Abstract
PR/SET domain-containing (PRDM) proteins are metazoan-specific transcriptional regulators that play diverse roles in mammalian development and disease. Several members such as PRDM1, PRDM14 and PRDM9, have been implicated in germ cell specification and homoeostasis and are essential to fertility-related processes. Others, such as PRDM14, PRDM15 and PRDM10 play a role in early embryogenesis and embryonic stem cell maintenance. Here, we describe the first PRDM family member with a maternal effect. Absence of maternal Prdm10 results in catastrophic failure of oocyte-to-embryo transition and complete arrest at the 2-cell stage. We describe multiple defects in oocytes, zygotes and 2-cell stage embryos relating to the failure to accumulate PRDM10 target gene transcripts in the egg. Transcriptomic analysis and integration of genome-wide chromatin-binding data reveals new and essential PRDM10 targets, including the cytoskeletal protein encoding gene Septin11. We demonstrate that the failure to express maternal Septin11, in the absence of maternal PRDM10, disrupts Septin-complex assembly at the polar body extrusion site in MII oocytes. Our study sheds light into the essentiality of maternal PRDM10, the requirement of the maternal Septin-complex and the likely evolutionary conservation of this regulatory axis in human female germ cells.
Similar content being viewed by others
Introduction
The oocyte-to-embryo transition (OET) involves activation and maturation of the oocyte, completion of meiosis, fertilization, and subsequent transformation into a developing embryo1. This transformation may arguably be the most dramatic change in mammalian development, even more so, considering the two highly specialized cell types, oocyte and sperm, involved2. Particularly the oocyte has been of great interest to research, as it provides nearly all gene products required for its successful culmination in form of maternally stored gene products, accumulated during oocyte growth3,4. This naturally puts a spotlight on the transcriptional regulators in charge of the deposition of these maternal transcripts, ultimately ensuring the smooth transition from germ cell to new life5. Through their study, and the study of their targets, we gain insights into the complex processes, which often, when deregulated, will lead to catastrophic failure and be responsible for female infertility, abortions, aneuploidies, or developmental syndromes3,6,7,8. Here we focus our attention on the maternal function of the transcriptional activator PRDM10, uncovering its essential function in preparation for OET.
PRDMs are highly conserved regulators of transcription with members shown to play crucial roles during development and homoeostasis. Characteristic to PRDMs are their N-terminal PR domain which has high homology to the SET domain of methyltransferases, and their highly variable C2H2-zinc finger repeats, which enable sequence-specific DNA-binding, making the 19 members of the family by and large non-redundant9,10. While some members such as PRDM1, PRDM14 and PRDM15 are well characterized and shown to control important cell-fate decisions, such as primordial germ cells or naïve pluripotency fates11,12, other members are still poorly understood.
We recently added a new family member, PRDM10 (aka tristanin), to the map of PRDMs essential for embryonic development13. In mice, zygotic loss of PRDM10 causes fully penetrant embryonic arrest at the early blastocyst stage, consequence of substantial transcriptional deregulation of essential genes. Amongst them we characterized the translation initiation factor, Eif3b, as a key downstream target, and demonstrate partial functional rescue of the PRDM10 KO in mouse Embryonic Stem Cells (mESCs)13.
Here we show that not only zygotic-, but also maternal loss of PRDM10 disrupts development. Preventing the accumulation of PRDM10-target genes in the fully grown oocyte (FGO) disrupts an array of processes in the egg and in the zygote, culminating in a fully penetrant 2-cell arrest. Through transcriptomic analysis and integration of genome-wide PRDM10 DNA binding analyses, we not only reveal Flcn14 and Eif3b13 as conserved targets of maternal PRDM10 but additionally characterize a full dependency of the cytoskeletal Septin-network15 on PRDM10-regulated transcription.
Results
Maternal Prdm family gene expression
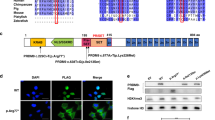
Several members of the PRDM family play important roles in the female and/or male germ line11,12. Others, such as maternal deletion of Prdm15, appear to be dispensable (unpublished observation). To gain insights into potential maternal functions of PRDMs we mined RNA-seq data of mouse16 and human GV oocytes17 for Prdm/PRDM family member transcripts (Fig. 1a, Supplementary Fig. 1a, b). We further confirmed the observations of Prdm/PRDM family member expression patterns across oocyte and early embryonic stages in recent transcriptomic data both in mouse18,19 and humans7 (Fig. 1b, Supplementary Fig. 1c, e). Lastly, we explored the expression of Prdm genes at oogenesis (from non-growing oocytes [NGOs] to fully grown oocytes [FGOs]20 (Fig. 1d). We find Prdm1, 2, 4, 6, 10 and 16 are consistently expressed in mouse GV oocytes which is also reflected in NGO, GO1 and GO2 oocytes. In humans only PRDM2, 4 and 10 show maternal expression. Interestingly, at least in myoblasts, Prdm4/10 have been suggested to be partially transcriptional-dependent on PRDM221. Prdm4 knock-out female mice are viable and fertile, excluding essential maternal requirements22. Similarly, Prdm2/Riz1 knock-out mice are viable and fertile but have an increased risk of tumour development in adults23. Expression levels of Prdm1, 6 and 16 are divergent between humans and mouse, being substantially activated only in the latter. Simultaneous deletion of maternal Prdm16 and Prdm3, which we find expressed at marginal levels in GV oocytes (Supplementary Fig. 1a) causes post-natal lethality for unknown reasons24.
a Expression heatmap (RNA-seq) of Prdm family members in mouse (n = 7, GV1-GV7) and human (n = 14, GV1-GV14) GV (germinal vesical) oocytes. (Prdm7 is primate-specific; Prdm10 is highlighted.) b Expression heatmap (RNA-seq) of Prdm family members in oocytes and early embryos. c RPF (ribosome-protected mRNA fragment) heatmap of Prdm family members in oocytes and early embryos. (FGO [fully grown oocytes], LPI [late prometaphase I oocyte], MII [Metaphase II oocyte], zygotes (PN3/5 [pronuclear stage 3/5]) and preimplantation stages (E2C/L2C [early/late 2-cell stage], 4C/8C [4-cell/8-cell stage], ICM [inner cell mass]). d Expression heatmap (RNA-seq) of Prdm family members in NGO (non-growing oocytes), GO1/2 (growing oocytes) and FGO. e Prdm10 expression in GV oocytes after Zp3-Cre-induced deletion (normalized to Trim28 and shown relative to Ctr expression levels). f Pups/litter born to Ctr and MatKO females mated to wildtype males (n = 15 and n = 8 observed mating plugs for Ctr and MatKO producing mothers, respectively; showing median, upper and lower quartiles; ****, p = 7.9*10−11, unpaired, two-tailed, parametric t-test. g In vitro culture of Ctr and MatKO zygotes (E0.5) to the blastocyst stage (E3.5) (scale bar: 100 μm). h Percentage of embryos developing across preimplantation stages to blastocyst in culture (n = 59/106 Ctr or MatKO embryos, respectively). i Representative still images from Supplementary Movie 1 (a) and Supplementary Movie 2 (b) of a Ctr and Prdm10 MatKO zygote, respectively, at various timepoints across first cleavage division. Injected cRNA for H2B-RFP (red) labels chromatin while membrane-targeted GFP (green) labels membranes (scale bar: 50 μm). j Symmetric vs asymmetric blastomere size in Ctr and MatKO 2-cell stage embryos respectively (red arrowheads indicate smaller blastomere, scale bar: 100 μm). k Quantification of skewed blastomere size ratios (****, p = 2.1*10−5, unpaired, two-tailed, parametric t-test). At least three experimental replicates for all embryo isolations and counts were performed.
Maternal expression levels of Prdm10 are conserved across humans and mice and found in the latter as early as in NGOs. While zygotic expression is essential in preimplantation embryos13 its maternal function remains unexplored. Translatomic profiling further substantiates the presence of maternal PRDM10 protein across all tested stages of oogenesis, OET and early development in humans and mice (Fig. 1c, Supplementary Fig. 1d, f).
Prdm10 is a maternal-effect gene
We used the established conditional knock-out mouse line13 to address maternal PRDM10 requirements using Zp3-Cre mediated recombination25 (Supplementary Fig. 2a). Deletion of exon five of Prdm10 gives rise to a frameshift mutation; the resulting truncation of the protein due to a premature stop codon occurs upstream of all annotated functional domains, such as the PR domain, the zinc fingers, and the (Q)-rich activation domain13 (Supplementary Fig. 2b, c). Deletion efficiency was confirmed by qPCR analysis in pooled GV oocytes derived from three mutant/control females, respectively (Fig. 1e).
Breeding pairs of maternal knock-out (MatKO) and control (Ctr) oocyte-producing females with wildtype males were observed over 6 months. Despite successful mating (by evidence of n = 15 and n = 8 vaginal plugs for Ctr and mutants respectively), no pups were obtained from MatKO oocytes (Fig. 1f), nor did we observe overt pregnancies. Control females delivered expected litter numbers and sizes (Fig. 1f). However, infertile MatKO females’ ovaries appeared normal and both GV and MII oocytes could readily be obtained (Supplementary Fig. 2d–f). We thus assessed the developmental potential of Prdm10 MatKO and Ctr zygotes (0.5 dpc) across preimplantation stages in vitro. MatKO-derived embryos (n = 106) displayed complete developmental arrest with nearly 70% of MatKO embryos arrested at the 2-cell stage and 30% failing to even undergo successful first cleavage division. In contrast, Ctr zygotes (n = 59) developed efficiently into blastocysts (94%) after three/four days in culture (Fig. 1g, h).
Live imaging of H2B-RFP/membrane-targeted GFP injected zygotes hints towards multiple defects at first cleavage division in mutant embryos, including the presence of multiple/fragmented pronuclei, incomplete abscission, chromosome segregation defects and severe membrane blebbing (Fig. 1i and Supplementary Movie 1–3). A consequence of these disturbances is the apparent deviation of the typically symmetrical blastomere size in MatKO embryos (n = 68) compared to Ctrs (n = 43) (Fig. 1j, k). In summary, we find Prdm10 is a maternal effect gene, with a fully penetrant 2-cell block or even earlier developmental arrest.
Loss of maternal Prdm10 causes pleiotropic phenotypes at OET
We set out to define the cause of the developmental block of MatKO-derived embryos at and before the 2-cell stage first by addressing egg quality and meiotic competence. MatKO GV and MII oocytes appeared normal compared to their wildtype counterparts based on presence of prominent nucleoli surrounded by a well-defined germinal vesicle (GV) or presence of the extruded first polar body, respectively (Supplementary Fig. 2e). We also recovered comparable numbers of MII oocytes from MatKO-producing females after super-ovulation relative to controls (Supplementary Fig. 2f).
Characteristic of MII eggs is the presence of metaphase arrest chromosomes alongside barrel-shaped spindles in the secondary oocytes, opposite the extruded first polar body. Immunofluorescence detection of α-tubulin and DNA staining revealed that, whilst approximately three-quarters of MatKO MII oocytes appeared normal, the remaining fourth displayed various metaphase spindle/plate abnormalities (Fig. 2a, b). These included multiple spindle tubulin formations, seemingly anaphase arrested-like spindles, or misalignment of chromosomes along spindles (Fig. 2b). Pole-to-pole distance of the metaphase spindles is usually consistent in each cell type. We measured spindle length-width ratios in MatKO MII oocytes without overt meiotic spindle defects (n = 20/29) and found a significant shortening compared to analyzed controls (n = 29), indicative of disturbed microtubule dynamics and/or its regulators (Fig. 2c, Supplementary Fig. 3a, b). The observed abnormalities in spindle formation and chromosomal misalignment in the MatKO MII eggs prompted us to investigate aneuploidies. A remarkable nearly 70% of MatKO MII oocytes showed some degree of aneuploidy compared to just nearly 20% of control MII oocytes with chromosome number aberrations observed in our analysis (Fig. 2d). Notably, aneuploid MatKO MII oocytes were hyperhaploid (Fig. 2e, Supplementary Fig. 3c, d), in some cases containing twice the normal chromosome complement.
a Quantification of abnormal spindle occurrence in control (n = 30) and MatKO (n = 29) MII oocytes. In mutants, 14% of MII oocytes showed spindles resembling anaphase, 10% had apolar or multipolar spindles and 7% had misaligned chromosome (b) Exemplary MatKO MII oocytes spindles stained for alpha-tubulin (grey) and DNA (blue). c Spindle width/length ratio in apparently normal MatKO (n = 20) and Ctr (n = 29) MII oocytes (****, p = 7*10−8, unpaired, two-tailed, parametric t-test) with exemplary images (stained for DNA and tubulin). d Mutant MII oocytes (n = 22) show significantly higher occurrence of aneuploidies compared to controls (n = 24) (****, p = 2.34*10−9, two-tailed Fisher’s exact test). e Quantification of kinetochores shows predominantly increased genomic content in MatKO vs Ctr oocytes. f Transmission light images of Ctr and MatKO zygotes isolated after natural mating. Spermatozoa in the perivitelline space and/or attached to the zona pellucida of MatKO zygotes indicated with black arrowheads. g Maximal projections of DNA stained Ctr and MatKO zygotes revealing multiple spermatozoa attached to MatKO zygotes (white arrowheads). Dotted, dashed, and solid circles delineate cell borders, pronuclei and polar bodies, respectively. h Quantification of MatKO (n = 57) and Ctr (n = 44) zygotes with overt accumulation of spermatozoa after natural mating. i Cortical granule labelling using FITC-labelled Lens culinaris agglutinin (LCA) in MatKO and Ctr GV or MII oocytes, respectively. j Still images from Supplementary Movie 1 (a) and Supplementary Movie 2 (b) with chromatin labelling (H2B-RFP) revealing multiple, pronuclear-like structures in Prdm10 MatKO zygotes. k Quantification of ‘poly-pronuclei’ phenotype (MatKO n = 38; Ctr n = 65; ****, p < 1*10−15, two-tailed Fisher’s exact test). l Maximal projections zygotes with labelled DNA (DAPI, blue), pronuclei (TRIM28, red) and maternal-derived chromatin (H3K9me3, green). Blue circles indicate paternal pronuclei (H3K9me3-negative), red circles indicate maternal-derived pro-/micronuclei, white circles indicate polar bodies [PB], asterisks indicate abnormally associated spermatozoa, dashed white circles indicate zygotes’ circumference. (all scale bars: 50 μm).
Polyspermy or pronuclear fragmentation?
In mice and humans, polyspermy (the fertilization of an oocyte by multiple sperm) is prevented by a combination of zona pellucida and membrane block26. Consequently, rarely more than one flagellum (if at all) is visibly attached to normal zygotes after isolation (Fig. 2f–h). Conversely, a remarkable 75% of isolated MatKO zygotes were found swarmed by multiple sperm, attached to zona pellucida or perivitelline space (Fig. 2f–h). The zona pellucida (ZP) block occurs after the first sperm entry into the MII oocytes. This triggers exocytosis of secretory granules (referred to as cortical granules [GCs]) from the just-fertilized egg and prevents the attachment to and penetration through the egg coat by additional spermatozoa. CGs line the inner edge of the egg’s membrane to which they are transported along actin fibres in either myosin Va or Rab11a-dependent myosin Vb powered manner, during oocyte growth and maturation27. When released into the perivitelline space their enzymatic cocktail causes impermeabilization of the zona pellucida. The accumulation of sperm around MatKO-derived zygotes suggests either absence of or failure to release these granules upon sperm entry. FITC-labelled lens culinaris agglutinin (LCA) staining28,29,30 revealed MatKO GV and MII oocytes to be completely devoid of cortical granules (Fig. 2i, right panels), while normal GC distribution was normal in controls (Fig. 2i, left panels).
Following fertilization, the haploid sperm chromatin quickly decondenses and forms the paternal pronucleus. Sperm entry triggers meiotic completion with extrusion of the second polar body with the remaining haploid set of chromosomes forming the maternal pronucleus. In contrast to normal zygotes with two distinct pronuclei of paternal and maternal origin respectively, we observed additional pronuclear-like structures of varying sizes in over 66% of MatKO-derived zygotes (n = 38) (Fig. 2j, k). Having established sperm block defects and multiple sperm attached to MatKO-derived zygotes but also meiotic defects in MatKO oocytes, we wanted to explore the parental origin of these additional pronucleus-like structures. We took advantage of H3K9me3 asymmetry in the mouse zygote to discern parental genomes31,32. Only one pronucleus of maternal (H3K9me3-positive) and paternal origin (H3K9me3-negative, respectively) is present in control zygotes (Fig. 2l, upper panels). Notably, despite a disrupted zona pellucida block and superfluous spermatozoa attached to the outside of the MatKO-derived zygotes consistently only one paternally originated (H3K9me3-negative) pronucleus was found. All other pronuclei and pronuclear-like structures were of maternal origin (i.e. H3K9me3-positive, Fig. 2l, lower panels) suggesting disrupted meiotic processes such as e.g. chromosome congression or cytokinesis (polar body extrusion/abscission). This further shows that despite the absence of cortical granules and disruption of the zona pellucida block, the membrane block appears unaffected and efficient in these mutants. All in all, we find Prdm10 MatKO oocytes and embryos display a spectrum of meiotic and other defects culminating in a 2-cell (or earlier) arrest. A combination of spindle and chromosome segregation defects, failed accumulation of cortical vesicles and failure of orderly cytokinesis is reminiscent of and may indicate substantial cytoskeletal disruptions33,34.
PRDM10 activates maternal genes
PRDM10 is an essential transcriptional activator regulating vital genes in mESCs and the preimplantation embryo13. Clinical data confirms this role and further shows functional conservation in humans14. In line with its expression/translation throughout oocyte growth (Fig. 1a–d) PRDM10 is likely to act as a regulator of the maternal transcriptome depositing essential maternal factors in growing oocytes, later required for successful OET. These target genes may further be context-dependent and only partially (or not at all) overlap between maternal and zygotic-regulated genes. Understanding these PRDM10-maternal targets will shed light into the essential processes of maternal-zygotic transition and novel players involved. We generated maternal transcriptomes from MatKO and Ctr GV oocytes (3 replicate samples per genotype, each a pool of ~10 oocytes derived from individual females) to identify deregulated genes. We find a clear segregation of genotypes (Supplementary Fig. 4a, b), with close to 3.3% of genes (924/27179) significantly (FDR ≤ 0.05) and substantially (baseMean≥10, FC≥2) deregulated. Of these 292 (1% of all genes) and 632 (2.3% of all genes) genes were down- or up-regulated respectively (Fig. 3a, Supplementary Data 1). Gene ontology (GO) analysis of significantly (FDR ≤ 0.005) up- and downregulated genes, point amongst others towards ‘DNA damage response’, ‘nuclear division’, ‘cell division’, ‘meiotic cell cycle’, ‘GTP-related processes’ and ‘cytoskeleton organization’ (Fig. 3b [upper panel], Supplementary Fig. 5a and Supplementary Data 3, Supplementary Data 6).
a Volcano plot of RNA-seq data from MatKO and Ctr GV oocytes (grey: FDR [false discovery rate]≤0.05 and baseMean≥10; black: genes FC [fold change]≥2; purple: genes FC ≥ 2 and promoter bound by PRDM10 in ESCs13). Genes of interest are highlighted. b Gene ontology analysis overview of regulated genes (FDR≤0.001) (top) and ‘bound®ulated’ genes (FDR≤0.05) (bottom). c Heatmap representation of PRDM10 binding signal (mESCs)13 of peak centres (+/−1kb) and associated chromatin features of active transcription/DNA accessibility in growing (GO1)(ATAC)36 and fully grown oocytes (FGO)(ATAC36, H3K4me335, H3K36me337). d Proportion of genes significantly (FDR≤0.05) up- or downregulated (FC ≥ 2) amongst PRDM10 bound/unbound fractions. Direct PRDM10 targets are predominantly downregulated. e Heatmap of all deregulated genes (FDR≤0.05, baseMean≥10, FC≥2) ranked from most down- to upregulated with PRDM10 promoter-binding indicated. ‘Bound®ulated’ genes defining GO categories ‘DNA damage response’ (pink) and ‘bacterial invasion of epithelial cells’ (blue) are highlighted. f PRDM10-dependent relative expression, RNA-seq track and promoter-binding peaks of formerly unknown target Sept11. Expression in 2 Ctr and 3 MatKO pools of oocytes is shown, respectively. Three overlayed tracks for PRDM10 ChIP in iKO (inducible knock out) and control mESCs at the Sept11 promoter are shown, respectively (GSE13502213). Motif location in respect to peak is indicated (red bar). g PRDM10 binding motif13 (top) and matching peak-underlying sequence in the Sept11 promoter in mouse with respective fully conserved motifs in humans.
PRDM10 was shown to predominantly associated with active gene promoters in ESCs13. Likewise, in growing and fully grown oocytes PRDM10 binding sites13 are predominantly associated with active (H3K4me3)35, accessible (ATAC-seq)36 gene promoter regions and transcriptional elongation (H3K36me3)37 (Fig. 3c). We integrated the PRDM10 binding sites with our maternal RNA-seq data sets to identify direct regulatory relationships. Of 540 genes with promoter-overlapping PRDM10 peaks 98 were found to be significantly and substantially deregulated (‘bound®ulated’; FDR≤0.05, baseMean≥10, FC≥2) in our RNA-seq data. The remaining putatively bound genes were either not expressed (80, baseMean<10) or just mildly/not changed in their expression (362, base mean≥10, FC < 2) (Fig. 3a, d, e; Supplementary Data 2). Underscoring the function of PRDM10 as a transcriptional activator, 86% (84/98) of ‘bound®ulated’ genes showed transcriptional down-regulation, including previously described targets, Eif3b13 and Flcn14 (Fig. 3a, Supplementary Fig. 4c, d) and only 16% (14/98) showed up-regulation (Fig. 3a, d, e). GO categories linked to ‘bound®ulated’ genes (FDR≤0.05, no FC cut off) included in addition to ‘DNA damage’ also ‘Bacterial invasion of epithelial cells’ (Fig. 3b [lower panel], Supplementary Fig. 5b, Supplementary Data 4, Supplementary Data 7). Secondary targets (unbound, >2-fold deregulated) showed an unexpected skewing towards upregulation of genes (618 [75%] up- vs. 208 [25%] down-regulated, respectively). GO analysis suggests unbound, up-regulated genes (Supplementary Data 5) to be predominantly involved in ‘cell homoeostasis’ and regulation of supramolecular fibre organization (Supplementary Fig. 5c, Supplementary Data 8).
Unimpaired zygotic genome activation (ZGA) in Prdm10 maternal KO embryos
In mouse, ZGA depends on maternal transcription factors such as OBOX family members, maternal loss of which results in 2-4 cell arrest6. Though PRDM10 is also found translated at stages of OET (Fig. 1c, Supplementary Fig. 1d), the PRDM10-binding motif (in contrast to the OBOX motif, is not significantly enriched at sites of open chromatin (ATAC-seq) of late 2-cell stage embryos, nor enriched in ZGA gene promoters (Supplementary Fig. 6a, b)6. None of the 633 genes associated with promoter-associated PRDM10 peaks in ESCs13 is minor ZGA genes (0/65)18 and only few (2.8%) are major ZGA genes18 (31/1107) (Supplementary Fig. 6c, d). Eight of the latter showed deregulation in Prdm10 matKO oocytes (four up- and down-regulated, respectively), none were deregulated in 8-cell stage zygotic KO embryos13 and only one showed reduced expression in KO ESCs13 (Supplementary Fig. 6e). Finally, three representative ZGA genes (Eif1a, Zfp352 and Zscan4d) were shown to be transcriptionally activated in Prdm10 MatKO 2-cell stage embryos by qPCR (Supplementary Fig. 6f). In conclusion, while maternal PRDM10 may be required to activate its target genes in early embryos after ZGA, it appears to be dispensable for triggering ZGA in the first place.
Sept11 is a PRDM10 target
Our integrated RNA-seq analysis reveals multiple ‘bound®ulated’ PRDM10 target genes potentially contributing to the observed 2-cell arrest. GO analyses point strongly either towards ‘DNA damage response’ (most prominent target Ube2a (Fig. 3e) or towards a less defined, category group of genes relating likely cytoskeletal processes (bacterial invasion, cell division, GTPases, etc.) (Fig. 3b, e and Supplementary Fig. 5) matching the observed pleiotropic defects culminating in a complete 2-cell arrest. Prominent amongst the severely deregulated targets in this category is Sept11 (Fig. 3e, f). Sept11 is highly expressed in wildtype oocytes yet reduced to almost undetectable levels in Prdm10 MatKO oocytes (Fig. 3a, f). PRDM10 binds the Sept11 promoter containing a strong binding motif13, which is furthermore highly conserved from mouse to human (Fig. 3f, g), as also observed in the known targets Flcn and Eif3b (Supplementary Fig. 4e). Septins are polymerizing GTP-binding proteins and considered ‘the forth element of the cytoskeleton’ 15. They are involved in a multitude of cellular processes including mitotic- (e.g. cytokinesis)38,39,40,41,42,43, as well as meiotic functions in mammals44,45,46. Perturbation of Septins causes pleiotropic defects in mitotic spindle organization, chromosome alignment and segregation, cleavage furrow formation and midbody abscission in mammals39,43,47,48,49. SEPTIN11 itself is linked to biological processes such as ‘cell division’ (GO:0051301), ‘cytokinesis’ (GO:0000910) and ‘chromosome segregation’ (GO:0007059) and is associated with cellular components such as the ‘cytoskeleton’ (GO:0005856), the ‘Septin complex’ (GO:0031105) and the ‘cleavage furrow’ (GO:0032154). As a member of the Septin family of GTP-binding proteins, SEPTIN11 has GO annotations for ‘GTP binding’ (GO:0005525) and ‘GTPase activity’ (GO:0003924), linked to its function in filament formation within the cytoskeleton. An oocyte-specific function for Sept11 has not been described to date.
Septin-network gene expression analysis in oocytes and early embryos
The Septin family has thirteen paralogs, classified into four groups: SEPT3 (SEPTIN3/9/12), SEPT2 (SEPTIN1/2/4/5), SEPT7 (sole member is SEPTIN7) and SEPT6 (SEPTIN6/8/10/11/14) (Fig. 4a). Septins assemble into heteromeric complexes between either 3 (SEPT2/SEPT7/SEPT6) or 4 (SEPT2/SEPT7/SEPT6/SEPT3) of the groups, respectively50,51,52,53,54,55. These complexes assemble further into highly organized filaments, in turn forming more complex structures (rings, gauzes, bundles) still50. Removal of any essential family member disrupts these structures, on the other hand, Septins have been shown experimentally to be largely redundant within their respective homology subgroups55,56.
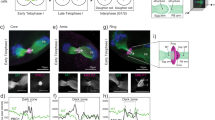
a Heat maps showing mRNA transcription of Septin family genes during oogenesis20 (RPKM), OET and preimplantation development19 (FPKM). b Heat map showing RPFs (ribosome protected fragments) of Septins in oocytes and preimplantation embryos19 (FPKM). c Heat map showing protein intensities of Septin family members in oocytes and preimplantation embryos by LC-MS/MS57. Septin members are clustered in functional homology groups. d Predicted Septin hexamer complex based on transcription and proteomic data in Ctr (left) and Prdm10 MatKO oocytes (right). e Expression heatmap from RNA-seq data from mouse Ctr/MatKO oocytes, Ctr/ZygKO 8-cell embryos and Ctr/iKO mESCs, respectively. Septin members are clustered in groups based on structural and functional similarity (column 1). PRDM10 promoter-binding as defined by ChIP-seq in mESCs is indicated (column 2). Only Sept11 is bound by PRDM10 and regulated by PRDM10 (red box/asterisk) in oocytes and mESCs. f Schematic illustration of oocyte activation and staining protocol to detect maternal Septin complex at polar body abscission site. g Immunofluorescence detection of core Septin members 2, 7 and 11 at the polar body abscission site in parthenogenically activated Ctr/MatKO MII oocytes, respectively (DNA blue; SEPTIN2/7/11 green; TUBULIN red). In Ctr parthenotes, SEPTIN2 (n = 22), 7 (n = 13) and 11 (n = 12) were all localized at the cytokinetic ring (white arrowheads) where abscission of the polar body will occur. In MatKO parthenotes only SEPTIN2 (n = 17) was detected at the cytokinetic ring (white arrowheads); SEPTIN11 (n = 12) and 7 (n = 11) were absent (scale bar 50 μm).
We mined RNA-seq, translatomic and proteomic data19,20,57 for Septin family member expression in oocytes and preimplantation embryos to understand the impact of Sept11 loss in Prdm10 MatKO embryos (Fig. 4a–c). Notably, maternal expression of Septins is restricted to unique or limited members per family, while at later stages (e.g. 8-cell stage or ICM/blastocyst or somatic cells55,56 a broader expression/translation can be observed (Fig. 4c). Consistent with its requirement in oocytes45, Sept2 is the only member of its class (SEPT2) expressed throughout oogenesis and OET. Sept7, also required for orderly meiosis44, is unique in its class and fittingly found in both maternal germline and soma. On maternal mRNA levels Sept11 and 14 are the only detectable members of their homology group (SEPT6). However, although also detectable in the translated RNA pool (RPF, Fig. 4b), Septin14 protein was not detectable by LC-MS/MS (Fig. 4c). In short, these data show that at time of meiotic completion and OET, the predominant (if not only) Septin core complex available to the cell consists of Septin2/7/11 (Fig. 4d [left]).
Amongst all Septin genes, Sept11 is the only ‘bound®ulated’ by PRDM10 and consequently reduced to near absence in Prdm10 MatKO oocytes (Fig. 4e, Supplementary Fig. 7a). Other maternal Septins (i.e. Sept2 and 7) are not affected. Notably, Sept11 levels are endogenously low-to-absent at the 8-cell stage, however, Sept8 and Sept10 are the expressed members of the SEPT6 class, instead (Fig. 4e). Deletion of Prdm10 has no effect on Septin expression at this stage. In mESCs, where Sept11 is endogenously expressed and repressed (though to a lesser degree) upon Prdm10 deletion, redundant Sept8 and Sept10 are also expressed (Fig. 4e, Supplementary Fig. 7b).
In essence, we reveal a highly dynamic Septin complex composition post-implantation and in mESCs, yet equally a high dependency of the maternally provided Septin complex on Sept11 (Fig. 4d [left]), which in turn is dependent on PRDM10 transcriptional activation in the oocyte.
We additionally mined human oocyte and preimplantation embryo expression and translation data7,17 (Supplementary Fig. 7c, d). As in mouse, SEPT2 is the unique expressed member of its class in human oocytes and early embryos. In contrast, a broader expression of SEPT6 class members (SEPT8, SEPT10 and SEPT11) is observed in oocytes and embryos in humans, better reflecting the findings in mESCs than mouse oocytes (Supplementary Fig. 7c, d).
SEPTIN11 is required for Septin complex assembly in oocytes
To shed light into the composition of the maternal Septin complex we attempted to identify Septin proteins in oocytes and zygotes. This worked best at the abscission point of the polar body of meiosis 2 following parthenogenic activation (Fig. 4h). Here we detected a distinct ring structure composed of SEPTIN2, SEPTIN7 and SEPTIN11 by immunofluorescence (Fig. 4i [top panels] and Supplementary Fig. 8a).
We next tested if this ring would also form in the maternal Prdm10 knock-out oocytes, despite the significant reduction of Sept11 transcript. This would further hint towards possible SEPTIN11/14 redundancy (Fig. 4i, Supplementary Fig. 8a). SEPTIN2, remains recruited to a ring-structure at the abscission site in mutants, comparable to controls. Conversely, SEPTIN11 was consistently undetectable in the mutant, in line with the near-complete absence of maternal transcripts shown by RNA-seq. Most remarkably, SEPTIN7 was also no longer detectable at the abscission site, even though its transcript expression is not affected by the PRDM10 deletion (Fig. 4i, Supplementary Fig. 8a). It thus appears that in the oocyte SEPTIN11 is essential to form a proper Septin-complex structure and mediates the recruitment of SEPTIN7 to the abscission point of the polar body. SEPTIN14 is either not able and/or insufficient to rescue this function.
Rescue of PRDM10-dependent Septin complex assembly
Recruitment of SEPTIN7 to the site of polar body abscission appears to be SEPTIN11 dependent, whose expression in turn is dependent on PRDM10. Consequently, we attempted to rescue this recruitment in Prdm10 MatKO oocytes by reconstituting SEPTIN11 expression and thus confirming this link. To this end we cloned Sept11 cDNA from oocytes to produce a Sept11-GFP fusion construct. cRNA was produced by in vitro transcription and subsequently used for injection. We first injected GV oocytes to allow cRNA translation during overnight maturation to MII oocytes. MII oocytes were then parthenogenically activated and fixed upon polar body extrusion for immunofluorescent analysis for SEPTIN2/7/11 (Fig. 5a). Double staining for GFP (exogenous SEPTIN11) and SEPTIN7 in injected Ctr embryos confirmed their co-localization, the undisturbed formation of the SEPTIN ring structure at the abscission point and thus functionality of the SEPTIN11/GFP fusion protein (Fig. 5b [top panels]). Remarkably, the reintroduction of SEPTIN11 in the Prdm10 MatKO oocytes reinstated SEPTIN7 recruitment and therefore at least rescues Septin-complex formation at the abscission site (Fig. 5b). This confirms essentiality of SEPTIN11 for the maternal Septin complex, at least at the site of polar body abscission in the oocyte.
a Schematic of Sept11-eGFP cRNA injection rescue strategy for SEPTIN7 recruitment: GV oocytes are microinjected, matured to MII oocytes, parthenogenically activated and fixed and stained upon polar body extrusion. b Sept11-eGFP Ctr and MatKO injected and mock injected parthenotes were stained for eGFP (green/arrowhead), SEPTIN7 (red/arrowhead) and DNA (blue), respectively (scale bar 50 μm). Rightmost column shows magnification of the polar body abscission site (scale bar 25 μm). c Schematic diagram for 2-cell block rescue strategy: Zygotes were injected with Sept11-eGFP-cRNA and cultured beyond the 2-cell stage, up to blastocyst formation. d Quantification of in vitro development post-injection: 80% of wildtype embryos developed beyond the 2-cell stage (n = 52/66). Full 2-cell block was observed in mock-injected Prdm10 MatKO embryos (n = 0/12, rescued). Sept11-eGFP-cRNA injection enabled 22% of MatKO embryos to overcome the 2-cell block (n = 11/51, rescued). e Percentual distribution of developmental stage arrests of rescued MatKO embryos overcoming the 2-cell block. f Brightfield images of representative rescue embryos arrested at the 3-cell, 8-cell (2x) and blastocyst stage, respectively (scale bar 50 μm).
The pleiotropic nature of the Prdm10 MatKO phenotype is likely caused by the deregulation of the broad PRDM10-regulated transcriptional programme and not limited to SEPTIN11 function alone. Nonetheless, given the variable severity of phenotypes observed in the Prdm10 MatKO zygotes, we reasoned that we may overcome the observed 2-cell block, if only in a subset of embryos. To this end we injected zygotes with Sept11-GFP cRNA and cultured them in vitro to score successful cleavage divisions beyond the 2-cell stage (Fig. 5d). Unsurprisingly, given the severe and pleiotropic phenotypic aberrations observed in many embryos, we still observe most embryos (>60%) arresting at or before first cleavage (Fig. 5d). Unlike the complete 2-cell block or even earlier arrest observed in un-injected embryos however, SEPTIN11-GFP expression allowed over 20% of embryos to undergo second or third cleavage division, and even rare blastocyst formation (Fig. 5d, f).
Discussion
Oocyte-to-embryo transition (OET) is the process through which fully grown, quiescent oocytes undergo maturation, fertilization, and transform into a developmentally active, mitotically dividing embryo. This transition depends entirely on maternal gene products deposited in the oocyte during its growth. However, our understanding of the transcription factor networks that regulate this critical accumulation of gene products remains incomplete. Here we have characterized the role of a PR/SET domain-containing (PRDM) protein (i.e. PRDM10) in contributing to this essential developmental step. While other family members such as BLIMP1/PRDM1 and PRDM14 have been characterized as crucial transcription factors in mammalian germ cell specification11,12, none has been linked specifically to oocyte growth, maturation and OET. While PRDM proteins share an overall structure comprised of a PR domain and several C-terminal Zn-fingers10, they have non-overlapping functions, due to their unique DNA-binding patterns, leading to the regulation of very specific downstream transcriptional programmes9,10. It is therefore unsurprising that PRDM10 plays a distinct, non-redundant role in regulating the transcription of target genes, among others essential for maintaining cell homoeostasis and organizing the cytoskeletal network in oocytes and early embryos.
Mechanistically, PRDM10 functions as a transcriptional activator by binding to highly conserved DNA motifs and recruiting transcriptional coactivators via its glutamine (Q)-rich C-terminal domain13, while other family members such as PRDM9 directly methylate histones, or BLIMP1/PRDM1 recruits transcriptional co-repressor to silence transcription58,59,60.
A transcription-independent role for PRDM10 has not been described to date. However, an increase of the ribosome-associated Prdm10 mRNA in translatomic data at transcriptionally inert stages (MII, zygote and 2-cell stage) (Fig. 1b, c and Supplementary Fig. 2b, c) may hint towards potential transcription-independent roles in OET, or merely reflect the maintenance/accumulation of PRDM10 protein required for resumption of its transcriptional activity after ZGA, the last step of successful OET. Interestingly ZGA itself is PRDM10 independent (Supplementary Fig. 6), unlike the requirement of other maternal transcription factors such as OBOX in mice6 or TPRX in humans7.
The role of BLIMP1/PRDM1 and PRDM14 in germ cell specification is evolutionary conserved61,62. In support of the evolutionarily conserved function of PRDM10, heterozygous missense variants in its zinc-finger domain have been recently identified in a family presenting with Birt-Hogg-Dube syndrome (BHD)14. BHD is linked to epigenetic silencing or loss-of-function variants in the FLCN gene, a PRDM10-target we previously described in mESCs13 and now confirmed in oocytes. Furthermore, are PRDM10 binding sites in oocyte target genes highly conserved from mouse to humans. These findings highlight the importance of PRDM10 as transcriptional activator and the evolutionary and functional conservation of PRDM10 in development and disease.
We have further uncovered and functionally characterized a previously unknown PRDM10 target, Sept11, and in the process shed light into the composition and requirement of the maternal Septin complex. Sept11 expression is fully dependent on PRDM10 in oocytes, and it is essential for the Septin complex formation at the polar body abscission site during oocyte maturation. Septins fulfil numerous functions in the cell and it is likely that other observed defects (such as Spindle defects, chromosome segregation defects, absence of vesicles38,39,44,45,63,64 etc.) in Prdm10 MatKOs are also Septin related. Remarkably, reintroduction of SEPTIN11 in Prdm10 MatKO oocytes/zygotes rescued Septin complex assembly and partially alleviated the developmental block caused by PRDM10 absence, respectively. The aforementioned evolutionary conservation of PRDM10 function also extends to SEPT11.
SEPT11 is highly expressed in human oocytes and likely regulated by PRDM10 given the full conservation of the PRDM10 motif at the promoter region, similar to other known targets such as Eif3b and Flcn (Fig. 3g and Supplementary Fig. 4e).
Analysis of maternal mouse and human transcriptomes, translatomes and proteomes revealed the presence of few Septin family members in oocytes and early embryos. Sept2 and Sept7 are the sole members expressed in their respective ‘Septin family member’ group and have previously been associated with meiotic phenotypes44,45. Here we add, through the genetic disruption of PRDM10 and consequential downregulation of Sept11, a third member to this list. While homozygous Sept11 deletion is embryonic lethal in mice at E13.565, we predict that maternal deletion would lead to a preimplantation lethality, given the demonstrated inability of SEPTIN14 to compensate for this loss of function. While redundancy of SEPTIN11 with SEPTIN6 (not maternally expressed) has been suggested66,67, and SEPTIN11 was shown to be able to form a functional trimeric complex with SEPTIN2 and 768, its functional redundancy with SEPTIN14 was not demonstrated. Whether this is because maternal SEPTIN14 protein was not detectable by LC-MS/MS despite the presence of maternal Sept14 mRNA in RNA-seq and Ribo-seq data or because SEPTIN14 cannot rescue SEPTIN11 function requires further investigation.
The rescue of the Prdm10 MatKO phenotype by SEPTIN11 is far from efficient. While the maternal Septin complex can be restored and in a subset of embryos cleavage capacity can be extended, most embryos arrest. Other facets of the pleiotropic phenotype are also not alleviated. We couldn’t, for example, restore cortical granules in MII oocytes obtained from Sept11-cRNA injected GV oocytes. Possibly the restoration of Septin complexes at the GV oocyte is too late for cortical granule establishment, normally occurring throughout oocyte growth. On the other hand, the pleiotropic nature of the Prdm10 MatKO phenotype could be attributed to additional targets we find substantially deregulated. FLCN, for instance, is a Rab11a-interacting protein and controls vesicle transport69. Cortical granules utilize various modes of transport, including hitching a ride on Rab11a vesicles, to reach the oocyte membrane27. Other targets, such as Ube2a are involved in DNA repair/damage response and themselves have severe maternal phenotypes70, yet apart from a 2-cell arrest, Prdm10 MatKO-like (meiotic/mitotic/cytokinesis) defects were not reported in maternal Ube2a knock-out oocytes.
Undoubtedly, disrupting transcriptional programmes in growing oocytes causes phenotypes that will require further dissection and target gene-specific analyses in the future. Yet, our data underscore an essential role for maternal PRDM10 in regulating the meiotic Septin complex and multiple other targets previously not linked to OET. Whether the disruption of these regulatory axes, as shown in BHD patients, could be of clinical relevance, contributing to human aneuploidies, infertilities and pregnancy abortions remains to be investigated.
Methods
Ethics statement and animal model
Female Prdm10f/f mice were mated to Zp3Cre[C57BL/6-Tg(Zp3-Cre)93Knw]25 males. The F1 male offspring with Prdm10+/f;Zp3Cre/ø) genotype were mated with Prdm10f/f females to generate an oocyte-specific Prdm10 knock-out (MatKO, Prdm10f/f Zp3Cre/ø) and control littermates (Ctr, Prdm10f/f Zp3ø/ø). Mice were house in pathogen-free conditions under 12 h light/dark cycle with ad libitum food and water (Biological Resources Centre (BRC), A*STAR, Singapore). All procedures conducted followed the Institutional Animal Care and Use Committee (IACUC) protocols #181393 and #221679. Genotyping of mice was performed as described13; primer sequences are listed in Supplementary Data 9.
Gamete and embryo isolation
Germinal vesicle (GV) oocytes and Metaphase II (MII) eggs were isolated from 4–8 weeks-old mice. To maintain GV arrest for bench handling and microinjection, GV oocytes were cultured in M2 medium (Merck) containing 5μM of milrinone (Sigma), a phosphodiesterase-3 inhibitor71. For egg and zygote collections, mice were super-ovulated with Pregnant Mare Serum Gonadotrophin (PMSG, Prospec Protein Specialists, 5-10IU), followed by human chorionic gonadotrophin (hCG, Sigma, 5-10IU) 48 h later. MII eggs were isolated 12–16 h after hCG injection. Zygotes are isolated from the ampulla of dissected oviducts either from naturally mated females after the presence of vaginal plug is noted the following morning or 19 h post-hCG. Both procedures are performed in M2 medium containing 0.3 mg/ml hyaluronidase (Sigma) to remove attached cumulus cells. 2-cell, 8-cell and blastocyst staged embryos were isolated ~24 h, 48 h and 72 h post-observation of vaginal plugs in naturally mated females. Embryos were flushed from the dissected oviducts and uterine horns in M2 medium. KSOM-AA (Merck) was used for all in vitro cultures of embryos that were incubated at 37 °C with 15% CO2 immersed by mineral oil (Millipore)72.
RNA extraction and qRT-PCR
Total RNA from pooled or individual gametes/embryos were isolated using PicoPure RNA Isolation Kit (Applied Biosystems). All eluted RNA was used for reversed transcription carried out using the High-Capacity cDNA Reverse Transcription kit and random hexamer primers (Applied Biosystems). qPCR assays were performed as previously described16. Primer sequences are listed in Supplementary Data 9.
Immunofluorescence (IF) microscopy of oocytes and embryos
Oocytes or embryos were fixed in 4% paraformaldehyde for 30 min followed by permeabilization with 0.1% Triton X-100 for 30 min at room temperature. Samples were blocked in 1% FBS/0.1% Triton X-100 and incubated overnight at 4 °C using the following primary antibodies and dilutions: α-tubulin (mouse, 1:5000, Sigma, T6199), CREST (human, 1:100, ACA, #15-234), TRIM28 (mouse, 1:100, Abcam, ab22553), H3K9me3 (rabbit, 1:100, Abcam, ab8899), SEPTIN2 (rabbit, 1:25, Proteintech, 11397-1-AP), SEPTIN7 (rabbit, 1:25, Proteintech, 13818-1-AP), SEPTIN11 (rabbit, 1:50, Abcam, ab183537), GFP (chicken, 1:1000, Abcam, ab13970). Samples were washed 3 times for 10 min in blocking buffer and incubated with Alexa Fluor-conjugated secondary antibodies (Alexa-405, Alexa-488, Alexa-568, Alexa-594 and Alexa-647; 1:500, Invitrogen) at room temperature for 1 h. Nuclei were counterstained using DAPI (Invitrogen, 12333553). FITC-labelled Lens culinaris agglutinin (LCA; 1:500, Vector, FL-1041) was used to stain cortical granules. Fixed and blocked specimens were incubated overnight at 4 °C and imaged immediately after 3 consecutive washes at room temperature. Images were acquired on either an Olympus Fluorview 1000 (60x oil immersion objective) or Zeiss LSM800 (63x oil immersion objective) confocal laser-scanning microscope and analyzed in Image J (FIJI, NIH, Bethesda).
Statistical analysis
Statistical analyses were carried out on GraphPad Prism version 9 (GraphPad Software, CA). Appropriate statistical tests were applied and are indicated with p-values and significances in the respective figure legends. Reproducibility of experiments was confirmed by at least three independent experiments and significant n numbers detailed in figures, graphs and figure legends.
Gene expression and RNA-seq analysis
For the maternal expression analysis of PRDM and Septin gene family members we used published RNA seq data sets for mouse and human GV oocyte. For the human samples we obtained normalized count files from NCBI GEO17 (GSE158802), randomly selected 14 GV oocytes (identifiers indicated in Supplementary Data 10) and curated for PRDM (MECOM was changed to PRDM3) or SEPTIN genes, respectively. For mouse expression analysis we performed featureCounts73 (Galaxy Version 2.0.3) with published NCBI37/mm9 aligned bam files16 (GSE126687) against the built-in gene annotation file73 and generated normalized count files using DeSeq274 (Galaxy Version 2.11.40.8).
For RNA-seq of Prdm10 MatKO and Ctr oocytes RNA from ~10 pooled mouse oocytes per replicate (isolated from individual females, respectively) was extracted with the Arcturus PicoPure RNA Isolation Kit (Thermo Fisher Scientific). A total of six libraries were prepared, three from MatKO oocytes and three from Ctrs, respectively. cDNA libraries were generated from total RNA using the SMARTer Ultra Low Input RNA Kit for sequencing (Clontech), followed by sequencing library preparation with the NexteraXT DNA Sample Preparation Kit (Illumina) according to manufacturer’s instructions. One Ctr library was identified as outlier in cDNA and sequencing library concentrations. Post-sequencing this library showed increased duplicate reads and low sequencing quality and was omitted from downstream analysis.
RNA-seq analysis was performed on the Galaxy platform75. In brief, sequence reads were mapped to the mouse genome build GRCm38.p6 (mm10) using STAR (Galaxy Version 2.7.11a), and gene-level transcript abundances were quantified by featureCounts73 (Galaxy Version 2.0.3) against the built-in gene annotation file (NCBI RefSeq)73. DeSeq274 (Galaxy Version 2.11.40.8) was used for differential expression analysis; genes were considered to be significantly differentially expressed at FDR≤0.05, and a minimum expression threshold (baseMean ≥ 10) was applied to exclude low-abundance genes. Plots and heatmaps were generated using GraphPad Prism version 10 (GraphPad Software, CA). GO enrichment analysis was performed using Metascape76 (http://metascape.org). Aligned reads with splice junctions were visualized on the IGV browser77.
Enrichment analysis
PRDM10 binding and chromatin state association were performed in easeq78. Data was clustered according to transcriptional state based on H3K36me3 (kmeans-clustering). Published datasets used: GSE13427936 for ATAC in GO and FGO, GSE11262237 for H3K36me3 in FGO, GSE7278435 for H3K4me3 in FGO, GSE13502213 for PRDM10 binding in mESCs.
Plasmid construction, cRNA synthesis, microinjection and live imaging
The coding sequence of Sept11 (Ensembl transcript ID: NM_001009818.2) was inserted by restriction cloning into pCS2+ expression plasmid containing EGFP. H2B-RFP and membrane-targeted-GFP in PCS2+ were used to label nuclei and membranes, respectively. Endotoxin-free plasmids were prepared using the Nucleobond Xtra Midi EF Kit (Macherey-Nagel). All constructs were verified by Sanger sequencing. In vitro, transcription was performed using the mMessage mMACHINETM SP6 kit (Invitrogen) according to manufacturer’s instructions from linearized plasmid as template. RNA was purified using the RNAeasy kit (Qiagen) according to manufacturer’s instructions. Embryos were injected with RNA diluted in injection buffer with a FemtoJet (Eppendorf). Embryos were cultured in LabTek chambers (Nunc) at 37 °C with 5% CO2 in a humidified environmental chamber adapted for the LSM880 microscope system (Carl Zeiss). Live imaging of embryos was performed at 6 min intervals. Image analysis was performed using ImageJ (FIJI, NIH, Bethesda) and Imaris software (Bitplane AG).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
RNA-seq data sets generated in this study have been deposited in NCBI’S Gene Expression Omnibus (GEO) under accession code GSE283055 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE283055]. Source data are shared via GEO (GSE283055) or provided within this paper in Supplementary Data. Published data sets used for enrichment analysis are: GSE13427936 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE134279] for ATAC in GO and FGO, GSE11262237 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE112622] for H3K36me3 in FGO, GSE7278435 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE72784] for H3K4me3 in FGO, GSE13502213 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE135022] for PRDM10 binding in mESCs. Source data are provided with this paper.
Code availability
This paper did not generate or use any custom code or software. All code and software packages utilized in the study are referenced in the Methods section and the Reporting Summary. All information is available from the corresponding authors upon request.
References
Schultz, R. M., Stein, P. & Svoboda, P. The oocyte-to-embryo transition in mouse: past, present, and future. Biol. Reprod. 99, 160–174 (2018).
Clift, D. & Schuh, M. Restarting life: fertilization and the transition from meiosis to mitosis. Nat. Rev. Mol. Cell Biol. 14, 549–562 (2013).
Mitchell, L. E. Maternal effect genes: Update and review of evidence for a link with birth defects. HGG Adv. 3, 100067 (2022).
Condic, M. L. The role of maternal-effect genes in mammalian development: are mammalian embryos really an exception? Stem Cell Rev. Rep. 12, 276–284 (2016).
Tora, L. & Vincent, S. D. What defines the maternal transcriptome? Biochem Soc. Trans. 49, 2051–2062 (2021).
Ji, S. et al. OBOX regulates mouse zygotic genome activation and early development. Nature 620, 1047–1053 (2023).
Zou, Z. et al. Translatome and transcriptome co-profiling reveals a role of TPRXs in human zygotic genome activation. Science 378, abo7923 (2022).
Messerschmidt, D. M. et al. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science 335, 1499–1502 (2012).
Mzoughi, S. et al. PRDM15 safeguards naive pluripotency by transcriptionally regulating WNT and MAPK-ERK signaling. Nat. Genet 49, 1354–1363 (2017).
Di Tullio, F., Schwarz, M., Zorgati, H., Mzoughi, S. & Guccione, E. The duality of PRDM proteins: epigenetic and structural perspectives. FEBS J. 289, 1256–1275 (2022).
Yamaji, M. et al. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat. Genet 40, 1016–1022 (2008).
Ohinata, Y. et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436, 207–213 (2005).
Han, B. Y. et al. Global translation during early development depends on the essential transcription factor PRDM10. Nat. Commun. 11, 3603 (2020).
van de Beek, I. et al. PRDM10 directs FLCN expression in a novel disorder overlapping with Birt-Hogg-Dube syndrome and familial lipomatosis. Hum. Mol. Genet 32, 1223–1235 (2023).
Mostowy, S. & Cossart, P. Septins: the fourth component of the cytoskeleton. Nat. Rev. Mol. Cell Biol. 13, 183–194 (2012).
Seah, M. K. Y. et al. The KRAB-zinc-finger protein ZFP708 mediates epigenetic repression at RMER19B retrotransposons. Development 146, https://doi.org/10.1242/dev.170266 (2019).
Llonch, S. et al. Single human oocyte transcriptome analysis reveals distinct maturation stage-dependent pathways impacted by age. Aging Cell 20, e13360 (2021).
Zhang, C., Wang, M., Li, Y. & Zhang, Y. Profiling and functional characterization of maternal mRNA translation during mouse maternal-to-zygotic transition. Sci. Adv. 8, eabj3967 (2022).
Xiong, Z. et al. Ultrasensitive Ribo-seq reveals translational landscapes during mammalian oocyte-to-embryo transition and pre-implantation development. Nat. Cell Biol. 24, 968–980 (2022).
Veselovska, L. et al. Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape. Genome Biol. 16, 209 (2015).
Di Zazzo, E., Porcile, C., Bartollino, S. & Moncharmont, B. Critical function of PRDM2 in the neoplastic growth of testicular germ cell tumors. Biology 5, https://doi.org/10.3390/biology5040054 (2016).
Bogani, D. et al. The PR/SET domain zinc finger protein Prdm4 regulates gene expression in embryonic stem cells but plays a nonessential role in the developing mouse embryo. Mol. Cell Biol. 33, 3936–3950 (2013).
Steele-Perkins, G. et al. Tumor formation and inactivation of RIZ1, an Rb-binding member of a nuclear protein-methyltransferase superfamily. Genes Dev. 15, 2250–2262 (2001).
Nishimura, H. et al. Maternal epigenetic factors in embryonic and postnatal development. Genes Cells 28, 422–432 (2023).
de Vries, W. N. et al. Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis 26, 110–112 (2000).
Evans, J. P. Preventing polyspermy in mammalian eggs-Contributions of the membrane block and other mechanisms. Mol. Reprod. Dev. 87, 341–349 (2020).
Cheeseman, L. P., Boulanger, J., Bond, L. M. & Schuh, M. Two pathways regulate cortical granule translocation to prevent polyspermy in mouse oocytes. Nat. Commun. 7, 13726 (2016).
Cherr, G. N., Drobnis, E. Z. & Katz, D. F. Localization of cortical granule constituents before and after exocytosis in the hamster egg. J. Exp. Zool. 246, 81–93 (1988).
Ducibella, T., Anderson, E., Albertini, D. F., Aalberg, J. & Rangarajan, S. Quantitative studies of changes in cortical granule number and distribution in the mouse oocyte during meiotic maturation. Dev. Biol. 130, 184–197 (1988).
Ducibella, T., Rangarajan, S. & Anderson, E. The development of mouse oocyte cortical reaction competence is accompanied by major changes in cortical vesicles and not cortical granule depth. Dev. Biol. 130, 789–792 (1988).
Arney, K. L., Bao, S., Bannister, A. J., Kouzarides, T. & Surani, M. A. Histone methylation defines epigenetic asymmetry in the mouse zygote. Int J. Dev. Biol. 46, 317–320 (2002).
Burton, A. & Torres-Padilla, M. E. Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat. Rev. Mol. Cell Biol. 15, 723–734 (2014).
Sun, Q. Y. & Schatten, H. Regulation of dynamic events by microfilaments during oocyte maturation and fertilization. Reproduction 131, 193–205 (2006).
Dunkley, S., Scheffler, K. & Mogessie, B. Cytoskeletal form and function in mammalian oocytes and zygotes. Curr. Opin. Cell Biol. 75, 102073 (2022).
Dahl, J. A. et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537, 548–552 (2016).
Zhang, C. et al. The chromatin remodeler Snf2h is essential for oocyte meiotic cell cycle progression. Genes Dev. 34, 166–178 (2020).
Brind’Amour, J. et al. LTR retrotransposons transcribed in oocytes drive species-specific and heritable changes in DNA methylation. Nat. Commun. 9, 3331 (2018).
Hall, P. A. & Russell, S. E. The pathobiology of the septin gene family. J. Pathol. 204, 489–505 (2004).
Spiliotis, E. T., Kinoshita, M. & Nelson, W. J. A mitotic septin scaffold required for Mammalian chromosome congression and segregation. Science 307, 1781–1785 (2005).
Hall, P. A., Jung, K., Hillan, K. J. & Russell, S. E. Expression profiling the human septin gene family. J. Pathol. 206, 269–278 (2005).
Fededa, J. P. & Gerlich, D. W. Molecular control of animal cell cytokinesis. Nat. Cell Biol. 14, 440–447 (2012).
Green, R. A., Paluch, E. & Oegema, K. Cytokinesis in animal cells. Annu Rev. Cell Dev. Biol. 28, 29–58 (2012).
Menon, M. B. et al. Genetic deletion of SEPT7 reveals a cell type-specific role of septins in microtubule destabilization for the completion of cytokinesis. PLoS Genet 10, e1004558 (2014).
Li, S. et al. Septin 7 is required for orderly meiosis in mouse oocytes. Cell Cycle 11, 3211–3218 (2012).
Zhu, J. L. et al. Septin2 is modified by SUMOylation and required for chromosome congression in mouse oocytes. Cell Cycle 9, 1607–1616 (2010).
Sharif, B., Fadero, T. & Maddox, A. S. Anillin localization suggests distinct mechanisms of division plane specification in mouse oogenic meiosis I and II. Gene Expr. Patterns 17, 98–106 (2015).
Kinoshita, M. et al. Nedd5, a mammalian septin, is a novel cytoskeletal component interacting with actin-based structures. Genes Dev. 11, 1535–1547 (1997).
Estey, M. P. et al. Mitotic regulation of SEPT9 protein by cyclin-dependent kinase 1 (Cdk1) and Pin1 protein is important for the completion of cytokinesis. J. Biol. Chem. 288, 30075–30086 (2013).
Estey, M. P., Di Ciano-Oliveira, C., Froese, C. D., Bejide, M. T. & Trimble, W. S. Distinct roles of septins in cytokinesis: SEPT9 mediates midbody abscission. J. Cell Biol. 191, 741–749 (2010).
Cavini, I. A. et al. The structural biology of septins and their filaments: an update. Front. Cell Dev. Biol. 9, 765085 (2021).
Kim, M. S., Froese, C. D., Estey, M. P. & Trimble, W. S. SEPT9 occupies the terminal positions in septin octamers and mediates polymerization-dependent functions in abscission. J. Cell Biol. 195, 815–826 (2011).
Sellin, M. E., Sandblad, L., Stenmark, S. & Gullberg, M. Deciphering the rules governing assembly order of mammalian septin complexes. Mol. Biol. Cell 22, 3152–3164 (2011).
Mendonca, D. C. et al. An atomic model for the human septin hexamer by cryo-EM. J. Mol. Biol. 433, 167096 (2021).
Benoit, B., Pous, C. & Baillet, A. Septins as membrane influencers: direct play or in association with other cytoskeleton partners. Front Cell Dev. Biol. 11, 1112319 (2023).
Kaplan, C., Steinmann, M., Zapiorkowska, N. A. & Ewers, H. Functional redundancy of septin homologs in dendritic branching. Front Cell Dev. Biol. 5, 11 (2017).
Schampera, J. N. & Schwan, C. Septin dynamics and organization in mammalian cells. Curr. Opin. Cell Biol. 91, 102442 (2024).
Zhang, H. et al. Stable maternal proteins underlie distinct transcriptome, translatome, and proteome reprogramming during mouse oocyte-to-embryo transition. Genome Biol. 24, 166 (2023).
Kim, S. et al. PRMT5 protects genomic integrity during global DNA demethylation in primordial germ cells and preimplantation embryos. Mol. Cell 56, 564–579 (2014).
Su, S. T. et al. Involvement of histone demethylase LSD1 in Blimp-1-mediated gene repression during plasma cell differentiation. Mol. Cell Biol. 29, 1421–1431 (2009).
Hayashi, K., Yoshida, K. & Matsui, Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438, 374–378 (2005).
Kojima, Y. et al. Evolutionarily distinctive transcriptional and signaling programs drive human germ cell lineage specification from pluripotent stem cells. Cell Stem Cell 21, 517–532.e515 (2017).
Sybirna, A. et al. A critical role of PRDM14 in human primordial germ cell fate revealed by inducible degrons. Nat. Commun. 11, 1282 (2020).
Blaser, S. et al. Human endothelial cell septins: SEPT11 is an interaction partner of SEPT5. J. Pathol. 210, 103–110 (2006).
Beites, C. L., Xie, H., Bowser, R. & Trimble, W. S. The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat. Neurosci. 2, 434–439 (1999).
Roseler, S. et al. Lethal phenotype of mice carrying a Sept11 null mutation. Biol. Chem. 392, 779–781 (2011).
Gozal, Y. M. et al. Aberrant septin 11 is associated with sporadic frontotemporal lobar degeneration. Mol. Neurodegener. 6, 82 (2011).
Ono, R. et al. Disruption of Sept6, a fusion partner gene of MLL, does not affect ontogeny, leukemogenesis induced by MLL-SEPT6, or phenotype induced by the loss of Sept4. Mol. Cell Biol. 25, 10965–10978 (2005).
Bartsch, I. et al. Human endothelial and platelet septin SEPT11: cloning of novel variants and characterisation of interaction partners. Thromb. Haemost. 104, 1201–1210 (2010).
Zhao, L. et al. FLCN is a novel Rab11A-interacting protein that is involved in the Rab11A-mediated recycling transport. J. Cell Sci. 131, https://doi.org/10.1242/jcs.218792 (2018).
Roest, H. P. et al. The ubiquitin-conjugating DNA repair enzyme HR6A is a maternal factor essential for early embryonic development in mice. Mol. Cell Biol. 24, 5485–5495 (2004).
Tsafriri, A., Chun, S. Y., Zhang, R., Hsueh, A. J. & Conti, M. Oocyte maturation involves compartmentalization and opposing changes of cAMP levels in follicular somatic and germ cells: studies using selective phosphodiesterase inhibitors. Dev. Biol. 178, 393–402 (1996).
Nagy, A. Manipulating the mouse embryo: a laboratory manual. 3rd edn, (Cold Spring Harbor Laboratory Press, 2003).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Galaxy, C. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 50, W345–W351 (2022).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Lerdrup, M., Johansen, J. V., Agrawal-Singh, S. & Hansen, K. An interactive environment for agile analysis and visualization of ChIP-sequencing data. Nat. Struct. Mol. Biol. 23, 349–357 (2016).
Acknowledgements
Research reported in this publication was supported in part by R01HD102385 and ISMMS seed fund to EG, a A*STAR Career Development Award (Project 202D800034) to MKYS, the NovoNordisk Young investigator award (NNF20OC0059301) to DM, Danish National Research Foundation (grant no 116), Novo Nordisk Foundation (NNF22OC0074308), and a Carlsberg Foundation grant CF22-1209 supporting JBR and AC. The authors gratefully acknowledge use of the services and facilities of the Tisch Cancer Institute supported by the NCI Cancer Center Support Grant (P30 CA196521). This work was supported in part by the Bioinformatics for Next Generation Sequencing (BiNGS) shared resource facility within the Tisch Cancer Institute at the Icahn School of Medicine at Mount Sinai, which is partially supported by NIH grant P30CA196521. This work was also supported in part through the computational resources and staff expertise provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai and supported by the Clinical and Translational Science Awards (CTSA) grant UL1TR004419 from the National Center for Advancing Translational Sciences. The authors acknowledge the support of the Freiburg Galaxy Team: Person X and Björn Grüning, Bioinformatics, University of Freiburg (Germany) funded by the German Federal Ministry of Education and Research BMBF grant 031 A538A de.NBI-RBC and the Ministry of Science, Research and the Arts Baden-Württemberg (MWK) within the framework of LIBIS/de.NBI Freiburg. We thank all current and former DM and EG lab members for fruitful scientific discussions and input to this work.
Author information
Authors and Affiliations
Contributions
D.M. conceptualized the project and designed experiments with inputs from E.G. M.K.Y.S., B.H., M.G., Y.H., J.B.R. and H.W. performed experiments and acquired the data. D.M., A.S.,. A.C. and L.R. performed computational and omics analysis. All authors provided scientific input. D.M. and E.G. wrote the manuscript with inputs from M.K.Y.S. and supervised research.
Corresponding authors
Ethics declarations
Competing interests
The Guccione laboratory received research funds from AZ and Prelude Therapeutics (for unrelated projects), E.G. is a cofounder and shareholder of Immunoa Pte.Ltd and cofounder, shareholder, consultant and advisory board member of Prometeo Therapeutics. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Seah, M.K.Y., Han, B.Y., Huang, Y. et al. Maternal PRDM10 activates essential genes for oocyte-to-embryo transition. Nat Commun 16, 1939 (2025). https://doi.org/10.1038/s41467-025-56991-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-56991-8