Abstract
Echinacoside (ECH), one of the most representative phenylethanoid glycosides (PhGs), has considerable neuroprotective effects and is an effective ingredient in numerous commercial drugs. Here, we elucidate the complete ECH biosynthetic pathway in the medicinal plant Cistanche tubulosa. In total, 14 related genes are cloned and functionally characterized. Two upstream pathways for tyrosol biosynthesis from L-tyrosine are identified: one includes separate decarboxylation, deamination and reduction steps; the other uses microbial-like transamination, decarboxylation and reduction steps. In addition, a distinct downstream assembly process from tyrosol to ECH is revealed that includes sequential glucosylation, acylation, hydroxylation, and rhamnosylation to form acteoside, and ends with a final glucosylation converting acteoside to ECH. Furthermore, the de novo synthesis of 23 PhG derivatives is achieved via the heterologous expression of different combinations of the functional genes in tobacco. Our findings provide insights into the biosynthesis of ECH and a platform for alternative production of complex PhGs.
Similar content being viewed by others
Introduction
Cistanche Herba (CH, Chinese name: Rou Cong Rong), often referred to the dried succulent stem of Cistanche plants (Fig. 1a), is a valuable traditional Chinese medicine known as the “ginseng of the desert”. It has a long history of use in Asian countries because of its pharmacological effects on treating kidney deficiency, impotence, and chronic renal diseases1,2. Phenylethanoid glycosides (PhGs) are the primary chemical components of Cistanche species3. These compounds possess a phenylethanol glucoside skeleton, substituted with various sugar moieties and acyl groups at different positions on the glucose residue (Fig. 1b). Echinacoside (ECH, Fig. 1c) is a representative PhG and one of the most well-studied. It processes pharmacological benefits in the prevention and treatment of neurodegenerative diseases, such as Alzheimer’s disease and Parkinson’s disease4,5. In nature, Cistanche tubulosa (Schenk) Wight, documented in the 2005 Edition of the Chinese Pharmacopoeia, contains up 30% (w/w) ECH5,6. The total glycosides of C. tubulosa, with ECH as the primary ingredient, have been certified by the National Medical Products Administration of China to be used in clinical trials for treating vascular dementia2.
a Cistanche tubulosa plant in desert. b Skeleton of PhG compounds. c Chemical structure of echinacoside (ECH). d Three possible upstream pathways for tyrosol or hydroxytyrosol biosynthesis, including the TyDC-TYO-ADH pathway (pathway 1), AAS-ADH pathway (pathway 2) and TAT-PPDC-ADH pathway (Ehrlich pathway, pathway 3) that are shown in different colours. e Downstream pathways involving the assembling of acyl, hydroxyl, rhamnosyl, and glucosyl modules in the biosynthesis of ECH.
As a holoparasitic medicinal plant grown in desert regions, wild CH resources are substantially impacted by the natural environment and are currently confronted with escalating constraints on both its supply and demand. On the other hand, as a triglycoside that has rhamnosyl, caffeoyl and glucosyl substituents at the 3′-OH, 4′-OH and 6′-OH positions of the hydroxysalidroside backbone, respectively, ECH (Fig. 1c) possesses one of the most complex PhG structures among natural products7. Many other bioactive PhGs, such as salidroside, osmanthuside A (osmA), osmanthuside B (osmB), syringalide A and acteoside, are predicted to be key intermediates in the ECH biosynthetic pathway8. However, the complete biosynthetic pathway of ECH remains unelucidated.
Biosynthesis of ECH is generally considered to involve three primary steps: the formation of tyrosol or hydroxytyrosol as the aglycone (annotated “upstream pathway” in Fig. 1d); the incorporation of the central glucose moiety; and the assembly of the 4′-acyl, 3-hydroxyl, 3′-rhamnosyl and 6′-glucosyl groups (annotated “downstream pathway” in Fig. 1e)9. 4-Hydroxyphenylacetaldehyde (4-HPAA) is the direct biosynthetic precursor to form tyrosol through reduction catalysed by alcohol dehydrogenase (ADH). However, reports on the upstream pathway of 4-HPAA biosynthesis are controversial, especially in PhGs-producing plants10. For many years, 4-HPAA was considered to be synthesized from tyrosine through separate decarboxylation (catalysed by tyrosine decarboxylase (TyDC)) and oxidative deamination (catalysed by tyramine oxidase (TYO)) reactions, referred as TyDC-TYO-ADH pathway (pathway 1 in Fig. 1d). However, only a limited number of TyDC genes involved in PhG biosynthesis have been identified thus far11,12,13, and no TYO genes have been functionally validated in PhGs-producing plants10,14. Torrens-Spence et al. reported a pyridoxal phosphate (PLP)-dependent 4-HPAA synthase (4-HPAAS) named Rr4HPAAS from Rhodiola rosea that can directly convert tyrosine into 4-HPAA through a combination of “decarboxylation-deamination” and then into tyrosol via ADH, thus providing the only fully elucidated upstream AAS-ADH pathway (pathway 2 in Fig. 1d)10. Additionally, in microbes, tyrosol can be synthesized from tyrosine through the sequential actions of tyrosine aminotransferase (TAT) and phenylpyruvate decarboxylase (PPDC), referred to as Ehrlich pathway (pathway 3 in Fig. 1d)15,16. Genes encoding TATs have also been identified17,18, however, 4-hydroxyphenylpyruvic acid (4-HPPA) decarboxylase genes have not yet been identified in plants19,20. Therefore, the roles of these three potential upstream pathways in PhG biosynthesis and which pathway occurs in Cistanche are still unknown. In terms of the downstream pathway (Fig. 1e), a sequential assembly process including p-coumaroylation, rhamnosylation and hydroxylation from salidroside to acteoside has been reported in Ligustrum robustum, Rehmannia glutinosa and Sesamum indicum21. An alternative route for introducing the dihydroxy groups of acteoside was also identified in S. indicum where an efficient glucosyltransferase that catalyses hydroxytyrosol to hydroxysalidroside and an acyltransferase that transfers the caffeoyl group to hydroxysalidroside to form calceolarioside A (calA) were discovered22, suggesting that multiple routes may be involved in the biosynthesis of PhGs within plants. Recently, Yao et al. reported the enzymatic synthesis of PhGs through multienzyme cascades using enzymes of different origin and engineered variants23. However, the complete ECH biosynthetic pathway, particularly several enigmatic processes such as the controversial upstream pathway and the unclear downstream assembly procedures, as well as the natural 6′-O-glucosyltransferase, remains to be elucidated.
In this work, we elucidate the complete ECH biosynthetic pathway from L-tyrosine in C. tubulosa. A total of 14 related enzymes are identified (Supplementary Table 1). Three potential upstream pathways for tyrosol and hydroxytyrosol biosynthesis are proposed using naturally identified or artificially modified enzymes. A distinct assembly mechanism that utilizes rhamnosylation as the last step for acteoside synthesis, which differs from what has been reported in L. robustum and R. glutinosa21, is elucidated. It indicates the presence of multiple parallel pathways for PhGs biosynthesis in different plants. Additionally, the natural glucosyltransferase responsible for the final glucosylation of acteoside to form ECH is reported. Furthermore, de novo synthesis of 23 structurally diverse PhGs is achieved in tobacco leaves, providing as an effective platform for the production of ECH and various PhGs.
Results and discussion
Identification of glycosyltransferases involved in downstream ECH biosynthetic pathways
Glycosyltransferases were initially explored to elucidate the biosynthetic pathway of ECH (Fig. 2a). A total of 31 candidate genes belonging to the UDP-glycosyltransferase (UGT) family with gene lengths >500 bp and FPKM values >20 were screened from the transcriptome of C. tubulosa. Phylogenetic analysis of their amino acid sequences was conducted using the previously reported “sugar-sugar UGTs” that catalyse sugar chain elongation21 and UGTs that catalyse the glucosylation of tyrosol to form salidroside (Supplementary Data 1)24,25,26,27,28. As shown in Fig. 2b, clades I, II and III contained most of the referenced sugar-sugar UGTs, and clade IV contains both sugar-sugar UGTs and UGTs involved in salidroside biosynthesis. In total, 11 C. tubulosa UGT genes were phylogenetically located in clades I and IV. These genes were selected as candidates for further heterologous expression in Escherichia coli (Supplementary Fig. 1) and subsequent in vitro enzymatic assays.
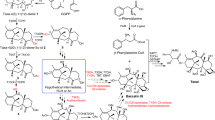
a The identified glycosyltransferases in the downstream biosynthesis pathway of ECH, including CtUGT85A191, UGT85AF12, UGT85AF13, UGT79G13 and UGT73EV1. b Phylogenetic analysis of candidate genes with reported “sugar-sugar UGTs”, which catalyse sugar chain elongation (distributed in clades I–IV labelled in black), and UGTs, which catalyse the glucosylation of tyrosol to form salidroside (located in clade IV labelled in black with green circles). Candidate genes located in clades I–IV for further functional analysis are labelled in red. Information on the referenced genes is summarized in Supplementary Data 1. c Glucosylation of tyrosol (9) to form salidroside (11) or hydroxytyrosol (10) to form hydroxysalidroside (12), catalysed by CtUGT85A191, UGT85AF12 and UGT85AF13 at 1 h or 12 h. d Kinetic parameter determination of CtUGT85A191 towards 9 or 10. e Kinetic parameter determination of UGT85AF12 towards 9 or 10. f Rhamnosylation of calA (18) to form acteoside (20) or osmA (13) to form osmB (14) by CtUGT79G13 at 0.5 h and 12 h. g Kinetic parameter determination of CtUGT79G13 towards 18. h, i Molecular docking of CtUGT79G13–UDP-rhamnose binary complex with calA (h) or osmA (i). j Glucosylation of acteoside (20) to form ECH (21) by CtUGT73EV1. k Kinetic parameter determination of CtUGT73EV1 towards 20. The detection wavelengths were set at 280 nm (c) and 330 nm (f, j), respectively. Kinetic assays were performed in independent triplicates, and data are presented as mean values ± SDs (d and e; g and k). Source data are provided as a Source Data file.
For the glycosylation of tyrosol to generate salidroside, three UGT candidates exhibited corresponding activities. They were further named CtUGT85A191, CtUGT85AF12 and CtUGT85AF13, respectively, by the UGT Nomenclature Committee. These three genes clustered in the same clade and were phylogenetically closely related to genes encoding tyrosol glucosyltransferases in R. rosea. In vitro enzymatic assays revealed that under the same conditions, CtUGT85A191 and CtUGT85AF12 effectively converted tyrosol (9) to salidroside (11, Fig. 2c) in >50% yield within 1 h, whereas CtUGT85AF13 required 12 h to provide a similar conversion rate (Fig. 2c and Supplementary Fig. 2). In addition, unlike salidroside, the aglycone of ECH harbours a hydroxytyrosol moiety. Therefore, we also examined the glucosylation activity of CtUGT85A191, CtUGT85AF12 and CtUGT85AF13 towards hydroxytyrosol (10). Both CtUGT85A191 and CtUGT85AF12 glucosylated hydroxytyrosol (10) to form hydroxysalidroside (12), whose structure was elucidated through nuclear magnetic resonance (NMR) analysis (Supplementary Figs. 3–6), whereas the catalytic activity of CtUGT85AF13 was relatively weak (Fig. 2c). Moreover, the expression profiles of CtUGT85A191 and CtUGT85AF12 were in accordance with the observed ECH accumulation patterns in PEG6000-treated C. tubulosa cell cultures, whereas that of CtUGT85AF13 was not. The apparent KM values of CtUGT85A191 and CtUGT85AF12 for tyrosol (or hydroxytyrosol) were further determined to be 137.70 ± 22.66 µM (288.50 ± 36.81 µM) and 9.92 ± 2.20 µM (75.74 ± 11.95 µM), respectively (Fig. 2d, e; kinetic parameters are summarized in Supplementary Table 2), under the identified optimal conditions (Supplementary Figs. 7 and 8). Both of these UGTs exhibited greater affinity for tyrosol than for hydroxytyrosol, and CtUGT85AF12 demonstrated greater substrate affinity and catalytic efficiency towards tyrosol substrates than CtUGT85A191 did.
Another crucial glycosylation step in ECH biosynthesis is the introduction of a rhamnose moiety into the central glucose through a rhamnose (1 → 3) glucose linkage. Bioinformatics analysis revealed that the enzyme encoded by Unigene8227 (later named CtUGT79G13) shares a close phylogenetic relationship with LrUGT79G7, a known rhamnosyltransferase responsible for converting osmA (13) (Supplementary Fig. 9) into osmB (14), which is further hydroxylated to form acteoside in L. robustum24. However, for the dihydroxyl-type substrate, calA (18), the probable direct precursor of acteoside, LrUGT79G7 showed deficient activity24. In contrast to LrUGT79G7, comprehensive enzymatic assays revealed that CtUGT79G13 could catalyse the rhamnosylation of both osmA (13) and calA (18) in the presence of UDP-rhamnose, with higher activity towards calA (Fig. 2f). When osmA was utilized as the substrate, only a minor product was detected within 30 min. When the reaction time was extended to 12 h, the peak of 14 was prominently observed. Using a reference standard for comparison, 14 was identified as osmB (Supplementary Figs. 10–13). Improved rhamnosylation activity of CtUGT79G13 was observed when calA (18) was used as the substrate, with a conversion rate of 35.7% within 30 min and up to 76.4% after 12 h under the same conditions, affording product 20, whose chemical structure was identified as acteoside through further NMR analysis (Supplementary Figs. 14–17). The kinetic parameters of CtUGT79G13 were further evaluated under the identified optimal conditions (Supplementary Fig. 18), which yielded a KM value of 77.82 ± 6.10 µM, and a kcat/KM value of 5.71 × 10−3 s−1 µM−1 for calA (Fig. 2g and Supplementary Table 2). To investigate the structural basis for substrate preference, we predicted the protein structure of CtUGT79G13 using AlphaFold2 (Supplementary Fig. 19) and further docked osmA and calA into the CtUGT79G13–UDP-rhamnose binary complex. The tyrosol (hydroxytyrosol) and central glucose moieties of osmA and calA exhibited similar conformations when interacting with CtUGT79G13 (Fig. 2h, i), whereas the 3-hydroxyl group of calA formed an extra hydrogen bond with K89 of CtUGT79G13. In addition, the conformation of the caffeoyl moiety of calA was flipped compared with that of osmA, thus facilitating the formation of a hydrogen bond between the 3″-hydroxyl group of calA and residue S82, as well as two hydrophobic interactions between the caffeoyl-aromatic ring and residues A18 and A81 (Fig. 2h and Supplementary Fig. 20), which benefit the binding of calA to CtUGT79G13.
On the basis of the identified activity of CtUGT79G13 in the production of acteoside, the final step in the biosynthesis of ECH was conclusively determined to be the glucosylation of acteoside at the 6′-OH position. All the UGT candidates in clades I and IV (Fig. 2b) were assayed with acteoside as substrate in the presence of UDP-glucose (UDP-Glc). The protein encoded by Unigene11946 specifically recognized acteoside to generate a single product 21 with the same retention time as that of ECH (Fig. 2j). The product peak exhibited an [M − H]− ion at m/z 785.2476 with the predicted formula of C35H46O20 (calcd 785.2510 [M − H]−), an MS2 fragment at m/z 623.2137 [M − H − 162]− and an MS3 fragment at m/z 477.1546 [M − H − 162 − 146]−, which are identical to those of ECH (Supplementary Fig. 21). The structure was further confirmed to be that of ECH based on NMR analysis (Supplementary Figs. 22 and 23). Unigene11946 was subsequently named CtUGT73EV1. Interestingly, CtUGT73EV1 is located in clade IV and is phylogenetically close to both UGT703E1 (a sugar-sugar UGT) and UGT73B6 (a tyrosol glucosylation-related UGT). The KM and kcat/KM values of CtUGT73EV1 with acteoside were further determined to be 135.3 ± 24.06 µM and 0.15 × 10−3 s−1 µM−1, respectively (Fig. 2k) under the identified optimal conditions (Supplementary Fig. 24).
Identification of acyltransferases involved in downstream ECH biosynthetic pathways
The PhGs in C. tubulosa usually feature acyl substituents such as p-coumaroyl, caffeoyl, or feruloyl groups, at the 4′-OH or 6′-OH group of the central glucose moiety29. In plants, such acylation is catalysed by acyltransferases (ATs), which can be divided into two families: BAHD-ATs (named after the first four biochemically characterized enzymes), which use acyl-CoA thioesters as donors30,31, and serine carboxypeptidase-like ATs (SCPL-ATs), which use 1-O-β-glucose esters as donors30,32 (Fig. 3a). Here, we employed multiple strategies to refine the candidate ATs. Initially, a local protein BLAST was conducted using known ATs with a preference for aromatic acyl donors as search templates (Supplementary Data 2). We subsequently assessed the expression levels of the obtained genes in PEG6000-treated C. tubulosa cell cultures through comparative transcriptome sequencing, and the upregulated genes were further screened since drought stress induced by PEG6000 could effectively increase the accumulation of ECH and acteoside in cell suspension cultures of C. tubulosa (Supplementary Fig. 25). Moreover, Murayama et al. discussed the key amino acids for acyl-CoA selectivity using Gt5,3′AT as a template and revealed that caffeoyl/p-coumaroyl-CoA-selective enzymes have Ala/Gly at position 179 and Gly/Ser at position 401 and include Arg neither at position 45 nor 182 (Fig. 3b)33. According to these criteria, six BAHD-AT candidate genes were further screened and named CtAT-A–F (Fig. 3b). These genes were subsequently amplified and heterologously expressed in E. coli. In vitro enzymatic assays were designed orthogonally using PhGs lacking acyl substitutions, including salidroside, forsythoside E and decaffeoylacteoside as substrates and aromatic CoAs, including cinnamoyl-CoA, p-coumaroyl-CoA, feruloyl-CoA and caffeoyl-CoA, as acyl donors. CtAT-E exhibited acylation activity towards salidroside when p-coumaroyl-CoA (Fig. 3c) or caffeoyl-CoA (Fig. 3d) was used as the acyl donor to form the corresponding products 13 and 15, respectively. Two additional products, 15′ and 15″, with the same molecular weight as 15 were also generated; these products were assumed to be formed due to the migration of the caffeoyl moiety22. More noticeable products 14 and 16 (Fig. 3c, d) were observed when UGT79G13 was added to catalyse the coupled rhamnosylation reaction, effectively preventing acyl group migration and increasing the substrate conversion rate. Compounds 13 and 14 were confirmed to be osmA and osmB, respectively, upon comparison with reference standards. Product 15 exhibited an m/z of 461.1455 [M − H]− with the predicted formula C23H26O10, which supported the occurrence of caffeoyl transfer to salidroside. Its rhamnosylated product 16 exhibited an [M − H]− ion at m/z 607.2035 and an MS2 fragment at m/z 445.1752 [M − H − 162]− (Fig. 3e), corresponding to cleavage of the caffeoyl group. This mass spectral information was identical to that of syringalide A-3′-rhamnoside34. Other candidate ATs did not show acylation activity towards the tested PhG substrates (Supplementary Figs. 26 and 27). The above results indicated that CtAT-E could recognize either p-coumaroyl-CoA or caffeoyl-CoA as an acyl donor to acylate salidroside to form osmA or syringalide A, respectively (Fig. 3f), both of which are important intermediates in the biosynthesis of ECH. In addition, acyl-CoA donor degradation was also observed in the reactions generating p-coumaric acid and caffeic acid, which are shown as the predominate peak in Fig. 3c and Fig. 3d, respectively.
a Phylogenetic analysis of the candidate acyltransferases CtAT-A–CtAT-G and reported acyltransferases belonging to the BAHD and SCPL families. Different clades are shown in different colours. The reported acyltransferases that prefer to utilize aromatic acyl donors are listed mainly in the red region. Information on the referenced genes is summarized in Supplementary Data 2. b Multiple sequence alignment of CtAT-A–CtAT-F with Gt5,3′AT. The key residues for acyl-CoA selectivity are highlighted by green triangles. c,d In vitro enzymatic assays of CtAT-E and “CtAT-E + CtUGT79G13” coupled reactions using salidroside (11) as the acyl acceptor and p-coumaroyl-CoA (c) or caffeoyl-CoA (d) as the acyl donor, respectively. The detection wavelength was set at 280 nm. e HRESI-MS and MS2 spectra of the acylated products (13, 15, 15′ and 15″) and their corresponding rhamnosylated products (14 and 16) in negative mode. f Reactions catalysed by CtAT-E and CtUGT79G13 using salidroside (11) as substrate.
Elucidation and identification of upstream biosynthetic pathways of ECH in C. tubulosa
Tyrosol is a common precursor of many important phenolic natural products in plants, including the representative PhGs35. Three potential pathways for tyrosol biosynthesis were predicted in plants, including TyDC-TYO-ADH pathway (pathway 1 in Fig. 1d), which occurs through sequential decarboxylation and tyramine oxidation; AAS-ADH pathway (pathway 2 in Fig. 1d), which involves aromatic acetaldehyde synthase (AAS) through a combined decarboxylation-deamination process10; and TAT-PPDC-ADH pathway (pathway 3 in Fig. 1d), through sequential tyrosine aminotransfer and phenylpyruvate decarboxylation. However, only AAS-ADH pathway has been thoroughly characterized in PhGs-producing plants10.
To elucidate the three potential upstream pathways, TyDC in pathway 1 and AAS in pathway 2 were firstly identified in C. tubulosa. These two enzymes both belong to the plant aromatic amino acid decarboxylase (AAAD) family36. They presented high sequence similarity with each other but were distinguished using two key residues differentiating their catalysis activity (decarboxylase activity or aldehyde synthase activity determined by a tyrosine or phenylalanine residue at the position marked with an asterisk in Supplementary Fig. 28a)10,37,38 and substrate selectivity (indolic substrate selectivity or phenolic substrate selectivity determined by a glycine or serine residue marked with an asterisk in Supplementary Fig. 28b)10,39. On the basis of transcriptome analysis, two AAAD genes were cloned from C. tubulosa. According to the residues that dictate substrate specificity and activity, one gene was predicted to encode a decarboxylase that used phenolic amino acid as substrate and was named CtTyDC. The other gene encodes a protein that indicated an aldehyde synthase activity using indolic substrate; therefore, this gene was named CtAAS (Supplementary Fig. 28). The expression profile of CtTyDC coincided with that of the identified downstream genes as well as with the ECH accumulation patterns, whereas that of CtAAS did not (Fig. 4a). Additionally, the protein sequences encoded by these genes were phylogenetically distinct (Supplementary Table 3 and Supplementary Fig. 29). In vitro enzymatic assays were subsequently designed accordingly. CtTyDC can catalyse the decarboxylation of tyrosine (1) to form tyramine (2) (Fig. 4b) with strict stereoselectivity for L-tyrosine (Supplementary Fig. 30). In contrast, CtAAS did not exhibit aldehyde synthase activity towards either tyrosine or tryptophan.
a Expression heatmap of candidate upstream genes and the identified downstream genes in C. tubulosa cell suspension cultures treated with 6% PEG6000. b Decarboxylation reaction catalysed by CtTyDC towards L-tyrosine (1) or L-dopa (3) to form tyramine (2) or dopamine (4), respectively. c Combined decarboxylation-deamination reaction catalysed by the mutant CtTyDCY347F in the presence of PLP towards L-tyrosine (1) to form 4-HPAA (7), which was further reduced to tyrosol (9) by NaBH4, or L-dopa (3) to 3,4-diHPAA (8), which was further reduced to hydroxytyrosol (10) by NaBH4. d The amino transfer reaction of L-tyrosine (1) to form 4-HPPA (5) or L-dopa (3) to form 3,4-diHPPA (6), respectively, catalysed by CtTAT in the presence of PLP and α-ketoglutarate. e With the addition of TPP, CtPPDC could decarboxylate 4-HPPA (5) to form 4-HPAA (7), which was further reduced to tyrosol (9) by NaBH4, or decarboxylate 3,4-diHPPA (6) to form 3,4-diHPAA (8), which was further reduced to hydroxytyrosol (10) by NaBH4. f Ct4HPAR could catalyse the reduction of 4-HPAA (7) or 3,4-diHPAA (8) to generate tyrosol (9) or hydroxytyrosol (10), respectively, using NADH as the cofactor. (b-f) HRESI-MS and MS2 spectra of the products are listed in Supplementary Fig. 36. g Heterologous transient expression of different combinations of upstream-related genes (Comb. 1–Comb. 7 annotated in Supplementary Table 8) in N. benthamiana. The detection wavelength was set at 280 nm. Source data are provided as a Source Data file.
To achieve aldehyde synthase activity, the tyrosine residue at position 347 of CtTyDC was mutated to phenylalanine. Then incubation of L-tyrosine (1) with the CtTyDCY347F mutant and the cofactor PLP led to the production of 4-HPAA (7). Since 4-HPAA is chemically unstable, the identity of 4-HPAA product was confirmed through a coupled reduction with NaBH4 to yield tyrosol (9) (Fig. 4c). In addition, the production of peroxide was detected during the reaction process (Supplementary Fig. 31), which provided more evidence for the occurrence of the decarboxylation-deamination process. However, when we used this activity-related residue as a clue to screen the 4-HPAAS gene in the C. tubulosa transcriptome (SRR31047969), no targeted gene was found, implying that the AAS-ADH pathway for tyrosol biosynthesis might be not present in C. tubulosa based on current transcriptome data.
Then, TAT and PPDC in pathway 3 were explored. Bioinformatics analysis revealed that the expression of one TAT gene in the C. tubulosa transcriptome was consistent with that of the identified downstream genes (Fig. 4a). This gene was subsequently cloned and named CtTAT. CtTAT is phylogenetically closely related to PsTyrAT from Opium poppy17, which uses ɑ-ketoglutarate and L-tyrosine as the preferred amino acceptor and amino donor, respectively (Supplementary Table 4 and Supplementary Fig. 32a). Conserved residues involved in PLP cofactor linkage were also observed in the CtTAT amino acid sequence (Supplementary Fig. 32b). Hence, an in vitro enzymatic assay was designed in the presence of PLP and ɑ-ketoglutarate. CtTAT effectively catalysed the amino transfer from L-tyrosine (1) to ɑ-ketoglutarate to yield L-glutamate and 4-HPPA (5), which was confirmed upon comparison with a reference standard, as shown in Fig. 4d. These findings confirmed the transamination activity of CtTAT.
Three candidate PPDC genes, namely CtPPDC1, CtPPDC2, and CtPPDC3, were identified by screening of C. tubulosa transcriptome data. All of their protein coding sequences contained an approximately 30-residues sequence motif common to thiamine pyrophosphate (TPP)-binding enzymes that begins with the highly conserved sequence “-GDG-” and ends with the conserved “-NN-” (Supplementary Table 5 and Supplementary Fig. 33)40. CtPPDC2 and CtPPDC3 presented increased expression levels in PEG6000-treated cell cultures (Fig. 4a). In vitro assays were performed using 4-HPPA (5) as a substrate in the presence of TPP. To prevent degradation of the 4-HPAA (7) product and facilitate product detection, NaBH4, which can directly convert the produced 4-HPAA (7) into tyrosol (9), was also added to the reaction mixture. The production of tyrosol (9) was detected only in the CtPPDC2-conducted assay based on high-performance liquid chromatography (HPLC) and high-resolution mass spectrometry (HRMS) analyses as well as upon comparison with the reference standard (Fig. 4e), demonstrating the decarboxylation activity of CtPPDC2, which was subsequently renamed CtPPDC.
The last step of tyrosol biosynthesis is the reduction of 4-HPAA by hydroxyphenylacetaldehyde reductase (HPAR), which belongs to a large family of NAD(P)H-dependent reductases. Using the previously reported gene encoding 4-HPAR from R. rosea as a query (Supplementary Table 6)10, one gene annotated as an alcohol dehydrogenase with a consistent expression pattern that correlated with the identified downstream genes was screened from the C. tubulosa transcriptome and named Ct4HPAR accordingly (Supplementary Fig. 34). With the addition of NADH as a cofactor, Ct4HPAR catalysed the reduction of 4-HPAA (7) to tyrosol (9) (Fig. 4f), revealing the universal last step for tyrosol biosynthesis in all three upstream pathways.
Finally, TYO in pathway 1 was investigated using the Nicotiana benthamiana expression system. The TyDC-TYO pathway has long been regarded as one of the primary routes for tyrosol biosynthesis. However, functional identification of TYO genes involved in PhG biosynthesis has been frustrated in many studies and these genes remain elusive. In particular, after Torrens-Spence et al. rectified the function of many previously mischaracterized TyDCs were actually 4-HPAAS on the basis of their actual function10,36,37,39,41, the participation of TYO in the upstream pathway of PhGs was considered doubtful10. Using the reported amine oxidases as templates (Supplementary Table 7), we obtained three TYO genes from the C. tubulosa transcriptome data and named them CtTYO1–CtTYO3. These genes all encode copper-containing amine oxidases with a consensus sequence of Asn-Tyr-Asp/Glu-Tyr (Supplementary Fig. 35)42,43,44,45. Although many efforts have been made towards the heterologous expression and functional identification of CtTYO1–CtTYO3, the targeted products have not been detected in in vitro enzymatic assays. Therefore, a plant expression system was subsequently used. The substrate tyramine (2) was initially injected into N. benthamiana leaves; however, these leaves tended to wilt, which was likely attributable to the toxicity of the injected tyramine. To solve this problem, we subsequently attempted to test the activity of candidate TYO genes by coexpressing the entire upstream pathway. This manipulation could allow the simultaneous verification of the functions of other upstream pathway-related genes in N. benthamiana. All candidate genes were inserted into the pCAMBIA1300 vector, and transiently expressed in N. benthamiana via different combinations (Combs. 1–7) as detailed in Supplementary Table 8. Interestingly, the coexpression of CtTyDC, CtTYO1 and Ct4HPAR (Comb. 1 in Fig. 4g) led to the formation of tyrosol (9), as determined by comparison with the reference standard, suggesting that CtTYO1 has the ability to oxidize tyramine; therefore, it was renamed CtTYO. The expression of CtTYO2 and CtTYO3 did not result in the detectable production of tyrosol but rather the obvious accumulation of tyramine (2), which is generated via the decarboxylation activity of CtTyDC (Combs. 2 and 3 in Fig. 4g). No products were detected with any of other combinations (Combs. 4–7 in Fig. 4g).
Biosynthesis of hydroxytyrosol from dopamine using the identified upstream genes
There are two phenolic hydroxyl substituents on the phenylethyl alcohol moiety of ECH. Since tyrosol has been well accepted as the crucial biosynthetic precursor of ECH, the introduction of the other hydroxy group is considered controversial and may occur at different stages. According to Yang et al., hydroxylation is the last step in acteoside biosynthesis using osmB as substrate in L. robustum and R. glutinosa21, whereas Yao et al. recently reported that polyphenol oxidase undergoes hydroxylation at the C3 position in the early stage of the downstream biosynthetic pathway for acteoside biosynthesis in Forsythia suspensa23. Based on the three possible upstream pathways for tyrosol biosynthesis, we further validated the catalytic activity of the upstream enzymes using L-dopa (3) as the initial precursor. As shown in Fig. 4b–f, CtTyDC in pathway 1 catalysed the decarboxylation of dopa (3) to dopamine (4) in 100% yield (Fig. 4b). CtTAT and CtPPDC in pathway 3 also catalysed amino transfer from dopa (3) to form 3,4-dihydroxyphenylpyruvic acid (3,4-diHPPA, 6) (Fig. 4d), followed by the decarboxylation of 3,4-diHPPA (6) to form 3,4-dihydroxyphenylacetaldehyde (3,4-diHPAA, 8) (Fig. 4e), with a conversion rate comparable to that of reactions using tyrosine as the initial precursor. The final step in the upstream biosynthesis of ECH catalysed by Ct4HPAR, which is shared by all three pathways, was also performed using 3,4-diHPAA (8) as the substrate, and the reduced product hydroxytyrosol (10) was effectively generated (Fig. 4f). The aldehyde synthesis ability of the CtTyDCY347F mutant towards dopa was also tested. 3,4-DiHPAA (8) was effectively generated through the one-step decarboxylation-deamination of dopa (3) by CtTyDCY347F (Fig. 4c). Therefore, enzymes involved in upstream biosynthesis that demonstrate catalytic activity towards tyrosine in vitro are also capable of catalysing corresponding reactions using dopa as a starting precursor (Supplementary Fig. 36), thereby offering additional alternative pathways for the incorporation of dihydroxyl groups.
In vivo functional identification of upstream biosynthesis genes of ECH in C. tubulosa
The transient expression of the upstream biosynthetic genes in N. benthamiana enabled the functional identification of CtTYO, which led to the identification of the predicted TyDC-TYO-ADH pathway for tyrosol biosynthesis (Fig. 4g and Supplementary Fig. 37). Moreover, CtAAS showed no activity in either in vitro enzymatic assays or in the N. benthamiana expression system, further excluding AAS-ADH pathway for tyrosol biosynthesis in C. tubulosa. Moreover, although CtTAT and CtPPDC in Ehrlich pathway showed obvious transamination and decarboxylation activity in vitro (Fig. 4d, e), when they were transiently expressed in N. benthamiana together with Ct4HPAR (Comb. 6 in Fig. 4g), no product was detected. Therefore, to further explore the roles of TyDC-TYO-ADH pathway-related genes and Ehrlich pathway-related genes in ECH biosynthesis in C. tubulosa (Supplementary Fig. 37), the in vivo functions of the genes involved in these two pathways were evaluated.
Initially, CtTyDC, CtTYO, CtTAT, CtPPDC and Ct4HPAR were expressed in C. tubulosa suspension cell cultures separately (Supplementary Fig. 38a) and in two different combinations (“CtTyDC + CtTYO + Ct4HPAR” and “CtTAT + CtPPDC + Ct4HPAR”, Supplementary Fig. 38b). The contents of ECH and acteoside in C. tubulosa suspension cells subjected to different treatments were subsequently determined. Overexpressing these genes individually resulted in only slight increases in the contents of ECH and acteoside (Fig. 5a), whereas clear increases in the ECH and acteoside contents were detected with both gene combinations (Fig. 5b), which indicated that overexpression of the entire pathway 1 or pathway 3 was beneficial for the accumulation of both ECH and acteoside.
a, b Contents of ECH and acteoside in C. tubulosa suspension cell cultures overexpressing the genes of CtTyDC, CtTYO, CtTAT, CtPPDC and Ct4HPAR individually (a) or in two combinations of “CtTyDC + CtTYO + Ct4HPAR” and “CtTAT + CtPPDC + Ct4HPAR”, respectively (b). c, d Contents of ECH and acteoside accumulated in C. tubulosa calli cultured with the knockdown of CtTyDC, CtTYO, CtTAT, and CtPPDC genes individually (c) or in three combinations of “CtTyDC + CtTYO”, “CtTAT + CtPPDC” and “CtTyDC + CtTYO + CtTAT + CtPPDC”, respectively (d). The data are presented as the means ± SDs (n = 3 biologically independent replicates). Statistical analysis was performed with unpaired two-tailed Student’s t tests. P value for each comparison from left to right in (d): 0.0484, 0.0045, <0.0001, 0.0206, 0.0022, and 0.0002. Source data are provided as a Source Data file.
In addition, the expression of CtTyDC, CtTYO, CtTAT, and CtPPDC in C. tubulosa calli was knocked down using RNAi (Supplementary Fig. 38c). The individual downregulation of these genes led to only a marginal reduction in the ECH and acteoside contents (Fig. 5c). When CtTyDC and CtTYO (pathway 1) or CtTAT and CtPPDC (pathway 3) were suppressed in together (Supplementary Fig. 38d), the contents of ECH and acteoside decreased moderately (Fig. 5d). This observation was consistent with the gene overexpression results and suggested that the biosynthesis of tyrosol was not solely dependent on either pathway. Thus, we downregulated the expression of CtTyDC and CtTYO in pathway 1 and CtTAT and CtPPDC in pathway 3 simultaneously (Supplementary Fig. 38d). The content of ECH in the treated callus culture showed a sharp decrease from 28.8% (control) to 10.1% (Fig. 5d). Based on these results, we speculated that both TyDC-TYO-ADH pathway and TAT-PPDC-ADH pathway are employed by C. tubulosa to generate the precursor tyrosol for ECH biosynthesis. Utilization of microbial-like pathways for the biosynthesis of natural products in plants has been revealed recently46. Our study demonstrated that plants could also utilize a microbial-like Ehrlich pathway to synthesize tyrosol in vivo.
Reconstruction of the ECH biosynthetic pathway in N. benthamiana
Given the desired de novo synthesis activity of CtTyDC, CtTYO and Ct4HPAR in N. benthamiana to produce tyrosol (Comb. 1 in Fig. 4g), all the functionally identified enzymes related to ECH biosynthesis were sequentially introduced into the N. benthamiana expression system. It was found that adding either CtUGT85A191 or CtUGT85AF12 to N. benthamiana led to the efficient de novo biosynthesis of salidroside, with CtUGT85AF12 exhibiting better activity, which is in agreement with the in vitro assays (Supplementary Fig. 39). On the other hand, we also attempted tyrosol infiltration with the transient expression of CtUGT85A191 or CtUGT85AF12. However, only minor salidroside products were detected (Supplementary Fig. 39). This observation highlights the advantage of utilizing the identified TyDC-TYO-ADH upstream pathway genes in the generation of PhGs. The identified acyltransferase gene CtAT-E was subsequently introduced into the N. benthamiana expression system, resulting in the production of osmA (13) and syringalide A (15) based on comparisons with the reference standard and HRESI-MSn analysis (Supplementary Figs. 40 and 41). These two compounds were further rhamnosylated to form osmB (14) and syringalide A-3′-rhamnoside (16), respectively, after CtUGT79G13 was coexpressed in N. benthamiana (Supplementary Figs. 40 and 42). These observations demonstrated that CtAT-E could transfer p-coumaroyl or caffeoyl groups and that CtUGT79G13 could catalyse the subsequent rhamnosylation for ECH biosynthesis in the N. benthamiana system, which are consistent with their in vitro activities.
The in vitro enzymatic assays demonstrated that, in comparison with osmA, calA serves as the optimal natural substrate for CtUGT79G13. Thus, the introduction of the phenolic hydroxy at the C3 position should occur before rhamnosylation. To elucidate the gene responsible for C3 hydroxylation, a comprehensive screening of 23 oxidase or hydroxylase genes was conducted using the transcriptome of C. tubulosa. Among them, a cytochrome P450 gene that is phylogenetically close to the CYP98A family (Supplementary Table 9 and Supplementary Fig. 43) and named CtCYP98A248 by the Cytochrome P450 Nomenclature Committee exhibited targeted hydroxylation activity when coexpressed with other genes. In the extracts of N. benthamiana transiently expressing the CtTyDC, CtTYO, Ct4HPAR, CtUGT85AF12, CtAT-E and CtCYP98A248 combination, the extracted ion chromatogram (EIC) for calA (18) revealed a product peak with the same retention time as the standard (Supplementary Fig. 44). The EIC for C23H25O10 [M − H]− showed a peak for 17 (Supplementary Fig. 44), which was further rhamnosylated to generate 19 after CtUGT79G13 was coexpressed (Supplementary Figs. 40 and 45). Compound 19 exhibited a different retention time from 16. Its molecular ion was detected at m/z 607.2002, with an MS2 fragment at m/z 461.1643, which corresponds to the mass spectrum of isosyringalide A 3′-rhamnoside (Supplementary Fig. 45). Owing to the weak activity of CtCYP98A248, all of these hydroxylated products were detected in trace amounts. Therefore, we further tested the catalytic activity of CtUGT79G13 and CtUGT73EV1 through substrate infiltration. The formation of acteoside was detected after the transient expression of CtUGT79G13 in N. benthamiana with calA coinfiltration (Supplementary Fig. 46). The formation of ECH was detected both in the CtUGT73EV1 transient expression system in N. benthamiana with acteoside coinfiltration and in the CtUGT79G13 and CtUGT73EV1 transient expression system in N. benthamiana with calA coinfiltration (Supplementary Fig. 47), confirming the last two steps of in vivo biosynthesis of ECH through the rhamnosylation of calA and the subsequent glucosylation of acteoside (Fig. 6).
De novo biosynthesis of various phenylethanoid glycosides in N. benthamiana
Most of the identified genes displayed desired activities in N. benthamiana, except CtCYP98A248, whose activity improvement was under investigation. The combinatorial transient expression of other identified genes paves a way for the efficient de novo synthesis of PhGs with mono-hydroxyl unit (Fig. 6). These genes were therefore coexpressed in different combinations in N. benthamiana (Fig. 7a). De novo synthesis of tyrosol and salidroside, in yields of 0.3% DW and 2% DW, respectively, was initially achieved through using gene Combs. 8 and 9 (Fig. 7b). Since the downstream modifications involve multiple glycosylation steps, we further identified a sucrose synthase (SuS) gene CtSuS from C. tubulosa to increase the supplementation of glycosyl donors (Supplementary Fig. 48). The incorporation of CtSuS in tobacco leaves harbouring CtTyDC, CtTYO, Ct4HPAR and CtUGT85AF12 effectively improved the de novo production yield of salidroside 11 from 2% DW to 3% DW (Fig. 7b). Subsequently, the combined transient expression of CtSuS in conjunction with CtTyDC, CtTYO, Ct4HPAR, CtUGT85AF12 and CtAT-E resulted in the generation of eleven additional PhG products labelled a1–a11 (Fig. 7c). These compounds were converted into their corresponding rhamnosylated products (b1–b11), as labelled in Fig. 7d, after transient coexpression of CtUGT79G13. According to the mass spectrometric patterns and diagnostic fragments summarized in Supplementary Figs. 49–71, as well as comparisons with the reference standards (for a2 and b2), the chemical structures of a1–a11 and b1–b11 were putatively identified (Fig. 7e). These compounds all harbour tyrosol moieties with different acyl substituents, including caffeoyl (a1 and b1; a3 and b3), coumaroyl (a2 and b2; a5 and b5; a7 and b7), feruloyl (a4 and b4; a6 and b6), cinnamoyl (a8 and b8; a10 and b10), and methoxycinnamoyl (a9 and b9; a11 and b11) substituents. Products with caffeoyl or coumaroyl substituents were predominant, which indicated the strong in vivo acylation ability of CtAT-E when unitizing the endogenous coumaroyl-CoA and caffeoyl-CoA in tobacco and is in accordance with the in vitro coumaroyl transferase and caffeoyl transferase activities of CtAT-E. Additionally, we hypothesized that other types of acylation reactions may be beneficial due to the improved activity of CtAT-E in the plant expression system; meanwhile, the possibility that the acyl group was methylated by native methyltransferases in N. benthamiana cannot be fully excluded. Moreover, the major peaks of a3, a5, a6, a7, a10 and a11 and their corresponding rhamnosylated products b3, b5, b6, b7, b10 and b11 all showed the characteristic sequential neutral loss of 44 Da for CO2 and 86 Da for the malonyl substituent, which is consistent with the spectrometric pattern of malonylated glycosides. We speculated that malonylation of the PhG products was conducted by an endogenous malonyltransferase that exists in tobacco since malonylation is recognized as a key reaction in the metabolism of xenobiotic phenolic glucosides in tobacco47, and several glycoside malonyltransferases have been identified previously48,49,50. Moreover, because the malonyl substituent occupied the 6′-OH position, which is the preferred glucosylation site of UGT73EV1, further incorporation of the UGT73EV1 gene in the N. benthamiana transient expression system did not generate new products. According to the residue content of 11 (~2% DW), the yields of a1–a11 and b1–b11 ranged from 0.02–1% DW (Fig. 7f). This finding indicated that N. benthamiana harbouring the functional genes identified in this study can serve as an efficient platform for the de novo synthesis of a wide range of natural and synthetic PhG derivatives. Further improvement of the C3 hydroxylation activity as well as inhibition of endogenous malonylation will offer a more advantageous approach for the de novo synthesis of ECH.
a De novo biosynthesis of PhGs using endogenous L-tyrosine in N. benthamiana harbouring functionally identified genes in different combinations (Combs. 8–14). b Yields of tyrosol (0.3% DW) or salidroside (2–3% DW) in tobacco harbouring gene Combs. 8–10. The data are presented as the means ± SDs; n = 3 biological replicates. Statistical analysis was performed with unpaired two-tailed Student’s t-tests. c HPLC chromatogram of tobacco extracts harbouring gene Comb. 12, which led to the production of salidroside and PhGs a1–a11. The detection wavelength was set at 330 nm. d HPLC chromatogram of tobacco extracts harbouring gene Comb. 14, which led to the production of salidroside and PhGs b1–b11. e Plausible identities of the generated products a1–a11 and b1–b11 based on their mass spectrometric patterns and diagnostic fragments (Supplementary Figs. 49–71), as well as through reference standard comparisons. Compounds b1–b11 are the corresponding rhamnosylated products of a1–a11, respectively. f The liquid chromatography mass spectrometry peak areas of products a1–a11 and b1–b11 from engineered tobacco harbouring different gene combinations. The data are presented as the means ± SDs; n = 3 biological replicates. Source data are provided as a Source Data file.
In summary, we have elucidated the complete biosynthetic pathway of ECH and various PhG intermediates from L-tyrosine in C. tubulosa. A total of 14 genes were functionally identified, including the separate decarboxylation and tyramine oxidation pathway i.e. TyDC-TYO-ADH pathway for tyrosol biosynthesis in PhG-producing plants. In addition, a microbial-like Ehrlich pathway for tyrosol biosynthesis was elucidated, which involves a separate tyrosine aminotransfer and phenylpyruvate decarboxylation pathway i.e. TAT-PPDC-ADH pathway. Furthermore, downstream assembly processes of tyrosol were elucidated, which involves sequential glucosylation to form salidroside, acylation to produce osmA and syringalide A, hydroxylation to produce calA, rhamnosylation to form acteoside, and ultimately glucosylation to yield ECH through the action of a specific 6’-O-glucosyaltransferase enzyme. Finally, de novo biosynthesis of salidroside, osmA, omsB, syringalide A, syringalide A-3’-rhamnoside, and 18 PhGs with to the best of our knowledge previously not reported structures was achieved in tobacco through expression of seven functional genes. The present study contributes to enhancing our understanding of the biosynthesis of ECH and various complex PhGs, and establishes a robust foundation for efficient and sustainable production of diverse bioactive PhGs through synthetic biology.
Methods
Chemicals and reagents
Chemicals and reference standards used in this study were listed in Supplementary Table 10. p-Coumaroyl-CoA and caffeoyl-CoA were prepared using a 4-coumarate coenzyme A ligase51. All solvents for preparative HPLC and MS analysis were of HPLC grade and MS grade, respectively, and purchased from Fisher Scientific.
Plant materials and comparative transcriptome sequencing
C. tubulosa plant was obtained from Hetian, Xinjiang Uygur Autonomous Region. Callus culture and cell suspension of C. tubulosa were induced and maintained in our laboratory for years52. 6% PEG6000 treatment was performed on cell suspension of C. tubulosa after subculture in MS liquid medium. Cells were harvested after 15 days’ treatment at 25 °C, 120 rpm in darkness. Total RNA of different C. tubulosa samples was extracted using OMEGA plant RNA kit (GA, USA). Comparative transcriptome sequencing and analysis were completed using the BGISEQ-500 sequencing platform (BGI, Wuhan, China).
HPLC and MS analysis methods
HPLC analyses were performed using Agilent 1260 Series HPLC system equipped with a diode array detector and a LC-20ADXR modular platform (Shimadzu, Kyoto, Japan). HRESI-MS data were recorded on an LCMS-IT-TOF system in negative ion mode with automatic multi-level MS1, MS2, MS3 full scans using ultrahigh-purity Ar as the collision gas, N2 as the atomizing gas. Ion accumulation time was 100 ms; CID collision energy was set to 50%. Ion trap was set to 1.9 × 10−2 Pa. The mobile phase and gradient programs for different detection purpose are listed in Supplementary Data 3.
Gene cloning, heterologous expression, and recombinant protein purification
Total RNA was extracted from the fresh stem of C. tubulosa using a plant RNA Extraction Kit (OMEQA Bio-Tek, USA) and used to synthesize the cDNA using M-MLV reverse transcriptase (Promega, USA). The coding sequences of genes of interest were amplified from cDNA using high fidelity KOD-Plus-Neo DNA polymerase (TOYOBO, Japan) through three steps or gradient touch down PCR methods. To achieve the soluble expression in E. coli, pET-28a vector, pET-24b vector, pColdTM I vector and pColdTM TF vector were used for recombinant strain construction. Protein expression in pET expression system was induced by addition of 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) and grown for 16 h at 25 °C, 200 rpm53. For pCold expression system, strains were induced via adding IPTG with the final concentration of 0.5 mM when the OD600 value reached 0.4–0.6, and the cells were grown for 24 h at 16 °C, 180 rpm. All primers used in this study are listed in Supplementary Data 4. SDS-PAGE analysis of all soluble proteins is shown in Supplementary Fig. 1.
Bioinformatic analysis
Hierarchical clustering for gene expression data analysis was performed in TBtools software54 based on the FPKM values of candidate genes. Phylogenetic analyses were performed using the Neighbour-Joining method based on the Poisson model by MEGA7 software. The reliability of the tree was measured by bootstrap analysis with 1000 replicates55. The amino acid sequences were aligned using Clustal W56.
In vitro enzymatic assays
In vitro enzymatic assays of different enzymes were performed in the reaction mixture as the following descriptions. Parallel reaction using boiled enzymes were set as control. The reactions were terminated by the addition of two-fold volume of ice-cold methanol. Afterward, the samples were centrifuged at 12,000 × g for 30 min at 4 °C. The obtained supernatants were analysed by HPLC and HRESI-MS using different methods summarized in Supplementary Data 3.
In vitro enzymatic assays of CtTyDC were performed using a mixed reaction solution (100 μL) containing 100 mM sodium phosphate (pH 8.0), 60 μg of the purified protein and 4 mM L-tyrosine, and incubated at 30 °C for 2 h.
In vitro enzymatic assays of CtPAAS and CtTyDCY347F were performed using a mixed reaction solution (100 μL) containing 50 mM sodium phosphate (pH 8.0), 0.2 mM PLP, 2 mM L-tyrosine, and 30 μg purified protein, and incubated at 30 °C for 3 h. After termination, NaBH4 was added to reduce the product, the obtained supernatants were analysed.
In vitro enzymatic assays of CtTAT were performed using a mixed reaction solution (100 μL) containing 100 mM HEPES (pH 8.5), 0.2 mM PLP, 2 mM L-tyrosine, 10 mM α-ketoglutarate and 30 μg purified protein, and incubated at 30 °C for 1 h.
In vitro enzymatic assays of CtPPDC were performed using a mixed reaction solution (100 μL) containing 50 mM Tris-HCl (pH 7.5), 2 mM 4-HPPA, 0.2 mM TPP and 30 μg purified protein, and incubated at 30 °C for 10 h. After termination, NaBH4 was added to reduce the product. The obtained supernatants were analysed.
In vitro enzymatic assays of Ct4HPAR were performed using a mixed reaction solution (100 μL) containing 50 mM Tris-HCl (pH 7.5), 4 mM 4-HPAA, 2 mM NADH and 50 μg purified protein, and incubated at 30 °C for 2 h.
In vitro enzymatic assays of CtUGT85A191, CtUGT85AF12 and CtUGT85AF13 were performed in the reaction mixture containing 0.8 mM UDP-Glc, 0.4 mM tyrosol or hydroxytyrosol, 10–60 µg of purified protein in 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM DTT at a final volume of 100 μL, respectively. The reactions were incubated at 30 °C for 1–12 h.
In vitro enzymatic assays of CtUGT79G13 were conducted in enzyme reaction mixtures containing 100 mM K2HPO4-KH2PO4 (pH 7.0), 0.4 mM osmA or calA, 0.8 mM UDP-rhamnose and 50 μg purified CtUGT79G13. The reactions were incubated at 30 °C for 0.5–12 h.
In vitro enzymatic assays of CtUGT73EV1 were performed in the reaction mixture containing 2 mM UDP-Glc, 0.4 mM acetoside, 100 µg of purified CtUGT73EV1 in 100 mM K2HPO4-KH2PO4 (pH 7.0), 50 mM NaCl, 1 mM DTT at a final volume of 100 μL. The reactions were incubated at 37 °C for 1.5 h.
In vitro enzymatic assays of CtATs were performed using a mixed reaction solution (100 μL) containing 50 mM Tris-HCl (pH 7.5), 0.4 mM salidroside, 0.6 mM aromatic acyl coenzyme A and 30 μg purified protein, and incubated at 30 °C for 12 h.
In vitro enzymatic assays of CtATs and CtUGT79G13 were performed in the reaction mixture containing 1 mM aromatic acyl coenzyme A, 1 mM UDP-rhamnose, 0.4 mM salidroside, 50 µg of purified CtATs and 30 µg of purified CtUGT79G13 in 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM DTT at a final volume of 150 μL. The reactions were incubated at 30 °C for 12 h.
Biochemical properties determination and kinetic studies
The time courses of the reactions were assayed by terminating the reactions at different time points (5 min, 15 min, 30 min, 1 h, 2 h, 4 h, 6 h, 8 h, 12 h and 24 h). To assay the optimal reaction temperature, the reactions were incubated at various temperatures ranging from 0 to 65 °C. To investigate the optimal pH, the enzymatic reactions were performed in various reaction buffers ranging in pH values from 4.0 to 6.0 (100 mM citric acid-sodium citrate buffer), 6.0 to 8.0 (100 mM K2HPO4-KH2PO4 buffer), 7.0 to 9.0 (50 mM Tris-HCl buffer) and 8.0 to 10.0 (100 mM Na2CO3-NaHCO3 buffer).
Kinetic characterization of CtUGT85A191, CtUGT85AF12, CtCTUGT79G13, CtUGT73EV1 was performed as listed in Supplementary Method 1–4. Kinetic constants were calculated by nonlinear Michaelis-Menten regression using GraphPad Prism 8. All the reactions were terminated by adding a double volume of methanol. After being centrifuged at 12,000 × g for 30 min, the samples were analysed by HPLC using methods in Supplementary Data 3.
Preparation, separation and structural elucidation of glycosylated products
For preparative enzymatic synthesis of glycosylated products, reactions were carried out in 50 mL total volume containing 50 mM of Tris-HCl buffer (pH 7.0), 0.6–1 mM glycosyl-acceptors, 3 mM glycosyl-donors and 50–100 mg of purified enzymes at 30 °C for 12 h. Then the reaction solutions were centrifuged at 12,000 × g for 30 min to obtain the supernatants which were subjected into a MCL Gel (Mitsubishi Chemical) using gradient elution (0%–100% methanol/H2O). The obtained fractions were analysed by HPLC-UV. Fractions contained targeted products were combined and evaporated to dryness before being redissolved in 2 mL of 50% methanol. The glycosylated products were subsequently purified by reversed-phase semi-preparative HPLC using a YMC-Pack ODS-A HPLC column (10 mm i.d. × 250 mm, 5 μm) at a flow rate of 3.0 mL/min. Water and acetonitrile were used as the mobile phase. The obtained glycosylated products were further structurally identified by HRESI-MS, 1H NMR and 13C NMR analyses (Supplementary Note 1).
AlphaFold2 protein structure prediction and molecular docking
The protein model for UGT79G13 was established using Alphafold257. Software AutoDock Vina v.1.2.0. was used for molecular docking58. Grid boxes for sugar donor and acceptor were calculated using the crystal structures of plant originated glycosyltransferases UGT89C1 (PDBID: 6IJA) and UGT72B1 (PDBID: 2VCE), respectively. Discovery Studio 2016 and Pymol 2.0.6 were used to visualize models and construct graphical illustrative figures.
Transient expression of candidate genes in N. benthamiana
Candidate biosynthetic genes were expressed in N. benthamiana via Agrobacterium mediated transient expression. Interested genes were constructed into pCAMBIA-1300-35S-EGFP vector using primers in Supplementary Data 4. Agrobacterium strains that contained the recombinant plasmid were grown in LB medium with antibiotics (25 μg/mL rifampicin, 50 μg/mL kanamycin) for 16 h at 28 °C. Then the cells were collected through centrifuge at 4000 × g for 6 min. The cell pellet was resuspended in infiltration buffer (10 mM MES, pH 5.6, 10 mM MgCl2, 150 μM acetosyringone) and centrifuged at 4000 × g for 6 min. The supernatant was removed and the cell pellet was resuspended in infiltration buffer to the optical density OD600 of 0.6. For individually tested strains, Agrobacterium suspensions were diluted. After incubation at 28 °C for 2–4 h, infiltration was performed using a 1 mL needle-free syringe on underside of four-week-old N. benthamiana leaves. For substrate infiltration, about 200 μL of aqueous substrate solution was infiltrated into underside side of previously Agrobacterium-infiltrated leaves with a needleless syringe. The infiltrated area was marked for further harvest at 48 hours post-infiltration.
Overexpression of interested genes in C. tubulosa cell suspension culture
The candidate genes were constructed into pCAMBIA1300 35s-EGFP binary vector and transformed into the A. tumefaciens strain EHA105. Strains harbouring empty vector were used as negative control. The transformed cells were cultured on LB medium containing selective antibiotics (50 μg L−1 kanamycin and 25 μg L−1 rifampicin) at 28 °C on a shaker rotating at 200 rpm until the OD600 reached 0.8–1.0. The cells were harvested by centrifugation at 4000 × g for 6 min and the pellets were resuspended in MS liquid medium supplemented with 100 mM acetosyringone to an OD600 reached 0.6–0.8. When gene constructs were tested in combination, strains were mixed in equal concentration such that each strain had an OD600 of 0.3. Each test had 3 parallels. The mixed cultures were added to the suspension cells of C. tubulosa growing in a stable phase and incubated at 28 °C with gentle shaking for 4 h. Subsequently, fresh MS liquid medium was added and incubation continued at 25 °C for 60 h in darkness. Then the suspension cells were harvested in duplicate, one was directly dried to constant weight for content determination, and the other one was frozen in liquid nitrogen and stored at −80 °C for RNA extraction.
Knockdown of gene expression through RNA interference in C. tubulosa calli
According to the sequence information of interested genes, the highly specific fragment about 200 bp was selected as the target and inserted into pBWA(V)HS-RNAi to generate the hairpin RNA vector with intron. Primers used for RNAi are summarized in Supplementary Data 4. The resulting recombinant vectors were transformed into A. tumefaciens GV3101, and the positive colonies carrying the recombinant plasmids were inoculated in 10 mL of LB medium with antibiotics (25 μg/mL rifampicin, 50 μg/mL kanamycin) and grown at 28 °C until the OD600 reached 0.6. Then the cells were centrifuged at 4 °C, 4000 × g for 6 min and the supernatant was removed. The Agrobacterium cells were washed with B5 medium twice and then resuspended in B5 medium for the infection of healthy C. tubulosa calli. The infected calli were cultured on B5 solid medium containing 300 μg/mL cefotaxime and 50 μg/mL hygromycin for 5 days in the dark at 25 °C. The collected calli were sampled in duplicate, one was directly dried to constant weight for content determination, and the other one was frozen in liquid nitrogen and stored at −80 °C for RNA extraction.
De novo synthesis of PhGs in N. benthamiana
For de novo synthesis of PhGs in N. benthamiana, gene constructs were expressed in different combination as shown in Supplementary Table 8. Agrobacterium strains that contained the recombinant plasmid were cultured and then collected by centrifuge at 4000 × g for 6 min. The cell pellets were resuspended in infiltration buffer and then centrifuged at 4000 × g for 6 min. For gene transient expression in combinations, the multiple constructs were infiltrated simultaneously. The corresponding A. tumefaciens cell cultures were mixed so that the final OD600 of each to be 0.3. The infiltration mixture was subsequently incubated in an incubator at 28 °C for 2–3 h for injection59. In a typical experiment, three individual leaves were used as replicates for each strain combination being tested, with each leaf belonging to a different plant to randomize any batch effects. The Agrobacterium infection solutions were infiltrated into the leaves of N. benthamiana with a 1 mL needle-free syringe. The infection area was marked and cultured overnight in dark, then cultured under normal conditions. After 120 h, the leaves were freeze-dried and crushed into powders. The powders were then ultrasonic extracted using 50% methanol (w:v = 1:5) for 30 min, The supernatants were obtained after being centrifuged at 12,000 × g, 15 min for HPLC and HRESI-MS analyses using method listed in Supplementary Data 3.
qRT-PCR
RNA retraction of C. tubulosa culture under different treatment and the subsequent cDNA preparation were performed as described above. The expression of CtTyDC, CtTYO, CtTAT, CtPPDC, Ct4HPAR were quantified through real-time PCR using specific primers in Supplementary Data 4 by TransStart Top Green qPCR SuperMix (TransGen Biotech). PCR procedures were performed on BIO-RAD CFX Connect Real-Time PCR Detection System using procedure as follows: 30 s at 94 °C, 5 s at 94 °C, 30 s at 60 °C. This procedure was repeated for 40 cycles followed by the dissociation stage. The relative expression of genes was analysed using the 2−ΔΔCt method and normalized to the expression level of the internal standard DNAj.
Content determination
The contents of salidroside, hydroxysalidroside, acteoside and echinacoside for kinetic studies, as well as contents of de novo biosynthesis of tyrosol and salidroside in tobacco leaves were determined using standard curves in Supplementary Table 11.
NMR analysis
NMR analysis was performed on 400 MHz, 500 MHz and 600 MHz Bruker Avance III spectrometers (Bruker Corp. Karlsruhe, Germany). Chemical shifts were reported in parts per million and coupling constants were recorded in hertz.
Statistical analysis
All numerical data are expressed as mean ± standard deviation (SD). Unpaired two-tailed Student’s t-tests was performed to evaluate the difference between groups. P value of less than 0.05 was statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Genes identified in this study have been deposited in GenBank database under following accession numbers: CtTyDC, PP911309; CtTYO, PP911310; CtTAT, PP911311; CtPPDC, PP911312; Ct4HPAR, PP911313; CtUGT85A191, PP911315; CtUGT85AF12, PP911316; CtUGT85AF13, PP911317; CtUGT79G13, PP911318; CtUGT73EV1, P911319; CtAT-E, PP911321; CtCYP98A248, PQ540720; CtSuS, PP911320. CtUGT85A191, CtUGT85AF12, CtUGT85AF13, CtUGT79G13, CtUGT73EV1 and CtCYP98A248 were named by the UGT Nomenclature Committee or the P450 Nomenclature Committee. The raw reads from the transcriptome analysis of C. tubulosa have been deposited in the NCBI Sequence Read Archive (SRA) database under the BioProject PRJNA1175030. Source data are provided with this paper.
References
Lei, H. et al. Herba Cistanche (Rou Cong Rong): a review of its phytochemistry and pharmacology. Chem. Pharm. Bull. 68, 694–712 (2020).
Song, Y., Zeng, K., Jiang, Y. & Tu, P. Cistanches Herba, from an endangered species to a big brand of Chinese medicine. Med. Res. Rev. 41, 1539–1577 (2021).
Jiang, Y. & Tu, P. F. Analysis of chemical constituents in Cistanche species. J. Chromatogr. A 1216, 1970–1979 (2009).
Wang, W., Jiang, S., Zhao, Y. & Zhu, G. Echinacoside: A promising active natural products and pharmacological agents. Pharmacol. Res. 197, 106951 (2023).
Li, J., Yu, H., Yang, C., Ma, T. & Dai, Y. Therapeutic potential and molecular mechanisms of echinacoside in neurodegenerative diseases. Front. Pharmacol. 13, 841110 (2022).
Tian, X. Y. et al. A review on the structure and pharmacological activity of phenylethanoid glycosides. Eur. J. Med. Chem. 209, 112563 (2021).
Morikawa, T. et al. A review of biologically active natural products from a desert plant Cistanche tubulosa. Chem. Pharm. Bull. 67, 675–689 (2019).
Hou, L. et al. De novo full length transcriptome analysis and gene expression profiling to identify genes involved in phenylethanol glycosides biosynthesis in Cistanche tubulosa. BMC Genomics 23, 698 (2022).
Alipieva, K., Korkina, L., Orhan, I. E. & Georgiev, M. I. Verbascoside-a review of its occurrence, (bio)synthesis and pharmacological significance. Biotechnol. Adv. 32, 1065–1076 (2014).
Torrens-Spence, M. P., Pluskal, T., Li, F. S., Carballo, V. & Weng, J. K. Complete pathway elucidation and heterologous reconstitution of Rhodiola salidroside biosynthesis. Mol. Plant 11, 205–217 (2018).
Yang, Y. H. et al. Functional characterization of tyrosine decarboxylase genes that contribute to acteoside biosynthesis in Rehmannia glutinosa. Planta 255, 64 (2022).
Li, Y. et al. Identification and functional characterization of tyrosine decarboxylase from Rehmannia glutinosa. Molecules 27, 1634 (2022).
Lan, X. et al. Engineering salidroside biosynthetic pathway in hairy root cultures of Rhodiola crenulata based on metabolic characterization of tyrosine decarboxylase. PloS one 8, e75459 (2013).
Zhou, Y., Zhu, J., Shao, L. & Guo, M. Current advances in acteoside biosynthesis pathway elucidation and biosynthesis. Fitoterapia 142, 104495 (2020).
Bai, Y. et al. Production of salidroside in metabolically engineered Escherichia coli. Sci. Rep. 4, 6640 (2014).
Hazelwood, L. A., Daran, J. M., Maris, A. J. A. V., Pronk, J. T. & Dickinson, J. R. The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 74, 2259–2266 (2008).
Lee, E. J. & Facchini, P. J. Tyrosine aminotransferase contributes to benzylisoquinoline alkaloid biosynthesis in opium poppy. Plant Physiol 157, 1067–1078 (2011).
Prabhu, P. R. & Hudson, A. O. Identification and partial characterization of an L-Tyrosine aminotransferase (TAT) from Arabidopsis thaliana. Biochem. Res. Int. 2010, 549572 (2010).
Hirata, H. et al. Seasonal induction of alternative principal pathway for rose flower scent. Sci. Rep. 6, 20234 (2016).
Xu, J. J., Fang, X., Li, C. Y., Yang, L. & Chen, X. Y. General and specialized tyrosine metabolism pathways in plants. aBIOTECH 1, 97–105 (2019).
Yang, Y., Xi, D., Wu, Y. & Liu, T. Complete biosynthesis of the phenylethanoid glycoside verbascoside. Plant Commun. 4, 100592 (2023).
Fuji, Y. et al. Molecular identification of UDP-sugar-dependent glycosyltransferase and acyltransferase involved in the phenylethanoid glycoside biosynthesis induced by methyl jasmonate in Sesamum indicum L. Plant Cell Physiol. 64, 716–728 (2023).
Yao, M. et al. Construct phenylethanoid glycosides harnessing biosynthetic networks, protein engineering and one‐pot multienzyme cascades. Angew. Chem. Int. Ed. Engl. 63, e202402546 (2024).
Yang, Y., Wu, Y., Zhuang, Y. & Liu, T. Discovery of glycosyltransferases involved in the biosynthesis of ligupurpuroside B. Org. Lett. 23, 7851–7854 (2021).
Mirmazloum, I. et al. Identification of a novel UDP-glycosyltransferase gene from Rhodiola rosea and its expression during biotransformation of upstream precursors in callus culture. Int. J. Biol. Macromol. 136, 847–858 (2019).
Xue, F. et al. Expression of codon-optimized plant glycosyltransferase UGT72B14 in Escherichia coli enhances salidroside production. Biomed Res. Int. 2016, 9845927 (2016).
Yu, H. S. et al. Characterization of glycosyltransferases responsible for salidroside biosynthesis in Rhodiola sachalinensis. Phytochemistry 72, 862–870 (2011).
Ma, L. Q. et al. Molecular cloning and overexpression of a novel UDP-glucosyltransferase elevating salidroside levels in Rhodiola sachalinensis. Plant Cell Rep. 26, 989–999 (2007).
Yan, Y. et al. Simultaneous determination of components with wide polarity and content ranges in Cistanche tubulosa using serially coupled reverse phase-hydrophilic interaction chromatography-tandem mass spectrometry. J. Chromatogr. A 1501, 39–50 (2017).
Wang, L. L., Chen, K., Zhang, M., Ye, M. & Qiao, X. Catalytic function, mechanism, and application of plant acyltransferases. Crit. Rev. Biotechnol. 42, 125–144 (2022).
D’Auria, J. C. Acyltransferases in plants: a good time to be BAHD. Curr. Opin. Plant Biol. 9, 331–340 (2006).
Bontpart, T., Cheynier, V., Ageorges, A. & Terrier, N. BAHD or SCPL acyltransferase? What a dilemma for acylation in the world of plant phenolic compounds. New Phytol. 208, 695–707 (2015).
Murayama, K. et al. Anthocyanin 5, 3′-aromatic acyltransferase from Gentiana triflora, a structural insight into biosynthesis of a blue anthocyanin. Phytochemistry 186, 112727 (2021).
Song, Q. Q. et al. Home-made online hyphenation of pressurized liquid extraction, turbulent flow chromatography, and high performance liquid chromatography, Cistanche deserticola as a case study. J. Chromatogr. A 1438, 189–197 (2016).
Chapple, C. C. S., Walker, M. A. & Ellis, B. E. Plant tyrosine decarboxylase can be strongly inhibited by L-α-aminooxy-β-phenylpropionate. Planta 167, 101–105 (1986).
Torrens-Spence, M. P. et al. Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins. Proc. Natl Acad. Sci. USA 117, 10806–10817 (2020).
Torrens-Spence, M. P. et al. Biochemical evaluation of the decarboxylation and decarboxylation-deamination activities of plant aromatic amino acid decarboxylases. J. Biol. Chem. 288, 2376–2387 (2013).
Bertoldi, M., Gonsalvi, M., Contestabile, R. & Voltattorni, C. B. Mutation of tyrosine 332 to phenylalanine converts dopa decarboxylase into a decarboxylation-dependent oxidative deaminase. J. Biol. Chem. 277, 36357–36362 (2002).
Torrens-Spence, M. P., Lazear, M., Von Guggenberg, R., Ding, H. Z. & Li, J. Y. Investigation of a substrate-specifying residue within Papaver somniferum and Catharanthus roseus aromatic amino acid decarboxylases. Phytochemistry 106, 37–43 (2014).
Hawkins, C. F., Borges, A. & Perham, R. N. A common structural motif in thiamin pyrophosphate-binding enzymes. FEBS Lett. 255, 77–82 (1989).
Torrens-Spence, M. P. et al. Biochemical evaluation of a parsley tyrosine decarboxylase results in a novel 4-hydroxyphenylacetaldehyde synthase enzyme. Biochem. Biophys. Res. Commun. 418, 211–216 (2012).
Janes, S. M. et al. A new redox cofactor in eukaryotic enzymes: 6-hydroxydopa at the active site of bovine serum amine oxidase. Science 248, 981–987 (1990).
Mydy, L. S., Chigumba, D. N. & Kersten, R. D. Plant copper metalloenzymes as prospects for new metabolism involving aromatic compounds. Front. Plant Sci. 12, 692108 (2021).
Mura, A. et al. Tyramine oxidation by copper/TPQ amine oxidase and peroxidase from Euphorbia characias latex. Arch. Biochem. Biophys. 475, 18–24 (2008).
Zarei, A. et al. Apple fruit copper amine oxidase isoforms: peroxisomal MdAO1 prefers diamines as substrates, whereas extracellular MdAO2 exclusively utilizes monoamines. Plant Cell Physiol. 56, 137–147 (2015).
Yoo, H. et al. An alternative pathway contributes to phenylalanine biosynthesis in plants via a cytosolic tyrosine: phenylpyruvate aminotransferase. Nat. Commun. 4, 2833 (2013).
Taguchi, G. et al. Malonylation is a key reaction in the metabolism of xenobiotic phenolic glucosides in Arabidopsis and tobacco. Plant J. 63, 1031–1041 (2010).
Taguchi, G., Shitchi, Y., Shirasawa, S., Yamamoto, H. & Hayashida, N. Molecular cloning, characterization, and downregulation of an acyltransferase that catalyzes the malonylation of flavonoid and naphthol glucosides in tobacco cells. Plant J. 42, 481–491 (2005).
Liu, Y. et al. Identification and functional application of a new malonyltransferase NbMaT1 towards diverse aromatic glycosides from Nicotiana benthamiana. RSC Adv. 7, 21028–21035 (2017).
Manjasetty, B. A. et al. Structural basis for modification of flavonol and naphthol glucoconjugates by Nicotiana tabacum malonyltransferase (NtMaT1). Planta 236, 781–793 (2012).
Mo, T. et al. Combinatorial synthesis of flavonoids and 4-hydroxy-δ-lactones by plant-originated enzymes. Chinese J. Org. Chem. 35, 1052–1059 (2015).
Liu, X. et al. Cell culture establishment and regulation of two phenylethanoid glycosides accumulation in cell suspension culture of desert plant Cistanche tubulosa. Plant Cell Tiss. Org. 134, 107–118 (2018).
Liu, X. et al. Molecular characterization and structure basis of a malonyltransferase with both substrate promiscuity and catalytic regiospecificity from Cistanche tubulosa. Acta Pharm. Sin. B 14, 2333–2348 (2024).
Chen, C. et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742 (2023).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Hong, B. et al. Biosynthesis of strychnine. Nature 607, 617–622 (2022).
Acknowledgements
This work was financially supported by National Natural Science Foundation of China (Grant No. 82173922, X.L.; Grant No. 81773832, P.T.; Grant No. 81402809, X.L.), National Key Research and Development Program Special Project of Synthetic Biology (Grant No. 2023YFA0914100-2023YFA0914103, S.-P.S.), Beijing Natural Science Foundation (Grant No. 7192112, X.L.), Fundamental Research Funds for the Central Universities (Grant No. 2023-JYB-JBQN-054, X.L.) and Young Elite Scientists Sponsorship Program by CAST (Grant No. CACM-2018-QNRC1-02, X.L.).
Author information
Authors and Affiliations
Contributions
P.T., X.L., S.-P.S. and J.L. conceived the project and designed the studies. W.H. performed experiments related to upstream pathways; Y.Y. and W.T. performed experiments related to downstream pathways; X.C. performed experiments related to heterologous expression in tobacco; Y.W., T.M., X.X., S.Z. and Y.L. contributed to the candidate genes screen and enzymatically functional identification work; Y.S., X.W., J.W. and Y.J. provide guidance to the experimental performance; X.L., W.H., Y.Y., W.T. and X.C. wrote the manuscript; P.T., S.-P.S. and J. L. revised the paper; P.T. and X.L. acquired fundings.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Chang-Jun Liu, Iman Mirmazloum and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Huang, W., Yan, Y., Tian, W. et al. Complete pathway elucidation of echinacoside in Cistanche tubulosa and de novo biosynthesis of phenylethanoid glycosides. Nat Commun 16, 882 (2025). https://doi.org/10.1038/s41467-025-56243-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-56243-9