Abstract
Triple-negative breast cancer (TNBC) is known for frequent copy number alterations (CNAs) and metabolic reprogramming. However, the mechanism by which CNAs of metabolic genes drive distinct metabolic reprogramming and affect disease progression remains unclear. Through an integrated analysis of our TNBC multiomic dataset (n = 465) and subsequent experimental validation, we identify copy number amplification of the metabolic gene flavin-adenine dinucleotide synthetase 1 (FLAD1) as a crucial genetic event that drives TNBC progression. Mechanistically, FLAD1, but not its enzymatically inactive mutant, upregulates the enzymatic activity of FAD-dependent lysine-specific demethylase 1 (LSD1). LSD1 subsequently promotes the expression of sterol regulatory element-binding protein 1 (SREBP1) by demethylating dimethyl histone H3 lysine 9 (H3K9me2). The upregulation of SREBP1 enhances the expression of lipid biosynthesis genes, ultimately facilitating the progression of TNBC. Clinically, pharmacological inhibition of the FLAD1/LSD1/SREBP1 axis effectively suppresses FLAD1-induced tumor progression. Moreover, LSD1 inhibitor enhances the therapeutic effect of doxorubicin and sacituzumab govitecan (SG). In conclusion, our findings reveal the CNA-derived oncogenic signalling axis of FLAD1/LSD1/SREBP1 and present a promising treatment strategy for TNBC.
Similar content being viewed by others
Introduction
The latest epidemiological statistics show that breast cancer now ranks as the most commonly diagnosed cancer for women globally1. TNBC is a subtype of breast cancer that is characterized by the absence of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) expression. TNBC accounts for approximately 10-15% of breast cancer cases and is linked to an unfavorable outcome, elevated rates of recurrence, heightened metastasis, and mortality2. Currently, chemotherapy remains the established mainstay treatment for TNBC, highlighting the urgent need for the exploration of novel and efficient therapeutic approaches for this aggressive subtype of breast cancer.
CNA is a vital characteristic of TNBC, occurring when there are structural changes in DNA that lead to the gain, amplification, loss, or deletion of copies of normal DNA segments. As one of the hallmarks of cancer, CNA is known to drive tumor growth and is associated with prognosis3. Previous study revealed that breast cancer is a disease influenced by CNAs4. With respect to breast cancer subtypes, TNBC shows a greater incidence of CNAs than other subtypes5,6. Apart from CNA, tumor metabolism is a promising focus for cancer treatment. Previous studies have reported that CNAs in certain oncogenes can influence tumor progression by reprogramming metabolic pathways. For instance, MYC activation driven by CNA promotes the expression of metabolic genes involved in glycolysis, glutaminolysis, and nucleotide and lipid synthesis, and targeting these metabolic pathways limits the growth of MYC-driven tumors7,8. In addition to that, amplification of EGFR activates mTORC2 and AKT to promote metabolic reprogramming9. Similarly, metabolic genes amplification could also induce tumor metabolic reprogramming. The amplification of serine and glycine biosynthetic genes promoted de novo synthesis of serine and glycine, thereby facilitating tumor progression through the secretion of various metabolites10. Our previous study identified that PDSS1 amplification promoted TNBC metastasis through the PDSS1/STAT3/CAMK2A pathway11. However, the precise mechanism through which CNAs regulate TNBC metabolic reprogramming requires further exploration.
In this work, we conducted an analysis of our TNBC multiomic cohort, aiming to identify CNA-driven metabolic genes in TNBC. We identified FLAD1, a CNA-driven metabolic gene involved in the riboflavin metabolism pathway. We aimed to delineate the oncogenic role of FLAD1, elucidate the underlying molecular mechanism by which FLAD1 shapes the metabolism of tumor cells, and explore its potential therapeutic value in TNBC patients.
Results
Integrated genomic and transcriptomic analysis revealed the potential oncogenic role of FLAD1 in TNBC
To screen and identify metabolic genes that were dysregulated at the genomic level and determine the progression of TNBC, we conducted an integrated analysis of our previously established multiomic dataset of TNBC patients from Fudan University Shanghai Cancer Center (FUSCC)12. First, we filtered metabolic genes that were frequently amplified and highly expressed in tumor tissues, resulting in the identification of 69 metabolic genes that were frequently amplified (amplification frequency > 15%) among the 302 samples with both CNA and RNA-seq data. Second, we examined the correlations between the CNA values and RNA expression of these 69 metabolic genes, identifying 20 metabolic genes for which amplifications are positively correlated with mRNA expression (Pearson r > 0.35). Third, we analysed the mRNA expression of these genes in TNBC tissues and paracancerous tissues. Notably, five candidate genes (NDUFB9, SQLE, PYCRL, CYC1, and FLAD1) were obtained. For NDUFB9, SQLE, PYCRL, and CYC1, previous studies have revealed the mechanisms underlying tumor progression13,14,15,16,17. Here, we decided to focus on FLAD1 for further study, as it is located in a frequently amplified chromosome region, 1q, which has been widely reported in TNBC (Fig. 1a)6,18. The detailed amplification frequency, correlation between CNA and RNA, and mRNA expression level of FLAD1 between tumor and normal breast tissues are illustrated. In primary TNBC tissues, 20.2% (61/302) of patients in the FUSCC TNBC cohort exhibited FLAD1 copy number amplification, which was positively correlated with its mRNA expression (Fig. 1b, upper; Supplementary Fig. 1a). These findings were further validated in TNBC samples from the TCGA cohort19, with copy number amplification detected in 21.1% of the samples (35/166), which was positively correlated with FLAD1 mRNA expression (Fig. 1b, lower; Supplementary Fig. 1b). In terms of RNA expression, FLAD1 mRNA expression was significantly higher in primary TNBC tissues than in adjacent normal tissues (Fig. 1c). In addition, higher FLAD1 expression was observed in TNBC samples with higher T stage and histologic grade (Fig. 1d). Moreover, we performed survival analysis to evaluate the prognostic value of FLAD1, and the results revealed that high FLAD1 mRNA and protein expression were associated with poorer prognosis in the FUSCC TNBC cohort and KMPlot dataset (https://kmplot.com/analysis/), respectively (Fig. 1e). The above results suggested that FLAD1, amplified in the 1q21.3 genomic locus and highly expressed in tumors, might play an important role in the progression of TNBC.
a Schematic diagram of the screening for the metabolic genes that were significantly amplified and highly expressed in TNBC. b Correlations between FLAD1 copy number and mRNA expression levels in the FUSCC TNBC and TCGA cohorts12,19. c FLAD1 mRNA expression between tumor tissues and adjacent normal tissues in the FUSCC and TCGA TNBC cohorts. d FLAD1 mRNA expression in the FUSCC TNBC and TCGA cohorts according to different T stages and histologic grades. e Kaplan-Meier survival analysis of FLAD1 mRNA expression in the FUSCC TNBC cohort and protein expression in the TNBC samples in the KMPlot database based on the optimal best cut-off. TNBC, triple-negative breast cancer. The center line represents the median, the box shows the interquartile range (IQR), and the whiskers extend to 1.5×IQR beyond the 25th and 75th percentiles. b–d P value was calculated by two-tailed Student’s t-test. e P value of the survival curve was calculated by log-rank test. Source data are provided as a Source Data file.
FLAD1 facilitates TNBC proliferation and migration
First, we examined the basal expression of FLAD1 in TNBC cells. The results showed FLAD1 is highly expressed in LM2-4175 and SUM159 cells, but is expressed at low levels in other TNBC cells (Supplementary Fig. 1c). We further validated the role of FLAD1 in TNBC progression. First, we employed siRNAs to knock down FLAD1 in the LM2-4175 and SUM159 cells. The results of cell counting kit-8 (CCK-8) assays showed that transient suppression of FLAD1 impeded cell proliferation (Supplementary Fig. 2a and b). To further confirm our findings, we used lentiviral-mediated delivery of shRNA constructs to establish stable FLAD1 knockdown in LM2-4175 and SUM159 cell lines (Fig. 2a). Subsequent CCK-8 and colony formation assays demonstrated that stable FLAD1 knockdown significantly inhibited cell proliferation and colony formation (Fig. 2b and c). To investigate the effect of FLAD1 on the migratory capacity of TNBC cells, we performed transwell assays and found that FLAD1 knockdown significantly attenuated the migratory capability of LM2-4175 and SUM159 cells (Fig. 2d). The oncogenic property of FLAD1 was further substantiated in vivo via orthotropic tumor transplantation models, and the results indicated that FLAD1 knockdown significantly reduced tumor weight and volume (Fig. 2e and f). Taken together, our data showed that FLAD1 promoted the malignant phenotype of TNBC cells.
a Knockdown of FLAD1 using two independent shRNAs in LM2-4175 and SUM159 cells was assessed by western blot analysis. b, c The effects of FLAD1 loss of function on the proliferation of the indicated TNBC cells were evaluated using CCK-8 (b) and colony formation (c) assays. d Transwell assays were performed to evaluate the effects of FLAD1 loss of function on the migration of the indicated TNBC cells. Scale bars, 100 μm. e, f LM2-4175 cells infected with lentivirus encoding control or FLAD1 shRNAs were injected orthotopically into the mammary fat pad of 6-week-old BALB/c female nude mice, with six mice per group. After 30 days, the tumors were collected, and the tumor volume and weight (e) were measured. Representative images (f) are presented. Graph bars represent mean ± SEM, and P value was calculated by two-tailed Student’s t-test. Results in (b) represent biologically independent experiments of n = 3. a, c, d, n = 3 independent experiments, a representative example is shown. Source data are provided as a Source Data file.
FLAD1 contributes to malignancy in an enzyme-dependent manner
We subsequently investigated whether the contribution of FLAD1 to TNBC progression depends on its metabolic pathway. FLAD1 participates in riboflavin metabolism and catalyzes the conversion of flavin mononucleotide (FMN) to flavin adenine dinucleotide (FAD)20,21. The FAD synthase domain of FLAD1 is located near its C-terminus, and previous research has demonstrated that the substitution of arginine with a cysteine at residue 530 (R530C) within the catalytic site leads to the inactivation of its enzymatic activity (Fig. 3a)22,23. Subsequently, HCC1806 and Hs578T cells were subjected to infection with vector, wild-type FLAD1 (WT FLAD1) or catalytically inactive mutant FLAD1 (FLAD1-R530C), in which the key amino acid residue (Arg530) crucial for FAD synthase was mutated (Fig. 3b). The results of the CCK-8 and colony formation assays revealed that the overexpression of WT FLAD1 significantly increased cell proliferation and colony formation, whereas FLAD1-R530C failed to induce these effects (Fig. 3c and d). In addition, transwell assays revealed that the overexpression of WT FLAD1 significantly increased the migratory capacity of HCC1806 and Hs578T cells. However, no such enhancement was observed when the FLAD1-R530C was expressed (Fig. 3e). To further validate our in vitro results, LM2-4175 cells were subjected to infection with vector, WT FLAD1 or FLAD1-R530C (Fig. 3f). The oncogenic property of FLAD1 was further substantiated in vivo using orthotropic tumor transplantation models, and the results revealed that the overexpression of WT FLAD1, but not FLAD1-R530C, significantly increased tumor weight and volume (Fig. 3g and h). In summary, our results suggested that FLAD1 promoted the malignant phenotype of TNBC cells in an enzymatic-dependent manner.
a Schematic of the riboflavin metabolic pathway and FLAD1 enzymatic activity site. b HCC1806 and Hs578T cells expressing the control vector, WT FLAD1 or FLAD1-R530C were assessed by western blot analysis. c, d The effects of the indicated TNBC cells expressing the control vector, WT FLAD1 or FLAD1-R530C on proliferation were evaluated using CCK-8 (c) and colony formation (d) assays. e Transwell assays were performed to evaluate the effects of the indicated TNBC cells expressing the control vector, WT FLAD1 or FLAD1-R530C on migration. Scale bars, 100 μm. f The overexpression efficiency of WT and R530C mutant FLAD1 was assessed by western blot in LM2-4175 cells. g, h LM2-4175 cells expressing the control vector, WT FLAD1 or FLAD1-R530C were injected orthotopically into the mammary fat pad of 6-week-old BALB/c female nude mice, with six mice per group. After 30 days, tumors were harvested, and both their volume and weight (g) were measured. Representative image (h) is presented. Graph bars represent mean ± SEM, and P value was calculated by two-tailed Student’s t-test. Results in (c) represent biologically independent experiments of n = 3. b, d, e, f n = 3 independent experiments, a representative example is shown. Source data are provided as a Source Data file.
FLAD1 deficiency attenuates lipid accumulation in TNBC cells
To determine the potential molecular mechanism of FLAD1-regulated signalling pathways, we performed transcriptome sequencing analysis of control and FLAD1 knockdown SUM159 cells. We identified 538 and 358 differentially expressed genes in two independent shRNA FLAD1 knockdown SUM159 cells respectively, compared to the control group (Fig. 4a). Subsequently, gene set enrichment analysis (GSEA) revealed that FLAD1 knockdown significantly suppressed cholesterol and fatty acid metabolism pathways (Fig. 4b and c).
a Schematic diagram of the differentially expressed genes between the control and FLAD1 knockdown SUM159 cells. b Enriched signaling pathways were identified by GSEA with the hallmark gene set in RNA-seq data from the control and FLAD1 knockdown SUM159 cells. c Individual GSEA plots of the cholesterol and fatty acid metabolism pathways in RNA-seq data from the control and FLAD1 knockdown SUM159 cells. d Cellular contents of cholesterol, fatty acid and triglyceride were detected by LC/MS-MS-based lipidomics in control and FLAD1 knockdown SUM159 cells. e, f Cellular contents of cholesterol (e) and triglyceride (f) were detected by high-throughput spectrophotometry in SUM159 and HCC1806 cells. g, h Neutral lipids stained with the BODIPY 493/503 probe in the control and FLAD1 knockdown SUM159 cells (g) or in the HCC1806 cells (h) expressing the control vector, WT FLAD1 or FLAD1-R530C. Scale bars, 20 μm. Graph bars represent mean ± SEM, and P value was calculated by two-tailed Student’s t-test. Results in (d, e, f) represent biologically independent experiments of n = 3. g, h n = 3 independent experiments, a representative example is shown. Source data are provided as a Source Data file. CE, cholesterol. FA, fatty acid. TAG, triglyceride.
To validate the results of the transcriptome analysis, we performed LC/MS-MS-based lipidomics to evaluate the impact of FLAD1 knockdown on lipid profiles in SUM159 cells. FLAD1 knockdown decreased various types of lipids, particularly cholesterol, fatty acids and triglycerides (Fig. 4d; Supplementary Fig. 3a). To evaluate whether FLAD1 knockdown influences the abundance of intermediates in the pathways of de novo fatty acid biosynthesis, we performed targeted metabolomics in control and FLAD1 knockdown SUM159 cells (Supplementary Table 1). The results showed that FLAD1 knockdown decreased the abundance of acetyl-CoA, butyryl-CoA, isobutyryl-CoA, oleoyl-CoA, palmitoyl-CoA and Propionyl-CoA (Supplementary Fig. 3b). Glucose is an important synthetic source for the de novo synthesis of lipids. Therefore, we used [U-13C] glucose isotope experiments to trace fatty acid synthesis derived from glucose (Supplementary Table 2). The results showed that FLAD1 knockdown significantly decreased the cholesterol and triglyceride labelling from glucose tracers (Supplementary Fig. 3c). Moreover, we measured total cholesterol and triglycerides by high-throughput spectrophotometry, and the results showed that FLAD1 knockdown decreased cholesterol and triglyceride accumulation, whereas overexpression of WT FLAD1, but not FLAD1-R530C, increased total cellular cholesterol and triglyceride accumulation (Fig. 4e and f). Additionally, BODIPY neutral lipid staining showed that FLAD1 knockdown decreased the level of neutral lipids in SUM159 and MDA-MB-453 cells (Fig. 4g; Supplementary Fig. 3d and e), whereas overexpression of WT FLAD1, but not FLAD1-R530C, increased the level of neutral lipids in HCC1806 cells (Fig. 4h). Collectively, these findings indicated that FLAD1 was an important activator of lipid metabolism.
FLAD1 promotes de novo lipogenesis via SREBP1-dependent upregulation of lipogenic enzymes
Elevated cellular lipid levels may result from enhanced lipid synthesis, elevated uptake of fatty acids, and reduced lipid breakdown24. Therefore, we assessed whether FLAD1 knockdown altered the expression of genes involved in fatty acid biosynthesis (FASN, ACC1 and SCD), cholesterol biosynthesis (HMGCR), fatty acid uptake (CD36), and fatty acid oxidation (CPT1A) (Fig. 5a and b). We further validated that FLAD1 knockdown significantly reduced the mRNA and protein levels of enzymes involved in the synthesis of fatty acids and cholesterol in LM2-4175 and SUM159 cells through RT-qPCR and western blot experiments, whereas CD36 and CPT1A mRNA expression was almost unaffected (Fig. 5c and e; Supplementary Fig. 4a). Conversely, the overexpression of WT FLAD1, but not the FLAD1-R530C mutant, significantly upregulated the mRNA and protein levels of enzymes involved in the synthesis of fatty acids and cholesterol in HCC1806 and Hs578T cells, whereas CD36 and CPT1A mRNA expression was almost unaffected (Fig. 5d and f, supplementary Fig. 4b). We also demonstrated that knockdown of FLAD1 reduced the concentrations of FASN and ACC1 (Fig. 5g). Conversely, overexpression of WT FLAD1, but not its mutant form, led to increase the concentrations of FASN and ACC1 (Fig. 5h). Collectively, these findings indicated that FLAD1 regulated genes involved in lipid biosynthesis.
a, b Schematic diagrams of fatty acid (a) and cholesterol (b) metabolic processes. c Relative mRNA levels of the indicated fatty acid and cholesterol biosynthesis genes following shRNA-mediated knockdown of FLAD1 in LM2-4175 and SUM159 cells. d Relative mRNA levels of the indicated fatty acid and cholesterol biosynthesis genes following the overexpression of WT FLAD1 and FLAD1-R530C in HCC1806 and Hs578T cells. e Relative protein levels of the indicated fatty acid and cholesterol biosynthesis genes following shRNA-mediated knockdown of FLAD1 in LM2-4175 and SUM159 cells. f Relative protein levels of the indicated fatty acid and cholesterol biosynthesis genes following the overexpression of WT FLAD1 and FLAD1-R530C in HCC1806 and Hs578T cells. g The concentrations of FASN and ACC1 were measured in control and FLAD1 knockdown SUM159 cells. h The concentrations of FASN and ACC1 were measured in Hs578T cells overexpressing the control vector, WT FLAD1 or FLAD1-R530C. Graph bars represent mean ± SEM, and P value was calculated by two-tailed Student’s t-test. Results in (c, d, g, h) represent biologically independent experiments of n = 3. e, f n = 3 independent experiments, a representative example is shown. Source data are provided as a Source Data file.
Past research has shown that lipogenic enzymes are regulated at the transcriptional level by sterol regulatory element-binding proteins 1 and 2 (SREBP1 and SREBP2). SREBP1 plays a crucial role in controlling the transcription of genes involved in cholesterol and fatty acid metabolism, whereas SREBP2 specifically regulates cholesterol metabolism25. Therefore, we further evaluated whether the expression of SREBP1 and SREBP2 was regulated by FLAD1. We observed that FLAD1 knockdown resulted in a decrease in SREBP1 mRNA and protein expression (Fig. 5c and e). Conversely, the overexpression of WT FLAD1, but not the FLAD1-R530C mutant, significantly upregulated the expression of SREBP1 (Fig. 5d and f). The SREBP2 mRNA and protein levels were not significantly affected by either FLAD1 knockdown or overexpression (Fig. 5e and f; Supplementary Fig. 4a and b). To further strengthen our conclusions, we depleted endogenous FLAD1 followed by the restoration of WT FLAD1 (Supplementary Fig. 5a). The results indicated that FLAD1 restoration rescued the impaired proliferation and migration ability, lipid content decrease and lipogenic genes expression downregulation (Supplementary Fig. 5b–k). Overall, our results suggested that SREBP1 played a crucial role in the regulation of lipogenic enzyme expression by FLAD1.
FLAD1 promotes SREBP1 expression through FAD-LSD1 signalling
We speculated that FLAD1 might regulate lipid metabolism by modulating FAD-dependent enzymes. LSD1 is an FAD-dependent histone demethylase26,27,28, and it can potentially function as a coactivator or corepressor by demethylating mono-methyl and dimethyl H3K4 or H3K9 (Fig. 6a)29,30. We wondered whether FLAD1 could regulate the enzymatic activity of LSD1. We observed that FLAD1 knockdown increased H3K4me2 and H3K9me2 protein expression levels which indirectly reflected LSD1 enzymatic activity (Fig. 6b)30,31. Conversely, the overexpression of WT FLAD1, but not FLAD1-R530C, significantly decreased H3K4me2 and H3K9me2 protein expression levels (Fig. 6c). We also verified that genetic LSD1 knockdown or pharmacological inhibition significantly upregulated the global expression of H3K4me2 and H3K9me2 while inhibited the expression of lipogenic genes at the mRNA and protein levels (Fig. 6d–g). To verify that LSD1 regulated the expression of lipogenic genes in an SREBP1-dependent manner. We generated SUM159 cells with LSD1 overexpression, SREBP1 knockdown, and a combination of LSD1 overexpression along with SREBP1 knockdown. The results indicated that LSD1 overexpression upregulated the expression of lipogenic genes, which was largely inhibited by simultaneous knockdown of SREBP1 (Fig. 6h). Next, we evaluated whether the upregulation of lipogenic enzyme expression induced by FLAD1 expression is LSD1 dependent. qRT-PCR and western blot analysis revealed that GSK-LSD1, an LSD1 inhibitor, rescued the increases in the mRNA and protein levels of cholesterol and fatty acid synthesis enzymes induced by FLAD1 overexpression (Fig. 6i and j). For ChIP-qPCR, primer sets were designed to cover the proximal promoter regions of SREBP1 near the transcriptional start site (TSS) to investigate the impact of LSD1 on FLAD1-induced upregulation of SREBP1 (Fig. 6k). ChIP-qPCR analysis revealed that the enrichment of H3K9me2, but not H3K4me2, in the SREBP1 promoter was markedly increased after FLAD1 knockdown (Fig. 6l). Additionally, FLAD1 overexpression effectively reduced the enrichment of H3K9me2 in the promoter of SREBP1, which was rescued by the addition of GSK-LSD1 (Fig. 6m). We also evaluated whether the H3K4me2 and H3K9me2 status of FASN, ACC1 and SCD was affected by LSD1 knockdown. As shown in supplementary Fig. 6, LSD1 knockdown increased the level of H3K9me2 at the FASN, ACC1 and SCD promoters. Conversely, the level of H3K4me2 at the FASN and ACC1 promoters was not significantly altered by LSD1 knockdown, whereas it was mildly upregulated at the SCD promoter. Overall, although FLAD1/LSD1 slightly promoted the expression of a few lipogenic genes (e.g., FASN, ACC1 and SCD) by demethylating H3K9me2, this regulation was, to a great extent, dependent on SREBP1 (Fig. 6h). In order to verify whether the effect on lipogenic genes of FLAD1 is secondary to an effect on the beta-oxidation process, we inhibited fatty acid oxidation through knockdown of CPT1A or using the CPT1A inhibitor etomoxir. The results showed that FLAD1 could regulate lipogenesis independently, even when fatty acid oxidation was inhibited through genetic or pharmacological CPT1A inhibition (Supplementary Fig. 7a and b). Collectively, these results suggested that FLAD1 upregulated the expression of SREBP1 via LSD1-mediated dimethyl H3K9 demethylation (Fig. 6n).
a Schematic diagram showing the FAD-dependent demethylation mechanism of mono-methylated and di-methylated lysine residues by LSD1. b Relative protein levels of H3K4me2 and H3K9me2 following shRNA-mediated knockdown of FLAD1 in LM2-4175 and SUM159 cells. c Relative protein levels of H3K4me2 and H3K9me2 following the overexpression of WT FLAD1 and FLAD1-R530C in HCC1806 and Hs578T cells. d The expression of H3K4me2 and H3K9me2 were evaluated by western blot in control and LSD1 knockdown LM2-4175 and SUM159 cells. e The expression of H3K4me2 and H3K9me2 were evaluated by western blot in control and pharmacologic LSD1 inhibition SUM159 cells. f The expression of lipogenic genes were detected by RT-qPCR in control and LSD1 knockdown LM2-4175 and SUM159 cells. g The protein expression of lipogenic genes were showed in the indicated LM2-4175 and SUM159 cells. h The protein levels of LSD1, SREBP1 and lipogenic genes in LSD1 overexpressing SUM159 cells after SREBP1 was knocked down. i, j Relative mRNA (i) and protein (j) levels of genes associated with fatty acid and cholesterol biosynthesis in HCC1806 and Hs578T cells expressing the control vector, WT FLAD1 and WT FLAD1 treated with 80 µM GSK-LSD1 for 24 h. k Schematic of the ChIP-qPCR primers designed for the SREBP1 promoter regions. l The enrichment levels of H3K4me2 and H3K9me2 in different regions of the SREBP1 gene promoter in SUM159 cells with or without FLAD1 knockdown were determined by ChIP-qPCR assay. m The enrichment levels of H3K9me2 in different regions of the SREBP1 gene promoter in Hs578T cells expressing control vector, WT FLAD1 and WT FLAD1 treated with 80 µM GSK-LSD1 for 24 h were determined by ChIP-qPCR assay. n Schematic model of proposed mechanisms of the FLAD1/LSD1/SREBP1 signaling pathway. Graph bars represent mean ± SEM, and P value was calculated by two-tailed Student’s t-test. Results in (f, i, l, m) represent biologically independent experiments of n = 3. b–e, g, h, j n = 3 independent experiments, a representative example is shown. Source data are provided as a Source Data file.
FLAD1 determines the therapeutic effects of LSD1 and SREBP1 inhibitors
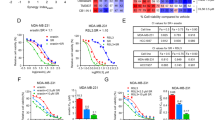
As FLAD1 upregulates the expression of SREBP1 via LSD1-mediated dimethyl H3K9 demethylation, we next investigated whether pharmacological inhibition of LSD1 or SREBP1 might be a suitable therapeutic approach for FLAD1-overexpressing TNBC. The control group, which had higher FLAD1 expression levels, exhibited greater sensitivity to GSK-LSD1 or Fatostatin compared to the FLAD1 knockdown group, resulting in significantly reduced growth following treatment with GSK-LSD1 or Fatostatin (Fig. 7a and b; Supplementary Fig. 8a, b and e). We also selected IMG-7289, a new-generation LSD1 inhibitor, used for patients with advanced myeloid malignancies in clinical trial32. The results of the drug sensitivity experiments revealed that FLAD1 knockdown reduced the sensitivity of TNBC cells to IMG-7289 (Supplementary Fig. 8c and d). Additionally, in organoids obtained from two TNBC patients with different FLAD1 protein expression levels, we noticed increased sensitivity of the organoids to GSK-LSD1 or Fatostatin, as well as an increase in FLAD1 expression (Fig. 7c; Supplementary Fig. 9a and b). Next, we evaluated the effectiveness of GSK-LSD1 or Fatostatin in suppressing TNBC growth in vivo. Following the establishment of the TNBC xenograft models, mice received treatment with either GSK-LSD1 or Fatostatin. The results showed a notable decrease in sensitivity to GSK-LSD1 and Fatostatin upon depletion of FLAD1 (Fig. 7d–f; Supplementary Fig. 9c). We also performed the immunohistochemistry and indicated that GSK-LSD1 treatment, Fatostatin treatment and FLAD1 knockdown significantly reduced the protein expression of lipogenic genes (Supplementary Fig. 9d). Our data collectively indicated that the FLAD1/LSD1/SREBP1 axis played a crucial role in enhancing TNBC proliferation, indicating that FLAD1 could serve as a biomarker and potential therapeutic target for TNBC.
a Left: dose–response curves and half maximal inhibition concentration values of GSK-LSD1 in LM2-4175 cells expressing control or FLAD1 shRNA. Right: Bar plot illustrating the mean cell survival rates in LM2-4175 cells expressing control or FLAD1 shRNA treated with 80 µM GSK-LSD1 for 48 h. b Left: dose–response curves and half maximal inhibition concentration values of Fatostatin in LM2-4175 cells expressing control or FLAD1 shRNA. Right: Bar plot illustrating the mean cell survival rates in LM2-4175 cells expressing control or FLAD1 shRNA treated with 10 µM Fatostatin for 48 h. c The results of the cell viability assay in two TNBC patient-derived organoid models treated with 100 µM GSK-LSD1 or 5 µM Fatostatin. Representative bright-field images and cell viability assays are shown. Scale bars, 100 μm. d–f Experimental protocols (d) for animal studies to observe whether FLAD1 expression is associated with GSK-LSD1 and Fatostatin sensitivity. Tumor volume (e) and weight (f) of shCtrl or shFLAD1 LM2-4175 cells in BALB/c nude mice treated with GSK-LSD1 or Fatostatin are presented (6 mice per group). g–i Experimental protocols (g) for animal studies to observe whether GSK-LSD1 could improve sensitivity to doxorubicin. Tumor volume (h) and weight (i) of LM2-4175 cells in BALB/c nude mice treated with GSK-LSD1 and/or doxorubicin are presented (6 mice per group). j–l Experimental protocols (j) for animal studies to observe whether GSK-LSD1 could improve sensitivity to SG. Tumor volume (k) and weight (l) of LM2-4175 cells in NOD-SCID mice treated with GSK-LSD1 or/and SG are presented (6 mice per group). Graph bars represent mean ± SEM, and P value was calculated by two-tailed Student’s t-test. Results in a and b represent biologically independent experiments of n = 5. c, n = 3 independent experiments, a representative example is shown. Source data are provided as a Source Data file. PBS, phosphate buffer saline. SG, sacituzumab govitecan. Doxo, doxorubicin. i.p, intraperitoneal injection. i.v, intravenous injection.
GSK-LSD1 combined with doxorubicin or sacituzumab govitecan enhanced the therapeutic effects
Chemotherapy is the main strategy for the treatment of TNBC. However, some TNBC patients still insensitive to chemotherapy33. Many factors affect chemotherapy efficacy. Among these, cancer stem cells (CSCs) are critical for chemoresistance34,35. It has been reported that LSD1 is necessary for maintaining the stemness of cancer cells36,37. These results provide strong justification for the use of chemotherapy in combination with LSD1 inhibitor. Hence, we aimed to analyze the impact of GSK-LSD1 when combined with the frequently utilized chemical drug doxorubicin to determine the possible synergistic effects of this medication duo in a live setting. Following the establishment of the orthotopic tumor transplantation models, the mice were administered either GSK-LSD1 or doxorubicin individually, or a combination of both GSK-LSD1 and doxorubicin (Fig. 7g). The findings indicated that the combined therapy was superior in inhibiting tumor advancement compared to individual treatments (Fig. 7h and i; Supplementary Fig. 9e). In addition, cotreatment with GSK-LSD1 and doxorubicin-induced higher expression of cleaved caspase 3 and lower expression of Ki-67 than each monotherapy (Supplementary Fig. 9f).
Human trophoblast cell surface antigen 2 (TROP2), a glycoprotein found on the surface of human trophoblast cells, is significantly more abundant in primary tumor tissues than in nearby normal tissues38,39. In a phase III study, SG, an antibody-drug conjugate that targets Trop2, has been shown to improve the progression-free survival (PFS) of patients with TNBC40. Currently, SG has been Food and Drug Administration (FDA) approved for treating metastatic TNBC41. Despite the proven effectiveness of SG, therapeutic resistance nevertheless continues to emerge, which can be partly attributed to the emergence of absence or mutation of Trop-2 expression42. Thus, combination therapy with SG and other drugs should be explored. Mechanistically, SG could induce cell-cycle arrest via p21 upregulation43, and p21 is also a direct downstream target of LSD144. Therefore, in our study, we sought to evaluate whether the combination of SG and GSK-LSD1 could show greater efficacy in vivo. Following the establishment of orthotropic tumor transplantation models, mice were administered either GSK-LSD1 or SG individually or a combination of both GSK-LSD1 and SG (Fig. 7j). While SG was somewhat effective at suppressing tumor progression, the therapeutic effect in vivo was further enhanced by cotreatment with SG and GSK-LSD1 (Fig. 7k and l; Supplementary Fig. 9g). In addition, cotreatment with GSK-LSD1 and SG induced higher expression of cleaved caspase 3 and lower expression of Ki-67 than either monotherapy did (Supplementary Fig. 9h). Moreover, we did not observe drug-related toxicity in mice at therapeutic doses of SG (Supplementary Fig. 10a–d).
Discussion
Comprehending metabolic reprogramming mechanisms is essential not only for obtaining mechanistic understandings of tumor advancement but also for the creation of pertinent treatments. In our study, we demonstrated that FLAD1, a key regulator driving TNBC progression, promoted malignant behaviours in TNBC through FLAD1/LSD1/SREBP1 signalling (Fig. 6n). The research we conducted offers more insight into the metabolic pathways that control the progression of TNBC, indicating that FLAD1 could be utilized as a promising indicator and target for the treatment of TNBC.
FLAD1 is involved in the riboflavin metabolism pathway and positively regulates the level of FAD. Cell bioenergetics, growth, and regulation are dependent on the essential role of FAD45. FLAD1 mutation leads to deficiency in FAD synthase, resulting in a muscle-specific disorder that impacts mitochondrial energy metabolism46. Previous studies have demonstrated that FLAD1 was upregulated in prostate cancer and gastric cancer and was associated with poor prognosis47,48. Furthermore, it has been reported that FLAD1 could maintain tumor stemness45. In addition, the function of FLAD1 in cancer has rarely been studied, and herein, we demonstrated that FLAD1 knockdown inhibited TNBC cell proliferation and migration, whereas FLAD1 overexpression promoted TNBC cell proliferation and migration. Our study suggests that FLAD1 acts as an oncogene in TNBC.
During the malignant transformation of tumors, metabolic reprogramming at the molecular level is often observed. These changes may be caused by mutations in metabolic enzyme-encoding genes or alterations in their expression levels, which in turn drive the progression of the tumor. Previous research suggested that some metabolic genes can drive tumor progression through an enzymatic-dependent fashion. For instance, PI3K/Akt signalling pathway activation is able to promote cell proliferation by increasing GLUT1‐mediated glucose transport49,50 The upregulated expression of ENO2 could increase the accumulation of acetyl-CoA, inducing resistance to anti-angiogenic agents by activating the Wnt/β‐catenin signalling pathway51. However, several metabolic genes could facilitate tumor progression in an enzymatic-independent manner. For instance, BBOX1 could bind with IP3R3, preventing the degradation of IP3R3, thereby inhibiting apoptosis52. PKM2 is moved to the mitochondria and phosphorylates the antiapoptotic protein bcl2 under conditions of oxidative stress, preventing its degradation by the proteasome and ultimately blocking tumor cell apoptosis caused by oxidative stress53,54,55. To investigate whether FLAD1 promotes TNBC progression in an enzymatic-dependent manner, we constructed FLAD1 and FLAD1-R530C overexpression plasmids. Our results revealed that the overexpression of FLAD1, but not FLAD1-R530C, promoted the malignant phenotype of TNBC cells. In summary, FLAD1 promoted TNBC progression in an enzymatic-dependent fashion.
Cholesterol and fatty acid synthesis play significant roles in driving tumor progression by promoting cell proliferation, inhibiting apoptosis, facilitating migration, maintaining stemness, and enabling immune evasion of tumors56,57,58,59,60,61,62,63. SREBPs play crucial roles in regulating the synthesis of cholesterol and fatty acids through transcriptional mechanisms64. SREBP1 is involved in the synthesis of fatty acids and cholesterol, whereas SREBP2 is specifically involved in cholesterol biosynthesis25,65,66,67. The expression of SREBP1 is subject to complex regulation at multiple levels: transcriptional, posttranscriptional and posttranslational. For example, Akt triggers the production of complete SREBP1 and leads to an increase in the level of intracellular lipids68. mTORC1 can activate SREBP1 transcription to regulate lipid synthesis69,70. Additionally, a prior investigation demonstrated that PRP19 could enhance the stability of SREBP1 mRNA through a m6A-dependent mechanism24. O-GlcNAc transferase, the most common posttranslational modification of proteins, was demonstrated to regulate SREBP1 protein expression levels and stability71. DAXX could bind to SREBP1 and SREBP2, thereby promoting the expression of lipogenic genes and tumor growth72. In our study, we identified FLAD1 as an upstream regulator of SREBP1 at the transcriptional level.
LSD1 is an FAD-dependent lysine-specific demethylase26,27,28, which was upregulated in various tumor types73,74,75,76. LSD1 promotes tumor progression through multiple mechanisms, including an accelerated cell cycle, immune escape, and metastasis77,78,79. Previous study indicated that LSD1 is a vital factor leading to metabolic reprogramming, Sakamoto et al. demonstrated that LSD1 depletion reduced glucose uptake and glycolytic activity by decreasing HIF-1α levels80. In our study, we revealed that LSD1 promoted TNBC progression through SREBP1-mediated lipid metabolism reprogramming. However, it has been reported that LSD1 can stimulate FASN expression and lipogenesis by demethylating dimethyl H3K981. Moreover, Abdulla et al. reported that LSD1 knockdown reduces the binding of SREBP1 to the FASN promoter. Therefore, we inferred that FLAD1-regulated lipogenic enzyme expression was not completely dependent on LSD1-mediated demethylation and SREBP1-dependent transcription, which needs further investigation in the future.
Our study has potential clinical and translational significance in TNBC. First, we proved that TNBC cells with high FLAD1 expression are more sensitive to GSK-LSD1 or Fatostatin. Therefore, FLAD1 expression may be a promising biomarker to stratify patients with TNBC for LSD1- or SREBP1-targeted therapy. Second, several studies have already reported that some drugs, such as poly (ADP-ribose) polymerase (PARP) inhibitors and some immune checkpoint inhibitors (ICIs), could be universal sensitizers to multiple chemotherapeutics82,83,84. Here, we demonstrated that the combination of GSK-LSD1 and doxorubicin could be a promising therapeutic strategy for TNBC patients. Third, we demonstrated that cotreatment with GSK-LSD1 and SG had a greater synergistic antitumour effect than single-drug treatment. Therefore, we propose that LSD1 inhibitors combined with antibody-drug conjugates (ADCs) is a promising TNBC treatment strategy.
In summary, our study provides a perspective on the role of the CNA driver gene FLAD1 in facilitating SREBP1-dependent lipid biosynthesis through LSD1-mediated H3K9me2 demethylation. These results suggest that targeting the FLAD1/LSD1/SREBP1 pathway represents a promising strategy for treating TNBC.
Methods
Study cohorts
The TNBC cohort used in this study was obtained from the FUSCC and consisted of 465 patients who were diagnosed with TNBC based on pathology. Our previous study has provided a description of this cohort12. Among the samples, 401 individuals had OncoScan microarray copy number data, while RNA sequencing data was available for 360 individuals, and 302 had both OncoScan microarray copy number and RNA sequencing data. None of the samples had received any prior treatment, and clinical information was available for all patients in the dataset. In addition to the FUSCC cohort, we also included data from the TCGA cohort19. The datasets derived from TCGA are available at the cBioPortal website (www.cbioportal.org/) and comprised 156 TNBC samples with both OncoScan microarray copy number data and RNA sequencing data.
Cell lines
The TNBC cell lines HCC1806 (cat no. CRL-2335) and Hs578T (cat no. HTB-126), both derived from female patients, along with HEK293T embryonic kidney cells (cat no. ACS-4500), were sourced from the American Type Culture Collection (ATCC). LM2-4175 cells were graciously provided by Professor Guohong Hu. SUM159 cells were acquired from Asterand in Detroit, Michigan. Each cell line was cultured in specific media: LM2-4175, SUM159, Hs578T, and HEK293T cells were grown in DMEM with 10% fetal bovine serum (FBS), while HCC1806 cells were cultured in RPMI 1640 with 10% FBS. Regular testing with a Mycoplasma Detection Kit from Vazyme verified the lack of mycoplasma contamination in all cell lines. Standard culture conditions, including 37 °C incubation with 5% CO2, were maintained for all cells. Furthermore, the cell lines’ authenticity was verified via STR profiling.
Transfection and virus infection
To transiently transfect siRNA, LM2-4175 and SUM159 cells were grown to 60% confluency and then transfected with control siRNA and FLAD1 siRNA utilizing Lipofectamine RNAiMAX (Invitrogen). The evaluation of knockdown efficiency was conducted 48-72 h post-transfection.
HEK293T cells were utilized in the generation of lentiviruses, in conjunction with the pLKO.1 vector and packaging plasmids. An oligonucleotide targeting FLAD1 was synthesized (Sangon Biotech), annealed, cloned, and then inserted into pLKO.1-Puro. Supernatants were harvested and filtered using a 0.45μm syringe filter. Lentiviral particles were added to target cells at a multiplicity of infection of 0.7 in the presence of polybrene (6 mg/ml; Sigma-Aldrich). Following this, TNBC cells were subjected to puromycin selection to establish stable transfection or infection. The efficiency and specificity of knockdown for all shRNAs were confirmed through qRT-PCR and immunoblotting. The top two shRNAs with the most effective knockdown were chosen for subsequent investigations. The specific target sequences for FLAD1 shRNAs are provided in Supplementary Table 3.
Plasmid, shRNA and siRNA construction
Human WT FLAD1 (NM 201398.2), FLAD1-R530C, and LSD1 plasmids were purchased from Jikai Genetic Company (Shanghai, China) and cloned and inserted into the pCDH-CMV-MCS-EF1-puro plasmid. The shRNA oligo sequences for FLAD1 were designed by Jikai Genetic Company (Shanghai, China) and inserted into the pLKO.1-Puro vector. FLAD1, LSD1, and CPT1A siRNAs were synthesized by Guangzhou RiboBio Co., Ltd. The sequences can be found in Supplementary Table 3.
RNA isolation and qRT-PCR
After cell isolation with TRIzol reagent (Invitrogen), cDNA was synthesized from the extracted RNA using the PrimeScript RT Reagent Kit (TaKaRa). Primers were designed via the PrimerBank website (https://pga.mgh.harvard.edu/primerbank). SYBR Premix Ex Taq (TAKARA) was employed for qRT-PCR analysis on an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems). The qRT-PCR results were standardized to the expression level of the ACTB (actin beta) gene, acting as an internal control. Detailed information on the gene-specific primers utilized for amplification is provided in Supplementary Table 4.
Western blotting
Following extraction with RIPA lysis buffer, proteins were quantified utilizing a BCA reagent kit (Solarbio). Subsequently, the protein specimens underwent separation via SDS-PAGE and transfer onto polyvinylidene difluoride membranes (Millipore). Incubation with primary antibodies specific to the target proteins, including anti-FLAD1 (1:100, Santa Cruz, mouse monoclonal, cat no. sc-376819), anti-GAPDH (1:5000, Proteintech, mouse monoclonal, cat no. 60004-1-lg), anti-SREBP1 (1:1000, Abcam, mouse monoclonal, cat no. ab3259), anti-SREBP2 (1:1000, Abcam, rabbit polyclonal, cat no, ab30682), anti-FASN (1:1000, Abcam, rabbit monoclonal, cat no, ab128856), anti-CPT1A (1:1000, Abcam, mouse monoclonal, cat no. ab128568), anti-ACC1 (1:1000, Abcam, rabbit monoclonal, cat no. ab45174), anti-SCD1 (1:1000, Abcam, rabbit monoclonal, cat no. ab236868), anti-HMGCR (1:2000, Abcam, rabbit monoclonal, cat no. ab174830), anti-Histong H3 (1:1000, Cell Signaling Technology, rabbit, cat no. 9715S), anti-H3K4me2 (1:2000, Abcam, rabbit monoclonal, cat no. ab32356), anti-H3K9me2 (1:1000, Abcam, mouse monoclonal, cat no. ab1220), anti-LSD1 (1:10000, Abcam, rabbit monoclonal, cat no. ab129195), was followed by secondary antibody treatment conjugated with HRP. Secondary antibodies against mouse and rabbit conjugated with horseradish peroxidase were obtained from Jackson ImmunoResearch. Enhanced chemiluminescence (Pierce Biotechnology) was employed for signal detection. Image capture was conducted utilizing the Molecular Imager ChemiDoc XRS system (Bio-Rad).
Lipidomics detection
To profile the effect of FLAD1 knockdown on lipid accumulation, 1 × 107 shCtrl or shFLAD1 SUM159 cells of each sample were used. Internal standards as well as quality control samples were utilized to assess the quality of the data. For details, Samples were prepared by homogenization and sonication in water, followed by extraction using a mixture of methyl tert-butyl ether (MTBE) and methanol (MeOH) in a 5:1 ratio. After centrifugation, the lipid extracts were reconstituted in 50% methanol/dichloromethane containing an internal standard. Lipidomics data were acquired on an ultra-high-performance liquid chromatography (UHPLC) system (Vanquish, Thermo Fisher) equipped with a C18 column and coupled to a quadrupole-Orbitrap mass spectrometer (QE-MS) in data-dependent acquisition mode. The mobile phases consisted of acetonitrile, isopropanol, and ammonium formate, applied in a gradient elution. Data processing was performed using the XCMS software for peak detection and alignment, and lipid species were annotated with the LipidBlast library.
Targeted metabolomics analysis
To evaluate whether FLAD1 knockdown could influence the abundance of intermediate in the pathways of de novo fatty acid biosynthesis. 1 × 107 shCtrl or shFLAD1 SUM159 cells of each sample were harvested and counted. Cells were rapidly frozen in liquid nitrogen for 30 s and then placed into a -80 °C freezer for standby. For details, Acyl-CoAs were extracted using methanol, chloroform, and water, followed by centrifugation, vacuum drying, and reconstitution in methanol/water (3:2, v/v). Calibration curves (R² > 0.99) were generated across 14 concentrations (10–2000 ng/mL), with the lower limit of detection (LLOD) and quantification (LLOQ) determined according to FDA bioanalytical validation guidelines. Separation was performed on a Vanquish Core HPLC system using a CSH C18 column with ammonium acetate and acetonitrile as mobile phases. Mass spectrometry (MS) data were acquired in electrospray ionization (ESI) mode. Quality control (QC) samples exhibited relative standard deviations (RSD) < 15% and recoveries within 85–115%. Target compounds were well-separated with reproducible retention times and consistent peak shapes.
Tracing with [U-13C] glucose
In order to investigate whether FLAD1 knockdown impacts the fatty acid form tracers. [U-13C] glucose was added into low-glucose DMEM (2 g/L final concentration of [U-13C] glucose) and cultured control and FLAD1 knockdown SUM159 cells for 24 h. After 24 h, 1 × 107 control and FLAD1 knockdown SUM159 cells were harvested by 1 ml pre-cold methanol: acetonitrile: water (2:2:1, v/v/v). Samples were placed into a -80 °C freezer for standby. For details, Liquid samples (100 μL) were extracted with Methyl tert-butyl ether (MTBE):Methanol (MeOH) (5:1, v/v), sonicated (10 min, 4 °C), incubated (-40 °C, 1 h), and centrifuged (3000 rpm, 15 min, 4 °C). Supernatants were dried, reconstituted in Dichloromethane (DCM): MeOH (1:1, v/v), sonicated, and centrifuged (12000 rpm, 15 min, 4 °C). Solid samples (tissues, soil, plants) were homogenized in MeOH:MTBE:Water (H2O) (5:10:2, v/v) and processed similarly. Bacterial and cell pellets (~107 cells) underwent freeze-thaw cycles and sonication, with fast-quenching for adherent cells using pre-cooled extraction buffer. QC samples were prepared by pooling unlabeled supernatants. Lipids were analyzed using a Vanquish UHPLC system (Thermo) with a Kinetex C18 column (2.1 × 100 mm, 2.6 μm) coupled to a Q Exactive Orbitrap MS. Mobile phases included Water (H2O): Acetonitrile (ACN) (6:4, v/v) and Isopropanol (IPA):ACN (9:1, v/v), both with 10 mM ammonium formate. Electrospray ionization (ESI) parameters were as follows: sheath gas 30 Arb, auxiliary gas 10 Arb, capillary temperature 320 °C, full MS resolution 70000, MS/MS resolution 17500, and spray voltage ± 4.5 kV.
Total triglyceride, cholesterol and neutral lipid quantification
Cholesterol levels were determined with a cholesterol assay kit (Solarbio, cat no. BC1985) and triglyceride levels were quantified using a triglyceride quantification kit (Solarbio, cat no. BC0625), respectively.
To quantify neutral lipids, 5 × 103 TNBC cells were seeded in a 24-well cell culture plate. After seeding, the cells were treated with 4% paraformaldehyde for 15 minutes to fix them. Subsequently, the cells were exposed to a 0.2 μg/ml BODIPY 493/503 (MedChemExpress, cat no. HY-W090090) solution in the dark at 37 °C for 30 minutes. Cell images were captured using microscope. The levels of neutral lipids’ fluorescence intensity were analyzed with ImageJ software.
Cell growth and colony formation assay
To assess cell proliferation, the CCK-8 from Vazyme was employed. Initially, 2 × 103 TNBC cells were plated in 96-well plates and incubated for 24 h. Subsequently, CCK-8 reagent was introduced to the plates and allowed to incubate for an extra 1.5 h at 37 °C among four consecutive days. Cell proliferation was measured by recording the absorbance at 450 nm and visualized the growth curve.
For the colony formation assay, 2 × 103 (LM2-4175, HCC1806, and Hs578T) and 1 × 103 SUM159 cells were plated into 6-well plates. TNBC cells were cultured for approximately two weeks, with the induction medium changing every other day. After two weeks, after fixing TNBC cells with methanol, they were treated with 1% crystal violet staining. The counting of colonies was performed with the assistance of ImageJ software.
Cell migration assay
In the cell migration assay, the lower chambers from Corning were filled with 500 μl of DMEM supplemented with 10% FBS. Cells (4-10 × 104) in 200 μl DMEM were seeded in the upper chamber. Migration was allowed for 12-24 h. Subsequently, non-migrated cells were eliminated by gently swabbing the upper chamber with a cotton swab. The cells that had migrated were subsequently stained with 0.1% crystal violet with paraformaldehyde. Images of the migrated cells were captured using an inverted microscope, and the quantification of migrated cells was conducted with ImageJ software.
ChIP assays
The ChIP-qPCR experiment was performed using the Simple ChIP kit from CST. The immunoprecipitation (IP) samples consisted of 4 × 106 cells. To start the experiment, the cells were fixed with 37% formaldehyde to crosslink the DNA and proteins. The chromatin was then fragmented into 200-500 bp fragments using sonication. Each IP sample was supplemented with anti-human H3K4me2 and anti-human H3K9me2 antibodies. Protein G Magnetic beads were used to capture the DNA-antigen-antibody complex, and the elution of chromatin was carried out from the beads. The cross-links were undone, and the DNA that had been immunoprecipitated was utilized for real-time PCR amplification. The primers used for ChIP-qPCR validation can be found in Supplementary Table 5.
Cell survival assay
Cells were plated in 96-well plates at a concentration of 5 × 103 cells per well. and exposed to varying concentrations of chemicals for 48 h. Following the incubation, 10 μl of CCK-8 solution was introduced to each well, and the cells were subsequently cultured at 37 °C for an additional 2 h. The cell survival rate was determined by dividing the cell density of the treated cells by the cell density of the untreated cells.
Animal studies
The animal trials were carried out at Shanghai Laboratory Animal Center, within a specific pathogen-free (SPF) environment. All animal-related experiments were conducted following the approved protocols of the FUSCC Institutional Animal Care and Use Committee (FUSCC-IACUC-2021550). The mice were kept in a regulated setting with temperatures ranging from 20–23 °C, a 12/12 h light/dark cycle, and humidity levels maintained at 50-60%. They were given unrestricted access to food and water. Tumor studies were performed exclusively in female mice. The investigators were blinded to group allocation during data collection and/or analysis. Tumor volume was calculated using the formula: (L × W ²)/2, where L represents the length and W the width. To comply with ethical guidelines and ensure data quality, animals were humanely euthanized before tumors reached the maximum permitted size (2000 mm³).
In order to assess the impact of FLAD1 knockdown on tumor growth in vivo, 18 female BALB/c-nude mice, aged six to eight weeks, were divided into 3 sets, each containing 6 mice. They were then orthotopically injected with either 1 × 106 shCtrl or shFLAD1 LM2-4175 cells suspended in a 50 μl PBS solution mixed with an equivalent amount of Matrigel (Corning, cat no. 356234) into the mammary fat pad. Tumor volume was measured using calipers, and then calculated the volume using the formula: (L × W ²)/2. After 30 days, the mice were euthanized through CO2 inhalation. The tumor weight was documented, and images of the tumor samples were captured.
In order to examine the impact of FLAD1 overexpression on tumor development in vivo, 18 female BALB/c-nude mice, aged six to eight weeks old, were divided into 3 sets, each consisting of 6 mice. They were then orthotopically injected with either 1 × 106 vector, WT FLAD1, or FLAD1-R530C LM2-4175 cells suspended in a 50 μl PBS solution mixed with an equivalent amount of Matrigel into the mammary fat pad. The remaining experimental procedures were identical to those described previously.
In order to evaluate the impact of FLAD1 on responsiveness to GSK-LSD1 and Fatostatin, 36 female BALB/c-nude mice, aged six to eight weeks, were randomly divided into 2 sets. They were then orthotopically injected with 5 × 106 shCtrl or shFLAD1 LM2-4175 cells (shFLAD1 #1) suspended in a 50 μl PBS solution mixed with an equivalent amount of Matrigel into the mammary fat pad. Following a 14-day period, the shCtrl group was divided into 3 subgroups at random: shCtrl, shCtrl + GSK-LSD1 (10 mg/kg/every other day), and shCtrl + Fatostatin (30 mg/kg/every other day). Similarly, the shFLAD1 group was divided into 3 subgroups: shFLAD1, shFLAD1 + GSK-LSD1 (10 mg/kg/every other day), and shFLAD1 + Fatostatin (30 mg/kg/every other day). The remaining experimental procedures were consistent with those described previously.
In order to assess the effectiveness of combining GSK-LSD1 with doxorubicin, 24 female BALB/c-nude mice, aged six to eight weeks, were orthotopically injected with 1 × 106 LM2-4175 cells suspended in a 50 μl PBS solution mixed with an equivalent amount of Matrigel into the mammary fat pad. Following a 14-day period, the mice were randomly divided into four groups: Ctrl, Ctrl + GSK-LSD1 (5 mg/kg/every other day), Ctrl + doxorubicin (2 mg/kg/twice per week), and the combination of GSK-LSD1 and doxorubicin. The remaining experimental procedures were consistent with those described previously.
In order to assess the effectiveness of combining GSK-LSD1 with SG, 24 female NOD-SCID mice, aged six to eight weeks, were orthotopically injected with 1 × 106 LM2-4175 cells suspended in a 50 μl PBS solution mixed with an equivalent amount of Matrigel into the mammary fat pad. Following a 14-day period, the mice were randomly divided into four groups: Ctrl, Ctrl + GSK-LSD1 (5 mg/kg/every other day), Ctrl + SG (10 mg/kg/twice per week), and the combination of GSK-LSD1 and SG. The remaining experimental procedures were consistent with those described previously.
Patient-derived organoids
In order to create TNBC organoids derived from patients, tissue samples were collected from surgical specimens of two female patients who had surgery at the FUSCC (Supplementary Table 6). The organoids derived from the patients were cultured using established methods as described previously85. Following several passages, the organoids were moved to separate wells in a 384-well plate. GSK-LSD1 (100 µM) or Fatostatin (5 µM) was introduced into each well in duplicate and allowed to incubate for a period of 7 days. Following the drug treatments, the organoids were visually photographed and examined. All clinical samples were acquired with informed consent, signing an informed consent form approved by FUSCC’s Ethics Committee (Protocol number: 1802-ZZK-17).
Histology and immunohistochemistry (IHC) staining
Formalin-fixed and paraffin-embedded sections were employed for histological examination (HE) and IHC with antibodies targeting specific proteins including SREBP1, FASN, ACC1, SCD, Ki-67 and cleaved caspase-3. The tissue sections underwent deparaffinization and rehydration through a series of xylene-ethanol baths. Antigen retrieval was carried out in citrate buffer at 95 °C for 20 minutes. Subsequently, the sections were exposed to primary antibodies at room temperature for 2 h. The SREBP1 (Abcam, cat no. ab3259), FASN (Abcam, cat no, ab128856), ACC1 (Abcam, cat no. ab45174) and SCD (Abcam, cat no. ab236868) antibodies were diluted at a ratio of 1:400. The Ki-67 (Abcam, cat no. ab15580) and cleaved caspase-3 (Cell Signaling Tech, cat no. 9661S) antibodies were diluted at a ratio of 1:100. Visualization was achieved using the Novolink Polymer Detection System from Leica Biosystems. The analysis was conducted in a double-blind manner, with the quantification of positive cells performed in each stained tissue section per high-power field (40×).
ELISA assay
We used the ELASA kit to measure the concentrations of FASN and ACC1. The ELISA kit (CSB-EL001119HU, CSB-EL008435HU) was purchased from Wuhan Huamei Biological Engineering Co., LTD. 1 × 107 cells were washed and resuspended with PBS, and then frozen at −20 °C. To disrupt cell membranes, the above procedure should be repeated twice. Biotin-antibody was added into standards and samples, and then incubated for 1 h at 37 °C. The supernatant was discarded and added HRP-avidin, incubating for 0.5 h at 37 °C. TMB substrate and stop solution were added, and then the optical density (OD450 nm) values were analyzed by utilizing a microplate reader.
Standard curves were plotted, and the concentrations of FASN and ACC1 were calculated according to OD values.
Chemical products
The subsequent chemical items were utilized: GSK-LSD1, a LSD1 inhibitor (MedChemExpress, cat no. HY-100546A); Fatostatin, a SREBP1 inhibitor (MedChemExpress, cat no. HY-14452); Doxorubicin, a chemotherapeutic agent (MedChemExpress, cat no. HY-15142); IMG-7289, a LSD1 inhibitor (MedChemExpress, cat no. HY-109169A); and Etomoxir, a CPT1A inhibitor (MedChemExpress, cat no. HY-50202).
RNA sequencing
RNA was extracted from SUM159 cells treated with either control or two distinct FLAD1-targeted shRNAs. Subsequently, the RNA samples underwent library preparation for RNA sequencing (RNA-seq) with the VAHTS mRNA-seq V2 Library Prep Kit for Illumina (#NR601-01, Vazyme). These libraries were then sequenced on the HiSeq-2500 platform (Illumina). The sequencing reads obtained were mapped to the hg38 genome assembly using HISAT2 software. Transcript abundance for each gene was calculated using Cufflinks in terms of transcripts per kilobase of exon model per million mapped reads. Differential gene expression (DEGs) was determined based on a fold change threshold of ≥ 1.5 or ≤ 0.67. Gene set enrichment analysis (GSEA) was performed to identify enriched biological pathways in our dataset. Functional gene sets were pulled from hallmark and C2 MsigDB pathways (https://www.gsea-msigdb.org/gsea/msigdb/human/genesets.jsp?collection=C2).
Statistical analysis
GraphPad Prism 9.3.0 and R (R version 4.2.1) was utilized for statistical analysis. Survival curves were generated using the Kaplan-Meier method and compared through log-rank tests. Recurrence-free survival (RFS) was calculated from the surgery date to recurrence identification. Overall survival (OS) was calculated from surgery to death date or last follow-up. Distant metastasis-free survival (DMFS) was defined as the period between surgery and metastasis or follow-up end. Patients without events were censored at the last follow-up date. Statistical significance was set at P < 0.05. Specific statistical methods are detailed in the figure legends. Experiments were conducted at least three times, and representative results are presented.
Study approval
The Ethical Committee of Fudan University Shanghai Cancer Center approved this study. Written informed consent was obtained from all participating patients before the study. Approval for all mouse experiments in this research was granted by the Institutional Animal Care and Use Committee of Fudan University Shanghai Cancer Center.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The FUSCC TNBC cohort can be viewed in The National Omics Data Encyclopedia (NODE) (http://www.biosino.org/node) by pasting the accession (OEP000155) into the text search box or through the URL: http://www.biosino.org/node/project/detail/OEP00015512. The RNA sequencing data for our study have been deposited into the Genome Sequence Archive (GSA) database under accession codes PRJCA030823 (https://bigd.big.ac.cn/gsa-human/browse/HRA008804). The raw LC-MS data for our study is NODE: OEP00005949. All data can be viewed in The National Omics Data Encyclopedia (NODE) (http://www.biosino.org/node) by pasting the accession (OEP00005949) into the text search box or through the URL: http://www.biosino.org/node/project/detail/OEP00005949. The datasets derived from TCGA are available at the cBioPortal website (www.cbioportal.org/)19. The model in Figs. 6n, 7d, 7g, 7j and supplementary Fig. 9a was created with BioRender.com. Source data are provided with this paper. All remaining data can be found in the Article, Supplementary and Source Data files. Source data are provided with this paper.
References
Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74, 229–263 (2024).
Bianchini, G., De Angelis, C., Licata, L. & Gianni, L. Treatment landscape of triple-negative breast cancer - expanded options, evolving needs. Nat. Rev. Clin. Oncol. 19, 91–113 (2022).
Steele, C. D. et al. Signatures of copy number alterations in human cancer. Nature 606, 984–991 (2022).
Ciriello, G. et al. Emerging landscape of oncogenic signatures across human cancers. Nat. Genet. 45, 1127–1133 (2013).
Natrajan, R. et al. Tiling path genomic profiling of grade 3 invasive ductal breast cancers. Clin. Cancer Res.: Off. J. Am. Assoc. Cancer Res. 15, 2711–2722 (2009).
Chin, K. et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer cell 10, 529–541 (2006).
Yuneva, M. O. et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 15, 157–170 (2012).
Altman, B. J., Stine, Z. E. & Dang, C. V. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Cancer 16, 619–634 (2016).
Ciminera, A. K., Jandial, R. & Termini, J. Metabolic advantages and vulnerabilities in brain metastases. Clin. Exp. metastasis 34, 401–410 (2017).
Geeraerts, S. L., Heylen, E., De Keersmaecker, K. & Kampen, K. R. The ins and outs of serine and glycine metabolism in cancer. Nat. Metab. 3, 131–141 (2021).
Yu, T. J. et al. PDSS1-Mediated Activation of CAMK2A-STAT3 Signaling Promotes Metastasis in Triple-Negative Breast Cancer. Cancer Res. 81, 5491–5505 (2021).
Jiang, Y. Z. et al. Genomic and transcriptomic landscape of triple-negative breast cancers: subtypes and treatment strategies. Cancer cell 35, 428–440.e425 (2019).
Li, L. D. et al. Down-Regulation of NDUFB9 Promotes Breast Cancer Cell Proliferation, Metastasis by Mediating Mitochondrial Metabolism. PloS one 10, e0144441 (2015).
Wang, D. et al. A comprehensive profile of TCF1(+) progenitor and TCF1(-) terminally exhausted PD-1(+)CD8(+) T cells in head and neck squamous cell carcinoma: implications for prognosis and immunotherapy. Int. J. oral. Sci. 14, 8 (2022).
Kazberuk, A., Chalecka, M., Palka, J., Bielawska, K. & Surazynski, A. NSAIDs Induce Proline Dehydrogenase/Proline Oxidase-Dependent and Independent Apoptosis in MCF7 Breast Cancer Cells. Int. J. Mol. Sci. 23, 3813 (2022).
Lu, T., Liu, H. & You, G. Long non-coding RNA C5orf66-AS1 prevents oral squamous cell carcinoma through inhibiting cell growth and metastasis. Int. J. Mol. Med. 42, 3291–3299 (2018).
Sun, L. et al. RNA-binding protein RALY reprogrammes mitochondrial metabolism via mediating miRNA processing in colorectal cancer. Gut 70, 1698–1712 (2021).
Pariyar, M., Johns, A., Thorne, R. F., Scott, R. J. & Avery-Kiejda, K. A. Copy number variation in triple negative breast cancer samples associated with lymph node metastasis. Neoplasia 23, 743–753 (2021).
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012).
Giancaspero, T. A. et al. Human FAD synthase is a bi-functional enzyme with a FAD hydrolase activity in the molybdopterin binding domain. Biochemical biophysical Res. Commun. 465, 443–449 (2015).
Balasubramaniam, S., Christodoulou, J. & Rahman, S. Disorders of riboflavin metabolism. J. Inherit. Metab. Dis. 42, 608–619 (2019).
Olsen, R. K. J. et al. Riboflavin-responsive and -non-responsive mutations in fad synthase cause multiple acyl-coa dehydrogenase and combined respiratory-chain deficiency. Am. J. Hum. Genet. 98, 1130–1145 (2016).
Wen, B. et al. A comparative study on riboflavin responsive multiple acyl-CoA dehydrogenation deficiency due to variants in FLAD1 and ETFDH gene. J. Human Genet. 69, 125–131 (2024).
Zhang, G. C. et al. PRP19 enhances esophageal squamous cell carcinoma progression by reprogramming srebf1-dependent fatty acid metabolism. Cancer Res. 83, 521–537 (2023).
Amemiya-Kudo, M. et al. Transcriptional activities of nuclear SREBP-1a, -1c, and -2 to different target promoters of lipogenic and cholesterogenic genes. J. lipid Res. 43, 1220–1235 (2002).
Vinyard, M. E. et al. CRISPR-suppressor scanning reveals a nonenzymatic role of LSD1 in AML. Nat. Chem. Biol. 15, 529–539 (2019).
Anand, R. & Marmorstein, R. Structure and mechanism of lysine-specific demethylase enzymes. J. Biol. Chem. 282, 35425–35429 (2007).
Hino, S. et al. FAD-dependent lysine-specific demethylase-1 regulates cellular energy expenditure. Nat. Commun. 3, 758 (2012).
Kim, J. et al. LSD1 is essential for oocyte meiotic progression by regulating CDC25B expression in mice. Nat. Commun. 6, 10116 (2015).
Metzger, E. et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437, 436–439 (2005).
Hirano, K. & Namihira, M. FAD influx enhances neuronal differentiation of human neural stem cells by facilitating nuclear localization of LSD1. FEBS open bio 7, 1932–1942 (2017).
Zhang, S., Liu, M., Yao, Y., Yu, B. & Liu, H. Targeting LSD1 for acute myeloid leukemia (AML) treatment. Pharmacol. Res. 164, 105335 (2021).
Sato, K. et al. Simultaneous CK2/TNIK/DYRK1 inhibition by 108600 suppresses triple negative breast cancer stem cells and chemotherapy-resistant disease. Nat. Commun. 12, 4671 (2021).
Ciccone, V. et al. Correction to: Stemness marker ALDH1A1 promotes tumor angiogenesis via retinoic acid/HIF-1α/VEGF signalling in MCF-7 breast cancer cells. J. Exp. Clin. cancer Res.: CR 38, 45 (2019).
Wang, T. et al. JAK/STAT3-regulated fatty acid β-oxidation is critical for breast cancer stem cell self-renewal and chemoresistance. Cell Metab. 27, 136–150.e135 (2018).
Lourenço, B. C. et al. Ki67 and LSD1 expression in testicular germ cell tumors is not associated with patient outcome: investigation using a digital pathology algorithm. Life (Basel, Switzerland) 12, 264 (2022).
Lu, Y. et al. Pan-cancer analysis revealed H3K4me1 at bivalent promoters premarks DNA hypermethylation during tumor development and identified the regulatory role of DNA methylation in relation to histone modifications. BMC genomics 24, 235 (2023).
Zhao, W. et al. Trop2 is overexpressed in gastric cancer and predicts poor prognosis. Oncotarget 7, 6136–6145 (2016).
Lin, H. et al. A novel human Fab antibody for Trop2 inhibits breast cancer growth in vitro and in vivo. Int. J. cancer 134, 1239–1249 (2014).
Bardia, A. et al. Sacituzumab govitecan in metastatic triple-negative breast cancer. N. Engl. J. Med. 384, 1529–1541 (2021).
Carey, L. A. Finding the positive in triple-negative breast cancer. Nat. cancer 2, 476–478 (2021).
Coates, J. T. et al. Parallel genomic alterations of antigen and payload targets mediate polyclonal acquired clinical resistance to sacituzumab govitecan in triple-negative breast cancer. Cancer Discov. 11, 2436–2445 (2021).
Cardillo, T. M., Govindan, S. V., Sharkey, R. M., Trisal, P. & Goldenberg, D. M. Humanized anti-Trop-2 IgG-SN-38 conjugate for effective treatment of diverse epithelial cancers: preclinical studies in human cancer xenograft models and monkeys. Clin. Cancer Res.: Off. J. Am. Assoc. Cancer Res. 17, 3157–3169 (2011).
Zhao, J. et al. Combination LSD1 and HOTAIR-EZH2 inhibition disrupts cell cycle processes and induces apoptosis in glioblastoma cells. Pharmacol. Res. 171, 105764 (2021).
Nisco, A. et al. Increased demand for FAD synthesis in differentiated and stem pancreatic cancer cells is accomplished by modulating FLAD1 gene expression: the inhibitory effect of Chicago Sky Blue. FEBS J. 290, 4679–4694 (2023).
Yıldız, Y. et al. Post-mortem detection of FLAD1 mutations in 2 Turkish siblings with hypotonia in early infancy. Neuromuscul. Disord.: NMD 28, 787–790 (2018).
Hu, P. et al. FLAD1 is up-regulated in Gastric Cancer and is a potential prediction of prognosis. Int. J. Med. Sci. 17, 1763–1772 (2020).
Kim, S. H. et al. OGT and FLAD1 genes had significant prognostic roles in progressive pathogenesis in prostate cancer. world J. men.’s. health 41, 712–723, (2023).
Morani, F. et al. PTEN regulates plasma membrane expression of glucose transporter 1 and glucose uptake in thyroid cancer cells. J. Mol. Endocrinol. 53, 247–258 (2014).
Ni, W. J. et al. Berberine regulates mesangial cell proliferation and cell cycle to attenuate diabetic nephropathy through the PI3K/Akt/AS160/GLUT1 signalling pathway. J. Cell. Mol. Med. 26, 1144–1155 (2022).
Wang, C. et al. ENO2-derived phosphoenolpyruvate functions as an endogenous inhibitor of HDAC1 and confers resistance to antiangiogenic therapy. Nat. Metab. 5, 1765–1786 (2023).
Liao, C. et al. Identification of BBOX1 as a therapeutic target in triple-negative breast cancer. Cancer Discov. 10, 1706–1721 (2020).
Wu, S. et al. Pyruvate Facilitates FACT-Mediated γH2AX Loading to Chromatin and Promotes the Radiation Resistance of Glioblastoma. Adv. Sci. (Weinh., Baden.-Wurtt., Ger.) 9, e2104055 (2022).
Yang, W. et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell 150, 685–696 (2012).
Liang, J. et al. Mitochondrial PKM2 regulates oxidative stress-induced apoptosis by stabilizing Bcl2. Cell Res. 27, 329–351 (2017).
Huang, B., Song, B. L. & Xu, C. Cholesterol metabolism in cancer: mechanisms and therapeutic opportunities. Nat. Metab. 2, 132–141 (2020).
Liu, Z., Liu, X., Liu, S. & Cao, Q. Cholesterol promotes the migration and invasion of renal carcinoma cells by regulating the KLF5/miR-27a/FBXW7 pathway. Biochemical biophysical Res. Commun. 502, 69–75 (2018).
Wang, Y., Liu, C. & Hu, L. Cholesterol regulates cell proliferation and apoptosis of colorectal cancer by modulating miR-33a-PIM3 pathway. Biochemical biophysical Res. Commun. 511, 685–692 (2019).
Ehmsen, S. et al. Increased cholesterol biosynthesis is a key characteristic of breast cancer stem cells influencing patient outcome. Cell Rep. 27, 3927–3938.e3926 (2019).
Novak, A. et al. Cholesterol masks membrane glycosphingolipid tumor-associated antigens to reduce their immunodetection in human cancer biopsies. Glycobiology 23, 1230–1239 (2013).
Menendez, J. A. & Lupu, R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer 7, 763–777 (2007).
Koundouros, N. & Poulogiannis, G. Reprogramming of fatty acid metabolism in cancer. Br. J. cancer 122, 4–22 (2020).
Dai, W. et al. PTPRO represses colorectal cancer tumorigenesis and progression by reprogramming fatty acid metabolism. Cancer Commun. (Lond., Engl.) 42, 848–867 (2022).
Goldstein, J. L., DeBose-Boyd, R. A. & Brown, M. S. Protein sensors for membrane sterols. Cell 124, 35–46 (2006).
Xiao, X. & Song, B. L. SREBP: a novel therapeutic target. Acta biochimica et. biophysica Sin. 45, 2–10 (2013).
Moslehi, A. & Hamidi-Zad, Z. Role of SREBPs in liver diseases: a mini-review. J. Clin. Transl. Hepatol. 6, 332–338 (2018).
Xue, L. et al. Targeting SREBP-2-regulated mevalonate metabolism for cancer therapy. Front. Oncol. 10, 1510 (2020).
Porstmann, T. et al. PKB/Akt induces transcription of enzymes involved in cholesterol and fatty acid biosynthesis via activation of SREBP. Oncogene 24, 6465–6481 (2005).
Bakan, I. & Laplante, M. Connecting mTORC1 signaling to SREBP-1 activation. Curr. Opin. Lipidol. 23, 226–234 (2012).
Ricoult, S. J., Yecies, J. L., Ben-Sahra, I. & Manning, B. D. Oncogenic PI3K and K-Ras stimulate de novo lipid synthesis through mTORC1 and SREBP. Oncogene 35, 1250–1260 (2016).
Sodi, V. L. et al. Nutrient sensor O-GlcNAc transferase controls cancer lipid metabolism via SREBP-1 regulation. Oncogene 37, 924–934 (2018).
Mahmud, I. et al. DAXX drives de novo lipogenesis and contributes to tumorigenesis. Nat. Commun. 14, 1927 (2023).
Fu, X., Zhang, P. & Yu, B. Advances toward LSD1 inhibitors for cancer therapy. Future medicinal Chem. 9, 1227–1242 (2017).
Zheng, Y. C. et al. Irreversible LSD1 inhibitors: application of tranylcypromine and its derivatives in cancer treatment. Curr. Top. medicinal Chem. 16, 2179–2188 (2016).
Duan, Y. C. et al. Design and synthesis of tranylcypromine derivatives as novel LSD1/HDACs dual inhibitors for cancer treatment. Eur. J. medicinal Chem. 140, 392–402 (2017).
Liu, C. et al. LSD1 Stimulates Cancer-Associated Fibroblasts to Drive Notch3-Dependent Self-Renewal of Liver Cancer Stem-like Cells. Cancer Res. 78, 938–949 (2018).
Escoubet-Lozach, L. et al. Pomalidomide and lenalidomide induce p21 WAF-1 expression in both lymphoma and multiple myeloma through a LSD1-mediated epigenetic mechanism. Cancer Res. 69, 7347–7356 (2009).
Shen, D. D. et al. LSD1 deletion decreases exosomal PD-L1 and restores T-cell response in gastric cancer. Mol. cancer 21, 75 (2022).
Zhang, J., Fan, X., Zhou, Y., Chen, L. & Rao, H. The PRMT5-LSD1 axis confers Slug dual transcriptional activities and promotes breast cancer progression. J. Exp. Clin. cancer Res.: CR 41, 191 (2022).
Kim, D., Kim, K. I. & Baek, S. H. Roles of lysine-specific demethylase 1 (LSD1) in homeostasis and diseases. J. Biomed. Sci. 28, 41 (2021).
Liu, Y. et al. Hepatic Slug epigenetically promotes liver lipogenesis, fatty liver disease, and type 2 diabetes. J. Clin. Investig. 130, 2992–3004 (2020).
Owonikoko, T. K. et al. Randomized phase II trial of cisplatin and etoposide in combination with veliparib or placebo for extensive-stage small-cell lung cancer: ECOG-ACRIN 2511 study. J. Clin. Oncol.: Off. J. Am. Soc. Clin. Oncol. 37, 222–229 (2019).
Liu, J. et al. Co-delivery of IOX1 and doxorubicin for antibody-independent cancer chemo-immunotherapy. Nat. Commun. 12, 2425 (2021).
Gong, J. et al. Combination systemic therapies with immune checkpoint inhibitors in pancreatic cancer: overcoming resistance to single-agent checkpoint blockade. Clin. Transl. Med. 7, 32 (2018).
Gong, Y. et al. Metabolic-pathway-based subtyping of triple-negative breast cancer reveals potential therapeutic targets. Cell Metab. 33, 51–64.e59 (2021).
Acknowledgements
We thank bioRENDER (biorender.com) for assisting in drawing graphics. This work was supported by the National Natural Science Foundation of China (Grant number 82373167 to X.Y., Grant number 82202883 to T.J.Y., and Grant number 92159301and 82341003 to Z.M.S.), the Shanghai Key Laboratory of Breast Cancer (Grant number 12DZ2260100 to Z.M.S.), Shanghai Medical Innovation Research Project (22Y11912700) and the Seed Program for Research and Translation of Innovative Medical Technologies of Shanghai Municipal Commission of Health (2024ZZ2023). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Z.M.S. and Y.X. conceptualized and supervised the study. X.Q.S. and T.J.Y. found the link between FLAD1 and tumor growth. X.Q.S., T.J.Y., and Y.X. designed the experiments. X.Q.S. and Y.OY. conducted data analysis and data visualization. J.H.D. assisted with the animal experiments. X.Q.S. wrote the original draft of the manuscript. X.Q.S., T.J.Y., and Y.X. contributed to writing the manuscript. All authors reviewed and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Anna Giudetti, Daiqing Liao, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Song, XQ., Yu, TJ., Ou-Yang, Y. et al. Copy number amplification of FLAD1 promotes the progression of triple-negative breast cancer through lipid metabolism. Nat Commun 16, 1241 (2025). https://doi.org/10.1038/s41467-025-56458-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-56458-w