Abstract
Immune checkpoint blockade (ICB) has become a standard anti-cancer treatment, offering durable clinical benefits. However, the limited response rate of ICB necessitates biomarkers to predict and modulate the efficacy of the therapy. The gut microbiome’s influence on ICB efficacy is of particular interest due to its modifiability through various interventions. However, gut microbiome biomarkers for ICB response have been inconsistent across different studies. Here, we identify TANB77, an uncultured and distinct bacterial clade, as the most consistent responder-enriched taxon through meta-analysis of ten independent ICB recipient cohorts. Traditional taxonomy fails to distinguish TANB77 from unrelated taxa, leading to its oversight. Mice with higher gut TANB77 abundance, either naturally or through transplantation, show improved response to anti-PD-1 therapy. Additionally, mice injected with TANB77-derived pilin-like protein exhibit improved anti-PD-1 therapy response, providing in vivo evidence for the beneficial role of the pilin-like protein. These findings suggest that pilins from the TANB77 order may enhance responses to ICB therapy across diverse cohorts of cancer patients.
Similar content being viewed by others
Introduction
Cancer immunotherapy, particularly immune checkpoint blockade (ICB) therapy, has marked a significant breakthrough in oncology, providing durable clinical benefits and transforming the treatment landscape for various malignancies. ICB therapies, which include antibodies targeting PD-1, PD-L1, and CTLA-4, function by blocking the interaction between immune checkpoint molecules and its ligand, leading to reactivation of immune system1,2. Despite these advancements, the overall response rate (ORR) to ICB therapy varies widely among patients, with a substantial proportion failing to achieve significant clinical benefit3,4. The limited response rate underscores the importance of identifying biomarkers to predict responders and developing combination therapy to enhance ICB efficacy.
Multiple studies have revealed associations between the gut microbiome and ICB response, identifying specific gut microbial taxa that are differentially abundant between responders and non-responders to ICB therapy5,6,7,8,9,10. State-of-the-art research includes clinical trials demonstrating that fecal microbiota transplantation (FMT) to patients can improve therapeutic outcomes11,12,13, as well as longitudinal studies observing shifts in the gut microbiome composition during ICB treatment14. Despite these promising developments, significant challenges remain. Results across studies have shown limited consistency, with different taxa being associated with ICB response. A comprehensive meta-analysis highlighted the complexity and cohort-dependency of gut microbiome associations with ICB response, revealing limited reproducibility of microbiome-based signatures across cohorts15. Furthermore, there is a lack of mechanistic understanding of how specific gut microbiota influence ICB efficacy, which hinders the development of reliable microbiome-based biomarkers and therapeutics.
In this study, we identified TANB77 order as a consistent metagenomic biomarker for ICB response, with its higher abundance correlating with better clinical outcomes after treatment across ten independent cohorts of patients. The TANB77 order is an uncultured and distinct bacterial clade often overlooked in previous analyses due to taxonomic discrepancies outside the Genome Taxonomy Database (GTDB) system16. We found that an exclusively conserved pilin-like protein derived from TANB77 can stimulate dendritic cell (DC) activation through the TLR4 signaling pathway in vitro and its direct injection enhanced ICB efficacy in mice. These findings highlight the potential of TANB77 order and its pilin-like proteins to serve as biomarkers and modulators of ICB response.
Results
TANB77 order was identified as a strong predictor of ICB response and prognosis
We collected fecal samples from non-small cell lung cancer (NSCLC) patients (n = 49) enrolled at Yonsei Cancer Center (YCC) in Korea before the initiation of anti-PD-1 therapy as a discovery cohort (hereafter referred to as YCC_1st cohort). The samples underwent ultra-deep whole metagenome sequencing (WMS) (>30Gbp on average), substantially deeper than similar studies (Supplementary Fig. 1). The patients were classified as responders (R, n = 18) or non-responders (NR, n = 31) based on the Response Evaluation Criteria in Solid Tumors, version 1.1 (RECIST 1.1)17 (Methods). The R and NR groups showed no significant differences in age, sex, smoking history, ECOG status, histology, and antibiotic treatment history (Supplementary Table 1).
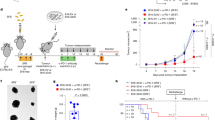
The taxonomic abundance of the patients’ gut microbiota was profiled based on GTDB taxonomy16 using Kraken218 with the HRGM database19, which is presented as the primary results in this paper, along with mOTUs320, and MetaPhlAn421 (Methods). No significant difference in alpha and beta diversity of the microbial community was detected between the R and NR groups (Supplementary Fig. 2a–f). Using a multivariable analysis, we found several differentially abundant taxa significantly enriched in either R or NR groups. Seven out of the top ten responder-enriched taxa (based on multivariable MaAsLin222 test P value) corresponded to the bacterial order TANB77 and its sub-taxa (Fig. 1a), while the relative abundance of the TANB77 order was 6.1-fold higher in R compared to NR patients (two-tailed Mann-Whitney U test, P < 0.01) (Fig. 1b). The enrichment of TANB77 in responders was also detected using mOTUs3 and MetaPhlAn4 taxonomic profiles (Supplementary Fig. 2g, h). Notably, taxa previously reported to be associated with ICB response in studies with similar designs5,6,7,8,9,10 showed limited reproducibility (Full analysis results are available, see Data availability).
a Volcano plot showing taxa enriched in responders (R, n = 18) or non-responders (NR, n = 31) in the YCC_1st cohort, produced by MaAsLin2 multivariable analysis. The horizontal dashed line indicates the unadjusted two-sided MaAsLin2 P = 0.05 threshold, and the vertical dashed line indicates the |MaAsLin2 coefficient | = 2 threshold. A positive MaAsLin2 coefficient means taxa are enriched in responders. TANB77 and its sub-taxa are highlighted in red. b Relative abundance of TANB77 in R and NR patients. Significance was evaluated by a two-tailed Mann-Whitney U test. The center line of the boxplot represents the median, the bounds of the box represent the 25th, and 75th percentiles, and the whiskers represent the minimum and maximum values within the 1.5 interquartile range from the lower and upper quartiles. c Receiver operating characteristic (ROC) curve illustrating the retrieval of responders based on the relative abundance of TANB77, with the area under the ROC curve (AUROC) indicated. d, e Kaplan-Meier plots representing progression-free survival (PFS, d) and overall survival (OS, e) of high- and low-TANB77 groups. Significance was evaluated by a two-tailed log-rank test. f, g Multivariate Cox regression models of PFS (f) and OS (g) in the YCC_1st cohort. Horizontal bars denote the 95% confidence interval (CI) of the hazard ratio (HR) for each category. Vertical marks indicate the HR. Significant categories are depicted by blue (decreasing HR) and red (increasing HR) bars, with their two-sided Cox regression test unadjusted P-values indicated (*P < 0.05, **P < 0.01, ***P < 0.001).
Based on the relative abundance of TANB77, receiver operating characteristic (ROC) analysis revealed an area under the ROC curve (AUROC) of 0.737 for predicting responders (Fig. 1c). We next tested whether TANB77 was predictive of prognosis in patients undergoing anti-PD-1 treatment. We stratified anti-PD-1 recipients into high- and low-TANB77 groups based on the TANB77 abundance threshold defined by the maximum Youden index in the ROC curve. Progression-free survival (PFS) and overall survival (OS) were significantly higher in the high- compared to the low-TANB77 group (two-tailed log-rank test, P < 0.05) (Fig. 1d, e). Furthermore, we performed Cox regression analysis to investigate whether the observed positive effect of TANB77 on clinical outcome was dependent on other factors known to be associated with immunotherapy response, such as oncogene mutations23, smoking history24, and PD-L1 expression25. Results showed that the abundance of TANB77 was a significant factor for the reduced hazard ratio of both PFS and OS (Fig. 1f, g, Supplementary Table 2). Notably, high expression of PD-L1 was a positive factor for PFS, whereas oncogenic mutation (EGFR or ALK) and antibiotic use were negative factors for PFS and OS, respectively.
Higher abundance of TANB77 consistently linked to better ICB response in diverse cohorts
Next, we examined whether the observed association between TANB77 and the ICB response could be reproduced in other independent cohorts. We established a validation cohort by performing WMS on fecal samples from an additional cohort of 51 NSCLC patients who underwent anti-PD-L1 therapy (hereafter referred to as YCC_2nd cohort, Methods Supplementary Table 1). Additionally, we collected publicly available WMS datasets on fecal samples from eight independent cohorts of ICB therapy recipients5,6,7,8,9,10,15. The datasets varied in immunotherapy type, cancer type, sequencing technology, and geographical location. We selected baseline samples before the first immunotherapy treatment and standardized the response evaluation criteria in accordance with the YCC cohort. The final dataset included 660 fecal WMS samples (265 R and 395 NR), from ten independent ICB recipient cohorts. (Methods Supplementary Table 3).
Consistent with the previous cross-cohort study, the gut communities differed significantly by cohort (PERMANOVA test, P < 0.0001) (Supplementary Fig. 3). SIAMCAT26 analysis reaffirmed that the variance in most taxa was explained more by cohort than by ICB response. However, we noted that the variance of TANB77 was explained more by ICB response than by cohort. Additionally, TANB77 showed the highest ratio of variance explained by ICB response to the variance explained by cohort consistently across different taxonomic profilers (Supplementary Fig. 4).
We next performed a meta-analysis using MMUPHin27 and identified consistent biomarkers by giving more weight to taxa that were enriched in the same direction (either toward R or NR) across multiple cohorts (Methods). We found that the TANB77 order was most consistently enriched in responders, while the Hungatella genus was most consistently enriched in non-responders (Fig. 2a). Although the enrichment of TANB77 was not statistically significant in all individual cohorts, the direction of enrichment was consistent across all cohorts and the enrichment in the pooled sample was significant (Benjamini-Hochberg corrected MMUPHin q-value < 0.001) (Fig. 2b, c). This high level of consistency in TANB77 enrichment toward responders was confirmed with different microbial taxonomic profilers (Supplementary Fig. 5a–d). To further examine the consistency of our findings, we compiled six datasets6,7,9,28,29,30 containing fecal 16S rRNA sequences of ICB therapy recipients, consisting of 125 R and 96 NR (Supplementary Table 4). Taxonomic profiling using a refined set of TANB77 16S rRNA sequences followed by batch correction27 and LEfSe analyses31 (Methods) revealed a major family of order TANB77, CAG-508, as a significant responder-enriched taxon (Supplementary Fig. 6). Notably, CAG-508 was the only taxon to be enriched for ICB responders by both WMS and 16S rRNA sequencing analyses. The enrichment of TANB77 in responders was also reproduced in cancer-type-specific and nivolumab recipient-specific analyses (Supplementary Figs. 7, 8).
a Gut microbial taxa showing consistent associations with ICB response in ten cohorts of immunotherapy recipients. The 50 taxa with the highest absolute consensus scores are shown, along with their consensus scores and unadjusted meta-analysis P-values (by MMUPHin, two-sided). Taxa are sorted by absolute consensus score. Taxa within the TANB77 and Hungatella clades are highlighted in blue and red, respectively. b MaAsLin2 effect size of TANB77 abundance as a predictor of immunotherapy response in individual cohorts and pooled samples. Center circle and horizontal line denote the MaAsLin2 coefficient and 95% confidence interval (CI), respectively. Statistically significant (two-sided unadjusted MaAsLin2 P < 0.05) cohorts are highlighted in blue. c Volcano plot representing MMUPHin coefficients and two-sided unadjusted P-values. A positive coefficient represents responder-enriched taxa. The x-axis scale was calculated using a symmetric log function. d Kaplan-Meier (KM) plots representing progression-free survival (PFS) of high- and low-TANB77 groups in the Gopalakrishnan, Peters, and YCC_2nd cohorts. Significance was evaluated by a one-tailed log-rank test. e Number of patients with PFS above or below certain months in the Routy, Lee-PRIMM-UK, and Lee-PRIMM-NL cohorts. Patients were categorized into high- and low-TANB77 groups. Significance was evaluated by a one-tailed Fisher’s exact test. f KM plots representing overall survival (OS) of high- and low-TANB77 groups in the Derosa, Peters, and YCC_2nd cohorts. Significance was evaluated by a one-tailed log-rank test. g Change in the tumor size after immunotherapy treatment in the Frankel and Matson cohorts. Significance was evaluated by a one-tailed Mann-Whitney U test. The center line of the boxplot represents the median, the bounds of the box represent the 25th, and 75th percentiles, and the whiskers represent the minimum and maximum values within the 1.5 interquartile range from the lower and upper quartiles. h Summary of the association between high TANB77 abundance in the gut and ICB therapy outcome in ten cohorts. Source data are provided as a Source Data file. (*P < 0.05, **P < 0.01, ***P < 0.001).
We next examined whether the higher abundance of TANB77 correlates with better clinical outcomes in ICB recipients. We divided the patients into high- and low-TANB77 groups based on the threshold defined by the maximum Youden index in ROC analysis in individual cohorts. We observed prolonged PFS in the Gopalakrishnan, Peters, and YCC_2nd cohorts (one-tailed log-rank test, P < 0.05) (Fig. 2d). In the Routy cohort, which provides categorical metadata on whether PFS is above or below 3 or 6 months, high-TANB77 patients were more frequent in the group with longer PFS at both the 3-month and 6-month thresholds (one-tailed Fisher’s exact test, P < 0.01). Similarly, both Lee-PRIMM-UK and Lee-PRIMM-NL cohorts showed a higher ratio of high-TANB77 patients in the group with PFS longer than 12 months, although the significance test was not passed for the Lee-PRIMM-NL cohort (Fig. 2e). Overall survival was significantly longer in the high-TANB77 group in the Derosa and YCC_2nd cohorts (one-tailed log-rank test, P < 0.05) (Fig. 2f). In the Frankel cohort, we observed that the tumor size growth was significantly lower in the high-TANB77 group (one-tailed Mann-Whitney U test, P < 0.01). However, in the Matson cohort, there was no significant difference (one-tailed Mann-Whitney U test, P > 0.05) (Fig. 2g). Overall, a significant correlation between higher TANB77 abundance and better ICB efficacy was observed in 8 out of 10 independent cohorts (Fig. 2h). Notably, even in the cohorts that did not reach statistical significance showed the same trend of correlation between higher abundance of TANB77 and better clinical outcomes. These results were also reproduced when classifying patients into TANB77 positive and negative groups based on MetaPhlAn4 and mOTUs3 profiles (Supplementary Fig. 9).
TANB77 is a distinct bacterial order with taxonomic discrepancies outside the GTDB system
The GTDB taxonomic classification system, which is based on 120 ubiquitous single-copy proteins, defines TANB77 as a bacterial order within the class Clostridia32. The TANB77 order has been consistently maintained throughout the updates of GTDB from r80 (November 2017) to r220 (April 2024) (Supplementary Fig. 10a). In contrast, non-GTDB taxonomies annotate TANB77 inconsistently. NCBI taxonomy places TANB77 genomes into several polyphyletic clades, many of which lack taxonomic resolution and include genomes not classified as TANB77 by GTDB taxonomy33 (Fig. 3a). MetaPhlAn421, a widely used taxonomic profiler in metagenomics, uses its own taxonomy of species genome bins (SGBs). In MetaPhlAn4, TANB77 is represented by 120 SGBs from 27 families, 24 orders, 21 classes, and 6 phyla (Supplementary Fig. 10b). Similarly, clades containing at least one TANB77 SGB also contain many non-TANB77 SGBs (Supplementary Fig. 10c). Such discrepancy is also observed in another widely used taxonomic profiler, mOTUs320 (Supplementary Fig. 10d). Furthermore, this issue persists in the 16S rRNA-based taxonomy, where the taxonomy corresponding to TANB77 order was inconsistent across SILVA34, RDP35, and EzBioCloud36 databases (Supplementary Data 1).
a Comparative taxonomic classification of TANB77 genomes. TANB77 genomes are classified into seven NCBI families shown on the right (the relationship is highlighted in red lines), and the genomes within these NCBI clades are classified into multiple GTDB orders on the left. b Comparison of GC content between genomes classified as TANB77 and non-TANB77 genomes within the top three NCBI families containing the most TANB77 genomes. Significance was evaluated by a two-tailed Mann-Whitney U test. (**P < 0.01, ***P < 0.001).
These discrepancies in the taxonomy of TANB77 may explain why previous studies failed to detect TANB77 as an ICB response marker. None of the clades (and even none of the combinations of clades) outside of GTDB can accurately represent TANB77. Consequently, the TANB77 signal has likely been dispersed or confounded with other taxa in previous works. We compared the meta-analysis results of the ten cohorts of ICB recipients using the MetaPhlAn4 or mOTUs3 taxonomy and those using the GTDB-converted taxonomy. The results showed that when using the MetaPhlAn4 or mOTUs3 taxonomy, the signals of features belonging to TANB77 were spread out, resulting in less prominent results compared to using the GTDB taxonomy (Supplementary Fig. 5e, f).
Despite discrepancies in taxonomy outside the GTDB, analyses using the entire genome contents confirmed that TANB77 is a distinct bacterial clade. In a previous study, where complete and circular genomes of TANB77 were assembled using long-read sequencing data37, genomes of TANB77 were exclusively clustered based on the KEGG pathway and KEGG ortholog profiles. We also observed that genomes of TANB77 possess a distinct GC content when compared to other genomes of clades commonly misclassified by NCBI (Fig. 3b). Analysis using traitar38 also revealed that TANB77 genomes exhibit trait profiles significantly different from their closest genomes outside of order TANB77, with the absence of many growth-related traits (Supplementary Fig. 11a).
To better understand in which ecosystems TANB77 can thrive, we assessed the prevalence of TANB77 in the planetary-scale microbiome resource SPIRE39 (Methods). TANB77 genomes are predominantly found in human oral, human gut, and non-human animal gut microbiomes (Supplementary Fig. 11b, c) and are infrequently detected in infants, suggesting they likely establish later in life (Supplementary Fig. 11d, e). Searches across various cultured bacterial genome databases40,41,42,43,44 revealed no TANB77 genomes, indicating that despite their prevalence in the human microbiome, TANB77 remains uncultured. TANB77 abundance showed a strong positive correlation with the RF39 order and CAG-272 family, both of which are also uncultured bacterial clades (Supplementary Fig. 11f, g). This suggests that these uncultured clades with reduced genome sizes thrive in specific gut environments. Among cultivated taxa, Coprococcus genus showed the strongest positive correlation with TANB77 (Supplementary Fig. 11f, h). In contrast, taxa showing a negative correlation included species like Ruminococcus_B gnavus, known to be enriched in diseases such as IBD and CRC45. Those taxa with positive and negative correlations with TANB77 were also observed as the most prominent high-TANB77 and low-TANB77 group enriched taxa in 660 ICB recipients, respectively (Supplementary Fig. 12).
The abundance of TANB77 in the mouse gut inversely correlates with tumor progression after anti-PD-1 treatment
We further investigated the relationship between TANB77 levels in the gut and ICB therapy response in a mouse model. Sivan et al. (2015) reported that mice from the Jackson Laboratory (JAX) showed slower tumor progression and better anti-PD-L1 therapy response than Taconic Farms (TAC) mice46. Moreover, TAC mice transplanted with JAX feces showed an enhanced response to ICB, demonstrating the causal effect of the gut microbiome on immunotherapy response. Here, we investigated TANB77 levels in the murine gut using 16S rRNA data from the Sivan et al. (2015) study. We first classified the animals into JAX and TAC phenotype mice (Supplementary Table 5). The JAX phenotype mice had significantly higher TANB77 levels in their gut microbiome compared to TAC phenotype mice (two-tailed Mann-Whitney U test, P < 0.0001) (Fig. 4a). To assess whether the altered ICB response in TAC mice after JAX feces transplantation was associated with TANB77 abundance, we examined data from a cross-transplantation experiment. Thus, we compared TANB77 abundance between TAC mice transplanted with JAX feces (JAX to TAC) and TAC mice transplanted with TAC feces (TAC to TAC). Interestingly, TANB77 abundance increased significantly during 14 days in JAX to TAC animals (two-tailed Mann-Whitney U test, P < 0.001), whereas TAC to TAC individuals showed no significant difference (two-tailed Mann-Whitney U test, P > 0.05) (Fig. 4b). While TANB77 abundance was similar between the two groups on day 0 (two-tailed Mann-Whitney U test P > 0.05), it ended up being significantly higher for JAX to TAC mice on day 14 (two-tailed Mann-Whitney U test P < 0.001) (Fig. 4b), where it coincided with that in JAX donor mice (two-tailed Mann-Whitney U test, P > 0.05) (Fig. 4c). This finding implies that the increased TANB77 originated from the JAX donor feces and could explain the improved ICB response in JAX mice.
a TANB77 abundance in JAX and TAC phenotype mice. Significance was evaluated by a two-tailed Mann Whitney U test. The center line of the boxplot represents the median, the bounds of the box represent the 25th, and 75th percentiles, and the whiskers represent the minimum and maximum values within the 1.5 interquartile range from the lower and upper quartiles. b Changes of TANB77 abundance in guts of “JAX to TAC” (blue, n = 6) and “TAC to TAC” (red, n = 6) mice after transplantation. The plot denotes average TANB77 abundance ± SEM. Statistical significance was calculated by a two-tailed Mann-Whitney U test. c Relative abundance of TANB77 in JAX donor (navy, n = 4) and TAC recipient mice on day 14 (D + 14). The plot denotes average TANB77 abundance ± SEM. Statistical significance was calculated by a two-tailed Mann-Whitney U test. d Schematic timeline of the mouse experiment relative to the tumor injection date. e Tumor growth curve of high- and low-TANB77 mice subjected to aPD-1 or isotype treatment. Data points and error bars represent the mean ± SEM of tumor volume from two independent experiments (aPD1 high-TANB77 = 9, aPD1 low-TANB77 = 12, Isotype high-TANB77 = 12, Isotype low-TANB77 = 9). Significance was evaluated by two-way ANOVA using Tukey’s multiple comparison post hoc test (all significant comparisons are shown). f Correlation between normalized tumor growth and relative abundance of the UMGS1663 genus in aPD-1 treated mice from day 11 (D + 11) to D + 14 (left), from D + 14 to D + 17 (center), and from D + 17 to D + 20 (right). The plots share y-axis. Solid lines denote linear regression. Source data are provided as a Source Data file. (n.s.: not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
To substantiate the correlation between baseline gut TANB77 abundance and anti-PD-1 response, we performed 16S rRNA sequencing on fecal samples of mice from two different vendors, followed by tumor injection and anti-PD-1 or isotype treatment (Fig. 4d). The composition of the gut microbiome differed between the two vendors (PERMANOVA, P < 0.0001) (Supplementary Fig. 13a), with higher alpha diversity in the Samtako mice (two-tailed Mann-Whitney U test, P < 0.0001) (Supplementary Fig. 13b), but similar TANB77 abundance (two-tailed Mann-Whitney U test, P > 0.05) (Supplementary Fig. 13c). Next, we examined the response to anti-PD-1 therapy. Mice from both vendors responded to anti-PD-1 treatment, with the Samtako mice displaying significantly reduced tumor growth (two-tailed Student’s t-test, P < 0.05) (Supplementary Fig. 13d) and longer survival (two-tailed log-rank test, P < 0.01) (Supplementary Fig. 13e) compared to isotype controls. In the Orient mice, tumor growth did not differ significantly between the two groups of mice (two-tailed Student’s t-test, P > 0.05) (Supplementary Fig. 13f), but the anti-PD-1-treated group survived longer than the isotype-treated control (two-tailed log-rank test, P < 0.01) (Supplementary Fig. 13g). Although mice from the two vendors had different gut microbiota, they shared similar baseline tumor growth, response to anti-PD-1 therapy (two-tailed Student’s t-test, P > 0.05) (Supplementary Fig. 13h), and survival duration (two-tailed log-rank test, P > 0.05) (Supplementary Fig. 13i, j). We then classified the animals into high-TANB77 and low-TANB77 mice based on the median abundance of species belonging to the TANB77 order. Interestingly, the high-TANB77 group mice showed a more significant reduction in tumor volume after anti-PD-1 therapy than the low-TANB77 group (two-way ANOVA followed by Tukey’s post hoc test) (Fig. 4e). To pinpoint which taxon contributed most to immunotherapy response, we investigated the gut commensals showing a negative correlation with tumor growth in anti-PD-1-treated mice. The genus UMGS1663, which belongs to the TANB77 order, displayed the strongest negative correlation with tumor growth (Spearman’s correlation, P < 0.05) from days 11 to 14, 14 to 17, and 17 to 20 (Fig. 4f). Moreover, half of the top ten taxa showing a negative correlation with the normalized tumor growth belonged to the TANB77 order, and the TANB77 itself was ranked 6th among 253 taxa (Supplementary Data 2).
FMT from a high-TANB77 human donor to mice improves the efficacy of anti-PD-1 treatment
We next conducted a human-to-mouse FMT experiment to investigate whether the abundance of gut TANB77 was an indicator of being a good donor for an improved ICB response (Fig. 5a). We first selected a high-TANB77 donor and a low-TANB77 donor from healthy individuals. The relative abundance of TANB77 from the high-TANB77 donor was 0.0334, placing it in the top 5.75% compared to 660 ICB recipients and in the top 14.75% compared to the global population. TANB77 was not detected in the low-TANB77 donor based on GTDB-converted mOTUs3 profiling (Fig. 5b). The two donor samples had different microbial compositions and alpha diversity (Supplementary Fig. 14a, full taxonomic profiles are available, see Data availability). We treated mice from the same vendor with an antibiotic cocktail to reduce the effect of intrinsic TANB77 or other resident gut microbiota and confirmed gut TANB77 to be below the detection limit (Supplementary Fig. 14b). Next, we performed FMT with the feces of high- and low-TANB77 human donors, injected the mice with tumor cells, and finally applied anti-PD-1 or isotype treatment.
a Timeline of human-to-mice FMT experiments. b Relative abundance of TANB77 in high- and low-TANB77 donors compared to global population (n = 20,900) and ICB recipients (n = 660). Blue and red horizontal lines indicate the relative abundance of high-TANB77 and low-TANB77 donor, respectively. c Tumor growth curve of high- and low-TANB77 FMT recipient mice subjected to anti-PD-1 or isotype treatment. Each treatment group had six to eight mice allocated per group (aPD1 high-TANB77 = 7, aPD1 low-TANB77 = 8, Isotype high-TANB77 = 6, Isotype low-TANB77 = 8). Data points and error bars represent the mean ± SEM. Significance was evaluated by two-way ANOVA using Tukey’s multiple comparison post hoc test (all significant comparisons are shown). d Survival curve for high- and low-TANB77 FMT recipient mice. Significance was evaluated by the pairwise two-tailed log-rank test (all significant comparisons are shown). e Proportion of Ki-67+ PD-1+ CD8+ T cells in mice PBMCs on D + 15. Significance was evaluated by a two-tailed Mann-Whitney U test and the error bars represent mean ± SEM f Schematic diagram of sequential filtration steps to identify candidate taxa responsible for improving the ICB response in high-TANB77 FMT recipient mice. g Relative abundance of the CAG-245 genus in high- (n = 13) and low-TANB77 (n = 16) FMT recipient mice feces. Significance was evaluated by two-tailed Mann-Whitney U test. b, g The center line of the boxplot represents the median, the bounds of the box represent the 25th, and 75th percentiles, and the whiskers represent the minimum and maximum values within the 1.5 interquartile range from the lower and upper quartiles. Source data are provided as a Source Data file. (n.s.: not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Notably, mice that received FMT from the high-TANB77 donor exhibited significantly better anti-PD-1 response in terms of tumor growth (two-way ANOVA followed by Tukey’s post hoc test, P < 0.05) (Fig. 5c) and survival duration (two-tailed log-rank test, P < 0.05) (Fig. 5d) than low-TANB77 FMT recipient mice. Moreover, we found a significantly higher proportion of Ki-67+ PD-1+ CD8+ T cells (two-tailed Mann-Whitney U test, P < 0.01) that are likely proliferating T cells in response to anti-PD-1 treatment47, among peripheral blood mononuclear cells (PBMCs) in high-TANB77 FMT recipient mice subjected to anti-PD-1 treatment compared to other mice (Fig. 5e, Supplementary Fig. 15a, Supplementary Fig. 16a–c).
Furthermore, we investigated murine gut microbiota using 16S rRNA sequencing data from fecal samples collected before tumor injection (D + 0). The taxonomic composition of the gut microbiome revealed a clear distinction between mice that received FMT from high- and low-TANB77 feces (PERMANOVA, P < 0.0001) (Supplementary Fig. 14c), with significantly higher alpha diversity in the former (two-tailed Mann-Whitney U test, P < 0.01) (Supplementary Fig. 14d). These results suggest that the taxonomic composition of fecal microbes from FMT donors contributes to the different anti-PD-1 responses between recipient groups. Accordingly, the elevated diversity of fecal microbiota may explain the improved ICB response. However, no significant difference in alpha diversity of fecal microbiota between responders and non-responders was detected for nine out of ten study cohorts (Supplementary Fig. 2e, f, Supplementary Fig. 14e). Even though the alpha diversity of gut microbiota was significantly higher in the Samtako mice than in the Orient mice (Supplementary Fig. 13b), both displayed a similar ICB response (Supplementary Fig. 13h–j). These findings suggest that the improved ICB response may not be caused by the higher alpha diversity. Therefore, we applied sequential filtering steps to identify candidate taxa that might specifically contribute to the improved ICB response observed upon fecal transplantation (Fig. 5f). The first filtering step in high-TANB77 FMT recipient mice yielded nine taxa (≥ 2-fold enrichment; two-tailed Mann-Whitney U test, P < 0.05; LEfSe LDA ≥ 3) (Supplementary Table 6). The second filtering step through taxa with greater abundance in high-TANB77 donors than in low-TANB77 donors (≥ 2-fold) retained only four taxa. Finally, we filtered out taxa that were not responder-enriched in the ten study cohorts of ICB recipients, which left a CAG-245 genus specific to the TANB77 lineage. CAG-245 was significantly more abundant in high-TANB77 FMT recipient mice (two-tailed Mann-Whitney U test, P < 0.01) (Fig. 5g) and 2.65-fold higher in high-TANB77 donors than in low-TANB77 donors. These results suggest that the improved anti-PD-1 response observed in high-TANB77 FMT mice may be attributed to CAG-245, a genus that belongs to the TANB77 lineage.
Sequence abundance of pilin-like proteins of TANB77 correlates with ICB response
TANB77 is an order-level taxonomic clade that includes largely heterogeneous bacterial species. We could not observe any subclade of TANB77, such as family, genus, or species, that is more predictive for immunotherapy response than TANB77 (Figs. 1a, 2a). Therefore, we hypothesized that gene products exclusively shared among the TANB77 bacterial species may be responsible for the increased responses to ICB treatment. We found a stretch of 10-14 genes that are highly conserved in the TANB77 genomes (Methods). Among the 129 species in the order TANB77, 122 ( ~ 95%) species carry a single copy of the conserved gene cluster in their representative genomes. We selected a genomic region for the gene cluster from HRGM_Genome_2725 (s_CAG-245 sp000435175) based on intactness and concordance and assigned gene identification numbers for the 12 conserved genes, from 927 through 938, based on the order of genomic loci (Fig. 6a, b). To confirm their exclusive presence within the order TANB77, we searched against the entire gut microbial proteins encoded in 5,414 HRGM species genomes. We found that all proteins encoded in the gene cluster except protein 930 have no or only a few homologous proteins outside the TANB77 lineage (Supplementary Fig. 17). Based on evidence from orthologous groups, protein domains, transmembrane topology (Supplementary Fig. 18), and 3D structural homology (Supplementary Fig. 19, Table 1, Methods), we hypothesized that the gene cluster encodes a pilus structure. Specifically, we propose that the first 5-6 genes encode the pilus assembly machinery, while the subsequent genes encode pilin proteins (Fig. 6b).
a Diagram of a pilus gene cluster conserved in TANB77. The tree on the left represents the maximum likelihood phylogenetic tree of 129 representative MAGs of TANB77 species. The center plot illustrates the length and relative location of genes. The arrows represent coding sequences, and arrows with the same color indicate homologous sequences, while gray arrows represent non-conserved genes. The black lines represent contigs. The ‘x’ mark represents the end of the contig. The heatmap on the right provides information on the source of the genome and family. b A representative gene cluster derived from HRGM_Genome_2725, which is highlighted in red (a). The arrows represent coding sequences. The products of genes 927 to 932 are potentially involved in assembly machinery, and products of genes 932 to 938 are pilin-like proteins. Black triangles indicate the predicted promoters. c, d Mouse BMDCs (c) and human monocyte derived dendritic cells (d) were stimulated with indicated agonists targeting TLR2 and TLR4 and pilin 938 at indicated concentration for 24 hours. e After an hour of pre-treatment with TAK-242 inhibitors, BMDC were stimulated for 24 hours with TLR agonists and pilin 938 at the indicated concentration. f BMDC created from WT and TLR4 knockout (KO) bone marrow were stimulated with LPS and pilin 938 to see the TLR4 dependency of pilin 938 activation. g BMDCs were activated with either LPS or pilin 938 before being pulsed with GP33-41, then, these cells were co-cultured with P14 CD8+ T cells. c–g The in vitro experiments were conducted in triplet (n = 3) and the error bars represent mean ± SEM. The data are representative of two independent experiments except for (d) and (f) which were conducted once. The statistical analysis was conducted using One-Way ANOVA Tukey’s multiple comparisons test other than (e) and (f) where student t-test was used for analysis. Source data are provided as a Source Data file. (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
We next investigated the abundance of metagenomic sequencing reads for the seven pilin-like proteins (from protein 932 to 938) derived from gut microbiomes of immunotherapy recipients. The sequence abundance of the pilin-like proteins exhibited a strong positive correlation with the relative abundance of TANB77 order (Pearson r = 0.943, P < 1e-300) (Supplementary Fig. 20). Of interest, it is worthwhile to note that some non-responders, despite having a high abundance of TANB77, had lower-than-expected levels of pilin-like protein sequences. We identified the most abundant TANB77 species in these non-responders to be CAG-492 sp900553225 in the Derosa cohort sample (SRR15373054), and CAG-492 sp000434335 in the Routy cohort sample (ERR2235228) and Derosa cohort sample (SRR15372921), which were among the few exceptional TANB77 species that lack the pilus gene complex (Fig. 6a).
A pilin-like protein of TANB77 can stimulate dendritic cells (DCs) via TLR4 signaling pathway
Based on the correlation between the sequence abundance of TANB77 species’ pilin-like proteins and ICB response, we hypothesized that the pilin-like proteins might function as pathogen-associated molecular patterns (PAMPs) and activate antigen-presenting cells (APCs), specifically DCs, which are known for recognizing microbial molecules and activating the adaptive immune system48. Among the seven pilin-like proteins, we selected the potential minor pilin, 938, as an experimental target and synthesized the protein because minor pilins are known to confer structural and functional uniqueness to the pilus49 (Fig. 6b, Supplementary Fig. 21, Methods). We observed that the treatment of both mouse bone marrow-derived DCs (BMDCs) and human monocyte-derived DCs with pilin 938 increased expression levels of known activation markers such as CD80 and CD86 in a dosage-dependent manner (Fig. 6c, d, Supplementary Fig. 15b, Supplementary Fig. 22a–d). The ability of pilin 938 to activate BMDCs was found to be mediated by TLR4 signaling, as both BMDCs pre-incubated with a TLR4 inhibitor (TAK-242) and TLR4 knockout BMDCs showed a dramatic decrease in response to pilin 938 (Fig. 6e, f, Supplementary Fig. 22e, f). However, blocking the TLR2 signaling did not reduce marker expression when pilin 938 was treated (Supplementary Fig. 22g, h), thus demonstrating that BMDC recognition and activation are induced through the TLR4 signaling pathway. When BMDCs were activated with pilin 938 and co-cultured with T cells, the T cells demonstrated proliferative and functional responses similar to those of BMDCs stimulated with LPS (Fig. 6g). Overall, our in vitro experiments revealed that pilin 938 can activate DCs via TLR4 cell signaling, leading to the proliferative and functional response of T cells.
In vivo administration of pilin 938 enhances the efficacy of anti-PD-1 treatment
Observing the capability of pilin 938 to activate DCs, we next examined whether direct treatment of mice with pilin 938 via intraperitoneal injection could enhance the immunotherapy response, similar to the effects observed with transferring high-TANB77 human fecal samples to tumor-bearing mice. Using the same tumor model and anti-PD-1 treatment schedule as the FMT experiments, pilin 938 was co-administered with anti-PD-1 (Fig. 7a). Compared to the outcome of anti-PD-1 treatment alone, combined treatment of pilin 938 and anti-PD-1 inhibited tumor progression and increased the survival rate (Fig. 7b, c). As in the FMT experiment, peripheral blood mononuclear cell (PBMC) analysis showed that mice co-administered with pilin 938 and anti-PD-1 exhibited an increased frequency of CD8+ T cells expressing Ki67+ PD-1+ compared to the frequency acquired from other treatment methods (Fig. 7d), indicating that the treatment with pilin 938 had a similar effect to the transplantation of high-TANB77 fecal samples.
a Schematic timeline of the mouse experiment relative to the tumor injection date. b, c MC38 tumor growth of mice injected with or without pilin 938 along with aPD-1 treatment (b) and its corresponding survival curves (c). The significance between each group was calculated using the Mantel-Cox test. Each treatment group had seven mice (n = 7) allocated per group. d Frequency of Ki67+ PD-1+ CD8+ T cells from peripheral blood mononuclear cell (PBMC). (aPD1+Pilin 938 = 9, aPD1+PBS = 10, Isotype+Pilin938 = 9, Isotype+PBS = 10) e, f Flow cytometry plot (e) and bar graph (f) showing the cytokine expression frequency of tumor infiltrated CD8+ T cells stimulated ex vivo with PMA. Each treatment group had five mice (n = 5) per group. b–e Data points and error bars represent mean ± SEM. g UMAP plot showing subclusters of T cells integrated from all treatment types. h Proportions of T cell subclusters in T cells for each treatment group. i Ratios of proliferating CD8+ T cells to Treg. j UMAP plots of T cells colored with TCR clonal expansion status. k Cell count of clonal types from proliferating CD8+ T cell subsets. l Shannon index for indicating TCR diversity for T cells. For (b), the significance was calculated using Two-Way ANOVA with Tukey’s multiple comparisons test (all significant comparisons are shown), and for (c), the indicated significance was calculated using pairwise two-tailed log-rank test (all significant comparisons are shown). The significance shown in (d) and (f) were calculated using One-Way ANOVA Tukey’s multiple comparisons test. Except for FACS and sequencing data of tumor infiltrated CD8+ T cells (e–l), which were conducted once, all other data (a–d) are representative of two independent experiments. Source data are provided as a Source Data file. (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Further analyses of CD8+ T cells retrieved from draining lymph nodes and tumors did not show significant phenotypic differences except for the PD-1 level expression in CD8+ T cells from lymph nodes (Supplementary Fig. 23a–c). However, re-stimulating these cells with PMA showed an increased frequency of CD8+ T cells expressing both IFNγ and TNFα (Fig. 7e, f, Supplementary Fig. 23d). Additionally, we conducted single-cell RNA sequencing on isolated lymphocytes from the tumor to investigate changes in the tumor immune microenvironment resulting from pilin 938 and anti-PD-1 treatment (Supplementary Fig. 23e). Focusing on CD8+ T cell subpopulations, we found that proliferating CD8+ T cell subsets were significantly increased in the combination treatment group (Fig. 7g–i). The CD8+ T cell proliferation was further supported by single-cell TCR sequencing data, which demonstrated larger clonal expansion of proliferating CD8+ T cell subsets in the combination treatment group (Fig. 7j, k). This clonal expansion mainly occurred in the combination treatment group (Supplementary Fig. 23f), consequently reducing TCR diversity (Fig. 7l). These data suggest that in vivo administration of the pilin from TANB77 can enhance responsiveness and proliferative ability of CD8+ T cells.
Discussion
In this study, we found that TANB77 is consistently enriched in responders across diverse cohorts, and patients with a higher abundance of TANB77 in their gut microbiome exhibit better responses to ICB therapy. We validated the correlation between the abundance of TANB77 and ICB efficacy through various mouse experiments. Furthermore, we identified a gene cluster for pilus assembly exclusively conserved within the TANB77 order and demonstrated the ability of one of its pilin-like proteins to enhance ICB efficacy. The consistent correlation of higher TANB77 abundance with better ICB efficacy, along with enhanced ICB efficacy from the administration of pilin 938, highlights the clinical impact of our findings in cancer immunotherapy.
Recent advancements in technologies and methodologies have enabled high-resolution profiling of the microbiome. Consequently, the current trend in microbiome research focuses on strain-level associations between the microbiome and host phenotypes50. Contrary to this trend, in this study, we identified an order-level taxon as a biomarker. However, in cases where a common functional feature among taxa of a monophyletic or paraphyletic group is responsible for the host phenotype, such as the case of pilin-like proteins conserved among the TANB77 order, the statistical signal for the association could be weakened at the species or genus level. This could result in a failure to identify the causal molecules. Our observations show that the TANB77 order is the most predictive metagenomic feature for the ICB responders. These results suggest that higher-level taxonomic features from metagenome-wide association studies are also noteworthy and should be considered.
TANB77 and its conserved pilin-like proteins may improve ICB therapy through various routes. First, as biomarkers for patient stratification, both TANB77 and its conserved pilin-like protein can enable the selection of patients with a high probability of responding to ICB therapy. Second, increasing TANB77 abundance in the gut microbiome may enhance ICB response. This can be achieved through the monoculture of a strain belonging to the TANB77 order or by using prebiotics favored by TANB77. Third, TANB77 may be useful in selecting FMT donors to enhance ICB efficacy. Lastly, TANB77 pilin-like proteins may be developed as adjuvants to enhance immunogenicity during ICB treatment. Given the challenges in cultivating TANB77, this represents a valuable therapeutic option.
This study has several limitations and areas for improvement. Firstly, the limited number of donors in the FMT experiments makes it difficult to rule out the possibility that the improved response in FMT recipient mice was influenced by factors other than TANB77. Secondly, interactions with other taxa need further consideration. Given the complexity of the gut microbiome, it is possible that TANB77 affects host physiology in conjunction with other taxa linked to ICB response in previous studies. Future research should explore the interactions between TANB77 and other taxa that showed consistent enrichment, especially the Hungatella genus, and other promising taxa identified in previous studies7,8,9,10,15. Indeed, we observed significant enrichment of some of the previously studied taxa in cohorts where TANB77’s significance was weaker (Supplementary Fig. 24). Finally, the association between TANB77 and the efficacy of ICB observed in this study is based on a meta-analysis that includes data from various cancer types and therapy types. Additionally, we confirmed that in human models, patients with high levels of TANB77 are associated with positive ICB responses in melanoma- and NSCLC-specific cohorts, as well as in cohorts of nivolumab recipients. In mouse models, we also observed that the abundance of TANB77 correlates with better efficacy of anti-PD-1 treatment. However, the association between TANB77 and ICB efficacy may still be specific to cancer type, treatment type, drug type, or host species, and the effect size may vary. As more data on ICB responses accumulate in the future, it will become possible to perform these detailed analyses and better understand the context-specificity of the microbiome in modulating ICB response.
Methods
Whole metagenomic shotgun (WMS) sequencing on non-small cell lung cancer (NSCLC) patients
For YCC_1st cohort, 49 patients with advanced NSCLC who started PD-1 inhibitor (nivolumab) treatment at Yonsei Cancer Center between February 2018 and January 2019, were enrolled (ClinicalTrials.gov identifier: NCT03486119). Clinical data, including patient characteristics, driver gene mutation status, metastatic locations, response to immunotherapy, and adverse events were prospectively collected and analyzed. The patients were treated intravenously with nivolumab at 3.0 mg/kg body weight every two weeks. This study was approved by the Severance Institutional Review Board (IRB No.4-2017-0788), and informed consent was obtained from all participants. We performed a prospective study investigating predictive biomarkers for atezolizumab in NSCLC patients who failed prior platinum-based chemotherapy (ClinicalTrials.gov identifier: NCT 04312308). Fifty-one patients who were able to provide fecal samples were included in YCC_2nd cohort. Patients were enrolled in the clinical trial from April 2020 to April 2022. Atezolizumab 1200 mg was administered once every three weeks, and all clinical data were collected prospectively. This study was approved by the Severance IRB (IRB No. 4-2019-0948), and informed consent was obtained from all participants.
Fecal samples were collected before initial PD-1 treatment. Feces were collected into a Stool Nucleic Acid Collection and Preservation Tube (NORGEN BIOTEK; Cat 45630, 45660) and transferred to Macrogen Inc. for WMS sequencing. For YCC_1st cohort, libraries were prepared according to the TruSeq Nano DNA Prep Reference Guide #15041110. DNA (100 ng) was fragmented using an LE220-plus Focused ultrasonicator (Covaris). The fragmented DNA was end-repaired, and the adapter sequences were ligated to ~350 bp fragments. After eight PCR cycles, libraries were quantified using the Kapa Illumina Library Quantification Kit (Kapa Biosystems; #KK4854). Next, 150 bp × 2 paired-end reads were sequenced on an Illumina HiSeq4000 machine. For YCC_2nd cohort, the sequencing libraries were prepared following the manufacturer’s protocol for the TruSeq Nano DNA High Throughput Library Prep Kit (Illumina). In brief, 100 ng of genomic DNA was sheared using adaptive focused acoustic technology (Covaris), and the fragmented DNA was end-repaired to generate 5’-phosphorylated, blunt-ended dsDNA molecules. After end-repair, DNA was size-selected using a bead-based method. These DNA fragments underwent the addition of a single ‘A’ base and ligation of the TruSeq DNA UD Indexing adapters. The products were then purified and enriched via PCR to create the final DNA library. The libraries were quantified using qPCR in accordance with the qPCR Quantification Protocol Guide (KAPA Library Quantification kits for Illumina Sequencing platforms) and assessed using the Agilent 4200 TapeStation D1000 ScreenTape (Agilent Technology). Subsequently, paired-end (2×150 bp) sequencing was performed by Macrogen Inc. using the Illumina NovaSeq6000 platform.
Assessments of response and clinical data
Patients were evaluated for treatment response using chest posterior-anterior radiography or chest computed tomography. Radiography was performed at every cycle and tomography every two or three cycles during treatment. In addition to regular follow-ups, images were acquired at the physician’s discretion and used to evaluate the clinical response to nivolumab according to the Response Evaluation Criteria in Solid Tumors version 1.117. The objective response rate was defined as the proportion of patients achieving complete response (CR) or partial response (PR), whereas the disease control rate was defined as the proportion of patients with CR, PR, or stable disease (SD). Progression-free survival (PFS) was defined as the time from the start of anti-PD-1 treatment to disease progression or death. Overall survival (OS) was defined as the time from the start of anti-PD-1 treatment to death from any cause. We divided patients into responders (R) and non-responders (NR) to anti-PD-1 treatment. According to RECIST criteria17, R were defined as those with CR, PR, or SD for more than 6 months, while the remaining patients were defined as NR, similar to another study51.
In most cases, tumor PD-L1 expression was determined immunohistochemically using the PD-L1 22C3 pharmDx antibody (Dako North America Inc.) or the Ventana PD-L1 SP263 antibody (Ventana Medical Systems) as companion diagnostic. PD-L1 expression levels in tumor cells were determined by the percentage of stained cells in each section, which was estimated in increments of 5%. Patients in whom at least 1% of the tumor cells were stained for PD-L1 were considered positive.
Collection of public metagenomic sequencing data for immunotherapy recipients
We compiled fecal WMS samples from eight cohorts of immunotherapy recipients5,6,7,8,9,10,15. We only used WMS data from baseline fecal samples collected before the initial immunotherapy treatment. We re-annotated the samples’ responses to standardize the response evaluation criteria with the YCC cohorts. Samples lacking metadata to annotate responses were removed from the analysis. We excluded datasets with insufficient patient numbers29,52 or insufficient metadata to standardize the criteria30.
We also compiled the fecal 16S rRNA amplicon sequencing data of checkpoint inhibitor therapy recipients from a public dataset. Compared to the WMS data, we applied relatively generous criteria to guarantee a sufficient number of cohorts and samples for further meta-analysis. Cohort information and detailed sample selection criteria are described in Supplementary Table 3 and Supplementary Table 4 for the WMS and 16S datasets, respectively.
Pre-processing of WMS data
For sequencing reads from the Illumina sequencing platform, we removed adapter sequences and trimmed low-quality ends with Trimmomatic v0.3953. We found that specific k-mers were abnormally enriched at the end of long reads obtained from the Ion Torrent sequencing machine. Therefore, we trimmed all reads to 280 bp. Next, all WMS reads were aligned to the human reference genome (GRCh38.p7) using Bowtie2 v2.3.554; those with positive alignment were considered human contaminants and were removed from further analysis.
Taxonomic profiling with WMS data and taxonomy conversion
To obtain a taxonomic abundance profile, we used Kraken2 v2.1.118, MetaPhlAn v4.0.621, and mOTUs v.3.0.120. For Kraken2, we used HRGM-based custom database19 and normalized the profiles according to genome length. Kraken2 mapped the reads to leaf features (5414 HRGM species) and internal nodes using the lowest common ancestor mapping algorithms. For leaf features, l, we normalized the abundance by genome length.
For internal nodes, t, with child nodes C, we calculated the abundance using the following equation:
Next, we obtained the relative abundance of taxon t by normalizing the abundance by the read abundance of the root node, sum of d__Bacteria and d__Archaea, and the classification rate of each sample.
For MetaPhlAn4, we performed profiling using the database version “mpa_vOct22_CHOCOPhlAnSGB_202212” with the default parameters. To convert MetaPhlAn4 results to the GTDB taxonomy, we used the “sgb_to_gtdb_profile.py” Python script provided by the tool’s authors. For mOTUs3, taxonomic profiling was conducted using the default parameters. Subsequently, we performed taxonomy conversion according to the mOTUs3 to GTDB taxonomy mapping file provided by the authors. Briefly, if more than 80% of the genomes within a single mOTU matched a consensus GTDB taxonomy, we converted it to that GTDB taxonomy. If no consensus taxonomy was available, we allocated the abundance according to the proportion of genomes possessing a GTDB taxonomy that was shared by at least 15% of the genomes.
Collection and refinement of 16S rRNA sequences of TANB77
The 16S rRNA sequence is relatively difficult to obtain from a metagenome-assembled genome because it contains highly conserved regions55. Therefore, to obtain a more inclusive 16S rRNA and perform 16S rRNA profiling of mouse TANB77, we compiled 16S rRNA sequences from three databases: HRGM, MRGM56, and GTDB32. We downloaded TANB77 16S rRNA sequences from HRGM, used barmap to predict 16S rRNA sequences from TANB77 genomes in MRGM, and collected small-subunit sequences from TANB77 genomes of GTDB r202 genomes. Sequences shorter than 600 bp were excluded.
The predicted 16S rRNA sequences from MAGs may contain false positives owing to contamination. To control the quality of 16S rRNA sequences, we ran SINA v1.2.1157 and removed sequences with a quality score <90. For the remaining 83 sequences, we generated a phylogenetic tree using FastTree v2.1.1058. Next, we removed eight more sequences according to the following criteria: annotation as Archaea or Bacilli instead of Clostridia (SILVA34 classification); large phylogenetic distance from the remaining sequences (average distance > 0.19); and phylogenetic patterns that could not be explained by genome-based classification. Finally, we selected a refined set of 75 TANB77 16S rRNA sequences and annotated them based on the SILVA34, RDP59, and EzBioCloud36 taxonomy with a 95% sequence match similarity threshold. All sequence information pertaining to the generated genomes and taxonomic classifications is available in Supplementary Data 1.
Taxonomic profiling with 16S rRNA sequencing data
We used the QIIME2 v2020.260 pipeline to obtain taxonomic profiles from 16S rRNA sequencing data. The sequences were denoised and merged using QIIME2-embedded DADA261. For profiling with the GTDB32 classification system, we generated custom 16S databases. For human fecal samples, we downloaded the 16S rRNA library from HRGM and replaced existing TANB77 16S rRNA sequences with the 75 re-defined ones. Similarly, for profiling of the mouse gut microbiome, we used the small-subunit sequences of GTDB genomes and replaced TANB77 16S rRNA sequences with 75 re-defined sequences.
Metagenomic data analysis
We performed a multivariate association study using MaAsLin222 v1.8.0 for the YCC cohorts. Clinical data (EGFR_ALK mutation status, PDL1 expression level, antibiotics treatment history, pathology, smoking history, age, and sex) of the therapy recipients were considered as confounders of microbial association. For the multiple cohort dataset, The amount of variance explained by cohort or response was measured by the ‘check.confounder’ function of the SIAMCAT v1.10.126 package. Meta-analysis and batch correction were carried out with the ‘lm_meta’ and ‘adjust_batch’ functions of the MMUPHin v1.4.027 library, respectively. For meta-analysis we used cohort as a random effect, immunotherapy response as the exposure variable, and sex and age as covariates. To identify features that are significantly and consistently enriched across multiple cohorts, we introduced the concept of a consensus score. In each cohort, responder-enriched taxa with a coefficient value greater than or equal to 1e-03 were assigned 3 points if the p-value was less than 0.01, 2 points if the p-value was less than 0.05, and 1 point otherwise. Similarly, non-responder enriched taxa with a coefficient value less than or equal to -1e-03 were assigned -3, -2, or -1 points based on the same p-value cutoffs. It is worth noting that taxa with an absolute coefficient value of 1e-03 or higher represent between 2.3% to 4.0% of all taxa, depending on the cohort. This indicates that the chosen threshold is sufficiently stringent to focus on the most relevant taxa. The taxa with the highest absolute sum of these scores were considered to show significant and consistent results across multiple cohorts. To use MMUPHin and SIAMCAT, we generated a taxonomic profile at every taxonomic level, from domain to species, and used each of them as an input to avoid redundant profiles.
Trait, habitat, and cultured status investigation of TANB77
To analyze the traits of TANB77 species, we compiled the representative genomes of 120 TANB77 species from the Human Reference Gut Microbiome (HRGM) and 9 complete and circular metagenome-assembled genomes (MAGs) of TANB77. To compare the characteristics of TANB77 with neighboring taxa, we also compiled the genomes of the 100 closest species based on the phylogenetic tree of HRGM. We analyzed their traits using Traitar v1.1.2.38. To study the habitats of TANB77, we analyzed 11,008 samples containing TANB77 genomes provided by SPIRE39. The habitat and age group information were analyzed through the microntology data provided by SPIRE. The mOTUs3 profiles of all SPIRE samples were converted to GTDB taxonomy to analyze abundance correlations between TANB77 and other taxa. Only human intestine samples annotated as adult and elderly (18 years and older) in microntology were included in the correlation analysis.
All HRGM 120 TANB77 species were annotated as not isolated based on HRGM metadata, indicating that no TANB77 genome originated from the PATRIC41/IMG42, HBC43, CGR44, and RefSeq40 isolated genomes. A total of 154 TANB77 genomes are listed in the GTDB r202 datasets32, and 116 have an assembly source annotation, such as “derived from environmental sample” or “derived from metagenome”. We further investigated the 115 isolated genomes from the recently expanded cultured bacteria in the human gut microbial biobank (hGMB)62. We annotated the 115 genomes with GTDB-TK63 but failed to find any TANB77 genome among them.
The identification and functional examination of proteins exclusively conserved among TANB77 lineage
To identify proteins exclusively conserved in TANB77, we used the 129 TANB77 MAGs analyzed in the aforementioned TANB77 genome characteristics investigation. We predicted their genes using Prodigal and performed all-versus-all pairwise sequence comparisons using DIAMOND v2.0.15.15364 blastp with ‘--ultra-sensitive’ parameter. Proteins with ≥80% of sequence similarity, ≥80% aligned regions, and an e-value ≤ 1e-20 were considered conserved.
If a conserved protein was present in ≥80% of TANB77 genomes (i.e., 103 of 120 genomes), we used the protein as an anchor and examined conservation of its neighboring protein coding genes on the genome. If more than five coding genes in a row shared the same genomic context, we define them as a conserved gene cluster. We chose representative coding gene sequences from HRGM_Genome_2725 by considering intactness (harboring both start and stop codons with no fragmentation) and concordance with others. We aligned the protein sequences of the representative gene cluster against the entire protein database of 5,414 HRGM species using DIAMOD blastp. We searched for homologous proteins with ≥50% similarity and ≥50% aligned regions. We generated a protein similarity network in which nodes and edges represent proteins and their homologous relationship, respectively.
To find their closest orthologous group, we used eggNOG-mapper65. Protein domain and cellular location were predicted by InterProScan66 and DeepTMHMM67, respectively. The 3D structure of the proteins was predicted by AlphaFold2 v2.2.368 using the monomer_ptm model. Proteins with homologous proteins were searched with the FoldSeek69 webserver, using all available databases70. Promoter of the pilus genes are predicted by the CNNProm71.
We observed sequence and structural homology between the protein 927 and a pilus assembly protein CpaB72. Protein 928 and 929 showed structural homology with other proteins for pilus assembly, TadZ and CpaE72,73, respectively. In addition, protein 930 showed both sequence and structural homologies with ATPase for pilus assembly, TadA and CpaF72. Each half of the protein 931 has structural homology with TadB and TadC72,73, which are known to form a platform of pilus as a dimer. About 25% N-terminus of the protein 938 has structural homology with known minor pilin, TadE, and pilV72. It was difficult to infer functions of the proteins from 932 to 937 using sequence or structural homology, partly because of their short sequence length. Nevertheless, we annotated them as pilin-like proteins because of the following reasons: (1) These proteins are encoded in the same gene cluster together with the pilus assembly machinery proteins, from 927 to 931. Since the TANB77 lineage belongs to the Clostridia, which consists mostly of gram-positive bacteria, it is worth noting that the conserved proteins from 927 to 931 appear to comprise most of the machinery part of Tad pili formation74; (2) In contrast to the proteins from 927 to 932, each of the protein 933, 934, 935, and 937 have their own promoter sequences, suggesting that they are subject to independent regulatory programs that control their expression (Fig. 5b); (3) The size of these proteins ranges from 143 to 236 amino acids, and transmembrane topology consists of a short cytoplasmic region followed by a transmembrane helix and a longer folded non-cytoplasmic region, which is similar to that of the known pilin proteins49.
Estimating sequence abundance of pilin-like proteins
To ensure identification of homologous proteins within the TANB77 lineage, only pilin-like proteins between positions 932 to 938 were selected for analysis. A total of 776 pilin-like protein sequences were identified from 122 genomes of the TANB77 lineage. Pre-processed metagenomic reads from 660 samples were aligned to the 776 pilin-like protein sequences using DIAMOND blastx, and only reads with ≥80% similarity and ≥80% aligned region were counted. For paired-end sequencing samples, pairs were considered aligned if at least one of the two reads aligned to the proteins. The relative abundance of each pilin-like protein sequence was determined by dividing the number of aligned reads by the total number of sequencing reads in the sample.
Mice
Six-week-old C57BL/6 female mice were purchased from Samtako Bio Inc. and ORIENT Bio Inc. P14 TCR transgenic mice expressing a TCR specific for the LCMV GP33-41 peptide were provided by Dr. Rafi Ahmed (Emory University School of Medicine, Atlanta, GA, USA). The TLR4 knockout mice bone marrows for the bone marrow-derived dendritic cell (BMDC) experiment were provided by Dr. Jong-Hwan Park (Chonnam National University, College of Veterinary Medicine, Republic of Korea). The mice were allowed to acclimatize and maintained in a specific pathogen-free facility under a 12 h light/dark cycle with unrestricted access to water and food (Cargill, Purina Lab Rodent Chow, 95135) at Yonsei University. The experiments were conducted after being reviewed and approved by the Institutional Animal Care and Use Committee of Yonsei University(permit number IACUC-A-202310-1735-01).
Cell culture and reagents
C57BL/6-derived colon carcinoma MC38 cells were cultured at 37 °C with 5% CO2 in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum (GE Healthcare UK Ltd.) and 1% penicillin/streptomycin (Gibco Laboratories).
Tumor induction and treatment
C57BL/6 mice were inoculated subcutaneously with 5× 105 MC38 cells on the flank. Once the tumors reached 30 to 50 mm3 in size, the mice were treated five times at 3-day intervals with anti-PD-1 mAb (200 μg/mouse; clone RMP1-14) or rat IgG2a isotype control (200 μg/mouse; clone 2A3) (both BioXcell). Tumor volume was routinely monitored using a caliper and was calculated according to the formula: 0.5 × length × width2.
Evaluating tumor growth rate and its correlation with microbiota abundance
We measured tumor growth fold-changes for intervals between PD-1 treatment dates (D + 8 to D + 11, D + 11 to D + 14, D + 14 to D + 17, and D + 17 to D + 20). For each anti-PD-1-treated mouse, we calculated the normalized tumor growth rate by dividing the growth fold-change by the average tumor growth fold-change of isotype-treated controls.
We profiled the murine gut microbiome using 16S rRNA sequencing data from baseline fecal samples. If taxa with inclusive relations shared the same profile across all mice, we only considered the profile of the more specific taxon. For instance, because the Pasteurellaceae family displayed an abundance profile identical to that of the Conservatibacter genus (belonging to the Pasteurellaceae family) across all mice, we used only the profiles of Conservatibacter to avoid redundant results. The Spearman correlation between microbial abundance and normalized tumor growth was calculated at each time interval in anti-PD-1-treated mice.
Collection and storage of mice fecal samples
After collecting fecal samples from individual mice, the samples were minced, mixed with PBS, and vortexed until they were fully homogenized. To exclude larger particles, homogenized samples were filtered through a 40-μm cell strainer (Falcon) and supplemented with 10% glycerol (Sigma). The amount of PBS and glycerol was calculated based on the weight of the collected fecal samples (30 mg/mL). Finalized samples were stored at −80 °C before sequencing analysis.
16S rRNA sequencing of mouse fecal samples
16S rRNA sequencing of murine fecal samples was conducted by Macrogen Inc. Sequencing libraries were prepared according to the 16S Metagenomic Sequencing Library Preparation Part # 15044223 Rev. B. Briefly, DNA was quantified using PicoGreen and quality was measured using a Nanodrop. The 16S rRNA sequences were PCR-amplified using V3-V4 primers:
16S Amplicon PCR Forward Primer:
5’ TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG 3’
16S Amplicon PCR Reverse Primer:
5’ GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC 3’
A limited-cycle amplification step was performed to add Illumina sequencing adapters and demultiplexing indices. Library size was verified on a TapeStation DNA screentape D1000 (Agilent). Sequencing was performed on an Illumina MiSeq platform.
Antibiotics treatment and FMT
An antibiotic solution was prepared by adding ampicillin (1 mg/mL), colistin (1 mg/mL), and streptomycin (5 mg/mL) (Goldbio) into sterile mice drinking water. The solution (200 μL) was administered daily for three days by oral gavage. FMT was performed by thawing human fecal samples and transferring 200 μL of the suspension by oral gavage to mice that had recently received antibiotic treatment.
Preparation of pilin-like protein 938
A codon-optimized 938 complementary DNA (cDNA) for Escherichia coli was synthesized and cloned into the pBT7-N-HIS vector (purchased from Bioneer, Inc., Republic of Korea). The recombinant plasmid was transformed into E. coli BL21 DE3 (NEB, C2527), which was grown in Lysogeny broth (LB) medium (BD DIFCO, 244620) supplemented with ampicillin at 37 °C until the culture reached an optical density at 600 nm of 0.5. Protein expression was induced with 0.5 M Isopropyl β-D-1-thiogalactopyranoside (IPTG, Gold Bio, I2481C5), and the cells were further incubated overnight (16 hours) at 37 °C. The cells were harvested by centrifugation (4000 g for 20 min), and the cell pellet was resuspended in lysis buffer consisting of 20 mM sodium phosphate (pH 8.0), 500 mM NaCl, and 1x protease inhibitor (ApexBio, K1008). The cells were sonicated on ice for 10 minutes (Sonics, VCX130) and centrifuged (15,000 g for 10 min). The cell lysate was incubated with Ni-NTA (nitrilotriacetic acid) agarose resin (QIAGEN, 30230) in a closed centrifuge column (Thermo Scientific, 89897) for 2 hours at 4 °C on a rotator mixer. The protein-bound NTA agarose resin was washed with a volume of 30 mM imidazole buffer equivalent to twice the resin bed volume. The proteins bound to the resin were detached and eluted with 250 mM imidazole buffer. The elution fraction containing protein 938 with a 6x His-tag was transferred to a spin column (Sartorius, VS04T31), and the protein was recovered and enriched following the manufacturer’s instructions.
Endotoxin was removed using a Triton X-114 based LPS (Lipopolysaccharides) extraction method as described here75. Briefly, TX-114 was added to the protein solution up to 1% v/v. The solution was incubated at 4 °C for 30 min on a rotator mixer. The sample was transferred to a water bath at 37 °C and incubated for 10 min, after which it was immediately centrifuged at 20,000 g for 20 min at 37 °C. This resulted in two separate phases: the upper part (aqueous phase containing 938 protein) and the lower part (detergent phase containing LPS). The upper part was carefully collected for further TX-114 removal. Bio-Beads SM-2 resin (Bio-Rad, 152-3920), which has a high affinity for Triton, was added to the solvent containing protein 938, and the mixture was incubated at 4 °C for 2 hours on a rotator mixer. The solution was then centrifuged using a 0.22 μm filter column (Costar, 8160) at 10,000 g at 37 °C for 4 min to remove the SM-2 resin. The collected solution containing protein 938 was further purified using FPLC (Cytiva, Akta go) with a size exclusion chromatography column (Cytiva, 28- 9909- 44), following the manufacturer’s manual. The purified protein 938 solution was enriched, and the buffer was exchanged with PBS using a spin column (Sartorius, VS04T31). The concentration and purity of the final protein 938 solution were evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and western blot with an anti-His tag antibody (Santa Cruz, sc-8036) (Supplementary Fig. 20).
Mouse BMDC and Human Monocyte-derived DC generation and pilin 938 treatment
To culture BMDCs, 7.5 × 105 bone marrow cells were seeded in non-tissue culture-treated 6-well plates in 2 mL of complete medium supplemented with GM-CSF (20 ng/mL, Creagen). On the third day of culture, 2 mL of fresh medium was added with the same concentration of GM-CSF. After seven days of culture, non-adherent cells present in the supernatant were collected gently. The cells were transferred to 96-well round-bottom culture plates and pre-treated with either DMSO or TLR inhibitors for one hour. A TLR4 inhibitor, TAK-242 or Resatorvid (Selleckchem, S7455), was used at a concentration of 1 μM, while the TLR2 inhibitor C29 (Selleckchem, S6597) was used at a concentration of 10 μM. TLR agonizts were treated with a concentration of 50 ng/mL and pilin 938 were treated as indicated in the Fig. 5 for 24 hours before flow cytometry analysis. For human monocyte-derived DCs(hMo-DCs), CD14+ cells were isolated from human PBMC following the protocol given from manufacturer. Then the cells were seeded in non-tissue culture-treated 24-well plates in 1 mL of Mo-DC differentiation medium (130-094-812, Miltenyi Biotec). On the second day of culture, 1 mL of fresh Mo-DC differentiation medium was added. After four days of culture, cells were collected and transferred to 96-well round-bottom culture plates for stimulation with LPS or pilin 938 for 24 hours before FACS analysis.
In vitro CD8+ T cell proliferation assay
CD8+ T cells were isolated from splenocytes of P14 mice using magnetic-activated cell sorting (MACS) CD8+ T cell isolation kit (Miltenyi Biotec, 130-104-075). Purified T cells were labeled with Cell Trace Violet dye (ThermoFisher, C34557) following the protocol provided by the manufacturer and cultured with activated BMDCs at a ratio of 5:1 at 37 °C for 60 hours. BMDCs were pulsed with GP33-41 peptide with 0.2ugmL−1 for 30 minutes and washed twice before starting the co-culture with CD8+ T cells. In order to measure cytokine production, the cells were supplemented with 0.2ugmL−1 of GP33-41 peptide along with GolgiPlug/GolgiStop 5 hours before analysis.
Cell isolation and MACS sorting for single cell-RNA sequencing
To retrieve mouse peripheral blood mononuclear cells (PBMCs), three to four drops of blood were collected by the eye-bleeding method. Histopaque-1077 (Sigma) was placed on blood samples prior to centrifugation. PBMCs were resuspended in PBS containing 2% fetal bovine serum and plated in each well of a 96-well plate. Draining lymph node, in this case, inguinal lymph node, were collected and meshed through a 40μm strainer to retrieve the cells. In ways to isolate lymphocytes from tumors, tumor tissue was digested with a type IV collagenase and DNase I mixture for 40 min at 37 °C shaking incubator. Lymphocytes were further isolated from the digested samples after removing the red blood cells by ACK lysis buffer (Gibco Laboratories). In order to purify the lymphocyte for single-cell RNA sequencing, CD45.2 MACS sorting was conducted following protocol provided by manufacturer. In brief, cells were first stained with CD45.2-FITC antibody which were later separated using a bead for anti-FITC (Miltenyi).
Staining and antibodies used for flow cytometry analysis
The cells were stained with antibodies against extracellular markers for 20 minutes at 4 °C, along with the LIVE/DEAD kit (Invitrogen) to remove dead cells through analysis. Intracellular staining was conducted using the Foxp3 Fixation/Permeabilization Kit (ThermoFisher) according to the manufacturer’s protocol. And for cytokines expression, intracellular cytokine staining was performed after cells were incubated with PMA (50 ng/ml) and Ionomycin (500 ng/mL) in the presence of GolgiPlug/GolgiStop at 37 °C for 5 hours. Surface staining was conducted first before using a BD Ctyofix/Cytoperm fixation/permeabilization kit (BD Biosciences) and followed the manufacturer’s instructions.
Moreover, for BMDC staining process, the cells were washed with PBS containing 2% fetal bovine serum and incubated with an anti-mouse CD16/CD32 (93; ThermoFisher) blocker for 20 minutes at 4 °C before proceeding to the staining process. All samples were acquired on either CytoFLEX or CytoFLEX LX (Beckman Coulter) and analyzed using FlowJo software. Antibodies against the following markers were used for mice samples: CD4 (RM4-5), CD8 (53-6.7), CD25 (PC61), CD44 (IM7), CD69 (H1.2F3), Ly108 (330-AJ), PD-1 (29 F.1A12), Tim3 (RMT3-23), Ki67 (16A8), PD-L1 (10 F.9G2), CD11c (N418), I-A/I-E (M5/114.15.2), and CD86 (GL-1) (all from Biolegend, clones indicated in parenthesis), as well as CD11b (M1/70), CD40 (3/23), CD62L (MEL-14), CD80 (16-10A1), CD90.2 (53-2.1), TNFa (MP6-XT22), and IFNγ (XMG1.2) from BD Biosciences and Foxp3 (FJK-16s) CD45.1 (A20), and CD101 (Moushi101) were from Invitrogen eBiosciences. For staining human Mo-DC, PD-L1 (29E.2A3), HLA-DR (L243), CD11c (S-HCL-3), CD86 (IT2.2), CD11b (ICRF44), and CD80 (2D10), were from Biolegend, CD14 (MφP9) from BD Biosciences were used.
Processing of single-cell RNA sequencing data
We utilized CellRanger70 (v8.0.0) count with CellRanger reference “GRCm39-2024-A” for quantifying raw expression counts for each sample. To remove possible ambient RNA, we used CellBender76 (v0.3.0) with “raw_feature_bc_matrix.h5” files from CellRanger outputs. As the initial raw count matrix, we used filtered count matrices from CellBender outputs and selected cells that are also present in filtered count matrices from CellRanger outputs. We used R-base (v4.3.1) and Seurat (v5.0.1) for all single-cell analysis unless otherwise stated. For initial quality control, we filtered cells with fewer than 200 genes, more than 7000 genes, more than 10% mitochondrial reads, or more than 40000 reads. We additionally filtered possible doublets with 5% expected doublet counts for each sample with scDblFinder77 (v1.14.0).
For initial read count processing and clustering, we normalized and scaled all genes with ‘NormalizeData’ and ‘ScaleData’ functions from Seurat with default parameters. We selected 3000 highly variable genes with ‘FindVariableFeatures’ function with vst method and performed linear dimensional reduction with PCA methods with ‘RunPCA’ function on the scaled data with the selected variable genes. We used 30 PC dimensions for constructing the k-nearest neighbor (kNN) graph with ‘FindNeighbors’ and for UMAP visualization with ‘RunUMAP’ function. For clustering, we used 0.5 resolution and calculated kNN graph for ‘FindClusters’ function. After initial clustering, we filtered clusters with cells with no PTPRC expression for removing non-immune cells. We re-processed and clustered cells after non-immune cell filtering with the same method above. We annotated cell clusters as “Macrophage”, “Neutrophil”, “Monocyte”, “DC”, “T”, and “NK” with their canonical markers and existence of cell matched TCR sequences.
We selected T cells for subclustering analysis. We processed and clustered the cells with the same parameters above except for choosing 2000 variable genes instead of 3000. After initial T cell clustering, we removed one cluster with NK cells (annotated as having to TCR sequences) and another cluster with non-T cell markers. We reperformed clustering after the initial filtering. We annotated cell clusters as “Naïve T”, “Proliferating CD8+ T”, “Exhausted CD8+ T”, and “Treg.”
Processing and analysis of single-cell TCR sequencing data
We utilized CellRanger78 (v8.0.0) vdj with CellRanger VDJ reference “GRCm38-alts-ensembl-7.0.0” for quantifying TCR sequences for each sample. We used filtered contig files from CellRanger VDJ outputs. For analysis of TCR sequences, we used scRepertoire79 (v2.0.0) and Seurat80 (v5.0.1). We combined TCR sequences for the processed cells with RNA sequencing profiles by matching cell barcodes from both single-cell RNA and TCR sequencing with combineTCR and combineExpression functions. For calling the TCR clones, we used combination of VDJC genes and CDR3 nucleotides (“strict” option from scRepertoire). We annotated T cells with one clonal frequency as having unique TCR clones and T cells with more than one clonal frequency as having expanded TCR clones.
Cell composition and ratio analysis
For cell composition analysis with immune cells and T cells, we calculated proportions of corresponding cell subtypes (clusters) for each sample by dividing cell counts for each cell subtype by total immune cells or total T cells from each sample. For cell subtype ratio, we divided one cell subtype with another cell subtype for each sample. All plots were visualized with ggplot2 (v3.4.3) and patchwork (v1.1.3).
Statistics and Reproducibility
The differences in microbial abundance between high- and low-TANB77 groups were analyzed using the MaAsLin2 test and the Mann-Whitney U test, while meta-analyses were conducted using MMUPHin. Progression-free survival or overall survival in patients was assessed using the log-rank test and Cox regression analysis. If survival data were provided categorically, they were analyzed using Fisher’s exact test. Survival data in mice were analyzed with the log-rank test, and changes in tumor size over time were compared using two-way ANOVA with Tukey’s multiple comparison post hoc test. Differences in biomarker measurements were tested using Student’s t-test. For mouse experiments involving different vendors, two independent experiments were conducted, yielding similar results. However, in the FMT experiments, replicating results was challenging due to the limited availability of identical human fecal samples. For the BMDC experiments, two independent experiments were performed with a sample size of n = 3 for each experiment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The sequencing data generated in this study have been deposited in the Sequence Read Archive under accession codes PRJNA678426 (fecal whole metagenomic shotgun sequencing (WMS) of YCC_1st cohort), PRJNA1130699 (fecal WMS of YCC_2nd cohort) and PRJNA771940 (murine gut microbiome 16S rRNA and fecal WMS of FMT donors). The taxonomic profile and raw data are Provided in the Supplementary Data and Zenodo [https://doi.org/10.5281/zenodo.12786069]. Source data are provided with this paper.
References
Pardoll, D. M. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 12, 252–264 (2012).
Waldman, A. D., Fritz, J. M. & Lenardo, M. J. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol 20, 651–668 (2020).
Robert, C. et al. Pembrolizumab versus Ipilimumab in Advanced Melanoma. N Engl J Med 372, 2521–2532 (2015).
Garon, E. B. et al. Pembrolizumab for the treatment of non–small-cell lung cancer. N Engl J Med 372, 2018–2028 (2015).
Frankel, A. E. et al. Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia 19, 848–855 (2017).
Peters, B. A. et al. Relating the gut metagenome and metatranscriptome to immunotherapy responses in melanoma patients. Genome Med 11, 61 (2019).
Gopalakrishnan, V. et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018).
Routy, B. et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97 (2018).
Matson, V. et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108 (2018).
Derosa, L. et al. Intestinal Akkermansia muciniphila predicts clinical response to PD-1 blockade in patients with advanced non-small-cell lung cancer. Nat Med 28, 315–324 (2022).
Davar, D. et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 371, 595–602 (2021).
Baruch, E. N. et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 371, 602–609 (2021).
Routy, B. et al. Fecal microbiota transplantation plus anti-PD-1 immunotherapy in advanced melanoma: a phase I trial. Nat Med 29, 2121–2132 (2023).
Bjork, J. R. et al. Longitudinal gut microbiome changes in immune checkpoint blockade-treated advanced melanoma. Nat Med 30, 785–796 (2024).
Lee, K. A. et al. Cross-cohort gut microbiome associations with immune checkpoint inhibitor response in advanced melanoma. Nat Med 28, 535–544 (2022).
Parks, D. H. et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36, 996–1004 (2018).
Eisenhauer, E. A. et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 45, 228–247 (2009).
Wood, D. E., Lu, J. & Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol 20, 257 (2019).
Kim, C. Y. et al. Human reference gut microbiome catalog including newly assembled genomes from under-represented Asian metagenomes. Genome Med 13, 134 (2021).
Ruscheweyh, H. J. et al. Cultivation-independent genomes greatly expand taxonomic-profiling capabilities of mOTUs across various environments. Microbiome 10, 212 (2022).
Blanco-Miguez, A. et al. Extending and improving metagenomic taxonomic profiling with uncharacterized species using MetaPhlAn 4. Nat Biotechnol 41, 1633–1644 (2023).
Mallick, H. et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput Biol 17, e1009442 (2021).
Miura, Y. & Sunaga, N. Role of immunotherapy for oncogene-driven non-small cell lung cancer. Cancers (Basel) 10, 245 (2018).
Norum, J. & Nieder, C. Tobacco smoking and cessation and PD-L1 inhibitors in non-small cell lung cancer (NSCLC): a review of the literature. ESMO Open 3, e000406 (2018).
Patel, S. P. & Kurzrock, R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther 14, 847–856 (2015).
Wirbel, J. et al. Microbiome meta-analysis and cross-disease comparison enabled by the SIAMCAT machine learning toolbox. Genome Biol 22, 93 (2021).
Ma, S. et al. Population structure discovery in meta-analyzed microbial communities and inflammatory bowel disease using MMUPHin. Genome Biol 23, 208 (2022).
Chaput, N. et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol 28, 1368–1379 (2017).
Lee, S. H. et al. Bifidobacterium bifidum strains synergize with immune checkpoint inhibitors to reduce tumour burden in mice. Nat Microbiol 6, 277–288 (2021).
Peng, Z. et al. The gut microbiome is associated with clinical response to anti-pd-1/pd-l1 immunotherapy in gastrointestinal cancer. Cancer Immunol Res 8, 1251–1261 (2020).
Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol 12, R60 (2011).
Parks, D. H. et al. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat Biotechnol 38, 1079–1086 (2020).
Federhen, S. The NCBI taxonomy database. Nucleic Acids Res 40, D136–D143 (2012).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41, D590–D596 (2013).
Cole, J. R. et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42, D633–D642 (2014).
Yoon, S. H. et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67, 1613–1617 (2017).
Kim, C. Y., Ma, J. & Lee, I. HiFi metagenomic sequencing enables assembly of accurate and complete genomes from human gut microbiota. Nat Commun 13, 6367 (2022).
Weimann, A. et al. From Genomes to Phenotypes: Traitar, the Microbial Trait Analyzer. mSystems 1, e00101–e00116 (2016).
Schmidt, T. S. B. et al. SPIRE: a Searchable, Planetary-scale mIcrobiome REsource. Nucleic Acids Res 52, D777–D783 (2024).
Pruitt, K. D., Tatusova, T., Brown, G. R. & Maglott, D. R. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res 40, D130–D135 (2012).
Gillespie, J. J. et al. PATRIC: the comprehensive bacterial bioinformatics resource with a focus on human pathogenic species. Infect Immun 79, 4286–4298 (2011).
Chen, I. A. et al. The IMG/M data management and analysis system v.6.0: new tools and advanced capabilities. Nucleic Acids Res 49, D751–D763 (2021).
Forster, S. C. et al. A human gut bacterial genome and culture collection for improved metagenomic analyses. Nat Biotechnol 37, 186–192 (2019).
Zou, Y. et al. 1520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat Biotechnol 37, 179–185 (2019).
Crost, E. H., Coletto, E., Bell, A. & Juge, N. Ruminococcus gnavus: friend or foe for human health. FEMS Microbiol. Rev. 47, fuad014 (2023).
Sivan, A. et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089 (2015).
Kamphorst, A. O. et al. Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1-targeted therapy in lung cancer patients. Proc Natl Acad Sci USA 114, 4993–4998 (2017).
Hayase, E. & Jenq, R. R. Role of the intestinal microbiome and microbial-derived metabolites in immune checkpoint blockade immunotherapy of cancer. Genome Med 13, 107 (2021).
Giltner, C. L., Nguyen, Y. & Burrows, L. L. Type IV pilin proteins: versatile molecular modules. Microbiol Mol Biol Rev 76, 740–772 (2012).
Podlesny, D. et al. Metagenomic strain detection with SameStr: identification of a persisting core gut microbiota transferable by fecal transplantation. Microbiome 10, 53 (2022).
Rizvi, N. A. et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015).
Zheng, Y. et al. Gut microbiome affects the response to anti-PD-1 immunotherapy in patients with hepatocellular carcinoma. J Immunother Cancer 7, 193 (2019).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359 (2012).
Miller, C. S., Baker, B. J., Thomas, B. C., Singer, S. W. & Banfield, J. F. EMIRGE: reconstruction of full-length ribosomal genes from microbial community short read sequencing data. Genome Biol 12, R44 (2011).
Kim, N. et al. MRGM: an enhanced catalog of mouse gut microbial genomes substantially broadening taxonomic and functional landscapes. Gut Microbes 16, 2393791 (2024).
Pruesse, E., Peplies, J. & Glockner, F. O. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–1829 (2012).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490 (2010).
Wang, Q., Garrity, G. M., Tiedje, J. M. & Cole, J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73, 5261–5267 (2007).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336 (2010).
Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13, 581–583 (2016).
Xie, F. et al. An integrated gene catalog and over 10,000 metagenome-assembled genomes from the gastrointestinal microbiome of ruminants. Microbiome 9, 137 (2021).
Chaumeil, P. A., Mussig, A. J., Hugenholtz, P. & Parks, D. H. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36, 1925–1927 (2019).
Buchfink, B., Reuter, K. & Drost, H. G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods 18, 366–368 (2021).
Huerta-Cepas, J. et al. Fast genome-wide functional annotation through orthology assignment by eggnog-mapper. Mol Biol Evol 34, 2115–2122 (2017).
Paysan-Lafosse, T. et al. InterPro in 2022. Nucleic Acids Res 51, D418–D427 (2023).
Hallgren, J. et al. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. https://doi.org/10.1101/2022.04.08.487609 (2022).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
van Kempen, M. et al. Fast and accurate protein structure search with Foldseek. Nat Biotechnol 42, 243–246 (2024).
Varadi, M. et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022).
Umarov, R. K. & Solovyev, V. V. Recognition of prokaryotic and eukaryotic promoters using convolutional deep learning neural networks. PLoS One 12, e0171410 (2017).
Singh, P. K., Little, J. & Donnenberg, M. S. Landmark Discoveries and Recent Advances in Type IV Pilus Research. Microbiol Mol Biol Rev 86, e0007622 (2022).
Cai, L. et al. Tad pilus-mediated twitching motility is essential for DNA uptake and survival of Liberibacters. PLoS One 16, e0258583 (2021).
Clock, S. A., Planet, P. J., Perez, B. A. & Figurski, D. H. Outer membrane components of the Tad (tight adherence) secreton of Aggregatibacter actinomycetemcomitans. J Bacteriol 190, 980–990 (2008).
Magalhaes, P. O. et al. Methods of endotoxin removal from biological preparations: a review. J Pharm Pharm Sci 10, 388–404 (2007).
Fleming, S. J. et al. Unsupervised removal of systematic background noise from droplet-based single-cell experiments using CellBender. Nat Methods 20, 1323–1335 (2023).
Germain, P. L., Lun, A., Garcia Meixide, C., Macnair, W. & Robinson, M. D. Doublet identification in single-cell sequencing data using scDblFinder. F1000Res 10, 979 (2021).
Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun 8, 14049 (2017).
Borcherding, N., Bormann, N. L. & Kraus, G. scRepertoire: An R-based toolkit for single-cell immune receptor analysis. F1000Res 9, 47 (2020).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat Biotechnol 42, 293–304 (2024).
Acknowledgements
This research was supported by the National Research Foundation funded by the Ministry of Science and ICT (2018R1A5A2025079, 2022R1A2C1092062, and 2022M3A9F3016364 to I.L.; RS-2024-00392705 to S-J.H.; 2021R1A2C2094629 to H.R.K.; RS-2023-00278521 to B.C.K.). This work was supported by the Technology Innovation Program (20022947) funded by the Ministry of Trade Industry & Energy (MOTIE), the Brain Korea 21 (BK21) FOUR Program, the Korea Drug Development Fund (RS-2021-DD121415), and the Korean Health Technology R&D Project (HV23C0090). This work was also supported by Samsung Science and Technology Foundation under project number SSTF-BA2001-05. Investigator initiated clinical trial were kindly supported by Ono and Roche. The funder had no role in the study design, data collection, analyses, and interpretation of data, in the writing of the report, or in the decision to submit the article for publication. This work was partially supported by the Yonsei Fellow Program, funded by Lee Youn Jae (HRK).
Author information
Authors and Affiliations
Contributions
C.Y.K., H.R.K., S-J.H. and I.L. conceived and designed the study. C.Y.K. performed the bioinformatics analysis on metagenomic data and formulated the study hypothesis. B-C.A., H.I.Y., M.H.H., and S.Y.P. collected and annotated the clinical data. D.J.P. performed the experimental validation. S.B. performed bioinformatics analysis on single-cell data. L.T.N. and S.H.H. prepared the protein 938. H.S.S. performed the pathological examination. N.K. assisted in the collection of 16S rRNA sequences of the murine gut microbiome. C.S., S.M.R., D.P., and A.O. contributed to bioinformatics analysis and taxonomy conversion. D.Y. provided fecal samples and related data for FMT experiments. M.H. contributed to sample collection. P.B. provided scientific advice and computing resources on bioinformatic analysis. B.C.K, H.R.K., S-J.H., and I.L. supervised the protein preparation, clinical analysis, experimental analysis, and bioinformatics analysis, respectively. All authors contributed to the writing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
I.L., S-J.H., H.R.K., C.Y.K., and B.C.A. are inventors of the patent (PCT/KR2021/008284) relevant to this work (Patent Applicant(s): Industry-academic cooperation foundation, Yonsei University; Inventors: Insuk Lee, Sang Jun Ha, Chan Yeong Kim, Hye Ryun Kim, Beung Chul Ahn). The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Bertrand Routy, and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kim, C.Y., Park, D.J., Ahn, B.C. et al. A conserved pilin from uncultured gut bacterial clade TANB77 enhances cancer immunotherapy. Nat Commun 15, 10726 (2024). https://doi.org/10.1038/s41467-024-55388-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-55388-3
This article is cited by
-
Drug resistance in cancer: molecular mechanisms and emerging treatment strategies
Molecular Biomedicine (2025)
-
The gut microbiome as a target in cancer immunotherapy: opportunities and challenges for drug development
Nature Reviews Drug Discovery (2025)