Abstract
GWASs have linked the 3p21.1 locus, which is associated with the expression levels of NEK4, to bipolar disorder. Here, we use integrative analyses of GWAS statistics and eQTL annotations to establish that elevated NEK4 expression in the hippocampus is associated with an increased risk of bipolar disorder. To further study this association, we generate transgenic male mice that conditionally overexpress NEK4 in the pyramidal neurons of the adult forebrain, or use AAV to overexpress NEK4 in the dorsal hippocampus. Compared to the control mice, male mice of both strains exhibit a shift from a diurnal anxiety state to a nocturnal normal or anxiolytic-like state. Overexpression of NEK4 also affects the circadian fluctuations in dendritic spine morphology and synaptic structure. Furthermore, we show that treatment with lithium ameliorates the effects of NEK4 overexpression in male mice. We then perform phosphoproteomic analyses to demonstrate that the diurnal and nocturnal phosphoproteomic profiles of male control and NEK4 overexpressing mice are different. These results suggest that male mice with different NEK4 expression levels may recapitulate some of the core features observed in patients with bipolar disorder, indicating that interruption of the homeostatic dynamics of synapses may underlie the emotional swings in bipolar disorder.
Similar content being viewed by others
Introduction
Bipolar disorder (BD) is a severe mental illness characterized by mood swings between manic and depressive states. Despite an over 80% heritability and multiple risk genomic loci identified by genome-wide association study (GWAS), the etiological and pathogenic mechanisms of BD remain largely opaque1. The most robust neurobiological evidence underlying BD lies in the discoveries of altered synaptic structures and functions in prefrontal cortex and hippocampus2, and many BD risk genes could modulate the structural dynamics of dendritic spines and synapses in those brain regions3,4,5,6. To further elucidate the mechanisms of BD, researchers have attempted to reproduce BD symptoms in model animals through manipulating GWAS risk genes7,8,9,10,11,12,13, or administration of environmental stimuli14. Nonetheless, majority of those model animals exhibit only either mania-like, anxiety-like, or depression-like behaviors during the experiment, whereas mechanisms underlying the fluctuations in mood episodes, a key characteristic of BD, remain less represented or explored15,16,17. Appropriate animal models that could mimic the fluctuations in mood episodes of BD are therefore urgently needed.
Chromosome 3p21.1 is one of the most prominent risk loci for BD, and genetic variations spanning multiple protein coding genes within this genomic region have been consistently highlighted in BD GWAS18,19,20,21,22. We noticed that multiple independent BD risk SNPs at 3p21.1 showed correlations with the structural indices of the hippocampus23, and these SNPs were significantly associated with NEK4 expression in the hippocampus24. We hence examined the physiological impact of increased hippocampal NEK4, which predicted a higher risk of BD, using genetic and pharmacological approaches in murine models, followed by assessment of behavioral and physiological phenotypes. We found that overexpression of this key BD risk gene in hippocampus led to concomitant presentations of disturbed circadian fluctuations in mood and synapses, which were ameliorated by the mood stabilizer lithium. Given that NEK4 encodes a serine/threonine kinase belonging to the NIMA kinases family, we analyzed the phosphoproteomic profiles of these mice, and confirmed that overexpression of NEK4 significantly disturbed the existing circadian pattern of the phosphorylation of numerous proteins involved in synapses. Furthermore, a DisGeNET gene set term “mood swings” was highlighted only in NEK4 overexpressing mice, suggesting that disturbance of the circadian homeostasis of synaptic structures might (at least in part) explain the emotion transitions observed in BD. Taken together, this study has identified a BD risk gene modulating the circadian swings in emotional behaviors in mice, providing a potential model for investigating emotion swings in BD and insights into the underlying biological mechanisms.
Results
BD genetic risk variants at 3p21.1 locus predicted higher NEK4 mRNA expression in hippocampus
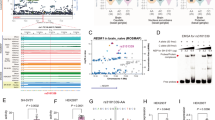
We carried out a detailed examination of SNPs spanning 3p21.1 (chr3: 52300000-53300000, hg19) for their associations with risk of BD using data from the PGC3 GWAS (including 41917 cases of clinical BD and biobank ascertained BD as well as 371549 controls of European ancestry)18, and found 368 SNPs in varied linkage disequilibrium (LD) showing genome-wide level of statistical significance (P ≤ 5.00 ⨯ 10–8), among which rs2336147 was the lead SNP (P = 3.62 ⨯ 10–13, Fig. 1A). Notably, two SNPs (rs7622851 and rs2276824) independent of rs2336147 (rs7622851-rs2336147: r2 = 0.001; rs2276824-rs2336147: r2 = 0.008) also exhibited genome-wide significance in the PGC3 GWAS (rs7622851: P = 1.83 ⨯ 10–11; rs2276824: P = 1.02 ⨯ 10–10, Fig. 1A), suggesting partially independent BD risk association signals in the 3p21.1 locus.
A Genetic associations of SNPs spanning 3p21.1 region with BD in European populations (41917 cases and 371549 controls). A physical map of the region is given and depicts known genes within the region, and three independent risk SNPs (rs7622851, rs2336147 and rs2276824) were marked. The linkage disequilibrium (LD) of the three SNPs were calculated in European individuals from 1000 Genomes Project, based on the r2 algorithm implemented in SHEsis program. B Associations of rs7622851, rs2336147 and rs2276824 with mRNA expression of NEK4 in hippocampus (n = 371) from BrainSeq Phase 2 dataset. The BD risk-associated genotypes at each SNP were marked. In box plot B, the lines from top to bottom represent maximum, 3rd quartile, median, 1st quartile, and minimum, while the middle area represents the interquartile range. Source data are provided as a Source data file.
Then, a PheWeb-based brain-wide imaging analysis using data from UK biobank (n = 32224)23,25 showed that the three independent SNPs (rs7622851, rs2336147 and rs2276824) were associated with structural indices of several brain regions including the hippocampus (Supplementary Fig. 1). We therefore conducted an expression quantitative trait loci (eQTL) analysis of the three SNPs using data from a hippocampus RNA-Seq sample (n = 371) in BrainSeq Phase 2 dataset24. There are three genes showing significant eQTL associations with the three SNPs in hippocampus, including NEK4 (rs7622851, P = 1.01 ⨯ 10–5; rs2336147, P = 6.88 ⨯ 10–11; rs2276824, P = 3.79 ⨯ 10–5, Fig. 1B), GNL3 (rs7622851, P = 5.63 ⨯ 10–5; rs2336147, P = 2.39 ⨯ 10–10; rs2276824, P = 1.56 ⨯ 10–6, Supplementary Fig. 2) and GLYCTK (rs7622851, P = 2.96 ⨯ 10–8; rs2336147, P = 1.83 ⨯ 10–5; rs2276824, P = 3.82 ⨯ 10–5, Supplementary Fig. 2), and the BD risk alleles at the three SNPs consistently predicted higher mRNA expression of NEK4, GNL3 and GLYCTK. Notably, these eQTL associations were successfully replicated in the same direction of allelic effects in an independent hippocampus RNA-Seq sample (n = 168) in MetaBrain dataset (Supplementary Data 1)26, including NEK4 (rs7622851, P = 1.08 ⨯ 10–5; rs2336147, P = 5.588 ⨯ 10–4; rs2276824, P = 6.24 ⨯ 10–3), GNL3 (rs7622851, P = 5.59 ⨯ 10–3; rs2336147, P = 2.98 ⨯ 10–5; rs2276824, P = 1.21 ⨯ 10–4) and GLYCTK (rs7622851, P = 7.86 ⨯ 10–4; rs2336147, P = 3.11 ⨯ 10–2; rs2276824, P = 1.80 ⨯ 10–2).
Furthermore, in a public BrainEXP dataset27 including differential expression analysis of BD cases versus controls in hippocampus—the data can be found in GSE53987 (18 cases and 18 controls)28—the mRNA expression of NEK4 was slightly increased in BD cases compared with controls (P = 0.031, mean ± S.D.: 7.519 ± 0.087 in cases and 7.447 ± 0.097 in controls, Supplementary Fig. 3), which was consistent with the eQTL analysis of this gene. However, the mRNA expression of GNL3 (P = 0.834, mean ± S.D.: 8.918 ± 0.068 in cases and 8.911 ± 0.120 in controls) or GLYCTK (P = 0.412, mean ± S.D.: 5.766 ± 0.099 in cases and 5.800 ± 0.146 in controls) was not altered between BD cases and controls (Supplementary Fig. 3). In another independent BD hippocampus expression dataset—GSE210064 (including 11 cases and 11 controls)29—the mRNA expression of NEK4 was also higher in BD cases compared with controls (P = 0.036, Supplementary Fig. 4). Therefore, we focused on NEK4 in the following functional explorations.
Forebrain conditional transgenic mice (H11-LSL-NEK4±; CaMKIIα-Cre) exhibited abnormal circadian fluctuations in emotional behaviors
We generated NEK4 conditional transgenic (cTG) mice (H11-LSL-NEK4±; CaMKIIα-Cre) through crossing mice harboring NEK4 transgene flanked by two loxP sites at H11 safe harbor locus with mice expressing Cre recombinase driven by the excitatory neuron-specific CaMKIIα promoter. These mice expressed NEK4 in the forebrain pyramidal neurons (predominantly in the CA1 region of the hippocampus), and the expression efficiency was verified through western blot (Supplementary Fig. 5A). Open filed, elevated plus maze and forced swimming tests were carried out to examine their emotional behaviors (Fig. 2A).
A Time scheme for mouse behavioral tests in NEK4 cTG mice. B Results of open field tests during the day and night of the NEK4 cTG mice compared to the control mice, including the total distance travelled, the distance spent in the center (day: P = 0.0393, df = 14, t = 2.272; night: P = 0.6931, df = 14, t = 0.403) and time spent in the center, and the time spent in the corner, were recorded and analyzed. C Results of the elevated plus maze during the day and night of the NEK4 cTG mice compared to the control mice, and their total distance traveled (day: P = 0.0008, df = 14, t = 4.280; night: P = 0.5681, df = 14, t = 0.585), distance spent in the open arms (day: P = 0.0019, df = 14, t = 3.801; night: P = 0.2647, df = 14, t = 1.162), time spent in the open arms (day: P = 0.0018, df = 14, t = 3.833; night: P = 0.0131, df = 14, t = 2.839) and time spent in the closed arms (day: P = 0.0006, df = 14, t = 4.363; night: P = 0.0238, df = 14, t = 2.535) were calculated. D The NEK4 cTG mice and the control mice underwent the forced swimming tests and monitored for 8 min, and the immobility time in the remaining 6 min was calculated for each mouse. N = 8 mice/group. All data were shown as mean ± SD, the two-sided student’s t test was used to perform the significance test. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P < 0.0001. The shaded part represents the behavioral experiment data carried out in the dark environment at night (21:00-24:00). Source data are provided as a Source data file.
In the open field, control mice were more active at night, and their emotional behaviors were not significantly altered between day and night. However, the NEK4 cTG mice traveled shorter distances in the central area than age and gender-matched controls during the daytime (control, 3132 ± 1266 mm; NEK4 cTG, 1862 ± 946.4 mm, P = 0.0393; Fig. 2B), while there was no significant difference at night. This phenomenon was also observed for the time spent in the central area. In the elevated plus maze, NEK4 cTG mice spent significantly less time in the open arms and more time in the closed arms than the control mice during the daytime (open arms time: control, 55.83 ± 26.52 s; NEK4 cTG, 17.10 ± 10.64 s, P = 0.0018; closed arms time: control, 205.0 ± 33.87 s; NEK4 cTG, 261.3 ± 13.74 s, P = 0.0006; Fig. 2C), whereas at night, the NEK4 cTG mice spent significantly more time in the open arms and less time in the closed arms than the control mice (open arms time: control, 28.77 ± 12.84 s; NEK4 cTG, 50.87 ± 17.89 s, P = 0.0131; closed arms time: control, 256.1 ± 19.93 s; NEK4 OE, 231.4 ± 19.03 s, P = 0.0238; Fig. 2C). In the forced swimming test, we also observed a trend of fluctuated depression-like behaviors of the NEK4 cTG mice between day and night, despite that the results were not statistically significant (Fig. 2D). Taken together, these results suggest that the NEK4 cTG mice shifted from an anxious state in the daytime to an anxiolytic-like state during the night.
AAV-mediated overexpression of NEK4 in the hippocampus led to aberrant circadian fluctuations in mood behaviors
To confirm the behavioral results observed in the NEK4 cTG mice, we then overexpressed NEK4 in the dorsal hippocampus of wild-type C57BL/6 J male mice through stereotaxic injections of adeno-associated virus (AAV) (Fig. 3A), and the expression efficiency was also verified through western blot (Supplementary Fig. 5B). For the control mice, we observed that they were more active at night, as indicated by a significant increase in their movement distances. However, their emotional behaviors showed no significant differences between day and night. Interestingly, we again found an obvious circadian fluctuation of the emotional behaviors in the NEK4 overexpression (NEK4 OE) mice compared with controls (Fig. 3B). Briefly, in the open field, NEK4 OE mice spent significantly less time in the central area (control, 26.33 ± 11.64 s; NEK4 OE, 18.53 ± 9.37 s, P = 0.0483; Fig. 3C) and more time in the corner area (control, 153.0 ± 17.79 s; NEK4 OE, 178.3 ± 34.49 s, P = 0.0166; Fig. 3C) compared with controls during the daytime, while the total traveled distances were similar between the two experimental groups (control, 17705 ± 2893 mm; NEK4 OE, 17671 ± 6012 mm, P = 0.984; Fig. 3C). By contrast, no statistically significant difference was seen in the time spent in the central area between NEK4 OE mice and the control mice (control, 27.80 ± 11.85 s; NEK4 OE, 31.57 ± 13.08 s, P = 0.409; Fig. 3C) during the night, while the NEK4 OE mice spent slightly more time in the center area. These results suggested presence of an anxious state in the NEK4 OE mice only in the daytime.
A Diagram showing the injection site and fluorescence expression in the dorsal hippocampus of mice (injection position [AP]: −2.06 mm; [ML]: ±1.50 mm; [DV]: −1.50 mm). B Time scheme for mouse behavioral tests in NEK4 OE mice. C Results of open field tests during the day and night of the NEK4 OE mice compared to the control mice. The total distance travelled, the time spent in the center (day: P = 0.0483, df = 29, t = 2.062; night: P = 0.4094, df = 29, t = 0.837), and the time spent in the corner (day: P = 0.0166, df = 29, t = 2.542; night: P = 0.4935, df = 29, t = 0.694) were recorded. Center area time during the day compared with the night in NEK4 OE group (P = 0.0029, df = 30, t = 3.241). N = 15 Control mice/group, n = 16 NEK4 OE mice/group. D Results of the elevated plus maze tests during the day and night of the NEK4 OE mice compared to the control mice. The total distance traveled, time spent in the open arms (day: P = 0.0299, df = 26, t = 2.297; night: P = 0. 0.0017, df = 28, t = 3.472), and distance traveled in the open arms (day: P = 0.0294, df = 26, t = 2.304; night: P = 0. 1056, df = 28, t = 1.672) were measured. Opened arm time and distance during the day compared with the night in NEK4 OE group (time: P < 0.0001, df = 27, t = 5.271; distance: P < 0.0001, df = 27, t = 4.090). Opened arm distance during the day compared with the night in control group (P = 0.0440, df = 27, t = 2.113). N = 14 mice/day group, n = 15 mice/night group. E Results of forced swimming tests during the day and night of the NEK4 OE mice compared to the control mice, the mice were placed in water for 8 min, and the lengths of their immobility time within the remaining 6 min were calculated (day: P = 0.909, df = 28, t = 0.116; night: P = 0.0337, df = 28, t = 2.234). N = 15 Control mice/group, n = 15 NEK4 OE mice/group. F The small animal metabolic and behavioral phenotype analysis system was used to monitor free activities of the mice for 2 days. N = 3 Control mice/group, n = 3 NEK4 OE mice/group. Data A-E were shown as mean ± SD, F was shown as mean ± SEM. The two-sided student’s t test was used to perform the significance test. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P < 0.0001. The shaded part represents the behavioral experiment data carried out in the dark environment at night (21:00-24:00). Source data are provided as a Source data file.
In the elevated plus maze, the NEK4 OE mice also showed significant reductions of both the time and distance in the open arms compared with controls during the daytime (spent time: control, 22.38 ± 13.05 s; NEK4 OE, 12.51 ± 9.40 s, P = 0.0299; distance: control, 487.8 ± 355.3 mm; NEK4 OE, 230.1 ± 220.9 mm, P = 0.0294; Fig. 3D). Nonetheless, NEK4 OE mice exhibited an opposite behavioral pattern during the night, i.e., they spent significantly more time (control, 20.64 ± 16.82 s; NEK4 OE, 44.74 ± 20.98 s, P = 0.0017; Fig. 3D) and traveled slightly more distance (control, 1011 ± 860.0 mm; NEK4 OE, 1583 ± 1008 mm, P = 0.106) in the open arms than the control mice, indicating elevated exploratory activities indicative of anxiolytic-like behaviors at night.
We then investigated whether depression-like behaviors were affected by NEK4 using the forced swimming test. We saw no difference in the immobility time between NEK4 OE mice and controls in the daytime (control, 134.3 ± 58.36 s; NEK4 OE, 131.8 ± 59.39 s, P = 0.909; Fig. 3E). Notably, the control mice showed slightly more immobility time during the night, whereas the NEK4 OE mice spent more time swimming (control, 171.6 ± 79.46 s; NEK4 OE, 109.9 ± 71.63 s, P = 0.0337; Fig. 3E). This result confirmed the disturbed circadian fluctuations in emotional behaviors caused by overexpressing NEK4 in hippocampus.
We then used the small animal metabolic and behavioral phenotype analytical system to monitor the circadian rhythm in NEK4 OE mice and the control mice, but no significant changes in the biological rhythm were seen (Fig. 3F). We also examined the results presented by CircaDB30, and the mRNA level of NEK4 was relatively stable between daytime and night, suggesting that NEK4 did not affect nor was its transcription affected by the biological rhythm in mice.
NEK4 overexpression disrupted circadian fluctuations in dendritic spine morphogenesis and synaptogenesis
Given that overexpression of NEK4 resulted in a circadian fluctuation of emotional behaviors in mice, we then explored the cellular phenotypes in the hippocampus of these mice during day and night respectively. Since the synapse is one of the most important elements in brain function, we examined the impact of NEK4 on synaptic structure in hippocampus through neuron sparse labeling (Fig. 4A), and noticed that overexpression of NEK4 led to abnormal circadian fluctuations in dendritic spine morphogenesis. We noticed that there was a “baseline” difference in the dendritic spines between day and night in the control mice, a significant increase of mushroom spines and a slight reduction of thin spines during the night compared with the day (Day: mushroom, 3.885 ± 0.6455 spines per 10 μm; Night: mushroom, 5.196 ± 0.8599 spines per 10 μm, P < 0.0001; Day: thin, 11.59 ± 1.867 spines per 10 μm; Night: thin, 10.250 ± 2.180 spines per 10 μm, P = 0.0675; Fig. 4B), probably reflecting the nocturnality of mice. There was no difference in the densities of total dendritic spines between NEK4 OE mice and the control mice either during day or night. However, compared with the control mice, NEK4 OE mice had higher densities of mushroom dendritic spines during the daytime (control, 3.885 ± 0.645 spines per 10 μm; NEK4 OE, 5.366 ± 0.973 spines per 10 μm, P = 0.0018; Fig. 4B) while lower densities of mushroom dendritic spines during the night (control, 5.196 ± 0.860 spines per 10 μm; NEK4 OE, 3.243 ± 0.823 spines per 10 μm, P = 0.0008; Fig. 4B), indicating a potent impact of NEK4 on dendritic spine morphogenesis in a circadian manner. Additionally, overexpression of NEK4 resulted in a significant decrease in the density of thin spines in the daytime (control, 11.587 ± 1.867 spines per 10 μm; NEK4 OE, 9.220 ± 2.054 spines per 10 μm, P < 0.0001; Fig. 4B), while this effect disappeared during the night (control, 10.25 ± 2.180 spines per 10 μm; NEK4 OE, 10.16 ± 2.049 spines per 10 μm, P = 0.998; Fig. 4B).
A Sparsely labeled neurons in the dorsal hippocampus CA1 region are shown in the left panel. The injection site of NEK4 overexpression AAV was identified by red fluorescence, and gray fluorescence indicates neurons in the CA1 region labeled with sparsely labeled AAV. Scale bars represent 100 μm. The right panel shows the second or third branch in the apical dendrites of sparsely labeled neurons in the dorsal hippocampus CA1 region, and scale bars represent 5 μm. B The density of various types of dendritic spines in the apical dendrites of neurons from NEK4 OE mice and the control mice during day and night were calculated and compared. Dendritic spines during the day compared with the night in control group (thin: P = 0.0675, df = 31, t = 1.895; mushroom: P < 0.0001, df = 31, t = 5.009; two-sided student’s t test). Dendritic spines during the day compared with the night in NEK4 OE group (mushroom: P < 0.0001, df = 27, t = 6.251; two-sided student’s t test). Dendritic spines of the control group compared with NEK4 OE group (Day: thin: P < 0.0001, df = 99, t = 5.663; night: thin: P = 0.998, df = 75, t = 0.172. Day: mushroom: P = 0.0018, df = 99, t = 3.541; night: mushroom: P = 0.0008, df = 75, t = 3.815; 2-way ANOVA with Bonferroni’s multiple comparisons test). N = 19 neurons were analyzed in the daytime control group, n = 14 neurons were analyzed in the night control group, n = 16 neurons were analyzed in the daytime NEK4 OE group, n = 13 neurons were analyzed in the night NEK4 OE group. C The density of total dendritic spines from these mice were analyzed, the two-sided student’s t test was used to perform the significance test. All data were shown as mean ± SD, and P < 0.05 after corrections was defined as significant. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P < 0.0001. Shadows represent data at night. Source data are provided as a Source data file.
Through the electron microscopy experiments (Fig. 5A), we found that the number of synapses formed per unit area in the hippocampal CA1 region were also significantly different between NEK4 OE mice and controls in a circadian manner. Consistent with our findings regarding the mushroom dendritic spines, the control mice had more synapses during the night than during the daytime due to their nocturnality. We also saw that the NEK4 OE mice had significantly more synapses than controls during the daytime (control, 8.421 ± 2.143; NEK4 OE, 11.790 ± 2.070, P < 0.0001; Fig. 5B), and significantly less synapses during the night (control, 10.74 ± 2.218; NEK4 OE, 8.400 ± 2.010, P = 0.0013; Fig. 5B). Therefore, both the dendritic spine and synaptic structure analyses confirmed the effects of NEK4 on synapses.
The number of synapses in NEK4 OE mice and the control mice during day and night were compared to assess the effects of NEK4 on synaptogenesis. A synapse was characterized based on the presence of the pre-protrusion (containing vesicles), synaptic cleft, and postsynaptic (PSD region) structures. The number of synapses in electron microscope images covering a same area was calculated and analyzed, and scale bars represent 2 μm. A Representative images of synaptic structures acquired by electron microscope. B The numbers of synapses in the CA1 region of dorsal hippocampus of the mice. Synapses during the day compared with the night in control group (P = 0.0022, df = 36, t = 3.301). Synapses during the day compared with the night in NEK4 OE group (P < 0.0001, df = 37, t = 5.187). Synapses of the control group compared with NEK4 OE group (Day: P < 0.0001, df = 36, t = 4.928; night: P = 0.0013, df = 37, t = 3.481). N = 19 images were analyzed in the daytime control group, n = 19 images were analyzed in the night control group, n = 19 images were analyzed in the daytime NEK4 OE group, n = 20 images were analyzed in the night NEK4 OE group. All data were shown as mean ± SD, the two-sided student’s t test was used to perform the significance test. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P < 0.0001. Shadows represent data at night. Source data are provided as a Source data file.
Lithium treatment ameliorated the circadian fluctuations in emotional behaviors and synaptic structures in NEK4 OE mice
Given that the unstable emotional behaviors are key clinical manifestations in BD patients, we then examined the effect of lithium, a common mood stabilizer in BD treatment, on the emotional fluctuations of the NEK4 OE mice to assess the potential correlation of this phenotype with BD. In the open field, we noticed that lithium treatment caused significant alleviation of the anxiety-like behaviors in the NEK4 OE mice during the daytime, i.e., NEK4 OE mice treated with lithium spent more time in the center and less time in the corner than their vehicle controls (Fig. 6A). Consistently, in the elevated plus maze, despite a reduction of the total distance, the diurnal anxiety-like and nocturnal anxiolytic-like behaviors of NEK4 OE mice were both significantly mitigated (Fig. 6B). Taken together, the aberrant fluctuations of emotional behaviors in NEK4 OE mice could be rescued by lithium treatment, suggesting that these mice likely mimicked the abnormal mood transitions in BD patients.
A Mice treated with either vehicle or lithium were allowed to explore for 6 min in the open field, and the total distance traveled (day: control-NEK4 OE, P = 0.0354, df = 19, t = 2.265; night: control-NEK4 OE, P = 0.0233, df = 18, t = 2.479), distance traveled in the center area (day: control-NEK4 OE, P = 0.0324, df = 19, t = 2.308), time spent in the center area and time spent in the corner area (day: control-NEK4 OE, P = 0.0244, df = 19, t = 2.445) were calculated. B The mice were allowed to explore the elevated plus maze for 5 min, and the total distance traveled (day: control-NEK4 OE, P = 0.0046, df = 18, t = 3.237), distance traveled in the open arms (day: control-NEK4 OE, P = 0.1122, df = 18, t = 1.670; night: control-NEK4 OE, P = 0.0154, df = 18, t = 2.678), time traveled in the open arms (day: control-NEK4 OE, P = 0.0953, df = 18, t = 1.760; night: control-NEK4 OE, P = 0.0144, df = 18, t = 2.709), time spent in the closed arms (day: control-NEK4 OE, P = 0.0146, df = 18, t = 2.702; night: control-NEK4 OE, P = 0.0070, df = 18, t = 3.040) were calculated. N = 10 mice in the daytime of control group, n = 11 mice in the daytime of NEK4 OE group, n = 9 mice in the daytime of control group treated with lithium, n = 10 mice in the daytime of NEK4 OE group treated with lithium; n = 10 mice in the night of control group, n = 10 mice in the night of NEK4 OE group, n = 9 mice in the night of control group treated with lithium, n = 10 mice in the night of NEK4 OE group treated with lithium. All data were shown as mean ± SD, the two-sided student’s t test was used to perform the significance test. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P < 0.0001. #P: Close to statistically significant. NEK4 OE: NEK4 overexpression. NEK4 OE-Li: Lithium treated NEK4 OE mice. Control-Li: Lithium treated control mice. The shaded part represents the behavioral experiment data carried out in the dark environment at night (21:00-24:00). Source data are provided as a Source data file.
We then examined the effect of lithium on the synaptic structures in the NEK4 OE mice. Consistent with previous results, vehicle-treated NEK4 OE mice showed alterations of the morphology of dendritic spines during the day and night, particularly in mushroom spines (Day: control, 3.354 ± 0.426 spines per 10 μm; Day: NEK4 OE, 4.553 ± 0.970 spines per 10 μm, P = 0.0085. Night: control, 4.684 ± 0.597 spines per 10 μm; Night: NEK4 OE, 2.463 ± 0.847 spines per 10 μm, P < 0.0001; Fig. 7A), and lithium treatment reduced such morphological changes (Day: control-Li, 3.557 ± 0.248 spines per 10 μm; Day: NEK4 OE-Li, 3.144 ± 0.615 spines per 10 μm, P = 0.6826. Night: control-Li, 4.392 ± 0.607 spines per 10 μm; Night: NEK4 OE-Li, 4.295 ± 0.646 spines per 10 μm, P = 0.8907; Fig. 7A). Similarly, lithium treatment also alleviated the alterations in synapse numbers caused by NEK4 overexpression (Day: control, 9.893 ± 2.149; Day: NEK4 OE, 11.760 ± 2.444, P = 0.0035. Night: control, 11.570 ± 2.373; Night: NEK4 OE, 9.214 ± 3.281, P = 0.0027. Day: control-Li, 10.90 ± 2.119; Day: NEK4 OE-Li, 11.18 ± 2.653. Night: control-Li, 11.57 ± 2.348; Night: NEK4 OE-Li, 11.34 ± 2.497; Fig. 7B).
A Densities of various types of dendritic spines in apical dendrites neurons of dorsal hippocampus CA1 from NEK4 OE mice and the control mice treated with either vehicle or lithium. Dendritic spines of the control group compared with NEK4 OE group (day: thin: P = 0.0306, df = 138, q = 3.941; night: thin: P = 0.0417, df = 147, q = 3.774. Day: mushroom: P = 0.0085, df = 138, q = 4.559; night: mushroom: P < 0.0001, df = 147, q = 8.667; 2-way ANOVA with Tukey’s multiple comparisons test). N = 12 neurons in control-day; 13 neurons in NEK4 OE-day; 12 neurons in control-Li-day; 13 neurons in NEK4 OE-Li-day; 13 neurons in control-night; 13 neurons in NEK4 OE-night; 13 neurons in control-Li-night; 14 neurons in NEK4 OE-Li- night. B Effect of lithium treatment on synaptic formation in mice. synaptogenesis during the day compared with the night in control group (P = 0.0068, df = 56, t = 2.809; two-sided student’s t test); synaptogenesis of the control group compared with NEK4 OE group without lithium treatment (day: P = 0.0035, df = 55, t = 3.056; night: P = 0.0027, df = 56, t = 3.144; two-sided student’s t test). N = 28 images in control-day; 29 images in NEK4 OE-day; 31 images in control-Li-day; 28 images in NEK4 OE-Li-day; 30 images in control-night; 28 images in NEK4 OE-night; 28 images in control-Li-night; 29 images in NEK4 OE-Li-night. C Effect of lithium treatment on PSD thickness in mice. PSD thickness during the day compared with the night in control group (P = 0.0126, df = 43, t = 2.604; two-sided student’s t test); PSD thickness of the control group compared with NEK4 OE group without lithium treatment (day: P = 0.0173, df = 38, t = 2.489; night: P < 0.0001, df = 45, t = 4.940; two-sided student’s t test). N = 20 images in control-day; 20 images in NEK4 OE-day; 21 images in control-Li-day; 22 images in NEK4 OE-Li-day; 25 images in control-night; 22 images in NEK4 OE-night; 21 images in control-Li-night; 21 images in NEK4 OE-Li-night. All data were shown as mean ± SD, P < 0.05 after corrections was defined as significant. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P < 0.0001. Shadows represent data at night. Source data are provided as a Source data file.
We then investigated the structure of postsynaptic dense (PSD) regions in these mice for further insights into the synaptic structure and function. We also noticed a slight increase of the PSD thickness during the night compared with that during the daytime in the control mice (Day: 26.71 ± 8.263 nm; Night: 31.81 ± 4.713 nm, P = 0.0126; Fig. 7C). As expected, we found that NEK4 OE mice exhibited increased thickness of the PSD during the daytime and decreased thickness at night compared with controls (Day: control, 26.71 ± 8.263 nm; NEK4 OE, 33.58 ± 9.178 nm, P = 0.0173. Night: control, 31.81 ± 4.713 nm; NEK4 OE, 23.99 ± 6.117 nm, P < 0.0001; Fig. 7C), and lithium treatment alleviated the abnormal changes caused by NEK4 overexpression (Day: control-Li, 28.59 ± 5.414 nm; Day: NEK4 OE-Li, 26.45 ± 4.810 nm. Night: control-Li, 33.43 ± 6.183 nm; Night: NEK4 OE-Li, 35.51 ± 8.283 nm; Fig. 7C).
NEK4 affected phosphorylation levels of synapse-associated proteins in a circadian manner
The aberrant circadian swing of emotional behaviors and synapses in NEK4 OE mice suggested different physiological outcomes of increase NEK4 expression between day and night. Since this gene encodes a serine/threonine kinase, we explored the phosphoproteomic profiles of these mice at different time of the day and compared them with those of the control mice. We isolated the hippocampal proteins of the NEK4 OE mice and the control mice during day and night respectively (n = 3 for each group), and performed quantitative phosphorylated proteomics analyses. After quality control, 5376 quantifiable phosphorylated peptides were identified in 2415 phosphorylated proteins.
As both the current study and previous investigations indicate light/dark cycle correlated synaptic structural changes31, we firstly compared the diurnal and nocturnal phosphoproteomic profiles in the control mice to characterize “baseline” differences in protein phosphorylation statuses between day and night. We found 109 peptides showing different phosphorylation levels in the control mice between day and night (|log2(fold change)| ≥ 1 and P≤0.005, Supplementary Data 2), among which 38 peptides (mapped to 38 proteins) had higher phosphorylation levels during the daytime while lower phosphorylation levels during the night (referred to as diurnal proteins in subsequent discussions), whereas additional 71 peptides (mapped to 71 proteins) had higher phosphorylation levels during the night but lower during the daytime (referred to as nocturnal proteins in subsequent discussions). Notably, proteins (such as Mecp2, Actb, Map1b) critical for synaptic structures and dendritic spines have varied phosphorylation levels between day and night in the control mice (Fig. 8), suggesting that these proteins likely mediated the circadian alterations of the spines and synapses.
Remarkably, the diurnal and nocturnal phosphoproteomic profiles were strikingly different between NEK4 OE mice and the control mice. In the NEK4 OE mice, there were only 6 diurnal proteins, whereas 83 nocturnal proteins (|log2(fold change)| ≥ 1 and P ≤ 0.005, Supplementary Data 3). There were no overlapped diurnal proteins between NEK4 OE mice and the control mice, and only 8 overlapped nocturnal proteins (Nsfl1c, Tprg1l, Pclo, Slc7a10, Map2, Trim28, Rpl18a, Ank2).
To gain insights into the pathways mediating the circadian emotional swings resulted from NEK4 overexpression, we carried out the gene set enrichment analysis (GSEA) of the nocturnal proteins in the NEK4 OE mice and the control mice (analyses of diurnal proteins were not performed since the number of proteins in NEK4 OE mice was too small). Specifically, Reactome gene set analysis32 in the control mice highlighted the terms of “Vesicle-mediated transport”, “Signaling by Receptor Tyrosine Kinases”, “Translocation of SLC2A4 (GLUT4) to the plasma membrane”, “RHO GTPase Effectors” and so on (Supplementary Data 4), while the terms “MAPK family signaling cascades”, “Post NMDA receptor activation events”, “Protein-protein interactions at synapses” and “Activation of NMDA receptors and postsynaptic events” were specifically highlighted in NEK4 OE mice (Supplementary Data 5). The two groups also shared some gene sets such as “Signaling by Rho GTPases”, “Axon guidance” and “Transmission across Chemical Synapses” (Fig. 8). We then conducted the DisGeNET gene set analysis33, which integrates gene-disease/phenotype association data. In such analysis, the nocturnal proteins in the control mice were significantly enriched for multiple diseases, and the most significant term was “mood disorders” (Supplementary Data 6), confirming potential link between biological rhythm and mood disorders34. Among the nocturnal proteins of NEK4 OE mice, the term “mood disorders” was also significant, despite the included proteins were largely different with the control mice (Fig. 8). Moreover, we also identified many enriched terms that were not highlighted in the control mice, such as “Moderate intellectual disability”, “Impulsive Behavior” and “Autistic behavior” (Supplementary Data 7), suggesting pleotropic impacts of NEK4. Remarkably, the term “Mood swings” (included proteins: ATP1A3, GRIN2A, NCAM1, MATR3) was highlighted only among the nocturnal proteins of NEK4 OE mice. Given the putative role of NEK4 in BD pathogenesis, the abnormal circadian fluctuations in emotional behaviors and synaptogenesis caused by altered expression of this gene might reminiscent the emotional swings in this disorder.
Discussion
BD patients exhibit cyclic extreme mood swings between (hypo) mania and depression separated by periods of normal affect. The biological processes underlying the mood swings, which is probably the most prominent clinical feature of BD, remains poorly explored. The extreme mood episodes in BD patients are usually concurrent with significant changes in sleep, activity, and energy levels35. For example, a cohort study have revealed that individuals with BD experience significant fluctuations in anxiety and depression over time36, and BD patients also exhibit significantly delayed circadian function compared with controls37,38. Notably, individuals with BD often experience highly disrupted sleep/wake patterns, and circadian phases might be advanced in mania episodes and delayed in depressive episodes39. These data support the hypothesis that mood episodes in BD are associated with circadian misalignment between an individual’s endogenous circadian rhythms and their physical environment39.
To date, animal models that can simulate mania-like behavior, including the modification of circadian rhythm genes (e.g., Clock-19)40,41 and sleep deprivation14, have been described and made tremendous contributions to the field. However, few animal models exhibited spontaneous emotional shifts reminiscent of BD patients. Two recent studies have shown mood transitions in mice with insufficient expression of BD risk genes. Briefly, one study found that mice lacking synaptotagmin-7 (Syt7), a BD risk gene identified in neurons derived from patient-derived iPSCs, exhibited emotional fluctuations during the day and at night42; the other study found that mice lacking a BD GWAS risk gene FADS1/2 showed emotional fluctuations with a multi-week interval43. These inspiring findings suggest that modulating BD risk genes in mice may produce the unique mood transitions in patients and provide opportunities to study the underlying mechanisms.
Chromosome 3p21.1 is one of the prominent BD GWAS loci, and we demonstrated that independent BD risk SNPs in this area consistently predicted higher NEK4 mRNA in hippocampus. Yang et al. previously showed that overexpressing NEK4 led to altered dendritic spine morphology, dendritic arborization and axon length in primary cultured neurons44,45, and here we found that NEK4 overexpression in hippocampus led to spontaneous fluctuations between diurnal anxiety-like behaviors and nocturnal anxiolytic-like behaviors in mice using both genetic and pharmacological approaches. However, although manipulation of Syt7, FADS1/2, or NEK4 all result in emotional fluctuations, their underlying mechanisms are unlikely the same. Previous studies showed that Syt7 linked BD and insulin/glucose metabolic syndrome42, whereas FADS1/2 likely contributed to BD-like episodic behavioral changes due to peripheral abnormalities43. While NEK4 mice exhibited circadian swings in emotion-related behaviors, this gene did not disrupt the circadian rhythm of the mice, but rather affect biological events aligned to the circadian rhythm. We also realized that light might be a driver in emotion transitions mediated by NEK4 overexpression, as mood symptoms may be associated with the amount, duration, and intensity of light46,47,48. Nonetheless, NEK4 cTG mice exhibited anxiety-like behaviors in the elevated plus maze under acute light-off conditions during the daytime (Supplementary Fig. 6), suggesting that light was unlikely the key mediator of its impact on emotional behaviors. Further studies assessing the circadian differences in the enzymatic activities or interactome of NEK4, with a focus in the involvement of the “inner-clock” machineries, might provide valuable clues.
Mice are active in the dark environment at night and are in a resting state in the light environment during the day. We herein noticed a concordance between the activities of mice and their synaptic structures in the control mice, with significant elevations of all these indices during the night. While this finding likely reflects the nocturnality of mice, it also highlights the existing homeostatic fluctuations of synaptic structures in a circadian manner, which has recently been discovered and brought revolutions to this field31,49. Intriguingly, we found that overexpression of NEK4 led to obvious interruptions of such homeostatic dynamics of synapses, i.e., NEK4 overexpression caused increased densities of mushroom spines, number of synapses and PSD thickness during the day, and reductions of those indices during the night. We hence speculated that disturbance of the “inner-clock” of synapses likely explained the impact of this gene on emotional behaviors. In consistency with our hypothesis, we found that lithium treatment, which balanced synaptic homeostasis, also ameliorated the behavioral abnormalities of NEK4 OE mice. It should be noticed that lithium treatment also resulted in slight trends of lower diurnal activities and higher nocturnal activities in the control mice, which is in line with the previous findings that lithium could enhance circadian amplitudes50. In addition, it has been reported that lithium has a regulatory effect on the structure of synapses and postsynaptic proteins51,52,53. Our phosphoproteomic analyses also found that NEK4 influenced a large number of postsynaptic proteins. Therefore, lithium may alleviate the behavioral fluctuations in NEK4 OE mice by influencing postsynaptic proteins, but the specific involved molecules remain to be characterized.
However, there are several limitations in the present study. NEK4 is widely expressed in many brain regions and multiple neural cells including excitatory neurons, inhibitory neurons, ciliated cells and astrocytes (https://www.proteinatlas.org/ENSG00000114904-NEK4/single+cell+type). In our study, we focused on the phenotypes caused by overexpressing NEK4 in pyramidal neurons of the adult forebrain or all cell types of the dorsal hippocampus. It is unclear whether manipulating NEK4 expression in distinct cell types, different brain regions or at different developmental stages will cause varied phenotypes, which should be analyzed in further studies. In addition, multiple GWAS have identified NEK4 as a shared risk gene between BD and schizophrenia44, therefore, the observed phenotypes in NEK4 mice might not be exclusive to the pathogenesis of BD. While the putative role of increased hippocampal NEK4 expression gains support from the eQTL and transcriptomic analyses in humans, whether the observed behavioral and synaptic disturbance caused by NEK4 were in the causative chain related to BD should be further validated in models with greater relevance to the human brain (e.g., additional human cohorts, brain organoids, non-human primates, etc.). Furthermore, although the genetic risk and BD diagnosis predicted higher expression of NEK4 in the hippocampus, the mRNA levels of NEK4 could be affected by other factors such as medications or particular stages of the illness; similarly, while GNL3 and GLYCTK were not examined in our murine experiments, their potential roles in BD are not denied. Further investigations in larger samples from patients will be helpful.
In summary, despite that many questions remain to be answered to fully elucidate all molecular mechanisms underlying mood swings in BD, the current study suggests that interruption of the homeostatic dynamics of synaptic structures might be part of the explanation. Likewise, we believe that other homeostatic dynamic processes, which also undergoes physiological fluctuations just like the circadian transitions of the synaptic structures, could also be interrupted in BD patients, leading to the clinically observed fluctuations in emotions. Further studies identifying such vital biological processes and their regulatory mechanisms are necessary to understand the pathogenesis of BD.
Methods
Ethics
All animal experiments were carried out according to the guidelines (developed by the National Advisory Committee for Laboratory Animal Research) for ethical conduct in the care and use of animals, and all protocols were approved by the Animal Ethic Committee of Kunming Institute of Zoology (NO: IACUC-RE-2021-11-001). Wild-type C57BL/6 J male mice (5–6 weeks old) were purchased from GemPharmatech Co., Ltd. (Nanjing, China) and groups of four animals were kept together in transparent, industry-standard Individually Ventilated Cages (IVC), with free access to food and water. The housing room had a temperature of 22 °C and a 12-hour light/dark cycle (lights on at 08:00 and lights off at 20:00).
The GWAS and eQTL data from human study participants were publicly accessible. No original data were collected from humans, and no ethical committee approval was requisite for the present study. Each original GWAS study obtained approval from their respective institutional ethics review committees.
Public GWAS statistics of bipolar disorder
The PGC3 BD GWAS included 41917 cases and 371549 controls of European ancestry18, and detailed information can be found in the original study (https://pgc.unc.edu/for-researchers/download-results/). The pairwise LD (r2) between the SNPs were calculated using an online program SHEsis (http://analysis.bio-x.cn)54. Regional association results of the 3p21.1 locus (chr3: 52300000-53300000, hg19) in PGC3 BD GWAS were plotted using LocusZoom (http://locuszoom.sph.umich.edu/locuszoom/)55.
Hippocampal expression quantitative trait loci (eQTL) dataset
We utilized eQTL data in hippocampus from BrainSeq Phase 2, a RiboZero RNA-Seq dataset of 371 individuals aged 13 or older24. Detailed information about the sample description and statistical methods can be found in the original study, and the eQTL was calculated based on the formula: log2(RPKM + 1) ~ SNP + diagnosis + sex + SNP PCs + expression PCs.
An independent hippocampus RNA-Seq eQTL dataset from MetaBrain (https://metabrain.nl/) was also utilized (n = 168). The hippocampus samples in MetaBrain were originally from Braineac (n = 101)56 and GTEx (n = 67)57, which did not have overlap in samples from BrainSeq24. Detailed description about the processing of multiple datasets, quality control and statistical analyses were shown in the original study, and the eQTL was calculated using Spearman correlations within each cohort first, followed by a sample size-weighted z-score meta-analysis.
Differential expression between BD cases and controls in the hippocampus
To compare the differential expression of NEK4, GNL3 and GLYCTK between BD cases and controls in hippocampus, we firstly utilized a public BrainEXP dataset27 including expression data of 18 BD cases versus 18 controls (data was from GSE53987)28. We also downloaded an independent hippocampus expression dataset including 11 BD cases and 11 controls (GSE210064 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE210064])29, and the normalized expression metrics were analyzed for linear models using limma in R software (version 4.3.1)58.
Generation of H11-LSL-NEK4±; CaMKIIα-Cre conditional transgenic mice
NEK4 (ENST00000233027.10) conditional transgenic (cTG) mice model was delegated to GemPharmatech Co., Ltd. (Nanjing, China). In brief, the CAG-LSL-NEK4-HA-WPRE-polyA gene fragment was site-directed inserted into the H11 locus of the mouse (NEK4± mice). F1 mice were tail-clipped at 5–7 days and genomic DNA was taken to identify transgene using PCR and sequencing. Following the creation of NEK4 cTG mice, female heterozygous CaMKIIα-Cre mice were crossed with male NEK4± mice to obtain animals with NEK4 conditional transgene, primarily in the hippocampal CA1 pyramidal cells and forebrain. Eight weeks following birth, behavioral testing was carried out using the following described behavioral paradigms and procedures. The anti-HA antibody (#901513, BioLegend) was used to detect NEK4 expression in NEK4 cTG mice via Western blot (Supplementary Fig. 5A).
AAV vector production and titration
The coding sequence of the human dominant NEK4 transcript (ENST00000233027) was constructed into the pAAV-CAG-tdTomato vector (#59462, Addgene) with BamHI and HindIII, the constructed plasmid was amplified using Stbl3 (#CC96134, Tolobio). Adeno-associated virus (AAV) was produced by co-transfection of pHelper, AAV-DJ and the recombinant pAAV-CAG-NEK4-FLAG or control vector (pAAV-CAG-tdTomato) into HEK293T cells (#KCB 200744YJ, Conservation Genetics CAS Kunming Cell Bank) using PEI Max (#23966, Polysciences). Three days post transfection, cells containing AAV were harvested by centrifugation at 3000 x g for 5 min at room temperature and were then lysed through rapid freezing with liquid nitrogen and thawing at 37 °C three times with 0.15 M NaCl/20 mM Tris PH 8.0 to release the AAV. The lysate was then added sodium deoxycholate (#D6750, Sigma-Aldrich) for a final concentration of 0.5% and Benzonase (#E1014, Millipore) and then incubated at 37 °C for 30 min, followed by centrifuging at 8,000 x g for 1 h at 4 °C. The lysate supernatant was layered onto an Iodixanol density step gradient and centrifuged at 350,000 x g for 2 h at 18 °C. The virus was obtained from the 40% iodixanol portion of the gradient, which was then transferred to 100 kDa centrifugal filter units for purification and concentration with precooling PBS.
To assess the AAV titration, a pAAV-CAG-tdTomato plasmid sample with known concentration was utilized as the standard samp, and the copy number was set as 1010, 109, 108, 107, 106, and 10^=5. 5 µL of each AAV sample was treated with DNaseI (#EN0529, Thermo Scientific) for 30 min at 37 °C followed by 10 min at 95 °C, and then 1:1000 diluted. Real-time quantitative PCR (RT-qPCR) was used to measure the AAV titers using primers specific to the ITR (forward primer: 5’-GGAACCCCTAGTGATGGAGTT-3’, reverse primer: 5’-CGGCCTCAGTGAGCGA-3’). The titer value was calculated according to the standard curve generated with the copy number and Ct value of the standard plasmid.
Intracranial stereotaxic injection of AAV in adult mice
Wild-type C57BL/6 J male mice (7-week-old) were placed in an induction anesthesia box and anesthetized with 3% isoflurane. The mice were then placed in the stereotaxic frame with 1% isoflurane, and an incision was gently made to create a small burr hole. We injected 0.2 µL NEK4 overexpressing AAV or control (5 × 1012 GC/mL) vectors to each site of the dorsal hippocampus (anteroposterior [AP]: −2.06 mm; mediolateral [ML]: ±1.50 mm; dorsoventral [DV]: −1.50 mm) with an infusion rate of 60 nL/min to prevent tissue damage. During surgery, erythromycin eye ointment was placed on the eye to avoid corneal dryness. After surgery, the incision was sutured, cleaned, and injected with buprenorphine (0.1 mg/kg) and carprofen (5 mg/kg) for analgesic and anti-inflammatory effects. Following these procedures, mice were placed on 37 °C heating pad until they regained normal mobility. The anti-flag antibody (#14793S, Cell Signaling Technology) was used to detect NEK4 expression in NEK4 OE mice via Western blot (Supplementary Fig. 5B).
Behavior paradigm
The onset age of BD usually starts from puberty, which corresponds to the age of 8–12 weeks in mice. Following previous studies59,60,61, we carried out the behavioral tests in NEK4 cTG mice during the ages of 8–12 weeks, and in the AAV-injected NEK4 OE mice the behavioral tests started at 11 weeks. The mice were transferred from the housing room to the behavioral room (environmental conditions were same as the housing room) and were handled daily by the researcher for a week prior to all behavioral tests to reduce the anxiety levels of the mice during the experiment. Diurnal (light phase) and nocturnal (dark phase) behavioral tests were respectively performed, from 10:00 to 17:00 during the day and from 21:00 to 24:00 at night. During nocturnal behavioral experiments, we tracked mouse movements with infrared lamps and cameras. The mice were moved to the behavioral room for about 1 h before the experiment. All equipment used in behavioral tests were bought from Shanghai Xinsoft Information Technology. Due to the possibility of erratic outcomes from varying hormone levels during the female estrous cycle, only male mice were used in this study. Before the behavioral tests, four randomly selected NEK4 cTG mice, NEK4 OE mice, and their corresponding controls were assessed for their body length, body weight, and motor activities. Specifically, the body length was measured with a standard ruler (Supplementary Fig. 7), the body weight was measured with an electronic weighing scale (Supplementary Fig. 7), and the motor abilities of the mice were assessed through the rotarod test (Supplementary Fig. 8). Mice from different experimental groups were comparable in these indices. The visual cliff test and vertical placement test were also performed to ensure that the vision of mice from each experimental group was intact (Supplementary Fig. 9 and Supplementary Movie 1). All mice were visibly healthy with smooth coat and normal ingestion of food and water. They were gently handled and kept away from any organic damage during the experiments.
Open field test. The mice were placed in the center of the apparatus surrounded by four square walls (40 × 40 × 40 cm) and allowed to explore freely for 5 min. The middle 20 × 20 cm portion of the field was defined as the center area region, the region of 10 ×10 cm in each of the four corners was defined as the corner area. The mice’s time and distance traveled in the center and corners, as well as the total mobility distance during the experiment were recorded.
Elevated plus maze. The Elevated plus maze comprises two cross-shaped open and closed arms that are 75 cm above the ground. Mice were placed in the center area of the maze, facing an open arm, and allowed to explore the apparatus freely for 5 min. The travelled distance and time spent in the open arms and closed arms were recorded.
Forced swim test. Mice were gently placed in an open, see-through cylinder (25 cm high, 10 cm in diameter) containing water with a depth of 18 cm and a temperature of 23 ± 1 °C. Their motion condition within 8 min were recorded, and the immobility time lengths during the final 6 min were analyzed. The water was entirely changed after each mouse.
Visual cliff test. The equipment for the visual cliff experiment consisted of a transparent open field (40 cm×40 cm x 40 cm) placed on a high platform (85 cm) with half of it extending outward (the safe area). A black and white cloth was positioned at the bottom of the open field and hanging vertically to the ground to create the illusion of a “cliff ” (the dangerous area). A rectangular platform (5 mm in height) was placed at the junction of the safe area and the dangerous area. Mice were placed on this platform to allow free exploration for 3 min, and the lengths of time they spent in the safe area and the dangerous area were respectively recorded. This experiment was carried out following a previous study with modifications62.
Vertical placement test. The vertical placement test was performed to assess the mice’s ability to respond to visual cues. Briefly, the experiment began by pinching the end of a mouse’s tail and dangling it 25 cm above the edge of a table, the mouse was then slowly put down while ensuring that its whiskers did not touch the table. When its eyes were almost level with the table, it was allowed three seconds to extend its front legs and climb onto the table. Mice with normal vision would lift their heads, arch their backs, and reach out with their front paws to grab the edge of the table during the descent. This experiment was performed following a previous study with modifications63.
Rotarod test. The rotarod test was performed to examine the motor function, coordination ability, and anti-fatigue capacity of mice. The mice were placed in each compartment of the rods, and they could run along with the rod rotation to maintain balance and avoid slipping. When a mouse falls, the instrument automatically records the time the mouse stays on the rod and the speed of the rod when the mouse falls. Before the experiment, the mice were trained at speeds of 8 rpm, 12 rpm, and 16 rpm for two consecutive days, three times a day for 5 min each, with a minimum interval of 30 min between training sessions. During the experiment, the rotation speed of the instrument increased steadily from 4 rpm to 40 rpm over a period of 5 min, and the time and speed at which the mice fell were recorded. The average of three tests for each mouse was recorded as the final result.
Fluorescent sparsely labeling of neurons, image capturing and statistical analyses
Mice receiving stereotaxic injections of NEK4 overexpressing AAV or control virus were allowed to recover for 3 weeks, and were then injected with another AAV (#BC-SL001, Brain Case) that sparsely labeled neurons at the same location in the dorsal hippocampus. Two weeks after the second injection, the mice were proceeded for slow trans-cardiac perfusion with 20 mL of PBS and 10 mL of per-cold 4% paraformaldehyde (PFA). Their brain tissues were harvested and fixed for around 24 h with PFA at 4 °C, and then dehydrated through incubation in 20% sucrose PBS for 24 h followed by 30% sucrose PBS for 24 h. The brain tissues were then freezed at –80 °C with an optimal cutting temperature compound (OCT) and sectioned at 50 μm. Free-floating brain slices were washed with PBS and mounted onto microscope slides with a drop of antifade mounting medium (#H-1000, Vector). Coverslips were carefully placed over the antifade solution to prevent air bubbles. To analyze the circadian alterations of the neurons, the mice were euthanized either at 11:00 am (daytime) or 11:00 pm (night).
Sparsely labeled neurons in the dorsal hippocampus CA1 were captured using a Zeiss LSM800 confocal microscope to produce z-stack pictures. The Image-J software was used to analyze the stacked images, and NeuronStudio was used to analyze dendritic spines. The dendritic spines in the second or the third branch of the apical dendrites of neurons were counted, and the numbers and proportions of different types of spines, including mushroom, thin, and stubby, were calculated and analyzed.
Analyses of synaptic structures with transmission electron microscopy
To analyze the synaptic structure, AAV overexpressing NEK4 and a virus expressing the tdTomato fluorescent protein were mixed at a 10:1 ratio and injected into the dorsal hippocampus of mice. Four weeks later, the dorsal hippocampus tissues were harvested during both daytime and night, and fluorescence signals were captured using electron microscopy to analyze the structural characteristics of synapses.
Lithium administration
To achieve a clinically relevant blood concentration of lithium, chronic lithium chloride administration using intraperitoneal injection followed by chronic ingestion was performed following previous studies64,65. In detail, an intraperitoneal loading dosage of 4 mmol/kg LiCl (#L0416, Sangon Biotech) or 0.9% NaCl was given to the mice 3 weeks after the AAV (control or NEK4 overexpressing virus) injection, following by adding 0.06% LiCl (#L0416, Sangon Biotech) to the drinking water for 15 days, both during the day and at night. Subsequently, animal behavioral tests, neuronal dendritic spines, and synaptic assessments were carried out.
Quantitative phosphoproteomic analysis
Hippocampus tissues (n = 3 in each group) were rapidly dissected at both daytime and night and lysed with SDT buffer (4% SDS, 100 mM Tris-HCl, 1 mM DTT, pH 7.6). The protein concentration of the lysates was determined using the BCA Protein Assay Kit (Bio-Rad, USA). Subsequently, 200 μg of proteins from each sample were used for further experiments. The proteins were digested by trypsin following filter-aided sample preparation (FASP) procedure66, and the digested peptides were processed for desalination, quantification and concentration. Next, phosphopeptides were enriched using the High-SelectTM Fe-NTA Phosphopeptides Enrichment Kit according to the manufacturer’s instructions (Thermo Scientific). After lyophilization, the phosphopeptides were resuspended in 20 µL loading buffer (0.1% formic acid) for liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS).
A timsTOF Pro mass spectrometer (Bruker) connected to Nanoelute (Bruker Daltonics) was used for the 60-minute LC-MS/MS. Buffer A (0.1% formic acid) was used to load peptides onto a handmade C18-reversed phase analytical column (25 cm length, 75 μm inner diameter, 1.9 μm, C18), and separated using a linear gradient of buffer B (84% acetonitrile and 0.1% formic acid) at a flow rate of 300 nL/min. Using a mass range of m/z 100-1700 and 1/k0 of 0.6 to 1.6, the mass spectrometer collected ion mobility MS spectra while operating in positive ion mode. After that, ten PASEF MS/MS cycles were run with a threshold of 2500 and a goal intensity of 1.5k. The release time for active exclusion was set to 0.4 min. The raw MS data for each sample were combined and analyzed using the MaxQuant software for identification and quantitation using Swissprot_mouse_17097_20220104 database with following related parameters and instructions (Modifications: Carbamidomethyl C, Variable modifications: Oxidation (M); Phospho (STY), Digestion: Trypsin, type: label-free, Database pattern: Reverse, Peptide/Site/Protein FDR ≤ 0.01, Time window: 2 min).
A good mass deviation was maintained during data collection, with the mass deviation of modified peptide ions predominantly within 10 ppm, indicating accurate and reliable identification results. Analysis of the MS map data using the Andromeda analysis tool showed that the score of modified peptide ions was ideal, with approximately 64.55% of the modified peptides scoring more than 20 points, and a median peptide score of 79.89 points. Peptide FDR < 0.01 and protein FDR < 0.01 were used as screening criteria in the qualitative analysis of the data (Supplementary Fig. 10 and Supplementary Data 8).
The phosphoproteomic profiles were analyzed and compared between tissues obtained at daytime and night in either the control mice or NEK4 OE mice. Briefly, proteins that had significantly different phosphorylation between day and night were characterized, and those having |log2(fold change)|≥ 1 and P ≤ 0.005 were considered proteins with circadian alteration of their phosphorylation statuses. These differentially phosphorylated proteins between day and night in either the control mice or NEK4 OE mice were then subjected for enrichment analyses of Reactome gene sets32 and DisGeNET gene sets33 using the Metascape dataset (https://metascape.org/)67.
Statistics and reproducibility
Two to three biological replicates were performed for each experiment, and no data were excluded for the analysis of dendritic spines and synaptic structures of neurons. For behavioral data, we first statistically assessed whether there were any apparent outliers in the open field test data. Outliers were calculated using the ROUT method in GraphPad Prism 9.0 software, with Q = 1%, and were excluded from the dataset. Mice that were isolated in the open field were not included in subsequent behavioral experiments. In addition, the mice in the experiment were in an unhealthy state, such as the loss of beard will also be eliminated. Data analysis was performed using GraphPad Prism 9.0. Unpaired two-tailed Student’s t test or two-way ANOVA was applied depending on the experimental groups, and specific statistical methods are indicated in the figure legends. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P < 0.0001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The mass spectrometry proteomics data in NEK4 mice generated in this study have been deposited in the ProteomeXchange Consortium database under accession code PXD052195. The processed mass spectrometry proteomics statistics are available within the Supplementary Data 8. The other mice related data generated in this study are provided in the Supplementary Materials. All datasets involving human participants used in this study are publicly accessible. Specifically, the GWAS summary statistics for bipolar disorder are available on Figshare [https://figshare.com/articles/dataset/PGC3_bipolar_disorder_GWAS_summary_statistics/14102594], the data for PheWeb-based brain-wide imaging analysis in UK biobank are available at https://open.win.ox.ac.uk/ukbiobank/big40/pheweb33k/ via inquiring specific SNPs, the eQTL statistics in BrainSeq Phase 2 dataset are available at http://eqtl.brainseq.org/phase2/eqtl/ through inquiring specific SNPs, the eQTL statistics in MetaBrain dataset are available at https://www.metabrain.nl/cis-eqtls.html through inquiring specific SNPs, the processed NEK4 mRNA data in bipolar disorder of GSE53987 are obtained from http://brainexpnpd.org:8088/BrainEXPNPD/boxresult.html?value = NEK4 = BD, the processed GNL3 mRNA data in bipolar disorder of GSE53987 are obtained from http://brainexpnpd.org:8088/BrainEXPNPD/boxresult.html?value = GNL3 = BD, the processed GLYCTK mRNA data in bipolar disorder of GSE53987 are obtained from http://brainexpnpd.org:8088/BrainEXPNPD/boxresult.html?value = GLYCTK = BD, and we also use processed mRNA data from GSE210064. Source data are provided with this paper.
Code availability
Analysis codes used in this manuscript can be found at https://doi.org/10.5281/zenodo.13938317.
References
Zhang, C., Xiao, X., Li, T. & Li, M. Translational genomics and beyond in bipolar disorder. Mol. Psychiatry 26, 186–202 (2021).
Lee, Y., Zhang, Y., Kim, S. & Han, K. Excitatory and inhibitory synaptic dysfunction in mania: an emerging hypothesis from animal model studies. Exp. Mol. Med. 50, 12 (2018).
Smith, K. R. et al. Psychiatric risk factor ANK3/ankyrin-G nanodomains regulate the structure and function of glutamatergic synapses. Neuron 84, 399–415 (2014).
Yoon, S. et al. Homer1 promotes dendritic spine growth through ankyrin-G and its loss reshapes the synaptic proteome. Mol. Psychiatry 26, 1775–1789 (2021).
Yoon, S. et al. Usp9X controls Ankyrin-repeat domain protein homeostasis during dendritic spine development. Neuron 105, 506–521 (2020).
Deans, P. J. M. et al. Psychosis risk candidate ZNF804A localizes to synapses and regulates neurite formation and dendritic spine structure. Biol. Psychiatry 82, 49–61 (2017).
Pappas, A. L. et al. Deficiency of Shank2 causes mania-like behavior that responds to mood stabilizers. JCI Insight 2, e92052 (2017).
Han, K. et al. SHANK3 overexpression causes manic-like behaviour with unique pharmacogenetic properties. Nature 503, 72–77 (2013).
Zhu, S. et al. Genetic disruption of ankyrin-G in adult mouse forebrain causes cortical synapse alteration and behavior reminiscent of bipolar disorder. Proc. Natl. Acad. Sci. USA 114, 10479–10484 (2017).
Miro, X. et al. Studies in humans and mice implicate neurocan in the etiology of mania. Am. J. Psychiatry 169, 982–990 (2012).
Dao, D. T. et al. Mood disorder susceptibility gene CACNA1C modifies mood-related behaviors in mice and interacts with sex to influence behavior in mice and diagnosis in humans. Biol. Psychiatry 68, 801–810 (2010).
Leussis, M. P. et al. The ANK3 bipolar disorder gene regulates psychiatric-related behaviors that are modulated by lithium and stress. Biol. Psychiatry 73, 683–690 (2013).
Sen, P. et al. A bipolar disorder-associated missense variant alters adenylyl cyclase 2 activity and promotes mania-like behavior. Mol. Psychiatry https://doi.org/10.1038/s41380-024-02663-w (2024).
Li, X. et al. A novel murine model of mania. Mol. Psychiatry 28, 3044–3054 (2023).
Cosgrove, V. E., Kelsoe, J. R. & Suppes, T. Toward a valid animal model of bipolar disorder: how the Research Domain Criteria help bridge the clinical-basic science divide. Biol. Psychiatry 79, 62–70 (2016).
Li, M. et al. Phenotypes, mechanisms and therapeutics: insights from bipolar disorder GWAS findings. Mol. Psychiatry 27, 2927–2939 (2022).
van Enkhuizen, J. et al. Investigating the underlying mechanisms of aberrant behaviors in bipolar disorder from patients to models: Rodent and human studies. Neurosci. Biobehav. Rev. 58, 4–18 (2015).
Mullins, N. et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat. Genet. 53, 817–829 (2021).
McMahon, F. J. et al. Meta-analysis of genome-wide association data identifies a risk locus for major mood disorders on 3p21.1. Nat. Genet. 42, 128–131 (2010).
Stahl, E. A. et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 51, 793–803 (2019).
Chen, D. T. et al. Genome-wide association study meta-analysis of European and Asian-ancestry samples identifies three novel loci associated with bipolar disorder. Mol. Psychiatry 18, 195–205 (2013).
Breen, G. et al. Replication of association of 3p21.1 with susceptibility to bipolar disorder but not major depression. Nat. Genet. 43, 3–5 (2011).
Gagliano Taliun, S. A. et al. Exploring and visualizing large-scale genetic associations by using PheWeb. Nat. Genet. 52, 550–552 (2020).
Collado-Torres, L. et al. Regional heterogeneity in gene expression, regulation, and coherence in the frontal cortex and hippocampus across development and schizophrenia. Neuron 103, 203–216 (2019).
Smith, S. M. et al. An expanded set of genome-wide association studies of brain imaging phenotypes in UK Biobank. Nat. Neurosci. 24, 737–745 (2021).
de Klein, N. et al. Brain expression quantitative trait locus and network analyses reveal downstream effects and putative drivers for brain-related diseases. Nat. Genet. 55, 377–388 (2023).
Jiao, C. et al. BrainEXP: a database featuring with spatiotemporal expression variations and co-expression organizations in human brains. Bioinformatics 35, 172–174 (2019).
Lanz, T. A. et al. Postmortem transcriptional profiling reveals widespread increase in inflammation in schizophrenia: a comparison of prefrontal cortex, striatum, and hippocampus among matched tetrads of controls with subjects diagnosed with schizophrenia, bipolar or major depressive disorder. Transl. Psychiatry 9, 151 (2019).
Nascimento, C. et al. Gene expression alterations in the postmortem hippocampus from older patients with bipolar disorder—a hypothesis generating study. J. Psychiatr. Res. 164, 329–334 (2023).
Pizarro, A., Hayer, K., Lahens, N. F. & Hogenesch, J. B. CircaDB: a database of mammalian circadian gene expression profiles. Nucleic Acids Res. 41, D1009–D1013 (2013).
Brüning, F. et al. Sleep-wake cycles drive daily dynamics of synaptic phosphorylation. Science 366, eaav3617 (2019).
Gillespie, M. et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 50, D687–d92 (2022).
Piñero, J. et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 48, D845–D855 (2020).
Dollish, H. K., Tsyglakova, M. & McClung, C. A. Circadian rhythms and mood disorders: time to see the light. Neuron 112, 25–40 (2024).
Walker, W. H. 2nd, Walton, J. C., DeVries, A. C. & Nelson, R. J. Circadian rhythm disruption and mental health. Transl. Psychiatry 10, 28 (2020).
Kim, H., McInnis, M. G. & Sperry, S. H. Longitudinal dynamics between anxiety and depression in bipolar spectrum disorders. J. Psychopathol. Clin. Sci. 133, 129–139 (2024).
McGowan, N. M., Goodwin, G. M., Bilderbeck, A. C. & Saunders, K. E. A. Circadian rest-activity patterns in bipolar disorder and borderline personality disorder. Transl. Psychiatry 9, 195 (2019).
Esaki, Y. et al. Association between circadian activity rhythms and mood episode relapse in bipolar disorder: a 12-month prospective cohort study. Transl. Psychiatry 11, 525 (2021).
Moon, J. H. et al. Advanced circadian phase in mania and delayed circadian phase in mixed mania and depression returned to normal after treatment of bipolar disorder. EBioMedicine 11, 285–295 (2016).
Roybal, K. et al. Mania-like behavior induced by disruption of CLOCK. Proc. Natl Acad. Sci. USA 104, 6406–6411 (2007).
Kristensen, M., Nierenberg, A. A. & Ostergaard, S. D. Face and predictive validity of the ClockDelta19 mouse as an animal model for bipolar disorder: a systematic review. Mol. Psychiatry 23, 70–80 (2018).
Shen, W. et al. Synaptotagmin-7 is a key factor for bipolar-like behavioral abnormalities in mice. Proc. Natl Acad. Sci. USA 117, 4392–4399 (2020).
Yamamoto, H. et al. GWAS-identified bipolar disorder risk allele in the FADS1/2 gene region links mood episodes and unsaturated fatty acid metabolism in mutant mice. Mol. Psychiatry 28, 2848–2856 (2023).
Yang, Z. et al. The genome-wide risk alleles for psychiatric disorders at 3p21.1 show convergent effects on mRNA expression, cognitive function and mushroom dendritic spine. Mol. Psychiatry 25, 48–66 (2020).
Zhang, C. et al. Unraveling NEK4 as a potential drug target in schizophrenia and bipolar I disorder: a proteomic and genomic approach. Schizophr. Bull. 50, 1185–1196 (2024).
Blume, C., Garbazza, C. & Spitschan, M. Effects of light on human circadian rhythms, sleep and mood. Somnologie (Berl.) 23, 147–156 (2019).
Young, J. W. & Dulcis, D. Investigating the mechanism(s) underlying switching between states in bipolar disorder. Eur. J. Pharm. 759, 151–162 (2015).
Burns, A. C. et al. Day and night light exposure are associated with psychiatric disorders: an objective light study in >85,000 people. Nat. Ment. Health 1, 853–862 (2023).
de Vivo, L. et al. Ultrastructural evidence for synaptic scaling across the wake/sleep cycle. Science 355, 507–510 (2017).
McCarthy, M. J. & Welsh, D. K. Cellular circadian clocks in mood disorders. J. Biol. Rhythms 27, 339–352 (2012).
Piguel N. H. et al. Lithium rescues dendritic abnormalities in Ank3 deficiency models through the synergic effects of GSK3beta and cyclic AMP signaling pathways. Neuropsychopharmacology 48, 1000–1010 (2022).
Tobe, B. T. D. et al. Probing the lithium-response pathway in hiPSCs implicates the phosphoregulatory set-point for a cytoskeletal modulator in bipolar pathogenesis. Proc. Natl. Acad. Sci. USA 114, E4462–E4471 (2017).
Gideons, E. S., Lin, P. Y., Mahgoub, M., Kavalali, E. T. & Monteggia, L. M. Chronic lithium treatment elicits its antimanic effects via BDNF-TrkB dependent synaptic downscaling. Elife 6, e25480 (2017).
Shi, Y. Y. & He, L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 15, 97–98 (2005).
Pruim, R. J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010).
Ramasamy, A. et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat. Neurosci. 17, 1418–1428 (2014).
GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
Smyth G. K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3 (2004).
Han, K. A., Yoon, T. H., Shin, J., Um, J. W. & Ko, J. Differentially altered social dominance- and cooperative-like behaviors in Shank2- and Shank3-mutant mice. Mol. Autism 11, 87 (2020).
Lee, S. et al. Anterior cingulate cortex-related functional hyperconnectivity underlies sensory hypersensitivity in Grin2b-mutant mice. Mol. Psychiatry https://doi.org/10.1038/s41380-024-02572-y (2024).
Ma, L. et al. Brain methylome remodeling selectively regulates neuronal activity genes linking to emotional behaviors in mice exposed to maternal immune activation. Nat. Commun. 14, 7829 (2023).
Su, J. et al. In vivo base editing rescues photoreceptors in a mouse model of retinitis pigmentosa. Mol. Ther. Nucleic Acids 31, 596–609 (2023).
Leamey, C. A. et al. Ten_m3 regulates eye-specific patterning in the mammalian visual pathway and is required for binocular vision. PLoS Biol. 5, e241 (2007).
Beaulieu, J. M. et al. A beta-arrestin 2 signaling complex mediates lithium action on behavior. Cell 132, 125–136 (2008).
Yin, Y. et al. Glycogen synthase kinase 3β inhibition as a therapeutic approach in the treatment of endometrial cancer. Int. J. Mol. Sci. 14, 16617–16637 (2013).
Wisniewski, J. R., Zougman, A., Nagaraj, N. & Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 (2009).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Acknowledgements
This study was supported by National Key Research and Development Program of China (2023YFA1800500 to M.L.); National Natural Science Foundation of China (82225016 to M.L., 82222024 to X.X., 82071534 to H.C., 82101592 to C.W.); Yunnan Fundamental Research Projects (202201AS070048 to M.L., 202401AS070080 to M.L., 202401AT070204 to X.C., 202301AW070009 to S.W.L., 202305AH340006 to H.C., 202305AH340007 to X.X.); Spring City Plan: the High-level Talent Promotion and Training Project of Kunming (2022SCP001 to M.L.); Municipal Key R&D Program of Ningbo (2022Z127 to C.W.); Open Program of Yunnan Key Laboratory of Animal Models and Human Disease Mechanisms (#AMHD-2024-2 and #AMHD-2024-6 to C.W.). Xiao Xiao was also supported by the CAS “Light of West China” Program, CAS Youth Innovation Promotion Association, and Yunnan Revitalization Talent Support Program Young Talent Project. Ming Li was also supported by the Yunnan Revitalization Talent Support Program Yunling Scholar Project. Hong Chang was also supported by the CAS “Light of West China” Program and Yunnan Revitalization Talent Support Program Innovation Team (202405AS350008). Xin Cai was supported by the CAS Special Research Assistant (SRA) Program.
Author information
Authors and Affiliations
Contributions
X.X. and M.L. conceived and designed the study. Z.H.Y. and X.C. performed the primary experiments, with the help of Q.Z., Z.L.D., Y.G., G.M. and C.H.Y. C.Y.Z. and M.L. conducted the bioinformatic analyses. L.G., H.C. and C.W. provided advice and discussion during the design and practice of the current study. M.L., Z.H.Y. and X.X. drafted the first version of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors report no competing interests.
Peer review
Peer review information
Nature Communications thanks Amanda Law, and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, ZH., Cai, X., Zhang, CY. et al. NEK4 modulates circadian fluctuations of emotional behaviors and synaptogenesis in male mice. Nat Commun 15, 9180 (2024). https://doi.org/10.1038/s41467-024-53585-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-53585-8