Abstract
Gene drives are alleles that can bias the inheritance of specific traits in target populations for the purpose of modification or suppression. Here, we construct a homing suppression drive in the major urban malaria vector Anopheles stephensi targeting the female-specific exon of doublesex, incorporating two gRNAs and a nanos-Cas9 to reduce functional resistance and improve female heterozygote fitness. Our results show that the drive was recessive sterile in both females and males, with various intersex phenotypes in drive homozygotes. Both male and female drive heterozygotes show only moderate drive conversion, indicating that the nanos promoter has lower activity in A. stephensi than in Anopheles gambiae. By amplicon sequencing, we detect a very low level of resistance allele formation. Combination of the homing suppression drive and a vasa-Cas9 line boosts the drive conversion rate of the homing drive to 100%, suggesting the use of similar systems for population suppression in a continuous release strategy with a lower release rate than SIT or fsRIDL techniques. This study contributes valuable insights to the development of more efficient and environmentally friendly pest control tools aimed at disrupting disease transmission.
Similar content being viewed by others
Introduction
Vector-bone diseases, including malaria, Dengue fever, and West Nile Virus, continue to pose a global health threat, causing numerous annual fatalities worldwide. Disease vector management is crucial for halting transmission. However, overuse of chemical-based treatments has accelerated the emergence of pesticide resistance1. In the ongoing effort to combat these diseases, various genetic engineering tools such as irradiation/chemosterilant-induced or transgene-based sterile insect technique (SIT)2,3,4, release of insects carrying a dominant lethal (RIDL)5,6,7, and Wolbachia-mediated incompatible insect technique (IIT)8,9,10 have been developed. Notably, gene drive technology stands out as a highly efficient control tool with the potential to affect a whole population with a minimal release.
The concept of homing gene drive, utilizing selfish genetic elements, was proposed two decades ago11, but its development was hindered by lack of ability to target specific sequences. The advent of CRISPR genome editing tools marked a significant breakthrough, enabling more sophisticated and practical gene drive strategies. A homing gene drive consists of a Cas9 endonuclease and gRNA cassette capable of cleaving the target site, generating a double-strand break in the wild-type allele, and utilizing homology-direct repair to copy the drive allele into the wild-type allele. This converts a heterozygote into homozygote in the germline, enabling the drive to bias its inheritance in the offspring, so-called Super-Mendelian inheritance. However, if end-joining repair takes place instead of homology-directed repair, the target site can be mutated. This is called a resistance allele because it can no longer be cleaved by Cas9/gRNA, hindering gene drive transmission.
While gene drive development has been explored in model species such as fruit flies12,13,14, mice15,16, and Arabidopsis17,18, as well as non-model organisms like yeast19, herpesviruses20, and agriculture pests21, the overall efficiency in insect species beyond possibly a few studies in Drosophila12,13,22, Aedes23 and especially Anopheles mosquitoes24,25,26 has been relatively lower due to varies factors such as low conversion rate, high fitness cost, parental effects and resistant allele formation. Several potential solutions have been proposed to address these issues. For instance, drive conversion and fitness of drive carriers can be promoted by improved regulatory elements or coding sequences22,24,27,28, while carefully selected targets24,27, “toxin-antidote” (or “Cleave and Rescue”) systems12,13,29 and gRNA multiplexing12,13,30 can help in removing resistant alleles.
Gene-drive strategies primarily aim at population modification or suppression. Population suppression gene drives have garnered attention for their potential to directly remove pests by disrupting reproductive capabilities or biasing the sex ratio of a population. Common targets include essential female fertility genes. In proof-of-principle studies in Drosophila melanogaster targeting yellow-G, only moderate drive equilibrium frequency was achieved in cage populations30, limiting suppressive power. This was caused by lack of high drive conversion rates, moderate fitness costs, and moderate rates of resistance allele formation from early embryo cleavage by maternally deposited Cas9 and gRNA. Drives utilizing improved germline promoters28 or decoupling of drive and fertility gene cleavage14 could mitigate these issues. Another promising target for population suppression is the sex determination pathway, with the female-specific exon of doublesex (dsx) emerging as a key target. While laboratory cage trials have shown successful suppression in Anopheles gambiae24,31, it is important to note that these results are not definitive for larger populations. Functional resistance and fitness costs remain potential issues that could affect the efficacy of this approach in broader populational or ecological contexts. A different interpretation of previous experimental data32 indicated that the nanos promoter could be have equal or better performance for drive conversion rate and especially fitness costs, which inspired our design in this study.
Although studies on modification drives have been published in A. stephensi, the major urban malaria vector in Asia that is becoming more invasive in East Africa, these efforts have met with challenges. The initial design targeting kh had high fitness costs and would tend to suffer from resistance or suppress the population33,34. A later rescue design improved fitness but lacked anti-malaria factors and still had some functional resistance25. Also, no suppression gene drive study has been reported for this species. Here, we construct a suppression drive (named HSDdsx) as well as a vasa-Cas9 line. The suppression drive exhibits intermediate drive inheritance rate and minimal resistance, indicating success of the 2-gRNA design but failure of the nanos-Cas9 allele to achieve high cut rates. Crosses of both lines result in significantly improved inheritance rate (to 100%) in HSDdsx but also female heterozygote sterility, suggesting their use in a continuous-release deployment for substantially more efficient population suppression compared to SIT and fsRIDL. Our study lays the groundwork for the further construction of highly efficient gene drives for population management of A. stephensi and provides valuable insights for other non-model organisms.
Results
Construction of a homing suppression drive HSDdsx
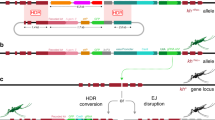
We built a suppression drive (HSDdsx) in A. stephensi, specifically disrupting the female exon of the haplosufficient gene dsx (Fig. 1A). It was expected that the female offspring inheriting two disrupted alleles would be sterile and eliminated from the population (Fig. 1B). Our suppression drive design was inspired by a previous study in A. gambiae24 but incorporated several enhancements. Firstly, we employed two gRNAs, targeting closely located sequences around the boundary, each regulated by distinct Pol III promoters (U6A and 7SK) and positioned in opposite directions to prevent recombination and deletion of one of the genes. The proximity of these gRNA target sites was intentional to maintain high rates of homology-directed repair35. Second, Cas9 expression was driven by a nanos promoter, which may reduce fitness costs32 caused by somatic disruption of dsx in heterozygous females. Following the injection of 1110 eggs, 58 adults survived, and upon crossing with wild-type, one EGFP-positive male was recovered. Subsequently, this male was crossed with wild-type females to establish the HSDdsx transgenic line. Morphological analysis revealed no observable differences between heterozygous drive carriers and wild-type mosquitoes. Additionally, molecular investigation confirmed the correct integration and integrity of our drive construct with successful PCR amplification and sequencing of fragments from genomic DNA spanning both sides of the homology arm in the original drive plasmid.
A HSDdsx is inserted into the female-specific intron 4-exon 5 boundaries of dsx. The drive element contains a nanos-Cas9 cassette, a 3xP3-EGFP-SV40 marker, and two gRNAs under the control of U6A and 7SK promoters. B A drive allele will express Cas9 to cut the wild-type allele in the germline, after which the wild-type allele is either converted to drive allele or disrupted via end-joining. Maternally deposited Cas9/gRNA can cut wild-type alleles in the embryo, followed by disruption through end-joining. Females carrying two nonfunctional alleles (drive or resistance) will be sterile. C Drive heterozygotes were outcrossed with wild-type, and their offspring were phenotyped for EGFP fluorescence, indicating the rate of drive inheritance. The size of the dots indicates the relative sample size of larval progeny from a single cross batch. “n” indicates the total number of offspring in each group. The mean and standard error of the mean (SEM) are displayed.
Drive efficiency test of HSDdsx
To assess the drive efficiency of HSDdsx, we initiated crosses between drive heterozygotes (G0) and wild-type. The generated heterozygous drive carriers (G1) were then either outcrossed with wild-type or intercrossed with heterozygous siblings. Their G2 larvae progeny were screened for fluorescence, and morphological phenotyping was conducted upon their emergence as adults. The drive inheritance rate was measured as the percentage of EGFP-expressing larvae. Our findings revealed similar drive inheritance rates for HSDdsx from male (74.6% ± 1.4%) and female (73.6% ± 2.6%) drive parents (p = 0.741, z-test). These correspond to drive conversion rates of 50% and 45%, respectively. Notably, both drive male and female groups exhibited drive inheritance rates significantly higher than the 50% Mendelian expectation (p < 0.0001, z-test). For the crosses between male and female heterozygotes, the drive carrier rate among offspring was 94.7% ± 1.0%, consistent with the inheritance rates from each heterozygote parent (Fig. 1C, Source Data). This result suggested that the expression of Cas9 driven by the nanos promoter and two gRNAs respectively driven by U6A and 7SK promoters were functional in causing biased inheritance of the drive allele, though the drive efficiency was lower than a similar suppression drive in the closely related species Anopheles gambiae24.
Fertility tests for drive females revealed no significant difference in hatch rates among eggs from drive and wild-type females (Supplementary Fig. 6, Source Data), regardless of whether the drive was maternally (p = 0.0608, z-test) or paternally (p = 0.7330, z-test) inherited. However, the standard errors of these hatch rates were relatively high, likely due to significant batch variations.
Phenotyping of offspring resulting from the outcross of heterozygotes and wild-type revealed normal sex morphological features, of which male had plumose antennae and downward-facing claspers in its external genitalia while female had pilose antennae and cerci (Fig. 2). We randomly collected 15 offspring for genotyping, which showed no resistance alleles in either drive or non-drive individuals. However, when drive heterozygotes were intercrossed, 60% of drive-carrying offspring showed three intersex phenotypes, which we termed as intersex (XX), intersex (XY), and intersex-90 (XY). Intersex (XX) was genetically female, showing less bushy antennae and upward rotated claspers. Intersex (XY) and intersex-90 (XY) were both genetically male and had male-like bushy antennae, but the former had upward claspers while the latter exhibited claspers that were twisted 90 degrees (Fig. 2 and S1). Genotyping revealed that all of the intersex (XX) and intersex-90 (XY) mosquitoes were drive homozygous females and males, respectively, and most intersex (XY) mosquitoes were also homozygous males. Note that two out of 14 intersex (XY) mosquitoes had mosaic wild-type/resistance alleles, with the wild-type sequence dominant (in addition to one drive allele). While it is unlikely that resistance alleles could disrupt male expression, this may be possible. More likely, these mosquitoes were misidentified because wild-type males also exhibit up-toward claspers shortly after emergence, but they eventually rotate 180 degrees within ~48 h post-eclosion. The upward claspers of these two individuals might have eventually been able to rotate in a more extended time window (though phenotyping was conducted five days post-eclosion), or they may have had a developmental defect. These male intersex mosquitoes were respectively pool-crossed to wild-type females, each with 5 males and 15–20 females and three cages each for intersex and intersex-90 males, but none of them produced any progeny. Our findings suggest that dsx is likely a haplosufficient gene in A. stephensi and that insertion of the drive construct into the female-specific intron 4-exon 5 boundary disrupted dsx expression and introduced sterility in both males and females.
To investigate the potential causes of intersex phenotypes, we analyzed the splicing patterns and relative expression levels of dsx in adult mosquitoes exhibiting different phenotypes. We identified two female-specific dsx transcripts: one containing the complete exon 5 (dsxF1) and another lacking part of its 5’ fragment (dsxF2), consistent with previous findings in A. stephensi36. Semi-quantitative RT-PCR and real-time PCR analysis indicated that intersex (XX) individuals expressed both dsxF2 and dsxM, albeit at lower relative expression levels compared to wild-type, suggesting a potential contribution to its intersex phenotype. Only dsxM was detected in intersex (XY) adults, which, surprisingly, showed no significant difference from wild-type males (Supplementary Fig. 2). However, it remains unclear whether differential protein expression at other developmental stages or in specific tissues influences intersex phenotypes.
Potential resistance in HSDdsx
To confirm if resistance emerged in our suppression drive line, deep sequencing was conducted around the closely spaced gRNA target sites. 100 drive and non-drive offspring from drive heterozygous parents were pooled for deep sequencing. The result showed a very low level of cleavage at both gRNA target sites, with only 0.657% and 1.38% reads modified in gRNA1 and gRNA2 targets, respectively. Probably only one mosquito had a resistance allele (with a cut at the second gRNA under control of the 7SK promoter) that was formed from the germline or at the early embryo stage (and thus inherited by many or all cells in the offspring). Most detected resistance alleles were found in a small proportion and were thus likely formed by somatic expression or mosaic cleavage from maternal Cas9 in a small number of cells at later developmental stages. None of these mutations seemed to be in-frame, making it unlikely that any would be functional (Supplementary Fig. 3). As expected from the low cut rates, the frequency of simultaneous cutting of both gRNAs was even lower (<0.00021%). This result reveals that both gRNAs were active and that resistance rates were overall very low, preventing a thorough assessment of functional versus nonfunctional resistance.
Modeling of HSDdsx
To assess HSDdsx performance at population level, we applied an SLiM mosquito model to analyze allele frequency and population size on a weekly basis following a 5% introduction of heterozygous drive males into the population. Drive performance parameters (Table S1) were based on experimental measurements of drive conversion and female fertility. Nonfunctional resistance alleles could only be formed by maternal deposition at a 1% rate, a pessimistic estimate (we would likely have found some resistance carriers based on phenotyping with this rate). We compared our drive performance in two scenarios where male homozygotes are fertile and sterile, respectively. In addition, two different low-density growth rates we used to simulate different potential population dynamics.
Our results demonstrated a consistent increase in drive allele frequency until reaching equilibrium (Fig. 3A). However, while all drives affected the population size, only the population with fertile male homozygotes and lower low-density growth rate was successful at eliminating the target population (Fig. 3B). The drive’s genetic load (suppressive power) was too weak to overcome a low-density density growth rate of 6, though with a reduced low-density density growth rate of 2, elimination could be achieved (such a low growth rate could still be realistic if it approximates the effects of a competing species and predators37). This was largely because of low drive conversion and female fitness costs, resulting in a balance of new drive alleles from conversion and removal of drive alleles in homozygous females. The small amount of nonfunctional resistance allele formation was detrimental to the drive as well, but it had only a minor effect on the drive’s suppressive power.
Drive heterozygous males were released into the population at 5% frequency. Drive performance parameters are based on experimental results (Table S1), with and without homozygous male sterility, as well as low and high low-density growth rates. A Allele frequencies and B average number of fertile females are displayed. Each simulation was repeated five times, and average values are shown in the figure. r2 indicates nonfunctional resistance alleles.
Furthermore, sterile homozygous males substantially reduced suppression power. This was because nonfunctional resistance alleles gained an advantage in this scenario over the drive allele (only drive homozygous males were sterile, while for females, any combination of drive and nonfunctional resistance alleles was sterile). Notably, the population size expanded when low-density growth rate was high (Fig. 3B). This was because the reduction in larvae competition (there were fewer eggs because of sterile females and reduced fertility females) allowed more larvae to survive, which has been seen in some natural environments38. However, the number of fertile females (the only type that can bite for this drive) did not increase.
Combination of HSDdsx and vasa-Cas9
In addition to HSDdsx, we generated another A. stephensi transgenic line, which was based on a “toxin-antidote” modification drive in a previous D. melanogaster study13. However, this line was not able to substantially bias its inheritance, likely due to low cleavage efficiency at its hairy target site. We thus employed it as a vasa-Cas9 line. To further investigate the performance of both lines, we first crossed HSDdsx with vasa-Cas9 (G0) to generate double heterozygous individuals (G1). Though both lines had the 3xP3-EGFP marker, their fluorescence patterns were different, enabling the reliable identification of each allele, even when both were present in the same individual (Fig. 4B). The inheritance rates of HSDdsx and vasa-Cas9 in G1 were 79.8% ± 1.4% and 50.6% ± 1.8%, respectively, when HSDdsx heterozygous females were crossed with vasa-Cas9 heterozygous males. This was consistent with the results of drive efficiency tests of each line. The vasa promoter has known activity in both somatic tissues and also has significant maternal deposition33. To avoid Cas9 deposition from a maternal vasa-Cas9 allele, only the double heterozygotes from HSDdsx females and vasa-Cas9 males were selected to cross with wild-type. Notably, all the double heterozygotes were confirmed as either intersex or morphological males, without any females being identified, indicating complete masculinization in drive females due to somatic cutting of dsx. Additionally, we genotyped seven of these double heterozygous males, six of which showed mosaic mutations in both the dsx gRNA target sites, indicating high somatic expression of Cas9 and cleavage activity from both gRNAs.
A HSDdsx females and vasa-Cas9 males were crossed to generate double heterozygous males, which were crossed to wild-type for assessing drive inheritance in offspring. Disrupted dsx can come from drive conversion and/or resistance allele formation in somatic cells. B The left panel picture was taken with an EGFP filter, and the right panel was under white light. Triangles with different colors show the differences in the fluorescence patterns between the lines. Wild-type (WT) mosquitoes are also displayed for comparison. C In the offspring of double heterozygous drive males and wild-type females, HSDdsx and vasa-Cas9 inheritance rates were measured.
Double heterozygous males were further crossed to wild-type to create G2 offspring. The phenotyping of G2 offspring revealed a significant boost in the HSDdsx drive inheritance to 100% (Wilsonian 95% confidence interval: 99.5%-100%), while the inheritance of vasa-Cas9 was 43.2% ± 2.36% (Fig. 4C). This showed that an extra Cas9 source was sufficient to induce higher germline cleavage and conversion of HSDdsx. The vasa-Cas9 inheritance was slightly reduced (even compared to previous crosses of vasa-Cas9 and wild-type), which could be potentially explained by a fitness cost of individuals carrying both alleles, leading to death in early egg or larval stages, or perhaps vasa-Cas9 carrying sperm were less competitive than wild-type sperm. However, phenotyping inaccuracy could also contribute to this apparent result.
To assess phenotyping accuracy, we conducted a small-scale genotyping of mosquitoes carrying different markers (Supplementary Fig. 7). Our result confirmed that most of our phenotyping was correct, including all identification of HSDdsx (both alone and together with vasa-Cas9). Identification of vasa-Cas9-only individuals was also successful. However, one sample was misidentified as HSDdsx-only when in fact it had both constructs. We found that part of the larval tail showing fluorescence in vasa-Cas9 was fragile and sometimes was removed during phenotyping, leading to misidentification of HSDdsx+vasa-Cas9 vs. HSDdsx only phenotype. This could also explain why the inheritance ratio of vasa-Cas9 was lower than the Mendelian expectation.
We observed that females with one HSDdsx and one vasa-Cas9 allele exhibited intersex phenotypes, but the extent of intersex characteristics varied. Females with vasa-Cas9 fathers and HSDdsx mothers displayed up-toward claspers (as shown in intersex XX), whereas female progeny of males heterozygous for both HSDdsx and vasa-Cas9 showed more pronounced abnormalities in external genitalia (Supplementary Fig. 4A). Genotyping of these latter individuals carrying both HSDdsx and vasa-Cas9 alleles confirmed their status as females that were heterozygous for HSDdsx and had mutated alleles at the dsx targets (Supplementary Fig. 4B).
Modeling of the combination of HSDdsx and vasa-Cas9 for SIT-like population suppression
Based on the results of the combination crosses, we propose a SIT-like strategy for population suppression and assessed its capacity via our SLiM mosquito model. This combination drive consists of two components located in unlinked loci, a gRNA-only homing allele targeting dsx and an unlinked Cas9 allele. This allows the system to be self-limiting, though use of our actual lines would allow slightly higher suppressive power at the cost of potential spread of the weak but unconfined HSDdsx into nontarget populations. Our modeling result showed that the population could be successfully suppressed when the release ratios were higher than 0.8 (Fig. 5A). When release ratio was 0.8, 20% of the simulated populations could be successfully suppressed. The drive female frequency and the number of fertile females eventually reached equilibrium when release ratios were lower (Fig. 5B and C). Having a higher release ratio sped up suppression, but this had decreasing returns (Fig. 5D). For comparison, we simulated the dynamics of SIT and late-acting fsRIDL using the same mosquito model. The results indicate that higher release ratios were required for effective population suppression: A release ratio of at least 6 for fsRIDL and at least 11 for SIT were required to eliminate the population (Supplementary Fig. 5). These findings demonstrated that our combination drive can be applied as an efficient self-limiting suppression strategy with substantially higher efficiency than SIT and fsRIDL strategies. If the vasa-Cas9 were integrated into the drive allele, then efficiency would be even higher39.
Mosquitoes heterozygous for the drive allele and homozygous for the Cas9 allele were released into the population every week. A, B show the number of fertile females and the drive female frequency, respectively, over time at different release ratios. C Number of fertile females at various release ratios when the population is not eliminated. D Actual time needed to suppress the population when elimination is possible. Note that for the release ratio of 0.8, population elimination occurred in four out of 20 simulations, so this release level is shown in (C, D) to assess both situations.
Discussion
In this study, we developed a 2-gRNA homing suppression gene drive (HSDdsx) in A. stephensi. Our experimental findings support several conclusions: 1) the nanos-Cas9 in HSDdsx was functional but not strong enough to induce high germline cut rate and conversion rate; 2) the multiplexed gRNA expression driven by U6 and 7SK promoters was effective, and modeling indicates that this is likely to substantially reduce functional resistance allele formation rates35; 3) resistance allele formation from germline-restricted nanos-Cas9 was minimal, though this could have been due to low expression in general; 4) the expression of vasa-Cas9 in the transgenic line was high, supporting 100% drive conversion efficiency in HDSdsx.
Our suppression drive was inspired by the previous study in A. gambiae, with improvements including gRNA multiplexing, the use of a different germline promoter nanos, and an insect codon-optimized Cas9 sequence. Suppression drives are generally more sensitive than modification drives to fitness costs from leaky somatic Cas9 expression or resistance allele formation from early embryonic activity of Cas9/gRNA, as reported in D. melanogaster28,30 and Anopheles mosquitoes27,40. Even though A. stephensi and A. gambiae are relatively closely related, the A. stephensi nanos promoter was substantially less effective in A. stephensi than the A. gambiae nanos promoter in A. gambiae, though one caveat to this is that the A. gambiae examples were at a different target site26,40. This difference could be attributed to lower expression of Cas9, which may have been due to the limited size of our nanos promoter (3489 bp, though it was larger than versions used in A. gambiae, which were 1642 ~ 2092 bp26,40), potentially lacking regulatory sequences. nanos may also simply have lower expression in A. stephensi than A. gambiae. Regulatory elements flanking the drive insertion site could also have affected expression. To enhance Cas9 germline expression and genetic load of the drive, future experiments can explore the activity of nanos promoters in different lengths, target different genes or loci, or test other germline promoters (e.g., the zpg promoter, which also has excellent performance in A. gambiae24,40). However, it is also possible that our gRNAs were simply lower activity (despite one being effective in A. gambiae with the zpg promoter24) and that a better gRNA would function well with nanos-Cas9. Similarly, while vasa-Cas9 offered better performance, it is not clear if vasa-Cas9 alone would have allowed 100% drive conversion because it was here combined with a nanos-Cas9 source (though it supported high drive conversion alone in previous studies25,33).
The existence of intersex drive homozygous males in our study (Fig. 2) can be potentially explained by the insertion of the large drive construct (with Cas9, gRNA, and fluorescent protein genes), affecting the transcription level and potentially the splicing of male-specific dsx exons, which are downstream of the gene drive. Although our RT-PCR and qPCR results did not show significant difference between intersex (XY) and wild-type male at adult stage, it is still unclear whether the expression at other developmental stages or tissues contributed to this morphological differentiation. Heterozygous males may be unaffected because the wild-type allele is haplosufficient. Similarly, we would expect male resistance allele homozygotes to be fertile because a resistance allele is unlikely to affect male transcription due to its smaller size and reduced complexity compared to the drive. Also of note is that our drive heterozygous males with vasa-Cas9 were fertile. The high level of somatic expression means that many or even nearly all wild-type alleles would have been converted to drive or resistance alleles. This could indicate that dsx expression timing plays a role if drive conversion was dominant in somatic cells (some male dsx expression could occur normally before somatic cell drive conversion), or it could indicate that resistance alleles do not affect male fertility if these occurred in a large enough fraction of somatic cells.
Based on the publication in A. gambiae, disrupting the female-specific exon of dsx converted only homozygous null-mutant females (either with or without drive) into an intersex phenotype causing sterility, while males remained healthy and fertile if homozygous for a fluorescent protein24. It remains unclear if drive males were fertile in this study. It is possible that there is no difference between this study and our own if A. gambiae drive homozygous males (as opposed to fluorescent protein homozygous males) are in fact sterile. If they are fertile, then the difference could be species-based or related to specific sequences in the drives. Perhaps nanos 3′ regulatory elements terminated male transcription at higher rates than zpg elements. A similar intersex phenotype in homozygous males has also been observed in Drosophila suzukii22, while other reports in D. suzukii and D. melanogaster showed dominant sterility in drive females, illustrating the complexity of sex-specific dsx expression22,41. These, together with the current study, demonstrate the necessity of more detailed assessments of both male and female genotypes in such suppression drives targeting dsx. Homozygous male sterility could substantially reduce overall drive efficiency (though genetic load would still be high with nearly 100% drive conversion) and make the drive more vulnerable to chasing42,43.
Resistance alleles could be classified into functional (r1) and non-functional (r2) alleles. While germline-restricted promoters can help in reducing total resistance allele formation, gRNA multiplexing is a useful method to reduce the fraction of r1 alleles, which is essential for suppression drive success. The separate gRNA expressing cassettes (with different promoters, U6 and 7SK) used in HSDdsx demonstrated strong gRNA expression capacity, providing an alternative tool for future gRNA multiplexing. Such additional promoters may be needed to avoid undesired recombination44 if multiple gRNAs cannot be expressed effectively from a single promoter. While increased numbers of gRNAs can eventually reduce drive conversion efficiency (though not total cutting rate)35, it is more important to minimize functional resistance. In this study, the reduced drive efficiency was not likely due to use of a second gRNA because the total resistance allele formation rate was very low, and failed drive conversion due to gRNA multiplexing is likely to produce resistance alleles35. Further, if resistance alleles produced by the multiple gRNAs turn out to be dominant sterile, as seen in dsx for D. melanogaster41 and for at least some alleles in A. gambiae45, then the drive may prove to have better performance than a standard homing suppression drive, even with male homozygous sterility.
Our research yields insights for the advancement of efficient and environmentally friendly pest control tools aimed for disrupting disease transmission. It suggests that constructing high-efficiency drives in A. stephensi could be achieved through modest modifications to our existing constructs. For the homing suppression drive, a stronger germline promoter can be used to drive Cas9 expression, which could increase drive efficiency and potentially suppress the population even with homozygous sterility in both sexes. It may also be possible to restore male homozygote fertility. Alternately, self-limiting suppression systems based on dsx with vasa-Cas9 could also provide improvements over existing methods.
Methods
Plasmid design and construction
Donor and helper plasmids were generated employing the Gibson assembly method. The donor plasmid designed for homing-based suppression denoted as HSDdsx, contained a Cas9 coding sequence controlled by the nanos promoter, a 3xP3-EGFP-SV40 fluorescence marker, two distinct gRNA cassettes under the control of the U6A and 7SK promoters, and flanking homology arms facilitating homology-directed repair-mediated integration. The U6-gRNA target (gRNA1: 5’-ttcaactacaggtcaagcgg-3’) was strategically positioned at the highly conserved intron 4-exon 5 boundary, consistent with a prior Anopheles gambiae study24. The target of 7SK-gRNA (gRNA2: 5’-cgcaataccacccgtcagag-3’) was situated within exon 5, 56 bp downstream of gRNA1.
An independent Cas9 helper plasmid was also constructed, featuring a Cas9 coding sequence driven by the vasa promoter, to facilitate knock-in for both the HSDdsx and vasa-Cas9 constructs. Additionally, two gRNA-expressing helper plasmids were developed exclusively for the transformation of vasa-Cas9, which contained a recoded hairy, a Cas9 coding sequence driven by vasa promoter, a 3xP3-EGFP-SV40 fluorescence marker and a U6-gRNA cassette. The target sites of these two gRNA plasmids (KI-gRNA1: 5’-cacacatccaaaatggtgac-3’; KI-gRNA2: 5’-ggccaccagccagataccgc-3’) were located around the translation start site of hairy, enabling successful integration of the vasa-Cas9 construct and subsequent translation of the recoded coding sequence for gene function rescue.
All the regulatory elements and target gene sequences were identified by reciprocal BLAST analysis of their homologs in D. melanogaster against the genome of A. stephensi through NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). gRNA target sites, their activity, and their potential off-target sites were analyzed by using the online tool CHOPCHOP.
All plasmids were constructed using Hifi DNA Assembly Cloning Kit (NEB, USA) and then miniprep with ZymoPure Midiprep Kit (Zymo Research, USA). Plasmid sequences were confirmed with Sanger sequencing by BGI. The final plasmid sequences are available at GitHub (https://github.com/jchamper/ChamperLab/tree/main/Anopheles-stephensi-dsx-Drive).
Mosquito rearing
All mosquitoes were maintained in a containment room at 27 ± 1 °C, 75% humidity, and a 12 h light/dark cycle. Larvae and adults were provided with fish food (Hikari, Japan) and a 10% sucrose solution, respectively. Adult mosquitoes were housed in 30 cm × 30 cm cages for mating, and females were blood-fed using the Hemotek blood-feeding system (Hemotek, UK) with defibrinated cow blood, except for injected females, which usually blood-fed on JC. All biosafety protocols were approved by Peking University.
Embryonic microinjection and germline transformation
Three to four days post blood meal, plastic cups covered with wet filter paper were put into the cage for a 30 min interval to collect eggs, which were subsequently lined up for microinjection. The injection mix for generating the HSDdsx line comprised 152 ng/μL of the donor plasmid, 300 ng/μL of the vasa-Cas9 helper plasmid, and 300 ng/μL of Cas9 protein. For the vasa-Cas9 line, the injection mix included 613 ng/μl of the donor plasmid, ~100 ng/μL of each gRNA plasmid, and 170 ng/μL of the vasa-Cas9 helper plasmid. Surviving G0 mosquitoes were subsequently mated with wild-type counterparts, and the resulting G1 larvae were screened for green fluorescence using the NIGHTSEA system (EMS, USA). Positive lines were maintained as heterozygous through outcrosses with wild-type (HSDdsx) or as homozygotes (vasa-Cas9) via intercrosses.
Crosses and phenotypes
To evaluate drive efficiency, heterozygous transgenic mosquitoes were crossed with their siblings or wild-type mosquitoes. Progeny was screened for fluorescence at the larval stage. Drive carriers (displaying fluorescence) and non-drive individuals (lacking fluorescence) were segregated and reared separately until adulthood. Subsequently, individuals were sexed using a stereo microscope (Olympus, USA).
Additionally, in the fertility test, either male or female drive heterozygous mosquitoes were crossed with wild-type of the opposite sex. Their drive daughters were subsequently crossed to wild-type mosquitos in a single-pair mating setup. The number of eggs laid and hatched larvae as well as drive inheritance rate were subsequently recorded.
In investigating the potential enhancement of HSDdsx drive efficiency with an additional vasa-Cas9 source, HSDdsx and vasa-Cas9 adults were crossed to generate double-heterozygous lines containing both drive alleles. The resulting double-heterozygous males were then crossed with wild-type females, and their offspring were phenotyped.
Statistics and reproducibility
Data were originally pooled (see Source Data) to calculate drive inheritance, drive conversion, and hatch rate. However, this approach did not account for batch variability because all offspring from different cages, parents, and experiments were combined together. To address this issue, we employed a “batch effect analysis” method using the R program, which accommodates variance between cross batches. It uses a generalized linear mixed model fit by maximum likelihood (Adaptive Gauss-Hermite Quadrature, nAGQ = 25). This approach usually results in slightly different parameter estimates and increased standard error estimates, but differences can be larger when batch effects are prominent. All reported p-values are derived from this method based on the z-test. The R program is available on GitHub (https://github.com/jchamper/ChamperLab/tree/main/Anopheles-stephensi-dsx-Drive).
No statistical method was used to predetermine sample size. No data were excluded from the analyses. The experiments were not randomized. The Investigators were not blinded to allocation during experiments and outcome assessment.
Genotyping
Genomic DNA was extracted using either DNAzol Reagent (Invitrogen, UK) or the Animal Genomic DNA Quick Extraction Kit (Beyotime, China). PCR reactions were performed with Q5 High-Fidelity DNA Polymerase (NEB, UK). Genomic integration of both constructs was confirmed through PCR and Sanger sequencing. For genotyping of each target gene, a primer pair covering the target region was designed to detect possible end-joining-induced resistant alleles. Primers (708: 5’- atcttgctcctcacttgccc −3’; 710: 5’- ggtgtcgcccactccttaaac −3’) were designed for amplifying a 539 bp region covering dsx target sites, and primers (711: 5’- tcaaagctgccacggatctc −3’; 714: 5’- aacccagactatgtgaaggatg −3’) were used to amplify a 655 bp fragment of GUY1 specifically present in Y chromosome. Another pair of primers (565: 5’- tcgtatcaacaactgtctgaacgagctg-3’; 39: 5’- tgaaggatggccggccaatc-3’) were designed to amplify 557 bp covering a fragment of hairy genomic sequence. In addition, primers (148: 5’-AGCACAAGATTAGCATGACTGAAGTG-3’; 149: 5’ATCAGCCATACCACATTTGTAGAGG-3’; 559: 5’-tgctgccatcttttgagcaacc3’; 560: 5’-TCCATAATGGGCTTATTCGAGCG-3’) were used for the genotyping of HSDdsx and vasa-Cas9 fragments.
Semi-quantitative RT-PCR and real-time PCR
Adult mosquitoes with various phenotypes were collected for total RNA extraction using the RNA Kit (Qiagen, Germany), with each biological replicate consisting of a single mosquito. The same quantity of RNA was utilized for cDNA synthesis with the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher, USA). RT-PCR and real-time PCR were subsequently performed with LightCycler 480 SYBR Green I Master (Roche, Switzerland), in order to compare the gene expression patterns of drive homozygous males and females against wild-type mosquitoes.
We first applied a pair of primers located in the common exon 4 and exon 6 (780: 5’-aatggctgttggagaagctcg-3’; 781: 5’-accagcgcttcgtacgtc-3’), yielding 1122 bp and 145 bp amplicons indicative of male and female dsx, respectively. Additionally, primers (807: 5’-ggagctactcattcgtgtgt-3’; 808: 5’-atgcaatcgtgagtattcgttga-3’) were designed to detect the 5’ region of the complete female-specific exon 5 (dsxF1). A 125 bp fragment of reference gene RpS7 (NCBI accession number: XM_036042790) was amplified using primers (793: 5’-tcaacaacaagaaggcgatcatc-3’; 794: 5’-aatgaacacgacgtgcttgc-3’). GraphPad Prism 10 was employed for figure generation and statistical analysis comparing drive homozygous males to wild-type males.
Amplicon sequencing
Heterozygous HSDdsx adults were crossed with each other, and their offspring were subsequently phenotyped. A total of 100 adults, encompassing both individuals with and without the drive allele were pooled for genomic DNA extraction. The target region was amplified using primers 581 (5’-agaagatgaggctcttgatcttgatc-3’) and 582 (5’-agaactatcgaagaattcggttcacc-3’). Subsequently, PCR products were purified using the Zymoclean Gel DNA Recovery Kit (Zymo Research, USA) to prepare for deep sequencing conducted by Genwiz. Sequencing data has been uploaded into GitHub (https://github.com/jchamper/ChamperLab/tree/main/Anopheles-stephensi-dsx-Drive).
Modeling
A previously developed mosquito-specific SLiM model32,37,46,47, which simulated various life stages of Anopheles mosquitoes, was applied to analyze population dynamics by releasing different combinations of HSDdsx into a wild population. Simulations were run for 317 weeks post release, with 3.167 weeks representing one generation. The wild-type population was allowed to equilibrate for 10 weeks before releasing drive carriers.
For HSDdsx, we set up a one-time release of male heterozygotes with various combinations of low-density growth rate and female heterozygote fitness. The low-density growth rate represents the multiplier of the offspring survival rate under optimal conditions without competition, while the fitness of female drive heterozygotes directly multiples female fecundity. Other default parameters were set up based on our experimental results. The data of its simulated drive allele frequency, nonfunctional (r2) resistance allele frequency, total fertile female number, and total adult number in each week were collected. Low-density growth rate has not been reliably measured in year-round ecological conditions to our knowledge, so our values were selected to show a range of plausible outcomes. We used a linear density growth curve to represent a robust population with intense competition at the larval stage47.
We used the same underlying model for our assessments of self-sustaining and self-limiting suppression systems. While the self-sustaining HSDdsx system has a single release, the self-limiting system has continuous releases. In the self-limiting system, we assumed a fitness-neutral vasa-Cas9 allele and a HSDdsx drive lacking Cas9. In these simulations, mosquitoes that were heterozygous for the split suppression drive allele and homozygous for the Cas9 allele were continuously released into the population every week. If the population was not eliminated, we recorded the number of fertile females and the drive frequency in females. If population elimination took place, we recorded the time that the population reached zero.
Considering that both drives used a gRNA multiplexing strategy to prevent the generation of functional r1 alleles35, only non-functional r2 alleles were modeled. Each simulation was independently run five times, and data was collected for figure generation with Python. The default parameters for both drives are listed in Table S1, and corresponding SLiM scripts and data can be found in GitHub (https://github.com/jchamper/ChamperLab/tree/main/Anopheles-stephensi-dsx-Drive).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All raw data is available in the Source Data file, provided with this paper. Plasmid sequences are available in https://github.com/jchamper/ChamperLab/tree/main/Anopheles-stephensi-dsx-Drive. A DOI link is https://doi.org/10.5281/zenodo.14245820. Source data are provided with this paper.
Code availability
All code is available for free and unlimited use at https://github.com/jchamper/ChamperLab/tree/main/Anopheles-stephensi-dsx-Drive. A DOI link is https://doi.org/10.5281/zenodo.14245820. The code repository is publicly accessible and open for free use.
References
Reid, M. C. & McKenzie, F. E. The contribution of agricultural insecticide use to increasing insecticide resistance in African malaria vectors. Malar. J. 15, 107 (2016).
Robinson, A. S. Genetic Basis of the Sterile Insect Technique. in Sterile Insect Technique: Principles and Practice in Area-Wide Integrated Pest Management (eds. Dyck, V. A., Hendrichs, J. & Robinson, A. S.) 95–114. https://doi.org/10.1007/1-4020-4051-2_4 (Springer Netherlands, Dordrecht, 2005).
Black, W. C., Alphey, L. & James, A. A. Why RIDL is not SIT. Trends Parasitol. 27, 362–370 (2011).
Li, M. et al. Suppressing mosquito populations with precision guided sterile males. Nat. Commun. 12, 5374 (2021).
Fu, G. et al. Female-specific flightless phenotype for mosquito control. Proc. Natl. Acad. Sci. USA 107, 4550–4554 (2010).
Alphey, L. Genetic control of mosquitoes. Annu. Rev. Entomol. 59, 205–224 (2014).
Marinotti, O. et al. Development of a population suppression strain of the human malaria vector mosquito, Anopheles stephensi. Malar. J. 12, 142 (2013).
Adams, K. L. et al. Wolbachia cifB induces cytoplasmic incompatibility in the malaria mosquito vector. Nat. Microbiol. 6, 1575–1582 (2021).
McNamara, C. J. et al. Transgenic expression of cif genes from Wolbachia strain wAlbB recapitulates cytoplasmic incompatibility in Aedes aegypti. Nat. Commun. 15, 869 (2024).
Zheng, X. et al. Incompatible and sterile insect techniques combined eliminate mosquitoes. Nature 572, 56–61 (2019).
Burt, A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc. Biol. Sci. 270, 921–928 (2003).
Champer, J. et al. A CRISPR homing gene drive targeting a haplolethal gene removes resistance alleles and successfully spreads through a cage population. Proc. Natl. Acad. Sci. USA 117, 24377–24383 (2020).
Champer, J. et al. A toxin-antidote CRISPR gene drive system for regional population modification. Nat. Commun. 11, 1082 (2020).
Faber, N. R. et al. Improving the suppressive power of homing gene drive by co-targeting a distant-site female fertility gene. Nat. Commun. 15, 9249 (2024).
Pfitzner, C. et al. Progress toward zygotic and germline gene drives in mice. CRISPR J. 3, 388–397 (2020).
Grunwald, H. A. et al. Super-Mendelian inheritance mediated by CRISPR-Cas9 in the female mouse germline. Nature 566, 105–109 (2019).
Oberhofer, G., Johnson, M. L., Ivy, T., Antoshechkin, I. & Hay, B. A. Cleave and Rescue gamete killers create conditions for gene drive in plants. Nat. Plants 10, 936–953 (2024).
Liu, Y., Jiao, B., Champer, J. & Qian, W. Overriding Mendelian inheritance in Arabidopsis with a CRISPR toxin-antidote gene drive that impairs pollen germination. Nat. Plants 10, 910–922 (2024).
DiCarlo, J. E., Chavez, A., Dietz, S. L., Esvelt, K. M. & Church, G. M. Safeguarding CRISPR-Cas9 gene drives in yeast. Nat. Biotechnol. 33, 1250–1255 (2015).
Walter, M. et al. Viral gene drive spread during herpes simplex virus 1 infection in mice. Nat. Commun. 15, 8161 (2024).
Xu, X. et al. Toward a CRISPR-Cas9-based gene drive in the diamondback moth Plutella xylostella. CRISPR J. 5, 224–236 (2022).
Yadav, A. K. et al. CRISPR/Cas9-based split homing gene drive targeting doublesex for population suppression of the global fruit pest Drosophila suzukii. Proc. Natl. Acad. Sci. USA 120, e2301525120 (2023).
Anderson, M. A. E. et al. A multiplexed, confinable CRISPR/Cas9 gene drive can propagate in caged Aedes aegypti populations. Nat. Commun. 15, 729 (2024).
Kyrou, K. et al. A CRISPR-Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol. 36, 1062–1066 (2018).
Adolfi, A. et al. Efficient population modification gene-drive rescue system in the malaria mosquito Anopheles stephensi. Nat. Commun. 11, 5553 (2020).
Carballar-Lejarazú, R. et al. Dual effector population modification gene-drive strains of the African malaria mosquitoes, Anopheles gambiae and Anopheles coluzzii. Proc. Natl. Acad. Sci. USA 120, e2221118120 (2023).
Hammond, A. et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 34, 78–83 (2016).
Du, J. et al. Germline Cas9 promoters with improved performance for homing gene drive. Nat. Commun. 15, 4560 (2024).
Oberhofer, G., Ivy, T. & Hay, B. A. Cleave and Rescue, a novel selfish genetic element and general strategy for gene drive. Proc. Natl. Acad. Sci. USA 116, 6250–6259 (2019).
Yang, E. et al. A homing suppression gene drive with multiplexed gRNAs maintains high drive conversion efficiency and avoids functional resistance alleles. G312, jkac081 (2022).
Simoni, A. et al. A male-biased sex-distorter gene drive for the human malaria vector Anopheles gambiae. Nat. Biotechnol. 38, 1054–1060 (2020).
Champer, S. E., Kim, I. K., Clark, A. G., Messer, P. W. & Champer, J. Anopheles homing suppression drive candidates exhibit unexpected performance differences in simulations with spatial structure. Elife 11, e79121 (2022).
Gantz, V. M. et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl. Acad. Sci. USA 112, E6736–E6743 (2015).
Pham, T. B. et al. Experimental population modification of the malaria vector mosquito, Anopheles stephensi. PLoS Genet 15, e1008440 (2019).
Champer, S. E. et al. Computational and experimental performance of CRISPR homing gene drive strategies with multiplexed gRNAs. Sci. Adv. 6, eaaz0525 (2020).
Weng, S.-C. et al. Establishing a male-positive genetic sexing strain in the asian malaria vector Anopheles stephensi. bioRxiv 2024.07.17.603997 https://doi.org/10.1101/2024.07.17.603997 (2024).
Liu, Y., Teo, W., Yang, H. & Champer, J. Adversarial interspecies relationships facilitate population suppression by gene drive in spatially explicit models. Ecol. Lett. 26, 1174–1185 (2023).
Bouyer, J. When less is more: accounting for overcompensation in mosquito SIT projects. Trends Parasitol. 39, 235–237 (2023).
Han, Y. & Champer, J. A comparative assessment of self-limiting genetic control strategies for population suppression. bioRxiv 2024.09.23.614516 https://doi.org/10.1101/2024.09.23.614516 (2024).
Hammond, A. et al. Regulating the expression of gene drives is key to increasing their invasive potential and the mitigation of resistance. PLoS Genet. 17, e1009321 (2021).
Chen, W., Guo, J., Liu, Y. & Champer, J. Population suppression by release of insects carrying a dominant sterile homing gene drive targeting doublesex in Drosophila. Nat. Commun. 15, 8053 (2024).
Champer, J., Kim, I. K., Champer, S. E., Clark, A. G. & Messer, P. W. Suppression gene drive in continuous space can result in unstable persistence of both drive and wild-type alleles. Mol. Ecol. 30, 1086–1101 (2021).
Zhang, X., Sun, W., Kim, I. K., Messer, P. W. & Champer, J. Population dynamics in spatial suppression gene drive models and the effect of resistance, density dependence, and life history. bioRxiv 2024.08.14.607913 https://doi.org/10.1101/2024.08.14.607913 (2024).
Green, E. I. et al. A population modification gene drive targeting both Saglin and Lipophorin impairs Plasmodium transmission in Anopheles mosquitoes. eLife 12, e93142 (2023).
Tolosana, I. et al. A Y chromosome-linked genome editor for efficient population suppression in the malaria vector Anopheles gambiae. Nat. Commun. 16, 206 (2025).
Haller, B. C. & Messer, P. W. SLiM 4: multispecies eco-evolutionary modeling. Am. Nat. 201, E127–E139 (2023).
Zhu, J., Chen, J., Liu, Y., Xu, X. & Champer, J. Population suppression with dominant female-lethal alleles is boosted by homing gene drive. BMC Biol. 22, 201 (2024).
Acknowledgements
This study was supported by grants from the National Science Foundation of China (32302455 to XX and 32270672 to J. Champer) and laboratory startup support from Peking University and the Center for Life Sciences to J. Champer. We thank Sam Champer for assistance with batch effect analysis.
Author information
Authors and Affiliations
Contributions
X.X. and J. Champer designed the research. X.X., J. Chen, Y.W., Y.L., Y.Z., J.Y., X.Y., and Z.H. performed the research. X.X. drafted the initial manuscript, and X.X., J. Chen, Y.W., Y.L., Y.Z., J.Y., X.Y., Z.H., B.C., and J. Champer made contributions to reviewing and editing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks John Marshall and Sebald Verkuijl for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xu, X., Chen, J., Wang, Y. et al. Gene drive-based population suppression in the malaria vector Anopheles stephensi. Nat Commun 16, 1007 (2025). https://doi.org/10.1038/s41467-025-56290-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-56290-2
This article is cited by
-
A suppression-modification gene drive for malaria control targeting the ultra-conserved RNA gene mir-184
Nature Communications (2025)