Abstract
Dysfunctional NF-κB signaling is critically involved in inflammatory bowel disease (IBD). We investigated the mechanism by which RIPK1 and TRADD, two key mediators of NF-κB signaling, in mediating intestinal pathology using TAK1 IEC deficient model. We show that phosphorylation of TRADD by TAK1 modulates RIPK1-dependent apoptosis. TRADD and RIPK1 act cooperatively to mediate cell death regulated by TNF and TLR signaling. We demonstrate the pathological evolution from RIPK1-dependent ileitis to RIPK1- and TRADD-co-dependent colitis in TAK1 IEC deficient condition. Combined RIPK1 inhibition and TRADD knockout completely protect against intestinal pathology and lethality in TAK1 IEC KO mice. Furthermore, we identify distinctive microbiota dysbiosis biomarkers for RIPK1-dependent ileitis and TRADD-dependent colitis. These findings reveal the cooperation between RIPK1 and TRADD in mediating cell death and inflammation in IBD with NF-κB deficiency and suggest the possibility of combined inhibition of RIPK1 kinase and TRADD as a new therapeutic strategy for IBD.
Similar content being viewed by others
Introduction
Inflammatory bowel disease (IBD), comprising Crohn disease and ulcerative colitis, is complex chronic inflammatory disorders of the gastrointestinal tract1. Both genetic and environmental factors are implicated in the causation of IBD. The intestine epithelial cells (IECs) constitute a large barrier surface that not only protects mammalian hosts from the external environment by physical separation but also allows the colonization by commensal bacteria which, in turn, exert an important influence on the development and function of the mucosal immune system2. Dysregulated epithelial cell turnover and apoptosis is associated with intestinal inflammation, barrier breaches, and dysbiosis of microbial populations and function in IBD. A breakdown in the regulatory constraints of mucosal immune responses to enteric bacteria leads to an exaggerated inflammatory response which in turn promotes damage to IECs. The success of anti-TNF therapy underscores the importance of the TNF pathway in IBD3. However, about 10–30% of IBD patients are unresponsive to anti-TNF, and 40–50% of patients eventually lose response to anti-TNF therapy4. Understanding the mechanisms that underlie the pathological evolution in IBD is critical for us to develop new therapeutics for the treatment of this disease.
RIPK1 and TRADD are two death domain-containing signaling mediators critically involved in mediating TNF and TLR signaling pathways, which are known to be involved in IBD5,6. Both TRADD and RIPK1 are quickly recruited to the intracellular death domain of TNFR1 upon TNF stimulation to form complex I. TRADD is involved in organizing the ubiquitination in complex I to recruit the key kinase TAK1 and its adapter proteins TAB1/2 to phosphorylate IKKs, which in turn activate the NF-κB pathway7. TAK1 is also implicated as a regulator of RIPK1 by direct phosphorylation8. TAK1 inhibition sensitizes cells to RIPK1 kinase-dependent apoptosis (RDA) upon TNF or LPS stimulation8,9,10. Blocking NF-κB pathway in cells stimulated by TNF can promote RIPK1-independent apoptosis (RIA) which can be blocked by TRADD deficiency11, but not by inhibition of RIPK1 kinase. The relationship between RDA and RIA, both of which can be activated by TNF, is unclear.
NF-κB pathway is the core regulator of the immune response involved in the dysregulated inflammatory response in IBD12. GWAS analysis has identified TAB1/2 as IBD-related genes13,14. Mice with TAB1/2 double knockout, specifically in IECs, develop spontaneous intestine pathology, which resembles TAK1 IEC-specific knockout mice15. Enterocyte-specific deletion of TAK1 promote severe TNF dependent intestinal pathology in newborn mice but TNF independent intestinal pathology in adult mice16,17. Thus, TAK1 IEC KO mice might provide an interesting model to investigate TNFR1 dependent and independent mechanisms in IBD.
Here, we demonstrate that TRADD and RIPK1 act cooperatively in mediating pathological progression in IECs under TAK1 deficient conditions. TAK1 directly modulates the activation of TRADD by phosphorylation. TRADD cooperates with RIPK1 kinase to promote ileitis and colitis in both neonatal and adult TAK1 IEC-specific knockout mice. We further reveal that gut microbiota contributes to TRADD-dependent caspase-8 activation in adult mice, and the contribution of RIPK1 and TRADD-dependent cell death and inflammation to microbiome dysbiosis. In sum, our work uncovers the cooperative effect and pathological progression from RIPK1-dependent to TRADD and RIPK1 co-dependent pathology in this IBD model.
Results
RIPK1 kinase inhibition protects early-stage ileitis and colitis in newborn Tak1 IEC-KO mice
To investigate the role of RIPK1 kinase in IBD using newborn Tak1IEC-KO mice (Tak1fl/fl;Villincre/+), we first generated Tak1IEC-KO Ripk1D138N/D138N mice which carried a kinase-dead knock-in substitution mutation D138N in the endogenous RIPK1 locus18. Newborn Tak1IEC-KO mice spontaneously developed severe ileitis and colitis, manifesting with epithelial erosion and increased neutrophil infiltration (Fig. 1a, b and Supplementary Fig. 1a, b). Inhibition of RIPK1 kinase in newborn Tak1IEC-KO Ripk1D138N/D138N mice at P0 displayed a normal intestine length and architecture without inflammation in both ileum and colon (Fig. 1a–c and Supplementary Fig. 1a, b). Increased mRNA levels of proinflammatory cytokines and chemokines, such as Ccl2, Ccl5, Cxcl1, Cxcl2, and Il-1β, were detected in the ileum, but not in the colon, of Tak1IEC-KO mice at P0, which was inhibited by Ripk1D138N/D138N (Supplementary Fig. 1c). Increased caspase-8 activation and IEC apoptosis, indicated by cleaved caspase-8(CC8) and caspase-3(CC3) staining, were detected in both ileum and colon of newborn Tak1IEC-KO mice at P0, which were blocked by the inactivation of RIPK1 kinase (Fig. 1a, b and Supplementary Fig. 1a, b). The loss of goblet cells, showed by AB-PAS staining, in the ileum and colon of Tak1IEC-KO mice at P0, was rescued by inhibition of RIPK1 in Tak1IEC-KO Ripk1D138N/D138N mice (Supplementary Fig. 1a, b). These results revealed the critical role of RIPK1 kinase in mediating ileitis and colitis in newborn mice with TAK1 deficiency.
a, b Representative images of ileum and colon sections from mice at P0 with indicated genotypes stained with H&E or immunostained for CC3 (a). Graphs showed the CC3+ signals in the ileum and colon of mice with the indicated genotypes (b). Scale bar, 100 μm. c Intestine length in mice with indicated genotypes at P0. d Kaplan–Meier survival curves of the indicated genotypes. e–h Representative images of the ileum (e) and colon (g) sections from mice at P4 with indicated genotypes stained with H&E or immunostained for CC3. Graphs showed the CC3+ signals in the ileum (f) and colon (h) of mice with indicated genotypes. Scale bar, 100 μm. i Intestine length in mice with indicated genotypes at P4. j The scheme of tamoxifen-induced TAK1 deletion. Created in BioRender. Yuan, J. (2025) https://BioRender.com/b69p319. k Kaplan–Meier survival curves of the mice with indicated genotypes. l Relative body weight curve of the mice with indicated genotypes. The body weight of each mouse was normalized to the weight of the day 0. m Intestine length of the mice at 3 dpi with indicated genotypes. n, o Representative images of ileum and colon sections from the mice at 3 dpi with indicated genotypes stained with H&E or immunostained for CC3 (n). Graphs showed the CC3+ signals in the ileum and colon of the mice with indicated genotypes (o). Scale bar, 150 μm for ileum; 100 μm for colon; p–s Representative images of the ileum (p) and colon (r) sections from the mice at 7 dpi with indicated genotypes stained with H&E or immunostained for CC3. Graphs showed the CC3+ signals in the ileum (q) and colon (s) of the mice with indicated genotypes. Anti-TNFα was treated in the indicated group from 1 day before tamoxifen injection every other day for a total of four times until sacrificed. Scale bar, 150 μm. Each dot represented one mouse. The results were shown as mean ± SEM. An unpaired two-tailed t-test was performed.
Inhibition of RIPK1 in Tak1IEC-KO Ripk1D138N/D138N mice increased the survival rate of Tak1IEC-KO mice (Fig. 1d). At P4, the ileum of Tak1IEC-KO Ripk1D138N/D138N mice was morphologically indistinguishable from that of Tak1fl/fl Ripk1D138N/D138N mice in the intestine architecture (Fig. 1e), although the evidence of inflammation, including low level increases in mRNA levels of Cxcl1, Cxcl2, Ccl2, Il-1β, and Tnfα, low levels of activated caspase-3 as well as infiltration of neutrophils in low numbers, were present in the ileum of P4 Tak1IEC-KO Ripk1D138N/D138N mice (Fig. 1e, f and Supplementary Fig. 1d–f). In contrast, Tak1IEC-KO Ripk1D138N/D138N mice at P4 began to demonstrate morphological evidence of colitis with epithelial erosion and increased neutrophil infiltration, although the intestinal length of Tak1IEC-KO Ripk1D138N/D138N mice was comparable with Tak1fl/fl Ripk1D138N/D138N mice (Fig. 1g–i and Supplementary Fig. 1g, h). Increased caspase-8 activation and apoptosis of IECs, as well as the loss of goblet cells, were also detected in the colon of Tak1IEC-KO Ripk1D138N/D138N mice (Fig. 1g, h and Supplementary Fig. 1g, h). The mRNA levels of proinflammatory cytokines, such as Cxcl2 and Tnfa, began to show elevation, with a few reaching high levels, in the colon of Tak1IEC-KO Ripk1D138N/D138N mice on P4 (Supplementary Fig. 1f). These results suggest that in newborn Tak1IEC-KO mice, RIPK1 kinase plays an important role in promoting ileitis and early-stage colitis.
Inhibition of RIPK1 kinase protects ileitis and early-stage colitis in adult Tak1 tamIEC-KO mice
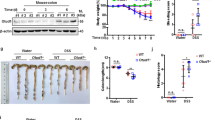
To investigate the role of TAK1 in maintaining intestinal homeostasis in adults, we generated mice with tamoxifen-inducible ablation of TAK1 in IECs by crossing Tak1fl/fl mice with Villin-creERT2 mice (hereafter referred to as Tak1tamIEC-KO). Adult Tak1tamIEC-KO mice were treated with tamoxifen for 3 consecutive days to induce TAK1 deletion at the age of 7–8 weeks (Fig. 1j). Rapid weight loss, diarrhea, and death were observed within 5 days after tamoxifen-induced TAK1 deletion (Fig. 1k, l). Inhibition of RIPK1 kinase in adult Tak1tamIEC-KO Ripk1D138N/D138N mice provided partial protection, including delayed death of Tak1tamIEC-KO mice to 9 days post-induction (dpi) (Fig. 1k), normal weight until 4 dpi (Fig. 1l), the rescue of intestine length to 3 dpi (Fig. 1m). At 3 dpi, both ileum and colon of adult Tak1tamIEC-KO Ripk1D138N/D138N mice exhibited normal architecture without infiltration of neutrophils, the activation of caspase-3/-8, or loss of goblet cells and Paneth cells (Fig. 1n, o and Supplementary Fig. 2a, b). RNA-SEQ analysis of ileum at 3 dpi showed that 1395 genes were upregulated, of which 1087 genes were rescued by RIPK1 kinase inhibition; while 2132 genes were downregulated, of which 1725 genes were rescued by RIPK1 kinase inhibition (Supplementary Fig. 2c, d). In addition, RNA-SEQ analysis of the colon showed that 678 genes were upregulated, of which 532 genes were rescued by RIPK1 kinase inhibition; while 1178 genes were downregulated, of which 763 genes were rescued by RIPK1 kinase inhibition (Supplementary Fig. 2e, f). GO analysis using these genes upregulated in Tak1tamIEC-KO mice and rescued by Ripk1D138N/D138N revealed RIPK1-dependent upregulation of Type-I and Type-II interferon pathways in both ileum and colon at 3 dpi (Supplementary Fig. 2g, h). Thus, the activation of RIPK1 kinase plays an important role in dysregulated innate immune response to promote ileitis and early-stage colitis in adult Tak1tamIEC-KO mice.
Tak1tamIEC-KO Ripk1D138N/D138N mice began to lose weight 5 days after tamoxifen-induced TAK1 deletion (Fig. 1l). Interestingly, the ileum of Tak1tamIEC-KO Ripk1D138N/D138N mice at 7 dpi showed no obvious signs of pathology with normal numbers of Paneth cells and goblet cells, no apoptosis of IECs, or aberrant neutrophil infiltration (Fig. 1p, q and Supplementary Fig. 2i, j). Thus, inhibition of RIPK1 kinase alone was sufficient to block ileitis, at least up to dip7, in adult mice after TAK1 IEC deletion. In contrast, at 7 dpi, Tak1tamIEC-KO Ripk1D138N/D138N mice began to demonstrate severe colitis manifested with epithelial erosion, caspase activation, and neutrophil infiltration with almost total loss of Goblet cells (Fig. 1r, s and Supplementary Fig. 2k, l). Thus, RIPK1 inactivation alone was insufficient to fully protect colitis in adult Tak1tamIEC-KO mice beyond 5 dpi.
We noted that mRNA levels of Tnfα were significantly increased in the colon of Tak1tamIEC-KO Ripk1D138N/D138N mice at 7 dpi (Supplementary Fig. 2m). Blocking TNF pathway via TNF neutralization antibody restored the colonic architecture in Tak1tamIEC-KO Ripk1D138N/D138N mice at 7 dpi (Fig. 1r). The number of goblet cells in the colon was partially, but not fully rescued by TNF neutralization (Supplementary Fig. 2k, l). TNF neutralization also attenuated inflammation indicated by reduced infiltration of Ly-6G+ neutrophils (Supplementary Fig. 2k, l). However, the activation of caspase-8 and caspase-3 in Tak1tamIEC-KO Ripk1D138N/D138N mice was not inhibited by TNF neutralization (Fig. 1r, s and Supplementary Fig. 2k, l). Collectively, these results suggest that TNF is a contributing, but not the only factor that mediates the RIPK1-independent colitis.
These results suggest that in adult Tak1tamIEC-KO mice, RIPK1 may play a dominant role in mediating intestinal pathology in the ileum and early-stage colitis, while additional factor(s) may be activated in the colon to mediate colitis in advanced disease stage. These observations raise interesting questions about the mechanisms that mediate the evolution of inflammatory processes during IBD and the potential difference(s) in the key pathological mediators for ileitis and colitis.
Combined loss of FADD and MLKL prevents IECs death in Tak1 IEC-KO mice
Since caspase-8 is a key downstream mediator of RIPK1-dependent apoptosis8, we next tested the role of caspase-8 by knocking out FADD, the adapter mediating the activation of caspase-819. Since FADD knockout promotes necroptosis, we introduced the MLKL knockout allele by generating Tak1IEC-KO Fadd−/− Mlkl−/− triple knockout (TKO) mice. Interestingly, 90% of Tak1IEC-KO Fadd−/− Mlkl−/− TKO mice survived normally without weight loss up to 50 days when sacrificed as Fadd−/− Mlkl−/− mice develop autoimmunity by the age of ~7 weeks20 (Fig. 2a–c). Notably, Tak1IEC-KO Fadd−/− Mlkl−/− TKO mice did not develop ileitis or colitis as that of Tak1IEC-KO Ripk1D138N/D138N mice at P4 (Fig. 2d, e and Supplementary Fig. 3a, b). Tak1IEC-KO Fadd−/− Mlkl−/− TKO mice had a similar number of goblet cells as that of control Tak1fl/fl Fadd−/− Mlkl−/− mice and without caspase activation or neutrophil infiltration in both ileum and colon (Fig. 2d, e and Supplementary Fig. 3a, b).
a Kaplan–Meier survival curves of the mice with indicated genotypes. b, c Body weight curve of the mice with indicated genotypes. d, e, g, h Representative images of ileum and colon sections from mice at P4 (d) or P0 (g) with indicated genotypes stained with H&E or immunostained for CC3. Graphs showed the CC3+ signals in the ileum and colon of the mice with indicated genotypes at P4 (e) or P0 (h). Scale bar, 100 μm. f Intestine length in the mice with indicated genotypes at P0. i Kaplan–Meier survival curves of the mice with indicated genotypes. j Relative body weight curve of the mice with indicated genotypes. The body weight of each mouse was normalized to the weight of day 0. k, l Representative images of ileum and colon sections from mice at 3 dpi and 7 dpi of the indicated genotypes stained with H&E or immunostained for CC3 (k). Graphs showed the CC3+ signals in the ileum and colon of mice with the indicated genotypes (l). Scale bar, 150 μm for ileum; 100 μm for colon; m Intestine length of the mice at 3 dpi with indicated genotypes. Each dot represented one mouse. The results were shown as mean ± SEM. An unpaired two-tailed t-test was performed.
MLKL deficiency alone was not sufficient to protect the lethality of Tak1IEC-KO mice (Fig. 2a). The shortened length of intestine in Tak1IEC-KO mice was normalized by double knockout of FADD and MLKL, but not MLKL knockout alone (Fig. 2f). Tak1IEC-KO Mlkl−/− mice had increased neutrophil infiltration in both ileum and colon, which was inhibited by FADD deletion (Supplementary Fig. 3c, d). Increased mRNA levels of Ccl2, Ccl5, Cxcl1, Cxcl2, and Tnfα were also detected in the ileum of Tak1IEC-KO Mlkl−/− mice, but not that of Tak1IEC-KO Fadd−/− Mlkl−/− TKO mice (Supplementary Fig. 3e). MLKL deficiency did not inhibit caspase-8 activation and IEC apoptosis in either ileum or colon (Fig. 2g, h and Supplementary Fig. 3c, d). Tak1IEC-KO Mlkl−/− mice still showed obvious loss of goblet cells in both the ileum and colon (Supplementary Fig. 3c, d).
Above data suggest FADD-caspase-8 mediated apoptosis, but not MLKL mediated necroptosis, is directly involved in mediating lethality and intestine pathology of newborn Tak1IEC-KO mice.
Combined loss of FADD and MLKL prevents intestinal pathology in adult Tak1 tamIEC-KO mice
Next, we tested the role of caspase-8 in adult Tak1tamIEC-KO mice. Tak1tamIEC-KO Fadd−/− Mlkl−/− mice, but not Tak1tamIEC-KO Mlkl−/− or Tak1tamIEC-KO Fadd+/− Mlkl−/− mice, survived to 21 dpi when the mice were sacrificed, with normal weight after TAK1 deletion induced by tamoxifen injection (Fig. 2i, j). Unlike Tak1tamIEC-KO Ripk1D138N/D138N mice, Tak1tamIEC-KO Fadd−/− Mlkl−/− mice displayed no signs of colitis when examined at 7 dpi (Fig. 2k, l and Supplementary Fig. 3f, g). The intestine of Tak1tamIEC-KO Fadd−/− Mlkl−/− TKO mice did not show increased neutrophil infiltration, caspase-3/8 activation, or loss of goblet cells and Paneth cells at 7 dpi (Fig. 2k, l and Supplementary Fig. 3f, g).
Heterozygosity of FADD was not sufficient to protect TAK1 IEC deficiency as Tak1tamIEC-KO Fadd+/− Mlkl−/− mice still developed severe ileitis and colitis including intestinal architecture damage (Fig. 2k, l and Supplementary Fig. 3f–h). Tak1tamIEC-KO Fadd+/− Mlkl−/− mice showed reduced body weight and shortened intestine length, and developed diarrhea (Fig. 2j, m). At 3 dpi, the ileum and colon of Tak1tamIEC-KO Fadd+/− Mlkl−/− mice had obvious epithelial erosion, activated caspase-8, apoptosis, and loss of goblet cells (Fig. 2k, l and Supplementary Fig. 3f, g). Aberrant inflammation demonstrated by increased neutrophil infiltration as well as increased mRNA levels of Ccl2, Cxcl2, and Il-1α was found in both the ileum and colon of Tak1tamIEC-KO Fadd+/− Mlkl−/− mice (Supplementary Fig. 3f–h).
Thus, similar to that of newborn TAK1 IEC-deficient mice, FADD/caspase-8 mediated apoptosis, rather than MLKL mediated necroptosis, is directly involved in driving the intestine pathology and lethality in adult mice with TAK1 IEC deficiency.
Cooperation of TRADD and RIPK1 in mediating intestinal pathology of Tak1 IEC-KO mice
TRADD, a DD-containing adapter protein, is directly involved in mediating caspase-8 activation in the TNFR1 signaling pathway21. We next examined the role of TRADD by generating Tak1IEC-KO TraddIEC-KO mice. Similar to that of RIPK1 inactivation with D138N mutation (Fig. 1d), TRADD-IEC knockout alone could also increase the survival of Tak1IEC-KO mice with normal length of intestine to 30% (Fig. 3a, b). TRADD-IEC knockout also prevented the intestine pathology of Tak1IEC-KO mice on P0, without epithelial erosion, loss of goblet cells, or the activation of caspase-8 and caspase-3 in both ileum and colon (Fig. 3c, d and Supplementary Fig. 4a, b). The numbers of neutrophils and mRNA levels of Ccl2, Cxcl1, Cxcl2, and Il-1β in the intestine of Tak1IEC-KO TraddIEC-KO mice were restored to that of the levels in control mice (Supplementary Fig. 4a–c). Collectively, these data identified that TRADD, as that of RIPK1 kinase, was also a critical factor in mediating the intestine pathology of newborn Tak1IEC-KO mice.
a Kaplan–Meier survival curves of the mice with indicated genotypes. b Intestine length in the mice with indicated genotypes at P0. c–h Representative images of ileum and colon sections from the mice at P0 (c) or P4 (e for ileum, g for colon) with indicated genotypes stained with H&E or immunostained for CC3. Graphs showed the CC3+ signals in the ileum and colon of the mice with indicated genotypes at P0 (d) or P4 (f for ileum, h for colon). i, j Body weight curve of the mice with indicated genotypes. Scale bar, 100 μm. Each dot represented one mouse. The results were shown as mean ± SEM. An unpaired two-tailed t-test was performed. k, l Tradd+/+ and Tradd−/− ileum (k) and colon (l) organoids were treated with 500 nM (5Z)-7-oxozeaeno and 20 ng/ml mTNF-α for 24 h (k) or 12 h (l) with or without 50 μM Nec-1s. The cell death in organoids was determined by PI staining. Scale bar, 50 μm. Each dot represented one organoid. The results were shown as mean ± SEM. An unpaired two-tailed t-test was performed. (m) Tradd+/+ and Tradd−/− MEFs were pretreated with 500 nM (5Z)-7-oxozeaeno, +/− Nec-1s (20 μM), +/− zVAD (50 μM) for 0.5 hr followed by 20 ng/ml mTNF-α treatment. The complex-II was isolated by FADD immunoprecipitated and detected by immunoblotting. n Phos-tag SDS-PAGE analysis of mTNF-α (100 ng/ml) stimulated WT MEFs with or without (5Z)-7-oxozeaeno (500 nM) pretreatment for 0.5 h. o HEK293T cells were co-transfected with expression vectors for Flag-mTRADD with different mutations and +/−HA-mFADD for 24 h. The cell lysates were then immunoprecipitated using anti-Flag resin. The binding of TRADD and FADD was analyzed by immunoblotting. p, q Tradd−/− MEFs were retrovirally reconstituted with HA-tagged TRADD with different mutations. Reconstituted cells were stimulated with 500 nM (5Z)-7-oxozeaeno for 0.5 h followed by 20 ng/ml mTNF-α. Cell death was measured by SytoxGreen, and the mean ± SEM from eight biological replicates was shown (p). The levels of Caspase-8, CC8, Caspase-3, CC3, RIPK1, p-RIPK1(S166), and HA were determined by immunoblotting (q).
Similar to that of Tak1IEC-KO Ripk1D138N/D138N mice, the ileum of Tak1IEC-KO TraddIEC-KO mice at P4 had normal architecture with a similar number of goblet cells as that of control mice, although increased caspase activation and neutrophil infiltration could be detected (Fig. 3e, f and Supplementary Fig. 4d, e). Tak1IEC-KO TraddIEC-KO mice at P4 also developed colitis with severe epithelial erosion, activation of caspase-3/-8, dramatic goblet cell loss, and increased neutrophil infiltration (Fig. 3g, h and Supplementary Fig. 4f, g).
Since these results suggest that both TRADD and RIPK1 kinase may be involved in mediating ileitis and colitis in newborn Tak1IEC-KO mice, we next generated Tak1IEC-KO TraddIEC-KO Ripk1D138N/D138N mice to investigate the interaction of RIPK1 and TRADD. Interestingly, all Tak1IEC-KO TraddIEC-KO Ripk1D138N/D138N mice survived with normal body weight compared to that of littermate control up to P60 when sacrificed (Fig. 3a, i, j). The ileum and colon of Tak1IEC-KO TraddIEC-KO Ripk1D138N/D138N mice displayed normal tissue architecture without evidence of cleaved caspase-8 or apoptosis, in contrast to that of Tak1IEC-KO Ripk1D138N/D138N mice and Tak1IEC-KO TraddIEC-KO mice (Fig. 3e–h and Supplementary Fig. 4d–g). The intestine of Tak1IEC-KO TraddIEC-KO Ripk1D138N/D138N mice displayed no evidence of inflammation, with comparable numbers of neutrophils (Supplementary Fig. 4d–g). The numbers of goblet cells were normal in both the ileum and colon of Tak1IEC-KO TraddIEC-KO Ripk1D138N/D138N mice (Supplementary Fig. 4d–g). In addition, Ripk1D138N/+ or TraddhetIEC-KO (Traddfl+ Villin-creTg/+) mice also increased survival rate from ~20–30% to ~60% compared to that of Tak1IEC-KO TraddIEC-KO or Tak1IEC-KO Ripk1D138N/D138N mice, respectively (Fig. 3a). Deletion of TAK1 in IECs of Tak1IEC-KO TraddhetIEC-KO Ripk1D138N/D138N mice, Tak1IEC-KO TraddIEC-KO Ripk1D138N/+ mice as well as Tak1IEC-KO TraddIEC-KO Ripk1D138N/D138N mice was confirmed (Supplementary Fig. 4h).
Thus, RIPK1 kinase and TRADD act cooperatively to mediate the intestine pathology and lethality of Tak1IEC-KO mice.
TAK1 directly phosphorylates TRADD to inhibit cell death in the TNF pathway
Since the data above demonstrate the cooperation of RIPK1 and TRADD in mediating the intestine pathology and lethality of Tak1IEC-KO mice, we next examined the interaction of TRADD and RIPK1 in the TNF pathway using organoid and cell models. We found that TRADD deficiency and RIPK1 kinase inhibitor R-7-Cl-O-necrostatin-1 (Nec-1s)22 showed an additive effect in protecting the death of intestinal organoids induced by TNF/5Z7 (5Z-7-Oxozeaenol, TAK1 inhibitor), while each alone had a partial protection (Fig. 3k, l). Similar results were obtained using MEFs (Supplementary Fig. 5a, b). TRADD deficiency alone could partially reduce the activation of RIPK1(p-S166), the cleavage of caspase-8, caspase-3, and RIPK1; while treatment with RIPK1 kinase inhibitor Nec-1s alone or RIPK1 kinase-dead mutation alone could inhibit RIPK1 activation and partially reduce the cleavage of caspase-8 and caspase-3 in MEFs induced with TNF/5Z7 (Supplementary Fig. 5c, d). Combined TRADD deficiency and RIPK1 kinase inhibition could completely prevent the activation of caspase-8 and caspase-3 (Supplementary Fig. 5c, d). In addition, TRADD deficiency could reduce the binding of FADD with RIPK1 and caspase-8 in the formation of complex IIa in MEFs treated with TNF/5Z7 in RDA (Fig. 3m). However, TRADD deficiency could not prevent the necroptosis or the binding of FADD with caspase-8 and RIPK3 in the formation of complex IIb upon TNF/5Z7/zVAD (zVAD-fmk, pan-caspase inhibitor) treatment in MEFs (Fig. 3m and Supplementary Fig. 5e).
Next, we investigated the mechanism by which TAK1 regulated TRADD. Since TAK1 could regulate the activation of RIPK1 by phosphorylation in cells stimulated by TNF8, we examined the possibility that TRADD might also be phosphorylated by TAK1. We found that using phospho-tag gel, the phosphorylation of TRADD was detectable within 10 min when cells were stimulated by TNF, which was inhibited by TAK1 inhibitor 5Z7, implicating the role of TAK1 in the phosphorylation (Fig. 3n). We re-introduced Flag-mTRADD into Tradd−/− MEFs, and identified the phosphorylation sites of TRADD which were induced by TNF and inhibited by TAK1 inhibitor 5Z7 using mass spectrometry (Supplementary Fig. 5f). We found that TAK1 could mediate the phosphorylation of multiple sites in TRADD upon TNF stimulation (Supplementary Fig. 5g). We next performed in vitro kinase assay using recombinant human TAK1 and human TRADD, followed by mass spectrometry to identify the sites directly phosphorylated by TAK1 (Supplementary Fig. 5h, i). These experiments led us to identify S55, S67, S186, S249(murine)/S251(human), Y252(murine)/Y254(human) and Y254(murine)/Y256(human) as potential sites in TRADD that were directly phosphorylated by TAK1 (Supplementary Fig. 5g, i). Y252(murine)/Y254(human) and Y254(murine)/Y256(human) were highly conserved during evolution and located in the death domain of TRADD (Supplementary Fig. 5j). Phosphorylation mimetic mutation of Y252D, Y252E, Y254D and Y254E in murine TRADD reduced its binding with FADD (Fig. 3o). We next re-introduced phosphorylation mimetic mutation of TRADD into Tradd−/− MEF to assess the effect of TRADD phosphorylation on TNF/5Z7 induced cell death. We found that phosphorylation mimetic mutation of TRADD, including Y252D, Y252E, Y254D, and Y254E reduced TNF/5Z7 induced cell death, as well as the activation of RIPK1 and caspases (Fig. 3p, q). These results suggest that phosphorylation of TRADD by TAK1 inhibits the formation of complex IIa by reducing its binding with FADD.
Cooperation of TRADD and RIPK1 in mediating intestinal pathology of adult Tak1 tamIEC-KO mice
Similar to that of TAK1 IEC deficiency in neonatal mice, we found that combined RIPK1 inhibition and TRADD knockout fully protected the reduction in body weight and lethality of adult Tak1tamIEC-KO mice after induced TAK1 IEC deletion (Fig. 4a, b). Histological analysis of the colon from Tak1tamIEC-KO TraddtamIEC-KO Ripk1D138N/D138N mice revealed normal architecture with similar numbers of goblet cells without CC3 or CC8 as that of control Tak1f/f Ripk1D138N/D138N mice (Fig. 4c, d and Supplementary Fig. 6a, b). RNA-SEQ analysis of the colons showed that 4137 genes were upregulated in Tak1tamIEC-KO Ripk1D138N/D138N mice at 7 dpi, of which 3515 genes were rescued by additional TRADD-IEC deficiency; while 4414 genes were downregulated, of which 3578 genes were rescued by additional TRADD deficiency (Supplementary Fig. 6c). RNA-SEQ analysis demonstrated that combined knockout TRADD and inhibition of RIPK1 led to effective inhibition of inflammation response of Tak1tamIEC-KO mice (Fig. 4e, f). Consistently, the numbers of intestinal neutrophils in Tak1tamIEC-KO TraddtamIEC-KO Ripk1D138N/D138N mice were also restored to that of Tak1fl/fl Tradd fl/fl Ripk1D138N/D138N mice (Supplementary Fig. 6a, b). These results suggest that TRADD and RIPK1 kinase are both important for driving colitis in adult TAK1 IEC-deficient mice.
a Kaplan–Meier survival curves of the indicated genotypes. ABX was pretreated for 14 days before and through tamoxifen injection of the mice with indicated genotypes until sacrificed. Survival data for Tak1tamIEC-KO Ripk1D138N/D138N mice from Fig. 1k were included for comparison. b Relative body weight of the mice with indicated genotypes at 7 dpi. The body weight of each mouse was normalized to the weight of day 0. Body weight data for Tak1fl/fl Ripk1D138N/D138N and Tak1tamIEC-KO Ripk1D138N/D138N mice at 7 dpi from Fig. 1l were included for comparison. c, d Representative images of colon sections from the mice at 7 dpi with indicated genotypes stained with H&E or immunostained for CC3 (c). Graphs showed the CC3+ signals in the colon of the mice with indicated genotypes (d). CC3+ signals for Tak1fl/fl Ripk1D138N/D138N mice and Tak1tamIEC-KO Ripk1D138N/D138N mice from Fig. 1s were included for comparison. e PCA on RNA-seq data from the colon of the mice with indicated genotypes at 7 dpi. PCA was based on genes differentially expressed between Tak1fl/fl Ripk1D138N/D138N mice and Tak1tamIEC-KO Ripk1D138N/D138N mice (cut-off: log2|fold change | ≥1, p ≤ 0.05). f GO analysis of genes upregulated in the colon of Tak1tamIEC-KO Ripk1D138N/D138N mice in TRADD-dependent fashion. g The mRNA levels of inflammatory cytokines in the colon of the mice with indicated genotypes at 7 dpi were measured by qRT-PCR. Scale bar, 150 μm. Each dot represented one mouse. The results were shown as mean ± SEM. An unpaired two-tailed t-test was performed.
Notably, TRADD-IEC knockout alone was not sufficient to protect the loss of body weight and lethality in adult Tak1tamIEC-KO mice after tamoxifen-induced TAK1 deletion (Supplementary Fig. 6d, e). Tak1tamIEC-KO TraddtamIEC-KO mice had shortened intestinal length as well as severe epithelial erosion in both the ileum and colon, similar to that of Tak1tamIEC-KO mice (Supplementary Fig. 6f, g). Tak1tamIEC-KO TraddtamIEC-KO mice lost most goblet cells as well as Paneth cells both in the ileum and colon (Supplementary Fig. 6g, h). Increased caspase-8 activation and IEC apoptosis, as well as neutrophils, were detected in both the ileum and colon of Tak1tamIEC-KO TraddtamIEC-KO mice (Supplementary Fig. 6g, h).
Collectively, above data suggest the cooperative role of RIPK1 kinase and TRADD mediating lethality and intestinal pathology both in newborn Tak1IEC-KO mice and adult Tak1tamIEC-KO mice; while RIPK1 kinase inhibition provides better protection than TRADD deficiency alone in adult TAK1 IEC KO mice.
Involvement of microbiota in RIPK1 kinase and TRADD-dependent intestinal pathology in adult Tak1 tamIEC-KO mice
Since our data above demonstrate that inhibition of RIPK1 and TRADD knockout provide complete protection against IBD and lethality of adult Tak1tamIEC-KO mice, we next investigated the interaction of RIPK1 and TRADD with microbiota which plays an important role in maintaining intestinal homeostasis23. We used antibiotics cocktails (ABX) to delete luminal microbiota and assess the role of microbiota in RIPK1 kinase and TRADD-mediated ileitis and colitis in adult Tak1tamIEC-KO mice. ABX treatment alone was only able to provide a limited extension for the survival of Tak1tamIEC-KO mice after induced TAK1 IEC deletion up to 12 dpi (Supplementary Fig. 7a). Tak1tamIEC-KO mice treated with ABX at 3 dpi had a comparable length of the intestine as that of control mice and normal villi architecture in both ileum and colon (Supplementary Fig. 7b, c), as well as reduced caspase-8 activation, IEC apoptosis and neutrophil infiltration, restored the number of goblet cells in both ileum and colon as well as the number of Paneth cells in ileum to the levels of control mice (Supplementary Fig. 7c, d). At 3 dpi, the elevated mRNA levels of Cxcl1, Cxcl2, Ccl2, Ccl5, Il-1α, Il-1β, Tnfα, Ifit1, Isg15 and Zbp1 were also blocked by ABX treatment in the ileum of Tak1tamIEC-KO mice (Supplementary Fig. 7e). Tak1tamIEC-KO mice treated with ABX at 7 dpi had normal villi architecture in the ileum (Supplementary Fig. 8a, b). No caspase-8 activation, IEC apoptosis, neutrophil infiltration, or loss of goblet cells, but the loss of Paneth cells were detected in the ileum of Tak1tamIEC-KO mice treated with ABX at 7 dpi (Supplementary Fig. 8a, b). These data suggest that lumen microbiota was involved in RIPK1 kinase-mediated ileitis and early-stage colitis in adult Tak1tamIEC-KO mice.
Tak1tamIEC-KO mice with ABX treatment died at 12 dpi and showed severe colitis characterized by nearly total loss of goblet cells and dramatic activation of caspase-8 and caspase-3 as well as neutrophil infiltration in the colon at 7 dpi. TNF neutralization had minimum effect to rescue the colon architecture damage, goblet cells loss, IEC apoptosis as well as neutrophil infiltration of Tak1tamIEC-KO mice treated with ABX, suggesting the involvement of factors beyond TNF (Supplementary Fig. 8c, d).
While inhibition of RIPK1 in Tak1tamIEC-KO Ripk1D138N/D138N mice allowed the survival to 7-9 dpi, ABX treatment was able to provide additional protection by preventing the loss of body weight and lethality of Tak1tamIEC-KO Ripk1D138N/D138N mice induced by TAK1 IEC deficiency (Fig. 4a, b). ABX-treated Tak1tamIEC-KO Ripk1D138N/D138N mice at 7 dpi had normal colon architecture, although slight caspase activation, goblet cell loss, and neutrophil infiltration were detected (Fig. 4c, d and Supplementary Fig. 8e, f). ABX treatment was able to significantly reduce caspase-8 activation and apoptosis in the colonic tissues of Tak1tamIEC-KO Ripk1D138N/D138N mice (Fig. 4c, d and Supplementary Fig. 8e, f). ABX administration also prevented the increased mRNA levels of Cxcl1, Cxcl2, Ccl2, Ccl5, Il-1α, Il-1β, and Tnfα as well as neutrophil infiltration in Tak1tamIEC-KO Ripk1D138N/D138N mice (Fig. 4g and Supplementary Fig. 8e, f). Thus, similar to that of TRADD knockout, ABX treatment was able to provide additive protection with inhibition of RIPK1 in Tak1tamIEC-KO Ripk1D138N/D138N mice. These results suggest that microbiota may be the major driver of TRADD-mediated colitis in Tak1tamIEC-KO Ripk1D138N/D138N mice.
Collectively, these data revealed the important role of microbiota in RIPK1 kinase and TRADD-mediated ileitis and colitis in adult Tak1tamIEC-KO mice.
Interaction of RIPK1 and TRADD in TLRs signaling pathways
Since the above data suggest the importance of microbiota, we next examined the role of TRADD and RIPK1 in TLR signaling using intestinal organoids. Colon organoids treated with Pam3csk4 (TLR1/2 ligands) or LPS (TLR4 ligands) were sensitized to TAK1 inhibition-induced cell death (Fig. 5a–d). TRADD deficiency or RIPK1 inhibition alone partially rescued colon organoid cell death, which can be effectively inhibited by the combination of TRADD deficiency and RIPK1 inhibition (Fig. 5a–d). As for treatment with flagellin (TLR5 ligand) and TAK1 inhibition, inhibition of RIPK1 kinase alone provided better protection than TRADD deficiency alone, and the combination of TRADD deficiency and inhibition of RIPK1 kinase effectively inhibited cell death in organoid culture (Fig. 5e, f). We also conducted above experiences using ileum organoids, however, ileum organoids were not that sensitive as that of colon organoids, which might be due to the low expression of TLRs in ileum organoids24.
a–f Tradd+/+ and Tradd−/− colon organoids were treated with 500 nM (5Z)-7-oxozeaeno and Pam3CSK4 (1 μg/ml) (a, b), LPS (1 μg/ml) (c, d) or Flagellin (500 ng/ml) (e, f) for 12 h with or without 50 μM Nec-1s. Cell death in organoids was determined by PI staining. Each dot represented one organoid. Scale bar, 50 μm. The results were shown as mean ± SEM. An unpaired two-tailed t-test was performed. g–l Primary BMDMs from the mice with indicated genotypes pretreated Pam3CSK4 (1 μg/ml) (g, h), LPS (100 ng/ml) (i, j) or Flagellin (100 ng/ml) (k, l) for 4 h and followed by 200 nM (5Z)-7-oxozeaeno. Cell death was measured by SytoxGreen, and the mean ± SEM from five biological replicates was shown (g, i, k). The levels of GSDMD, Caspase-1, Caspase-8, CC8, Caspase-3, CC3, RIPK1, and p-RIPK1(S166) were determined by immunoblotting (h, j, l).
Nevertheless, combined inhibition of RIPK1 kinase and TRADD knockout was also able to reduce the cell death of ileum organoids induced by TLR ligands and TAK1 inhibition (Supplementary Fig. 9a–f).
We also examined the response of primary bone marrow-derived macrophages (BMDMs), which are commonly used to study the TLR signaling pathway, to TAK1 inhibition. Primary BMDMs priming with Pam3csk4 or LPS were sensitized to TAK1 inhibition-induced cell death and mature IL-1β release, which were partially reduced by TRADD deficiency or RIPK1 inhibition (D138N) alone and effectively inhibited with a combination of TRADD deficiency and RIPK1 inhibition (D138N) (Fig. 5g–j and Supplementary Fig. 9g, h). The biomarkers of apoptosis, including the cleavage of caspase-8 and caspase-3, induced by Pam3csk4 and LPS with TAK1 inhibitor 5Z7, were partially reduced by TRADD deficiency or RIPK1 inhibition (D138N) alone, and effectively inhibited with a combination of TRADD deficiency and RIPK1 inhibition (D138N) (Fig. 5h–j). The activation of RIPK1 (p-S166) in primary BMDMs induced by Pam3csk4 and LPS with TAK1 inhibitor 5Z7 was inhibited by RIPK1 inhibition (D138N), but not by TRADD deficiency alone (Fig. 5h, j). The cleavage caspase-1 and GSDMD, which could form pores on a cytoplasmic membrane for the release of mature IL-1β25, induced by Pam3csk4 and LPS with TAK1 inhibitor 5Z7 in primary BMDMs, were effectively inhibited by combined TRADD deficiency and RIPK1 inhibition (D138N) (Fig. 5h, j).
RIPK1 kinase inhibition alone was able to protect against apoptosis and inflammation in BMDMs induced by flagellin (TLR5 ligand) with TAK1 inhibition, while TRADD deficiency reduced mature IL-1β release but had minimum effect on cell death (Fig. 5k, l and Supplementary Fig. 9i). In BMDMs stimulated by flagellin with TAK1 inhibition, the cleavage of caspase-1, GSDMD as well as caspase-3 and caspase-8 was inhibited by RIPK1 inhibition alone, but combined TRADD deficiency and RIPK1 inhibition was still more effective than that of RIPK1 inhibition alone (Fig. 5l). Thus, RIPK1 inhibition can selectively reduce certain signaling by TLR ligand (such as TLR5) while combined TRADD KO and RIPK1 inhibition are more effective in blocking cell death and inflammation mediated by ligands for TLR1/2 and TLR4.
Collectively, combined TRADD deficiency and RIPK1 inhibition is able to effectively inhibit both apoptosis upon activation of different TLR signaling with TAK1 inhibition.
Interaction of microbiota and RIPK1 in ileitis of Tak1 tamIEC-KO mice
We next assessed the influence of RIPK1 kinase on the ileum microbiota community using 16S rDNA sequencing, as inhibition of RIPK1 kinase alone was sufficient to effectively prevent ileitis in Tak1tamIEC-KO mice at 3 dpi. The Chao1 alpha-diversity, a measure for the numbers of distinct type microbes in a community, was higher in the ileum fecal of Tak1tamIEC-KO mice at 3 dpi than that of Tak1fl/fl mice (Fig. 6a). The Chao1 alpha-diversity of Tak1tamIEC-KO Ripk1D138N/D138N mice showed a shift towards that of Tak1fl/fl mice (Fig. 6a). Principle component analysis (PCoA) of beta diversity based on the genera levels differentiated Tak1tamIEC-KO mice from Tak1fl/fl mice, suggesting an altered microbiota communities after induced TAK1 IEC deletion, which was restored in Tak1tamIEC-KO Ripk1D138N/D138N mice (Fig. 6b, c and Supplementary Data 1).
a Chao1 alpha-diversity from mice of the indicated genotypes at 3 dpi. Each dot represented one sample. The results were shown as mean ± SEM. An unpaired two-tailed t-test was performed. b Principal coordinate analysis (PCoA) of 16S sequencing of ileum fecal at 3 dpi were analysed with Bray-Curtis distances (p = 0.001, PERMANOVA by Adonis) on the taxonomic profile (at the ASV level). c Average relative abundance of the top 30 abundant genera of ileum fecal at 3 dpi of mice with the indicated genotypes. d Cluster heat map of average relative microbiota abundance at genus level in the ileum of the mice with indicated genotypes at 3 dpi (cut-off: Tak1fl/fl V.S. Tak1tamIEC-KO p ≤ 0.05). * indicated Tak1tamIEC-KO V.S. Tak1tamIEC-KO Ripk1D138N/D138N p ≤ 0.05. Z scores are shown. e Relative abundance of Enterococcus faecalis in ileum fecal of the mice with indicated genotypes at 3 dpi. Each dot represented one sample. The results were shown as mean ± SEM. An unpaired two-tailed t-test was performed. f Cluster heat map of AMP expression level in the ileum of the mice with indicated genotypes at 3 dpi (log2|fold change | ≥1, p ≤ 0.05). Z scores are shown. Each column represented one mouse. g Ileal IECs from the mice with indicated genotypes at 3 dpi were isolated and immunoblotting for TAK1, RIPK1, Caspase-8, CC8, Caspase-3, CC3, p-ERK, ERK, p-P65, and P65.
We identified 40 increased and 7 decreased genera in the ileum fecal of Tak1tamIEC-KO mice that were different from that of Tak1fl/fl mice, and RIPK1 kinase inhibition restored the levels of 18 increased genera and 2 decreased genera in the ileum of Tak1tamIEC-KO mice (Fig. 6d). Among 2 decreased genera, Bifidobacterium contributes to sphingolipid metabolism as well as the maintenance of barrier function26, the level of which was reported to be positively correlated with successful outcome of anti-TNF treatment27.
Among 18 upregulated genera, the levels of Enterococcus in the ileum of Tak1tamIEC-KO mice were increased more than 200 folds compared to that of Tak1fl/fl mice, which was also restored to that of levels upon RIPK1 inactivation in the ileum fecal of Tak1tamIEC-KO Ripk1D138N/D138N mice (Fig. 6c, d). Enterococcus faecalis, one species of the Enterococcus genus, was also increased in the ileum fecal of Tak1tamIEC-KO mice in a RIPK1 kinase-dependent manner (Fig. 6e). Enterococcus faecalis contributes to the intestine barrier impairment and intestinal inflammation by producing metalloprotease GelE28. Increased levels of Helicobacter and Mucispirillum, mediated by RIPK1 kinase in Tak1tamIEC-KO mice (Fig. 6d), were also associated with human IBD29,30. We also assessed the microbiota interaction using idopNetwork which was developed to measure the interaction networks among the luminal bacteria31. Compared with that of Tak1fl/fl mice, the microbiota in Tak1tamIEC-KO mice showed substantially reduced interactions, which could be restored by RIPK1 kinase inhibition (Supplementary Fig. 10a). Thus, the activation of RIPK1 kinase is a major driving force in ileum microbiota dysbiosis of Tak1tamIEC-KO mice.
Since the above data suggest that activation of RIPK1 kinase disturbs the microbiota community in the ileum of Tak1tamIEC-KO mice, we considered the contribution of RIPK1-mediated loss of IECs to the expression of the antimicrobial peptides (AMP) genes. The expression levels of α-defensin 17/23/24/31/40, Lysozyme-1, and Angiogenin-4, secreted by Paneth cells, were decreased in the ileum of Tak1tamIEC-KO mice and restored by RIPK1 inhibition (Fig. 6f and Supplementary Fig. 2a, b). Ccl6 and Ccl28, which were chemokines with antibacterial activity and highly expressed in IECs32,33, were downregulated in the ileum of Tak1tamIEC-KO mice in RIPK1-dependent manner (Fig. 6f). CCL28 was reported to kill Enterococcus faecalis in vitro34. Thus, the reduction in the levels of CCL28 may contribute to the dramatically increased level of Enterococcus faecalis in the ileum fecal of Tak1tamIEC-KO mice. Increased levels of cleaved caspase-8 and cleaved caspase-3 in the ileum IECs of Tak1tamIEC-KO mice was inhibited by RIPK1 inhibition (Fig. 6g). Collectively, these results suggest that the RIPK1-mediated death of IECs may contribute to the microbiota dysbiosis in the ileum of Tak1tamIEC-KO mice.
In addition, among these upregulated genes in the ileum of Tak1tamIEC-KO mice, S100a8/935, Gbp3/6/736,37, Cxcl9/10 38 and Ltf 39 that were of neutrophil origin and with broad-spectrum antimicrobial activity, were increased in RIPK1 kinase-dependent manner (Fig. 6f). Increased neutrophils were detected in the ileum of Tak1tamIEC-KO mice at 3 dpi, which were protected by RIPK1 kinase inhibition (Supplementary Fig. 2a, b). Cxcl1440,41 and Ccn142,43, with direct antimicrobial effects and expressed mainly by IECs, were found upregulated in the ileum of Tak1tamIEC-KO mice and rescued by RIPK1 kinase inhibition (Fig. 6f). We also found that the IECs of Tak1tamIEC-KO mice demonstrated increased levels of p-P65 and p-ERK, suggesting the activation of NF-κB and ERK pathways, which was restored in the IECs of Tak1tamIEC-KO Ripk1D138N/D138N mice to that of mice (Fig. 6g). RIPK1 activation mediated increased transcription may contribute to increased levels Cxcl14 and Ccn1 (Fig. 6f, g). CAMP, a member of Cathelicidins, was stored in the secretory granules of neutrophils and macrophages44. Camp was found to express in epithelial cells and was influenced by ERK pathway45. Lcn2 is expressed in IECs and myeloid cells to enhance phagocytic bacterial clearance46,47. Thus, RIPK1 regulates increased neutrophil infiltration as well as the activation of NF-kB and MAPK pathways which act cooperatively to promote cytokines with antimicrobial properties such as Camp and Lcn2 expression in Tak1tamIEC-KO mice (Fig. 6f, g). Collectively, RIPK1 activation can promote the transcription and immune cell infiltration to disrupt the normal homeostatic levels of the genes that encode antimicrobial peptides or proteins to promote microbiota dysbiosis in the ileum Tak1tamIEC-KO mice.
Cooperation of RIPK1 and TRADD in the evolution of colonic microbiota
Our data above demonstrate RIPK1 kinase mediated early-stage colitis while RIPK1 and TRADD together mediated late-stage colitis with TAK1 IEC deletion. We next collected the stool samples from the colon of Tak1fl/fl mice, Tak1tamIEC-KO mice as well as Tak1tamIEC-KO Ripk1D138N/D138N mice at 3 dpi. We also collected the stool samples from the colon of Tak1fl/fl Ripk1D138N/D138Nmice, Tak1tamIEC-KO Ripk1D138N/D138N mice as well as Tak1tamIEC-KO Ripk1D138N/D138N TraddIEC-KO mice at 7 dpi. We submitted these stool samples for microbiome profiling using 16S rDNA sequencing. PCoA based on the genera levels revealed the dramatic differences of microbiota community in the colon between Tak1fl/fl mice and Tak1tamIEC-KO mice at 3 dpi, which was restored by RIPK1 kinase inhibition in Tak1tamIEC-KO Ripk1D138N/D138N mice (Fig. 7a, b and Supplementary Data 1). At 7 dpi, TAK1 deletion still disrupted the microbiota community in Tak1tamIEC-KO Ripk1D138N/D138N mice, which was further restored by TRADD deficiency (Fig. 7c, d and Supplementary Data 1). This analysis revealed the time-dependent dynamics of microbiota dysbiosis transitioned from RIPK1-dependent to RIPK1- and TRADD-codependent after TAK1 deletion.
a, c Principal coordinate analysis (PCoA) of 16S sequencing of colon fecal at 3 dpi (a) or 7 dpi (c) were analysis with Bray-Curtis distances (p = 0.001, PERMANOVA by Adonis) on the taxonomic profile (at the ASV level). b, d Average relative abundance of the top 30 abundant genera of colon fecal at 3 dpi (b) or 7 dpi (d) of the mice with indicated genotypes. e Cluster heat map of average relative microbiota abundance at genus level in colon of the mice with indicated genotypes at 3 dpi and 7 dpi (including the genus fit either Tak1fl/fl V.S. Tak1tamIEC-KO p ≤ 0.05 or Tak1fl/fl Ripk1D138N/D138N V.S. Tak1tamIEC-KO Ripk1D138N/D138N p ≤ 0.05). Z scores are shown. * on Tak1tamIEC-KO (3 dpi) indicated Tak1fl/fl V.S. Tak1tamIEC-KO p ≤ 0.05. * on Tak1tamIEC-KO Ripk1D138N/D138N (3 dpi) indicated Tak1tamIEC-KO V.S. Tak1tamIEC-KO Ripk1D138N/D138N p ≤ 0.05. * on Tak1tamIEC-KO Ripk1D138N/D138N (7 dpi) indicated Tak1fl/fl Ripk1D138N/D138N V.S. Tak1tamIEC-KO Ripk1D138N/D138N p ≤ 0.05. * on Tak1tamIEC-KO Ripk1D138N/D138N TraddtamIEC-KO (7 dpi) indicated Tak1tamIEC-KO Ripk1D138N/D138N V.S. Tak1tamIEC-KO Ripk1D138N/D138N TraddtamIEC-KO p ≤ 0.05. f Relative abundance of Escherichia coli in colon fecal of the mice with indicated genotypes at 7 dpi. Each dot represented one sample. The results were shown as mean ± SEM. An unpaired two-tailed t-test was performed. g, h Cluster heat map of AMP expression level in the colon of the mice with indicated genotypes at 3 dpi (g) or 7 dpi (h) (log2|fold change | ≥1, p ≤ 0.05). Z scores are shown. Each column represented one mouse. i, j Colonic IECs from the mice with indicated genotypes at 3 dpi (i) or 7 dpi (j) were isolated and immunoblotting for TAK1, RIPK1, CC8, CC3, p-ERK, p-P65, p-JNK, and p-P38. k, l Chao1 alpha-diversity from the mice with indicated genotypes at 3 dpi (k) or 7 dpi (l). Each dot represented one sample. The results were shown as mean ± SEM. An unpaired two-tailed t-test was performed.
We observed that TAK1 deficiency changed the colon microbiota community in two major patterns (Fig. 7e). In the first pattern, the overgrowth of microbiota including Uncultured, Helicobacter, Bacteroides, Odoribacter, Rodentibacter, Parabacteroides, and Mucispirillum or the reduced growth of Muribaculaceae, Lachnospiraceae NK4A136 group, Clostridia UCG-014, A2, NK4A214 group, Monoglobus, Gordonibacter, Christensenellaceae, and Family XIII UCG-001, were restored by RIPK1 kinase inhibition at 3 dpi. However, these genera still changed in Tak1tamIEC-KO Ripk1D138N/D138N mice at 7 dpi, which were rescued by further TRADD deficiency. These data suggest the evolution from early RIPK1-dependent to late RIPK1- and TRADD-co-dependent regulation of these genera (Fig. 7e). In the second pattern, the overgrowth of microbiota including Escherichia-Shigella, Rodentibacter as well as Bacteroides was not obvious at 3 dpi. While these genera increased dramatically at 7 dpi in Tak1tamIEC-KO Ripk1D138N/D138N mice which was mediated by TRADD (Fig. 7e). Notably, Escherichia-Shigella which dominated in the colon microbiome of Tak1tamIEC-KO Ripk1D138N/D138N mice at 7 dpi, was reduced to low levels in Tak1tamIEC-KO Ripk1D138N/D138N TraddtamIEC-KO mice (Fig. 7d).
Increased levels of Escherichia-Shigella have been found to be positively correlated with the levels of inflammatory markers in IBD patients48. We further identified Escherichia coli, one of the species in Escherichia-Shigella, was dramatically increased in the colon fecal of Tak1tamIEC-KO Ripk1D138N/D138N mice at 7 dpi and reduced to that of normal levels in the colon of Tak1tamIEC-KO Ripk1D138N/D138N TraddtamIEC-KO mice (Fig. 7f). Increased levels of Escherichia coli are highly associated with human IBD49. Adherent-invasive Escherichia coli can utilize fucose fermentation product 1,2-propanediol to promote T cell-mediated intestine inflammation50. The upregulation of Escherichia-Shigella was specifically regulated by TRADD after prolonged TAK1 IEC deficiency with RIPK1 inhibition. These results suggest that the activation of TRADD can drive colon microbiota dysbiosis beyond inhibition of RIPK1 in Tak1tamIEC-KO Ripk1D138N/D138N mice.
TAK1 deletion at 3 dpi increased Gbp3/6/7/10, Cxcl9/10, Lcn2 as well as Saa251, attributable to increased neutrophil infiltration in the colon of Tak1tamIEC-KO mice, which were reduced upon inhibition of RIPK1 in Tak1tamIEC-KO Ripk1D138N/D138N mice (Fig. 7g). At 7 dpi, immune cell infiltration including neutrophils mediated by TRADD increased S100a8/9, Lcn2, Ltf, lyz252, Camp2 as well as Saa2 in Tak1tamIEC-KO Ripk1D138N/D138N mice (Fig. 7h). The increased AMPs suppressed microbiota growth in RIPK1 kinase-dependent manner at early stage and TRADD dependent manner at late stage.
We observed that reduced levels of Reg4, mostly expressed in enteroendocrine cells and known to have antibacterial activity53,54, and increased caspase activation in Tak1tamIEC-KO mice at 3 dpi were rescued in Tak1tamIEC-KO Ripk1D138N/D138N mice (Fig. 7g, i). However, reduced levels of Reg4 and increased caspase activation were still found in Tak1tamIEC-KO Ripk1D138N/D138N mice at 7 dpi, which were blocked in Tak1tamIEC-KO Ripk1D138N/D138N TraddtamIEC-KO mice (Fig. 7h, j). Thus, the reduction in Reg4 may facilitate microbiota overgrowth in RIPK1 kinase-dependent manner at an early stage and RIPK1/TRADD co-dependent manner at a late stage. Unlike RIPK1 activation in the colon IECs at 3 dpi, TRADD activation in the colon IECs at 7 dpi inhibited P65, ERK, JNK as well as P38 activation (Fig. 7i, j). Thus, TAK1 deficiency further reduced Chga/b and Muc13 in the colon of Tak1tamIEC-KO Ripk1D138N/D138N mice at 7 dpi in a TRADD-dependent manner (Fig. 7h). Chga or Chgb were produced by enteroendocrine cells and were found decreased in IBD patients55,56. Muc13 were found on the apical surface of intestinal enterocytes and goblet cells. The polymorphisms of Muc13 in IBD patients were also found to be significantly associated with susceptibility to Escherichia coli infection in pigs57. The reduced levels of Muc13 may explain the extreme overgrowth of Escherichia coli in the colon of Tak1tamIEC-KO Ripk1D138N/D138N mice at 7 dpi (Fig. 7f). TRADD mediated transcription inhibition as well as IECs death including enteroendocrine cells and goblets cells contributed to the decreased levels of the Reg4, Chga/b and Muc13 (Fig. 7h, j).
Chao1 alpha-diversity was downregulated in the colon microbiome of Tak1tamIEC-KO mice relative to that of Tak1fl/fl mice due to the overall upregulated AMPs, which was also rescued by RIPK1 kinase inhibition (Fig. 7g, k). However, Chao1 alpha-diversity at 7 dpi was upregulated in the colon microbiome of Tak1tamIEC-KO Ripk1D138N/D138N mice compared to that of Tak1fl/fl Ripk1D138N/D138N mice (Fig. 7l). Chao1 alpha-diversity in the colon of Tak1tamIEC-KO Ripk1D138N/D138N TraddtamIEC-KO mice was corrected towards that of Tak1fl/fl Ripk1D138N/D138N mice (Fig. 7l). We observed some microbiota genera of Tak1tamIEC-KO mice, including Escherichia-Shigella, which were not obviously changed at 3 dpi, were significantly increased in the colon of Tak1tamIEC-KO Ripk1D138N/D138N mice at 7 dpi (Fig. 7e). This may be attributable to the TRADD activation induced such as Chga/b and Muc13 reduction and consequently elevated Chao1 alpha-diversity at 7 dpi in the colon of Tak1tamIEC-KO Ripk1D138N/D138N mice.
In conclusion, our data demonstrated the cooperative role of RIPK1 kinase and TRADD in the evolution of colonic microbiota dysbiosis.
Discussion
The intestinal epithelium provides the first-line defense for the integrity of the mucosa system. We explored the interaction of intestinal epithelial cells with the mucosa microbiota using TAK1 IEC-deficient mice. TAK1 has been established to be a key signaling mediator for the NF-κB pathway and a modulator of RIPK17,8. TAK1 inhibition promotes apoptosis and necroptosis in the TNF signaling pathway. Our study demonstrates that TAK1 can directly phosphorylate TRADD to modulate its interaction with signaling mediators in the TNFR1 pathway. Thus, TAK1 can phosphorylate not only IKKs to mediate the NF-κB pathway and RIPK1 to modulate its activation, but also TRADD to control its interaction with the components of complex IIa to regulate apoptosis. Our results demonstrate that blocking IEC apoptosis in TAK1 IEC KO mice by FADD/MLKL DKO is sufficient to maintain the intestinal health of TAK1 IEC KO mice, supporting the idea that death of IECs is a key driver for pathology in IBD. However, since FADD/MLKL DKO mice eventually develop lethal autoimmunity as adults20, simultaneous loss of FADD and MLKL cannot be considered as a therapeutic strategy for IBD.
Our study reveals the interactive relationship between RIPK1-dependent apoptosis (RDA) and RIPK1-independent apoptosis (RIA) in cells, organoids, and in vivo. TAK1 inhibition sensitizes cells to RIPK1 kinase-dependent apoptosis (RDA) upon TNF or LPS stimulation8,9,10. In the TAK1 IEC-deficient model, RDA is activated at an early stage to mediate apoptosis and inflammation, which eventually evolved into RDA- and RIA-co-dependent apoptosis and inflammation, which can be blocked by co-inhibition of RIPK1 kinase and TRADD, but not by either alone. In contrast to that of autoimmunity in adult FADD/MLKL DKO mice, combined inhibition of RIPK1 and TRADD KO was able to provide full protection against the lethality and intestinal pathology in TAK1 IEC-deficient mice without promoting autoimmunity. It is possible that reduced NF-κB activation with TRADD deficiency might help to restrain immune response to prevent autoimmunity.
Our results highlight the role of microbiota in driving the pathological progression from early to late stage IBD. Since inhibition of RIPK1 is sufficient to delay the onset of early-stage intestinal pathology and lethality in newborn and adult TAK1 IEC-deficient mice, RIPK1-mediated apoptosis may be the primary contributor to ileitis and early-stage colitis. Our results also demonstrate that the genes responded to type I and II interferon, such as GBP family, are upregulated in RIPK1-dependent manner. Since the intestine of newborn mice are yet to be fully colonized by microbiota and the ileum is less populated by microbiota than that of colon58,59, our data suggest that RIPK1 may play a dominant role in mediating chronic and low level inflammation in gut. In addition, ileum has higher expression of TLR5 than that of TLR1/2 or TLR424, and we show that apoptosis and inflammation mediated by the ligand for TLR5 can be effectively blocked by RIPK1 inhibition alone, which may explain why RIPK1 inhibition alone was sufficient to block ileitis in adult TAK1 IEC mice. These results may shed light as to why a RIPK1 inhibitor failed to demonstrate efficacy in human clinical trials for the treatment of ulcerative colitis which are most likely involved patients with advanced disease in colon60. The roles of RIPK1 and TRADD in mediating intestinal pathology are functionally different as inhibiting RIPK1 kinase, but not TRADD knockout alone, can protect against intestinal pathology in early-stage of TAK1 IEC KO mice. TRADD is involved in mediating the signaling for the TLR3 or TLR4 pathway by regulating the production of cytokines11,61,62. The critical role of TRADD in mediating TLR signaling likely explains the importance of TRADD in colitis since the colon is subjected to routine higher levels of microbiota presence than that of the ileum. The differential chronic inflammatory medium in colon vs. ileum may not only provide an adaptive selection paradigm for microbiota species that can survive in such an environment but also determine the signaling mechanisms that may be activated in response to pathological stimulus. The higher TLR expression in the colon than that of ileum24 may also help to explain why the cooperative role of TRADD and RIPK1 kinase is involved in mediating colitis in adult TAK1 IEC KO mice.
Antibiotic treatment or anti-TNF treatment cannot fully rescue the intestine phenotype of Tak1tamIEC-KO mice, while the combination of TRADD KO and RIPK1 kinase inhibition strongly protects the intestine pathology and lethality. Thus, inhibition of RIPK1 and TRADD deficiency might inhibit proinflammatory factors beyond microbiota. Since inhibition of RIPK1 has been shown to be safe in humans60 and Ripk1D138N/D138N or Tradd−/− mice are normal11,61,62,63,64, our data suggest that simultaneous inhibition of RIPK1 kinase and TRADD might provide a new strategy to module the cell death, inflammation, as well as microbiota dysbiosis in IBD patients where both TNF and TLR signaling pathways are involved. Moreover, our finding that combined inhibition of RIPK1 kinase and TRADD to module the microbiota dysbiosis might suggest new perspectives for the treatment of IBD to overcome obstacles of lost response to anti-TNF treatment.
Methods
Ethics statement
Animal experiments were conducted according to the protocols approved by the Institutional Animal Care and Use Committee (IACUC) of the Institute of Interdisciplinary Research Center on Biology and Chemistry and the experiments involving mice were performed according to the IACUC’s guidelines (Approval no. ECSIOC2021-04).
Mice
Tak1fl/fl mice were kindly provided by Shizuo Akira of Osaka University, Japan65. Villin-cre mice were purchased from the SHANGHAI MODEL ORGANISMS company. Villin-creERT2 mice were gifts kindly provided by Dr. Wei Mo66. Ripk1D138N/D138N mice were generated via CRISPR/Cas9 system as described previously18. Traddfl/fl mice were purchased from Cyagen Biosciences. Fadd−/− Mlkl−/− mice were gifts kindly provided by Dr. Haibing Zhang67. Newborn mice were designated P0 on the day their birth was detected. Male and female Tak1IEC-KO mice with or without Ripk1D138N/D138N, Tradd IEC-KO, Fadd−/−, and Mlkl−/− ranging in age from P0 to P4 were analyzed. Mice of 7–8 weeks with or without Villin-creERT2 were treated with 3 daily intraperitoneal administrations of 80 mg/kg tamoxifen dissolved in corn oil/ethanol. Only male mice were used for Villin-creERT2 recombinase activity induction. For the TNF blockade experiment, 125 μg anti-mouse TNFα antibody (Bioxcell, BE0058) was administered intraperitoneally every other day at -1, 1, 3, 5 dpi. All animals (C57/B6J) were maintained in a specific pathogen-free environment and housed with no more than five animals per cage under controlled light (12 h light and 12 h dark cycle), temperature (24 ± 2 °C), and humidity (50 ± 10%) conditions, and provided with ad libitum access to food (Jiangsu Xietong Pharmaceutical Bio-Engineering Co.,Ltd., XTI01FZ) and water throughout all experiments.
Administration of antibiotics
Antibiotics including ciprofloxacin hydrochloride (0.2 mg/ml), ampicillin sodium salt (1 mg/ml), metronidazole (1 mg/ml), vancomycin hydrochloride (0.5 mg/ml), and kanamycin sulfate (0.5 mg/ml) were dissolved in sterile water66. Tak1tamIEC-KO mice, or Tak1tamIEC-KO Ripk1D138N/D138N mice and their littermates involved in the experiments were pretreated with antibiotics water two weeks before tamoxifen injection.
Generation and immortalization of MEFs
MEFs were isolated from E11-13 embryos using trypsin/EDTA and sieved through a 70-micron filter. Primary MEFs were immortalized by transfection with SV40 large T antigen-expressing plasmid. Immortalized MEFs were maintained at 37 °C and 5% CO2 in DMEM (Thermo Fisher, C11995500BT) supplemented with 10% (vol/vol) fetal bovine serum (FBS, Gibco) and 100 units ml−1 penicillin/streptomycin.
Organoid culture
Mouse organoids were established from isolated crypts collected from the ileum and colon according to the manufacturer’s instructions (StemCell Technologies, 06005). Organoids were passaged at least twice and grown using IntestiCult organoid growth medium (StemCell Technologies, 06005). Propodium iodide (PI) staining was used to quantitate the levels of cell death. Images were acquired using the PerkinElmer Operetta CLS™ high content analysis system. The percentage of PI+ organoid area was calculated by HALO (v4.0.5107.318) image analysis software.
Isolation of primary BMDMs
BMDMs were differentiated from whole bone marrow-derived from flushed tibia and femurs from C57BL/6 J mice. Differentiation was carried out in complete DMEM supplemented with 30% L929 supernatants containing macrophage colony-stimulating factor.
Analysis of cell death in culture
MEFs or BMDMs were seeded the day before in a 384-well plate (25000 cells per well for BMDMs). The cells were pretreated on the next day with indicated compounds or TLRs ligands for 30 min or 4 h and then stimulated with mouse TNF or 5Z7 in the presence of 5 mM SytoxGreen (Invitrogen). SytoxGreen intensity was measured at intervals of 0.5 h using a SYNERGY H1 microplate reader (BioTek), with an excitation filter of 488 nm and an emission filter of 523 nm. Data were collected using Gen5 software version 3.08.01 (BioTek). Cell death (%) was expressed as percentages of cell death per well after deducting the background signal in non-induced cells and compared to that of the maximal cell death with 100% Lysis Reagent (0.1% Triton X-100).
Immunoblotting
Cell lysates were lysed in 1% NP-40 buffer supplemented with protease and phosphatase inhibitors, followed by denaturation in LDS sample buffer. Protein lysates were then subjected to western blotting. Primary antibodies against the following proteins were used for western blotting: TAK1 (Abcam, ab109526, 1:1,000), p-S166 RIPK1 (Biolynx, BX60008, 1:1,000), RIPK1 (homemade, 1:1,000), TRADD (homemade, 1:1,000), Cleaved Caspase-3 (CST, 9661, 1:1,000), Cleaved Caspase-8 (CST, 8592, 1:1,000), Caspase-3 (CST, 9662, 1:1,000), Caspase-8 (CST, 4790, 1:1,000), GSDMD (Abcam, ab209845, 1:1,000), Caspase-1 (p20) (AdipoGen, AG-20B-0042-C100, 1:1,000), p-ERK1/2 (CST, 4370, 1:1,000), p-p38 (CST, 4511, 1:1,000), p-JNK (CST, 4668, 1:1,000), p-NF-κB p65 (CST, 3033, 1:1,000), ERK1/2 (CST, 9102, 1:1,000), p38 (CST, 8690, 1:1,000), p-JNK (CST, 9252, 1:1,000), NF-κB p65 (CST, 8242, 1:1,000), FADD (Abcam, ab124812, 1:1,000), RIPK3 (ProSci, 2283, 1:1,000), MLKL (Abgent, AP14272b, 1:1,000), p-MLKL (Abcam, ab196436, 1:1,000), p-RIPK3 (Abcam, ab195117, 1:1,000), HA (CST, 3724, 1:1,000), β-Tubulin (MBL, PM054, 1:2000), β-actin (TransGen Biotech, HC201, 1:2000).
Complex-II purification
For complex-II purification, cells were seeded in 15 cm dishes and treated as indicated. Cells were lysed on ice in 1% NP-40 lysis buffer. The lysates were incubated with FADD antibody (Santa Cruz Biotechnology, sc-6036, 1:100) in rotation overnight at 4 °C. The immunocomplex was captured by 20 μl of protein A/G agarose (Invitrogen) 4 h at 4 °C. The samples were then washed three times in 1% NP-40 buffer and eluted by boiling in 40 μl 1 × SDS loading buffer.
RNA extraction, cDNA synthesis, and quantitative real-time PCR
Total RNA was extracted from intestine tissues using RNAiso Plus (Takara, 9109) according to the manufacturer’s instructions. The quantity and quality of RNA were measured by using a nanodrop (Thermo Scientific). RNA was used to synthesize cDNA with Reverse Transcription Regents [Takara, Recombinant RNase Inhibitor 2313A, Reverse Transcriptase M-MLV (RNaseH-) 2641 A, Oligo(dT)18 Primer 3806, Random Primer 3801, dNTP Mixture 4030]. Then cDNA was used to run the quantitative real-time PCR assay with the QuantStudioTM 7 Flex Real-Time PCR system (Life Technologies) to measure the expression of interested genes. Actin was used as an internal control.
Mouse Actin
forward: ATGGAGGGGAATACAGCCC,
reverse: TTCTTTGCAGCTCCTTCGTT;
Ccl2
forward: GGGATCATCTTGCTGGTGAA,
reverse: AGGTCCCTGTCATGCTTCTG;
Ccl5
forward: CCAATCTTGCAGTCGTGTTTGT,
reverse: CATCTCCAAATAGTTGATGTATTCTTGAAC;
Cxcl1
forward: CTGGGATTCACCTCAAGAACATC,
reverse: CAGGGTCAAGGCAAGCCTC;
Cxcl2
forward: ACAGAAGTCATAGCCACTCTC,
reverse: TTAGCCTTGCCTTTGTTCAG;
Ifit1
forward: TACAGGCTGGAGTGTGCTGAGA,
reverse: CTCCACTTTCAGAGCCTTCGCA;
Il1a
forward: CGCTTGAGTCGGCAAAGAAATC,
reverse: GTGCAAGTCTCATGAAGTGAGC;
Il1β
forward: ACTCATTGTGGCTGTGGAGA,
reverse: TTGTTCATCTCGGAGCCTGT;
Isg15
forward: CATCCTGGTGAGGAACGAAAGG,
reverse: CTCAGCCAGAACTGGTCTTCGT;
Tnf
forward: CCCTCACACTCAGATCATCTTCT,
reverse: GCTACGACGTGGGCTACAG;
Zbp1
forward: TTGAGCACAGGAGACAATCTG,
reverse: TTCAGGCGGTAAAGGACTTG;
ELISA
The levels of IL-1β in the culture medium were determined using ELISA kits according to the manufacturer’s instructions (Thermo Fisher, 88-7013A-88).
Immunohistochemistry and histology
Tissue samples from mice were fixed in 4% paraformaldehyde and embedded in paraffin. Sections of 5 µm in thickness were subjected to hematoxylin and eosin (H&E) staining. For immunohistochemical analysis, slides were rehydrated and antigen retrieval was performed by boiling with Tris-EDTA buffer (10 mM Tris, 1 mM EDTA, 0.05% Tween-20, pH 9.0) for CC3 (CST, 9661, 1:200) and CC8 (CST, 8592, 1:200), or digestion with trypsin solution (0.05% Trypsin, 0.1% CaCl2, pH 7.8) for lysozyme-1(Invitrogen, PA5-16668, 1:200) and Ly-6G (Biolegend, 127608, 1:200). Slides were washed and incubated with peroxidase blocking buffer (0.3% H2O2) for 15 min. Slides were washed and blocked in 1.5% goat serum for 20 min, and incubated with primary antibody for 6 h at 4 °C. Sections were incubated with biotinylated anti-rat secondary antibody (Vector Laboratories, BA-9401, 1:500) or anti-rabbit secondary antibody (Vector Laboratories, BA-1000, 1:500) for 60 min at room temperature. Staining was visualized using the Vectastain Elite ABC-HRP kit (Vector Laboratories, VEC-PK-6100) and DAB substrate (Vector Laboratories, SK-4100). Sections were then counterstained with hematoxylin for staining of the nuclei, dehydrated, and mounted with neutral balsam (Solarbio, G8590). Histological sections were scanned using 3DHistech PANNORAMIC® 250 Flash III DX.
In vitro kinase assay
Kinase assays were performed with 75 µg recombinant 6*HIS hTRADD (Proteintech, Ag7768) and 150 μg hTAK1(15-303)-TAB1(468-504) in a reaction containing 25 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM beta-glycerophosphate, 2 mM dithiothreitol (DTT), 0.1 mM Na3VO4, 10 mM MgCl2, 200 μM ATP at 30 °C for 1 h68.
Mass spectrometry and data analysis
The protein samples were resolved in 8 M urea and 500 mM Tris-HCl (pH 8.5). Disulfide bridges were reduced by adding Tris (2-carboxyethyl) phosphine (TCEP) at a final concentration of 5 mM for 20 min. Reduced cysteine residues were then alkylated by adding 10 mM iodoacetamide (IAA) and incubating for 15 min in the dark at room temperature. The urea concentration was reduced to 2 M by adding 100 mM Tris-HCl (pH 8.5) and 1 mM CaCl2. Then, the protein samples were digested overnight at 37 °C with trypsin (V5111, Promega) at an enzyme-to-substrate ratio of 1:100 (w/w). The resulting peptides were enriched for phosphorylated peptides using TiO2 and then analyzed using a Bruker TimsTOFTM mass spectrometer. The peptides were separated on an analytical column (250 mm × 75 μm 1.6 μm C18 resin, IonOpticks). The dual TIMS analyzer was operated at a fixed duty cycle with a ramp time of 100 ms, and the total cycle time was 1.16 s. Data-dependent acquisition (DDA) was performed in PASEF mode with ten PASEF scans per acquisition cycle in a mass range from 100 m/z to 1700 m/z with charge states from 0 to 5+. The ion mobility was scanned from 0.6 to 1.6 Vs/cm2. Precursors that reached a target intensity of 20,000 were selected for fragmentation and dynamically excluded for 0.4 min (mass width 0.015 m/z, 1/K0 width 0.015 Vs/cm2). The protein identification and quantification were done by FragPipe version 21.169. The tandem mass spectra were searched against the UniProt human and mouse protein database. Trypsin was set as the enzyme, and the specificity was set to both N and C terminal of the peptides. The peptide length was set as 7 to 50 amino acids. The maximum missed cleavage was set to 2. The cysteine carbamidomethylation was set as a static modification, and the methionine oxidation and phosphorylation on serine, threonine, and tyrosine were set as variable modifications. The precursor and fragment mass tolerance were set as 20 ppm. The false discovery rate at the peptide spectrum match level and protein level was controlled to be <1%. The minimal localization probability was set to 0.75. The “match between runs” option was applied.
Construction and transfection of plasmids
pMSCV-blasticidin was used as the plasmid backbone for all of the constructs used for the reconstitution study. Full-length cDNAs for mouse TRADD were PCR-amplified from the plasmid library using Q5® Hot Start High-Fidelity DNA Polymerase (NEB, M0493) and cloned into pMSCV vector using ClonExpress® MultiS One Step Cloning Kit (Vazyme, C113) with Flag or HA tag. Mutant mTRADD (Y252D, Y252E, Y254D, Y254E) were generated using Q5® Hot Start High-Fidelity DNA Polymerase and cloned into the XhoI/EcoRI sites in the pMSCV vector using ClonExpress® MultiS One Step Cloning Kit (Vazyme, C113). All plasmids were verified by DNA sequencing, and details of the plasmid’s sequences are available upon request. For viral packaging, HEK293T cells were transfected with different vectors using PEI (Polysciences, Cat#23966-2), and collected at 48 h post-transfection. The virus-containing supernatant was then used to infect Tradd−/− MEFs with 8 μg/ml−1 polybrene for 48 h.
RNA-seq
Total RNA was extracted using the TRIzol reagent (Invitrogen, CA, USA) according to the manufacturer’s protocol. RNA purity and quantification were evaluated using the NanoDrop 2000 spectrophotometer (Thermo Scientific, USA). RNA integrity was assessed using Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The libraries were constructed using VAHTS Universal V6 RNA-seq Library Prep Kit according to the manufacturer’s instructions. The transcriptome sequencing and analysis were conducted using OE Biotech Co., Ltd. (Shanghai, China). The libraries were sequenced on an llumina Novaseq 6000 platform, and 150 bp paired-end reads were generated. About ~50 million reads for each sample were generated. Raw reads of fastq format were firstly processed using fastp and the low-quality reads were removed to obtain the clean reads70. About ~48 million clean reads for each sample were retained for subsequent analyses. The clean reads were mapped to the reference genome using HISAT271. FPKM of each gene was calculated and the read counts of each gene were obtained by HTSeq-count72,73.
16S rDNA seq
Ileum and colon samples were snap-frozen and stored at −80 °C after collection. Ileum fecal were collected from the mice with indicated genotypes at 3 dpi for 16S sequencing. We collected two biological replicates from the ileum of each mouse. In total, 24 ileum stool samples from 12 Tak1fl/fl mice, 23 ileum stool samples from 12 Tak1tamIEC-KO mice and 20 ileum stool samples from 10 Tak1tamIEC-KO Ripk1D138N/D138N mice were obtained and submitted for microbiome profiling using 16S rDNA sequencing. Colon fecal were collected from the mice with indicated genotypes at 3 dpi and 7 dpi for 16S sequencing. We collected two biological replicates from the colon of each mouse. In total, 24 colon stool samples from 12 Tak1fl/fl mice, 24 colon stool samples from 12 Tak1tamIEC-KO mice and 20 colon stool samples from Tak1tamIEC-KO Ripk1D138N/D138N mice at 3 dpi, while 22 colon stool samples from 11 Tak1fl/fl Ripk1D138N/D138N mice, 21 colon stool samples from 11 Tak1tamIEC-KO Ripk1D138N/D138N mice and 20 colon stool samples from 10 Tak1tamIEC-KO Ripk1D138N/D138N TraddtamIEC-KO mice were obtained and submitted for microbiome profiling using 16S rDNA sequencing. Bacterial DNA was isolated using a DNeasy PowerSoil kit (Qiagen) following the manufacturer’s instructions. DNA concentration and integrity were measured using NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific) and agarose gel electrophoresis. PCR amplification of the V3-V4 hypervariable regions of bacterial 16S rRNA gene was carried out in a 25 μl reaction using universal primer pairs (343 F: 5′-TACGGRAGGCAGCAG-3′; 798 R: 5′-AGGGTATCTAATCCT-3′). The reverse primer contained a sample barcode and both primers were connected with an Illumina sequencing adapter. The Amplicon quality was visualized using gel electrophoresis. The PCR products were purified with Agencourt AMPure XP beads (Beckman Coulter Co., USA) and quantified using a Qubit dsDNA assay kit. The concentrations were then adjusted for sequencing. Sequencing was performed on an Illumina NovaSeq 6000 with two paired-end read cycles of 250 bases each (Illumina Inc., San Diego, CA; OE Biotech Company; Shanghai, China). Raw sequencing data were in FASTQ format. Paired-end reads were then preprocessed using cutadapt software to detect and cut off the adapter. After trimming, paired-end reads were filtered low-quality sequences, denoised, merged, detected, and cut off the chimera reads using DADA2 with the default parameters of QIIME274,75. The software outputs the representative reads and the ASV abundance table. The representative read of each ASV was selected using the QIIME2 package. All representative reads were annotated and blasted against Silva database Version 138 (or Unite) (16 s/18 s/ITS rDNA) using q2-feature-classifier with the default parameters. The 16S rRNA gene amplicon sequencing and analysis were conducted using OE Biotech Co., Ltd. (Shanghai, China).
Inferring context-specific microbial interactions
We consider each part of a mouse as an ecosystem inhabited by the microbiota (containing m genera). Let yij denote the microbial abundance of genus j for an ecosystem i. We calculate \({N}_{i}={\sum }_{i=1}^{m}{y}_{{ij}}\) and define it as the niche index (HI) of ecosystem i. The abundance level of an individual genus i allometrically scales as HI across ecosystems, which obeys the power law, expressed as
where \({\alpha }_{j}\) is the scale-depending constant and \({\beta }_{j}\) is the scaling exponent, both of which determine the shape of the power curve. As a function of Ni, we denote yij by yi(Nj).
Chen et al. proposed a statistical mechanics model to reconstruct interaction networks from abundance data76. This model is the combination of elements of different disciplines, from evolutionary game theory and ecosystem ecology theory through mathematical and statistical principles. Specifically, this model is founded on a system of quasi-dynamic mixed ordinary differential equations (mODE), expressed as
where the overall abundance change of genus j per HI change is decomposed into two components. The first component is the independent abundance change that is due to the self-regulation of genus j, which occurs when this genus is assumed to be in isolation. The magnitude of the independent change depends on the intrinsic capacity of this genus. The second component is the dependent abundance change that results from the regulation of all possible other genus \(j{\prime}\), whose sign and magnitude depend on how and how strongly this genus is affected by genus \(j{\prime}\). Based on these notions, the independent and dependent component changes are expressed as a function of the abundance of genus j and \(j{\prime}\), specified by parameters, \({\Theta }_{j}\) and \({\Theta }_{{jj}{\prime} }\), respectively. Here, two points are considered to fit the independent and dependent component curves. First, the fitted values of abundance of each genus from the power Eq. (1) are used as an exploratory variable. This treatment allows a universal physical law to underpin the basis of network reconstruction. Second, because no explicit forms exist, a nonparametric function, such as Legendre Orthogonal Polynomials, is used to smooth independent and dependent component curves.
Equation (2) is a full mODE model under which each microbe is linked with every other microbe. However, for an ecosystem composed of extremely many microbes, i.e., m is large, this full model forms a fully interconnected network, which is neither computationally feasible nor biologically justifiable. In living systems, a stable and robust ecosystem tends to be sparse. Thus, we need to reconstruct sparsely interconnected networks in which one microbe is only linked with a small set of other microbes. To obtain these small set of microbes, we implement regularity-based variable selection based on the mODE model of Eq. (2) to choose a set of the most significant genera (as predictors) (say dj) that affect a given genus (as a response). After then, the full model is reduced to a sparse model in which the summation of the dependent components is made over dj genera rather than m genera.
We implement the fourth-order Runge–Kutta algorithm to solve the reduced mODE equation. Following graph theory, we encode the independent components for individual genera as codes and the dependent components for individual genus pairs as edges into informative (capturing all interaction properties, such as sign, strength, and causality), dynamic (interaction properties as a function of HI), omnidirectional (characterizing all possible interactions each and every microbes may has), and personalized (illustrating interaction architecture specific to individual subjects) networks (idopNetworks). idopNetworks provides a tool to reveal how microbial interactions respond to the change of genetic background and identify the microbial cause of disease formation77.
Statistics
Data shown in the graphs were the mean ± SEM. Comparisons between the two groups were performed using the unpaired Student’s t-test. All statistical analyses were performed with GraphPad Prism 9.5.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The mass spectrometry proteomics data generated in this study have been deposited in the ProteomeXchange Consortium via the iProX partner repository78,79 under accession PXD056588. The RNA-seq data generated in this study have been deposited at Gene Expression Omnibus (GEO) database under accessions GSE279034, GSE279700, and GSE279702 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE279702]. The 16S rDNA seq data generated in this study are available in Supplementary Data 1. Source data are provided as a Source Data file. Uncropped western blot scans are shown in the Source Data File. Source data are provided with this paper.
References
Honap, S. et al. Navigating the complexities of drug development for inflammatory bowel disease. Nat. Rev. Drug Discov. 23, 546–562 (2024).
Peterson, L. W. & Artis, D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 14, 141–153 (2014).
Neurath, M. F. Strategies for targeting cytokines in inflammatory bowel disease. Nat. Rev. Immunol. 24, 559–576 (2024).
Danese, S. & Fiocchi, C. Ulcerative colitis. N. Engl. J. Med. 365, 1713–1725 (2011).
Holbrook, J. et al. Tumour necrosis factor signalling in health and disease. F1000Res. 8, F1000 (2019).
Ofengeim, D. & Yuan, J. Regulation of RIP1 kinase signalling at the crossroads of inflammation and cell death. Nat. Rev. Mol. Cell Biol. 14, 727–736 (2013).
Wang, C. et al. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412, 346–351 (2001).
Geng, J. et al. Regulation of RIPK1 activation by TAK1-mediated phosphorylation dictates apoptosis and necroptosis. Nat. Commun. 8, 359 (2017).
Morioka, S. et al. TAK1 kinase switches cell fate from apoptosis to necrosis following TNF stimulation. J. Cell Biol. 204, 607–623 (2014).
Orning, P. et al. Pathogen blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin D and cell death. Science 362, 1064–1069 (2018).
Chen, N. J. et al. Beyond tumor necrosis factor receptor: TRADD signaling in toll-like receptors. Proc. Natl Acad. Sci. USA 105, 12429–12434 (2008).
Mukherjee, T. et al. The NF-kappaB signaling system in the immunopathogenesis of inflammatory bowel disease. Sci. Signal 17, eadh1641 (2024).
Jostins, L. et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012).
Liu, J. Z. et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 47, 979–986 (2015).
Omori, E. et al. Epithelial transforming growth factor β-activated kinase 1 (TAK1) is activated through two independent mechanisms and regulates reactive oxygen species. Proc. Natl Acad. Sci. USA 109, 3365–3370 (2012).
Kajino-Sakamoto, R. et al. Enterocyte-derived TAK1 signaling prevents epithelium apoptosis and the development of ileitis and colitis. J. Immunol. 181, 1143–1152 (2008).
Kajino-Sakamoto, R. et al. TGF-beta-activated kinase 1 signaling maintains intestinal integrity by preventing accumulation of reactive oxygen species in the intestinal epithelium. J. Immunol. 185, 4729–4737 (2010).
Tan, S. et al. Hepatocyte-specific TAK1 deficiency drives RIPK1 kinase-dependent inflammation to promote liver fibrosis and hepatocellular carcinoma. Proc. Natl Acad. Sci. USA 117, 14231–14242 (2020).
Dillon, C. P. et al. Developmental checkpoints guarded by regulated necrosis. Cell Mol. Life Sci. 73, 2125–2136 (2016).
Alvarez-Diaz, S. et al. The pseudokinase MLKL and the kinase RIPK3 have distinct roles in autoimmune disease caused by loss of death-receptor-induced apoptosis. Immunity 45, 513–526 (2016).
Hsu, H. et al. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity 4, 387–396 (1996).
Degterev, A. et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat. Chem. Biol. 4, 313–321 (2008).
Plichta, D. R. et al. Therapeutic opportunities in inflammatory bowel disease: mechanistic dissection of host-microbiome relationships. Cell 178, 1041–1056 (2019).
Price, A. E. et al. A map of Toll-like receptor expression in the intestinal epithelium reveals distinct spatial, cell type-specific, and temporal patterns. Immunity 49, 560–575.e6 (2018).
Xia, S. et al. Gasdermin D pore structure reveals preferential release of mature interleukin-1. Nature 593, 607–611 (2021).
Brown, E. M., Clardy, J. & Xavier, R. J. Gut microbiome lipid metabolism and its impact on host physiology. Cell Host Microbe 31, 173–186 (2023).
Yilmaz, B. et al. Microbial network disturbances in relapsing refractory Crohn’s disease. Nat. Med. 25, 323–336 (2019).
Steck, N. et al. Enterococcus faecalis metalloprotease compromises epithelial barrier and contributes to intestinal inflammation. Gastroenterology 141, 959–971 (2011).
Fraschilla, I. et al. Immune chromatin reader SP140 regulates microbiota and risk for inflammatory bowel disease. Cell Host Microbe 30, 1370–1381.e5 (2022).
Herp, S. et al. The human symbiont Mucispirillum schaedleri: causality in health and disease. Med. Microbiol. Immunol. 210, 173–179 (2021).
Cao, X. et al. Modeling spatial interaction networks of the gut microbiota. Gut Microbes 14, 2106103 (2022).
Kotarsky, K. et al. A novel role for constitutively expressed epithelial-derived chemokines as antibacterial peptides in the intestinal mucosa. Mucosal Immunol. 3, 40–48 (2010).
Mohan, T., Deng, L. & Wang, B. Z. CCL28 chemokine: an anchoring point bridging innate and adaptive immunity. Int. Immunopharmacol. 51, 165–170 (2017).
Matsuo, K. et al. CCL28-deficient mice have reduced IgA antibody-secreting cells and an altered microbiota in the colon. J. Immunol. 200, 800–809 (2018).
Wang, S. et al. S100A8/A9 in inflammation. Front. Immunol. 9, 1298 (2018).
Kim, B.-H. et al. A family of IFN-γ–inducible 65-kD GTPases protects against bacterial infection. Science 332, 717–721 (2011).
Kirkby, M. et al. Guanylate-binding proteins: mechanisms of pattern recognition and antimicrobial functions. Trends Biochem. Sci. 48, 883–893 (2023).
Marshall, A. et al. Antimicrobial activity and regulation of CXCL9 and CXCL10 in oral keratinocytes. Eur. J. Oral. Sci. 124, 433–439 (2016).
Cao, X. et al. Lactoferrin: a glycoprotein that plays an active role in human health. Front. Nutr. 9, 1018336 (2022).
Meuter, S. & Moser, B. Constitutive expression of CXCL14 in healthy human and murine epithelial tissues. Cytokine 44, 248–255 (2008).
Lu, J. et al. CXCL14 as an emerging immune and inflammatory modulator. J. Inflamm. 13, 1 (2016).
Choi, J. S., Kim, K. H. & Lau, L. F. The matricellular protein CCN1 promotes mucosal healing in murine colitis through IL-6. Mucosal Immunol. 8, 1285–1296 (2015).
Jun, J.-I. & Lau, L. F. CCN1 is an opsonin for bacterial clearance and a direct activator of Toll-like receptor signaling. Nat. Commun. 11, 1242 (2020).
Kościuczuk, E. M. et al. Cathelicidins: family of antimicrobial peptides. A review. Mol. Biol. Rep. 39, 10957–10970 (2012).
Myszor, I. T. et al. Bile acid metabolites enhance expression of cathelicidin antimicrobial peptide in airway epithelium through activation of the TGR5-ERK1/2 pathway. Sci. Rep. 14, 6750 (2024).
Devireddy, L. R. et al. A cell-surface receptor for lipocalin 24p3 selectively mediates apoptosis and iron uptake. Cell 123, 1293–1305 (2005).
Toyonaga, T. et al. Lipocalin 2 prevents intestinal inflammation by enhancing phagocytic bacterial clearance in macrophages. Sci. Rep. 6, 35014 (2016).
Wang, X. et al. Characteristics of fecal microbiota and machine learning strategy for fecal invasive biomarkers in pediatric inflammatory bowel disease. Front. Cell. Infect. Microbiol. 11, 711884 (2021).
Mirsepasi-Lauridsen, H. C. et al. Escherichia coli pathobionts associated with inflammatory bowel disease. Clin. Microbiol. Rev. 32, e00060-18 (2019).
Viladomiu, M. et al. Adherent-invasive E. coli metabolism of propanediol in Crohn’s disease regulates phagocytes to drive intestinal inflammation. Cell Host Microbe 29, 607–619.e8 (2021).
De Buck, M. et al. Structure and expression of different serum amyloid A (SAA) variants and their concentration-dependent functions during host insults. Curr. Med. Chem. 23, 1725–1755 (2016).
Markart, P. et al. Comparison of the microbicidal and muramidase activities of mouse lysozyme M and P. Biochem. J. 380, 385–392 (2004).
van Beelen Granlund, A. et al. REG gene expression in inflamed and healthy colon mucosa explored by in situ hybridisation. Cell Tissue Res. 352, 639–646 (2013).
Wang, W. et al. Reg4 protects against Salmonella infection-associated intestinal inflammation via adopting a calcium-dependent lectin-like domain. Int. Immunopharmacol. 113, 109310 (2022).
Arijs, I. et al. Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PLoS ONE 4, e7984 (2009).
Eissa, N. et al. Chromofungin (CHR: CHGA(47-66)) is downregulated in persons with active ulcerative colitis and suppresses pro-inflammatory macrophage function through the inhibition of NF-κB signaling. Biochem. Pharm. 145, 102–113 (2017).
Zhang, B. et al. Investigation of the porcine MUC13 gene: isolation, expression, polymorphisms and strong association with susceptibility to enterotoxigenic Escherichia coli F4ab/ac. Anim. Genet. 39, 258–266 (2008).
Barker-Tejeda, T. C. et al. Comparative characterization of the infant gut microbiome and their maternal lineage by a multi-omics approach. Nat. Commun. 15, 3004 (2024).
Martinez-Guryn, K., Leone, V. & Chang, E. B. Regional diversity of the gastrointestinal microbiome. Cell Host Microbe 26, 314–324 (2019).
Weisel, K. et al. A randomised, placebo-controlled study of RIPK1 inhibitor GSK2982772 in patients with active ulcerative colitis. BMJ Open Gastroenterol. 8, e000680 (2021).
Ermolaeva, M. A. et al. Function of TRADD in tumor necrosis factor receptor 1 signaling and in TRIF-dependent inflammatory responses. Nat. Immunol. 9, 1037–1046 (2008).
Pobezinskaya, Y. L. et al. The function of TRADD in signaling through tumor necrosis factor receptor 1 and TRIF-dependent Toll-like receptors. Nat. Immunol. 9, 1047–1054 (2008).
Newton, K. et al. Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science 343, 1357–1360 (2014).
Polykratis, A. et al. Cutting edge: RIPK1 Kinase inactive mice are viable and protected from TNF-induced necroptosis in vivo. J. Immunol. 193, 1539–1543 (2014).
Sato, S. et al. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat. Immunol. 6, 1087–1095 (2005).
Wang, R. et al. Gut stem cell necroptosis by genome instability triggers bowel inflammation. Nature 580, 386–390 (2020).
Zhang, X. et al. MLKL and FADD are critical for suppressing progressive lymphoproliferative disease and activating the NLRP3 inflammasome. Cell Rep. 16, 3247–3259 (2016).
Sun, W. et al. Small molecule activators of TAK1 promotes its activity-dependent ubiquitination and TRAIL-mediated tumor cell death. Proc. Natl Acad. Sci. USA 120, e2308079120 (2023).
Kong, A. T. et al. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat. Methods 14, 513–520 (2017).
Chen, S. et al. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Roberts, A. et al. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 12, R22 (2011).
Anders, S., Pyl, P. T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019).
Chen, C. et al. An omnidirectional visualization model of personalized gene regulatory networks. NPJ Syst. Biol. Appl 5, 38 (2019).
Dong, A. et al. idopNetwork: a network tool to dissect spatial community ecology. Methods Ecol. Evol. 14, 2272–2283 (2023).
Ma, J. et al. iProX: an integrated proteome resource. Nucleic Acids Res. 47, D1211–d1217 (2019).
Chen, T. et al. iProX in 2021: connecting proteomics data sharing with big data. Nucleic Acids Res. 50, D1522–d1527 (2022).
Acknowledgements
The authors thank Dr. Shizuo Akira of Osaka University, Japan, Dr. Wei Mo of Zhejiang University, China, and Dr. Haibing Zhang of Shanghai Institute of Nutrition and Health, China, for kindly providing Tak1fl/fl mice, Villin-creERT2 mice, and Fadd−/− Mlkl−/− mice, respectively. This project was supported (to Junying Y.) in part by the China National Natural Science Foundation (82188101 and 92049303), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB39030200), Shanghai Science and Technology Development Funds (22JC1410400), the Shanghai Municipal Science and Technology Major Project (2019SHZDZX02), the Shanghai Basic Research Pioneer Project, the Shanghai Key Laboratory of Aging Studies (19DZ2260400), the Shanghai Municipal Science and Technology Major Project, the Basic Research Pioneer Project of the Science and Technology Commission of Shanghai Municipality (STCSM), (to B.S.) by the China National Natural Science Foundation (32370796), (to H.P.) by the China National Natural Science Foundation (32170754), (to R.W.) by the Interdisciplinary Project at the Shanghai Institute for Mathematics and Interdisciplinary Sciences (SIMIS-ID-2024-WN) and (to HY. J.) a fellowship (2022T150677) from the China Postdoctoral Science Foundation.
Author information
Authors and Affiliations
Contributions
This project was conceptualized and directed by Junying Y. The experiments were designed by ZY. S., Junying Y., and J.Y. Most of the experiments were performed and data analyzed by Z.S. and J.Y. H.P. helped with the project coordination. W. S. helped with in vitro kinase assay and western blot. L.J. and R.W. performed an idopNetwork analysis. C.X., J.X., and W.L. helped with the microbiota analysis. B.S. and M.Z. conducted mass spec analysis. J.L. helped with phosphorylation site analysis. H.J., T.Z., and M.H. helped with mouse care during the pandemic. The manuscript was written by J.Y., Z.S., and Junying Y.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sun, Z., Ye, J., Sun, W. et al. Cooperation of TRADD- and RIPK1-dependent cell death pathways in maintaining intestinal homeostasis. Nat Commun 16, 1890 (2025). https://doi.org/10.1038/s41467-025-57211-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-57211-z