Abstract
The sinoatrial node regulates the heart rate throughout life. Failure of this primary pacemaker results in life-threatening, slow heart rhythm. Despite its critical function, the cellular and molecular composition of the human sinoatrial node is not resolved. Particularly, no cell surface marker to identify and isolate sinoatrial node pacemaker cells has been reported. Here we use single-nuclei/cell RNA sequencing of fetal and human pluripotent stem cell-derived sinoatrial node cells to reveal that they consist of three subtypes of pacemaker cells: Core Pacemaker, Sinus Venosus, and Transitional Cells. Our study identifies a host of sinoatrial node pacemaker markers including MYH11, BMP4, and the cell surface antigen CD34. We demonstrate that sorting for CD34+ cells from stem cell differentiation cultures enriches for sinoatrial node cells exhibiting a functional pacemaker phenotype. This sinoatrial node pacemaker cell surface marker is highly valuable for stem cell-based disease modeling, drug discovery, cell replacement therapies, and the targeted delivery of therapeutics to sinoatrial node cells in vivo using antibody-drug conjugates.
Similar content being viewed by others
Introduction
The sinoatrial node (SAN) is the primary pacemaker of the heart located at the boundary of the right atrium and the superior vena cava. The pacemaker cells of the SAN generate electrical impulses that initiate each of the three billion heartbeats that a human heart experiences in a lifetime. SAN diseases can lead to a life-threatening decrease in heart rate, a condition known as bradyarrhythmia. The current standard treatment for SAN dysfunction is implantation of an electronic pacemaker device which, while lifesaving, has multiple downsides1. To improve treatment options, a better understanding of the SAN pacemaker cells and their molecular identifiers is needed.
The SAN is a small structure that is comprised of about 10,000 pacemaker cells and has therefore been hard to access and study. Additionally, it is a heterogeneous tissue that contains multiple cell types, including fibroblasts, smooth muscle cells, endothelial cells, and neurons from the innervating autonomic nervous system, as well as different subtypes of pacemaker cardiomyocytes. In the adult human heart, these include the Core SAN pacemaker cardiomyocytes which express well-established pacemaker genes TBX3, TBX18, ISL1, HCN4, while lacking the expression of the pan-cardiomyocyte marker NKX2–52,3,4. These core pacemaker cells are responsible for initiating the heartbeat. In addition, a transitional cell type, also referred to as paranodal cells, that expresses both pacemaker and atrial cardiomyocyte genes has been described at the border of the Core SAN and atrial myocardium5,6,7. These cells are thought to facilitate the conduction of the electrical signals from the SAN tissue into the atrial myocardium.
Bulk RNA sequencing has been performed to gain a better understanding of the molecular identity of the SAN pacemaker cardiomyocytes, but tissue resident non-cardiomyocytes resulted in the detection of off-target genes that are expressed in neurons or fibroblasts4,8,9. Single-cell-based sequencing technologies have revolutionized our ability to study tissue compositions at the transcriptome level. Single-cell RNA sequencing (scRNA-seq) of the mouse SAN uncovered several new SAN pacemaker cardiomyocyte-specific marker genes9,10,11. Most recently, Kanemaru et al. dissected SAN tissue from adult hearts to generate a single-cell transcriptome atlas of the human SAN, but a single-cell-based dataset of the developing human SAN has not yet been established12.
Studies of the human SAN are hampered by the limited access to healthy primary tissue. To overcome this, investigators have turned to human pluripotent stem cells (hPSCs) as an alternative source of SAN pacemaker cells13. Accessing these cells from patient-specific, induced pluripotent stem cells (iPSCs) enables modeling of SAN diseases to gain deeper insights into disease mechanisms. Beyond that, hPSC-derived SAN cardiomyocytes are a source of functional pacemaker cells for the development of biological pacemakers that could overcome the downsides of electronic pacing. Several groups, including our own, have developed protocols for the directed differentiation of hPSCs into SAN-like pacemaker cells (SANLPCs)14,15,16,17,18. To isolate enriched populations of SANLPCs from the differentiation cultures, transgenic reporters have been employed, including NKX2–5:GFP for the negative selection of NKX2–5− SANLPCs or SHOX2:GFP for the positive selection of SHOX2+ SANLPCs14,15,18. Identification of a cell surface antigen that is specifically expressed on SANLPCs would eliminate the need to generate reporter lines, enabling straightforward identification and isolation of SANLPCs from differentiation cultures of any hPSC line. To date, a SANLPC-specific cell surface marker has not been identified.
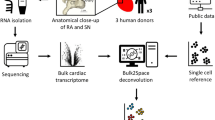
To gain a better understanding of the cellular and molecular composition of the human SAN and identify potential SAN-specific cell surface markers, we performed single-nuclei/single-cell RNA sequencing (sn/scRNA-seq) of human fetal SAN tissue and hPSC-derived SANLPCs. This analysis showed that hPSC-derived SANLPCs closely resemble fetal SAN pacemaker cells on the transcriptome level. Our analysis identified three subtypes of SAN pacemaker cells, including Core SAN, Sinus Venosus, and Transitional Cells. Combining the fetal and hPSC-derived datasets, we established a shared list of Core SAN pacemaker cell-specific genes. This list contains a host of SAN markers, most notably the surface antigen CD34. We show that CD34 is specifically expressed by the pacemaker cardiomyocytes in human SAN tissue and by hPSC-derived SANLPCs, but not by other cardiomyocyte subtypes. Finally, we demonstrate that sorting for CD34+ cells from hPSC differentiation cultures enriches for SANLPCs with a functional pacemaker phenotype.
Results
Single-cell transcriptomic analysis of the human fetal SAN
To assess the molecular heterogeneity of the cells in the developing human SAN, we dissected the SAN tissue of a fetal heart (gestation week 19) and performed snRNA-seq (Fig. 1a). Unsupervised clustering identified 19 cell clusters representing cell types expected to be present in the heart, including cardiomyocytes as well as non-cardiomyocytes such as fibroblasts, smooth muscle, endothelial, endocardial, epicardial cells, neurons, and macrophages9,19 (Fig. 1b, Supplementary Table 1). To obtain a better resolution of the cardiomyocyte subtypes, we performed subcluster analysis on the cardiomyocyte cell clusters. This resulted in eight cell clusters that expressed high levels of TNNT2, indicative of cardiomyocytes (Fig. 1b, c). To identify the cardiomyocytes representing SAN pacemaker cells, we analyzed the expression of established SAN pacemaker genes (TBX3, TBX18, SHOX2, ISL1, HCN4)2,3,4,5. We detected one cell cluster expressing high levels of all these genes that we annotated as Core SAN cells. We identified a second cell cluster expressing these SAN genes but with lower expression levels of TBX3 and ISL1. This expression pattern is consistent with a Sinus Venosus cardiomyocyte phenotype, a cardiomyocyte population developing together with the Core SAN from the TBX18+ mesenchyme at the venous pole of the heart2,20,21,22. Of note, most of the Core SAN and Sinus Venosus cells did not express the cardiac transcription factor NKX2–5, which is a well-established characteristic of SAN pacemaker cells and the Sinus Venosus2,3. In addition, three cell clusters representing Atrial cells were identified based on the expression of NKX2–5, the atrial marker NPPA, and SCN5A4,19. As expected, we also identified a cluster of cells expressing both atrial and pacemaker genes, representing the myocytes of the Transition Zone between the pacemaker and atrial tissues6,7,9. Finally, two clusters of proliferating MKI67-expressing cells were detected (Supplementary Fig. 1 a-c). Differential gene expression analysis showed that the Transition Zone cells were marked by the expression of LRRC4C (netrin-G1 ligand), MYO16 (myosin 16), and SLC24A3 (sodium/potassium/calcium exchanger 3), while Sinus Venosus cells were marked by the expression of the long non-coding RNA LINC01880, PDEA1 (phosphodiesterase 1A), and CDH13 (cadherin 13) (Fig. 1d, Supplementary Data 1). We next focused our analysis on the Core SAN cells to identify markers of this population. The Core SAN cells expressed the highest levels of the established pan-pacemaker marker TBX32. In addition, the expression of other well-known pacemaker genes (TBX18, SHOX2, ISL1, RGS6, VSNL1, CACNA1D, HCN1, HCN4) was significantly enriched in this cell cluster, confirming a pacemaker phenotype4,8,9,22. Gene ontology (GO) analysis for biological processes of the genes enriched in the Core SAN cells resulted in terms including regulation of heart rate, cardiac pacemaker cell differentiation, sinoatrial node development, and regulation of SAN cell action potential, further supporting the SAN pacemaker phenotype of these cells (Fig. 1e). Importantly, the top ten differentially expressed genes (DEGs) within the Core SAN cluster contained several genes not previously reported to be involved in SAN pacemaker cell function or development, including TENM2 (teneurin transmembrane protein 2), PPFIA2 (protein tyrosine phosphatase receptor type F polypeptide interacting protein alpha 2), CNTN4/5 (contactin 4/5) PRKG1 (protein kinase cGMP-dependent 1), SLIT2 (slit guidance ligand 2), DGKB (diacylglycerol kinase beta), and KCNIP4 (potassium voltage-gated channel interacting protein 4) (Fig. 1 d, f). Furthermore, the guanylate kinase-associated protein DLGAP1, recently described as a SAN-specific gene in the mouse, was also contained in our top ten gene list10.
a Schematic overview of SAN tissue dissection and processing for snRNA-seq on the 10x Genomics Chromium platform. b Uniform manifold approximation and projection (UMAP) of gestation week 19 fetal heart SAN tissue sample showing 19 cell clusters (left). Subclustering of the TNNT2+ cardiomyocytes showing 8 sub-clusters (right). c UMAPs of the subclustered cardiomyocytes showing the expression of the indicated genes. d Heatmap of the top 5 differentially expressed genes (DEGs) within the indicated cardiomyocyte subclusters. e Gene Ontology (GO) analysis (biological processes) of all genes enriched in the Core SAN cluster. f UMAPs showing the top 10 DEGs of the core SAN cluster. Schematics in (a) were generated using Biorender (https://biorender.com).
To get a better understanding of the potential functional differences between the three pacemaker cell clusters we compared their expression of key cardiac ion channels (Supplementary Fig. 1d and Supplementary Data 2, 3). For most of the ion channels, expression was comparable between Core SAN and Sinus Venosus cardiomyocytes except for the working cardiomyocyte genes SCN5A, CACNA1C, GJA5, ITPR1, and RYR2 that were higher expressed by Sinus Venosus cells. Comparison between Core SAN and Transition Zone myocytes showed lower expression of pacemaker ion channels such as HCN1, HCN4, and CACNA1D and higher expression working cardiomyocyte genes such as SCN5A, CACNA1C, GJA5, GJA1 in Transition Zone cells. This agrees with the notion that Transition Zone cells have a mixed phenotype between atrial and pacemaker cells.
SANLPCs transcriptionally resemble fetal SAN cells
We next wanted to compare the expression profiles of the fetal SAN cells to hPSC-derived SANLPCs generated using our previously reported protocol14 (Fig. 2a). This protocol generates a mixed population, containing both NKX2–5− SANLPCs (50%) and NKX2–5+ cardiomyocytes (30%), that we used for scRNA-seq analysis. As shown in Fig. 2b, 11 cell clusters were identified, including fibroblasts, epithelial, and epicardial cells, as well as cardiomyocytes. To specifically compare the expression between the cardiomyocyte subtypes, we subclustered the TNNT2+ cardiomyocytes. Analysis of the expression of established SAN pacemaker markers TBX3, TBX18, SHOX2, ISL1, and HCN42,3,4,5 identified a Core SAN cluster that was mostly negative for NKX2–5 expression (Fig. 2c). Like in the fetal SAN tissue, a second NKX2–5− cardiomyocyte cluster was identified that expressed all the pacemaker markers, but lower levels of TBX3, that we annotated as Sinus Venosus cells. Analysis of expression of the atrial marker NPPA together with working cardiomyocyte markers NKX2–5 and SCN5A identified a cluster containing Atrial cardiomyocytes. The hPSC cultures also contained Transition Zone cells that expressed both pacemaker and atrial genes. In addition, two clusters of proliferating MKI67-expressing cardiomyocytes were identified (Supplementary Fig. 2a–c). To further confirm the identity of the hPSC-derived cardiomyocyte clusters, we scored the cells in the hPSC-derived dataset with the DEG lists of the fetal tissue dataset23 (Fig. 2d). Applying the top 200 DEGs from the fetal Core SAN cluster identified the Core SAN cluster in the hPSC dataset. Using the same approach, the identities of the Sinus Venosus, Transition Zone, and Atrial cell clusters were confirmed. These findings indicate that the NKX2–5+ cells within the hPSC-derived population represent Atrial and Transition Zone cells while the NKX2–5− cells represent Core SAN and Sinus Venosus cells.
a Schematic overview of SANLPC differentiation protocol and sample processing for scRNA-seq on the 10x Genomics Chromium platform. b UMAP of day 25 HES2-derived SANLPCs showing 11 cell clusters (left). Subclustering of the TNNT2+ cardiomyocytes showing 6 sub-clusters (right). c UMAPs of the subclustered cardiomyocytes showing the expression of the indicated genes. d UMAPs showing signature score distribution for the DEGs of the indicated fetal SAN cell types. e Heatmap of the top 5 DEGs within the indicated cardiomyocyte subclusters. f GO analysis (biological processes) of all genes enriched in the Core SAN cluster. g UMAPs showing the top 10 DEGs of the core SAN cluster. h UMAPs showing Harmony integration of the fetal SAN snRNA-seq data and the hPSC scRNA-seq data labeled by source dataset (left), cluster number (center), and assigned cell types (right). i Stacked bar graph showing the frequency of each cell type from both fetal and hPSC datasets in the indicated cell clusters. j UMAPs of integrated fetal SAN and hPSC datasets showing the expression of the indicated genes. k Spearman correlation between selected clusters from the fetal SAN and hPSC datasets. p < 0.05 for all correlations (asymptotic t approximation). Schematics in (a) were generated using Biorender (https://biorender.com).
Differential gene expression analysis identified expression patterns that distinguish the different hPSC-derived cardiomyocyte subtypes. Transition Zone cells were marked by the expression of ANKRD1 (ankyrin repeat domain 1, cardiac muscle), CSRP3 (cysteine and glycine-rich protein 3, cardiac LIM protein), and MYL3 (myosin light chain 3), while Sinus Venosus cells were marked by the expression of TRH (thyrotropin-releasing hormone), CKM (creatine kinase, muscle) and DES (desmin) (Fig. 2e, Supplementary Data 4). The Core SAN cells were enriched in the expression of previously established conduction system and SAN pacemaker markers (TBX3, VSNL1, LBH, CPNE5)4,8,9,24. GO term analysis identified an enrichment of genes involved in regulation of heart contraction, cardiac conduction system development, and sinoatrial node development, further supporting the SAN pacemaker phenotype of the cells contained in the Core SAN cluster (Fig. 2f). Interestingly, the top ten DEGs of the Core SAN cells contained genes not previously reported to be specifically expressed in SAN pacemaker cardiomyocytes including ACTA2 (actin alpha 2, smooth muscle), ALDH1A1 (aldehyde dehydrogenase 1a1), HAMP (hepcidin antimicrobial peptide), MYLK (myosin light chain kinase, smooth muscle), and ALDOC (aldolase c, brain-type) (Fig. 2g). The list also contained the C-X-C motif chemokine ligand CXCL12, the smooth muscle myosin heavy chain isoform MYH11, the BMP signaling ligand BMP4, and the annexin family member ANXA2 which have been previously detected in transcriptomic analysis of mouse and human SAN cells4,8,12,22.
To further compare the hPSC-derived cells and the fetal SAN cells, we used Harmony integration and combined the two datasets (Fig. 2h–j, Supplementary Fig. 2d–g)25. This analysis showed that the hPSC-derived cardiomyocytes clustered together with the fetal cardiomyocytes. Within the integrated cardiomyocytes we identified clusters of Core SAN, Sinus Venosus, Transition Zone, and Atrial cardiomyocytes as described in the individual datasets. Importantly, the Core SAN cluster contained both hPSC-derived and fetal tissue-derived Core SAN cells (Fig. 2i, Supplementary Fig. 2e). The vast majority of the Core SAN cells from the hPSC and fetal datasets (74% and 94%) clustered together in this merged Core SAN cluster. The Sinus Venosus, Transition Zone, and Atrial clusters similarly contained both hPSC-derived and fetal-derived cells of the respective cardiomyocyte subtype. However, along with the matching subtypes, a more heterogenous contribution of other cardiomyocyte subtypes was detected in these clusters. Spearman correlation analysis further demonstrated that the hPSC-derived Core SAN cells are most similar to the cells of the fetal Core SAN (Fig. 2k, Supplementary Fig. 2g). Taken together, this molecular analysis indicates that the hPSC-derived Core SAN pacemaker cells closely resemble the Core SAN pacemaker cells found in the developing human heart.
Identification of shared Core SAN pacemaker markers
To identify a list of marker genes that can be used to identify SAN pacemaker cells in vitro and in vivo, we next compared the DEGs of the Core SAN cells from the hPSC and fetal datasets. This analysis identified 36 genes that were significantly enriched in both populations (Fig. 3a–d, Supplementary Fig. 3a–c). Signature scoring analysis showed that these 36 genes clearly identify the Core SAN cell clusters in the hPSC and fetal data. The gene list contains the established cardiac conduction system and SAN markers TBX3, TBX5, VSNL1, LBH, CPNE2,4,8,9,24 as well as GNAO, a subunit of the G-protein signal-transducing complex, that has been recently described to be expressed in hPSC-derived SAN-like cells22. In addition, this shared list contains a number of genes that have not been previously associated with human SAN function or development including; (i) contractile apparatus and cytoskeletal genes: MYH11 (myosin heavy chain 11), ACTB (actin beta), WIPF3 (WAS/WASL interacting protein family member 3); (ii) genes involved in BMP signaling: BMP4 (bone morphogenetic protein 4), BMP5 (bone morphogenetic protein 5), ID3 (inhibitor of DNA binding 3); (iii) genes encoding for calcium binding proteins involved in cellular signaling: ANXA2 (annexin 2), NECAB1 (N-terminal EF-hand calcium binding protein 1), SPOCK1 (proteoglycan 1); (iv) genes encoding for regulators of transmembrane calcium ion-currents: RRAD (Ras related glycolysis inhibitor and calcium channel regulator), PRKG1 (protein kinase cGMP dependent 1); (v) and genes classically known to be expressed in neurons and glia cells that play a role in axon growth and guidance: BASP1 (brain abundant membrane attached signal protein 1), PPFIA2 (protein tyrosine phosphatase receptor type F polypeptide interacting protein alpha 2), EFR3B (EFR3 homolog B), SLIT2 (slit guidance ligand 2), LYPD6 (LY6/PLAUR domain containing 6). Importantly, the list also contains five genes encoding membrane-spanning proteins that could serve as SAN pacemaker cell surface markers that have not been previously associated with SAN cells: CD34 (CD34 antigen), CADM1 (cell adhesion molecule 1), EFNB2 (ephrin B2), NTRK2 (neurotrophic receptor tyrosine kinase 2), and ELAPOR2 (endosome-lysosome associated apoptosis and autophagy regulator family member 2).
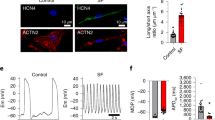
a Venn diagram of the Core SAN markers identified in the fetal and hPSC datasets. b UMAPs of TNNT2+ cardiomyocytes showing the assigned cell types (top row) and the signature score distribution (bottom row) of the shared 36 Core SAN marker genes in the fetal (left) and hPSC datasets (right). c, d Dot plots showing the expression of the conserved core SAN markers in the fetal (c) and hPSC (d) datasets. *indicates genes encoding for membrane-spanning proteins. e–h Immunofluorescent staining of day 25 hPSC-derived SANLPCs, atrial-like cardiomyocytes (ALCMs), and ventricular-like cardiomyocytes (VLCMs) for NKX2–5 and SAN pacemaker transcription factor SHOX2, ventricular contractile apparatus protein MLC2V and pacemaker ion channel HCN4, NKX2–5:GFP and atrial protein NPPA (e), Core SAN marker MYH11 (f), Core SAN marker BMP4 (g), and Core SAN marker CD34 (h). Cells were counterstained with cTNT to identify cardiomyocytes and Hoechst to visualize all cells (n = 3) independent differentiations. Images were denoised using nikons denoise.ai. Scale bars, 100 μm.
To assess whether the 36 Core SAN marker genes that we identified in fetal/hPSC-derived SAN pacemaker cells are maintained during maturation, we analyzed their expression in the recently published snRNA-seq data of the adult human SAN from Kanemaru et al.12. Subclustering of the SAN cell cluster identified Atrial and Transition Zone cardiomyocytes similar to our fetal dataset. (Supplementary Fig. 3d–f). In addition, two Core SAN cell clusters were identified, based on the expression of high levels of TBX3, SHOX2, HCN4 and very low levels of NKX2–5. These adult Core SAN cells expressed significantly higher levels of 22 of the 36 Core SAN genes compared to adult Atrial cardiomyocytes, suggesting that the majority of the Core SAN markers are maintained during maturation (Supplementary Fig 3g–i, and Supplementary Data 5).
To determine if the 36 Core SAN markers are conserved between species, we compared our list to the previously published scRNA-seq data of the embryonic (E16.5) mouse SAN from Goodyer et al.9,26 (Supplementary Fig. 4a-c). For the mouse dataset, we used the same cluster annotation of SAN cells, Transition zone cells, and Atrial cells as originally published by the authors. As expected, the established SAN markers TBX3, TBX5, VSNL1, LBH, and CPNE5 were conserved between human and mouse. However, only half of the human Core SAN genes from our list were specifically expressed in mouse SAN pacemaker cells. The other half was higher expressed in atrial cardiomyocytes of the mouse or expressed at very low levels. These observations prompted us to assess whether the SAN-specific genes identified in the mouse SAN scRNA-seq dataset are conserved in the human SAN9. Out of the 50 genes that we analyzed, the majority (56%) were either not detected in the human Core SAN cells or expressed at higher levels in human atrial cardiomyocytes. Notably, the list of genes also contained mouse SAN pacemaker cell surface markers26, and only 13 out of 25 were conserved in the human SAN. Taken together, this analysis shows that, while key markers are conserved between species, there are numerous markers that are specific to the human SAN. This highlights the importance of analyzing human cells and tissues.
MYH11, BMP4, and CD34 are SAN pacemaker markers
We selected three Core SAN genes identified in our study to further validate them as SAN markers. We chose the top two differentially expressed genes MYH11 and BMP4, which have previously been detected in transcriptomic studies of SAN pacemaker cells but have not been further validated on the protein level4,8,12,22. In addition, we chose the surface antigen CD34 due to its promising potential application as a SAN cell surface marker and the fact that its expression is maintained in the adult SAN (Supplementary Fig 3h). We first focused on hPSC-derived cardiomyocytes and used previously reported protocols to generate SANLPCs, atrial-like cardiomyocytes (ALCMs), and ventricular-like cardiomyocytes (VLCMs)14,27. RT-qPCR analysis and immunofluorescence staining confirmed the phenotype of the different cardiomyocyte subtypes. As expected, SANLPCs expressed SHOX2 and HCN4, ALCMs expressed NKX2–5 and NPPA, and VLCMs expressed NKX2–5 and MLC2V (Fig. 3e, Supplementary Fig. 5a, b). Importantly, immunostaining for MYH11, BMP4, and CD34 showed that these proteins are specifically expressed by SANLPCs but not ALCMs and VLCMs (Fig. 3f-h). Of note, a nuclear staining for BMP4 was detected in the VLCMs. This is most likely an unspecific signal as RT-qPCRs confirmed significantly lower BMP4 expression in VLCMs compared to SANLPCs (Supplementary Fig. 5c).
To validate the expression of these SAN markers in primary tissue, we dissected the SAN of fetal hearts (gestation week 17–20) and prepared longitudinal sections for immunostaining (Fig. 4a, Supplementary Fig. 6a)28. We used the expression of TBX3, SHOX2, and HCN4 to identify fetal Core SAN pacemaker cells, expression of NPPA to identify Atrial cardiomyocytes, and expression of cTNT to distinguish cardiomyocytes from non-cardiomyocytes (Fig. 4a). Using these markers we also detected TBX3−SHOX2+HCN4+NPPA+ cardiomyocytes adjacent to the Core SAN representing Transition Zone cardiomyocytes (Supplementary Fig. 6a, cI & 7a, bI-II). Furthermore, a region of TBX3lowSHOX2+HCN4+NPPA− cardiomyocytes was detected cranial to the SAN. These cardiomyocytes did not express NKX2–5 suggesting that they represent the Sinus Venosus cardiomyocytes. Immunostaining for MYH11 labeled the SAN pacemaker cardiomyocytes but did not label Atrial, Transition Zone, or Sinus Venosus cardiomyocytes (Fig. 4bI, Supplementary Fig. 6cII & 7bIII). As expected, MYH11 was also detected in the smooth muscle cells of blood vessels contained in the sections. BMP4 expression was detected in HCN4+cTNT+ SAN pacemaker cells and in some of the HCN4−cTNT− non-cardiomyocytes of the SAN (Fig. 4bII, Supplementary Fig. 6cIII). BMP4 expression was higher in the SAN cardiomyocytes than in the adjacent Atrial and Transition Zone cardiomyocytes. CD34 specifically labeled cTNT+ SAN cardiomyocytes but not Atrial, Transition Zone, or Sinus Venosus cardiomyocytes (Fig. 4bIII, c, Supplementary Fig. 7bIII, IV). In agreement with our transcriptional analysis most of the SAN cardiomyocytes were CD34+NKX2–5− (white arrowheads, Fig. 4cIV). In addition, we detected a few CD34+NKX2–5+ (green arrowheads) and CD34−NKX2–5− (yellow arrowheads) pacemaker cardiomyocytes in the SAN. Of note, CD34 is also expressed by endothelial cells, which most likely explains the detection of some CD34+ cells in the atrial tissue. Co-staining for CD31 and cTNT confirmed that these CD34+ cells are indeed CD31+ endothelial cells and that only SAN cardiomyocytes but not atrial cardiomyocytes stain positive for CD34 (Supplementary Fig. 6b).
Immunofluorescent staining of gestation week 19 fetal human SAN for: (a) pacemaker transcription factor TBX3 and pacemaker ion channel HCN4 (aI), SAN pacemaker transcription factor SHOX2 and atrial protein NPPA (aII); (b) core SAN marker MYH11 and HCN4 (bI), Core SAN marker BMP4 and HCN4 (bII), Core SAN marker CD34 and HCN4 (bIII); (c) CD34 and SHOX2 (cI), CD34 and TBX3 (cII), CD34 and MYH11 (cIII), and CD34 and NKX2–5 (cIV). White dashed line outlines the SAN. Yellow and green boxes indicate location of high magnification insets shown on the right marked with *. White arrows in bI* indicate MYH11+ smooth muscle cells of a blood vessel. Arrowheads in cIV* indicate the following cell types: white, CD34+NKX2–5−; green, CD34+NKX2–5+; yellow, CD34−NKX2–5−. Tissue sections were counterstained with cTNT to identify cardiomyocytes and DAPI to visualize all cells (n = 3 independent stains each of sections from 3 heart samples). Images were denoised using nikons denoise.ai. Scale bars, 500 μm (left) and 50 μm in the high magnification insets (right). RA right atrium.
Our computational analysis suggested that CD34 is specific to human SAN pacemaker cells, because its expression was not detected in mouse SAN pacemaker cells (Supplementary Fig. 4b)9. To further validate this, we performed immunostaining for CD34 on sections of mouse SAN tissue (postnatal days 0–3). These experiments confirmed that SHOX2+TBX3+HCN4+ mouse SAN cardiomyocytes do not express CD34 (Supplementary Fig. 8a, b).
To determine if expression of MYH11, BMP4, and CD34 is specific to the primary pacemaker, the SAN, or whether these proteins are also expressed by the secondary pacemaker, the atrioventricular node (AVN), we analyzed sections of the atrioventricular junctional region of fetal hearts (gestation week 17–20) (Supplementary Fig. 9a). AVN pacemaker cardiomyocytes were identified based on the expression of the AVN markers TBX3, MSX2, and the lack of expression of the SAN marker SHOX2 (Fig. 5aI, IV, Supplementary Fig. 9bI). NPPA and MLC2V expression identified adjacent atrial and ventricular tissues, respectively (Supplementary Fig. 9bI, II). Neither MYH11 nor BMP4 were expressed in AVN cardiomyocytes (Fig. 5aII, III). Similarly, no CD34+cTNT+ cells were found in AVN tissue (Fig. 5aIV, Supplementary Fig. 9bIII). The CD34+cTNT− cells that were detected likely represent a CD34+ mesenchymal/fibroblast cell population that has previously been reported in multiple studies19,29,30. Taken together, these findings show that MYH11, BMP4, and CD34 are specifically expressed by the primary pacemaker cardiomyocytes of the SAN, but not by atrial, ventricular, nor AVN cardiomyocytes.
Immunofluorescent staining of gestation week 17 fetal human AVN for: AVN transcription factor MSX2 and HCN4 (aI), Core SAN marker MYH11 and HCN4 (aII), Core SAN marker BMP4 and HCN4 (aIII), and Core SAN marker CD34 and TBX3 (aIV). Images represent consecutive sections of AVN tissue. White dashed line outlines the AVN. Yellow box indicates location of high magnification insets shown on the right marked with *. Tissue sections were counterstained with cTNT to identify cardiomyocytes and DAPI to visualize all cells (n = 3 independent stains each of sections from 1 heart sample). Images were denoised using nikons denoise.ai. Scale bars, 500 μm (left) and 50 μm in the high magnification insets (right). IVS interventricular septum; RA right atrium.
Isolation of SANLPCs based on CD34 expression
To determine if CD34 could be used for the isolation of SANLPCs from hPSC differentiation cultures, we first carried out flow cytometric analysis of CD34 expression. The HES3 NKX2–5egfp/w reporter line was used for these experiments which allowed us to identify NKX2–5:GFP− cardiomyocytes as SANLPCs and NKX2–5+ cardiomyocytes as VLCMs or ALCMs14,31. Analysis within the fraction of SIRPA+CD90− cardiomyocytes32 at day 25 of differentiation showed that only SANLPCs are marked by CD34 while VLCMs and ALCMs do not express CD34 (Fig. 6a, b, Supplementary Fig. 10a). Within the SANLPC differentiation cultures, the majority of the NKX2–5− cells expressed CD34 (54 ± 4%). Notably, a small fraction of NKX2–5+ cells contained in the SANLPC cultures also expressed CD34 (17 ± 2%), an observation consistent with our findings in the fetal heart tissue sections. To identify the earliest timepoint of CD34 expression in SANLPC progenitors, we performed a time course analysis (Fig. 6c, d). The first SIPRA+CD90− cardiomyocytes detected at day 8 of differentiation did not express CD34. By day 10, a small population of NKX2–5− cardiomyocytes started to express CD34, and by day 16, most of the NKX2–5− cardiomyocytes expressed CD34. The proportion of CD34+NKX2–5− cardiomyocytes continued to increase until day 50 at which point 81 ± 4% of NKX2–5− cardiomyocytes expressed CD34 (Supplementary Fig. 10b, c). The first NKX2–5+CD34+ cardiomyocytes were detected at day 16. The proportion of NKX2–5+CD34+ cardiomyocytes also increased over time, but expression levels of CD34 remained lower than in the NKX2–5− cardiomyocytes at all time points analyzed. Accordingly, analysis at day 50 showed that 64 ± 6% of NKX2–5− cells and 20 ± 3% of NKX2–5+ cells expressed high levels of CD34 (CD34++), similar to the proportions of CD34+ cells observed at day 25. Importantly, day 50 VLCM and ALCM populations remained CD34 negative, confirming that CD34 is exclusively expressed on SAN pacemaker cardiomyocytes (Supplementary Fig. 10d, e).
a Flow cytometric analyses at day 25 of CD34 and NKX2–5:GFP expression in SIRPA+CD90− cardiomyocytes in VLCMs, ALCMs, and SANLPCs. b Bar graphs summarizing the expression of CD34 in myocytes as shown in (a) in the indicated differentiation cultures (left) and within the NKX2–5− and NKX2–5+ fraction of SANLPC differentiation cultures (right) (n = 6 independent differentiations). c Flow cytometric analyses of CD34 and NKX2–5:GFP expression in SIRPA+CD90− cardiomyocytes at indicated time points throughout the differentiation. d Bar graph summarizing the expression of CD34 in myocytes as shown in (c) (n = 6 independent differentiations). e Flow cytometric analyses of CD34 and NKX2–5:GFP expression in SIRPA+CD90− cardiomyocytes before and after FACS at day 25. Teal shading indicates CD34+ and blue shading indicates CD34− FACS sorting gates. f Bar graphs summarizing the proportion of NKX2–5− and NKX2–5+ cells in presort, CD34+, and CD34− FACS sorted samples (n = 7 independent differentiations). g RT-qPCR analysis of the expression of the indicated genes in presort, CD34+, and CD34− FACS sorted samples (n = 7 independent differentiations, n = 4 independent differentiations (genes BMP4)). Values represent expression relative to the housekeeping gene TBP. h Flow cytometric analyses of CD34 and NKX2–5:GFP expression in SIRPA+CD90− cardiomyocytes before and after MACS at day 25. Teal shading indicates CD34+ and blue shading indicates CD34− MACS sorting gates. i Bar graphs summarizing the proportion of NKX2–5− and NKX2–5+ cells in presort, CD34+, and CD34− MACS sorted samples (n = 8 independent differentiations). j RT-qPCR analysis of the expression of the indicated genes in presort, CD34+, and CD34− MACS sorted samples (n = 6 independent differentiations, n = 5 independent differentiation (MYH11, BMP4)). Values represent expression relative to the housekeeping gene TBP. Statistical analysis was performed using two-sided paired t-test when comparing two samples (b, f, i) and one-way ANOVA followed by Bonferroni’s post hoc test when comparing >2 samples (b, g, j). Datasets that failed the normality test ((g): SCN5A, (j): SCN5A) were analyzed using Friedman test with Dunn’s post hoc test: *p < 0.05, **p < 0.01, ***p < 0.001 vs indicated sample. Error bars represent SEM. Violin plot elements: center line, median; lower line, first quartile; upper line, third quartile. Source data are provided as a Source Data file.
To test if it is possible to enrich for NKX2–5− SANLPCs based on CD34 expression, we isolated SIRPA+CD90−CD34+ and SIRPA+CD90−CD34− cardiomyocytes by fluorescence-activated cell sorting (FACS) and analyzed the populations for NKX2–5:GFP expression (Fig. 6e, f). We performed these experiments at day 25 of differentiation because the majority of NKX2–5− cells expressed CD34 while the majority of NKX2–5+ cells were still CD34 negative at this timepoint. As shown in Fig. 6e and f, the CD34+ sorted population was significantly enriched for NKX2–5− cells and significantly depleted of NKX2–5+ cells compared to pre-sort. The opposite trend was observed for the CD34− sorted population. RT-qPCR analysis showed significantly enriched expression of SAN genes (TBX3, SHOX2, HCN4, MYH11, BMP4) and decreased expression of atrial/working cardiomyocyte genes (SCN5A, NPPA) in the CD34+ sorted cells compared to pre-sort and CD34− sorted cells (Fig. 6g). To determine if the CD34+NKX2–5− phenotype is stable over time, we cultured the cells for an additional 25 days. Flow cytometric analyses at days 1, 10, and 25 post-sort revealed that the cells maintained CD34 expression and did not upregulate NKX2–5 expression (Supplementary Fig. 10f, g). These experiments also showed that a sizeable proportion of CD34− cells upregulated CD34 expression by day 50. To further assess the fate of the NKX2–5−CD34− cells specifically, we isolated this population on day 25 and analyzed their phenotype on day 50 (Supplementary Fig 11a, b). 35 ± 2% of these NKX2–5−CD34− cells did upregulate CD34 expression by day 50 and showed higher expression of TBX3 compared to the day 25 and day 50 NKX2–5−CD34− cells. Taken together, our data suggests that the day 25 NKX2–5−CD34− cells represent a progenitor population and that the majority of NKX2–5− SANLPCs do express CD34 upon extended culture.
For disease modeling and cell therapy applications, large cell numbers of SAN pacemaker cells will be required, thus prompting us to test whether magnetic-activated cell sorting (MACS) for CD34+ cells can also enrich for SANLPCs. Interestingly, MACS for CD34+ cells enriched for SIRPA+CD90− cardiomyocytes without the need for additional antibodies (Fig. 6h-j, Supplementary Fig. 11c). Notably, the CD34+ sorted cells were also significantly enriched in NKX2–5− cardiomyocytes and depleted of NKX2–5+ cells compared to pre-sort. Accordingly, RT-qPCR analysis showed increased expression of SAN pacemaker genes in the MACS-sorted CD34+ cells and decreased expression of working cardiomyocyte genes compared to pre-sort and CD34− sorted cells (Fig. 6j). The CD34+ population contained a low frequency (<5%) of CD31+ endothelial cells that are known to contaminate hPSC-derived cardiomyocyte differentiations (Supplementary Fig. 11d, e). These cells were no longer detectable following 5 days of culture, likely because our culture conditions did not support their survival.
To confirm that this CD34-based isolation of NX2–5− SANLPCs can be applied to other hPSC lines, we repeated the experiments using the HES2 hESC line (Supplementary Fig. 12a–f). First, we confirmed that CD34 is specifically expressed by HES2-derived SANLPCs but not by ALCMs and VLCMs. Next, we showed that MACS sorting for CD34+ cells results in a significant enrichment of NKX2–5− cardiomyocytes and depletion of NKX2–5+ cardiomyocytes. In addition, CD34+ sorted cells showed higher expression of SAN pacemaker genes and reduced expression of working cardiomyocyte genes, thus confirming the enrichment for SANLPCs. Taken together, these experiments demonstrate that CD34 can be universally used to identify NKX2–5− SANLPCs and isolate them from cardiac hPSC-differentiation cultures.
Previously, CD166 has been reported to label SAN progenitors in the mouse33. We therefore analyzed the expression of CD166 during SANLPC and VLCM differentiations. We found that both SANLPCs and VLCMs do express CD166 between days 8–25, with higher proportions of CD166+ cells detected in VLCMs (Supplementary Fig. 13a–c). This is in agreement with previous reports showing that CD166 is expressed by hPSC-derived cardiomyocytes34. Furthermore, only a small fraction of CD166+ cardiomyocytes co-expressed CD34 (12 ± 2%) and the majority of CD34+ SANLPCs did not express CD166. Taken together this data suggests that CD166 does not specifically mark human SAN pacemaker cells.
CD34+NKX2–5+ and CD34−NKX2–5− cells are SAN pacemaker cells
In addition to the CD34+NKX2–5− SAN pacemaker cells, we detected some CD34+NKX2–5+ and CD34−NKX2–5− cardiomyocytes in the fetal SAN (Fig. 4cIV) and hPSC-derived SANLPC cultures (Fig. 6a, b). Furthermore, sorting for CD34+ cells also isolated a small proportion of CD34+NKX2–5+ cells (Fig. 6e–i). To characterize the identity of both of these cardiomyocyte populations in more detail, we performed Cellular Indexing of Transcriptomes and Epitopes by sequencing (CITE-seq) on day 25 of SANLPC differentiation using a barcoded CD34 antibody (Supplementary Fig. 14a, b)35. This enabled us to read out protein-level CD34 expression in parallel to the whole transcriptome of the cells. Unsupervised clustering identified 11 cell clusters with the majority representing cardiomyocytes. Small clusters of fibroblasts, epithelial cells, and epicardial cells were also detected with a similar distribution as in our original hPSC dataset (Fig. 7a). Subclustering of TNNT2+ cardiomyocytes identified the same cardiomyocyte subtypes as before, including NKX2–5−TBX3high Core SAN cells, NKX2–5−TBX3mid Sinus Venosus cells, NKX2–5+NPPA+ Atrial cells, and Transition Zone cells that express both pacemaker and atrial genes (Fig. 7a, b, Supplementary Data 6). Signature scoring analysis using the top 200 DEGs of the fetal SAN dataset as a reference confirmed the identity of these cardiomyocyte subclusters (Fig. 7c).
a UMAP of the day 25 HES2-derived SANLPC CITE-seq dataset showing 11 cell clusters (left). Subclustering of the TNNT2+ cardiomyocytes showing 7 sub-clusters (right). b UMAPs of the subclustered cardiomyocytes showing the expression of the indicated genes. c UMAPs showing signature score distribution for the DEGs of the indicated fetal SAN cell types. d UMAPs showing the expression of CD34 on the transcript (RNA) or protein level (antibody-derived tag (ADT)) (left) and co-expression with NKX2–5 transcript (right). e Stacked bar graph quantifying the number of cells expressing CD34 by transcript (RNA) or protein (ADT) grouped based on NKX2–5 expression by transcript. f UMAPs showing the distribution of the selected cell populations indicated above. g Box plots showing the expression of the indicated genes on the transcript level and CD34 on the protein level (ADT) in the selected cell populations indicated below (CD34+NKX2–5+ n = 78; CD34+NKX2–5− n = 1465; CD34−NKX2–5− n = 2294; Atrial CD34−NKX2–5+ n = _97 single cells). h Spearman correlation between the selected cell populations indicated on the axes. p < 0.05 for all correlations (asymptotic t approximation). Blue labels indicate the data that is showing protein level (ADT)-based CD34 expression. Box plot elements: center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range. Source data are provided as a Source Data file.
We next analyzed the expression of CD34 at the transcript (RNA) and protein (antibody-derived tag, ADT) levels. A larger number of CD34+ cells was detected by ADT counts compared to transcript counts (1552 vs. 387) (Fig. 7d, e). Accordingly, we also identified a larger number of CD34+NKX2–5+ cells (85 vs. 24) when using the antibody-based detection of CD34+ cells. To assess the identity of the different cell populations, we isolated CD34+NKX2–5+, CD34−NKX2–5−, and CD34+NKX2–5− cells based on their expression of CD34 (ADT) and NKX2–5 (RNA) (Fig. 7f, Supplementary Fig. 14c, d). The majority of the CD34+NKX2–5− cells (62%) segregated into the Core SAN cell cluster. The distribution of NKX2–5+CD34+ and CD34−NKX2–5− cells was more scattered with about half of these cells (47% and 46% respectively) segregating into the Core SAN cell cluster.
To further characterize the CD34+NKX2–5+ and CD34−NKX2–5− cells we compared the expression of typical pacemaker and working myocyte markers in these cells with the CD34+NKX2–5− SAN pacemaker cells. We included CD34−NKX2–5+ myocytes from the Atrial cluster as an additional expression reference in this analysis. Both the CD34+NKX2–5+ and CD34−NKX2–5− cells expressed comparable levels of the SAN pacemaker genes TBX18, SHOX2, ISL1, HCN4, MYH11 and BMP4 like the CD34+NKX2–5− cells (Fig. 7g). Lower expression of these SAN genes was detected in the Atrial myocytes. Interestingly, expression of the pan-pacemaker marker TBX3 was high in the CD34+NKX2–5+ and CD34+NKX2–5−cells, but lower in the CD34−NKX2–5− cells. Expression of the atrial genes NPPA, SCN5A, GJA5, and KCNA5 was comparable between the CD34+NKX2–5+, CD34−NKX2–5−, and CD34+NKX2–5− cells and lower than in the Atrial myocytes. In addition, we performed Spearman correlation analysis using the DEGs of each of these cell populations, which showed that the CD34+NKX2–5+ and CD34−NKX2–5− cells most closely correlate with the CD34+NKX2–5− SAN pacemaker cells but not with the CD34−NKX2–5+ Atrial myocytes (Fig. 7h).
Collectively, these data indicate that the CD34+NKX2–5+ and CD34−NKX2–5− cells are transcriptionally comparable to the CD34+NKX2–5− SAN pacemaker cells and have a SAN pacemaker phenotype. The lower expression of TBX3 in the CD34−NKX2–5− cells suggests that they might be a SAN pacemaker progenitor population. This is consistent with the observation that the CD34− sorted NKX2–5− cardiomyocytes upregulate CD34 and TBX3 expression upon extended culture (Supplementary Fig. 10f, g and Supplementary Fig. 11a, b).
CD34+ cardiomyocytes have a functional pacemaker phenotype
To assess if the hPSC-derived CD34+ cardiomyocytes also show the electrophysiological characteristics of pacemaker cells we recorded optical action potentials (Fig. 8a). SIRPA+CD90−NKX2–5− FACS sorted SANLPCs, which we have previously shown to functionally represent pacemaker cells, were included as positive control14. In addition, ALCMs and VLCMs were included as reference cell types. We recorded fast spontaneous beating rates in MACS-sorted CD34+ cells and SANLPC controls that were significantly faster than beating rates of ALCMs and VLCMs14,17 (Fig. 8b). As expected for pacemaker cells, CD34+ cells and SANLPC controls displayed action potentials with significantly slower maximum upstroke velocities than ALCMs and VLCMs14,36. Additional parameters that distinguish pacemaker cells from working cardiomyocytes are their action potential durations14,27,36. Both CD34+ cells and SANLPC controls showed significantly shorter action potential durations at 30% (APD30) and 90% (APD90) repolarization compared to VLCMs. The APD30/90 ratios for CD34+ cells and SANLPC controls were significantly larger compared to ALCMs which clearly distinguishes them from ALCMs. Finally, we analyzed the diastolic depolarization during phase 4 of the action potential, which is driven by funny currents specifically present in pacemaker cells36,37. Accordingly, we observed pronounced diastolic depolarization in CD34+ cells and SANLPC controls that was significantly faster than in ALCMs and VLCMs. Of note, both CD34+ and CD34− cells, that were isolated from the SANLPC cultures, displayed pacemaker-like action potentials. This could be due to the fact that the samples were MACS sorted on day 25 when the CD34− population still contained a sizable fraction of NKX2–5− cardiomyocytes (41 ± 3%, Fig. 6h, i), in addition to the NKX2–5+ Atrial and Transition Zone cells. This is consistent with the idea that the CD34−NKX2–5− cardiomyocytes represent an immature pacemaker cell population that upregulates CD34 expression upon extended culture.
a Representative optical action potential traces of day 25 MACS sorted CD34+ and CD34− cells, FACS sorted SIRPA+CD90−NKX2–5− SANLPCs, as well as ALCMs and VLCMs generated from the HES3 NKX2–5eGFP/w hPSC line. b Optical action potential parameters analyzed in the indicated cell types (MACS pre-sort, n = 46; MACS CD34+, n = 47; MACS CD34−, n = 43; SANLPC, n = 57; ALCM, n = 79; VLCM, n = 51 cell aggregates from three biological replicates (independent differentiations) for each cell type). Statistical analysis was performed using Kruskal-Wallis test followed by Dunn’s post hoc test: *p < 0.05, **p < 0.01, ***p < 0.001 vs. indicated sample. c Schematic overview of the experimental design used to assess the pacemaker capacity of the CD34+ cardiomyocytes in vitro. d Representative bright field images (left) and color-coded activation maps (right) of a VLCM monolayer before and ~4 weeks after the addition of a VLCM control aggregate (n = 12 independent experiments from three biological replicates (independent differentiations)). e Representative bright field images (left) and color-coded activation maps (right) of a VLCM monolayer before and ~4 weeks after the addition of a MACS-sorted CD34+ cardiomyocyte (CM) aggregate (n = 10 independent experiments from three biological replicates (independent differentiations)). Scale bars, 1000 μm. Violin plot elements: center line, median; lower line, first quartile; upper line, third quartile. APD30/90, Action potential duration at 30%/90% repolarization. Source data are provided as a Source Data file. Schematics in (c) were generated using Biorender (https://biorender.com).
To further confirm the functional pacemaker phenotype of the CD34+ cells, we tested their capacity to pace other cardiomyocytes. For these experiments, monolayers of VLCMs were prepared (Fig. 8c). Once the VLCMs had formed an electrical syncytium, aggregates of CD34+ MACS sorted cardiomyocytes or control VLCMs were added. Using optical mapping, we observed that the CD34+ cell aggregate initiated the electrical activation and thereby paced the monolayers in 10 out of 14 of these co-cultures (Fig. 8d, e). In contrast, only 1 out of 13 VLCM aggregates was the source of electrical activation (P = 0.0013 Fisher’s exact test, CD34+ cardiomyocyte aggregates vs VLCM aggregates). In summary, our functional analysis clearly distinguished CD34+ cardiomyocytes from ALCMs and VLCMs and showed that they have a functional pacemaker phenotype comparable to NKX2–5− SANLPCs.
CD34 does not impact SANLPC differentiation and function
To assess whether CD34 is just a marker or whether it plays a role in SAN pacemaker differentiation and function, we used CRISPR-Cas9 to generate two pools of CD34 knockout (CD34 KO) hPSCs from the HES3 NKX2–5egfp/w reporter cell line (Supplementary Fig. 15a–d). To control for any potential gene editing-related effects, we generated control HES3 NKX2–5egfp/w hPSCs with edits in the AAVS1 safe harbor locus. CD34 knockout efficiency was assessed to be 96% (CD34 KO 1) and 24% (CD34 KO 2) using endothelial cell differentiations (Supplementary Fig. 15e). When we applied the SANLPC differentiation protocol, we observed a significant reduction in CD34+ myocytes for CD34 KO 1 but not CD34 KO 2, most likely due to the lower knockout efficiency of CD34 KO 2 (Fig. 9a, b, Supplementary Fig. 15f). Importantly, the proportion of NKX2–5− and NKX2–5+ cardiomyocytes was unchanged in both CD34 KO pools compared to AAVS1 Control-derived SANLPC differentiations. Further experiments were performed using the CD34 KO 1 hPSCs due to their superior knockout efficiency. RT-qPCR analysis on SIRPA+CD90−NKX2–5− sorted cardiomyocytes showed that the expression of key pacemaker genes (TBX3, SHOX2, HCN4, MYH11, BMP4) and atrial genes (SCN5A, NPPA) was unchanged in CD34 KO 1 cells (Fig. 9c). Functional assessment using optical action potential recordings showed a small decrease in action potential durations in CD34 KO 1 cells but no significant changes in key pacemaker characteristics including beating rates, maximum upstroke velocity, and diastolic depolarization compared to AAVS1 Control cells (Fig. 9d). The reduced action potential durations are most likely caused by the small, but not significant, increase in beating rates in the CD34 KO 1 cells and the fact that a faster beating rate results in shorting of the action potential38. Taken together, these data support the notion that CD34 marks SAN pacemaker cells but does not play a role in SAN pacemaker lineage specification and function.
a Flow cytometric analyses at day 25 of CD34 and NKX2–5:GFP expression in SIRPA+CD90− cardiomyocytes in AAVS1 Control, CD34 KO 1, and CD34 KO 2 SANLPC differentiations. b Bar graphs summarizing the percentage of NKX2–5− myocytes (left) and NKX2–5+ myocytes (right) in the indicated differentiation cultures (n = 3 independent differentiations). c RT-qPCR analysis of the expression of the indicated genes in SIRPA+CD90−NKX2–5− FACS sorted cells from day 25 AAVS1 Control and CD34 KO 1 differentiations (n = 3 independent differentiations). Statistical analysis was performed using two-sided unpaired t-test. Datasets that failed the normality test (MYH11) were analyzed using two-sided Mann-Whitney test: *p < 0.05 vs indicated sample. d Optical action potential parameters recorded in day 25 FACS sorted SIRPA+CD90−NKX2−5− cardiomyocytes from AAVS1 Control and CD34 KO 1 differentiations (AAVS1 Control, n = 48 (n1 = 16, n2 = 16, n3 = 16) and CD34 KO 1, n = 38 (n1 = 12, n2 = 13, n3 = 13) cell aggregates from three biological replicates (independent differentiations)). Statistical analysis was performed using two-sided unpaired t-test. Datasets that failed the normality test (APD90) were analyzed using two-sided Mann-Whitney test: *p < 0.05, **p < 0.01 vs indicated sample. Error bars represent SEM. Violin plot elements: center line, median; lower line, first quartile; upper line, third quartile. APD30/90, Action potential duration at 30%/90% repolarization, KO knockout. Source data are provided as a Source Data file.
Discussion
The SAN is the primary pacemaker of the heart that initiates every single heartbeat. SAN failure is treated with the implantation of an electronic pacemaker device, which is associated with multiple downsides. Despite the important function of the SAN, its cellular and molecular composition is not completely resolved. This limits our ability to develop novel treatment options for SAN diseases. To date, no surface markers for SAN pacemaker cells have been identified, which represents a major bottleneck for disease modeling and the advancement of biological pacemaker cell therapy approaches. To expand our knowledge on the cellular and molecular makeup of the human SAN, we performed snRNA-seq of human fetal SAN tissue and scRNA-seq of hPSC-derived SANLPCs. Our findings establish that the fetal SAN is composed of three distinct subtypes of pacemaker cells, including Core SAN, Sinus Venosus, and Transition Zone cells. Notably, the hPSC-derived cells closely resemble these fetal SAN pacemaker cells. Combining the fetal and hPSC-derived datasets allowed us to establish a shared list of Core SAN-specific genes. This list contains a host of SAN markers, most importantly, the cell surface antigen CD34. We provide evidence that CD34 is specifically expressed by SAN pacemaker cells and that sorting for CD34+ cells from hPSC differentiations enriches for SANLPCs with a functional pacemaker phenotype.
Previous studies of the human SAN describe a core pacemaker structure that expresses TBX3, TBX18, ISL1, HCN4 and lacks the expression of NKX2–53,4,5,6,7,12. In addition, a transitional structure, also known as the paranodal area, located at the border zone between the SAN and atrial tissues, has been described. This transitional structure expresses a mix of pacemaker and atrial myocyte genes. Consistent with this, we identified Core SAN pacemaker cells and Transition Zone cells in our fetal SAN and hPSC-derived SANLPC sn/scRNA-seq datasets. We also detected a second NKX2–5 negative myocyte cell cluster that expressed pacemaker genes TBX18, SHOX2, HCN4 but low levels of TBX3 and ISL1, resembling Sinus Venosus cardiomyocytes2,20,21,22. Sinus Venosus cardiomyocytes develop from the same TBX18+ progenitor population as the Core SAN at the venous pole of the heart. At early developmental stages, they contribute to the pacing of the heart until the Core SAN is fully established and takes over pacing2. The expression of key cardiac ion channels including pacemaker channels was comparable between fetal Core SAN and Sinus Venosus myocytes except for the higher expression of a few working cardiomyocyte ion channels in the Sinus Venosus cells. This is consistent with the literature suggesting that Sinus Venosus cardiomyocytes upregulate atrial genes at later developmental stages and eventually molecularly and functionally integrate with the atrial myocardium in the adult39. Accordingly, we did not identify a Sinus Venosus cell cluster when we subclustered the snRNA-seq data of the adult SAN published by Kanemaru et al12.
Studies of the SAN in the mouse heart identified an NKX2–5−TBX3+TBX18+ SAN head, located at the boundary between the superior vena cava and the right atria, and an NKX2–5+TBX3+TBX18− SAN tail that extends along the terminal crest towards the inferior vena cava7,9,40,41,42,43. In the human heart, a SAN head and tail can be anatomically defined6,44. However, a clear molecular distinction between a SAN head and SAN tail, like in the mouse, has not been shown. In agreement with that, we did not identify a SAN tail cell population in our fetal nor hPSC-derived datasets. Together our molecular analysis provides detailed insights into the composition of the developing human SAN. In addition, our study corroborates that hPSC differentiations can replicate SAN development including the development of the different subtypes of pacemaker cells.
To identify markers that can be used in vivo and in vitro to detect, target, and isolate the rare pacemaker cells of the Core SAN, we combined the fetal snRNA-seq and hPSC-derived scRNA-seq datasets. This analysis resulted in a list of 36 shared SAN marker genes. Amongst the most highly ranked markers was MYH11, the gene encoding for the smooth muscle-specific myosin heavy chain 11. MYH11 is expressed in smooth muscle cells of the digestive system, bladder, aorta, and coronary arteries of the heart. Previous transcriptomic studies have detected MYH11 in SAN myocytes4,8,10,18,22. We extended these findings to show that MYH11 protein is specifically expressed by hPSC-derived SANLPCs and fetal SAN pacemaker cells but not AVN, atrial, nor ventricular cardiomyocytes. Our analysis of the adult snRNA-seq data12 confirmed that specific expression of MYH11 is maintained in adult human SAN pacemaker cells. We and others also found that additional smooth muscle genes (ACTA2, MYLK) are specifically expressed in SAN myocytes4,8,10,18. It remains to be explored whether these genes play an important role in SAN physiology. As a second gene from our Core SAN marker list, we validated the expression of the TGF-β signaling ligand BMP4 specifically in SAN pacemaker cardiomyocytes. BMP signaling has previously been shown to play an important role during SAN development in both animal models as well as hPSC-differentiations, where BMP4 is used to induce the specification of SAN pacemaker cells from cardiogenic mesoderm4,8,14,22,45. Interestingly, our analysis of the adult snRNA-seq data12 showed that BMP4 is not expressed by adult human SAN pacemaker cells, suggesting that BMP4 is not relevant for the maintenance of the SAN phenotype.
The marker list further contained several genes involved in calcium binding and calcium channel regulation (ANXA2, NECAB1, SPOCK1, RRAD, PRKG1). This is consistent with the well-established role of calcium ions in the regulation of the spontaneous action potential firing of the SAN (Ca2+-clock)46. In addition, we identified several genes commonly expressed in neurons as Core SAN markers (BASP1, PPFIA2, EFR3B, SLIT2, LYPD6). These genes play a role in axon growth and guidance which is in keeping with the finding that Neuron Projection Guidance/Development were identified as top GO terms for the fetal/hPSC-derived Core SAN cells. The SAN tissue is highly innervated by the autonomic nervous system which led to the detection of neuronal genes in past bulk RNA-seq experiments4,8. Here we analyzed gene expression on the single-cell level within TNNT2+ cardiomyocytes making it highly unlikely that gene expression from neuronal cells was detected. Similarly, multiple neuronal genes were detected in SAN pacemaker cells in recent sc/snRNA-Seq studies of the mouse and human SAN9,10,12,47. Collectively, these findings suggest that SAN pacemaker cells express a subset of neuronal genes, potentially utilize comparable transcriptional and signaling networks as neurons, and actively participate in guiding the axons of the innervating autonomic nervous system. These are interesting concepts that need to be further explored in the future.
In addition to the above set of genes we also identified a number of cell surface markers that were specifically expressed in Core SAN pacemaker cells, including CD34, CADM1, EFNB2, NTRK2, and ELAPOR2. We focused our analysis on the single-pass transmembrane glycoprotein CD34, because of its routine use as surface marker for cell isolations. Classically, CD34 is known as a selection marker for hematopoietic stem and progenitor cells, as well as for its expression in endothelial cells48,49,50. In recent years, CD34 has also been described to mark tissue-resident progenitors such as muscle satellite cells and mesenchymal stem cells51. Additionally, there have been reports of CD34 expression in telocytes, an interstitial cell type that can be found throughout the body, including the heart29. Here we extend these findings and show expression of CD34 in human fetal and adult SAN pacemaker myocytes and in hPSC-derived SAN pacemaker cells.
Interestingly, CD34 labeled a subset of NKX2–5+ cells in the hPSC differentiation cultures and in the fetal SAN tissue. This contradicts the notion that SAN pacemaker cells lack NKX2–5 expression2,3,14,15. Recent single nuclei transcriptome analysis of the adult human SAN also showed that a subset of SAN pacemaker cells do express NKX2–512. Furthermore, our CITE-Seq analysis confirmed that the CD34+NKX2–5+ cells have a SAN pacemaker phenotype, rather than an Atrial or Transition Zone phenotype. Taken together, this data suggests that a subset of NKX2–5+ SAN pacemaker cells do exist in the human SAN.
Although CD34 is a well-established marker, especially in the hematopoietic system, its functional role is not well-established52. Here we applied a CRISPR-Cas9 knockout approach to test for the functional role of CD34 in hPSC-derived SAN-like pacemaker cells. We did not find any effect of the CD34 knockout on SAN lineage specification or key pacemaker action potential parameters. Interestingly, multiple genome-wide association studies have implicated genetic variants in the CD34 gene in heart rate changes53,54,55, suggesting that CD34 might regulate SAN function in vivo. Assessing CD34 function in vivo is complicated by the fact that it is not expressed by mouse SAN pacemaker cells. In the hematopoietic system, CD34 has been implicated in cell migration and adhesion via binding of selectins by its extracellular mucin domain52. Further studies are needed to address whether CD34 expression on developing SAN cells regulates their migratory behavior and overall SAN tissue organization and thereby affects SAN function.
We used FACS and MACS sorting for CD34+ cells to demonstrate that this approach enables transgene-independent and straightforward isolation of SAN-like pacemaker cells from multiple hPSC lines. Our analysis showed that these CD34+ sorted cells display molecular and functional properties of SAN pacemaker cells. This SAN cell surface marker will be highly valuable for future SAN-specific disease modeling and drug screening projects using patient-derived iPSCs, to gain a better understanding of the mechanisms of sinus node diseases and to develop novel therapies. It will further enable the enrichment of SAN pacemaker cells for toxicology and safety pharmacology screens. From a cell therapy perspective, MACS will allow efficient isolation of the large numbers of CD34+ SAN pacemaker cells from hPSC-differentiation cultures that will be required for future biological pacemaker applications14,56. Conversely, MACS-based depletion of CD34+ cells can be used to remove unwanted pacemaker cells from populations of hPSC-derived ventricular cardiomyocytes. This could reduce the risk of unwanted arrhythmias in cell replacement therapies for patients who suffered a myocardial infarction57,58. Beyond that, CD34-specific antibodies conjugated to a drug or other therapeutic cargo could be used in the future to specifically deliver therapeutics to the SAN in patients with too slow or too fast sinus rhythm26,59. To avoid cross-reaction to other CD34+ cells in the body, bispecific antibodies that target both CD34 and a myocyte-specific epitope such as SIRPA or HCN4 could be employed60.
When we compared our human Core SAN marker list to previously published mouse scRNA-seq data9,26, we found that only half of the markers were conserved. Specifically, CD34 expression was not detected in mouse SAN pacemaker cells. Likewise, the majority of the previously reported mouse SAN marker genes were not specifically expressed by human SAN pacemaker cells. These species differences are important when choosing markers for the characterization and isolation of hPSC-derived SAN-like pacemaker cells as well as for the clinical translation of SAN cell surface marker-based therapeutics. Hence, the insights that our study of the human SAN provides will be a valuable resource for the scientific community.
In summary, our study provides a transcriptomic profile of the developing human SAN at single-cell resolution. Using this data, we showed that hPSC-derived SANLPCs closely resemble the SAN pacemaker cells in the fetal heart and identified a shared set of 36 markers for the identification of SAN pacemaker cells in vitro and in vivo. Most notably this maker list contains the cell surface antigen CD34. We show that CD34 is specifically expressed by human SAN pacemaker cardiomyocytes and that it can be used to isolate SANLPCs from hPSC differentiation cultures. Our findings have important implications for future iPSC-based SAN disease modeling studies as well as cell therapy approaches using hPSC-derived biological pacemakers. The markers identified in our study will also advance our ability to specifically deliver therapeutics to SAN cells in vivo, using antibody-based approaches. Taken together, our findings will contribute to a better understanding of SAN diseases and to improved future treatment options for patients.
Methods
The research presented in the article complies with all relevant ethical regulations and has been approved by the Research Ethics Board of the University Health Network (UHN), Toronto.
Mice
Wild-type, P0-3 CD1 mice were obtained from the Animal Resource Centre at UHN. All animal work was performed in accordance with the Animal Use and Care Committee at UHN. Animals were housed under standard conditions (22 degrees, 45–60% humidity, and 12 h light/dark cycle, Teklad Irradiated LM-485 Mouse/Rat diet) at UHN’s Animal Resources Centre. Neonatal mice were euthanized by cervical dislocation. Both female and male mice were used.
Human fetal heart samples
Human fetal heart samples gestation week 17–20 were obtained from the Mount Sinai Hospital Research Centre for Women’s and Infants’ Health BioBank, Toronto. Work with the human fetal tissue was approved by the Research Ethics Boards of the Mount Sinai Hospital, Toronto, and the University Health Network (UHN), Toronto. Samples were collected from voluntary pregnancy terminations. After patients did sign the standard consent form for the procedure, they were approached by the Mount Sinai Hospital Research Centre for Women’s and Infants’ Health BioBank with the request to consent for the donation of the fetal tissue. Donors were informed about the use of samples for research purposes and do not receive compensation for donated material. To ensure sampling from healthy heart samples, fetuses with chromosomal abnormalities or cardiovascular defects as well as fetuses from mothers with congenital heart defects, arrhythmias, diabetes (type 1, 2 or gestational), or Systemic Lupus Erythematosus were excluded from this study. The fetal sex was not specified for this study and not included in the health information shared with the study team.
hPSC lines
Human pluripotent stem cell lines HES3-Nkx2–5egfp/w (karyotype: 46, XX)31, and HES2 (karyotype: 46, XX)61 were used in this study. Work with hPSCs was approved by the Stem Cell Oversight Committee of the Canadian Institutes of Health Research.
Generation of CD34 knockout (CD34 KO) hPSC lines
Single guide RNA (sgRNA) targeting the AAVS1 (intron 1) or CD34 locus (exon 1 or 5, Supplementary Table 2) were cloned into the lentiCRISPRv2 transfer vector (gift from Prof. Feng Zhang, Addgene plasmid #52961) via BsmBI restriction enzyme cloning (NEB, R0739S)62. All plasmids were propagated in the Stbl3 E. coli strain (ThermoFisher, C737303), and transfection-quality plasmid DNA was obtained using the QIAprep Spin Miniprep Kit (QIAGEN, 27104). HEK293T packaging cells were co-transfected with the lentiCRISPRv2, pMD2.G (gift from Prof. Didier Trono, Addgene plasmid #12259) and psPAX2 (gift from Prof. Didier Trono, Addgene plasmid #12260) plasmids in a 1:1:1 molar ratio using Opti-MEM (ThermoFisher, 31985070) and X-tremeGENE 9 (MilliporeSigma, 6365779001) in standard HEK293T growth medium consisting of DMEM high glucose (ThermoFisher, 11995065) + 10% FBS (FisherScientific, SH3039603) + 0.1% Pen/Strep (ThermoFisher, 15070-063), and lentiviral particles were harvested 48 h and 72 h later in high-BSA medium consisting of DMEM high glucose + 100 g/L BSA (Sigma, A1470) + 1% Pen/Strep63. To generate control AAVS1-KO and CD34-KO hPSC pools, HES3 NKX2–5egfp/w hPSCs were transduced in suspension at MOI > 1 and seeded onto DR4 multidrug-resistant MEFs (ThermoFisher, A34966). 24 h later, 0.5 μg/mL puromycin (ThermoFisher, A1113803) was applied for 72 h to select for transduced cells, and surviving cells were passaged 3 times before cryopreservation. Pluripotency was confirmed by flow cytometry as described below using anti-OCT4 and anti-SOX2 antibodies. Knockout efficiency was measured using Synthego’s ICE tool to analyze Sanger sequencing traces of PCR amplicons spanning the sgRNA cut sites (Supplementary Table 3). Genomic DNA was isolated using the DNeasy Blood & Tissue Kit (QIAGEN, 69504), PCRs were performed using Q5 High-fidelity DNA Polymerase (NEB, M0491S), and purified using the QIAquick PCR Purification Kit (QIAGEN, 28104) before submission for sequencing with The Centre for Applied Genomics (TCAG) at SickKids.
Maintenance and directed differentiation of hPSCs
The human pluripotent stem cell lines were maintained on irradiated mouse embryonic fibroblasts in DMEMF/12 (ThermoFisher, MT-10-092-CV) supplemented with 20% KnockOut serum replacement (ThermoFisher, 10828028), 2 mM L-glutamine (ThermoFisher, 25030081), 1x non-essential amino acids (ThermoFisher, 11140050), 55 μM β-Mercaptoethanol (ThermoFisher, 21985023) and 1% penicillin/streptomycin (ThermoFisher, 15070-063), and 20 ng/ml recombinant human (rh)-bFGF (Biotechne, 233-FB/CF) as described previously64. All cell lines used in this study had a normal karyotype and tested negative for mycoplasma contamination.
Cardiomyocyte differentiations
For directed differentiations into the different cardiomyocyte subtypes, we used our previously reported embryoid body (EB)-based protocols with the following modifications14,27. StemPro-34 Media (ThermoFisher, 10639011) supplemented with 1% penicillin/streptomycin, 2 mM L-glutamine, 50 μg/mL ascorbic acid (MilliporeSigma, A4544), 75 μg/mL transferrin (MilliporeSigma, 10652202001) and 50 μg/mL monothioglycerol (MilliporeSigma, M6145) was used as a base media for all differentiation steps. hPSCs grown to ~80% confluence were dissociated into single cells using TrypLE (ThermoFisher, 12605010). Cells were suspended in base media containing 20% StemPro-34 diluted with IMDM (ThermoFisher, 12440061) supplemented with 0.01x ITSX (ThermoFisher, 51500056), 1 ng/mL rhBMP4 (Biotechne, 314-BP/CF), 10 μM ROCK inhibitor Y27632 (Selleckchem, S1049), and 250 U/ml DNase I (MilliporeSigma, 260913) at a concentration of 1 × 106 cells/mL and cultured for 18 h on an orbital shaker at 80 rpm in low attachment 6 cm dishes for the formation of EBs. EBs were cultured in a low oxygen environment (5% CO2, 5% O2, 90% N2) until day 11 and then in a 5% CO2 ambient air environment for the remainder of the culture period. Tissue culture plastic was pre-coated with 5% (w/v) poly-HEMA (MilliporeSigma, P3932) to generate low-adherence plates for the maintenance of EBs in suspension culture.
Ventricular-like cardiomyocyte (VLCM) differentiation
At day 1 of differentiation the EBs were moved into ventricular mesoderm induction media consisting of base media containing 100% StempPro-34 supplemented with 1 μM CHIR99021 (Biotechne, 4423), 2.5 ng/ml rhbFGF, 10 ng/ml rhBMP4, and 8 ng/ml rhActivinA (Biotechne, 338-AC/CF). At day 3 the EBs were washed once with IMDM and transferred to cardiac induction media that consisted of base media containing 100% StemPro-34 supplemented with 1 uM IWP2 (Biotechne, 3533) and 10 ng/ml rhVEGF (Biotechne, 293-VE/CF). At day 5 media was changed to cardiac maintenance media that consisted of base media containing 20% StemPro-34 diluted with IMDM supplemented with 5 ng/ml rhVEGF. Media was changed every 3–4 days and starting at day 11 EBs were cultured in maintenance media without rhVEGF.
Atrial-like cardiomyocyte (ALCM) differentiation
At day 1 of differentiation the EBs were moved into atrial mesoderm induction media consisting of base media containing 100% StempPro-34 supplemented with 1 μM CHIR99021 (Biotechne, 4423), 2.5 ng/ml rhbFGF, 4 ng/ml rhBMP4, and 3 ng/ml rhActivinA. At day 3 the EBs were washed once with IMDM and transferred to cardiac induction media as above (VLCM differentiation) that was additionally supplemented with 0.5uM retinoic acid (MilliporeSigma, R2625). From day 5 onwards the differentiation was continued as described for VLCMs.
Sinoatrial Node-like pacemaker cell (SANLPC) differentiation
At day 1 of differentiation the EBs were moved into pacemaker mesoderm induction media consisting of base media containing 100% StempPro-34 supplemented with 1 μM CHIR99021 (Biotechne, 4423), 2.5 ng/ml rhbFGF, 4 ng/ml rhBMP4, and 3 ng/ml rhActivinA. At day 3 the EBs were washed once with IMDM and transferred to pacemaker induction media that consisted of base media containing 100% StemPro-34 supplemented with 0.5uM IWP2 (Biotechne, 3533), 10 ng/ml rhVEGF, 2.5 ng/ml rhBMP4, 1.5 uM SB-431542 (MilliporeSigma, S4317), and 0.125 uM retinoic acid. 500–1000 nM PD173074 (Biotechne, 3044) were added to the media on either day 3 (HES3-Nkx2–5egfp/w line) or day 4 (HES2 cell line). At day 6 media was changed to base media containing 100% StemPro-34 supplemented with 5 ng/ml rhVEGF. At day 8 media was changed to cardiac maintenance media and differentiations were continued as described above for VLCMs.
Endothelial cell differentiation
To assess the CD34 knockout efficiency CD34 KO hPSCs were differentiated into endothelial cells using the following protocol. hPSCs were differentiated into ventricular mesoderm as described above. At day 3 EBs are dissociated using TrypLE for 3 min at 37 °C and plated in 24 well plates coated with Matrigel (25% v/v, FisherScientific, CB40230) at 400k cells per well. The monolayers were cultured in base media containing 100% StemPro-34 supplemented with 30 ng/ml rhbFGF, 30 ng/ml rhVEGF, and 9uM SB-431542 from day 3 to day 9. Media was changed every 2 days.
Flow cytometry and cell sorting
Day 6–8 EBs were dissociated using TrypLE for 3–6 min at 37 °C. Day 10–50 EBs were dissociated using Collagenase type 2 (1 mg/ml, Worthington, 4176) in HANKs buffer for 16 h at room temperature, followed by treatment with TrypLE as above if further dissociation was required. Endothelial monolayers were dissociated using TrypLE for 5 min at 37 °C. The cells were stained using the following antibodies: anti-CD172a/b-PE-Cy7 (SIRPa) (Biolegend, 323808, 1:1000), anti-CD90-BV421 (BD, 562556, 1:300), anti-CD31-PE (BD, 555446, 1:50), anti-CD34-APC (BD, 340441, 1:300), anti-CD34-PE-Cy7 (ThermoFisher, 25-0349-42, 1:100), anti-CD166 (BD, 560903, 1:100), anti-Cardiac Troponin T (cTNT) (ThermoFisher, MA5-12960, 1:2000), anti-NKX2–5 (Cell Signaling, 8792S, 1:1000), anti-OCT4-PE (BD, 561556, 1:300), anti-SOX2-PerCP-Cy5.5 (BD, 561506, 1:100). To detect unconjugated primary antibodies the following secondary antibodies were used: goat anti-mouse IgG-PE (Jackson ImmunoResearch, 115-115-164, 1:500), donkey anti-rabbit IgG-A647 (ThermoFisher, A31573, 1:500)
For cell-surface markers, cells were stained PBS containing 5% FCS (FisherScientific, SH3039603) and 0.02% sodium azide (Sigma, S2002) for 30 min at 4 °C. For intracellular staining (cTNT, NKX2–5, OCT4, and SOX2), cells were fixed using 4% PFA for 10 min at 4 °C and stained in PBS containing 0.3% BSA (Sigma, A1470), 0.3% Triton X-100 (Sigma, X100) overnight at 4 °C. Stained cells were analyzed using the LSRFortessa flow cytometer (BD) or Cytoflex flow cytometer (Beckman Coulter). For FACS, the cells were kept in IMDM containing 0.5% FCS and sorted at a concentration of 10 million cells/ml using a FACSAria (BD) sorter at the SickKids-UHN Flow Cytometer Facility. For MACS, the PSC-Derived Cardiomyocyte Isolation Kit (Miltenyi, 130-110-188) and the CD34 MicroBead Kit (Miltenyi, 130-046-702) were used according to the manufacturer’s instructions. All data was analyzed using FlowJo software (BD). The gating strategies used for data analysis are shown in Supplementary Fig. 16.
Immunohistochemistry
For cultured cells, day25 EBs were dissociated, and MACS sorted using the PSC-Derived Cardiomyocyte Isolation Kit to enrich for cardiomyocytes as described above. Cells were plated on 12 mm cover glasses (FisherScientific, 22293232 P) precoated with Matrigel (25% v/v, FisherScientific, CB40230) and cultured for 3–5 days until confluent monolayers were obtained. Cells were fixed with 4% PFA in PBS for 10 min at 4 °C and permeabilized using PBS containing 0.3% Triton X-100, 200 mM glycerin (MilliporeSigma, G2289) for 20 min at room temperature (RT). Samples were blocked with PBS containing 10% donkey serum, 2% BSA, and 0.3% Triton X-100 (blocking buffer) for 30 min at RT. Primary antibodies were incubated in PBS containing 1% donkey serum and 0.3% Triton X-100 (staining buffer) overnight at 4 °C. The samples were washed three times with PBS containing 0.1% BSA, 0.1% Triton X-100 (wash buffer), before applying secondary antibodies together with 10ug/ml Hoechst (FisherScientific, H3570) in staining buffer for 1 h at RT. The samples were washed three times with wash buffer before mounting with ProLong Diamond Antifade Mounting Media (FisherScientific, P36965). Note, staining of cells for CD34 was performed on fresh, unfixed cultures in PBS and 5% FCS using the anti-CD34-APC antibody (BD, 340441, 1:300) together with a donkey anti-mouse IgG-A555 (ThermoFisher, A31570, 1:500) secondary antibody.
To prepare tissue sections of human gestation week 17–20 SAN and AVN we followed a previously described dissection protocol (Supplementary Fig. 6a)28. Briefly, the hearts were placed with the anterior side facing up and the ventricular chambers were dissected away leaving a small rim of ventricular tissue apical to the mitral and tricuspid valves. Next, the right atria was opened by cutting through the tricuspid valve towards the superior vena cava on the anterior side of the heart. Next, the atrial free wall was cut open, flattened, and pinned down revealing the view of the crista terminalis, the SAN, and the AVN. Sections were prepared from the posterior (epicedial) side of the dissected tissue. To obtain tissue sections of P0-3 mouse SAN whole hearts were isolated, and cross sections were prepared from the base of the hearts.
All tissue preparations were fixed overnight in 4% formalin, transferred to 70% ethanol, and sent for paraffin embedding and preparation of 5um thick sections at the Pathology Research Program at UHN. Sections were re-hydrated using decreasing xylene/ethanol solutions before heat-induced antigen retrieval in citrate buffer (pH 6). Sections were blocked in PBS containing 0.5% donkey serum and incubated with primary antibodies overnight at 4 °C. The samples were washed three times with PBS before applying secondary antibodies together with 10ug/ml DAPI (FisherScientific, D1306) in blocking buffer for 1 h at RT. The samples were washed three times with PBS and once with distilled water before mounting with ProLong Diamond Antifade Mounting Media.
The following primary antibodies were used: anti-BMP4 (Abcam, ab124715, 1:50), anti-Cardiac Troponin T (cTNT)-FITC (Miltenyi, 130-119-575, 1:100), anti-Cardiac Troponin T (cTNT) (ThermoFisher, MA5-12960, 1:200), anti-CD31 (DAKO, M0823, 1:50), anti-CD34 (Abcam, ab81289, 1:300), anti-CD34 (BD, 340441 1:100), anti-CD34 (Abcam, ab8536 1:100), anti-GFP (Abcam, ab13970, 1:500), anti-HCN4 (Antibodies Inc, 75–150, 1:100), anti-MLC2V (Abcam, ab79935, 1:100), anti-MSX2 (Abcam, ab223692, 1:300), anti-MYH11 (Abcam, ab224804, 1:100), anti-MYH11 (Sigma, M7786 1:100), anti-NKX2–5 (Cell Signaling, 8792S, 1:1000), anti-NPPA (Abcam, ab209232, 1:300), anti-SHOX2 (Abcam, ab55740, 1:300), and anti-TBX3 (ThermoFisher, 424800, 1:100). The following secondary antibodies were used: Donkey anti-rabbit IgG-A647 (ThermoFisher, A31573, 1:500), Donkey anti-rabbit IgG-A555 (ThermoFisher, A31572, 1:500), Donkey anti-mouse IgG-A647 (ThermoFisher, A31571, 1:500), Donkey anti-mouse IgG-A555 (ThermoFisher, A31570, 1:500), and Donkey anti-chicken IgY-A488 (ThermoFisher, A78948, 1:500). Masson’s Trichrome staining was performed by the UHN Pathology Research Program.
All samples were imaged at UHN’s Advanced Optical Microscopy Facility using the Nikon A1R confocal microscope and associated NIS-Elements software (Nikon). Images were denoised using Nikon’s denoise.ai. Whole-slide images were scanned using the 20X objective lens on the Scan-scope AT2 brightfield scanner (Aperio). Images were processed and prepared using ImageJ (FIJI)65.
Optical action potential recordings
All cell types analyzed by optical action potential recording were generated from the HES3 NKX2–5eGFP/w hPSC line. Day 25 EBs were dissociated into single cells as described above and re-aggregated into smaller aggregates (~50–100 μm diameter) by plating into 96 well ultra-low attachment plates (FisherScientific, 07-200-603) at 200,000 cells/well. To obtain enriched populations of SANLPCs SIRPA+CD90−NKX2–5− cells were isolated by FACS. CD34+ and CD34− cells were isolated from SANLPC differentiation cultures by MACS. Cells were allowed to recover for 3–5 days and stained at 37 °C, 5% CO2 in salt-adjusted IMDM (final concentrations in mmol/l: 137 NaCl, 4.4 KCl, 1.5 CaCl2, 25 HEPES, and 25 D-glucose) (ThermoFisher, 12440061) using the FluoVolt Membrane Potential Kit (ThermoFisher, F10488) according to the manufacturer’s instructions. Imaging of optical action potentials was performed in salt-adjusted IMDM at 37 °C using either a pco.edge 4.2Q sCMOS camera mounted on a Nikon Ti2-E inverted microscope with a CFI60 10X objective or a Photometrics Evolve 128 EMCCD camera mounted on an Olympus IX71 inverted microscope with a 20x objective. Recordings of at least 10 s were acquired at 400 frames/second using NIS-Elements (Nikon) or Metamorph (Molecular Devices) software. Optical action potential characteristics (frequency, maximal upstroke velocity, and action potential duration) were analyzed using custom Python scripts and standard techniques previously described66,67. Briefly, signals were pre-processed to correct for staining-related artifacts, and filtered to remove high-frequency noise, using a low-pass Butterworth filter with a cut-off frequency of 100 Hz. Recordings were segmented into individual action potentials that were then normalized in intensity and aligned for ensemble averaging (n ≥ 5 action potentials were averaged per recording). To determine the diastolic depolarization, the maxima of the second derivative of an over-filtered representation of the original signal was used, to detect the point of transition from the action potential phase 4 depolarization to action potential phase 0 depolarization. The phase 4 portion of the action potential was extracted, and linear regression analysis was used to determine the slope of depolarization.
In vitro assessment of pacemaker capacity
Day 25 VLCM EBs were dissociated into single cells as described above and replated into 24 well plate wells. Cultures were incubated for 3–10 days to form an electrical syncytium and analyzed by optical mapping. For optical mapping the same experimental conditions as described above for optical action potential recordings were applied using a CFI60 4X objective. To assess the electrical activation patterns in the monolayers color coded activation maps were generated using a procedure similar to previous implementations67. Recordings were segmented into repeating temporal units corresponding to individual cardiac cycles. Within a cycle, recordings were binned into 3 × 3 pixel windows, and processed using baseline subtraction, signal filtering, and action potential normalization. The time of half activation was determined within each individual pixel bin. These half-activation times were used to generate smoothened activation maps, with the first-pixel bin to reach half-activation denoted 0 ms activation time, and all other depolarizations scaled referentially. After the baseline electrical activation pattern of the monolayers was recorded, large aggregates (~300–500 μm diameter) of either control VLCMs or MACS-sorted CD34+ cardiomyocytes were seeded on top of the monolayer. After ~4 weeks these co-cultures were analyzed again by optical mapping to assess the electrical activation patterns and whether the cell aggregates are pacing the VLCM monolayers.
Quantitative reverse transcription PCR
Total RNA from hPSC-derived cardiomyocytes was isolated using the RNAqueous-Micro Total RNA isolation kit including RNase-free DNase treatment (ThermoFisher, AM1931). cDNA was prepared by reverse transcribing the isolated RNA using oligo(dT) primers and random hexamers with Superscript III Reverse Transcriptase (ThermoFisher, 18080044). RT-qPCRs were performed using the QuantiNova SYBR Green PCR kit (Qiagen, 208057) according to the manufacturer’s instructions on the QuantStudio5 RT-qPCR machine (ThermoFisher). Each experiment included a tenfold dilution series ranging from 25 ng/mL to 2.5 pg/mL of human genomic DNA standards to evaluate PCR efficiency and to calculate the copy number of each gene relative to the housekeeping gene TBP as described previously32. Primer sequences are listed in Supplementary Table 3.
Sample processing for single-nucleus RNA sequencing
Single-nucleus RNA-seq was performed on a gestation week 19 fetal heart. To capture the tissue containing the SAN, we dissected the heart and isolated the tissue at the boundary of the right atria with the superior vena cava (Fig. 1a). The tissue was immediately frozen using liquid nitrogen and stored at −80 °C. The frozen tissue was retrieved and dissociated using lysis buffer containing: 0.32 M sucrose (Sigma, S0389), 5 mM CaCl2 (Fluka, 21114), 3 mM Mg(AC)2 (Sigma, 228648), 20 mM Tris-HCl (ThermoFisher, 15567-027), 0.1%Triton X-100 (MilliporeSigma, X100), 0.1 mM EDTA (ThermoFisher, AM9260G) in DNase/RNase free water (Qiagen, 129115). A douncer was used to mechanically homogenize the tissue until single nuclei were obtained. Homogenate was washed twice with PBS containing 1% BSA and 0.2 U/ul RNAseOUT (ThermoFisher, 10777019) and filtered through a 40 μm cell strainer. Nuclei were stained with DAPI (10μg/ml) and sorted for DAPI+ single-nuclei on a FACSAria (BD) sorter at the SickKids-UHN Flow Cytometer Facility.
Sample processing for single-cell RNA sequencing and CITE sequencing
Single-cell RNA-seq was performed on day 25 SANLPC differentiation cultures of the HES2 cell line (Fig. 2a). The EB’s were dissociated and stained with DAPI as described above before being sorted for DAPI− single live cells using a FACSAria (BD) sorter at the SickKids-UHN Flow Cytometer Facility. For CITE-seq, day 25 SANLPC differentiation cultures of the HES2 cell line were processed as described above. Before sorting, samples were stained with DAPI, and the hCD34 TotalSeqTM-B0054 antibody (Biolegend, 343539) and DAPI− single cells were collected as detailed above. The optimal concentration of the hCD34 TotalSeqTM antibody was determined using flow cytometric analysis with an anti-mouse AF647 secondary antibody (ThermoFisher, A-31571). The hCD34 TotalSeqTM antibody concentration that matched to the proportion of CD34+ seen when using the anti-CD34 APC antibody (BD, 340441 1:300) was chosen for the CITE-Seq experiment (Supplementary Fig. 14a).
Single-cell/nuclei RNA sequencing, raw data processing, quality control, clustering, and differential gene expression analysis
Single-cell and single-nuclei suspensions were processed for sequencing on the 10x Genomics platform using the Chromium Single Cell 3’ v3 reagent kit. Single-cell/nuclei libraries were sequenced on the Illumina NovaSeq 6000 with a sequencing depth of > 45,000 reads per cell. Raw data processing was performed with the 10x Genomics CellRanger pipeline by UHN’s Princess Margaret Genomics Centre. Single-cell RNA-seq data and single-nuclei RNA-seq data were mapped to the human reference Genome GRCh38 and GRCh38-premrna respectively. Downstream data analysis was performed in R using the Seurat toolkit (version 4.0.2, https://satijalab.org/seurat/)68. First, data was filtered to remove low-quality cells and potential doublets by removing cells with low library size (<1500) and high library size (>50,000). In addition, a cut-off for mitochondrial gene expression was applied to remove damaged cells (fetal snRNA seq: 1%, hPSC scRNA-seq: 31.512%, hPSC CITE-seq 21%). Data was normalized using ScTransform, principal components analysis was performed using RunPCA, and the top 25 principal components were used for clustering into distinct cell clusters (FindClusters function). The scClustViz tool69 was used to determine optimal cluster resolution by increasing resolution until the minimal number of differentially expressed genes between each cluster reached 0–10. The following final cluster resolutions were used in this study: fetal snRNA-seq: 0.8, hPSC scRNA-seq: 0.8, hPSC CITE-seq: 0.4. The FindAllMarkers function (only.pos = TRUE, min.pct = 0.1, logfc.threshold = 0.25, adjusted p-value < 0.05) was used to identify differentially expressed genes (DEGs) within each cluster or between subsets of selected cells. The top DEGs were identified after sorting the gene lists based on average log-fold expression changes. The genes used to identify which cell type each cluster represents are listed in Supplementary Table 1. To generate datasets that only contain cardiomyocytes, cell clusters expressing TNNT2 were selected for subclustering. The genes used to identify the cardiomyocyte subtype clusters are discussed in the main text. Uniform manifold approximation and projection (UMAP) was used for dimensionality reduction to visualize the data in two dimensions. To read out CD34 expression on the protein level in the CITE-seq dataset, raw counts from the antibody-derived tag (ADT) were normalized with NormalizeData function (method = CLR), and a cut-off of < 0.84 was set to match CD34 positive expression in flow cytometric data (Supplementary Fig. 14). For selection of cells by RNA expression, a cut off of >0 was used for both NKX2–5 and CD34.
Gene signature score and gene ontology analysis
Gene signature scores were obtained using UCell23 for the top 200 positive differentially expressed genes from the comparison groups. Signature score distributions were visualized using UMAPs. Gene ontology analyses were performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) Bioinformatics Resources (version Dec. 2021, https://david.ncifcrf.gov/summary.jsp). Analysis was focused on biological processes (GOTERM_BP_all).
Harmony data integration of the fetal snRNA-seq and hPSC scRNA-seq datasets
The fetal and hPSCs datasets were normalized and clustered using Seurat. Each dataset was normalization using ScTransform. Next, both samples were scaled separately as it has been shown to facilitate integration of single-cell and single-nucleus RNA-seq datasets in prior studies70. The two datasets were then concatenated to generate a merged gene expression matrix. The merged dataset was scaled and principal component analysis was applied to the data for dimension reduction. We then applied Harmony (version 1.0) integration on the principal components to remove the technical batch variations. UMAP was next applied to Harmony-adjusted top components for visualization.
Spearman correlation analysis
To compare the cell types in the fetal and hPSC datasets correlation analysis was performed. The list of DEGs for each fetal cluster was sorted based on fold change and the top 200 genes were selected as the feature set for the correlation analysis. We then restricted the comparison to cell clusters of interest from each dataset (fetal: Core SAN, SAN, Transition Zone, Atrial, Epithelial cells, Epicardial cells, Fibroblasts; hPSC: Core SAN, SAN, Transition Zone, Atrial, Epithelial cells, Epicardial cells, Fibroblasts). Duplicate features and the genes not detected in the hPSC sample were removed. The resulting genes were used to calculate Spearman correlation between the fetal and hPSC clusters (Fig. 2k). To compare the CD34+ NKX2–5+, CD34+NKX2–5−, CD34−NKX2–5−, and CD34−NKX2–5+ Atrial cells in the CITE-Seq dataset a similar correlation analysis was performed with the following modifications. The positive DEGs from each of the selected cell types were selected as the feature set for the correlation analysis. The resulting genes were used to calculate Spearman correlation between the selected cell types (Fig. 7h).
Comparison of adult human and fetal human datasets
The raw data of the single-nuclei RNA-seq of the adult SAN deposited by Kanemaru et al.12 was downloaded and analyzed using Seurat with the same settings as described in the original publication. Their original annotations labeled four clusters as atrial and one cluster as SAN pacemaker cells. We further subclustered the SAN pacemaker cells into four cardiomyocyte subclusters: Core SAN 1, Core SAN 2, Transition Zone, and Atrial. For further analysis, cells from Core SAN 1 and Core SAN2 were combined and compared against all atrial cells in the original dataset. DEGs were determined using FindAllMarkers function (min.pct = 0, logfc.threshold = 0) and compared against the list of 36 Core SAN marker genes. The P value was corrected using the Benjamini-Hochberg method (adjusted p-value < 0.05).
Comparison of mouse and human datasets
The raw data deposited by Goodyer et al.9 for the single-cell RNA-seq dataset of the mouse SAN was downloaded and analyzed using Seurat with the same settings as described in the original publication. For the comparison of gene expression in mouse and human SAN cells, the 36 Core SAN DEGs contained in both human fetal and hPSC datasets were selected. In addition, the top 25 SAN DEGs9 and the top 25 SAN surface marker DEGs26 from the mouse dataset were selected (sorted based on fold change). To visualize the gene expression in each dataset, bar graphs were generated in R. Genes from our human datasets were categorized as conserved if their expression in the mouse Core SAN cells was higher than in the Atrial cells. Genes from the mouse dataset were categorized as conserved if their expression in the human Core SAN cells was higher than in the Atrial cells in at least one of the human datasets, fetal or hPSC. If gene expression in the Core SAN was below 0.1 the gene was considered not expressed and listed as not conserved. These low-expressing genes were also visually confirmed looking at the gene expression in UMAPs.
Statistics and reproducibility
All data are presented as mean ± standard error of the mean (SEM). Indicated sample sizes (n) represent biological replicates (cell culture replicates or primary tissue samples). Sample sizes from single-cell datasets represent the number of cells analyzed as indicated in the individual figures. Statistical significance was determined using GraphPad Prism 9 (version 9.5.1). Datasets were tested for normal (Gaussian) distribution using the Shapiro-Wilk test (alpha = 0.05). The following statistical tests were applied for normally distributed data: For the comparison between two conditions Student’s t-test (two-tailed) was applied. For the comparison between multiple conditions one-way ANOVA with Bonferroni post hoc test was applied. The following nonparametric tests were applied for datasets that failed the normality test: For the comparison between two conditions Mann-Whitney test (unpaired samples) and Wilcoxon matched-pairs signed rank test (paired samples) were applied. For the comparison between multiple conditions Kruskal-Wallis test with Dunn’s post hoc test (unpaired samples) and Friedman test with Dunn’s post hoc test (paired samples) were applied. Results were considered significant at p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***). All statistical parameters are detailed in the respective figures and figure legends. Due to the nature of the study, the experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment. No statistical methods were used to pre-determine sample sizes.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data supporting the findings of this study are available within the article and the supplementary information files. Additional information is available from the corresponding author upon request. Source data are provided with this paper (Source Data file). Raw sc/snRNA-seq data generated in this study have been deposited in the GEO database under accession codes: GSE279630, GSE279631, and GSE279632. Source data are provided with this paper.
Code availability
We did not generate any original code in this study and all applied software packages are detailed and referenced in the method section.
References
Cantillon, D. J. et al. Complications and health care costs associated with transvenous cardiac pacemakers in a nationwide assessment. JACC Clin. Electrophysiol. 3, 1296–1305 (2017).
Christoffels, V. M., Smits, G. J., Kispert, A. & Moorman, A. F. Development of the pacemaker tissues of the heart. Circ. Res. 106, 240–254 (2010).
Sizarov, A. et al. Molecular analysis of patterning of conduction tissues in the developing human heart. Circ. Arrhythm. Electrophysiol. 4, 532–542 (2011).
van Eif, V. W. W. et al. Transcriptome analysis of mouse and human sinoatrial node cells reveals a conserved genetic program. Development 146, dev173161 (2019).
Chandler, N. J. et al. Molecular architecture of the human sinus node: insights into the function of the cardiac pacemaker. Circulation 119, 1562–1575 (2009).
Chandler, N. et al. Computer three-dimensional anatomical reconstruction of the human sinus node and a novel paranodal area. Anat. Rec. 294, 970–979 (2011).
Csepe, T. A. et al. Human sinoatrial node structure: 3D microanatomy of sinoatrial conduction pathways. Prog. Biophys. Mol. Biol. 120, 164–178 (2016).
Vedantham, V., Galang, G., Evangelista, M., Deo, R. C. & Srivastava, D. RNA sequencing of mouse sinoatrial node reveals an upstream regulatory role for Islet-1 in cardiac pacemaker cells. Circ. Res. 116, 797–803 (2015).
Goodyer, W. R. et al. Transcriptomic profiling of the developing cardiac conduction system at single-cell resolution. Circ. Res. 125, 379–397 (2019).
Liang, D. et al. Cellular and molecular landscape of mammalian sinoatrial node revealed by single-cell RNA sequencing. Nat. Commun. 12, 287 (2021).
Linscheid, N. et al. Quantitative proteomics and single-nucleus transcriptomics of the sinus node elucidates the foundation of cardiac pacemaking. Nat. Commun. 10, 2889 (2019).
Kanemaru, K. et al. Spatially resolved multiomics of human cardiac niches. Nature 619, 801–810 (2023).
Protze, S. I., Lee, J. H. & Keller, G. M. Human pluripotent stem cell-derived cardiovascular cells: from developmental biology to therapeutic applications. Cell Stem Cell 25, 311–327 (2019).
Protze, S. I. et al. Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker. Nat. Biotechnol. 35, 56–68 (2017).
Birket, M. J. et al. Expansion and patterning of cardiovascular progenitors derived from human pluripotent stem cells. Nat. Biotechnol. 33, 970–979 (2015).
Liang, W. et al. Canonical Wnt signaling promotes pacemaker cell specification of cardiac mesodermal cells derived from mouse and human embryonic stem cells. Stem cells 38, 352–368 (2020).
Li, J. et al. Molecular and electrophysiological evaluation of human cardiomyocyte subtypes to facilitate generation of composite cardiac models. J. Tissue Eng. 13, 20417314221127908 (2022).
Ghazizadeh, Z. et al. A dual SHOX2:GFP; MYH6:mCherry knockin hESC reporter line for derivation of human SAN-like cells. iScience 25, 104153 (2022).
Litvinukova, M. et al. Cells of the adult human heart. Nature 588, 466–472 (2020).
Blaschke, R. J. et al. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation 115, 1830–1838 (2007).
Cai, C. L. et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell 5, 877–889 (2003).
Wiesinger, A. et al. A single cell transcriptional roadmap of human pacemaker cell differentiation. Elife 11, e76781 (2022).
Andreatta, M. & Carmona, S. J. UCell: robust and scalable single-cell gene signature scoring. Comput. Struct. Biotechnol. J. 19, 3796–3798 (2021).
Aminu, A. J. et al. Further insights into the molecular complexity of the human sinus node—the role of ‘novel’ transcription factors and microRNAs. Prog. Biophys. Mol. Biol. 166, 86–104 (2021).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Goodyer, W. R. et al. In vivo visualization and molecular targeting of the cardiac conduction system. J. Clin. Invest 132, e156955 (2022).
Lee, J. H., Protze, S. I., Laksman, Z., Backx, P. H. & Keller, G. M. Human pluripotent stem cell-derived atrial and ventricular cardiomyocytes develop from distinct mesoderm populations. Cell Stem Cell 21, 179–194 e174 (2017).
Lang, D. & Glukhov, A. V. High-resolution optical mapping of the mouse sino-atrial node. J. Vis. Exp. 2, 54773 (2016).
Bei, Y. et al. Cardiac telocytes and fibroblasts in primary culture: different morphologies and immunophenotypes. PLoS One 10, e0115991 (2015).
Farbehi, N. et al. Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. Elife 8, e43882 (2019).
Elliott, D. A. et al. NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat. Methods 8, 1037–1040 (2011).
Dubois, N. C. et al. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat. Biotechnol. 29, 1011–1018 (2011).
Scavone, A. et al. Embryonic stem cell-derived CD166+ precursors develop into fully functional sinoatrial-like cells. Circ. Res. 113, 389–398 (2013).
Rust, W., Balakrishnan, T. & Zweigerdt, R. Cardiomyocyte enrichment from human embryonic stem cell cultures by selection of ALCAM surface expression. Regen. Med. 4, 225–237 (2009).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
He, J. Q., Ma, Y., Lee, Y., Thomson, J. A. & Kamp, T. J. Human embryonic stem cells develop into multiple types of cardiac myocytes: action potential characterization. Circ. Res. 93, 32–39 (2003).
Liu, J., Laksman, Z. & Backx, P. H. The electrophysiological development of cardiomyocytes. Adv. Drug Deliv. Rev. 96, 253–273 (2016).
Ravens, U. & Wettwer, E. Electrophysiological aspects of changes in heart rate. Basic Res. Cardiol. 93, 60–65 (1998).
Faber, J. W. et al. Sinus venosus incorporation: contentious issues and operational criteria for developmental and evolutionary studies. J. Anat. 234, 583–591 (2019).
Wiese, C. et al. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ. Res. 104, 388–397 (2009).
Liu, J., Dobrzynski, H., Yanni, J., Boyett, M. R. & Lei, M. Organisation of the mouse sinoatrial node: structure and expression of HCN channels. Cardiovasc. Res. 73, 729–738 (2007).
Boyett, M. R., Honjo, H. & Kodama, I. The sinoatrial node, a heterogeneous pacemaker structure. Cardiovasc. Res. 47, 658–687 (2000).
Liang, X., Evans, S. M. & Sun, Y. Development of the cardiac pacemaker. Cell Mol. Life Sci. 74, 1247–1259 (2017).
Li, N. et al. Molecular mapping of sinoatrial node HCN channel expression in the human heart. Circ. Arrhythm. Electrophysiol. 8, 1219–1227 (2015).
Puskaric, S. et al. Shox2 mediates Tbx5 activity by regulating Bmp4 in the pacemaker region of the developing heart. Hum. Mol. Genet. 19, 4625–4633 (2010).
Lakatta, E. G., Maltsev, V. A. & Vinogradova, T. M. A coupled SYSTEM of intracellular Ca2+ clocks and surface membrane voltage clocks controls the timekeeping mechanism of the heart’s pacemaker. Circ. Res. 106, 659–673 (2010).
Liang, D. et al. Sinoatrial node pacemaker cells share dominant biological properties with glutamatergic neurons. Protein Cell 12, 545–556 (2021).
Civin, C. I. et al. Antigenic analysis of hematopoiesis. III. A hematopoietic progenitor cell surface antigen defined by a monoclonal antibody raised against KG-1a cells. J. Immunol. 133, 157–165 (1984).
Majeti, R., Park, C. Y. & Weissman, I. L. Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell 1, 635–645 (2007).
Fina, L. et al. Expression of the CD34 gene in vascular endothelial cells. Blood 75, 2417–2426 (1990).
Sidney, L. E., Branch, M. J., Dunphy, S. E., Dua, H. S. & Hopkinson, A. Concise review: evidence for CD34 as a common marker for diverse progenitors. Stem Cells 32, 1380–1389 (2014).
Hughes, M. R. et al. A sticky wicket: defining molecular functions for CD34 in hematopoietic cells. Exp. Hematol. 86, 1–14 (2020).
Eijgelsheim, M. et al. Genome-wide association analysis identifies multiple loci related to resting heart rate. Hum. Mol. Genet. 19, 3885–3894 (2010).
den Hoed, M. et al. Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat. Genet. 45, 621–631 (2013).
Eppinga, R. N. et al. Identification of genomic loci associated with resting heart rate and shared genetic predictors with all-cause mortality. Nat. Genet. 48, 1557–1563 (2016).
Chauveau, S., Brink, P. R. & Cohen, I. S. Stem cell-based biological pacemakers from proof of principle to therapy: a review. Cytotherapy 16, 873–880 (2014).
Liu, Y. W. et al. Human embryonic stem cell-derived cardiomyocytes restore function in infarcted hearts of non-human primates. Nat. Biotechnol. 36, 597–605 (2018).
Romagnuolo, R. et al. Human embryonic stem cell-derived cardiomyocytes regenerate the infarcted pig heart but induce ventricular tachyarrhythmias. Stem Cell Rep. 12, 967–981 (2019).
Liu, R., Wang, R. E. & Wang, F. Antibody-drug conjugates for non-oncological indications. Expert Opin. Biol. Ther. 16, 591–593 (2016).
Szijj, P. & Chudasama, V. The renaissance of chemically generated bispecific antibodies. Nat. Rev. Chem. 5, 78–92 (2021).
Reubinoff, B. E., Pera, M. F., Fong, C. Y., Trounson, A. & Bongso, A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat. Biotechnol. 18, 399–404 (2000).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Mair, B. et al. Essential gene profiles for human pluripotent stem cells identify uncharacterized genes and substrate dependencies. Cell Rep. 27, 599–615 e512 (2019).
Kennedy, M., D’Souza, S. L., Lynch-Kattman, M., Schwantz, S. & Keller, G. Development of the hemangioblast defines the onset of hematopoiesis in human ES cell differentiation cultures. Blood 109, 2679–2687 (2007).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Laughner, J. I., Ng, F. S., Sulkin, M. S., Arthur, R. M. & Efimov, I. R. Processing and analysis of cardiac optical mapping data obtained with potentiometric dyes. Am. J. Physiol. Heart Circ. Physiol. 303, H753–H765 (2012).
Tomek, J., Wang, Z. J., Burton, R. B., Herring, N. & Bub, G. COSMAS: a lightweight toolbox for cardiac optical mapping analysis. Sci. Rep. 11, 9147 (2021).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 e3529 (2021).
Innes, B. T. & Bader, G. D. scClustViz—single-cell RNAseq cluster assessment and visualization. F1000Res 7, ISCB Comm J-1522 (2018).
Andrews, T. S. et al. Single-cell, single-nucleus, and spatial rna sequencing of the human liver identifies cholangiocyte and mesenchymal heterogeneity. Hepatol. Commun. 6, 821–840 (2022).
Acknowledgements
We would like to thank the members of the Protze laboratory for experimental advice and critical comments on the manuscript. We thank Prof. A. Elefanty and Prof. E. Stanley (Monash University, Victoria, AU) for providing the HES3 NKX2–5egfp/w reporter line, Prof. Feng Zhang (MIT, Boston, US) for sharing the lentiCRISPRv2 plasmid, and Prof. Didier Trono (EPFL, Lausanne, CH) for sharing the pMD2.G and psPAX2 plasmids. We would also like to thank the core facilities that supported this project including, the SickKids-UHN Flow Cytometry Facility for assistance with cell sorting, the Advanced Optical Microscopy Facility at UHN for assistance with confocal microscopy, the Princess Margaret Genomics Centre at UHN for assistance with sc/snRNA-seq and data processing, the Centre for Applied Genomics (TCAG) at SickKids for Sanger sequencing. We thank the donors, the Research Centre for Women’s and Infants’ Health BioBank Program, the Lunenfeld-Tannenbaum Research Institute, and the Department of Obstetrics & Gynecology of Sinai Health System for the human specimens used in this study. This work was supported by grants from the Canadian Stem Cell Network (IRP-ECI-19, S.P.& Z.L.), the Canadian Institutes of Health Research (CIHR PJT 169090, S.P.), the Canadian Foundation for Innovation (John R. Evans Leaders Fund, 38651, S.P.), the Government of Canada’s New Frontiers in Research Fund (NRFT-2022-00447, M.A.L.)”, the Canada Research Chairs Program (CRC-2020-00245, M.A.L.), and funding from BlueRock Therapeutics LP (S.P.). A.A.L. and M.L. were supported by Ontario Graduate Scholarships, B.M.M., and M.L.C. were supported by Canadian Institutes of Health Research Doctoral Fellowships, and F.J.A. was supported by a Canadian Institutes of Health Research Postdoctoral Fellowship.
Author information
Authors and Affiliations
Contributions
A.A.L. designed the project, performed experiments, analyzed data, and wrote the manuscript. A.L., D.P., T.T., G.D.B., and S.P., generated and analyzed the sc/snRNA-seq data. M.A., K.H., F.J.A., M.A.L., and Z.L., designed, performed, and analyzed the optical action potential/optical mapping experiments. M.L., M.L.C., M.K., and B.M.M., performed and analyzed tissue culture, flow cytometry, and immunohistochemistry experiments and provided valuable input on the manuscript. B.M.M. generated the CD34-knockout hPSC lines. A.M. assisted with the dissection of fetal heart tissues. S.P. designed the project and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
S.P. is a paid consultant for BlueRock Therapeutics LP. M.A.L. is a scientific founder and paid consultant for BlueRock Therapeutics LP. G.D.B. is a paid advisor of Adela, Inc.. A.A.L., M.L., and S.P. declare a patent titled “Use of CD34 as a marker for sinoatrial node-like pacemaker cells” (PCT/IB2021/053646) related to this study. All other authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lim, A.A., Pouyabahar, D., Ashraf, M. et al. Single-cell transcriptome analysis reveals CD34 as a marker of human sinoatrial node pacemaker cardiomyocytes. Nat Commun 15, 10206 (2024). https://doi.org/10.1038/s41467-024-54337-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-54337-4