Abstract
Blood brain barrier-crossing molecules targeting transferrin receptor (TfR) and CD98 heavy chain (CD98hc) are widely reported to promote enhanced brain delivery of therapeutics. Here, we provide a comprehensive and unbiased biodistribution characterization of TfR and CD98hc antibody transport vehicles (ATVTfR and ATVCD98hc) compared to control IgG. Mouse whole-body tissue clearing reveals distinct organ localization for each molecule. In the brain, ATVTfR and ATVCD98hc achieve enhanced exposure and parenchymal distribution even when brain exposures are matched between ATV and control IgG in bulk tissue. Using a combination of cell sorting and single-cell RNAseq, we reveal that control IgG is nearly absent from parenchymal cells and is distributed primarily to brain perivascular and leptomeningeal cells. In contrast, ATVTfR and ATVCD98hc exhibit broad and unique parenchymal cell-type distribution. Finally, we profile in detail brain region-specific biodistribution of ATVTfR in cynomolgus monkey brain and spinal cord. Taken together, this in-depth multiscale characterization will guide platform selection for therapeutic targets of interest.
Similar content being viewed by others
Introduction
Disorders of the central nervous system (CNS) comprise a large area of unmet medical need. A major limitation to the success of many CNS treatments is the high selectivity and restrictiveness of the blood-brain barrier (BBB), which severely limits the effective delivery of antibodies and other large molecules into the brain1,2,3,4,5,6. Despite numerous clinical trials for neurodegenerative diseases using large molecule therapeutics over the past decades, only a few recent efforts have shown clinical efficacy7,8,9. Given that only around 0.01–0.1% of systemically introduced antibodies reach the brain, there is a great need to further improve the target engagement potential of CNS therapeutics by enhancing their brain uptake capacity and biodistribution10,11.
The entry route of systemically delivered protein therapeutics into the brain remains incompletely understood. Brain endothelial cells (BECs) possess many unique cellular properties, including limited vesicular trafficking and preferential lysosomal degradation, both of which leads to very low levels of immunoglobulin (IgG) transport across BECs10,12. Initial studies examining CNS biodistribution described endogenous serum proteins as distributing primarily to the leptomeninges and perivascular spaces in addition to the circumventricular organs13,14,15, yielding distribution patterns similar to that resulting from exogenous protein administration into the CSF16. Subsequent work has demonstrated a clear molecular size-dependent access of circulating proteins to the CSF under normal conditions, a finding historically interpreted as consistent with serum proteins primarily accessing the CSF across choroid plexus epithelial cells and nonchoroidal sites such as the circumventricular organs17. Taken together, this suggests systemic and even direct CSF delivery of large molecules may often result in limited brain parenchymal distribution16. Approaches for large molecule CNS delivery have sought to target highly expressed luminal proteins on BECs to enable receptor-mediated transcytosis or potentially other trafficking pathways. The most well-characterized of these BBB proteins are the transferrin receptor (TfR1 or TfR) and the more recently described CD98 heavy chain (CD98hc, also known as 4F2)11,18,19,20,21,22,23,24,25,26,27. There has been an increase in the development of BBB-targeted large molecule platforms28,29,30,31,32,33,34,35,36,37,38,39 as well as approvals of standard non-BBB targeted CNS antibody therapeutics7,8,9 yet an unbiased and comprehensive characterization of the precise pathways and cell type these BBB-targeting molecules distribute to in the CNS has not yet been explored. Additionally, a detailed biodistribution comparison of BBB-enabled molecules to traditional IgG has been lacking; it remains unclear whether the parenchymal biodistribution limitations of standard antibodies may be overcome simply with higher doses, or if their limited biodistribution is an inherent consequence of their specific route of entry into the brain.
We have previously described two BBB-crossing transport vehicles (TVs) that facilitate increased brain uptake and biodistribution by directly engineering the huIgG Fc to bind to either TfR or CD98hc (TVTfR, TVCD98hc)11,18. The TV platform is highly modular, with applications for antibodies (ATVs), enzymes (ETVs), other proteins (PTVs), and oligonucleotide conjugates (OTVs)11,18,28,34,35,40. The TV platform is engineered for distinct advantages compared to other brain delivery platforms, e.g., no unnatural linkers or appended sequences, retention of the native IgG architecture, and optimized TV affinity to maximize exposure11,18. Several different TV fusion proteins have demonstrated efficacy in preclinical animal models of disease11,18,28,34,35,40 and evaluations in clinical trials are on-going41. Although these prior preclinical reports have consistently described significant increases in brain exposure with TfR- and CD98hc-targeted TV molecules compared to non-targeted control IgG, a more comprehensive description of whole-body distribution and cell-type specific distribution of TVs throughout the brain has yet to be reported. Given that TfR and CD98hc are also expressed across peripheral organs, a more granular multilevel characterization is particularly important for optimized TV platform selection for various types of therapeutic targets. This knowledge would help to broaden the window between efficacy and safety, as well as to identify potential for targeting peripheral organs. Finally, more information is needed to better appreciate how distribution patterns and mechanisms revealed in rodent studies translate to primates.
Here, we address several of these questions using multiple technologies to characterize ATV and non-targeted control IgG biodistribution at a multiscale level from whole body to brain cellular subtypes. Whole-body tissue clearing identified organ-specific uptake of ATVs that was previously not appreciated, as well as enhanced ATV brain and spinal cord biodistribution to both vasculature and parenchyma. In contrast, control IgG predominantly localized to CSF-associated compartments, even at substantially higher doses, suggesting TV-enabled distribution via the vasculature is superior for widespread brain delivery. Brain cell-specific distribution using fluorescence activated cell sorting (FACS) and single cell RNA sequencing (scRNA-seq) demonstrate that ATVTfR and ATVCD98hc can be found in BECs and pericytes as they traffic through the BBB, with unique and consistent uptake in most parenchymal cell types. In contrast, control IgG predominantly distributed to CSF-bordering and perivascular cells associated with cerebral blood vessels, with minimal localization to BECs or parenchymal cell types. Importantly, we demonstrate that the vascular and parenchymal biodistribution of TVTfR-enabled molecules obtained in our mouse studies translated to larger non-human primate brain and spinal cord.
Results
ATVTfR and ATVCD98hc exhibit distinct peripheral biodistribution patterns
Previous reports of peripheral distribution of TV-enabled molecules have provided useful insights, yet most approaches utilized to date have been focused upon preselected organs without spatial biodistribution information18,34. We used whole-body tissue clearing and light sheet fluorescence microscopy (LSFM) to enable a more comprehensive and unbiased understanding of peripheral biodistribution42,43,44. To isolate the contribution of TfR and CD98hc binding alone on brain exposure and peripheral biodistribution, ATVTfR and ATVCD98hc were engineered with non-targeting Fabs as well as with Fc mutations to eliminate binding to Fc gamma receptor and complement proteins (L234A/L235A/P329G)45 (Fig. 1a). Control IgG, ATVTfR, or ATVCD98hc were pre-conjugated with Alexa Fluor 647 (AF647) and systemically administered into wild-type, or previously described homozygous chimeric TfRmu/hu KI11 or CD98hcmu/hu KI mice18 followed by whole body tissue clearing and 3D LSFM (Fig. 1b). Using a series of in vitro and in vivo experiments we validated that the AF647 fluorophore does not alter the ATV biochemical properties or its brain biodistribution profile, and the fluorophore remains attached to the molecule in vivo (Supplementary Fig. 1). A single terminal time point, prioritized for when maximum brain concentrations are achieved with each platform, was selected to characterize brain and whole body biodistribution. Based on previous studies, this corresponds to 1 day post-dose for control IgG and ATVTfR, and around 5 days post-dose for ATVCD98hc11,18. Despite the different terminal timepoints selected for control IgG and ATVCD98hc, we have not previously observed significant differences in control IgG brain distribution over time18. To further validate this, we assessed the cellular biodistribution of control IgG by IHC over 1, 3, and 7 days, and observed undetectable signal at all timepoints (Supplementary Fig. 2a).
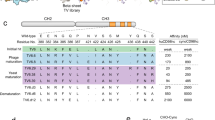
a Schematic of the molecules used with non-targeting control Fabs. The orange patch in ATVTfR Fc region binds to TfR and blue patch in ATVCD98hc binds to CD98hc. b Schematic of the experimental paradigm. TfRmu/hu KI, CD98hcmu/hu KI, or WT mice were dosed with 30 mg/kg AF647-conjugated control IgG and ATVTfR for 1 day and ATVCD98hc for 5 days. c Ventral and dorsal view 3D immunofluorescence images AF647-conjugated control IgG, ATVTfR, or ATVCD98hc in the whole mouse. Representative immunostaining from n = 3/group. Quantification of mean fluorescence intensity of AF647-ATVTfR (d) or AF647-ATVCD98hc (e) normalized to AF647-conjugated control IgG. n = 3/group, mean ± sem, two-tailed t-test with Benjamini–Hochberg false discovery rate, vertebrae adjusted p value 0.045, brain cerebrum adjusted p value 0.045, brain hippocampus adjusted p value 0.045, mean fluorescence intensity of ATV compared to control IgG. 3D reconstructed images of select peripheral organs with high ATVTfR (f) or ATVCD98hc (g) uptake. VNO vomeronasal organ, intra. l.g. intraorbital lacrimal gland, extra. l.g. extraorbital lacrimal gland. Representative images from n = 3/group.
Dorsal and ventral 3D views, as well as rotating and sectional fly-through movie, revealed unique biodistribution profiles for each molecule (Fig. 1c and Supplementary Movie 1). Raw mean fluorescence intensity (MFI) measurements highlighted a few organs with particularly high levels of uptake for each molecule, with most of the remaining organs showing similar levels of uptake (Supplementary Fig. 3). Interestingly, all molecules including the control IgG, prominently distribute to nasal structures appearing to correspond to the nasal respiratory mucosa and nasal associated lymphoid tissue (NALT) in the rostrolateral nasal passage, as well as to the vomeronasal organ (VNO) of Jacobson in the rostromedial nasal passage (Fig. 1 and Supplementary Fig. 3). These areas appear to correspond to regions in the rat we previously demonstrated to be fenestrated (based on dextran and BSA permeability measurements), with large pores that would allow a molecule the size of IgG to easily extravasate from the circulation46. It is likely that the mouse also exhibits fenestrated nasal vasculature in these same regions47, supporting the significant uptake observed for all three molecules. Since a certain level of IgG uptake will be non-specific, we normalized the MFI values of each organ to that of control IgG to provide a semi-quantitative survey of ATVTfR- and ATVCD98hc-specific uptake (Fig. 1d, e). Although the terminal time points are different between control IgG (1 day) and ATVCD98hc (5 days), we previously observed that the trends of biodistribution across peripheral organs from 1 to 5 post-treatment remain relatively similar for ATVCD98hc and control IgG, albeit with higher peak concentrations for kidney and testes where among the highest uptake is observed (Supplementary Fig. 3)18. Overall, this analysis revealed that both ATVs are taken up at higher levels across numerous peripheral organs with differentiated distribution patterns (Fig. 1d, e). ATVTfR preferentially localized to bones (e.g., tibia, shoulder bone, vertebra, and skull), consistent with high TfR expression in erythroid precursor cells within the bone marrow (Fig. 1d, f and Supplementary Movie 1)48,49. The brain exhibited the second highest level of ATVTfR uptake, with some slight regional differences observed. Additional organs that showed pronounced uptake of ATVTfR included the large intestine, kidney (cortex and medulla), lacrimal glands, lung, and spleen. Intriguingly, the organ that showed the highest level of ATVCD98hc uptake was the lacrimal gland which, in addition to the parotid gland, and sciatic nerve, have not been previously identified as peripheral targets for CD98hc-binding constructs (Fig. 1e, g and Supplementary Movie 1). Consistent with previous reports, enhanced uptake of ATVCD98hc was also observed in the large intestine, kidneys, pancreas, and testes, in addition to the brain18. As with ATVTfR, the brain exhibited widespread and significant uptake of ATVCD98hc18. These peripheral distribution data suggest that TVs may not only serve as brain delivery platforms but could potentially also be exploited in certain cases to provide enhanced delivery to specific peripheral organs.
3D imaging reveals enhanced brain biodistribution of ATVTfR and ATVCD98hc
Whole-body tissue clearing also provided the opportunity to further evaluate the biodistribution of ATVTfR and ATVCD98hc compared to control IgG throughout the brain, meninges, and other associated tissues in the intact mouse head. We segmented the brain from the whole-body tissue cleared mice using AI-based tools and generated 3D-reconstructed views of the brains, including the meninges. 3D reconstructed images revealed striking differences in the overall distribution of ATVs compared to control IgG. The latter exhibited a clear signal primarily at the brain surface, corresponding to leptomeningeal tissue and associated blood vessels on the brain surface (Fig. 2a). Weaker IgG signal was also evident in the lateral ventricles. In contrast, the ATVTfR and ATVCD98hc groups exhibited strong and distinct patterns of signal intensities throughout the brain (Fig. 2a and Supplementary Movies 2–4). Quantification of the AF647 signal from the brain surface extending through deeper cortical regions revealed a consistently elevated signal for both ATVs, in stark contrast to the low level of signal for control IgG (Fig. 2b).
a Representative images from AI-segmented 3D-reconstructed brains obtained from the whole-body tissue cleared mice dosed with AF647-conjugated control IgG, ATVTfR, or ATVCD98hc. arc. hypoth nuc arcuate hypothalamic nucleus, ventromed. hypoth nuc. ventromedial hypothalamic nucleus, median em. median eminence. Representative images from n = 3/group. b Mean fluorescence intensity (a.u.) of control IgG, ATVTfR, and ATVCD98hc as a function of brain depth from the cortical surface (0 μm) moving into deeper cortical tissues (1350 μm). n = 3/group, mean ± sem, two-tailed t-test with Benjamini–Hochberg false discovery rate control per depth bin, *p < 0.05, **p < 0.01, compared to control IgG, exact values provided in Source Data. Mean fluorescence intensity (a.u.) of AF647-conjugated control IgG (c), ATVTfR (d), or ATVCD98hc (e) across different mouse brain regions from the whole-body tissue cleared mice. n = 3/group, mean ± sem, two-tailed t-test with Benjamini–Hochberg false discovery rate control, all regions nonsignificant p value > 0.05 (mean fluorescence intensity of each brain region compared to whole brain average per treatment group). Top ten regions with highest fold change in fluorescence intensity compared to brain average fluorescence intensity for AF647-conjugated IgG (f), ATVTfR (g), or ATVCD98hc (h). Dotted line denotes the average brain intensity. n = 3/group, mean ± sem, two-tailed t-test with Benjamini–Hochberg false discovery rate control, subgeniculate nucleus adjusted p value 0.043, stria terminalis adjusted p value 0.024, arcuate hypothalamic nucleus adjusted p value 0.012 (mean fluorescence intensity of each brain region compared to whole brain average per treatment group). Optical slice longitudinal (i) or transverse (j) views of fluorescence images from mouse spinal cord from whole-body tissue cleared mice. Representative images from n = 3/group.
As many neurological disorders affect distinct brain regions, we next determined whether the two ATV platforms exhibited differential uptake across brain regions that could be informative for targeting of specific therapeutics. As expected, control IgG administration yielded a consistently low signal across all AI (artificial intelligence)-segmented brain regions, in marked contrast to the higher intensities across all regions associated with both ATV molecules (Fig. 2c–e). A global assessment revealed a relatively homogenous exposure for ATVTfR across the whole brain, albeit with slightly more enhanced uptake in the hippocampus and cortex (Fig. 2d). ATVCD98hc also exhibited a relatively homogenous exposure and revealed regions with elevated exposure such as the thalamus, hypothalamus, and hippocampus (Fig. 2e). Since this initial quantification approach covered large brain areas, an unbiased approach identified additional smaller subregions with the highest exposure levels relative to the average whole brain signal (Fig. 2f–h). Subregion-specific enhancement was found to be modest for control IgG and ATVTfR. In contrast, ATVCD98hc distribution revealed several subregions with IgG concentrations more than two-fold above the average signal in brain, including the arcuate hypothalamic nucleus, the ventromedial hypothalamic nucleus, area postrema, and median eminence, among others (Fig. 2h). The high uptake of ATVCD98hc observed in these circumventricular organs (arcuate hypothalamic nucleus, ventromedial hypothalamic nucleus, area postrema, and median eminence) is consistent with high expression in these regions of CD98hc and one of its binding partners, LAT1 (L-type amino acid transporter)50,51,52.
Previous studies have demonstrated that TfR is expressed on endothelial cells in both mouse and rat spinal cord, as well as in neurons in the mouse53,54, suggesting this is another expected site for uptake of ATVTfR. Given this, we assessed uptake of both ATV molecules in the spinal cord, a region particularly affected in CNS diseases such as amyotrophic lateral sclerosis, multiple sclerosis, and spinal muscular atrophy, among others. Both longitudinal (Fig. 2i) and cross-sectional (Fig. 2j) views of the spinal cord showed clear and enhanced exposure of both ATVTfR and ATVCD98hc compared to control IgG, which did not show detectable signal. Additionally, the cross-sectional images revealed that within the ATV groups, uptake was mostly evident in the spinal gray matter (Fig. 2j). Taken together, these findings further support the higher exposure and distribution of ATVTfR and ATVCD98hc across CNS regions.
ATVTfR and ATVCD98hc exhibit enhanced brain exposure and biodistribution
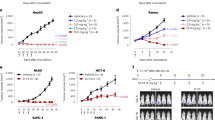
Although higher total brain concentration can be achieved with control IgG using very high systemic doses, it remains unclear whether dosing higher can result in a homogeneous brain biodistribution achieved with ATVs. To address this, we first compared brain exposure of control IgG, ATVTfR, or ATVCD98hc that were pre-conjugated with AF647 and systemically administered at the same dose of 50 mg/kg into wild-type, TfRmu/hu KI, or CD98hcmu/hu KI mice, respectively (Fig. 3a). Bulk brain exposure levels for ATVTfR and ATVCD98hc were significantly higher compared to control IgG when each molecule was administered at the same dose (Fig. 3b). Consistent with our whole-body tissue clearing, widefield fluorescence imaging of whole sagittal brain sections by IHC revealed widespread vascular and parenchymal localization of ATVTfR and ATVCD98hc across the whole brain, while control IgG was primarily localized to the choroid plexus and leptomeningeal tissues at the brain surfaces (Fig. 3c). We next increased the dose of control IgG to 100 mg/kg to match total brain concentration achieved with 15 mg/kg of ATVTfR to determine whether a higher dose could overcome the limited brain distribution of control IgG. These dose levels resulted in comparable bulk brain concentrations between the two molecules (Fig. 3d). However, despite a more than 6-fold higher dose of control IgG compared to ATVTfR the biodistribution pattern of both molecules remained similar to what was observed in the dose-matched experiment (Fig. 3c, e and Supplementary Fig. 4a–d). That is, control IgG exposure remained predominantly limited to the pial brain surface and appeared remarkably similar to what has been reported when IgG is applied directly into the CSF by intracisternal administration16. High signal was also observed in association with the choroid plexus, particularly in the stroma on the blood-side (basolateral aspect) of the polarized epithelial cells (Supplementary Fig. 4a). In contrast, ATVTfR signal was also observed in the choroid plexus, but with a more diffuse pattern and relatively lower intensity compared to control IgG (Supplementary Fig. 4a). Putative perivascular signal was observed with control IgG, particularly prominent in the colliculi, hindbrain, and the thalamus adjacent to the lateral ventricle (Fig. 3f) and appeared predominantly associated with αSMA+ vessels (arterioles; Fig. 3e–g). We observed a prominent signal in the fimbria of the hippocampus bordering the ventricle and a gradient of diffuse parenchymal signal in the thalamus which appeared to arise from the lateral ventricle (Fig. 3f). Control IgG signal was also prominently observed around the olfactory nerve layer of the ventral olfactory bulb, near a known drainage pathway for CSF (Fig. 3e and Supplementary Fig. 4b)16,55,56,57. Despite the high dose of control IgG, the parenchyma appeared to have little signal relative to ATVTfR. Notably, ATVTfR localized to most of the vasculature but was largely absent from αSMA-positive vessels (arterioles), suggesting potential differences in uptake across the arterio-venous spectrum (Fig. 3f and Supplementary Fig. 4e–g).
a Schematic of the experimental design. WT, TfRmu/hu KI, and CD98hcmu/hu KI mice were dosed with 50 mg/kg of AF647-conjugated control IgG and ATVTfR for 1 day and ATVCD98hc for 5 days. b Brain concentration of AF647-conjugated control IgG, ATVTfR, or ATVCD98hc as measured by huIgG ELISA in bulk brain lysates after a single 45 mg/kg IV dose. n = 4/ group, mean ± SEM, one-way ANOVA ****p < 0.0001 compared to control IgG, 95% confidence interval of ATVTfR vs. control IgG −34.76 to −23.5, 95% confidence interval of ATVCD98hc vs. control IgG −24.72 to −13.47. c Representative immunofluorescence in sagittal brain sections of AF647-conjugated control IgG, ATVTfR, and ATVCD98hc in mouse. Representative immunostainings from n = 2/group. d Brain lysate concentration measured by ELISA 1 day after a single 100 mg/kg IV dose of control IgG or 15 mg/kg of ATVTfR (n = 4 mice/group), unpaired two-tailed t-test, nonsignificant (n.s.) p value > 0.05. e Immunodetection of huIgG in whole sagittal brain sections by widefield imaging. Experiment conducted once with n = 4/group. f Magnified examples of brain regions from (e) including the superior colliculus, neocortex, and thalamus. Examples of putative perivascular huIgG signal indicated by arrowheads; notably darker alpha-smooth muscle actin (αSMA) + arteriole indicated by arrows (see Supplementary Fig. 4e–g). Insets show immunodetected huIgG (magenta) and vascular marker caveolin-1 (green), scalebar 50 μm. g Ventral volume view of immunodetected huIgG in tissue-cleared whole mouse hemibrain (n = 6 mice/group). Arrowheads indicate huIgG associated with the large surface arteries, asterisk indicates the median eminence, arrow indicates cranial nerves (region of trigeminal, facial, and vestibulocochlear nerves). h Higher magnification light sheet imaging of immunodetected huIgG and vascular marker lectin around the middle cerebral artery (MCA) and anterior cerebral artery (ACA) branching from the circle of Willis. Circumferential banding pattern around putative smooth muscle cells indicated by arrowheads; arrow indicates putative perivascular profile of a penetrating vessel. For micrographs, display settings were optimized independently for each region of interest and were identical for both treatment groups.
We next took advantage of the prominent signal achieved with the high 100 mg/kg dose of control IgG to better visualize distribution within the leptomeninges and perivascular spaces of surface vessels in 3D by brain tissue clearing and LSFM imaging. These regions are often difficult to evaluate spatially by 2D IHC but may have particular safety implications for certain classes of CNS antibody therapeutics58,59,60. Perfusion with lectin was used to enable visualization of the entire brain vasculature. Control IgG showed a clear and substantial association with the brain surface (presumably associated with leptomeningeal cells of the arachnoid and pia), leptomeningeal vessels (putative perivascular signal), the median eminence (a circumventricular organ lacking a blood-brain barrier), and hindbrain cranial nerves (putative routes for CSF clearance), while ATVTfR did not strongly localize to any of these compartments (Fig. 3g). Higher resolution imaging of the middle cerebral artery as it branches from the circle of Willis revealed control IgG localization on the abluminal (CNS) side of the vasculature (Fig. 3h). The signal appeared to be perivascular, with a circumferential banding pattern around smooth muscle cells that suggests control IgG could be accessing these spaces from the CSF. In contrast, ATVTfR was minimally associated with these leptomeningeal and perivascular compartments. A lateral volume view of the middle cerebral artery and the underlying parenchyma showed a stark drop-off in control IgG signal within the parenchyma, suggesting inefficient penetration from the CSF and leptomeninges, aside from occasional perivascular signal evident around penetrating vessels (Supplementary Fig. 4h, i).
ATVTfR and ATVCD98hc localize to brain vascular and parenchymal cells while control IgG localizes to blood-CSF barrier and perivascular BBB cells
Obtaining unbiased brain cell-specific biodistribution patterns of ATVTfR, ATVCD98hc, and control IgG by IHC faces numerous challenges, particularly detection in less abundant cell types. We sought to address this by devising a highly sensitive and semiquantitative approach that allowed characterization of brain cellular biodistribution in an unbiased and comprehensive manner. Mice were dosed with AF647-conjugated ATVTfR, ATVCD98hc, or control IgG, and brains were dissociated and sorted by flow cytometry based on AF647 intensity. Terminal time points were selected based on previously reported brain Cmax for each platform (1 day for control IgG, ATVTfR, or 5 days for ATVCD98hc)11,18 (Fig. 4a). Similar to the whole body biodistribution study, we selected the timepoints at which brain Cmax is reached in order to maximize the number of captured cells for each molecule. To further validate our selection of these single timepoints, we assessed the cellular biodistribution of ATVTfR and ATVCD98hc versus control IgG by IHC over 1, 3, and 7 days across parenchymal cell types that have previously been reported to exhibit localization of ATVTfR (i.e., neurons, using NeuN) and ATVCD98hc (i.e., astrocytic endfeet, using AQP4)11,18 (Supplementary Fig. 2a–c). Consistent with our previous reports, we observe that once the ATVs cross the cerebrovasculature, their parenchymal biodistribution profiles are not altered across time except in terms of the magnitude of localization. Flow cytometry revealed that only around 1% of the live brain cells from the control IgG group were AF647-positive in contrast with substantially higher percentages for ATVTfR and ATVCD98hc (Fig. 4b and Supplementary Fig. 5). To generate semiquantitative distribution data, we divided the AF647-positive into low, medium, and high bins by dividing the total signal from the ATVTfR group equally into thirds, such that each AF647-positive bin contained approximately 33% of the total AF647-positive cells (Fig. 4b). We then applied these gating cutoffs to the remaining treatment groups to enable comparisons across groups. Single cell RNA-seq was performed on all four bins (i.e., AF647-low, -medium, and -high bins, along with a negative bin denoting negligible signal) allowing us to identify and quantify AF647-labeled molecules within cell types that constitute each bin.
a Schematic of the experimental design: single cells obtained from dissociated brains of WT, TfR mu/hu KI, and CD98hcmu/hu KI mice dosed with 45 mg/kg AF647-conjugated antibodies were sorted based on AF647 signal and then loaded onto the 10x Genomics platform for scRNA sequencing. b Representative flow cytometry plots showing the percentage of AF647-positive cells and their percent distribution across the negative, low, mid, and high AF647 intensity bins. c UMAP of cell clusters captured from the dissociated brains of mice from all three treatment groups and across all four bins. d Bar graphs represent mean percent of total huIgG distribution across indicated cell types captured per treatment group across fluorescence intensity bins. Points represent individual mice. Numbers displayed represent the total number of cells captured for the respective cell type and fluorescent intensity bin. Control IgG (n = 2/group); ATVTfR and ATVCD98hc (n = 4/group).
Dimensionality reduction and predictive cell type labeling across all samples revealed that our approach captured not only major cell types of the brain such as neurons, microglia, astrocytes, and oligodendrocytes, but also cells of the neurovascular unit such as endothelial cells, pericytes, and vascular smooth muscle cells (VSMC), along with other border-associated cell types including perivascular lymphoid and myeloid cells, choroid plexus epithelial cells (CPECs) and two types of fibroblast cells (Fig. 4c and Supplementary Fig. 6b). We divided fibroblasts into two broad types for ease of evaluation given the parameters of our dataset: type 1 fibroblasts predominantly expressed markers of the recently reported brain fibroblast (BFB) subtypes including dural border and arachnoid barrier fibroblasts (BFB5, BFB4 and BFB3), while type 2 fibroblasts expressed markers associated with the inner arachnoid, pia, perivascular compartments, and choroid plexus (BFB3, BFB2, BFB1b, BFB1a, and BFB6) (Supplementary Fig. 6c)61. Notably, no control IgG was observed in any major parenchymal cell types including astrocytes, neurons, microglia, and oligodendrocytes, as these cell types were only found in the AF647-negative bin (Fig. 4d). The vast majority (97%) of vascular endothelial cells (i.e., BECs), pericytes, and vascular smooth muscle cells (VSMCs) also contained negligible levels of control IgG, suggesting minimal trafficking across the BBB. The only cell types where any appreciable control IgG was detected were the choroid plexus epithelial cells (inner blood-CSF epithelial barrier), type 1 and 2 fibroblasts associated with the meninges and perivascular compartments, and perivascular lymphoid and myeloid cells (Fig. 4d and Supplementary Fig. 7a).
ATVTfR and ATVCD98hc biodistribution exhibited a markedly different cellular biodistribution pattern. A significant percentage of ATVTfR and ATVCD98hc localized to both vascular and parenchymal cell types, where the vast majority of BECs and pericytes in both ATVTfR and ATVCD98hc-treated groups fell in the AF647-mid and -high bins (Fig. 4d and Supplementary Fig. 7a). In addition, ATVs displayed localization to cells of the blood-CSF barriers (choroid plexus epithelial cells), fibroblasts and immune cell populations (Fig. 4d). Each ATV exhibited unique parenchymal biodistribution characteristics as well. ATVTfR exhibited strong localization to astrocytes and neurons, along with minimal but measurable localization to microglia and oligodendrocytes. Despite the very low cell numbers of neurons captured (279 cells in total across treatments), we were able to discern distinct localization patterns between ATVTfR and ATVCD98hc in this cell type, broadly consistent with previous reports by IHC11,18. Specifically, higher localization of ATVTfR was found in neurons, whereas ATVCD98hc instead exhibited appreciably more localization to microglia compared to ATVTfR (Fig. 4d). One caveat of our approach was an expected loss of astrocytic processes due to the single-cell dissociation protocol. As we previously observed high protein expression of CD98hc on aquaporin 4-positive astrocytic endfeet and processes18, it is likely that our current approach underestimates localization of ATVCD98hc to astrocytes. Additional analysis evaluating the cellular distribution per fluorescence intensity bin for each treatment revealed that among the few cells captured in the A647-high bin for the control IgG group, there were some fibroblasts type 2 and microglial cells in addition to peripheral myeloid and choroid plexus epithelial cells (Supplementary Fig. 7b). Overall, significant positive associations between receptor expression and ATV uptake were observed only in a few cell types, suggesting either there is a lack of strong correlation between mRNA and protein expression, and/or mRNA levels does not reliably reflect the amount of accessible cell surface antigen available for TV binding (Supplementary Fig. 8a, b). Taken together, our application of a unique method for evaluation of brain cell biodistribution revealed a highly specific cell-type distribution pattern of each ATV, compared to the limited parenchymal cell distribution of control IgG.
ATVTfR and ATVCD98hc exhibit localization to BECs across the arterio-venous spectrum
Distinct expression patterns of transporters along the arterio-venous axis have been previously described in both mice and humans62,63. To determine if ATVs are differentially taken up by BEC subtypes, we next subclustered BECs into arterial, capillary, and venous populations based on markers established in previous mouse single-cell studies64,65 (Supplementary Fig. 9a, b) and determined the distribution of molecules to these subtypes across the intensity bins (Fig. 5). Consistent with active trafficking across the BBB, ATVTfR localized strongly to capillary and venous cells, with greater than 80% of AF647-positive cells falling within the high bin for these BEC subtypes, and a lower proportion (~60%) falling within the high bin for arterial cells (Fig. 5b). Based on the lack of arterial localization by IHC (Fig. 3f and Supplementary Fig. 3e–g), the arterial ATVTfR signal we observed in the medium and high bins was unexpected (Fig. 5b). We sought to determine the localization of ATVTfR with more granularity by overlaying the BEC subtypes captured per bin over the total BEC UMAP (Uniform Manifold Approximation and Projection) (Supplementary Fig. 9b, d). This additional analysis revealed that the BECs falling within the negative bin appear to be arterial cells at the most proximal end of the arteriovenous spectrum, i.e., marker phenotype indicative of arteries and larger caliber arterioles (Supplementary Fig. 9a, b). In contrast, BECs in the low, mid, and high bins appear to display an increasingly mixed phenotype indicative of smaller caliber arterioles gradually transitioning to capillaries (Supplementary Fig. 9b, d). Indeed, this uptake pattern is consistent with Tfrc expression pattern across the arteriovenous axis, where we observe little to very low expression of Tfrc for the proximal arterial cells versus arterial cells closer to capillaries (Supplementary Fig. 9a). Taken together, we hypothesize that the apparent discrepancy between our IHC and FACS/scRNAseq data is likely due to: (1) the inability of αSMA staining by IHC to fully capture all arterial endothelial cells across this zonation and (2) the inability of the FACS-scRNAseq approach to fully distinguish between arteriolar and capillary brain endothelial cells along the zonal continuum.
a–c Bar graph represent mean percent of total huIgG distribution across arterial, capillary, and venous endothelial cells captured per treatment group across fluorescence intensity bins. Points represent individual mice. Numbers displayed represent the total number of BEC subtypes captured. Control IgG (n = 2/group); ATVTfR and ATVCD98hc (n = 4/group).
ATVCD98hc exhibited a more similar distribution across all three BEC subtypes, with ~50% of AF647-positive cells falling within both mid and high bins (Fig. 5c). There was minimal localization of control IgG to BECs, and among the ~2% found in the AF647-low bin, nearly all signal belonged to the venous population (Figs. 4d, 5a and Supplementary Fig. 9c). Overall, these data are largely consistent with their expression along the arterio-venous axis as previously reported in both mice and human transcriptomics datasets, and suggests ATVTfR and ATVCD98hc are actively trafficked across the BBB through multiple BEC subtypes62,63.
Enhanced vascular and parenchymal biodistribution patterns of ATVTfR in cynomolgus monkey brain and spinal cord
Our previous work established significant increases in bulk brain exposure ATV in non-human primates, though a more granular biodistribution analysis across the entire brain and within more discrete brain regions has yet to be evaluated11. We sought to examine whole brain biodistribution of ATVTfR versus control IgG using tissue clearing and 3D LSFM imaging on the brains and spinal cords of cynomolgus monkeys. We only pursued the comparison with ATVTfR due to the limited availability of cynomolgus monkeys, associated risk of tissue clearing and imaging technology for primate brains, and because TfR-based approaches are the most clinically advanced and widely used brain delivery platforms. Animals were dosed systemically with AF647-conjugated control IgG or ATVTfR and brain vasculature were simultaneously labeled using wheat germ agglutinin (Fig. 6a)66. A global 3D view of the hemibrain revealed higher signal for ATVTfR throughout the entire brain compared to control IgG (Fig. 6b, c). Coronal slice views and higher magnification images further revealed clear vascular and parenchymal signals for ATVTfR within both the smallest microvessels (i.e., capillaries) as well as larger caliber vessels, while control IgG was primarily localized only to the perivascular space of larger penetrating vessels (Fig. 6d–g and Supplementary Fig. 10a, b). This biodistribution pattern was evident across multiple brain regions (Supplementary Fig. 10a–d), in coronal sections throughout the whole brain (Supplementary Fig. 11), and sectional fly-through 3D movie (Supplementary Movies 5 and 6). In all areas and regions of interest (ROIs) examined, delivery of ATVTfR to the cynomolgus monkey brain clearly exceeded that of control IgG. The mean fluorescence of ATVTfR was higher than control IgG in both segmented brain vessels as well as in the non-vessel parenchymal fraction (Fig. 6h, i). Indeed, the ATVTfR distribution reached deep into the tissue as evidenced by the persistence of signal intensity in deeper brain regions (Fig. 6i). Significantly higher ATVTfR signal was found in smaller caliber vessels (<40 µm) compared to larger vessels (>60 µm), consistent with the predominant capillary and venule TfR BEC distribution patterns that have been reported in mice as well as our BEC subtype cell distribution (Figs. 6j and 5)62,67. ATVTfR signal was higher than control IgG across all brain regions measured, with the largest differences observed in the cortex, diagonal subpallium, amygdala, hypothalamus, and pons (Fig. 6k). Isolated cervical spinal cords also revealed striking levels of ATVTfR uptake in both the vasculature and parenchyma, with notably higher signal in the spinal gray matter (Fig. 6m). In contrast, control IgG signal in the spinal cord was notably faint, with prominent signal primarily observed only in spinal nerve roots (Fig. 6l), comprising another putative CSF drainage pathway and similar to what has been observed following intrathecal antibody administration16,55. Taken together, these findings in cynomolgus monkey brain and spinal cord illustrate how ATVTfR enables both higher CNS exposure as well as significantly broader and homogenous biodistribution throughout the parenchyma compared to non-BBB targeted IgG in larger primate brains.
a Schematic of the in vivo experimental design. Immunofluorescence images of 3D reconstructed hemibrains (1x objective) (b, c), coronal slice view of hemibrain (200 μm thick, 1x objective) (d, e), and cortical slice view of temporal lobe (ventral cortex) (200 μm thick, 4x objective) (f, g) from cynomolgus monkeys dosed with 23 mg/kg AF647-conjugated control IgG or ATVTfR (IV) for 2 days. Arrow heads in (f) highlight localization of control IgG to large penetrating vessels, while arrow heads in (g) highlight localization of ATVTfR within large vessels as well as capillaries. Experiment conducted once with from n = 1/group. Mean fluorescence intensity of ATVTfR and control IgG in vessels (h) and non-vessels (i) of cynomolgus monkey brains across regions of different depths from surface of the brain. j Mean fluorescence intensity of ATVTfR and control IgG within vessels of different radii in the cynomolgus monkey brains. k Mean fluorescence intensity of ATVTfR and control IgG across different brain regions in cynomolgus monkeys. 3D Immunofluorescence images of spinal cords from cynomolgus monkeys dosed with AF647-conjugated control IgG (l) or ATVTfR (m). Immunostaining image from n = 1/group.
Discussion
BBB-crossing molecules are emerging as a new class of therapeutics with huge potential for CNS indications. In this study, we provide a comprehensive and unbiased examination of the biodistribution of non-targeted control IgG antibody, ATVTfR, and ATVCD98hc from whole body to single-cell resolution. Our approach combining fluorescent tagging of molecules with FACS-sorting and single cell RNAseq reveals that ATV distributes robustly to numerous parenchymal cell types and blood-brain barrier endothelial cells, whereas control IgG localization was limited to choroid plexus epithelial cells, meningeal, perivascular cells. At an organ level, we leveraged the enhanced 3D imaging capabilities of light sheet fluorescence microscopy and find distinct organ-specific biodistribution of ATVTfR, ATVCD98hc, and control IgG throughout the whole mouse body. Widespread brain distribution was observed for both ATVTfR and ATVCD98hc compared to control IgG. Finally, 3D imaging in tissue-cleared cynomolgus monkey brains reveal widespread and enhanced distribution of ATVTfR throughout the superficial and deep regions of both the brain and spinal cord, where ATVTfR distribution had a striking resemblance to previously published patterns of TfR immunostaining in human spinal cord68. Altogether, these data shed further light on how the pathways governing IgG uptake and distribution enables broad delivery with ATV but severely limits distribution of control IgG antibodies in the CNS of both rodents and primates.
The peripheral distribution of each ATV platform was largely consistent with previous reports using more traditional methods11,18, but the unbiased nature of the 3D whole-body imaging methodology reported here provide a more holistic view of biodistribution and reveals important insights. These findings from the present study also highlights the potential for ATVTfR and ATVCD98hc to treat CNS diseases in which peripheral pathology might also be present, as well as provide additional biological considerations for platform-therapeutic pairings where there is a desire to target or avoid certain organs. For example, tissues where both platform and target antigen are localized may require additional safety evaluation. The current data is especially informative for some of the previously unappreciated organs identified as tissues of localization that may have otherwise been overlooked (e.g., ATVCD98hc to the lacrimal glands, parotid gland, and sciatic nerve). The addition of targeting Fabs and/or cargo, as well as the binding affinity of TfR and CD98hc will undoubtedly impact both the peripheral biodistribution as well as tissue uptake kinetics of each TV-enabled therapeutic molecule and will thus need to be characterized on a case-by-case basis. Stronger affinity TfR and CD98hc TVs would likely drive even greater cell- and organ-specific uptake, as well as faster peripheral pharmacokinetics; the opposite would be expected for weaker affinity variants. Overall, the current data set provides a starting point for understanding how these platforms distribute across peripheral organs to help guide platform selection and tissues for further investigation.
The contrast between TV-enabled and control IgG biodistribution reveals several major limitations for standard antibody therapeutics for CNS diseases. Both our IHC and scRNAseq data strongly suggest non-BBB targeted IgG has a predominant route into the brain via the CSF, and not across the BBB. These data bear remarkable similarity to previously reported patterns for endogenous IgG10,12,13 as well as intracisternal administered IgG16,61. This plasma-to-CSF-to-brain trafficking route dominates for systemically administered IgG, with lowest concentrations in the deeper brain regions far from the ventricular and pial brain surfaces. Such patterns are expected to ultimately result in inefficient therapeutic target engagement, particularly in species with larger brain volumes69. One example where more homogenous biodistribution has the potential to further improve efficacy are for amyloid antibodies for the treatment of Alzheimer’s disease, some of which have been clinically approved in the last several years37,38,70. Amyloid antibodies have some amount of access the CNS after peripheral administration (similar to what we observe with control IgG in the current study), leading to modest improvements in clinical outcomes. However, this is a highly inefficient process, likely due to sub-optimal target engagement and slow and/or incomplete amyloid removal. It has been recently reported that ATVTfR can greatly improve brain biodistribution and target engagement of an amyloid antibody in a preclinical mouse model of Alzheimer’s disease71. Indeed, interim clinical data from of a TfR-enabled amyloid antibody exhibited a much more rapid removal of plaque compared to standard antibodies, and it was achieved with a significantly lower dose level72. Together, these data suggest the biodistribution patterns observed in preclinical species is readily translatable to humans. Interestingly, we noted ATVTfR or ATVCD98hc had relatively higher exposures in several brain regions implicated in neurodegenerative diseases (e.g., cortex, hippocampus, caudoputamen, thalamus) and thus may further benefit from based drug delivery for these diseases.
The cellular distribution profile of control IgG is consistent with the interpretation that non-BBB targeted antibodies predominantly enter the CSF compartment from the circulation, rather than cross the BBB. This distribution pattern may account for the typically higher exposures of standard antibodies detected in the CSF compared to brain parenchyma. Strikingly, less than 3% of BECs and pericytes had any control IgG signal. Importantly, control IgG yielded almost no association with other parenchymal CNS cells. Rather, the cells with the highest control IgG signal were located at the inner and outer blood-CSF barriers, suggesting these cells are likely involved in blood-to-CSF transport of non-targeted control IgG. This distribution agrees well with imaging results showing strongest signal in the choroid plexus and leptomeninges. Additionally, the perivascular signal on surface vessels and penetrating vessels points to subsequent CSF-to-brain entry of control IgG, supported by the cellular data showing localization with type 2 fibroblasts, perivascular lymphocytes, and macrophages. This pattern of leptomeningeal and perivascular signal strikingly matches well with that previously reported for exogenous antibodies administered into the CSF as well as that of endogenous serum proteins, further supporting a mechanism of circulating IgG distribution first into the CSF and then into brain13,16. Similar biodistribution patterns have also been observed when comparing intrathecal delivery of antisense oligonucleotides versus intravenous delivery of ATVTfR-enabled ASO40, suggesting limited brain biodistribution likely poses a consistent challenge to other non-protein therapeutics when not enabled by a BBB-targeting platform. Importantly, even when bulk brain exposures were made equivalent by increasing the dose of control IgG by almost 7-fold over that of ATVTfR, IgG biodistribution to the parenchyma remained severely limited. Together, these observations underscore the critical importance of the brain entry route in shaping robust brain cellular biodistribution, with BBB entry greatly favored over CSF entry if the goal is to obtain widespread biodistribution to both superficial and deeper brain.
Consistent with active trafficking across the BBB as the predominant route of entry into the brain, ATVTfR and ATVCD98hc are highly localized to arteriovenous endothelial cells with lower levels in arterial compared to venous and capillary BECs. This pattern is also consistent with previously reported zonal expression of TfR and CD98hc (i.e., highest in capillaries and post-capillary venules) and direct blood-to-brain transport across the BECs62,67. These distribution patterns translated to the cynomolgus monkey, where we observe greater ATVTfR signal in smaller diameter capillaries and venules compared to larger diameter arteries. Each ATV also exhibit a unique cellular distribution profile to other CNS cells. ATVTfR highly localizes to neurons, consistent with previous reports of brain TfR expression11. Interestingly, ATVTfR localizes to cell types that have not been previously appreciated, including astrocytes, and to a lower extent, to microglia and oligodendrocytes. Since these molecules were modified to lack effector function, it is entirely plausible that the addition of Fcγ receptor binding could alter the cellular distribution of the molecules. Indeed, we previously observed that ATVCD98hc localization to Iba1-positive microglia is enhanced when the ATVCD98hc is on a wild-type huIgG Fc (i.e., effector-positive), compared to effector-attenuated molecules in both mice and NHP18. Furthermore, addition of effector function to ATVTfR with anti-amyloid beta Fabs resulted in robust microglia activation and subsequent plaque phagocytosis71, suggesting that although minimal microglia localization is observed with the effectorless ATVTfR from this current study, addition of effector function can result in a molecule that can readily engage with microglia.
ATVCD98hc localizes to both astrocytes and microglia but no detectable localization was found with neurons (albeit in a limited sampling) or oligodendrocytes. Like ATVTfR, additional target binding of a therapeutic would likely impact the cellular biodistribution for ATVCD98hc as well. We previously demonstrated that the addition of anti-BACE1 to ATVCD98hc led to neuronal localization not normally observed with ATVCD98hc molecules, as well as reduced brain exposure, likely due to an increase in internalization and degradation in neurons driven by BACE1 binding18. This suggests that cell-specific targeting, as well as the trafficking biology of the target, could impact not just the biodistribution, but also the overall brain exposure profile as well.
Although this work provides important insights, the biodistribution of ATV-enabled therapeutic molecules will need to be further assessed independently, as both CNS cell-specific as well as peripheral distribution will be driven not only by TfR and CD98hc binding, but also by the therapeutic cargo (e.g., Fab targets, enzymes, ASOs, etc). Additionally, TV binding affinity, dose, and timepoint will likely all impact the distribution of molecules both in brain and periphery. While these results are relevant for the fundamental understanding of receptor-mediated BBB transport, each BBB technology will need to be evaluated separately, given that differences in architecture of the molecules could have significant impacts on CNS exposure and distribution.
Finally, the route of entry of therapeutics may have additional important therapeutic implications that warrants further investigation. For example, it will be interesting for future studies to evaluate whether ATV-enabled delivery of anti-amyloid antibodies can reduce engagement with the vascular amyloid commonly present on leptomeningeal arteries and perivascular drainage pathways58,59,60. Avoiding these pathways with a BBB-dominant route of entry into the CNS could potentially mitigate risks such as immunotherapy-induced vascular inflammation and microhemorrhages60,73,74. Nevertheless, our results provide a thorough foundational understanding of the brain cell types and tissues targeted by each TV platform, which will enable optimal platform selection to efficiently drive desired distribution and target engagement profiles of a variety of TV-enabled therapeutics for neurological diseases.
Methods
All animal procedures were performed in adherence to ethical regulations and protocols approved by Denali Therapeutic Institutional Animal Care and Use Committee (IACUC). We used the following IACUC protocols 2023.04.001 Tissue Harvest for Biomarker Development & Primary Culture (protocol number 2023.04.002) and 2023.10.001 PK/PD and PK Studies in Rodents (protocol number 2023.10.001). For the cynomolgus monkey studies we used the IACUC protocol ID 2087590 approved by the IACUC committee at Charles River Laboratories, Inc. at Reno Nevada.
Animal care
Mice were housed under a 12 h light/dark cycle and had access to water and a standard rodent diet (LabDiet 5LG4, Irradiated) ad libitum. Temperature and humidity in all animal rooms were monitored daily by Thermo ScientificTM InSight. The normal temperature range was 18.3–23.3 °C and the normal humidity range was 30–70%.
In vivo mouse studies
Mice used in this experiment included C57BL/6J and homozygous TfRmu/hu KI, and CD98hcmu/hu KI (on a C57Bl6J background) (The Jackson Laboratory)11,18. For the brain uptake study of fluorophore-labeled ATVTfR 12-week-old TfRmu/hu KI females were used, and for the wild-type plasma PK study 12-week-old C57Bl6J were used. For the whole body biodistribution and brain imaging experiments, we used 12-week-old male mice. For the FACS-scRNA-seq experiment, 12-week-old mixed sex mice were used with 2 males and 2 females per group. For the brain concentration matching experiments, female TfRmu/hu KI mice were used at 18–19 weeks of age. Mice were IV dosed via the tail vein with the test articles and then anesthetized deeply via IP injection of 2.5% Avertin at the terminal timepoints. For PK studies, mice were transcardially perfused with ice cold PBS at a rate of 5 mL/min and brains were collected.
We tried to include both females and males mice when possible. Yet since the cost of conducting each of our experiments (whole body tissue clearing and cyno brain and spinal cord tissue clearing) was very high, we could not include an additional group to assess both females and males in our experiment thus we selected one sex for each.
TV engineering
ATVTfR and ATVCD98hc platforms were engineered as previously described using a huIgG1 backbone11,18. To isolate the biodistribution attributed to TfR and CD98hc binding only, the control huIgG1 antibody, ATVTfR, and ATVCD98hc molecules were engineered with bivalent non-targeting control Fabs that bind a small molecule, the hapten dinitrophenol (DNP), not present in vivo75. The L234A/L235A/P329G (LALAPG) mutations were made to the Fc region to abolish FcγR and C1q binding and consequently eliminate effector function activity45. For the experiments involving the validation of our fluorophore conjugation approach, whole body biodistribution, and FACS-scRNAseq studies, we used a monovalent form of ATVTfR variant with 100 nM affinity to human TfR, as well as the monovalent form of ATVCD98hc with 170 nM affinity to human CD98hc. For the experiment involving the assessment of brain biodistribution of control IgG versus ATVTfR using brain-exposure matched doses we used an ATVTfR variant with 620 nM affinity to human TfR. For the cynomolgus monkey studies, we used a monovalent form of ATVTfR variant with 390 nM affinity to cynomolgus monkey TfR.
Fluorescent antibody conjugation
To fluorescently conjugate our antibodies, we used the Thermo Fisher Scientific SiteClick Labeling Kits (S10911, S10901) which allow the attachment of Alexa-647 to the heavy chain N-linked glycans. The main advantage of this approach is that the conjugation is far from the antigen-binding domain and the TV location which reduces the potential for interference with molecule biochemical properties. On average we achieved a degree of labeling between 1–2 AF647 fluorophores per antibody. This was determined by the absorbance of the fluorophore and antibody on the nanodrop and followed by the calculation using the kit’s recommendation. Quality control steps including in vitro (cell binding and SPR (Surface Plasmon Resonance)) and in vivo (wild-type plasma clearance and brain uptake) were conducted to ensure the fluorophore does not interfere with the binding and biodistribution properties of the ATV platform (Supplementary Fig. 1).
Cell binding of fluorophore-labeled molecules
Cell binding was assessed similarly as described18. Briefly, HEK293T (ATCC, CRL-3216) and CHO(ATCC, CCL-61) were plated in triplicates at 15,000 cells/well in a 96-well Poly-D lysine-coated plate (Fisher Scientific, PerkinElmer LLC 6055302) and incubated overnight at 37 °C. Cells were treated with ATVTfR or AF647-ATVTfR for 1 h at 37 °C, fixed with 4% paraformaldehyde, and blocked with 1xPBS containing 5% BSA and 0.3% Triton. Cells were then incubated with anti-human IgG AlexaFluor®488 (1:1000, Jackson ImmunoResearch, 109-545-003 lot 151904), DAPI, and Deep Red Cell Mask (1:5000, Thermo Fisher, C10046). A minimum of 20 field of views were acquired for each replicate at ×40 using the Opera Phenix High Content imaging system (PerkinElmer). Images were analyzed using the Harmony Software (PerkinElmer version 4.9).
Surface plasmon resonance (SPR) of fluorophore-labeled molecules
Affinities of unlabeled and A647-labeled ATVTfR were determined by surface plasmon resonance using a Biacore 8K instrument. Samples were immobilized on a Cytiva Series S CM5 sensor chip (Cytiva, #29149603) using a Cytiva Human Fab capture kit (Cytiva, #28958325) at 10 ug/mL using a flow rate of 10 uL/min for 60 s. Three-fold serial dilutions of Full Length human TfR at concentrations of 300, 100, 33.3, 11.11, 3.70 and 0 nM were injected at a flow rate of 30 ul/min for 60 s, followed by a 300-s dissociation in a 1x HBS-EP+ running buffer (Cytiva, #BR100826). After injection, CM5 sensor chip was regenerated using 10 mM Glycine-HCl (pH 2.0) at the end of each cycle. Data analysis was conducted using Biacore Insight Evaluation software (version 2.0.15.12933). Kinetic analysis was performed with a 1:1 Langmuir kinetic binding model for evaluation of K on, K off and K D.
Wild-type plasma PK of fluorophore-labeled molecules
C57Bl6J (n = 3) were subject to a 3 mg/kg tail vein IV injection of unlabeled or AF647-labeled ATVTFR and in-life bleeds were collected in EDTA tubes mice at 0.5, 4, 24, and 48 h post-treatment. At 72 h post-dose mice were deeply anesthetized via an IP injection of 2.5% and terminal blood collected in EDTA tubes. From each timepoint the collected whole blood was stored temporarily on ice and then processed to plasma by centrifugation (18,213 × g for 7 min at 4 °C) within 30 min of collection. Plasma was then transferred into cluster tubes and stored at −80 °C until ready for huIgG analysis by ELISA.
Brain uptake PK of fluorophore-labeled molecules
TfRmu/hu KI mice (n = 4) were subject to a 10 mg/kg tail vein IV injection of unlabeled or AF647-labeled ATVTFR and in-life bleeds were collected in EDTA tubes mice at 0.5 h post-treatment. At 24 h post-treatment mice were deeply anesthetized via an IP injection of 2.5% and terminal blood collected in EDTA tubes. The frontal third of the left hemibrain was collected in 1.5 ml Eppendorf tube and fresh-frozen on dry ice and stored at −80 °C until ready for huIgG analysis by ELISA.
Brain cellular biodistribution timecourse by IHC
ATVTfR, ATVCD98hc, and Ctrl IgG were intravenously administered at 50 mg/kg to CD98hcmu/hu, TfRmu/hu double KI mice (n = 5/group). At 1, 3, and 7 days post dose, animals were deeply anesthetized with an intraperitoneal injection of 2.5% Avertin, and mice were transcardially perfused with cold PBS. Following transcardial perfusion with PBS, hemi-brains were drop-fixed in 4% paraformaldehyde overnight at 4 °C.
Brain and plasma huIgG quantification
The left side of the frontal brains were fresh-frozen and homogenized using a Qiagen TissueLyser with 5 mm steal beads for 6 min at 30 Hz in 10x volume/tissue weight of lysis buffer containing 1% NP-40 in PBS with protease inhibitors. Homogenate was centrifuged at 18,213 × g for 20 min at 4 °C and supernatant collected. Brain lysates were diluted 1:2 and 1:20 for analysis of huIgG concentration.
HuIgG concentrations from brain or plasma were quantified using a generic anti-human IgG sandwich-format ELISA. Briefly, plates were coated overnight at 4 °C with donkey anti-human IgG (JIR #709-006- 098) at 1 μg/mL in sodium bicarbonate solution (Sigma #C3041- 50CAP) with gentle agitation. Plates were then washed 3x with wash buffer (PBS + 0.05% Tween 20). Assay standards and samples were diluted in PBS + 0.05% Tween 20 and 1% BSA. Standard curve preparation ranged from 0.41 to 1500 ng/mL or 0.003 to 10 nM (BLQ < 0.03 nM). Standards and diluted samples were incubated with agitation for 2 h at room temperature. After incubation, plates were washed 3× with wash buffer. The detection antibody, goat anti-human IgG (JIR #109-036-098), was diluted in blocking buffer (PBS + 0.05% Tween-20 + 5% BSA) to a final concentration of 0.02 μg/mL and plates were incubated with agitation for 1 h at room temperature. After a final 3× wash, plates were developed by adding TMB substrate and incubated for 5–10 min. Reaction was quenched by adding 4 N H2SO4 and read using 450 nm absorbance.
Mouse immunohistochemistry
For the experiment involving dose-matched 2D imaging, after perfusing the mice with PBS as described above, the hemibrains were drop fixed in 4% PFA overnight. Sagittal brain sections (40 μm) were cut using a microtome (MultiBrain® Technology by NeuroScience Associates), blocked in 5% BSA + 0.3% Triton X-100. Sections were mounted in Prolong glass (Thermo Fisher P36984) before imaging at ×20 magnification using a widefield epifluorescence slide scanner (Axio Scan Z1; Carl Zeiss; 20x/0.8 NA air objective; each channel was acquired independently with the appropriate filter cube for that fluorophore).
Perfused hemibrains were immersion fixed in 4% PFA for approximately 24 h at 4 °C before transferring to PBS + 0.01% sodium azide followed by sectioning sagittal at 40 μm thickness (MultiBrain® Technology by NeuroScience Associates). Serial sections were stained free-floating by incubating in blocking buffer (PBS/TBS + 1% BSA + 1x fish gelatin (BioWorld 21761058) + 0.5% Triton X-100 + 0.1% sodium azide) at room temperature for 2 h, incubation with primary antibodies (in PBS + 1% BSA + 0.3% Triton x-100 + 0.01% sodium azide) overnight at 4 °C, three 15 min washes in PBS + 0.3% Triton X-100, incubation in secondary antibody (in PBS + 1% BSA + 0.3% Triton x-100 + 0.01% sodium azide with DAPI) for 4–5 h, and three 15 min washes in PBS + 0.3% Triton x-100. Sections were mounted onto slides and sealed with a coverslip with Prolong Glass (Invitrogen, P36984) For the dose- and brain-concentration-matched 2D imaging, antibodies used were anti-huIgG AlexaFluor 647 (Jackson Immunoresearch, 709-606-149 lot 168098), aSMA alpha-Smooth Muscle Actin Antibody (1A4/asm-1) AF750 (Novus Biological NBP2-33006AF750 lot D149482), Caveolin-1 (Cell Signaling 3267S lot 9, 1:500), and donkey anti-rabbit IgG (H + L) AF488 (Invitrogen A21206 lot 2072687, 1:500). For the brain uptake of unlabeled and AF647-ATVTfR molecules, antibody used to anti-human IgG AlexaFluor®488 (1:1000, Jackson ImmunoResearch, 109-545-003 lot 151904). Slides were imaged at ×20 magnification using a widefield epifluorescence slide scanner (Axio Scan Z1; Carl Zeiss; 20x/0.8 NA air objective; each channel was acquired independently with the appropriate filter cube for that fluorophore). For the characterization of brain cellular biodistribution over time by IHC, anti-AQP4 (Millipore, AB2218, 1:500), anti-NeuN (Millipore, MAB377, 1:500), and anti-human IgG-647 (Jackson ImmunoResearch, 709-606-149, 1:500) were used in addition to the secondary antibodies donkey anti-rabbit Alexa Fluor 488 (Invitrogen, A21206, 1:500) and goat anti-mouse IgG1-568 (Invitrogen, A-21124, 1:500). For each tissue section, a small region of the cortex directly superior to the hippocampus was imaged using a Leica SP8 scanning confocal with a 25x water objective (0.95 NA) followed by Lightning superresolution post processing with default settings for the “adaptive” algorithm.
Mouse 3D brain tissue clearing and imaging
After 24 h of dosing mice with 100 mg/kg control IgG or 15 mg/kg ATVTfR through tail vein IV injection (brain-concentration-matched study), mice were deeply anesthetized via IP injection of 2.5% Avertin and immediately transcardially perfused using ice cold PBS containing 5 mg/L LEL tomato (DyLight 649 conjugated; Invitrogen, L32472 lot32472) at a flow rate of 2.5 mL/min for 6 min, followed by perfusion of room temperature 4% PFA for 25 min. Spinal cords and brains were extracted, the brains bisected, and each tissue post-fixed in 4% PFA for 24 h, rinsed in PBS, and then transferred to PBS + 0.01% sodium azide and stored at 4 °C, protected from light throughout the process. One animal in the control IgG group was not well fixed (body not rigid) due to an air bubble in the line during perfusion-fixation and was excluded from further analysis.
Samples were brought to RT and washed in 1xPBS before being dehydrated in a 200-proof methanol/milliq water gradient of 20%, 40%, 60%, 80% and 100% at 1 h while on a rocker at gentle speed. After washing once more in 100% methanol at room temperature, samples were incubated overnight in 67% dichloromethane/33% methanol, after which they were washed twice in 100% methanol for 30 min (RT), cooled to 4 °C and bleached in chilled fresh 5% hydrogen peroxide in methanol overnight at 4 °C. After rehydration in a room-temperature methanol/PBST series (1 h each at 80%, 60%, 40%, 20% methanol in PBS with 0.2% TX-100), samples were washed in PBST twice for an hour before incubating in permeabilization solution (2.3% glycine, 20% DMSO, 0.02% sodium azide in PBST) at 37 °C for 4 days. Once permeabilized, samples were blocked at 37 °C for 3 days in a solution of 6% donkey serum, 10% DMSO, 0.02% sodium azide, and 0.2% Triton x-100 in PBS. After blocking, samples were incubated with primary antibody in a buffer of 0.2% gelatin, 0.5% Trion x-100 and 0.1% saponin in PBS for 14 days at room temperature, while being rocked gently. After washing (in PBS with 0.1% saponin, 0.2% tween20, and 0.1% of a 10 mg/ml heparin solution) for 1 × 10 min, 1 × 20 min, 1 × 30 min, 1 × 1 h, 1 × 1.5 h (at RT) and 1x 3 days (at 4 °C), samples were incubated in secondary antibody for 7 days. The secondary incubation took place in antibody dilution buffer (0.2% Tween 20, 0.1% of 10 mg/ml heparin stock solution, 0.1% saponin, 0.5% Triton X-100, 0.2% gelatin) at RT with gentle rocking. Samples were then washed again (as after primary antibody incubation), and dehydrated in a methanol/milliq water gradient as before. After a final overnight incubation in 100% methanol, samples were washed for an hour in fresh 100% methanol and incubated in 66%DCM (Dichloromethane)/33% methanol at room temperature and finally washed in 100% DCM 15 min twice (with shaking) to remove traces of methanol. Stained and delipidated tissue samples were cleared in DiBenzyl Ether in airtight vials for 24 h, and in ECi for another 24 h (RT and protected from light) before imaging in ECi. Antibodies used were anti-huIgG, Dylight800 (1:500 Thermo Fisher SA510132 lot 4D3898685), Goat anti-CD31 (1:300 RD systems AF3628 lot YZU0223011), Goat anti-Podocalyxin (1:1000 RD systems AF1556 lot JPC0122111) and donkey anti-gt Cy3 (1:500 Jackson ImmunoResearch 705-166-147 lot 167205).
Fully cleared samples were imaged in the Miltenyi Ultramicroscope blaze with the 1x objective (1x zoom), and again with 12x objective (1x zoom). For both imaging runs, photographs were taken with 0.186 numerical aperture, with two-sided illumination, adaptive focus and adaptive blending. All images of one magnification were taken with the same optimized settings. A total of 1x images were taken at 70% sheetwidth, at 3.86 µM steps. Excitation (Emission) as follows: 785(805): 10%, 640 (680) 7%, 561(620) 5%, and 488 (525) 40%. A total of 12x images (of the medial branch of the circle of Willis) were taken at 50% sheetwidth, at 0.3 µM steps. Excitation (Emission) as follows: 785(805): 8%, 640 (680) 9%, 561(620) 6%, and 488 (525) 77%.
FACS-scRNA-seq
For each treatment group (Ctrl IgG, ATVTfR, ATVCD98hc), 2 male and 2 female mice were perfused with 1x PBS and their whole brain were collected (excluding olfactory bulbs as well as cerebellum, pons, and medulla). For downstream processing, brains from 1 male and 1 female per treatment were pooled and processed together for single cell dissociation in order to reduce the number and cost of single cell RNA-seq libraries. Cells from individual animals were then demultiplexed based on sex-specific gene expression allowing us to analyze cells from all 4 animals per treatment group.
To dissociate the brains into single cells we used the Neural Tissue Dissociation Kit (Miltenyi Biotec, cat. #130-092-628) as previously described76. Briefly, the brains were chopped using a razor, resuspended with HBSS, and centrifuged. The pellet was resuspended and incubated with the enzyme P mix at 37 °C for 10 min along with the transcriptional and translational inhibitors Actinomycin D (Cell Signaling Technology, cat. #15021, 5 uM final concentration) and Anisomycin (Cell Signaling Technology, cat. #2222, 2 uM final concentration) respectively. Samples were then homogenized by trituration using a 5 ml serological pipette and incubated with Enzyme A at 37 °C for another 10 min. Following incubation, the brains were triturated with a 2 ml serological pipette, washed with DPBS, and passed through a 100 uM filters and spun at 300 × g 10 min. To remove the excess myelin, cells were resuspended in 0.9M sucrose and spun at 850 × g for 35 min. Cells were then washed with FACS buffer (1x PBS, 1% BSA, 2 mM EDTA) and stained with SYTOX™ blue (Thermo Fisher) to distinguish live cells.
On the FACS sorter (BD FACSAria™ III Cell Sorter), the gates were set to capture single live cells, and using the naïve mouse sample the Alexa-647 positive gate was drawn (Supplementary Fig. 5). We used ATVTfR samples to draw the gates for the low, mid, and high samples (ensuring no overlap) such that each gate would capture 33% of the cells from the AF647-positive gate for that group. To have a fair comparison across treatments, these same gates were applied to all other groups. Additionally, since the AF647-negative bin accounts for 99% of the cells in the IgG control group, sorting cells from this bin for the same duration of time as the low, mid, and high AF647-bins (total of ~1% for the IgG control group) would have exceeded the capacity of the collection tubes for the AF647-ve bin. Thus, we decided to sort only 60,000 cells for the AF647-ve bin for all treatment groups and record the time required to obtain this cell number. Afterward, we simultaneously sorted the cells from the AF647-low, -mid, and -high bins as well as recorded the time required. Then to obtain the theoretical number for the AF647-ve bin that we would have obtained, we extrapolated this number from the total time it took to sort the AF647-low, mid, and high bins for that group (further described in “Single-cell RNA-seq data processing and analysis”). Even though we started out with an n = 4/group (2 males, 2 females) for all groups, we unfortunately lost n = 2 samples (1 female, 1 male) for the control group AF647-negative bin due to a technical malfunction in the 10x Genomics instrument that was used to conduct the scRNAseq experiment.
Single-cell RNA-seq library construction and sequencing
Six single-cell suspensions of pooled male/female dissociated brain tissues were FACS sorted into four different bins based on AF647 signal intensity resulting in 24 samples for single-cell capture. Up to 60,000 cells were sorted into each 1.5 mL centrifuge tube containing the 24 samples consisting of the 3 treatment groups with 4 bins each and 2 pooled animals per treatment group (1 male, 1 female). To concentrate the single-cell suspensions, each sample was pelleted by centrifugation at 200 × g for 10 min in a swing bucket rotor at 4 °C, and supernatant was removed leaving only 43.3 µL volume for resuspension. The entire volume of cells was then added to the RT master mix and loaded onto Chromium Next GEM Chip G microfluidic channels per manufacturer’s protocol (10X Genomics, CG000315 Rev C). One sample (IgG1- bin 0) was lost due to a fluidic wetting issue, leaving 23 samples for library preparation and sequencing analysis.
Post GEM–RT Cleanup was performed using a Dynabeads cleanup mix via magnetic purification, and cDNA was generated using 12 cycles of amplification followed by cDNA purification using SPRIselect reagent. A quarter volume of the cDNA generated was used as input for 3’ gene expression library construction starting with fragmentation, end repair and A-tailing, followed by Illumina adapter ligation and sample index PCR, with SPRIselect bead purification performed in between each step.
Library quantity and quality were assessed with High Sensitivity D1000 ScreenTapes (Agilent 5067-5584) and then pooled in equimolar ratios for shallow sequencing on an Illumina MiSeq Reagent Kit v3 (Illumina, MS-102-3001) to determine cell capture rate. With the cell capture number information from the MiSeq sequencing result, libraries were pooled targeting 20,000 reads per captured cell. RNA-seq data for analysis was generated on an Illumina NovaSeq 6000 instrument, S2 cartridge, Paired end (28X10X10X90) by SeqMatic (Fremont, CA, USA).
Single-cell RNA-seq data processing and analysis
After sequencing, fastq files were processed using Cell Ranger (v7.1.0). To increase the number of samples without generating additional libraries, each library consisted of one male and one female mouse which could then be computationally demultiplexed using gene expression data. For every cell, male/female identity was predicted using a random forest classifier trained on mouse single-cell data from the Linnarsson Adolescent Mouse Brain Atlas77. The model was trained on 6 genes that have strong differential expression between males and females: Xist, Tsix, Ddx3y, Kdm5d, Uty, and Eif2s3y. After applying a 90% confidence threshold, the model predicted sex with ~99% accuracy. This accuracy was supported by observed separation of X and Y chromosome gene expression between predicted male and female mice (Supplementary Fig. 6a). Normalization, variable feature selection, PCA, UMAP dimensionality reduction, and louvain clustering were all carried out in R using Seurat v5.1.078. Broad cell types were predicted using the Seurat label transfer methodology and a single-nucleus atlas of mouse brain cell types. Predictive cell typing was supported by observation of top marker genes in each predicted class of cells (Supplementary Fig. 6b). Following cell typing, putative doublets were identified and removed using the scDblFinder R package (version 1.18.0)79. Fine cell typing of immune and endothelial populations was performed using iterative subclustering and manual cell typing through marker gene analysis. Briefly, the populations were isolated and clustered, obvious doublet clusters were removed, and cluster identities were mapped to known cell types by examination of canonical marker gene expression. These detailed cell type labels were projected back onto the full object as seen in Fig. 2.
FACS was performed for different amounts of time, e.g., longer run times were required to capture sufficient numbers of rarer cells in the A647+ gates. To compare cell populations across fluorescent bins, we calculated capture rates (cells captured per second) by dividing the total number of cells collected in each bin by the duration of the FACS run.
To examine the expression of marker genes, including TfR and Slc3a2, single-cell counts were aggregated into pseudobulk expression values for each sample and cell type using the AggregateAcrossCells function in the scuttle R package (version 1.14.0). For visualization, log2 CPMs (counts per million) were calculated using a pseudocount of 1.
Associations between log2 (pseudobulk) expression and fluorescent intensity bins were assessed by fitting polynomials of degree two using the voomLmFit function from the edgeR R package (version 4.2.1) for each gene, allowing for a monotonous but non-linear increase in intensity between the low, mid and high fluorescent bins. In addition, the sex of each animal was modeled as a fixed term.
Whole body tissue clearing
Mice were transcardially perfused with ice cold PBS at a rate of 4 mL/min followed by 20 min perfusion with 4% PFA. They were then post-fixed for 12 h with 4% PFA then washed with PBS and stored in PBS with 0.05% sodium azide until ready for shipment. Upon arrival to Deep Piction, they were skinned, washed in PBS and set up for whole-body perfusion as described in the vDISCO protocol, omitting the nanobody staining step43. In brief, the mice were stored in a glass staining jar and a stainless-steel oral gavage needle was attached to the left ventricle of the heart and secured with superglue. Subsequently, the mice were perfused with the following solutions at room temperature: PBS (1x 12 h), CUBIC-I solution for decolorization (4x 12 h), PBS (3x 3 h), 20% EDTA for decalcification (3d), PBS (3x 3 h), Permeabilization solution (3.5d). Subsequently, the perfusion needle was removed, and the stomach and intestines were cleaned of chyme and feces by small incisions every 2–3 cm and careful rinsing with PBS via a syringe. The mice were then washed 2x 2 h in PBS and dehydrated at RT on a shaker in a series of Tetrahydrofuran (THF)/dH2O mixes with increasing THF concentrations as follows: 50% THF (12 h), 70% THF (12 h), 90% THF (12 h), 100% THF (2x 12 h). They were then dilapidated in Dichloromethane (DCM) for 6 h and refractive index matched in a 1:2 mixture of Benzyl Alcohol/Benzyl Benzoate (BABB). We replaced the BABB after the first 12 h for better clearing performance.
Whole body tissue clearing imaging and processing
The mice were scanned in a customized Ultramicroscope Blaze (Miltenyi Biotec, Germany) outfitted with a custom whole-mouse imaging chamber and a custom stage. Samples were imaged with a 1.1x objective optimized for BABB imaging (LaVision-Miltenyi BioTec MI PLAN ×1.1/0.1 NA, WD 17 mm) and recorded with a scientific sCMOS camera (Miltenyi Biotec, resolution 2048 × 2048 px/frame, 16-bit depth, effective pixel size 5.8 µm/px). Samples were superglued to the sample holder and first imaged along the ventral side with z-step size of 6 µm/step (ca 1700 z-planes) and a 3 × 9 tiling layout in X/Y (25% overlap), The light-sheet thickness was set to 11.4 µm (sheet NA: 0.03) and sheet width of 100%, which evenly covered the entire field of view of the 1.1x objective. The exposure time was set to 80 ms/frame for imaging of the background tissue autofluorescence using a 488 nm laser (emission filters 525/50, 12% laser power) and AlexaFluor-647 conjugated antibody signal with a 640 nm laser (emission filters 680/30, 14% laser power). Following ventral imaging, mice were flipped to the dorsal position, re-glued, and the dorsal half of the animal was imaged using the same parameters.
Imaged tiles were stitched with Fiji (ImageJ) Stitch Sequence of Grids of Images function. For the organ-wise region of interest (ROI) analysis, the whole-body image stack (signal channel only) was loaded as a virtual stack in Fiji and placed a set of markers on the respective organs (minimum two per organ). Regions Of Interest (ROIs) were then processed by cutting out a 10 × 10 × 10 voxel cube centered around the marker. The median, standard deviation, minimum and maximum values were first calculated from all voxels. These were then grouped by ROI type and treatment group, normalized the means to the mean of the control condition, and log-transformed the results.
In addition, the whole-body image stacks were segmented into organs of interest, brain, kidney and lungs, using a custom machine learning pipeline.
Mouse brain regional analysis
Segmented brains were resampled from their initial resolution of 5.80 μm × 5.80 μm × 6.00 μm to 10 μm isotropic volumes with 3rd order b-spline interpolation using the ResampleImageFilter in the Insight Toolkit (ITK) version 5.3.080. Using this resampled volume, an image pyramid with isotropic resolutions 10 μm, 25 μm, 50 μm, and 100 μm was built for each brain image, corresponding to the voxel pitches of the Allen Institute of Brain Science common coordinate framework version 3 brain atlas (CCFv3)81.
The CCFv3 atlas defines a nested hierarchy of over 1300 brain regions, with the most finely delineated layers at or below the resolution of the segmented whole brain data. To enhance the robustness of our region-based quantification, we collapsed the CCFv3 annotations into 14 high-level brain regions by combining the parent region mask(s) and all child region mask(s) into a single region of interest (ROI) as described in Supplementary Table 1.
Individual brain volumes were registered in stages to the Nissl-stained template using the ElastixImageFilter class in itk-elastix v0.17.182. iDISCO cleared brains were uniformly flatter along the dorsal-ventral (D/V) axis compared to the template image, so the 25 μm brain volumes were each expanded by 50% along the D/V axis, then rigidly registered to the 25 μm Nissl-stained atlas image using parameters given by the GetDefaultParameterMap(“rigid”) function in itk-elastix, coarsely aligning the two in space. Next, the rigidly aligned brains were further registered to the Nissl template with an affine transform using parameters from the GetDefaultParameterMap(“affine”) function. Finally, each brain was nonlinearly warped with a B-spline warp using parameters from the GetDefaultParameterMap(“bspline”) function, except with a final grid spacing of 1600 μm, 9 rounds of optimization, and a grid spacing schedule of 8x, 8x, 8x, 4x, 4x, 4x, 2x, 2x, 2x for each round respectively. Warped brains were visually overlaid on the atlas Nissl template image to assess registration quality using napari v0.5.483. While gross region alignment was good across all brains, fine structures were visibly misaligned. To improve alignment of fine structures, all 9 b-spline warped brain images and the 25 μm Nissl atlas template were simultaneously aligned using the parameters from the GetDefaultParameterMap(“groupwise”) function, except with a final grid spacing of 800 μm, an increased maximum number of iterations to 512 steps, and a 10-step grid spacing schedule with 2 steps each of 32x, 24x, 16x 12x and 8x optimization. To transfer ROIs onto the brains, both the high level merged ROIs described above, and the lowest level annotations for each voxel of the 25 μm Allen Brain Atlas were warped through the rigid, affine, b-spline and groupwise sequence using the TransformixImageFilter with a final b-spline interpolation order of 0 (nearest neighbor interpolation).
A brain mask was defined on the 25 μm isotropic fluorescence image by thresholding the image to include the entire visible brain volume as well as any surface vessels. Voxel-wise brain depth was then quantified inside this brain mask using the distance_transform_edt function from scipy v1.14.184. Mean intensity values were quantified as the mean huIgG intensity within the intersection of each warped brain atlas ROI and the whole brain mask. For hierarchically defined brain regions, mean intensity was calculated for all voxels annotated with that region id or any descendent region ids. Normalized mean intensity was calculated for each region by dividing the regional mean intensity by the regional mean intensity for Brain Atlas region id #8, basic cell groups and regions. Annotations from the fused atlas were warped back onto the original image space using the numerical inverses of the bspline, affine, then rigid transforms above, then overlaid onto the 10 μm isotropic brains in native space. Depth-wise mean intensity was calculated by binning depth from surface into 10 μm bins, then calculating mean signal within the intersection of each inverse warped brain atlas ROI and the depth bin.
In vivo cynomolgus monkey study
Female cynomolgus monkeys (2.6–3.6 years old, Macaca fascicularis, n = 1/group; Charles River Laboratories) were housed at 64–84 °F, 30–70% humidity. Housing set-up is as specified in the USDA Animal Welfare Act (Code of Federal Regulations, Title 9) and as described in the Guide for the Care and Use of Laboratory Animals. Cynomolgus monkeys were IV administered with a single 23 mg/kg dose of appropriate test article, 2 days post-dose the monkeys were anesthetized with ketamine HCl combined with isoflurane/O2 and perfused with 4 ml phosphate buffer saline (PBS) containing heparin, followed by 12.5 mg/kg of WGA-Alexa555 (wheat germ agglutinin) 1.6 mg/ml to label blood vessels, and then 3ml of 1x PBS. After euthanasia, the brain and spinal cord were collected and drop fixed in 4% PFA overnight and then stored in 1xPBS with sodium azide.
Cynomolgus monkey brain and spinal cord clearing
Hemi-brains were washed twice in a 10x volume of 1xPBS for a total of 24 h at room temperature with gentle rocking, protected from light. The tissue was then dehydrated through a series of 200 proof fresh ethanol (LC/MS grade) and milliq water solutions: first at 50% ethanol for 48 h (37 °C), followed by 4 days at 70% ethanol (RT), 48 h at 80% (37 °C), 48 h at 90% (37 °C), 72 h at 100% (RT), and finally 48 h at fresh 100% (37 °C). After dehydration, tissue was incubated in a mixture of 50% EtOH and 50% THF for 24 h at 37 °C, followed by a 4-day incubation in 100% THF (37 °C), and finally a 6-day incubation in 100% DCM (37 °C).
Hemi-brains were then cleared in 99.9% ECi at RT in the dark without agitation for 3 days, before being switched to fresh ECi and allowed to clear further for another 5 days before imaging in ECi in the Miltenyi Blaze Ultramicroscope.
Spinal cord segments were chosen to be comparable between the two animals, taken from the first three, non-enlargement cervical spinal segments. After bringing samples to room temperature, tissue was washed once in PBS for an hour on gentle rocking at RT before dehydrating in similar manner to the brain samples, in 15 ml of an ethanol/water series. First, tissue was incubated in 50% ethanol for 3 h (RT), followed by 70% ethanol overnight (RT), 80% for 3 h (RT), 90% 3 h (RT), 100% overnight (37 °C), and finally fresh 100% for 3 h (37 °C). After dehydration, tissue was moved to a 10 ml glass vial filled completely with 50% EtOH & 50% THF, and rocked gently for 3 h at 37 °C, before incubating in 100% THF overnight (37 °C). The following day, samples were rocked gently in 100% THF for 3 h (37 °C) before moving to 100% Dichloromethane overnight (37 °C). Finally, delipidated cord segments were cleared in 99% Ethyl cinnamate (ECi) for 3 days, followed by switching to fresh ECi for 24 h before imaging in ECi in the Miltenyi Blaze Ultramicroscope.
Samples from the two animals were treated identically and imaged using the same parameters, the details of which varied with the region of interest (hemi-brain or specific ROI), tissue (hemi-brain or spinal cord), and objective used.
Cynomolgus monkey brain imaging
All samples were imaged in ethyl cinammate (ECi), with a retail Ultramicroscope Blaze microscope using the standard 2048 × 2048 px/frame 16-bit depth sCMOS camera, with the exposure time set to 650 ms. For overview scans, the samples were imaged using the standard organic-solvent-optimized 1.1x objective and a 0.6x zoom-out setting in the ImSpector (v7.5.3) software using the 640 laser (emission filter 680/30 16% laser power) in a 2 × 4 tiling grid with 35% overlap and dual-sided illumination. The samples were imaged sagitally, first glued on the medial plane so that the lateral cortex was closest to the objective (lateral scan), then they were repositioned so that the medial sulcus was closest to the objective (medial scan). The z-step size was set to 13 µm. Detailed volume imaging was done with the standard organic-solvent-optimized 4x objective without additional zoom in the software, by using the 561 nm and 640 nm lasers (emission filters 620/60 and 680/30 respectively and 14% laser power) in a 2 × 3 tiling grid with 33% overlap and single sided illumination. The z-stepping was set to 2.71 µm. The samples were positioned in an oblique plane, glued on the medial plane so that the ventral hippocampal region was closest to the objective and the dorsal cortex was next to the sample holder bar, below the laser’s height.
Cynomolgus monkey spinal cord imaging
All samples were imaged in ethyl cinnamate (ECi), with a retail Ultramicroscope Blaze microscope using the standard organic-solvent-optimized 4x objective and the 0.6x zoom-out setting in the ImSpector acquisition software. We used a standard 2048 × 2048 px/frame 16-bit depth sCMOS camera with 264 ms and 650 ms exposure times for the 561 nm and 640 nm lasers respectively. We used the 561 nm and 640 nm lasers (emission filters 620/60 and 680/30 respectively and 14% laser power) for dual-sided illumination. The z-stepping was set to 2.71 µm.
Cynomolgus monkey vessel processing
The produced image stacks were stitched using TeraStitcher. The lateral and medial overview scans were fused together using Vision4D’s landmark-based Volume Fusion function to a single volume. The detailed scans were segmented using VesSAP’s vascular segmentation in the channel highlighting the brain blood vessels. This mask was then expanded using Fiji’s Maximum 3D filter using 2, 4 and 6 voxels in XYZ. To analyze the distribution of parenchymal intensity, we first removed the vascular mask from the 2vx expanded mask, resulting in a concentric shell surrounding the blood vessels. Then, we removed the 2vx mask from the 4vx mask, forming a larger shell, and then removed the 4vx shell from the 6vx mask to create the outermost shell surrounding the vasculature. These masks were applied to the signal channel only. Next, a 5-step tissue depth binning was produced in Fiji using polygon selections, each bin encompassing a 135 µm (50vx) depth. The histogram of each vascular and parenchymal shell (in each depth bin) was extracted using the standard Fiji function and the weighted mean was compared.
To extract the local vessel radius, the vessel mask was first processed with an iterative quasi-3D hole closing filter. For each possible Z plane, the XY vessel mask was extracted then filtered using the remove small holes function from scikit-image with a maximum area threshold of 7344 µm2 (1000 px2). This filter was repeated for each possible Y plane filtering the XZ mask, followed by each possible X plane filtering the YZ mask. The vessel mask was next processed with a binary opening operation to remove axis-aligned stripe artifacts. The morphological skeleton was then calculated using a 3D skeletonization algorithm85 and the distance from the edge of the hole filled vessel mask to the skeleton was calculated using a Euclidean distance transform86. Each point along the skeleton was assigned an index and the distance from the edge at that index was used as the estimate for the local radius at that point.
Each voxel of the hole-filled vessel mask was assigned to the nearest point on the vessel skeleton using watershed segmentation. This segmentation was then masked so only voxels in the original segmentation were considered, and any voxels not assigned to a corresponding point on the vessel skeleton were filtered (~1.0–1.5% of the vessel mask). The mean huIgG was then calculated for each label in the segmentation, resulting in a measurement of radius and mean intensity at each point in the vessel skeleton. The radii were binned into 20 buckets from 2.71 µm (1 voxel) to 54.2 µm (20 voxels) and the mean intensity within each bin was calculated.
Cynomolgus monkey brain atlas registration
1x magnification tiled scans of the cynomolgus brains were first stitched using TeraStitcher then converted to Imaris image pyramid volumes using ImarisFileConverter. The 2x down sampled volume was then loaded and flipped along the y-axis to bring it into rough alignment with the T1 weighted template image for the 24-month-old cynomolgus atlas87. The brain volume was N4 corrected using a shrink factor of 32x, with 5 levels of correction and 50 iterations at each level. The corrected brain was thresholded followed by morphological filtering to remove mask fragments smaller than 0.00292 mm3 (500 vx) and to remove holes smaller than 0.292 mm3 (50,000 vx), then all values outside the brain mask were set to 0.
The T1 weighted template image was split along the midline to produce a hemibrain image, then trimmed 1.9 mm (5 voxels) on each side to match the approximate imaging area of the light sheet image. The template image was then upsampled, and the corrected brain image down sampled each to 0.25 mm × 0.25 mm × 0.25 mm isotropic using linear resampling with the ResampleImageFiler from ITK. The corrected brain image was then registered to the trimmed T1 template image using ANTsPy (antspyx v0.5.4)88. To calculate a robust initial transform, the corrected brain was first coarsely aligned to the template using the “Rigid” transform preset, then this initial transform was further refined using the “Affine” transform preset. The resulting affine initial transform was used to initialize the “SyNAggro” transform preset with no additional refining affine iterations, SyN sampling of 128 per iteration, and a series of six levels of registration with 80, 80, 80, 40, 20, and 10 iterations per level respectively. This resulted in a set of forward and inverse affine and warp parameters mapping between the input volume and the atlas template space.
The SARM and CHARM annotations for level 2 of the cynomolgus 48-month atlas were transformed back to the 0.25 mm isotropic image space using the apply_transforms method from ANTsPy with the “genericLabel” interpolator. The labeling was manually inspected on the original images using napari to ensure alignment with the light sheet anatomy. To enable comparison with the mouse atlas, the CHARM cortical parcellation was combined into a single “cortex” region, while the SARM labels were not modified. Mean intensity was calculated within each region inside the brain mask using the (UNCORRECTED | N4 CORRECTED) images.
Statistical analysis
Unless where labeled otherwise, unpaired t-tests, one-way, and two-way ANOVAs were performed using Prism 9 software (GraphPad 9.5.1). The raw data and results of the statistical analysis found in the Source Data file. Statistical comparisons between Mean Fluorescence Intensities in whole body images and depthwise were conducted using unpaired two-tailed t-tests with the ttest_ind function in scipy. Where multiple comparisons were performed, the false discovery rate was controlled using the Benjamini/Hochberg method as implemented using the false_discovery_control function in scipy. Mean Fluorescence Intensities within fused brain regions were compared to whole brain mean intensity using paired two-tailed t-tests using the ttest_rel function in scipy. Mean Fluorescence Intensities within all regions of level 8 of the Allen Brain Atlas were compared to the mean brain intensity using ttest_rel, p values for all regions were corrected using the false_discovery_control function, and then the 10 regions on average most different from the mean brain intensity were presented. Comparisons between cell proportions detected by single-cell RNA-seq were performed using unpaired two-tailed t-tests assuming unequal variance with R’s t.test function.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The scRNA-seq raw and processed data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database under accession code GSE262436. Due to the terabyte sizes of the raw imaging data and the high cost associated with depositing it, we have not provided this data in a publicly accessible repository but can still provide it upon request. The remaining data generated in this study are provided in the Source Data file. Source data are provided with this paper.
Code availability
scRNAseq and image analysis code is available via Zenodo https://doi.org/10.5281/zenodo.13935844; https://zenodo.org/records/13935844.
References
Daneman, R. & Prat, A. The blood-brain barrier. Cold Spring Harb. Perspect. Biol. 7, a020412 (2015).
Wu, D. et al. The blood-brain barrier: structure, regulation, and drug delivery. Signal Transduct. Target Ther. 8, 217 (2023).
Neuwelt, E. A. et al. Engaging neuroscience to advance translational research in brain barrier biology. Nat. Rev. Neurosci. 12, 169–182 (2011).
Profaci, C. P., Munji, R. N., Pulido, R. S. & Daneman, R. The blood-brain barrier in health and disease: important unanswered questions. J. Exp. Med. 217, e20190062 (2020).
Banks, W. A. From blood-brain barrier to blood-brain interface: new opportunities for CNS drug delivery. Nat. Rev. Drug Discov. 15, 275–292 (2016).
Terstappen, G. C., Meyer, A. H., Bell, R. D. & Zhang, W. Strategies for delivering therapeutics across the blood-brain barrier. Nat. Rev. Drug Discov. 20, 362–383 (2021).
Wu, W. et al. The FDA-approved anti-amyloid-β monoclonal antibodies for the treatment of Alzheimer’s disease: a systematic review and meta-analysis of randomized controlled trials. Eur. J. Med. Res. 28, 544 (2023).
Gueorguieva, I. et al. Donanemab population pharmacokinetics, amyloid plaque reduction, and safety in participants with Alzheimer’s disease. Clin. Pharm. Ther. 113, 1258–1267 (2023).
Cohen, S. et al. Lecanemab clarity AD: quality-of-life results from a randomized, double-blind phase 3 trial in early Alzheimer's disease. J. Prev. Alzheimers Dis. 10, 771–777 (2023).
St-Amour, I. et al. Brain bioavailability of human intravenous immunoglobulin and its transport through the murine blood-brain barrier. J. Cereb. Blood Flow. Metab. 33, 1983–1992 (2013).
Kariolis, M. S. et al. Brain delivery of therapeutic proteins using an Fc fragment blood-brain barrier transport vehicle in mice and monkeys. Sci. Transl. Med. 12, eaay1359 (2020).
Villaseñor, R. et al. Trafficking of endogenous immunoglobulins by endothelial cells at the blood-brain barrier. Sci. Rep. 6, 25658 (2016).
Broadwell, R. D. & Sofroniew, M. V. Serum proteins bypass the blood-brain fluid barriers for extracellular entry to the central nervous system. Exp. Neurol. 120, 245–263 (1993).
Kozlowski, G. P., Sterzl, I. & Nilaver, G. Localization patterns for immunoglobulins and albumins in the brain suggest diverse mechanisms for their transport across the blood-brain barrier (BBB). Prog. Brain Res. 91, 149–154 (1992).
Schmidt-Kastner, R. et al. A one-step immunohistochemical method for detection of blood-brain barrier disturbances for immunoglobulins in lesioned rat brain with special reference to false-positive labelling in immunohistochemistry. J. Neurosci. Methods 46, 121–132 (1993).
Pizzo, M. E. et al. Intrathecal antibody distribution in the rat brain: surface diffusion, perivascular transport and osmotic enhancement of delivery. J. Physiol. 596, 445–475 (2018).
Felgenhauer, K. Protein size and cerebrospinal fluid composition. Klin. Wochenschr. 52, 1158–1164 (1974).
Chew, K. S. et al. CD98hc is a target for brain delivery of biotherapeutics. Nat. Commun. 14, 5053 (2023).
Zuchero, Y. J. Y. et al. Discovery of novel blood-brain barrier targets to enhance brain uptake of therapeutic antibodies. Neuron. 89, 70–82 (2016).
Kanodia, J. S. et al. Prospective design of anti-transferrin receptor bispecific antibodies for optimal delivery into the human brain. CPT Pharmacomet. Syst. Pharm. 5, 283–291 (2016).
Yu, Y. J. et al. Therapeutic bispecific antibodies cross the blood-brain barrier in nonhuman primates. Sci. Transl. Med. 6, 261ra154 (2014).
Bien-Ly, N. et al. Transferrin receptor (TfR) trafficking determines brain uptake of TfR antibody affinity variants. J. Exp. Med. 211, 233–244 (2014).
Yu, Y. J. et al. Boosting brain uptake of a therapeutic antibody by reducing its affinity for a transcytosis target. Sci. Transl. Med. 3, 84ra44 (2011).
Gehrlein, A. et al. Targeting neuronal lysosomal dysfunction caused by β-glucocerebrosidase deficiency with an enzyme-based brain shuttle construct. Nat. Commun. 14, 2057 (2023).
Weber, F. et al. Brain shuttle antibody for Alzheimer’s disease with attenuated peripheral effector function due to an inverted binding mode. Cell Rep. 22, 149–162 (2018).
Pornnoppadol, G. et al. Bispecific antibody shuttles targeting CD98hc mediate efficient and long-lived brain delivery of IgGs. Cell Chem. Biol. https://doi.org/10.1016/j.chembiol.2023.09.008 (2023).
Edavettal, S. et al. Enhanced delivery of antibodies across the blood-brain barrier via TEMs with inherent receptor-mediated phagocytosis. Med. 3, 860–882.e15 (2022).
Arguello, A. et al. Molecular architecture determines brain delivery of a transferrin receptor-targeted lysosomal enzyme. J. Exp. Med. 219, e20211057 (2022).
Faresjö, R., Sehlin, D. & Syvänen, S. Age, dose, and binding to TfR on blood cells influence brain delivery of a TfR-transported antibody. Fluids Barriers CNS 20, 34 (2023).
Faresjö, R. et al. Transferrin receptor binding BBB-shuttle facilitates brain delivery of anti-Aβ-affibodies. Pharm. Res. 39, 1509–1521 (2022).
Zhou, Z., Austin, G. L., Shaffer, R., Armstrong, D. D. & Gentry, M. S. Antibody-mediated enzyme therapeutics and applications in glycogen storage diseases. Trends Mol. Med. 25, 1094–1109 (2019).
Roshanbin, S. et al. Reduction of αSYN pathology in a mouse model of PD using a brain-penetrating bispecific antibody. Pharmaceutics 14, 1412 (2022).
Barker, S. J. et al. Targeting transferrin receptor to transport antisense oligonucleotides across the blood-brain barrier. BioRxiv https://doi.org/10.1101/2023.04.25.538145 (2023).
van Lengerich, B. et al. A TREM2-activating antibody with a blood-brain barrier transport vehicle enhances microglial metabolism in Alzheimer’s disease models. Nat. Neurosci. 26, 416–429 (2023).
Logan, T. et al. Rescue of a lysosomal storage disorder caused by Grn loss of function with a brain penetrant progranulin biologic. Cell 184, 4651–4668.e25 (2021).
Ullman, J. C. et al. Brain delivery and activity of a lysosomal enzyme using a blood-brain barrier transport vehicle in mice. Sci. Transl. Med. 12, eaay1163 (2020).
Sevigny, J. et al. The antibody aducanumab reduces Aβ plaques in Alzheimer’s disease. Nature 537, 50–56 (2016).
van Dyck, C. H. et al. Lecanemab in early Alzheimer’s disease. N. Engl. J. Med. 388, 9–21 (2023).
Grimm, H. P. et al. Delivery of the BrainshuttleTM amyloid-beta antibody fusion trontinemab to non-human primate brain and projected efficacious dose regimens in humans. MAbs 15, 2261509 (2023).
Barker, S. J. et al. Targeting the transferrin receptor to transport antisense oligonucleotides across the mammalian blood-brain barrier. Sci. Transl. Med. 16, eadi2245 (2024).
NCT05371613. A Study to Determine the Efficacy and Safety of Tividenofusp Alfa (DNL310) vs Idursulfase in Pediatric Participants With Neuronopathic (nMPS II) or Non-Neuronopathic Mucopolysaccharidosis Type II (nnMPS II) (COMPASS). https://clinicaltrials.gov/study/NCT05371613#publications (2022).
Mai, H. et al. Whole-body cellular mapping in mouse using standard IgG antibodies. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01846-0 (2023).
Cai, R. et al. Whole-mouse clearing and imaging at the cellular level with vDISCO. Nat. Protoc. 18, 1197–1242 (2023).
Cai, R. et al. Panoptic imaging of transparent mice reveals whole-body neuronal projections and skull-meninges connections. Nat. Neurosci. 22, 317–327 (2019).
Schlothauer, T. et al. Novel human IgG1 and IgG4 Fc-engineered antibodies with completely abolished immune effector functions. Protein Eng. Des. Sel. 29, 457–466 (2016).
Kumar, N. N. et al. Relative vascular permeability and vascularity across different regions of the rat nasal mucosa: implications for nasal physiology and drug delivery. Sci. Rep. 6, 31732 (2016).
Lochhead, J. J. & Thorne, R. G. Intranasal delivery of biologics to the central nervous system. Adv. Drug Deliv. Rev. 64, 614–628 (2012).
Chen, K. et al. Resolving the distinct stages in erythroid differentiation based on dynamic changes in membrane protein expression during erythropoiesis. Proc. Natl Acad. Sci. USA 106, 17413–17418 (2009).
Marsee, D. K., Pinkus, G. S. & Yu, H. CD71 (transferrin receptor): an effective marker for erythroid precursors in bone marrow biopsy specimens. Am. J. Clin. Pathol. 134, 429–435 (2010).
Sato, H. et al. Distribution of cystine/glutamate exchange transporter, system x(c)-, in the mouse brain. J. Neurosci. 22, 8028–8033 (2002).
Kageyama, T., Imura, T., Matsuo, A., Minato, N. & Shimohama, S. Distribution of the 4F2 light chain, LAT1, in the mouse brain. Neuroreport 11, 3663–3666 (2000).
Nakamura, E. et al. 4F2 (CD98) heavy chain is associated covalently with an amino acid transporter and controls intracellular trafficking and membrane topology of 4F2 heterodimer. J. Biol. Chem. 274, 3009–3016 (1999).
Jefferies, W. A. et al. Transferrin receptor on endothelium of brain capillaries. Nature 312, 162–163 (1984).
Moos, T. Immunohistochemical localization of intraneuronal transferrin receptor immunoreactivity in the adult mouse central nervous system. J. Comp. Neurol. 375, 675–692 (1996).
Koh, L., Zakharov, A. & Johnston, M. Integration of the subarachnoid space and lymphatics: is it time to embrace a new concept of cerebrospinal fluid absorption? Cerebrospinal Fluid Res. 2, 6 (2005).
Spera, I. et al. Open pathways for cerebrospinal fluid outflow at the cribriform plate along the olfactory nerves. EBioMedicine 91, 104558 (2023).
Jacob, L. et al. Conserved meningeal lymphatic drainage circuits in mice and humans. J. Exp. Med. 219, e20220035 (2022).
Weller, R. O. et al. Cerebral amyloid angiopathy: amyloid beta accumulates in putative interstitial fluid drainage pathways in Alzheimer’s disease. Am. J. Pathol. 153, 725–733 (1998).
Keable, A. et al. Deposition of amyloid β in the walls of human leptomeningeal arteries in relation to perivascular drainage pathways in cerebral amyloid angiopathy. Biochim. Biophys. Acta 1862, 1037–1046 (2016).
Greenberg, S. M. et al. Cerebral amyloid angiopathy and Alzheimer disease—one peptide, two pathways. Nat. Rev. Neurol. 16, 30–42 (2020).
Pietilä, R. et al. Molecular anatomy of adult mouse leptomeninges. Neuron 111, 3745–3764.e7 (2023).
Vanlandewijck, M. et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480 (2018).
Yang, A. C. et al. A human brain vascular atlas reveals diverse mediators of Alzheimer’s risk. Nature 603, 885–892 (2022).
Chen, M. B. et al. Brain endothelial cells are exquisite sensors of age-related circulatory cues. Cell Rep. 30, 4418–4432.e4 (2020).
Jeong, H.-W. et al. Single-cell transcriptomics reveals functionally specialized vascular endothelium in brain. Elife 11, e57520 (2022).
Todorov, M. I. et al. Machine learning analysis of whole mouse brain vasculature. Nat. Methods 17, 442–449 (2020).
Kucharz, K. et al. Post-capillary venules are the key locus for transcytosis-mediated brain delivery of therapeutic nanoparticles. Nat. Commun. 12, 4121 (2021).
Morris, C. M., Candy, J. M., Bloxham, C. A. & Edwardson, J. A. Distribution of transferrin receptors in relation to cytochrome oxidase activity in the human spinal cord, lower brainstem and cerebellum. J. Neurol. Sci. 111, 158–172 (1992).
Wolak, D. J. & Thorne, R. G. Diffusion of macromolecules in the brain: implications for drug delivery. Mol. Pharm. 10, 1492–1504 (2013).
Mintun, M. A. et al. Donanemab in early Alzheimer’s disease. N. Engl. J. Med. 384, 1691–1704 (2021).
Pizzo, M. E. et al. Engineering anti-amyloid antibodies with transferrin receptor targeting improves safety and brain biodistribution. bioRxiv https://doi.org/10.1101/2024.07.26.604664 (2024).
Kulic, L. et al. Latest interim results from the BrainshuttleTMAD study, a phase Ib/IIa study of Trontinemab in people with Alzheimer’s disease. Preprint at https://medically.roche.com/content/dam/pdmahub/restricted/neurology/ctad-2024/CTAD-2024-presentation-kulic-latest-interim-results-from-brainshuttle-ad-study.pdf (2024).
Withington, C. G. & Turner, R. S. Amyloid-related imaging abnormalities with anti-amyloid antibodies for the treatment of dementia due to Alzheimer’s disease. Front. Neurol. 13, 862369 (2022).
Solopova, E. et al. Fatal iatrogenic cerebral β-amyloid-related arteritis in a woman treated with lecanemab for Alzheimer’s disease. Nat. Commun. 14, 8220 (2023).
Leahy, D. J., Rule, G. S., Whittaker, M. M. & McConnell, H. M. Sequences of 12 monoclonal anti-dinitrophenyl spin-label antibodies for NMR studies. Proc. Natl Acad. Sci. USA 85, 3661–3665 (1988).
Yousef, H., Czupalla, C. J., Lee, D., Butcher, E. C. & Wyss-Coray, T. Papain-based single cell isolation of primary murine brain endothelial cells using flow cytometry. Bio Protoc. 8, e3091 (2018).
Zeisel, A. et al. Molecular architecture of the mouse nervous system. Cell 174, 999–1014.e22 (2018).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021).
Germain, P.-L., Lun, A., Garcia Meixide, C., Macnair, W. & Robinson, M. D. Doublet identification in single-cell sequencing data using scDblFinder. F1000Res 10, 979 (2021).
McCormick, M., Liu, X., Jomier, J., Marion, C. & Ibanez, L. ITK: enabling reproducible research and open science. Front. Neuroinform. 8, 13 (2014).
Wang, Q. et al. The Allen Mouse Brain Common Coordinate Framework: a 3D reference atlas. Cell 181, 936–953.e20 (2020).
Ntatsis, K. et al. itk-elastix: Medical image registration in Python. 101–105 https://doi.org/10.25080/gerudo-f2bc6f59-00d (2023).
Ahlers, J. et al. napari: a multi-dimensional image viewer for Python. zenodo (2023).
Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020).
Lee, T. C. et al. Building skeleton models via 3-D medial surface axis thinning algorithms. CVGIP Graph. Models Image Process. 56, 462–478 (1994).
Felzenszwalb, P. F. & Huttenlocher, D. P. Distance transforms of sampled functions. Theory Comput. 8, 415–428 (2012).
Zhong, T. et al. Longitudinal brain atlases of early developing cynomolgus macaques from birth to 48 months of age. Neuroimage 247, 118799 (2022).
Tustison, N. J. et al. The ANTsX ecosystem for quantitative biological and medical imaging. Sci. Rep. 11, 9068 (2021).
Acknowledgements
The authors would like to thank Rene Meisner for her input on the anatomical interpretation of whole-body tissue clearing imaging results; Shan V. Andrews, Alexander Schuth, Mitesh Shridhar, and Ryan Watts for providing helpful comments on the manuscript, Butch Benitez, Kevin Rebadulla, and Dominic Sobrepena for their help with mice care at Denali Therapeutics.
Author information
Authors and Affiliations
Contributions
Conceptualization: N.K., M.E.P., R.T., M.E.K.C., J.L., A.E., and Y.J.Y.Z. Data generation and or analysis: N.K., M.E.P., C.B.D., D.J., D.T., M.I.T., M.N., C.H., G.L.D.M., L.S., M.J.S., D.C., R.C., K.C., J.C., A.C., Y.R.C., J.C.D., J.D., D.K., H.K., A.L., E.L., A.M., E.R., T.S., M.T., K.X., Y.Z., and M.E.K.C. Writing and main edits: N.K., M.E.P., R.G.T., M.E.K.C., and Y.J.Y.Z. Supervision: J.L., A.E., R.G.T., M.E.K.C., and Y.J.Y.Z.
Corresponding authors
Ethics declarations
Competing interests
N.K., M.E.P., C.B.D., D.J., D.T., C.H., G.L.D., L.S., M.J.S., D.C., R.C., J.C., A.C., Y.R.C., J.C.D., J.D., H.K., A.L., E.L., A.M., E.R., T.S., M.T., K.X., Y.Z., J.L., R.G.T., M.E.K.C., and Y.J.Y.Z. are currently or were previously paid employees of Denali Therapeutics Inc. M.I.T., M.N., and D.K. are paid employees of Deep Piction and A.E. is the chief executive officer of Deep Piction.
Peer review
Peer review information
Nature Communications thanks Dag Sehlin, Peter Tessier and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Khoury, N., Pizzo, M.E., Discenza, C.B. et al. Fc-engineered large molecules targeting blood-brain barrier transferrin receptor and CD98hc have distinct central nervous system and peripheral biodistribution. Nat Commun 16, 1822 (2025). https://doi.org/10.1038/s41467-025-57108-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-57108-x
This article is cited by
-
Bridging the blood-brain barrier: strategies to improve delivery of biologics to tumors in the brain
Fluids and Barriers of the CNS (2026)
-
Complement in brain and eye disease: shared mechanisms, convergent pathologies, and common therapeutic opportunities
Mammalian Genome (2026)
-
Transferrin receptor-binding blood-brain barrier shuttle enhances brain delivery and plaque-clearing efficacy of a therapeutic anti-Aβ antibody
Fluids and Barriers of the CNS (2025)
-
Brain drugs can now cross the once impenetrable blood–brain barrier
Nature (2025)
-
Disruption of the cerebrospinal fluid–plasma protein balance in cognitive impairment and aging
Nature Medicine (2025)