesis of glaucoma.
Similar content being viewed by others
Introduction
Primary open angle glaucoma (POAG) is the most common form of glaucoma. Worldwide, the condition affects more than 3% of the population aged 40 and above1, although the prevalence varies widely by ethnicity2. The primary risk factor for POAG is increasing age1. POAG is usually asymptomatic in the early stage, suggesting its prevalence may be underestimated3. As well as being highly prevalent, the visual impairment due to POAG is irreversible, therefore the condition has a major adverse impact on health-related quality of life4. POAG is heritable, and previous studies have explored its genetic architecture extensively5,6,7,8,9. The largest genome-wide association study (GWAS) for POAG to date identified 127 loci associated with disease risk10. Despite this success, our knowledge of the disease mechanisms underlying the vision loss caused by POAG remains incomplete.
Intraocular pressure (IOP) is the only known modifiable risk factor for POAG and IOP-lowering medication is currently the mainstay treatment for POAG. IOP can be measured non-invasively in population-based samples, which has facilitated GWAS analyses of IOP and the identification of more than 100 genetic variants associated with this trait9,11,12. However, a substantial proportion of patients with POAG have IOP within the normal range—so called, normal tension glaucoma (NTG)13. In addition, the genetic correlation between IOP and POAG ranges from 0.49 to 0.80, which suggests that IOP alone cannot comprehensively account for the development of POAG11,14,15. In animal models, glaucoma can be induced by non-IOP-related factors16. Together, these sources of evidence suggest that both IOP-dependent and IOP-independent (non-IOP-dependent) mechanisms contribute to the pathobiology of POAG16,17.
Past research into the IOP-dependent component of POAG has highlighted the role of mechanical compression or stretching forces exerted on the retina, especially at the laminar cribrosa of the optic nerve head (ONH), where retinal ganglion cell (RGC) axons may be particularly vulnerable18. In animal models, long-term exposure to elevated IOP-induced mechanical compression leads to hallmark signs of glaucomatous optic neuropathy (GON), such as thinning of the retinal nerve fiber layer (RNFL) and excavation (cupping) of the optic disc, and finally progressive visual field loss19. Research focusing on non-IOP-dependent mechanisms underlying POAG has emphasized the role of the retinal vascular system. Deficient ocular blood perfusion (OBF) results in the degeneration of RGCs13,20,21,22. Vasospasm, autoregulatory dysfunction, endothelial cell dysfunction, and morphological changes in retinal vessels have been shown to disrupt OBF and thus may contribute to the non-IOP-dependent component of POAG23.
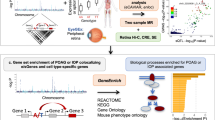
As with POAG, existing evidence supports a role for both non-IOP-dependent and IOP-dependent mechanisms in the etiology of NTG20,24. Hence, GWAS analyses of NTG cases and healthy controls may not be the most effective way to delineate the genetic architecture of the non-IOP-dependent component of POAG. Here, we applied a genomic structural equation model (GSEM) approach called GWAS-by-Subtraction to gain insight into the non-IOP-dependent component of POAG (Fig. 1a). In the first stage, we derived the genome-wide genetic effects corresponding to the non-IOP-dependent component of POAG by subtracting IOP-related genetic effects from those of POAG in general. The findings were validated using a GWAS-by-Subtraction analysis with different, independent input GWAS samples (Fig. 1b). We then constructed separate genetic risk scores (GRS) quantifying an individual’s susceptibility to IOP-dependent and non-IOP-dependent POAG disease mechanisms. We found the GRS for the non-IOP-dependent component of POAG was associated with retinal microvascular features (RMF), providing strong evidence supporting the role of the vascular system in non-IOP-dependent POAG disease mechanisms.
Schematic diagram of the genomic SEM implemented in the GWAS-by-subtraction method. a The measured (observed) variables SNP, IOPcc, and POAG, indicated as rectangles, correspond to the SNP genotype, the observed IOPcc and the observed POAG case-control status for a sample population. Two latent variables, labeled IOP and non-IOP, are indicated as filled circles. The latent variables correspond to hypothetical (unobserved) factors that mediate the effect of the SNP on IOPcc and POAG. Straight arrows depict causal paths, while circular arrows indicate variances used to constrain the SEM. b Flowchart of the analysis process.
Results
GWAS-by-subtraction identifies novel variants associated with the non-IOP-dependent component of POAG and replicates known NTG loci
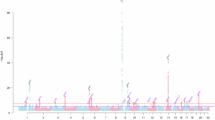
In the discovery stage, genetic effects associated with corneal-compensated IOP (IOPcc) (n = 97,644) were subtracted from those of POAG (14853 cases vs. 106544 controls; reported by Gharahkhani et al.10 that excluded participants in the FinnGen study) (Supplementary Data 1 and 2). GWAS-by-subtraction identified 17 independent genome-wide significant SNPs (P < 5 × 10−8) from 15 separate regions that were associated with the non-IOP-dependent component of POAG (Fig. 2; Table 1). The SNP heritability (\({h}_{{SNP}}^{2}\)) of the non-IOP-dependent component of POAG estimated by LDSC was 5.7%, with no evidence of inflation of test statistics due to population structure (LDSC intercept = 1.03). Four of the lead genome-wide significant SNPs in the 15 regions corresponded to known NTG variants (rs2790049 at the TMCO1 locus, rs6475604 at the CDKN2B-AS1 locus, rs2472494 at the ABCA1-SLC44A1 locus and rs34935520 at the RPL37P5-SIX1 locus)7,10. In addition, four SNPs were reported in a Multi-Trait analysis of GWAS for NTG described in a recent preprint25 (rs6845653 at the AFAP1 locus, rs10740731 at the BICC1 locus, rs2667477 at the TMTC2 locus and rs6117318 at the CASC20 locus), while the results for 7 regions were not reported as NTG loci in previous studies. Notably, variants rs113894504 (ANAPC1) and rs7803764 (TMEM196) were not significant in the GWAS for POAG used as input for the GWAS-by-subtraction analysis (Table 1; Supplementary Data 3). This demonstrated that our approach went beyond simply separating known POAG loci into IOP-dependent and non-IOP-dependent categories26.
Plot of the −log10(P-value) associated with Wald’s test (two-sided) of the beta coefficient for the non-IOP-dependent component of POAG for all SNPs, ordered by chromosome and base position. The threshold for genome-wide significance after considering of multiple tests is indicated by the red dashed line (P = 5.0 × 10−8).
Sensitivity analyses in which the genetic correlation between the IOP-dependent and non-IOP-dependent latent factors for POAG was less tightly constrained (rg = 0.1 or 0.2) during the GWAS-by-subtraction analysis led to progressively more SNPs being associated with the non-IOP-dependent component of POAG at genome-wide significance (Supplementary Fig. 1 and Supplementary Data 4).
Lead SNPs for the non-IOP-dependent component of POAG were replicated
Three validation analyses were carried out to verify that the GWAS-by-subtraction results were not reliant on either of the input GWAS datasets utilized in the discovery analysis (Supplementary Data 1). In validation analysis 1, GWAS-by-subtraction was performed with a different IOP sample as input, namely an International Glaucoma Genetic Consortium (IGGC) GWAS for IOP carried out by Bonnemaijer et al.12 that did not include participants from UK Biobank. This IGGC IOP-based GWAS-by-subtraction analysis identified 20 independent SNPs, all of which were significant (P < 0.05) in the main analysis, while 16 of the 17 SNPs reported in the discovery phase showed evidence of replication (P < 0.05) (Supplementary Figs. 2 and 3, Supplementary Data 4). In validation analysis 2, GWAS-by-subtraction was performed with a different POAG sample as input, namely, a GWAS for POAG in FinnGen participants (release 9; 7756 cases and 358375 controls) that had no overlap with the POAG sample used in the discovery analysis (Supplementary Figs. 2 and 3, Supplementary Data 4). Of the 17 loci reported in the discovery phase, the association results for 15 SNPs were available in validation analysis 2. An additional 2 SNPs were selected as proxies for those not present; both were in perfect linkage disequilibrium (LD) r2 = 1 with the target SNP. Of these 17 SNPs, there were 14 that showed evidence of replication (P < 0.05), including loci in known NTG regions as well as newly identified loci in ANAPC1, CADM2, MIR3925, and PKH1D (Supplementary Data 4). The lead SNP at the TMEM196 locus had the same direction of effect and approach nominal significance (P = 0.07). In validation analysis 3, GWAS-by-subtraction was performed with completely independent input samples: the GWAS for POAG in FinnGen participants and the IGGC GWAS for IOP reported by Bonnemaijer et al.12. Of the 14 SNPs present in both the discovery analysis and validation analysis 3, 11 SNPs showed evidence of replication, while for the 3 other variants for which a proxy SNP was assessed, 2 showed evidence of replication (Supplementary Figs. 2 and 3, Supplementary Data 4).
The IOP-dependent component of POAG does not include novel loci
There were 75 independent genome-wide significant SNPs from 48 regions associated with the IOP-dependent component of POAG (Supplementary Data 5, Supplementary Fig. 4). Once again, there was minimal evidence of inflation due to population stratification (LDSC intercept = 1.04). The correlation between the initial GWAS for IOPcc and the GWAS-by-subtraction results for the IOP-dependent component of POAG was high (rg = 1.0, P < 0.001). The \({h}_{{SNP}}^{2}\) for IOPcc was 14.8%, whereas for the IOP-dependent component of POAG the \({h}_{{SNP}}^{2}\) was 11.9%, which tentatively suggested that the IOP-dependent and non-IOP-dependent risk mechanisms for POAG partially overlap.
The IOP-dependent and non-IOP-dependent components of POAG have distinct patterns of association with other traits
Fifteen of the 17 genome-wide significant variants associated with the non-IOP-dependent component of POAG were found to have been linked to POAG or a glaucoma-related trait in previous GWAS analyses (Supplementary Data 6). Furthermore, SNPs from 13 of the 15 regions where the SNPs were located had pleiotropic effects on other traits. The lead variant at the ANAPC1 locus, rs113894504, was in strong LD with a known IOP-associated variant, rs2030500, suggesting that ANAPC1 or a nearby gene influences susceptibility to POAG through pleiotropic mechanisms that are both dependent and independent of IOP. Many of the lead non-IOP-dependent component SNPs were also associated with a variety of glaucoma endophenotypes, such as optic cup area, optic disc area, vertical cup-disc ratio (VCDR), and RNFL thickness (Fig. 3a). By contrast, only a few IOP-dependent component SNPs were associated with glaucoma endophenotypes (Fig. 3b; Supplementary Data 7). Instead, the majority of the IOP-dependent component SNPs had only been associated with either glaucoma or IOP in previous GWAS analyses.
The genetic correlation between POAG and various glaucoma endophenotypes12 was moderate-to-high: optic cup area, rg = 0.57 (95% CI: 0.45–0.69, P = 1.67 × 10−20), optic disc area, rg = 0.22 (95% CI: 0.10–0.34, P = 5.32 × 10−4), VCDR, rg = 0.53 (95% CI:0.41– 0.65, P = 1.18 × 10−17). Consistent with the results described above, genetic correlations between the non-IOP-dependent component of POAG and these glaucoma endophenotypes were also moderate-to-high (Fig. 4; Supplementary Data 8). By contrast, the genetic correlations between the IOP-dependent component of POAG and these glaucoma endophenotypes were notably lower, at about half the magnitude observed for the non-IOP-dependent component (Fig. 4; Supplementary Data 8). For example, in the case of VCDR, the rg = 0.54 (95% CI: 0.41–0.68, P = 2.50 × 10−15) for the non-IOP-dependent component of POAG and rg = 0.19 (95% CI: 0.10–0.29, P = 4.86 × 10−5) for the IOP-dependent component of POAG.
The dots represent genetic correlations estimated using LDSC. Correlations with the non-IOP-dependent component of POAG are shown in red and with the IOP-dependent component of POAG in dark blue. Error bars represent 95% confidence intervals (CIs). Closed circles indicate statistically significant genetic correlation (LDSC uncorrected P < 0.05, two-sided test). Full results, study details and sample size are reported in Supplementary Data 8. Body mass index (BMI); systolic blood pressure (SBP), diastolic blood pressure (DBP); pulse pressure (PP); low-density lipoprotein (LDL); high-density lipoprotein (HDL); coronary artery disease (CAD); chronic kidney disease (CKD). Source data are provided on figshare (https://doi.org/10.6084/m9.figshare.25045220).
Relationship with systolic blood pressure and other clinical traits
Further genetic correlation analyses were performed to identify relationships between the two GWAS-by-subtraction traits and other clinically relevant traits. POAG itself demonstrated a low, but statistically significant, positive genetic correlation with systolic blood pressure (SBP) (rg = 0.05, 95% CI: 7.1e-04–0.1, P = 0.046)27. Interestingly, the IOP-dependent component of POAG had higher, positive genetic correlations with blood pressure traits: SPB, rg = 0.10 (95% CI: 0.05–0.15, P = 2.42 × 10−5); pulse pressure (PP)27, rg = 0.10 (95% CI: 0.05–0.14, P = 8.76 × 10−5). By contrast, the non-IOP-dependent component of POAG was not correlated with blood pressure traits to an appreciable extent (Fig. 4; Supplementary Data 8). For the remaining traits investigated28,29,30,31,32,33,34,35,36, none of the genetic correlations was significant after accounting for multiple testing.
eQTL, gene, pathway, and tissue-specific enrichment analyses
First, the function of the SNPs associated with the non-IOP-dependent component of POAG was investigated by examining their roles in the regulation of gene expression. Thirteen of the 17 lead non-IOP-dependent SNPs (including those in LD r2 > 0.8) were known expression quantitative trait loci (eQTLs; false discovery rate [FDR] <0.05 across various tissues; Supplementary Data 9). A Bayesian-based colocalization analysis between the 17 SNPs associated with the non-IOP-dependent component of POAG and gene expression signatures in blood (GTEx v7.0) identified 3 loci with evidence that the GWAS-by-subtraction SNP was a causal variant regulating gene expression (Colocalization Posterior Probability [CLPP] > 0.01; Supplementary Data 10).
Gene-based enrichment analysis with MAGMA prioritized 23 genes associated with the non-IOP-dependent component of POAG (Supplementary Data 11). Among these 23 genes, ALDH9A1, TMCO1, AFAP1, CDKN2A, CDKN2B, BICC1, ABCA1, PPM1A, C14orf39, and SIX6 were located in regions previously associated with NTG7,10,25, while ETV7, PXT1, KCTD20, TWISTNB, TMEM196, and C9orf53 genes were previously known to be associated with ONH morphology37. Gene-based enrichment analysis also prioritized 115 genes associated with the IOP-dependent component of POAG (Supplementary Data 12).
GO pathway analysis for the non-IOP-dependent component of POAG yielded a single significantly enriched pathway, Apolipoprotein A-1 binding (P = 3.24 × 10−7). The GO pathway, Negative regulation of vascular permeability, was the most strongly enriched pathway in the MAGMA analysis for the IOP-dependent component of POAG (P = 3.90 × 10−7; Supplementary Data 13). This enrichment was largely driven by the association of the ANGPT1 and PTPRJ genes (Supplementary Data 13). GENE2FUNC analysis with FUMA prioritized pathways related to cell cycle or immune reaction for the non-IOP-dependent component of POAG (Supplementary Fig. 5) and several pathways related to the sensory system for the IOP-dependent component of POAG (Supplementary Fig. 6).
We performed cell-type-specific enrichment analyses with WebCSEA to identify tissues or cell types enriched for genes identified in the GWAS-by-subtraction analysis. Skeletal muscle satellite stem cells were prioritized for the non-IOP-dependent component of POAG (Supplementary Figs. 7 and 8). In the visual system (eye, retina, and visual cortex tissues), genes relevant to the non-IOP-dependent component of POAG were relatively enriched in photoreceptor cells, retinal progenitors and Muller glia cells and visual cortex cells (Supplementary Figs. 9). Consistent with the pathway analysis, CSEA of the IOP-dependent component of POAG prioritized vascular-related endothelial cells when considering overall tissue and cell types, suggesting a potential role for vessel permeability in POAG (Supplementary Figs. 10 and 11). When focusing on the visual system, genes were predominantly enriched in cells of the juxtacanalicular region of the ocular trabecular meshwork and retina fibroblast cells (Supplementary Fig. 12).
Capacity to predict POAG and its subtypes
A subset of UK Biobank participants could be classified as NTG cases, high-tension glaucoma (HTG) cases or non-POAG controls (see Methods subsection “Assessment of IOP, glaucoma, fundus images, and covariates”). When comparing POAG cases (n = 7205) vs. controls (n = 191,710), a GRS for the non-IOP-dependent component of POAG was modestly predictive of glaucoma risk (OR = 1.22 per one standard deviation increase in the GRS; P < 0.001), as was the GRS for the IOP-dependent component of POAG (OR = 1.27; P < 0.001). As regards POAG subtypes, both GRSs demonstrated significant association with the risk of NTG and HTG. Specifically, in the case of NTG (2384 cases compared to 191,710 controls), the GRS for the non-IOP-dependent component demonstrated an OR of 1.08 (P = < 0.001), while the GRS for the IOP-dependent component yielded an OR of 1.07 (P < 0.001). Similarly, in the context of HTG risk (4821 cases vs. 191,710 controls), the GRS for the non-IOP-dependent component resulted in an OR of 1.30 (P < 0.001), and the GRS for the IOP-dependent component produced an OR of 1.38 (P < 0.001). A polygenic risk ratio (PRR)—defined as the GRS of the non-IOP-dependent component of POAG divided by that for the IOP-dependent component—was assessed for its capacity to distinguish NTG cases (n = 2384) from HTG cases (n = 4821). After adjusting for the GRS of POAG, a 10% increase in the PRR was associated with 5% increase in the risk of NTG (Table 2). Since these analyses predicting POAG and its subtypes were carried out in samples of UK Biobank participants, a sensitivity analysis was conducted in which SNP effect sizes were derived from validation analysis 3 (a GWAS-by-subtraction analysis that was fully independent of UK Biobank). In the sensitivity analysis, the GRSs for both the IOP-dependent and non-IOP-dependent components of POAG demonstrated similar predictive effects as observed in the main analysis (Table 2).
Associations with retinal microvascular features
Retinal-based Microvascular Health Assessment System (RMHAS) analysis quantified a total of 91 distinct RMFs (Supplementary Data 14) in the sample of 50,624 participants whose data passed quality control filters. After adjusting for covariates, lower central retinal artery equivalent (CRAE) caliber, lower arterial complexity, greater artery length diameter ratio and longer artery length were associated with an increased risk of ICD-10-inferred glaucoma (583 cases vs. 48497 controls) (Supplementary Data 15). The GRS for the IOP-dependent component of POAG was associated with 4 of the 91 RMFs after accounting for multiple testing (P < 5.49 × 10−4; corresponding to 0.05/91 features) (Supplementary Data 16), and they were all highly correlated RMFs related to vessel complexity. However, the GRS for the non-IOP-dependent component of POAG was significantly associated with 26 different RMFs (P < 5.49 × 10−4) (Supplementary Data 17). The most significantly associated RMF was the number of vessels within the optic disc area (termed ntree_a for arterioles and ntree_v for venules). A higher GRS was associated with fewer vessels passing across the optic disc area (ntree_a: beta = −3.56 × 10−03, P = 1.28 × 10−19; ntree_v: beta = −1.51 × 10−03, P = 5.26 × 10−6). Four RMFs related to vascular network complexity—artery and total fractal dimension (fda and fdt) and number of vessel segments (nseg_a and nseg_v)—were also negatively associated with the GRS (fda, beta = −8.23 × 10−05, P = 2.85 × 10−13; fdt, beta = −4.62 × 10−05, P = 2.07 × 10−5; nseg_a, beta = −4.59 × 10−2, P = 1.33 × 10−13; nseg_v, beta = −0.03, P = 4.15 × 10−8). These findings suggest that reduced retinal vessel complexity might be a sign of POAG, especially in patients whose IOP is within the normal range. In addition, a further 13 highly correlated RMFs indicative of vessel width were positively associated with increasing GRS (Supplementary Fig. 13). There were also associations with indices of vessel tortuosity (curveangle_mean_a and curveangle_mean_v). A higher GRS was associated with reduced tortuosity, quantified as lower values of curveangle_mean_a or curveangle_mean_v (curveangle_mean_a: beta = −2.58 × 10−3, P = 6.84 × 10−9; curveangle_mean_v: beta = −1.37 × 10−3, P = 1.14 × 10−6; Fig. 5, Supplementary Data 17).
The x-axis lists vessel features, the y-axis is the Z-score of association. Circles represent the GRS of the IOP-dependent component of POAG, while triangles represent the GRS of the non-IOP-dependent component. Symbols in yellow were significant after Bonferroni correction. Source data are provided on figshare (https://doi.org/10.6084/m9.figshare.25045220).
Discussion
In this study we used a genomic structural equation modeling approach, GWAS-by-subtraction, to dissect the genetic risk of POAG into an IOP-independent component and an IOP-dependent component. Unlike single-trait GWAS, genomic SEM considers multiple traits in a single model, incorporating their intricate interrelationships, thereby enhancing the likelihood of uncovering mechanism-specific associations. We hypothesized both the IOP-dependent and non-IOP-dependent biological processes contribute to the etiology of POAG. By isolating the impact of IOP-dependent effects on POAG through GSEM, the residual genetic effects represent a component of POAG not influenced by IOP. GWAS summary statistics for the non-IOP-dependent component revealed 17 independent SNPs located in 15 separate regions that were genome-wide significantly associated with POAG through mechanisms likely to be unrelated to elevated IOP. The GWAS summary statistics for the IOP-dependent component yielded 75 independent variants from 48 distinct regions associated with POAG, through mechanisms likely to be associated with raised IOP. The genetic insights gained from these two components of POAG risk provided us with a unique opportunity to explore the mechanisms underlying POAG.
A wide range of glaucoma endophenotypes have been discovered, including VCDR, optic disc size, cup size, and RNFL thickness38,39. The IOP-dependent and non-IOP-dependent components of POAG displayed very different patterns of association with these endophenotypes. For instance, while the IOP-dependent component of POAG was predominantly associated with IOP itself, rather than with morphological endophenotypes, the non-IOP-dependent component shared a greater genetic architecture with ONH-related endophenotypes. Thus, for VCDR and optic cup area, where both components were associated, the magnitude of association was approximately two-fold higher for the non-IOP-dependent component of POAG.
The non-IOP-dependent genetic component of POAG prioritized two variants—rs113894504 and rs7803764—that had not previously been associated with POAG at genome-wide significance. These two loci were associated with a reduced risk of POAG and yet, surprisingly, with higher IOPcc. Our GWAS-by-subtraction model implemented a theoretical framework in which SNPs influence POAG through either an IOP-dependent or non-IOP-dependent pathway. By assuming zero genetic correlation between the IOP and non-IOP components, the IOP-dependent component is excluded from the genetic risk of POAG. The IOP-associated variants rs113894504 and rs7803764 would be expected a priori to be associated with an increased risk of POAG via their effect on IOP. However, the GWAS-by-subtraction analysis was able to reveal that this was not the case and, instead, suggested that these variants were associated with a reduced risk of POAG through a non-IOP-dependent mechanism. Our work suggests antagonistic effects of these loci; an increased risk of POAG via an association with higher IOP balanced by a reduced risk of POAG via a non-IOP-dependent pathway. Variant rs113894504 (and its r2 = 1.0 proxy, rs80233695) resides within the ANAPC1 gene. This variant was strongly associated with the non-IOP-dependent component of POAG (1.99 × 10−8) as well as the IOP-dependent component of POAG (P = 5.17 × 10−30) in the discovery analysis and is an eQTL for ANAPC1 (Supplementary Data 3 and 9). In tissues from the GTEx catalog, the C allele of rs80233695 is associated with higher expression of ANAPC1 in skeletal muscle (P = 9.3 × 10−7), yet reduced expression in tibial nerve (P = 4.0 × 10−3). The variant is not listed in the ClinVar database, but other ANAPC1 variants have been reported to regulate specific features of corneal structure11,40,41,42,43, such as corneal endothelial cell density and central corneal thickness. ANAPC1 variants are also associated with retinitis pigmentosa (RP)44, hence it will be of interest for future studies to investigate if ANAPC1 (or another gene at the locus) increases the risk of POAG through a mechanism related to either the anterior or the posterior segment. rs7803764 is situated within the TMEM196 gene, as well as within the LOC107986774 long non-coding RNA (lncRNA) transcript. However, rs7803764 is an expression quantitative trait locus (eQTL) for the TWIST1 gene, which is located over 600 kb away (specifically, the C allele of rs7803764 is associated with increased expression of TWIST1 in GTEx tibial nerve samples; P = 1.7 × 10−6). TMEM196 has not been linked to glaucoma in previous studies, however, TWIST1 has been implicated in trabecular meshwork cell-initiated remodeling of the extracellular matrix45. Nevertheless, a GWAS based on artificial intelligence (AI)-graded ONH features identified an association between VCDR and variant rs57584385, which is located nearby the TMEM196 and TWISTNB (POLR1F) genes37. This VCDR-associated variant and our lead variant at the locus, rs7803764, are in moderate LD in Europeans (r2 = 0.34). The risk allele (G) of the known VCDR-associated variant rs57584385 was associated with the non-IOP-dependent component of POAG (beta = 0.06, P = 0.015) but not with the IOP-dependent component of POAG (beta = −0.02, P = 0.10). Additional PheWAS analysis supported its association with RMFs (Supplementary Data 18). Thus, our current findings support the earlier findings linking TMEM196—or potentially TWIST1 or TWISTNB—to POAG via degeneration of the ONH or trabecular meshwork remodeling.
A noteworthy feature of the non-IOP-dependent GWAS component of POAG was its replication of several loci discovered in GWAS analyses for NTG. However, IOP-related mechanisms are still part of the pathogenic process of NTG, as evidenced by IOP lowering medication serving as the key therapeutic intervention24,46,47. Thus, whereas a GWAS for NTG may disclose genetic associations with a contribution from IOP, our GWAS-by-subtraction-derived non-IOP-dependent component should, by design, identify variants that influence POAG risk independently of IOP. Two findings supported this line of reasoning. First, when GWAS-by-subtraction was repeated with a less stringent genetic correlation between the IOP-dependent and IOP-independent latent factors, the resulting non-IOP-dependent component showed greater similarity to a GWAS of NTG10 (Fig. 2, Supplementary Fig. 1). Second, among the 6 known SNPs associated with NTG10, 4 are also significantly associated with IOP (1 SNP with 5 × 10−8 < P < 0.05, 3 SNPs with P < 5 × 10−8). By contrast, only 6 out of the 17 SNPs identified in the non-IOP-dependent component of POAG were associated with IOP at a genome-wide significant level.
This study provided abundant evidence to support the ‘vascular theory’ of glaucoma, with both the IOP-dependent and non-IOP-dependent components of POAG yielding complementary evidence. Thus, first, we found that the IOP-dependent component of POAG had a shared genetic architecture with blood pressure (Supplementary Data 8). Numerous epidemiological studies have reported a positive association between hypertension and glaucoma48. Acting via the autonomic nervous system, increased blood pressure might lead to excess production of aqueous humor and/or a decrease in aqueous outflow, resulting in increased IOP49,50. This could promote a feedback cycle in which elevated IOP increases resistance to blood flow in the ocular tissues, reducing OBF. On the contrary, there are also studies suggesting that the risk of POAG is higher in individuals with low SBP due to the reduced ocular perfusion pressure51 and a Mendelian Randomization study found little support for a causal relationship between blood pressure and POAG52. In summary, the relationship between blood pressure and POAG may be complex, with hypertension linked to elevated IOP and an increased risk of glaucoma, but simultaneously causing increased OBF and a reduced risk of POAG. Second, we found evidence that vascular cell properties, such as vascular permeability, vascular endothelial cell function and smooth muscle cells, were associated with the IOP-dependent component of POAG (Supplementary Data 13, Supplementary Figs. 10–12). Vascular cells are already known to play an important role in the etiology of glaucoma. Experimental studies have demonstrated that endothelial nitric oxide synthase (eNOS) can regulate vascular contractility17 and that dysfunction of the endothelial cells lining Schlemm’s canal may lead to an increase in IOP17,53,54. Moreover, impaired functioning of the vascular system may compromise the aqueous humor outflow pathway, which would directly lead to ocular hypertension55. Dysregulation of arteriole vascular muscle and consequent ischemia of the optical nerve may impair delivery of nutrients to the retina56,57. Third, we also identified links between the non-IOP-dependent component of POAG and the retinal vascular system (Fig. 5, Supplementary Data 17). Specifically, we identified multiple RMFs associated with the non-IOP-dependent component of POAG. These RMFs included indices of vessel density, complexity, caliber, and tortuosity. We speculate that increased vessel density may result in a wider area of the ONH being supplied with blood and nutrients, and hence confer a lower susceptibility to ischemia. Our findings linking vessel complexity to glaucoma are consistent with previous studies58,59. Higher indices of RMFs for ‘fractal dimensions’, which correspond to vessel complexity, are indicative of a better blood supply to the retina (thus, hypertension and diabetic microvasculopathy60 are associated with lower fractal dimensions). We also found that RMFs indicative of less steeply curved retinal vessels were associated with a greater genetic risk of glaucoma. Consistent with this finding, Wu et al.58 and Rudnicka et al.61 have reported that less steeply curved vessels were associated with an increased risk of POAG, although Chiquet62 found limited evidence of an association between vessel tortuosity and POAG or NTG. Thus, in general, our findings support the hypothesis that vascular morphology might be associated with OBF, and that these morphological features are less likely to affect, or be influenced by, IOP. Also, we observed a negative association between the vessel caliber (CRAE) and ICD-10 code-inferred glaucoma, consistent with previous findings63,64,65,66. However, these associations were non-significant in our subsequent GRS analyses. Instead, other RMFs relating to vessel caliber were positively associated with the GRS for the non-IOP-dependent component of POAG. A key difference between the current study and previous work was that our RMFs for vessel caliber were derived form a wide range of vessels, not just from the CRAE and CRVE (likewise, our RMF indices for vessel width may not be directly comparable with those used in previous studies). No previous study has identified associations between the genetic risk of glaucoma and RMFs, which was also the case for our non-IOP-dependent component of POAG. These findings are summarized in Table 3 and Fig. 6.
This summary figure highlights the key distinctions between the IOP-dependent and non-IOP-dependent components of POAG, including differences in SNP pleiotropy, genetic correlation, pathway enrichment, and single-cell enrichment analyses. The genetic risk scores for each component are associated with distinct retinal microvascular features. Furthermore, these genetic risk scores can distinguish between NTG and HTG. This figure is created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license (https://creativecommons.org/licenses/by-nc-nd/4.0/deed.en).
Another noteworthy finding in the current work was that the genetic correlation between glaucoma traits and CKD was negative, while recent epidemiology studies have reported a positive phenotypic association for these two diseases67,68. This discrepancy may result from bias in the genetic correlation estimation, as precedents exist for such bias (in a scenario where non-genetic confounding factors exist for two binary traits and the genetic correlation is estimated from GWAS summary statistics that did not account for covariates69,70).
We acknowledge several limitations of the current study. For example, the GWAS for POAG had a relatively modest sample size, which would have led to limited statistical precision in estimating the associations between SNPs and glaucoma risk. Furthermore, both HTG and NTG cases were included in the GWAS for POAG, yet the SNP heritability for these two traits could be different. Although, GWAS-by-subtraction is expected in theory to be robust to such a difference in SNP heritability71, we could not rule out this potential source of imprecision. Our GWAS for IOPcc relied on a single measurement of IOP in UK Biobank participants, whereas IOP is known to fluctuate over the diurnal cycle such that a single assessment will not capture its full effects. Furthermore, despite our use of a ‘corneal compensated’ measure of IOP, any such compensation will be error-prone and thus will not fully account for differences in corneal biomechanical properties that influence IOP measurement. The GWAS for POAG and the GWAS for IOP were performed in separate samples, therefore any differences in genetic and/or environmental risk factors for POAG in the two samples may have introduced errors into the GWAS-by-subtraction model. We used GWAS summary statistics as input for the GWAS-by-subtraction analysis; ideally, it would have been preferable to study the causal variants for each trait, since the tagging of causal variants in the two GWAS samples may have been subtly different. Finally, the GWAS-by-subtraction model assumes a simple set of causal relationships involving just two latent risk factors for POAG, one of which is fully IOP-dependent and the other fully independent of IOP. The reality is probably a much more complex set of interrelationships that includes pleiotropic effects. Despite these limitations, we found that dissecting the genetic risk of POAG into an IOP-dependent and a non-IOP-dependent component yielded further insight into the mechanism through which these two distinct mechanisms increase the risk of POAG.
Methods
Ethics statement
The UK Biobank resource received approval from the North West Centre for Research Ethics Committees under reference number 11/NW/0382, with all participants providing (digital) signed informed permission. UK Biobank participants are volunteers and do not receive compensation for their involvement. Data from the UKB resource were accessed under approved application numbers 86091. The study followed the principles of the Declaration of Helsinki.
Study cohort
The UK Biobank is a longitudinal study that focuses on how lifestyle and genetic factors affect health and well-being. Approximately 500,000 participants, aged 37 to 73, underwent physical examinations and completed health-related questionnaires at baseline. Recruitment took place between 2006 and 2010; examinations were conducted at 22 assessment centers in Scotland, Wales, and England.
Genotype data
We analyzed imputed genotype data from the UK Biobank July 2017 release72. Briefly, the UK Biobank Axiom array or the BiLEVE Axiom array was used to genotype DNA samples. Genotype data for 805,426 markers in 488,377 participants were imputed using a joint Haplotype Reference Consortium and UK 10,000-Genomes Project (UK10K) reference panel, followed by stringent quality control, as described72.
Assessment of IOP, glaucoma, fundus images, and covariates
During the UK Biobank baseline assessment visit, IOP was measured once for each eye using an Ocular Response Analyzer noncontact tonometer (Reichert Corporation). The instrument calculates a corneal-compensated IOP (IOPcc) value, which is derived from a linear combination of the inward and outward applanation tensions. Participant-level IOP values were calculated by averaging the IOPcc values from the right and left eye of each participant. If data were available for only one eye, then we used that eye’s IOP value as the participant’s IOP. A digital color fundus image of each eye of participants was obtained with a Topcon 3D OCT 1000 Mk2 instrument (Topcon Corporation).
Glaucoma cases were identified from UK Biobank via different sources including: hospitalization records (ICD-10 codes: H40.X, H42.X; and ICD-9 codes: 365.X); operation procedures (OPCS-4 codes: C60-C62, C65; and OPCS-3 codes: 1574, 1512); self-reported non-cancer illness (data field 20002, code:1277); self-reported medical conditions about eye disorders (data field 6148, Supplementary Data 19) and prescribing of eye drops indicating glaucoma (data field 20003). The earliest date identified from the above sources was defined as the date of diagnosis of glaucoma (Supplementary Fig. 14, Supplementary Data 19). Glaucoma cases were defined as having normal tension glaucoma if: (1) they were not prescribed glaucoma eye-drops or did not receive an operation procedure during the first 5 years after glaucoma diagnosis; (2) having at least one IOP measure (corneal compensated IOP or Goldmann Applanation Tonometry [IOPg], both recorded in UK Biobank, data fields 5263,5255,5262 and 5254) within the first 5 years of glaucoma diagnosis; and (3) their maximum IOP (of IOPcc and IOPg), measured within the first 5 years, was less than 21 mmHg. High-tension glaucoma (HTG) cases were defined as those identified as POAG cases from hospitalization records (ICD codes: H40.1, H40.8, H40.9, and 365.1) but not meeting the NTG criteria. We did not require the identified NTG or HTG cases to have fundus images available. Glaucoma cases that could not be confirmed as NTG or HTG cases were excluded from the comparisons of NTG versus HTG, NTG versus non-glaucoma controls, and HTG versus non-glaucoma controls. The non-glaucoma controls were defined as individuals who underwent eye disorder screening and had confirmed they had no history of any kind of glaucoma (Supplementary Fig. 14).
Clinical measurements, including SBP, PP, BMI, and cholesterol (HDL, LDL, triglycerides, and total cholesterol) were selected as covariates. At the baseline visit, SBP was measured twice using a digital sphygmomanometer (Omron 705 IT; OMRON Healthcare Europe), and the average of the two measurements was used in the analysis. Body weight was measured by a Tanita BC-418MA body composition analyzer (Tanita Corporation) and body height was measured in a barefoot standing position using a Saca 202 device. BMI (in units kg/m2) was calculated by dividing body weight by height-squared. Cholesterol levels were measured by a direct enzymatic method (Konelab, Thermo Fisher Scientific). Refractive error was measured using an autorefractor (Tomey RC 5000, Tomey Corporation); participants were assigned a refractive error based on the value for their right eye. Age, gender, Townsend deprivation score and smoking status were collected by a touchscreen questionnaire at the baseline visit.
Summary statistics from published GWASs for POAG
Gharahkhani et al.10 reported a multi-ethnic investigation in which GWAS summary statistics for POAG were meta-analyzed across 21 independent studies. These 21 studies included age, sex, and genetic principal components as covariates in the GWAS analyses. In the discovery stage GWAS-by-subtraction, we used as input the summary statistics reported by Gharahkhani et al.10 from a meta-analysis restricted to participants of European ancestry (16,677 POAG cases and 199,580 controls). Details of the GWAS for POAG in the individual samples are described in the original article10. However, to avoid sample overlap with our FinnGen-based validation sample, we first reverse-meta-analyzed the FinnGen summary statistics from the GWAS for POAG reported by Gharahkhani et al.10. FinnGen summary statistics for POAG (release 2; 1824 cases vs. 93,036 controls) were downloaded from https://storage.googleapis.com/finngen-public-data-r2/summary_stats/finngen_r2_H7_GLAUCPRIMOPEN.gz. Genetic variants with minor allele frequency (MAF) < 1% and without valid rsIDs were removed, leaving 7,691,441 SNPs for reverse-meta-analysis. MetaSubtract V1.673,74 was used to performed the reverse-meta-analysis. SNPs with a high level of heterogeneity in the reverse meta-analysis (heterogeneity test, P < 0.001) were excluded from further analysis.
In the replication stage of GWAS-by-subtraction, we used as input the summary statistics from a GWAS for POAG reported by FinnGen (release 9; 7756 cases vs. 358,375 controls). There was no sample overlap between the discovery and replication GWAS for POAG. Demographic information for the samples is provided in Supplementary Data 2.
GWAS for IOPcc
We performed a GWAS for IOPcc using PLINK 2.075 in a sample of 97,644 unrelated participants of European ancestry in the UK Biobank cohort who had this trait measured, for a set of 7,458,361 genetic variants with minor allele frequency (MAF) ≥1%, missing genotype call rate <1.5% and Hardy-Weinberg equilibrium P < 1.0 × 10−6. European ancestry was defined based on the self-reported ethnic background (data field 21000, code: 1, 1001, 1002, 1003) and a sample of unrelated participants was obtained by excluding those who were third-degree relatives or closer. To reduce bias from outlier IOP values, participants with measurements in the top or bottom 0.5 percentile were excluded52. Given the impact of glaucoma surgery on IOP, we excluded participants who had a history of glaucoma laser therapy or surgery. Each SNP was tested for association with IOPcc in a linear regression model, with age, sex, genotyping array, BMI and the first 5 ancestry principal components (PCs) included as covariates.
Summary statistics from a published GWAS for IOP
As a sensitivity analysis, we used summary statistics from a published GWAS for IOP carried out by the International Glaucoma Genetic Consortium (IGGC) and reported by Bonnemaijer et al.12. The IGGC study included 31,269 participants of European ancestry and identified 16 SNPs associated with IOP at P < 5 × 10−8. Age, sex, and five genetic principal components were included covariates in the GWAS analysis.
GWAS-by-subtraction
The principle of GWAS-by-subtraction is outlined in Fig. 1. Starting from a set of GWAS summary statistics for POAG and a set of GWAS summary statistics for IOP, the method ‘subtracts’ the influence on IOP from each SNP’s effect on POAG to reveal the SNP’s influence on POAG independent of IOP. The GWAS-by-subtraction model outputs an alternative set of summary statistics that approximate those expected from a GWAS for POAG with IOP included as a covariate. The theoretical basis of the GWAS-by-subtraction method is explained in the original article by Demange et al.71. We performed GWAS-by-subtraction with the R package Genomic-SEM (0.0.5c), using the summary statistics from the Gharahkhani et al.10. GWAS for POAG (excluding FinnGen participants) and the summary statistics from the GWAS for IOPcc in UK Biobank participants as input sources (Supplementary Data 1). From the output summary statistics, SNPs with P < 5 × 10−8 were considered as genome-wide significant. The lead SNP in each region was identified by clumping the genome-wide significant SNPs using a threshold r2 < 0.1 for pairs of SNPs located within a distance of 250 kb. For the main GWAS-by-subtraction analysis, the genetic correlation (rg) between the IOP-dependent and non-IOP-dependent latent factors was set as zero. In sensitivity analyses, rg was set as 0.1 or 0.2 to explore how robust the results were to this parameter.
The cross-trait effects of the IOP-dependent and non-IOP-dependent were further investigated in the GWAS catalog by retrieving all association results for SNPs in high LD (r2 ≥ 0.8, European ancestry) with the leading SNPs and were genome-wide significant at P < 5.0 × 10−8.
Gene-based pathway analysis
MAGMA76 analyses of GWAS summary statistics were used to identify genes and pathways with potential roles in IOP-dependent and non-IOP-dependent POAG mechanisms. For these analyses, the mean of the χ2 statistic for all SNPs within the gene was estimated after applying a window of 50 kb at the 5’ and 3’ ends of the gene. For the MAGMA gene-set analysis, we performed a ‘competitive’ analysis, with the Molecular Signatures Database (MSigDB)77 as the reference.
The FUMA GENE2FUNC78 feature was implemented to map genes to functional pathways and provide deeper biological insights. Gene sets from MsigDB, KEGG, WikiPathways and the GWAS Catalog were considered. P-values were corrected for multiple-testing using the Benjamini–Hochberg FDR method, based on the number of data sources of the tested gene sets. We took forward gene sets with an FDR-adjusted P < 0.05 and a gene set overlap of at least one gene.
eQTL analysis
For SNPs significantly associated with the non-IOP-dependent component of POAG in the discovery sample, variants annotation was based on the Genome Reference Consortium Human genome build 37 derived from the University of California Santa Cruz (UCSC) Genome resource79. eQTL mapping was performed using EyeGEx80, eQTL catalog81 and GTEx82 database implemented in FUMA. Colocalization analysis between GWAS signal and eQTLs in blood was conducted via ezQTL under the eCAVIAR framework83.
Cell-type-specific enrichment analysis
Cell-type-specific enrichment analyses (CSEA) of the target genes were undertaken with WebCSEA (https://doi.org/10.1093/nar/gkac392). This approach calculates the raw P-value across 1355 tissue-cell types. To overcome the potential bias due to the different lengths of signature genes and input gene lists, WebCSEA applies a permutation-based method to adjust the raw P-values. We considered target genes with a permutation-adjusted P < 0.001 as suggestive and a permutation-adjusted P < 3.7 × 10−5 (corresponding to 0.05/1355 tissue-cell types) as experiment-wide significant. As suggested by the WebCSEA authors, the experiment-wide significance threshold may be too stringent, hence we report both raw and corrected P-values.
Genetic correlations
SNP heritability (the phenotype variance explained by a specific set of GWAS variants) and genetic correlations were estimated by LDSC84. These analyses took the GWAS summary statistics from the Gharahkhani et al.10. GWAS for POAG excluding FinnGen participants, or the summary statistics for the IOP-dependent or non-IOP-dependent component of POAG generated by GWAS-by-subtraction as input. Genetic correlations between the POAG traits and a series of anthropometric, clinical ocular and systemic traits and diseases were calculated. Details of the traits tested are listed in Supplementary Data 6.
Retinal microvascular features
Fundus images were available for the right eyes of 66,500 UK Biobank participants. We previously developed a fast, automated convolutional neural network (CNN) system to identify and quantify retinal microvascular features (RMFs), which we designated ‘Retinal-based Microvascular Health Assessment System’ (RMHAS)85. We applied RMHAS to the fundus images of UK Biobank participants, to segment the retinal artery, vein, and optic disc. For quality control, images were excluded if they had less than 6 identifiable arteries or veins in around the optic disc region or less than 2 detectable arteries or veins over the whole fundus. RMFs including the caliber, complexity, length, tortuosity, and branching angle of the retinal arteries and veins were quantified.
Generation of the genetic risk scores (GRSs)
Separate GRSs were derived for the IOP-dependent component and non-IOP-dependent component of POAG, using the summary statistics generated by GWAS-by-subtraction. For each trait, significant (P < 5.0 × 10−8), independent SNPs (r2 < 0.1) were used to generate weighted genetic risk scores (GRSs) for each participant using the --score function implemented in PLINK 2.075. In the main analysis, the magnitude of the association with IOP-dependent POAG or non-IOP-dependent POAG (GWAS Z-score) in the discovery analysis was used as the weighting factor for each variant included in the GRS. In the sensitivity analysis, the weighting factors were obtained from validation analysis 3, which was independent of UK Biobank. In the comparative analysis between individuals with NTG and HTG, a polygenic risk ratio (PRR) was formulated. The PRR is expressed as:
wherein GRS_non-IOP-dependent denotes the GRS associated with the non-IOP-dependent component of POAG, and GRS_IOP-dependent represents the GRS for the IOP-dependent component of POAG. The GRS for POAG was computed based on independent POAG SNPs reported by Gharahkhani et al.10. This GRS was adjusted as a covariate to control for POAG genetic background variation in the differentiation analysis between NTG and HTG cases.
PheWAS analysis for retinal vascular features
The R package ‘PheWAS’ was used to identify RMFs associated with either of the two GRSs, after adjusting for age, sex, SBP, Townsend deprivation score and refractive error. The PheWAS analysis encompassed individuals who had retinal images and validated RMFs, thereby yielding a sample size of n = 50,624. Analogously, logistic regression was carried out to identify RMFs associated with POAG case-control status, after adjusting for the same covariates. GRS-RMF and POAG-RMF associations P < 5.49 × 10−4 were considered significant (corresponding to 0.05/91 RMFs).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The summary statistics for (a) the non-IOP-dependent component of POAG, (b) the IOP-dependent component of POAG generated in this study, and (c) the GWAS for IOPcc in UK Biobank participants, along with the summary statistics for the sensitivity analysis, have been deposited in Zenodo [https://doi.org/10.5281/zenodo.13765063]. The UK Biobank data are available under restricted access for health-related research that is in the public interest. Access can be obtained by submitting an application to https://www.ukbiobank.ac.uk/, where applications are considered from bona fide researchers. The summary statistics for the GWAS for POAG reported by Gharahkhani et al.10 and for the GWAS of IOP reported by Bonnemaijer et al.12 are available on the GWAS Catalog under accession codes GCST90011766 and GCST009413. The summary statistics for the GWAS for POAG in the FinnGen release 9 and 2 are available via FinnGen database (release 9: https://storage.googleapis.com/finngen-public-data-r9/summary_stats/finngen_R9_H7_GLAUCOMA_POAG.gz; release 2: https://storage.googleapis.com/finngen-public-data-r2/summary_stats/finngen_r2_H7_GLAUCPRIMOPEN.gz). All summary genetic association statistics datasets used in the genetic correlation analysis are publicly available at GWAS Catalog and can be accessed by entering the identifiers provided in the ‘GWAS_catalog_access_number’ column in Supplementary Data 8 at https://www.ebi.ac.uk/gwas/. Source data are provided on Figshare [https://doi.org/10.6084/m9.figshare.25045220].
References
Tham, Y. C. et al. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology 121, 2081–2090 (2014).
Quigley, H. A. & Broman, A. T. The number of people with glaucoma worldwide in 2010 and 2020. Br. J. Ophthalmol. 90, 262–267 (2006).
Kroese, M., Burton, H., Vardy, S., Rimmer, T. & McCarter, D. Prevalence of primary open angle glaucoma in general ophthalmic practice in the United Kingdom. Br. J. Ophthalmol. 86, 978–980 (2002).
Varma, R., Lee, P. P., Goldberg, I. & Kotak, S. An assessment of the health and economic burdens of glaucoma. Am. J. Ophthalmol. 152, 515–522 (2011).
Gharahkhani, P. et al. Common variants near ABCA1, AFAP1 and GMDS confer risk of primary open-angle glaucoma. Nat. Genet. 46, 1120–1125 (2014).
Bailey, J. N. et al. Genome-wide association analysis identifies TXNRD2, ATXN2 and FOXC1 as susceptibility loci for primary open-angle glaucoma. Nat. Genet 48, 189–194 (2016).
Choquet, H. et al. A multiethnic genome-wide association study of primary open-angle glaucoma identifies novel risk loci. Nat. Commun. 9, 2278 (2018).
Shiga, Y. et al. Genome-wide association study identifies seven novel susceptibility loci for primary open-angle glaucoma. Hum. Mol. Genet. 27, 1486–1496 (2018).
Khawaja, A. P. et al. Genome-wide analyses identify 68 new loci associated with intraocular pressure and improve risk prediction for primary open-angle glaucoma. Nat. Genet. 50, 778–782 (2018).
Gharahkhani, P. et al. Genome-wide meta-analysis identifies 127 open-angle glaucoma loci with consistent effect across ancestries. Nat. Commun. 12, 1258 (2021).
MacGregor, S. et al. Genome-wide association study of intraocular pressure uncovers new pathways to glaucoma. Nat. Genet 50, 1067–1071 (2018).
Bonnemaijer, P. W. M. et al. Multi-trait genome-wide association study identifies new loci associated with optic disc parameters. Commun. Biol. 2, 435 (2019).
Flammer, J. The vascular concept of glaucoma. Surv. Ophthalmol. 38, S3–S6 (1994).
Charlesworth, J. et al. The path to open-angle glaucoma gene discovery: endophenotypic status of intraocular pressure, cup-to-disc ratio, and central corneal thickness. Invest Ophthalmol. Vis. Sci. 51, 3509–3514 (2010).
Aschard, H. et al. Genetic correlations between intraocular pressure, blood pressure and primary open-angle glaucoma: a multi-cohort analysis. Eur. J. Hum. Genet. 25, 1261–1267 (2017).
Pang, I. H. & Clark, A. F. Inducible rodent models of glaucoma. Prog. Retin Eye Res. 75, 100799 (2020).
Reina-Torres, E. et al. The vital role for nitric oxide in intraocular pressure homeostasis. Prog. Retin Eye Res. 83, 100922 (2021).
Yan, D. B. et al. Deformation of the lamina cribrosa by elevated intraocular pressure. Br. J. Ophthalmol. 78, 643–648 (1994).
Levkovitch-Verbin, H. Animal models of optic nerve diseases. Eye 18, 1066–1074 (2004).
Flammer, J. et al. The impact of ocular blood flow in glaucoma. Prog. Retin Eye Res. 21, 359–393 (2002).
Chan, K. K. W., Tang, F., Tham, C. C. Y., Young, A. L. & Cheung, C. Y. Retinal vasculature in glaucoma: a review. BMJ Open Ophthalmol. 1, e000032 (2017).
Yanagi, M. et al. Vascular risk factors in glaucoma: a review. Clin. Exp. Ophthalmol. 39, 252–258 (2011).
Flammer, J., Konieczka, K. & Flammer, A. J. The primary vascular dysregulation syndrome: implications for eye diseases. EPMA J. 4, 14 (2013).
Shields, M. B. Normal-tension glaucoma: is it different from primary open-angle glaucoma? Curr. Opin. Ophthalmol. 19, 85–88 (2008).
Torres, S. D. et al. Genome-wide Multi-ethnic Meta-analysis Identifies 22 Independent Risk Loci for Normal Tension Glaucoma and Predominantly Overlaps with High Tension Glaucoma (Research Square, 2023).
Gao, X. R., Huang, H., Nannini, D. R., Fan, F. & Kim, H. Genome-wide association analyses identify new loci influencing intraocular pressure. Hum. Mol. Genet. 27, 2205–2213 (2018).
Evangelou, E. et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat. Genet. 50, 1412–1425 (2018).
Yengo, L. et al. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum. Mol. Genet. 27, 3641–3649 (2018).
Willer, C. J. et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45, 1274–1283 (2013).
Wheeler, E. et al. Impact of common genetic determinants of Hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: A transethnic genome-wide meta-analysis. PLoS Med. 14, e1002383 (2017).
van der Harst, P. & Verweij, N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ. Res. 122, 433–443 (2018).
Malik, R. et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat. Genet. 50, 524–537 (2018).
Nielsen, J. B. et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat. Genet. 50, 1234–1239 (2018).
Kunkle, B. W. et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat. Genet. 51, 414–430 (2019).
Wuttke, M. et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat. Genet. 51, 957–972 (2019).
Xue, A. et al. Genome-wide association analyses identify 143 risk variants and putative regulatory mechanisms for type 2 diabetes. Nat. Commun. 9, 2941 (2018).
Han, X. et al. Automated AI labeling of optic nerve head enables insights into cross-ancestry glaucoma risk and genetic discovery in >280,000 images from UKB and CLSA. Am. J. Hum. Genet. 108, 1204–1216 (2021).
Asefa, N. G., Neustaeter, A., Jansonius, N. M. & Snieder, H. Heritability of glaucoma and glaucoma-related endophenotypes: Systematic review and meta-analysis. Surv. Ophthalmol. 64, 835–851 (2019).
Craig, J. E. et al. Multitrait analysis of glaucoma identifies new risk loci and enables polygenic prediction of disease susceptibility and progression. Nat. Genet. 52, 160–166 (2020).
Khawaja, A. P. et al. Genetic variants associated with corneal biomechanical properties and potentially conferring susceptibility to keratoconus in a genome-wide association study. JAMA Ophthalmol. 137, 1005–1012 (2019).
Ivarsdottir, E. V. et al. Sequence variation at ANAPC1 accounts for 24% of the variability in corneal endothelial cell density. Nat. Commun. 10, 1284 (2019).
He, W. et al. Association of novel loci with keratoconus susceptibility in a multitrait genome-wide association study of the UK Biobank database and Canadian longitudinal study on aging. JAMA Ophthalmol. 140, 568–576 (2022).
Ahonen, S. J. et al. Increased expression of MERTK is associated with a unique form of canine retinopathy. PLoS ONE 9, e114552 (2014).
Rodriguez-Munoz, A. et al. Expanding the clinical and molecular heterogeneity of nonsyndromic inherited retinal dystrophies. J. Mol. Diagn. 22, 532–543 (2020).
Pattabiraman, P. P., Maddala, R. & Rao, P. V. Regulation of plasticity and fibrogenic activity of trabecular meshwork cells by Rho GTPase signaling. J. Cell Physiol. 229, 927–942 (2014).
Caprioli, J. & Coleman, A. L. Blood flow in glaucoma, D. Blood pressure, perfusion pressure, and glaucoma. Am. J. Ophthalmol. 149, 704–712 (2010).
Kamal, D. & Hitchings, R. Normal tension glaucoma-a practical approach. Br. J. Ophthalmol. 82, 835–840 (1998).
Leeman, M. & Kestelyn, P. Glaucoma and blood pressure. Hypertension 73, 944–950 (2019).
Gherghel, D., Hosking, S. L. & Orgul, S. Autonomic nervous system, circadian rhythms, and primary open-angle glaucoma. Surv. Ophthalmol. 49, 491–508 (2004).
Cui, Y. K., Pan, L., Lam, T., Wen, C. Y. & Do, C. W. Mechanistic links between systemic hypertension and open angle glaucoma. Clin. Exp. Optom. 105, 362–371 (2022).
Flammer, J. et al. The eye and the heart. Eur. Heart J. 34, 1270–1278 (2013).
Plotnikov, D. et al. High blood pressure and intraocular pressure: a Mendelian randomization study. Invest Ophthalmol. Vis. Sci. 63, 29 (2022).
Nakabayashi, M. Review of the ischemia hypothesis for ocular hypertension other than congenital glaucoma and closed-angle glaucoma. Ophthalmologica 218, 344–349 (2004).
Nakabayashi, M. Ischemic hypertension of pigeon eye. Jpn J. Ophthalmol. 45, 128–136 (2001).
Carreon, T., van der Merwe, E., Fellman, R. L., Johnstone, M. & Bhattacharya, S. K. Aqueous outflow—a continuum from trabecular meshwork to episcleral veins. Prog. Retin Eye Res. 57, 108–133 (2017).
Buckley, C., Hadoke, P. W., Henry, E. & O’Brien, C. Systemic vascular endothelial cell dysfunction in normal pressure glaucoma. Br. J. Ophthalmol. 86, 227–232 (2002).
Su, W. W. et al. Glaucoma is associated with peripheral vascular endothelial dysfunction. Ophthalmology 115, 1173–1178 e1 (2008).
Wu, R. et al. Retinal vascular geometry and glaucoma: the Singapore Malay Eye Study. Ophthalmology 120, 77–83 (2013).
Ciancaglini, M. et al. Fractal dimension as a new tool to analyze optic nerve head vasculature in primary open angle glaucoma. Vivo 29, 273–279 (2015).
Grauslund, J. et al. Retinal vascular fractals and microvascular and macrovascular complications in type 1 diabetes. Ophthalmology 117, 1400–1405 (2010).
Rudnicka, A. R. et al. Retinal vasculometry associations with glaucoma: findings from the European Prospective Investigation of Cancer-Norfolk Eye study. Am. J. Ophthalmol. 220, 140–151 (2020).
Chiquet, C. et al. Retinal vessel phenotype in patients with primary open-angle glaucoma. Acta Ophthalmol. 98, e88–e93 (2020).
Rong, X. et al. Peripapillary retinal artery in first diagnosed and untreated normal tension glaucoma. BMC Ophthalmol. 19, 203 (2019).
Mitchell, P. et al. Retinal vessel diameter and open-angle glaucoma: the Blue Mountains Eye Study. Ophthalmology 112, 245–250 (2005).
Kawasaki, R. et al. Retinal vessel caliber is associated with the 10-year incidence of glaucoma: the Blue Mountains Eye Study. Ophthalmology 120, 84–90 (2013).
Lin, T. P. H. et al. Assessment of risk of normal tension glaucoma progression via deep-learning based retinal-vessel caliber measurement. Invest. Ophthalmol. Vis. Sci. 62, 1027–1027 (2021).
Wong, C. W., Wong, T. Y., Cheng, C. Y. & Sabanayagam, C. Kidney and eye diseases: common risk factors, etiological mechanisms, and pathways. Kidney Int. 85, 1290–1302 (2014).
Ng, F. Y. C. et al. Bidirectional association between glaucoma and chronic kidney disease: a systematic review and meta-analysis. EClinicalMedicine 49, 101498 (2022).
Weissbrod, O., Flint, J. & Rosset, S. Estimating SNP-based heritability and genetic correlation in case-control studies directly and with summary statistics. Am. J. Hum. Gene.t 103, 89–99 (2018).
Holmes, J. B., Speed, D. & Balding, D. J. Summary statistic analyses can mistake confounding bias for heritability. Genet. Epidemiol. 43, 930–940 (2019).
Demange, P. A. et al. Investigating the genetic architecture of noncognitive skills using GWAS-by-subtraction. Nat. Genet. 53, 35–44 (2021).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Nolte, I. M. Metasubtract: an R-package to analytically produce leave-one-out meta-analysis GWAS summary statistics. Bioinformatics 36, 4521–4522 (2020).
Peyrot, W. J. & Price, A. L. Identifying loci with different allele frequencies among cases of eight psychiatric disorders using CC-GWAS. Nat. Genet. 53, 445–454 (2021).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219 (2015).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
Speir, M. L. et al. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 44, D717–D725 (2016).
Ratnapriya, R. et al. Retinal transcriptome and eQTL analyses identify genes associated with age-related macular degeneration. Nat. Genet. 51, 606–610 (2019).
Kerimov, N. et al. A compendium of uniformly processed human gene expression and splicing quantitative trait loci. Nat. Genet. 53, 1290–1299 (2021).
Consortium, G. T. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
Zhang, T., Klein, A., Sang, J., Choi, J. & Brown, K. M. ezQTL: a web platform for interactive visualization and colocalization of QTLs and GWAS Loci. Genom. Proteom. Bioinf. 20, 541–548 (2022).
Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Shi, D. et al. A deep learning system for fully automated retinal vessel measurement in high throughput image analysis. Front. Cardiovasc. Med. 9, 823436 (2022).
Acknowledgements
This research was conducted using the UK Biobank resource. We thank the participants of the UK Biobank. We also want to acknowledge the participants and investigators of the FinnGen study. Y.H. receives support from the Research Foundation of Medical Science and Technology of Guangdong Province, China (A2022323); NSFC Incubation Project of Guangdong Provincial People’s Hospital, China (KY0120220051), NSFC Young Scientist Found (82301246) and Science and Technology Program of Guangzhou, China (202002020049); D.P. was supported by a grant from Kazan State Medical University (2/22-2 01.08.2022); C.L. receives support from the National Natural Science Foundation of China (81870663, 82171075), the Outstanding Young Talent Trainee Program of Guangdong Provincial People’s Hospital (KJ012019087), Guangdong Provincial People’s Hospital Scientific Research Funds for Leading Medical Talents and Distinguished Young Scholars in Guangdong Province (KJ012019457), Talent Introduction Fund of Guangdong Provincial People’s Hospital (Y012018145). X.S. receives support from the University of Melbourne at Research Accelerator Program and the CERA Foundation. The Centre for Eye Research Australia receives Operational Infrastructure Support from the Victorian State Government.
Author information
Authors and Affiliations
Contributions
Y.H., H.W., D.P., H.Y., H.Z., J.G., and M.H. conceived and designed the study. Y.H., D.P., H.W., and J.G. performed data analysis and drafted the initial manuscript. D.P., D.S., and X.S. performed data curation. C.L., X.l.Z., X.y.Z., S.T., Y.j.H., H.Y., H.Z., and M.H. made a critical revision to the manuscript for important intellectual content. All authors read the manuscript and approved the final draft. The corresponding authors attest that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Huang, Y., Plotnikov, D., Wang, H. et al. GWAS-by-subtraction reveals an IOP-independent component of primary open angle glaucoma. Nat Commun 15, 8962 (2024). https://doi.org/10.1038/s41467-024-53331-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-53331-0