Abstract
Herpes simplex encephalitis (HSE) caused by HSV-1 is the most common non-epidemic viral encephalitis, and the neuropathogenesis of HSE remains elusive. This work describes a 3D human neurovascular unit (NVU) model that allows to explore the neuropathogenesis of HSE in vitro. This model is established by co-culturing human microvascular endothelial cells, astrocytes, microglia and neurons on a multi-compartment chip. Upon HSV-1 infection, this NVU model exhibited HSE-associated pathological changes, including cytopathic effects, blood-brain barrier dysfunction and pro-inflammatory cytokines release. Besides, significant innate immune responses were observed with the infiltration of peripheral immune cells and microglial activation. Transcriptomic analysis revealed broadly inflammatory and chemotactic responses in host cells. Mechanistically, we found HSV-1 could induce severe suppression of autophagic flux in glial cells, especially in microglia. Autophagy activators could effectively inhibit HSV-1 replication and rescue neurovascular injuries, indicating the utility of this unique platform for studying neurological diseases and new therapeutics.
Similar content being viewed by others
Introduction
Herpes simplex encephalitis (HSE) is the most common sporadic encephalitis with high mortality1, and over 90% of HSE cases in adults are caused by HSV-12,3,4. Although the administration of antiviral acyclovir has greatly reduced the mortality, most survivors still suffer from permanent neurological sequelae4,5. Besides, increasing evidence implicates that HSV-1 infection is a potential risk factor for Alzheimer’s disease1,6,7. So far, an effective vaccine against HSV-1 has not yet been developed, and the only antiviral drug (acyclovir) is facing the increasing problem of viral drug resistance.
Clinically, HSE is characterized by severe cerebral damage, which is considered to be attributed to a combination of viral replication and excessive inflammatory responses1,3,8. In vivo, the brain is the most complex organ, with multiple cell types and complicated interactions between distinctive cell types in a 3D brain microenvironment. In the last two decades, the concept of neurovascular unit (NVU) was formalized, which refers specifically to nerve tissue and its neighboring blood vessels, consisting of neurons, glial cells, basement membrane and vascular cells (such as brain microvascular endothelial cells and pericytes)9,10. NVU emphasized the close connections between brain cells and their neighboring microvasculature, thus facilitating to study the various brain disease as a whole and to find targeted therapy11,12,13,14,15. It has been reported that NVU injuries occur in a variety of neurological disorders, including ischemic stroke, Alzheimer’s disease, and multiple sclerosis11,12,13,14. In particular, some pathological studies revealed neurovascular injuries and dysfunctional blood-brain barrier (BBB) occurred in the affected brain region of HSE patients16,17,18,19. However, the basis underlying HSV-1-induced neurovascular injuries remains poorly defined.
At present, HSV-1 research mainly relies on mouse models. Due to the inter-species differences, mouse models often showed inconsistent findings with HSV-1-infected human cells or human clinical trials20,21. In recent years, brain organoids, as a new type of 3D organ or tissue models, have been utilized for the neuropathological studies of HSV-1 infection, such as the studying neurodevelopmental disorders associated with neonatal HSV-1 infection22,23, modeling HSV-1 encephalitis and drug discovery24, and testing AD pathogenesis association with HSV-1 infection25. However, these brain organoids more resemble the embryonic human brain and lack the vasculatures and immune cells that are vital for constituting brain microenvironment. These features make them not suitable for investigating the complex neurovascular injuries and host immune responses underlying HSV-1 encephalitis. Organ-on-a-chip, also called microphysiological system, is a form of engineered microfluidic 3D culture system, that allows to recapitulate the basic structure and key function of various human organs or tissues in vitro26,27,28. Based on this technology, some types of NVU or BBB chips have been developed for the study of brain infection, such as fungal brain infection29, SARS-CoV-2-induced neuroinflammation30, Venezuelan equine encephalitis31 and bacterial meningitis32.
In this work, we microengineered a new in vitro 3D NVU model that allows to reconstitute the BBB interface and NVU structure by co-culturing brain endothelial cells, microglia, astrocytes and neurons in a biomimetic brain microenvironment. This model can not only emulate the features of NVU in normal condition but also enables to monitor the multi-stage intercellular interactions in pathological condition dynamically, such as the responses of peripheral immune cells and microglia. Upon HSV-1 infection, significant HSE-associated pathological changes were detected on the NVU model. Mechanistically, we found HSV-1 infection induced severe disturbance of autophagic flux in glial cells. While autophagy activators could effectively inhibit virus proliferation and rescue neurovascular injuries, revealing the therapeutic potential of autophagy activators against HSV-1 encephalitis.
Results
Construction of a microengineered human 3D NVU model
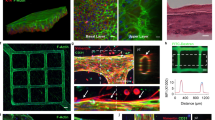
The human brain is an organ of vast complexity in terms of the cell types it comprises. In order to study brain infection associated with HSV-1 (Fig. 1a), we initially designed and engineered a biomimetic neurovascular unit device (also called NVU chip) with multi-compartment chambers and different functional regions (Fig. 1b). The NVU chip consisted of a top layer (named as blood side) and a bottom layer (named as brain side), separated by a transparent porous membrane (8 µm pore diameter, 7 µm thick) (Fig. 1c; Supplementary Fig. 1). The top blood side included an open reservoir for seeding the brain microvascular endothelial cells (HBMECs). The bottom brain side consisted of five parallel channels for co-culturing of human glial cells and neurons to mimic the in vivo brain microenvironment, as shown in Supplementary Fig. 2. The astrocytes and neurons (astrocytes: neurons = 10:1) mixed in 3D Matrigel were seeded in the central compartment, and microglia were seeded into the two lateral channels. The central and lateral culture compartments were connected via 12 trapezoidal channels (6 channels on each side) to enable the intercellular communication of astrocytes/neurons and microglia. In this way, the migration trajectories of microglia in the culture region can be tracked visually under a microscope. Using this device, the four types of cells (HBMECs, astrocytes, neurons, and microglia) were co-cultured in an orderly manner on the chip to mirror the in vivo-like 3D NVU structure. According to this design, responses of distinctive cell types to pathological stimuli can be monitored and recorded conveniently.
a Schematic diagram of the human neurovascular unit. b Image of a real chip. c 3D schematic diagram showing cell culture on the NVU chip. The yellow area indicates the HBMECs culture compartment of the blood side (1). The dark gray area indicates the porous PCTE membrane between the blood side and brain side (2). The red area indicates the astrocytes/neurons culture compartment of the brain side (3). The blue area indicates the microglia culture compartment of the brain side (4). d 3 days after seeding cells on the chip, confocal micrographs showing the HBMECs on the chip, immunostained for VE-cadherin, ZO-1 and PECAM-1. e Confocal micrographs showing the astrocytes on the chip, immunostained for S100β and GFAP. f Confocal micrographs showing the neurons on the chip, stained for Tuj-1 and MAP2. g A confocal micrograph showing the co-culture of astrocytes (GFAP) and neurons (Tuj-1) in Matrigel. h Confocal micrographs showing the microglia on the chip, immunostained for CD11b and IBA1. i A 3D configuration image showing BBB interface formed by HBMECs (VE-cadherin) and astrocytes (S100β) after co-culture for 3 days. d–i Representative images were from 3 biological replicates. All experiments were repeated at least 3 times. j FITC-dextran permeability assays for BBB formed by HBMECs alone, or HBMECs and astrocytes, after culture for 3 days (n = 6 for biological replicates; n = 3 for technical replicates). Data are presented as the mean ± SD and were analyzed using an unpaired two-sided Student’s t-test.
Prior to the study of HSV-1 infection on the NVU model, we initially characterized the structure and function of NVU chip by immunofluorescent imaging after co-culture for 3 days. In the blood side, an intact endothelium was formed with strong expression of adherens junction protein VE-cadherin, tight junction protein ZO-1 and endothelial marker PECAM-1 on the cell periphery (Fig. 1d). In the brain side, astrocytes (S100β, GFAP) and neurons (Tuj-1, MAP2) were co-cultured in 3D Matrigel exhibiting specific markers of the individual cell type in the middle channels (Fig. 1e–g). Quantification of GFAP+ astrocytes and Tuj-1+ neurons showed the ratio of astrocytes to neurons is ~2.33 (Fig. 1g, Supplementary Fig. 3). While, microglia exhibited polygonal shape with expressing microglial markers (CD11b and IBA1) in the lateral channels (Fig. 1h). As we know, in human NVU, HBMECs, astrocytes, pericytes and basement membrane work in concert to form BBB in vivo, which strictly controls the metabolites transport between blood circulation and brain parenchyma, and prevents invasion of toxic substances and pathogens. Here, 3D confocal configuration images revealed an intact BBB interface formed by co-culturing HBMECs (VE-cadherin) and astrocytes (S100β) on the porous membrane (Fig. 1i). Aquaporin-4 (AQP4) is a vital water channel protein which showed concentrated expression at the end-feet of astrocytes near vasculature in human brain33. Confocal micrographs showed many fluorescent puncta of AQP4 were detected facing toward the brain endothelium (Supplementary Fig. 4a), and some of the AQP4 puncta were located at the end of end process of astrocytes (Supplementary Fig. 4b), indicating the partially polarized localization of AQP4 on the NVU model. Also, the barrier function of BBB was evaluated by permeability assessment using dextran-FITC with various molecular weights (4 kDa and 40 kDa). It appeared the co-culture of astrocytes and HBMECs significantly enhanced the barrier function of BBB with integrated interface (Fig. 1j). Collectively, these data indicated the formed human 3D NVU model enables to recapitulate the major features of NVU in vitro at both structural and functional levels.
HSV-1 induced severe BBB dysfunction and neurovascular injuries on the NVU model
To probe the neuropathogenesis with HSV-1, we then tested HSV-1 infection on the NVU model. The virus (MOI = 0.5) was inoculated in the bottom brain side (Fig. 2a), and viral infection was examined by immunostaining for viral gG protein (HSV-1 gG). Immunofluorescence analysis revealed obvious HSV-1 infection in the lower brain side (13.68 ± 3.98% at 3 days post-infection (dpi), 20.73 ± 11.47% at 5 dpi and 31.05 ± 8.14% at 7 dpi for astrocytes/neurons; 22.25 ± 5.72% at 3 dpi, 44.78 ± 8.46% at 5 dpi and 61.50 ± 8.33% 7 dpi for microglia) (Fig. 2b, c). Meanwhile, significant cytopathies were detected in the glial cells and neurons, such as syncytia, cell shrinkage, and cell debris (Fig. 2b). Although no obvious viral infection was found, marked downregulation of VE-cadherin was observed in the brain endothelium (Fig. 2b). Permeability assay (4 kDa FITC-dextran) revealed increased permeability on the infected-NVU model at 7 dpi, indicating the loss of BBB integrity following HSV-1 infection (Fig. 2d).
a Schematic diagram of HSV-1 inoculation on the NVU model. b Confocal micrographs showing the HBMECs, astrocytes, microglia and neurons following HSV-1 infection at 7 dpi on the chip by detecting for viral gG protein (red) (n = 3 for biological replicates; n = 3 for technical replicates). c Quantification of the infected cells for HBMECs, astrocytes/neurons and microglia and at 3, 5 and 7 dpi (n = 3 for biological replicates; n = 3 for technical replicates). Three fields were quantified for each chip. Data are presented as the mean ± SD. d BBB permeability assay using 4 kDa FITC-dextran for Mock or HSV-1-infected NVU chips at 7 dpi (n = 6 for biological replicates; n = 3 for technical replicates). Data are presented as the mean ± SD and were analyzed using an unpaired Student’s t-test. e LDH assay for Mock or HSV-1-infected NVU chips at 3, 5, and 7 dpi, respectively (n = 3 for biological replicates; n = 3 for technical replicates). Data are presented as the mean ± SD and were analyzed using an unpaired two-sided Student’s t-test. f Confocal micrographs of HBMECs, astrocytes/neurons, and microglia, stained with Annexin V-FITC/PI dyes following HSV-1 infection at 7 dpi (n = 4 for biological replicates; n = 3 for technical replicates). g Quantification of Annexin V positive or PI-positive cells for HBMECs, astrocytes/neurons, and microglia following HSV-1 infection, based on (f) (n = 4 for biological replicates; n = 3 for technical replicates). Data are presented as the mean ± SD and were analyzed using an unpaired two-sided Student’s t-test. h ELISA results of medium from brain side for Mock or HSV-1-infected chips at 3 dpi (n = 3 for biological replicates; n = 3 for technical replicates). Data are presented as the mean ± SD and were analyzed using an unpaired two-sided Student’s t-test.
Furthermore, cell viability was examined by LDH (lactate dehydrogenase) activity assay for the medium, and it was observed LDH activity was significantly increased following viral infection in both blood side and brain side at 3, 5, and 7 dpi (Fig. 2e). Cell death assay (Annexin V/PI kit staining) showed, at 7 dpi, no obvious cell death was detected in the brain endothelial cells, whereas marked cell death was found in the glial cells and neurons (Fig. 2f, g). As increasing evidences implicate that HSV-1 infection is a potential risk factor for Alzheimer’s disease1,6,7, and several studies reported HSV-1 infection can induce Aβ accumulation or deposition in vitro25,34,35. We then determined the amyloid pathological responses following HSV-1 infection on the NVU model. By immunostaining for amyloid-β (Aβ) in astrocytes/neurons compartment, we found HSV-1 infection had no obvious effect on Aβ40 accumulation at 4 dpi, while led to a significant increase in Aβ42 level (Supplementary Fig. 5), indicating this NVU chip can recapitulate the amyloid pathology caused by HSV-1 infection in vitro. Moreover, cytokine levels in the medium of brain side were examined by ELISA experiment, and results showed IL-1β, IL-6, TNF-α, CXCL1, and IFN-β were significantly released following HSV-1 infection at 3 dpi (Fig. 2h).
Taken together, these studies reflected this NVU model could recapitulate the neuropathological features associated with HSV-1 infection in vitro, including BBB dysfunction, viral infection, cell injuries, Aβ42 accumulation and pro-inflammatory cytokines release, which indicated it can be further utilized for studies of disease pathogenesis and development of new therapeutics.
HSV-1 infection induced microglial response on the NVU model
In the central nervous system, microglia are the resident immune cells and function as the first line of defense against pathogen invasion36,37. When pathogens invade the brain tissue, microglia respond quickly and participate in a range of immune responses, such as migration to the infected area, recognition and phagocytosis of pathogens and dying cells, antigen presentation, or secretion of a variety of bioactive substances38,39. To track the microglial responses on the NVU model following HSV-1 infection, microglia were labeled with RFP by lentivirus-RFP vector and were visualized in real time. HSV-1 (MOI = 0.1) was inoculated in the right channel of astrocytes/neurons compartment, and 2 h later, microglia labeled with RFP were seeded in the left microglial channel. At 0, 1, 3 and 5 dpi, microglial migration was recorded under a fluorescent microscope. The results showed more microglia migrated to the astrocytes/neurons compartment on the HSV-1-infected model (Fig. 3a, b; Supplementary Fig. 6), which is reminiscent of the pathological changes of microglia in HSE patient samples18,19. It was reported that pro-inflammatory cytokines, such as IL-1β and TNF-α, can stimulate microglia proliferation40,41, which is a key component of the responses to brain injuries42. The ELISA experiments showed, HSV-1 infection triggered a release of IL-1β and TNF-α on the NVU model (Fig. 2h). We then examined the proliferation state of microglia by immunostaining for Ki67, and the result showed more proliferating microglia were detected on the HSV-1-infected NVU model (Fig. 3c, d). Based on these findings, we speculated that enhanced proliferation and increased migration might jointly lead to more appearance of microglia in the astrocytes/neurons compartment following HSV-1 infection.
a Fluorescent images showing microglial migration (red) following HSV-1 infection on the chip at 0, 1, 3, and 5 dpi (n = 3 for biological replicates; n = 3 for technical replicates). The dotted white line indicates the interface between astrocytes-neurons compartment and microglia compartment. b Quantification of the migrated microglia at 0, 1, 3 and 5 dpi (n = 3 for biological replicates; n = 3 for technical replicates). Data are presented as the mean ± SD and were analyzed using an unpaired two-sided Student’s t-test. c Confocal micrographs of microglia (red) immunostained for Ki67 at 4 dpi (n = 3 for biological replicates; n = 3 for technical replicates). The dotted white line indicates the interface between astrocytes-neurons compartment and microglia compartment. d Quantification of Ki67+ microglia based on c. Three fields were quantified for each chip. Data are presented as the mean ± SD and were analyzed using an unpaired two-sided Student’s t-test. e Confocal micrographs of astrocytes/neurons immunostained for viral gG protein (green) and MAP2 (red) at 4 dpi, with or without microglia (n = 4 for biological replicates; n = 3 for technical replicates). f Quantification of the infected neurons based on (c) (n = 4 for biological replicates; n = 3 for technical replicates). Three fields were quantified for each chip. Data are presented as the mean ± SD and were analyzed using an unpaired two-sided Student’s t-test.
Microglia are known to phagocytose damaged cells or cell debris under a number of pathological conditions, including viral infections43,44. We then examined microglial phagocytosis on the NVU model 5 days after HSV-1 infection (MOI = 0.1). As shown in Supplementary Fig. 6, HSV-1 infection resulted in more cell debris in the astrocytes/neurons compartment compared with Mock-infected NVU model. Intriguingly, some cell debris was engulfed by the microglia (Supplementary Fig. 7), indicative of microglial phagocytosis induced by HSV-1 infection. To further determine the antiviral effect of microglia, we conducted a microglia ablation study. To exclude the possibility that less cells affect the infection efficiency, microglia were replaced with the same quantity of astrocytes-neurons on the microglia-deprived NVU model (w/o microglia). After 4 days infection (MOI = 0.5), more infected neurons were detected on the microglia-deprived NVU model, compared with NVU model with microglia (w microglia) (Fig. 3e, f), indicating the protective roles of microglia against acute HSV-1 infection.
HSV-1 infection elicited infiltration of peripheral immune cells on the NVU model
To gain a deeper understanding of the molecular signatures underlying HSV-1-associated encephalitis, we performed RNA-sequencing (RNA-seq) analysis of cells on the NVU model at 3 dpi. Genes differentially expressed with fold changes of >2.0 and P < 0.05 were identified as differentially expressed genes (DEGs). Transcriptomic analysis revealed that HSV-1 infection induced global perturbations in all three types of cells (1049 DEGs in HBMECs, 5093 DEGs in astrocytes, and 5610 DEGs in microglia) (Fig. 4a; Supplementary Fig. 8). Consistent with ELISA results in Fig. 2h, RNA-seq data revealed many cytokine genes (GO: 0005125) elevated in HBMECs following HSV-1 infection (Fig. 4b), some of which are enriched in “TNF signaling pathway” according to KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment analysis (Supplementary Fig. 9). Gene set enrichment analysis (GSEA) indicated “TNF signaling pathway” was significantly activated in HBMECs following HSV-1 infection (Fig. 4c), and upregulated genes included chemokines (CXCL1, CXCL2, CCL2, CXCL3, CX3CL1 and CCL20), pro-inflammatory cytokine (IL-6) and cellular adhesion molecule (ICAM1) (Supplementary Fig. 10). These findings implicated HSV-1 infection induced an inflammatory state in HBMECs, which might trigger recruitment of peripheral immune cells to further participate in inflammation.
a Scatter plots showing the differentially expressed genes in HBMECs following HSV-1 infection at 3 dpi. Genes differentially expressed with fold changes of >2.0 and P < 0.05 are marked in color. P-values were calculated using a two-sided, unpaired Student’s t-test with equal variance assumed (n = 3). b Heatmap indicating the expression levels of upregulated genes annotated to cytokine activity (GO: 0005125) in HBMECs (n = 3). c GSEA analysis reveals the correlation between HSV-1 infection and genes involved in the TNF signaling pathway in HBMECs. The analyses were all one-sided and adjustments were made for multiple comparisons. NES normalized enrichment score, FDR false discovery rate. Gene sets were considered significant when P < 0.05 and FDR < 0.25. d Schematic diagram of adding PBMCs following HSV-1 inoculation on the NVU model. e Fluorescent micrographs of PBMCs (red) attached to the HBMECs (green) following HSV-1 infection at 3 dpi (n = 4 for biological replicates; n = 3 for technical replicates). f Quantification of the attached PBMDs per filed for the Mock or HSV-1-infected chips (n = 4 for biological replicates; n = 3 for technical replicates). Two fields were quantified for each chip. Data are presented as the mean ± SD and were analyzed using an unpaired two-sided Student’s t-test. g Fluorescent micrographs of migrating PBMCs through the brain endothelium on the HSV-1-infected chip (n = 4 for biological replicates; n = 3 for technical replicates). h Heatmap indicating the expression levels of upregulated genes annotated to chemokine activity (GO: 0008009) in astrocytes (n = 3). i Heatmap indicating the expression levels of upregulated genes annotated to chemokine activity (GO: 0008009) in microglia (n = 3). j Fluorescent micrographs of infiltrated PBMCs in the astrocytes culture compartment 3 h after PBMCs addition (n = 3 for biological replicates; n = 3 for technical replicates). k Quantification of the infiltrated PBMCs per filed for the Mock or HSV-1-infected chips (n = 3 for biological replicates; n = 3 for technical replicates). Two fields were quantified for each chip. Data are presented as the mean ± SD and were analyzed using an unpaired two-sided Student’s t-test. l Fluorescent micrographs showing infiltrated T cells (CD45+CD3+) in the astrocytes culture compartment 24 h after PBMCs addition (n = 4 for biological replicates; n = 3 for technical replicates). m Fluorescent micrographs showing infiltrated B cells (CD45+CD19+) in the astrocytes culture compartment 24 h after PBMCs addition (n = 4 for biological replicates; n = 3 for technical replicates). n Fluorescent micrographs showing infiltrated macrophages (CD45+CD68+) in the astrocytes culture compartment 24 h after PBMCs addition (n = 4 for biological replicates; n = 3 for technical replicates). o Quantification of the infiltrated T cells, B cells and macrophages per field for the Mock or HSV-1-infected chips (n = 4 for biological replicates; n = 3 for technical replicates). Two fields were quantified for each chip. Data are presented as the mean ± SD and were analyzed using an unpaired two-sided Student’s t-test.
To probe the responses of peripheral immune cells to HSV-1 infection, peripheral blood mononuclear cells (PBMCs) were added to the HBMECs compartment to monitor the recruitment of immune cells (Fig. 4d). Two hours later, more PBMCs were found to attach to the brain endothelium of the HSV-1-infected group (Fig. 4e, f). Moreover, transmigration of PBMCs through the brain endothelium was further detected on the HSV-1-infected group (Fig. 4g). Next, we analyzed the transcriptome data of glia cells and found more chemokine genes were upregulated in both astrocytes and microglia in the infected group (Fig. 4h, i). In line with the RNA-seq analysis, obvious infiltration of peripheral immune cells was observed in the astrocytes/neurons compartment of the HSV-1-infected models, while almost no peripheral immune cells were detected in the Mock-infected models (Fig. 4j, k). Twenty-four hours after addition of PBMCs, the types of infiltrated immune cells were determined by immunostaining for specific markers of immune cells, including T cells (CD45+CD3+), B cells (CD45+CD19+) and macrophages (CD45+CD68+). The results showed the main infiltrated immune cells are T cells and macrophages, while no obvious infiltration of B cells was detected following HSV-1 infection (Fig. 4l–o), which was consistent with the components of parenchymal inflammatory infiltrates identified in the HSE pathological samples8,18,19. Intriguingly, the infiltrated immune cells exhibited in vivo-like morphology, such as spherical T cells (enlarged images in Fig. 4l) and irregularly shaped macrophages with many pseudopodia (enlarged images in Fig. 4n).
Collectively, it appeared this NVU model can reflect the host immune responses in brain to HSV-1 infection via brain-resident microglia and peripheral immune cells, indicating it can be further utilized to explore the roles of immune cells in the pathogenesis of HSV-1 encephalitis.
Blockage of autophagic flux in glial cells following HSV-1 infection
As above, HSV-1 infection induced obvious dysfunctions of NVU, we then tried to identify the molecular basis of the host responses to the viral infection by KEGG enrichment analysis (Fig. 5a, b). Intriguingly, we noticed some vital biological processes were commonly modulated in both astrocytes and microglia, especially the autophagic process (e.g., autophagy or mitophagy). Next, we examined the NVU models by transmission electron microscope (TEM), and discovered more autophagic vacuoles in the microglia and astrocytes of HSV-1 infected models at 3 dpi (Fig. 5c; Supplementary Fig. 11), especially in the microglia. Further, the microglia were examined by immunofluorescence for autophagic markers (LC3B and p62), and the p62 and LC3B positive dots were significantly increased in both rapamycin (a potent autophagy activator) treated and HSV-1 infected NVU models at 3 dpi (Fig. 5d).
a KEGG pathway enrichment analysis of upregulated genes and downregulated genes in astrocytes following HSV-1 infection at 3 dpi. b KEGG pathway enrichment analysis of upregulated genes and downregulated genes in microglia following HSV-1 infection at 3 dpi. a, b Autophagy-related terms were marked with red boxes. The analyses were all one-sided and adjustments were made for multiple comparisons. c TEM images showing microglia on Mock or HSV-1-infected NVU models at 3 dpi (n = 3 for biological replicates; n = 3 for technical replicates). The autophagic vacuoles were indicated by red asterisks. d Confocal micrographs showing the microglia immunostained for p62 (green) and LC3B (red) following HSV-1 infection at 3 dpi on the chip (n = 3 for biological replicates; n = 3 for technical replicates). The chip treated with rapamycin was used as a positive control group. e Confocal micrographs showing autophagic flux in microglia following HSV-1 infection at 3 dpi (n = 3 for biological replicates; n = 3 for technical replicates). The red dots indicate autolysosome, and the yellow dots indicate autophagosome. The chip treated with rapamycin was used as a positive control. f Quantification of autolysosome and autophagosome numbers in microglia following HSV-1 infection based on (e) (n = 3 for biological replicates; n = 3 for technical replicates). Ten cells were quantified for each chip. Data are presented as the mean ± SD and were analyzed using an unpaired two-sided Student’s t-test.
As autophagic flux is a dynamic process, it is difficult to distinguish whether the increased autophagosomes are due to increased autophagosome formation or suppressed autophagosome clearance. We then used GSEA as a targeted method to analyze the potential correlation between HSV-1 infection and the autophagic process in all three types of cells. The results showed that the pathway related to autophagy exhibited significant differences between the Mock group and HSV-1 group in both astrocytes and microglia, while not in HBMECs (Supplementary Fig. 12). Especially, we found this difference was more significant in the microglia (P = 0.299 in HBMECs; P = 0.048 in astrocytes; P = 0.020 in microglia), in which HSV-1 infection was negatively correlated with the autophagic process (Supplementary Fig. 12). To verify this GSEA analysis, autophagic flux was examined in microglia using an adenovirus-mRFP-GFP-LC3 tool, which can directly reflect the level of autophagic flux by the fluorescent dots45. The red dots indicate the autolysosomes and the yellow dots (overlay of GFP and RFP) indicate the autophagosomes. The NVU model treated with rapamycin was used as a positive control (increased RFP dots) (Fig. 5e). The results showed the number of autophagosomes was significantly increased following HSV-1 infection at 3 dpi, while the number of autolysosomes was decreased at the meanwhile (Fig. 5e, f). These findings reflected that HSV-1 infection caused excess accumulation of autophagosomes in the microglia, by blocking the fusion of autophagosomes with lysosomes.
Autophagy activators ameliorated HSV-1-induced neurovascular injury by inhibiting viral replication
As HSV-1 infection caused severe blockage of the autophagic process in host cells, we then tried to test whether autophagy could be a therapy target for HSV-1 encephalitis. To verify the hypothesis, we performed a drug screening by testing autophagy activators or inhibitors on the HSV-1 encephalitis model (Fig. 6a). In this drug screening test, acyclovir (Acyc) which is a common antiviral drug to treat HSV-1 infection, was used a positive control drug. Autophagy activators include rapamycin (Rapa) and two natural compounds (Euphpepluone K (EupK) and Munronin V (MunV)). Euphpepluone K was isolated from Euphorbia peplus Linn46, and Munronin V was isolated from Munronia henryi Harms47. Previous studies reported that both of the two compounds significantly activated autophagic flux in cells46,47. As a comparison, 3-Methyladenine (3-MA) was chosen as an autophagy inhibitor in this test. Cell viability assay showed, similar to acyclovir, all 3 autophagy activators (rapamycin, Euphpepluone K and Munronin V) could effectively rescue the cell death caused by HSV-1 infection, while autophagy inhibitors (3-MA) significantly exacerbated the cell death (Fig. 6b). Besides, BBB permeability assay and LDH assay were performed, and the results showed acyclovir and all 3 autophagy activators could restore the BBB injury and LDH release caused by HSV-1 infection to different extend (Fig. 6c, d).
a Experimental flow diagram showing the drug testing on the HSV-1 encephalitis model. b Cell viability assay showing the therapeutic effects of different drugs on HBMECs, Astrocytes/Neurons, and microglia following HSV-1 infection on the model at 7 dpi (n = 3 for biological replicates; n = 3 for technical replicates). Acyclovir (Acyc) was used as a positive control drug. Rapa rapamycin, 3-MA 3-methyladenine, Euphpepluone K EupK, Munronin V MunV. Cells were stained with LIVE/DEAD® Viability/Cytotoxicity Kit, and red color indicates the dead cell. c 4 kDa FITC-dextran assay showing the effects of different drugs on the BBB permeability following HSV-1 infection at 7 dpi (n = 3 for biological replicates; n = 3 for technical replicates). Data are presented as the mean ± SD and were analyzed using a one-way analysis of variance (ANOVA) followed by the Bonferroni post hoc test. d LDH assay showing the therapeutic effects of different drugs on the model following HSV-1 infection at 7 dpi (n = 3 for biological replicates; n = 3 for technical replicates). Data are presented as the mean ± SD and were analyzed using a one-way ANOVA followed by the Bonferroni post hoc test.
Next, we continued to investigate whether the autophagy activators could rescue HSV-1-caused neurovascular injury by inhibiting HSV-1 replication. Immunofluorescent analysis showed the infected cells were dramatically decreased following treatment with acyclovir or 3 autophagy activators at 7 dpi, while were significantly increased following treatment with autophagy inhibitor (3-MA) (Fig. 7a). TCID50 assay was performed to determine the viral load in medium of brain side at 7 dpi, and the results showed the viral load was decreased following treatment with autophagy activators, while was increased following treatment with autophagy inhibitor (Fig. 7b), indicating that the autophagy activators rescued neurovascular injury by inhibiting HSV-1 replication. Furthermore, autophagic flux assay was also performed, and the results showed that autophagy activators effectively decreased the excess accumulation of autophagosomes and restored the blockade of autophagic process on the HSV-1-infected NVU model (Fig. 7c, d).
a At 7 dpi, confocal micrographs showing the HSV-1 infection (red, immunostained for viral gG protein) in astrocytes/neurons and microglia after adding the indicated drugs (n = 3 for biological replicates; n = 3 for technical replicates). b TCID50 assay showing the antiviral effects of different drugs on the brain side (n = 3 for biological replicates; n = 3 for technical replicates). c Confocal micrographs showing autophagic flux in microglia with the addition of indicated drugs following HSV-1 infection at 3 dpi n = 3 for biological replicates; n = 3 for technical replicates. The red dots indicate autolysosome, and the yellow dots indicate autophagosome. d Quantification of autolysosome and autophagosome numbers in microglia of the HSV-1-infected chips after adding indicated drugs (n = 3 for biological replicates; n = 3 for technical replicates). Ten cells were quantified for each chip. Data are presented as the mean ± SD and were analyzed using a one-way ANOVA followed by the Bonferroni post hoc test.
Discussion
This work described a 3D human NVU model containing a functional BBB interface and neural components, which allows to simulate the brain microenvironment in normal and pathophysiology of HSE associated with HSV-1 infection. Upon HSV-1 infection, this model revealed obvious pathological changes, such as cytopathic effects, BBB dysfunction, pro-inflammatory cytokines release, microglial response and infiltration of peripheral immune cells. These results are consistent with the findings of pathological studies in SAE patients16,17,18,19. Intriguingly, it was found that HSV-1 induced severe suppression of autophagic flux in glial cells, and autophagy activators could effectively rescue the neurovascular injuries induced by HSV-1.
In vivo, the brain has the largest number of cell types and the most complex physiological microenvironment. The pathogenesis of HSE still remains elusive, partially due to the lack of models that accurately mimic the complex physiological structure of the brain and reflect the human-relevant responses to HSV-1 infection. Although mouse models are widely used in HSV-1 studies, mice are not natural hosts for this virus, which leads to host responses different from those in humans. For example, HSV-1 infection leads to inhibition of necroptosis in human cells, but activation of necroptosis in mouse cells20. Many anti-HSV-1 candidate drugs that showed promise in mouse models, all failed in human clinical trials, such as vaccines. In addition, organoids have recently been utilized in the neuropathological studies of HSV-1 infection22,23,24,25. Although the brain organoid models show great potential in the study of brain HSV-1 infection, they still face some challenges at this stage, such as a lack of vasculatures or immune cells, which makes them not suitable for investigating neurovascular injuries and the immune cells-mediated inflammatory responses underlying HSV-1 encephalitis. Till now, the model that can accurately reflect the pathogenesis of HSV-1-associated encephalitis is still lacking.
In this work, to probe the neuropathogenesis with HSV-1 in a human-relevant manner, we created a human NVU-on-a-chip model, including brain endothelial cells, neurons, astrocytes and microglia in 3D microenvironment. A multi-compartment co-culture chip was designed for the evaluation of host responses to HSV-1 infection and enabled real-time tracking of distinctive types of cells. Following viral infection in the brain side, we detected prominent viral replication in the brain cells, including glial cells and neurons, while no visible HSV-1 replication was observed in HBMECs. As we know, brain endothelium is the core component of BBB, which line the brain capillaries. Although it is not the direct target for HSV-1 infection, brain endothelium still suffered from severe injuries, such as disruption of cell junctions, endothelial cell death, increased BBB permeability and more LDH release. This indicates the injuries of HBMECs are caused in an indirect manner, and this might be mediated by the cross-talk between infected brain cells and endothelial cells.
In addition to the direct injury of brain cells by viral infection, inflammatory response was considered another cause for severe cerebral injury in HSE. We then evaluated the host immune responses to HSV-1 infection on the NVU model. By real-time recording of microglial responses following HSV-1 infection, it appeared more microglia migrated to the infected area and phagocytosed cell debris. Ablation of microglia significantly exacerbated HSV-1 infection in the astrocytes/neurons compartment, and this result indicated the protective role of brain-resident immune cells against viral infection in HSE. Furthermore, we also probed the responses of peripheral immune cells to HSV-1 infection. This NVU model contains a continuous layer of brain endothelial cells that, together with extracellular matrix and astrocytes, form a functional BBB interface. It is thus convenient to record the adhesion and infiltration of peripheral immune cells from the blood side into the brain side. The results indicated HSV-1 infection triggered the secretion of chemokines from HBMEs and glial cells, which might cause peripheral immune cells (mainly T cells and macrophages) to adhere to the brain endothelial cells, transmigrating through the endothelium, followed by infiltration into brain side. These findings demonstrated the unique advantages of this model to emulate the complex inflammatory responses mediated by resident immune cells (microglia) or peripheral immune cells in HSE18,19.
To better understand the molecular basis underlying HSE, we performed RNA-seq analysis on the NVU at 3 dpi. Consistent with pathological studies, transcriptomic analysis revealed the upregulation of many immune-related genes, including pro-inflammatory cytokines and chemokines. Further GO enrichment analysis of upregulated genes indicated several vital inflammation-associated pathways were activated following HSV-1 infection, such as the TNF signaling pathway, IL-17 signaling pathway, NF-kappa B signaling pathway, etc. However, type I interferon (INF-1) signaling pathway, which plays pivotal roles in defense against viral infection48,49, is not significantly activated by HSV-1 infection on the NVU model, according to the RNA-seq analysis. This might be due to the immune evasion mechanism exploited by HSV-1. Previous studies reported HSV-1 has evolved several strategies to interfere with IFN responses at multiple levels50. For example, HSV-1 ICP0 protein was reported to degrade the host deubiquitinase BRCC36 and further downregulate IFN-I receptor IFNAR151, and HSV-1 ICP27 protein was found to interfere with STAT-1 activation and hamper the downstream transcription of ISGs (interferon-stimulated genes)52. Moreover, another study reported the levels of IFN antiviral responses to HSV-1 are different among distinctive tissues, and IFN induction is more robust in skin than in brain53. Taken together, we suggested that the immune evasion mechanism of HSV-1 might be responsible for the failure to trigger a significant IFN response on the NVU model, which in turn increased the degree of brain damage.
Intriguingly, we found HSV-1 infection induced severe suppression of autophagic flux in glial cells. Autophagy has been shown to act as a vital cellular defense mechanism against pathogens by eliminating intracellular microorganisms54,55. Several studies reported HSV-1 infection causes autophagy inhibition, by which the virus evades the innate immune immunity to promote its propagation56,57,58. In support of this view, deeper studies revealed HSV-1 neurovirulence factor ICP34.5 reverses PKR-mediated eIF2a phosphorylation and translational shutoff in host cells59, and it could also bind to BECN1 and inhibits BECN1-mediated autophagy57,60. Other studies, however, reported HSV-1 infection induces autophagy in some types of cells, such as macrophages, fibroblast, neurons and glial cells, which contributes to antigen presentation or promoting cell survival61,62,63. With regard to the controversial findings, further studies indicated that HSV-1 might modulate autophagy in a cell-type-specific manner55,64. For example, autophagy plays a critical role in limiting HSV-1 replication in terminally differentiated neurons, while it is not required for HSV-1 control in mucosal epithelial cells and other mitotic cells65. In this work, we found HSV-1 infection caused excess accumulation of autophagosomes and decreased autolysosomes in glial cells, which indicated HSV-1 infection interfered with the fusion of autophagosomes with lysosomes and blocked the autophagic flux. In addition, the addition of autophagy inhibitor promoted HSV-1 replication and exacerbated neurovascular injuries, while treatment of three types of autophagy activators obviously rescued the injuries caused by HSV-1 infection. Intriguingly, we found the degrees of autophagic blockage caused by HSV-1 infection were different in three cell types (HBMECs, astrocytes and microglia), and this effect was particularly pronounced in the microglia. These data supported that HSV-1 can suppress autophagic flux in host cells and promote viral replication, which indicates the autophagy pathway might be a valuable therapeutic target for HSV-1 encephalitis.
The brain is a complex organ with various cell types, and each cell type has different functions. Therefore, it is necessary to consider the proportion of different cells, especially the ratio of astrocytes to neurons. Previous studies reported, the glia/neurons ratio in the whole human brain is close 166,67,68. However, the glia/neuron ratio varies dramatically among distinctive brain regions, and the ratio is ~3.7 in cerebral cortex, ~0.2 in cerebellum, and ~11.3 in the rest of brain66. Clinical studies reported HSV-1 mainly attacks cortical areas of the brain in HSE patients, including temporal lobe, frontal lobe and limbic system69,70. Besides, considering that astrocytes are the most abundant cells among glial cells71,72, we set the cell seeding ratio of astrocyte/neuron to 10 in the NVU chip. As SH-SY5Y cells have a higher proliferation rate than HA cells, the final ratio of astrocytes to neurons reaches ~2.33 after co-culture for 3 days.
Despite the advantage of this NVU model for the study of HSV-1 encephalitis, there are still some limitations that need to be improved. First of all, immortalized cell lines were used to represent neurons and microglia in this NVU model. Especially for neurons, SH-SY5Y cells lack some vital neuronal features, such as mature synapses (Supplementary Fig. 13) and electrophysiological characteristics. Neurons and microglia differentiated from iPSCs could be a better choice for the construction of a more biomimetic NVU model. Second, a vascular channel design with an open reservoir was applied in our model, without fluidic flow acting on the brain endothelium. It is known that dynamic fluid flow and shear stress significantly contribute to the normal structure and function of blood vessels. Third, pericyte is also a vital cell type contributing to a functional BBB, which should be incorporated in the future to improve the overall functions of the NVU model. Finally, although Matrigel is widely used for the establishment of BBB or NVU models33,73,74, it is limited in the ill-defined composition and high laminin (~60%) content75, which is different from the real composition of extracellular matrix in the human brain76. Thus, it is suggested to reconstitute the human NVU model using an extracellular matrix with brain-rich and chemical defined substances in further studies, such as collagen IV or fibrin/fibrinogen.
Taken together, we established a new 3D NVU model that allows to recapitulate the features of NVU in normal and pathological condition of HSV-1 infection in vitro. The novelty of this work lies in its ability to mimic the physiologically relevant microenvironment of NVU and the host immune responses to viral infection in a human-organ context. In particular, it enables the visualization of interactions between the diverse brain cells (astrocytes, neurons) and immune cells (peripheral immune cells, microglia) dynamically, which is not easily achieved with animal models. This work provides a unique platform for the study of pathophysiology in HSV-1 encephalitis and the development of new therapeutics for this devastating disease.
Methods
Cell culture
Human microvascular endothelial cells (HBMECs) were purchased from ScienCell Corporation (no. 1000) and cultured in an endothelial cell medium (ECM; ScienCell; no. 1001) supplemented with 5% FBS. Human astrocytes (HA cells) were purchased from ScienCell Corporation (no. 1800) and cultured in AM medium (ScienCell; no. 1801). Human neuroblastoma cell line SH-SY5Y was purchased from Procell Corporation (no. CL-0208) and maintained in DMEM/F-12 medium (Gibco; no. C11330500BT) supplemented with 10% FBS and 1% P/S. HMC3 cells were purchased from Procell Corporation (no. CL-0620) and maintained in MEM medium containing NEAA (Procell; no. MP150410) supplemented with 10% FBS and 1% P/S. HMC3-RFP cells were transfected with a lentivirus-GFP vector for 48 h, and RFP-positive cells were selected using 10 μg/ml puromycin dihydrochloride (Beyotime; no. ST551). PBMCs were isolated from fresh human peripheral blood using Ficoll (GE Healthcare) density gradient centrifugation and cultured in RPMI 1640 medium containing 10% FBS, 1% P/S, and 50 IU/ml IL-2. All cells were cultured at 37 °C in a humidified atmosphere of 5% CO2.
Chip fabrication
The microfluidic NVU model device was composed of two polydimethylsiloxane (PDMS) layers and a PCTE membrane (Whatman; no. 10417512; 1 × 105 pores/cm2). First, SU-8 molds with different heights were prepared by standard photolithography technology as previously described77. The base and curing agent (Sylgard 184, Dow Corning company) were mixed at a 10:1 ratio (wt/wt) and poured on the molds, and cured for 45 min at 80 °C. Peeled off the cured PDMS replica and punched a rounded rectangle (10 mm long × 5 mm wide) in the center of the upper layer. Punched round holes (1.6 mm diameter for 6 middle pores; 3.5 mm diameter for 4 lateral pores) at both ends of PDMS channels for the inlets and outlets of cell-containing compartments. Using uncured PDMS as glue, a porous polycarbonate track-etched (PCTE) membrane (7 μm thick, 8 μm pore size) was glued onto the central hole in the upper layer. Then, two PDMS layers were bonded using oxygen plasma treatment (250 W, 30 s, TS-PL30MA, Plasma Systems). The assembled devices were sterilized by ultraviolet irradiation overnight and then were pre-coated with 100 μg/ml collagen I (1:100, Corning) at 37 °C for 12 h to promote cell adhesion.
Cell culture on the NVU chip
Before seeding cells on the chips, SH-SY5Y cells were treated with retinoic acid (10 µM) for 5 days and differentiated to a neuron-like phenotype. Astrocytes and SH-SY5Y cells were mixed at a ratio of 10:1, then were mixed with half volume of Matrigel on ice to get a final density of 5 × 105 cells/ml. 60 μl cell-Matrigel suspension was injected into the astrocytes/neurons culture compartment of the lower brain side, and the chips were flipped immediately. The height of the middle three channels was designed to be lower than that of two lateral channels. Height difference between two channels could generate surface tension in the junction while cell loading. The mixture flowed into the middle three channels, stayed within the junction of the two compartments and formed a well-defined interface after curing. Thirty minutes later, the Matrigel was solidified and the chips were flipped back. Then, 5 × 104 HBMECs (in 100 μl endothelial medium) were seeded in the central compartment of the upper blood side. Next, 2.5 × 104 microglia were seeded in each lateral channel of the lower brain side. Two hours later, 100 μl mixed medium (AM: DMEM/F-12: MEM = 1:1:1) was infused into each lateral channel of the lower layer. Medium was changed every day, and the chips were maintained in an incubator for three days with 5% CO2 at 37 °C.
Virus
HSV-1 virus was provided by Professor Jumin Zhou’s lab (Kunming Institute of Zoology, CAS). The HSV-1 infection experiments were performed in a biosafety level-2 (BSL-2) laboratory.
Direct viral infection
For HSV-1 infection on the 3D NVU model (except for microglial migration assay), 20 μl medium containing indicated multiplicity of virus (MOI = 0.5) was infused in the lateral channels of the lower brain side. Two hours after virus inoculation, cells on the NVU model were washed twice with PBS and kept in a fresh medium for continued culture.
Immunofluorescent imaging
Cells on the chips were washed twice with PBS and fixed with 4% PFA at 4 °C overnight. The fixed cells were permeabilized and blocked with 0.2% Triton X-100 in PBS (PBST) buffer containing 5% normal goat serum for 30 min at room temperature. Antibodies were diluted with PBST buffer and added into the upper and lower channels, respectively. Cells were stained with the primary antibodies at 4 °C overnight and corresponding secondary antibodies at room temperature for 2 h. The chips were disassembled after counterstaining with DAPI, and the porous membrane and bottom layer were mounted on slides. Images were acquired using an Olympus FV3000 confocal fluorescent microscope. Image processing was done using ImageJ software (NIH).
PBMC adhesion and infiltration assay
PBMC adhesion and infiltration assay on NVU model was performed at 2 dpi following HSV-1 infection (MOI = 0.5). Briefly, PBMCs were collected from fresh peripheral blood and stained with CellTracker red dye (Invitrogen, Cat no. C34552) in RPMI 1640 medium at the concentration of 5 µM at 37 °C for 30 min. Meanwhile, cells in NVU model were stained with CellTracker green dye (Invitrogen, Cat no. C7025). After washing with medium twice, the PBMCs were resuspended in ECM medium to 3 × 106 cells/ml. 100 µl cell suspension was added in the central compartment of the upper blood side and incubated at 37 °C for 3 h. Then, the central compartment of the upper layer was washed with PBS three times to wash out unattached PBMCs, and the whole chip was fixed with 4% PFA. Finally, the attached PBMCs in the upper blood side and infiltrated PBMCs in the bottom astrocytes/neurons compartment were imaged using a fluorescent microscope, respectively.
BBB permeability assay
Permeability was assessed by detecting the diffusion of FITC-dextran from the upper blood channel to the bottom brain channel. Briefly, medium containing 50 μg/ml FITC-dextran was infused into the upper blood channel of the chip. Two hours later, the medium in the upper blood channel and the bottom brain channel were collected, respectively. The FITC-dextran concentration was determined by FITC fluorescence intensity using a microplate reader system at 488 nm (excitation) and 525 nm (emission). The apparent permeability coefficient (Papp) was calculated according to the equation below78,79: Papp = (Vb*Cb)/(A*Cv*t)
where Vb (ml) and Cb (ng/ml) indicate the volume (200 μl) and concentration of FITC-dextran in the bottom brain channel, respectively; A (cm2) indicates the contact area between upper layer and bottom layer (0.27 cm2); Cv (ng/ml) is the FITC-dextran concentration in the upper channel; and t (s) is the diffusion time of FITC-dextran (7200 s).
ELISA assay
Culture supernatant was harvested from the lower brain side at 3 dpi. Cytokines in the culture supernatants, including IL-1β, IL-6, TNF-α, CXCL-1 and IFN-β were analyzed using ELISA kits (Beyotime Biotechnology, China) according to the manufacturer’s instructions.
Microglial migration assay
For microglial migration assay, HBMECs, astrocytes, neurons and microglia were seeded on the NVU model for co-culture without microglia seeded on the right lateral channel of the lower layer. Three days later, HSV-1 (MOI = 0.1) was inoculated in the left lateral channel of the lower layer, and 2.5 × 104 microglia labeled with RFP (microglia-RFP) were seeded in the right lateral channel of the lower layer. From 0 to 5 dpi, migration of microglia-RFP was recorded by fluorescent microscope every day.
TCID50 assay
The tissue culture infectious dose 50 (TCID50) was determined as described previously with minor modifications80. Briefly, 10,000 HMC3 cells were seeded per well in 96-well plates in 100 μl medium and incubated overnight. HSV-1 containing medium was serially diluted 10-fold with fresh medium, then the diluted medium was used to inoculate HMC3 cells, resulting in final HSV-1 dilutions of 1:102 to 1:108 on the cells in sextuplicates. Seventy-two hours later, the cytopathic effect (CPE) was recorded under a microscope, and the TCID50 was calculated using the Reed and Muench method.
Autophagic flux detection
Autophagic flux in microglia was evaluated using an HBAD-mRFP-GFP-LC3 adenovirus (Hanbio Co. Ltd., Shanghai, China). Briefly, microglia on the chips were transfected with the BAD-mRFP-GFP-LC3 adenovirus (MOI = 10) for 4 h, then half of the medium containing adenovirus was renewed with fresh normal medium. Forty-eight hours later, HSV-1 virus (MOI = 0.5) was inoculated in the lateral channels of the lower layer. Three days later, cells in NVU model were fixed with 4% PFA and counterstained with DAPI. Images were acquired using a confocal fluorescent microscope (Olympus FV3000). The number of yellow dots (overlay of RFP and GFP) and mRFP dots was determined by manual counting of fluorescent puncta in ten cells from three chips.
For evaluation assay of autophagy activators, 2 h after HSV-1 inoculation, medium containing virus in lower brain side was replaced with fresh medium containing indicated chemicals for another 3-day culture. The subsequent detection procedure was consistent with the above experiment.
Drug testing
To test the drugs on the 3D NVU model after viral infection, 20 μl medium containing HSV-1 (MOI = 0.5) was infused in the lateral channels of the lower brain side. Two hours after virus inoculation, cells on the NVU model were washed twice with PBS and kept in fresh medium containing indicated chemicals (50 µM Acyclovir, 2 μM Rapamycin, 50 µM 3-Methyladenine, 20 μM Euphpepluone K, 20 μM Munronin V) for continued culture. At 7 dpi, BBB permeability was examined by 4 kDa FITC-dextran assay. Medium supernatants of the lower brain side were analyzed for LDH and TCID50 assays. Cell viability was examined using LIVE/DEAD® Viability/Cytotoxicity Kit, and viral infection was detected by immunostaining for HSV-1 gG protein.
RNA extraction, library preparation, and sequencing
For RNA-seq experiments, HBMECs, astrocytes and microglia were seeded on vascular channel, central compartment and lateral channels, respectively. Three days after HSV-1 infection (MOI = 0.5) in the brain side, HBMECs, astrocytes and microglia were collected, respectively. Total RNAs of three cell types were extracted using TRIzol (Invitrogen) following the methods of Chomczynski et al.81. RNA samples were treated with DNaseI for DNA digestion and then determined by examining A260/A280 using a Nanodrop OneC spectrophotometer (Thermo Fisher Scientific Inc). RNA integrity was confirmed by 1.5% agarose gel electrophoresis. Finally, qualified RNAs were quantified by Qubit 3.0 with a Qubit RNA Broad Range Assay kit (Life Technologies).
Samples with 2 μg total RNA were used for stranded RNA-sequencing library preparation using a KC-Digital Stranded mRNA Library Prep Kit for Illumina (Catalog NO. DR08502, Wuhan Seqhealth Co., Ltd. China), following the manufacturer’s instructions. The kit eliminates duplication bias in PCR and sequencing steps by using a unique molecular identifier (UMI) of 8 random bases to label the pre-amplified cDNA molecules. The library products corresponding to 200-500 bps were enriched, quantified, and finally sequenced on a Hiseq X 10 sequencer (Illumina).
RNA-Seq data analysis
Raw sequencing data were first filtered by Trimmomatic (version 0.36). Low-quality reads were discarded, and the reads contaminated with adapter sequences were trimmed. Clean reads were further treated with in-house scripts to eliminate duplication bias introduced in library preparation and sequencing. Briefly, clean reads were first clustered according to the UMI sequences, in which reads with the same UMI sequence were grouped into the same cluster, resulting in 65,536 clusters. Reads in the same cluster were compared by pairwise alignment, and then reads with sequence identities of over 95% were extracted to a new sub-cluster. Finally, sub-clusters were generated, and multiple sequence alignment was performed to obtain one consensus sequence for each sub-cluster. These steps eliminated any errors or biases introduced by PCR amplification or sequencing.
The de-duplicated consensus sequences were used for standard RNA-seq analysis. They were mapped to the reference genome of Homo sapiens from the Ensembl database (ftp://ftp.ensembl.org/pub/release-87/fasta/homo_sapiens/dna/) using STAR software (version 2.5.3a) with default parameters. Reads mapped to the exon regions of each gene were counted by featureCounts (Subread-1.5.1; Bioconductor), and then RPKMs were calculated. Genes differentially expressed between groups were identified using the edgeR package (version 3.12.1). An FDR-corrected P-value cutoff of 0.05 and a fold-change cutoff of 2 were used to judge the statistical significance of gene expression differences. KEGG enrichment analysis of differentially expressed genes were both implemented using KOBAS software (version: 2.1.1) with a corrected P-value cutoff of 0.05 to judge statistically significant enrichment. The gene set enrichment analysis (GSEA) was performed via clusterProfiler (version 3.14.3) to determine the KEGG signaling pathways. Annotated gene sets (c2.cp.v7.2.symbols.gmt) were selected for the GSEA analysis. The gene list input for the GSEA was ranked by values of log2 fold-change. The R package ggplot 2 (version 3.3.3) was used for data visualization.
Transmission electron microscopy
Chip samples were fixed in PBS buffer containing 4% PFA (Electron Microscopy Sciences) and 2.5% glutaraldehyde (Electron Microscopy Sciences) at 4 °C overnight. After being washed with PBS three times and fixed in 1% OsO4 buffer for 2 h, the samples were dehydrated with graded ethanol solutions and then embedded in Epon812 resin (SPI). Then, 70-nm ultrathin sections were stained with 2% uranyl acetate for 30 min, followed by lead citrate for 10 min. Images were acquired using a JEM-1400PLUS electron microscope.
Ethics
This study does not involve experiments involving animals, human participants, or clinical samples.
Statistical analyses
Data were recorded using Excel (Microsoft) software. GraphPad Prism 8 software was used for statistical analysis. Differences between the two groups were analyzed using unpaired two-sided Student’s t-test. Multiple comparisons were performed using a one-way analysis of variance (ANOVA) followed by the Bonferroni post hoc test. Data are presented as the mean ± SD, and P < 0.05 was considered significant. P-values are presented in the figures.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the findings of this study are available within the article and its supplementary files. The RNA-seq data generated in this study have been deposited in the Sequence Read Archive (SRA) database under accession code PRJNA1027066. Every request can be directed to the corresponding author. Source data are provided with this paper.
References
Marcocci, M. E. et al. Herpes simplex virus-1 in the brain: the dark side of a sneaky infection. Trends Microbiol. 28, 808–820 (2020).
Stahl, J. P. & Mailles, A. Herpes simplex virus encephalitis update. Curr. Opin. Infect. Dis. 32, 239–243 (2019).
Gnann, J. W. Jr. & Whitley, R. J. Herpes simplex encephalitis: an update. Curr. Infect. Dis. Rep. 19, 13 (2017).
Bradshaw, M. J. & Venkatesan, A. Herpes simplex virus-1 encephalitis in adults: pathophysiology, diagnosis, and management. Neurotherapeutics 13, 493–508 (2016).
Liu, H. et al. Proteomics analysis of HSV-1-induced alterations in mouse brain microvascular endothelial cells. J. Neurovirol. 25, 525–539 (2019).
Ball, M. J. Herpesvirus in the hippocampus as a cause of Alzheimer’s disease. Arch. Neurol. 43, 313 (1986).
Itzhaki, R. F. et al. Herpes simplex virus type 1 in brain and risk of Alzheimer’s disease. Lancet 349, 241–244 (1997).
Sobel, R. A. et al. The in situ cellular immune response in acute herpes simplex encephalitis. Am. J. Pathol. 125, 332–338 (1986).
Kugler, E. C., Greenwood, J. & MacDonald, R. B. The “neuro-glial-vascular” unit: the role of glia in neurovascular unit formation and dysfunction. Front. Cell Dev. Biol. 9, 732820 (2021).
Naranjo, O. et al. In vivo targeting of the neurovascular unit: challenges and advancements. Cell Mol. Neurobiol. 42, 2131–2146 (2022).
Cai, W. et al. Dysfunction of the neurovascular unit in ischemic stroke and neurodegenerative diseases: an aging effect. Ageing Res. Rev. 34, 77–87 (2017).
Zarekiani, P. et al. The neurovascular unit in leukodystrophies: towards solving the puzzle. Fluids Barriers CNS 19, 18 (2022).
van Vliet, E. A. & Marchi, N. Neurovascular unit dysfunction as a mechanism of seizures and epilepsy during aging. Epilepsia 63, 1297–1313 (2022).
Cashion, J. M., Young, K. M. & Sutherland, B. A. How does neurovascular unit dysfunction contribute to multiple sclerosis? Neurobiol. Dis. 178, 106028 (2023).
Dong, Y. et al. Cerebrospinal fluid efflux through dynamic paracellular pores on venules as a missing piece of the brain drainage system. Exploration 4, 20230029 (2024).
Thakur, K. T. et al. COVID-19 neuropathology at Columbia University Irving Medical Center/New York Presbyterian Hospital. Brain 144, 2696–2708 (2021).
Taskinen, E., Koskiniemi, M. L. & Vaheri, A. Herpes simplex virus encephalitis. Prolonged intrathecal IgG synthesis and cellular activity in the cerebrospinal fluid with transient impairment of blood-brain barrier. J. Neurol. Sci. 63, 331–338 (1984).
Lellouch-Tubiana, A. et al. Immunocytochemical characterization of long-term persistent immune activation in human brain after herpes simplex encephalitis. Neuropathol. Appl. Neurobiol. 26, 285–294 (2000).
Nicoll, J. A., Maitland, N. J. & Love, S. Autopsy neuropathological findings in ‘burnt out’ herpes simplex encephalitis and use of the polymerase chain reaction to detect viral DNA. Neuropathol. Appl. Neurobiol. 17, 375–382 (1991).
He, S. & Han, J. Manipulation of host cell death pathways by herpes simplex virus. Curr. Top. Microbiol. Immunol. 442, 85–103 (2023).
Johnston, C., Koelle, D. M. & Wald, A. Current status and prospects for development of an HSV vaccine. Vaccine 32, 1553–1560 (2014).
Qiao, H. et al. Herpes simplex virus type 1 infection leads to neurodevelopmental disorder-associated neuropathological changes. PLoS Pathog. 16, e1008899 (2020).
Krenn, V. et al. Organoid modeling of Zika and herpes simplex virus 1 infections reveals virus-specific responses leading to microcephaly. Cell Stem Cell 28, 1362–1379.e7 (2021).
Rybak-Wolf, A. et al. Modelling viral encephalitis caused by herpes simplex virus 1 infection in cerebral organoids. Nat. Microbiol. 8, 1252–1266 (2023).
Qiao, H. et al. Cerebral organoids for modeling of HSV-1-induced-amyloid β associated neuropathology and phenotypic rescue. Int. J. Mol. Sci. 23, 5981 (2022).
Shuler, M. L. Organ-on-a-chip systems: translating concept into practice. Lab Chip 20, 3072–3073 (2020).
Ingber, D. E. Human organs-on-chips for disease modelling, drug development and personalized medicine. Nat. Rev. Genet. 23, 467–491 (2022).
Wang, Y., Wang, P. & Qin, J. Microfluidic organs-on-a-chip for modeling human infectious diseases. Acc. Chem. Res. 54, 3550–3562 (2021).
Kim, J. et al. Fungal brain infection modelled in a human-neurovascular-unit-on-a-chip with a functional blood-brain barrier. Nat. Biomed. Eng. 5, 830–846 (2021).
Wang, P. et al. Blood-brain barrier injury and neuroinflammation induced by SARS-CoV-2 in a lung-brain microphysiological system. Nat. Biomed. Eng. 8, 1053–1068 (2024).
Boghdeh, N. A. et al. Application of a human blood brain barrier organ-on-a-chip model to evaluate small molecule effectiveness against Venezuelan equine encephalitis virus. Viruses 14, 2799 (2022).
Rauti, R. et al. Insight on bacterial newborn meningitis using a neurovascular-unit-on-a-chip. Microbiol. Spectr. 11, e0123323 (2023).
Ahn, S. I. et al. Microengineered human blood-brain barrier platform for understanding nanoparticle transport mechanisms. Nat. Commun. 11, 175 (2020).
Cairns, D. M. et al. A 3D human brain-like tissue model of herpes-induced Alzheimer’s disease. Sci. Adv. 6, eaay8828 (2020).
Wozniak, M. A. et al. Herpes simplex virus infection causes cellular β-amyloid accumulation and secretase upregulation. Neurosci. Lett. 429, 95–100 (2007).
Zhang, W. et al. Microglial targeted therapy relieves cognitive impairment caused by Cntnap4 deficiency. Exploration 3, 20220160 (2023).
Xu, L., He, D. & Bai, Y. Microglia-mediated inflammation and neurodegenerative disease. Mol. Neurobiol. 53, 6709–6715 (2016).
Borst, K., Dumas, A. A. & Prinz, M. Microglia: immune and non-immune functions. Immunity 54, 2194–2208 (2021).
Yu, F. et al. Phagocytic microglia and macrophages in brain injury and repair. CNS Neurosci. Ther. 28, 1279–1293 (2022).
Mander, P. K., Jekabsone, A. & Brown, G. C. Microglia proliferation is regulated by hydrogen peroxide from NADPH oxidase. J. Immunol. 176, 1046–1052 (2006).
Ganter, S. et al. Growth control of cultured microglia. J. Neurosci. Res. 33, 218–230 (1992).
Vela, J. M. et al. Time course of proliferation and elimination of microglia/macrophages in different neurodegenerative conditions. J. Neurotrauma 19, 1503–1520 (2002).
Fu, R. et al. Phagocytosis of microglia in the central nervous system diseases. Mol. Neurobiol. 49, 1422–1434 (2014).
Quick, E. D. et al. Activation of intrinsic immune responses and microglial phagocytosis in an ex vivo spinal cord slice culture model of West Nile virus infection. J. Virol. 88, 13005–13014 (2014).
Hariharan, N., Zhai, P. & Sadoshima, J. Oxidative stress stimulates autophagic flux during ischemia/reperfusion. Antioxid. Redox Signal 14, 2179–2190 (2011).
Yan, Y. et al. Jatrophane diterpenoids from Euphorbia peplus Linn. as activators of autophagy and inhibitors of tau pathology. Int. J. Mol. Sci. 24, 1088 (2023).
Yan, Y. et al. Munronin V with 7/7/6 tricarbocyclic framework from Munronia henryi harms inhibits tau pathology by activating autophagy. Org. Biomol. Chem. 21, 514–519 (2023).
Zhao, Q. et al. Ubiquitination network in the type I IFN-induced antiviral signaling pathway. Eur. J. Immunol. 53, e2350384 (2023).
McNab, F. et al. Type I interferons in infectious disease. Nat. Rev. Immunol. 15, 87–103 (2015).
Tognarelli, E. I. et al. Herpes simplex virus evasion of early host antiviral responses. Front. Cell Infect. Microbiol. 9, 127 (2019).
Zhang, L. et al. HSV-1-encoded ICP0 degrades the host deubiquitinase BRCC36 to antagonize interferon antiviral response. Mol. Immunol. 135, 28–35 (2021).
Johnson, K. E., Song, B. & Knipe, D. M. Role for herpes simplex virus 1 ICP27 in the inhibition of type I interferon signaling. Virology 374, 487–494 (2008).
Tsalenchuck, Y., Steiner, I. & Panet, A. Innate defense mechanisms against HSV-1 infection in the target tissues, skin and brain. J. Neurovirol. 22, 641–649 (2016).
Deretic, V. & Levine, B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe 5, 527–549 (2009).
Cavignac, Y. & Esclatine, A. Herpesviruses and autophagy: catch me if you can! Viruses 2, 314–333 (2010).
Gobeil, P. A. & Leib, D. A. Herpes simplex virus γ34.5 interferes with autophagosome maturation and antigen presentation in dendritic cells. mBio 3, e00267–12 (2012).
Orvedahl, A. et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 1, 23–35 (2007).
Talloczy, Z., Virgin, H. W. T. & Levine, B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy 2, 24–29 (2006).
He, B., Gross, M. & Roizman, B. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1α to dephosphorylate the α subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl Acad. Sci. USA 94, 843–848 (1997).
O’Connell, D. & Liang, C. Autophagy interaction with herpes simplex virus type-1 infection. Autophagy 12, 451–459 (2016).
Alexander, D. E. et al. Analysis of the role of autophagy in replication of herpes simplex virus in cell culture. J. Virol. 81, 12128–12134 (2007).
McFarlane, S. et al. Early induction of autophagy in human fibroblasts after infection with human cytomegalovirus or herpes simplex virus 1. J. Virol. 85, 4212–4221 (2011).
Tovilovic, G. et al. mTOR-independent autophagy counteracts apoptosis in herpes simplex virus type 1-infected U251 glioma cells. Microbes Infect. 15, 615–624 (2013).
Petrovski, G. et al. Herpes simplex virus types 1 and 2 modulate autophagy in SIRC corneal cells. J. Biosci. 39, 683–692 (2014).
Yordy, B. et al. A neuron-specific role for autophagy in antiviral defense against herpes simplex virus. Cell Host Microbe 12, 334–345 (2012).
Azevedo, F. A. et al. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J. Comp. Neurol. 513, 532–541 (2009).
von Bartheld, C. S., Bahney, J. & Herculano-Houzel, S. The search for true numbers of neurons and glial cells in the human brain: a review of 150 years of cell counting. J. Comp. Neurol. 524, 3865–3895 (2016).
Herculano-Houzel, S. The glia/neuron ratio: how it varies uniformly across brain structures and species and what that means for brain physiology and evolution. Glia 62, 1377–1391 (2014).
Sun, S. et al. Case report: Overlapping anti-AMPAR encephalitis with anti-IgLON5 disease post herpes simplex virus encephalitis. Front. Immunol. 14, 1329540 (2023).
Fazekas, C. et al. Depressive symptoms following herpes simplex encephalitis—an underestimated phenomenon? Gen. Hosp. Psychiatry 28, 403–407 (2006).
Campos, A. C. P. et al. What do we know about astrocytes and the antidepressant effects of DBS? Exp. Neurol. 368, 114501 (2023).
Blanco-Suarez, E., Caldwell, A. L. & Allen, N. J. Role of astrocyte-synapse interactions in CNS disorders. J. Physiol. 595, 1903–1916 (2017).
Bolden, C. T. et al. Validation and characterization of a novel blood-brain barrier platform for investigating traumatic brain injury. Sci. Rep. 13, 16150 (2023).
Blanchard, J. W. et al. Reconstruction of the human blood-brain barrier in vitro reveals a pathogenic mechanism of APOE4 in pericytes. Nat. Med. 26, 952–963 (2020).
Aisenbrey, E. A. & Murphy, W. L. Synthetic alternatives to Matrigel. Nat. Rev. Mater. 5, 539–551 (2020).
Xu, L., Nirwane, A. & Yao, Y. Basement membrane and blood-brain barrier. Stroke Vasc. Neurol. 4, 78–82 (2019).
Liu, T., Lin, B. & Qin, J. Carcinoma-associated fibroblasts promoted tumor spheroid invasion on a microfluidic 3D co-culture device. Lab Chip 10, 1671–1677 (2010).
Wang, P. et al. Malignant melanoma-derived exosomes induce endothelial damage and glial activation on a human BBB chip model. Biosensors 12, 89 (2022).
Oddo, A. et al. Advances in microfluidic blood-brain barrier (BBB) models. Trends Biotechnol. 37, 1295–1314 (2019).
Conzelmann, C. et al. An enzyme-based immunodetection assay to quantify SARS-CoV-2 infection. Antivir. Res. 181, 104882 (2020).
Chomczynski, P. & Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159 (1987).
Acknowledgements
We thank Ms. Yingqi Guo (Kunming Institute of Zoology, CAS) for helping with the preparation of electron microscopy samples. We thank Dr. Wenping Zeng in the Biomedical Platform (Suzhou Institute for Advanced Research of USTC) for technical support in bioimaging experiments. This research was supported by the National Key R&D Program of China (No. 2022YFA1104700) warded to J.Q., National Key R&D Program of China (No. 2022YFA1205000) awarded to M.Z., National Key R&D Program of China (No. 2024YFA0919800) awarded to P.W., Manned Space Station Engineering Space Science and Application Project (No. KJZ-YY-NSM0S05) awarded to J.Q. and P.W., and National Natural Science Foundation of China (No. 32171406) awarded to J.Q.
Author information
Authors and Affiliations
Contributions
J.Q., M.Z. and P.W. conceived the study. M.Z. and P.W. performed the experiments and analyzed the results. L.J. performed the HSV-1 infection in BSL-2 lab. Y.W. performed TCID50 assay. X.C. performed the ELISA assay. Y.G., J.L., P.D. and X.Z. performed chip fabrication. R.L. helped perform autophagic flux. Y.Y. and Y.D. provided the autophagy activators. P.W., M.Z. and J.Q. wrote and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Ann-Na Cho, Silvia Vidal, and the other anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, M., Wang, P., Wu, Y. et al. A microengineered 3D human neurovascular unit model to probe the neuropathogenesis of herpes simplex encephalitis. Nat Commun 16, 3701 (2025). https://doi.org/10.1038/s41467-025-59042-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-59042-4