Abstract
GWAS of Alzheimer’s disease have been predominantly based on European ancestry cohorts with clinically diagnosed patients. Increasing the ancestral diversity of GWAS and focusing on imaging brain biomarkers for Alzheimer’s disease may lead to the identification of new genetic loci. Here, we perform a GWAS on cerebral β-amyloid deposition measured by PET imaging in 3,885 East Asians and a cross-ancestry GWAS meta-analysis with data from 11,816 European participants. Our GWAS analysis replicates known loci (APOE4, CR1, and FERMT2) and identifies a novel locus near SORL1 that is significantly associated with β-amyloid deposition. Single-nucleus expression analysis shows that SORL1 is differentially expressed according to β-amyloid positivity in microglia. Our joint association analysis using the SORL1 lead variant (rs76490923) and the APOE4 allele demonstrates that the risk of β-amyloid deposition is reduced by up to 43.5% in APOE4 non-carriers and up to 55.6% in APOE4 carriers, according to the allelic dosage of the rs76490923 T allele. Our findings suggest that SORL1 may play an important role in the pathogenesis of Alzheimer’s disease, particularly in relation to β-amyloid deposition.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) is a complex neurodegenerative disease with a high genetic heritability estimated to be 60–80%1. Large genome-wide association studies (GWAS) have identified genetic variants associated with AD and highlighted important genes and pathways in AD pathology2,3. However, while over 70 genetic loci were identified by leveraging up to 1,126,563 samples, they only explained about 15% of the phenotypic variance in the diagnosis of AD3. This may be partially due to the failure of outcome phenotypes of GWAS to match the biology of the disease and the lack of sufficient ancestral diversity in study populations.
Over the past decades, the paradigm of AD diagnosis has shifted from clinical diagnosis to biomarker-based diagnosis. The recently revised criteria for diagnosis and staging of AD emphasize abnormalities on core biomarkers for the diagnosis of AD, of which amyloid positron emission tomography (PET) is the best validated4. However, most large GWAS on AD used clinically diagnosed AD or proxy AD using family history as the outcome phenotype2,3. Recently, GWAS analyses using imaging or fluid biomarkers to identify genetic factors that are more biologically relevant have been reported5,6,7,8,9. For instance, a GWAS meta-analysis using amyloid PET identified genome-wide significant loci in genes previously known to be associated with clinical AD, such as APOE, CR1, FERMT2, and ABCA7, along with a novel African-specific variant in PTDSS1. This observation supports their involvement in AD pathology via amyloid-related mechanisms and suggests the importance of cross-ancestry biomarker-based research in expanding our understanding of AD genetics5.
The ethnic diversity of study populations is also an essential factor that could contribute to significant discoveries. The majority of GWAS to date have been conducted primarily in European (EUR) populations, not only limiting the generalization of the findings in non-EUR populations but also inevitably missing many variants that are absent or rare in EUR. Given that only a small fraction of variants is common across different populations10, many biologically relevant but rare variants may not have reached genome-wide significance due to the insufficient statistical power. Therefore, multi-ethnic GWAS including non-EUR populations may identify novel genetic loci.
In this study, we performed a cross-ancestry GWAS meta-analysis of cerebral β-amyloid (Aβ) deposition in EUR and East Asian (EAS) populations. We identified variants associated with a core pathological biomarker of AD using quantified uptake on amyloid PET. We demonstrated that the lead variant in the SORL1 region is associated with SORL1 expression specifically in microglia, based on single-nucleus RNA sequencing (snRNA-seq) data from the ROSMAP (Religious Orders Study/Memory and Aging Project)11. Finally, we investigated whether SORL1 was differentially expressed according to Aβ pathology in single-cell transcriptome data of postmortem brain samples.
Results
β-amyloid genetic association analysis and meta-analysis in EAS population (stage 1)
For the discovery GWAS analysis (stage 1), Aβ and genome-wide genotype data for 3387 EAS samples were obtained from the Korea-Registries to Overcome and Accelerate Dementia research (K-ROAD) cohort from 2017 to 2023. The stage 1 participants had a mean age of 72.0 years (standard deviation [SD] = 8.4), and 63.4% were female (Supplementary Data 1). After stringent quality control (QC) and genotype imputation, we performed a GWAS for Aβ with overlapped 5,651,782 variants in 2049 GWAS microarray chip data and 1338 whole-genome sequencing (WGS) data (Supplementary Fig. 1). In addition, we obtained external summary statistics-level GWAS data in EAS, including 498 samples and 5,619,153 variants from a previous multi-ethnic GWAS performed by Ali et al.5. We then performed a genome-wide fixed-effect meta-analysis for Aβ between K-ROAD (chip array and WGS data) and the external GWAS summary statistics, which retained the association results for 5,110,277 autosomal variants with minor allele frequency (MAF) > 0.5% in both cohorts. A total of 3885 samples and 5,110,277 variants (stage 1) were included in the meta-analysis of EAS population (Fig. 1a).
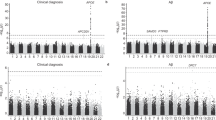
a Manhattan plot of the GWAS meta-analysis for Aβ deposition in EAS (stage 1, n = 3855). b Manhattan plot of the cross-ancestry GWAS meta-analysis for Aβ deposition in EAS and EUR populations (n = 15,701). p values for GWASs were calculated using a fixed-effect inverse variance-weighted meta-analysis. The x-axis represents chromosomal position, and the y-axis represents the −log10(p value) for the association of variants with Aβ deposition. Reported p values are two-sided and not corrected for multiple testing. The light green dots represent the genome-wide significant lead variants. The red text highlights previously unreported loci along with their mapped genes, while the black text indicates previously reported loci. The pink and blue horizontal dashed lines indicate the genome-wide significance level (p value = 5.00 × 10−8) and the genome-wide suggestive level (p value = 1.00 × 10−5), respectively. EAS East Asian, EUR European, GWAS genome-wide association study.
We identified three genome-wide significant loci (p < 5.00 × 10−8), comprising a total of 87 variants, from the GWAS meta-analysis for Aβ in EAS. The most significant association was observed with rs429358, a single-nucleotide polymorphism (SNP) involved in determining APOE4 status, which demonstrated a strong association with Aβ levels (beta = 0.5589, standard error [SE] = 0.0244, p = 8.70 × 10−116). Moreover, 73 variants were in high linkage disequilibrium (LD) with rs429358 and located near the APOE region on chromosome 19. On chromosome 11, a set of 12 variants, led by rs76490923 (beta = −0.1661, SE = 0.0279, p = 2.46 × 10−9), exceeded genome-wide significance and had not been previously reported in the largest Aβ GWAS5. These variants were located within the intron region of the SORL1 gene and demonstrated high LD with each other. In addition, the rs4851914 single variant located intron region of the ECRG4 gene was identified on chromosome 2 (beta = 0.3926, SE = 0.0712, p = 3.47 × 10−8) (Table 1). The λGC was 1.093, and the SNP-based heritability for Aβ was estimated to be 14.0% in the EAS GWAS meta-analysis.
Replication analysis for novel associations in EAS population (stage 2)
To replicate the significantly associated SNPs in the SORL1 and ECRG4 regions with Aβ level, we genotyped an additional 753 samples in the next phase of K-ROAD from 2023 to 2024. The stage 2 participants had a mean age of 70.8 years (SD = 8.8), and 62.4% were female (Supplementary Data 1). We estimated genetic relationships and selected 655 unrelated samples (stage 2) with the discovery samples (stage 1) that were eligible for replication analysis. We then performed linear regression analyses adjusted for age, sex, and five genetic principal components (PCs) with Aβ as the dependent variable using the significantly associated 12 SNPs in SORL1 and the rs4851914 variant in ECRG4.
In the replication analysis, we found that the 11 SNPs identified in the SORL1 region from the stage 1 GWAS were associated with Aβ levels. The lead SNP, rs76490923, was significantly associated with Aβ levels (beta = −0.1527, SE = 0.0561, p = 6.67 × 10−3), showing a similar effect size to the discovery results (beta = −0.1661 [stage 1] vs. −0.1527 [stage 2]). The detailed results are shown in Supplementary Data 2. In contrast, the variant rs4851914 in the ECRG4 region was not significantly associated with Aβ levels in the replication sample (beta = 0.0874, SE = 0.1100, p = 0.427).
Comparison of SORL1 variants for β-amyloid between EAS and EUR
To investigate ethnic differences in the identified SORL1 variants, we obtained the summary statistics of EUR GWAS for Aβ, including 11,816 samples and 5,622,864 variants from a large-scale EUR GWAS performed by Ali et al.4 and then compared these results with our GWAS signals.
In the EUR GWAS for Aβ, we found that rs76490923 had a marginally significant association (beta = −0.1240, SE = 0.0404, p = 2.20 × 10−3) with Aβ in EUR population. rs76490923 was associated with decreased Aβ levels and exhibited the same effect directionality (beta coefficient, −0.1240 [EUR] vs. −0.1661 [EAS]) as in our EAS GWAS results (Fig. 2). In contrast, the lead SNP, rs76490923, showed a notable difference in MAF between the two populations. According to the gnomAD database12, the MAF for rs76490923 differed nearly tenfold between EUR (non-Finnish, MAF: 0.021) and EAS (MAF: 0.210).
a Regional plot for SORL1 signals from the East Asian GWAS. b Regional plot for SORL1 signals from the European GWAS. c Regional plot for SORL1 signals from the cross-ancestry meta-GWAS. p values for GWASs were calculated using a fixed-effect inverse variance-weighted meta-analysis. Each dot is colored by r2 of linkage disequilibrium with the purple-colored lead SNPs indicated with texts. Reported p values are two-sided and not corrected for multiple testing. The x-axis represents chromosomal position, and the y-axis represents the −log10(p value) for the association of variants with Aβ deposition. EAS East Asian, EUR European, GWAS genome-wide association study, SE standard error, MAF minor allele frequency, gnomAD Genome Aggregation Database.
Cross-ancestry GWAS meta-analysis in EAS and EUR populations
To maximize the power of gene discovery, we conducted a cross-ancestry meta-analysis with Aβ GWAS summary statistics in EAS (stage 1 [n = 3885] in this study) and EUR (n = 11,816 from Ali et al.4) using METAL13.
A total of 15,701 samples and 6,648,274 variants were included in the cross-ancestry GWAS meta-analysis. The λGC was 1.035. We identified four genome-wide significant loci associated with Aβ levels (Fig. 1b). The APOE allele, rs429358, demonstrated the most significant association with Aβ levels (beta = 0.5982, SE = 0.0122, p < 1.00 × 10−320). The SORL1 signals on chromosome 11 (rs76490923, beta = −0.1526, SE = 0.0230, p = 3.09 × 10−11), initially identified in the EAS GWAS (stage 1), demonstrated even a greater significance level in the cross-ancestry meta-analysis (Fig. 2 and Supplementary Fig. 2). The remaining two loci had been previously reported in EUR GWAS (21 variants near the CR1 gene and 16 near the FERMT2 gene) and also exhibited greater significance levels in the cross-ancestry analysis than the EUR GWAS (Supplementary Data 3 and Supplementary Figs. 3–5). However, the ABCA7 locus on chromosome 19, previously reported in EUR GWAS4, was not significantly associated (rs12151021, beta = 0.0054, SE = 0.0317, p = 0.864) with Aβ in our EAS GWAS (stage 1).
At the suggestive level of association (p < 1.00 × 10−5), we identified an additional 24 independent genome-wide suggestive loci associated with Aβ (Supplementary Data 4). We then conducted a pathway enrichment analysis using the 28 lead SNPs and their corresponding mapped genes and gene ontology (GO) database (biological process 2023)14. This analysis revealed 10 significant GO pathways that met the Bonferroni correction threshold (p < 0.05/619) and 55 pathways that passed the false discovery rate (FDR) < 0.05 using Benjamini–Hochberg correction (Table 2 and Supplementary Data 5). Notably, pathways associated with Aβ formation and amyloid precursor protein (APP) catabolism were among the most significant, alongside those implicated in tau protein kinase activity and neuronal death.
SORL1 signals in AD case-control GWAS
To investigate whether the Aβ-associated SORL1/rs76490923 variant is also associated with AD risk, we performed a case-control association analysis for the SORL1/rs76490923 ± 2 Mb region using individual-level data from 4042 EAS participants across stages 1 and 2. After excluding individuals with mild cognitive impairment (MCI), 2043 participants were eligible for the association test, comprising 961 AD cases and 1082 cognitive unimpaired (CU) controls. The analysis revealed a significant protective association between the SORL1/rs76490923 variant and AD risk (odds ratio [OR] = 0.7657, 95% confidence interval [CI] = 0.6119–0.9196, p = 6.79 × 10−4) (Supplementary Fig. 6).
We also examined the association between SORL1/rs76490923 and clinical AD risk in previously established large-scale EUR AD or AD proxy GWASs with sample sizes ranging from 63,920 to 788,989 individuals2,3,15,16. Although the significance levels varied slightly, the SORL1/rs76490923 variant consistently showed a protective association against AD in the EUR population. Additionally, we identified rs11218343, an LD proxy variant for SORL1/rs76490923 (r² = 0.62), which exhibited a strong association with the EUR-based AD GWAS. rs11218343 (MAF = 0.039 in EUR) was a more frequent variant than rs76490923 (MAF = 0.021 in EUR) (Supplementary Fig. 7).
Joint association of SORL1 and APOE 4 with Aβ deposition
We performed a joint association analysis to investigate the complementary effects of the identified SORL1 signal and APOE4 on Aβ deposition risk using the 4042 participants (combined stage 1 and 2 datasets). This analysis involved SORL1/rs76490923 genotype dosage (0 [CC], 1 [TC or CT], or 2 [TT]) and APOE4 status (carrier or non-carrier) (Fig. 3).
a Aβ deposition risk between APOE4 carriers and non-carriers. b Aβ deposition risk according to APOE4 status and SORL1/rs76490923 genotype. c Aβ deposition risk according to SORL1/rs76490923 genotype stratified by APOE4 status. The joint association analyses were conducted on 4042 study samples, combining data from stage 1 and stage 2 datasets. The number of Aβ-positive cases represents participants classified as Aβ-positive according to the CERAD neuropathological category. Boxes represent the adjusted odds ratio, with horizontal lines around the boxes indicating 95% confidence intervals. The odds ratios were estimated using logistic regression models, adjusted for age, sex, cohort type, and the first five principal components of ancestry. p values were determined using a Wald test. p values for the trend were calculated using the Cochran-Armitage trend test. All p values were derived from two-sided tests. Exact statistical values are provided as a Source Data file. OR odds ratio, CI confidence interval, CERAD Consortium to Establish a Registry for Alzheimer’s disease.
We observed a gradual decremental association between increasing rs76490923 T allele dosage and a decreased risk of Aβ deposition. Non-carriers for APOE4 with rs76490923 alternative homozygote (TT) had the lowest risk for Aβ deposition (OR = 0.092, 95% CI = 0.059–0.143, p = 1.80 × 10−26) (Fig. 3b). In stratified analyses by APOE4 status, rs76490923 was associated with up to a 55.6% reduction in the risk of Aβ deposition among APOE4 carriers (OR = 0.444, 95% CI = 0.256–0.770, p = 3.85 × 10−3) and up to a 43.5% reduction among non-carriers (OR = 0.565, 95% CI = 0.370–0.862, p = 8.06 × 10−3) (Fig. 3c). All trend analyses showed significant (p < 0.001), but no significant interactions between SORL1 and APOE4 genotypes were identified (p = 0.391).
Brain single-nucleus RNA sequencing eQTL and colocalization analyses
To further investigate the link between SORL1 expression levels in the brain and the identified GWAS signals, we conducted an expression quantitative trait loci (eQTL) analysis using brain snRNA-seq expression data from Columbia ROSMAP17. Our results revealed that the lead SNP (rs76490923) was significantly associated with SORL1 expression levels in microglia (beta = 0.393, SE = 0.087, p = 8.81 × 10−6) among seven cell types in the dorsolateral prefrontal cortex (Table 3 and Supplementary Fig. 8). Furthermore, we found that GWAS signals near the SORL1 gene also overlapped with the microglia-specific eQTL signals of SORL1 (posterior probability of H4 = 0.56).
Brain single-nucleus RNA sequencing data and DEG analysis
We additionally investigated whether the SORL1 gene was differentially expressed according to Aβ positivity using cell-type specific differentially expressed genes (DEG) in the brain using the snRNA-seq data derived from 15 EAS individuals. The expression levels of SORL1 (log fold change = −2.94, SE = 0.121, FDR corrected p = 0.017) were lower in Aβ-positive patients in microglia cell type than in controls (Fig. 4 and Table 4).
a UMAP colored by major brain single-cell types. b UMAP plot colored by Aβ positivity across single-cell types. The red represents Aβ-positive and blue represents Aβ-negative. c SORL1 gene expression across single-cell types colored by z-scored expression levels. The red indicates higher gene expression, and the blue indicates lower expression. d Box plots for differential gene expression of SORL1 by Aβ positivity in single-cell types. Aβ positivity was defined based on the CERAD neuropathological category. Differential gene expression analysis was performed using MAST (v1.16.0) and brain cell-type-specific gene expression data from 15 Korean individuals (nine Aβ-positive and six Aβ-negative individuals). The asterisk indicates a significant result obtained from the likelihood ratio test that meets the threshold of a false discovery rate-corrected p value < 0.05, calculated using the Benjamini–Hochberg correction from two-sided tests. This box plot illustrates the differential expression of SORL1 across major brain cell types, grouped by Aβ positivity (positive in red and negative in blue). On the box plots, the horizontal line indicates the median, the box indicates the first to third quartile of expression and whiskers indicate 1.5 × the interquartile range. The x-axis represents various cell types, and the y-axis represents SORL1 log normalization expression. Source data are provided as a Source Data file. UMAP uniform manifold approximation, and projection, UMI unique molecular identifiers, MG microglia, OD oligodendrocytes, IN inhibitory neurons, OPC oligodendrocyte precursor cells, End endothelial cells, Ast astrocytes, Ext excitatory neurons, ns non-significant, CERAD Consortium to Establish a Registry for Alzheimer’s disease.
Discussion
We performed a cross-ancestry GWAS using our Korean genotype-phenotype cohort to identify genetic variants associated with amyloid pathology. By including a substantial proportion of EAS samples with amyloid PET data, we identified four loci with genome-wide significant association and 24 additional suggestive loci. The SNPs identified in our analysis were enriched in pathways related to Aβ formation and APP catabolism, as well as pathways implicated in tau protein kinase activity and neuronal death—key processes in AD pathogenesis. Ancestral diversity and precise phenotype enabled us to identify SORL1 as the most significant locus besides the well-known APOE gene. Our joint association analysis using the SORL1 lead variant and APOE4 allele showed that the risk of Aβ positivity was significantly diminished by carrying the SORL1 variant.
Clinical diagnosis of AD is made based on clinical and neuropsychological examination and the absence of other diseases on structural brain imaging. However, because it takes more than 15 years from the deposition of Aβ in the brain to the manifestation of cognitive impairment18, many individuals with AD pathophysiology can be included in the control group. Also, individuals with the same amount of pathology can either be symptomatic or asymptomatic depending on their cognitive reserve19. Furthermore, AD can present with atypical symptoms and non-AD neurodegenerative diseases can mimic AD20, making accurate clinical diagnosis challenging. Therefore, GWAS using clinical diagnosis labels can be limited and biased by the heterogeneity of study participants in both case and control groups. As a promising alternative, genetic studies using phenotypes based on imaging biomarkers are needed.
Previous GWAS meta-analysis using amyloid PET reported four genome-wide significant loci associated with cerebral Aβ deposition. Our GWAS of the largest EAS individuals to date identified significant associations at a previously unreported locus near the SORL1 gene. While this locus was not identified in previous EUR amyloid PET GWAS, the lead SNP showed an association with amyloid PET uptake at a nominal significance with the same effect direction (beta = −0.1240, SE = 0.0404, p = 2.20 × 10−3). In line with this, the association level was further strengthened in meta-analyses with EAS and EUR GWAS. This suggests that variants in SORL1 may have shared pathophysiological effects between EAS and EUR but their associations were not identified in the EUR GWAS partly due to insufficient statistical power because of their low MAF.
We further investigated whether the identified SORL1/rs76490923 was associated with AD risk. Our findings confirmed that the SORL1/rs76490923 variant or its LD proxy variant may have a potential protective effect against AD risk across populations. Despite variations in allele frequency, this protective association was consistently observed in both EAS and EUR populations. These results suggested that the identified SORL1/rs76490923 variant plays an important role in the pathogenic mechanism of AD following that of Aβ and exemplifies the necessity of studying ancestry-specific variants and the rationale for conducting genetic studies in diverse non-EUR populations.
We confirmed that the SORL1/rs76490923 T allele was significantly associated with the upregulation of SORL1 expression levels in microglia, among seven cell types in the dorsolateral prefrontal cortex using brain snRNA-seq. In addition, DEG analysis revealed that SORL1 expression levels were significantly lower in microglia from Aβ-positive individuals than in those from controls. Our results aligned with this previous knowledge in that the levels of SORL1 expression were decreased in Aβ-positive individuals21,22. Since SORL1 is implicated in the reduction of amyloidogenic processing of APP23,24, this upregulation is particularly significant. Our snRNA-seq DEG analysis supports this, demonstrating that higher SORL1 expression correlates with a reduced Aβ burden. Therefore, the protective effect of the rs76490923 T allele on amyloid positivity may be mediated through enhanced SORL1 expression in microglia, leading to decreased Aβ accumulation. This underscores the critical role of microglial SORL1 in modulating Aβ pathology and highlights the potential of targeting microglial pathways to develop therapeutic strategies for AD.
Our joint association analysis using the SORL1 lead variant (rs76490923) and the APOE4 allele demonstrated that the risk of Aβ positivity was reduced by up to 43.5% in APOE4 non-carriers and up to 55.6% in APOE4 carriers, according to the allelic dosage of the rs76490923 T allele. In the current clinical setting, the routine use of amyloid PET is constrained by financial considerations and accessibility. Among the tests currently performed in clinical settings, genotyping APOE4 is considered the most important for predicting the risk of Aβ positivity. However, given that the APOE4 genotype alone still does not explain a substantial portion of variability in Aβ positivity, the identification of additional genetic risk factors may enable more precise risk evaluation. These genetic risk factors may be ancestry-specific and may need to be applied in real-life practice under consideration of ethnicity.
Although SORL1 has not been identified in previous amyloid PET GWAS, variants in SORL1 have been identified in relation to familial and sporadic AD. Prior studies showed that rare variants in SORL1 can be the major cause of familial AD without the well-known APP, PSEN1, and PSEN2 mutations25,26. Also, a recent whole-exome sequencing study showed that the gene-based burden of rare damaging variants in SORL1 had significant impact on AD27. In terms of sporadic AD, SORL1 has been identified in large-scale case-control GWASs3,16. In EAS GWASs, association signals at SORL1 tended to be more pronounced despite relatively small sample sizes28,29. The SORL1 gene encodes a transmembrane protein SORLA which is involved in the trafficking and recycling of endosomal cargoes including APP, which is a key protein in the development of cerebral amyloidosis30,31,32,33. Specifically, depletion of SORL1 leads to longer residence of APP in the early and recycling endosomes, which is a favorable environment for amyloidogenic cleavage of APP by BACE134.
There are several limitations in our study. First, while we used temporal validation scheme using samples having difference in calendar time from the discovery set, replication in an independent cohort is needed to confirm our finding regarding the SORL1 variant. Second, the ROSMAP data used in the snRNA-seq eQTL and colocalization analyses are derived from only EUR while our main analysis was performed in EAS data. This may have contributed to rather marginal results in our colocalization analysis. Despite the discrepancy, the significant eQTL result in microglia was in line with the differential expression analysis using Korean autopsy samples. Lastly, the autopsy data used in differential expression analysis is relatively small in sample size. Nevertheless, SORL1 showed significant differential expression in microglia despite small sample size even when using the more conservative DEG analysis tool demonstrating the biological impact of this gene.
In summary, by utilizing the EAS genotype-phenotype cohort with quantified amyloid PET measurements, our cross-ancestry GWAS meta-analysis identified a previously unreported association of SORL1 variants with Aβ and replicated known associations. SORL1 demonstrated significant differential expression in microglia between Aβ-positive and Aβ-negative individuals. Our findings suggest that SORL1 may have potential value in the diagnosis and treatment of AD, particularly considering the growing importance of early diagnosis and aggressive treatment of amyloidosis.
Methods
Ethics
This study complied with all relevant ethical regulations for research involving human participants and was conducted in accordance with the criteria set by the Declaration of Helsinki. This study received approval from the institutional review board (IRB) of Samsung Medical Center, and written informed consent was provided by all participants. WGS and genotyping using microarray were conducted using blood samples obtained from the participants. The collection, storage, and analyses of biospecimens, genetic data, and data as part of the K-ROAD were approved under the Samsung Medical Center; IRB No. 2022-07-092. All data were handled in accordance with relevant data protection and privacy regulations.
Study population
A total of 4381 individuals of Korean descent with either WGS or GWAS microarray chip data were recruited from the Korean dementia hospital-based cohort named K-ROAD. As an open cohort with ongoing data accumulation, K-ROAD aims to develop a genotype–phenotype cohort to accelerate the development of advanced diagnostic and therapeutic techniques for AD and related other dementias. Overall, 25 university-affiliated hospitals in South Korea participated in the K-ROAD cohort. Eligible participants were individuals on the spectrum of Alzheimer’s clinical syndrome—CU, MCI, and DAT—who underwent amyloid PET imaging. We used genomic data generated up to 2022 for the discovery analysis (stage 1) and data generated from 2023 for the replication analysis (stage 2). Detailed characteristics of the participants by stage are shown in Supplementary Data 1.
Phenotype definitions
All participants were assessed through clinical interviews, neurological examinations, neuropsychological testing, and brain magnetic resonance imaging (MRI). After these evaluations, clinical diagnoses were established by consensus among a multidisciplinary team. CU was defined to have (1) no medical or psychiatric history that is likely to affect cognitive function (2) no objective cognitive impairment observed after a comprehensive neuropsychological test on any cognitive domain (above the −1.0 SD of age- and education-matched norms in memory and below −1.5 SD in other cognitive domains). MCI was diagnosed as having: (1) subjective cognitive complaints by the participants or caregiver; (2) objective cognitive impairment in any cognitive domain (below the −1.0 SD of age- and education-matched norms in memory or below −1.5 SD in other cognitive domains); and (3) no significant impairment in activities of daily living. The participants with dementia of the Alzheimer type (DAT) met the NIA-AA diagnostic criteria for AD dementia35.
All K-ROAD participants underwent Aβ PET with either 18F-florbetaben (FBB) or 18F-flutemetamol (FMM). To quantify the Aβ burden upon PET scans as Centiloids (CL), we followed the methods described in our previous study36. In brief, T1-weighted MR images were co-registered onto the MNI-152 template, followed by co-registration of individual PET images to the corresponding MRI images. The PET and MRI images were spatially normalized using the transformation parameters of the SPM8 unified segmentation method of T1-weighted MRIs. A whole cerebellum mask downloaded from the GAAIN website (http://www.gaain.org/) was used as the reference region. The common cortical target volume of interest (VOI) for FMM and FBB PET was defined by subtracting the average image of 18 Aβ-negative old controls from the average image of Aβ-positive DAT participants in a head-to-head study cohort37. After calculating the individual standardized uptake values of the common cortical target VOI in all PET images, we converted the values into CL units using the equation from the original paper by Klunk et al.38:
Here, SUVRind refers to the individual standardized uptake value ratio (SUVR) values, whereas SUVRYC and SUVRDAT denote the group mean SUVR values from Aβ-negative young controls and Aβ-positive DAT patients, respectively.
All imaging analyses for the K-ROAD study were conducted at the laboratory of Samsung Medical Center, which served as a core center. While centiloid values were used as the main outcome variable, Aβ positivity as determined by expert visual assessment was used in our joint association analysis. Visual assessment was performed by board-certified nuclear medicine physicians according to the guidelines of each radiotracer manufacturer39,40.
Genotyping and imputation
DNA used for genotyping K-ROAD participants was collected from whole blood. The majority of samples were genotyped using the Illumina Asian Screening Array (ASA) BeadChip (Illumina, CA, USA) (n = 2314). Additionally, 496 participants were genotyped on the customized Korea Biobank array (KBA) chip (Affymetrix, CA, USA).
We performed QC for both types of SNP data. SNPs were removed using the following criteria: (1) call rate of <98%, (2) MAF of <1%, or (3) genotype frequencies significantly deviating from the Hardy–Weinberg equilibrium (HWE) with a p value of <10−6. After QC, the genotype data were imputed to estimate genotypes for variants that were not directly genotyped and to combine datasets of different genotyping arrays (ASA chip and KBA chip). Imputation was conducted using the Minimac4 software with all available reference haplotypes from the Trans-Omics for Precision Medicine (TOPMed) at the Michigan Imputation Server. Consequently, we performed post-imputation QC with (1) an MAF of <1% or (2) a low imputation quality (R2 < 0.8 for imputed SNPs). Genome annotations were generated using the GRCh38 assembly.
Based on the genotype data, participants were excluded in accordance with the following criteria: (1) call rate of <95%, (2) sex mismatch, (3) heterozygosity excess (±5 SDs from the mean), or (4) one of the related pairs of individuals with second-degree or closer relationships estimated using the KING software41. After QC, 2049 samples remained available for genetic association analyses in stage 1, and 526 samples remained in stage 2.
WGS data alignment and variant calling
Genomic DNA was extracted from blood samples using the QIAmp DNA Mini Kit (QIAGEN). For sequencing, library preparation was performed with the TruSeq® DNA PCR-Free Library Prep Kit (Illumina) with size selection DNA performed with Covaris ultrasonication using 1 µg of input DNA for an average insert size of 350 bp and sequencing was performed at an average depth of 30X with paired-end sequencing using NovaSeq™ 6000 instrument with an S4 flow cell.
The paired-end raw sequencing data were initially processed by quality trimming, adapter trimming, removing short sequences, and hard-trimming using trim galore software (RRID:SCR_011847) (https://github.com/FelixKrueger/TrimGalore). Subsequently, the sequenced reads were aligned to the hg38 reference genome using BWA-MEM software42,43. After alignment, duplicates were removed using GATK MarkDuplicate44,45,46. Base quality score recalibration was conducted using BaseRecalibrator with WGS interval contig, insertions and deletions (Indels) from Mills and 1000 G gold standard, known Indels from Homo sapiens assembly41, and high confidence SNPs from 1000 G phase1. Next, germline SNPs and Indels calling were performed using HaplotypeCaller, and base quality score recalibration was conducted using ApplyBQSR. GenotypeGVCFs was used to produce gVCFs, and a DB folder was created using genomicsdbimport. Finally, gVCFs from the DB folder were combined using CombineGVCFs.
WGS quality control
QC of variants and samples was mainly performed by hail (https://github.com/hail-is/hail). Firstly, we conducted pre-filtering and genotype QC. Pre-filtering included splitting multi-allelic variants, Variant Quality Score Recalibration (VQSR) filtering, allele count greater than 0, removing low complexity regions (LCRs), and removing a duplicated sample (n = 1). Genotype QC was performed using criteria including genotype quality (GQ) (GQ ≥ 20), allele balance (AB) (hetero-variants AB ≥ 0.2 and ≤0.8, homo-variants AB ≥ 0.9), and read depth (DP) (autosomal DP ≥ 10 and ≤200, chrX DP (female) ≥10 and ≤200, chrX DP (male) ≥5 and ≤200, chrY DP ≥ 5 and ≤200).
For sample QC, we used high-confidence variants based on the criteria including bi-allelic variants, high call rates (>0.95), and common single-nucleotide variants (allele frequency >0.1%). We excluded samples with low coverage (mean depth ≥15) and low sample-level call rate (missingness ≥0.9). Samples with unmatched sex (f stat for female <0.2, f stat for male >0.8) or ambiguous sex (f stat >0.3 and <0.8) were excluded. For the next steps, we applied different variant QC criteria including only autosomal variants, bi-allelic variants, high call rate (>0.95), and allele frequency (>5%). Relatedness was calculated using KING39, and samples up to the second degree were excluded, keeping only one sample. The sample removal process followed a specific prioritization order. Samples with a diagnosis of AD were prioritized, followed by those of older age. Additionally, samples from earlier sequencing batches were prioritized for inclusion. After removing related samples, principal component analysis (PCA) was performed using PLINK 2.047.
After sample QC, we removed samples that failed sample QC from the raw VCF file. We repeated pre-filtering and genotype QC, including VQSR, LCRs, allele balance, GQ, and DP. We excluded variants with excess heterozygosity (inbreeding coefficient < −0.3), high missing rate (call rate <0.9), and high HWE of control samples (HWE >1e−09). We divided variants into SNPs and Indels, and QC procedures were conducted separately. SNPs were filtered based on QD ≥ 2, SOR ≤ 3, FS ≤ 60, MQ ≥ 50, MQRankSum ≥ −12.5, and ReadPosRankSum ≥ −8.0. Indels were filtered based on QD ≥ 2, FS ≤ 200, ReadPosRankSum ≥ −20, MQ ≥ 50, MQRankSum ≥ −12.5.
In the final step, after merging SNP and Indel variants, we conducted the final sample QC. Samples exceeding 5 SDs from criteria, such as the number of SNPs, Indels, transition/transversion (Ti/Tv), hetero/homo-variants, and the ratio of Indels, hetero/homo-variants, and Ti/Tv, were excluded. After QC, 1338 samples remained available for genetic association analyses in stage 1, and 129 samples remained in stage 2.
Identification of the genetic ancestry group
To confirm the ancestral background of our study sample, we conducted PCA for the 1000 Genomes Project samples and projected 4042 participants (K-ROAD) from stages 1 (n = 3387) and 2 (n = 655) onto the PCA plot to check the ancestral distribution.
Genome-wide association study and meta-analysis
We conducted GWASs for Aβ using a linear regression model with age, sex, and the first five PCs of ancestry as covariates. To account for the non-normal distribution of each amyloid PET level and to ensure the robustness of our results, the rank-based inverse normal transformation was applied to each amyloid PET level. An inverse variance-weighted fixed-effects meta-analysis of each EAS and meta-GWAS result was performed using METAL13.
Cis-eQTL mapping and colocalization
To test for cis-eQTLs, we used the tensorQTL v.1.0.2 cis_nominal mode with genotypes and gene expression matrix with cis-eQTL mapping software (https://github.com/RajLabMSSM/QTL-mapping-pipeline). We filtered out the individuals under 10 nuclei by each major cell type. Pseudo-bulk gene expression matrices were averaged in all counts for each gene in each individual by each cell type. As input covariates to analysis, we included PEER48 factors from 30 to 70 by each brain cell type and the first four PCA of genotypes. Each SNP–gene pair used a 1-Mb window within the transcription start site of a gene. We used the tensorQTL cis permutation pass per gene with 1000 permutations. To test whether SNPs from the GWAS colocalized with bulk RNA-seq or single-nucleus RNA-seq expression QTLs14,49, we used the COLOC package (v3.2-1). We extracted a significant genome-wide locus within 1 Mb on either side of the lead SNP (2 Mb-wide region total) in the GWAS. In each QTL dataset, we filtered all SNPs by each gene matched with a significant genome-wide locus within 100 kb to test colocalization. Missing MAF was used by reference values from the 1000 Genomes (phase 3) EUR superpopulations. Matching sets of SNPs colocalized by comparing the p value distributions.
Korean single-nucleus RNA sequencing
Korean single-cell transcriptomics data were generated using the dorsolateral prefrontal cortex from 15 individuals who underwent autopsy at Samsung Medical Center. All collected tissue samples were frozen and stored at −80 °C. The tissue pieces were homogenized and sorted using a FACSMelody cell sorter (BD) via flow cytometry (Supplementary Fig. 9). The sorted nuclei were used in the library construction following Chromium 3’ v3.1 protocol (10x Genomics, Cat-No.100268). The quality of library was confirmed using Agilent TapeStation (Agilent) and Qubit fluorometer (Thermo Fisher Scientific). Sequencing was performed on a NovaSeq6000 platform (Illumina) under conditions specified in the 10x Genomics protocol. Of the individuals, 9 were Aβ-positive and 6 were Aβ-negative by Consortium to Establish a Registry for Alzheimer’s disease (CERAD) neuropathological category (none to sparse: negative, moderate to frequent: positive). To obtain the gene count, we used the Cell Ranger software50 (v.6.1.2) (10x Genomics) with the GRCh38 assembly. The Cell Ranger count pipeline, including the pre-mRNA, was processed to account for unspliced nuclear transcripts. The gene-count matrix of all libraries was generated by the Cell Ranger aggr pipeline with Cell Ranger 3.0 default parameters to call cell barcodes.
To process snRNA-seq, we used SCANPY (v1.9.8)51. First, we excluded the outlier nuclei (the range [Q1 − 3(Q3 − Q1), Q3 + 3(Q3 − Q1)], with Q1 as the lower quartiles and Q3 as the upper quartiles) in terms of the number of genes, total counts, and percentage of mitochondrial genes. Second, we removed the doublets labeled cells using Scrublet (v.0.2.3)52. After filtering out the 11,780 nuclei, 88,622 nuclei were retained. The integration method to remove single-nucleus platform, dataset-specific batch effects was performed by Harmony53, which is the external function of SCANPY using individual and batches with normalized gene expression. After that, to annotate major brain cell types based on published snRNA-seq data, we projected annotations of brain cell types defined by the Allen Brain Institute (https://portal.brain-map.org/atlases-and-data/rnaseq/human-multiple-cortical-areas-smart-seq) onto this study.
Differential gene expression analysis
Differential gene expression of genes comparing Aβ positivity by CERAD neuropathological category was done using the MAST algorithm (v.1.16.0)48, which implements a two-part hurdle model. The genes with expression in greater than 10% of nuclei were required to be considered for differential gene expression analysis by each cell type. The fixed-effect covariates included batch, age, sex, and status (Aβ positivity) on each cell type. The individuals are a random-effect covariate in the model for the interindividual correlation of expression data. We tested differential expression analysis across all major cell types to estimate the effect of disease on expression. The significant differential express genes filtered by FDR < 0.05 using Benjamini–Hochberg correction. Using the discrete coefficient of MAST, we defined the coefficient estimate of the effect of Aβ positivity on expression.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The full summary statistics of Korean Aβ GWAS (stage 1) are publicly available at the NHGRI-EBI GWAS Catalog under the accession number GCST90483382. The raw genotype and single-nuclei RNA-seq data for Koreans are accessible for collaborative research under restricted conditions to protect participant privacy. For inquiries regarding data access, please contact the corresponding authors (S.W.Seo (sangwonseo@empas.com) and H.-H.Won (wonhh@skku.edu)). Data access requests will be reviewed and responded to within 2 to 4 weeks of receipt. The genotype data for ROSMAP are available under restricted access to protect participant privacy and can be accessed by submitting a data Data Use Certificate (DUC) through Synapse at https://www.synapse.org/#!Synapse:syn10901595. Similarly, the single-nuclei RNA-seq data for ROSMAP are available under restricted access and can be accessed via Synapse at https://www.synapse.org/#!Synapse:syn31512863. Instructions for submitting a DUC can be found at https://adknowledgeportal.synapse.org/Data%20Access. Source data are provided with this paper.
Code availability
All software used in the study is publicly available as described in the “Methods” and “Reporting summary”. The single-cell processing pipeline, differentially expressed gene analysis, and other custom scripts for colocalization are available at https://doi.org/10.5281/zenodo.14791596.
References
Gatz, M. et al. Role of genes and environments for explaining Alzheimer disease. Arch. Gen. Psychiatry 63, 168–174 (2006).
Wightman, D. P. et al. A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer’s disease. Nat. Genet. 53, 1276–1282 (2021).
Bellenguez, C. et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet. 54, 412–436 (2022).
Jack, C. R. Jr. et al. Revised criteria for diagnosis and staging of Alzheimer’s disease: Alzheimer’s Association Workgroup. Alzheimers Dement. 20, 5143–5169 (2024).
Ali, M. et al. Large multi-ethnic genetic analyses of amyloid imaging identify new genes for Alzheimer disease. Acta Neuropathol. Commun. 11, 68 (2023).
Nho, K. et al. CYP1B1-RMDN2 Alzheimer’s disease endophenotype locus identified for cerebral tau PET. Nat Commun 15, 8251 (2024).
Stevenson-Hoare, J. et al. Plasma biomarkers and genetics in the diagnosis and prediction of Alzheimer’s disease. Brain 146, 690–699 (2023).
Bradley, J. et al. Genetic architecture of plasma Alzheimer disease biomarkers. Hum. Mol. Genet. 32, 2532–2543 (2023).
Jansen, I. E. et al. Genome-wide meta-analysis for Alzheimer’s disease cerebrospinal fluid biomarkers. Acta Neuropathol. 144, 821–842 (2022).
Biddanda, A., Rice, D. P. & Novembre, J. A variant-centric perspective on geographic patterns of human allele frequency variation. Elife 9, e60107 (2020).
Bennett, D. A. et al. Religious Orders Study and Rush Memory and Aging Project. J. Alzheimers Dis. 64, S161–S189 (2018).
Gudmundsson, S. et al. Variant interpretation using population databases: lessons from gnomAD. Hum. Mutat. 43, 1012–1030 (2022).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Milacic, M. et al. The reactome pathway knowledgebase 2024. Nucleic Acids Res. 52, D672–D678 (2024).
Kunkle, B. W. et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 51, 414–430 (2019).
Jansen, I. E. et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 51, 404–413 (2019).
Fujita, M. et al. Cell subtype-specific effects of genetic variation in the Alzheimer’s disease brain. Nat. Genet. 56, 605–614 (2024).
Villemagne, V. L. et al. Amyloid β deposition, neurodegeneration, and cognitive decline in sporadic Alzheimer’s disease: a prospective cohort study. Lancet Neurol. 12, 357–367 (2013).
Stern, Y. Cognitive reserve in ageing and Alzheimer’s disease. Lancet Neurol. 11, 1006–1012 (2012).
Shim, Y. S., Roe, C. M., Buckles, V. D. & Morris, J. C. Clinicopathologic study of Alzheimer’s disease: Alzheimer mimics. J. Alzheimers Dis. 35, 799–811 (2013).
Dodson, S. E. et al. LR11/SorLA expression is reduced in sporadic Alzheimer disease but not in familial Alzheimer disease. J. Neuropathol. Exp. Neurol. 65, 866–872 (2006).
Scherzer, C. R. et al. Loss of apolipoprotein E receptor LR11 in Alzheimer disease. Arch. Neurol. 61, 1200–1205 (2004).
Caglayan, S. et al. Lysosomal sorting of amyloid-β by the SORLA receptor is impaired by a familial Alzheimer’s disease mutation. Sci. Transl. Med. 6, 223ra20 (2014).
Rogaeva, E. et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat. Genet. 39, 168–177 (2007).
Pottier, C. et al. High frequency of potentially pathogenic SORL1 mutations in autosomal dominant early-onset Alzheimer disease. Mol. Psychiatry 17, 875–879 (2012).
Nicolas, G. et al. SORL1 rare variants: a major risk factor for familial early-onset Alzheimer’s disease. Mol. Psychiatry 21, 831–836 (2016).
Holstege, H. et al. Exome sequencing identifies rare damaging variants in ATP8B4 and ABCA1 as risk factors for Alzheimer’s disease. Nat. Genet. 54, 1786–1794 (2022).
Shigemizu, D. et al. Ethnic and trans-ethnic genome-wide association studies identify new loci influencing Japanese Alzheimer’s disease risk. Transl. Psychiatry 11, 151 (2021).
Miyashita, A. et al. SORL1 is genetically associated with late-onset Alzheimer’s disease in Japanese, Koreans and Caucasians. PLoS ONE 8, e58618 (2013).
Mishra, S. et al. The Alzheimer’s gene SORL1 is a regulator of endosomal traffic and recycling in human neurons. Cell Mol. Life Sci. 79, 162 (2022).
Hansen, D. V. et al. Microglia in Alzheimer’s disease. J. Cell Biol. 217, 459–472 (2018).
Barthelson, K., Newman, M. & Lardelli, M. Sorting out the role of the sortilin-related receptor 1 in Alzheimer’s disease. J. Alzheimers Dis. Rep. 4, 123–140 (2020).
Andersen, O. M. et al. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc. Natl. Acad. Sci. USA 102, 13461–13466 (2005).
Sun, J. & Roy, S. The physical approximation of APP and BACE-1: a key event in Alzheimer’s disease pathogenesis. Dev. Neurobiol. 78, 340–347 (2018).
McKhann, G. M. et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 7, 263–269 (2011).
Kim, S. J. et al. Development and clinical validation of CT-based regional modified Centiloid method for amyloid PET. Alzheimers Res. Ther. 14, 157 (2022).
Cho, S. H. et al. A new Centiloid method for 18 F-florbetaben and 18 F-flutemetamol PET without conversion to PiB. Eur. J. Nucl. Med. Mol. Imaging 47, 1938–1948 (2020).
Klunk, W. E. et al. The Centiloid Project: standardizing quantitative amyloid plaque estimation by PET. Alzheimers Dement. 11, 1–15 (2015).
Collij, L. E. et al. Visual assessment of [(18)F]flutemetamol PET images can detect early amyloid pathology and grade its extent. Eur. J. Nucl. Med. Mol. Imaging 48, 2169–2182 (2021).
Barthel, H. et al. Cerebral amyloid-β PET with florbetaben (18F) in patients with Alzheimer’s disease and healthy controls: a multicentre phase 2 diagnostic study. Lancet Neurol. 10, 424–435 (2011).
Manichaikul, A. et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Van der Auwera, G. A. et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinforma. 43, 11.10.1–11.10.33 (2013).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Finak, G. et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015).
Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014).
Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291.e9 (2019).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Acknowledgements
This research was supported by the Future Medicine 2030 Project of the Samsung Medical Center (SMX1250081 awarded to S.W.Seo and H.-H.Won); the National Research Foundation of Korea (NRF) (RS-2023-00262527 and RS-2023-00223277 awarded to H.-H.Won, RS-2019-NR040057 awarded to S.W.Seo, RS-2023-00247408 awarded to J.P.Kim); the Korea Dementia Research Project through the Korea Dementia Research Center (KDRC), funded by the Ministry of Health & Welfare and Ministry of Science and ICT, Republic of Korea (RS-2022-KH125557 awarded to H.-H.Won, RS-2020-KH106434 awarded to S.W.Seo); This research was supported by the “Korea National Institute of Health” research project (2023-ER1001-02 awarded to J.P.Kim, 2024-ER1003-01 awarded to S.W.Seo); This work was partly supported by Institute of Information & communications Technology Planning & Evaluation (IITP) grant funded by the Korea government (MSIT) (RS-2021-II212068, Artificial Intelligence Innovation Hub, awarded to S.W.Seo); This work was supported by the Postdoctoral Research Program of Sungkyunkwan University (2023, awarded to S.-H.Jung); This study utilized BeauBrain Amylo’s image processing technology to quantify amyloid uptakes using PET-CT.
Author information
Authors and Affiliations
Contributions
J.P.Kim, S.-H.Jung, S.W.Seo, and H.-H.Won conceived and designed the study. J.P.Kim and S.-H.Jung performed the statistical analyses. J.P.Kim and S.-H.Jung wrote the manuscript. S.-H.Jung performed the genetic data analyses. B.Jang, M.Cho, M.Song, J.Kim, and B.Kim conducted data preprocessing and interpretation. J.P.Kim, H.Lee, D.Shin, E.H.Lee, H.Jang, B.-H.Kim, and H.Ham curated the data. D.Kim, T.Raj, C.Cruchaga, H.J.Kim, and D.L.Na have read and critically revised the manuscript for intellectual content. S.W.Seo and H.-H.Won supervised the study. All authors agreed to the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks M. Ilyas Kamboh, and Aniket Mishra for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kim, J.P., Jung, SH., Jang, B. et al. Cross-ancestry genome-wide association study identifies implications of SORL1 in cerebral beta-amyloid deposition. Nat Commun 16, 3150 (2025). https://doi.org/10.1038/s41467-025-57751-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-57751-4