Abstract
Proinsulin translation and folding is crucial for glucose homeostasis. However, islet β-cell control of Proinsulin translation remains incompletely understood. Here, we identify OSGEP, an enzyme responsible for t6A37 modification of tRNANNU that tunes glucose metabolism in β-cells. Global Osgep deletion causes glucose intolerance, while β-cell-specific deletion induces hyperglycemia and glucose intolerance due to impaired insulin activity. Transcriptomics and proteomics reveal activation of the unfolded protein response (UPR) and apoptosis signaling pathways in Osgep-deficient islets, linked to an increase in misfolded Proinsulin from reduced t6A37 modification. Osgep overexpression in pancreas rescues insulin secretion and mitigates diabetes in high-fat diet mice. Osgep enhances translational fidelity and alleviates UPR signaling, highlighting its potential as a therapeutic target for diabetes. Individuals carrying the C allele at rs74512655, which promotes OSGEP transcription, may show reduced susceptibility to T2DM. These findings show OSGEP is essential for islet β-cells and a potential diabetes therapy target.
Similar content being viewed by others
Introduction
Diabetes is an age-related chronic disease characterized by hyperglycemia and severe complications that caused by absolute or relative insulin deficiency1,2. As diet-induced obesity or ageing progresses, islet β-cells need augment insulin production and secretion in response to increased metabolic demand to maintain normoglycaemia3. The abundant production of insulin in islet β-cells imposes a significant burden on the endoplasmic reticulum (ER), where Proinsulin undergoes its initial folding, including the formation of three evolutionarily conserved disulfide bonds4. Normally, up to 20% of Proinsulin may fail to reach its proper protein conformation, and islet β-cells can handle a certain amount of misfolded Proinsulin molecules through ER stress response mechanisms5,6. However, under certain pathological conditions, mistranslation of the Proinsulin can further exacerbate the extent of misfolding7,8. Once the threshold is reached, the accumulation of misfolded Proinsulin in the ER would trigger unfolded protein response (UPR) signaling, leading to the loss of function and survival of β-cells9. Therefore, further exploration into the mechanism of insulin mistranslation and misfolding is critical for diabetes treatment.
O-sialoglycoprotein endopeptidase (OSGEP) is a N6-threonyl-carbamoylation (t6A) modification enzyme of the 37th adenosine in ANN-type (N = A, T, G, C) tRNA. OSGEP is the catalytic subunit of Kinase, Endopeptidase, and Other Proteins of Small size (KEOPS) complex, and mutations in OSGEP are connected with Galloway-Mowat syndrome (GAMOS), characterized by the combination of early onset nephrotic syndrome and microcephaly10,11. There is accumulating evidence that t6A37 stabilizes correct codon-anticodon pairing by preventing an interloop hydrogen bond of U33:A37, and mediates base-stacking interactions at the ribosomal decoding site to prevent codon +1 frameshifting during translation in yeasts and bacteria12,13,14,15. As early as 2004, scientists listed OSGEP as the first of Top 10 Conserved hypothetical genes, highly conserved from archaea to mammals, and likely to possess an essential function, making it a priority target for researchers in the respective field16. Recently, studies have reported that knockdown of OSGEP results in impaired protein synthesis, ER stress, activation of UPR signaling, and increased apoptosis17,18,19, which are the potential molecular and cellular pathophysiological mechanisms underlying for islet β-cell dysfunction. Despite advances in understanding the role of OSGEP in regulating t6A37 modification in bacteria, yeast and archaea, the mechanism of OSGEP in mammalian cells has not been unearthed. Interestingly, one case report described a 13-year-old Turkish GAMOS syndrome patient with OSGEP genetic mutations who was diagnosed with T2DM (Type 2 diabetes mellitus)17. Therefore, it would be of great interest to investigate the function of OSGEP in the islet-β cell.
In this study, we found that Osgep expression is decreased in diabetic human and mouse islets. We show that Osgep plays a role in regulating Proinsulin translation and ameliorating ER stress in islet β cells, thereby maintaining blood glucose homeostasis. We used inducible global knockout Osgep mice, islet β-cell-specific knockout Osgep mice, and Adeno-associated virus (AAV)-mediated Osgep overexpression in the pancreas of mice. Furthermore, we found an OSGEP functional variant, rs74512655, that may contribute to individual differences in susceptibility to T2DM. This study may provide a promising therapeutic target for diabetes in the future.
Results
Osgep expression is lower in diabetic patients and mice
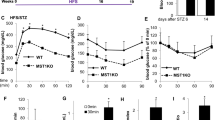
To identify whether OSGEP expression is associated with diabetes, we analyzed several transcriptomic datasets. We examined a human islet tissue bulk RNA-seq dataset GSE76896 (116 healthy control, 55 T2DM)20, as well as scRNA-seq datasets E-MTAB-5061 (6 healthy control with 171 β cells; 4 T2DM with 99 β cells)21 and GSE86473 (5 healthy control with 168 β cells; 3 T2DM with 96 β cells)22. In all three datasets, the OSGEP expression level was lower in individuals diagnosed with type 2 diabetes compared to in the non-diabetic individuals (Fig. 1a–c). Next, we found significantly lower Osgep protein and mRNA levels in islets of db/db diabetic mice (Fig. 1d, e, i). Consistent results were obtained from high-fat diet (HFD) mice in comparison to mice fed a chow diet (CD) (Fig. 1f–h, j). Taken together, these findings suggest that Osgep may play an important function in islet.
The OSGEP expression level compared between non-T2DM and T2DM individuals in dataset GSE76896 (healthy control n = 116, T2DM n = 55) (a), E-MTAB-5061 (β-cells of healthy control n = 171, β-cells of T2DM n = 99) (b), GSE86473 (β-cells of healthy control n = 168, β-cells of T2DM n = 96) (c). Protein of Osgep in the islets of WT and db/db mice (d) or CD and HFD-fed mice (f). The experiment was repeated three times with similar results. mRNA of Osgep in the islets of WT and db/db mice (e) or CD and HFD-fed mice (g) (n = 6). h Fasting blood glucose of CD and HFD mice (n = 6). Mice were fed at HFD for 16 weeks. i, j The representative images of immunofluorescence staining (IF) for insulin (green), Osgep (red), and nucleus (blue) in sections from mice. Results are repeated three times independently with similar results. a–c, e, g, h Data are presented as the mean ± SD. a–c The p value was obtained by Benjamini-Hochberg false discovery rate correction. e, g, h Unpaired two-tailed Student’s t-test. *p < 0.05, *** p < 0.001; **** p < 0.0001. Source data are provided as a Source Data file. CD Chow diet. HFD High fat diet.
Global Osgep knockout mice develop glucose intolerance
To explore whether Osgep regulates blood glucose homeostasis, we generated global Osgep knockout mice. Osgep-floxed mice (WT) and global Osgep knockout mice (Osgep-Ubc-CreERT, Osgepflox/flox mice) were born at the expected Mendelian ratio. At 6 weeks of age, 2 genotyped mice were injected with tamoxifen (Supplementary Fig. 1a-c). We found that homozygous knockout of Osgep caused lethality (Supplementary Fig. 1d). Therefore, heterozygous global Osgep deletion mice (Osgep-Ubc-CreERT, Osgepflox/- mice) were chosen for further study. Two weeks after tamoxifen induction, Osgep protein levels were reduced in multiple organs of Osgep-Ubc-CreERT mice, as assessed by Western blot (Supplementary Fig. 1e). Notably, Glucose tolerance was significantly impaired in Osgep-Ubc-CreERT mice, when challenged in an intraperitoneal glucose tolerance test (IPGTT) (Supplementary Fig. 1g). Whereas 2 groups of mice had no significant differences in fasting blood glucose and insulin sensitivity (Supplementary Fig. 1f, h). Taken together, these findings suggest that Osgep involves in blood glucose metabolism.
Islet β-cell-specific Osgep knockout mice have impaired glucose homeostasis
To further dissect the physiological function of Osgep in the islet, we generated islet β-cell-specific Osgep knockout mice by crossing Osgep-floxed mice with Ins2-Cre mice (Supplementary Fig. 2a). Two genotypes of mice were verified by PCR (Supplementary Fig. 2b). Osgep knockout mice were born at Mendelian frequencies and were morphologically indistinguishable with their control littermates in neonatal pups. Western blot and qPCR experiments showed that Osgep expression was significant reduced in the islets of Osgep-Ins2-Cre mice, but remained unchanged in other tissues (Supplementary Fig. 2c, d). Immunofluorescence (IF) straining of Insulin and Osgep further confirmed the specific depletion of Osgep in β-cells of Osgep-Ins2-Cre mice (Supplementary Fig. 2e). We monitored the body weight of the male mice from 12 to 40 weeks. Body weight curve showed that Osgep-Ins2-Cre male mice exhibited a significant increase (11 g) compared to their WT counterparts (Fig. 2a). Dual energy X-ray absorptiometry (DEXA) analysis was applied to examine the development of 20-week-old mice. Osgep-Ins2-Cre mice displayed a significant increase in lean mass, fat mass, and percentage of body fat mass, while reduced in bone mineral content (BMC) and no difference on body length and bone mineral density (BMD) (Supplementary Fig. 2f–k). Furthermore, the spleen and kidney index of 40-week-old Osgep-Ins2-Cre mice were decreased as compared with WT, while their liver index was increased (Supplementary Fig. 2l–n). These findings demonstrate that Osgep deficiency in islet β-cells can affect mice development.
a Body weight curve of male (WT n = 6, Osgep-Ins2-Cre n = 5) mice was monitored weekly till 40 weeks. b Fasting blood glucose of male WT (n = 7) and Osgep-Ins2-Cre (n = 5) mice were monitored in 5, 15, and 30-week-old. IPGTT (c–e) in male mice at 8 (WT n = 7, Osgep-Ins2-Cre n = 6), 12 (WT n = 6, Osgep-Ins2-Cre n = 5), and 36-week-old (WT n = 5, Osgep-Ins2-Cre n = 5). ITT (f–h) in male mice at 8 (WT n = 6, Osgep-Ins2-Cre n = 5), 12 (WT n = 6, Osgep-Ins2-Cre n = 5), and 36-week-old (WT n = 6, Osgep-Ins2-Cre n = 6). Fasting serum insulin levels (WT n = 7, Osgep-Ins2-Cre n = 5) i, j and in vivo GSIS (WT n = 6, Osgep-Ins2-Cre n = 5) k, l were monitored at 12, and 36-week-old. m Islets number count of male WT and Osgep-Ins2-Cre mice (n = 6). n The representative images of immunofluorescence staining (IF) for insulin (green), Glucagon (red), and nucleus (blue) in sections from male WT and Osgep-Ins2-Cre mice (n = 6 biological replicates), and the proportion of α-cells and β-cells in the total islet cells. a–n Data are presented as the mean ± SD. a, c–h, k, l One-way ANOVA with Tukey’s post-test and Unpaired two-tailed Student’s t-test. b, i, j, m, n Unpaired two-tailed Student’s t-test. *p < 0.05, ** p < 0.01; *** p < 0.001. Source data are provided as a Source Data file.
To assess the function involvement of Osgep in glucose homeostasis, we assayed the metabolic phenotype of Osgep-Ins2-Cre mice and their WT littermates at different weeks. Fasting blood glucose levels in Osgep-Ins2-Cre male mice showed an increase at 5 weeks, a significant addition at 15 weeks, and an even greater difference at 30 weeks (Fig. 2b). IPGTT revealed a severe glucose intolerance phenotype in 8- to 36-week-old Osgep-Ins2-Cre male mice (Fig. 2c–e). Notably, the difference in glucose intolerance between the two genotypes of mice became more pronounced with age, as indicated by the increasing AUC of the IPGTT, from 139% of the WT value in 12-week-old mice to 149% in 36-week-old mice. These results suggest that the diabetic phenotype in the knockout mice worsened over time. No difference in insulin sensitivity was found between WT and Osgep-Ins2-Cre male mice aged 8-12 weeks, indicating that peripheral insulin sensitivity was not affected. Insulin sensitivity of knockout mice slightly decreased at 36 weeks (Fig. 2f–h). Fasting insulin levels and glucose-stimulated insulin secretion levels were not different between groups at 12 and 36 weeks (Fig. 2i–l). Additionally, islet count did not reveal any difference in islet number between the 2 genotypes of mice (Fig. 2m) and islet specific IF staining showed similar islet morphology and cell compositions in Osgep-Ins2-Cre and WT mice (Fig. 2n). Consistent with the metabolic phenotype in male mice, Osgep-Ins2-Cre female mice also showed significantly weight gain and impaired glucose tolerance, but no significant discrepancy in insulin sensitivity or insulin secretion levels (Supplementary Fig. 3). These contradictory results of Osgep in glucose regulation lead us to the hypothesis that the same amounts of insulin are secreted, but the activity to sweep glucose is dysfunction in Osgep knockout mice, which results in hyperglycemia and glucose intolerance.
Osgep deletion in islet β-cells triggers aggravated hyperglycemia, glucose intolerance, and hypoinsulinemia in HFD-fed mice
HFD animal models are widely used in the study of metabolic diseases, including diabetes, fatty liver disease, and obesity23. In general, HFD induces metabolic stress leading to weight gain, hyperglycemia, hyperinsulinemia, insulin resistance, inflammatory cytokine secretion and ectopic lipid accumulation. We subjected WT and Osgep-Ins2-Cre mice to an HFD-fed and assessed the related metabolic parameters at the indicated weeks (Supplementary Fig. 4a). During the 16-week diet, deletion of Osgep resulted in a significant increase in body weight gain compared to HFD-WT mice, despite the same food intake, which is consistent with the larger liver and white adipose tissues (WAT) (Fig. 3a, Supplementary Fig. 4b–d). Lipid profiles showed serum triglycerides (TG) and high-density lipoprotein cholesterol (HDL-C) levels were comparable, while serum cholesterol (CHO) and low-density lipoprotein cholesterol (LDL-C) levels were significantly increased in HFD-fed Osgep-Ins2-Cre mice (Supplementary Fig. 4e). H&E and oil red O staining of insulin target tissue sections revealed greater lipid deposition in the liver and significant enlargement of the adipocyte size in HFD-fed Osgep-Ins2-Cre mice compared to the WT mice (Supplementary Fig. 4f). These results indicate that Osgep-Ins2-Cre mice are more susceptible to obesity under metabolic stress conditions.
Body weight gain (a), fasting blood glucose (b), IPGTT (c–e), and ITT (f) were conducted in HFD-fed mice (HFD-WT n = 6, HFD-Osgep-Ins2-Cre n = 5). g Protein of p-Akt and total Akt were detected in liver and WAT of HFD-feed mice. The protein samples derive from the same experiment and that blots were processed in parallel. In vivo GSIS (h) and fasting serum insulin levels (i) were conducted in HFD feeding mice (HFD-WT n = 6, HFD-Osgep-Ins2-Cre n = 5). In vitro GSIS assays (HFD-WT n = 4, HFD-Osgep-Ins2-Cre n = 4) (j), and islets number count (HFD-WT n = 6, HFD-Osgep-Ins2-Cre n = 8) (k) were performed in islets isolated from HFD-fed mice. l Hematoxylin and eosin (H&E) stain analysis of the pancreatic sections and quantification of islet area (HFD-WT n = 6, HFD-Osgep-Ins2-Cre n = 5). m Representative immunofluorescence staining (IF) images showing major islet hormones anti-insulin (red), anti-glucagon (green), and anti-DAPI (blue) in pancreatic sections from HFD-fed mice (n = 6), and the proportion of α-cells and β-cells in the total islet cells. n Electron microscopy analysis of mice islet, arrowheads denoted insulin granules (n = 6). a–f, h–m Data are presented as the mean ± SD. a, b, i–m Unpaired two-tailed Student’s t-test. c–f, h One-way ANOVA with Tukey’s post-test and Unpaired two-tailed Student’s t-test. *p < 0.05, **p < 0.01; ***p < 0.001; ****p < 0.0001. Source data are provided as a Source Data file. HFD High fat diet, WAT White adipose tissue.
Obesity is usually accompanied by blood glucose metabolic disorders24,25. Osgep-Ins2-Cre mice displayed more severe hyperglycemia than WT mice when exposed to HFD diets (Fig. 3b). IPGTTs performed at 10- 14- and 19-week-old showed that Osgep-Ins2-Cre mice were more severely glucose intolerant than WT mice over time following the dietary intervention (Fig. 3c-e). Unlike the results in normal diet-fed mice, the HFD-fed Osgep-Ins2-Cre mice exhibited a statistical reduction in insulin sensitivity, and p-Akt expression levels in liver ang WAT were significantly decreased (Fig. 3f–g). In response to a glucose challenge, the HFD-fed Osgep-Ins2-Cre mice secreted lower levels of insulin compared to HFD-fed WT mice (Fig. 3h), and their circulating insulin levels were markedly reduced (Fig. 3i). To examine whether this phenotype was associated with defective GSIS in vitro, we performed 30-min incubations of islets isolated from the mice. While the basal level of insulin secretion (2.8 mM glucose) was equivalent, the high glucose (16.7 mM)-induced insulin secretion was markedly reduced in the HFD-fed Osgep-Ins2-Cre mice β-cells (Fig. 3j). Impaired insulin sensitivity and secretion may result from reduced islet mass and morphology. We calculated the islets of 2 genotype mice by divided the size of islets into 3 groups: small islets (0-5,000 μm2), medium islets (5,001-10,000 μm2), and large islets (>10,000 μm2). In Osgep-Ins2-Cre mice, the total islet number and each size of islet mass was significantly reduced (Fig. 3k). The phenotypes of reduced islet number were further demonstrated by H&E stain (Fig. 3l). Furthermore, IF staining for insulin and glucagon showed that HFD-fed Osgep-Ins2-Cre mice had atrophied islet architecture and reduced β-cell proportion (Fig. 3m). Ultrastructural analyses revealed that Osgep-deficient β-cells had smaller insulin granules with reduced halo and core area (Fig. 3n). These data indicate that islet β-cell-specific deletion of Osgep exacerbates HFD-induced hyperglycemia and glucose intolerance. Meanwhile, after HFD-feeding, deletion of Osgep impaires insulin secretion and islets morphology.
Islet β-cell-specific deletion of Osgep leads to defective insulin activity
To investigate the contradictory results of impaired glucose metabolism and unchanged insulin level in CD-fed Osgep-Ins2-Cre mice, we purified insulin from the mice to determine its activity. The insulin was extracted from the pancreas referring to pharmacochemistry method for producing animal insulin, and insulin standard was used as a positive control. The same concentration of insulin solution was injected intraperitoneally into 3 groups of 6-h-fasted mice with similar age and weight. The insulin extracted from the pancreas could significantly decrease blood glucose level, consistent with the insulin standard (Fig. 4a). However, the drop in blood glucose levels was significantly weaker in mice injected with insulin-extracted from Osgep-Ins2-Cre pancreas, suggesting a deficiency in glucose clearance ability of insulin. We further conducted the same experiments in HFD-fed mice. Under the HFD condition, the insulin extracted from the Osgep-Ins2-Cre mice had significantly lower activity compared to the WT mice (Fig. 4b). Additionally, HFD-fed WT mice had worse ability to lower glucose compared to the insulin from CD-fed mice (Fig. 4c). Collectively, these data demonstrate that the deletion of Osgep in islet β-cells can reduce the insulin activity to sweep glucose, thereby significantly increase blood glucose level. This impairment in insulin activity was more progressive in the Osgep-Ins2-Cre mice under the HFD condition compared to the CD condition.
ITT assay detected the activity of insulin extracted from WT and Osgep-Ins2-Cre mice pancreas at CD (a) or HFD (b)-fed, and insulin standard as positive control (n = 4). c AUC of ITT curve (n = 4). d Schematic of the universal t6A37 biosynthesis pathway in mammalian cell. Kinase, Endopeptidase, and Other Proteins of Small size (KEOPS) compound, with OSGEP as the active catalytic enzyme, utilizes threonylcarbamoyl adenylate (TC-AMP) to catalyze the formation of t6A37 modification in ANN-decoding tRNA substrates. e HPLC-MS/MS chromatograms of t6A37 in islet tRNA and relative t6A37 levels were showed in WT and Osgep-Ins2-Cre mice islet (n = 4). f Circular dichroism spectra of protein extracted from WT and Osgep-Ins2-Cre islet at CD. Proinsulin protein was analyzed by reducing western blots, and misfolded Proinsulin protein was analyzed by non-reducing Western blots in islets of mice under CD (g) and HFD (h)-fed condition. The ratios of Monomer to total Proinsulin were calculated. The ratios of Dimers, Trimers plus High-Molecular-Weight (HMW) to total Proinsulin were calculated (n = 3 biological replicates). Results are repeated three times independently with similar results. i Experimental procedure of in vitro transcription and translation for proinsulin protein. j Western blots analyzed the proinsulin product in addition of tRNAs from islets of HFD-fed WT and Osgep-Ins2-Cre mice in vitro reticulocytes translation system (n = 3). a–c, e, g, h, j Data are presented as the mean ± SD. a, b One-way ANOVA with Tukey’s post-test. c, e, g, h, j Unpaired two-tailed Student’s t-test. *p < 0.05, **p < 0.01; ***p < 0.001; ****p < 0.0001. Source data are provided as a Source Data file. CD Chow diet, HFD High fat diet, KEOPS Kinase, Endopeptidase, and Other Proteins of Small size, TC-AMP threonylcarbamoyl adenylate, RRL Rabbit Reticulocyte Lysate.
Loss of Osgep impairs oxidative folding and reduces Proinsulin protein content via downregulated t6A37 level of tRNA
Previous studies demonstrated that Osgep is an NNU-t6A37 modifying enzyme (Fig. 4d). Out of the total proinsulin protein, 11 amino acids are encoded by ANN (N = A, T, G, C) codon type tRNAs, accounting for 12.79% of the total peptide chain (Supplementary Fig. 5a). Dysregulation of tRNA modification can increase the possibility of the translation error and reduced the translation efficiency26. We isolated mouse islets to analyze the t6A37 level using HPLC-MS/MS detection. The results suggested that Osgep knockout islets exhibited about 60% lower t6A37 level compared to WT islets (Fig. 4e, Supplementary Fig. 5b). We then compared the secondary structure of total protein from WT and Osgep-Ins2-Cre islets using circular dichroism spectrum (Fig. 4f). The spectra showed similar band intensity in 180-200 nm, but the Osgep-Ins2-Cre protein was completely disordered, as seen from the presence of an increased circular dichroism band at 220 nm. This indicates that Osgep-Ins2-Cre islets displayed abnormal protein folding state. Proinsulin is the most abundantly synthesized protein in β-cells. We further examined proinsulin oxidative folding under non-reducing conditions. Although the total amount of proinsulin was unchanged in CD-fed Osgep-Ins2-Cre islets as measured under reducing conditions, intermolecular disulfide-linked complexes of proinsulin (Dimers, Trimers, and High-Molecular-Weight proinsulin-associated complexes) were markedly increased in Osgep-Ins2-Cre islets analyzed under non-reducing conditions (Fig. 4g). We also analyzed the expression levels of proinsulin protein in islets of HFD-fed mice. In contrast to the CD-fed mice, the proinsulin as measured by reducing Western blot was significantly decreased under the HFD condition (Fig. 4h). Additionally, the intermolecular disulfide-linked complexes of proinsulin were increased to a higher degree, and the monomer proinsulin was markedly decreased in the HFD-fed Osgep-Ins2-Cre islets. Furthermore, in the HFD-fed WT mice, the intermolecular disulfide-linked complexes of proinsulin were also significantly higher compared to the CD-fed WT mice. These results indicated that deficiency of Osgep impairs proinsulin oxidative folding.
In vitro Min6 cell experiments also showed that Osgep regulated Proinsulin translational and secretion, but not transcription (Supplementary Fig. 5c–e). To further verify tRNA as the mediator of Osgep affecting insulin translation, we applied in vitro translation of Proinsulin protein assay with rabbit reticulocyte lysates system (Fig. 4i). Luciferase mRNA was used to verify the endogenic tRNA eliminated efficiency. The luciferase activity in filtered-RRL (Rabbit Reticulocyte Lysate) obviously decreased by ~90% compared with untreated-RRL, which proved the feasibility of experimental model (Supplementary Fig. 5f). Preproinsulin mRNA as templates was synthesized by in vitro transcription (Supplementary Fig. 5g). tRNA as materials was purified from different Osgep expression level of islets and Min6 cells (Supplementary Fig. 5h). Western blot analysis showed that the level of the Proinsulin protein was significantly lower when added with the tRNA isolated from HFD-fed Osgep-Ins2-Cre islets (Fig. 4j). The same tendency was verified in the group of shOsgep-Min6 cells (Supplementary Fig. 5i). Taken together, we demonstrate that Osgep deficiency impairs the oxidative folding of Proinsulin protein and decreases its translation level, which are mediated by the defective t6A37 level of tRNA.
Osgep deficiency alters the islet transcriptome and proteome profiles, affecting Unfolded Protein Response and Apoptosis Pathways
To investigate the networks regulated following the deletion of Osegp expression in islet β-cells, we explored how transcriptome and proteome profiles were altered in islets isolated from 12-week-old male WT and Osgep-Ins2-Cre mice. The Partial Least-Squares (PLS) analysis plot clearly separated the 2 groups for both RNA-seq and proteins (Supplementary Fig. 6a, b). Combined analysis of all samples yielded the identification of 16383 annotated RNAs and 5932 proteins, of which 5621 overlapped. Protein counts positively correlated with mRNA counts, with Spearman correlation coefficient of 0.358 in WT and 0.369 in Osgep-Ins2-Cre (Supplementary Fig. 6c, d).
GSEA (Gene Set Enrichment Analysis) revealed 33 biological pathways in RNA-seq results and 11 pathways in proteome results that were differentially enriched in WT relative to Osgep-Ins2-Cre with NOM p value < 0.05. Of which, 8 pathways overlapped (Supplementary Data 1-3). Among the enriched pathways, the hallmark_unfolded_protein_response was upregulated in Osgep-Ins2-Cre mice in the RNA-seq data, and it was top-ranked and formed a large cluster in the enrichment map (Fig. 5a). Meanwhile, the hallmark_apoptosis pathway showed a similar trend in the proteome data (Fig. 5b). To explore more reliable enrichment analysis considering the effects of multi-gene expression patterns, we conducted k-means clustering analysis on 1895 differentially expressed (DE) RNAs and 108 DE proteins, successfully identifying 9 RNA expression clusters (RNA_A-RNA_I) and 3 protein expression clusters (pro_A-pro_C) (Fig. 5c). To explore the biological functions of these expression patterns, we carried out a KEGG analysis and organized the identified networks between the enriched pathways and related DE genes, excluding pathways specific to diseases unrelated to diabetes, pathways pointing to general biological mechanisms, and pathways with no known connection to diabetes, glucolipid metabolism, or protein translation and processing. The comprehensive analysis showed that the most noticeable pathways enriched in multiple clusters could be divided into three categories: 1) Associated with diabetes (Associated with diabetes, Diabetic cardiomyopathy, Maturity onset diabetes of the young); 2) Associated with protein synthesis (Protein processing in endoplasmic reticulum, Protein export, Protein digestion and absorption, Arginine and proline metabolism, Glycine, serine and threonine metabolism, Apoptosis); and 3) Associated with insulin (Insulin secretion, PI3K-Akt signaling pathway, MAPK signaling pathway). Various circumstances, such as increased protein folding demand, accumulation of misfolded proteins, and excess metabolic demand, can induce endoplasmic reticulum (ER) stress in islet-β cells. The production of large quantities of insulin in β cells imposes a significant burden on the ER. In turn, adaptation to ER stress by sophisticated and highly dynamic UPR signaling network activation is proposed to play a protective role in increased insulin production, whereas the failure of adaptation has been implicated in islet-β cell dysfunction and apoptosis. This result was aligned with the role of Osgep-dependent tRNA involved in protein translation and implied that Osgep regulated ER homeostasis and UPR signaling.
Enrichment plots of unfolded protein response (UPR) pathway of the transcriptome (a) and apoptosis pathway of the proteome (b) enriched in Gene set enrichment analysis (GSEA), showing the profile of the running ES Score and positions of gene set members on the rank-ordered list (WT n = 3, Osgep-Ins2-Cre n = 3). c Circular heatmap showed expression clustering analysis of DE genes. First, counts of 1895 DE RNAs and 108 DE proteins were transformed into z-scores. Next, the k-means clustering method was used to classify the genes into 9 RNA and 3 protein clusters, displayed as a circular figure. KEGG enrichment analyses of the genes within each pattern were carried out by using clusterProfiler and enrich plot package in R software. The enriched functional pathways are selectively showed by networks between enriched pathways and related DE genes. Source data are provided as a Source Data file.
Osgep knockout causes ER stress and induces cell apoptosis
To explore how Osgep altered ER stress and UPR signaling, we first observed the ER ultrastructure of islet-β cells in Osgep-Ins2-Cre mice. Electron microscopy revealed that Osgep-deficiency islets showed typical dilated ER cisternae with degranulation, a classical feature of ER stress (Fig. 6a). Next, we explored the expression level of UPR markers in islets of WT and Osgep-Ins2-Cre mice. Both mRNA and protein levels of Bip (a master molecular chaperone regulating the UPR) were increased along with Osgep-deficiency. The UPR comprises 3 signaling arms, inositol-requiring enzyme 1 (IRE1α), protein kinase R-like ER kinase (PERK), and activating transcription factor 6 (ATF6), that sense ER stress and are activated through release from Bip. IRE1α, ATF6 and phosphorylated PERK showed significantly higher expression levels in the Osgep-Ins2-Cre mice islets on a HFD (Fig. 6b, e, Supplementary Fig. 7a). As a main substrate of PERK signaling, phospho-eIF2α was activated in the same way. Phosphorylation of eIF2α, while reducing global protein synthesis, increases ATF4 expression. Consistent with the increased phospho-eIF2α, the mRNA and protein levels of ATF4 were higher in HFD-fed Osgep-Ins2-Cre mice. Meanwhile, CHOP, which contributes to ER stress induced β-cell apoptosis, was prominently activated by ATF4. We also constructed an in vivo acute ER stress mouse model by intraperitoneal injected of 1 μg/g tunicamycin (Tm) solution twice27. Western blot and qPCR similarly revealed that UPR signaling was activated in Tm-induced Osgep knockout mouse islets (Fig. 6c, f, Supplementary Fig. 7b). Additionally, isolated islets were incubated with 1 μM thapsigargin (Tg) for 6 h to establish an in vitro ER stress model, and the same trends were confirmed in Tg-induced Osgep-Ins2-Cre mice (Fig. 6d, Supplementary Fig. 7c). Furthermore, Osgep-knockdown in Min6 cells increased the number of PI+ cells triggered by ER stress agonist Tg, indicating that Osgep deletion promoted ER stress-induced cell apoptosis (Fig. 6g, h). Collectively, our results suggest a role for Osgep in the orchestration of UPR and apoptosis signaling.
a Electron micrograph showing ER membrane changes in β-cell of WT and Osgep-Ins2-Cre mice. Arrowheads indicated ER. Western blot (n = 3) b and qPCR (n = 6) e analyzed UPR-related markers of WT and Osgep-Ins2-Cre islets after 16 weeks HFD-fed. Western blot (n = 3) c and qPCR (n = 6) f analyzed UPR-related markers from WT and Osgep-Ins2-Cre islets i.p. injected twice 0.67 mg/kg Tunicamycin (Tm). d Western blot analyzed UPR-related markers in the islets treated with 1 μM Thapsigargin (Tg) for 6 h isolated from WT and Osgep-Ins2-Cre mice. Immunofluorescence staining (IF) (n = 6 biological replicates) (g) and quantifications (h) of PI+ apoptotic cells in the presence or absence of Osgep Min6 cell treatment with DMSO or 1 μM Tg for 6 h (n = 3). b–d The protein samples were derived from the same experiment and the blots were processed in parallel. e, f, h Data are presented as the mean ± SD. e, f, h Unpaired two-tailed Student’s t-test. **p < 0.01; ***p < 0.001; ****p < 0.0001. Source data are provided as a Source Data file. HFD High-fat diet, Tm Tunicamycin, Tg Thapsigargin.
AAV-Osgep delivery into the pancreas rescues the diabetic phenotype in diabetic mice
AAV-based gene therapy has shown potential in clinical applications due to its low toxicity and long-term transgene expression. This approach has been actively applied for chronic diseases by replenishment single gene expression28. Since we had established Osgep as a regulator of insulin translation in islet β-cell, and observed lower Osgep expression in HFD-fed and db/db diabetic mice, we sought to test whether Osgep expression via AAV2/Pan delivery could be an effective approach to accomplish the desired outcome of treating diabetes. To achieve an adequate and specific efficiency of Osgep expression in pancreas, we applied the AAV2/Pan and CMV promoter systems to deliver Osgep in CD- or HFD-fed mice and measured the relative metabolic phenotype as illustrated in Fig. 7a. IF and Western blot analysis were confirmed the effective delivery of the adeno-associated virus (Fig. 7b, c), and Western blot of Osgep expression in several tissues showed the specificity of the AAV2/Pan system for targeting and transducing the pancreas, without off-target effects in other tissues (Supplementary Fig. 8a). We found that body weight, the weight of glucose metabolism-effected organ and lipid content were upregulated in HFD-fed mice overexpressing Osgep (Fig. 7d, Supplementary Fig. 8c–f). Moreover, increasing the Osgep level in the pancreas could significantly reduce blood glucose and improve glucose tolerance (Fig. 7e, f). The HFD-fed, pancreas-specific Osgep overexpressing mice exhibited improved insulin sensitivity, and higher level of GSIS, while no change in CD-fed mice (Fig. 8g–i). We also found AAV-Osgep did not exhibit changes in glucagon levels (Supplementary Fig. 8b). To explore the underlying reasons for the improved glucose homeostasis in CD- AAV-Osgep, we assessed the insulin activity in the collected pancreas. The results revealed that the insulin extracted from AAV-Osgep pancreas exhibited significantly higher activity compared to the control group (Fig. 8j). Additionally, the HFD-fed pancreas-specific Osgep overexpressing mice exhibited a decrease in the expression of UPR signaling markers (Fig. 8k–m). Collectively, these data demonstrated that a single-dose AAV vector- mediated delivered of Osgep expression in the pancreas significantly ameliorated the diabetic phenotype and alleviated UPR signaling, suggesting it could be an effective diabetes therapeutic.
a Study design. C57BL/6 J mice were fed CD or HFD (n = 6) starting at 5 weeks old, and i.p. injected with AAV-GFP or AAV-Osgep at 9 weeks old with frequent metabolic phenotyping analysis. b In vitro tissue fluorescence activity was assessed following i.p. injection of adenovirus for 12 weeks. c Protein expression of Osgep in the pancreas of CD or HFD-fed mice assessed by Western blots. Body weight change (d), fasting blood glucose (e), IPGTT (f), ITT (g), fasting serum insulin levels (h), and in vivo GSIS (i) were conducted in CD or HFD feeding of AAV-GFP or AAV-Osgep mice. (n = 6; *, comparison between CD-AAV-GFP and CD-AAV-Osgep; #, comparison between HFD-AAV-GFP and HFD-AAV-Osgep in line charts). j ITT assay detected the activity of insulin extracted from AAV-GFP or AAV-Osgep mice pancreas at CD-fed, and insulin standard as positive control (n = 4; insulin-extracted from CD-AAV-GFP pancreas versus insulin-extracted from CD-AAV-Osgep pancreas: *p < 0.05, **p < 0.01). QPCR analysis (k) and Western blot analysis (l) of UPR-related markers in the pancreas from AAV-GFP or AAV-Osgep mice after HFD-fed. m Quantitation of the immunoblotting of l. c, l The protein samples were derived from the same experiment and the blots were processed in parallel. d–k, m Data are presented as the mean ± SD. d, f, g, i, j One-way ANOVA with Tukey’s post-test and Unpaired two-tailed Student’s t-test. e, h, k, m Unpaired two-tailed Student’s t-test. d, f, g, i *p < 0.05, **p < 0.01; ***p < 0.001; ****p < 0.0001; #p < 0.05, ##p < 0.01; ###p < 0.001; ####p < 0.0001. Source data are provided as a Source Data file. CD Chow diet. HFD High-fat diet. AAV Adeno-associated virus. FBG Fasting blood glucose. IPGTT Intraperitoneal glucose tolerance test. ITT Insulin tolerance test. GSIS Glucose stimulated insulin secretion. PA pancreas, Co colon, Ce cecum, Ile ileum, Je jejunum, Du duodenum. St stomach. Sp spleen; K kindey, L liver, Lu lung, Ht heart.
a Overview of the pGL3-rs74512655 reporter plasmid constructs used for dual luciferase assays. b The relative firefly luciferase activities were compared between pGL3-G and pGL3-C group (n = 3). c Overview of the OSGEP promoter-rs74512655 reporter plasmid constructs used for dual luciferase assays. (d) Relative firefly luciferase activities in constructs containing OSGEP promoter driven by DNA fragment harboring rs74512655 (n = 3). b, d Data are presented as the mean ± SD. b, d Unpaired two-tailed Student’s t-test. **p < 0.01; ****p < 0.0001. Source data are provided as a Source Data file.
Regulatory role of rs74512655 on OSGEP transcription is associated with T2DM
The above study confirmed that Osgep deficiency caused diabetes in mice, and we next wanted to explore the association between OSGEP and T2DM at a clinical level. As shown in Table 1, we recruited 234 control and 285 T2DM subjects as discovery cohort to genotype 13 OSGEP single-nucleotide polymorphisms (SNPs). Only rs74512655 (G > C) was found to be associated with T2DM (p = 0.016, Supplementary Data 6). However, after strict correction for multiple testing, we found this SNP cannot reach the corrected threshold of p_correct = 0.003. To further validate this result, we included an independent replication cohort of 228 T2DM and 228 control individuals. The genotype results verified that C allele of rs74512655 was associated with a decreased T2DM risk (OR = 0.636 (0.435-0.928), p value = 0.019). Finally, when we combined the data from the two stages, the association between OSGEP rs74512655 and T2DM risk persisted (Table 2). We also explored a genome-wide association study GWAS summary from the GWASATLAS29, which analyzed 385,420 European individuals (Ncase = 18,483, Ncontrol = 366,937) and containing 10,598,160 genetic variants. By searching the list of OSGEP SNPs, only rs74512655 was validated, and it showed the same genetic effect (Table 2, OR = 0.939 (0.886-0.995), p value = 0.033) as in our study. Those results collectively suggest that OSGEP rs74512655 was the risk locus of T2DM.
To investigate the effect of rs74512655 on the transcriptional activity of OSGEP, we created promoter-luciferase constructs containing the G or C allele and performed dual luciferase assays (Fig. 8a). As shown in Fig. 8b, the group transfected with pGL3-C presented higher luciferase activity than the group transfected with pGL3-G, indicating that the mutation site may have a regulatory function. Given that the variant rs74512655 is located in the third intron of OSGEP gene, it might activate the gene transcription activity through a long-range interaction with the OSGEP promotor. This hypothesis was further confirmed by the observation that the group transfected with the vector containing the OSGEP promoter region (Fig. 8c). The group transfected with OSGEP-promoter-C displayed an enhanced luciferase activity compared to the OSGEP-promoter-G group (Fig. 8d). The results suggest that the protective C allele of rs74512655 influences the interaction with the OSGEP promoter, facilitates OSGEP transcriptional activity, and may reduce susceptibility to T2DM.
Discussion
In summary, our results uncover a role for Osgep-regulated glucose metabolism. We show that loss of Osgep-dependent tRNA modification impairs Proinsulin translation and oxidative folding, resulting in severe hyperglycemia and glucose intolerance. Indeed, the observed accumulation of misfolded proteins further activates of UPR signaling and β-cell apoptosis. Given the importance of Proinsulin translation, we infer that Osgep is essential for maintaining islet β-cell function. In addition, we identified a SNP rs74512655 (G > C), located in the third intron region and C allele enhancing the transcription of OSGEP, which may be associated with a lower susceptibility to T2DM.
The main function of islet β-cells is to rapidly synthesize and store large amounts of insulin, approximately 6000 insulin molecules per second, in response to high glucose signals in peripheral circulation26,30,31,32. In this process, mistranslation and misfolded Proinsulin proteins lead severe proteotoxic ER stress and activation of the UPR signaling pathway. The diabetes of youth (MIDY) caused by misfolded Proinsulin has been well studied, as mutant INS gene exhibit perturbed proinsulin disulfide bond formation to block misfolded insulin in the ER33. OSGEP is evolutionarily conserved across three species, responsible for catalyzing the t6A37 modification of tRNANNU 34. tRNA modifications have a crucial role in protein translation by guaranteeing translational fidelity and efficiency, and position 37 presents the largest variety of modified nucleoside derivatives, which corresponds to about 70% of all modifications35,36. The proinsulin peptide chain contains totally 86 amino acids, of which 11 amino acids (2 of Isoleucines, 2 Lysines, 2 Asparagines, 1 Serine, 1 Methionine, and 3 Threonines) are encoded by ANN codon type tRNAs, accounting for 12.79% of the total peptide chain. We observe that the lysine and arginine residues at the two ends of the C-peptide may potentially impacting the correct cleavage of the C-peptide. Additionally, the Threonine-Serine-Isoleucine residues located within the 2 disulfide bond formation sites may affect the proper formation of the disulfide bonds. Those sites may affect the function of proinsulin protein under Osgep deficiency condition. Under CD feeding conditions, islet β-cell-specific Osgep knockout mice displayed impaired insulin activity but maintained normal insulin levels. However, under HFD feeding conditions, both insulin activity and levels were reduced. This suggests that abnormalities in proinsulin folding can occur before a decrease in insulin quantity, indicating a high degree of adaptive plasticity in β-cells37. This finding opens up opportunities for detecting insulin folding states, which could facilitate early diabetes diagnosis aid and enhance cure rates.
Structural prediction suggests OSGEP performs a vital function in the organism, as well our study showed global Osgep knockout mice lethal after tamoxifen-induced. We found that Osgep deficiency in β-cells is associated with impaired oxidative folding of Proinsulin by diminished t6A37 modification. Meanwhile, both our in vivo and in vitro studies demonstrated that Osgep deficiency in β-cells caused a marked reduction of insulin activity and content with severe deterioration on islet morphology. Here, we show that loss of Osgep causes misfolded Proinsulin in islet β-cell, thus representing a unique mechanism of Osgep, which is consistent with the findings of Cdkal1 in islet β-cells. Cdkal1 catalyzes the biosynthesis of ms2t6A37 for tRNALys (UUU), required for the accurate translation of lysine codons in Proinsulin38,39. Functional loss of Cdkal1 may cause misread of lysine residues, located at the cleavage site of the A-chain and C-peptide, potentially leading to cleavage-resistant Proinsulin40. Furthermore, the other subunits of the KEOPS complex (GON7, LAGE3, TP53RK, and TPRKB) and YRDC also participate in the process of t6A37 synthesis, which may play roles in regulating proinsulin synthesis and folding in β-cells. Those aspects require further investigation.
Relative and/or absolute insulin deficiency leads to a greater need for insulin in the peripheral circulation, forcing islet β-cells to accelerate the process of protein translation, further produces more mistranslated proteins, including Proinsulin. Deficiency of Osgep expression impairs the t6A37 modification. Previously, studies in Drosophila, yeast, and bacteria have found that the absence of t6A leads to translation infidelity due to incorrect codon-anticodon pairing during decoding, thus resulting in protein aggregates. The excessive incorrect protein synthesis induces the accumulation of misfolded and unfolded proteins in the lumen of endoplasmic reticulum (ER), leading to ER stress, and triggering the Unfolded Protein Response (UPR)41,42. In the present work, one of the most-significant change we observed in Osgep knockout islet β-cells was an ultrastructural hallmarks of ER stress, showing swollen ER and ribosome shedding evident. This finding is consistent with GSEA and KEGG pathway analysis of transcriptomics and proteomics data, suggest that Osgep may execute its function through the interconnected pathways linked to protein processing in ER and insulin secretion. Meanwhile, it has been reported that Osgep may be involves in homeostatic regulation of the ER stress19. In support of this general possibility, we found that Osgep knockout causes an upregulation of the ER chaperone BiP at the mRNA and protein levels. Bip is an ER chaperone protein that binds with ER stress sensors (IRE1α, PERK, and ATF6) under normal conditions. Increased misfolding/unfolding proteins competitively bind with Bip and release sensors to activate ER stress. Depending on the extent of ER stress, this response can be adaptive or maladaptive, restoring ER homeostasis or activating the UPR signaling further promoting β-cellular death43,44,45. We observe all three transmembrane proteins are activated to different extents. The activated PERK pathway promoted the phosphorylation of eIF2α to reduce the protein influx into the ER. The PERK-eIF2α axis suppresses of canonical translation but enhances the translation of ATF4, then resulting in transcriptional activation of CHOP, a direct regulator of ER stress-drove β-cells apoptosis. Up-regulated expression of UPR were investigated in the Osgep deficiency islets at HFD-fed. What’s more, in TM-induced in vivo- and TG-induced in vitro-ER stress model mouse islet, the activation trends of the three UPR pathways were also validated, suggesting that Osgep deficiency caused ER stress and subsequent activated UPR signaling pathway.
In particular, active ER stress and dysregulated UPR signaling are significant causes of the dysfunction and death of β-cells in diabetes. Therefore, improvement of ER function and intervention of the UPR in β-cells may be useful therapeutic strategies for diabetes. Several studies in humans have provided some strategies. However, known targets may not adequately reduce ER stress and improve islet β-cells function. Meanwhile, Osgep protein was lower expression in diabetic islets both in humans and rodents. Our results illustrate the potential of Osgep as targets for relieving ER stress in diabetes. Currently, protein-based biopharmaceuticals are difficult to achieve, owing to the large molecule, complicated production and purification, and in certain instances non-native post-translational modification of the protein. Given these limitations, nucleic acid-based therapy as an alternative way demonstrates its superiorities, which has been extensively evaluated for both therapeutic proteins and prophylactic vaccines. The most advanced nucleic acid drugs including AAV vectors vaccine and mRNA vaccine are currently being used for vaccination purposes for COVID-19 before46,47. Here, we specifically deliver Osgep to the pancreas using an AAV2/Pan vector in HFD-induced diabetic mice. Our results showed that increasing Osgep could significantly reduce blood glucose levels and increase glucose-stimulated insulin secretion. Meanwhile, adenovirally facilitated Osgep expression in mouse islets reduced the expression levels of ER stress-related genes. However, this type of AAV cannot be specifically expressed in islet β-cell48,49. Therefore, the therapeutic translatability of our current study remains to be further investigated.
In conclusion, our results suggest that functional loss of Osgep in β-cells affects the accuracy and efficiency of protein translation via t6A37 modification of tRNA, resulting in the synthesis of misfolded Proinsulin protein, which triggers the UPR and diabetes. Together, Osgep upregulation could be considered as a potential treatment to improve UPR-dependent diabetes.
Methods
Statement of ethical regulations
All animal care protocols and experiments were reviewed and approved by the Animal Care and Use Committees of the Laboratory Animal Research Center of Central South University (Changsha, China; Permit No. 2019sydw0126).
All human samples were approved by the Medical Ethics Committee of Xiangya Hospital, Central South University (ID: 2019040116). All individuals provided informed consent prior to participating in the study.
Analysis of human islet RNA-seq datasets
The public human islet bulk RNA-seq dataset GSE7689620 (116 healthy control; 55 T2DM), and scRNA-seq dataset GSE8647322 (5 healthy control with 168 β cells; 3 T2DM with 96 β cells), E-MTAB-506121 (6 healthy control with 171 β cells; 4 T2DM with 99 β cells) were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/) and ArrayExpress database (https://www.ebi.ac.uk/biostudies/arrayexpress). For the scRNA-seq datasets, we downloaded the pre-processed fastq files of β cells. Using the Bioconductor limma package, we performed statistical analysis to identify differentially expressed genes (DEGs) between the two groups, The p values were adjusted by Benjamini-Hochberg false discovery rate correction.
Animal studies
C57BL/6 J mice were obtained from Gempharmatech Co., Ltd. To generate Osgep-floxed mice, two loxP sequence fragment were inserted at the two terminals of Osgep exon 2-3 using CRISPR/Cas9 technology. Ubc-creERT mice and Ins2-cre mice were obtained as previously described. For genotyping, genomic DNA was extracted from tail tips and PCR was performed using the designed primers (Supplementary Table 1). Based on the principle of the cre-loxP system, Osgep-floxed mice were interbred with 2 types of cre mice respectively to generate global Osgep knockout mice and islet β cell-specific Osgep knockout mice. The Osgep-floxed littermates were used as controls.
All mice were housed in the Laboratory Animal Research Center of Central South University, where is a pathogen-free animal facility at a controlled temperature under standard laboratory conditions (12 h light/dark cycle, temperature kept at 21–24 °C and 40-70% humidity) with food (Cat#1016706714625204224, Beijing Keao Xieli Feed Co., Ltd.) and water provided ad libitum.
All animal care protocols and experiments were reviewed and approved by the Animal Care and Use Committees of the Laboratory Animal Research Center of Central South University (Changsha, China; Permit No. 2019sydw0126).
Intraperitoneal glucose tolerance test (IPGTT)
For IPGTT, mice were fasted overnight (5:30 PM to 8:00 AM) and then intraperitoneally injected with 2 g/kg body weight of 20% D-glucose (Solarbio, Beijing, China) in sterile saline solution by standard methods. Blood glucose levels was measured from the tail vein using a glucometer (AUUC-CHEK, Roche) at 0, 15, 30, 60, and 120 min post-injection.
Insulin tolerance test (ITT)
For ITT, mice were fasted for 6 h (8:00 AM to 2:00 PM). Blood glucose levels were measured from the tip of the tail vein prior to an intraperitoneal injection 0.75U/kg insulin (Beyotime, Beijing, China) in sterile saline solution. Blood glucose levels were measured at 0, 15, 30, 60, and 90 min after insulin administration using a glucometer.
Glucose stimulated insulin secretion (GSIS)
For in vivo GSIS, mice were fasted overnight, then injected intraperitoneally with glucose (2 g/kg). After 15 min, blood samples were obtained from retro-orbital plexus of the mice. The blood samples were centrifuged at 1000 g for 15 minutes, and the serum insulin levels were measured using a mouse insulin ELISA kit (10-1247-01, Mercodia, Sweden), according to the manufacturer’ instructions. For in vitro GSIS, 10 size-matched medium islets were loaded on a 24 well plate in RPMI-1640 with 10% FBS for 24 h after isolation. The islets were then equilibrated with Krebs Ringer Buffer (KRBH; 128 mM NaCl, 4.8 mM KCl, 1.2 mM KH2PO4, 1.2 mM MgSO4, 2.5 mM CaCl2, 5 mM NaHCO3, 10 mM HEPES, and 0.1% BSA) plus 2.8 mM glucose for 30 min. The buffer was then replaced with KRBH supplemented with either 2.8 or 16.7 mM glucose for 30 min. The supernatant was collected and used to measure insulin levels by ELISA as described above. All steps were performed at 37 °C in a tissue culture incubator.
Fasting serum insulin and glucagon concentrations
After fasting for 6 h, blood samples were collected from the mice and the serum was separated. The serum insulin concentration was determined by an ELISA kit (10-1247-01, Mercodia, Sweden). The serum glucagon concentrations were determined by an ELISA kit (ZC-38937, Zcibio, China). following the manufacturer’s instructions.
Pancreas digestion and islet isolation
Overnight fasted mice were anesthetized by inhalation of isoflurane. After cervical dislocation resulting in euthanasia, the mouse the mouse abdomen was disinfected with ethanol, and the abdominal cavity was opened to allow isolation of the pancreatic tissue. Next, 3 mL of a collagenase Type V solution (C9263, Sigma-Aldrich, St. Louis, MO) was slowly injected into the bile duct after occlusion of the proximal end. The distended pancreas was then excised and digested at 37 °C for approximately 8 min to allow complete digestion. The digestion was stopped by the addition of 20 mL RPMI-1640 with 10% FBS, followed by centrifugation (1000 g, 2 min). The pancreas samples were then washed three times with 20 mL of RPMI-1640 medium. The isolated islets were stained reddish-brown by Dithizone (Solarbio, Ch. Beijing) to facilitate the harvest and then hand-picked with the aid of a pipette under a stereoscopic microscope (Olympus, Japan).
Mice insulin purification
To preserve the activity of insulin, pancreases (n = 8) were removed and immediately frozen in liquid nitrogen. Insulin was extracted by mechanical homogenization of the frozen pancreases in iced acid ethanol (0.7 M HCl/ethanol 25:75 with pH = 2.0). After stirring the mixture for 3 h at 8 °C, the samples were centrifuged and the supernatant was collected. Additional iced acid ethanol was added to the precipitate to extract insulin in the same manner. The combined extracts were then neutralized by slowly adding ammonia solution dropwise until the pH reached 7.8, and the supernatant was collected by vacuum filtration. Next, the pH was adjusted to 3.5 using sulfuric acid, which caused acidic proteins to precipitate for 4 h at 4 °C. The supernatant was then concentrated to 1/10 of the original volume under reduced pressure, followed by the steps: 50 °C for 5 min; 4 °C for 4 h. Finally, the insulin was extracted via salting out and stored tightly sealed after vacuum drying. The activity of the extracted insulin was measured following the method described in the Chinese Pharmacopeia 2020.
Morphometric evaluation
For immunofluorescence staining, formalin-fixed, paraffin-embedded mice pancreas samples were sectioned and immunostained. The primary antibodies used were anti-insulin (Proteintech, 15848-1-AP, 1:500 dilution in blocking buffer), anti-glucagon (Proteintech, 15954-1-AP, 1:200 dilution in blocking buffer), and Osgep (SANTA CRUZ, sc-393199, 1:50 dilution in blocking buffer) antibodies. The second antibodies used were the CoraLite488-conjugated Donkey Anti-Rabbit IgG(H + L) (Proteintech, SA00013-6, 1:500 dilution in blocking buffer) and CoraLite594-conjugated Goat Anti-Mouse IgG (H + L) (Proteintech, SA00013-3, 1:500 dilution in blocking buffer).
For hematoxylin and eosin (H&E) staining, paraffin-embedded sections of the pancreas, liver, white adipose tissue (WAT), and brown adipose tissue (BAT) were stained with hematoxylin and eosin.
For Oil red O staining of liver tissues, frozen sections of mouse liver were stained with 0.5% Oil red O working solution to examine intracellular lipid accumulation. All slides were imaged using a 3DHISTECH Pannoramic MIDI fluorescent microscope.
Transmission Electron Microscopy
Isolated islets were fixed in 2.5% glutaraldehyde overnight. After washed three times with 0.1 M phosphate buffer (pH 7.2), the samples were fixed in 1% osmic acid at 4 °C for 2 h. The samples then underwent a series of alcohol dehydration steps (30, 50, 70, 80, 95, and 100% ethanol). Following dehydration, the samples were embedded in epoxy resin. Ultrathin sections (70 nm thick) were cut and stained with uranyl acetate. The stained sections were examined using a HT7800 transmission electron microscope.
Cell culture
The mouse pancreatic β-cell line, Min6 cells (Cellosaurus, CVCL_0431), were cultured in DMEM medium supplemented with 10% fetal bovine serum (FBS) and 50 μM β-mercaptoethanol. The cells were maintained in a humidified incubator at 37 °C with 95% air and 5% CO2. To generate Osgep stable knockout Min6 cell, Osgep-shRNAs in the pLKO-puro lentiviral vector were packaged in Hek293T cells (Takara Bio, Cat. 632180). The packaging was done by transient transfection, in combination with the packaging constructs pRSV-Rev, pCMV-VSVG-G, pCgpV. The viral supernatants were collected 48 h and 72 h after transfection. Min6 cell were infected with the lentiviral supernatants in the presence of 8 μg/mL polybrene. Infected cells were selected with 10 μg/mL puromycin for 48 h. The knockdown efficacy was confirmed by qPCR and western blot analysis. The HEK293T cells were cultured in DMEM supplemented with 10% FBS.
Cell apoptosis assay
Min6 cells with different Osgep expression levels were plated at a density of 1 × 104 cells/well in 96-well plates. The cells were then incubated with DMSO or 1 μM Thapsigargin (Tg) for 6 h. Cell apoptosis was measured using the nucleic acid dye propidium iodide (PI) and Hoechst. Briefly, the cells were incubated with 10 μL of PI and Hoechst reagent per well for 2 h. The absorbance was then measured at 636 and 460 nm. ImageJ was used to count the number of PI+ cells and total cells.
Isolation of RNA and qPCR
Total RNA was extracted from the cultured cells or the tissues of mice using Trizol reagent (Takara, Japan) according to the manufacturer’s protocol. 1 μg RNA was reverse transcribed to cDNA using the Prime Script RT reagent kit with gDNA Eraser (Takara, Japan). Quantitative PCR (qPCR) was conducted using a SYBR-based real-time PCR assay on a QuantStudio 5 Real-Time PCR system (Thermo Fisher Scientific) with 2 × SYBR Green qPCR Master Mix (Bimake.cn China). The relative expression of the genes of interest was calculated using comparative Ct approach with ACTB as an endogenous control. The primer sequences were summarized in Supplementary Table 2. All experiments were performed a minimum of three times.
Western blot analysis
Total protein from the cells or tissues were separated by SDS-PAGE electrophoresis and then transferred to a PVDF membrane. After blocking with 5% skim milk, the membrane was incubated overnight at 4 °C with one of the following primary antibodies: Osgep (SANTA CRUZ, sc-393199, 1:500 dilution in blocking buffer), β-Tubulin (Proteintech, 10094-1-AP, 1:20000 in blocking buffer), Insulin (Proteintech, 66198-1-Ig, 1:2000 in blocking buffer), Proinsulin, (Novus, NB100-73013, 1:1000 in blocking buffer), IRE1α (Cell Signaling Technology, 3294, 1:1000 in blocking buffer), Bip (Proteintech, 66574-1-Ig, 1:1000 in blocking buffer), PERK (Sigma, P0074, 1:2000 in blocking buffer), Phospho-PERK (Sigma, SAB5700521, 1:1000 in blocking buffer), eIF2α (Cell Signaling Technology, 5324, 1:2000 in blocking buffer), Phospho-eIF2α (Cell Signaling Technology, 3398, 1:1000 in blocking buffer), ATF4 (Cell Signaling Technology, 11815, 1:1000 in blocking buffer), CHOP (Cell Signaling Technology, 2895, 1:1000 in blocking buffer), ATF6 (Proteintech, 66563-1-Ig, 1:1000 in blocking buffer), AKT (Proteintech, 60203-2-Ig, 1:2000 in blocking buffer), p-Akt1/2/3 (SANTA CRUZ, SC-514032, 1:500 in blocking buffer) or Gapdh (Proteintech, 60004-1-Ig, 1:50000 in blocking buffer). The membranes were then incubated for 1 h at room temperature with goat anti-rabbit-HRP antibody (Sigma, A0545, 1:20000 dilution in blocking buffer) or goat anti-mouse-HRP antibody (Sigma, A6154, 1:2000 dilution in blocking buffer). The signals were detected by chemiluminescence assay using a ChemiDoc Touch (Bio-Rad). Uncropped scans of all blots are supplied as Source Data files.
Analysis of t6A37 modification of tRNA
Total RNA from 3 WT and 3 Osgep-Ins2-Cre mice was extracted from mouse islet cells, and the tRNA was then purified from this total RNA sample. To quantify the t6A37 level of tRNAs, they were enzymatically hydrolysed into individual nucleosides50,51. Approximately 100 μg of the bulk tRNA was digested with 0.01 units of snake venom phosphodiesterase (Sigma), 10 units of nuclease P1 (Sigma), and 3 μL of alkaline phosphatase (Sigma). The resulting nucleosides were lyophilized, resuspended in 100 μL of water, and then subjected to High-Performance Liquid Chromatography–Mass Spectrometry Analysis (HPLC-MS). The nucleosides were resolved using an ExionLC AD instrument (AB SCIEX, USA) equipped with a Discovery C18 (250 mm, 4.6 mm, 5 μm) reverse-phase column (Waters) and maintained at a column temperature of 30 °C. The elution gradient consisted of 10 mM ammonium acetate (A) and 40% acetonitrile (B), with the following the steps: 0–7 min 99% A, 7–30 min 95% A, 30–40 min 85% A, 40–50 min 95% A, and 50–60 min 99% A. The nucleosides were then analyzed using a mass spectrometer (Triple Quad 6,500, AB SCIEX, USA) operated in positive ion mode using Multiple/Selected reaction monitoring (MRM/SRM) mode with a collision energy of 10 eV. The mass transitions monitored were m/z 268-m/z 136 for adenosine (A), m/z 413.2-m/z 281 for t6A. Then calculated ratio of t6A to A to accurately reflect the differences in t6A content by normalized to the respective A content. Data acquisition and quantitative spectral analysis was conducted using the Analyst Software 1.6.3.
Circular dichroism analysis
The 6 μg protein was dissolved in 300 μL of water. The protein solution was then scanned with a circular dichroism spectrometer (MOS-500 circular dichroism chromatograph, Bio-Logic Science Instruments, USA) with a 0.1 cm path-length quartz cuvette. The spectra were recorded in a step size of 1 nm and a bandwidth of 2.0 nm, collecting ellipticity data from 190 to 260 nm at a 1 nm data pitch. A background value for each test was subtracted from the corresponding value of each sample, and the data was analyzed using the Jasco software.
In vitro transcription system
Plasmids (pT7-PreProinsulin) for in vitro translation were prepared by Genechem (Shanghai, China), which was linearized by dissecting with Hind III to serve as an in vitro transcription DNA template (Supplementary Fig. 7A). In vitro transcription was performed using the T7 High Yield Capped RNA Transcription Kit (Thermo Fisher) in a 20 μL rection containing the following components: 10 μL of 2 × NTP/CAP, 2 μL of 10 × Reaction Buffer, 2 μL of Enzyme Mix, 5 μL of Nuclease-free water and 1 μg of liner template. This process yielded the Proinsulin mRNA, which along with purified tRNA were assayed by denaturing polyacrylamide gels electrophoresis (Supplementary Fig. 7B). In vitro translation reactions were performed according to the manufacturer’s instructions in the Flexi® Rabbit Reticulocyte Lysate System (Promega). To identify the differences of exogenous tRNAs, we depleted endogenous tRNAs from RRL (Flexi® Rabbit Reticulocyte Lysate) by ethanolamine-sepharose column chromatography52. The luciferase activity was detected to quantify the in vitro translation system’s output, and the Proinsulin protein product was measured by Western blot analysis.
RNA-seq
Total RNA was extracted from islets of 15-week-old male WT and Osgep-Ins2-Cre mice. RNA expression profiling was performed using the Illumina NovaSeq 6000. The clean reads were mapped to the reference genome sequence using the Hisat2 software. Differential expression analysis between the two groups was performed using the DESeq2 software.
TMT-based proteomics analysis
Protein was extracted from the isolated islets using SDT lysis buffer (containing 100 mM NaCl and 1/100 DTT). Sufficient iodoacetamide (IAM) were added to the samples, and they were incubated for 1 h at room temperature in the dark. After a quality test by Bradford quantitative method, the proteins were digested with trypsin and incubated overnight at 37 °C. The next day, TMT labeling reagent (Thermo Fisher Scientific) dissolved in acetonitrile was added, and all the labeled samples were mixed in equal volumes, desalted, and lyophilized. The protein powder was then dissolved in 2% acetonitrile, and fractionated using a C18 column (Waters BEH C18, 4.6 × 250 mm, 5 μm) on a Rigol L3000 HPLC system. Finally, the eluate was split into 10 equal-volume aliquots and lyophilized by vacuum centrifugation.
Mobile phase A (100% water, 0.1% formic acid) and liquid B (80% acetonitrile, 0.1% formic acid) were prepared for liquid chromatography (LC). Samples of 1 μg from each fraction were injected into the LC-MS/MS system for detection. The EASY-nLCTM 1200 nano-upgraded UHPLC system was connected to an Q ExactiveTM HF-X mass spectrometer, fitted with a Nanospray Flex™ (ESI) ion source. The following gradient was employed for peptide separation: 2 min, 15% B; 48 min, 40% B; 50 min, 50% B; 51 min, 55% B; 60 min, 100% B. The mass spectrometry data-dependent acquisition mode was used with the detection mode set to positive ion, a parent ion scan range of 350–1500 m/z, a resolution of 60000 (at m/z 200), a maximum ion injection time of 20 ms, and a dynamic exclusion parameter of 20 s.
Data analysis and visualization of TMT data were conducted using the Proteome Discoverer (PD 2.2, Thermo Fisher Scientific) platform and R statistical framework. The search parameters were set as follows: mass tolerance for precursor ion was 10 ppm and mass tolerance for production was 0.02 Da. To improve data quality, Peptide Spectrum Matches (PSMs) with a credibility of more than 99% were identified and the identified protein contains at least 1 unique peptide. The identified PSMs and protein were retained and processed with an FDR rate no more than 1.0%. Data normalization was then performed by dividing each protein ratio by the respective sample median.
Bioinformatics analysis
Both mRNA and protein expressions levels were submitted to perform the Gene Set Enrichment Analysis using GSEA 4.3.2 software. Differentially expressed (DE) genes were assessed based on two criteria: 1) the p value criterion, namely adjusted p value is less than 0.05; 2) the fold change (FC) criterion, namely that the fold up- or downregulation in the islet of Osgep-KO mice is greater than 1.2 or less than 0.83. The k-means clustering algorithm was used to classify the DE gene expression patterns, implemented using the ConsensusClusterPlus package53. The results were visualized as circular heatmaps using the TBtools v2.019 software54. Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathway enrichment analysis was carried out using the clusterProfiler and enrichplot package in R software55.
Adenoviral vector construction and infection
The 9-week-old C57BL/6 J mice were injected with AAV2/Pan-CMV-Osgep-EF1-GdGreen-WPRE virus to obtain pancreas-specific Osgep overexpression mice, or with the control virus AAV2/Pan-CMV-MCS-EF1-GdGreen-WPRE (OBiO Technology). The viral solution (5 × 1011 v.g. per mouse) was thawed at 25 °C before injection. The desired amount of virus was diluted with saline to a final volume of 100 μL per mouse, then introduced to the mice through intraperitoneal injection. The pancreas-specific Osgep overexpression in the mice was confirmed by IF and Western blot analyses.
Human subjects
The T2DM diagnosis was based on fasting blood glucose (FBG) levels ≥ 7.0 mmol/L or a 2 h post-glucose level ≥ 11.1 mmol/L. Patients with any type of cancer, autoimmune diseases, nephropathy, or severe liver disease history were excluded. Medical data, including age, sex, body mass index (BMI) and blood glucose, were collected for the subjects. The BMI was calculated based on height and weight using the formula BMI = weight (kg)/height2 (m2). The control group was no family history of diabetes mellitus or any type of cancer or chronic inflammatory disorders. 234 T2DM and 285 control individuals were recruited for the discovery cohort, while 228 T2DM and 228 control individuals were recruited for the validation cohort.
The study was approved by the Medical Ethics Committee of Xiangya Hospital, Central South University (ID: 2019040116). All individuals provided informed consent prior to participating in the study.
The subjects of T2DM GWASs have been reported previously, which comprised 18,483 cases and 366,937 controls29. The GWAS summary dataset was deposited in the database of GWASaATLAS (https://atlas.ctglab.nl) with GWAS Atlas ID 3328.
OSGEP SNP Genotyping
After collecting 5 mL of peripheral venous blood from all enrolled individuals, genomic DNA was extracted from hemocyte using the Wizard® Genomic DNA Purification Kit (Promega, USA) according to the manufacturer’s instructions. The OSGEP gene is located on chromosome 14 with 11 exons. All SNPs were selected using Haploview 4.1 with default settings (pairwise r2 ≥ 0.8) and a minor allele frequency (MAF) ≥ 0.05). Sequenom MassARRAY system was used to genotype for candidate SNPs. The relevant information was in Supplementary Data 4.
OSGEP Reporter Constructs and luciferase reporter assay
The sequence (chr14:20,449,457 - 20,450,034) in intron 3 of OSGEP containing rs74512655 was first PCR amplified with primers: forward: 5’-CTATCGATAGGTACC GAGCTCTGGCCTTAAGTGAATTGCCATT-3’; reverse: 5’-GGCTAGCACGCGTAA GAGCTCGGATTCCAGGAACAGATTCCTAGA-3’. This PCR product and the pGL3 vector (Promega) were both digested with SacI restriction enzyme and subsequently ligated. Site-directed mutagenesis was then used to change the allele at rs74512655 from G to C, which was performed using the Fast Mutagenesis System (TransGen Biotech, Beijing). Another sequence of OSGEP promoter (fragment corresponding to -1500 to the ATG start site of the OSGEP gene) was PCR amplified with the following primers: forward: 5’-ATTTCTCTATCGATAGGTACCGAAATACAACTGGCAAGA CTGGG-3’; reverse: 5’-CACGCGTAAGAGCTCGGTACCACTCCTGGCAATGTCA GGAGC-3’. These PCR products, along with the pGL3 vector, were digested with KpnI restriction enzyme and subsequently ligated. The correct sequences and insert orientations of all plasmids were verified by Sanger sequencing. 2.5 μg of reporter plasmid and 50 ng of phRG-TK Renilla luciferase internal control plasmid were co-transfected into HEK293T cells using the Lipofectamine 2000 reagent (Invitrogen, USA). After 24 h, the luciferase activities were measured using the dual-luciferase reporter assay system (Promega, USA). Data were presented as the ratio of firefly to renilla luciferase activity (FLuc/RLuc).
Statistics & reproducibility
The statistical analyses were performed using GraphPad Prism 8, Microsoft Excel, and SPSS software 26. All samples represent biological replicates, and the statistical measurements are presented as mean ± SD as specified in each figure. No statistical method was used to predetermine sample size. No data were excluded from the analyses. The experiments were randomized. The Investigators were blinded to allocation during experiments and outcome assessment. An unpaired Student’s t-test (two-tailed) was conducted to assess the statistical significance of one observed parameter between two experimental groups. For comparisons of more than two groups, the data were analyzed using one-way analysis of variance (ANOVA) with Tukey’s test. Chi-square test was used to assess the relationship between the binary categorical variable (sex) and 2 groups of population, while the nonparametric Mann-Whitney U test was used to analyze the continuous variables (age, BMI, fasting plasma glucose). Single-factor binomial Logistic Regression Analysis was used to calculate the influences of sex, age and BMI (Supplementary Data 5). Then, we defined age and sex as confounders in the discovery and combined cohort, as well as age in validation cohort. Multi-factor binomial Logistic Regression Analysis with adjusted for the confounders mentioned previously to analyze the influences of genetic variants on the T2DM risk. p value < 0.05 was considered to indicate a statistically significant difference. The P values and n values are given in the figures or corresponding legends. All representative experiments (such as Western blot, IF, H&E and oil red O staining, and Electron microscopy) are repeated at least three times independently with similar results.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNA-seq datasets of human islet (GSE76896, E-MTAB-506, and GSE86473) have been previously published. The transcriptomic data was obtained from the SRA website with accession number PRJNA1132596. Proteomics data have been deposited to the ProteomeXchange Consortium via the iProX partner repository56,57 with the dataset identifier PXD053834. All data are included within the article or Supplementary Data. Source data are provided with this paper.
References
Wang, P. et al. Diabetes mellitus–advances and challenges in human β-cell proliferation. Nat. Rev. Endocrinol. 11, 201–212 (2015).
Gurzov, E. N. et al. Novel strategies to protect and visualize pancreatic β cells in diabetes. Trends Endocrinol. Metab. 31, 905–917 (2020).
Weir, G. C., Gaglia, J. & Bonner-Weir, S. Inadequate β-cell mass is essential for the pathogenesis of type 2 diabetes. Lancet Diabetes Endocrinol. 8, 249–256 (2020).
Kalwat, M. A. & Cobb, M. H. Mechanisms of the amplifying pathway of insulin secretion in the β cell. Pharm. Ther. 179, 17–30 (2017).
Eizirik, D. L., Cardozo, A. K. & Cnop, M. The role for endoplasmic reticulum stress in diabetes mellitus. Endocr. Rev. 29, 42–61 (2008).
Ghosh, R., Colon-Negron, K. & Papa, F. R. Endoplasmic reticulum stress, degeneration of pancreatic islet β-cells, and therapeutic modulation of the unfolded protein response in diabetes. Mol. Metab. 27s, S60–s68 (2019).
Laybutt, D. R. et al. Endoplasmic reticulum stress contributes to beta cell apoptosis in type 2 diabetes. Diabetologia 50, 752–763 (2007).
Wilinski, D. & Dus, M. N(6)-adenosine methylation controls the translation of insulin mRNA. Nat. Struct. Mol. Biol. 30, 1260–1264 (2023).
Scheuner, D. & Kaufman, R. J. The unfolded protein response: a pathway that links insulin demand with beta-cell failure and diabetes. Endocr. Rev. 29, 317–333 (2008).
Arrondel, C. et al. Defects in t(6)A tRNA modification due to GON7 and YRDC mutations lead to Galloway-Mowat syndrome. Nat. Commun. 10, 3967 (2019).
Wang, J. T. et al. Commonality and diversity in tRNA substrate recognition in t6A biogenesis by eukaryotic KEOPSs. Nucleic Acids Res 50, 2223–2239 (2022).
Zhou, J. B. et al. Molecular basis for t6A modification in human mitochondria. Nucleic Acids Res 48, 3181–3194 (2020).
Su, C., Jin, M., & Zhang, W. Conservation and Diversification of tRNA t(6)A-Modifying Enzymes across the Three Domains of Life. Int. J. Mol. Sci., 23, 10.3390/ijms232113600 (2022).
Thiaville, P. C. et al. Essentiality of threonylcarbamoyladenosine (t(6)A), a universal tRNA modification, in bacteria. Mol. Microbiol 98, 1199–1221 (2015).
Thiaville, P. C. et al. Global translational impacts of the loss of the tRNA modification t(6)A in yeast. Micro. Cell 3, 29–45 (2016).
Galperin, M. Y. & Koonin, E. V. ‘Conserved hypothetical’ proteins: prioritization of targets for experimental study. Nucleic Acids Res 32, 5452–5463 (2004).
Braun, D. A. et al. Mutations in KEOPS-complex genes cause nephrotic syndrome with primary microcephaly. Nat. Genet 49, 1529–1538 (2017).
Ng, A. P. et al. Cleavage of misfolded nuclear receptor corepressor confers resistance to unfolded protein response-induced apoptosis. Cancer Res 66, 9903–9912 (2006).
Krausel, V. et al. The transcription factor ATF4 mediates endoplasmic reticulum stress-related podocyte injury and slit diaphragm defects. Kidney Int 103, 872–885 (2023).
Solimena, M. et al. Systems biology of the IMIDIA biobank from organ donors and pancreatectomised patients defines a novel transcriptomic signature of islets from individuals with type 2 diabetes. Diabetologia 61, 641–657 (2018).
Segerstolpe, Å. et al. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 24, 593–607 (2016).
Lawlor, N. et al. Single-cell transcriptomes identify human islet cell signatures and reveal cell-type-specific expression changes in type 2 diabetes. Genome Res 27, 208–222 (2017).
Stott, N. L. & Marino, J. S. High fat rodent models of type 2 diabetes: from rodent to human. Nutrients, 12, 10.3390/nu12123650 (2020).
Lu, Y. et al. Obesity-induced excess of 17-hydroxyprogesterone promotes hyperglycemia through activation of glucocorticoid receptor. J. Clin. Invest 130, 3791–3804 (2020).
Nagarajan, S. R. et al. Dysregulation of hepatic metabolism with obesity: factors influencing glucose and lipid metabolism. Proc. Nutr. Soc. 81, 1–11 (2022).
Wei, F. Y. & Tomizawa, K. tRNA modifications and islet function. Diabetes Obes. Metab. 20, 20–27 (2018). p.
Abdullahi, A. et al. Modeling acute ER stress in vivo and in vitro. Shock 47, 506–513 (2017). p.
Naso, M. F. et al. Adeno-associated virus (AAV) as a vector for gene therapy. BioDrugs 31, 317–334 (2017).
Watanabe, K. et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet 51, 1339–1348 (2019).
Sun, J. et al. Proinsulin misfolding and endoplasmic reticulum stress during the development and progression of diabetes. Mol. Asp. Med 42, 105–118 (2015).
Lee, J. H. & Lee, J. Endoplasmic reticulum (ER) stress and its role in pancreatic β-cell dysfunction and senescence in type 2 diabetes. Int. J. Mol. Sci. 10.3390/ijms23094843 (2022).
Morris, G. E. & Korner, A. The effect of glucose on insulin biosynthesis by isolated islets of Langerhans of the rat. Biochim Biophys. Acta 208, 404–413 (1970).
Haataja, L. et al. Distinct states of proinsulin misfolding in MIDY. Cell Mol. Life Sci. 78, 6017–6031 (2021). p.
Beenstock, J. & Sicheri, F. The structural and functional workings of KEOPS. Nucleic Acids Res 49, 10818–10834 (2021).
Machnicka, M. A. et al. Distribution and frequencies of post-transcriptional modifications in tRNAs. RNA Biol. 11, 1619–1629 (2014).
de Crécy-Lagard, V. et al. Matching tRNA modifications in humans to their known and predicted enzymes. Nucleic Acids Res 47, 2143–2159 (2019).
Chen, C. W. et al. Adaptation to chronic ER stress enforces pancreatic β-cell plasticity. Nat. Commun. 13, 4621 (2022). p.
Arragain, S. et al. Identification of eukaryotic and prokaryotic methylthiotransferase for biosynthesis of 2-methylthio-N6-threonylcarbamoyladenosine in tRNA. J. Biol. Chem. 285, 28425–28433 (2010).
Wei, F. Y. et al. Deficit of tRNA(Lys) modification by Cdkal1 causes the development of type 2 diabetes in mice. J. Clin. Invest 121, 3598–3608 (2011).
Kautz, S. et al. Early insulin therapy prevents beta cell loss in a mouse model for permanent neonatal diabetes (Munich Ins2(C95S). Diabetologia 55, 382–391 (2012).
Pollo-Oliveira, L. et al. Loss of elongator- and keops-dependent trna modifications leads to severe growth phenotypes and protein aggregation in yeast. Biomolecules 10, 10.3390/biom10020322 (2020).
Rojas-Benítez, D., Eggers, C., & Glavic, A. Modulation of the proteostasis machinery to overcome stress caused by diminished levels of t6A-Modified tRNAs in Drosophila. Biomolecules, 7, 10.3390/biom7010025 (2017).
Eizirik, D. L. & Cnop, M. ER stress in pancreatic beta cells: the thin red line between adaptation and failure. Sci. Signal 3, pe7 (2010).
Herrema, H. et al. FKBP11 rewires UPR signaling to promote glucose homeostasis in type 2 diabetes and obesity. Cell Metab. 34, 1004–1022.e8 (2022).
Lee, H., et al. Stress-induced β cell early senescence confers protection against type 1 diabetes. Cell Metab. 10.3390/ijms232113600 (2023).
Walsh, E. E. et al. Safety and immunogenicity of two rna-based covid-19 vaccine candidates. N. Engl. J. Med 383, 2439–2450 (2020).
Tong, D. et al. Single-dose AAV-based vaccine induces a high level of neutralizing antibodies against SARS-CoV-2 in rhesus macaques. Protein Cell 14, 69–73 (2023).
Zheng, Y. et al. Activation of pancreatic acinar fxr protects against pancreatitis via osgin1-mediated restoration of efficient autophagy. Res. (Wash. D. C.) 2022, 9784081 (2022).
Chen, M. et al. Efficient gene delivery and expression in pancreas and pancreatic tumors by capsid-optimized AAV8 Vectors. Hum. Gene Ther. Methods 28, 49–59 (2017).
Pomerantz, S. C. & McCloskey, J. A. Analysis of RNA hydrolyzates by liquid chromatography-mass spectrometry. Methods Enzymol. 193, 796–824 (1990).
Srinivasan, M. et al. The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. Embo j. 30, 873–881 (2011).
Guo, J. et al. YRDC mediates the resistance of lenvatinib in hepatocarcinoma cells via modulating the translation of KRAS. Front Pharm. 12, 744578 (2021).
Wilkerson, M. D. & Hayes, D. N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 26, 1572–1573 (2010).
Hou, J. et al. Temporal transcriptomic and proteomic landscapes of deteriorating pancreatic islets in type 2 diabetic rats. Diabetes 66, 2188–2200 (2017).
Yu, G. et al. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16, 284–287 (2012).
Ma, J. et al. iProX: an integrated proteome resource. Nucleic Acids Res 47, D1211–d1217 (2019).
Chen, T. et al. iProX in 2021: connecting proteomics data sharing with big data. Nucleic Acids Res 50, D1522–d1527 (2022).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant no. 82373963 to H. Zhou, 82171467 and 82371477 to Z. Ye), as well as the Provincial Natural Science Foundation of Hunan (grant no. 2024JJ5569 to Q. Li). We thank A. Long (Tsinghua University) for guidance with islet isolation and X. Le (Central South University) for support in the pharmaceutical chemistry laboratory platform. Additionally, we acknowledge the authors from published studies to share their GWAS and scRNA-seq sequencing data of T2DM trials. We are grateful for resources from the Public Platform for Advanced Medical Research Equipments, Xiangya Hospital, Central South University.
Author information
Authors and Affiliations
Contributions
Y.L. and Q.L. provided the study concept and design. Y.L., X.Y., J.Z., R.Y., and H.Y., contributed to data acquisition and drafting of the manuscript. Y.L., P.Z., R.Z., T.W., Y.G., Z.Y., R.L., W.Z., H.Z. X.L. H.Z. and Q.L. participated in critical data analysis and discussion of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Johan Palmfeldt, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, Y., Yang, X., Zhou, J. et al. OSGEP regulates islet β-cell function by modulating proinsulin translation and maintaining ER stress homeostasis in mice. Nat Commun 15, 10479 (2024). https://doi.org/10.1038/s41467-024-54905-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-54905-8