Abstract
Yes-associated protein (YAP) activation confers resistance to chemotherapy and targeted therapy. Methionine participates in cellular processes by converting to methyl donor for the methylation of DNA, RNA and protein. However, it remains unclear whether methionine affects drug resistance by influencing YAP activity. In this study, we report that methionine deprivation remarkably suppresses the transcriptional activity of YAP–TEAD in cancer cells. Methionine promotes PRMT1-catalyzed asymmetric dimethylation at R124 of YAP (YAP R124me2a). Mimicking of YAP methylation abolishes the reduction effect of methionine-restricted diet on YAP-induced drug resistance. YAP activates the transcription of SLC43A2, the methionine transporter, to increase methionine uptake in cancer cells. Knockdown of SLC43A2 decreases the level of YAP R124me2a. BCH, the inhibitor of SLC43A2, sensitizes tumors to anticancer drugs. Thus, our results unravel the positive feedback between YAP R124 methylation and SLC43A2 that contributes to anticancer drug resistance. Disrupting this positive feedback could be a potential strategy for cancer therapy.
Similar content being viewed by others
Introduction
Methionine is a sulfur-containing essential amino acid, and its metabolism and biological function have been extensively explored in metabolic disorders, aging, and particularly in cancer1,2. Emerging evidence has demonstrated that multiple cancer cells depend on exogenous sources of methionine compared to normal cells, which is called the Hoffman effect. Given that an adequate amount of methionine must be obtained from food, dietary methionine intervention has emerged as a promising strategy for cancer treatment3. Dietary methionine restriction has been shown to impede tumor growth and sensitize tumors to chemotherapy, radiation therapy, and immunotherapy in multiple tumor types4,5,6,7,8. Methionine contributes to therapy resistance by participating in a variety of biological processes, including nucleotide synthesis, redox homeostasis, as well as converting to S-adenosylmethionine (SAM), the methyl donor for the methylation of DNA, RNA, and protein5,7. The roles of DNA or RNA methylation in drug resistance have been intensively investigated9,10. However, the mechanism of methionine-induced protein methylation in drug resistance remains to be illustrated.
The oncogenic transcriptional coactivator Yes-associated protein (YAP) is the primary effector of the Hippo signaling pathway, which plays a critical role in organ size control, tissue homeostasis, tumorigenesis, and drug resistance11,12,13,14. Post-translational modifications (PTMs) are crucial modifiers involved in the subcellular localization and transcriptional activity of YAP. Phosphorylation of YAP at S127 inhibits its activity by retaining YAP in the cytoplasm and enhances the therapeutic sensitivity of hepatocellular carcinoma (HCC) to lenvatinib treatment15. YAP-K342 methylation enhances the nuclear retention of YAP and promotes tumorigenesis and sorafenib resistance16,17. In contrast, methylation of YAP at K494 is critical for its cytoplasmic retention18. Recently, we revealed that protein arginine methyltransferase 1 (PRMT1) catalyzes asymmetric dimethylation at R124 of YAP (YAP R124me2a)19. YAP R124me2a facilitates YAP nuclear translocation, enhances YAP–TEAD transcriptional activity, and is associated with poor patient prognosis in multiple cancers. However, whether methionine affects the methylation of YAP and the role of YAP R124me2a in drug resistance remains unclear.
YAP is involved in metabolic reprogramming and serves as a critical coordinator of nutrient availability with the development and progression of cancer20. YAP increases glycolysis via augmenting the transcription of glucose transporter 3 (GLUT3)21 and several key enzymes in glycolysis, including hexokinase 2 (HK2)22 and phosphofructokinase B3 (PFKFB3)23. YAP also upregulates the transcription of glutaminase1 (GLS1) and glutamine transporters to promote glutaminolysis by interacting with TEAD24,25,26. In addition, YAP activates polyamine biosynthesis and cholesterol biogenesis in cancers27,28. Previous metabolomic analyses have found that overexpression of YAP led to an increase, whereas knockdown of YAP resulted in a decrease in the level of methionine-related metabolites27,29. Nevertheless, the mechanism by which YAP regulates methionine metabolism has not been reported.
In the present study, we find that methionine induces resistance to sorafenib and oxaliplatin by elevating PRMT1-catalyzed methylation of YAP R124. We also demonstrate that YAP activates the transcription of SLC43A2, the major methionine transporter, to sustain high methionine levels in cancer cells by interacting with TEAD4. We reveal positive feedback between YAP R124me2a and SLC43A2 during drug resistance. Furthermore, the disruption of this positive feedback with BCH, a pharmacologic inhibitor of SLC43A2, results in the sensitization of YAP-related drug resistance.
Results
Methionine is essential for YAP–TEAD activation and PRMT1-mediated YAP R124me2a
Previous studies have found that methionine deficiency sensitizes cancer cells to drug therapy5. In this study, we found that methionine deprivation significantly enhanced the effect of anticancer drugs on the cell viability and the apoptosis of Huh-7 and HCT 116 cells, although methionine deprivation alone did not inhibit the cell viability and induce the apoptosis of Huh-7 cells at 24 h and HCT 116 cells at 48 h (Supplementary Fig. 1a–e). We also generated sorafenib-resistant Huh-7 cells and oxaliplatin-resistant HCT 116 cells and found methionine deprivation also enhanced the drug sensitivity of these cells (Supplementary Fig. 1f, g). These data further confirmed that methionine confers potent drug resistance to HCC and CRC cells.
Methionine participates in a variety of biological processes, including protein synthesis, redox homeostasis, as well as converting to SAM. We found that short-term methionine deprivation showed no obvious effect on protein synthesis and redox homeostasis in Huh-7 and HCT 116 cells (Supplementary Fig. 1h–k), suggesting that the conversion of methionine to SAM may be involved in drug resistance. It is known that the methylation of proteins modulates the activity of signal transduction. Thus, we conducted dual-luciferase assays to investigate the activities of canonical oncogenic signaling pathways in Huh-7 and HCT 116 cells cultured with methionine-depleted medium. The results showed that the activity of YAP–TEAD, but not other tested pathways, was remarkably suppressed by methionine deprivation (Fig. 1a, b). As methionine is converted into SAM, the major methyl donor for protein methylation (Supplementary Fig. 2a), we then investigate whether methionine promoted drug resistance by improving the methylation of YAP. Our recent study has demonstrated that PRMT1-mediated asymmetric arginine dimethylation of YAP at R124 (YAP R124me2a) enhanced YAP–TEAD transcriptional activity19. Consistently, we confirmed that PRMT1 knockdown reduced YAP–TEAD activity and downregulated the expression of YAP target genes (Supplementary Fig. 2b, c). Bioinformatic analyses of the Cancer Genome Atlas (TCGA) data revealed increased expression of YAP target genes in HCC tissues with high PRMT1 levels compared to those with low PRMT1 levels (Supplementary Fig. 2d). PRMT1 expression showed a positive correlation with YAP target genes, such as CDC20, FOXM1, TGFB2, and ANKRD1 (Supplementary Fig. 2e). Moreover, we found that PRMT1 knockdown significantly decreased the cell viability upon sorafenib administration in Huh-7 cells and upon oxaliplatin administration in HCT 116 cells (Supplementary Fig. 2f, g). In addition, MS023, an inhibitor of PRMT1, exhibited similar effects on enhancing drug sensitivity and hampering YAP–TEAD transcriptional activity in vivo and in vitro (Supplementary Fig. 3a–g).
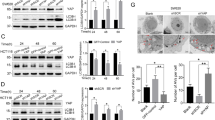
a, b The relative luciferase activity of YAP-TEAD, Wnt, TGF-β, NF-κB, and Hedgehog signaling pathways in Huh-7 cells (a) and HCT 116 cells (b) cultured in completed medium (CM) or methionine-deprived medium (MD) for 48 h. c, d The level of YAP R124 methylation in Huh-7 cells (c) and HCT 116 cells (d) cultured in MD for 72 h. The data represents the relative ratio of YAP R124me2a to YAP. The red arrow indicates the brand of YAP R124me2a. e, f The transcriptional expression of YAP, CTGF, CYR61, and AREG in Huh-7 cells (e) and HCT 116 cells (f) cultured in MD and supplemented with methionine for 72 h. g The S-adenosylmethionine (SAM) concentration in Huh-7 cells cultured in MD and supplemented with SAM (50 μM) was detected by UPLC-MS/MS. h, i The level of YAP R124me2a in Huh-7 cells (h) and HCT 116 cells (i) cultured in MD for 24 h and then supplemented with SAM (50 μM) for another 24 h. The data represents the relative ratio of YAP R124me2a to YAP. j, k The transcriptional expression of YAP, CTGF, CYR61, and AREG in Huh-7 cells (j) and HCT 116 cells (k) cultured in MD for 24 h and then supplemented with SAM (50 μM) for 36 h. l The level of YAP R124me2a in Huh-7 cells transfected with si-PRMT1 and cultured in MD for 48 h. The data represents the relative ratio of YAP R124me2a to YAP and PRMT1 to GAPDH. m The viability of Huh-7 cells transfected with si-PRMT1 and cultured in MD for 6 days. Statistical analyses were performed using two-sided unpaired Student’s t-test (a, b), one-way ANOVA with multiple comparisons (e–g, j, k), or two-way ANOVA with multiple comparisons (m). Data were presented as mean ± SD. n = 3 biologically independent samples (a, b, e–g, j, k, m). The data presented in (c, d, h, i) are representative of three independent experiments, and those in (l) are representative of two independent experiments. Source data are provided as a Source Data file.
Then, we evaluated the effect of methionine on YAP R124me2a. Supplementation with methionine elevated the level of YAP R124me2a (Fig. 1c, d), accompanied by enhanced mRNA levels of YAP target genes in a dose-dependent manner in Huh-7 and HCT 116 cells (Fig. 1e, f). UPLC-MS/MS results indicated that the intracellular level of SAM in Huh-7 cells was decreased in methionine-depleted medium, and it was significantly elevated after SAM supplementation, which was also confirmed by ELISA in Huh-7 cells and HCT 116 cells (Fig. 1g and Supplementary Fig. 4a, b). Supplementation with the methyl donor SAM blocked the inhibitory effect of methionine deprivation on the level of YAP R124me2a and the expression of YAP target genes (Fig. 1h–k). Moreover, PRMT1 knockdown reduced the level of YAP R124me2a and inhibited cell growth in the complete medium, but not in methionine-depleted medium in Huh-7 cells (Fig. 1l, m). Furthermore, PRMT1 knockdown sensitized Huh-7 cells to sorafenib in a completed medium, but not in methionine-deprived medium (Supplementary Fig. 5). Taken together, these results suggested that methionine is important for PRMT1-mediated methylation of YAP R124 and YAP–TEAD activity through providing methyl donor.
R124 asymmetric dimethylation of YAP enhances YAP-related drug resistance
It is known that YAP activation contributes to drug resistance in a variety of cancers30. As expected, treatment with sorafenib and oxaliplatin increased YAP–TEAD transactivation in a time- and dose-dependent manner (Fig. 2a, b). In addition, sorafenib and oxaliplatin markedly upregulated the transcription of YAP target genes (Fig. 2c, d). Interestingly, western blot analysis showed that the level of YAP R124me2a was significantly increased in sorafenib-treated Huh-7 cells and oxaliplatin-treated HCT 116 cells (Fig. 2e, f). Similar effects were also observed in human lung cancer A549 cells treated with gemcitabine and gastric cancer SGC7901 cells treated with 5-fluorouracil (5-Fu) (Supplementary Fig. 6a–e). These observations suggested that YAP R124 methylation may be involved in drug resistance.
a, b The relative YAP-TEAD activity in Huh-7 cells treated with the indicated concentration of sorafenib (a) and HCT 116 cells treated with the indicated concentration of oxaliplatin (b). c, d The relative expression of YAP target genes in Huh-7 cells treated with sorafenib (0.75 μM) (c) and HCT 116 cells treated with oxaliplatin (4 μM) (d) for 72 h. e, f The level of YAP R124me2a in Huh-7 cells treated with sorafenib (0.75 μM) (e) and HCT 116 cells treated with oxaliplatin (4 μM) (f). The data represents the relative ratio of YAP R124me2a to YAP. g The cell viability of YAP-, YAP-R124K-, YAP-S127A-, or YAP-R124F-overexpressed Huh-7 cells treated with sorafenib for 24 h. h The cell viability of YAP-, YAP-R124K-, YAP-S127A-, or YAP-R124F-overexpressed HCT 116 cells treated with oxaliplatin for 48 h. i Kaplan–Meier analysis of overall survival in a cohort of 191 hepatocellular carcinoma patients presenting a low level of YAP R124me2a (left panel) or high level of YAP R124me2a (right panel), who were treated with or without sorafenib. Statistical analyses were performed using one-way ANOVA with multiple comparisons (a–d, g, h) or Kaplan–Meier analysis (Gehan-Breslow-Wilcoxon test, i). Data were presented as mean ± SD. n = 4 (a) or 3 (b–d, g, h) biologically independent samples (a–d, g, h). The data presented in (e, f) are representative of three independent experiments. Source data are provided as a Source Data file.
To clarify the role of YAP R124 methylation in drug resistance, we replaced R124 with lysine (K) to block methylation, and with phenylalanine (F) to mimic methylation31 (Supplementary Fig. 6f). Of note, both R124 and S127 residues are located in a 14-3-3 binding motif (RX1-2SX2-3S), which is well-known for suppressing YAP transcriptional activity upon phosphorylation of Ser127 (Supplementary Fig. 6f). This prompted us to evaluate the impact of R124 methylation on S127 phosphorylation. In line with a previous study32, the YAP-R124K mutation resulted in a modest decrease in S127 phosphorylation in YAP (Supplementary Fig. 6g). However, YAP-R124F mutation completely blocked S127 phosphorylation (Supplementary Fig. 6g). To eliminate the potential bias of S127 phosphorylation on drug resistance, we employed YAP-S127A, a mutation that prevents phosphorylated while maintaining YAP activation, as a control for YAP-R124F. Compared with YAP, YAP-R124K mutation reduced the transactivation of the YAP–TEAD reporter, while YAP-S127A and YAP-R124F mutations enhanced its activity (Supplementary Fig. 6h). Moreover, the promoting effect of YAP-R124F on YAP–TEAD transcriptional activity was stronger than that of YAP-S127A (Supplementary Fig. 6h). Similarly, YAP-R124K mutation abolished the drug resistance induced by YAP, while YAP-S127A and YAP-R124F mutations enhanced drug resistance compared with YAP-overexpressed Huh-7 and HCT 116 cells (Fig. 2g, h). Furthermore, YAP-R124F-overexpressed cells exhibited greater resistance to drug therapy than YAP-S127A-overexpressed cells (Fig. 2g, h).
To further investigate the functional contribution of YAP R124me2a in sorafenib resistance, the levels of YAP R124me2a in HCC tissues were detected with immunohistochemistry staining. Patients from HCC tissue arrays were stratified into groups with either low or high levels of YAP R124me2a. The Kaplan–Meier survival analysis showed that patients with HCC presenting low levels of YAP R124me2a benefited from sorafenib administration, while patients with high levels of YAP R124me2a did not (Fig. 2i). Collectively, these data suggested that R124 asymmetric dimethylation of YAP promotes the drug resistance induced by YAP.
Dietary methionine restriction enhances drug efficacy by reducing YAP R124me2a
Next, we explored whether methionine deprivation enhanced the efficacy of sorafenib and oxaliplatin by blocking YAP R124me2a. Compared with YAP-overexpressed cells, YAP-R124K mutation decreased survival and increased apoptosis in the complete medium upon sorafenib and oxaliplatin treatment (Supplementary Fig. 7a–d), confirming that methylation of R124 is involved in the anti-drug capability of YAP. Moreover, methionine deprivation significantly enhanced the effect of sorafenib and oxaliplatin on the cell viability and the apoptosis of the YAP- or YAP-S127A-overexpressed cells, but not of the YAP-R124F-overexpressed cells (Fig. 3a, b and Supplementary Fig. 7e, f). Similarly, methionine deprivation significantly reduced YAP–TEAD transcriptional activity in YAP-, YAP-S127A-overexpressed cells but not of the YAP-R124F-overexpressed cells (Supplementary Fig. 6h). Taken together, our data indicated that methionine deprivation could improve sensitivity to drug therapy by diminishing YAP R124me2a in vitro.
a The cell viability of YAP-, YAP-S127A-, or YAP-R124F-overexpressed Huh-7 cells cultured in methionine-deprived medium (MD) and treated with sorafenib for 24 h. b The cell viability of YAP-, YAP-S127A-, or YAP-R124F-overexpressed HCT 116 cells cultured in MD and treated with oxaliplatin for 48 h. c Schematic representation of the experimental design in Balb/c nude mice to measure the combination effect of dietary methionine restriction and sorafenib in stable YAP-R124F- or YAP-S127A-overexpressed cells. d Representative image of tumor nodules in different conditions described in Fig. 3c. e, f The statistical analysis of tumor volume (e) and tumor weight (f) in different groups described in Fig. 3c. g, h Representative images (g) and quantitation (h) of ki-67 staining in tumors from different groups were shown. scale bar = 50 μm. i, j The relative expression of CTGF (i) and CYR61 (j) was analyzed using real-time PCR. Statistical analyses were performed using one-way ANOVA with multiple comparisons (a, b, f, h–j) or two-way ANOVA with multiple comparisons (e). Data were presented as mean ± SEM for e and mean ± SD for a, b, f, h, i, j. n = 3 (a, b) or 6 (e, f, h, i, j) biologically independent samples. Source data are provided as a Source Data file.
To further verify this view in vivo, we established subcutaneous tumor models via inoculating stable Huh-7 cells overexpressing YAP-R124F or YAP-S127A into Balb/c nude mice, followed by treatment with sorafenib and methionine-restricted diet (Fig. 3c). Methionine-restricted diet significantly reduced serum methionine in mice (Supplementary Fig. 7g). Eight days of dietary methionine restriction alone showed little effect on tumor growth, tumor volumes, and tumor weights in both YAP-R124F- and YAP-S127A-overexpressed cells (Fig. 3d–f). Dietary methionine restriction effectively sensitized YAP-S127A-overexpressed tumors to sorafenib, resulting in a considerable inhibition of tumor growth (Fig. 3d–f). IHC staining showed that the combination treatment of methionine restriction and sorafenib reduced cell proliferation in YAP-S127A-overexpressed tumors (Fig. 3g, h). Moreover, the combination treatment decreased the level of YAP R124me2a and the expression of CTGF, CYR61 in YAP-S127A-overexpressed tumors (Supplementary Fig. 7h and Fig. 3i, j). However, overexpression of YAP-R124F blunted the sensitizing effect of dietary methionine restriction on YAP-induced sorafenib resistance (Fig. 3d–j). Consistently, similar results were observed in subcutaneous CRC tumor models injected with HCT 116 cells and treated with oxaliplatin (Supplementary Fig. 8a–e).
It is known methionine-restricted diet played an antitumor role by enhancing adaptive immunity6. Thus, we subcutaneously injected Hepa1-6 cells into immunocompetent C57BL mice. Interestingly, the combination of methionine-restricted diet and sorafenib significantly inhibited tumor growth, tumor weight, and the proliferation of tumor cells in both YAP-S127A-overexpressed tumors and YAP-R124F-overexpressed tumors (Supplementary 8f–k). However, the combination therapy exhibited a greater effect on inhibiting tumor growth in YAP-S127A-overexpressed tumors compared to YAP-R124F-overexpressed tumors (Supplementary Fig. 8f–k). These data implied that apart from its impact on immunity, methionine restriction still improved drug sensitivity through modulating YAP R124me2a in tumor cells within a syngeneic cancer model. Altogether, these findings further indicate that dietary restriction of methionine enhances the therapeutic effect of sorafenib and oxaliplatin by diminishing YAP R124me2a.
YAP promotes methionine uptake by upregulating SLC43A2 expression
It has long been recognized that YAP plays a vital role in regulating various cellular metabolic processes, including glycolysis, glutaminolysis, and polyamine biosynthesis, which contribute to tumorigenesis and tumor progression20. However, it remains unknown whether YAP regulates methionine metabolism. Previous metabolomic analyses found that overexpression of YAP led to an increase, whereas knockdown of YAP resulted in a decrease in the levels of methionine-related metabolites27,29. Consistent with previous reports, we found intracellular methionine was elevated by about 1.5-fold in YAP-overexpressed Huh-7 and HCT 116 cells (Fig. 4a, b).
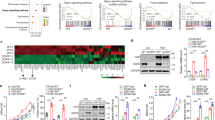
a, b The methionine concentration in Huh-7 cells (a) and HCT 116 cells (b) infected with YAP lentivirus for 96 h. c Schematic diagram for RNA sequence analysis. Seven-week-old mice were injected with AAV8-TBG-YAP-S127A through tail vein. 8 weeks later, the primary hepatocytes were isolated for RNA sequence analysis. d Heatmap from the RNA sequence analysis depicts different gene expression of transporters mediating methionine uptake and enzymes involving in methionine metabolism. e, f The mRNA levels of SLC family in Huh-7 cells (e) and HCT 116 cells (f) infected with YAP lentivirus for 96 h. g The methionine concentration in Huh-7 cells transfected with indicated siRNAs for 48 h. h The protein level of SLC43A2 in Huh-7 cells and HCT 116 cells infected with YAP lentivirus for 96 h. The data represents the relative ratio of SLC43A2 to GAPDH. i Spearman correlation between YAP and SLC43A2 in HCC tumors and normal tissues in TCGA dataset. j Kaplan–Meier overall survival analysis of SLC43A2 for HCC patients in TCGA dataset. k The mRNA expression of SLC43A2 in Huh-7 cells transfected with YAP or YAP-S94A plasmids for 72 h. l The mRNA expression of SLC43A2 in Huh-7 cells transfected with YAP and/or si-TEAD4 for 72 h. m, n ChIP-qPCR assays in Huh-7 cells with YAP (m) and TEAD4 (n) antibody showed the binding of YAP-TEAD4 to the promoter of SLC43A2. o Luciferase activity in 293 T cells transfected with YAP. The SLC43A2 luciferase reporters expressing wild-type or mutated TEAD binding sites (Mut1, Mut2, Mut1 + 2) were used. Statistical analyses were performed using two-sided unpaired Student’s t-test (a, b, e, f), one-way ANOVA with multiple comparisons (g, k, l–o), two-tailed Spearman’s correlation test (i) or Kaplan–Meier analysis (Mantel–Cox test, j). Data were presented as mean ± SD. n = 3 (a, b, d–g, k, l) or 4 (m–o) biologically independent samples. The data presented in (h) are representative of three independent experiments. Source data are provided as a Source Data file.
To understand how YAP improves intracellular methionine levels, seven-week-old wild-type mice were injected with AAV8-TBG-control or AAV8-TBG-YAP-S127A. The primary hepatocytes were isolated for RNA sequence analysis (Fig. 4c). A total of 4028 transcripts were differentially expressed in YAP-S127A-overexpressed hepatocytes versus control hepatocytes (Supplementary Fig. 9a). KEGG analysis indicated that metabolic pathways, including lipid, carbohydrate, nucleotide, and amino acid, were strongly enriched in YAP-S127A-overexpressed hepatocytes (Supplementary Fig. 9b). We compared the expression of key enzymes involved in methionine metabolism and the transporters from the solute carrier (SLC) family that mediates methionine uptake. The expression of most key enzymes involved in methionine metabolism was not significantly altered, except for Mat1a, which is the primary enzyme responsible for converting methionine to SAM in the liver (Fig. 4d). However, most methionine-related transporters (Slc1a5, Slc7a5, Slc7a6, Slc7a8, Slc38a1, Slc43a2, and Slc43a3) were upregulated in mouse hepatocytes overexpressing YAP-S127A, while Slc38a2 and Slc43a1 were downregulated (Fig. 4d). Consistently, YAP overexpression markedly increased the expression of SLC1A5, SLC7A5, SLC38A1, and SLC43A2 in Huh-7 and HCT 116 cells (Fig. 4e, f). Thus, YAP increased methionine uptake by upregulating the expression of methionine-related transporters to improve the level of intracellular methionine.
Next, we evaluated the level of cellular methionine in Huh-7 cells transfected with siRNA for SLC1A5, SLC7A5, SLC38A1, or SLC43A2, respectively. The knockdown efficiency of the indicated siRNAs was confirmed using real-time PCR (Supplementary Fig. 9c). Interestingly, knockdown of SLC43A2 markedly decreased the cellular methionine level, while silencing the other SLC family genes only slightly reduced the methionine level in Huh-7 cells (Fig. 4g). Therefore, we further investigated the correlation between YAP and SLC43A2 in detail. YAP overexpression enhanced the mRNA and protein levels of SLC43A2 (Fig. 4e, f, h), whereas knockdown of YAP downregulated SLC43A2 expression in Huh-7 and HCT 116 cells (Supplementary Fig. 9d, e). In addition, knockout of YAP (YAP-KO) in 293 A cells resulted in a remarkable reduction of SLC43A2 mRNA levels (Supplementary Fig. 9f). Analyses of human HCC and CRC data from TCGA revealed that the SLC43A2 mRNA level was significantly elevated in tumors and positively correlated with YAP mRNA levels (Fig. 4i and Supplementary Fig. 9g–i). Additionally, SLC43A2 was also identified as a prognostic biomarker for HCC patients, where higher expression levels of SLC43A2 were associated with shorter patient survival (Fig. 4j). These observations implicated SLC43A2 as a transcription target of YAP in human cells.
As a transcriptional coactivator, YAP needs to engage with the TEAD family in order to activate target gene expression33. YAP-S94A, a mutant defective in TEAD binding, failed to stimulate SLC43A2 expression (Fig. 4k). Similarly, silencing TEAD4 with siRNA prevented YAP-induced upregulation of SLC43A2, implying that YAP needs to collaborate with TEAD4 to regulate SLC43A2 expression (Fig. 4l and Supplementary Fig. 9j). We then utilized the JASPAR database to analyze the putative binding sequences of TEAD4 in the region 2 kb upstream of the SLC43A2 gene and identified 2 potential TEAD4-binding sequences (Supplementary Fig. 9k). Chromatin immunoprecipitation (ChIP)—quantitative PCR (qPCR) assays demonstrated that both YAP and TEAD4 bound to these two binding sites in Huh-7 cells (Fig. 4m, n). Luciferase reporter assays further showed that YAP overexpression strongly elevated luciferase activity in 293 T cells transfected with SLC43A2 reporter plasmids containing the wild-type TEAD4-binding sites, but not in cells transfected with the mutation reporter plasmids (Fig. 4o). Collectively, these data confirmed that YAP–TEAD4 activates the transcription of SLC43A2 by directly binding to specific responsive elements.
Next, we proceeded to investigate the relationship between YAP R124 methylation and the expression of SLC43A2. Compared with YAP-overexpressed cells, the expression of SLC43A2 and the level of methionine were decreased in YAP-R124K-overexpressed Huh-7 cells (Supplementary Fig. 9l, m). Compared with YAP-S127A-overexpressed cells, the expression of SLC43A2 and the level of methionine were more effectively upregulated in YAP-R124F-overexpressed cells (Supplementary Fig. 9l, m). Furthermore, western blot analysis revealed that the YAP-R124K mutation abolished the stimulatory effect of YAP on SLC43A2 expression (Supplementary Fig. 9n). These results suggested that R124 asymmetric dimethylation of YAP further promotes YAP-induced SLC43A2 expression and methionine uptake.
The positive feedback between YAP R124 methylation and SLC43A2 persists in drug resistance
As SLC43A2 is an important methionine transporter, we then evaluated the effect of SLC43A2 on YAP R124 methylation. The loss-of-function screening data showed that knockdown of SLC43A2 markedly diminished the level of YAP R124me2a stimulated by anticancer drugs, whereas knockdown of other SLC family genes only slightly decreased the level of YAP R124me2a (Fig. 5a, b). IHC evaluation of paraffin-embedded HCC tissues also showed a strong positive correlation between the levels of YAP R124me2a and SLC43A2 proteins, with R2 = 0.67 by Spearman’s coefficient analysis (Fig. 5c). We also found SLC43A2 knockdown decreased the mRNA levels of CTGF and CYR61 (Fig. 5d, e). In addition, SLC43A2 levels positively correlated with the transcripts of YAP downstream genes in HCC patients from TCGA (Fig. 5f).
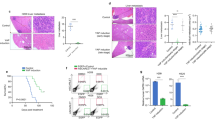
a, b The level of YAP R124 methylation in Huh-7 cells transfected with indicated siRNA and treated with sorafenib (0.75 μM) for 48 h (a) and HCT 116 cells (b) transfected with indicated siRNA and treated with oxaliplatin (4 μM) for 48 h. The data represents the relative ratio of YAP R124me2a to YAP. c Representative images of the level of SLC43A2 and YAP R124me2a in HCC tissues (left panel) and the clinical relevance between the level of SLC43A2 and YAP R124me2a was determined (right panel). n = 28. Scale bar = 50 μm. d, e The transcriptional levels of SLC43A2, YAP, and YAP target genes in Huh-7 cells (d) and HCT 116 cells (e) transfected with si-SLC43A2 for 72 h. f Heatmap from TCGA dataset analysis depicts differential expression of YAP target genes in HCC patients with high and low expression of SLC43A2. g, h The level of SLC43A2 in Huh-7 cells treated with sorafenib (0.75 μM) (g) and HCT 116 cells treated with oxaliplatin (4 μM) (h). The data represents the relative ratio of SLC43A2 to GAPDH. i The cell viability of YAP-, YAP-S127A-, or YAP-R124F-overexpressed Huh-7 cells treated with sorafenib and transfected with si-SLC43A2 for 24 h. j The cell viability of YAP-, YAP-S127A-, or YAP-R124F-overexpressed HCT 116 cells treated with oxaliplatin and transfected with si-SLC43A2 for 48 h. Statistical analyses were performed using two-tailed Spearman’s correlation test (c), two-sided unpaired Student’s t-test (d, e, g, h), or one-way ANOVA with multiple comparisons (i, j). Data were presented as mean ± SD. Data were presented as mean ± SD. n = 3 (d, e, g, h, i, j) or 28 (c) biologically independent samples. The data presented in (a, b, g, h) are representative of three independent experiments. Source data are provided as a Source Data file.
Of note, the expression of SLC43A2 at both mRNA and protein levels was increased 2–3-fold after treatment with sorafenib and oxaliplatin (Fig. 5g, h). Knockdown of SLC43A2 markedly increased the sensitivity of Huh-7 cells to sorafenib and HCT 116 cells to oxaliplatin (Fig. 5i, j). A similar effect was observed in the tumor cells with overexpression of YAP or YAP-S127A (Fig. 5i, j). In contrast, overexpression of YAP-R124F abolished the effect of SLC43A2 knockdown on drug sensitivity in tumor cells (Fig. 5i, j). Taken together, these data suggested that positive feedback between YAP R124 methylation and methionine transporter SLC43A2 enhances drug resistance.
Disrupting the positive feedback between YAP R124 methylation and SLC43A2 improves drug sensitivity
SLC43A2 is one of the system L-type transporters. Given that no specific SLC43A2 inhibitor is available, we cultured cells with BCH, an inhibitor of system L transporters, to inhibit SLC43A2 function34. Similar to SLC43A2 knockdown, BCH blocked YAP R124me2a and downregulated the mRNA expression of YAP target genes (Fig. 6a–c). BCH enhanced the sensitivity of control cells, as well as cells overexpressing YAP and YAP-S127A to sorafenib and oxaliplatin (Fig. 6d, e). However, overexpression of YAP-R124F notably reduced the effect of BCH on drug sensitivity in tumor cells (Fig. 6d, e). We also treated mice bearing YAP-S127A- or YAP-R124F-overexpressed Huh-7 tumors with BCH and sorafenib (Fig. 6f). Treatment with sorafenib alone partially inhibited tumor growth, whereas the combination of BCH and sorafenib markedly inhibited tumor growth in YAP-S127A-overexpressed tumors (Fig. 6g, h). Moreover, the combination treatment decreased the level of YAP R124me2a and the expression of YAP target genes (Supplementary Fig. 10a and Fig. 6i, j), as well as the proliferation of tumor cells in YAP-S127A-overexpressed tumors (Fig. 6k and Supplementary Fig. 10b), suggesting that targeting SLC43A2 in combination with anticancer drugs may be an effective anticancer approach. However, neither sorafenib alone nor the combination of BCH and sorafenib presented any effect on YAP-R124F-overexpressed tumors (Fig. 6g–k), indicating that BCH improves drug sensitivity by disrupting the positive feedback between YAP R124 methylation and SLC43A2.
a The level of YAP R124 methylation in Huh-7 cells and HCT 116 cells treated with BCH (1 mM) for 72 h. The data represents the relative ratio of YAP R124me2a to YAP. b, c The transcriptional levels of YAP and YAP target genes in Huh-7 cells (b) and HCT 116 cells (c) were treated with BCH (1 mM) for 72 h. d The cell viability of YAP-, YAP-S127A-, or YAP-R124F-overexpressed Huh-7 cells treated with sorafenib and BCH (1 mM) for 24 h. e The cell viability of YAP-, YAP-S127A-, or YAP-R124F-overexpressed HCT 116 cells treated with oxaliplatin and BCH (1 mM) for 48 h. f Schematic representation of the experimental design in Balb/c nude mice to measure the combination effect of BCH and sorafenib in stable YAP-R124F- or YAP-S127A-overexpressed cells. n = 5 per group. g Representative image of tumor nodules in different conditions described in Fig. 6f. h The statistical analysis of tumor weight in different conditions is described in Fig. 6f. i, j The expression of CTGF (i) and CYR61 (j) was analyzed in different conditions described in Fig. 6f. k The quantitation of ki-67 staining in tumors from different groups was analyzed. Statistical analyses were performed using two-sided unpaired Student’s t-test (b, c) or one-way ANOVA with multiple comparisons (d, e, h–k). Data were presented as mean ± SD. n = 3 (b–e) or 5 replicates (h–k) biologically independent samples. The data presented in (a) are representative of three independent experiments. Source data are provided as a Source Data file.
Discussion
The potential of methionine to enhance drug resistance has been verified in many tumor models. The high SAM consumption rate in tumor cells is the primary reason for methionine addiction35. However, there remains little understanding of the mechanism by which methionine-induced protein methylation affects drug resistance. In this study, we found methionine induces resistance to sorafenib and oxaliplatin by elevating PRMT1-catalyzed methylation of YAP R124. We also identified a target gene of YAP, SLC43A2, which is responsible for the uptake of methionine in cancer cells. Furthermore, we reveal positive feedback between YAP R124me2a and SLC43A2 during drug resistance. Disruption of this positive feedback results in the sensitization of YAP-related drug resistance.
Previous studies have indicated that phosphorylation and methylation of YAP contribute to drug resistance. Researchers found that the phosphorylation level of YAP S127 was decreased after sorafenib intervention36, and inhibition of YAP phosphorylation induced castration resistance in prostate cancer cells and lenvatinib resistance in HCC15,37. YAP-K342 methylation catalyzed by SET domain containing 1A (SET1A) promotes sorafenib resistance and is reversely correlated with cancer survival16,17. In this study, we found methionine-induced YAP R124me2a promoted YAP-related drug resistance in HCC and CRC. Indeed, YAP-R124F-overexpressed cells showed greater resistance to drug therapy than YAP-S127A-overexpressed cells. Moreover, methionine deprivation markedly reduced YAP-S127A-induced drug resistance but failed to alter YAP-R124F-induced drug resistance in tumor cells. These findings imply that YAP R124 methylation plays a more crucial role than S127 phosphorylation in the process of drug resistance.
Previous studies have indicated YAP is involved in amino acid metabolism in cancer cells. YAP stimulates the metabolism of glutamine, leucine, tryptophan, threonine, and phenylalanine by enhancing the expression of crucial enzymes, including GLS1, GOT1, and PSAT124,38, as well as related transporters, including SLC1A5, SLC7A5, and SLC38A125,26,39,40. Metabolomics data suggest that YAP may be involved in methionine metabolism27,29, but the specific mechanism has not been reported. Our RNA sequencing analysis suggested that YAP increases methionine levels by promoting methionine uptake rather than altering the process of methionine synthesis or its consumption. Moreover, our identification of YAP as a direct upstream regulator of SLC43A2, the major transporter for methionine import, provides important insights into the regulation of methionine metabolism in tumors.
Previous studies have demonstrated that SLC43A2, the major methionine transporter in tumor cells, is highly expressed in multiple human cancers and plays a crucial role in protein methylation34,41. In this study, we found that SLC43A2 was highly expressed and associated with the level of YAP R124me2a in HCC and CRC. By integrating the results that methionine increases the level of YAP R124me2a and YAP elevates intracellular methionine levels through upregulation of the methionine transporter SLC43A2, our study revealed positive feedback between YAP R124 methylation and methionine transporter SLC43A2. Indeed, we found that disruption of this positive feedback by BCH downregulated the level of YAP R124me2a, inhibited the expression of YAP target genes and enhanced YAP-related drug sensitivity. However, BCH is an inhibitor of system L-type transporters, which include SLC7A5, SLC7A8, SLC43A1, and SLC43A242. Among these, SLC7A5 and SLC43A2 have been demonstrated to be upregulated by YAP. Considering that SLC7A5 has minimal effect on YAP R124me2a, we proposed that BCH inhibits YAP R124me2a and alleviates YAP-induced drug resistance by suppressing the activity of SLC43A2. Nevertheless, it is valuable to develop an approach that specifically targets SLC43A2 to enhance the drug sensitivity. A nanoplatform has been established by encapsulating a CRISPR plasmid into Mn/Zn bimetallic metal-organic framework nanoparticles to downregulate SLC43A2 expression43, yet it remains far from clinical application.
Apart from its biological significance, our work may have relevant applications in the clinical management of cancer patients. There is a growing fascination with the therapeutic potential of a methionine-restricted diet for cancer treatment. However, no current cancer clinical trials combine drug therapy with a methionine-restricted diet. The feasibility of achieving beneficial effects on drug resistance through a methionine-restricted diet without inducing systemic toxicities remains a topic of debate3. So, what kind of patients are suitable for the combination of methionine-restricted diet and drug therapy? Our study has important translational implications as it highlights that the combination strategy of methionine restriction with sorafenib or oxaliplatin treatment based on the difference in YAP R124me2a level or SLC43A2 expression for patient selection may produce promising clinical benefits.
In summary, we found methionine promotes sorafenib and oxaliplatin resistance by improving the level of YAP R124me2a in cancer cells. The intracellular methionine levels are elevated in cancer cells due to increased methionine import by YAP-induced SLC43A2 expression. Furthermore, silencing the expression of SLC43A2 decreased YAP R124 methylation. Our work demonstrated positive feedback between YAP R124 methylation and the methionine transporter SLC43A2 during drug resistance. On this basis, disruption of this positive feedback has potential application value for improving tumor treatment outcomes (Supplementary Fig. 11).
Methods
All experiments were conducted in accordance with the guidelines of the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and were approved by Scientific Investigation Board of Naval Medical University. Every precaution was taken to minimize suffering, and the number of animals used in the experiments.
Mouse models
The 6–7-week-old C57BL/6J mice and 4–5-week-old Balb/c nude mice, purchased from Shanghai BK/KY Biotechnology Company (Shanghai, China), were housed under specific pathogen-free conditions. To avoid the influence of estrogen, all experiments used male mice.
To establish the xenograft model, tumor cells suspended in 100 μl of phosphate-buffered saline (PBS) were injected into the axilla of each mouse. Once the tumor volumes reached approximately 100 mm3, mice were randomly assigned to different experimenting groups, with no significant differences in tumor volume among the groups. The length and width of the tumors were measured every 2 days, and the tumor volumes were calculated using the formula: (length × width2)/2. TGI (tumor growth inhibition value) calculation was based on tumor weight, and the formula is as follows: TGI% = (1 − Tumor Weight of Treated Group/Average Tumor Weight of Control Group) × 100%. In cases where the tumor volume exceeded 2000 mm3, the mice were humanely euthanized for welfare reasons. At the experimental endpoint, the mice were humanely sacrificed, and their tumors were collected and weighed.
To investigate the role of MS023 in sorafenib resistance in vivo, Huh-7 cells (1 × 106) were injected into the right axilla of each mouse. The mice received daily intraperitoneal injections of sorafenib (MCE, USA, 30 mg/kg/day) and MS023 (MCE, 30 mg/kg/day) for 14 days. To assess whether methionine deprivation or BCH enhanced the efficacy of sorafenib by blocking YAP R124me2a, YAP-R124F-overexpressed Huh-7 cells (1.5 × 106) were injected into the right axilla, while YAP-S127A-overexpressed Huh-7 cells (1.5 × 106) were injected into the left axilla. The methionine-restricted diet containing 0.12% methionine (w/w, catalog no. A11051301) and the control diet containing 0.86% methionine (w/w, catalog no. A11051302) were purchased from Xietong Pharmaceutical Bioengineering (Jiangsu, China). BCH (Targetmol, USA) was administered via tail vein injection at a dose of 100 mg/kg every other day. Sorafenib was given via intraperitoneal injection at a dose of 20 mg/kg daily. To assess whether methionine deprivation enhanced the efficacy of oxaliplatin by blocking YAP R124me2a, YAP-R124F-overexpressed HCT 116 cells (5 × 106) were injected into the right axilla of mice and YAP-S127A-overexpressed HCT 116 cells (5 × 106) were injected into the left axilla. Oxaliplatin was administered via intraperitoneal injection at a dose of 5 mg/kg daily. In a syngeneic cancer model, YAP-R124F-overexpressed Hepa1-6 cells and YAP-S127A-overexpressed Hepa1-6 (1 × 106) were subcutaneously injected into C57BL/6J mice. Sorafenib was administered via intraperitoneal injection at a daily dose of 10 mg/kg.
Cell lines and cell culture
The Huh-7 (TCHu182) and SGC7901 (TCHu 46) cell lines were purchased from the National Collection of Authenticated Cell Cultures (Shanghai, China). The HCT 116 (CCL-247), Hepa1-6 (CRL-1830), A549 (CRM-CCL-185), and HEK 293A/293T (CRL-3216) cell lines were obtained from American Type Culture Collection (ATCC, Manassas, VA). Huh-7, Hepa1-6, A549, SGC7901, and HEK 293A/293T cells were cultured in Dulbecco’s Modified Eagle Medium (GIBCO, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (GIBCO). HCT 116 cells were cultured in RPMI-1640 medium (GIBCO) with 10% FBS. Cells were maintained at 37 °C in a humidified atmosphere containing 5% CO2. Regular testing for mycoplasma contamination was conducted every 2 weeks using a Mycoalert detection kit (Lonza, Swiss).
Human HCC tissues
HCC tissues from patients with or without sorafenib treatment (n = 191, cohort 1) and HCC tissues for the analysis of the correlation between SLC43A2 and YAP R124me2a (n = 28, cohort 2) were obtained with informed consent and in accordance with guidelines of Institutional Review Board and the Declaration of Helsinki at Eastern Hepatobiliary Surgery Hospital (Shanghai, China). The clinical characteristics of Cohort 1 and Cohort 2 are presented in Supplementary Data 1 and Supplementary Data 2, respectively. Samples from both females and males were used in this study. Sex and gender information was not taken into consideration for the statistical analyses of this study, as it was not the focus of the research.
Methionine restriction treatment
Medium without methionine was prepared using methionine-deficient DMEM (Bioagrio, USA) or RPMI-1640 (Bioagrio) medium supplemented with 10% dialyzed FBS (Bioagrio). Medium with methionine was prepared by adding 200 μM L-Methionine (Sigma-Aldrich, USA) to the DMEM medium and 100 μM methionine to the RPMI-1640 medium, according to the complete medium formula. Cells were seeded with a complete medium and then treated with a methionine-deficient medium or control medium for 24 to 72 h. To explore the effects of SAM on YAP methylation, cells were cultured in a methionine-deficient medium and then treated with 50 μM SAM (Sigma-Aldrich) for 48 h.
Gene overexpression, silencing, and transfection
The plasmids were synthesized and cloned into appropriate vectors at the Beijing Genomics Institute (Beijing, China). Small interfering RNAs (siRNAs) were synthesized from GenePharma (Suzhou, China). For transfection with plasmid or siRNA, cells were seeded at 50% confluence in six-well plates one day ahead and transfected with LipofectamineTM 2000 (Thermo Fisher Scientific, 11668019, USA) following the manufacturer’s protocol. The final concentration of all siRNAs utilized in this study was 20 nM. The sequences of siRNAs used in this study were provided in Supplementary Table 1.
Lentivirus
The full-length coding sequences of wild-type or mutant YAP were inserted into the pCDH-CMV-MCS-EF1-copGFP vector (System Biosciences) in frame with a v5 tag. We generated lentivirus particles by co-transfecting HEK 293T cells with the packaging plasmid psPAX2 (Addgene, USA), envelope plasmid pMD2.G (Addgene), and the expression plasmid following the manufacturer’s instructions. The lentiviral supernatant was harvested at 48 and 72 h after transfection and filtered through a 0.45 μm filter (Sigma-Aldrich). The filtered viruses were used immediately or stored in cryovials at −80 °C until further use.
Cell counting kit-8 (CCK-8) assay
Cells were plated at a density of 5000 cells per well in 100 μl of medium in 96-well plates. Once the cells had been attached, they were subjected to different treatments using medium containing 2% FBS for 24–72 h. Subsequently, 10 μl of CCK-8 solution (Dojindo, Kumamoto, Japan) was applied to each well, and the plates were incubated at 37 °C for 1 h. The absorbance at 450 nm was recorded to evaluate cell viability.
Apoptosis staining
Apoptosis was examined by flow cytometric analysis. Cells were washed with 1× binding buffer and stained with Annexin V and PI (BioLegend, San Diego, USA) in 1× binding buffer in the dark for 15 min, following the manufacturer’s protocol. All samples were acquired on Aligent NovoExpress (Aligent, California, USA) and analyzed using FCS express V7.22 (De Novo Software, Pasadena, USA).
RNA isolation and qPCR analysis
Total RNA was extracted from cells or tissues using RNAiso Plus Reagent (Takara, Japan). 1 μg of total RNA was reversely transcribed to generate first-strand cDNA using SuperScript III reverse transcriptase (Invitrogen). Real-time PCR was conducted with TB Green Premix Ex Taq™ II (Takara) on an ABI Step One Real-Time Detection System (Life Technologies) according to manufacturer instructions. The relative expressions of different genes were normalized to GAPDH. The primer sequences are provided in Supplementary Table 1.
SDS-PAGE and immunoblot analysis
The western blot was performed using the method previously described44. In brief, the whole proteins were extracted using 2× sodium dodecyl sulfate (SDS) protein lysis buffer (4% SDS, 20% glycerol, 100 mM Tris-HCl [pH 6.8]) containing PMSF, and the concentration was quantified using BCA Protein Assay Kit (Beyotime, Shanghai, China). 60 μg of proteins were separated by 10% SDS-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes (Millipore, Hercules, CA). The membranes were blocked in 5% skim milk for 1 h and then incubated with the primary antibodies at 4 °C overnight. After incubation with fluorescent-conjugated secondary antibodies for 1 h at room temperature, signals were detected using an Odyssey Infrared Imaging System (LI-COR). The gray values of immunoblots were measured using Image J software (version 2.1). The antibodies used are provided in Supplementary Table 2.
Dual-luciferase reporter assay
The transcriptional activity of YAP mutations was evaluated in HEK 293T cells infected with wild-type or mutant YAP lentivirus and transfected with YAP–TEAD responsive reporter (8×GTIIC, 100 ng) (RIKEN, Japan). The binding site of YAP–TEAD4 on SLC43A2 promoter was evaluated in HEK 293T cells transfected with wild-type or mutant SLC43A2 promoter vectors (100 ng) in a 96-well tissue culture plate. All cells were co-transfected with the pRL-SV40 vector (Promega, USA, 10 ng) to normalize the data. The assays were conducted using the Dual-Luciferase Reporter Assay System (Promega). Each experiment was repeated at least three times.
Determination of methionine and SAM
To determine methionine levels in the peripheral blood of mice fed with dietary methionine restriction, 500 μl of blood was collected and centrifuged at 1000 × g for 10 min to obtain plasma. To assess the effect of YAP on methionine, Huh-7 cells, and HCT 116 cells were infected with YAP lentivirus for 96 h. To assess the effect of SLC on methionine uptake, Huh-7 cells were transfected with siRNAs for 24 h, cultured in methionine-deprived medium for another 24 h, then exposed to 200 μM methionine for 90 min. Cells were washed with cold PBS and lysed through three freeze-thaw cycles. Methionine concentrations (mlBio, Shanghai, China) were evaluated by ELISA kit according to the manufacturer’s instructions. SAM concentrations were detected by ELISA kit (Jianglai Biology, Shanghai, China) and ultra-performance liquid chromatography coupled with tandem mass spectrometry (UPLC-MS/MS) on Shanghai Metabo-Profile Biotechnology. In brief, to extract SAM from cells for UPLC-MS/MS, 100 μl cold methanol was added to each sample (1 × 107 cells). The samples were vigorously vortexed, ultrasonicated twice for 10 min at 4 °C to ensure the cell pellet was completely disrupted, and centrifuged at 14,000 × g for 20 min at 4 °C. 50 μl supernatant of each sample was transferred to a 96-well plate and mixed with 75 μl Vitamin C solution for UPLC-MS/MS. The samples were analyzed using an ACQUITY UPLC-Xevo TQ-S (Waters Corp., MA, USA) coupled with MassLynx system (version 4.1, Waters). Each group has 3 biological replicates.
GSH detection
The GSH Content Assay Kit (Solarbio, Beijing, China) was used to evaluate the GSH level. The measurements were carried out according to the manufacturer’s protocols. Briefly, 1 × 107 cells were collected and lysed in PBS. The absorbance was measured at 412 nm, and the GSH content was calculated based on the GSH standard curve.
Immunohistochemistry
For immunohistochemistry, deparaffinization of 4-μm thick paraffin-embedded tissue sections was followed by treatment with EDTA buffer (ph = 8.0) at 100 °C for 15 min to retrieve the antigen. The antibodies used are provided in Supplementary Table 2. Staining was visualized using DAB chromogen. The representative image was acquired by microscope (Olympus, Tokyo, Japan). The proportion of ki-67 positive cells among the total cell count was quantified as the ki-67 positive cell rate. Protein expression levels were evaluated based on the extent and intensity of staining using ImageJ Pro Plus (version 6.0).
ChIP-qPCR
Cells were initially cross-linked with 2 mM disuccinimidyl glutarate for 30 min, followed by the addition of 1% formaldehyde for another 15 min. The cross-linking reaction was then terminated by the addition of 1.25 M glycine for 10 min. Subsequently, sonication was performed to shear DNA into 200–1000-bp fragments. Protein-DNA complexes were then precipitated using an indicated antibody (Supplementary Table 2). The DNA extraction was carried out using the phenol–chloroform–isoamyl alcohol method. The CYR61 primer was used as a positive control. The primers are listed in Supplementary Table 1.
Bioinformatics analyses
The TCGA Liver hepatocellular carcinoma (LIHC) and Colon adenocarcinoma (COAD) data, represented as transcripts per million (TPM), were obtained from UCSC Xena (https://xenabrowser.net/datapages/). The gene tissue expression (GTEx) dataset was integrated with the TCGA data for normal samples. The webtool GEPIA was utilized to conduct a Spearman correlation analysis between YAP and SLC43A2, as well as an overall survival analysis of SLC43A2. The correlation between PRMT1 or SLC43A2 with YAP target genes in the TCGA dataset was generated using R Bioconductor (https://www.bioconductor.org/Bioconductor).
Statistics and reproducibility
No methods were used to predetermine sample size, and no blinding or randomization was employed for data analysis. No data were excluded from the analyses. The data were presented as the mean ± standard deviation (SD) or the mean ± standard error of the mean (SEM). Inter-group data were analyzed using an unpaired two-tailed t-test or one-way ANOVA test, and tumor growth and growth curves were analyzed using two-way ANOVA. The expression of SLC43A2 in LIHC and COAD was calculated using the Wilcoxon test. The correlation coefficient was calculated using a two-tailed Spearman’s correlation test. Survival data were analyzed using the Kaplan–Meier statistical method. The detailed statistical methods and the sample sizes (n) are indicated in the figure legends. All data analyses were performed in GraphPad Prism version 9.5 (GraphPad Inc., CA, USA). Significant differences were considered at P-values below 0.05 and were indicated in the figures.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNA sequencing data generated in this study have been deposited in the GEO database under accession code GSE263889. The target region sequencing data of HCC driver genes mutation in this study of peer review file has been deposited in the SRA database under accession code PRJNA1163846. The raw data of UPLC-MS/MS in this study have been deposited in the MetaboLights database under accession code MTBLS11903. All the other data which support the findings of this study are available with in the article, supplementary information and source data. Source data are provided in this paper.
References
Parkhitko, A. A., Jouandin, P., Mohr, S. E. & Perrimon, N. Methionine metabolism and methyltransferases in the regulation of aging and lifespan extension across species. Aging Cell 18, e13034 (2019).
Li, Z. et al. Methionine metabolism in chronic liver diseases: an update on molecular mechanism and therapeutic implication. Signal Transduct. Target Ther. 5, 280 (2020).
Sanderson, S. M., Gao, X., Dai, Z. & Locasale, J. W. Methionine metabolism in health and cancer: a nexus of diet and precision medicine. Nat. Rev. Cancer 19, 625–637 (2019).
Cunningham, A. et al. Dietary methionine starvation impairs acute myeloid leukemia progression. Blood 140, 2037–2052 (2022).
Gao, X. et al. Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature 572, 397–401 (2019).
Fang, L. et al. Methionine restriction promotes cGAS activation and chromatin untethering through demethylation to enhance antitumor immunity. Cancer Cell 41, 1118–1133.e1112 (2023).
Li, T. et al. Methionine deficiency facilitates antitumour immunity by altering m6A methylation of immune checkpoint transcripts. Gut 72, 501–511 (2023).
Xue, Y. et al. Intermittent dietary methionine deprivation facilitates tumoral ferroptosis and synergizes with checkpoint blockade. Nat. Commun. 14, 4758 (2023).
Wang, N., Ma, T. & Yu, B. Targeting epigenetic regulators to overcome drug resistance in cancers. Signal Transduct. Target Ther. 8, 69 (2023).
Zhuang, H. et al. The role of m6A methylation in therapy resistance in cancer. Mol. Cancer 22, 91 (2023).
Hong, A. W., Meng, Z. & Guan, K.-L. The Hippo pathway in intestinal regeneration and disease. Nat. Rev. Gastroenterol. Hepatol. 13, 324–337 (2016).
Fu, M. et al. The Hippo signalling pathway and its implications in human health and diseases. Signal Transduct. Target Ther. 7, 376 (2022).
Ciamporcero, E. et al. YAP activation protects urothelial cell carcinoma from treatment-induced DNA damage. Oncogene 35, 1541–1553 (2015).
Garcia-Rendueles, M. E. R. et al. Yap governs a lineage-specific neuregulin1 pathway-driven adaptive resistance to RAF kinase inhibitors. Mol. Cancer 21, 213 (2022).
Yan, Y.-C. et al. Diacylglycerol lipase alpha promotes hepatocellular carcinoma progression and induces lenvatinib resistance by enhancing YAP activity. Cell Death Dis. 14, 404 (2023).
Fang, L. et al. SET1A-mediated mono-methylation at K342 regulates YAP activation by blocking its nuclear export and promotes tumorigenesis. Cancer Cell 34, 103–118.e109 (2018).
Wu, J., Chai, H., Li, F., Ren, Q. & Gu, Y. SETD1A augments sorafenib primary resistance via activating YAP in hepatocellular carcinoma. Life Sci. 260, 118406 (2020).
Oudhoff, M. J. et al. Control of the Hippo pathway by set7-dependent methylation of Yap. Dev. Cell 26, 188–194 (2013).
Qian, H. et al. SRY-Box transcription factor 9 triggers YAP nuclear entry via direct interaction in tumors. Signal Transduct. Target Ther. 9, 96 (2024).
Koo, J. H. & Guan, K.-L. Interplay between YAP/TAZ and Metabolism. Cell Metab. 28, 196–206 (2018).
Kuo, C.-C. et al. Metastatic colorectal cancer rewrites metabolic program through a Glut3-YAP-dependent signaling circuit. Theranostics 9, 2526–2540 (2019).
Gao, Y. et al. TNFα-YAP/p65-HK2 axis mediates breast cancer cell migration. Oncogenesis 6, e383–e383 (2017).
Feng, Y. et al. YAP promotes ocular neovascularization by modifying PFKFB3-driven endothelial glycolysis. Angiogenesis 24, 489–504 (2021).
Bertero, T. et al. Vascular stiffness mechanoactivates YAP/TAZ-dependent glutaminolysis to drive pulmonary hypertension. J. Clin. Invest 126, 3313–3335 (2016).
Edwards, D. N. et al. The receptor tyrosine kinase EphA2 promotes glutamine metabolism in tumors by activating the transcriptional coactivators YAP and TAZ. Sci. Signal. 10, eaan4667 (2017).
Park, Y. Y. et al. Yes‐associated protein 1 and transcriptional coactivator with PDZ‐binding motif activate the mammalian target of rapamycin complex 1 pathway by regulating amino acid transporters in hepatocellular carcinoma. Hepatology 63, 159–172 (2015).
Li, H. et al. YAP/TAZ drives cell proliferation and tumour growth via a polyamine–eIF5A hypusination–LSD1 axis. Nat. Cell Biol. 24, 373–383 (2022).
Pan, Q. et al. The ZMYND8-regulated mevalonate pathway endows YAP-high intestinal cancer with metabolic vulnerability. Mol. Cell 81, 2736–2751.e2738 (2021).
Murakami, S. et al. A Yap-Myc-Sox2-p53 regulatory network dictates metabolic homeostasis and differentiation in Kras-driven pancreatic ductal adenocarcinomas. Dev. Cell 51, 113–128.e119 (2019).
Nguyen, C. D. K. & Yi, C. YAP/TAZ signaling and resistance to cancer therapy. Trends Cancer 5, 283–296 (2019).
Zhao, Z. et al. CARM1-mediated methylation of ASXL2 impairs tumor-suppressive function of MLL3/COMPASS. Sci. Adv. 8, eadd3339 (2022).
Hao, Y., Chun, A., Cheung, K., Rashidi, B. & Yang, X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 283, 5496–5509 (2008).
Zhao, B. et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971 (2008).
Bian, Y. et al. Cancer SLC43A2 alters T cell methionine metabolism and histone methylation. Nature 585, 277–282 (2020).
Wang, Z. et al. Methionine is a metabolic dependency of tumor-initiating cells. Nat. Med. 25, 825–837 (2019).
Sun, T., Mao, W., Peng, H., Wang, Q. & Jiao, L. YAP promotes sorafenib resistance in hepatocellular carcinoma by upregulating survivin. Cell Oncol. 44, 689–699 (2021).
Li, Q. et al. MYBL2 disrupts the Hippo-YAP pathway and confers castration resistance and metastatic potential in prostate cancer. Theranostics 11, 5794–5812 (2021).
Yang, C. S. et al. Glutamine‐utilizing transaminases are a metabolic vulnerability of TAZ/YAP‐activated cancer cells. EMBO Rep. 19, e43577 (2018).
Hansen, C. G., Ng, Y. L. D., Lam, W.-L. M., Plouffe, S. W. & Guan, K.-L. The Hippo pathway effectors YAP and TAZ promote cell growth by modulating amino acid signaling to mTORC1. Cell Res. 25, 1299–1313 (2015).
Ong, Y. T. et al. A YAP/TAZ-TEAD signalling module links endothelial nutrient acquisition to angiogenic growth. Nat. Metab. 4, 672–682 (2022).
Pandit, M. et al. Methionine consumption by cancer cells drives a progressive upregulation of PD-1 expression in CD4 T cells. Nat. Commun. 14, 2593 (2023).
Wang, W. & Zou, W. Amino acids and their transporters in T cell immunity and cancer therapy. Mol. Cell 80, 384–395 (2020).
Huang, Y. et al. A bimetallic nanoplatform for STING activation and CRISPR/Cas mediated depletion of the methionine transporter in cancer cells restores anti-tumor immune responses. Nat. Commun. 14, 4647 (2023).
Cai, L.-Y. et al. Targeting p300/CBP attenuates hepatocellular carcinoma progression through epigenetic regulation of metabolism. Cancer Res. 81, 860–872 (2021).
Acknowledgements
We thank Prof. Fa-Xing Yu at Children’s Hospital of Fudan University for providing YAP-KO 293A cells and Prof. Jin Ding at Naval Medical University for providing the HCC tissue array with sorafenib administration. This work was supported by the National Natural Science Foundation of China (grant 82072641 to X.Z., grant 82030021 to W.F.X., grant 82070620, 82370598 to H.Q.), the National Key R&D Program of China (grant 2023YFC2507500 to W.F.X.). This work was also supported by the Ministry of Education's Key Laboratory on Signaling Regulation and Targeting Therapy of Liver Cancer and Shanghai's Key Laboratory of Hepato-biliary Tumor Biology.
Author information
Authors and Affiliations
Contributions
X.H. conducted the experiments, analyzed the data, and drafted the manuscript; C.H. performed animal experiments and analyzed the data; H.Q., C.D., and F.L. performed the experiments; H.H. and S.W. provided assistance in animal experiments; S.L. provided assistance in IHC; X.Z. and W.X. conceived, designed, and supervised the study and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Xiaoling Li, Fengyuan Tang, Ping Wang, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hong, XL., Huang, CK., Qian, H. et al. Positive feedback between arginine methylation of YAP and methionine transporter SLC43A2 drives anticancer drug resistance. Nat Commun 16, 87 (2025). https://doi.org/10.1038/s41467-024-55769-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-55769-8
This article is cited by
-
Posttranslational modifications of YAP/TAZ: molecular mechanisms and therapeutic opportunities
Cellular & Molecular Biology Letters (2025)