Abstract
Technical limitations in spatial and single-cell omics sequencing pose challenges for capturing and describing multimodal information at the spatial scale. To address this, we develop SIMO, a computational method designed for the Spatial Integration of Multi-Omics datasets through probabilistic alignment. Unlike previous tools, SIMO not only integrates spatial transcriptomics with single-cell RNA-seq but expands beyond, enabling integration across multiple single-cell modalities, such as chromatin accessibility and DNA methylation, which have not been co-profiled spatially before. We benchmark SIMO on simulated datasets, demonstrating its high accuracy and robustness. Further application on biological datasets reveals SIMO’s ability to detect topological patterns of cells and their regulatory modes across multiple omics layers. Through comprehensive analysis of real-world data, SIMO uncovers multimodal spatial heterogeneity, offering deeper insights into the spatial organization and regulation of biological molecules. These findings position SIMO as a powerful tool for advancing spatial biology by revealing previously inaccessible multimodal insights.
Similar content being viewed by others
Introduction
The evolution of spatial omics and single-cell sequencing technologies has transformed our ability to study tissues and diseases at unprecedented resolution1,2,3,4,5,6,7. However, current spatial omics sequencing primarily focuses on a single modality, mainly transcriptome and proteome, making it challenging to characterize multiple modalities on the same tissue section and achieve cellular-level resolution. On the other hand, single-cell omics sequencing methods can provide detailed snapshots of cellular identity and states across various modalities, such as gene expression, chromatin accessibility, and DNA methylation1,2,3,4,8. However, tissue dissociation steps lead to the loss of spatial information, which is crucial for understanding cell states and the cellular microenvironment6,9,10,11.
Current computational methods can integrate spatial transcriptomics (ST) data with single-cell RNA sequencing (scRNA-seq) data12,13,14,15, or map paired single-cell multi-omics data onto spatial tissues16. By integrating these datasets, researchers can explore gene expression profiles within the spatial context, uncovering the distribution and functional roles of specific cell types in tissues. Furthermore, combining spatial transcriptomics with epigenetic data enhances our understanding of spatial gene regulation, such as the activation of regulatory elements during development. Additionally, there are some tools that can achieve multi-omics integration to meet the needs of multi-omics analysis of single-cell data17,18,19. However, these methods either focus solely on transcriptomics, rely on paired data or fail to effectively incorporate spatial information. As a result, there remains a significant gap in tools that can map diverse single-cell data across multiple modalities within a spatial context, hindering a comprehensive understanding of tissue biology.
To address these challenges, we introduce SIMO, a computational tool for spatial transcriptomics with multiple non-spatial single-cell omics data, such as RNA, ATAC, and DNA methylation. SIMO uses these data sets to perform precise spatial mapping of single-cell data in different modalities, construct detailed spatial patterns of cell clusters, and conduct an in-depth analysis of gene regulatory networks in the spatial dimension. We comprehensively benchmark SIMO using simulated datasets containing complex spatial patterns as well as multiple biological datasets, and further apply it to real-world data ranging from mouse brains to human myocardial infarction cases, aiming to reveal the organization’s multimodal spatial structure. These results not only verify SIMO’s excellent performance on the task of spatial integration of multi-omics single-cell data but also demonstrate its potential as a powerful tool for analyzing tissue physiological and pathological states.
Results
Overview of SIMO
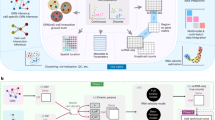
SIMO is a state-of-the-art computational tool designed for spatial mapping and integration of single-cell data from various modalities. Specifically, SIMO addresses the challenge of spatial integration of multi-omics data by breaking down the task into a sequential mapping process for each modality (Fig. 1).
a SIMO takes ST and multi-omics single-cell data as input. SIMO assigns individual cells from various modalities to specific spots, refining the spatial coordinates of single cells grounded on either the similarity of gene expression or the congruence of low-dimensional embedding representations. This process yields a comprehensive spatial multimodal dataset. By assigning coordinates to cells from different modality data, SIMO enables multidimensional deconvolution of spots and reconstruction of spatial omics patterns. Moreover, the downstream functions of SIMO can realize gene regulation analysis and spatial regulation analysis. b By leveraging the fused Gromov-Wasserstein optimal transport algorithm and taking into consideration the gene expression, as well as spatial and modal graphs constructed through k-NN, SIMO computes a probabilistic alignment between cells and spots. c For a second modality, SIMO integrates it with the already mapped transcriptomic data, utilizing Unbalanced Optimal Transport (UOT) to facilitate cluster-based label transfer. Following this, SIMO calculates cell alignment using Gromov-Wasserstein (GW) based on k-NN graphs.
Initially, SIMO integrates ST data with transcriptomics data based on the premise that these two data types originate from the same modality, aiming to minimize the interference caused by modal differences. This approach has been validated by prior research, underscoring the importance of shared modalities in precise data integration16. In the transcriptomics mapping step of SIMO, we borrowed the computational strategy of the previously developed tool SpaTrio. Specifically, the process uses the k-nearest neighbor (k-NN) algorithm to construct a spatial graph (based on spatial coordinates) and a modality map (based on low-dimensional embedding of sequencing data), and uses the fused Gromov-Wasserstein optimal transport to calculate the mapping relationship between cells and spots. We retain the key hyperparameter α for balancing the significance of transcriptomic differences and graph distances. We fine-tune the cell coordinates based on the transcriptome similarity between the mapped cells and their surrounding spots.
Next, SIMO targets the integration of non-transcriptomic single-cell data, such as single-cell epigenetic data, through a new sequential mapping process. For single-cell ATAC sequencing (scATAC-seq) data, SIMO first preprocesses both mapped scRNA-seq and scATAC-seq data, obtaining initial clusters via unsupervised clustering. To bridge RNA and ATAC modalities, gene activity scores are used as a key linkage point, calculated as a gene-level matrix based on chromatin accessibility (Methods). SIMO calculates the average Pearson Correlation Coefficients (PCCs) of gene activity scores between cell groups, facilitating label transfer between modalities using an Unbalanced Optimal Transport (UOT) algorithm. Subsequently, for cell groups with identical labels, SIMO constructs modality-specific k-NN graphs and calculates distance matrices, determining the alignment probabilities between cells across different modal datasets through Gromov-Wasserstein (GW) transport calculations. Finally, based on the cell matching relationship, SIMO precisely allocates scATAC-seq data to specific spatial locations (spots) and further adjusts cell coordinates based on the modality similarity between the mapped cells and their neighboring spots. This stepwise strategy enhances SIMO’s spatial integration compatibility across omics data. By modifying the UOT cost matrix construction, SIMO achieves spatial mapping of various omics types, irrespective of their positive or negative biological relationship with the transcriptome.
SIMO’s downstream analysis capabilities encompass both gene regulation analysis and spatial regulation analysis, collectively illuminating the complexities of gene regulation. In gene regulation analysis, depending on specific analytical needs, data are transformed into matrices with gene names as features, such as motif activity matrices calculated from ATAC data. Correlations and regulatory patterns between different cell populations were analyzed by calculating PCCs between fold changes in motif activity and gene expression. Spatial regulation analysis involves integrating data from both modalities and their spatial information. Applying a spatial smoothing algorithm to reduce data noise and using cross-modal smoothing to supplement information between modalities, the ratio of feature pairs is calculated as a regulatory score. A kernel matrix based on spatial location information is further constructed, and feature modules with similar spatial regulation patterns are identified through weighted correlation analysis and Consensus Clustering (CC).
Evaluation on simulated datasets
We first used multi-omics data from the mouse cerebral cortex, including both single-nucleus chromatin accessibility and mRNA expression sequencing (SNARE-seq) and in situ sequencing hetero RNA–DNA-hybrid after assay for transposase-accessible chromatin-sequencing (ISSAAC-seq)2,20 to construct simulated spatial datasets with varying degrees of spatial complexity, aiming to evaluate the performance of the SIMO tool and optimize the key parameter α (Supplementary Fig. 1a–c). The simulation of ST data involved sample extraction, data merging, and coordinate allocation. At the same time, we introduced pseudocount δ and resampled the readings to introduce varying degrees of noise into the spatial transcriptomics data, thereby assessing the robustness of the tool. To comprehensively evaluate the accuracy of spatial mapping for each modality, we employed several key evaluation metrics, including cell mapping accuracy (the percentage of cells correctly matched to their types), the Root Mean Square Error (RMSE) of deconvoluted cell type proportions, and two measures based on the Jensen-Shannon Distance (JSD): JSD of spot and JSD of type. JSD of spot focuses on the accuracy of cell-type distribution at spatial locations, evaluating by comparing the differences between actual and expected distributions. JSD of type assesses the accuracy of predicting proportions of each cell type across the entire sample, determined by calculating the difference between actual and expected proportions. These metrics were calculated for each modality and averaged to comprehensively reflect the tool’s overall performance in handling multimodal data (Fig. 2 and Supplementary Fig. 2).
The Root Mean Square Error (RMSE) for SIMO, across settings α = 0 (solely gene expression), α = 1 (exclusively graph data), and α = 0.1 (a combination of both) with a pseudocount δ in Pattern 1–6, was derived from twenty simulated trials per configuration. Each parameter was repeated 20 times (n = 20). The RMSE was calculated and averaged across omics. Source data are provided as a Source Data file.
In scenarios with simpler spatial distributions (Patterns 1 and 2), SIMO (α = 0.1) demonstrated greater stability as δ increased. In contrast, relying solely on gene expression data (α = 0) led to a faster decline in performance. When predictions were based only on graphical data (α = 1), only 21.0%-43.0% of cells in Pattern 1 were correctly mapped. Even under conditions of high noise (δ = 5), SIMO (α = 0.1) was able to accurately recover the spatial positions of more than 91% of cells in Pattern 1 and over 88% in Pattern 2, achieving the lowest RMSE, JSD of spot, and JSD of type values. Furthermore, we also simulated scenarios with more complex cell distributions. In Pattern 3, 15.4% of spots contained multiple cell types; this proportion increased to 67.8% in Pattern 4. Even in the presence of significant noise, SIMO (α = 0.1) showed exceptional stability in these complex scenarios. Specifically, Pattern 3 achieved 83% mapping accuracy, with an RMSE of 0.098, JSD (spot) of 0.056, and JSD (type) of 0.131; in Pattern 4, the accuracy was 73.8%, with an RMSE of 0.205, JSD (spot) of 0.222, and JSD (type) of 0.279. The simulated data Patterns 5 and 6 have more cell types (10 cell types). In Pattern 5, 61% of the spots contain multiple cell types, while Pattern 6 is more complex, with 91% of the spots containing only multiple cell types (Supplementary Fig. 1c). SIMO (α = 0.1) achieves 62.8% accuracy in Pattern 5, with an RMSE of 0.179, JSD (spot) of 0.300, and JSD (type) of 0.564, and 55.8% accuracy in Pattern 6, with an RMSE of 0.182, JSD (spot) of 0.419, and JSD (type) of 0.607 in high noise scenes. These results indicate that setting parameter α to 0.1 generally yields the best performance.
Comparison with existing tools
To further highlight the advantages of SIMO, we compared it with several other integration algorithms, including those specifically designed for ST data, such as CARD and Tangram, as well as integration methods for scRNA-seq, like Seurat, LIGER, and Scanorama14,17,18,19,21. In addition to simulated data, we collected three sets of biological datasets, from which matched ST and multi-omics single-cell data were prepared by splitting and merging, to test the tools11.
The results show that under noise-free conditions (pseudocount = 0), SIMO significantly outperformed other tools across all simulated datasets (Patterns 1-6), achieving the lowest RMSE values (0.000, 0.152, 0.049, 0.112, 0.050, 0.061) and its JSD values were also significantly lower than other methods (Fig. 3a). In tests on more complex real datasets, SIMO also displayed superior performance (RMSE = 0.156, 0.119, 0.123) (Fig. 3a and Supplementary Fig. 3), leading other methods except for a slight underperformance in Dataset 2 compared to Tangram (RMSE = 0.101). When noise was introduced, SIMO’s advantages became even more apparent, achieving the best results across all testing indicators across all datasets (Fig. 3b and Supplementary Fig. 4). In Dataset 1, SIMO reached an RMSE of 0.236, with JSD of spot and JSD of type at 0.489 and 0.575, respectively. In Dataset 2, SIMO’s RMSE decreased to 0.198, significantly outperforming the second-placed LIGER (RMSE = 0.232) and third-placed Tangram (RMSE = 0.238). In Dataset 3, SIMO led with an RMSE of 0.186, ahead of the second-placed LIGER (RMSE = 0.221), with a reduction in error of 15.84%.
a, b SIMO demonstrates superior performance compared to the other tools. The RMSE of the deconvolved cell-type proportions was calculated by comparing them with the ground truth. The indicators were calculated and averaged across omics. The pseudocount is set to 0 (a) and 5 (b) respectively. In the box plots, the range of each box extends from the first to the third quartile, with the horizontal line representing the median. The whiskers extend to 1.5 times the interquartile range beyond the lower and upper bounds of the box. Dataset 1: mouse embryo data (Spatial ATAC–RNA-seq); Dataset 2: mouse brain data (Spatial ATAC–RNA-seq); Dataset 3: mouse brain data (Spatial CUT&Tag–RNA-seq, H3K27me3). Each parameter was repeated 5 times (n = 5). Source data are provided as a Source Data file. c Spatial map of the original data and spatial maps reconstructed using various tools for the CUT&Tag assay (H3K27me3), with spatial pixels or cells colored according to their respective classifications.
On the biological datasets, we further analyzed the capacity of all tools to reconstruct cell-type spatial patterns under noise-free conditions. It was observed that the tools generally performed well in the spatial mapping of transcriptomic data but showed weaker performance in the spatial mapping of epigenetic data. In contrast, SIMO’s mapping results across each modality were relatively stable, particularly excelling in the second modality over other tools (Fig. 3c and Supplementary Figs. 5–7). For example, in reconstructing the spatial distribution of A3 in Dataset 1 and C11 in Dataset 2, SIMO revealed a clearer spatial distribution pattern (Supplementary Figs. 5, 6). Particularly noteworthy is SIMO’s performance in Dataset 3, which involves H3K27me3 (repressing loci) data from the mouse brain, where transcriptomic and epigenetic signals show opposite correlations. Unlike other integration tools that only integrate spatial and non-spatial data based on transcriptomic data similarity, thereby overlooking potential inconsistencies in signal strength across modalities, SIMO was capable of accurately merging transcriptomic and epigenetic data simultaneously (Fig. 3c and Supplementary Fig. 7). This underscores SIMO’s advanced and unique capability in effectively identifying and reconciling inter-modality differences when processing multimodal data.
In addition to comparing the same computational tools in each mapping, we can also combine different methods (Supplementary Fig. 8). Specifically, we applied spatial deconvolution tools (such as Tangram and CARD) in the first mapping and data integration tools (such as Seurat, LIGER, and Scanorama) in the second mapping, comparing their results with those of SIMO. The results show that across both simulated and biological datasets, SIMO outperforms other approaches in all metrics.
Evaluation of biological data
To assess SIMO’s capability to map complex spatial cell arrangements in real-world biological scenarios, we evaluated its performance on real-world spatial multi-omics datasets. Specifically, we tested SIMO across three distinct datasets: the Spatial ATAC-RNA-seq dataset from mouse embryos (Dataset 1), the Spatial ATAC-RNA-seq dataset from mouse brain tissue (Dataset 2), and the Spatial CUT&Tag-RNA-seq dataset from mouse brain tissue (Dataset 3). In addition to SIMO’s demonstrated ability to accurately reconstruct the spatial distribution of multi-omics cell types, its precise reconstruction of various omics features was also notable (Supplementary Figs. 9–13). In Dataset 1, Six6, a key gene involved in eye development, showed the highest gene expression and accessibility in the eye region. Genes such as Sox2, Myt1l, and Pax6, which are highly accessible in certain regions of the embryonic brain, exhibited relatively low RNA expression. This observation suggests that these genes may be involved in lineage specification during embryonic brain development22. For Dataset 2, multi-omics features in different mouse brain regions revealed high signals corresponding to specific cell types: striatum (Pde10a, medium spiny neurons), corpus callosum (Sox10, Mbp, and Tspan2, oligodendrocytes), cortex (Mef2c, Neurod6, and Cux2, excitatory neurons), and lateral ventricle (Dlx1, ependymal/neural progenitor cells). Overall, the spatial distribution patterns of gene expression and accessibility in Dataset 2 were relatively consistent, whereas this was not the case in Dataset 3. For instance, genes like Pde10a, Sox10, and Tspan2 showed high gene expression in regions with lower H3K27me3 signaling. SIMO accurately reconstructed the spatial distribution of multi-omics features across all datasets, preserving biological relationships between features, while other tools showed considerable errors, particularly in Dataset 3.
We further analyzed Dataset 1 to measure the preservation of biological relationships between different omics layers. We focused on radial glia and postmitotic premature neurons, which are known to have spatially graded differentiation relationships22. Cells in these regions were divided into three main regions (Region 1, 2, and 3), and we calculated the signal correlation of different omics features across these regions (Fig. 4a and Supplementary Fig 14a). Clustering based on these correlations identified three categories: C1, C2, and C3, with C1 genes highly expressed in Region 3, C2 genes in Region 2, and C3 genes in Region 1. All genes with high accessibility were found in C3 (Fig. 4b). These spatial distribution differences reflect distinct spatial omics regulation patterns. Gene Ontology (GO) pathway analysis of genes across the three omics layers revealed associations with different biological functions (Supplementary Fig 14c). For example, pathways related to development and forebrain development were linked to C2 and C3, while C1 was associated with negative regulation of neurogenesis, aligning with its presence in the terminal differentiation regions. SIMO’s accurate reconstruction of these spatial omics regulatory patterns, and the high correlation between reconstructed and actual results, demonstrated its superior performance compared to other methods (Fig. 4c and Supplementary Fig 14b).
a Spatial regions of radial glia and postmitotic premature neurons. b Spatial distribution of features in different clusters (Spatial-ATAC-RNA-seq). Clustering is based on multi-omics correlations of features, representing multi-omics regulatory patterns. c Correlation between the multi-omics regulatory relationships reconstructed by the tool and the actual situation. Source data are provided as a Source Data file. d Correlation of H3K27me3 signaling and RNA gene expression. Fold change was calculated by differential analysis of omics between spatial regions. e The count of regulatory relationships with correlation directions (positive or negative) consistent with those measured by Spatial CUT&Tag-RNA-seq. Source data are provided as a Source Data file.
In Dataset 3, the correlation between multi-omics features across different regions was crucial. Strong anti-correlation between H3K27me3 and RNA was observed in the original sequencing data. Low H3K27me3 levels in Mal, Mag, and Car2 corresponded to high RNA expression (quadrant IV), while high H3K27me3 levels in Syt1, Grin2b, and Mef2c were associated with low RNA expression (quadrant II). SIMO effectively captured the correlations between these two omics features, outperforming other methods that often showed errors or omissions in correlation (Fig. 4d and Supplementary Fig 14d). Specifically, SIMO reconstructed 611 feature associations, the highest among all compared methods (Fig. 4e).
Spatial integration of the mouse cortex
First, we applied SIMO to a publicly available mouse brain multimodal single-cell dataset. This dataset integrates gene expression, DNA methylation, and 10x Visium ST data8,23 (Supplementary Fig. 15a–c). Through preliminary analysis of single-cell transcriptomics data, we discovered that SIMO accurately reconstructs the stratified characteristics of excitatory neuron subtypes, arranged in the order of layers L2/3, L5, and L6, perfectly aligning with existing prior knowledge of cortical structure (Fig. 5a–c and Supplementary Fig. 15d, e). Additionally, we delved into the spatial distribution patterns of specific marker genes, for instance, finding Otof and Cux2 predominantly expressed in the upper layers (L2/3), Rorb and Fezf2 mainly concentrated in the middle layers, and Sulf2 and Foxp2 significantly distributed in the deeper layers. The spatial distribution of DNA methylation data also clearly reflected the stratification of layers L2/3, L4, L5, and L6, with markers such as the methylation markers (mCH and mCG) of Cux2 being more pronounced in the deeper layers, while the signals of Fezf2 and Sulf2 were stronger in the superficial layers (Supplementary Fig. 16a, b). Notably, compared to existing tools, SIMO demonstrated a significant advantage in the spatial mapping capability of DNA methylation signals (Supplementary Fig. 17a–d), emphasizing its unique advantage in compatibility with multimodal integration.
a Spatial integration results of mouse cerebral cortex data. Spots and cells are colored according to different categories. b, c The box plot shows the distance between the main cell groups in the single-cell transcriptome data (b) and single-cell DNA methylation data (c) after spatial mapping and the innermost C6 group in the ST data. Different cell types are included, with varying sample sizes for each cell type (n = 50 for Layer2/3, n = 351 for Layer5a, n = 120 for Layer5, n = 265 for Layer5b, n = 454 for Layer6, n = 237 for mL2/3, n = 92 for mL4, n = 94 for mL5-2, n = 50 for mL6-1 and n = 333 for mL6-2). In the box plots, the range of each box extends from the first to the third quartile, with the horizontal line representing the median. The whiskers extend to 1.5 times the interquartile range beyond the lower and upper bounds of the box. Source data are provided as a Source Data file. d, e SIMO mappings of Layer5a cells, with cells color-coded based on DNA methylation cell types (d) and RNA clusters (e).
Moreover, the integrative analysis facilitated by SIMO enriches our insight into cell typing across various omics layers from a spatial perspective. Notably, the Layer5a cell type spans a broad spatial extent within the transcriptomics landscape. Our integrated assessment reveals that these cells predominantly align with mL4 and mDL-2 cell types identified in DNA methylation profiles. Unsupervised clustering further uncovers the intrinsic heterogeneity within Layer5a cells (Fig. 5d, e). Additionally, we examined the expression patterns of key markers (including Cux2, Rorb, Rora, Sox5, Rbfox1, and Bcl11a)24,25,26 among Layer5a subtypes, observing expression variability across modalities and distinctive regional distribution (Supplementary Fig. 18a–d). These observations underscore SIMO’s robust capability in elucidating the spatial heterogeneity inherent in multi-omics single-cell datasets.
Next, we combined gene expression and DNA methylation data to deeply explore the gene regulatory mechanism in the mouse cerebral cortex. It is generally believed that DNA methylation has a profound regulatory impact on gene activity, and it affects cell fate decisions during aging and development by regulating gene expression4,9,10,11,27,28. We explored the interaction between gene expression and corresponding DNA methylation levels by analyzing population-level PCCs between them (Fig. 6a). The results showed that among the gene-methylation pairs with significant correlations, most genes showed negative correlations with their DNA methylation marks, which is consistent with previous research results. For example, when the Cux2 gene reaches its peak expression in the L2/3 layer, its DNA methylation level is relatively low; conversely, the high methylation level of the Rorb gene is consistent with its distribution in low-expression regions in the deeper layers (Supplementary Fig. 16a, b).
a The average DNA methylation activity, specific to each cell type, is graphically represented alongside the mean log-fold change in expression of the corresponding gene. Statistical analysis was performed using the two-sided PCC test. These plots uncover relationships that display significant correlations. b The dot plot shows the results of module analysis, and the color of the dots is defined according to the module to which they belong. c Spatial regulation modules of Layer5a cells were identified using SIMO. d SIMO maps of the activity score of the gene expression within modules of Layer5a cells. e Violin plots of the activity score of the gene expression within modules of Layer5a cells. f SIMO maps of the activity score of the DNA methylation within modules of Layer5a cells. g Violin plots of the activity score of the DNA methylation within modules of Layer5a cells.
To conduct further in-depth analysis of the Layer5a cell population, we explored its multimodal spatial regulation pattern. Through spatial module analysis of gene expression and DNA methylation signals, we identified two main modules, each exhibiting its unique spatial distribution pattern (Fig. 6b, c). The first module is mainly located in the outer layer of the cerebral cortex and covers genes such as Cux2, Rorb, and Rora and their corresponding DNA methylation signals, which is consistent with cluster 1 in the transcriptome data and DNA methylation data. The mL4 class matched, showing high gene expression and low DNA methylation levels in the outer region (Fig. 6d–g and Supplementary Fig. 18a–d). The second module is mainly distributed in the inner layer, including Sox5, Rbfox1, Bcl11a, and other genes and their corresponding DNA methylation signals, which is related to cluster 2 in the transcriptome data and mDL-2 class in the DNA methylation data. Correspondingly, high gene expression and low DNA methylation levels in the inner layer were demonstrated (Fig. 6d–g and Supplementary Fig. 18a–d). These findings further emphasize the heterogeneity of Layer5a cells at the multi-omics level from the perspective of spatial regulation.
Spatial integration of the human myocardial infarction
We apply SIMO to datasets covering human myocardial infarction events, including scRNA-seq, scATAC-seq data, and ST dataset29 (Supplementary Fig. 19a–f). The analysis results show that single cells in both scRNA-seq and scATAC-seq data are successfully mapped to spatial slices, involving key cell types such as cardiomyocytes (RYR2), endothelial cells (PECAM1), fibroblasts (COL12A1)30, myeloid cells (CD14), and vascular Smooth Muscle Cells (vSMCs) (MYH11). These cell types exhibited similar spatial distribution characteristics in different modalities (Fig. 7a and Supplementary Fig. 20a, b). We selected marker genes specific to multiple cell types to calculate the abundance of major cell types at each location in the ST data and compared these data with cell type proportions plotted using the SIMO tool. From a spatial distribution perspective, there is a clear consistency between the abundance of cell types and their spatial distribution proportions, and the spatial distribution calculated by cell differential genes is also similar to the original data (Fig. 7b and Supplementary Fig. 20c).
a Spatial integration results of human myocardial infarction data. Spots and cells are colored according to different categories. b Scaled PCCs between the expression of cell markers and the proportion of each cell type. c SIMO mappings of cardiomyocyte cells within RNA data, with cells color-coded based on RNA cluster. d Boxplots show the distance of cardiomyocyte subpopulations from the infarct area (cluster 0: n = 1799, cluster 1: n = 1247 and cluster 7: n = 687). In the box plots, the range of each box extends from the first to the third quartile, with the horizontal line representing the median. The whiskers extend to 1.5 times the interquartile range beyond the lower and upper bounds of the box. p-values were calculated using the two-sided Wilcoxon test. Source data are provided as a Source Data file. e SIMO mapping of cardiomyocytes in ATAC data, with cells color-coded according to grouping. This grouping is based on the distance of ATAC subpopulations from the infarct area. f The violin plot illustrates the scoring of biological processes for the GO pathway “Wound Healing” (Adjacent: n = 1903 and Distant: n = 664). In the box plots, the range of each box extends from the first to the third quartile, with the horizontal line representing the median. The whiskers extend to 1.5 times the interquartile range beyond the lower and upper bounds of the box. p-values was calculated using the two-sided Wilcoxon test.
Given the importance of cardiomyocytes among cell types, we performed an in-depth analysis of them. Single-cell transcriptome data revealed that cardiomyocytes are mainly divided into three unsupervised clustering subpopulations: clusters 0, 1, and 7 (Fig. 7c and Supplementary Fig. 21a). There are significant differences in the distance between these clusters and the myocardial infarction area (Fig. 7d). Among them, cluster 7 is closest to the infarction area, and cluster 0 is the farthest to the infarction area. Through pathway enrichment analysis, we found that cluster 0 is mainly related to normal heart functions such as cardiac contraction, muscle contraction, and blood circulation; cluster 1 is related to tissue morphology construction; cluster 7 is associated with pathways involved in wound healing, myofibril assembly, and the assembly of cell binding sites, these pathways may be involved in cardiac repair and recovery after myocardial infarction31,32 (Supplementary Fig. 21b–d). This suggests that cardiomyocytes adjacent to the infarcted area exhibit characteristics of post-infarction repair. At the same time, we also analyzed the ATAC data of cardiomyocytes and divided all unsupervised clusters into adjacent groups and distant groups according to the distance from the infarct area, which are spatially different distribution patterns (Fig. 7e and Supplementary Fig. 21e). By calculating the scores of biological pathways, we found that the wound healing scores in adjacent regions were significantly higher than those in distant regions, indicating that cardiomyocytes in adjacent regions activated accessible regions of relevant genes (Fig. 7f). These findings reveal the spatial heterogeneity of cardiomyocytes in function and gene regulation after myocardial infarction, providing valuable clues for in-depth exploration of the pathophysiological process of myocardial infarction.
Subsequently, by integrating gene expression data with motif activity insights from ATAC sequencing, we meticulously explored the multimodal distribution patterns within cardiomyocytes (Supplementary Fig. 22a). Our modularity analysis identified two principal gene-motif modules with distinct spatial distributions: module 1 is situated closer to the infarct zone, whereas module 2 proximates farther (Fig. 8a, b). Spatial analysis underscored a negative correlation between module 1 activity and the proximity to myeloid cells and the infarct region, underscoring the potential significance of module 1-associated subgroups in the immunomodulatory processes of myocardial infarction (Fig. 8c and Supplementary Fig. 22b). Utilizing the disparity in module scores, we categorized cardiomyocytes into two subgroups aligned with module 1 (high score group) and module 2 (low score group) (Supplementary Fig. 22c). By conducting Gene Set Enrichment Analysis (GSEA) to delve into the functional characteristics of these two subgroups, we discovered that module 1 is involved in platelet function, intraplatelet calcium levels, and cytokine-related pathways, which may play a crucial role in the onset and progression of myocardial infarction (Fig. 8d). Additionally, module 1 encompasses several transcription factors intimately linked to myocardial infarction; for instance, ATF6 mitigates myocardial ischemia/reperfusion (I/R) injuries by inducing genes associated with oxidative stress responses, and IRF2 modulates apoptosis by activating the cell pyroptosis pathway via Gasdermin-D (GSDMD)33,34 (Supplementary Fig. 22d). These insights intimate that subgroups related to module 1 may be pivotal in the development and progression of myocardial infarction.
a Spatial regulation modules of cardiomyocyte cells were identified using SIMO. b SIMO maps of the activity score of gene-motif pairs within modules of cardiomyocytes. c The activity scores of gene-motif pairs within module 1 and their correlation with the distance to the infarct area (left) and myeloid cells (right). Statistical analysis was performed using a two-sided linear regression model (ordinary least squares, OLS). d GSEA analysis results of module1-related genes. The Benjamini-Hochberg (BH) method was applied for multiple comparison adjustment. e SIMO maps of the activity score of gene-motif pairs within modules of fibroblasts. f GO analysis results of module2-related cell genes, including Biological Process (BP) and Molecular Function (MF) categories. The BH method was applied for multiple comparison adjustment.
Analysis of fibroblasts also yielded two main gene-motif modules, of which module 1 is closer to myeloid cells and module 2 is located near the infarct area (Fig. 8e and Supplementary Fig. 22e, f). The differential scores of these two modules further divided fibroblasts into two subgroups (Fig. 8g). GO pathway enrichment analysis showed that module 2 is related to pathways related to cell junction assembly and structural components of the extracellular matrix. These pathways may play an important role in the repair of cardiac tissue after injury35 (Fig. 8f). Transcription factors involved in module 2, such as KLF15, OVOL2, RUNX1, SMAD3, etc., may play a regulatory role in cardiac fibrosis and tissue repair36,37,38,39. The transcription factors involved in module 1, such as KLF2 and ZEB1, are involved in the regulation of inflammatory and immune responses40,41 (Supplementary Fig. 22h). Interestingly, RUNX1 and RUNX2, as structurally similar transcription factors, belong to different modules, reflecting their different functions and biological roles in cardiac tissue. Early studies have shown that the activity and stability of ETV2 can be regulated through its interaction with OVOL2, which has a significant impact on the methylation/demethylation process of DNA and the expression of downstream target genes37. Given the critical role of ETV2 in cardiovascular regeneration and cardiac repair after myocardial infarction, these findings highlight the possibility of OVOL2 as a potential target for the treatment of myocardial infarction.
Discussion
We introduced SIMO, a computational tool that utilizes optimal transport algorithms to integrate single-cell data from various single modalities through sequential spatial mapping, enabling spatial integration of multi-omics single-cell data. With its downstream analysis capabilities, SIMO can accurately perform spatial analysis of gene regulatory patterns. Compared to existing computational methods, SIMO displays several advantages. First, it is one of the most advanced tools available, capable of simultaneously reconstructing the spatial distribution of data from multiple modalities, enabling spatial mapping of two or more types of omics data (Supplementary Fig. 23). Second, SIMO excels in handling modal data with opposing biological signals, demonstrating strong compatibility in multimodal integration tasks. Furthermore, SIMO not only possesses gene regulation analysis capabilities but also allows for in-depth exploration of complex gene regulatory spatial patterns within tissues. Lastly, with the continuous advancement of single-cell omics sequencing technologies, SIMO is theoretically compatible with any omics data that can display features associated with the transcriptome, providing more diversified and in-depth omics insights for tissue studies.
Initially, we conducted benchmark tests on SIMO using simulated datasets to assess its accuracy and robustness across different spatial patterns, noise levels, and hyperparameter settings, ultimately determining the optimal hyperparameters. Compared to other integration tools, SIMO demonstrated superior performance, especially in integrating non-transcriptomic data, where it significantly outperformed other tools. We then applied SIMO to two real datasets. In the analysis of mouse cerebral cortex data, SIMO not only accurately identified different cortical layers but also revealed multimodal gene regulatory relationships at a spatial resolution. Additionally, SIMO resolved high-resolution substructures of cell populations and specific modal spatial heterogeneities. In studies of human myocardial infarction, SIMO, through spatial regulation analysis, revealed multimodal heterogeneity between cardiomyocytes and fibroblasts. Based on spatial regulatory analysis, SIMO also proposed potential therapeutic targets for the diagnosis and treatment of myocardial infarction.
As single-cell omics sequencing technology continues to evolve, we will be able to understand the complex biological states of individual cells from a more detailed omics perspective. By efficiently integrating key gene regulatory data, SIMO could provide a more comprehensive landscape of gene regulatory networks.
Currently, single-cell analysis has entered a new era centered on multi-omics, with single-cell omics and ST technologies becoming foundational to biological research. Therefore, as these technologies merge and innovate, we expect SIMO to become an important tool for exploring the physiological and pathological states of tissues in the future. It has the potential to assist scientists in examining spatiotemporal dynamics and whole-genome gene regulation mechanisms within tissue environments, and it may play a crucial role in building disease models and precisely analyzing pathological states, providing scientific bases for discovering new therapeutic targets and developing treatment strategies.
Methods
SIMO toolkit
Alignment of modality1
The input scRNA-seq and ST data were used for the first integration and processed before calculating the optimal probabilistic alignment and integration. We recommend using the top 100 genes per cell type/cluster shared between the two datasets for subsequent analysis under default parameters. Gene expression matrices were normalized and scaled in preparation for analysis. Complementary representations were used to capture the distinct characteristics of each dataset to facilitate integration. For scRNA-seq, the data were summarized as \(X\in {{\mathbb{R}}}^{p\times n}\), capturing the expression levels of \(p\) genes in \(n\) cells, and \(E\in {{\mathbb{R}}}^{h\times n}\), a reduced-dimensional representation that highlights critical cellular features using techniques like PCA. For spatial transcriptomics, the data were represented by \(Y\in {{\mathbb{R}}}^{p\times m}\), which captures the expression levels of \(p\) genes in \(m\) spots, and \(Z\in {{\mathbb{R}}}^{2\times m}\), which retains the spatial location of spots. To address the inherent differences between the dimensionally reduced embedding representations \(E\) and the spatial coordinates \(Z\), we adopted a graph-based strategy to harmonize their scales. Specifically, we constructed a k-NN graph for each dataset, leveraging the local neighborhood relationships within the embedding space and spatial domain. Distances between nodes were refined using type- or region-specific adjustments to reflect biological and spatial heterogeneity. Unconnected nodes were assigned the graph’s maximum distance to ensure completeness. The entire matrix was subsequently normalized to make spatial and embedding distances comparable across datasets. These resulting distance matrices, representing the relationships among cells and spots respectively, are defined as \(G\in {{\mathbb{R}}}^{n\times n}\) and \(G^{\prime} \in {{\mathbb{R}}}^{m\times m}\).
To integrate scRNA-seq and ST data, we adopted the previous algorithm strategy16, describing input datasets as triplets \(\left(X,G,w\right)\) and \(\left(Y,G^{\prime},w^{\prime} \right)\).Here, \(X\in {{\mathbb{R}}}^{p\times n}\) and \(Y\in {{\mathbb{R}}}^{p\times m}\) are gene expression matrices for \(n\) cells and \(m\) spots, \(G\in {{\mathbb{R}}}^{n\times n}\) and \(G^{\prime} \in {{\mathbb{R}}}^{m\times m}\) are pairwise graph distance matrices, and \(w/w^{\prime}\) are distributions over cells and spots, which can be user-defined based on biological priors or set to uniform by default. The alignment is represented by a mapping matrix \(\Pi=[{\pi }_{{ij}}]\in {{\mathbb{R}}}_{+}^{n\times m}\), where \({\pi }_{{ij}}\) indicates the probability of mapping cell \(i\) to spot \(j\). The alignment is achieved by minimizing a composite cost function \(F(\Pi )\), which combines two key objectives:
-
1.
Expression similarity cost: This term quantifies the overall dissimilarity between the gene expression profiles of cells and spots, defined as \({C}_{\exp }={\sum}_{i,j}d({x}_{{\cdot i}},{y}_{{\cdot j}}){\pi }_{{ij}}\), where \(d\) is the expression cost function and \(d({x}_{{\cdot i}},{y}_{{\cdot j}})\) quantifies the expression-level divergence between cell \(i\) and spot \(j\).
-
2.
Graph pairwise distance cost: Quantifies the difference in pairwise graph distances defined as \({C}_{{graph}}={\sum}_{i,j,k,l}{({g}_{{ik}}-{g}_{{jl}}^{{\prime} })}^{2}{\pi }_{{ij}}{\pi }_{{kl}}\), where \({g}_{{ik}}\) represents the graph pairwise distance between cell \(i\) and \(k\), and \({g}_{{jl}}^{{\prime} }\) represents the graph pairwise distance between spot \(j\) and \(l\). This term ensures that the pairwise relationships within the spatial and embedding graphs are preserved during the alignment process.
The overall objective function is a weighted combination of these two components:
where \(\alpha \in [{{\mathrm{0,1}}}]\) is a tunable parameter that determines the relative importance of gene expression similarity versus distance preservation in the alignment, a higher \(\alpha\) emphasizes spatial consistency, while a lower \(\alpha\) prioritizes expression similarity.
To determine the cell composition of each spot, we first rank cells based on their mapping probabilities, excluding those with zero probability. The top-ranked cells are selected as potential candidates for further analysis. Using the Non-Negative Least Squares (NNLS) method, we deconvolved the cell types contributing to each spot. Candidate cells that align with the deconvolution outcomes are prioritized, while any remaining gaps in the expected number of mapped cells are filled by selecting additional cells with the highest probabilities.
Coordinates assignment of modality1
The spatial coordinates of cells are assigned through a two-step process. Initially, cells are positioned based on the spatial location of the spot they belong to. These initial coordinates are then refined by accounting for the correlation between the cell’s gene expression and the expression profiles of nearby spots. For example, consider a spot \(j\) with coordinates \(({x}_{j},{y}_{j})\), surrounded by neighboring spots \({j}_{1}\), …, \({j}_{n}\). The similarity between cell \(i\) in spot \(j\) and its neighboring spots is defined as \({p}_{1}\), …, \({p}_{n}\), which are the PCCs of gene expression scaled to the range \([{{\mathrm{0,1}}}]\). Using these similarity scores, the coordinates of cell \(i\) in spot \(j\) are computed as follows:
To cope with the insufficient number of adjacent spots, we added pseudo spot \({j}_{{pseudo}}\), which inherits the gene expression profile of spot j\(,\) and is assigned spatial coordinates calculated as:
The pseudo-spot will participate in the coordinate correction process as a nearby spot. To ensure cells remain within the boundaries of their respective spots, we adjust their distances from the spot center. This scaling ensures that the farthest cells are positioned exactly at the edge of the spot, maintaining their spatial distribution within the defined spot area.
Alignment of modality2
The data of scRNA-seq and another modality (taking scATAC-seq data as an example) all go through a preprocessing process, using the corresponding modality low-dimensional representation to build a proximity network, and using the Leiden algorithm to assign initial cell cluster labels. Next, we use the gene-level matrix (gene activity score for ATAC) for subsequent analysis. The gene-level matrix can be calculated using the ArchR package or the Signac package. The top 10 differential genes of each cluster were used for subsequent label migration. Specifically, for each modality, we identified the top 10 genes with the highest differential expression in each cluster, thereby capturing the key marker genes that distinguish clusters. We then retained the intersection of key genes between different modalities. Datasets contain matrix \(X\) and \(Y\), where \(X\) represents the gene expression level, and \(Y\) gene activity score. We generate expression profile for each cell population based on the cell label, denoted as \(X^{\prime}\), where \({x^{\prime} }_{i}\) represent the average expression profile of i th cell population. Similarly, we create an average activity profile \(Y^{\prime}\), with \({y^{\prime} }_{j}\) representing the average activity profile of the j th cell population. We then calculated the PCC between \({x^{\prime} }_{i}\) and \({y^{\prime} }_{j}\), storing the results in the correlation matrix \(M\):
To facilitate the next step of label transfer, we scaled all correlation coefficients to a range of 0 to 1 and calculated the difference from 1, resulting in the correlation-based distance matrix, which is used for the next step of label transfer. If two modalities display opposite biological activity trends, such as transcriptional activity versus inhibitory epigenomic signals, we instead use the original correlation matrix \(M\) directly in the label transfer step.
We assign each cell population a weight greater than zero in each modality. By default, we apply a uniform distribution and normalize these weights. We define the alignment matrix between clusters as \(\Pi=[{\pi }_{{jl}}]\in {{\mathbb{R}}}_{+}^{{c}_{1}\times {c}_{2}}\) where \({c}_{1}\) and \({c}_{2}\) mean the cluster number of modalities. Given marginal relaxation term \({{reg}}_{m}\) use the following cost to find a mapping:
The obtained probability map is subjected to threshold processing to obtain the transfer relationship between cell populations.
For cell populations from two data sets that are successfully paired, we extract their low-dimensional embedding representation and use the previous method to construct a k-NN graph and calculate the distance matrix. Finally, the Gromov-Wasserstein transport was used to calculate the pairing probability between cells and determine the pairing relationship between cells based on the probability.
Cordinates assignment of modality2
After obtaining the matching relationship between cells between modality1 and 2, we allocate the cells of modality2 to the corresponding spot based on the information about the spot where the cells in modality are located and correct the coordinates. The correction process is similar to the previous method. The difference is that gene expression is no longer used for PCC calculations. Instead, low-dimensional embedding representation and cosine similarity are used to measure the relationship between cells and surrounding spots.
Gene regulation analysis
Gene regulation analysis integrates information from both modality 1 and modality 242. Before this, data from modality 2 must be converted into a matrix format with gene names as features and different types of data are selected according to analysis requirements. For instance, the RunChromVAR function in Signac43 (version 1.9.0), based on the JASPAR2020 database44, can estimate transcription factor activity as an input for analysis. For DNA methylation data, the average signal value of different DNA methylation sites of the same gene is calculated as an input for the analysis. Taking ATAC data as an example, the FindMarkers function calculates the fold change in transcription factor activity and gene expression, with a false discovery rate (FDR) threshold set to less than 0.05. Then, the PCCs between the activity of transcription factors and the fold change in corresponding gene expression is used to assess the correlation between the two modalities. Based on the strength and direction of these correlations, transcription factors are categorized into three groups. Those showing a strong positive correlation are inferred to act as transcriptional activators, enhancing gene expression. Conversely, transcription factors with negative correlations are likely to function as transcriptional repressors, reducing gene expression. For transcription factors with negligible or no correlation, their regulatory roles remain uncertain.
Spatial regulation analysis
Spatial regulation analysis integrates data from modality 1 and modality 2 along with their corresponding spatial location information. Initially, expression data and spatial information are extracted from both datasets. To reduce measurement noise and prepare for subsequent modular analysis, spatial smoothing is applied to the data within each dataset45. Essentially, this means that the expression data for each cell is adjusted based on the average expression of its surrounding neighboring cells. Moreover, cross-modal smoothing is performed to complement missing information in one modality, revealing potential interactions between the two datasets. Specifically, for a cell in modality 1, we use information from neighboring cells in modality 2 to estimate its missing data. The smoothed expression data are then merged to create a comprehensive data framework encompassing both modalities and minimum-maximum normalization ensures data consistency. Subsequently, for each feature pair (gene) across the two datasets, we calculate the expression ratio, defined as the expression level of the feature in dataset 2 divided by that in dataset 1. This ratio serves as the regulatory score to assess the strength of gene regulation between the features. To facilitate weighted correlation analysis, we construct a kernel matrix based on spatial location information. This matrix reflects the spatial proximity of cells, calculated by determining the pairwise Euclidean distances between cells and applying a Gaussian function to convert these distances into weights:
Where \({d}_{{ij}}\) represents the distance between cell \(i\) and \(j\), and \(\sigma\) is a parameter that controls the smoothness of the kernel. The resulting kernel matrix is crucial for capturing the influence of spatial relationships during the weighted correlation analysis. CC is applied to the weighted correlation matrix to categorize feature pairs into groups, identifying sets of features with similar regulatory patterns46. The results are further refined by setting specific criteria (such as upper and lower limits for feature numbers, average connectivity strength, and average correlation threshold). Module scores are calculated using the score_genes function in Scanpy47. In summary, this analysis, by merging and comparing data from two modalities along with their spatial information and through detailed preprocessing and analysis of the data, can identify feature pairs with significant regulatory roles at the spatial level. This provides important perspectives and tools for a deeper understanding of spatial biology.
Simulation data
To comprehensively evaluate the performance of SIMO, we adopted the simulation strategy used in our previous research16, which is based on SNARE-seq data from the mouse cerebral cortex to construct simulated datasets. The gene activity matrix was computed using the GeneActivity function within Signac (v1.9.0). After preprocessing and annotating cell types, we identified three key subgroups: L2/3 IT, L4, and L5 IT, labeled as Cell types 1, 2, and 3. From each type, 250 cells were randomly selected, divided into 50 groups representing spatial locations, and their average expression levels were calculated for each location. After assigning spatial coordinates to these locations, we created an ST dataset that includes the transcriptome and epigenome information for each spatial position to test the performance of SIMO. The original single-cell multi-omics data were split into two sets of single-omics data and input into the SIMO algorithm along with the constructed ST data. Patterns 5 and 6, which contain the 10 most important cell types in the ISSAAC-seq brain cortex data, are used to construct simulated data.
To increase the realism of the data and simulate the noise introduced by technical limitations, we introduced a pseudocount parameter δ. Specifically, before mapping cells to specific locations, new transcript counts were generated based on the total count at each location using a negative binomial distribution and then adjusted using a multinomial distribution for individual gene counts, thereby simulating randomness and adding a level of noise to the data. Moreover, to eliminate the potential impact of tissue slice angles, we randomly rotated the slices before aligning the data.
Biological data
To comprehensively evaluate the effectiveness of SIMO, we utilized three biological datasets as benchmarks: mouse embryonic Spatial ATAC-RNA-seq data, mouse brain Spatial ATAC-RNA-seq data, and mouse brain Spatial CUT&Tag–RNA-seq (H3K27me3) data. These datasets, derived from sequencing experiments, provide a realistic foundation for assessing the performance of tools. We use the gene activity score and chromatin silencing score in the original data as part of the input data.
Mouse Embryonic Spatial ATAC-RNA-seq Dataset (Dataset 1)
By merging every four adjacent pixels into one, we preserved the spatial transcriptomics data for input into the ST dataset while removing the original positional information and segmenting the data to create multi-omics single-cell datasets for input. The final scRNA-seq data included 2187 cells and 17058 genes; the scATAC-seq data comprised 2187 cells and 32437 peaks, corresponding to 24017 genes’ activity scores. The resultant ST dataset contained 576 spots and 17058 genes. We retained only the highly variable genes common between the expression data and gene activity scores, and the input datasets were subject to standard preprocessing steps, including normalization, PCA analysis (for RNA assay), LSI analysis (for ATAC assay), and nonlinear dimension reduction using UMAP. The original data’s clustering information was used as a basis for grouping in specific computational steps, and the omics data for mapping spots were obtained by merging the contained single-cell data. We manually delineated the primary regions of radial glia and postmitotic premature neurons and divided these into three regions (Region 1, 2, and 3) based on the distance between cells and radial glia. Subsequently, we selected differential features from the RNA and ATAC assays for comparison. We calculated the average gene expression levels and average gene activity scores for these three regions, normalizing them to a 0-1 scale. We then computed the Spearman correlation of the same features between different omics (RNA-ATAC) to summarize their multi-omics regulatory patterns. Using the correlation of each feature, we performed Hierarchical clustering, resulting in three distinct clusters. We computed spatial distribution scores for each feature cluster using AddModuleScore. To evaluate the accuracy of tools, we compared the reconstructed multi-omics regulatory patterns with the actual data. Specifically, we calculated the Spearman correlations between different omics features for each tool and then assessed these correlations against the true results using Pearson correlation.
Mouse Brain Spatial ATAC-RNA-seq Dataset (Dataset 2)
Following the same pixel merging strategy as with the embryonic dataset, we obtained scRNA-seq data containing 9215 cells and 22914 genes; scATAC-seq data included 9215 cells and 121068 peaks, corresponding to 24027 genes’ activity scores. The corresponding ST dataset contained 2315 spots and 22914 genes. All preprocessing steps were consistent with those mentioned for the embryonic dataset, and all computational steps of SIMO were executed with default parameters.
The Mouse Brain Spatial CUT&Tag–RNA-seq (H3K27me3) Dataset (Dataset 3)
After data segmentation and merging, the scRNA-seq portion includes 9752 cells and 25881 genes; the scATAC-seq segment covers 9752 cells with 70470 peaks, and the corresponding gene-cell chromatin silencing score matrix encompasses 24023 genes. The ST dataset contains 2441 spatial spots and 25881 genes. The preprocessing methods are consistent with those applied to the previous datasets. When integrating the second modality (CUT&Tag) data, the alignment_2 function’s modality2_type parameter is set to “neg”. We used transcriptomic clusters to define regional groupings, and based on these regional groupings, we calculated the log2(fold change) for features using FindAllMarkers function across two omics (RNA and H3K27me3) as the basis for final visualization. For the tool-generated mapping data, each cell’s regional grouping was assigned based on the location. We then applied the same calculations and processing steps. We classified omics log2 (fold change) values as either positive or negative relationships, depending on whether the signs were the same (positive correlation) or different (negative correlation). We then counted the number of correctly constructed feature relationships across all tools to assess their performance.
Performance evaluation
Mapping accuracy
The precision of cell mapping was evaluated by determining the percentage of cells accurately placed within their corresponding regions. For a given spot \(j\), where \(n\) cells were allocated and \(n^{\prime}\) of those cells shared the same cell type as that of spot \(j\), the precision of cell placement was quantified as the ratio of \(n^{\prime}\) to \(n\). To compute the overall allocation precision \(A\) for a slice containing m spots, we used the formula below:
RMSE
The RMSE was calculated to assess the discrepancy between the deconvolved proportions and the actual proportions of precise labels for each spot within the ST dataset. Specifically, for each spot, the RMSE is computed as the square root of the average of the squared differences between the deconvolved proportions and the actual proportions across all cell types. The calculation is performed using the equation:
Here, \(M\) represents the total number of spots, \(N\) represents the total number of cell types, \({y^{\prime} }_{n,j}\) denotes the deconvolved proportion of cell type \(n\) within the spot \(j\), and \({y}_{n,j}\) indicates the actual proportion of cell type \(n\) within spot \(j\).
JSD
The calculation process of JSD between cell type proportions and reference proportions in ST data can be performed for two different metrics: “spot” and “type”.
For the “spot” metric, JSD calculations are performed across spots to assess the similarity between cell type distributions within each spot, comparing observed cell ratios to a reference standard. For each spot \(j\), the JSD is caculated as follows:
Where \({P}_{j}\) represents the observed cell type proportions in spot \(j\), \({Q}_{j}\) represents the reference cell type proportions in spot \(j\), \({M}_{j}=\frac{1}{2}({P}_{j}+{Q}_{j})\) is the average of the two distributions and \({D}_{{KL}}\) is the Kullback-Leibler divergence.
In the case of the “type” metric, the JSD calculation across cell types measures the consistency of the overall distribution of each cell type at all spots, comparing the deconvoluted proportions to the true proportions in the reference data. For each cell type \(k\), the JSD is caculated as follows:
Where \({P}_{k}\) represents the observed cell type proportions across all spots for cell type \(k\), \({Q}_{k}\) represents the summed reference cell type proportions across all spots for cell type \(k\) and \({M}_{k}=\frac{1}{2}\left({P}_{k}+{Q}_{k}\right)\) is the average of the two distributions.
Baseline methods processing
To assess the performance of SIMO, we compared it against several existing integration tools, including CARD (v1.0), Tangram (v1.0.4), Seurat (v4.3.0), LIGER (v1.0.1), and Scanorama (v1.7.3). Before integration, ST data were randomly rotated and supplemented with a predetermined amount of pseudo-counts to simulate data variability in real-world applications. Using CARD, we inferred single-cell resolution gene expression for each spatial position. With Tangram, we executed cell-to-spot mapping using default parameters to determine the alignment probabilities between spots and cells. Data were integrated using Seurat’s IntegrateData function, and distance matrices between spots and cells were computed using PCA embedding, then inverted and divided by the maximum value to obtain the alignment matrix. The optimizeALS function of LIGER was utilized for data integration, calculating distance matrices between spots and cells based on Nonnegative Matrix Factorization (NMF) embedding. Scanorama integrated datasets to calculate the matching relationships between spots and cells. The resulting matching matrix was then inputted into the assign_coord_2 function to allocate cells to their respective spots. Finally, we compared the performance of these tools using the metrics mentioned above. In performing multimodal spatial integration, we followed SIMO’s strategy of sequentially mapping multiple single-omics data, using gene activity scores or transformed gene-cell signal matrices as input for non-transcriptomic modalities.
Mouse cerebral cortex data analysis
Single-cell transcriptomic data of the mouse cerebral cortex, measured through Drop-seq technology, revealed 8 major cell types. The single-cell DNA methylation data of the same region, obtained using snmC-seq technology, displayed 16 major cell types. For the 10x Visium ST data, only spots located in the cortical layers were retained, and based on the transcriptomic data’s heterogeneity, the brain sections were divided into five areas corresponding to the five main cortical layers (Astro, L2/3, L4, L5, L6, and Oligo). Data were processed using SIMO’s preprocessing workflow. Differential genes calculated using unsupervised clustering information from ST data and cell type labels from single-cell transcriptomic data were retained for further analysis, with only genes common to both kept. Five single cells were allocated to each spot for this analysis. When mapping DNA methylation data, differential genes were identified using unsupervised clustering information from mapped single-cell transcriptomic data and cell type labels from single-cell DNA methylation data, with common genes retained for further analysis. The alignment_2 function’s modality2_type parameter was set to “neg” for integrating DNA methylation data, with three single cells allocated to each spot. Gene regulation analysis was conducted using default settings, and spatial regulation analysis only retained gene-DNA methylation features with significant correlation, with parameters set to sigma = 140, mink = 2, maxK = 8, avg_con_min = 0.5, avg_cor_min = 0.5, min_feature = 20, max_feature = 200.
Human myocardial infarction data analysis
Gene expression matrices and 10x Visium data (patient_region_id: RZ_P3) were downloaded from https://cellxgene.cziscience.com/collections/8191c283-0816-424b-9b61-c3e1d6258a77, while peak count matrices were obtained from https://zenodo.org/records/6578553 and https://zenodo.org/record/6578617. Following the original study’s protocol, we processed the scATAC-seq data to derive gene activity scores. To enhance processing efficiency, we randomly selected scRNA-seq and scATAC-seq data for 20,000 cells. The data were processed using SIMO’s standardized preprocessing workflow. Differential genes shared between the original annotations of ST data and unsupervised clustering labels from single-cell transcriptomic data were calculated and retained. Up to three cells were allocated per spot. For mapping ATAC data, unsupervised clustering information from both mapped single-cell transcriptomic data and scATAC-seq data was used to identify differential genes, with three cells allocated per spot. According to the staining map and the spatial distribution of cell types (Fig. 6 and Supplementary Fig. 12a), we took the location of vSMC with an ordinate greater than 0.5 as the core of the infarct area. The gene regulation analysis was conducted with default parameters. Spatial regulation analysis for cardiomyocytes was set with parameters sigma = 140, mink = 2, maxK = 8, avg_con_min = 0.5, avg_cor_min = 0.5, min_feature = 20, max_feature = 200, while for fibroblasts, parameters were adjusted to avg_con_min = 0.6 and avg_cor_min = 0.6.
Pathway and biological process enrichment analysis
To identify pathways within gene clusters, the clusterProfiler package48 (version 4.6.2) in R was utilized for conducting enrichment analysis on genes from each cluster. The analysis focused on the BP and MF categories, selecting results that surpassed a predefined significance level (q-value cutoff = 0.05). For analyzing pathway enrichment among different cell groups, the FindAllMarkers function was employed to identify differentially expressed genes (DEGs) in each cell group relative to the others, applying criteria (only.pos = TRUE, min.pct = 0.2, logfc.threshold = 0.2) to filter for genes with an adjusted p-value (via the Wilcoxon test) below 0.05. Additionally, GSEA49 was carried out on a ranked list of genes to uncover significantly enriched pathways and biological processes, drawing upon gene sets from the Molecular Signatures Database (MSigDB, http://www.gsea-msigdb.org/gsea/msigdb) to interpret the gene signatures linked to these pathways and processes.
Scoring of biological processes
We generated scores for individual cells based on gene signatures representing biological functions, with the scores for biological processes defined as the average normalized expression of the associated genes. These functional signatures were collected from the GO database, comprising differentially expressed genes identified with an adjusted p-value cutoff of 0.05 using the Wilcoxon test. For the “Wound Healing” pathway (GO:0042060), scores were calculated based on the genes enriched in the corresponding pathway.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All relevant data supporting the key findings of this study are available within the article and its Supplementary Information files. The original data used in this paper can be accessed through the following links: (1) Mouse cerebral cortex SNARE-seq data: GEO accession: GSE126074; (2) ISSAAC-seq data of mouse brain cortex: https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-11264 (3) Spatial ATAC–RNA-seq data of mouse embryo and brain, Spatial CUT&Tag–RNA-seq (H3K27me3) data of mouse brain: https://web.atlasxomics.com/visualization/Fan; (4) Mouse cerebral cortex Drop-seq data: http://dropviz.org/; (5) Mouse cerebral cortex snmC-seq data: https://brainome.ucsd.edu/annoj/brain_single_nuclei/; (6) Mouse cerebral cortex 10x Visium data: https://satijalab.org/seurat/articles/spatial_vignette; (7) Human myocardial infarction scRNA-seq, scATAC-seq and 10x Visium data: https://cellxgene.cziscience.com/collections/8191c283-0816-424b-9b61-c3e1d6258a77, https://zenodo.org/record/6578553 and https://zenodo.org/record/6578617. Source data are provided with this paper.
Code availability
The SIMO toolkit is available at GitHub: https://github.com/ZJUFanLab/SIMO under GPL-3.0 license. It is also deposited at Zenodo: https://doi.org/10.5281/zenodo.1449825750.
References
Ma, S. et al. Chromatin Potential Identified by Shared Single-Cell Profiling of RNA and Chromatin. Cell 183, 1103–1116.e20 (2020).
Chen, S., Lake, B. B. & Zhang, K. High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nat. Biotechnol. 37, 1452–1457 (2019).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
Park, J. et al. Spatial omics technologies at multimodal and single cell/subcellular level. Genome Biol. 23, 256 (2022).
Liao, J., Lu, X., Shao, X., Zhu, L. & Fan, X. Uncovering an Organ’s Molecular Architecture at Single-Cell Resolution by Spatially Resolved Transcriptomics. Trends Biotechnol. 39, 43–58 (2021).
Schueder, F. & Bewersdorf, J. Omics goes spatial epigenomics. Cell 185, 4253–4255 (2022).
Perkel, J. M. Single-cell analysis enters the multiomics age. Nature 595, 614–616 (2021).
Luo, C. et al. Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science 357, 600–604 (2017).
Deng, Y. et al. Spatial-CUT&Tag: Spatially resolved chromatin modification profiling at the cellular level. Science 375, 681–686 (2022).
Deng, Y. et al. Spatial profiling of chromatin accessibility in mouse and human tissues. Nature 609, 375–383 (2022).
Zhang, D. et al. Spatial epigenome–transcriptome co-profiling of mammalian tissues. Nature 616, 113–122 (2023).
Longo, S. K., Guo, M. G., Ji, A. L. & Khavari, P. A. Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nat. Rev. Genet 22, 627–644 (2021).
Moncada, R. et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat. Biotechnol. 38, 333–342 (2020).
Ma, Y. & Zhou, X. Spatially informed cell-type deconvolution for spatial transcriptomics. Nat. Biotechnol. 40, 1349–1359 (2022).
Kleshchevnikov, V. et al. Cell2location maps fine-grained cell types in spatial transcriptomics. Nat. Biotechnol. 40, 661–671 (2022).
Yang, P. et al. Revealing spatial multimodal heterogeneity in tissues with SpaTrio. Cell Genom. 3, 100446 (2023).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021).
Welch, J. D. et al. Single-Cell Multi-omic Integration Compares and Contrasts Features of Brain Cell Identity. Cell 177, 1873–1887.e17 (2019).
Hie, B., Bryson, B. & Berger, B. Efficient integration of heterogeneous single-cell transcriptomes using Scanorama. Nat. Biotechnol. 37, 685–691 (2019).
Xu, W. et al. ISSAAC-seq enables sensitive and flexible multimodal profiling of chromatin accessibility and gene expression in single cells. Nat. Methods 19, 1243–1249 (2022).
Biancalani, T. et al. Deep learning and alignment of spatially resolved single-cell transcriptomes with Tangram. Nat. Methods 18, 1352–1362 (2021).
Fan, R. et al. Spatially Resolved Epigenome-Transcriptome Co-Profiling of Mammalian Tissues at the Cellular Level. https://www.researchsquare.com/article/rs-1728747/v1. https://doi.org/10.21203/rs.3.rs-1728747/v1 (2022).
Saunders, A. et al. Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 174, 1015–1030.e16 (2018).
Lai, T. et al. SOX5 Controls the Sequential Generation of Distinct Corticofugal Neuron Subtypes. Neuron 57, 232–247 (2008).
Zhang, Z. et al. Epigenomic diversity of cortical projection neurons in the mouse brain. Nature 598, 167–173 (2021).
Muñoz-Castañeda, R. et al. Cellular anatomy of the mouse primary motor cortex. Nature 598, 159–166 (2021).
Liu, Y. et al. High-Spatial-Resolution Multi-Omics Sequencing via Deterministic Barcoding in Tissue. Cell 183, 1665–1681.e18 (2020).
Lu, T., Ang, C. E. & Zhuang, X. Spatially resolved epigenomic profiling of single cells in complex tissues. Cell 185, 4448–4464.e17 (2022).
Kuppe, C. et al. Spatial multi-omic map of human myocardial infarction. Nature 608, 766–777 (2022).
Guerrero-Juarez, C. F. et al. Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds. Nat. Commun. 10, 650 (2019).
Chalise, U., Becirovic‐Agic, M. & Lindsey, M. L. The cardiac wound healing response to myocardial infarction. WIREs Mech. Dis. 15, e1584 (2023).
Machackova, J., Barta, J. & Dhalla, N. S. Myofibrillar remodelling in cardiac hypertrophy, heart failure and cardiomyopathies. Can. J. Cardiol. 22, 953–968 (2006).
Jin, J.-K. et al. ATF6 Decreases Myocardial Ischemia/Reperfusion Damage and Links ER Stress and Oxidative Stress Signaling Pathways in the Heart. Circ. Res 120, 862–875 (2017).
Li, Y., Wang, Y., Guo, H., Wu, Q. & Hu, Y. IRF2 contributes to myocardial infarction via regulation of GSDMD induced pyroptosis. Mol. Med Rep. 25, 40 (2021).
McCain, M. L., Lee, H., Aratyn-Schaus, Y., Kléber, A. G. & Parker, K. K. Cooperative coupling of cell-matrix and cell–cell adhesions in cardiac muscle. Proc. Natl Acad. Sci. USA. 109, 9881–9886 (2012).
Wang, B. et al. The Kruppel-like factor KLF15 inhibits connective tissue growth factor (CTGF) expression in cardiac fibroblasts. J. Mol. Cell. Cardiol. 45, 193–197 (2008).
Kim, J. Y. et al. OVOL2 is a critical regulator of ER71/ETV2 in generating FLK1+, hematopoietic, and endothelial cells from embryonic stem cells. Blood 124, 2948–2952 (2014).
McCarroll, C. S. et al. Runx1 Deficiency Protects Against Adverse Cardiac Remodeling After Myocardial Infarction. Circulation 137, 57–70 (2018).
Bujak, M. et al. Essential Role of Smad3 in Infarct Healing and in the Pathogenesis of Cardiac Remodeling. Circulation 116, 2127–2138 (2007).
Tang, X. et al. KLF2 regulates neutrophil activation and thrombosis in cardiac hypertrophy and heart failure progression. J. Clin. Investig. 132, e147191 (2022).
Plaschka, M. et al. ZEB1 transcription factor promotes immune escape in melanoma. J. Immunother. Cancer 10, e003484 (2022).
Muto, Y. et al. Single cell transcriptional and chromatin accessibility profiling redefine cellular heterogeneity in the adult human kidney. Nat. Commun. 12, 2190 (2021).
Stuart, T., Srivastava, A., Madad, S., Lareau, C. A. & Satija, R. Single-cell chromatin state analysis with Signac. Nat. Methods 18, 1333–1341 (2021).
Fornes, O. et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Research gkz1001 https://doi.org/10.1093/nar/gkz1001 (2019).
Kartha, V. K. et al. Functional inference of gene regulation using single-cell multi-omics. Cell Genom. 2, 100166 (2022).
Wei, R. et al. Spatial charting of single-cell transcriptomes in tissues. Nat. Biotechnol. 40, 1190–1199 (2022).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS: A J. Integr. Biol. 16, 284–287 (2012).
Subramanian, A. et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 102, 15545–15550 (2005).
Penghui Yang. Spatial integration of multi-omics single-cell data with SIMO. Zenodo https://doi.org/10.5281/ZENODO.14498256 (2024).
Acknowledgements
This work is supported by the National Natural Science Foundation of China (U23A20513), Ningbo Top Medical and Health Research Program (No. 2022030309). The authors thank the High-Performance Computing Cluster of Zhejiang University Innovation Center of Yangtze River Delta for their technical support.
Author information
Authors and Affiliations
Contributions
X.F. and X.L. conceived the study. P.Y. drafted the manuscript. K.J. and X.S. collected and analyzed the multi-omics single-cell data and ST data. Y.Y. and C.L. implemented the algorithm of SIMO. P.Y. developed the package of SIMO. K.J. provided important advice on cell-type annotation and regulation analysis of brain tissue. Y.Y. and L.J. provided important advice on the application of algorithms in human myocardial infarction. All authors wrote the manuscript, read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, P., Jin, K., Yao, Y. et al. Spatial integration of multi-omics single-cell data with SIMO. Nat Commun 16, 1265 (2025). https://doi.org/10.1038/s41467-025-56523-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-56523-4
This article is cited by
-
Single-cell multi-omics in cancer immunotherapy: from tumor heterogeneity to personalized precision treatment
Molecular Cancer (2025)
-
Medical laboratory data-based models: opportunities, obstacles, and solutions
Journal of Translational Medicine (2025)