Abstract
Intestinal epithelial cells are segregated into proliferative crypts and differentiated regions. This organization relies on specific signals, including Wnt3a, which regulates cell proliferation within crypts, and Eph/Ephrin, which dictates cell positioning along the crypt-villus axis. However, studying how the spatial distributions of these signals influences crypt-villus organization is challenging both in vitro and in vivo. Here we show that micropatterns of Wnt3a can govern the size, shape and long-range organization of crypts in vitro. By adjusting the spacing between Wnt3a ligand patterns at the microscale over large surfaces, we override endogenous Wnt3a to precisely control the distribution and long-range order of crypt-like regions in primary epithelial monolayers. Additionally, an agent-based model integrating Wnt3a/BMP feedback and Eph/Ephrin repulsion effectively replicates experimental tissue compartmentalization, crypt size, shape, and organization. This combined experimental and computational approach offers a framework to study how signaling pathways help organize intestinal epithelial tissue.
Similar content being viewed by others
Introduction
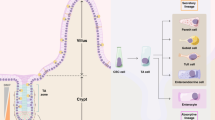
The small intestinal epithelium is organized in specialized structures: evaginations called villi, populated by terminally differentiated cells, and invaginations named crypts, populated by proliferative cells1. Adult intestinal stem cells (ISCs) reside at the bottom of these crypts2, interspersed between secretory Paneth cells3. In there, ISCs undergo self-renewal, generating transit-amplifying (TA) cells. These TA cells differentiate and migrate up towards the villus tips, where they are extruded ensuring tissue renewal2. Changes in this compartmentalization are associated to pathologies such as inflammatory bowel disease and cancer4,5. Maintaining this cell compartmentalization amid dynamic process of cell proliferation, differentiation, and extrusion relies on a set of signaling pathways along the crypt-villus axis, including the Wnt and Bone Morphogenetic Protein (BMP) pathways6 and the repulsive EphB2/EphrinB1 receptor/ligand interaction7,8. Wnt and Eph decrease in a gradient fashion from the bottom to the top of the crypts. Conversely, BMP and Ephrin increase towards the villi7,9,10,11.
Understanding these signaling pathways is crucial to comprehend intestinal epithelial organization, and often involves genetic approaches conducted in vivo. However, these approaches have some limitations. For example, while Paneth cells, located adjacent to Lgr5+ ISCs in the crypts, are a major Wnt source within the intestinal epithelium, it is known that other cell types such as intestinal fibroblasts also secrete Wnt3,12,13. This indicates that Paneth cells are not the only Wnt source in the stem cell niche11,14,15. Therefore, with redundant signaling sources11,14, determining the specific role of each source in regulating epithelial compartmentalization becomes a challenge. Hence, there is a need for in vitro systems that allow for the controlled supply of individual signaling factors and the precise control over their spatial distribution. These systems would be instrumental in gaining insights into how the various signal sources collectively influence the organization of crypt-villus structures.
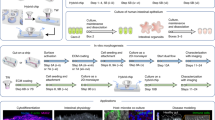
In vitro, single cells derived from intestinal organoids form epithelial monolayers that spontaneously self-organize in crypt- and villus-like domains that retrieve in vivo compartmentalization6,16,17. This simplified system offers a simple and robust approach to gain valuable insights into tissue organization and properties. The self-organization and the maintenance of its homeostasis in these compartmentalized intestinal monolayers are governed by a feedback loop involving intrinsic Wnt and BMP signals within the epithelium6. However, understanding how multiple sources of signals impact on the crypt-villus organization requires in vitro systems where individual signaling factors can be externally supplied, allowing the precise control over their spatial distribution. Several in vitro models have been developed to replicate the architecture and signaling gradients of the intestinal epithelium, but controlling the spatial distribution of specific molecules remains a challenge18,19,20,21. Previous attempts to localize signaling factors involved the culture of intestinal epithelial monolayers derived from colon organoids (mouse22 or human23) on impermeable collagen-coated membranes patterned with microholes22. These studies demonstrated that localized growth factors, such as Wnt, Noggin, and R-Spondin (Rspo) from the basal compartment, concentrate proliferative cells above the microholes, thereby localizing the crypt domains22,23. However, the influence of the signal source size and spacing on intestinal compartmentalization remains unclear. Additionally, exploring the impact of spatially distributed signaling factors found in the villi, such as EphrinB1, has been challenging using this approach. Here, we employ 2D intestinal epithelial monolayers16 and micropatterning techniques24,25 to engineer a system that precisely controls both the molecular signaling and its spatial localization (Fig. 1a, b). This innovative approach allows us to investigate how exogenously supplied Wnt3a and EphrinB1 molecules pattern the intestinal epithelium. Our experimental and computational models reveal that circular patterns of Wnt3a effectively regulate the size, shape, and long-range order of intestinal crypt-like compartments.
a Schematic of small intestine crypt, showing a decreasing Wnt3a gradient toward the crypt top and increasing EphrinB1 expression toward the villus. b Simplified model with on/off Wnt3a and EphrinB1 patterns in an intestinal monolayer culture, top view and cross-section. Yellow cells indicate the area covered by Wnt3a patterns, green cells indicate the area covered with EphrinB1, and stem cells are marked in light green. c Representative fluorescence images of micropatterns of EphB2 receptors and EphrinB1 ligands printed on treated rBM-coated substrates. Images were immunostained for EphB2, EphrinB1 and laminin (Ln). Scale bars: 50 µm. d Representative fluorescence images of micropatterns of Wnt3a and EphrinB1 printed on treated rBM-coated substrates. Images were immunostained for Wnt3a, EphrinB1, and laminin. Scale bars: 2 mm (left-panels) and 50 µm (right-panels). e Left-panel and middle-panel (zoom): Representative images of organoid-derived single cells seeded on treated rBM at 24 h of culture. Cells were immunostained for Ki67, GFP (Lgr5+ stem cells), F-Actin and cell nuclei (DNA). Scale bars: 50 μm. Right-Panel: Graph plotting the percentage of positive cells (from the total cell number counted) for GFP at 24, 72 and 120 h. Mean ± SEM. N = 3 experiments. f Representative images of monolayers of organoid-derived single cells seeded on treated rBM. Cells were immunostained for Ki67, EphB2 receptor and F-Actin. Scale bars: 50 μm. g Left-panel and middle-panel (zoom): Representative images of organoid-derived single cells seeded on treated rBM at 48 h. Cells were immunostained for Ki67, Lysozyme (Lyz), F-Actin and DNA. Scale bars: 50 μm. Right-panel: Graph plotting the number of Lysozyme+ cells per crypt at 48 h of culture. Mean ± SEM. N = 3 experiments. h Representative images of organoid-derived single cells seeded on treated rBM at 48 h. Cells were immunostained for Ki67, Wnt3a and EphrinB1. Scale bars: 20 μm. i Graph plotting the probability of hosting a crypt for Wnt3a and EphrinB1 patterns. Mean ± SEM. NWnt3a = 3 and NEphrinB1 = 4 experiments. Source data are provided as Source Data file.
Results
Crypt-like regions are spatially controlled by Wnt3a and EphrinB1 micropatterns
Microcontact printing has been successfully adapted to transfer proteins onto soft and adhesive substrates like reconstituted basement membranes (rBM)24,25. This involved a freeze-drying method applied to rBM-coated substrates (Supplementary Fig. 1a). By using this approach, we were able to produce designs featuring paracrine signaling molecule Wnt3a and juxtracrine signaling molecules EphB2/EphrinB1 receptor/ligand pair in the form of dots and hole patterns (Fig. 1c, d). Notably, these patterns were highly uniform and reproducible over large substrate areas (∼20 mm2) (Fig. 1d). This micropatterning process effectively preserved the integrity of the rBM coatings, as evidenced by the even distribution of laminin (Fig. 1c and Supplementary Fig. 1b). The positive staining for laminin indicates stable adhesion of the rBM coating to the substrate after freeze-drying, reconstitution (rehydration), and immunostaining procedures (Supplementary Fig. 1b). The samples exhibited a punctate protein distribution, and there were no significant differences observed in terms of both average intensities and thickness when comparing the freeze-dried and reconstituted (treated) samples to the non-freeze-dried (fresh) samples (Supplementary Fig. 1c, d). In addition, the fluorescence intensity of Wnt3a and the EphrinB1 micropatterns demonstrated dimensions consistent with those of the stamps employed, and were unaffected by the reconstitution procedure (Supplementary Fig. 1e).
To ensure that the reconstituted freeze-dried rBM did not adversely affect single cells derived from organoids, we thoroughly examined the formation and self-organization of the intestinal epithelial monolayers into crypt- and villus-like compartments using this substrate. We first checked whether organoid-derived single cells could form monolayers that expanded over time. By culturing these single cells on both freeze-dried and fresh rBM-coated substrates (Supplementary Fig. 1f), we observed the successful monolayer formation and growth. The surface area covered by cells doubled from day 1 (24 h) to day 5 (120 h) (Supplementary Fig. 1g, h). This growth pattern remained consistent across both substrates, as indicated by similar growth ratios. Next, we confirmed the spontaneous self-organization of intestinal epithelial monolayers derived from organoids into distinct crypt- and villus-like compartments, as previously observed6,16. The analysis of the distribution of ISCs and differentiated cells within these monolayers revealed that Lgr5+ stem cells formed densely packed zones of actively dividing cells (Ki67 positive) (Fig. 1e), which expressed EphB2 (Fig. 1f). Likewise, secretory Paneth cells (Lysozyme, Lyz positive cells) were found within these proliferative clusters (Fig. 1g). In contrast, differentiated cells (Ck20 positive) were located in the surrounding regions, displaying clear spatial segregation (Supplementary Fig. 1i). Notably, while a few non-proliferative cells showed positive Ck20 staining at the 24 h mark, this proportion substantially increased over time (Supplementary Fig. 1j). Thus, in summary, primary intestinal epithelial monolayers on reconstituted freeze-dried rBM substrates exhibited the ability to spontaneously self-organize into crypt- and villus-like compartments, displaying a behavior akin to homeostasis.
Subsequently, we seeded organoid-derived single cells onto freeze-dried rBM that had been patterned with two distinct biomolecules: circular regions with Wnt3a or holes depleted of EphrinB1. These biomolecules play critical roles in two primary signaling pathways responsible for establishing crypt-villus compartmentalization in vivo6,7,8. Upon reconstitution, cells effectively adhered and formed a monolayer. After immunostaining for Ki67, a marker indicative of cell proliferation, we observed that crypt-like compartments were located precisely on the exogenous Wnt3a regions and co-localized with the EphrinB1 holes, respectively (Fig. 1h). To quantify this phenomenon, we computed the probability of finding areas containing crypts on patterned regions if both events were independent and compared it to the experimental probability of the presence of crypt areas on Wnt3a patterned circles and EphrinB1 holes. Remarkably, we found that the presence of Wnt3a significantly increased the probability of locating a crypt by ∼2.3 times, while regions depleted of EphrinB1 enhanced the probability of hosting a crypt by ∼1.3 times (Fig. 1i). These results provide strong evidence that the spatial location of crypt-like compartments can be precisely controlled through micropatterns of Wnt3a and EphrinB1 biomolecules.
Exogenous Wnt3a micropatterns regulate crypt-like domain size and shape
We then hypothesized that exogenous Wnt3a micropatterned signal could potentially modulate not only the spatial localization, but also the size and shape of the crypt-like compartments, as it does in vivo26. To investigate this, we designed an experiment to generate Wnt3a micropatterns with varying sizes and evaluate their influence on the size and shape of the crypt-like compartments. First, we compared mRNA expression levels of typical crypt (Lgr5, stem cells, and Ki67, proliferation) and villus (Ck20, epithelial differentiation, Muc2, goblet cells, Fabp1, absorptive enterocytes, and Anpep, epithelial brush border enzymes) markers27,28 from monolayers grown on non-patterned and patterned rBMs (Supplementary Fig. 2a–f). Notably, there were no significant differences in the mRNA levels of these markers obtained by reverse-transcription polymerase chain reaction (RT-qPCR), suggesting that monolayers growing on non-patterned and patterned rBMs are alike in terms of cell populations.
We then characterized the size and shape of these crypt-like compartments on non-patterned reconstituted freeze-dried rBM, which we called control samples. In these regions, characterized by cell proliferation and the presence of ISCs, cell density was significantly higher compared to the non-proliferative villus-like regions of the monolayers. Using custom image-processing algorithms (see methods section), we quantified the individual cell areas and correlated them with their proliferative status (Supplementary Fig. 3a–f). We found that cells within the crypt-like compartments, which included Lgr5+ cells and Paneth cells, had significantly smaller areas compared to those in the villus-like compartments (Fig. 2a, b). This approach allowed for the easy identification of crypt- and villus-like compartments in cell area maps, eliminating the need for immunostaining. We were then able to compute shape descriptors for the crypt-like domains present on Wnt3a patterns (Fig. 2c) and compare them with those observed in control monolayers. To define the crypt-like compartment characteristics, we measured their area and circularity on non-patterned surfaces, where the crypts exhibited a well-defined size of 3400 µm² and a circularity index of 0.73 (Supplementary Fig. 4). Interestingly, crypt-like compartments that formed on Wnt3a patterns with a diameter of 100 µm were more than two-fold larger than their counterparts on non-patterned substrates (3400 µm² for control crypts vs. 9000 µm² for crypts on Wnt3a patterns, which, in turn, were around 8000 µm², Fig. 2d). We then fitted each crypt-like domain to an ellipse to quantify its circularity. Crypts exhibited a relatively circular shape on non-patterned substrates, with a circularity index of around 0.7. However, on substrates with exogenous Wnt3a patterns, crypts displayed even higher circularity values, approaching approximately 0.9 (Fig. 2e). Remarkably, this increase in both area and circularity was converging toward the dimensions and shape of the Wnt3a patterns themselves.
a Representative images of cells immunostained for Lysozyme (Lyz), GFP, F-Actin and nuclei (DNA) (upper panels), cell area segmentation and contour map (lower panels). Scale bars: 50 μm. b Cell area distributions in the crypt- and villus-like domains. Symbols correspond to experimental measurements, and solid lines are the Gaussian fits. Mean ± SEM. N = 4 experiments. c Representative images of Wnt3a micropatterns after cell seeding and growth (upper panels), cell area segmentation and cell area contour map (lower panels). Scale bars: 50 μm. d Bars correspond to experimental measurements of crypts’ area on Wnt3a patterns, and the dashed line corresponds to the distribution on control surfaces (Lognormal fit). Mean ± SEM. N = 3 experiments. e Bars correspond to experimental measurements of crypts’ circularity on Wnt3a patterns, and the dashed line corresponds to the distribution on control surfaces (Gaussian fit). Mean ± SEM. N = 3 experiments. f Representative images of EphrinB1 micropatterns after cell seeding and growth (left panels), cell area segmentation and area contour map (right panels). Scale bars: 50 μm. g Bars correspond to experimental measurements of crypts’ area on EphrinB1 patterns, and the dashed line corresponds to the distribution on control surfaces (Lognormal fit). Mean ± SEM. N = 3 experiments. h Bars correspond to experimental measurements of crypts’ circularity on EphrinB1 patterns, and the dashed line corresponds to the distribution on control surfaces (Gaussian fit). Mean ± SEM. N = 3 experiments. i Organoid-derived single cells were seeded on Wnt3a micropatterns of different sizes after 48 h of culture. Representative fluorescence images of F-Actin, DNA and Wnt3a (upper panels). Representative cell area contour maps (lower panels). The diameters of the Wnt3a micropatterns are 100 μm (middle panel) and 200 μm (right panel). Scale bars: 100 μm. j Crypt-like domain areas. Mean ± SEM. One-way ANOVA. Ncontrol = 6, N100 = 3 and N200 = 3 experiments. k One-way ANOVA. Ncontrol = 6, N100 = 3 and N200 = 3 experiments. Source data are provided as Source Data file.
Next, considering that proliferative crypt cells express EphB2 receptors in a gradient fashion from the center to the periphery of the crypts (Fig. 1f), we explored the possibility that creating EphrinB1-depleted regions (holes) surrounded by EphrinB1 ligands, could enable control over the localization of crypt-like compartments through repulsion (Fig. 1h, i). In particular, we wondered whether 100 µm diameter EphrinB1-depleted regions could influence the dimensions and shape of crypt-like compartments. Using a procedure analogous to that used for Wnt3a patterns, we confirmed that the repulsive effect resulting from the interaction of EphB2/EphrinB1 receptors and ligands could indeed confine the crypt-like compartments within the EphrinB1-free regions (Fig. 2f). In this scenario, the crypt areas were larger than when patterns were absent (Fig. 2g), but the compartments did not expand to the same extent as observed on Wnt3a patterns. Additionally, the circularity of the crypts was less pronounced compared to those cultured on Wnt3a patterns (Fig. 2h). This observation shed light on the differing effects of Wnt3a and EphrinB1 patterns on the dimensions and shape of crypt-like compartments, providing insights into how distinct signaling pathways influence their organization.
To investigate the influence of pattern size on crypt area, we employed larger microcontact-printed Wnt3a patterns with a diameter of 200 µm. Following the formation of the monolayers, we conducted immunostaining for Ki67, F-actin, cell nuclei (DNA), and Wnt3a (Fig. 2i). Qualitatively, the crypt-like compartments on 200 µm patterns appeared notably larger than those observed on non-patterned surfaces and on patterns with a diameter of 100 µm. Crypt-like areas increased according to the increasing size of the Wnt3a micropattern features (Fig. 2j). Additionally, their shapes exhibited a slightly more circular configuration compared to cases where patterns were absent (Fig. 2k). In summary, we successfully controlled the size and shape of the crypt-like domains through the simple microcontact printing of exogenous Wnt3a ligands.
Exogenous Wnt3a micropatterns serve as the main source of Wnt3a in the formation of crypt-like regions
Paneth cells are intermingled with Lgr5+ stem cells within the crypt regions of the small intestinal epithelium, both in vivo and in vitro3. These Paneth cells secrete factors that promote growth and stem cell maintenance, including epidermal growth factor (EGF), Notch, and Wnt ligands. Therefore, the number of Paneth cells per crypt is a key factor that regulates crypt homeostasis and consequently determines crypt size26.
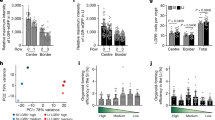
We quantified the number of lysozyme-positive cells per crypt in control monolayers and observed that crypts with more Paneth cells were larger (Fig. 3a). This suggests that crypt size may indeed be regulated by the number of Paneth cells in our experimental model as well. Next, we analyzed the number of lysozyme-positive cells in crypts formed on exogenous Wnt3a micropatterns (Fig. 3b). We found a reduction in the number of Paneth cells per crypt on Wnt3a micropatterns compared to those on control surfaces (Fig. 3c). Interestingly, these crypts required fewer lysozyme-positive cells to reach larger sizes (Fig. 3d). This indicates that the presence of exogenous Wnt3a determines crypt size independently of the Wnt signals provided by Paneth cells.
a Average crypt area as a function of the number of Lysozyme+ cells present in that crypt on freeze-dried rBM. Dots represent individual crypts and lines correspond to Mean ± SEM. N = 4 experiments. b Representative image of a crypt growing on a microcontact-printed Wnt3a dot immunostained for Lysozyme, Wnt3a, F-Actin and DNA. Scale bar 50 µm. Inset corresponds to the Lysozyme signal in the squared area. Scale bar 20 µm. c Number of Lysozyme+ cells per crypt in monolayers grown on non-patterned (control) and patterned (Wnt3a) freeze-dried rBM. Mean ± SEM. Two-tailed Mann-Whitney test. Ncontrol = 4 and NWnt3a = 3 experiments. d Number of Lysozyme+ cells per crypt as a function of crypt area for monolayers grown on non-patterned (gray) and patterned (yellow) freeze-dried rBM. Dots represent individual crypts from Ncontrol = 4 and NWnt3a = 3 experiments. e Representative image of Lysozyme+ cells stained also for Wnt3a in non-patterned (control) and patterned (Wnt3a) freeze-dried rBM. Scale bar 20 µm. f Percentage of Lysozyme+ cells which are also Wnt3a+ in monolayers grown on non-patterned (control) and patterned (Wnt3a) freeze-dried rBM. Mean ± SEM. Unpaired two-tailed t-test. N = 3 experiments. g Lyz1 mRNA relative expression in monolayers grown on non-patterned (control) and patterned (Wnt3a) freeze-dried rBM. Mean ± SEM. Paired two-tailed t-test N = 3 experiments. h Wnt3a mRNA relative expression in monolayers grown on non-patterned (control) and patterned (Wnt3a) freeze-dried rBM. Mean ± SEM. Paired two-tailed t-test N = 3 experiments. Source data are provided as Source Data file.
Immunostaining for Lysozyme and Wnt3a in control monolayers revealed a subset of cells positive for both markers, located adjacent to Wnt3a-positive/Lysozyme-negative cells. This pattern likely reflects the reported short-range Wnt3a gradient within crypts9 and suggests that, in our model, the Wnt3a signal may similarly be transmitted from Paneth cells to Lgr5+ stem cells. However, crypts on Wnt3a micropatterns showed a reduction in the number of Lysozyme-positive/Wnt3a-positive cells (Fig. 3f), and the Wnt3a signal appeared more diffuse throughout the crypt (Fig. 3e). This suggests that the intestinal epithelium may have the ability to respond to elevate Wnt concentration by downregulating the source of epithelial Wnt, as previously indicated6. In our case, Paneth cells may secrete less Wnt3a when exposed to exogenous Wnt3a from the patterns. RT-qPCR analysis further supported this, showing a complete loss of Wnt3a mRNA in crypts cultured on Wnt3a micropatterns, while Lyz1 mRNA levels remained comparable to those of monolayers grown on control surfaces (Fig. 3g, h). These findings indicate that crypt-like regions formed on exogenous Wnt3a micropatterns rely primarily on this external Wnt3a signal, rather than on endogenous Wnt3a produced by Paneth cells.
External control of the long-range organization of crypt-like domains
In vivo, crypts are distributed in the space in between villi following a honeycomb-like order29. This spatial arrangement implies that, beyond the crypt-villus axis organization, there is an inherent long-range order governing the distribution of crypts and villi. The capability of Wnt3a patterns to confine crypt-like domains and regulate their size and shape suggests that we could explore the long-range organization of the epithelial compartmentalization using arrays of microcontact printed patterns.
For this purpose, we first characterized the long-range organization of intestinal epithelial monolayers as they grew on non-patterned substrates. By examining immunostaining results and cell area maps, we could easily identify the spatial distribution of proliferative and differentiated cells (Supplementary Fig. 3). We then used Delaunay triangulation30,31 to establish crypt neighbor distances and the corresponding angles between them for each crypt (Fig. 4a and Supplementary Fig. 5). In the absence of microcontact printed patterns, neighboring crypts were found to be, on average, approximately 225 µm apart (Fig. 4b). However, these values displayed significant variability, spanning over one order of magnitude. In terms of the Delaunay angles, no distinct order was observed, with an average angle measuring approximately 55 degrees (Fig. 4c). This distribution closely resembled what one would obtain from a random distribution of points (Supplementary Fig. 5a, c). Hence, in the absence of external signals, crypt-like domains exhibited a random spatial distribution but with distances constrained within a specific range, typically ranging from 50 to 500 µm.
a Delaunay triangulation. Immunofluorescence image of the epithelial monolayer stained for Ki67, F-Actin and cell nuclei (DNA). Scale bar: 200 μm. b Distribution of the distance between crypts, dashed line corresponds to a Gaussian fit. Mean ± SEM. N = 6 experiments. c Angle distribution, dashed line corresponds to the fitting using a Gaussian distribution function. Mean ± SEM. N = 6 experiments. d Organoid-derived single cells seeded on treated rBM with the Wnt3a micropattern at 48 h of culture. Representative fluorescence images of F-Actin, cell nuclei (DNA) and Wnt3a (left panel), cell area segmentation (middle panel), and cell area contour map (right panel). Scale bars: 200 μm. e Upper panel: Distribution of the distance between crypts, dashed line corresponds to the Gaussian fit of the control distribution (in b). Mean ± SEM. N = 3 experiments. Lower panel: Angle distribution, dashed line corresponds to the Gaussian fit of the control distribution (in c). Mean ± SEM. N = 3 experiments. f Organoid-derived single cells seeded on treated rBM with the EphrinB1 micropattern at 48 h of culture. Representative fluorescence images of F-Actin, cell nuclei (DNA) and EphrinB1 (left panel), cell area segmentation (middle panel), and cell area contour map (right panel). Scale bars: 200 μm. g Upper panel: Distribution of the distance between crypts, dashed line corresponds to the Gaussian fit of the control distribution (in b). Mean ± SEM. N = 4 experiments. Lower panel: Angle distribution, dashed line corresponds to the Gaussian fit of the control distribution (in c). Mean ± SEM. N = 4 experiments. Source data are provided as Source Data file.
Next, we produced micropatterns consisting of dots of 100 μm in diameter of Wnt3a ligands, arranged in a squared lattice fashion with 200 μm center-to-center spacing across the entire surface of freeze-dried rBM-coated substrates. Following reconstitution and cell seeding, cells were cultured for 48 h and formed monolayers. These cells were then subjected to immunostaining for F-actin, segmented, and cell area contour maps were generated, as previously described (Fig. 4d). In the resulting color-coded cell area contour maps, the crypt-like domains appeared to align perfectly with the Wnt3a micropatterned regions. Differentiated cells, on the other hand, were predominantly situated in the regions between the Wnt3a patterns. Employing Delaunay triangulation, we observed that the presence of the Wnt3a micropattern had a significant impact on the distances between neighboring crypts. In this case, the distances between neighbors were less spread and their distribution was narrower than in the absence of Wnt3a pattern (Fig. 4e). Two peaks appeared in the distribution of neighbor distances: one around 210 µm and a second, smaller peak around 270 µm (Fig. 4e). These values corresponded to the sides and diagonal of a 200 µm square lattice, respectively. The angular distribution of the Delaunay triangles also underwent significant changes in response to the presence of Wnt3a patterns. They triggered the appearance of one peak at 90° and a second peak at 45°, twice as frequently (Fig. 4e). Thus, crypt-like domains exhibited long-range organization following the characteristic parameters of a square lattice as the one set by the micropattern (Fig. 4e and Supplementary Fig. 5b, c).
We then tested whether the repulsive effects mediated by EphB2/EphrinB1 could also induce long-range organization of the crypt- and villus-like compartments. Employing the same square lattice arrangement as before, we seeded cells on EphrinB1 patterned rBM, let them form the monolayer, immunostained, and plotted the cell area maps (Fig. 4f). Crypts displayed less uniformity in size compared to those on Wnt3a patterns, yet they appeared to order following the square lattice pattern established by the EphrinB1 holes. Interestingly, the areas positive for EphrinB1 were predominantly occupied by differentiated cells (Fig. 4f). The EphB2/EphrinB1 repulsive effect led to a narrowing of the distribution of neighbor distances, with a peak observed at approximately 210 µm (Fig. 4g). However, the frequency of this peak was lower compared to what was observed with Wnt3a patterns, and the secondary peak at 270 µm was less pronounced (Supplementary Fig. 6a). Analysis of the angles derived from the Delaunay triangulation indicated the presence of a peak around 45°, aligning with one of the expected values for a square lattice (Fig. 4g). Nevertheless, the peak at 90° was less distinct (Supplementary Fig. 6b). Altogether, these findings show that the long-range spatial distribution of ISCs and differentiated cells could be engineered by patterning one of the signaling pathways involved in setting the crypt-villus axis in vivo.
Effect of pattern dimensions on long-range epithelia organization
To better understand the capability of micropatterns to dictate the long-range distribution of crypt- and villus-like compartments, we introduced variations in the distances between circular patterns (or holes) while preserving the square lattice arrangement and the size of these circular patterns (or holes). We first varied the distance between Wnt3a patterns, resulting in dots spaced at distances of 150, 200, and 400 µm apart (Fig. 5a). We observed that the probability of finding a crypt on the pattern was increased compared to the expected occurrence, while the probability of finding differentiated cells on the pattern was decreased, irrespective of the spacing between Wnt3a patterns (Fig. 5b). This shows the capacity of localized exogenous Wnt3a to induce the presence of proliferative cells. Interestingly, Wnt3a patterns spaced 200 µm decreased the probability of finding proliferative clusters outside them. However, this probability was less altered when the distance between dots was decreased to 150 µm or increased to 400 µm. This distance-dependent effect was also reflected in the size and shape of the crypts: patterns spaced 200 µm apart triggered the development of larger crypts (Fig. 2d), while patterns that were more closely spaced (150 µm) or farther apart (400 µm) resulted in average crypt areas similar to those observed on non-patterned substrates (Supplementary Fig. 7a, b).
a Organoid-derived single cells seeded on treated rBM with the Wnt3a micropattern spaced 150 µm, 200 µm and 400 µm respectively at 48 h of culture. Representative fluorescence images of F-Actin, cell nuclei (DNA) and Wnt3a (upper panels). Representative cell area contour map (lower panel). Scale bars: 200 μm. b Graph plotting the probability of Wnt3a dots of hosting crypt and villus as a function of dots distance. Mean ± SEM. N = 3 experiments. c Distribution of the distance between crypts on Wnt3a patterns, dashed line corresponds to the Gaussian fit of the control distribution. Mean ± SEM. N = 3 experiments. d Distribution of the angles between crypts on Wnt3a patterns, dashed line corresponds to the Gaussian fit of the control distribution. Mean ± SEM. N = 3 experiments. e Organoid-derived single cells seeded on treated rBM with the EphrinB1 micropattern spaced 150 µm, 200 µm and 400 µm respectively at 48 h of culture. Representative fluorescence images of F-Actin, cell nuclei (DNA) and EphrinB1 (upper panels). Representative cell area contour map (lower panel). Scale bars: 200 μm. f Graph plotting the probability of EphrinB1 holes of hosting crypt and villus as a function of holes distance. Mean ± SEM. N = 3 experiments. g Distribution of the distance between crypts on EphrinB1 patterns, dashed line corresponds to the Gaussian fit of the control distribution. Mean ± SEM. N = 3 experiments. h Distribution of the angle between crypts on EphrinB1 patterns, dashed line corresponds to the Gaussian fit of the control distribution. Mean ± SEM. N = 3 experiments. Source data are provided as Source Data file.
In terms of long-range order, when the distance between Wnt3a dots was decreased to 150 µm, the distribution of distances and angles between neighboring crypts closely resembled that of non-patterned surfaces (Fig. 5c, d). On the contrary, when the distance between Wnt3a dots was increased to 400 µm, the distribution of distances and angles between neighboring crypts showed a convoluted behavior. On one hand, peaks at 300 µm and 450 µm, along with an angle of 45°, suggested the existence of some degree of square-lattice order (Fig. 5c, d). However, we also observed the presence of neighboring distances falling below the range imposed by the Wnt3a pattern, corresponding to crypts that formed in the spaces between Wnt3a circular patterns (Fig. 5a, c, d). Thus, when the imposed distance between circular patterns fell within the range of neighboring crypts on non-patterned surfaces, crypts enlarged reaching the Wnt3a pattern size and shape. However, when Wnt3a dots were spaced large distances, crypts were growing in between Wnt3a patterns analogously to the crypts on non-patterned surfaces (Supplementary Fig. 7a, b).
Next, we used holes in EphrinB1 surfaces at varying distances, with the aim of inhibiting the formation of crypts in the spaces between these holes. We incrementally increased the distance between the holes from 200 µm to 300 µm and 400 µm (Fig. 5d). The presence of EphrinB1 on the surface did not significantly reduce the probability of finding EphB2-positive proliferative compartments, nor increased the probability of finding differentiated cells, at any of the tested distances (Fig. 5e). Likewise, the probability of a proliferative compartment locating itself inside the EphrinB1 holes exhibited a slight enhancement, but this was independent of the distance between the holes. Moreover, crypt size and shape closely resembled that of non-patterned crypts in both the 300 µm and 400 µm cases (Supplementary Fig. 7c, d). In both scenarios, the distributions of neighbor distances resembled those observed on non-patterned substrates (Fig. 5g). Notably, long-range order was entirely absent, and angular distributions mirrored those observed on non-patterned substrates (Fig. 5h). These findings suggest that the repulsion effect exerted by EphrinB1 on EphB2-expressing crypts was not potent enough to prevent the appearance of crypts on EphrinB1-coated regions. These results show that, in this configuration, Wnt3a is more efficient than EphrinB1/EphB2 interaction in controlling (i) the position, (ii) size, and (iii) long-range order of 2D intestinal epithelial crypts in vitro.
ya | |a modelling framework recapitulates epithelial compartmentalization, dimensions and long-range order
To investigate the relationship between arrays of exogenous Wnt3a and long-range spatial distribution of ISCs and differentiated cells, we developed a particle-based model of the intestinal epithelia using the ya | |a modelling framework32 (Fig. 6a and Supplementary Notes). We considered individual cells (particles) seeded on a high-friction surface. Cell-cell forces were modeled by exclusion and adhesion as a function of cell-cell distance. We incorporated Eph/Ephrin interaction by introducing differential repulsion forces between cells based on whether their interactions were homotypic or heterotypic (\({{{{\bf{F}}}}}_{{{{\rm{homotipic}}}}}^{{{{\rm{repulsion}}}}} < {{{{\bf{F}}}}}_{{{{\rm{heterotipic}}}}}^{{{{\rm{repulsion}}}}}\))33. The model considered three distinct cell types: proliferative cells, Wnt3a-secreting Paneth cells and BMP-secreting differentiated cells6. The capacity of proliferative cells to undergo differentiation was regulated by an internal variable, \({{{{\rm{diff}}}}}_{i}\), within each cell, which evolved following the equation:
a Schematic description of the particle-based model. b Monolayer forming a segregated distribution of proliferative and differentiated cells. c Upper panel. Distance between crypts. Experimental in vitro distribution and fitting of the in silico results using a Gaussian distribution function in red. Lower panel. Angles between crypts. Experimental in vitro distribution and fitting of the in silico results using a Gaussian distribution function in red. Mean ± SEM for experimental data and Mean ± 95% confidence interval for the simulation. N = 6 experiments and N = 10 simulations. d Monolayer forming a squared lattice distribution of proliferative and differentiated cells on Wnt3a micropatterns in silico. e Left panel. Distribution of the in vitro experimental distance between crypts and fitting of the in silico results using a sum of two Gaussian distribution functions in red, in the presence of Wnt3a pattern. Right panel. Distribution of the in vitro experimental angles between crypts and fitting of the in silico results using a sum of two Gaussian distribution functions in red, in the presence of Wnt3a pattern. Mean ± SEM for experimental data and Mean±95% confidence interval for the simulation. N = 3 experiments and N = 10 simulations. Note that experimental data (in grey or orange) is compared to the best fitting of distributions obtained from 10 simulations performed for each condition (in red). Source data are provided as Source Data file.
Here, \({t}_{{{{\rm{diff}}}}}\) sets the time scale of cell differentiation, [BMP]i and [Wnt3a]i are the local concentrations of these signals, and \({k}_{{{{\rm{diff}}}}},{k}_{{{{\rm{stem}}}}}\) determine the magnitude of their effect on the differentiation dynamics. Thus, cells located close to a Wnt3a source would keep low values of \({{{{\rm{diff}}}}}_{i}\) and remain undifferentiated, while cells that move away from the Wnt3a source and into the high BMP regions would increase \({{{{\rm{diff}}}}}_{i}\) over time. Once \({{{{\rm{diff}}}}}_{i}\) is equal to one, the cell would differentiate and stop proliferating. Cell motility was modeled by a constant traction force (\({{{{\bf{F}}}}}^{{{{\rm{traction}}}}}\)) oriented along a 2D vector that randomly changed direction at intervals of \({t}_{{{{\rm{persistence}}}}}\). Cells were initially distributed randomly within a circle with an average separation of \({r}_{{{{\rm{seed}}}}}\), and the initial quantity of Paneth cells and proliferative cells corresponded to experimental conditions (Fig. 1). The system evolved until reaching confluency (Fig. 6b). Proliferative cells divided at a constant rate; however, cell proliferation ceased if the average distance between a cell and its nearest neighbors fell below 30% of the resting cell radius (\({r}_{{{{\rm{eq}}}}}\)). This feature emulated the global reduction in cell proliferation observed in monolayers after confluence34.
The confluent monolayer exhibited a segregated distribution of proliferative and differentiated cells such as the one observed in the experiments (Supplementary Movie 1). After optimizing the parameters (Supplementary Notes), the model successfully replicated the size of the proliferative regions, as well as the distances and angles between them (Fig. 6b, c and Supplementary Fig. 8a). Since the maximum cell size was set to be 10 µm in diameter in the simulations, comparisons with experimental data can be directly derived. The distances between neighboring crypts, obtained from Delaunay triangulation in the simulations, followed a distribution similar to that observed in vitro experiments (Fig. 6c), with values ranging from 0 to 600 µm and centered around the average values seen in the experiments (300 µm for simulations and 225 µm for experiments). Likewise, the angular distribution resulting from the Delaunay triangulation closely matched that of a random distribution, mirroring the experimental finding (Fig. 6c). Thus, our particle-based model featuring segregated populations of proliferative and differentiated cells, recapitulates key characteristics of our in vitro intestinal epithelial model, including compartmentalization into crypt- and villus-like domains, crypt size, inter-crypt distances, and spatial distribution of the crypts.
The Wnt3a gradient was critical in setting the spatial distribution of proliferative and differentiated cells (see Supplementary Notes). So, we sought to study whether exogenous Wnt3a signals could govern crypt distribution in our computational model similarly to experiments. We generated a Wnt3a pattern in our in silico model to simulate the microcontact-printing experiments, aligning its dimensions with those employed in the actual experiments. Upon reaching confluency, we observed the formation of regions containing proliferative cells encircled by differentiated cells in locations corresponding to the defined Wnt3a regions (Fig. 6d). We then analyzed the sizes and distribution of these proliferative regions using Delaunay triangulation. Similar to the in vitro findings, the introduction of Wnt3a dots in silico resulted in an enlargement of crypts (Supplementary Fig. 8b). Distances and angles between proliferative regions closely resembled those observed in the experimental data, the distribution of distances and angles showed two peaks in both cases (Fig. 6e), corresponding to a square lattice organization. Thus, our particle-based computational model successfully replicated the distributions of both distances and angles characterizing the long-range order of proliferative regions established by the Wnt3a microcontact-printed patterns in vitro.
By varying the spacing between Wnt3a dots, we managed to reproduce the crypt distribution observed in the experiments (Supplementary Fig. 8c, d). When Wnt3a dots were spaced 150 µm, we observed fewer crypts than the number of Wnt3a dots printed, both in vitro and in silico (Supplementary Fig. 8c), thus the monolayer was not able to follow the organization imposed by the dots. At 200 µm distance between dots, we found a one-to-one correspondence between Wnt3a dots and crypts (Supplementary Fig. 8c), resulting in the long-range order of the compartmentalized monolayer in simulations and experiments (Fig. 6d, e). Finally, increasing the distance between Wnt3a dots up to 400 µm led to the formation of more crypts than the number of Wnt3a dots (Supplementary Fig. 8c). While proliferative crypts primarily localized on the Wnt3a dots, they also appeared in the spaces between (Supplementary Fig. 8d). Altogether, the particle-based model was able to capture the main features of the experimental system, including (i) the generation of compartmentalized monolayers with well-defined crypt size and spatial distribution, (ii) the establishment of a long-range order through a spatial pattern of exogenous Wnt3a signal, and (iii) the effects of pattern dimension on crypt distribution.
Exogenous Wnt3a patterns reduce the size of proliferative crypts in Apc-/- monolayers
Loss of Apc in intestinal stem cells drives adenoma formation in mice through enhanced Wnt signaling, which leads to hypertrophic crypt formation and increased cellular proliferation35. Our results show that exogenous Wnt3a patterns can modulate the size, shape, and organization of intestinal crypts in 2D monolayers by overriding endogenous Wnt3a signals from Paneth cells. This raised the question of how intestinal epithelia with an altered Wnt pathway –characterized by enlarged crypts36,37 –would respond to these Wnt3a patterns.
To investigate this, we stablished an in vitro epithelial monolayer model using single cells derived from intestinal organoids of Apc-/- mice. Because the binding of Wnt3a to the surface of Apc-/- cells depends on Rspo9, we cultured organoids and generated monolayers on non-patterned rBM substrates, both with and without Rspo (Supplementary Fig. 9a). Indeed, Rspo had a significant effect on Wnt3a binding in Apc-/- monolayers, with increased Wnt3a signal when Rspo was added to the culture medium (Supplementary Fig. 9b–d). We next examined the proliferative state of these Apc-/- monolayers. In the absence of Rspo, a high proportion of Ki67 positive cells was observed, and the crypt-villus compartmentalization typical of wild-type (WT) monolayers was lost (Fig. 7a, b). In contrast, Apc-/- monolayers cultured with Rspo were less proliferative, with fewer Ki67 positive cells that clustered together, resembling the crypt-like regions of WT monolayers (Fig. 7a, b). Thus, we successfully generated Apc-/- intestinal epithelial monolayers, which showed increased proliferation and altered compartmentalization compared to WT, consistent with in vivo observations37,38.
a Immunofluorescence images of Apc-/- monolayers cultured with and without Rspo. Cells were stained for F-actin, cell nuclei (DNA), and Ki67. Scale bars: 50 µm. b Quantification of Ki67+ cells in Apc-/- monolayers. Mean ± SEM. Unpaired two tailed t-test with Welch’s correction. N = 3 experiments. c Crypt area distribution of Apc-/- crypt-like monolayers + Rspo. Black line corresponds to a Lorentzian fit. Mean ± SEM. N = 3 experiments. d Delaunay triangulation measuring the distance between crypt-like regions in Apc-/- monolayers + Rspo. Black line corresponds to a Gaussian fit. Mean ± SEM. N = 3 experiments. e Angular distribution of crypt-like regions in Apc-/- monolayers + Rspo. Black line corresponds to a Gaussian fit. Mean ± SEM. N = 3 experiments. f Immunofluorescence images of Apc-/- monolayers cultured with Rspo on freeze-dried rBM substrates micropatterned with Wnt3a (100 µm diameter dots spaced 200 µm apart in a square arrangement). Cells were stained for Wnt3a, F-actin, and cell nuclei (DNA) on the left panel, and for Wnt3a and Ki67 on the right panel. Dashed circles highlight Wnt3a patterns. Scale bars: 100 µm. g Characterization of crypt-like regions area in Apc-/- monolayers cultured with Rspo on Wnt3a micropatterns. Dashed black line corresponds to the Lorentzian fit of the Apc-/- crypts on non-patterned substrates. Mean ± SEM. N = 3 experiments. h Angular distribution of Delaunay triangles between crypt-like regions in Apc-/- monolayers cultured with Rspo on Wnt3a micropatterns. Dashed black line corresponds to the Gausian fit of the monolayers on non-patterned substrates. Mean ± SEM. N = 3 experiments. i Distribution of the distances between crypt-like regions in Apc-/- monolayers cultured with Rspo on Wnt3a micropatterns. Dashed black line corresponds to the Gausian fit of the monolayers on non-patterned substrates. Mean ± SEM. N = 3 experiments. Source data are provided as Source Data file.
Next, we characterized the crypt-like domains of Apc-/- monolayers cultured with Rspo on non-patterned rBM substrates by measuring their area and applying Delaunay triangulation. Consistent with in vivo findings37,38,39, Apc-/- crypt-like regions were larger than those in WT monolayers (Fig. 7c). Furthermore, the distance between crypt-like regions was significantly increased compared to WT (Fig. 7d), and the angular distribution followed that of randomly distributed crypts (Fig. 7e). We then cultured Apc-/- monolayers with Rspo on rBM substrates micropatterned with Wnt3a (100 µm diameter dots, spaced 200 µm and following a square arrangement) (Fig. 7f). Analysis of crypt area and Delaunay triangulation revealed that exogenous Wnt3a micropatterns impacted Apc-/- crypt-like regions. The crypts were smaller than those on non-patterned substrates (Fig. 7g), and although the crypts remained randomly distributed (Fig. 7h), the distance between them was reduced compared to their non-patterned counterparts (Fig. 7i). Therefore, in Apc-/- monolayers, where increased endogenous Wnt signaling leads to enlarged crypt-like regions, the exogenous Wnt3a signal from micropatterns effectively reduced crypt size and influenced their distribution by decreasing the distance between them.
Discussion
We have introduced a method to induce the long-range organization of crypt-like compartments of 2D primary intestinal epithelial monolayers, which would otherwise randomly self-organize in vitro. In vivo, epithelial and stromal cell-derived signals exert their effects through paracrine mechanisms involving the diffusion of cell-secreted molecules through the tissue’s extracellular matrix, as well as through direct cell-cell contacts (juxtracrine signals). Instead, our approach involved the immobilization of paracrine signaling molecules such as Wnt3a and juxtracrine signals like EphrinB1 on rBM coatings to guide the spontaneous compartmentalization of 2D intestinal epithelial monolayers. We showed that the use of immobilized micropatterns of these signaling factors can effectively govern the size, shape, and long-range arrangement of crypt-like compartments.
In vivo, these signals orchestrate tissue development, maintain homeostasis, and induce regeneration after damage by a set of complex inter-dependent mechanisms such as feedback loops6 or repulsive interactions along the crypt-villus axis7,8. Here, we propose an approach utilizing spatially localized surface-bound ligands created via micropatterning techniques to simulate in vivo signaling. Although the in vivo Wnt3a and EphrinB1 gradients were simplified in here using an on/off signal, micropatterns were able to clearly impact on crypt size, shape and organization. It is worth considering the use of other patterning technologies such as microfabricated scaffolds18,20, inkjet printing or microfluidics40,41, or photopatterning (PRIMO)42 to obtain more in vivo-like complex shapes of micropatterned proteins. These methodologies could also be adapted to allow to precisely control and manipulate these gradients. However, their compatibility with soft and sticky rBM layers can be challenging. Alternatively, the use of soft collagen coatings on photoresist films containing arrays of microholes was successfully used to position intestinal crypts22,23. But, this approach was limited to growth factors located in the crypt regions and could not address the role of signals specific to the villus region. In contrast, our technique allows the easy transfer of relevant biomolecules to organize the small intestinal epithelium, whether in crypts or villi regions, such as Wnt3a or EphrinB1 ligands and their EphB2 receptors, at precise locations on thin rBM coatings.
This simple approach allowed us to study the impact of Wnt3a and EphrinB1 ligands in dictating epithelial compartmentalization architecture. We showed that both signals increased the probability of guiding the position of a crypt. However, when it came to changing the size and shape of crypt-like domains, this could only be achieved by manipulating the size of exogenous Wnt3a patterns, not by using EphrinB1 micropatterns. These differences could be related to the fact that both ligand concentration and cellular density can impact the strength of the repulsive effect triggered by microcontact printed Eph-Ephrin signaling25. But also, because the observed expansion in the area of crypt-like compartments seems linked to an increase in the proliferative capacity of the cells, caused by the role of Wnt3a in the regulation of the intestinal proliferation. This appears to occur at the expense of endogenous Wnt3a secretion, likely through the downregulation of the Paneth cell population under Wnt conditions6. On one hand, while the crypts were enlarged by the Wnt3a patterns, the number of Paneth cells per crypt was slightly reduced. And in the other hand, the levels of endogenous Wnt3a expression were decreased when the cells were exposed to the exogenous Wnt3a. Moreover, we demonstrated the potential to further enlarge the size of proliferative crypts by expanding the area of Wnt3a patterns, effectively controlling the spatial distribution of Wnt3a while keeping its concentration constant. Our approach thus provides control of the local concentration of ligand (short-range diffusion), which has been shown critical in the growth of organoids3,43.
In addition to determining the size and shape of crypt-like compartments, Wnt3a micropatterns were effectively guiding the long-range localization of crypts onto these patterns. Interestingly, this effect could also be achieved by using similar templates of EphrinB1. In this case, the use of micropatterns featuring EphrinB1 ligands across the entire surface except within circular holes demonstrated that the EphB2/EphrinB1 receptor/ligand interaction could confine the crypt-like domains inside the regions devoid of EphrinB1. Indeed, when the dimensions of the EphrinB1 or Wnt3a arrays fell within a range close to the spontaneous crypt-crypt distances, proliferative compartments were redistributed following the signals imposed by the micropatterned template. Both signals acted at the crypt level, either by affinity or repulsion, essentially creating niche-like regions. Remarkably, their confining effect weakened when the spacing between domains was decreased or increased. This suggests that the size of the villus-like compartment also played a role in the organization of the monolayer. It is reasonable to assume that the villus compartment, predominantly composed of differentiated cells, has a defined size that cannot expand unlimitedly. Since cells proliferate in the crypt-like regions and are pulled towards the villus-like domain while differentiating17, larger villus-like regions should be achieved by boosting proliferation. Therefore, it is not surprising that when distance between domains is expanded above a given length, villus-like regions drag crypt-like domains out of the patterned niche-like regions in order to maintain the structural integrity of the epithelium. In this case, it is worth considering that the behavior observed is the convolution of the micropatterns localizing large crypts on Wnt3a dots together with the endogenous Wnt3a signal driving the formation of crypts in the regions in between dots. This behavior was indeed observed experimentally and by a particle-based model that we implemented. By using the Wnt3a/BMP pathway to control proliferation and differentiation6 and the Eph/Ephrin signaling to account for heterotypic cell-cell repulsion33, our theoretical model was capable of generating compartmentalized monolayers in silico that replicated the primary experimental characteristics, including the distribution of sizes and distances, as well as the long-range order induced by exogenous Wnt3a patterns. Moreover, our theoretical model also captured that by increasing the distance between the exogenous Wnt3a compartments, the distances between crypts could be similarly expanded, ultimately causing crypts to emerge in between the Wnt3a patterns.
Finally, we demonstrated the applicability of our method to clinically relevant models, specifically intestinal epithelia with Apc gene mutations. We successfully established in vitro 2D monolayers from single cells derived from Apc-/- mouse organoids. This Apc mutation leads to increased Wnt signaling, resulting in altered crypt-villus architecture, enhanced cellular proliferation, and changes in epithelial cell differentiation2,37,38,39. In vitro, this mutation produced a monolayer phenotype that was dependent on Rspo, consistent with prior reports for organoids9. In the absence of Rspo, Wnt3a signaling was reduced, most cells were Ki67-positive, and the typical crypt-villus organization was lost. However, upon adding Rspo, both Wnt3a signaling and the proportion of Ki67-positive cells decreased, and the monolayers became compartmentalized, with large, proliferative, crypt-like regions. This enabled us to examine the effect of exogenous Wnt3a patterns on these enlarged crypts. Wnt3a patterns successfully reduced both the size of the proliferative crypt-like regions and the distance between them in Apc-/- monolayers, although they did not establish long-range order. In WT epithelia, long-range order was typically achieved when the dimensions of the micropatterns matched the intrinsic size and spacing of the monolayer. Therefore, further optimization of the pattern dimensions may be necessary to induce long-range organization in the Apc-/- model. Nevertheless, these results highlight the suitability and potential of our method for studying disease mechanisms.
In summary, we have used a modified version of microcontact printing to investigate the self-organization of intestinal epithelial cells derived from organoids. This approach allowed us to systematically examine the effects of individual Wnt3a and EphrinB1 ligands on this process. We successfully controlled the position, size, and shape of crypt-like domains by microcontact printing exogenous Wnt3a, which in turn downregulated Paneth cells’ production of endogenous Wnt3a. Furthermore, we showed that, within an optimal range of pattern dimensions, ordered arrays of crypt-like regions could be established through exogenous signals. Notably, we extended this method to clinically relevant models, including Apc-/- intestinal epithelial monolayers, where increased Wnt signaling disrupts crypt-villus architecture and cell proliferation. Our approach effectively reduced crypt-like domain size and modified their distribution in these mutant monolayers, showcasing its broad applicability. Finally, we replicated the key experimental outcomes using a particle-based model that incorporated the Wnt3a-BMP signaling feedback loop and Eph/Ephrin-mediated heterotypic repulsion. In the long term, this system holds significant potential for providing insights into individual signaling pathways involved in intestinal epithelium organization, which could enhance our understanding of both physiological and pathological processes within this tissue in vivo.
Methods
Ethical statement
The research performed in this study complies with all relevant ethical regulations. The use of animals for the obtention of intestinal crypts from Lgr5-EGFP-IRES-creERT2 mice was approved by the Animal care and Use Committee of Barcelona Science Park (CEEA-PCB) and the Catalan government and performed by their relevant guidelines and regulations.
Intestinal crypt isolation and organoid culture
Intestinal crypts from Lgr5-EGFP-IRES-creERT2 male and female mice (3–6 month old) (WT) and from Apcfl/fl-Villin-CreERT2 male and female mice (3-6 month old) (Apc-/-) from E. Batlle lab were isolated as previously described44,45. Briefly, small intestines were flushed with PBS and cut longitudinally. Intestinal villi were removed mechanically, and the intestinal crypts were isolated by incubating the tissue with PBS containing 2 mM EDTA (Sigma-Aldrich) for 30 min at 4 °C. The digested material was filtered through a 70 µm pore cell strainer (Biologix Research Co.), and the so-called crypt fraction was obtained. Then, crypts were plated within drops of the commercial reconstituted basement membrane Matrigel (BD Bioscience), and supplemented with medium: advanced DMEM/F12 (Invitrogen) plus 1% Glutamax (Gibco), 1% HEPES (Sigma-Aldrich), Normocin (1:500, Invitrogen), 2% B27 (Gibco), 1% N2 (Gibco), and 1.25 mM N-acetylcysteine (Sigma-Aldrich). The medium was supplemented with recombinant murine epidermal growth factor (EGF) (100 ng mL−1, Gibco), recombinant murine Noggin (100 ng mL−1, Peprotech), and recombinant human R-Spondin 1 (200 ng mL−1, R&D Biosystems) to obtain what we called ENR-medium or EN if R-Spondin was not added46. For the experiments, WT organoid cultures were enriched in their stem cell amount by supplementing the ENR-medium with two small molecules, CHIR99021 (3 µM) and valproic acid (1 mM), leading to ENR_CV-medium46, while Apc-/- organoids were maintained and passaged with EN or ENR medium alone. For the maintenance of the organoid culture, outgrowing crypts were passaged twice a week. To obtain crypt pieces homogeneous in size, full-grown organoids were subjected to a mild digestion protocol as previously described16. Briefly, Matrigel drops containing organoids were mechanically disrupted using a syringe with a 23 G 1” needle (BD Microlance 3). The resulting solution was centrifuged at 70 g for 3 min. Crypt pieces were then plated again in Matrigel drops. Organoid stocks were maintained for up to 4 months at 37 °C in a humidified incubator under a 5% CO2 atmosphere.
Preparation and characterization of micropatterns on soft substrates
Microcontact printing was performed following standard procedures. First, silicon masters fabricated by UV-photolithography and reactive ion etching (D + T Microelectrónica AIE) were silanized to facilitate the demolding of PDMS replicas47. PDMS replicas were freshly prepared mixing Sylgard 184 Silicon Elastomer and curing agent (DowCorning) at a ratio of 10:1 w/w. PDMS was directly casted on the silicon master forming thick replicas (∼ 3 mm) and cured for 3 h at 65 °C. Then, PDMS replicas were incubated with 10 µg mL−1 Wnt3a (Abcam), 100 µg mL−1 recombinant mouse EphB2/Fc receptor (R&D Systems) or 69 µg mL−1 recombinant mouse EphrinB1/Fc ligand (R&D Systems) in a cluster fashion for 45 min at room temperature. Clusters were prepared conjugating ligand (EphrinB1 fused to Fc, named EphrinB1/Fc) at 2:5 molar ratio for 30 min under constant shaking. The EphrinB1 concentration for stamping was optimized based on prior work25, and the Wnt3a concentration was initially set according to previously reported values48, and then progressively increased until the patterns were transferred satisfactorily.
Matrigel was previously subjected to a freeze-drying process to make it mechanically resistant to sustain the PDMS stamp pressure, following a process developed by our research group24,25. In short, polymer well plates (Ibidi μ-Slide 8 Well, Ibidi GmbH) with an area of 1 cm2/well and stiffness of 3 GPa were coated with 200 µL cm-2 of Matrigel diluted at 0.25 mg mL−1. After gelation (1 h at 37 °C), the excess of the solution was removed, and samples were carefully rinsed first with PBS and then with Milli-Q water to avoid the presence of salts. Subsequently, samples were freeze-dried by immersion in liquid nitrogen and vacuum dried for 24 h at −50 °C and 0.06 mbar of pressure (1-4 LD-2, Christ Alpha) and sterilized with 10 min of ultraviolet (UV) light.
Matrigel thin films were characterized by immunofluorescence using antibodies against laminin, as this is its major component49. To minimize losing Matrigel material in the immunostaining process, the fixation step was eliminated, low temperatures were avoided, and detergents (such as Triton X-100) were removed from all solutions. Thus, Matrigel-coated substrates were blocked with a blocking buffer containing 1% BSA (Sigma-Aldrich) and 3% donkey serum (Millipore) in PBS for 30 min. Antibodies were diluted in PBS containing 0.1% BSA, and 0.3% donkey serum. Hydrogels were incubated with anti-laminin (1:200, Abcam) primary antibody for 30 min at room temperature (RT). Alexa Fluor® 647 donkey anti-rabbit (Jackson ImmunoResearch) diluted at 1:500 was used as secondary antibody, and it was incubated for 45 min at RT. Finally, samples were mounted with Fluoromount® (Thermo Fisher) for imaging. Images of the samples were acquired at randomly selected locations with a confocal laser scanning microscope (LSM 800, Zeiss) equipped with a 100x oil objective (NA = 1.40, WD = 0.17). The pinhole diameter was set to 1 Airy Unit (AU). For all images acquired, a z-step of 0.1 µm was used. To characterize the surface protein distribution, projection images of the confocal z-stack were obtained by adding the intensities of each z-slice. Then, average intensities of the resulting projection images were measured as follows: 10 × 10 µm ROIs (regions of interest) were defined and their average intensity was obtained. Finally, the average intensity of each ROI was normalized by the maximum intensity of the corresponding projection image. Three different ROIs of each projection image were measured for three independent samples. Finally, average intensity of all the ROIs was computed. On the other hand, Matrigel thickness was estimated by a method based on confocal microscopy, following a previously described procedure50. Briefly, we extracted all the orthogonal views of the confocal laminin images and added them up by summing their intensities. This generated an intensity profile of the film cross-sections. To mitigate the noise effects, intensity across the z-profile was obtained by averaging the intensity values of 10 μm width regions. The obtained profiles x were fitted by a Gaussian distribution:
where the coefficient \(a\) sets the Gaussian baseline, \(\left(b-a\right)\) is the Gaussian amplitude, \(c\) sets the peak position, and \(\sigma\) is the Gaussian width parameter. Then, for each profile the full width at half maximum (FWHM) of the peak was calculated as \(2\surd \left(2{{\mathrm{ln}}}2\right)\sigma\), and this value was attributed to the Matrigel layer thickness. The measurements were conducted using a custom-made ImageJ macro (http://rsb.info.nih.gov/ij, NIH).
The micropatterns were visualized by immunofluorescence following the protocol described above. The following antibodies were employed: as primary antibodies anti-laminin (1:200, Abcam), anti-EphB2 (1:200, R&D Systems), anti-Wnt3a (1:200, Abcam) and anti-Fc (560 µg mL−1, Jackson ImmunoResearch). As secondary antibodies, Alexa Fluor® 488 donkey anti-goat, Alexa Fluor® 488 donkey anti-mouse, Alexa Fluor® 488 donkey anti-rabbit, and Alexa Fluor® 647 donkey anti-rabbit (all from Jackson ImmunoResearch) diluted at 1:500 were employed. Finally, samples were mounted with Fluoromount® (Thermo Fisher) for their observation under the microscope. The success of the printing procedure (uniformity, reproducibility, and stability of the biochemical micropatterns) was checked using an epifluorescence inverted microscope (Axio Observer 7, Zeiss) with a 63x oil objective (NA = 1.4). Images were analyzed using ImageJ software. The protein coverage was quantified by measuring the fluorescence intensity of the antibodies. For comparison of the intensity measurements, all images were acquired using the same microscope settings.
Cell culture
To obtain organoid-derived single cells, full-grown WT or Apc-/- organoids were enzymatically and mechanical disrupted. When seeded in culture for our experiments, these organoid-derived single cells were cultured with ENR_CV-medium (WT organoids) or with EN o ENR medium (Apc-/- organoids) plus ROCK inhibitor Y-27632 (10 µM, Sigma-Aldrich) to prevent anoikis51. Organoid-derived single cells were then seeded onto micropatterns at a density of 105 cells per drop. The cells were cultured for different periods of time depending on the experiment. In all the cases, the medium was changed at 24 h after seeding, and then every 48 h.
Cells were fixed with 10% neutralized formalin (Sigma-Aldrich) for 40 mins at 4 °C, permeabilized with 0.5% Triton X-100 (Sigma-Aldrich) for 30 min at room temperature (RT) and blocked for 2 h at RT with a blocking buffer containing 1% BSA (Sigma-Aldrich), 3% donkey serum (Millipore), and 0.2% Triton X-100 in PBS. All primary antibodies were diluted in 0.1% BSA, 0.3% donkey serum and 0.2% Triton X-100 in PBS, and incubated on cells overnight at 4 °C. Primary antibodies used, which depended on the cell type and experimental layout, were: anti-GFP (1:100, Life Technologies), anti-GFP (1:100, Abcam), anti-Ki67 (1:100, Abcam), anti-Ki67 (1:100, BD Biosciences), anti-Lysozyme (1:100, Dako), anti-Cytokeratin 20 (1:100, Dako), anti-EphB2 (1:200, R&D Systems). Secondary antibodies were diluted in 0.1% BSA and 0.3% donkey serum in PBS and incubated on cells 1 h at RT. Secondary antibodies used were: Alexa Fluor® 488 donkey anti-goat, Alexa Fluor® 488 donkey anti-mouse, Alexa Fluor® 488 donkey anti-rabbit, Alexa Fluor® 647 donkey anti-goat, Alexa Fluor® 647 donkey anti-mouse, Alexa Fluor® 647 donkey anti-rabbit (all from Jackson ImmunoResearch) diluted at 1:500. Alexa Fluor® 568 phalloidin was used to stain F-Actin. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (1:1000) diluted in 0.1% BSA and 0.3% donkey serum in PBS and incubated on cells 40 min at RT. Finally, samples were mounted with Fluoromount® (Thermo Fisher). Fluorescence images (or tiles) were acquired using an epifluorescence inverted microscope (Axio Observer 7, Zeiss) with 5x air (NA = 0.16), 10x air (NA = 0.30) or 20x air (NA = 0.40) objectives. Images were acquired at randomly selected locations. The tiles were made up of several individual images to inspect the whole sample. The tiles used a 10% spatial overlap to stitch the single images into a map of the full screened-surface by the ZEN software (Zeiss). In addition, fluorescence images were acquired with a confocal laser scanning microscope (LSM 800, Zeiss) with a 10x air objective (NA = 0.3, WD = 2.0) or 20x air objective (NA = 0.8, WD = 0.55). The laser excitation and emission light spectral collections were optimized for each fluorophore, especially for the four-color scans, where the emission bands were carefully adjusted to avoid overlapping channels. The pinhole diameter was set to 1 Airy Unit (AU). For all images acquired, the optimal z-step was used as indicated by the equipment software. The images were used to characterize several features of the cultures such as cell markers, surface epithelial coverage and cell morphology, among others.
Quantification of cell markers
Monolayers were characterized for proliferative cells through Ki67 marker (nuclear marker), for Paneth cells through Lysozyme marker, the presence of Lgr5+ ISC by positive GFP signal and epithelial markers such as Cytokeratin 20 (Ck20) (cytoskeleton marker for terminally differentiated enterocytes and goblet cells). To allow quantifications, cell contour was defined through F-Actin and cell nuclei were located through DNA identification. To quantify the different cell markers, a semi-automatize routine implemented in the Imaris software (version 9.1.0; Bitplane) was used. First, cell nuclei were detected as spots using the smallest values of their diameters previously measured for representative images. Spot number detection was adjusted based on the signal quality (differences in intensity and shape) to improve nuclei detection. Then, F-Actin channel was used as a filter to define the segmentation area of the nuclei. To quantify the number of cells positive for a specific marker relative to the total cell population, the mean intensity of that specific marker was filtered by the total nuclei signal found in each F-Actin positive area. When the mean intensity of the marker was above a threshold, we counted the nucleus as positive for that particular marker. This process was performed for Ki67, Ck20, Lysozyme and GFP markers. Alternatively, a Fiji/ImageJ custom-made macro can be used to perform this quantification.
Quantification of cell surface coverage
To quantify the cell surface coverage from immunostained images, the fluorescence image tiles of F-Actin were used. The tiles were pre-processed and analyzed using ImageJ software. First, we applied a mean filter with a radius of 15 μm to smoothen the surface edges. The filtered images were then segmented using the default ImageJ threshold to obtain a binary mask where the area occupied by the cells appeared in white, and cell-free regions or holes in the cell monolayers appeared in black. To remove the background noise, the binary mask was twice eroded followed by twice dilated using ImageJ specific functions. The area was the filled region that describes the projected area. Then, from the binary image, the area occupied by the cell monolayer (including holes) and cell aggregates were determined and, by inverting the binary image, the areas of the holes (cell-free areas) were obtained. Subtracting both, the projected area effectively occupied by cell structures was evaluated. This area was then normalized by the seeding area. Also, in some experiments, cell surface coverage over culture time was alternatively quantified by analyzing brightfield images with ImageJ software. Briefly, the projected area occupied by cellular structures was measured by outlining its shape and normalizing it to the total area of the image.
Quantitative reverse-transcription polymerase chain reaction (RT-qPCR)
Total RNA was extracted from 4 wells of a Ibidi®µ-Slide 8 Well plate for each condition with 800 µL of TRI Reagent Solution® (Invitrogen™). The RNA extraction was achieved following the manufacturer’s instructions of the reagent, and the RNA was resuspended in DEPC treated water. The concentration and purity of the extracted RNA was measured using the Nanodrop 1000 (Thermo Scientific). For RNA-complementary DNA (cDNA) synthesis, 200 ng of total RNA were first treated with DNAse I Amplification Grade® (Invitrogen™) to remove all genomic DNA. The reverse transcription of the RNA was performed using the iScript™ cDNA first-strand Synthesis kit (BioRad) following the manufacturer’s protocol.
The mRNA transcript levels of the genes of interest were analysed by real-time quantitative PCR using the StepOne Plus real time PCR system (Applied Biosystems). The primers used for each gene are shown in Supplementary Table 1. The specificity of the primers to recognize correctly the gene sequence, and the absence of non-specific primer bindings were tested in silico. For each sample and gene studied, the analyses were performed in triplicate wells in a 96-well plate. The PCR reactions were carried out in a final volume of 10 µL containing 5 µL of Fast SYBR™ Green Master Mix (ThermoFisher), 0.5 µL of a mixture of forward and reverse primers (250 nM), 3.5 µL of DEPC water plus 1 µL of diluted cDNA. In addition, three negative controls, a non-template control, a non-reverse transcriptase control and a PCR control (water control), were included and ran in duplicate. The expression level of each gene was calculated with the ΔΔCT method and was analysed relative to the reference genes gapdh and rps18.
Analysis of crypt- and villus-like domains on the micropatterned substrates
2D monolayers of organoid-derived single cells were organized in crypt- and villus-like domains6,16. To define the crypt- and villus-like domains, we developed a method to identify, segment and characterize these domains on non-patterned and micropatterned substrates based on the cell areas. Fluorescence images were employed. The area of each cell was first identified by the segmentation of the cell monolayers. F-Actin fluorescence images were pre-processed with ImageJ, as previously described52. Briefly, the background noise of the F-Actin image was reduced with the subtract background function by using a rolling ball radius of 10 μm. The resulting image was then subjected to band-pass filter with upper and lower limits of 15 μm and 1 μm, respectively. The background noise from this output was reduced by using the subtract background command again with a rolling ball radius of 10 μm. The F-Actin pre-processed images from each experiment were then loaded to Tissue analyzer53 for edge detection and 2D monolayer segmentation. The segmented 2D monolayer was used to quantify various parameters such as cell area, and cell centroid positions \(\left(x,y\right)\). 2D cell area contour maps were performed using a custom-made code in Matlab using the cell area, and cell centroid positions obtained from the segmented 2D monolayers. The contour map is represented in logarithmic scale to enhance the visualization. To identify crypt-like domains, the cell area contour maps were correlated with the segmented epithelial cell marker Ki67 images (Ki67 stains positive for proliferative cells). For the Ki67 segmentation, we followed the protocol described above. To identify villus-like domains, the cell area contour maps were correlated with the inverted of the segmented Ki67 images. From these two correlations, the distributions of the crypt- and villus-like domains were plotted. These distributions were fitted with two independent Gaussian functions and were characterized by their mean value and the standard deviation (SD): \({{{{{\rm{mean}}}}}_{{{{\rm{area}}}}}}_{{{{\rm{Ki}}}}67+}\) and \({{{{{\rm{SD}}}}}_{{{{\rm{area}}}}}}_{{{{\rm{Ki}}}}67+}\), respectively.
Cell areas below threshold were counted as proliferative cells. These proliferative cells generate a binary image with proliferative domains. In addition, the area of a proliferative domain should be above \({10{{{{\rm{mean}}}}}_{{{{\rm{area}}}}}}_{{{{\rm{ki}}}}67+}\) threshold to be consider a crypt-like domain. Binary crypt-like domains images were treated as single object images. So, from them, we extracted the area, \(\left(x,y\right)\) centroid position and circularity index
from the crypt-like domains to define their morphology.
The organization of crypt-like domains was analyzed by Delaunay triangulation30,31. Binary images of crypt-like domains and the patterns were used for these analyses. These were processed using ImageJ to extract the center position \(\left(x,y\right)\) of each crypt-like domain and each pattern. These center coordinates were used to determine the sides (distances between crypts defining a triangle) and the angles in between by using a custom-made code in Matlab.
Quantification of the impact of Wnt3a and EphrinB1 on crypt positioning
To determine the impact of Wnt3a and EphrinB1 on crypt positioning, we evaluated the experimental conditional probability of finding a pixel with both crypt and pattern signals, \({P\left({{{\rm{crypt}}}}|{{{\rm{pattern}}}}\right)}_{\exp }\), in a given area \(A.\) We compared this with the expected conditional probability assuming the two events are independent, \({P\left({{{\rm{crypt}}}} |{{{\rm{pattern}}}}\right)}_{{{{\rm{ind}}}}}\).
The experimental conditional probability of finding a pixel with both crypt and pattern signals can be expressed as:
where:
and
Thus,
In contrast, the conditional probability assuming independence is given by:
where
We can then define a preference parameter as:
When this parameter is close to 1, the positioning of patterns and crypts appears independent. If it is greater than 1, the presence of a pattern signal increases the likelihood of finding a crypt signal at that location.
Statistical analysis
Statistical analysis was conducted using GraphPad Prism software (GraphPad Software Inc.). Results are expressed as mean±error, either standard deviation (SD), confidence interval (CI) or standard error of the mean (SEM), as noted in the figure captions. The number of experiments used for data extraction is specified for each quantitative analysis. Normality of the datasets was tested by the d’Agostino-Pearson normality test. Statistical comparisons were performed using parametric (e.g., t-test, ANOVA) or non-parametric tests (e.g., Mann-Whitney), depending on data distribution, as indicated in the corresponding figure legends. Exact p-values are provided in the plots when applicable. Images shown are representative of multiple independent experiments conducted by different researchers, consistently yielding similar results.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data are available within the Article, Supplementary Information or Source Data file. The source data underlying Figs. 1e, g, i, 2b, d, e, g, h, j, k, 3a, c, f–h, 4b, c, e, g, 5b–d, f–h, 6c, e, 7b–e, g–i and Supplementary Figs. 1c–e, h, j, 2a–f, 3f, 4, 5c, 6a, b, 7a–d, 8a–c, 9c, d are provided as a Source Data file. Source data are provided with this paper.
Code availability
Custom-made Fiji/Image macros and MATLAB scripts generated for this study can be found at: https://github.com/BiomimeticsLab/Larranaga_et_al_2024. The source code used to run the ABM simulations of this paper can be found at: https://github.com/mmarinriera/crypt_patterning_simulation.
References
Bjerknes, M. & Cheng, H. Intestinal Epithelial Stem Cells and Progenitors. Methods Enzymol. 419, 337–383 (2006).
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
Sato, T. et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418 (2011).
Radtke, F. & Clevers, H. Self-Renewal and Cancer of the Gut: Two Sides of a Coin. Science 307, 1904–1909 (2005).
Maloy, K. & Powrie, F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature 474, 298–306 (2011).
Thorne, C. A. et al. Enteroid Monolayers Reveal an Autonomous WNT and BMP Circuit Controlling Intestinal Epithelial Growth and Organization. Dev. Cell 44, 624–633.e4 (2018).
Batlle, E. et al. β-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/EphrinB. Cell 111, 251–263 (2002).
Holmberg, J. et al. EphB Receptors Coordinate Migration and Proliferation in the Intestinal Stem Cell Niche. Cell 125, 1151–1163 (2006).
Farin, H. F. et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature 530, 340–343 (2016).
Qi, Z. et al. BMP restricts stemness of intestinal Lgr5+ stem cells by directly suppressing their signature genes. Nat. Commun. 8, 13824 (2017).
Meran, L., Baulies, A. & Li, V. S. W. Intestinal Stem Cell Niche: The Extracellular Matrix and Cellular Components. Stem Cells Int. 2017, 7970385 (2017).
Gregorieff, A. et al. Expression Pattern of Wnt Signaling Components in the Adult Intestine. Gastroenterology 129, 626–638 (2005).
Crosnier, C., Stamataki, D. & Lewis, J. Organizing cell renewal in the intestine: Stem cells, signals and combinatorial control. Nat. Rev. Genet 7, 349–359 (2006).
Farin, H. F., Van Es, J. H. & Clevers, H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of paneth cells. Gastroenterology 143, 1518–1529.e7 (2012).
Durand, A. et al. Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1). Proc. Natl. Acad. Sci. USA 109, 8965–8970 (2012).
Altay, G. et al. Self-organized intestinal epithelial monolayers in crypt and villus-like domains show effective barrier function. Sci. Rep. 9, 1–14 (2019).
Pérez-González, C. et al. Mechanical compartmentalization of the intestinal organoid enables crypt folding and collective cell migration. Nat. Cell Biol. (2021) https://doi.org/10.1038/s41556-021-00699-6.
Wang, Y. et al. A microengineered collagen scaffold for generating a polarized crypt-villus architecture of human small intestinal epithelium. Biomaterials 128, 44–55 (2017).
Wang, Y. et al. Bioengineered Systems and Designer Matrices That Recapitulate the Intestinal Stem Cell Niche. CMGH 5, 440–453.e1 (2018).
Altay, G. et al. Modelling Biochemical Gradients in Vitro to Control Cell Compartmentalization in a Microengineered 3D Model of the Intestinal Epithelium. (2021).
Cameron, O., Neves, J. F. & Gentleman, E. Listen to Your Gut: Key Concepts for Bioengineering Advanced Models of the Intestine. Adv. Sci. Adv. Sci. 11, e2302165 (2024).
Kim, R. et al. Formation of arrays of planar, murine, intestinal crypts possessing a stem/proliferative cell compartment and differentiated cell zone. Lab Chip 18, 2202–2213 (2018).
Wang, Y., Sims, C. E. & Allbritton, N. L. Human 2D Crypt Model for Assaying Intestinal Stem Cell Proliferation and Differentiation. Anal. Chem. 94, 9345–9354 (2022).
Castaño, A. G., Hortigïela, V., Lagunas, A., Cortina, C. & Samitier, J. Protein patterning on hydrogels by direct microcontact printing: application to cardiac differentiation. RSC Adv. 4, 29120–29123 (2014).
Larrañaga, E., Fernández-Majada, V., Ojosnegros, S., Comelles, J. & Martinez, E. Ephrin Micropatterns Exogenously Modulate Cell Organization in Organoid-Derived Intestinal Epithelial Monolayers. Adv. Mater Interf. 9, (2022).
Mei, X., Gu, M. & Li, M. Plasticity of Paneth cells and their ability to regulate intestinal stem cells. Stem Cell Res Ther 11, 349 (2020).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Sato, T. & Clevers, H. Growing Self-Organizing Mini-Guts from a Single Intestinal Stem Cell: Mechanism and Applications. Science 340, 1190–1194 (2013).
Gehart, H. & Clevers, H. Tales from the crypt: new insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 16, 19–34 (2019).
Franz Aurenhammer, Rolf Klein & Der-tsai Lee. Voronoi Diagrams And Delaunay Triangulations. (World Scientific Publishing Company, 2013).
Okabe, A., Boots, B., Sugihara, K. & Chiu, S. N. Spatial Tessellations. (J. Wiley & Sons, 2000).
Germann, P., Marin-Riera, M. & Sharpe, J. ya||a: GPU-Powered Spheroid Models for Mesenchyme and Epithelium. Cell Syst. 8, 261–266.e3 (2019).
Kindberg, A. A., Srivastava, V., Muncie, J. M., Weaver, V. M. & Gartner, Z. J. EPH / EPHRIN regulates cellular organization by actomyosin contractility effects on cell contacts. J. Cell Biol. 220, e202005216 (2021).
Streichan, S. J., Hoerner, C. R., Schneidt, T., Holzer, D. & Hufnagel, L. Spatial constraints control cell proliferation in tissues. Proc. Natl. Acad. Sci. USA 111, 5586–5591 (2014).
Barker, N. et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457, 608–611 (2009).
Miyoshi, H. J. B. Special Review - Wnt Signaling: Biological Functions and Its Implications in Diseases: Wnt-expressing cells in the intestines: Guides for tissue remodeling. J. Biochem 161, 19–25 (2017).
Sansom, O. J. et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 18, 1385–1390 (2004).
Fischer, J. M., Schepers, A. G., Clevers, H., Shibata, D. & Liskay, R. M. Occult progression by Apc-deficient intestinal crypts as a target for chemoprevention. Carcinogenesis 35, 237–246 (2014).
Dow, L. E. et al. Apc Restoration Promotes Cellular Differentiation and Reestablishes Crypt Homeostasis in Colorectal Cancer. Cell 161, 1539–1552 (2015).
Rodríguez-Seguí, S. A., Ortuño, M. J., Ventura, F., Martínez, E. & Samitier, J. Simplified microenvironments and reduced cell culture size influence the cell differentiation outcome in cellular microarrays. J. Mater. Sci. Mater. Med. 24, 189–198 (2013).
Comelles, J., Hortigüela, V., Samitier, J. & Martínez, E. Versatile gradients of covalently bound proteins on microstructured substrates. Langmuir 28, 13688–13697 (2012).
Melero, C. et al. Light-induced molecular adsorption of proteins using the primo system for micro-patterning to study cell responses to extracellular matrix proteins. J. Visualized Exp. 2019, 1–19 (2019).
Snippert, H. J. et al. Intestinal Crypt Homeostasis Results from Neutral Competition between Symmetrically Dividing Lgr5 Stem Cells. Cell 143, 134–144 (2010).
Fernández-Majada, V. et al. The tumour suppressor CYLD regulates the p53 DNA damage response. Nat. Commun. 7, 12508 (2016).
Barriga, F. M. et al. Mex3a Marks a Slowly Dividing Subpopulation of Lgr5+ Intestinal Stem Cells. Cell Stem Cell 20, 801–816.e7 (2017).
Yin, X. et al. Niche-independent high-purity cultures of Lgr5 + intestinal stem cells and their progeny. Nat. Methods 11, 106–112 (2014).
Caballero, D. et al. Atomic Force Microscopy Characterization of a Microcontact Printed, Self‐Assembled Thiol Monolayer for Use in Biosensors. Anal. Lett. 39, 1721–1734 (2006).
Ahmad, A. A., Wang, Y., Sims, C. E., Magness, S. T. & Allbritton, N. L. Optimizing Wnt-3a and R-spondin1 concentrations for stem cell renewal and differentiation in intestinal organoids using a gradient-forming microdevice. RSC Adv. 5, 74881–74891 (2015).
Hughes, C. S., Postovit, L. M. & Lajoie, G. A. Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics 10, 1886–1890 (2010).
Brismar, H., Patwardhan, A., Jaremko, G. & Nyengaard, J. Thickness estimation of fluorescent sections using a CSLM. J. Microsc 184, 106–116 (1996).
Watanabe, K. et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 25, 681–686 (2007).
Comelles, J. et al. Epithelial colonies in vitro elongate through collective effects. Elife 10, e57730 (2021).
Aigouy, B. et al. Cell Flow Reorients the Axis of Planar Polarity in the Wing Epithelium of Drosophila. Cell 142, 773–786 (2010).
Acknowledgements
We thank the Martinez Lab members for discussions and help, and A.L. Godeau for critical reading of the manuscript. Funding for this project was provided by: European Union Horizon 2020 ERC grant (agreement no. 647863 – COMIET (E.M.)), CERCA Programme/Generalitat de Catalunya (2017-SGR-1079 (E.M.)), Spanish Ministry of Economy and Competitiveness (TEC2017-83716-C2-1-R (E.M.), and the Severo Ochoa Programme for Centres of Excellence in R&D 2016-2019). The results presented here only reflect the views of the authors; the European Research Commission is not responsible for any use that may be made of the information it contains.
Author information
Authors and Affiliations
Contributions
E.L., V.F-M., J.C. and E.M. conceived the project. E.L. and J.C. performed experiments and analyzed data. V.F.-M performed experiments, and contributed technical expertise and discussion. A.A.-L and D.B.-C. performed experiments with J.C. and contributed technical expertise and discussion. A.O. performed RT-qPCR experiments and analysis. M.M.-R developed the ABM and contributed technical expertise and discussion. E.B., J.S. and S.O. contributed technical expertise, materials and discussion. E.L. and J.C. wrote the manuscript. All authors revised the completed manuscript. J.C. and E.M. supervised the project. J.C. and E.M. have contributed equally to this work and share corresponding authorship.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Ying Chen, Pak-Wing Fok, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Larrañaga, E., Marin-Riera, M., Abad-Lázaro, A. et al. Long-range organization of intestinal 2D-crypts using exogenous Wnt3a micropatterning. Nat Commun 16, 382 (2025). https://doi.org/10.1038/s41467-024-55651-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-55651-7
This article is cited by
-
Physics-informed displacement control for variable pattern printing with V-shaped PDMS stamps in roll-to-roll microcontact printing
Communications Engineering (2025)