Abstract
Fibrolamellar Hepatocellular Carcinoma (FLC) is a rare liver cancer characterized by a fusion oncokinase of the genes DNAJB1 and PRKACA, the catalytic subunit of protein kinase A (PKA). A few FLC-like tumors have been reported showing other alterations involving PKA. To better understand FLC pathogenesis and the relationships among FLC, FLC-like, and other liver tumors, we performed a massive multi-omics analysis. RNA-seq data of 1412 liver tumors from FLC, hepatocellular carcinoma, hepatoblastoma and intrahepatic cholangiocarcinoma are analyzed, obtaining transcriptomic signatures unrestricted by experimental processing methods. These signatures reveal which dysregulations are unique to specific tumors and which are common to all liver cancers. Moreover, the transcriptomic FLC signature identifies a unifying phenotype for all FLC tumors regardless of how PKA was activated. We study this signature at multi-omics and single-cell levels in the first spatial transcriptomic characterization of FLC, identifying the contribution of tumor, normal, stromal, and infiltrating immune cells. Additionally, we study FLC metastases, finding small differences from the primary tumors.
Similar content being viewed by others
Introduction
Fibrolamellar Hepatocellular Carcinoma (FLC) is a rare liver cancer1,2 that occurs in patients without a history of viral hepatitis, cirrhosis, or other known risk factors3,4,5. FLC presents with non-specific and vague symptoms2,6,7,8, and there are no serum biomarkers. The diagnosis of FLC, even from pathology slides, can be problematic9, contributing to misdiagnosis and underdiagnosis10,11. Thus, FLC has frequently metastasized by the time of diagnosis, which is lethal7,12,13. FLC is classified as a subtype of Hepatocellular Carcinoma (HCC)14. However, the transcriptome and proteome of FLC tumors are distinct from those of adjacent non-transformed (called “normal”) tissue and of HCC15,16. Characterizing the molecular changes occurring in FLC tumors and the different cell types included in surgical samples could provide markers for diagnosis, prognosis, and therapeutic interventions10.
In FLC17, a heterozygous deletion of ~ 400 Kb in chromosome 19p13.12 connects exon 1 of DNAJB1 with the exons 2–10 of PRKACA (the catalytic subunit of Protein Kinase A)18,19. Producing this deletion20,21, or just expression of the DNAJB1::PRKACA transcript20, is sufficient to create the tumor in mouse liver or human hepatocytes, and elimination of the fusion transcript is sufficient to kill the tumor22,23. No other recurrent mutations have been observed in FLC17. These results demonstrate that FLC is the result of a somatic mutation. It is not genetically inherited, and FLC is both triggered by and driven by the DNAJB1::PRKACA fusion.
There are numerous unresolved questions: What are the pathways of pathogenesis; what is the relation between primary and metastatic tumors; what are the contributions of DNA methylation; how similar or different is FLC from other liver tumors, especially those considered FLC-like? Interestingly, tumors with FLC-like histopathology have been reported in patients missing expression of PRKAR1A (an inhibitory regulatory subunit of PKA)24, or with alternate fusions between ATP1B1 and a catalytic subunit of PKA25. Moreover, fibrotic tumors with histological features of both HCC and FLC have been observed in patients with mutations inactivating BAP1, with a chromosomal gain of PRKACA or loss of PRKAR2A (another inhibitory regulatory subunit)26.
The clean genetic background of FLC facilitates addressing these questions. Determining a transcriptomic signature for a disease has been useful for its diagnosis prognosis, gaining mechanistic insights for understanding pathogenesis or developing new treatments, classifying tumors and identifying cancer subtypes27, studying tumor evolution and metastasis, and validating biological disease models28,29,30. Previous RNA-seq studies of FLC15,26,31,32,33 analyzed small datasets, had multiple limitations, and yielded inconclusive characterization of FLC tumors.
Here, we sequenced the whole transcriptome of 127 FLC and 2 FLC-like frozen tissue samples and reprocessed 73 FLC and 18 FLC-like samples from external datasets26,31,32,33,34,35, making a total of 220 samples. We generated a transcriptomic FLC signature and studied it through multiomics. We distinguished which dysregulations are present in tumor, stromal, immune infiltrating cells, or normal hepatocytes, using single-cell spatial transcriptomics. The signature helped elucidate FLC pathogenesis and demonstrated that FLC and FLC-like tumors with diverse dysregulations of PKA are a single disease with a common transcriptome, rather than a collection of diverse diseases with similar pathologic features. Moreover, primary tumors and metastases were found to be highly similar, with differential expression of only 0.6% of transcripts. This opens directions for understanding tumor proliferation, maintenance, and immune evasion. The transcriptomic signature is valuable for interrogating and classifying potential FLC samples from different experiments and laboratories, and for validating in vitro (organoids29) and in vivo (PDX30) models. In addition, through analysis of 1192 tumor and normal samples of HCC, hepatoblastoma (HBL), and intrahepatic cholangiocarcinoma (iCCA), we determined their transcriptomic signatures and studied the commonalities and differences between these and FLC (Fig. 1).
This includes 1412 RNA-seq samples (FLC: 220, iCCA: 139, HBL: 148, HCC: 905. Some of these FLC samples were also studied through whole-genome and whole-exome sequencing (n = 39), targeted bisulfite methylation sequencing (n = 31), proteome sequencing (n = 15), and spatial single-cell transcriptomics (n = 6). We also developed FLC models, including 15 different PDX, 18 genetically engineered mice, 27 organoids, and primary human hepatocytes expressing the chimeras DNAJB1::PRKACA and ATP1B1∷PRKACA.
Results
Finding the transcriptomic signature of FLC
We analyzed 73 samples from 7 independent studies15,26,31,32,33,34,35 (Supplementary Data 1) and found little overlap among them in the differentially expressed genes in FLC tumors compared to paired normal samples (Supplementary Fig. 1). This low consistency may be the result of FLC being a mixture of different diseases, the consequence of analytical problems, or ambiguity in determining what is FLC. The initial studies relied on histopathology to diagnose FLC, which has low reproducibility9. Since FLC is rare, some studies were limited to small datasets without paired tumors and normal or used surrogate normal samples36. In addition, some studies used older methods and software for quantifying gene expression, which can distort or even reverse dysregulation trends37, or inappropriate integration of different batches, which may lead to underpowered and biased results38.
To address these issues, we restricted our focus to samples that were validated to be FLC with a molecular test, then did RNA-seq with state-of-the-art analysis tools to generate an “FLC signature”. This signature was next refined on independent libraries, and then tested on the samples from the external independent studies.
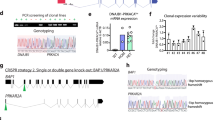
We performed RNA-seq of 6 libraries, comprising 127 patient samples of freshly frozen FLC tumors and normal tissue samples from the Fibrolamellar Tissue Repository at Rockefeller University (Supplementary Data 2). To confirm that the samples were FLC, we tested for reads mapping to both the first exon of DNAJB1 and the second exon of PRKACA, demonstrative of the DNAJB1::PRKACA fusion. Of these 127 FLC samples, we selected 81 matched tumor and normal samples from 5 libraries to calculate a transcriptomic FLC signature. These libraries were distributed into exploration and refinement datasets (see details in the “Methods”). For exploration, we selected the three largest libraries (RU-A, -B, and -C, with 67 matched tumor-normal samples). Unsupervised clustering showed a separation between tumor and normal samples but a strong batch effect among libraries (Supplementary Fig. 2). Batch effects can result from many factors, including different RNA extraction, library preparation or ribosomal depletion methods, or sequencing machines38. The batch effect in libraries RU-A, -B, and -C was not fully mitigated by adding a variable representing the experimental processing group or using batch effect removal tools39,40. Therefore, each library was analyzed independently, obtaining genes differentially expressed between FLC tumors versus normal tissue (Fig. 2A). Their intersection was filtered following detectability, consistency, and refinement filters (using 14 matched tumor and normal samples from libraries RU-D and -E). This resulted in a collection of 287 up- and 406 down-regulated genes, which we named the “transcriptomic FLC signature” (Fig. 2B and Supplementary Data 3).
A Differentially expressed genes obtained by the library (FDR < 0.05 and |log2(FC)| > 1). In (B) filters were applied to the intersection of the exploration datasets to obtain the transcriptomic FLC signature (287 up- and 406 down-regulated genes), as detailed in the Methods. C Validation using three external datasets. In these, we calculated the dysregulation trends of the FLC signature genes. In all cases, we confirmed that they matched with the trends obtained in panel (B). The libraries used correspond to the human tissue samples sequenced in RU-A: Simon et al.15, RU-B and RU-C: this study, RU-D: Lalazar et al.30, RU-E: Narayan et al.29. We used as validation three external datasets of patients’ samples: Sorenson et al.32, Francisco et al.33 and the TCGA-LIHC study35. In these datasets, we calculated the dysregulation trend of each of the genes in the FLC signature. For all genes, we confirmed that these trends matched those obtained in panel (B). The raw reads and normalized read counts for this figure are deposited in their corresponding dbGAP, GEO, and EGA repositories, as detailed in the data availability section. Access can be requested directly to these repositories under their privacy and confidentiality terms.
As a validation test, we checked the dysregulation trend of each of these 693 genes in the FLC signature against FLC transcriptomes from independent external studies. The samples were processed as were the samples we sequenced, excluding samples with reads that bridged the fusion of DNAJB1::PRKACA in the “normal” samples, or that did not have such reads in the “tumor”. We selected 3 studies with 28 matching tumor and normal samples32,33,35. Remarkably, all the dysregulation trends observed in the FLC signature were confirmed in these external samples (Fig. 2C). Further, with the transcriptomic FLC signature, unsupervised clustering cleanly distinguished tumor and normal (Fig. 3B, UMAP with HDBSCAN), with no signs of the batch effect observed with the whole transcriptome (Fig. 3A). All the validated DNAJB1::PRKACA FLC tumors, from different sequencing libraries and laboratories, clustered together. A similar result is observed in a heatmap (Supplementary Fig. 3). This consistency affirms the importance of using only samples validated by a molecular test and of remapping all the data using a consistent set of modern tools. Further, the result provides a tool that can give important insights into FLC.
Panels (A, B) Unsupervised clustering using UMAP with HDBSCAN of FLC and FLC-like samples. We analyzed the samples of Requena et al. (this study), and the samples deposited in public databases from the studies of Francisco et al.33, Xu et al.31, Robinson et al.34, Sorenson et al.32, Simon et al.15, Hirsch et al.26, and the TCGA-LIHC study35 (N = 185 samples). In the legend (upper right) the symbol “-” indicates that the corresponding dataset does not have normal samples. A Plot using all the genes, showing a strong batch effect. B Plot using only the genes of the FLC signature, showing no batch effect. All the FLC tumors included have the fusion transcript DNAJB1::PRKACA, but the FLC-like samples do not. Instead, they either have the chimera ATP1B1::PRKACA, mutations in BAP1, or missing R1A activity (∆R1A). Panels (C–E) Histopathological assessment of a pair of samples from a patient, classified by the hospital as a tumor and a normal sample. Surprisingly, the normal sample clustered with the FLC tumors (in B, a light blue square with a black arrowhead pointing to it). Blind pathological assessment with 300 additional de-identified slides resulted in the identification of normal regions (panel C), but also other regions with large eosinophilic cells and fibrous bands (panel D), characteristic of FLC tumors. These regions look histologically like the paired tumor piece (panel E). The scale bar for panels (C–E) is 100 μM. The specific sample IDs are detailed in the tables in Supplementary Data 1 and 2. The raw reads and normalized read counts for this figure are deposited in their corresponding dbGAP, GEO, and EGA repositories, detailed in the data availability section. Access can be requested directly to these repositories under their privacy and confidentiality terms.
The FLC signature is a useful diagnostic tool
One illustrative example of the utility of the signature is from the analysis of a pair of tissue blocks from a patient which the surgery hospital categorized as an FLC tumor and its adjacent normal sample. However, RNA-seq of the normal, sequenced in library F (which was not used to generate the FLC signature), clustered with the FLC tumors (Fig. 3B, light blue square with a black arrowhead). We extracted RNA of this presumptively normal sample and, with RT-PCR detected the chimeric DNAJB1::PRKACA. In parallel, the sample was analyzed as part of a deidentified collection of 300 histopathology slides by a board-certified pathologist (MST). The presumptive “normal” was determined to have not only normal regions (Fig. 3C) but also other regions (Fig. 3D) that were indistinguishable from the FLC tumor (Fig. 3E). Thus, the transcriptomic signature detected a mixed tumor-normal sample wrongly curated as normal.
FLC is a single disease
There are tumors that do not express the DNAJB1::PRKACA but are considered “FLC-like”, due to their histopathology. These are positive for arginase, CK7, and CD-68 and display large eosinophilic cells with abundant cytoplasm and prominent nucleoli, pale bodies in the cytosol, and lamellar bands of fibrosis traversing the tumor3. As the histopathological diagnosis of FLC has previously proven problematic9 (e.g., fibrous bands could be the consequence of inflammation of the liver41), we used the FLC signature to examine the extent to which they are “FLC-like” at the transcriptomic level.
One such tumor lacks detectable transcript and protein of the R1 regulatory subunit of PKA (PRKAR1A)24. The absence of R1 removes a constraint on PKA activity. Using the transcriptomic FLC signature, we found this sample clusters with the rest of the FLC tumors (in cyan, Fig. 3 and Supplementary Fig. 3).
A second set of FLC-like tumors has been found in the ductal cells of the liver and pancreas. These express either DNAJB1::PRKACA, ATP1B1::PRKACA, or ATP1B1::PRKACB. When we analyzed a tumor expressing ATP1B1::PRKACA25 in the cholangiocytes using the transcriptomic FLC signature, it clustered with the FLC tumors (hepatocytes with DNAJB1::PRKACA) (in navy blue in Fig. 3 and Supplementary Fig. 3). The ATP1B1::PRKACA and DNAJB1::PRKACA tumors have two different driver mutations and occur in two different cell types, yet share an increase of the catalytic subunit of protein kinase A, and display a common transcription profile.
A third set of FLC-like liver tumors have mutations in the deubiquitinating enzyme BRCA1 Associated Protein 1 (BAP1) and increased PKA activation26. However, the transcriptome of these does not cluster with the FLC tumors (Fig. 3B). A few factors may contribute to these differences. These patients are older (27–54 years old) than FLC patients (11–30 years old). Alternatively, this may be a different class of tumor, because these samples have genomic alterations additional to the mutations affecting PKA subunits. Inactivation of BAP1 is present in all these samples. This tumor suppressor gene42,43,44 has numerous effects on the development of stem cell pluripotency and controls the stability of different proteins45,46.
Small differences between the metastases and primary tumors of FLC
Of the 54,289 transcripts assessed, 6.5% (n = 3506) had a statistically significant (FDR < 0.05) and detectable alteration in primary tumors compared to paired normal adjacent tissue (1758 over- and 1748 under-expressed genes). In contrast, only 0.6% (n = 310) present alterations between metastases and primary tumors: 76 over and 113 under-expressed (Supplementary Data 4). Progressive dysregulation trends were observed comparing normal liver tissue (N) to primary (P) and metastatic (M) tumors. Some genes showed increasing expression (N < P < M, n = 14), while others showed the opposite pattern (N > P > M, n = 54) (Fig. 4A). Three of these genes were downregulated, not only on average but in every patient studied: CYP3A4, APOF, and APOM. Additionally, there were genes without statistically significant change between normal and primary tumors but showing a significant increase (n = 52) or decrease (n = 45) in the metastases (Fig. 4B).
The transcriptome of 51 samples from 13 patients with concurrent resections of at least one Normal (N), one Primary (P), and one metastatic (M) tumor samples were compared. A Differentially expressed genes with progressive dysregulation (N < P < M: n = 14, N > P > M, n = 54) from normal samples (N) to primary tumors (P) and metastases (M) of FLC patient samples. B Box-scatter-violin plots of the normalized counts (in log2 scale) of the top 39 genes with differential expression in metastases. The box plot spans the Q1, Q2 (median, red line), and Q3 quartiles, with the whiskers extending to 1.5 in the interquartile range. The heatmap and violin-box-scatter plots were generated using all the FLC patients with triplets of Normal-Primary-Metastastic samples. The specific sample IDs are detailed in the Supplementary Data 2. The raw reads and normalized read counts for the samples in these figures are deposited in their corresponding dbGAP, GEO, and EGA repositories (details in the data availability section). Access can be requested directly to these repositories under their privacy and confidentiality terms.
The transcriptomic FLC signature through different omics
The changes in the transcriptomic FLC signature were mostly reflected at the protein level (R2 = 0.8, see Fig. 5A). We explored upstream events (somatic genomic mutations or methylation events) that could lead to the dysregulations apparent in the FLC signature. Differential methylation analysis between 19 matching tumor and normal samples resulted in 1607 CpG sites altered in 979 genes (diff: 15%, FDR < 0.05), including only 41 genes of the FLC signature (see Fig. 5B and Supplemental Data 5). Notably, we found hypermethylation of 22 genes of the Protocadherin family cluster (PCDHGA1 to 8, PCDHGB1 to 5, PCDHA1 to 9). They are associated with cell adhesion and epigenetic silencing and have been previously reported in different cancers47,48. In addition, a few genomic alterations in genes of the FLC signature were found, although they have a low frequency among patients (see Fig. 5C).
A Fold Change (in log2 scale) of the genes in the transcriptomic FLC signature and the differentially regulated proteins in tumors versus normal samples16 (N = 238 genes). A positive correlation is observed (R = 0.73). B Differentially methylated genes in tumor versus normal samples in the transcriptomic FLC signature (N = 41 genes). Red: overexpressed, Blue: underexpressed. In (A, B), the gray-shaded region indicates the range of possible values for the linear regression fit with 95% confidence. C Circos plot representing the genes of the transcriptomic FLC signature at different omic levels, from external to internal circle: chromosome, transcriptome, proteome, methylome, and genome/exome. The Log2(Fold Change) of FLC tumor versus normal samples at transcriptional level is shown in a red (overexpressed) white (no change), and blue scale (underexpressed). The log2(Fold Change) at the proteome level is presented in a yellow (down) to blue (up) scale. The percentage of differentially hyper- and hypo-methylation in FLC tumors versus normal samples in a yellow (down) to blue (up) scale. For the proteome and methylome, white = undetected. The genomic variants detected in the WGS/WES data of FLC tumors versus normal samples are represented as bars, where the height represents the number of tumors mutated (from 0 to 10), and the grayscale intensity represents the number of mutations. The raw reads and normalized read counts of the transcriptome are deposited in dbGAP phs003643. Access can be requested directly from dbGAP under their privacy and confidentiality terms. The methylation data is provided in the Supplementary Data 5 in the present article. The proteome data is from Supplementary Table 1, 2 of Levin et al.16.
We explored the association between PKA and transcription factors (TFs) in the hTFtarget49 database on liver ChIP-seq data. We found 445 genes of the transcriptomic FLC signature associated with 59 TFs (see Supplementary Fig. 4A). No family was found predominant among these TFs, and no recurrent missense mutations were identified in their DNA-binding domain or effector domains. Some of them are related to many genes in the transcriptomic FLC signature (CTCF: 273 genes, CREB1: 253, FOXA2: 246, GABPA: 226, MAZ: 219, FOXA1: 193). After normalizing by each TF’s overall number of targets and adjusting for multiple-hypothesis testing, we found that seven TFs (NFIC, CEBPB, ARID3A, ZEB1, TCF12, HNF4G, TEAD4) preferentially target the FLC signature (FDR < 0.05). In the BioGrid50 data-base of protein-protein interactions, PRKACA is not reported to interact directly with any of these TFs. However, two were one interaction away: TEAD4 through RELA, and CEBPB through HDAC1 (Supplementary Fig. 4B, C). Interestingly, HDAC1 exhibited the highest connectivity between the TFs and PRKACA. Notably, a recent in vitro and in vivo massive drug screening showed that HDAC inhibitors are among the most effective drugs against FLC30. We also identified FOXA1 and FOXA2, which are downregulated in FLC and have been previously associated with migration and proliferation of HepG2 cells and HCC51,52.
Studying the cellular contribution to the transcriptomic FLC signature
The FLC signature was derived from tumor resections, which are composed of tumor cells, non-transformed hepatocytes, stellate cells, reticular-endothelial cells, and immune cells. We used standard single-cell and single-cell spatial transcriptomics to characterize the contributions of these different cells to the FLC signature. Standard single-cell transcriptomics, which requires treatment to dissociate single-cell populations from tumor samples, showed a weak correlation (Pearson’s R = 0.18) with bulk transcriptomics. This could be the consequence of the adverse effects of digesting the tissue or the loss of specific cell populations depleted by sorting (see Fig. 6A). Therefore, we turned to single-cell spatial transcriptomics, designing a panel containing 56 over- and 56 under-expressed genes in FLC (see “Methods”). The total gene counts from the single-cell spatial transcriptomics of FLC tumors showed a much better correlation with the quantification obtained by bulk RNA-seq (Pearson’s R = 0.86) than the standard single-cell transcriptomics (Fig. 6A).
A Changes in expression (as log2 fold change) in tumor vs normal samples of 140 genes screened using spatial transcriptomics (Sp), with their corresponding change in single-cell (Sc) transcriptomics and bulk RNA-seq. B Tissue section imaged using MERFISH. C Dimensionality reduction and unsupervised clustering of the gene expression by cell resulted in 3 Leiden clusters in three different patient samples (RU12, RU21, and RU26). D Top 3 differentially expressed genes among the 3 clusters identified in (C) across the three samples. The color represents the gene expression normalized from 0 (white) to 1 (dark red), and the size of the circle represents the percentage of cells expressing the gene. E, F 100X zoom of the pink rectangle in (B), showing morphological features that allow depicting stromal (left), normal (center), and tumor (right) cells. When the genes identified in (D) were mapped in (E), they matched with these three cell types, as shown in (F). G Paired normal and tumor sections from three different FLC patients (RU12, RU21, and RU26), colored using the markers selected in (D). The spatial transcriptomics data and the single cell reads are available under dbGAP accession code phs003643. Access can be requested directly from dbGAP under their privacy and confidentiality terms.
As spatial transcriptomics preserves histological features (Fig. 6B, E), this allowed us to map the signal to tumor, normal, stromal, and immune cells. We thus identified a distinctive expression pattern among these cell types, serving as cell type classifiers (Fig. 6C, D). This led to two important observations. First, the transcriptional profile was different in each of the cell types (Fig. 6C, F). Second, from patient to patient and even in different samples of the same patient, there were different proportions of each of these cell types (Fig. 6G). Thus, the overall transcriptional profile will vary not only among patients but between different samples of the same patient and is dependent on the relative distribution of cell types in the sample.
Almost all the genes follow the same trend, comparing tumors to normal tissue, in bulk RNA-seq and single cell spatial transcriptomics. COL1A1 and COL11A1 deviated from this pattern. In bulk transcriptomics, they were higher in tumors than normal, consistent with the observation of high levels of collagen that form the fibrolamellar bands. In contrast, in single-cell spatial transcriptomics, they were higher in normal than tumor cells. However, they were many times higher in the stromal cells of the tumor tissue (Col1A1 20x; Col11A1, 4x, see Fig. 6D). Therefore, these two genes are upregulated at the bulk tumor tissue level, because of the stromal cell content of the tumor blocks.
In the panel for spatial transcriptomics, we included markers of T cells (CD27, CD4, CD8A), B cells (CD19, CD40LG, TNFRSF8), stromal cells (ICAM1, MCAM, VAM1, ACTA2), macrophages (CD1D, CD14, CD163), neutrophils (ITGAM), and immune checkpoint blockade immunotherapy PD-1/PD-L1 (PDCD1 and CD274) and CTLA4 (CTLA4 and CD86). We observed a variable signal from B-cell, T-CD8+, and neutrophil markers among FLC tumors. However, there was a recurrent lower signal of monocytes, macrophages, and T-CD4 + cells in FLC tumors. Consistent with the low signal from immune cells in the tumors, the signal from immunotherapy markers was overall low. However, CTLA4 had a higher signal in tumors compared to normal samples. These observations agree with our bulk RNA-seq transcriptomics screening (Supplementary Fig. 5). Moreover, the low immunogenicity observed in FLC could explain the lack of success of immune checkpoint inhibitors53.
Using the transcriptomic signature to validate FLC models
The FLC transcriptomic signature was used to assess patient-derived xenografts (PDX) that were made directly from surgical resections without passage in plastic54. Using the transcriptomic FLC signature, the PDX segregate close to their originating tumor samples, away from the cluster of normal cells (Fig. 7A, B).
UMAP plots of the PDX and the normal and tumor patient samples (N = 25) presented by Lalazar et al.30, using (A) all the genes and (B) using only the genes of the transcriptomic FLC signature. Each color represents a different patient, and the shape indicates if the sample is normal (circle), patient tumor (square), or tumor PDX (rhombus) tissue sample. The letter inside the square indicates if the sample is a primary (P), recurrence (R), or metastatic (M) tumor. The number inside the rhombus indicates the PDX passage number. C Comparison of the log2(fold change) in FLC PDX relative to normal samples from patients (y-axis) and the log2(fold change) in patient tumors relative to normal samples (x-axis), using only the differentially expressed genes (FDR < 0.05) in the FLC signature, obtaining a high correlation (R2 = 0.95). D Revisiting the stemness of FLC. Violin plots (in yellow) of the log2 normalized gene counts of AHR and the stem/progenitor markers screened by Oikawa et al.54 We analyzed RNA-seq data of 143 FLC patient tumor and normal tissue samples from different FLC studies15,26,31,32,33,34,35 (in variations of green) and the tumor line and the biliary tree stem cells studied by Oikawa (magenta). The color of the significance bar represents the variation of each of the groups in the X-axis compared to patient normal samples (red: overexpressed, blue: underexpressed), and the symbols on top represent if that variation was significant (*: 0.01 < FDR ≤ 0.05, **: 0.001 < FDR ≤ 0.01, ***: 0.0001 < FDR ≤ 0.001, ****: FDR ≤ 0.0001) or not (n.s.) in the Wald two-sided test performed by DESeq2. The box plots (vertical rectangles in gray) span the Q1, Q2 (median, black line), and Q3 quartiles, with the whiskers extending to 1.5 in the interquartile range. We used all the FLC RNA-seq data available: Requena et al. (this study), and the samples deposited in public databases from the studies of Francisco et al.33, Xu et al.31, Robinson et al.34, Sorenson et al.32, Simon et al.15, Hirsch et al.26 and the TCGA-LIHC study. We included the tumor model data of Oikawa et al.54. Their accession numbers are provided in the Data Availability section. Access can be requested directly to dbGAP, GEO and EGA under their privacy and confidentiality terms.
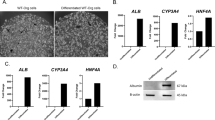
Moreover, we obtained a strong correlation (R2 = 0.95) between the expression change (in log2 scale) of patient tumors relative to normal tissue and the expression change in PDX relative to normal tissue (Fig. 7C). This suggests that these PDX reflect characteristics of their originating tumor, which is also supported by their histology and drug profile54.
Revisiting the stemness of FLC
Many cancers have been proposed to have an origin in stem cells55. Oikawa et al.54 reported eight stem/progenitor markers detected by immunohistochemistry in FLC tumors, biliary tree cells, and in a tumor line generated from ascites of an FLC patient passaged in plastic. The authors screened for and did not detect six of these genes in normal tissue (SOX9, SOX17, PDX1, POU5F1, SALL4, and SHH), concluding thereby a stem cell origin of FLC, which they associated with biliary tree cells by comparing their transcriptomes54. We analyzed 143 FLC patient tumors and normal tissue samples from different studies along with the RNA-seq samples from Oikawa et al.54 (4 tumor lines and 3 biliary tree stem cell samples). We observed in the patient tumor, relative to normal, an increase in only 2 of 8 genes: POU5F1 and PDX1. We found no change in 3 of 8 genes, SOX17, BMI, and SLC5A5 in the patient tumor and a decrease of SOX9, SALL4, and SHH. Moreover, in a previous mass spectrometric analysis, we found no statistical difference in the levels of these eight proteins in FLC tumors compared to normal, as quantified by TMT and LFQ proteomics16. To determine whether the discrepancy was the result of the specific biological sample used by Oikawa, or instead by how we processed these samples, we inspected the expression of these 8 genes in the RNA-seq data of Oikawa’s samples54. We found the reported increase in only three of the eight genes: POU5F1, PDX1, and SOX9 (which is not increased in patient samples). However, we found a substantial decrease of SOX17 and SALL4 and no change in SHH, SLC5A5, or BMI1 (Fig. 7D). We broadened our examination for stemness signal using Gene Set Enrichment Analysis (GSEA), screening gene sets of the Molecular Signatures Database (MSigDB). The FLC whole transcriptome showed no statistically significant enrichment in the gene sets “Stemness up” (MSigDB: M9473, 189 genes)56, “Curated Stemness Markers” (MSigDB: M30411, 21 genes)57, the “Liver Cancer Stem Cell Up” (MSigDB: M16956, 47 genes)58, and the “Liver Development Up” (MSigDB: M17163, 166 genes)59. Thus, even with a broader set of genes, there was no sign of a “stemness” signature for FLC (see Supplementary Fig. S9 and Supplementary Data 8).
An increased expression of the aryl hydrocarbon receptor (AHR) was also reported54, and the authors hypothesized a potential impact of environmental factors from the plastic industries. However, we found AHR to be decreased in FLC patient tumor samples relative to normal in the transcriptome and unaltered in the proteome, indicating that this is not a recurrent feature of FLC. The reported increased expression in their tumor line may be a consequence of an adaptation to grow in plastic. Moreover, when we analyzed Oikawa's data, we could not observe the reported increase in AHR (Fig. 7D).
The transcriptomic signature of other liver cancers
We collected and reprocessed RNA-seq data of 1192 tumor and normal samples of HCC, hepatoblastoma (HBL), and intrahepatic cholangiocarcinoma (iCCA), from which we selected 858 matching tumor and normal samples (Supplementary Data 6). Unsupervised clustering using the whole transcriptome showed batch effect (Supplementary Fig. 6). With application of the transcriptomic FLC signature, a clear discrimination was observed between FLC and the different liver tumors (see heatmap in Fig. 8 and Supplementary Fig. 7). Some genes of the transcriptomic FLC signature are dysregulated in the same direction (up or down) in other liver cancers, whereas some other genes present a unique distinctive pattern exclusive to FLC (Fig. 8).
Paired tumor-normal samples from hepatocellular carcinoma (HCC)35,83,84,85,86,87,88,89, hepatoblastoma (HBL)76,77,78, intrahepatic cholangiocarcinoma (iCCA)79,80,81,82, fibrolamellar carcinoma (FLC)15,26,31,32,33,34,35 and FLC-like tumors with the chimera ATP1B1::PRKACA, mutations in BAP1, or missing R1A activity (∆R1A) are represented in the columns (N = 986). They are categorized by sample type in: Normal sample, Primary tumor, Recurrence, Metastasis, or Uncategorized tumor. Using only genes of the FLC signature, hierarchical clustering separated the normal samples apart from the tumors, and FLC tumors in its own branch. The rows represent the genes of the transcriptomic FLC signature, which were hierarchically clustered in a dendrogram. It shows branches with expression patterns exclusive of FLC, distinct from other liver tumors. We used all the paired FLC tumor and normal samples with RNA-seq data available: Requena et al. (this study), and the samples deposited in public databases from the studies of Francisco et al.33, Xu et al.31, Robinson et al.34, Sorenson et al.32, Simon et al.15, Hirsch et al.26, and the TCGA-LIHC study. Also, the FLC-like samples with R1A and ATP1B1::PRKACA mutations from Requena et al. (this study) and the BAP-1 FLC-like samples from Hirsch et al.26. Their accession numbers are provided in the Data Availability section. Access can be requested directly to dbGAP, GEO and EGA under their privacy and confidentiality terms.
We determined the transcriptomic signatures of HCC, iCCA, and HBL (Supplementary Data 3, 7). Comparing them with FLC, we identified genes dysregulated in common in these four cancers: 1 up (CDCA7) and 18 downregulated. Also, there were 198 up- and 103 down-regulated genes exclusive to FLC (see Supplementary Fig. 8). Through gene set enrichment analysis, we identified pathways enriched in these four liver cancers (see Supplementary Fig. 9). Pathways enriched across FLC, HCC, and iCCA sets included mitotic spindle assembly hallmarks. HCC and iCCA showed enrichment in E2F targets and G2M checkpoints. Significant enrichment of stemness-related markers was found in HCC and iCCA, but not in FLC and HBL. We also identified pathways enriched only in FLC but not in HBL, iCCA, or HCC. They include positive enrichment in the proton-transporting ATPase complex and GalNAc transferase activity, while negative enrichment in fibroblast growth factor signaling (via PI-3K activated by FGFR4), protein acetylation and demethylation, RNA processing, cellular contact, nuclear receptor binding (thyroid hormone) and lamin binding (Supplementary Data 8).
Discussion
FLC is a powerful system for studying oncogenesis: It is driven by a single genetic event, exhibits no other recurrent genomic alterations17,18, and has a low mutational burden with a highly consistent transcriptome and proteome15,16,60. This provides a favorable scenario for multiomics exploration. Other cancers usually present with many mutations, which complicates molecular-level understanding. The study of FLC could, therefore, elucidate general mechanisms of oncogenesis.
FLC has been considered a subtype of HCC, despite its different age of incidence, histology, and transcriptomic and proteomic profiles10,61. This misclassification results in some patients receiving drug therapy standard for HCC, which has proven ineffective against FLC10. Defining the molecular identity of a cancer is critical for developing and administering proper drug treatment. Current RNA-seq studies of FLC15,26,31,32,33 had limited agreement in their differentially expressed genes. This lack of congruency is likely the result of diverse limitations. These include:
Tumors are categorized only by histopathology, which can be inconsistent among pathologists9. Some transcriptomic studies even had 30% of their samples mistakenly deemed to be FLC. Here we started by studying FLC tumors having DNAJB1::PRKACA, which allowed selecting a reliable group.
Small datasets using unpaired tumors and surrogate normal samples, some with samples from other tumor types as “surrogate normal control” or mostly unpaired tumors (Supplementary Data 1). A high proportion of unpaired tumors (> 10%) in the analysis can result in an incorrect estimation of the expression change36. Here, we sequenced 127 frozen FLC and 2 FLC-like patient samples, which included 98 matching FLC tumors and adjacent normal samples. This was complemented with 73 FLC and 18 FLC-like samples (30 FLC matching tumor and normal) from other studies26,31,32,33,34,35.
Gene expression estimated with older software, like ht-seq62 (also included in STAR63), featureCounts64, or equivalent, which count reads aligned to features (i.e., transcripts or genes). When a read matches with multiple features, it is either not counted; or counted multiple times in all matching features, distorting the biology37. We used Salmon65, which avoids this problem by assigning fractions of a read to the multiple features it matches based on a probability distribution that depends on factors like the abundance, start position, length, and orientation of the fragment.
Inappropriate integration of datasets. Combining RNA-seq datasets usually results in batch effects, introduced by the use of different RNA extraction, library preparation or ribosomal depletion methods, sequencing platforms, and sample ascertainment differences, among other unknown sources of variability38. It has been shown that adding variables accounting for processing groups or dates might not be sufficient to address batch effects, which can lead to mistaken, even opposite conclusions38. We utilized different datasets and performed unsupervised clustering, revealing batch-driven clusters. ComBat39 and RUV40, popular approximations for compensating batch effect, were used, although ineffectively. We opted to perform differential expression analysis in each dataset individually, and then intersect the significantly dysregulated genes obtained.
By collecting a large dataset and devising a methodology including exploration, refinement, and subsequent external validation, we found a distinct and reproducible transcriptomic FLC signature. This captures dysregulations characteristic of all FLC tumors, primary, recurrences, or metastases, independent of the laboratory, collection, or experimental processing methods. This FLC transcriptomic signature is thus inherent to FLC tumors and reflects a very specific driver program.
The transcriptomic FLC signature has shown to be useful for validating animal and cellular models of FLC, which is critical for the study of a very rare cancer with scarce patient experimentation material. The FLC signature allowed the inclusion of new samples from different experiments and laboratories, regardless of having a paired normal or not. Thus, it could be used to add future FLC tumor samples and to interrogate samples under suspicion of being FLC. Further studying the transcriptomic FLC signature may advance the knowledge of the biology of FLC and, more broadly, other cancers.
The transcriptional signature revealed that some diverse molecular changes in liver tumors converge on a common disease, FLC. Several FLC-like tumors do not have the DNAJB1::PRKACA fusion in hepatocytes. One set of tumors has no fusion to the PKA catalytic subunit; instead, it lacks activity of the regulatory subunit R1A. The transcriptome of these tumors clusters with the DNAJB1::PRKACA tumors. Seemingly related tumors, found in the ductal cells of the liver or pancreas, express a chimeric form of the catalytic subunit of protein kinase A in the fusions DNAJB1::PRKACA, ATP1B1::PRKACA or ATP1B1::PRKACB. When present in the ductal cells of the liver, these fusions produce a cholangiocarcinoma, and the ductal cells of the pancreas produce an intraductal oncolytic pancreatic neoplasm (IOPN). Application of the transcriptomic FLC signature to these FLC-like samples demonstrates that they cluster together with FLC samples with the canonical DNAJB1::PRKACA fusion. These common transcriptomes suggest a common pathogenesis, whether expressed in hepatocytes or the ductal cells of the liver or pancreas. This is consistent with the demonstration that the drug-response profile of cholangiocarcinoma from patients expressing ATP1B1::PRKACA in cholangiocytes is indistinguishable from the response profile of FLC tumors expressing DNAJB1::PRKACA in hepatocytes66.
They all share an increase in the amount of catalytic subunit to the regulatory subunit of PKA, whether the consequence of increased expression of the catalytic subunit of protein kinase A from a fusion of DNAJB1::PRKACA, ATP1B1::PRKACA or ATP1B1::PRKACB or due to R1A deficiency. These conclusions are consistent with the observation that primary human hepatocytes experimentally manipulated to express DNAJB1::PRKACA or ATP1B1::PRKACA produce similar transcriptomes, and these share the top altered transcripts with patient FLC tumors66. The transcriptomic FLC signature, by capturing dysregulations independent of the experimental processing and characteristic of FLC tumors, empowers identifying which FLC-like tumors cluster with DNAJB1::PRKACA and which, such as the BAP1-driven tumors, do not. This is important for determining what is exclusive and distinct among FLC and other FLC-like tumors. We have found that the FLC-like tumors that either overexpress the catalytic subunit (as a consequence of being expressed as a fusion protein) or are missing the regulatory subunit have a common transcriptome (this publication), histopathology24, and drug response profile66. Further, the transcriptome of FLC patients can be recapitulated just by increasing the ratio of catalytic to regulatory subunits in primary human hepatocytes. These data are consistent with PKA activation as the unifying causal mechanism of FLC. Thus, we suggest FLC be considered a single disease, rather than a mix of different pathologies with a similar phenotype24,66,67.
We studied which elements of the transcriptomic FLC signature were exclusive to FLC or shared with the transcriptomic signatures of other liver cancers (HBL, iCCA, and HCC) by analyzing 1412 bulk RNA-seq samples. We found only 1 upregulated and 18 downregulated genes in common. In FLC, we found 198 upregulated and 103 downregulated genes that were not in the others. Studying these genes will promote a better understanding of the biology of FLC, and, in the broader context of liver tumors, evaluating what is common and what is unique. To explore potential clinical uses, we selected a reduced list of 35 genes with the strongest upregulation and abundance (Supplementary Data 9).
Liver cancers shared common enrichment in pathways like mitotic spindle assembly and E2F targets in FLC, HCC, and iCCA, highlighting the role of cell cycle control dysregulation in liver oncogenesis. Despite originating from the same organ, they exhibit distinct activated pathways, suggesting exclusive oncogenic mechanisms not shared with other liver cancers. Evidence suggests that PKA-mediated phosphorylation regulates proton-transporting ATPase complex elements68, which we found enriched in FLC.
Cancers are frequently thought to have an origin in stem cells55. Such a claim has been made for FLC in a study that characterized the ascites tumor of a single patient, which was first passaged for weeks on plastic in a medium selective for endodermal stem cells and progenitors and then implanted in mice for four serial transplantations54. In that study its transcriptome was then compared to that of stem cells, but not to the adjacent normal or to the original patient tumor sample. In contrast, we found most of these same stem-cell markers54 to be decreased or unaltered in FLC patient tumors compared to paired normal samples at the transcriptional and proteomic level, even in those authors’ original publications. We expanded our analysis to four sets of stemness markers of the MSigDB, finding no enrichment in the transcriptome of FLC. This challenges the hypothesis of stem cell-driven oncogenesis for FLC. The stem hypothesis is further challenged by the recent demonstration that transducing primary human hepatocytes with the fusion oncotranscript is sufficient to recapitulate the transcriptome of FLC patients66. Further experiments are required to definitively test the stem cell hypothesis.
Genomic screening from various cancers has led to the conclusion that metastases are different from primary tumors69. Two previous studies32,70 compared the transcriptome of FLC metastases to paired primary tumors, but neither reported nor discussed the differentially expressed genes. We showed that the metastases differ in 0.6% of the transcriptome from primary FLC tumors, consistent with a low mutation rate of FLC17,53. Small differences between metastases and primary tumors have been reported in other cancers71. We identified a set of genes consistently dysregulated, related to tumor proliferation, maintenance, and immune evasion, like COLEC10 and COLEC11. It has been found that decreased expression of COLEC10 is prognostic of poor overall survival in patients with HCC72. COLEC11 participates in apoptosis, binding the DNA at the surface of apoptotic cells and activating the complement in response73. Also, it has been found that people affected by the 3MC syndrome (characterized by facial dysmorphic traits and other developmental problems) present genetic loss-of-function mutations in COLEC1174. Zebrafish morphants of COLEC11 exhibited dose-dependent pigmentary defects and cranial abnormalities74. In addition, CL-K1 (the protein encoded by COLEC11) acts during embryonic development as a guidance cue to the migration of neural crest cells. Therefore, COLEC10 and COLEC11 could play a role in the metastatic migration of FLC. It will require further experimental studies to understand better the role of these genes and the other candidates in the development and maintenance of the metastases of FLC.
The FLC signature also served as a tool to validate in vivo (PDX30) and in-vitro (organoids29) experimental models for FLC, by assessing how closely they reflect the transcriptional changes observed in patient tumors. Differences between the originating tumor and the models may arise for several reasons: the original tumor is a mixture of many cell types, whereas the model contains predominantly tumor cells. The tumor cells may evolve in adapting to the new environment. The present findings augment our confidence in these models, which had previously been verified by histopathology, proteomics, and drug susceptibility.
A surgical tumor sample of an FLC patient is usually composed of different cell types: FLC tumor cells, stromal cells (stellate cells, reticular endothelial cells), normal hepatocytes, and infiltrating immune cells (macrophages, T-cells, B-cells, myeloid-derived suppressor cells). With spatial transcriptomics, we identified differential expression patterns that distinguish these cell types. The different relative levels of these cell types in each tumor sample likely account for some variations in transcriptome and proteome observed across samples and models. Spatial single-cell transcriptomics also revealed that some of the phenotypic characteristics of FLC, such as the collagen in the fibrous bands, reflect contributions of stromal rather than tumor cells.
Integration of multiomic data layers may help to elucidate the biology of FLC. Thus, we identified transcription factors that could be involved in regulating the transcriptomic FLC signature. To facilitate further exploration, we developed an interactive web portal (Shiny app) aggregating all the data processed in this study. It can generate plots not requiring programming knowledge, integrating the genomic, proteomic, methylomic, and transcriptomic data from FLC and other liver cancers.
The transcriptional signature for FLC is useful for addressing problems of general interest such as: resolving ambiguous diagnoses, a task that challenges histopathology for FLC9; assessing the mechanistic basis of tumorigenesis, which requires not merely identifying tumor drivers but understanding also their diverse or uniform downstream consequences; improving the accuracy of classifying diverse tumors, for which the transcriptome provides an informative benchmark; determining the extent of deviation of metastases from primary tumors, to resolve the sources of tumor progression and dispersion, and to guide therapeutic investigation; testing the validity of biologic cancer models, an important investigational resource, but one that must adequately reflect the properties of its source; and determining the cells of origin of the tumors. Our work in defining the transcriptional signature presents a methodology with state-of-the-art tools and filters that we have extended to other liver cancers and which can be more broadly applied to oncogenesis.
Methods
A schematic representation of the methods described below is presented in Supplementary Fig. 10.
Ethics statement and demographics
Under the supervision of our Institutional Review Board approval (Rockefeller IRB #SSI 0797, SSI 0798) consent was obtained from patients scheduled for tumor resection. Since this is a rare disease, we accepted samples from all patients, without regard to age or sex. For patients not of the age of consent, we consented to both parents and, if the patient was older than 7, assented the patient. For each patient, the diagnosis of FLC was confirmed both by histopathology, demonstration of the DNAJB1::PRKACA fusion transcript by RT-PCR, and presence of the DNAJB1::PRKACA fusion protein by western blot18. This is a rare disease which means that inclusion of almost any data can be used to identify the patients. We have recently published an aggregated analysis of the age, sex, and outcomes of our patients75. All studies of the transcriptomes of patient-derived xenografts were from previously published mouse studies20,30, which were performed with the approval of the Institutional Animal Care and Use Committee at Rockefeller University (protocol 23011-H). For those studies, animals were monitored three times a week for changes in activity or weight loss. Tumors were not allowed to grow beyond 2 cm.
Statistics and reproducibility
This study was devised as an exploratory analysis of a very rare cancer. Therefore, no statistical method was used to predetermine sample size. From our cohort, we only used tumor samples validated as FLC as described above. From external cohorts, we only used samples in which we verified the presence of reads spanning the DNAJB1::PRKACA fusion transcript. In both cases, we verified the absence of the chimera in the normal samples. The experiments were not randomized, we used all the samples available in each comparison. All the statistical methods used are described in their corresponding sections below and were performed in R v4.3.3 and RStudio 2023.12.1 + 402.
Whole RNA extraction and sequencing
A total of 127 FLC and 2 FLC-like patient samples, collected by the Simon Laboratory at Rockefeller University, were sequenced from 2010 to 2023 in six different libraries (named RU-A to RU-F), representing the dataset of Requena et al. (this study). Libraries A, D, and E contain the human tissue samples studied in Simon et al.15, Lalazar et al.30, and Narayan et al.29, respectively. These 129 samples were processed using different combinations of library preparation and ribosomal depletion methods (details in Supplementary Data 1). After the RNA extraction, aliquots were used to assess the 260/280 ratio and RNA concentration using a Nanodrop 2000c (Thermo Fisher), and the RNA quality by estimating the RNA integrity number (Agilent BioAnalyzer and TapeStation). All samples had at least a concentration of 100 ng/µl and RIN ≥ 7. RNA-seq was performed at an average of 60 million raw reads per sample with Q30 > 90%.
Collection of external datasets
Bulk RNA-seq data from every transcriptomic study, including FLC patients and available metadata was collected and reprocessed. This comprised 73 FLC and 18 FLC-like samples from the studies of Xu et al.31, Robinson et al.34, Sorenson et al.32, the TCGA-LIHC study35, Hirsch et al.26, and Francisco et al.33 (see Supplementary Table 2).
In addition, 1192 bulk RNA-seq samples from other liver cancers were collected and reprocessed. This includes 148 HBL samples from the studies of Carrillo-Reixach et al.76, Hooks et al.77, and Wagner et al.78; 139 iCCA samples from the studies of Ahn et al.79, Gao et al.80, Sia et al.81, and the TCGA-CHOL study82; and 905 HCC samples from the studies of Long et al.83, Liu et al.84, Huang et al.85, Jin et al.86, Wang et al.87, Yoo et al.88, Yang et al.89, and the TCGA-LIHC study35.
RNA-seq mapping and quantification
Sambamba90 v1.0.0 and bamToFastq (part of bedtools91 v2.27.1) were used to covert.bam to.fastq files. Subsequently, quality control was performed using FastQC v0.11.9 and MultiQC92 v1.15, followed by adapter trimming by BBDuk (included in BBMap v39.01). The trimmed reads were mapped to the annotated Human Genome GRCh38.103 using STAR63 v2.7.10b, generating.bam files, which were indexed using SAMtools v1.10 in.bai files. These files were loaded into IGV v2.16.0. to visualize alterations in PKA subunits in Sashimi plots. The presence of reads in the junction of the chimera DNAJB1::PRKACA in all FLC tumors and its absence in all the normal samples was confirmed in all the samples from FLC patients used in this study. Expression by transcript was quantified using Salmon65 v1.10.0 with a gentrome generated connecting the human genome hg38 and its annotation GRCh38.103.
Dimensionality reduction and Unsupervised clustering
The samples from all the studies were inspected through unsupervised clustering to explore batch effect and covariables using four different approaches. These included dimensionality reduction by PCA93, t-Distributed Stochastic Neighbor Embedding (tSNE)94, and the Uniform Manifold Approximation and Projection (UMAP)95, followed by density-based hierarchical clustering using HDBSCAN96. Different random seeds and perplexity values (number of neighbors) were explored in tSNE and UMAP for robustness. And we used heatmaps with dendrograms of hierarchical clustering, using the Ward297 agglomerative clustering method. These methods are implemented in the R libraries OmicsKit v1.0.0 (PCA, tSNE, and UMAP with HDBSCAN), tsne v0.1-3.1 (tSNE), umap v0.2.10.0 (UMAP), dbscan v1.1.12 (HDBSCAN), pheatmap v1.0.12 (heatmap), and stats 4.3.3 (functions prcomp for PCA and hclust for Ward.D2 clustering).
Differential expression analysis
In each library, differential expression analysis was conducted using R v4.3.3 and RStudio 2023.12.1 + 402. We used the gene counts by transcript quantified by Salmon65, which were condensed by gene using tximport98 v1.30.0. These counts were analyzed in DESeq299 v1.42.1, performing the variance stabilizing transformation and fitting the data to a generalized linear model. Only those genes with |log2(Fold Change)| > 1 and FDR < 0.05 were considered differentially expressed. We developed an R library with the functions we developed for data manipulation and visualization of multi-omics data and differential expression analysis. It is available on GitHub (https://github.com/BigMindLab/OmicsKit) and can be installed by executing the following line of code: remotes::install_github(repo = ‘BigMindLab/OmicsKit’).
Calculating the transcriptomic signature of FLC tumors
Matching tumor and normal samples of five RNA-seq libraries sequenced at Rockefeller University were selected for the determination of the transcriptomic FLC signature. Of these, three libraries (RU-A, RU-B, and RU-C) were used for exploration. Libraries RU-D and RU-E were used to refine the results obtained in the first three. These two libraries were not used for exploration because they were smaller and had fewer tumor-normal pairs than the first three. The data of patient R17 in library RU-C was omitted in this analysis to prevent overrepresentation, because samples of this patient are already included in library RU-B. Library F was not used to determine the FLC signature because it has mostly unpaired samples.
Differential expression analysis was performed independently in each library, filtering by significance and fold-change as described in the “Differential expression analysis” section. The sets of differentially expressed genes obtained by the library were further reduced using the following filters:
Detectability
This consists of first discarding the genes with baseMean < 50. The baseMean of a gene (generated by DESeq2) represents the mean of its normalized gene counts across all the samples in the library. Of the remaining genes, we selected those with at least 50 normalized gene counts in the highest experimental condition. This means, for upregulated genes, the mean normalized gene counts in the tumor samples should be greater than 50. And for downregulated genes, the mean normalized gene counts in the normal samples should be greater than 50.
Trend consistency
This filter consists of verifying in every patient if the dysregulation trend observed (up- or down-regulation) is the same dysregulation trend obtained in the differential expression analysis of all tumors relative to their corresponding matching normal samples (called “group level”). With this filter, we only keep genes in which the group level trend is also observed in the tumor versus normal samples of every patient. The “trend consistency” filter was applied as follows: First, for each patient in a library, we calculated the average expression by gene in the tumors (\({\bar{X}}_{T}\)) and in the normal samples (\({\bar{X}}_{N}\)). Then, for every gene upregulated at the group level, the gene was discarded if at least one patient in the library shows signal in the opposite direction (\({\bar{X}}_{N}/{\bar{X}}_{T} > 1.1\)). And, for every gene downregulated at the group level, the gene was discarded if at least one patient in the library showed signal in the opposite direction (\({\bar{X}}_{T}/{\bar{X}}_{N} > 1.1\)).
These two filters were applied in all the libraries used for exploration and refinement, as follows:
Exploration
The differentially expressed genes obtained in each of the libraries for exploration (RU-A, RU-B, and RU-C) were intersected. We reduced the intersection by applying the filters of detectability and consistency. Thus, we selected a set of detectable genes with a consistent dysregulation trend.
Refinement
Libraries RU-D and RU-E were filtered by detectability. The resulting genes in each of these libraries were used to verify the trend consistency in the genes obtained in the exploration step, discarding those who do not pass this filter. We named the resulting genes the “transcriptomic FLC signature” (see Fig. 2).
Validating the FLC signature
Three of the six external RNA-seq studies selected FLC for validation (Francisco et al.33, Sorenson et al.32, and the TCGA-LIHC study35) because they studied at least 3 patients and contained at least one tumor-normal pair by the patient. These datasets were re-analyzed as described in the RNA-seq mapping, quantification, and differential expression analysis sections, using only patients verified as FLC tumors and normal samples. The results were filtered by detectability as described above.
As a validation test, for each gene, we evaluated if the dysregulation trend obtained in the transcriptomic FLC signature was observed as well in each of the validation datasets. The agreement between the FLC signature and an external dataset was calculated as the percentage of genes in the FLC signature that agree in the dysregulation trend obtained in the external dataset.
Metastasis compared to primary tumors
Metastases with matching primary tumors and normal samples from libraries RU-A, RU-B and RU-C were analyzed adjusting by library. Pairwise differential expression comparisons by sample type (metastasis, primary tumor, or normal) were performed, filtering by detectability and ranking by trend consistency.
DNA extraction, quantification, and quality control
DNA was extracted from OCT-embedded frozen tissue, using 10 curls (10 mm each) by patient sample. They were dissolved in 1 mL PBS, and the DNA was isolated using the DNeasy Blood & Tissue Kit (QIAGEN catalog N° 69504), according to the manufacturer’s protocol modified by replacing AW2 buffer with 80% ethanol. DNA was eluted in 50–55 µL 0.5X Buffer AE heated to 55 °C. DNA concentration was quantified by PicoGreen (ThermoFisher catalog N° P7589), and quality control was assessed in an Agilent BioAnalyzer.
Exome sequencing
Sequencing libraries were prepared using 100–113 ng of DNA and the KAPA Hyper Prep Kit (Kapa Biosystems N° KK8504), with 8 cycles of PCR amplification. After sample barcoding, 100 ng of the library were captured by hybridization using the xGen Exome Research Panel v2.0 (IDT) according to the manufacturer’s protocol, followed by 12 cycles of PCR amplification. Samples were sequenced in a PE150 run on a NovaSeq 6000 using S4 Reagent Kit (Illumina) for 300 Cycles, obtaining an average coverage of 124X.
Mapping and variant calling
All samples were processed following the GATK Best Practices100. Reads from WGS and WES were mapped to the Human Genome GRCh38 using BWA-MEM v0.7.17101. The resulting.bam files were tagged for read duplication, sorted, and calibrated using GATK v4.2102 and Picard v2.26 (https://broadinstitute.github.io/picard). Somatic variant calling in matching tumor and normal samples was performed with Mutect2 (GATK), Strelka2 v2.9.10103, and Lancet v1.1104. From the WES data, variants within regions of the xGen Exome Research Panel v2.0 (IDT) panel were called, keeping only those tagged with “PASS”. To capture more potential variants, the.vcf files from the same tumor and normal comparison but generated by different software were merged using bcftools (SAMTools)105. The resulting non-redundant variants were annotated using the Ensembl Variant Effect Predictor (VEP)106 v103.
Targeted bisulfite methylation sequencing
Approximately 500 ng of genomic DNA by sample were sheared using a LE220-plus Focused-ultrasonicator (Covaris catalog N° 500569). Sequencing libraries were prepared using the KAPA Hyper Prep Kit (Kapa Biosystems N° KK8504) without PCR amplification. Post-ligation cleanup proceeded with 110 µL of the Sample Purification Mix from the TruSeq Methyl Capture EPIC LT Library Prep Kit (Illumina catalog N° FC-151-1002) according to the manufacturer’s instructions. After purification, 2–3 samples were pooled to capture the methylome regions using EPIC oligos, which were bisulfite-converted and amplified in 12 PCR cycles. Then, sequenced on a NovaSeq 6000 in a PE100 or PE150 run, using NovaSeq 6000 S1 or S4 Reagent Kit (200/300 Cycles) (Illumina), obtaining an average of 88 million read pairs per sample.
Methylation mapping and variant calling
The targeted bisulfite sequencing data was processed with the methylseq v2.3.0 pipeline of nf-core v1.6.1 (https://nf-co.re/methylseq)107, using Docker v20.10.12, Nextflow v22.10.4.5836, python v3.10.6, yaml v6.0, fastqc v0.11.9, preseq v3.1.1, qualimap v2.2.2, cutadapt v3.4, Trim Galore! v0.6.7, and Bismarck v0.24.0. Briefly, quality control of the raw reads was performed using FastQC, and the adapters trimmed using Trim Galore! Next, Bismark108 was used for indexing the reference genome and read alignment. Duplicates were marked with Picard. Then, the methylation events were quantified.
Differential methylation analysis
The tables with methylation events were analyzed in R using the library MethylKit109 v1.22.0 and the human genome GRCh38 as a reference. CpG sites with a minimum coverage of 10, bases above the 99.9th percentile of coverage in each sample, and standard deviations larger than 2% were selected for further analysis. PCA, dendrograms, and correlation plots were used to explore the data. Differentially methylated sites in with aa percentual methylation difference of at least 15% between tumors and normal samples, and a q-value less than 0.05, were selected. These sites were annotated using the human genome annotation GRCh38.103 and visualized in a Volcano plot.
Gene set enrichment analysis
We selected 18 of the liver cancer datasets collected containing matching tumor-normal samples and at least 5 normal samples: 2 FLC, 3 HBL, 4 iCCA, and 9 HCC (see Supplementary Fig. 9). We tested the enrichment and statistical significance of different gene sets in tumors compared to normal samples. All the gene sets in seven collections from the MSigDB110 database v2023.2 were tested: Hallmarks (H: 50 gene sets), Curated Gene Sets (C2-CGP: 3438 gene sets, C2-CP: 619 gene sets of KEGG_MEDICUS and 1692 gene sets of REACTOME), Curated Cancer Cell Atlas (C4-3CA: 149 gene sets), Ontology gene sets (C5-GOBP: 7647 gene sets, C5-GOCC: 1015 gene sets, C5-GOMF: 1799 gene sets, C5-HPO: 5,547 gene sets), Oncogenic Signatures (C6: 189 gene sets), Immunologic signature gene sets (C7-ImmuneSigDB: 4872 gene sets), and Cell type signatures (C8: 830 gene sets). Collection C8 was supplemented with 42 gene sets from the scientific literature, containing markers of different liver cells57,111,112,113,114,115,116,117,118,119,120. In each liver cancer dataset, we tested each collection independently, performing Gene Set Enrichment Analysis using the software GSEA121 v4.3.2. It was run using the parameters: permutation = phenotype, statistic = weighted enrichment, metric = signal-to-noise, and permutations = 10,000 with a statistically significance cut-off of FDR < 0.25. The resulting gene sets were compared to find pathways shared and distinctive among liver cancers. These were visualized in balloon plots generated using the R package ggplot2 v3.4.3 (see Supplementary Fig. 9).
Multiomic assessment of the FLC signature
Multiomic comparisons were performed in R. Pearson correlations between transcriptome, proteome16, and methylome scores were performed using ggplot2 3.4.3. Standard error and regression lines for each plot were obtained using ‘lm’ as smoothing method. A Circos plot of the 693 genes of the FLC signature was generated using the package circlize 0.4.15. Each track was generated using the circos.rect function. The chromosome track was adjusted to represent the number of genes rather than the chromosome size.
Analysis of transcription factors
We searched for liver ChIP-seq data of transcription factors (TFs) associated with the genes of the transcriptomic FLC signature in the hTFtarget49 database. TFs were reported for 445 of the 693 genes, making a total of 59 TFs.
These 59 TFs were categorized according to their DNA-binding domain families using the Human TFs database122. We also annotated the DNA-binding and Effector domains of the TFs based on their canonical isoforms using the CIS-BP123 v2.0 and TFRegDB124 databases, respectively. The protein level of each TF was obtained from LFQ and TMT proteome screenings of FLC tumors16, which have complementary detection ranges. In the cases where the protein was detected in both the LFQ and TMT experiments, the value obtained in the LFQ experiment was used.
To identify TFs associated to genes in the transcriptomic FLC signature in an increased ratio than to any other set of genes of the same size, we generated a probability distribution as follows: First, the genes associated with least one TF in the hTFtarget database were collected. Excluding those in the FLC signature, we obtained 27,790 genes. Second, we generated 10,000 random samples of 445 genes from these 27,790 genes. Third, for each of the 59 TFs, we calculated the number of genes a TF targets in each of the 10,000 random sets, generating a probability distribution with these numbers. Fourth, for the same TF, we calculated the number of genes it regulates in the transcriptomic FLC signature. By locating this number in the distribution previously generated, we calculated the Z-score and p-value of enrichment (one-sided Z-test). Fifth, these p-values were adjusted for multiple-hypothesis testing using the Benjamini-Hochberg125 correction, calculating the q-values.
Having identified these TFs increasingly associated with FLC, we studied their interactions to explore associations with PRKACA. We retrieved evidence of their Protein-Protein Interactions (PPI) from the BioGrid50 database and selected only those with at least 2 sources of physical evidence between human proteins (ID: 9606). Then, from the list of proteins interacting with PRKACA and the TFs, we selected those connections with a maximum of two intermediaries (PRKACA-Protein1-Protein2-TF). This network was plotted using CytoScape126 v2.0 (Supplementary Fig. 4).
Spatial single-cell RNA sequencing
A personalized library of 140 probes was designed for MERFISH single-cell spatial transcriptomics in 16-bit encoding. For this library, we selected the genes with: |Log2FC | > 2 in bulk RNA-seq of tumor versus normal, average normalized read counts of at least 100, a transcript length of 1,500nt or more (because the probes are 30nt long, and approx. 50 per transcript are needed), and similar FPKM (because the protocol requires that the sum of the FPKMs of the genes in the library must be less than 8000). Thus, the following 140 genes were selected: 112 genes of the transcriptomics FLC signature (56 up- and 56 down-regulated); 10 of the least variable genes in FLC tumors versus normal samples obtained from the analysis of libraries RU-A, RU-B and RU-C (KMT2C, SEC23A, COPB1, HNRNPL, SUN1, SNX6, THUMPD1, TM9SF4, PIGN and RTF1); 4 targets of immunotherapy (CD86, CTLA4, PDCD1 and CD274); 3 B-cell markers (CD40LG, CD19 and TNFRSF8); 3 T-cell markers (CD4, CD8A and CD27); 4 markers of Stromal cells (MCAM, VCAM1, ICAM1 and ACTA2); 3 markers of monocytes and macrophages (CD1D, CD163 and CD14); and a marker of neutrophils (ITGAM).
For three different patients, we selected a slice of the tumor block presenting a mixture of tumor, normal, and stromal regions (Fig. 6G) and processed it along with a corresponding slice of a normal tissue block from the same patient resection. These samples were prepared for MERFISH spatial transcriptomics following Vizgen’s protocol for frozen liver tissue samples127. Briefly, it consists of 16 cycles of sequential hybridization, imaging, and photobleaching. The signal intensity of each probe was quantified and used as a proxy of gene expression, obtaining gene expression values with spatial coordinates. This was followed by co-staining with DAPI, polyA RNA, and the Vizgen Cell boundary Staining Kit (Cat. no.: 10400009) as recommended for MERFISH128. Cell segmentation was performed using the deep-learning-based Cellpose algorithm129. Samples were visualized in Vizgen’s MERSCOPE Visualizer software v2.1.2589.1.
The signal from the single cells was analyzed in Python 3.12.3 using scanpy v1.9.2. Gene expression counts were filtered by a minimum barcode count of 20 and cell volume between 100 and 2500. These counts were normalized by cell volume and total counts across all genes. Then, they were log-transformed and normalized to mean = 0 and variance = 1. We explored the presence of clusters in the data through UMAPs applying the Leiden algorithm (package leidenalg v0.8.10), exploring resolution values between 0.01 and 0.15, and identifying the main clusters in each sample. The differentially expressed genes among clusters were calculated and the top 3 genes in each cluster, along with the histological features, were used to identify the cell type corresponding to each cluster.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw data (RNA-seq, WGS, bisulfite methylation sequencing, and spatial transcriptomics) of the FLC samples sequenced in this study has been deposited in dbGAP (phs000709 [https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000709.v1.p1], phs002435 [https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs002435.v1.p1], phs002439 [https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs002439.v1.p1], and phs003643 [https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs003643.v1.p1]). This data corresponds to FLC human subjects from a very rare cancer, including some underage patients. Therefore, we chose this NIH-controlled repository in compliance with our IRB protocols to protect the patient’s privacy. Access can be requested directly from dbGAP under their privacy and confidentiality terms. We have no input into the process of granting access. However, deidentified grouped visualizations can be obtained from our Shiny app: https://simonlab.shinyapps.io/FLCdb/. The data accessed from other studies are under the following GEO accession codes. From HBL: GSE133039 (Carrillo-Reixach et al.76), GSE104766 (Hooks et al.77), GSE151347 (Wagner et al.78). From iCCA: GSE107943 (Ahn et al.79), GSE119336 (Gao et al.80), GSE63420 (Sia et al.81). From HCC: GSE214846 (Long et al.83), GSE77314 (Liu et al.84), GSE105130 (Jin et al.86), GSE207435 (Wang et al.87), GSE94660 (Yoo et al.88), GSE77276 (Yang et al.89). From FLC: GSE181922 (Francisco et al.33), GSE63018 (Sorenson et al.32). The rest of FLC samples were collected from the European Genome Phenome Archive EGAD00001005308 (Hirsch et al.26) and dbGAP codes phs000828 [https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000828.v1.p1] (Xu et al.31) and phs000673 [https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000673.v5.p1] (Robinson et al.34). The datasets LIHC (containing HCC and FLC samples) and CHOL (containing iCCA samples) of the TCGA study35 are available under dbGAP code phs000178 [https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000178.v11.p8]. Oikawa et al.54 tumor cell line is under GEO code GSE73114. The raw reads and normalized read counts for Figs. 3, 4, 7A, B, D, and 8 are deposited in their corresponding dbGAP, GEO, and EGA repositories, as detailed above. For Supplementary Fig. 2, UMAP plots of libraries RU-A, -B, and -C, are from dbGAP: phs000709 and phs003643 using all the genes. For Supplementary Fig. 3, all of the data are in dbGAP (phs000709, phs002435, phs002439, phs003643, phs000828, phs000673, phs000178), GEO (GSE181922 and GSE63018) and EGA (EGAD00001005308). For Supplementary Fig. 5 the data is in Requena, this study (dbGAP: phs003643), and the samples deposited in public databases from the studies Francisco et al.33 (GEO: GSE181922), Xu et al.31 (dbGAP: phs000828), Robinson et al.34 (dbGAP: phs000673), Sorenson et al.32 (GEO: GSE63018), Simon et al.15 (dbGAP: phs000709), Hirsch et al.26 (EGA: EGAD00001005308), and the TCGA-LIHC study (dbGAP: phs000178). For Supplementary Fig. 6 UMAP plots of samples from 15 RNA-seq studies: intrahepatic cholangiocarcinoma (iCCA)79,80,81,82 (GEO: GSE107943, GSE119336, GSE63420), hepatoblastoma (HBL)76,77,78 (GEO: GSE133039, GSE104766, GSE151347), and hepatocellular carcinoma (HCC)35,83,84,85,86,87,88,89 samples (GEO: GSE214846, GSE77314, GSE105130, GSE207435, GSE94660, GSE77276; and dbGAP: phs000178). For Supplementary Fig. 7 UMAP plots of tumor-normal paired samples of cholangiocarcinoma (iCCA)79,80,81,82 (GEO: GSE107943, GSE119336, GSE63420), hepatoblastoma (HBL)76,77,78 (GEO: GSE133039, GSE104766, GSE151347), and hepatocellular carcinoma (HCC)35,83,84,85,86,87,88,89 (GEO: GSE214846, GSE77314, GSE105130, GSE207435, GSE94660, GSE77276; and dbGAP: phs000178). The remaining data are available within the Article, Supplementary Information, or Source Data file. Source data are provided in this paper.
Code availability
The functions developed for manipulation and visualization of multi-omics data have been packaged into an R library, available on GitHub (https://github.com/BigMindLab/OmicsKit).
References
Edmondson, H. A. Differential diagnosis of tumors and tumor-like lesions of liver in infancy and childhood. AMA J. Dis. Child 91, 168–186 (1956).
Craig, J. R., Peters, R. L., Edmondson, H. A. & Omata, M. Fibrolamellar carcinoma of the liver: a tumor of adolescents and young adults with distinctive clinico-pathologic features. Cancer 46, 372–379 (1980).
Torbenson, M. Fibrolamellar carcinoma: 2012 update. Scientifica 2012, 743790 (2012).
Kassahun, W. T. Contemporary management of fibrolamellar hepatocellular carcinoma: diagnosis, treatment, outcome, prognostic factors, and recent developments. World J. Surg. Oncol. 14, 151 (2016).
Kaseb, A. O. et al. Prognostic indicators and treatment outcome in 94 cases of fibrolamellar hepatocellular carcinoma. Oncology 85, 197–203 (2013).
Darcy, D. G. et al. Prognostic factors in fibrolamellar hepatocellular carcinoma in young people. J. Pediatr. Surg. 50, 153–156 (2015).
El-Gazzaz, G. et al. Outcome of liver resection and transplantation for fibrolamellar hepatocellular carcinoma. Transpl. Int 13, S406–S409 (2000).
Ang, C. S. et al. Clinicopathologic characteristics and survival outcomes of patients with fibrolamellar carcinoma: data from the fibrolamellar carcinoma consortium. Gastrointest. Cancer Res. 6, 3–9 (2013).
Malouf, G. et al. Is histological diagnosis of primary liver carcinomas with fibrous stroma reproducible among experts? J. Clin. Pathol. 62, 519–524 (2009).
Lalazar, G. & Simon, S. M. Fibrolamellar Carcinoma: Recent Advances and Unresolved Questions on the Molecular Mechanisms. Semin. Liver Dis. 38, 51–59 (2018).
Zack, T. et al. Defining incidence and complications of fibrolamellar liver cancer through tiered computational analysis of clinical data. NPJ Precis. Oncol. 7, 29 (2023).
Yamashita, S. et al. Prognosis of fibrolamellar carcinoma compared to non-cirrhotic conventional hepatocellular carcinoma. J. Gastrointest. Surg. 20, 1725–1731 (2016).
Berkovitz, A. et al. Clinical and demographic predictors of survival for fibrolamellar carcinoma patients-A patient community, registry-based study. Hepatol. Commun. 6, 3539–3549 (2022).
Fritz, A. G. International Classification of Diseases for Oncology: ICD-O. 3rd edition, First revision. edn, (World Health Organization, 2013).
Simon, E. P. et al. Transcriptomic characterization of fibrolamellar hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 112, E5916–E5925 (2015).
Levin, S. N. et al. Disruption of proteome by an oncogenic fusion kinase alters metabolism in fibrolamellar hepatocellular carcinoma. Sci. Adv. 9, eadg7038 (2023).
Darcy, D. G. et al. The genomic landscape of fibrolamellar hepatocellular carcinoma: whole genome sequencing of ten patients. Oncotarget 6, 755–770 (2015).
Honeyman, J. N. et al. Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science 343, 1010–1014 (2014).
Graham, R. P. et al. DNAJB1-PRKACA is specific for fibrolamellar carcinoma. Mod. Pathol. 28, 822–829 (2015).
Kastenhuber, E. R. et al. DNAJB1-PRKACA fusion kinase interacts with beta-catenin and the liver regenerative response to drive fibrolamellar hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 114, 13076–13084 (2017).
Engelholm, L. H. et al. CRISPR/Cas9 Engineering of adult mouse liver demonstrates that the Dnajb1-Prkaca gene fusion Is sufficient to induce tumors resembling fibrolamellar hepatocellular carcinoma. Gastroenterology 153, 1662–1673 (2017).
Neumayer, C. et al. DNAJB1-PRKACA is specific for fibrolamellar carcinoma. Clin. Cancer Res. 29, 271–278 (2023).
Neumayer, C. et al. GalNAc-conjugated siRNA targeting the DNAJB1-PRKACA fusion junction in fibrolamellar hepatocellular carcinoma. Mol. Ther. J. 32, 140–151 (2024).
Graham, R. P. et al. Fibrolamellar carcinoma in the Carney complex: PRKAR1A loss instead of the classic DNAJB1-PRKACA fusion. Hepatology 68, 1441–1447 (2018).
Singhi, A. D. et al. Recurrent rearrangements in PRKACA and PRKACB in intraductal oncocytic papillary neoplasms of the pancreas and bile duct. Gastroenterology 158, 573–582 (2020).
Hirsch, T. Z. et al. BAP1 mutations define a homogeneous subgroup of hepatocellular carcinoma with fibrolamellar-like features and activated PKA. J. Hepatol. 72, 924–936 (2020).
Aggarwal, R. et al. Clinical and genomic characterization of treatment-emergent small-cell neuroendocrine prostate cancer: A multi-institutional prospective study. J. Clin. Oncol. 36, 2492–2503 (2018).
Saltsman, J. A. et al. A Human Organoid Model of Aggressive Hepatoblastoma for Disease Modeling and Drug Testing. Cancers 12, 2668 (2020).
Narayan, N. J. C. et al. Human liver organoids for disease modeling of fibrolamellar carcinoma. Stem Cell Rep. 17, 1874–1888 (2022).
Lalazar, G. et al. Identification of novel therapeutic targets for fibrolamellar carcinoma using patient-derived xenografts and direct-from-patient screening. Cancer Discov. 11, 2544–2563 (2021).
Xu, L. et al. Genomic analysis of fibrolamellar hepatocellular carcinoma. Hum. Mol. Genet. 24, 50–63 (2015).
Sorenson, E. C. et al. Genome and transcriptome profiling of fibrolamellar hepatocellular carcinoma demonstrates p53 and IGF2BP1 dysregulation. PloS ONE 12, e0176562 (2017).
Francisco, A. B. et al. Multiomic analysis of microRNA-mediated regulation reveals a proliferative axis involving miR-10b in fibrolamellar carcinoma. JCI Insight 7, https://doi.org/10.1172/jci.insight.154743 (2022).
Robinson, D. R. et al. Integrative clinical genomics of metastatic cancer. Nature 548, 297–303 (2017).
Cancer Genome Atlas Research Network. Electronic address, w. b. e. & cancer genome atlas research, N. comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 169, 1327–1341.e1323 (2017).
Stevens, J. R., Herrick, J. S., Wolff, R. K. & Slattery, M. L. Power in pairs: assessing the statistical value of paired samples in tests for differential expression. BMC Genomics 19, 953 (2018).
Trapnell, C. et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46 (2013).
Leek, J. T. et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 11, 733–739 (2010).
Johnson, W. E., Li, C. & Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007).
Gagnon-Bartsch, J. A. & Speed, T. P. Using control genes to correct for unwanted variation in microarray data. Biostatistics 13, 539–552 (2012).
Wang, S. & Friedman, S. L. Hepatic fibrosis: A convergent response to liver injury that is reversible. J. Hepatol. 73, 210–211 (2020).
Harbour, J. W. et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 330, 1410–1413 (2010).
Bott, M. et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat. Genet. 43, 668–672 (2011).
Pena-Llopis, S. et al. BAP1 loss defines a new class of renal cell carcinoma. Nat. Genet. 44, 751–759 (2012).
Scheuermann, J. C. et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465, 243–247 (2010).
Artegiani, B. et al. Probing the tumor suppressor function of BAP1 in CRISPR-engineered human liver organoids. Cell Stem Cell 24, 927–943 (2019).
Novak, P. et al. Agglomerative epigenetic aberrations are a common event in human breast cancer. Cancer Res. 68, 8616–8625 (2008).
Dallosso, A. R. et al. Frequent long-range epigenetic silencing of protocadherin gene clusters on chromosome 5q31 in Wilms’ tumor. PLoS Genet. 5, e1000745 (2009).
Zhang, Q. et al. hTFtarget: A comprehensive database for regulations of human transcription factors and their targets. Genom. Proteom. Bioinforma. 18, 120–128 (2020).
Oughtred, R. et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 30, 187–200 (2021).
Wang, J. et al. FOXA2 suppresses the metastasis of hepatocellular carcinoma partially through matrix metalloproteinase-9 inhibition. Carcinogenesis 35, 2576–2583 (2014).
He, S., Zhang, J., Zhang, W., Chen, F. & Luo, R. FOXA1 inhibits hepatocellular carcinoma progression by suppressing PIK3R1 expression in male patients. J. Exp. Clin. Cancer Res. 36, 175 (2017).
Chen, K. Y. et al. Clinical outcomes in fibrolamellar hepatocellular carcinoma treated with immune checkpoint inhibitors. Cancers 14, https://doi.org/10.3390/cancers14215347 (2022).
Oikawa, T. et al. Model of fibrolamellar hepatocellular carcinomas reveals striking enrichment in cancer stem cells. Nat. Commun. 6, 8070 (2015).
Sell, S. On the stem cell origin of cancer. Am. J. Pathol. 176, 2584–2494 (2010).
Ramalho-Santos, M., Yoon, S., Matsuzaki, Y., Mulligan, R. C. & Melton, D. A. Stemness”: transcriptional profiling of embryonic and adult stem cells. Science 298, 597–600 (2002).
Malta, T. M. et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell 173, 338–354 (2018).
Yamashita, T. et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology 136, 1012–1024 (2009).
Cairo, S. et al. Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell 14, 471–484 (2008).
Farber, B. A. et al. Non coding RNA analysis in fibrolamellar hepatocellular carcinoma. Oncotarget 9, 10211–10227 (2018).
O’Neill, A. F. et al. Fibrolamellar carcinoma: An entity all its own. Curr. Probl. Cancer 45, 100770 (2021).
Anders, S., Pyl, P. T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Shirani, M. et al. Increased protein kinase a activity induces fibrolamellar hepatocellular carcinoma features independent of DNAJB1. Cancer Res. 84, 2626–2644 (2024).
Simon, S. M. Fighting rare cancers: lessons from fibrolamellar hepatocellular carcinoma. Nat. Rev. Cancer 23, 335–346 (2023).
Garcia-Bermudez, J. et al. PKA Phosphorylates the ATPase inhibitory factor 1 and inactivates its capacity to bind and inhibit the mitochondrial H(+)-ATP synthase. Cell Rep. 12, 2143–2155 (2015).
Zehir, A. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 23, 703–713 (2017).
Kannangai, R., Vivekanandan, P., Martinez-Murillo, F., Choti, M. & Torbenson, M. Fibrolamellar carcinomas show overexpression of genes in the RAS, MAPK, PIK3, and xenobiotic degradation pathways. Hum. Pathol. 38, 639–644 (2007).
Huang, H. et al. Multi-omics analyses reveal spatial heterogeneity in primary and metastatic oesophageal squamous cell carcinoma. Clin. Transl. Med. 13, e1493 (2023).
Zhang, B. & Wu, H. Decreased expression of COLEC10 predicts poor overall survival in patients with hepatocellular carcinoma. Cancer Manag. Res. 10, 2369–2375 (2018).
Hansen, S. et al. Collectin 11 (CL-11, CL-K1) is a MASP-1/3-associated plasma collectin with microbial-binding activity. J. Immunol. 185, 6096–6104 (2010).
Rooryck, C. et al. Mutations in lectin complement pathway genes COLEC11 and MASP1 cause 3MC syndrome. Nat. Genet. 43, 197–203 (2011).
Berkovitz, A. et al. Clinical and demographic predictors of survival for fibrolamellar carcinoma patients – A patient community registry-based study. Hepatol. Commun. 6, 3539–3549 (2022).
Carrillo-Reixach, J. et al. Epigenetic footprint enables molecular risk stratification of hepatoblastoma with clinical implications. J. Hepatol. 73, 328–341 (2020).
Hooks, K. B. et al. New insights into diagnosis and therapeutic options for proliferative hepatoblastoma. Hepatology 68, 89–102 (2018).
Wagner, A. E. et al. SP8 Promotes an aggressive phenotype in hepatoblastoma via FGF8 activation. Cancers 12, https://doi.org/10.3390/cancers12082294 (2020).
Ahn, K. S. et al. Prognostic subclass of intrahepatic cholangiocarcinoma by integrative molecular-clinical analysis and potential targeted approach. Hepatol. Int. 13, 490–500 (2019).
Gao, C. et al. The A-to-I editing of KPC1 promotes intrahepatic cholangiocarcinoma by attenuating proteasomal processing of NF-kappaB1 p105 to p50. J. Exp. Clin. Cancer Res. 41, 338 (2022).
Sia, D. et al. Massive parallel sequencing uncovers actionable FGFR2-PPHLN1 fusion and ARAF mutations in intrahepatic cholangiocarcinoma. Nat. Commun. 6, 6087 (2015).
Farshidfar, F. et al. Integrative genomic analysis of cholangiocarcinoma identifies distinct IDH-Mutant molecular profiles. Cell Rep. 18, 2780–2794 (2017).
Long, M. et al. A novel risk score based on immune-related genes for hepatocellular carcinoma as a reliable prognostic biomarker and correlated with immune infiltration. Front. Immunol. 13, 1023349 (2022).
Liu, G. et al. Potential diagnostic and prognostic marker dimethylglycine dehydrogenase (DMGDH) suppresses hepatocellular carcinoma metastasis in vitro and in vivo. Oncotarget 7, 32607–32616 (2016).
Huang, H. et al. Integrated analysis of microbiome and host transcriptome reveals correlations between gut microbiota and clinical outcomes in HBV-related hepatocellular carcinoma. Genome Med. 12, 102 (2020).
Jin, Y. et al. Comprehensive analysis of transcriptome profiles in hepatocellular carcinoma. J. Transl. Med. 17, 273 (2019).
Wang, K. et al. PHGDH arginine methylation by PRMT1 promotes serine synthesis and represents a therapeutic vulnerability in hepatocellular carcinoma. Nat. Commun. 14, 1011 (2023).
Yoo, S. et al. A pilot systematic genomic comparison of recurrence risks of hepatitis B virus-associated hepatocellular carcinoma with low- and high-degree liver fibrosis. BMC Med. 15, 214 (2017).
Yang, Y. et al. Recurrently deregulated lncRNAs in hepatocellular carcinoma. Nat. Commun. 8, 14421 (2017).
Tarasov, A., Vilella, A. J., Cuppen, E., Nijman, I. J. & Prins, P. Sambamba: fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034 (2015).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Ewels, P., Magnusson, M., Lundin, S. & Kaller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016).
Pearson, K. On lines and planes of closest fit to systems of points in space. Philos. Mag. 2, 559–572 (1901).
van der Maaten, L. & Hinton, G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Healy, J. & McInnes, L. Uniform manifold approximation and projection. Nat. Rev. Methods Primers 4, 82 (2024).
Campello, R. J. G. B., Moulavi, D. & Sander, J. Density-Based Clustering Based on Hierarchical Density Estimates. In Advances in Knowledge Discovery and Data Mining. PAKDD 2013. Lecture Notes in Computer Science(), (eds Pei, J., Tseng, V.S., Cao, L., Motoda, H. & Xu, G.) vol 7819. (Springer, Berlin, Heidelberg, 2013).
Murtagh, F. & Legendre, P. Ward’s hierarchical agglomerative clustering method: Which algorithms implement ward’s criterion? J. Classif. 31, 274–295 (2014).
Soneson, C., Love, M. I. & Robinson, M. D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 4, 1521 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Van der Auwera, G. A. et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 43, 11 10 11–11 10 33 (2013).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Kim, S. et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat. Methods 15, 591–594 (2018).
Narzisi, G. et al. Genome-wide somatic variant calling using localized colored de Bruijn graphs. Commun. Biol. 1, 20 (2018).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, https://doi.org/10.1093/gigascience/giab008 (2021).
McLaren, W. et al. The ensembl variant effect predictor. Genome Biol. 17, 122 (2016).
Ewels, P. A. et al. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 38, 276–278 (2020).
Krueger, F. & Andrews, S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011).
Akalin, A. et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13, R87 (2012).
Liberzon, A. et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Brosch, M. et al. Epigenomic map of human liver reveals principles of zonated morphogenic and metabolic control. Nat. Commun. 9, 4150 (2018).
Hu, H. et al. Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell 175, 1591–1606 (2018).
MacParland, S. A. et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 9, 4383 (2018).
Cari, L. et al. Identification of 15 T cell restricted genes evaluates T cell infiltration of human healthy tissues and cancers and shows prognostic and predictive potential. Int. J. Mol. Sci. 20, https://doi.org/10.3390/ijms20205242 (2019).
Franzen, O., Gan, L. M. & Bjorkegren, J. L. M. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database 2019, https://doi.org/10.1093/database/baz046 (2019).
Fultang, N., Li, X., Li, T. & Chen, Y. H. Myeloid-derived suppressor cell differentiation in cancer: Transcriptional regulators and enhanceosome-mediated mechanisms. Front. Immunol. 11, 619253 (2020).
Verstegen, M. M. A. et al. Human extrahepatic and intrahepatic cholangiocyte organoids show region-specific differentiation potential and model cystic fibrosis-related bile duct disease. Sci. Rep. 10, 21900 (2020).
Sampaziotis, F. et al. Cholangiocyte organoids can repair bile ducts after transplantation in the human liver. Science 371, 839–846 (2021).
Yu, S. et al. Significance of liver zonation in hepatocellular carcinoma. Front. Cell Dev. Biol. 10, 806408 (2022).
Hu, C. et al. CellMarker 2.0: an updated database of manually curated cell markers in human/mouse and web tools based on scRNA-seq data. Nucleic Acids Res. 51, D870–D876 (2023).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Lambert, S. A. et al. The human transcription factors. Cell 172, 650–665 (2018).
Weirauch, M. T. et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158, 1431–1443 (2014).
Soto, L. F. et al. Compendium of human transcription factor effector domains. Mol. Cell 82, 514–526 (2022).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300 (2018).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Lu, Y. et al. Spatial transcriptome profiling by MERFISH reveals fetal liver hematopoietic stem cell niche architecture. Cell Discov. 7, 47 (2021).
Moffitt, J. R. et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 362, https://doi.org/10.1126/science.aau5324 (2018).
Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021).
Acknowledgements
We would like to thank the fibrolamellar patients and their caregivers for their contributions to The Fibrolamellar Registry, to our Fibrolamellar Tissue Repository, through work at the bench and contributions in too many ways to enumerate. We would also like to thank for their financial support, critical seed funds from private foundations and the NIH/NCI P50CA210964 (S.M.S.); NIH/NCI U54CA243126 (S.M.S.); Center for Basic and Translational Research on Disorders of the Digestive System through the generosity of the Leona M. and Harry B. Helmsley Charitable Trust (S.M.S.); The Rally Foundation (S.M.S.); The Bear Necessities (S.M.S.); The Truth365 (S.M.S.). Supported in part by grant NIH/NCATS UL1 TR001866 from a Clinical and Translational Science Award (CTSA). We acknowledge the use of the Integrated Genomics Operation Core of the Memorial Sloan Kettering Cancer Center, funded by the NCI Cancer Center Support Grant (CCSG, P30 CA08748), Cycle for Survival, and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology.
Author information
Authors and Affiliations
Contributions
D.R. and S.M.S. devised the study. D.R. and J.A.S. collected and conducted RNA-seq of our samples. M.S.T. carried out the histopathological assessment of the samples. D.R. performed the whole-genome and bisulfite methylation sequencing. D.R. and M.S. processed the samples for single-cell spatial transcriptomics. J.A.M. and D.R. collected and curated the external datasets and conducted the differential variant calling and gene set enrichment analysis. D.R. performed the differential expression and methylation analyses and calculated the transcriptomic signatures. L.F.S. and D.R. did the multiomics and transcription factor analysis. D.R. and J.A.M. generated the data for the Shiny App, and L.F.S. developed the user interface. D.R. prepared the manuscript. S.M.S., P.C., M.S., and J.A.M. provided critical reviews of the manuscript. All authors approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Sandrine Imbeaud, and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Requena, D., Medico, J.A., Soto-Ugaldi, L.F. et al. Liver cancer multiomics reveals diverse protein kinase A disruptions convergently produce fibrolamellar hepatocellular carcinoma. Nat Commun 15, 10887 (2024). https://doi.org/10.1038/s41467-024-55238-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-55238-2
This article is cited by
-
Advancing liver cancer diagnosis and treatment with multi-omics approaches: a systematic review
Discover Oncology (2025)