Abstract
Pupil dynamics has emerged as a critical non-invasive indicator of brain state changes. In particular, pupillary-light-responses (PLR) in Alzheimer’s disease (AD) patients show potential as biomarkers for brain degeneration. To investigate AD-specific PLR and its underlying neuromodulatory sources, we combine high-resolution awake mouse fMRI with real-time pupillometry to map brain-wide event-related correlation patterns based on illumination-driven pupil constriction (\({P}_{c}\)) and post-illumination pupil dilation recovery (amplitude, \({P}_{d}\), and time, T). The \({P}_{c}\)-driven differential analysis reveals altered visual signal processing and reduced thalamocortical activation in AD mice in comparison with wild-type (WT) control mice. In contrast, the post-illumination pupil dilation recovery-based fMRI highlights multiple brain areas associated with AD brain degeneration, including the cingulate cortex, hippocampus, septal area of the basal forebrain, medial raphe nucleus, and pontine reticular nuclei (PRN). Additionally, the brain-wide functional connectivity analysis highlights the most significant changes in PRN of AD mice, which serves as the major subcortical relay nuclei underlying oculomotor function. This work integrates non-invasive pupil-fMRI measurements in preclinical models to identify pupillary biomarkers based on brain-wide functional changes, including neuromodulatory dysfunction coupled with AD brain degeneration.
Similar content being viewed by others
Introduction
Pupil diameter changes reflect cognitive processing1,2,3,4, presenting a unique, non-invasive index of brain state fluctuation in both normal and degenerative conditions, e.g., Alzheimer’s Disease (AD)5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23. Although several studies associate pupillary responses with the risk for AD, it remains highly speculative given the lack of understanding of its mechanistic linkage24,25,26,27,28. Multiple neuronal sources drive the spontaneous pupil dynamics, as well as pupillary light responses (PLR), through autonomic neuromodulation to control pupil dilation and constriction9,10,11,29,30,31,32,33,34,35,36. Besides evoked visual pathways, widespread neuromodulatory pathways, including those involving acetylcholine37,38,39, norepinephrine39,40, and serotonin41, are recruited to regulate PLR. In addition, several task-related studies have reported that PLR is mediated by higher-order brain function with either direct cortical micro-stimulation42 or loss-of-function by brain lesion43. Given that AD-related cognitive decline is linked to impaired neuromodulatory circuits, particularly cholinergic deficits44,45,46,47, an intriguing hypothesis proposes that altered PLR could serve as an indirect indicator of brain-wide dysfunction resulting from neuromodulatory impairment in AD.
Combining fMRI with real-time pupillometry provides a non-invasive mapping approach that bridges altered pupillary responses with corresponding brain-wide functional changes in AD brains. Existing studies have primarily focused on analyzing spontaneous pupil diameter changes with resting-state (rs) fMRI signals. Human fMRI studies with pupillometry have revealed that pupil dynamic changes are associated with the activation of subcortical brain nuclei along the ascending arousal network, as well as the salience network including the cingulate cortex and insula32,35,48,49,50,51,52,53,54,55. Elman et al. have linked the locus coeruleus (LC)-related functional network with the task-related pupil responses, indicating potential LC malfunction-based pupil dynamic biomarkers for AD risk22. Whereas, the noradrenergic regulation of pupil-fMRI interactions, e.g., the noradrenergic cell group 5 (A5) nuclei at the brainstem, was detectable only at a certain brain state of rat brains56. By measuring the axonal activity of neuromodulatory neurons, it has been shown that pupillary responses to locomotion are more closely coupled with sustained cholinergic activity, but less with noradrenergic phasic activity57. This observation is further confirmed in the AD transgenic mice, which exhibit pupil dilation coupled with locomotion but less associated with the diminished astrocytic activity controlled by noradrenergic inputs58. Besides cholinergic and noradrenergic systems, other neuromodulatory pathways are also reported to regulate pupil dynamics at resting state41. A recent work demonstrates the highly varied spectral distribution of spontaneous pupil dynamics according to different activation patterns of neuromodulatory systems57. These studies have illustrated the challenges of dissecting the neuromodulatory contribution to spontaneous pupil dynamics based on rs-fMRI correlation analysis, which would be insufficient when bridging the AD-specific pupil dynamics changes with brain-wide functional alteration due to neuromodulatory dysfunction in AD brains.
Instead of relying on spontaneous pupil dynamics, light-triggered PLR responses can be controlled with fixed interstimulus durations, allowing for phase-locked analysis of fMRI and pupillary changes. In this study, an approach combining awake mouse fMRI with simultaneous real-time pupillometry was developed to elucidate altered brain function and potential neuromodulatory dysfunction underlying AD-specific PLR. Using an implantable radiofrequency (RF) surface coil, which also served as a headpost for head-fixation during scanning, robust high-resolution brain-wide BOLD functional maps (100 × 100 × 200 µm resolution) were acquired from awake mice undergoing visual stimulation. Epoch-specific PLR responses were measured during awake mouse fMRI, including the illumination-induced pupil constriction amplitude (\({P}_{c}\)) and post-illumination pupil dilation recovery time (T) and amplitude (\({P}_{d}\)). Using these time-varied PLR dynamic features, the \({P}_{c}\)-based fMRI differential maps between AD and wild-type (WT) mice were presented to identify altered brain activation in the visual pathways. In contrast, post-illumination pupil dilation features (T, \({P}_{d}\))-based fMRI differential maps highlighted several cortical (e.g., cingulate and retrosplenial cortex (RSP)) and subcortical regions (e.g., septal areas in the basal forebrain, hippocampus, and pontine reticular nucleus (PRN)) involved in the altered neuromodulation underlying the AD-specific PLR. Meanwhile, the region of interest (ROI)-based correlation analysis also identified altered functional connectivity among these PLR-related neuromodulatory nuclei, highlighting the most significant connectivity changes originating from the PRN between AD and WT mice. These results establish a pupil-fMRI mapping scheme for identifying AD-specific PLR dynamic features, revealing altered correlation patterns in functional nuclei potentially affected during brain degeneration.
Results
Distinguishing altered PLR dynamics of AD and WT mice during fMRI scanning
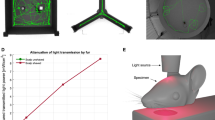
The real-time pupillometry was simultaneously performed inside the 14 Tesla scanner with high-resolution echo-planar-imaging (EPI) scans of awake mice (Fig. 1A). The awake mouse real-time pupillometry setup includes i. MR-compatible miniaturized complementary metal-oxide-semiconductor (CMOS) sensor with a focus adjustable lens to maintain at least a 2 cm distance from the mouse to avoid B0 field distortion, ii. A plane mirror to reflect the pupil toward the camera mounted on the animal holder, iii. The optical fibers to deliver the switchable blue/green light pulses (530 nm and 490 nm, flashed at 5 Hz and 5.1 Hz, respectively) for visual stimulation and the red light (660 nm) to illuminate the eye for pupillometry, as mice are 1000 times less sensitive to red light with wavelengths over 660 nm59. Mice were implanted with an RF surface coil that served as the headpost for head-fixation while inside the animal holder during scanning (the detailed setup was described in our previous work60). Figure 1B showed a representative trace of the PLR from an awake mouse, measured using DeepLabCut61. To improve the accuracy and efficiency of PLR measurements, we also implemented a grayscale-based pupil size measurement to verify different PLR dynamic features across different pupillometry recording trials (Supplementary Fig. 1). The mean PLRs of two groups of mice (AD vs. WT) were plotted in Fig. 1C, showing the dynamic PLR features during constriction (\({P}_{c}\)) and dilation recovery phases (\({P}_{d}\), T). The recovery time (T) was quantified based on exponential fitting of the pupillary dilation curve from each stimulation on/off epoch. Figure 1D showed the quantitative comparison of mean PLR features, presenting a significantly shorter T in AD mice compared to WT mice, but no significant difference in \({P}_{c}\) and \({P}_{d}\) between the two groups. These results provided robust PLR dynamic measurements of head-fixed awake mice during fMRI scanning. However, the altered PLR dynamic features detected in AD mice still need to be defined as potential biomarkers of AD. Instead, combining PLR measurements with simultaneous fMRI to identify the brain-wide functional changes underlying AD-specific PLR features may provide a promising approach to identifying bioimaging markers of AD.
A The snapshot of the awake mouse fMRI setup with real-time pupillometry and the visual stimulation setup through optical fibers. B The representative pupillometry results of an awake mouse with time-varied images of the pupil recorded during fMRI scanning (colored dots define the pupil size based on the DeepLabCut; green trace showed the pupil dynamic changes with visual stimulation in a block design; cyan lines showed stimulation on periods). C The normalized curves showed averaged pupil light response from AD and WT mice (the bracket indicates the \({P}_{c}\); dotted box highlights the dilation recovery period for an exponential fitting function to extract T and \({P}_{d}\)). D The PLR feature-specific box plots of \({P}_{c}\), \({P}_{d}\), and T between AD and WT mice. (WT: n = 9 mice, AD: n = 9 mice, the average feature value denoted by black dots; * means two-sided t-test p < 0.005.) Each box shows the median (red central mark), the lower and upper quartiles (gray box edges), and the whiskers extend to non-outlier extremes. Source data are provided as a Source Data file.
Brain-wide BOLD-fMRI of awake AD and WT mice with visual stimulation
High-resolution fMRI was applied to acquire EPI images of awake mice with 100x100x200µm resolution using a horizontal 14 T MRI scanner (Supplementary Fig. 2). During the awake mouse fMRI scanning, motion artifacts caused by the voluntary movement of the mice led to distorted images (Supplementary Fig. 2E), which can be removed from the 3D time series for regression analysis (Supplementary Fig. 2F, Movie 1). Figure 2 showed the visual stimulation-evoked brain-wide activation in the visual cortex (VC), superior colliculus (SC), lateral geniculate nucleus (LGN), retrosplenial cortex (RSP), and anterior cingulate cortex (ACA) in both WT and AD mice. Robust positive BOLD signals were detected in both groups from the time course data of specific ROIs segmented based on the Allen mouse brain atlas62 (Fig. 3), demonstrating the reliable visual stimulation-evoked fMRI signals in awake WT and AD mice.
The functional maps of awake mice show significant BOLD activation in the visual cortex (VC), superior colliculus (SC), retrosplenial cortex (RSP), lateral geniculate nucleus (LGN), and anterior cingulate area (ACA) from both WT (upper panel) (n = 13 mice with 58 trials) and AD (lower panel) mice (n = 9 mice with 60 trials; two-sided t-test p < 0.001, FDR < 0.01, minimum cluster size = 200 voxels). Source data are provided as a Source Data file.
A The functional maps overlaid with the brain atlas highlight activated brain regions: visual cortex (VC), superior colliculus (SC), retrosplenial cortex (RSP), lateral geniculate nucleus (LGN), and anterior cingulate area (ACA) from Alzheimer’s disease (AD) and wild-type (WT) mice. (Two-sided t-test p < 0.001, FDR < 0.01, minimum cluster size=200 voxels.) B The averaged time course based on the ROIs from brain atlas, demonstrating the evoked positive BOLD signal changes with the 8 s visual stimulation (5 Hz 530 nm and 5.1 Hz 490 nm 20 ms light pluses). Each graph displays the average of 116 sets of 5 stimulation epochs for 13 WT mice and 120 sets of 5 stimulation epochs for 9 AD mice. Shaded regions represent standard error. Cyan shadows represent the 8 s stimulation duration (WT: n = 13 mice with 58 trials; AD: n = 9 mice with 60 trials). Source data are provided as a Source Data file.
To investigate the difference of functional maps between the two groups, we first integrated the \({P}_{c}\)-based amplitude modulated (AM) hemodynamic response function (HRF) as the regressor across different trials. Since the varied pupil constriction at each stimulation on/off epoch altered retinal illumination, specific visual pathway activation would be better quantified by integrating the \({P}_{c}\)-based AM regression scheme. Quantitative group analysis was performed to produce voxel-wise differential maps between WT and AD mice. Figure 4 showed significantly higher BOLD responses in WT mice compared to AD mice in the primary and secondary visual cortices and LGN, but little difference was detected in the SC and cingulate cortex. In particular, well-separated higher visual cortical areas, including the adjacent RSP, were highlighted in the differential maps (Fig. 4B). These results indicate that the higher-order thalamocortical processing of visual signals is impaired in AD mice based on the \({P}_{c}\)-fMRI AM regression analysis.
A The brain-wide Pc-based fMRI differential maps (WT-AD) show significantly higher BOLD signals detected along visual pathways in WT mice compared to AD mice (WT: n = 9 mice with 20 sessions; AD: n = 9 mice with 19 sessions; two-sided t-test p < 0.001, Permutation-based FDR < 0.001, minimum cluster size=200 voxels). B The enlarged functional maps overlaid with the brain atlas highlight the higher-order visual pathways: visual cortex (V1, V2), retrosplenial cortex (A30, A29), and lateral geniculate nucleus (LGN). Source data are provided as a Source Data file.
Identify neuromodulation-related brain-wide functional changes coupled to the post-illumination pupil dilation recovery
Brain-wide functional changes associated with post-illumination pupil dilation were investigated using the pupil-fMRI mapping scheme. The pupil dilation features \({P}_{d}\) and T were quantified for each stimulation on/off epoch with awake mouse fMRI, enabling a PLR-based fMRI regression analysis. Figure 5 showed the \({P}_{d}\)-based fMRI correlation patterns in both WT and AD mice. In both groups, a negative correlation was observed in subcortical midbrain regions. In WT mice, this negative correlation was located in the serotonergic raphe nuclei, while in AD mice it was predominantly detected in sparse pontine areas adjacent to the LC, laterodorsal tegmental area (LDT), and the mesencephalic pedunculopontine tegmental nuclei. Also, \({P}_{d}\)-based negative correlation with the cingulate cortex, hippocampus, and lateral septal (LS) area were detected in AD mice. Group differential analysis revealed the most salient differences in the MnR, the cingulate/retrosplenial cortex, the hippocampus (dentate gyrus (DG) and CA1), and the LS of the basal forebrain regions.
A The averaged brain-wide \({P}_{d}\)-based correlation maps of WT and AD mice (left panel: WT, n = 9 mice with 20 sessions; middle panel: AD, n = 9 mice with 19 sessions), and the differential maps between WT and AD mice (right panel: WT-AD) (two-sided t-test p < 0.04, Permutation-based FDR < 0.04, minimum cluster size = 200 voxels). B The differential maps overlaid with the brain atlas, highlighting brain regions with significant differences: midbrain raphe nuclei (MnR), hippocampus (CA1, DG), retrosplenial cortex (RSP), and lateral septal nucleus (LS). Source data are provided as a Source Data file.
Figure 6 showed the T-based fMRI correlation patterns, highlighting the negative correlations in the PRN, the cingulate cortex, and the hippocampus in WT mice. Furthermore, negative correlations were observed in the midbrain reticular formation and central thalamic nuclei in WT mice. In contrast, AD mice showed positive correlations across the hippocampus and cingulate cortex, and the hypothalamus. The statistical differential maps between WT and AD mice highlighted the significantly different brain regions located at the PRN, the hippocampus, the cingulate/retrosplenial cortex, and central thalamic nuclei, including the paraventricular (PVP) and dorsal thalamus.
A The averaged brain-wide \({P}_{d}\)-based correlation maps of WT and AD mice (left panel: WT, n = 9 mice with 20 sessions; middle panel: AD, n = 9 mice with 19 sessions), and the differential maps between WT and AD mice (right panel: WT-AD) (two-sided t-test p < 0.04, Permutation-based FDR < 0.04, minimum cluster size = 180 voxels). B The differential maps overlaid with the brain atlas, highlighting brain regions with significant differences: pontine reticular nucleus (PRN), hippocampus (CA1, DG), paraventricular (PVP), and retrosplenial cortex (RSP). Source data are provided as a Source Data file.
To further identify the neuromodulatory nuclei responsible for the altered pupil-fMRI coupling in AD brains, we also performed ROI-based correlation network analysis given the reiterated visual stimulation paradigms. Figure 7A illustrated the spatial correlation network of functional connectivity among the highlighted nuclei from PLR-based differential maps between WT and AD mice. By comparing the ROI-based correlation between WT and AD mice, significantly altered connectivity among paired functional nuclei was highlighted in the spatial correlation network maps, showing increased connectivity between hippocampus-PRN, LS-PRN, and decreased connectivity between LS-VC/SC, ACA-VC/SC in AD mice. Figure 7B showed the corresponding correlation matrices, summarizing the paired functional nuclei with significantly different connectivity between the two groups. It should be noted that we also conducted permutation controls using randomly selected brain-wide regions of interest (ROIs) for differential analysis. This approach revealed a significantly reduced number of different connections compared to those identified in the nuclei selected based on the PLR-based fMRI analysis (Fig. 7C). Meanwhile, brain-wide correlation analysis of ROIs, independent of the PLR-based fMRI mapping results, was performed to identify the ROIs with the most significantly altered connectivity in AD mice. The top 20 paired ROIs with significantly altered connectivity between WT and AD mice highlighted that over 40% of the connectivity changes originated from PRNc/PRNr in AD mice (Fig. 7D), as identified by the PLR-based fMRI differential analysis. These results further confirm that the PLR-based fMRI differential analysis identified a critical set of neuromodulatory nuclei most responsible for AD-specific PLR.
A Colored-coded Pearson correlation network through line projections in 3D mouse brain contours, which is based on the ROIs defined by Pupillary-light-responses (PLR)-based fMRI analysis (pontine reticular nuclei (PRN), median raphe nucleus (MnR), superior colliculus (SC), visual cortex (VC), retrosplenial cortex (RSP), hippocampus (CA1), anterior cingulate cortex (ACA), and lateral septal area (LS)). The color-coded line projections present the paired ROIs with statistically stronger based on one-sample t-test of wild-type (WT) and Alzheimer’s disease (AD) mice (WT: n = 9 mice with 21 sessions, AD: n = 9 mice with 21 sessions, two-sided t-test p < 0.01), and with significantly different correlation coefficients between WT and AD mice (WT-AD, two-sample two-sided t-test, p < 0.01), B The correlation matrices of WT and AD mice, as well as the differential matrix (WT-AD), presenting an alternative way to show the correlation networks based on the PLR-fMRI specific ROIs. C The permutation test histogram shows the distribution of number of significantly different connections between WT and AD mice across 5000 permutation tests with randomly selected ROIs, which was fitted with a Gaussian distribution profile (red line, μ = 2.25, σ = 5). Red star indicates the number of significantly different correlation based on the PLF-fMRI specific ROIS, which is ~μ + 7σ of the Gaussian distribution. D A pie chart highlights the ROIs presenting the most different correlation patterns (i.e., the top 20 paired ROIs with the largest coefficient difference at two-sided t-test p < 0.01) between WT and AD mice, of which the PRN accounts for 45%. Source data are provided as a Source Data file.
Discussion
The present study combined awake mouse fMRI with real-time pupillometry to identify brain-wide functional changes associated with AD-specific PLR dynamics. This awake mouse pupil-fMRI mapping scheme provides a proof-of-concept method for identifying non-invasive PLR linked to altered brain function, particularly within neuromodulatory pathways, as potential biomarkers of AD brain degeneration.
PLR in AD patients has been reported to show altered pupil constriction phases24,25,26. These studies have suggested the potential of pupil dynamics as biomarkers of AD degeneration. However, a few studies have failed to reproduce altered PLR features when comparing AD patients with control groups27,28. Numerous efforts have been made to link pupil dynamics with various behavioral variables such as the subject’s arousal fluctuation5,6,7, designated tasks5,8,9,10,11,12, or specific cognitive processes13,14,15,16,17,18,19,20,21. The richness of behavioral correlates of pupil dynamics, especially in AD patients with cognitive impairments, could lead to confounding PLR results given the complex controlling mechanisms. In the 5xFAD mouse model, altered PLR features, such as the significantly shortened post-illumination dilation recovery time (T) (Fig. 1), could serve as alternative behavioral readouts reflecting brain functional changes associated with cognitive impairments, which have been extensively studied using behavioral tests in this transgenic mouse line63,64,65,66,67. Besides the well-documented behavioral impairments, detailed studies have also identified various neuroanatomical abnormalities in 5xFAD mice68,69,70,71. Our work addresses the gap in bridging PLR features with brain-wide functional changes using fMRI in 5xFAD mice, which could directly contribute to the reported cognitive impairments in this mouse model.
One intriguing technological development of this work is the high-resolution awake mouse fMRI at 14 T. Hike et al. developed a novel implantable RF coil, which also serves as the headpost for head-fixation during awake mouse fMRI scanning60. In contrast to the inductively coupled wireless RF coil72,73, this new design has enabled ultra-high-resolution EPI-based fMRI of awake mice with 100 × 100 × 200 um resolution, allowing the characterization of activated functional nuclei in mice with only a few hundred-micron size. Both AD and WT mice have gone through the acclimatization procedure with fMRI as described previously60,74, enabling the reliable detection of visual stimulation-evoked fMRI maps (Figs. 2 and 3). It should be noted that the functional maps showed brain-wide activation not only in the visual pathway but also included the thalamic areas and ACA in both groups. This demonstrated extended high-level cortical function involved in the cognitive processing of awake mice. This advanced awake mouse fMRI allowed the brain-wide functional characterization of AD-specific PLR features.
We have performed PLR-based fMRI correlation analysis based on the stimulation on/off epoch across different trials. In contrast to the direct comparison of evoked functional maps between WT and AD groups, the \({P}_{c}\)-based AM regression analysis enabled the characterization of evoked brain function based on illumination-driven retinal activity. The BOLD signal difference detected along the central visual pathways indicated significantly reduced visual cortical activity from primary and associated cortices in AD mice, as well as in the LGN, but no signal difference was detected in the central thalamic areas and cingulate cortex (Fig. 3A). This altered visual pathway activity in AD mice could be directly linked with impaired retina function due to vascular and neuronal degenerative effects75,76,77,78. The differential maps highlighted V1 and associated visual areas with LGN, indicating impaired high-order visual signal processing. Previous studies have reported altered laminar distribution of neurofibrillary tangles (NFTs) in the visual cortex79 and decreased long synaptic projection from area V2 to V1 in AD patients80. The \({P}_{c}\)-based fMRI provides a bottom-up mapping scheme to characterize the malfunction of visual signal processing in AD brains.
In contrast to the \({P}_{c}\)-based fMRI correlations, the post-illumination pupil dilation reflected the top-down neuromodulatory processes. The \({P}_{d}/T\)-based fMRI revealed altered coupling with multiple functional nuclei in AD mice. First, the posterior cingulate cortex and hippocampus of AD mice showed altered coupling features with \({P}_{d}/T\) compared to WT mice. Since both the posterior cingulate cortex and hippocampus are impaired in AD due to the neurodegeneration81,82,83,84,85, it is plausible that the reduced neuronal activation in these areas was directly involved in the altered pupil dilation features, especially the shortened dilation recovery time (T) that presented the most significant changes in PLR measurements (Fig. 1C). This observation is consistent with reports, showing that hippocampal damage in AD patients is related with impaired oculomotor behavior and networks86,87,88 as well as altered pupil response89,90,91,92. From the ROI-based correlation analysis, we observed fewer or weaker connections related to the hippocampus (CA1) and cingulate cortex in AD mice compared to WT mice. This finding is consistent with other functional connectivity studies in AD mice93,94,95.
Another intriguing observation is the identification of subcortical brain regions underlying the altered pupil dilation recovery in AD mice. First, the PLR (T)-based fMRI coupling in PRN was reduced in AD mice compared to WT mice. The PRN is the major relay nuclei in the brainstem controlling oculomotor function96,97,98. By performing the brain-wide ROI network connectivity analysis based on the reiterated visual stimulation (Fig. 7D), significant changes in PRN-based network connectivity in AD mice further confirmed the PLR (T)-based PRN coupling as a crucial estimate of the reliable pupil-fMRI biomarker for AD brains.
For the \({P}_{d}\)-based fMRI correlation, negative correlations at median raphe nuclei (MnR) were observed in WT mice but were less pronounced in AD brains, indicating impaired serotonergic regulation of pupil dilation. The pupil dilation and constriction can be controlled by serotonergic agonists and antagonists, respectively99,100,101, and significant neuronal reduction is also reported in the MnR of AD patients’ brains102. In contrast, the LS showed more negative correlations in AD than WT mice in the \({P}_{d}\)-based fMRI maps. The septal area of the basal forebrain is composed of glutamatergic, GABAergic, and cholinergic neurons and is reciprocally connected to multiple subcortical regions103, including the medial septal area (MS) which sends cholinergic projections throughout the cortex104. Cholinergic hypofunction is a hallmark of AD24,105, characterized by impaired cholinergic projections from septal areas to widespread brain regions, as observed in various AD transgenic models106,107,108,109,110,111, including 5xFAD mice112. Since cholinergic function directly influences pupil dilation57, the increased negative correlation in LS suggests that LS-mediated septal cholinergic function in AD brains contributes more significantly to regulate post-illumination pupillary dilation compared to healthy brains. Our work points to a future research direction on investigating the causal relationship between cholinergic deficits and altered PLR dynamics in AD. In particular, the multi-modal fMRI platform, which combines simultaneous fMRI and fiber photometry to record acetylcholine signals, can be integrated with chemogenetic tools to regulate the cholinergic neuronal activity in the septal areas of awake AD mice.
A counterintuitive observation in this work is the primary negative correlation patterns from multiple nuclei for the \({P}_{d}\)-based fMRI maps. The pupil-based fMRI negative correlation has been previously reported in anesthetized rodent brains56. And brain-wide negative BOLD signal was also coupled with the vigilant brain state changes based on eye open/close arousal indices113. The \({P}_{d}\)-based negative BOLD fMRI correlation could be mediated by the neurovascular coupling event underlying arousal state changes in awake mice involving multiple neuromodulatory pathways. By incorporating the resting-state spatiotemporal pupil-fMRI correlation patterns, principal component analysis (PCA) has been implemented to highlight different neuromodulatory pathways based on clustered pupil dynamic spectra114. To further elucidate the neuronal basis of the negative coupled pupil-fMRI relationship, cellular-specific fiber photometry Ca2+ recordings56 need to be implemented with awake mouse fMRI and real-time pupillometry. In addition, the brain state-dependent spontaneous pupil dynamics and locomotion-coupled pupil dynamics of awake mice could provide further insights into the coupled neuromodulatory dysfunction in AD mice.
In contrast to existing awake mouse fMRI studies115, we implemented a 5-week training scheme that has reported significantly reduced signs of stress-related behaviors, e.g., eye movements and spontaneous pupil dynamic changes60. Specifically, eye movements of head-fixed mice are much less frequent than in free-moving mice116, and these movements have shown gradual recovery during the mock-MRI acoustic training60. In addition, stress-related pupillary dilation predominates in the spontaneous pupil dynamics during the early phase of training74 but diminishes as acclimation progressed and leads to low-frequency fluctuations corresponding to brain state changes of normal behaving mice60. This work reported the highly comparable illuminance-induced pupil constriction between WT and AD mice, presenting similar acclimated states of both groups. And reduced corticosterone level of head-fixed mice has been reported with acclimation training in awake mouse fMRI studies117. However, corticosterone levels in head-fixed mice might still be higher than those in free-moving mice117, with variations depending on the duration of training and the length of the scanning session. Thus, the impact of stress-related confounding factors on the fMRI-pupil relationship of awake AD mice requires further investigation in future studies.
In conclusion, our study demonstrates that the epoch-specific post-illumination PLR features, i.e., the pupil dilation recovery time (T) and amplitude (\({P}_{d}\)), correlate with specific neuromodulatory nuclei that exhibit impaired functionality in AD brains. These nuclei also show dramatically altered network connectivity during visual stimulation in AD mice (Fig. 7), suggesting that AD-specific PLR features are linked to underlying changes in neuromodulatory function. To further explore the relationship between PLR-related neuromodulatory dysfunction and cognitive impairments, future research will need to connect these altered functional maps with behavioral changes in AD mice across different stages of neurodegeneration. Eventually, this pupil-fMRI mapping could offer non-invasive measurements for identifying pupillary biomarkers of brain degeneration in AD patients.
Methods
Animal surgical procedures
A total of nine female 5xFAD transgenic AD mice and thirteen matched genetic background WT (C57BL/6 J) mice (6 females and 7 males) at 6–12 months old were used, similar to previous studies60. The age distribution of the mice during scanning is shown in Supplementary Fig. 3. For real-time pupillometry with fMRI, only nine (4 females and 5 males) of the thirteen WT mice were used because of the poor quality of the pupil recordings from inside the 14 T MRI scanner. For the AD mice, only females were used since female 5xFAD mice display more pronounced molecular changes, including cortical and hippocampal Aβ accumulation and other inflammatory gene induction, than male 5xFAD mice64,65,66,118,119. Also, female 5xFAD mice exhibit significant Alzheimer’s-related behavioral and physiological changes by the age of 6 months63,64,65,66,67. Mice were group-housed (2–4/cage) under a 12-h light/dark cycle with food and water ad libitum. All animal procedures were conducted in accordance with protocols approved by the Massachusetts General Hospital (MGH) Institutional Animal Care and Use Committee (IACUC), and animals were cared for according to the requirements of the National Research Council’s Guide for the Care and Use of Laboratory Animals.
All mice underwent surgery under anesthesia (1–2% isoflurane with 1 L/min of medical air and 0.2 L/min additional O2 flow) to implant the RF coil to the skull60. Briefly, anesthetized mouse heads were stabilized in a stereotaxic stage, and the skull was exposed after the aseptic treatment. Before the coil was implanted, both 0.3% H2O2 and PBS was applied to clean the skull. It is critical to wait until the skull is fully dried before the coil implantation. The coil was secured with cyanoacrylate glue and dental cement to prevent movement and ensure stability. Analgesic (Buprenorphine, 0.1–0.3 mg/kg) was administrated before the surgical procedure and was also injected periodically (once per 12–24 h) for 3 days after the surgery. Mice were allowed at least a week of recovery in their home cages for neck strengthening to resume normal head movements before training.
Animals were acclimated to the MRI environment through a modified training program spanning five consecutive weeks. The training duration increased progressively increasing from 10 min to 60 min, transitioning from head-fixed mock MRI sessions to in-bore training with actual MRI scanning60,74. The stress levels of head-fixed mice during the training procedure were previously reported based on the monitored eye movements and pupil dynamics60. The stress-related confounding issue is discussed in the Discussion section.
MRI methods
MRI data was acquired with the 14 T horizontal MRI scanner (Magnex Sci, UK) located at the Athinoula A. Martinos Center for Biomedical Imaging in Boston, MA. The magnet is equipped with a Bruker Avance Neo Console (Bruker-Biospin, Billerica, MA) and is operated using ParaVision 360 V.3.3. A microimaging gradient system (Resonance Research, Inc., Billerica, MA) provides a peak gradient strength of 1.2 T/m within a 60-mm diameter. The MRI data for anatomical registration was obtained with a multi-slice T1-weighted 2D gradient echo fast low angle shot (FLASH) sequence (TE: 3 ms, TR: 475 ms, flip angle: 30°, 4 averages for an approximate acquisition time of 4.5 min, 100 × 100 × 200 µm resolution). The awake mouse fMRI images were obtained with a multi-slice 2D EPI (TE: 6.2 ms, TR: 1 s with two segments, BW: 278 kHz; Matrix size: 144 × 96 × 36, in-plane resolution: 100 × 100 µm, thickness: 200 µm). The 6.2 ms TE was chosen based on the measured T2* values of the cortex (~7–8 ms) of awake mice at 14 T. A total of 205 repetitions are acquired per trial for a total of 6 min 50 s. For each mouse, three trials of visual stimulation were conducted per scanning session. Within each trial, a block-design paradigm consisting of 10 stimulation on/off epochs was applied. For each epoch, the visual stimulation lasted for 8 s, followed by 32 s of inter-stimulus intervals. The optical stimulation during the “8 s on” period included two LED lights flashing at specific frequencies: 490 nm at 5.1 Hz and 530 nm at 5 Hz, with light pulse duration at 20 ms.
Pupillometry setup during fMRI scanning
The pupil recordings were obtained using a 3D-printed animal cradle with mounts for a camera, a mirror, and three fiber optic cables used as light sources as shown in Fig. 1A. The pupil dynamics were recorded through the mirror. The red light (660 nm) was used for video illumination since mice are 1000 times less sensitive to red light with wavelengths over 600 nm59, while the green and blue lights (530 nm and 490 nm) were used for visual stimulation.
To correlate the whole-brain fMRI signals with specific pupillary response features of normal and AD mice, the detection of pupil dynamics is first needed. Figure 1B shows an example of pupil dynamic recording. As shown in Fig. 1B, the edge of the mouse pupil was detected and tracked using DeepLabCut61. To further confirm the PLR results, an alternative pupil size detection method based on ocular and global grayscales was proposed. By comparing the average grayscale of the eye with the average grayscale of the whole image, the size of the pupil could be well characterized based on the signal intensity (Supplementary Fig. 1). Eventually, the normalized pupil dynamics data P can be calculated as:
Where \({g}_{{eye}}\) is the grayscale of the eye area and \({g}_{{all}}\) is the grayscale of the whole image.
PLR features extraction
To further map the correlation between pupil and fMRI, we extracted different features of the PLR in AD and WT mice with MATLAB R2023b (MathWorks, Natick, MA). The PLR time courses were divided into two components (constriction and dilation) for phase-dependent analysis of PLR. The constriction component represents the immediate reaction of the pupil to illumination, primarily regulated by the parasympathetic nervous system. To quantify pupil constriction, we used the difference of the trial-specific z-scored pupil dynamic data before and after constriction, denoted as \({P}_{c}\). For each epoch, the pre-constriction pupil size is calculated by averaging the pupil dynamic recording 1 s before the stimulation. The constricted pupil size is calculated by averaging the pupil recording below 75% of the epoch-specific minimum z-score. The dilation component, mainly controlled by the sympathetic nervous system, represents automatic pupil modulation and can provide insights into associated brain activity. Regarding the dilation component, we used an exponential function to accurately model the changes in pupil size.
Where \(P(t)\) is the pupil dilation defined from above 75% of the epoch-specific minimum z-score after stimulation to 75% of the epoch-specific maximum z-score. Through this modeling approach, we obtained two parameters: \({P}_{d}\), indicating the difference in pupil size before and after dilation; T, characterizing the shape of the pupil dilation curve and the potential full dilation time.
BOLD fMRI mapping with visual stimulation
To generate the visual-stimulation BOLD map, the fMRI images were processed using Analysis of Functional Neuroimages (AFNI_23.0.02) software120,121. A detailed awake mouse fMRI imaging processing procedure has been described previously60. First, the “to3d” function was used to transform the fMRI dataset into AFNI BRIK format. For group analysis, the images were registered to a mouse brain template. The mouse brain template acquired from the Australian Mouse Brain Mapping Consortium (AMBMC)122 was resampled at 100 µm isotropic resolution. For each scanning session, the anatomical FLASH image was registered to the AMBMC template using the “3dAllineate” function, creating a transformation matrix (TM1). Meanwhile, averaged EPI data was registered to the anatomical image, creating a second transformation matrix (TM2). By integrating the two transformation matrices, EPI time courses from multiple trials per scanning session will be registered to the brain template for concatenation. After the registration, several pre-processing procedures were implemented before the general linear regression using commands in “afni_proc.py”. The time series for the scanning session was first concatenated using the “3dTcat” command. The “despike” command was used to truncate spikes in each voxel’s time series before each EPI time point was registered to the atlas via a 6-degree transformation with the “volreg” command. The “mask” command was then applied to separate the brain from the background and other tissues. The “blur” command was used to smooth the transformed data by applying a Gaussian filter to average signal over nearby voxels before the “scale” command was applied to ensure compatibility and consistency across the analysis pipeline. In addition, the motion parameters were characterized and regressed out in the final general linear regression. The “censor_motion” parameter was set at 0.5 and the “censor_outlier” parameter was set at 0.02 based on the acquired awake mouse fMRI datasets (e.g., Supplementary Fig. 2F). The “3dDeconvolve” function was used to create the final statistical β coefficient maps with a “BLOCK” function.
PLR feature-driven fMRI map
To characterize PLR feature-driven fMRI maps, we applied the stimulation on/off epoch-specific PLR features to modulate the regression function. To improve the stability of regression, we concatenated the three trials from each scanning session and then obtained the PLR feature-driven fMRI maps. For group analysis, voxel-wise Student’s t-tests were performed to identify the differential PLR-based fMRI correlation maps between AD and WT mice. The permutation-based false discovery rate (FDR) was calculated to verify the statistical results. We compared the observed statistic to the null distribution by shuffling the labels to estimate the permutation-based p-value.
To analyze the constriction component, the pupil constriction parameter \({P}_{c}\) was applied to the amplitude-modulated pupil-fMRI correlation analysis. The \({P}_{c}\) was used as the auxiliary behavioral information to modulate the amplitude of the hemodynamic response function. The AM1 function (Supplementary Fig. 4A) of AFNI was applied with a regressor calculated as the following:
Where \({\sum }_{k=1}^{K}h(t-{\tau }_{k})\) is the ideal HRF, \({P}_{c}(k)\) is the value of the \({P}_{c}\) at time k. K is the time length of the fMRI data.
For the dilation component, pupil dilation parameters \({P}_{d}\) and T were used as the auxiliary behavioral information which was expected to be modulated by activation in specific brain areas. Those brain areas were expected to be associated with automatic pupil modulation and to exhibit variation between AD and WT groups. The AM2 function (Supplementary Fig. 4B) of AFNI was applied to incorporate the ideal HRF, as well as two regressors representing the epoch-specific modulation for pupil dilation parameters (\({P}_{d}\) and T), which were calculated as the following:
Where \(a(k)\) is the value of the pupil dilation parameter at time k. \(\bar{a}\) is the average pupil dilation parameter.
For the statistical differential maps, significant voxels were presented based on the p-values ranging from less than 0.04 to 0.001, according to different comparison schemes. The minimal cluster size was set at 180–200 voxels according to different comparison schemes.
Two-sample t-tests on the three pupil-fMRI correlation maps (\({P}_{c}\), \({P}_{d}\), and \(T\)) were also performed to compare male and female WT mice, aiming to characterize the gender-related differences. The threshold for p-value and cluster size were set the same as in the WT-AD analysis. No significant difference was observed for PLR-based fMRI differential maps between male and female WT mice in the ROIs highlighted from the PLR-based fMRI differential maps between AD and WT mice (Supplementary Fig. 5).
Functional network connectivity analysis
For the correlation network analysis, we defined all the ROIs based on the Allen mouse brain atlas given the brain-wide delineation of the functional nuclei. This Allen mouse brain atlas was registered to the previously chosen AMBMC brain template. For each ROI, we averaged the time course from the preprocessed fMRI datasets based on the 21 sessions of 9 WT mice and 21 sessions of 9 AD mice. Pearson correlation analysis was performed to calculate the correlation coefficients among paired ROIs that were identified based on the PLR-feature-driven BOLD differential maps between WT and AD mice (a total of 13 ROIs were selected). A one-sample Student’s t-test was performed to identify the significantly correlated paired ROIs in two groups of animals. A two-sample Student’s t-test was performed to compare the correlation difference of the paired ROIs between the two groups. To verify the differential correlation networks of PLR-specific ROIs, permutation control tests were performed by randomly selecting the same number of ROIs (excluding those pupil feature-related ROIs) 5000 times. For each permutation test, the number of randomly paired ROIs with significantly different correlations between the two groups was calculated. Gaussian fitting was applied to verify the normal distribution for the permutation tests. Alternatively, Pearson correlation analysis was also performed across all the ROIs based on the Allen mouse brain atlas (a total of 1086 ROIs), and the two-sample Student’s t-tests were performed to compare correlation coefficients of paired ROIs between the two groups. Among the paired ROIs showing significant correlation differences between the two groups (p < 0.01), the top 20 ROIs with the largest coefficient difference were chosen to plot the pie chart, which shows the individual ROIs’ contribution to the altered correlation network between WT and AD mice. Supplementary Fig. 6 also showed the pie charts from the top 10 to the top 200 verified paired ROIs.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw fMRI data are available at OpenNeuro: https://doi.org/10.18112/openneuro.ds005497.v1.0.0. The processed data generated in this study have been deposited in Zenodo: https://doi.org/10.5281/zenodo.13225145. Source data are provided with this paper.
Code availability
Scripts for running all analyses and visualizations are available at Zenodo (https://doi.org/10.5281/zenodo.13225145).
References
Beatty, J. & Lucero-Wagoner, B. In Handbook of Psychophysiology 2nd ed. 142–162 (Cambridge University Press, 2000).
Eckstein, M. K., Guerra-Carrillo, B., Miller Singley, A. T. & Bunge, S. A. Beyond eye gaze: What else can eyetracking reveal about cognition and cognitive development? Dev. Cogn. Neurosci. 25, 69–91 (2017).
Laeng, B., Sirois, S. & Gredebäck, G. Pupillometry: a window to the preconscious? Perspect. Psychol. Sci. https://doi.org/10.1177/1745691611427305 (2012).
Wilhelm, H. & Wilhelm, B. Clinical applications of pupillography. J. Neuro Ophthalmol. 23, 42–49 (2003).
McGinley, M. J., David, S. V. & McCormick, D. A. Cortical membrane potential signature of optimal states for sensory signal detection. Neuron 87, 179–192 (2015).
Yoss, R. E., Moyer, N. J. & Hollenhorst, R. W. Pupil size and spontaneous pupillary waves associated with alertness, drowsiness, and sleep. Neurology 20, 545–545 (1970).
McCormick, D. A., Nestvogel, D. B. & He, B. J. Neuromodulation of brain state and behavior. Annu. Rev. Neurosci. 43, 391–415 (2020).
Hakerem, G. A. D. & Sutton, S. Pupillary response at visual threshold. Nature 212, 485–486 (1966).
Stringer, C. et al. Spontaneous behaviors drive multidimensional, brainwide activity. Science 364 https://doi.org/10.1126/science.aav7893 (2019).
Salkoff, D. B., Zagha, E., McCarthy, E. & McCormick, D. A. Movement and performance explain widespread cortical activity in a visual detection task. Cereb. Cortex 30, 421–437 (2020).
Musall, S., Kaufman, M. T., Juavinett, A. L., Gluf, S. & Churchland, A. K. Single-trial neural dynamics are dominated by richly varied movements. Nat. Neurosci. 22, 1677–1686 (2019).
Reimer, J. et al. Pupil fluctuations track fast switching of cortical states during quiet wakefulness. Neuron 84, 355–362 (2014).
Hess, E. H. & Polt, J. M. Pupil size in relation to mental activity during simple problem-solving. Science 143, 1190–1192 (1964).
Kahneman, D. & Beatty, J. Pupil diameter and load on memory. Science 154, 1583–1585 (1966).
Alnæs, D. et al. Pupil size signals mental effort deployed during multiple object tracking and predicts brain activity in the dorsal attention network and the locus coeruleus. J. Vis. 14, 1–1 (2014).
O’Doherty, J. P., Dayan, P., Friston, K., Critchley, H. & Dolan, R. J. Temporal difference models and reward-related learning in the human brain. Neuron 38, 329–337 (2003).
Satterthwaite, T. D. et al. Dissociable but inter-related systems of cognitive control and reward during decision making: evidence from pupillometry and event-related fMRI. NeuroImage 37, 1017–1031 (2007).
Nassar, M. R. et al. Rational regulation of learning dynamics by pupil-linked arousal systems. Nat. Neurosci. 15, 1040–1046 (2012).
Richer, F. & Beatty, J. Contrasting effects of response uncertainty on the task-evoked pupillary response and reaction time. Psychophysiology 24, 258–262 (1987).
Gee, J. W. D., Knapen, T. & Donner, T. H. Decision-related pupil dilation reflects upcoming choice and individual bias. Proc. Natl. Acad. Sci. USA 111, E618–E625 (2014).
Sheng, F. et al. Decomposing loss aversion from gaze allocation and pupil dilation. Proc. Natl. Acad. Sci. USA 117, 11356–11363 (2020).
Elman, J. A. et al. Task-evoked pupil dilation and BOLD variance as indicators of locus coeruleus dysfunction. Cortex 97, 60–69 (2017).
Scinto, L. F. et al. Focal pathology in the Edinger-Westphal nucleus explains pupillary hypersensitivity in Alzheimer’s disease. Acta Neuropathol. 97, 557–564 (1999).
Frost, S. et al. Evaluation of cholinergic deficiency in preclinical Alzheimer’s disease using pupillometry. J. Ophthalmol. 2017, 7935406 (2017).
Nowak, W., Nakayama, M., Kręcicki, T. & Hachoł, A. Detection procedures for patients of Alzheimer’s disease using waveform features of pupil light reflex in response to chromatic stimuli. EAI Endorsed Trans. Pervasive Health Technol. 6, 167656 (2020).
Lustig-Barzelay, Y. et al. Machine learning for comprehensive prediction of high risk for Alzheimer’s disease based on chromatic pupilloperimetry. Sci. Rep. 12, 9945 (2022).
Bittner, D. M., Wieseler, I., Wilhelm, H., Riepe, M. W. & Müller, N. G. Repetitive pupil light reflex: potential marker in Alzheimer’s disease? J. Alzheimer’s Dis. 42, 1469–1477 (2014).
Gregory, P. V. S., Ling, B., Ying-Bo, S., Julie, H. & Mae, G. Pupillary light reaction in preclinical Alzheimer’s disease subjects compared with normal ageing controls. Br. J. Ophthalmol. 103, 971 (2019).
Bonvallet, M. & Zbrozyna, A. Les commandes réticulaires du système autonome et en particulier de l’innervation sympathique et parasympathique de la pupille. Arch. Italiennes de. Biol. 101, 174–207 (1963).
McDougal, D. H. & Gamlin, P. D. Autonomic control of the eye. Compr. Physiol. 5, 439–473 (2015).
Yüzgeç, Ö., Prsa, M., Zimmermann, R. & Huber, D. Pupil size coupling to cortical states protects the stability of deep sleep via parasympathetic modulation. Curr. Biol. 28, 392–400.e393 (2018).
Yellin, D., Berkovich-Ohana, A. & Malach, R. Coupling between pupil fluctuations and resting-state fMRI uncovers a slow build-up of antagonistic responses in the human cortex. Neuroimage 106, 414–427 (2015).
Joshi, S., Li, Y., Kalwani, R. M. & Gold, J. I. Relationships between pupil diameter and neuronal activity in the locus coeruleus, colliculi, and cingulate cortex. Neuron 89, 221–234 (2016).
Wang, C.-A., Boehnke, S. E., White, B. J. & Munoz, D. P. Microstimulation of the monkey superior colliculus induces pupil dilation without evoking saccades. J. Neurosci. 32, 3629–3636 (2012).
Schneider, M. et al. Spontaneous pupil dilations during the resting state are associated with activation of the salience network. Neuroimage 139, 189–201 (2016).
Ranson, S. W. & Magoun, H. Respiratory and pupillary reactions: induced by electrical stimulation of the hypothalamus. J. Nerv. Ment. Dis. 29, 1179–1194 (1933).
Mukherjee, S. & Vernino, S. Dysfunction of the pupillary light reflex in experimental autoimmune autonomic ganglionopathy. Auton. Neurosci. 137, 19–26 (2007).
Diamond, J. The pupil. Anatomy, physiology and clinical applications. BMJ Publishing Group Ltd. (2001).
Spector, R. H. Chapter 58. The Pupils. Clinical Methods: The History, Physical, and Laboratory Examinations 3rd Edition. Boston: Butterworths (1990).
Pereira, A., Ledent, E., Bervoets, L., Khan, A. & Colzi, A. PII‐32: pupillary light reflex effect of a new selective norepinephrine reuptake inhibitor assessed during the first human dose study. Clin. Pharmacol. Ther. 79, P44–P44 (2006).
Szabadi, E. Functional organization of the sympathetic pathways controlling the pupil: light-inhibited and light-stimulated pathways. Front. Neurol. 9 https://doi.org/10.3389/fneur.2018.01069 (2018).
Ebitz, R. B. & Moore, T. Selective modulation of the pupil light reflex by microstimulation of prefrontal cortex. J. Neurosci. 37, 5008–5018 (2017).
Dal Monte, O., Costa, V. D., Noble, P. L., Murray, E. A. & Averbeck, B. B. Amygdala lesions in rhesus macaques decrease attention to threat. Nat. Commun. 6, 10161 (2015).
Cheyuo, C. et al. Connectomic neuromodulation for Alzheimer’s disease: a systematic review and meta-analysis of invasive and non-invasive techniques. Transl. Psychiatry 12, 490 (2022).
Kocahan, S. & Doğan, Z. Mechanisms of Alzheimer’s disease pathogenesis and prevention: the brain, neural pathology, N-methyl-D-aspartate receptors, tau protein and other risk factors. Clin. Psychopharmacol. Neurosci. 15, 1–8 (2017).
Nava-Mesa, M. O., Jimenez-Diaz, L., Yajeya, J. & Navarro-Lopez, J. D. GABAergic neurotransmission and new strategies of neuromodulation to compensate synaptic dysfunction in early stages of Alzheimer’s disease. Front. Cell Neurosci. 8, 167 (2014).
Cunliffe, G., Lim, Y. T., Chae, W. & Jung, S. Alternative pharmacological strategies for the treatment of Alzheimer’s disease: focus on neuromodulator function. Biomedicines 10 https://doi.org/10.3390/biomedicines10123064 (2022).
Lloyd, B., de Voogd, L. D., Mäki-Marttunen, V. & Nieuwenhuis, S. Pupil size reflects activation of subcortical ascending arousal system nuclei during rest. ELife 12, e84822 (2023).
Murphy, P. R., O’Connell, R. G., O’Sullivan, M., Robertson, I. H. & Balsters, J. H. Pupil diameter covaries with BOLD activity in human locus coeruleus. Hum. Brain Mapp. 35, 4140–4154 (2014).
DiNuzzo, M. et al. Brain networks underlying eye’s pupil dynamics. Front. Neurosci. 13, 965 (2019).
Leuchs, L., Schneider, M., Czisch, M. & Spoormaker, V. I. Neural correlates of pupil dilation during human fear learning. Neuroimage 147, 186–197 (2017).
Schneider, M., Leuchs, L., Czisch, M., Sämann, P. G. & Spoormaker, V. I. Disentangling reward anticipation with simultaneous pupillometry/fMRI. NeuroImage 178, 11–22 (2018).
Mäki-Marttunen, V. Pupil-based states of brain integration across cognitive states. bioRxiv, 2020.2012.2015.422870 https://doi.org/10.1101/2020.12.15.422870 (2020).
Fietz, J. et al. Pupillometry tracks cognitive load and salience network activity in a working memory functional magnetic resonance imaging task. Hum. Brain Mapp. 43, 665–680 (2022).
He, H., Hong, L. & Sajda, P. Pupillary response is associated with the reset and switching of functional brain networks during salience processing. PLOS Comput. Biol. 19, e1011081 (2023).
Pais-Roldán, P. et al. Indexing brain state-dependent pupil dynamics with simultaneous fMRI and optical fiber calcium recording. Proc. Natl. Acad. Sci. USA 201909937 (2020).
Reimer, J. et al. Pupil fluctuations track rapid changes in adrenergic and cholinergic activity in cortex. Nat. Commun. 7, 13289 (2016).
Åbjørsbråten, K. S. et al. Impaired astrocytic Ca2+ signaling in awake-behaving Alzheimer’s disease transgenic mice. eLife 11, e75055 (2022).
Peirson, S. N., Brown, L. A., Pothecary, C. A., Benson, L. A. & Fisk, A. S. Light and the laboratory mouse. J. Neurosci. Methods 300, 26–36 (2018).
Hike, D. et al. High-resolution awake mouse fMRI at 14 Tesla. eLife 13, RP95528 (2024).
Mathis, A. et al. DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci. 21, 1281–1289 (2018).
Wang, Q. et al. The Allen mouse brain common coordinate framework: a 3D reference atlas. Cell 181, 936–953.e920 (2020).
Nam, Y., Kim, S., Kim, J., Hoe, H.-S. & Moon, M. Mesoscopic mapping of visual pathway in a female 5XFAD mouse model of Alzheimer’s disease. Cells 11, 3901 (2022).
Sil, A. et al. Sex differences in behavior and molecular pathology in the 5XFAD model. J. Alzheimer’s Dis. 85, 755–778 (2022).
Poon, C. H. et al. Sex differences between neuronal loss and the early onset of amyloid deposits and behavioral consequences in 5xFAD transgenic mouse as a model for Alzheimer’s disease. Cells 12, 780 (2023).
O’Leary, T. P. & Brown, R. E. Visuo‐spatial learning and memory impairments in the 5xFAD mouse model of Alzheimer’s disease: Effects of age, sex, albinism, and motor impairments. Genes Brain Behav. 21, e12794 (2022).
Oblak, A. L. et al. Comprehensive evaluation of the 5XFAD mouse model for preclinical testing applications: a MODEL-AD study. Front. Aging Neurosci. 13, 713726 (2021).
Tsui, K. C. et al. Distribution and inter-regional relationship of amyloid-beta plaque deposition in a 5xFAD mouse model of Alzheimer’s disease. Front. Aging Neurosci. 14, 964336 (2022).
Zhang, J. et al. Whole-brain three-dimensional profiling reveals brain region specific axon vulnerability in 5xFAD mouse model. Front. Neuroanat. 14, 608177 (2020).
Girard, S. D. et al. Onset of hippocampus‐dependent memory impairments in 5XFAD transgenic mouse model of Alzheimer’s disease. Hippocampus 24, 762–772 (2014).
Girard, S. D. et al. Evidence for early cognitive impairment related to frontal cortex in the 5XFAD mouse model of Alzheimer’s disease. J. Alzheimer’s Dis. 33, 781–796 (2013).
Chen, X. et al. Sensory evoked fMRI paradigms in awake mice. Neuroimage 204, 116242 (2020).
Qian, C. et al. Wireless amplified nuclear MR detector (WAND) for high-spatial-resolution MR imaging of internal organs: preclinical demonstration in a rodent model. Radiology 268, 228–236 (2013).
Zeng, H., Jiang, Y., Beer-Hammer, S. & Yu, X. Awake mouse fMRI and pupillary recordings in the ultra-high magnetic field. Front Neurosci. 16, 886709 (2022).
Iseri, P. K., Altinas, Ö., Tokay, T. & Yüksel, N. Relationship between cognitive impairment and retinal morphological and visual functional abnormalities in Alzheimer disease. J. Neuro Ophthalmol. 26, 18–24 (2006).
Moschos, M. M. et al. Structural and functional impairment of the retina and optic nerve in Alzheimer’s disease. Curr. Alzheimer Res. 9, 782–788 (2012).
Querques, G. et al. Functional and morphological changes of the retinal vessels in Alzheimer’s disease and mild cognitive impairment. Sci. Rep. 9, 63 (2019).
Polo, V. et al. Visual dysfunction and its correlation with retinal changes in patients with Alzheimer’s disease. Eye (Lond.) 31, 1034–1041 (2017).
Lewis, D. A., Campbell, M. J., Terry, R. D. & Morrison, J. H. Laminar and regional distributions of neurofibrillary tangles and neuritic plaques in Alzheimer’s disease: a quantitative study of visual and auditory cortices. J. Neurosci. 7, 1799–1808 (1987).
Callahan, L. M. & Coleman, P. D. Neurons bearing neurofibrillary tangles are responsible for selected synaptic deficits in Alzheimer’s disease. Neurobiol. Aging 16, 311–314 (1995).
Zhou, Y. et al. Abnormal connectivity in the posterior cingulate and hippocampus in early Alzheimer’s disease and mild cognitive impairment. Alzheimers Dement 4, 265–270 (2008).
Choo, I. H. et al. Posterior cingulate cortex atrophy and regional cingulum disruption in mild cognitive impairment and Alzheimer’s disease. Neurobiol. Aging 31, 772–779 (2010).
Huang, C., Wahlund, L. O., Svensson, L., Winblad, B. & Julin, P. Cingulate cortex hypoperfusion predicts Alzheimer’s disease in mild cognitive impairment. BMC Neurol. 2, 9 (2002).
Scheff, S. W. et al. Synaptic change in the posterior cingulate gyrus in the progression of Alzheimer’s disease. J. Alzheimers Dis. 43, 1073–1090 (2015).
Ali, D. G. et al. Amyloid-PET levels in the precuneus and posterior cingulate cortices are associated with executive function scores in preclinical Alzheimer’s disease prior to overt global amyloid positivity. J. Alzheimers Dis. 88, 1127–1135 (2022).
Hannula, D. E., Ryan, J. D., Tranel, D. & Cohen, N. J. Rapid onset relational memory effects are evident in eye movement behavior, but not in hippocampal amnesia. J. Cogn. Neurosci. 19, 1690–1705 (2007).
Liu, Z. X., Shen, K., Olsen, R. K. & Ryan, J. D. Visual sampling predicts hippocampal activity. J. Neurosci. 37, 599–609 (2017).
Hannula, D. E. & Ranganath, C. The eyes have it: hippocampal activity predicts expression of memory in eye movements. Neuron 63, 592–599 (2009).
Pajkossy, P., Szőllősi, Á. & Racsmány, M. Pupil size changes signal hippocampus-related memory functions. Sci. Rep. 10, 16393 (2020).
Zhao, A. et al. Visual abnormalities associate with hippocampus in mild cognitive impairment and early Alzheimer’s disease. Front. Aging Neurosci. 12, 597491 (2021).
Jiménez, E. C. et al. Altered vergence eye movements and pupil response of patients with Alzheimer’s disease and mild cognitive impairment during an oddball task. J. Alzheimer’s Dis. 82, 421–433 (2021).
Granholm, E. L. et al. Pupillary responses as a biomarker of early risk for Alzheimer’s disease. J. Alzheimer’s Dis. 56, 1419–1428 (2017).
Kesler, S. R., Acton, P., Rao, V. & Ray, W. J. Functional and structural connectome properties in the 5XFAD transgenic mouse model of Alzheimer’s disease. Netw. Neurosci. 2, 241–258 (2018).
Mondragón-Rodríguez, S., Gu, N., Fasano, C., Peña-Ortega, F. & Williams, S. Functional connectivity between hippocampus and lateral septum is affected in very young Alzheimer’s transgenic mouse model. Neuroscience 401, 96–105 (2019).
Green, C. et al. Functional networks are impaired by elevated tau-protein but reversible in a regulatable Alzheimer’s disease mouse model. Mol. Neurodegener. 14, 1–13 (2019).
Mangold, S. A. & Das, J. M. In StatPearls [Internet] (StatPearls Publishing, 2022).
Cohen, B. & Komatsuzaki, A. Eye movements induced by stimulation of the pontine reticular formation: evidence for integration in oculomotor pathways. Exp. Neurol. 36, 101–117 (1972).
Horn, A. K. E. In Progress in Brain Research Vol. 151 (ed Büttner-Ennever, J. A.) 127-155 (Elsevier, 2006).
Vitiello, B. et al. Cognitive and behavioral effects of cholinergic, dopaminergic, and serotonergic blockade in humans. Neuropsychopharmacology 16, 15–24 (1997).
Schmid, Y. et al. Acute effects of lysergic acid diethylamide in healthy subjects. Biol. Psychiatry 78, 544–553 (2015).
Cazettes, F., Reato, D., Morais, J. P., Renart, A. & Mainen, Z. F. Phasic activation of dorsal raphe serotonergic neurons increases pupil size. Curr. Biol. 31, 192–197.e194 (2021).
Chen, C. P. et al. Immunocytochemical study of the dorsal and median raphe nuclei in patients with Alzheimer’s disease prospectively assessed for behavioural changes. Neuropathol. Appl Neurobiol. 26, 347–355 (2000).
Rizzi-Wise, C. A. & Wang, D. V. Putting together pieces of the lateral septum: multifaceted functions and its neural pathways. eNeuro 8 https://doi.org/10.1523/ENEURO.0315-21.2021 (2021).
Woolf, N. J., Eckenstein, F. & Butcher, L. L. Cholinergic systems in the rat brain: I. projections to the limbic telencephalon. Brain Res. Bull. 13, 751–784 (1984).
Fotiou, D. F. et al. Cholinergic deficiency in Alzheimer’s and Parkinson’s disease: evaluation with pupillometry. Int. J. Psychophysiol. 73, 143–149 (2009).
Kim, S. et al. Alteration of neural pathways and its implications in Alzheimer’s disease. Biomedicines 10 https://doi.org/10.3390/biomedicines10040845 (2022).
Hara, Y. et al. Involvement of the septo-hippocampal cholinergic pathway in association with septal acetylcholinesterase upregulation in a mouse model of tauopathy. Curr. Alzheimer Res. 14, 94–103 (2017).
Belarbi, K. et al. Early Tau pathology involving the septo-hippocampal pathway in a Tau transgenic model: relevance to Alzheimer’s disease. Curr. Alzheimer Res. 6, 152–157 (2009).
Rubio, S. E. et al. Accelerated aging of the GABAergic septohippocampal pathway and decreased hippocampal rhythms in a mouse model of Alzheimer’s disease. FASEB J. 26, 4458–4467 (2012).
Soler, H. et al. The GABAergic septohippocampal connection is impaired in a mouse model of tauopathy. Neurobiol. Aging 49, 40–51 (2017).
Jeon, S. G., Kim, Y. J., Kim, K. A., Mook-Jung, I. & Moon, M. Visualization of altered hippocampal connectivity in an animal model of Alzheimer’s disease. Mol. Neurobiol. 55, 7886–7899 (2018).
Kim, S. et al. Topographical visualization of the reciprocal projection between the medial septum and the hippocampus in the 5XFAD mouse model of Alzheimer’s disease. Int. J. Mol. Sci. 20 https://doi.org/10.3390/ijms20163992 (2019).
Chang, C. et al. Tracking brain arousal fluctuations with fMRI. Proc. Natl. Acad. Sci. USA 113, 4518–4523 (2016).
Sobczak, F., Pais-Roldán, P., Takahashi, K. & Yu, X. Decoding the brain state-dependent relationship between pupil dynamics and resting state fMRI signal fluctuation. eLife 10, e68980 (2021).
Mandino, F., Vujic, S., Grandjean, J. & Lake, E. M. R. Where do we stand on fMRI in awake mice? Cerebral Cortex 34 https://doi.org/10.1093/cercor/bhad478 (2024).
Meyer, A. F., O’Keefe, J. & Poort, J. Two distinct types of eye-head coupling in freely moving mice. Curr. Biol. 30, 2116–2130.e2116 (2020).
Juczewski, K., Koussa, J. A., Kesner, A. J., Lee, J. O. & Lovinger, D. M. Stress and behavioral correlates in the head-fixed method: stress measurements, habituation dynamics, locomotion, and motor-skill learning in mice. Sci. Rep. 10, 12245 (2020).
Bundy, J. L., Vied, C., Badger, C. & Nowakowski, R. S. Sex‐biased hippocampal pathology in the 5XFAD mouse model of Alzheimer’s disease: a multi‐omic analysis. J. Comp. Neurol. 527, 462–475 (2019).
Manji, Z. et al. 5xFAD mice display sex-dependent inflammatory gene induction during the prodromal stage of Alzheimer’s disease. J. Alzheimer’s Dis. 70, 1259–1274 (2019).
Cox, R. W. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 29, 162–173 (1996).
Cox, R. W. & Hyde, J. S. Software tools for analysis and visualization of fMRI data. NMR Biomed. 10, 171–178 (1997).
Janke, A. L. & Ullmann, J. F. Robust methods to create ex vivo minimum deformation atlases for brain mapping. Methods 73, 18–26 (2015).
Acknowledgements
We thank Zeping Xie, Bei Zhang, and Biyue Zhu for their technical support, and Grace Yu for editing. We thank the AFNI group for its support of fMRI image processing. The research is supported by funding sources, including NIH grants (RF1NS113278 X.Y., RF1NS124778 X.Y., R01NS122904 X.Y., R01NS120594 X.Y., and R21NS121642 X.Y.), S10 instrument grants (S10OD028616 X.Y., S10OD036211 X.Y.) to Martinos Center, Massachusetts General Hospital, and NSF grant (2123970 X.Y.).
Author information
Authors and Affiliations
Contributions
X.L. and X.Y. designed the research. X.L., D.H., Y.J., and X.Y. performed the research. X.L., S.C., and W.M. analyzed data. C.R. provides animal models. X.A.Z. performed surgeries. D.H., X.Y. built coils. D.H. and X.L. designed the animal cradle. X.L., D.H., and X.Y. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
Xin Yu co-founded MRIBOT LLC. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Tracy Farr and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, X., Hike, D., Choi, S. et al. Identifying the bioimaging features of Alzheimer’s disease based on pupillary light response-driven brain-wide fMRI in awake mice. Nat Commun 15, 9657 (2024). https://doi.org/10.1038/s41467-024-53878-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-53878-y
This article is cited by
-
Probing locus coeruleus functional network in healthy aging and its association with Alzheimer’s disease biomarkers using pupillometry
Alzheimer's Research & Therapy (2025)