Abstract
Chemotherapy is an important treatment for glioblastoma (GBM) and a key component of comprehensive GBM therapy. However, the blood-brain barrier (BBB) and complex tumor microenvironment (TME) restrict the diffusion of drugs, which greatly reduces the chemotherapeutic effect on GBM. Single strategies, such as cell-based nanobots to cross the BBB or enzymatic nanobots propelled by enriched substrates in the TME for deep tumor penetration, remain inadequate to address multiple barriers and achieve precise targeting. Here, we develop a Trojan horse-inspired enzymatic nanobot-in-neutrobot system (Trojanbot) to greatly enhance targeted GBM therapy. Trojanbots traverse the BBB by leveraging positive chemotaxis in response to tumor-derived chemokine gradients, after which the released catalase-driven nanobots (CatNbot) undergo directional movement along the H2O2 gradients in TME, facilitating deep tumor penetration. This multi-stage targeting strategy improves drug delivery efficiency, providing considerable potential as a clinical approach for brain tumor treatment.
Similar content being viewed by others
Introduction
Glioblastoma (GBM) is one of the most common and aggressive primary brain tumors, owing to a high mortality rate and poor prognosis1,2,3. Despite the widespread use of chemotherapy in clinics, the successful treatment of GBM remains a significant challenge, primarily due to low target delivery efficiency and severe adverse effects4,5. The blood–brain barrier (BBB), a crucial protective structure of the central nervous system composed of tightly connected endothelial cells, a basal lamina, and astrocytes, severely limits the transport of therapeutics into brain tumor tissues6,7,8,9. Consequently, most drugs struggle to accumulate adequately in tumor tissues after passing through the BBB, leading to significantly reduced therapeutic efficacy10,11. Additionally, the tumor microenvironment (TME) is marked by high levels of hydrogen peroxide (H₂O₂)12, severe hypoxia13, and a dense extracellular matrix (ECM)14, which further undermines drug efficacy while restricting penetration into the tumor core.
Swimming nanobots capable of self-propulsion and programmable navigation have emerged as a promising solution for targeted drug delivery15,16,17,18,19,20,21,22. By utilizing endogenous chemical fuels or harnessing external energy sources from various physical fields, these nanobots can move efficiently and navigate complex environments23,24,25,26,27,28,29,30, including the circulatory system, gastrointestinal tract, and tumors31,32,33. Current strategies for targeted drug delivery to cancers using nanorobots primarily focus on chemical/biological recognition and TME-responsive approaches34,35,36,37,38. These methods leverage target specificity and the unique conditions within tumors, offering higher adaptability and potential biosafety compared to physical-field navigation39,40.
Chemical recognition strategies typically involve modifying specific transporters that target endothelial cells, such as penetrating peptides and lactoferrin, as well as utilizing tumor-recognizing molecules like folic acid and L-arginine to ensure precise drug delivery41. However, the effectiveness of this strategy can vary significantly due to individual differences, particularly in the expression levels of BBB components and tumor cell surface receptors across different individuals and tumor types. Biological recognition strategies employ live cells, such as neutrophils, to carry nanoparticles, taking advantage of their natural ability to penetrate barriers and home in tumors42,43,44. Studies have shown that neutrophil-loaded nanoparticles can effectively cross the BBB and target tumor regions, while also extending drug circulation time and reducing the risk of immune clearance45,46. However, this approach primarily relies on passive diffusion, limiting nanoparticle delivery due to the dense ECM present in solid tumors, which hampers adequate drug distribution to deeper tumor areas47.
Furthermore, TME-responsive strategies use nanomotors that react to specific conditions, such as acidic pH or reactive oxygen species (ROS), to enhance drug penetration48. For example, pH-responsive nanomotors can release drugs in low-pH environments49, and ROS-responsive nanomotors can trigger propulsion in the presence of high concentrations of ROS, thereby improving diffusion into deep tissues50. Despite these advancements, significant challenges remain in achieving effective BBB crossing and delivering drugs to the deeper regions of tumors. Against this backdrop, combining chemical/biological recognition with TME-responsive strategies offers the potential for multi-stage targeted delivery. This synergistic approach not only improves the precision of drug localization in complex biological environments but also significantly enhances drug-delivery efficiency in deep tumor tissues, addressing the challenges of achieving both precise navigation and deep tissue penetration with a single approach.
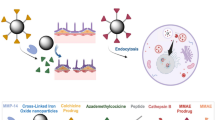
Here, we develop an enzymatic nanobot-in-neutrobot system, termed Trojanbot for targeted therapy of GBM. This innovative system employs bacterial membrane vesicles (EMVs) for surface camouflage and leverages the innate phagocytic ability of neutrophils to assemble catalytically driven nanorobots (CatNbots, referred to as soldiers) into chemotactic hybrid neutrophils (neutrobots, acting as Trojan Horse). This creates a multifunctional drug delivery platform. As illustrated in Fig. 1, we encapsulate doxorubicin (DOX) in collagen nanoparticles (GeNPs), then coat them with EMVs, and modify the catalase to form CatNbots. Thanks to EMV-mediated camouflage, the CatNbots are effectively internalized by neutrophils without losing their functionality. By taking advantage of the inflammation-driven chemotaxis of neutrophils, Trojanbots can actively travel across the BBB following the gradient of inflammation, allowing for targeted drug delivery. Once they reach the TME, inflammation signals trigger the release of CatNbots from the Trojanbots. These nanorobots utilize the catalytic decomposition of H₂O₂ present in the TME to generate propulsion, significantly improving deep tissue penetration and facilitating drug delivery to the tumor’s core. Experimental results show that Trojanbots efficiently traverse the BBB in vitro and accumulate significant amounts of the drug in glioma regions. In a mouse glioma model, Trojanbots demonstrate superior drug delivery efficiency and therapeutic effectiveness, highlighting their potential for future applications in treating central nervous system diseases.
Schematic illustration of the enzymatic nanobot-in-neutrobot system, inspired by the “Trojan Horse” strategy, which leverages neutrophil phagocytosis, chemotactic movement, BBB crossing, and the response of CatNbots to H2O2 in the TME to enhance the tumor penetration of DOX, thereby achieving targeted drug delivery to GBM.
Results
Preparation and characterization of Trojanbots
As depicted schematically in Fig. 2a, the construction of Trojanbots involves the following process: synthesis of DOX-loaded gelatin nanoparticles (GeNPs), surface camouflage of GeNPs using EMVs to obtain EMV@GeNPs, attachment of catalase onto EMV@GeNPs to prepare catalase-driven nanobots (CatNbots), and phagocytosis of CatNbots by activated neutrophils. Briefly, the GeNPs were synthesized through a desolvation method using gelatin as precursors and loaded with DOX, a broad-spectrum anticancer agent51. The transmission electron microscopy (TEM) and scanning electron microscopy (SEM) images reveal the presence of as-synthesized GeNPs exhibiting a spherical morphology, characterized by an average diameter of approximately 200 nm (Fig. 2b and Supplementary Fig. 1). The UV absorption spectra of both DOX and DOX-GeNPs exhibit identical absorption bands at 480 nm, indicating the successful loading of DOX (Supplementary Fig. 2a). Furthermore, typical emission peaks at 570 nm (λex = 488 nm) also confirms the presence of DOX in GeNPs, as shown in the fluorescence spectra (Supplementary Fig. 2b). To ensure the preservation of Trojanbot’s bioactivity and chemotactic capability following particle phagocytosis, a bio-camouflaging strategy was employed52, wherein freshly purified E. coli outer membranes were reassembled into E. coli membrane vesicles (EMVs). These EMVs were subsequently fused onto the surface of GeNPs using a 200-nm porous polycarbonate membrane, forming EMV-coated GeNPs (EMV@GeNPs). The TEM image shows that the spherical EMV@GeNPs were monodispersed, with a mean particle diameter of 220 nm (Fig. 2c). The final step for obtaining the CatNbots required the attachment of the catalase onto the particles through the cross-linker molecule, 1-(3-dimethylaminopropyl)−3-ethylcarbodiimide (EDC). The catalase was combined with EMV@GeNPs in a 1:1 ratio and immersed in a 2 mM EDC solution at 20 °C for 2 h to facilitate the enzyme attachment onto the surface of EMV@GeNPs. Notably, the TEM image in Fig. 2d shows the morphological features of as-synthesized CatNbots (~329.5 nm in diameter), where the catalase molecules were grafted asymmetrically at the edges. Further, TEM-assisted elemental mapping results display the presence of Fe elements in the CatNbots in addition to N, O, S, and P elements, providing evidence for the existence of catalase (Fig. 2e). We followed the sequential functionalization steps by dynamic light scattering (DLS) to determine the hydrodynamic radius and surface charge of the GeNPs, EMV@GeNPs, and CatNbots (Fig. 2f, g). The synthesized bare GeNPs exhibited a characteristic negative surface charge (−31.6 ± 1.2 mV) and displayed a single-population peak in the DLS, suggesting the particles were not aggregated. After the application of EMVs coating, a distinct reduction in the surface charge was observed for the EMV@GeNPs (−35.1 ± 1 mV), which closely resembled that of pure EMVs (−40.1 ± 1 mV). When the surface of EMV@GeNPs was further modified with catalase, the resulting surface charge of CatNbots was further decreased (−35.6 ± 1.6 mV) due to the isoelectric point of catalases at pH 5.4, thus showing a negative charge at pH 7.4. The hydrodynamic radius of the CatNbots increased with the addition of functional molecules to their surface. Moreover, the coating of the EMVs was confirmed by conducting an enzyme-linked immunosorbent assay to measure the lipopolysaccharide (LPS) content of CatNbots. Notably, the approximate LPS content of CatNbots (20 mg mL−1) and pure EMVs (21.2 mg mL−1) proved the translocation of EMVs onto the outer surface of CatNbots (Fig. 2h). The enzymatic activity test of CatNbots confirmed the successful surface modification with catalases, showing no significant impact on enzyme activity during the grafting process (Supplementary Fig. 3). We also verified the enzymatic reactions of the CatNbots by dissolved O2 level, as depicted in Fig. 2i. We initially assessed the catalytic efficacy of surface-bound catalases on CatNbots in neutral conditions for H2O2 decomposition. Our results demonstrate a gradual increase in O2 levels upon the addition of CatNbots, with no significant decline even at pH 6.5. These findings suggest that the catalytic activity of catalases on the surface of CatNbots remains stable under weakly acidic conditions similar to tumor microenvironments. Furthermore, we investigated the dependence of loading capacity on the DOX concentrations to optimize the drug loading dose of the CatNbots (Supplementary Fig. 4a). With the increase of DOX concentration, the loading dose gradually increased and reached the maximum value of 45 μg mg−1 at a DOX concentration of 100 μg mL−1. The CatNbots and GeNPs exhibited comparable DOX loading capacities, suggesting that the gelatin networks predominantly contribute to the adsorption of DOX. We further measured the drug loading capacity over time (Supplementary Fig. 4b) and determined that the optimal loading capacity of CatNbots was achieved at a DOX concentration of 100 μg mL−1 with a 1-h incubation period. All these results suggest that CatNbots can be synthesized on demand.
a Schematic outlining the fabrication of the Trojanbots. b TEM images of GeNPs. Inset: Magnified TEM image of GeNPs. c TEM images of EMV@GeNPs. Inset: Magnified TEM image of EMV@GeNPs. d TEM image of CatNbots. e The mapping spectrum of CatNbot, Fe element indicates that catalase is successfully connected to EMV@GeNPs with random asymmetry. f DLS analyzes the particle diameter of GeNPs, EMV@GeNPs, and CatNbots. g Zeta potential values of GeNPs, EMVs, EMV@GeNPs, and CatNbots (n = 3). The Zeta analysis represented data obtained from three independent experiments. h LPS content of GeNPs, EMVs, EMV@GeNPs, and CatNbots (n = 3). The LPS content analysis represented data from three independent experiments. i Changes in the dissolved O2 level of the reaction medium treated with CatNbots under different pH conditions represent the enzymatic activity of CatNbots. j Movie snapshot capturing neutrophils undergoing phagocytosis of CatNbots. The trajectory of a representative neutrophil was depicted in green, while the typical neutrophil and CatNbot were marked in purple and yellow, respectively. k Quantitative analysis of neutrophil phagocytosis efficiency (n = 3). l The confocal laser scanning microscopy (CLSM) image of a Trojanbot. The nucleus of the neutrophil was stained with Hoechst 33342 (blue); the cell membrane of the neutrophil was stained with Dio (green), and the fluorescence of CatNbot was red. Data are presented as mean ± SD of three independent experiments (g, h, k). P-values are determined by a two-tailed Student’s t-test in (k). Source data are provided as a Source Data file.
To prepare Trojanbots, we employed an ex vivo strategy by isolating neutrophils from murine bone marrow. The successful isolation of natural neutrophils was confirmed using both scanning electron microscopy (SEM) and fluorescence microscopy (Supplementary Figs. 5 and 6). Furthermore, flow cytometry analysis of surface markers CD11b and Ly6G showed that the purity of the isolated neutrophils was determined to be >83% (Supplementary Fig. 7). To observe the phagocytosis process of CatNbots by neutrophils, we incubated the resulting CatNbots with neutrophils and visualized the uptake process in real-time by CLSM (Fig. 2j and Supplementary Movie 1). The time-lapse images show that neutrophils rapidly migrated toward and phagocytosed CatNbots, potentially driven by the EMVs on their surface, which mimic specific chemotactic signals secreted by inflammatory sources. This mimicry induces directional migration of neutrophils along chemokine gradients, promoting efficient uptake of EMVs53. Using flow cytometry, we further monitored cellular uptake profiles of fluorescein isothiocyanate (FITC)-labeled CatNbots and GeNPs at distinct time intervals following a 30-min preculture with neutrophils. Taking advantage of the EMVs disguise, neutrophil phagocytosis of CatNbots was significantly higher than that of the original bare GeNPs, exhibiting a 1.38-fold increase (Fig. 2k and Supplementary Fig. 8). The time-lapse fluorescence microscopy images also indicate that more CatNbots were taken up by neutrophils in 30 min in contrast with bare GeNPs (Supplementary Fig. 9). To evaluate the stability after phagocytosis, we assessed the cell viability of neutrophils and those that had engulfed CatNbots using the CCK8 assay (Supplementary Fig. 10). Freshly isolated neutrophils and Trojanbots showed over 90% viability, demonstrating that neutrophils maintain their viability and function after engulfing CatNbots. Furthermore, the CLSM image reveals that the CatNbots predominantly localize within the neutrophils (Fig. 2l). Taken together, these findings demonstrate the successful encapsulation of CatNbots within neutrophils, thereby highlighting the feasibility of our approach in developing Trojanbots for targeted drug delivery.
Motion analysis of Trojanbots
Generally, neutrophils employ numerous cell surface receptors to detect and respond to chemokine gradient concentrations, which guide the chemotactic motion54. Generally, neutrophils utilize multiple cell surface receptors to sense and respond to chemokine gradient concentrations, which guide their chemotactic motion. Chemokines such as IL-8, CXCL1, and C5a bind to receptors like CXCR1 and CXCR2, activating intracellular signaling pathways that trigger cell polarization and direct migration toward the source of the chemokine gradient55,56. In this work, the Trojanbots inherit the chemotactic ability of natural neutrophils, enabling the migration toward chemokine gradients. As shown in Fig. 3a, upon activation by inflammatory stimuli (phorbol myristate acetate, PMA), Trojanbots release neutrophil extracellular traps (NETs) along with encapsulated CatNbots. The as-released CatNbots with asymmetrically distributed catalases can be further propelled due to the catalytic decomposition of localized H2O2, enabling much deeper tumor penetration.
a Schematic illustration of the chemotactic motion of Trojanbots and the release of CatNbots under PMA stimulation, and the catalytic propulsion of CatNbots. b Time-lapse images of the chemotactic motion of Trojanbots along a chemokine gradient. c Trajectories (n = 7) and d direction distribution (n = 7) of Trojanbots in response to a fMLP gradient (10 pM µm−1). The duration of chemotactic motion is 10 min. e Dependence of chemotaxis velocity of neutrophils and Trojanbots on the chemokine gradient of fMLP (n = 3). f Dependence of CI (ratio of distance to path length) of Trojanbots on the chemokine gradient of fMLP (n = 6). The center line of the box plots represents the median, the box bounds indicate the 25th and 75th percentiles and the whiskers extend to the minimum and maximum values within the 1.5× interquartile range (IQR). Individual data points are overlaid. No outliers are present. g TAD of Trojanbots under chemokine gradient (n = 600). h TAD at 0° of Trojanbots under different gradients of fMLP (n = 6). The center line of the box plots represents the median, the box bounds indicate the 25th and 75th percentiles and the whiskers extend to the minimum and maximum values within 1.5× IQR. Individual data points are overlaid. No outliers are present. i Time-lapse images of CatNbots releasing process under PMA stimulation. The resting Trojanbots (i) were activated by PMA stimulation (ii), and the as-released CatNbots (iii) exhibited self-propelled motion in H2O2 solution (iv). j The effect of different concentrations of PMA on the release rate of Trojanbots (n = 3). k Dependence of Trojanbot release percentage on time with or without PMA (100 nM) stimulation. (n = 3). l CLSM images of CatNbots released from Trojanbots with (i) and without 100 nM PMA (h) after 8 h of in vitro culturing. The CatNbots were loaded with Cou6 (green), and the released DNA segments were stained with PI (red). m Representative tracking trajectories of the CatNbots (n = 20) in H2O2 solutions with different concentrations (0, 2, 4, 6, 8 µM) within 1 s. n MSD analysis (Δt = 1 s, n = 20) of CatNbots at different H2O2 concentrations (0, 2, 4, 6, and 8 µM). o The effective diffusion coefficient (Deff) (left) and self-propelled velocity of CatNbots (right) at different H2O2 concentrations (0, 2, 4, 6, and 8 µM) based on MSD. (n = 20). p The instantaneous velocity of CatNbots at different H2O2 concentrations (0, 2, 4, 6, and 8 µM) (n = 20). Data are presented as mean ± SD of three independent experiments (e, j, k), six independent experiments (f, g, h), seven independent experiments (c, d), and 20 independent experiments (o, p). Source data are provided as a Source Data file.
To assess the chemotactic behavior of Trojanbots, we established an artificial chemokine gradient on a microfluidic chip, where an agarose hydrogel containing 1 µM N-Formylmethionine-leucyl-phenylalanine (fMLP) was incorporated as the source (Supplementary Fig. 11). In this way, a chemokine gradient formed naturally. The time-lapse images (Fig. 3b and Supplementary Movie 2) provide visual evidence for the directional movement of Trojanbots towards the fMLP source, indicating the chemotactic motion ability of the Trojanbots. To further study the dynamic behavior of the Trojanbots, the chemotactic motion in different chemokine gradients was evaluated. Figure 3c shows the trajectories of 7 Trojanbots over 20 min, normalized to a common origin (chemokine gradient of 10 pM µm−1). The Trojanbots demonstrated clear directional movement toward regions of higher chemokine concentration. As shown in Fig. 3d, the chemotactic motion of Trojanbots exhibited a centralized distribution angle between −60° and +30°, indicating their directional migration ability along the chemokine gradient. We further investigated the response of Trojanbots to chemokine stimulation in a 30-s timeframe by documenting alterations in their morphologies. In the beginning, the Trojanbot retained its spherical shape, indicating an inactive state. Within 30 s, the Trojanbot underwent a structural transformation from the initial spherical shape to a flattened configuration with extended pseudopods, while demonstrating displacement relative to its original position (Supplementary Fig. 12).
To optimize the chemotactic conditions, the relationship between the movement behavior of Trojanbots and the chemokine concentration gradients has been investigated and compared with that of neutrophils. The chemotactic velocity of both neutrophils and Trojanbots increased with the chemokine gradient. The velocity of the Trojanbots can reach 0.24 µm s−1 in a chemokine gradient of 100 pM μm−1, demonstrating comparable chemotactic motion ability to normal neutrophils (Fig. 3e). We further assessed the migration persistence by employing the chemotactic index (CI), which quantifies the ratio of total displacement to path length. As illustrated in Fig. 3f, the CI of Trojanbots was influenced by the fMLP concentrations. To gain deep insights into the motion orientation of Trojanbots concerning the chemokine gradient, we investigated the turning angles distribution (TAD) that represents the angle between two consecutive displacements during chemotactic movement (Supplementary Fig. 13). In the absence of chemokine, the Trojanbots exhibited a homogeneous distribution of turning angles. When the chemokine gradient formed, the motion of Trojanbots became regular (Fig. 3g). Moreover, higher chemokine concentrations can lead to a more focused distribution of turning angles (Fig. 3h). The curve of TAD at 0° versus chemokine concentration shows a typical ballistic behavior with a chemotactic response toward the chemokine gradient, similar with that of natural neutrophils (Supplementary Fig. 14). These results suggest that the velocity of Trojanbots can be influenced by the intensity of chemokine gradient.
To characterize the release dynamics of Trojanbots, PMA was applied as a stimulus, and the release process was recorded using a microscope (Fig. 3i and Supplementary Movie 3). Time-lapse images show the resting, activation, release, and movement of Trojanbots, demonstrating the release of CatNbots in response to PMA. By adjusting the PMA concentration in the range of 10–300 nM, the initial release time of Trojanbots varied from 12 to 2 h (Fig. 3j). Consequently, a PMA concentration of 100 nM was selected as the treatment condition for subsequent experiments. To address resolution limitations in conventional microscopy, we performed super-resolution imaging using amino fluorescent quantum dots-labeled CatNbots (emission: 605 nm, pseudo-colored red) under structured illumination microscopy (SIM). Time-resolved SIM revealed that upon stimulation with PMA (100 nM), CatNbot expulsion occurred within 1–3 s, with individual CatNbots dispersing up from rupture sites while maintaining structural integrity (Supplementary Movie 4; Supplementary Fig. S15). Furthermore, we quantified the percentage of CatNbots released from Trojanbots and determined the release rate under different conditions. In the untreated group, <2% of CatNbots were released in 4 h (Fig. 3k). In contrast, in the group treated with 100 nM PMA, a modest release of CatNbots (<5%) was observed in the first 2 h, followed by a significant increase to ~90% in the following 4 h. The CLSM images show the encapsulation of Cou6-labeled CatNbots in the Trojanbots for 6 h (Fig. 3l left). However, in the presence of PMA, enhanced red fluorescence was observed after 6 h in the PI channels (Fig. 3l right), because the NETs destroyed the neutrophil plasma membrane, leading to the subsequent staining. Moreover, the distribution of as-released CatNbots was also observed in the red fluorescence region, confirming the release of CatNbots along with NETs from the Trojanbots.
To further investigate the catalytic propulsion of the CatNbots released from Trojanbots, we observed the motion track of individual CatNbots in phosphate-buffered saline (PBS) solution of different H2O2 concentrations (Fig. 3m). The released CatNbots exhibited typical Brownian motion in pure PBS, whereas in the presence of H2O2, they demonstrated enhanced diffusion. As the H2O2 concentration increased, the motion range increased significantly, indicating enhanced diffusion behavior (Supplementary Fig. 16 and Supplementary Movie 4). Then, the mean square displacement (MSD) versus time was used to study the enhanced motion of CatNbots governed by Eq. (1),
with the diffusion coefficient D, the time interval Δt, the rotational time τR, the CatNbots propulsion speed V, and the anomalous diffusion exponent α57. The MSD was calculated as a function of the time interval (∆t) under different H2O2 concentrations. As shown in Fig. 3n, the MSD increases slightly with time following a quasi-linear dependence in low H2O2 concentration (0–2 µM). In an H2O2 solution of higher concentrations (4–8 µM), the MSD curve demonstrated a parabolic increase, indicating the environment-dependent motion abilities of CatNbots. Similarly, corresponding to Eq. (2)
Figure 3o (blue curve) shows the highest effective diffusion coefficient Deff of the CatNbots under 8 µM of H2O2, corresponding to the fastest speed of about 0.93 μm2/s, which is ~18 times that at 0 µM of H2O2 (0.05 μm2/s, Brownian motion). A similar trend was observed in propulsion velocity (Fig. 3o, purple curve), the CatNbots in H2O2 of 8 µM exhibited a moving velocity of 5.57 µm/s, 7-fold that without H2O2. These results demonstrate the dynamic self-propulsion of CatNbots in the presence of H2O2. Considering the enrichment of H2O2 in the TME, the H2O2-powered self-propulsion would significantly enhance the deep diffusion of CatNbots into tumors. As shown in Fig. 3p, the CatNbots in 8 µM H2O2 exhibited the highest instantaneous velocity compared to those in 0, 2, 4, and 6 µM H2O2. To evaluate whether the released CatNbots could maintain optimal motility, we conducted motility performance assessments on the pre-phagocytic CatNbots. The results showed that these pre-phagocytic CatNbots exhibited similar motility trends across different concentrations of H2O2 solution (Supplementary Figs. 17, 18, and Supplementary Movie 5), compared to the released CatNbots. This indicates that neutrophil phagocytosis did not significantly impair the motility of the CatNbots. Critically, this preserved motility ensures that CatNbots can actively navigate toward tumor regions, where their therapeutic payload is selectively released.
Under physiological pH (7.4), GeNPs maintain a compact structure due to limited swelling of the gelatin matrix, which slows molecular diffusion and minimizes premature drug release. However, in the acidic tumor microenvironment (pH 6.5–6.8), the protonation of amino groups in the gelatin matrix reduces electrostatic repulsion. This swelling disrupts the nanoparticle structure, which in turn accelerates the diffusion of DOX and facilitates its release from the GeNPs58. Building on this pH-responsive mechanism observed in GeNPs, we explored whether the structurally optimized CatNbots could achieve enhanced controlled release dynamics. Our drug release studies demonstrated that CatNbots, similar to GeNPs, exhibit enhanced drug release under tumor-mimicking conditions (Supplementary Figs. 19 and 20). To directly evaluate the controlled drug delivery capability of CatNbots, high-performance liquid chromatography (HPLC) was conducted to investigate the pH-dependent release kinetics of DOX from CatNbots over 72 h under both physiological (pH 7.4) and simulated acidic tumor conditions (pH 6.5) The cumulative release of DOX from CatNbots at pH 7.4 increased from 4.7% at 1 h and 9.7% at 4 h to 73.3% at 72 h. In contrast, under acidic conditions (pH 6.5), the release was significantly accelerated, reaching 7.5% at 1 h, 15.7% at 4 h, and 90.4% at 72 h (Supplementary Fig. 21). This pronounced pH sensitivity confirms the controlled release capability of CatNbots, ensuring minimal premature leakage during systemic circulation (pH 7.4) while enabling efficient drug release in the tumor microenvironment (pH 6.5). This environmental sensitivity aligns with previous reports on gelatin-based systems, where acidic pH and MMP activity synergistically promote carrier degradation and payload release.
Cross the BBB
To evaluate the BBB penetration capability of Trojanbots, a monolayer Transwell system was established as an in vitro BBB model. Briefly, murine-brain microvascular endothelial (bEnd.3) cells were cultured in the upper chambers, while mice GBM tumor cells (GL261) were cultured in the bottom chamber (Supplementary Fig. 22). Additionally, serum-free culture medium without fetal bovine serum (FBS) was added to the bottom chamber. As a chemotactic peptide that attracts neutrophils, 10 nM fMLP was introduced to facilitate BBB penetration. After 6 days, the bEnd.3 cells formed a dense layer without apparent gaps (Supplementary Fig. 23). To functionally validate tight junction integrity, transendothelial electrical resistance (TEER) was monitored using a RE1600 epithelial voltohmmeter equipped with STX-2 chopstick electrodes (Jingong Hongtai Technology Co., Ltd., China) at 37 °C. The TEER values progressively increased and plateaued at 100 ± 25 Ω cm² by Day 6 (Supplementary Fig. 24), aligning with established in vitro BBB benchmarks59. These combined morphological and biophysical validations confirm functional tight junction formation, thereby supporting the model’s utility for mechanistic studies of Trojanbot penetration.
In the control experiments, GeNPs, CatNbots, neutrophils, and Trojanbots were individually added to the upper chambers for comparison (Fig. 4a). Under the influence of the chemokine gradient, both neutrophils and Trojanbots exhibited active downward migration, enabling them to traverse the model BBB and reach the tumor cells. To investigate the BBB penetration performance of different groups, we labeled the bEnd.3 cells membrane with Dio, while 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (Dil) was used to label GeNPs, CatNbots, neutrophils, and Trojanbots. As shown in Fig. 4b–d, GeNPs and CatNbots groups exhibited negligible penetration into the BBB, while neutrophils and Trojanbots demonstrated considerable BBB penetration ability. Notably, in the presence of fMLP, the Trojanbots displayed the strongest fluorescence signals on the GL261 layers in the bottom chamber, indicating the highest penetration rate. This trend was consistent across the bEnd.3 and BBB layers (Fig. 4e–g), suggesting that the Trojanbots enable effective BBB penetration in response to the fMLP gradient.
a Schematic illustration of the in vitro BBB model used to evaluate the potential BBB penetration abilities of GeNPs, CatNbots, Neutrophils, and Trojanbots, both in the presence and absence of fMLP. b–d CLSM images of the bEnd.3 cell layer (b), BBB layer (c), and GL261 cells (d). In the images, bEnd.3 cells were labeled with Dio, while GeNPs, CatNbots, Neutrophils, and Trojanbots were labeled with Dil. The nuclei of the GL261 cells were stained with Hoechst 33342 (blue). e–g The fluorescence statistics of the bEnd.3 monolayer, BBB layer, and GL261 cells layer, respectively (n = 3) (exact P value of e: GeNP vs. Trojanbot + fMLP P = 1.51E−8; exact P value of f: GeNP vs. Trojanbot + fMLP P = 2E−10, CatNbot vs. Trojanbot + fMLP P = 3E−10, Neutrophil vs. Trojanbot + fMLP P = 9.666E−7, Trojanbot vs. Trojanbot + fMLP P = 1.17148E−5; exact P value of g: GeNP vs. Trojanbot + fMLP P = 4.2E−9, CatNbot vs. Trojanbot + fMLP P = 1.25964E−5.) h The penetration ratio of different groups for 6 h (n = 3). (exact P value: GeNP vs. Trojanbot + fMLP P = 1.484E−7, CatNbot vs. Trojanbot + fMLP P = 1.731E−7, Trojanbot vs. Trojanbot + fMLP P = 1.484E−7). i–l The time-dependent penetration ratios of GeNPs (i), CatNbots (j), Neutrophils (k), and Trojanbots (l) with and without fMLP (10 nM, n = 3). Data are presented as mean ± SD of three independent experiments (e–l). P-values are determined by one-way ANOVA and Tukey multiple comparisons tests in (e–h). Source data are provided as a Source Data file.
The efficiency of active BBB penetration was further quantitatively assessed in vitro for 6 h. As shown in Fig. 4h, the control groups, which included GeNPs, CatNbots, and red blood cells, exhibited very low penetration ratios. In contrast, the Trojanbots demonstrated a significantly higher penetration ratio of 31.1%, comparable to that of neutrophils. Time-dependent quantification data indicated that the penetration ratios of GeNPs and CatNbots increased slightly over time, with <10% growth after 6 h, suggesting a poor ability to penetrate the BBB (Fig. 4i and j). Interestingly, the penetration ratios of both neutrophil and Trojanbot exhibited a similar trend (Fig. 4k and l). In the first 4 h, the penetration ratio of neutrophils gradually increased to 9.1%, following a quasi-linear pattern. Subsequently, this ratio went up dramatically to ~32.2% over the next 2 h. In the presence of fMLP, the penetration ratio after 6 h could reach about 54.1%, likely due to the surface adhesion and migratory mechanisms that neutrophils employ to target brain endothelial cells60. Similarly, the penetration ratio of Trojanbots followed a comparable trend, indicating that the Trojanbots, loaded with CatNbots, effectively retained their adhesion and migration capabilities.
Deep penetration of tumor in vitro
In addition to BBB, the dense and viscous ECM in GBM poses another substantial physical barrier that hinders the delivery of therapeutic agents to tumor tissue, limiting the efficacy of antitumor treatments61,62. Therefore, there is a strong need for innovative methods that can facilitate the deep penetration of therapeutic agents into tumor sites. Our developed Trojanbots address this challenge by traversing the BBB and then strategically releasing CatNbots within the TME. The catalytic self-propulsion ability of the CatNbots, released by Trojanbots, allows for deeper infiltration and enhanced internalization into tumor tissues in the presence of H2O2. This method not only increases the penetration depth significantly but also reduces H2O2 levels in tumor cells, modulating the intracellular oxidative stress state and thereby optimizing the microenvironmental conditions (Fig. 5a). To validate this mechanism, we examined the chemotactic migration behavior of CatNbots in response to an H2O2 concentration gradient. To mimic the H2O2 gradient in vitro, agarose gel containing GL261 cancer cell lysate was introduced into the upper right chamber of a “Y-shape” microfluidic chip, forming an H2O2 gradient from left to right. For comparison, agarose gel containing bEnd.3 cell lysate was added to the bottom right chamber (Fig. 5b). The concentrations of H2O2 at different positions of the Y channel were further evaluated (Supplementary Fig. 25a). The results show that the concentration of the H2O2 was higher at the site containing GL261 lysate (location 4), reaching ~8.4 µM, whereas at location 1 (the region furthest from the H2O2 inducer), the concentration was about 0.93 µM (Supplementary Fig. 25b). These findings suggest that H2O2 gradually diffuses outward along the Y-type channel, thereby establishing a concentration gradient over time. In this experiment, CatNbots were introduced into the left chamber, and their movement was continuously tracked using CLSM. As shown in Fig. 5c, the fluorescence signals in both the GL261 chamber and the bEnd.3 chamber indicates selective migration of CatNbots towards the upper right chamber that contains GL261 cancer cell lysate. After 60 min, the fluorescence intensity of the upper right chamber was ~10 times higher than that contained bEnd.3 cell lysate (Fig. 5d). To further verify the robustness of this chemotactic behavior under more physiologically relevant conditions, we replaced cell lysates with live GL261 cells and bEnd.3 cell aggregates embedded in collagen gel. These 3D cellular clusters (~100 μm diameter) generated sustained H₂O₂ gradients (8 μM at proximity) through active metabolic activity. When introduced into the Y-chip system, CatNbots exhibited analogous directional migration toward the live tumor aggregates, with the target chamber accumulating ~15-fold higher fluorescence intensity than control chambers after 90 min (Supplementary Fig. 26). Specifically, the different diffusion rates along the surface of CatNbots would induce a phoretic torque, which persistently orients their motion toward the H2O2 fuel gradient, thereby facilitating chemotactic motion, in good agreement with other previous studies reported elsewhere63. These in vitro results confirm that the targeted self-propulsion ability of CatNbots holds great promise for deep penetration toward tumors.
a Schematic diagram of the chemotaxis of CatNbots toward tumor tissue and the internalization by tumor cells. b Schematic of chemotactic motion of CatNbots along the concentration gradient of the H2O2. c Fluorescent images of the different chambers. d The corresponding fluorescence quantification values at different time points (n = 3). e In vitro penetration results in 3D tumor spheroids. The bright-field images and Z-stack CLSM images were taken from the top to the equatorial plane in 160 µm thickness of 3D cellular spheroids after treatment with GeNPs, EMV@GeNPs, and CatNbots for 4 h. f The corresponding fluorescent spectra and g the mean fluorescence intensity of the GL261 tumor spheroids treated with different samples (n = 3) (exact P value: GeNP vs. CatNbot P = 8.16414E−5). h Tumor spheroid morphology and TUNEL apoptosis assay of GL261 3D spheroids treated with various groups for 3 days. Spheroid slices, 160 µm thick, were subjected to the TUNEL assay, with positive cells identified using CLSM imaging. i Volume statistics of tumor spheroids before and after treatment across different experimental groups. j Cytotoxicity assessment of GeNPs, EMV@GeNPs, and CatNbots on 3D tumor spheroids (n = 3). k Cellular uptake CLSM images for different samples (Green: labeled with Cou6, Red: cell membranes stained with Dil, Blue: cell nuclei stained with Hoechst 33342). l The corresponding fluorescence semi-quantitative analysis (n = 3) (exact P values: GeNP vs. CatNbot P = 3.77E−7, EMV@GeNP vs. CatNbot P = 1.2024E−6). m Cytotoxicity of GeNPs, EMV@GeNPs, and CatNbots on GL261 cell lines in a 2D culture (n = 3). n Mechanism diagram of CatNbots regulating the TME. o Typical CLSM images of intracellular H2O2 (labeled with the H2O2 fluorescent probe, ROSGreenTM H2O2 Probe) in cancer cells after CatNbots treatment for 12 h (n = 3). p Relative H2O2 level in cancer cells after treatment with different samples for 12 h (n = 3) (exact P-value: 3 h Control vs. CatNbot P = 1.51132E−5, 3 h GeNP vs. CatNbot P = 3.32681E−5, 9 h Control vs. CatNbot P = 3.9146E−6, 9 h GeNP vs. CatNbot P = 2.219E−6, 9 h EMV@GeNP vs. CatNbot P = 3.617E−7, 12 h Control vs. CatNbot P = 3.18E−8, 12 h GeNP vs. CatNbot P = 2.9E−8, 12 h EMV@GeNP vs. CatNbot P = 1.72E−8). Data are presented as mean ± SD of three independent experiments (d, g, j, l, m, p). P-values are determined by one-way ANOVA and Tukey multiple comparisons tests in (g, l, p). Source data are provided as a Source Data file.
To gain deeper insight into the chemotactic behavior of CatNbots, we utilized 3D cellular spheroid models composed of GL261 tumor cells to explore their permeability. After one week of culture, the tumor cells formed complete spherical shapes (Supplementary Fig. 27). Following incubation with CatNbots, we assessed the fluorescence intensity within the 3D cellular spheroids using CLSM (Fig. 5e). In the passive GeNPs and EMV@GeNPs groups, fluorescence signals were only detected around the periphery, while the center was dark. In contrast, the CatNbots group exhibited detectable fluorescence at various depths, indicating significant penetration of CatNbots into the spheroids. The fluorescence intensity and mean fluorescence intensity across the 160 μm section further confirmed the deep penetration ability of CatNbots, demonstrating the chemotactic response and the enhanced penetration of CatNbots in the intricately cellular structure of GBM (Fig. 5f and g).
To clarify the antitumor efficacy following extensive tumor penetration, we systematically evaluated the superior cytotoxicity of CatNbots against tumor spheroids through morphological changes and an apoptosis assay (Fig. 5h). After treatment with CatNbots, the tumor spheroids exhibited a noticeable transformation in morphology, changing from a smooth, spherical shape to rough, irregular, and collapsed structures within 10 h (Supplementary Fig. 28 and Supplementary Movie 6). Significant ablation can be observed after 3 days (Fig. 5h and i). Compared to the GeNPs, EMV@GeNPs, and control groups, treatment with CatNbots resulted in more apoptotic cells, even in the central region of the spheroids. This indicates the exceptional cytotoxicity of CatNbots against GL261 tumor cells (Fig. 5j). Additionally, to investigate the precise targeting and anti-tumor capability of CatNbots toward glioma cells, the cellular uptake of Cou6-labeled CatNbots was analyzed by CLSM (Fig. 5k). Compared with the GeNPs and EMV@GeNPs groups, the cells incubated with CatNbots showed more obvious green fluorescence signals, three times higher than that of the other two groups (Fig. 5l). This is attributed to the autonomous movement of CatNbots, which not only facilitates enhanced cellular uptake but also amplifies cytotoxic effects on tumor cells by effectively delivering encapsulated anti-cancer drugs to eliminate tumor cells.
Generally, the development of GBM is associated with elevated levels of ROS, with H₂O₂ playing a pivotal role in tumor progression. The accumulation of H₂O₂ not only leads to significant oxidative stress and cellular damage but also accelerates tumor malignant transformation64,65. By selectively targeting and reducing excessive H₂O₂, it is possible to effectively diminish oxidative stress within the tumor microenvironment TME, thereby inhibiting tumor cell proliferation and slowing disease progression. We have demonstrated that CatNbots are capable of generating self-propulsion within tumor tissues by catalyzing the decomposition of H2O2 in the TME. Based on this observation, we hypothesized that CatNbots could not only mitigate excess H₂O₂ during the catalytic process but also potentially regulate oxidative stress levels in glioma cells (Fig. 5n). To test this hypothesis, glioma cells were incubated with GeNPs, EMV@GeNPs, and CatNbots for 12 h, and intracellular H₂O₂ levels were continuously monitored using fluorescence probes. The results demonstrated that CatNbots significantly reduced intracellular H₂O₂ levels, achieving a 3.1-fold reduction compared to the control group (Fig. 5p). Additionally, the presence of CatNbots led to increased O₂ production within the cancer cell environment, whereas GeNPs and EMV@GeNPs exhibited minimal O₂ generation. This enhanced O₂ production by CatNbots may further inhibit tumor cell invasion and metastasis by alleviating the hypoxic TMZ (Supplementary Fig. 29).
Active target of Trojanbots
As shown in Fig. 6a, to explore the targeting ability of Trojanbots in vivo, we established an orthotopic glioma model by inoculating Gl261-luciferase cells into the brains of mice (C57BL/6J, male, aged 8 weeks). After 9 days, the established GBM model was confirmed through H&E tissue staining (Supplementary Fig. 30). We then assessed the tumor-targeting ability of Trojanbots by injecting therapeutic agent samples labeled with 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindotricarbocyanine iodide (DiR) into the tail veins of glioma-bearing mice and the subsequent tracking of fluorescence signal using near-infrared fluorescence microscopy. As shown in Fig. 6b, the mice treated with Trojanbots displayed a significantly higher brain fluorescence signal than the other three control groups (EMV@GeNPs, CatNbots, and NE@EDGNs, which refers to the engulfment of EMV@GeNPs by neutrophils). Quantitative fluorescence analysis indicated that the fluorescence intensity in the mouse brain reaches its maximum after 8 h. Mice injected with Trojanbots exhibited the highest fluorescence intensity among the four groups (Fig. 6c). This enhancement can be attributed to the dual chemotactic response facilitated by external neutrophils and internal CatNbots.
a The schematic representation of the construction of the brain glioma model. b In vivo fluorescence images of orthotopic GL261-Luc mouse models with GBM tumors following a single tail vein injection of potassium fluorescein (n = 3 mice per group). c Fluorescence intensity measurements of glioma mouse brains treated with various therapeutic agents (n = 3 mice per group). d Fluorescent images (IVIS imaging) of the brain and major organs, taken 48 h after injection (n = 3 mice per group). e Quantitative ROI analysis of DiR signals in the brain and organs (n = 3 mice per group) (exact P value: Spleen CatNbot vs. Trojanbot P = 5.89774E−5, Lung EMV@GeNP vs. Trojanbot P = 3.86669E−5, Brain EMV@GeNP vs. Trojanbot P = 3.98E−8, Brain CatNbot vs. Trojanbot P = 1.084E−7, NE@EDGN vs. Trojanbot P = 1.62003E−5). f Fluorescence images (IVIS imaging) of brain slices from four groups of mice, labeled with DiR (8 h post tail vein injection). g Quantitative analysis of the fluorescence images (IVIS imaging) of brain slices from the four groups (n = 3 mice per group). (exact P value: EMV@GeNP vs. Trojanbot P = 6.055E−7). h Representative fluorescence images of glioma tissue slices from GBM mice. In the images, EMV@GeNPs, CatNbots, EMV@GeNPs within NE@EDGN, and CatNbots within Trojanbots were labeled with FITC (green), while DOX exhibits intrinsic autofluorescence properties (red). The nuclei of the GL261 cells were stained with Hoechst 33342 (blue). Data are presented as mean ± SD of three independent experiments (e, g). P-values are determined by one-way ANOVA and Tukey multiple comparison tests in (e, g). Source data are provided as a Source Data file.
To substantiate the in vivo results, ex vivo fluorescence imaging of brain tumors and major organs was further performed (Fig. 6d and Supplementary Fig. 31). Quantitative fluorescence analysis shows that the fluorescence signal accumulated at the tumor tissue concerning the Trojanbots group was 5.1 times of the CatNbots group and 1.8 times of the NE@EDGNs group (Fig. 6e), confirming the superior BBB penetration and GBM tissue accumulation of Trojanbots. Fluorescence images (IVIS imaging) of brain slices from four groups of mice labeled with DiR were collected 8 h after tail vein injection (Supplementary Fig. 32). As depicted in Fig. 6f, Trojanbots show the highest accumulation at the brain tumor sites. Quantitative analysis results (Fig. 6g) indicate that the fluorescence signal intensity of the Trojanbot group is 5.2-fold higher than that of the CatNbot group, suggesting that utilizing neutrophils to cross the BBB significantly enhances tumor-targeting efficacy. Moreover, compared with the NE@EDGN group, the fluorescence intensity of the Trojanbot group was 2-fold greater, confirming that the CatNbot in Trojanbot achieves deeper tumor penetration upon release in response to the TME. These findings support the hypothesis that the design strategy combining neutrobots with CatNbots can more effectively target the interior of glioma.
To comprehensively compare the BBB penetration and targeted aggregation properties among different groups, we labeled EMV@GeNPs, CatNbots, NE@EDGNs, and Trojanbots with FITC. We then evaluated their fluorescence intensity in tumor sections taken 8 h after injection. As depicted in Fig. 6h, the tumor tissue treated with Trojanbots displayed a substantial green fluorescence signal that permeated nearly the entire tumor, suggesting the superior tissue penetration capability of Trojanbots. In contrast, only a minimal green fluorescence signal was observed in the NE@EDGNs group that had been loaded with EMV@GeNPs. H&E staining analysis revealed that the released CatNbots were able to enter brain tumors (Supplementary Fig. 33). This significant difference can be attributed to the self-propelling, deep penetration ability of CatNbots released from Trojanbots. Furthermore, the therapeutic drug DOX showed extensive distribution within the tumor tissues in the Trojanbots group, confirming the targeted drug delivery behavior of the Trojanbots.
Therapeutic efficacy of Trojanbots
To further validate the therapeutic efficacy of Trojanbots, in vivo antitumor experiments were conducted (Fig. 7a). We initially assessed circulating levels of Trojanbots in healthy mice at different time points after intravenous injection of FITC-labeled Trojanbots by flow cytometry. The results showed that Trojanbots remained at a relatively high level in the blood within 72 h after injection, demonstrating their excellent blood retention ability (Supplementary Fig. 34). Subsequently, 8-week-old male C57BL/6J mice with established orthotopic GL261-Luc tumors were randomly divided into 5 groups, with 5 tumor-bearing mice per experimental group. The groups were treated with PBS, EMV@GeNPs, CatNbots, NE@EDGNs, and Trojanbots (starting from the 9th day of model establishment, tail vein administration was performed every other day for a total of three times). Except for the PBS group, the DOX dose for the other groups was 100 mg/kg. Next, we conducted bioluminescence imaging, body weight measurement, survival status analysis, and histological examination at different stages for each group of mice. Bioluminescence imaging revealed progressive brain tumor growth in the control group of PBS, confirming success in the model establishment (Fig. 7b). The luminescence intensity of GBM of mice treated with EMV@GeNPs, CatNbots, and NE@EDGNs increased to 1.32, 1.25 and 1.2 times of the initial value at the end of treatment, respectively. Conversely, following treatment with Trojanbots, the fluorescence signal diminished to 75% of its initial value (Fig. 7c). Additionally, mice treated with Trojanbots exhibited stable body weight, indicating minimal side effects. In contrast, the PBS-treated group experienced significant weight loss (Fig. 7d). In the survival rate experiment, Trojanbots demonstrated the longest median survival time of 38 days due to its tumor growth inhibition effect, which was significantly longer than that of PBS (18 days), EMV@GeNPs (25 days), CatNbots (26 days), and NE@EDGNs (34 days) (Fig. 7e). Moreover, H&E staining of the entire brain revealed the smallest tumor volume in mice treated with Trojanbots (Fig. 7f). TUNEL staining analysis confirmed the increased tumor cell apoptosis induced by Trojanbots (Fig. 7f and g). To further evaluate the effect of Trojanbots on glioma treatment, immunohistochemical staining analyses of Ki-67, CD31, HIF-1α and N-cadherin at the tumor site were performed (Fig. 7f). The expression of the proliferation marker Ki-67 was significantly lower in mice treated with Trojanbots, reaching only 0.33 times that of the PBS control group (Fig. 7h). Conversely, the expression level of CD31, a marker of blood vessels, was markedly reduced in the tumor sections of the Trojanbot-treated group, indicating a significant decrease in tumor-associated vasculature (Fig. 7i). Immunofluorescent staining revealed a substantial reduction in HIF-1α levels in the Trojanbot group, demonstrating effective alleviation of hypoxia in the TME following Trojanbot treatment (Fig. 7j). Additionally, the expression of N-cadherin, which is associated with tumor invasion and metastasis, was significantly decreased in the Trojanbot group compared to the PBS control group (Fig. 7k). Collectively, these findings underscore the exceptional antitumor efficacy of Trojanbots in GBM models.
a The schematic diagram of intravenous injection treatment. b Time-course bioluminescence images of orthotopic GL261 tumor-bearing mice treated with PBS, EMV@GeNPs, CatNbots, NE@EDGNs, and Trojanbots (n = 5 mice per group). c Quantified luminescence levels of mice using the Lumina IVIS III system (n = 5 mice per group). d Body weight profiles (n = 5 mice per group). e Survival curves of mice (n = 5 mice per group) (exact P value: PBS vs. Trojanbots, P = 0.0003, EMV @ GeNP vs. Trojanbot, P = 0.0005, CatNbot vs. Trojanbot, P = 0.0022, NE @ EDGN vs. Trojanbot, P = 0.2517). f H&E images of the whole brains excised from the representative mice in treatments as described above, Histological analysis using the TUNEL (Green: apoptotic cells; Blue: Hoechst-stained cell nuclei) and Ki67 assay (Green: proliferating cells; Blue: Hoechst-stained cell nuclei), CD31, HIF-1α, and N-cadherin. The histological analyses were representative of data from three mice. g–k The fluorescence statistics derived from the histological analyses shown in (f) (n = 3 mice per group) (exact P value of h: PBS vs. Trojanbot P = 5.65E−8, exact P value of j: PBS vs. Trojanbot P = 6.91E−8, EMV@GeNP vs. Trojanbot P = 1.258E−7, exact P value of k: PBS vs. Trojanbot P = 9.00532E−5). l Representative flow cytometry results showing different groups of CD45+CD3+T cells in tumor tissues of GBM. For the gating strategy for T-cell analysis, refer to Supplementary Fig. 35. m Quantitative analysis of CD3+CD8+ cytotoxic Helper T cells (n = 3 mice per group). n Quantitative analysis of CD3+CD4+ Helper T cells (n = 3 mice per group). Data are presented as mean ± SD of three independent experiments (g–k, m, n) and five independent experiments (c–e). P-values are determined by one-way ANOVA and Tukey multiple comparisons tests in (c, e, g–k, m, n). Source data are provided as a Source Data file.
To further investigate the immune activation mechanism triggered by Trojanbots, we assessed the infiltration of anti-tumor immunity cells, specifically cytotoxic T cells (CD3+CD8+) and Helper T cells (CD3+CD4+), in the tumor tissues on day 16 post-treatment. Notably, in the Trojanbot treatment group, the abundance of CD8+ T cells was significantly increased, with values 1.52 and 1.68 times higher than those in the Ctrl (wild-type mice) and tumor-bearing group, respectively (Fig. 7l, m and Supplementary Fig. 35). Conversely, the relative proportion of CD4+ T cells decreased (Fig. 7n). The observed increase in immune cell infiltration could be attributed to Trojanbot’s ability to reduce ROS (particularly H2O2) levels in the tumor environment, coupled with the release of DOX, which induces immunogenic cell death66. The shift in immune composition may reflect either the direct expansion of CD8+ T cells or a conversion from CD4+ T cells, enhancing the anti-tumor response and promoting more effective tumor targeting and killing67.
Furthermore, to achieve a more comprehensive evaluation of the biocompatibility of Trojanbots treatment, mice were administered with EMV@GeNPs, CatNbots, NE@EDGNs, and Trojanbots, respectively. Major organs were harvested and subjected to H&E staining analysis to assess potential systemic toxic side effects. Notably, no discernible organ damage was observed post-treatment (Supplementary Fig. 36, thereby confirming the superior in vivo biocompatibility of Trojanbots. Consequently, these findings validate that the rationally designed enzymatic nanobot-in-neutrobot systems exhibit favorable antitumor efficacy without inducing additional systemic toxicity, offering insights into the active therapy of GBM.
Discussion
Two critical challenges should be addressed to achieve efficient drug delivery for GBM. First, the BBB exhibits highly selective permeability, blocking most exogenous drugs from entering the central nervous system (CNS). This significantly limits drug delivery efficiency and reduces the effective duration of therapeutic agents within the body. Second, the TME is characterized by elevated oxidative stress and hypoxic conditions, which not only impede drug diffusion but also enhance tumor cell survival and resistance to therapy, thereby complicating treatment68. To address these challenges, we have demonstrated a Trojanbot by integrating active hybrid neutrophils (neutrobot) with enzymatic nanobots, creating an efficient tumor-targeting delivery system.
This system leverages the natural chemotactic properties of neutrophils, enabling them to sense inflammatory factor gradients and achieve efficient chemotactic movement with an average speed of 0.24 µm s−1. In an in vitro BBB model, Trojanbots demonstrated a penetration efficiency of ~50%, which is tenfold higher than that of passive nanoparticles (GeNPs). After crossing the BBB, the enzyme-driven nanorobots are fully released and utilize hydrogen peroxide enriched in the TME as a fuel source, generating catalytic propulsion with an average speed of 5.57 µm s−1. This significantly enhances their ability to penetrate tumor tissues deeply. In a 3D tumor spheroid model, CatNbots released by Trojanbots achieved a penetration depth of 416 μm, which was 2.8 times greater than that of GeNPs. Notably, the released nanorobots not only exhibited high uptake efficiency by tumor cells but also catalytically degraded intracellular H2O2 (reducing levels by approximately 3.5-fold), effectively alleviating oxidative stress in the TME while significantly improving hypoxic conditions by generating oxygen. This synergistic effect further enhances anti-tumor efficacy, demonstrating the potential of Trojanbots in modulating the metabolic landscape of tumors.
In vivo, experimental results further validated the superior therapeutic performance of Trojanbots. By combining the BBB-penetration capability of active neutrophils with the deep tissue penetration of enzyme-driven nanorobots, Trojanbots improved targeted delivery efficiency by 33.5% compared to the control group and extended the median survival time of mice to 45 days. Additionally, while immune clearance is an inherent challenge for most exogenous nanoparticles, Trojanbots are endowed with natural immune tolerance-based incorporation of neutrophils. As innate immune cells, neutrophils are inherently recognized as “self”, allowing Trojanbots to evade rapid clearance by macrophages and other immune surveillance mechanisms. This immune evasion capability not only prolongs systemic circulation but also enhances targeted delivery efficiency, thereby improving the therapeutic efficacy of Trojanbots in vivo44,46. Moreover, literature reviews highlight the limitations of passive targeting strategies, where only 0.7% (median) of administered nanoparticles successfully reach solid tumors69. While our previous design, which combined neutrophils with passive nanoparticles, enabled BBB penetration, the internal nanoparticles’ passive nature restricted overall delivery efficiency43. Building upon this, we optimized the design to incorporate active targeting and deep penetration capabilities within the internal nanoparticles, achieving a nearly fivefold improvement in delivery efficiency for the optimized Trojanbots.
In conclusion, by integrating the natural chemotactic properties of active neutrophils with hydrogen peroxide-responsive nanorobots, Trojanbots successfully overcome the BBB and achieve precise delivery to deep tumor regions. This strategy not only demonstrates significant advantages in GBM therapy but also offers promising potential for application in the treatment of other cancers and neurological diseases, such as Alzheimer’s and Parkinson’s disease. Moreover, the versatility of Trojanbots, driven by their dual-targeting mechanism, enables them to adapt to various disease microenvironments, making them an attractive platform for personalized treatment strategies. The modular design further enhances their potential by allowing the flexible loading of different therapeutic agents, from chemotherapeutic drugs to gene-editing tools, thus broadening their application to a wide range of pathological conditions.
Methods
This research complies with all relevant ethical regulations. All animal experimental operations were approved by the Animal Experimentation Ethics Committee of Harbin Institute of Technology, in compliance with all relevant ethical regulations (approval number: IACUC-2024087). To comply with the SAGER guidelines, only male animals were used for the in vivo experiments. The sex of the animals was not considered a variable in this study design. The mice were kept in a barrier environment with a constant temperature of 24 °C and a relative humidity of 50%. The mice were maintained under a 12-h light and 12-h dark cycle. All experimental measurements are provided as Source data inside an Excel file named “Source Data”.
Materials
All chemicals were used without additional purification, and deionized water (resistance > 18.2 MΩ cm) obtained from a Milli-Q® integral water purification system was used for all solution preparations. Doxorubicin (DOX, >99%)), 1-(3-Dimethylaminopropyl)−3-ethylcarbodiimide (EDC) (≥95.0%), and Catalase were provided by Macklin. Hoechst 33342, Hoechst 33342/PI, Red Blood Cell Lysis Buffer, and DAPI were obtained from Solarbio. Gelatin, Penicillin–streptomycin mixture, and Sucrose were purchased from Biotopped. Percoll, PMA, 4% paraformaldehyde, and Triton X-100 were purchased from Biosharp. Ltd. Coumarin 6 (Cou6) was purchased from Sigma, 1,1’-dioctadecyl-3,3,3’,3’-tetramethylindocarbocyanine perch-lorate (DiI), 3,3’-dioctadecyloxacarbocyanine perchlorate (Dio), 1,1-dioctadecyl-3,3,3,3-tetramethylindotricar-bocyaine iodide (DiR), and Hoechst 33342 were provided by Beyotime Institute of Biotechnology. Dulbecco’s modified Eagle medium (DMEM) was purchased from Gibco, DMEM/High Glucose was purchased from HyClone, anti-fluorescence decay sealing tablets (BOSTER), PBS(Meilun), ROSGreenTM H2O2 Probe (Baiaolaibo). FITC-conjugated anti-CD11b (dilution 1:100, catalog number: 101205) and APC-conjugated anti-Ly6G (dilution 1:100, catalog number: 127613) were purchased from BioLegend. Anti-Ki67 antibody (dilution 1:200, catalog number: ab15580), anti-CD31 antibody (dilution 1:100, catalog number: ab222783), anti-N-Cadherin antibody (dilution 1:100, catalog number: ab76011), and anti-HIF-1 alpha antibody (dilution 1:100, catalog number: ab308433) were purchased from Abcam. CoraLite488-conjugated goat anti-rabbit IgG(H + L) (dilution 1:100, catalog number:SA00013-2) and multi-rAb CoraLite® Plus 594-Goat anti-rabbit recombinant secondary antibody (H + L) (dilution 1:200, catalog number: RGAR004) were purchased from Proteintech. Anti-CD3-PerCP-Cy5.5 (dilution 1:20, catalog number: 100218), anti-CD45-FITC (dilution 1:100, catalog number: 202205), anti-CD4-APC-Cy7 (dilution 1:100, catalog number: 201509), and anti-CD8-PE (dilution 1:50, catalog number: 201706) were purchased from BioLegend.
Instruments
Transmission electron microscopy (TEM) was performed on a Hitachi H-7650 transmission electron microscope; the acceleration voltage was 100 kV. The hydration diameter and zeta potential of the samples were analyzed by Zeta PALS™ Zeta potential analyzer from Brookhaven Instrument Co., Ltd, in PBS buffer. The fluorescence spectrum was analyzed using a HORIBA JOBIN YVON FluoroMax-4 spectrofluorometer. Oxygen content generated by CatNbots dissolving hydrogen peroxide was observed by Asmik with a dissolved oxygen analyzer. CLSM was performed on a Leica TCS SP5 II CLSM system. An Olympus BX31 normal microscope with a DP74 scientific camera and HIS-SIM intelligent ultra-sensitive super-resolution microscope (V3.0, China) was used to capture fluorescence images. in vivo fluorescence images were collected by IVIS Lumina III in vivo Imaging System from PerkinElmer Inc. The cell flow data were all collected by a FACS Calibur from Becton Dickinson Co., Ltd. (FACSAria™ Fortessa), and analyzed by FlowJo_V10. The desktop high-speed centrifuge used in the synthesis experiments was H-1650 from Hunan Xiangyi Centrifuge Device Co., Ltd. Deionized water was produced by the ELGA Lab Water MICRI + CLASSIC UV water purification system.
Preparation of GeNPs
The preparation of GeNPs was conducted utilizing a solvent-based method. Initially, 1.25 g of gel powder was dissolved in 25 mL of water at a temperature of 50 °C. Upon cooling the gel solution to room temperature, an additional 25 mL of acetone was introduced while maintaining agitation with a magnetic stirrer. After a duration of 3 min, stirring was halted, and the supernatant was carefully removed using a disposable dropper, thereby isolating the precipitate and eliminating the low molecular weight gel from the supernatant. Subsequently, 25 mL of water was added to the beaker containing the remaining gel pellet, which was then placed in a 50 °C water bath to ensure complete redissolution of the gel pellet. The pH of the gel solution was adjusted to 3.0 using 1 M hydrochloric acid. While stirring at 40 °C, 75 mL of acetone was added. Following this, 0.2 mL of 50% glutaraldehyde was added at a controlled rate of 0.05 mL per minute to facilitate complete crosslinking of the GeNPs. After an additional hour of agitation, the GeNP solution was allowed to stand overnight at room temperature. Upon achieving complete crosslinking, the GeNPs solution was subjected to rotation and evaporation at 25 °C. The resultant particle solution was then filtered through a 0.2 µm filter membrane after the removal of acetone, and the prepared GeNPs solution was subsequently stored at 4 °C.
Drug loading ability of GeNPs
GeNPs were synthesized according to the previously described method in the section titled “Preparation of GeNPs.” Subsequently, the loading capacity of DOX was assessed. Various concentrations of DOX solutions were prepared, and GeNPs of consistent mass were immersed in the DOX solution for a duration of 24 h. Following this incubation period, the samples were subjected to centrifugation to separate the supernatant. The concentration of DOX in the supernatant was quantified using UV–visible spectroscopy, thereby facilitating the calculation of the drug loading capacity.
Bacterial culture and collection of bacterial (E. coli) outer membrane vesicles (EMVs)
E. coli was cultivated in Luria broth medium using a rotary shaker at a temperature of 37 °C for 24 h until the optical density at 600 nm (OD600) reached ~1.0. Initially, 250 mL of the bacterial culture was subjected to centrifugation at 3000×g for 10 min to separate the bacterial cells from the medium. The resulting supernatant was subsequently collected and filtered through a 0.45 µm filter. Following this, the filtrate was concentrated utilizing Amicon centrifugal filters with a molecular weight cutoff of 100 kDa (Millipore). The concentrated medium was then centrifuged at 150,000×g for 2 h at 4 °C. The resultant outer membrane vesicle (OMV) pellet was resuspended in PBS and stored at −80 °C for future use.
Preparation and characterization of the EMV@GeNPs
After the aforementioned preparatory steps, the EMVs were subjected to extrusion through a 200 nm polycarbonate (PC) membrane utilizing an Avanti mini extruder, a process that was repeated 30 times to obtain the EMVs. Following this, the EMVs were combined with GeNPs and extruded through the 200 nm PC membrane an additional 100 times. The resultant particle mixture was then centrifuged at 11,180×g for three cycles of 10 min each to eliminate any unbound E. coli membranes.
Lipopolysaccharide (LPS) measurement
The enzyme-linked immunosorbent assay (ELISA) was employed to quantify the levels of LPS present in E. coli. The experimental setup involved the use of EM vesicles, GeNPs, EMV@GeNPs, and CatNbots. A standard LPS solution was incubated in separate wells of a 96-well plate that had been coated with LPS-specific antibodies. The substrate 3,3’,5,5’-tetramethylbenzidine (TMB) was utilized, which, upon catalysis by peroxidase, resulted in a blue coloration, while an acidic environment induced a yellow hue. TMB is a hydrophobic compound and is largely insoluble in water. Therefore, TMB was first dissolved in dimethyl sulfoxide (DMSO) to create a stock solution (10 mg/mL: 10 mg of TMB in 1 mL of DMSO). This stock solution was then diluted into an acetic acid–sodium acetate buffer (pH = 5.0−5.5) to achieve the desired final concentration for the assay (0.1 mg/mL). The intensity of the color observed in the E. coli samples was found to be positively correlated with the concentration of LPS. Consequently, absorbance measurements (optical density, OD) were taken for GeNPs, EM vesicles, EMV@GeNPs, and CatNbots, followed by appropriate calculations.
Preparation and characterization of the CatNbots
The chemical cross-linker 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) was employed to achieve covalent attachment of catalase to the surface of bacterial membranes. This methodology promotes the formation of an amide bond between the enzyme’s amino group and the carboxyl group present on the bacterial surface. Specifically, a PBS (pH 5.5) solution containing 2 mM 1-(3-dimethylaminopropyl)−3-ethylcarbodiimide hydrochloride (EDC) was added to the 2 mL EMV@GeNPs solution (0.5 mg mL−1). Following 6 h of agitation, the supernatant was removed by centrifugation at 7000 × g for 10 min. The resulting EMV@GeNPs were then dispersed in a PBS (pH 5.5) solution containing catalase (CAT), and after 4 h of agitation, the supernatant was again removed by centrifugation at 7000 × g for 10 min, yielding the CatNbots. Morphology of GeNPs and EMV@GeNPs, and CatNbots was characterized by TEM. GeNPs, EMV@GeNPs, and CatNbots were stained with 2% phosphotungstic acid.
Isolation of neutrophils
Neutrophils were isolated from the bone marrow of mice utilizing the Percoll gradient method. Specifically, tibiae and femora were extracted from 8-week-old mice, thoroughly rinsed in Roswell Park Memorial Institute 1640 (RPMI 1640) medium, and placed in a sterile dish. In a controlled sterile environment, the ends of the bones were cut, and the marrow cavity was flushed with culture medium using a sterile syringe. Percoll solutions at concentrations of 71% and 61% (v/v) were prepared in PBS. A 15 mL sterile centrifuge tube was sequentially layered with 3 mL of each Percoll solution. Bone marrow cells were then added dropwise to the tube and subjected to centrifugation at 1000 × g for 30 min. The neutrophils were subsequently collected from the interface between the 71% and 61% Percoll layers using a sterile syringe. Following the lysis of erythrocytes with a lysis buffer, the neutrophils were washed and cultured in DMEM supplemented with 10% FBS in a humidified atmosphere containing 5% CO2 at 37 °C.
Fabrication and characterization of Trojanbots
Trojanbots were synthesized through the incubation of neutrophils with CatNbots. Specifically, neutrophils were incubated with CatNbots (1 mg) containing DOX at a concentration of 50 µg in a sterile tube, at a density of 5 × 105 cells per mL, for 30 min at 37 °C. Following incubation, the resultant Trojanbots were isolated by washing with PBS. The phagocytosis of neutrophils was visualized using confocal laser scanning microscopy (CLSM, Leica TCS SP5 II). Additionally, flow cytometry was employed to assess the cellular uptake of gold nanoparticles (GeNPs) and CatNbots by neutrophils. For this analysis, neutrophils (5 × 105 cells per mL) were incubated with either GeNPs or CatNbots (1 mg/mL) for 30 min at 37 °C. After the incubation period, any unbound GeNPs or CatNbots were removed. The neutrophils loaded with GeNPs or CatNbots were subsequently collected for fluorescence analysis. The fluorescence intensity observed was compared to that of the original neutrophils, which served as a control group, allowing for the calculation of the loading percentage of neutrophils. CLSM images of the Trojanbots were captured, with CatNbots being specifically labeled with DOX.
Purity analysis of neutrophils
Freshly isolated murine neutrophils were resuspended in PBS supplemented with 2% FBS and subsequently stained with FITC-conjugated anti-CD11b (catalog number 101205, clone M1/70, Biolegend, diluted 1:100) and APC-conjugated anti-Ly6G (catalog number 127613, clone 1A8, Biolegend, diluted 1:100) antibodies for 30 min at 4 °C in the absence of light. Following staining, the cells were washed twice with cold PBS to eliminate unbound antibodies, after which red blood cell lysis was performed using ACK lysing buffer for 5 min at room temperature. The cells were then subjected to centrifugation at 300×g for 5 min, resuspended in PBS, and analyzed using a BD FACSAria™ Fortessa flow cytometer equipped with 488 and 640 nm lasers. Neutrophils were identified based on their forward and side scatter (FSC/SSC) characteristics, and the quantification of double-positive cells for CD11b and Ly6G was conducted using FlowJo_V10 software. Unstained and single-color controls were incorporated to ensure proper compensation and accuracy in gating.
The establishment of the chemokine gradient
A stable chemokine gradient of 10 pM µm⁻¹ was generated utilizing 10 nM fMLP within a microfluidic chamber. The formation of this gradient adheres to Fick’s second law of diffusion under quasi-steady-state conditions. The gradient slope, denoted as ∇C, was determined as follows:
In this context, ∆c signifies the concentration differential between the source channel, which is loaded with fMLP, and the sink channel, which contains a buffer. The variable L represents the length of the migration channel. With a concentration difference of ∆c = 10 nM and a channel length of L = 1000 μm, the calculated gradient slope is determined to be ∇C = 10 pM µm⁻¹.
Motion behavior study of the Trojanbots
A self-constructed cultivation apparatus equipped with a circulating water temperature control system was utilized, which was filled with RPMI 1640 medium supplemented with 10% FBS. All experimental procedures involved the combination of Trojanbots with H2O2 solutions at specified concentrations. PMA was added to the solution at a concentration of 100 nM, and the resultant movement of the Trojanbots, as well as the motion of the released CatNbots, was observed and documented using an inverted optical microscope (Olympus BX31). Furthermore, the TrackMate plugin of ImageJ was employed for tracking and quantifying the motion of the Trojanbots. The coordinate data obtained were subsequently utilized for mathematical statistical analysis and motion assessment. The trajectory data coordinates (x, y) for each frame were extracted through processing with ImageJ software. The MSD can be calculated using the following formula:
The velocity is determined by the formula:
In vitro assessment of BBB crossing
A Transwell cell culture system (3412, Corning) was employed to assess the ability of Trojanbots to traverse a model BBB. This model was established using bEnd.3 cells and GL261 within the Transwell cell culture framework. Specifically, a polycarbonate membrane from Corning was positioned in a 24-well plate, with a porous membrane affixed at the center of each well (as depicted in Fig. S22). bEnd.3 cells (1 × 105 cells per well) were introduced into the upper chamber of the Transwell and subsequently cultured in a medium supplemented with 10% FBS for a duration exceeding 6 days until a monolayer barrier was successfully developed (as illustrated in Fig. S23).
In the control group, which consisted of neutrophils or Trojanbots, a fetal bovine serum (FBS)-free DMEM was introduced into the lower chamber. Conversely, for the fLMP experimental groups (neutrophils + fLMP and Trojanbots + fLMP), an FBS-free DMEM supplemented with 10 nM fMLP was utilized in the lower chamber. The Transwell system was maintained in a 5% CO2 atmosphere at 37 °C. Neutrophils or Trojanbots, at a concentration of 1 × 106 cells per well, were introduced into the upper chamber of each group, using 100 µL of FBS-free DMEM medium. Observations of the bottom of the Transwell for each group were conducted using an optical microscope at intervals of 1–6 h. At each time point, the average cell count was determined from five images, and subsequently, the total number of cells in the lower chamber was calculated. Each experimental condition was replicated three times to ensure the reliability of the results.
Multimodal super-resolution imaging of released CatNbots
CatNbots were fluorescently labeled through incubation with carboxyl-functionalized carbon quantum dots (Amino fluorescent quantum dots, Macklin, emission: 605 nm) at a mass ratio of 1:10 in PBS at pH 7.4 for a duration of 2 h at 25 °C. Subsequently, unbound fluorescent quantum dots were eliminated by performing three cycles of centrifugation (800 × g for 10 min) followed by resuspension in fresh PBS. The fluorescent quantum dots-labeled CatNbots were then encapsulated within unlabeled Trojanbots.
The release process was monitored utilizing an HIS-SIM intelligent ultra-sensitive super-resolution microscope (V3.0, China). Trojanbots containing quantum dots-labeled CatNbots were immobilized on glass-bottom dishes coated with poly-L-lysine. The release was initiated by the addition of 100 nM PMA to the PBS medium at 37 °C. Time-lapse structured illumination microscopy (SIM) imaging was conducted using a ×100 oil-immersion objective (numerical aperture 1.46), with a 640 nm laser employed for the excitation of quantum dots. Images were acquired in 3D-SIM mode, achieving a lateral resolution of ~60 nm, at intervals of 5 s.
Chemotactic evaluation of the CatNbots
The dimensional specifications of the Y-shaped channel are as follows: the main body of the channel measures 1 cm in length and 0.4 cm in width, while the two branch channels of the Y-shaped glass substrate each measure 1 cm in length and 0.3 cm in width. A solution of agarose (5 mg) was prepared by dissolving it in 500 µL of PBS and subsequently heated to 90 °C. Upon cooling to ambient temperature, 200 µL of GL261-Luc cell lysate (5 × 106 cells/mL) was incorporated and subsequently transferred to a gelatin maintained at 4 °C. For comparative purposes, an equivalent concentration of bEnd.3 cell lysate was introduced under the same conditions to replicate an environment devoid of a chemotactic gradient. The resultant agarose gel, which contained the various cell lysates (~8 mm3), was positioned in both the upper and lower reservoirs of the Y-channel. An additional 300 µL of PBS was introduced into the Y-channel to sustain the water level and ensure complete coverage of the channel. The mixture solidified at room temperature, yielding a gel that encapsulated the cell lysate. Over time, hydrogen peroxide (H2O2) diffused through the Y-channel, establishing a chemotactic gradient. The concentration gradient of H2O2, extending from the center of the cancer cell lysate to its periphery, was quantified at five distinct locations utilizing a ROS Green TM H2O2 Probe (Baiaolaibo, KM0103). Concurrently, the fluorescence variations of the nanomotors within the Y-channel were monitored for 60 min. Subsequently, 10 µL of CatNbots solution was carefully introduced to the left terminus of the Y-channel, and fluorescence images of the upper and lower reservoirs were captured at predetermined time intervals (0, 15, 30, and 60 min) using an inverted fluorescence microscope. The trajectories of n = 50 particles were tracked and normalized to a common origin, with analyses conducted on chemotactic speed and index. The fluorescence intensity pseudo-color images were processed utilizing ImageJ (Version 2.0.0).
Cell culture
The GL261-Luc glioma cell line (catalog number: CTCC-DZX-0027) and the mouse-derived brain endothelial cells (bEnd.3, catalog number: CTCC-400-0303) were acquired from Hangzhou Meisen Life Science & Technology Co., Ltd. The cells were maintained in DMEM enriched with 10% fetal bovine serum, 100 U mL−1 of penicillin G sodium, and 100 µg mL−1 of streptomycin sulfate, under a humidified environment containing 5% CO2 at a temperature of 37 °C. All cell lines underwent testing and were confirmed to be free of mycoplasma contamination.
Cell invasion and ablation assays
To evaluate the invasion and ablation of three-dimensional tumor spheres, GL261 cells (5 × 103 per well) were cultured in low-adherence 96-well plates and incubated for a duration of 72 h, as illustrated in Fig. S15. Following the formation of tumor spheres, CatNbots were introduced into the culture. During this period, images were systematically captured at designated intervals to facilitate the analysis of both invasion (refer to Fig. 5b) and the ablation potential of the engineered tumor cells. High-resolution images of these co-cultures were acquired using confocal microscopy (Leica TCS SP5 II, Germany) to evaluate their invasion and ablation capabilities.
Cellular uptake performance assessment
GL261-Luc cells (1 × 105) were cultured in 12-well plates, followed by the addition of various samples and a subsequent incubation period of 12 h. After this incubation, the cells were fixed and stained with 200 µL of Dil and 200 µL of DAPI. The staining process lasted for 10 min allowing for the visualization of the cell membrane and nucleus, respectively. The uptake of the materials by the cells was then assessed using confocal laser scanning microscopy (CLSM).
Animal model
In the conducted experiments, C57BL/6J male mice, aged 8 weeks and weighing between 18 and 20 g, were procured from Liaoning Changsheng Biotechnology Co. Ltd., located in Liaoning, China. To create a glioblastoma (GBM) model, the mice were anesthetized through inhalation of a mixture of 1–2% isoflurane and oxygen. Subsequently, GL261-Luc cells, suspended in 5 µL of phosphate-buffered saline (PBS) at a concentration of 1 × 106 cells per mouse, were meticulously injected into the right hemisphere of the brain at a depth of 1.5 m, −0.5, and 3 mm, utilizing a stereotactic fixation device from RWD Life Science Co. in Shenzhen. Control mice received PBS injections under identical surgical conditions. The progression of the brain tumor was assessed by administering 100 µL of D-luciferin potassium salt (150 mg kg−1, Meisen, China) via intraperitoneal injection, followed by the measurement of luminescence intensity from the GL261-Luc cells in the brain using an in vivo imaging system (IVIS Lumina III, PerkinElmer, USA) as an indicator of tumor size. Following the guidelines set forth by the ethics committee, the animal shows obvious anxiety, restlessness, or the weight of a tumor exceeds 10% of the animal’s body weight (for example, a subcutaneous tumor in a 25 g mouse should not exceed 20 mm in maximum diameter), should result in euthanasia of the experimental animal. At the same time, it should be ensured that the animal’s body weight does not lose 20% or more from the pre-experimental weight and no obvious skin lesions of the body or signs of purulent infection. Tumor weight was measured at the end of each animal’s intervention cycle. Before euthanization, the body weight of each animal was calculated. After euthanasia, the glioma tumors were carefully excised and weighed. The specific method for calculating tumor weight is as follows:
-
1.
Tumor volume calculation: The tumor volume is first calculated by measuring the maximum length (d₁), width (d₂), and height (d₃) of the tumor. The volume is then estimated using the ellipsoid formula:
$$V=\frac{4}{3}{{{\rm{\pi }}}}\times \frac{{d}_{1}}{2}\times \frac{{d}_{2}}{2}\times \frac{{d}_{3}}{2}$$ -
2.
Tumor weight estimation: The tumor weight is then estimated by multiplying the volume by the tumor’s density (typically 1.05 g/cm³):
$$W=V\times {{{\rm{Density}}}}$$ -
3.
Comparison with animal body weight: Finally, the estimated tumor weight is compared with 10% of the animal’s body weight to ensure that the tumor weight remains within the acceptable limit.
All animal procedures complied with institutional ethical guidelines. Throughout the study, tumor growth was strictly monitored, and no tumors exceeded the ethical limits of 20 mm in diameter or 10% of the animal’s body weight.
In vivo biodistribution of Trojanbots
For in vivo fluorescence imaging, nine days following tumor implantation, glioma-bearing mice (C57BL/6J, male, aged 9 weeks, n = 3 per group) were randomly assigned to groups and administered DiR-labeled materials, including EMV@GeNPs, CatNbots, NE@EDGNs, and Trojanbots (100 µg kg−1 DiR), via tail vein injection. Subsequently, the mice were anesthetized with a mixture of 1–2% isoflurane and oxygen, and imaging was conducted at 1, 2, 4, 8, and 24 h post-injection utilizing a near-infrared fluorescence imaging system (IVIS Lumina III, PerkinElmer, USA), which was equipped with specific fluorescent filter sets (Excitation: 750 nm, Emission: 780 nm). For ex vivo fluorescence imaging, the brains and other organs (heart, liver, spleen, lungs, kidneys) of glioma-bearing mice (C57BL/6J, male, aged 9 weeks, n = 3 per group) were harvested, washed, and imaged at 8 h post-injection. Image acquisition and analysis were performed using the IVIS Lumina III and Living Image System 4.5.5 software. Additionally, brain tissues were preserved in cryo-embedding media (OCT) at −80 °C and sectioned into 20 μm slices. Following DAPI staining, the sections were examined using a confocal laser scanning microscope (Leica, Germany), and the fluorescence intensity of the images was quantified using Image J software (Version 2017).
In vivo antitumor effect
Mice with tumors of comparable size (C57BL/6J, male, aged 9 weeks, n = 5 per group) were randomly assigned to five groups: PBS, EMV@GeNPs, CatNbots, NE@EDGN, and Trojanbots (50 µg mg−1 DOX). Subsequently, the glioma-bearing mice received intravenous injections of each formulation on days 9, 12, and 15 post-tumor implantation. To assess tumor progression, bioluminescence imaging of the mouse brain tumors was conducted at various time points following the injections, utilizing the IVIS Imaging System. Additionally, the body weights of the mice were monitored throughout the treatment period, and the survival rates for each group were documented.
Detection and analysis of T cell infiltration in murine brain tumor
On day 16 following treatment, each cohort of mice (C57BL/6J, male, aged 10 weeks, n = 3 per group) was euthanized. Tumor tissues were harvested, sectioned into small fragments, and subsequently homogenized using a glass grinder in cold phosphate-buffered saline (PBS) to create a cell suspension. This suspension was then subjected to centrifugation at 1000×g for 5 min at a temperature of 4 °C, after which the pellet was collected. The cells were resuspended in PBS and filtered through a 70 µm nylon cell filter to ensure uniformity. A staining buffer was added to facilitate the thorough mixing of the cells. To perform the analysis of CD3+CD4+ and CD3+CD8+ T-cells, the following antibodies were utilized: anti-CD3-PerCP-Cy5.5 (diluted 1:20), anti-CD45-FITC (diluted 1:100), anti-CD4-APC-Cy7 (diluted 1:100), and anti-CD8-PE (diluted 1:50). Flow cytometric data were acquired using a BD FACSAria™ Fortessa flow cytometer and subsequently analyzed with FlowJo software (Version 10). The gating strategy employed for T-cell analysis is illustrated in Supplementary Fig. 35.
Histological analysis
Twenty-four hours post-final treatment, the mice (C57BL/6J, male, aged 10 weeks, n = 3 per group) were euthanized, and their brains were meticulously extracted. The extracted brains were then fixed in 4% paraformaldehyde, embedded in paraffin for the preparation of immunohistochemical sections, and then sectioned for further measurements. H&E-stained tissue sections were observed using a Slide Scanner (SCN400, Leica), and TUNEL staining was performed to detect apoptosis in tissue sections. In addition, the brain slices were further stained with Ki67 (dilution of 1:200, catalog number: ab15580, Abcam) to evaluate tumor cell proliferation and then incubated with coraLite488-conjugated Goat Anti-Rabbit IgG (H + L) (dilution of 1:100, Cat: SA00013-2, Proteintech). To further explore the Trojanbot-mediated effect on TME, sections were stained with antibodies specific for CD31 (dilution of 1:100, catalog number: ab222783, Abcam), HIF-1α (dilution of 1:100, catalog number: ab308433, Abcam) and N-cadherin (dilution of 1:100, catalog number: ab76011, Abcam), respectively, followed by incubation of labeled secondary antibodies with Multi-rAb CoraLite® Plus 594-goat anti-rabbit recombinant secondary antibody (H + L) (dilution of 1:200, Cat: RGAR004, Proteintech). As a control, cell nuclei were stained with DAPI for visualization, and all fluorescence images were captured using a fluorescence microscope.
Biosafety evaluation in vivo
The healthy C57BL/6J mice without tumors (C57BL/6J, male, aged 8 weeks, 3 animals per group) were injected with various materials (EMV@GeNPs, CatNbots, NE@EDGNs, and Trojanbots) to assess in vivo biological safety. At the end of injection, the cortical tissues of the major organs (brain, heart, liver, spleen, lung, and kidney) were harvested separately, fixed in paraformaldehyde in PBS (4%), and stained with H&E. The histological sections were observed using a Slide Scanner (SCN400, Leica).
Statistical information
Microsoft Excel (2020), GraphPad Prism (Version 8), and Origin 2018 software are used for numerical processing and drawing. All data are presented as the mean ± standard deviation (SD). The samples/animals were allocated to experimental groups and processed randomly. General statistical data were analyzed by GraphPad Prism 8 software. These experiments have been conducted three times or with enough mice to determine statistically significant differences in mean values. One-way analysis of variance (ANOVA) and unpaired two-tailed t-tests were used to calculate P values for different experimental purposes. Unless otherwise specified, all experiments were repeated at least three times. In addition, two-sided tests were used for all tests. Significant differences are indicated as P < 0.05, P < 0.01, P < 0.001, and P < 0.0001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data generated or analyzed during this study are available within the Article, its Supplementary Information file, and the Source data file. Source data are provided with this paper. The full image dataset is available from the corresponding author upon request. Source data are provided with this paper.
References
Aldape, K. et al. Challenges to curing primary brain tumours. Nat. Rev. Clin. Oncol. 16, 509–520 (2019).
Di Mascolo, D. et al. Conformable hierarchically engineered polymeric micromeshes enabling combinatorial therapies in brain tumours. Nat. Nanotechnol. 16, 820–829 (2021).
Weller, M. et al. Glioma. Nat. Rev. Dis. Prim. 10, 33 (2024).
Lu, G. H. et al. Engineered biomimetic nanoparticles achieve targeted delivery and efficient metabolism-based synergistic therapy against glioblastoma. Nat. Commun. 13, 4214 (2022).
Gao, J. J. et al. Precision drug delivery to the central nervous system using engineered nanoparticles. Nat. Rev. Mater. 9, 567–588 (2024).
Tang, W. et al. Emerging blood–brain-barrier-crossing nanotechnology for brain cancer theranostics. Chem. Soc. Rev. 48, 2967–3014 (2019).
Arvanitis, C. D., Ferraro, G. B. & Jain, R. K. The blood–brain barrier and blood–tumour barrier in brain tumours and metastases. Nat. Rev. Cancer 20, 26–41 (2020).
Mao, M., Wu, Y. J. & He, Q. Recent advances in targeted drug delivery for the treatment of glioblastoma. Nanoscale 16, 8689–8707 (2024).
Gao, C. Y. et al. Biomedical micro-/nanomotors: from overcoming biological barriers to in vivo imaging. Adv. Mater. 33, 2000512 (2021).
Wang, X. J. et al. Bioinspired adaptive microdrugs enhance the chemotherapy of malignant glioma: beyond their nanodrugs. Adv. Mater. 36, 2405165 (2024).
Chen, S. Q. et al. Enhanced tumour penetration and prolonged circulation in the blood of polyzwitterion-drug conjugates with cell-membrane affinity. Nat. Biomed. Eng. 5, 1019–1037 (2021).
Li, S. L., Jiang, P., Jiang, F. L. & Liu, Y. Recent advances in nanomaterial-based nanoplatforms for chemodynamic cancer therapy. Adv. Funct. Mater. 31, 2100243 (2021).
Meng, W. R., Hao, Y. Y., He, C. S., Li, L. & Zhu, G. Q. Exosome-orchestrated hypoxic tumor microenvironment. Mol. Cancer 18, 57 (2019).
Huang, J. C. et al. Extracellular matrix and its therapeutic potential for cancer treatment. Signal Transduct. Target. Ther. 6, 153 (2021).
Kim, J. et al. Advanced materials for micro/nanorobotics. Chem. Soc. Rev. 53, 9190–9253 (2024).
Zhou, H. J., Mayorga-Martinez, C. C., Pané, S., Zhang, L. & Pumera, M. Magnetically driven micro and nanorobots. Chem. Rev. 121, 4999–5041 (2021).
Han, H. et al. Imaging-guided bioresorbable acoustic hydrogel microrobots. Sci. Robot. 9, 3593 (2024).
Wu, Z. G., Chen, Y., Mukasa, D., Pak, O. S. & Gao, W. Medical micro/nanorobots in complex media. Chem. Soc. Rev. 49, 8088–8112 (2020).
Lu, H. J. et al. A bioinspired multilegged soft millirobot that functions in both dry and wet conditions. Nat. Commun. 9, 3944 (2018).
Chen, J. Y. et al. Enhancing aortic valve drug delivery with PAR2-targeting magnetic nano-cargoes for calcification alleviation. Nat. Commun. 15, 557 (2024).
Wang, S. H. et al. Hydrogen-powered microswimmers for precise and active hydrogen therapy towards acute ischemic stroke. Adv. Funct. Mater. 31, 202009475 (2021).
Yang, Z. L. et al. Ultrasmall enzyme-powered janus nanomotor working in blood circulation system. ACS Nano 17, 6023–6035 (2023).
Simo, C. et al. Urease-powered nanobots for radionuclide bladder cancer therapy. Nat. Nanotechnol. 19, 554–564 (2024).
Serra-Casablancas, M. et al. Catalase-powered nanobots for overcoming the mucus barrier. ACS Nano 18, 16701–16714 (2024).
Gu, Z. Q. et al. Autonomous nanorobots with powerful thrust under dry solid-contact conditions by photothermal shock. Nat. Commun. 14, 7663 (2023).
Villa, K. & Pumera, M. Fuel-free light-driven micro/nanomachines: artificial active matter mimicking nature. Chem. Soc. Rev. 48, 4966–4978 (2019).
Ma, Z. C. et al. Femtosecond laser programmed artificial musculoskeletal systems. Nat. Commun. 11, 4536 (2020).
Liang, Q. L. et al. The softness of tumour-cell-derived microparticles regulates their drug-delivery efficiency. Nat. Biomed. Eng. 3, 729–740 (2019).
Gao, C. et al. Treatment of atherosclerosis by macrophage-biomimetic nanoparticles via targeted pharmacotherapy and sequestration of proinflammatory cytokines. Nat. Commun. 11, 2622 (2020).
Zou, Y. et al. Cancer cell-mitochondria hybrid membrane coated Gboxin loaded nanomedicines for glioblastoma treatment. Nat. Commun. 14, 4557 (2023).
Gao, C. Y., Zhou, C., Lin, Z. H., Yang, M. C. & He, Q. Surface wettability-directed propulsion of glucose-powered nanoflask motors. ACS Nano 13, 12758–12766 (2019).
Fan, R. R. et al. Engineering MMP-2 activated nanoparticles carrying B7-H3 bispecific antibodies for ferroptosis-enhanced glioblastoma immunotherapy. ACS Nano 17, 9126–9139 (2023).
Li, Z. X. et al. Biohybrid microrobots regulate colonic cytokines and the epithelium barrier in inflammatory bowel disease. Sci. Robot. 9, eaat2007 (2024).
Soto, F., Wang, J., Ahmed, R. & Demirci, U. Medical micro/nanorobots in precision medicine. Adv. Sci. 7, 2002203 (2020).
Chen, B. et al. Cell-based micro/nano-robots for biomedical applications: a review. Small 20, 2304607 (2024).
Ye, J. M. et al. Biomimetic dual-driven heterojunction nanomotors for targeted catalytic immunotherapy of glioblastoma. Adv. Funct. Mater 35, 2416265 (2024).
Sun, J. W., Mathesh, M., Li, W. & Wilson, D. A. Enzyme-powered nanomotors with controlled size for biomedical applications. ACS Nano 13, 10191–10200 (2019).
Mathesh, M., Sun, J. W., van der Sandt, F. & Wilson, D. A. Supramolecular nanomotors with “pH taxis” for active drug delivery in the tumor microenvironment. Nanoscale 12, 22495–22501 (2020).
Zhang, F. Y. et al. Nanoparticle-modified microrobots for in vivo antibiotic delivery to treat acute bacterial pneumonia. Nat. Mater. 21, 1324–1332 (2022).
de Avila, B. E. F. et al. Hybrid biomembrane-functionalized nanorobots for concurrent removal of pathogenic bacteria and toxins. Sci. Robot. 3, eaat0485 (2018).
Yong, J. L., Mellick, A. S., Whitelock, J., Wang, J. S. & Liang, K. A Biomolecular toolbox for precision nanomotors. Adv. Mater. 35, 2205746 (2023).
Xue, J. W. et al. Neutrophil-mediated anticancer drug delivery for suppression of postoperative malignant glioma recurrence. Nat. Nanotechnol. 12, 692–700 (2017).
Wu, M. Y. et al. MR imaging tracking of inflammation-activatable engineered neutrophils for targeted therapy of surgically treated glioma. Nat. Commun. 9, 4777 (2018).
Chang, Y. et al. CAR-neutrophil mediated delivery of tumor-microenvironment responsive nanodrugs for glioblastoma chemo-immunotherapy. Nat. Commun. 14, 2266 (2023).
Zhang, H. Y. et al. Dual-responsive biohybrid neutrobots for active target delivery. Sci. Robot. 6, eaaz9519 (2021).
Li, M. et al. Chemotaxis-driven delivery of nano-pathogenoids for complete eradication of tumors post-phototherapy. Nat. Commun. 11, 1126 (2020).
Wu, J. F. et al. Dual-responsive nanorobot-based marsupial robotic system for intracranial cross-scale targeting drug delivery. Adv. Mater. 36, 2306876 (2024).
Chang, X. W. et al. Enhanced manipulation of tumor microenvironments by nanomotor for synergistic therapy of malignant tumor. Biomaterials 290, 121853 (2022).
de Avila, B. E. F. et al. Nanomotor-enabled pH-responsive intracellular delivery of caspase-3: toward rapid cell apoptosis. ACS Nano 11, 5367–5374 (2017).
Chen, H. et al. A nitric-oxide driven chemotactic nanomotor for enhanced immunotherapy of glioblastoma. Nat. Commun. 14, 941 (2023).
Gao, C. Y. et al. Stem cell membrane-coated nanogels for highly efficient in vivo tumor targeted drug delivery. Small 12, 4056–4062 (2016).
Xuan, M. J., Shao, J. X., Lin, X. K., Dai, L. R. & He, Q. Self-propelled janus mesoporous silica nanomotors with sub-100 nm diameters for drug encapsulation and delivery. Chemphyschem 15, 2255–2260 (2014).
Shao, J. X. et al. Chemotaxis-guided hybrid neutrophil micromotors for targeted drug transport. Angew. Chem.-Int. Ed. 56, 12935–12939 (2017).
Michael, M. & Vermeren, S. A neutrophil-centric view of chemotaxis. Essays Biochem. 63, 607–618 (2019).
Kolaczkowska, E. & Kubes, P. Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 13, 159–175 (2013).
de Oliveira, S., Rosowski, E. E. & Huttenlocher, A. Neutrophil migration in infection and wound repair: going forward in reverse. Nat. Rev. Immunol. 16, 378–391 (2016).
Howse, J. R. et al. Self-motile colloidal particles: from directed propulsion to random walk. Phys. Rev. Lett. 99, 048102 (2007).
Li, J. W. et al. Water-soluble and degradable gelatin/polyaniline assemblies with a high photothermal conversion efficiency for ph-switchable precise photothermal therapy. ACS Appl. Mater. Interfaces 14, 52670–52683 (2022).
Helms, H. C. et al. In vitro models of the blood-brain barrier: an overview of commonly used brain endothelial cell culture models and guidelines for their use. J. Cereb. Blood Flow. Metab. 36, 862–890 (2016).
Hyun, Y. M., Choe, Y. H., Park, S. A. & Kim, M. LFA-1 (CD11a/CD18) and Mac-1 (CD11b/CD18) distinctly regulate neutrophil extravasation through hotspots I and II. Exp. Mol. Med. 51, 1–13 (2019).
Chauhan, V. P. & Jain, R. K. Strategies for advancing cancer nanomedicine. Nat. Mater. 12, 958–962 (2013).
Cho, Y. & Doh, J. The extracellular matrix in solid tumor immunotherapy. Trends Immunol. 45, 705–714 (2024).
Zhou, C. et al. Torque-driven orientation motion of chemotactic colloidal motors. Angew. Chem.-Int. Ed. 61, e202116013 (2022).
Yan, Y. L. et al. Suppression of ITPKB degradation by Trim25 confers TMZ resistance in glioblastoma through ROS homeostasis. Signal Transduct. Target. Ther. 9, 58 (2024).
Zaher, A., Petronek, M. S., Allen, B. G. & Mapuskar, K. A. Balanced duality: H2O2-based therapy in cancer and its protective effects on non-malignant tissues. Int. J. Mol. Sci. 25, 8885 (2024).
Wang, H. et al. Synergistic chemoimmunotherapy augmentation via sequential nanocomposite hydrogel-mediated reprogramming of cancer-associated fibroblasts in osteosarcoma. Adv. Mater. 36, 2309591 (2024).
Robins, E. et al. Conversion of effector CD4+ T cells to a CD8+ MHC II-recognizing lineage. Cell. Mol. Immunol. 18, 150–161 (2021).
Hameedat, F. et al. Engineering nanomaterials for glioblastoma nanovaccination. Nat. Rev. Mater. 9, 628–642 (2024).
Wilhelm, S. et al. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 1, 16014 (2016).
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (Nos. 22193033, and U23A20342), the Key R&D Program of Heilongjiang Province, China (No. 2022ZX02C23), China Postdoctoral Science Foundation (No. 2024M764205), the Postdoctoral Fellowship Program of CPSF (GZC20233468), and the start-up grant of Wenzhou Institute, University of Chinese Academy of Sciences.
Author information
Authors and Affiliations
Contributions
Y.Y.G. and M.M. conceived the project, designed the experiments, and prepared the manuscript. Y.L. and M.J.X. contributed to movie processing, and Y.J.W. assisted in manuscript preparation. Q.H. commented on and reviewed the manuscript. All authors participated in discussions throughout the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Motilal Mathesh and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gao, Y., Mao, M., Li, Y. et al. A self-directed Trojanbot-enzymatic nanobot in neutrobot for active target therapy of glioblastoma. Nat Commun 16, 5263 (2025). https://doi.org/10.1038/s41467-025-60422-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-60422-z