Abstract
Pancreatic cancer is known for its lethal condition, with most cases being diagnosed at advanced stage. Recently, liquid biopsy has emerged as a promising tool in cancer detection. Here we develop both an early detection model and a prognostic model for pancreatic cancer using cell-free DNA (cfDNA) end motif, fragmentation, nucleosome footprint (NF), and copy number alteration (CNA) features from plasma cfDNA. A total of 975 individuals were enrolled in our study. We developed an integrated model that demonstrated superior performance in distinguishing patients with early-stage pancreatic cancer from non-cancer controls. Moreover, we find that cfDNA features are associated with prognostic outcomes among pancreatic cancer patients. In this study, a cfDNA-based liquid biopsy signature is established for the early detection and prognostic prediction of pancreatic cancer. CfDNA may become a valuable tool for enhancing early diagnosis and prognosis assessment in this challenging disease.
Similar content being viewed by others
Introduction
Pancreatic cancer is one of the most aggressive cancers in China with only a 10% 5-year survival rate1,2. With its high malignancy, pancreatic cancer remains a major cause of cancer-related mortality3,4. Pancreatic ductal adenocarcinoma (PDAC) accounts for 90% of all pancreatic cancer cases2,5. Smoking, nonhereditary or chronic pancreatitis, chronic diabetes mellitus, obesity, nontype O blood group, and age could be risk factors for PDAC3. Besides, germline mutations in genes such as BRCA2, BRCA1, CDKN2A, ATM, STK11, PRSS1, MLH1, and PALB2 are associated with pancreatic adenocarcinoma (PAAD)3. To date, there’s no reliable screening test for pancreatic cancer, while most patients with pancreatic cancer do not have evident symptoms until the advanced stage6. Surgical resection remains the main therapeutic method for the treatment of pancreatic cancer, but only 10–20% of patients are eligible for surgical resection6. Carbohydrate antigen 19-9 (CA19-9), and carcinoembryonic antigen (CEA), are considered biomarkers of pancreatic cancer7. CA19-9 also plays an important role in guidance of surgery decisions, the use of adjuvant therapy, and detection of post-operative tumor recurrence, but its effect is limited because 10% of patients do not secrete the antigen8. As for biomarker, CA19-9 is lack of sensitivity and specificity, and is elevated in pancreatic benign diseases and other gastrointestinal malignancies9. CEA is also neither sensitive nor specific, and it is elevated in alcoholic cirrhosis, hepatitis, and biliary disease10,11.
Cell-free DNA (cfDNA) is fragmented (approximately 150–350 bases), and typically double-stranded12. Most of the cfDNA is released from hematopoietic cells, and there is also a portion of cfDNA released from cancer cells13,14. It was found that cfDNA was more abundant in patients with gastrointestinal cancer than in healthy controls, and the level of cfDNA in the malignant group was higher than in the benign group15. CfDNA features are closely related to the early genesis of cancer16, therefore, multiple studies indicated that cfDNA can be utilized for early detection of cancer, including liver cancer17,18,19,20, lung cancer21,22, breast cancer23, urothelial bladder carcinoma24, colorectal cancer25, Hodgkin’s lymphoma26, and pancreatic cancer27,28,29.
CfDNA levels are elevated in pancreatic cancer16, providing a potential diagnostic biomarker for diagnosing pancreatic cancer. CfDNA offers several advantages: its detection technology is well established, and its relative stability enables consistent testing30,31,32. Several studies have investigated various cfDNA-based features, such as fragmentomics, mutations, and methylation, to develop diagnostic models for pancreatic cancer. For instance, one study developed a cancer diagnostic model using cfDNA fragmentation profiles, achieving sensitivities ranging from 57% to 99%, with a specificity of 98%33. Additionally, copy number alterations (CNAs) detected via cfDNA have been applied to identify various cancers, including pancreatic cancer34. In methylation-based approaches, leveraging cfDNA 5-hydroxymethylcytosine (5hmC) features has shown strong performance in identifying early-stage pancreatic cancer29. Combining circulating tumor DNA (ctDNA) with protein biomarkers has yielded high diagnostic accuracy for detecting PDAC35. These findings highlight the potential of cfDNA-based approaches as valuable tools for the early detection and diagnosis of pancreatic cancer. However, relying solely on a single biomarker for diagnosis presents inherent limitations. Integrating multiple cfDNA-based features has the potential to significantly enhance diagnostic accuracy and mitigate these constraints.
In this work, we performed a multi-center, large-scale cohort study and employed a state-of-the-art next generation sequencing (NGS) technology to acquire plasma cfDNA end motif36, nucleosome footprint (NF)13, fragmentation33,37,38 profiles, and copy number alteration of cfDNA from all enrolled cases. Predictive features were filtered out using the least absolute shrinkage and selection operator (LASSO). Based on these features, we developed a weighted diagnostic model (PCM score) and a prognostic evaluation model (PCP score).
Results
Characteristic signatures of cfDNA
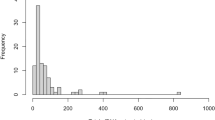
CfDNA fragment size was measured in plasma samples from patients with pancreatic cancer, pancreatic benign tumor (PBT), chronic pancreatitis (CP), and healthy controls (HC). The fragmentation profiles showed consistency among non-cancer cases (PBT, CP, and HC), but exhibited significant variability in patients with pancreatic cancer (Fig. 1a, Supplementary Fig. 1a). Notably, cfDNA fragments in pancreatic cancer patients were shorter compared to those in PBT, CP, and healthy controls, and the median cfDNA fragment size of pancreatic cancer were 175 bp (range 154 bp to 197 bp) while in CP + PBT and healthy controls were 182 bp (range 165 bp to 198 bp) and 186 bp (range 160 bp to 203 bp) (Fig. 1a, Supplementary Fig. 1a). Among patients in pancreatic cancer, fragment size was not influenced by age, gender, or level of CA125, CA19-9, and CEA but showed significant associations with AJCC stage (Supplementary Fig. 1b). In the PBT group, cfDNA fragment size remained unaffected by age, gender, level of CA125, CA19-9, and CEA (Supplementary Fig. 1c).
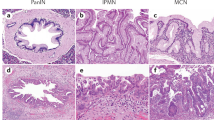
a Size distributions of cfDNA fragments in participants of healthy controls, CP + PBT and pancreatic cancer (The Z-score indicates the ratio of short fragments to long fragments); b KEGG pathway analysis of NF difference between healthy controls and pancreatic cancer. Hypergeometric test was used to detect whether a specific gene set is significantly enriched; c Plasma cfDNA end motif features distribution in healthy controls, CP + PBT and pancreatic cancer. d Size distributions of cfDNA fragments in different subtypes of pancreatic cancer, PBT, CP, and healthy controls. Box plots indicate median (middle line), 25%, 75% percentile (box) and minimum and maximum (whiskers) as well as outliers (single points). e CNA features in participants of healthy controls, CP + PBT, and pancreatic cancer. Source data are provided as a Source Data file. PDAC pancreatic ductal adenocarcinoma, ASCP adenosquamous carcinoma of the pancreas, IPMN intraductal papillary mucinous neoplasm, PNET pancreatic neuroendocrine tumor, SCN serous cystic neoplasm.
KEGG pathway analysis revealed that differentially expressed NF gene were enriched in several cancer-related pathways, including the hedgehog signaling pathway, VEGF signaling pathway, MAPK signaling pathway, TGF-β signaling pathway, and Wnt signaling pathway (Fig. 1b). Unsupervised hierarchical clustering demonstrated a clear distinction between healthy controls, CP, PBT, and pancreatic cancer (Fig. 1c). Fragment lengths were observed to decrease progressively with increasing malignancy (Fig. 1d). Additionally, CNA analysis showed that pancreatic cancer patients exhibited a higher number of CNAs compared to PBT and CP patients, with healthy individuals displaying the lowest CNA numbers (Fig. 1e).
Patients and cohorts
All cases were divided into 4 cohorts, including Training cohort (432 cases), Testing cohort (267 cases), External Validation cohort 1 (129 cases), and External Validation cohort 2 (139 cases) (Fig. 2). Training cohort was designed for the construction of PCM and PCP scoring System. Among 422 patients with pancreatic cancer or PBT, five subtypes were included: PDAC, ade nosquamouscarcinoma of the pancreas (ASCP), intraductal papillary mucinous neoplasm (IPMN), pancreatic neuroendocrine tumor (PNET), serous cystic neoplasm (SCN). Pancreatic cancer cases comprised PDAC and ASCP, while PBT cases included IPMN, PNET, and SCN. We used computer-generated random numbers to assign patients from Changhai Hospital to Training cohort (n = 272) and Testing cohort (n = 98). External Validation cohort 1 consists of patients from the Affiliated Hospital of Qingdao University, and External Validation cohort 2 consists of patients from The Second Affiliated Hospital of Shandong University and The Second Hospital, Cheeloo College of Medicine, Shandong University. The healthy controls were randomly distributed into the Training cohort, the Testing cohort, and the Validation cohorts. The levels of CA19-9, CA125, and CEA across different patient groups are presented in Supplementary Fig. 2, while the TNM stage distribution for all patients is detailed in Supplementary Fig. 3.
Establishment of PCM score
The workflow for constructing the diagnosis model was shown in Fig. 2. All participants were divided into four cohorts: Training cohort, Testing cohort, and two External Validation cohorts. In Training cohort, cfDNA was analyzed using low-pass whole-genome sequencing (WGS), and the PCM score were constructed with CNA, fragment signatures, motif signatures, and NF signatures. We constructed 4 models to identify malignant pancreatic cancer from non-cancer patients (PBT, CP, and healthy individuals). In the Training cohort, the combined model (PCM score) showed an AUC of 0. 975 (95% CI: 0.961–0.988), compared with NF (AUC: 0.973, 95% CI: 0.959–0.986), motif (AUC: 0.858, 95% CI: 0.823–0.894), fragment (AUC: 0.968, 95% CI: 0.952–0.983) (Fig. 3a). In the Testing cohort, the combined model showed an AUC of 0.979 (95% CI: 0.961–0.998) (Fig. 3b). In the External Validation cohort 1 and the External Validation cohort 2, our combined model showed AUC of 0.992 (95% CI: 0.983–1) and 0.986 (95% CI: 0.97–1) (Fig. 3c, d). The combined model outperformed the individual feature models across all four cohorts. The detailed information of performance of CNA in distinguishing different types of groups were shown in Supplementary Table 1.
Our combined model (PCM score) could distinguish pancreatic cancer from healthy controls (HC) with an AUC of 0.990 (95% CI: 0.983–0.997) in the Combined cohort (Testing cohort plus two External Validation cohorts) (Fig. 4a), and resectable stage (stage I/II) from healthy controls, with an AUC of 0.994 (95% CI: 0.989–0.999) in the Combined cohort (Fig. 4b). Fig. 4c shows that the PCM score was able to distinguish pancreatic cancer from PBT, with an AUC of 0.886 (95% CI: 0.835–0.936), compared with CA19-9 with an AUC of 0.819 (95%CI: 0.755–0.883). The model distinguished CA19-9 negative pancreatic cancer from HC with an AUC of 0.990 (95%CI: 0.977–1) (Fig. 4d).
a ROC curve analysis for the NF, motif, fragment or combined model (PCM score) in distinguishing pancreatic cancer and HC. b ROC curve analysis for the NF, motif, fragment or combined model (PCM score) in distinguishing early stage (stage I,II) of pancreatic cancer and HC. c ROC curve analysis for the NF, motif, fragment, combined model (PCM score), and CA19-9 in distinguishing pancreatic cancer and PBT. d ROC curve analysis for the NF, motif, fragment, and combined model (PCM score) in distinguishing CA19-9 negative pancreatic cancer and HC. Source data are provided as a Source Data file. HC healthy control.
The performance of the PCM score for staged pancreatic cancer versus non-cancer (including PBT, CP, and HC) and pancreatic cancer versus healthy is summarized in Table 1. Additionally, Table 2 compares the performance of the PCM score and CA19-9 in differentiating staged pancreatic cancer from benign pancreatic diseases (PBT and CP). As shown in Table 2, the PCM score outperformed CA19-9 across both the Testing cohort and the two External Validation cohorts. Notably, the PCM scoring system demonstrated a superior ability to accurately differentiate early-stage pancreatic cancer compared to CA19-9, highlighting its potential as a more reliable diagnostic tool.
The PCM score demonstrated high sensitivity in detecting pancreatic cancer, with positive detection rates of 92% for PDAC patients and 100% for ASCP patients. In contrast, the positive detection rates for PBT subtypes and HC were below 40% (Supplementary Fig. 4). Additionally, the PCM score was significantly higher in pancreatic cancer cases compared to non-cancer groups (Supplementary Fig. 5). Plasma samples from patients with other cancer types revealed that the Logistic score was notably elevated in pancreatic cancer compared to both other cancers and HC (Supplementary Fig. 6). When combining CA19-9 with the PCM score, the diagnostic performance improved further. The PCM score and CA19-9 combination distinguished pancreatic cancer from PBT and CP with AUCs of 0.936, 0.968, and 0.864 in the Training, Testing, and External Validation cohorts, respectively, compared to AUCs of 0.888, 0.942, and 0.841 for the PCM score alone (Supplementary Fig. 7a). This combination also exhibited superior performance in identifying early-stage (stage I and II) pancreatic cancer from PBT and CP (Supplementary Fig. 7b) and in distinguishing pancreatic cancer from CP (Supplementary Fig. 7c).
Establishment of PCP score
We investigated the relationship between cfDNA features and prognosis in pancreatic cancer using both the Training cohort and the Combined cohort (which included the Testing cohort and two External Validation cohorts). Utilizing end motif, fragment, and nucleosome footprint features, we developed a prognostic model and introduced the Pancreatic Cancer Prognostic (PCP) score. Kaplan–Meier survival analyses were conducted for both cohorts based on the PCP score. The results demonstrated a significant difference in median overall survival between the high and low PCP score groups in both the Training cohort (p < 0.0001) and the Combined cohort (p < 0.0001) by the log-rank test (Fig. 5a, b). Similarly, recurrence-free survival was significantly longer in the low PCP score group compared to the high PCP score group in both the Training cohort (p < 0.0001) and the Combined cohort (p < 0.0001) (Fig. 5c, d). If we defined patients who experienced death or recurrence within 1 year as high-risk patients, then in External validation cohort 1, there were 19 patients in high risk, of whom 17 were correctly identified based on our threshold, resulting in an identification accuracy of 89.5%. In External validation cohort 2, there were 18 high-risk patients, and all patients were correctly identified using our threshold, yielding an accuracy of 100%. The association of PCP score, clinicopathological characteristics with overall survival and recurrence-free survival was shown in Supplementary Tables 2 and 3.
a Kaplan–Meier analyses of PCP score with overall survival in Training cohort. Log-rank test was used to compare the survival distributions of two groups b Kaplan–Meier analyses of PCP score with overall survival in Combined cohort (Testing cohort + External validation cohort 1 + External validation cohort 2). Log-rank test was used to compare the survival distributions of two groups. c Kaplan–Meier analyses of PCP score with recurrence-free survival in Training cohort. Log-rank test was used to compare the survival distributions of two groups. d Kaplan–Meier analyses of PCP score with recurrence-free survival in Combined cohort. Log-rank test was used to compare the survival distributions of two groups. Source data are provided as a Source Data file.
Discussion
Pancreatic cancer is notorious for its high malignancy and poor prognosis39, with the majority of patients diagnosed at an advanced stage due to the lack of early symptoms. As such, early detection is crucial for reducing mortality rates. Currently, the blood-based marker CA19-9 is the most widely used biomarker for pancreatic cancer diagnosis. However, its relatively low sensitivity (79%–81%) and specificity (82%–90%) limit its effectiveness, particularly in early-stage detection40.
CfDNA levels are elevated in pancreatic cancer16, providing a potential diagnostic biomarker for diagnosing pancreatic cancer. CfDNA offers several advantages: its detection technology is well established, and its relative stability enables consistent testing30,31,32. Several studies have investigated various cfDNA-based features, such as fragmentomics, mutations, and methylation, to develop diagnostic models for pancreatic cancer. For example, Bie et al. adapt an enzyme-mediated methylation sequencing method and developed a genome-wide cfDNA methylation, fragmentation, and copy number alteration (CNA) characteristics integrated model for cancer detection41. Ju et al. investigated the cfDNA fragmentomic characteristics against nucleosome positioning patterns in hematopoietic cells and developed a cancer diagnostic model based on the cfDNA fragmentomic metrics42. Christopher et al. developed A-plus, which can enhanced sensitivity over that achieved for aneuploidy alone at matched specificities43. DNA methylation can also affect the length of cfDNA fragments, An et al. found that DNA methylation might regulate cfDNA fragmentation, then they developed a cfDNA end-preference-based metric for cancer diagnosis44. Another study using methylation-based cfDNA features constructed a four-gene methylation panel, with a sensitivity of 100% and specificity of 90%27. Additionally, Liu, M.C et al. enrolled more than 50 types of cancer (including pancreatic cancer), through using methylation signatures in cfDNA, achieved high sensitivity in detecting early stage of pancreatic cancer45. Another study used methylation signature of cfDNA, achieved sensitivity of 83.7% in detecting pancreatic cancer46. Combining cfDNA methylation markers with protein biomarkers, such as CA19-9 and TIMP1, significantly improved diagnostic accuracy47. Zill et al. conducted a prospective analysis of five genes (KRAS, TP53, APC, FBXW7, and SMAD4) in tumor tissues and ctDNA from 26 pancreatic cancer patients, and the diagnostic accuracy of ctDNA sequencing was 97.7%, with an average sensitivity of 92.3% and a specificity of 100% for the five genes48.
In this study, we developed a cfDNA-based diagnostic and prognostic model using four different cfDNA features: fragment length, nucleosome footprint, end motif, and CNA. These features demonstrated significant differences among groups, with shorter cfDNA fragment lengths observed in pancreatic cancer patients compared to those with benign pancreatic tumors, suggesting increased cfDNA fragmentation with tumor malignancy. The PCM score effectively distinguished between pancreatic cancer and PBT, as well as early-stage pancreatic cancer from healthy individuals. Importantly, the cfDNA features correlated with prognosis, with a high PCP score indicating high risk.
Previous studies on early pancreatic cancer diagnosis have focused primarily on PDAC, excluding other benign pancreatic tumors. However, distinguishing between malignant and benign pancreatic tumors is challenging using imaging techniques, often requiring pathological confirmation. Traditional liquid biopsy methods, including CA19-9, show poor performance in distinguishing pancreatic cancer from benign tumors. In our analysis, the AUC of CA19-9 for differentiating pancreatic cancer from PBT was 0.819, with 26.7% of pancreatic cancer patients testing negative and 19.1% of chronic pancreatitis patients testing positive for CA19-9. Misdiagnosis based on CA19-9 alone is a significant concern, as elevated levels are observed in many benign conditions. By incorporating cfDNA features, our model achieved an AUC of 0.886 for distinguishing pancreatic cancer from PBT, representing a promising approach for differentiating pancreatic cancer from other pancreatic diseases.
Although cfDNA has shown promise in early pancreatic cancer detection, other biomarkers, such as circulating tumor cells (CTCs) and ctDNA, have also been explored. However, the low abundance of CTCs in early-stage cancer and the lack of validated biomarkers for cell selection limit their utility49. Similarly, ctDNA is unstable and present in low concentrations in early-stage cancer, further constraining its diagnostic potential15. This leads to the current techniques for using ctDNA as a standalone diagnostic marker for early-stage pancreatic cancer being insufficiently developed50. Additionally, others have utilized various biomarkers for the diagnosis of pancreatic cancer. For instance, some studies have investigated extracellular vesicle long RNA51 and exosomal microRNAs for pancreatic cancer diagnosis52. Based on extracellular vesicles long RNA profiling, Shulin Yu et al. developed a d-signature model for PDAC detection, the d-signature was able to identify early stage of pancreatic cancer (stage I/II) with an AUC of 0.94951. Compared with other studies, our study has the following advantage: 1. While others often focus on PDAC in their research, we have collected some pancreatic benign tumor, and we not only detect pancreatic cancer but also differentiate between cancer and non-cancer cases. 2. We validate our model in multicenter cohort; 3. PCP score is associated with overall survival, allowing for prognostic prediction; 4. Our study utilized four different types of features of cfDNA, allowing for a more comprehensive reflection of the differences in cfDNA among different populations. However, there are limitations, first of all, our study was a retrospective study, lack of perspective cohort; Secondly, although we have multicenter cohort that covered patients from different regions of China, extending validation to other countries or ethnic populations would enhance the model’s applicability.
Pancreatic cancer patients generally have poor prognosis. Our PCP score, based on cfDNA features, was associated with survival outcomes, with higher scores indicating worse prognosis. which confirms that cfDNA features are related to prognosis. However, we did not investigate which specific features contribute most to poor outcomes, that is an avenue for future research.
In conclusion, we developed a cfDNA-based diagnostic and prognostic model for pancreatic cancer, validated across multiple independent cohorts. Our PCM score system, integrating CNA, NF, fragmentation, and end motif features, demonstrated high accuracy in distinguishing malignant from benign conditions and was predictive of patient outcomes. Additionally, combining PCM score with CA19-9 significantly improved diagnostic performance, reinforcing the importance of CA19-9 as a biomarker in pancreatic cancer diagnosis.
Methods
Patients
From April 2021 through November 2021, we retrospectively collected a total of 975 cases for this study. Eight cases were excluded from the study according to eligibility criteria (Fig. 2). Finally, 967 cases were analyzed in our study. Including 422 pancreatic cancer or PBT, 47 CP, and 498 healthy controls. Among them, 370 patients were recruited from Changhai Hospital (Shanghai, China), 45 patients were recruited from The Affiliated Hospital of Qingdao University (Shandong province, China), 54 patients were recruited from The Second Affiliated Hospital of Shandong University and The Second Hospital, Cheeloo College of Medicine, Shandong University (Shandong Province, China). Healthy controls were recruited in five geographically centers for regular physical examination and had no history of pancreatic or other systematic diseases. The size of the training cohort was determined to have a power of 80% at a two-sided type I error rate of 0.05, which required at least 174 participants per group (actual enrollment: 432 participants). Detailed information of all patients and healthy controls was listed in Supplementary Data 1. Institutional review board at all participating hospitals reviewed and approved the study protocol. We conducted follow-up on all pancreatic cancer patients until January 2023, with a median follow-up duration of 443 days.
Plasma sample collection and cfDNA isolation
Blood samples were collected from patients and healthy controls in 10 ml EDTA-coated Vacutainer tubes. For all patients enrolled in our study, blood samples were collected before treatment. The plasma sample was centrifuged at the speed of 1600 × g for 10 mins and then at 16,000 × g for 10 mins (Eppendorf 5810 R/5427 R, Germany). The plasma samples were stored at −80 °C. The MagMAX Cell-Free DNA Isolation Kit (Thermo Fisher Scientific, USA) was used to isolate cfDNA according to the product instructions with the help of DNA purification instrument (Thermo Kingfisher FLEX, USA). The concentration of DNA product was then measured with Qubit3 Fluorometers (Thermo, USA). The size of DNA fragments was detected by fragment analyzer (Agilent, USA). The research protocol was approved by Shanghai Changhai Hospital Ethics Committee (CHEC2018-112) and Research Ethics Committee of The Second Hospital of Shandong University (KYLL2024446), and written informed consent was provided by every participant. Institutional review board at all participating hospitals reviewed and approved the study protocol.
Whole-genome sequencing and data processing
Sequencing libraries were prepared using 5 ng DNA. DNA samples were then subjected to end-repair/dA-tailing (5X ER/A-Tailing Enzyme Mix) and adaptor ligation (WGS Ligase). The adaptor sequence was specifically designed for Illumina CN500 platform. After purified by Agencourt AMPure XP beads (Beckman Coulter, USA), Libraries were quantified by the KAPA Library Quantification Kit (Kapa Biosystems, USA) and size was confirmed using Bioanalyzer (Agilent, USA). Sequencing libraries were pooled at equal amount. WGS at an average coverage of 1.5X was performed on Illumina CN500 platform using 2 × 36 bp paired-end sequencing.
Fastq files were processed by fastp software (https://github.com/OpenGene/fastp) to remove adaptor and end sequence together with sequences below 25 bp to acquire clean data. Clean data were aligned to human reference genome GRCh37 using bwa-aln (https://github.com/lh3/bwa). Duplicate reads were marked by sambamba (https://github.com/biod/sambamba/). Samtools (http://samtools.sourceforge.net/) was used to calculate mapping rate, duplicate rate and genome coverage. Reads with mapping rate above 90%, duplicate rate below 25% and coverage above 50% passed the quality control. The bam files were further filtered by Samtools, removing unmapped reads, low quality reads, marked duplicates and sequences with no perfect match between read1 and 2.
We conducted low-pass whole-genome sequencing with all collected samples. The sequencing data allowed us to analyze multiple features, including CNA, nuclear footprint, end motif, and fragmentation. Individuals from each cohort were randomly assigned into training and testing cohorts. In the training process, we performed LASSO regression algorithm for each genomic feature to reduce dimensionality and extract markers, and further employed SVM algorithm to build the optimal model of each genomic features. At last, a logistic model was used to integrate three genomic features CNA.
Procedure of feature selection was as follows; all steps were conducted in Training cohort.
Procedure for quantifying fragments33,53
-
(1)
The whole genome was divided to 3055 regions; the length of every region is 1 Mbp.
-
(2)
After aligning with the reference genome, the genomic location of each DNA fragment is identified and corrected.
-
(3)
Fragments between 90 and 150 bp are defined as short fragments, and those between 151 and 220 bp are defined as long fragments.
-
(4)
Calculate the ratio of short fragment to long fragment.
Feature selection results: Regions on the Y chromosome and regions with no detected DNA fragment coverage were removed from the initial 3055 regions, leaving 2890 regions. LASSO was then used for feature selection, resulting in 154 regions, which were used to construct the model.
Procedure for quantifying end motif36,54
-
(1)
Align the DNA fragments with the reference genome to determine the start and end positions of each fragment, and perform correction.
-
(2)
Count the 4-mer nucleotide sequence at the 5′ end of each fragment.
-
(3)
There are 256 possible types of 4-mer sequences; calculate the proportion of each type.
Feature selection: LASSO was used for feature selection, resulting in 33 motifs, which were used to construct the model.
Procedure for quantifying nucleosome footprint 13,55
-
(1)
Acquisition of the promoter regions of the whole genome: Promoter regions were identified using transcription start sites (TSS) of the main transcripts of reference genes published in the UCSC database are used (https://genome.ucsc.edu/), with 2500 bp extended upstream and downstream as the promoter region of the gene;
-
(2)
Defining the Central and Peripheral Regions: The central region of the promoter is defined as the 250 bp immediately adjacent to the transcription start site of the gene, while the peripheral region was extended 2500 bp on either side of the central region. This classification is based on the observation that actively transcribed genes tend to have sparser nucleosome distribution near the TSS, making them more susceptible to degradation once in the bloodstream. As a result, sequencing depth is expected to be lower in the central region compared to the peripheral region;
-
(3)
Obtaining Region Coverage and Sequencing Depth: The software “Bedtools” (v1.6.2) was used to calculate region coverage, while “featureCounts” (v2.19.1) was employed to determine sequencing depth for each region, which was then converted to FPKM;
-
(4)
Quantifying the Differences in Nucleosome Distribution for Each Gene: The nucleosome distribution difference score is calculated as the sequencing depth of the peripheral region (FPKM) minus the central region (FPKM). This score represents the distribution of nucleosomes and the transcriptional activity of the gene;
-
(5)
Filter Genes and Model Construction: In the Training cohort, after removing housekeeping genes and silenced genes from the whole genome, 20315 genes remained. Genes covered in at least 90% of the samples were retained, and the rank-sum test was applied to calculate p-values. Genes with p ≤ 0.01 were further filtered, resulting in 428 genes. LASSO was then used for dimensionality reduction, ultimately selecting 102 genes for model construction.
Procedure for CNA score calculation56
The human genome was divided into numerous 20Kbp regions. In order to avoid the high variations of CNAs related to small bins, we have connected adjacent small bins that meet the requirements. A certain margin of error is allowed during the connection process, and the final reported length is at least 2Mbs. Any length below 2Mbs will be filtered out. The average sequencing depth of each was counted and GC content corrected (the GC correction process was calculating the average depth of bins for each GC content, then computing the overall average depth of all bins to correct the sequencing depth). A baseline threshold was established for each region with the mean and variance of the average sequencing depth from the data of healthy population in the training cohort. Each region used the above calculated mean and variance to calculate the Zscore. According to the distribution of the Zscore of healthy people in the training cohort in the region, defined that Zscore greater than 2 or less than -2 was the baseline threshold with significant difference. Those with Zscore greater than 2 were copy number amplification, and those with Zscore less than −2 were copy number deletion. Adjacent regions with the same copy number alteration direction will be connected. When adjacent regions were connected, the tolerance was set for regions that were not covered by sequencing data. The CNA region should contain at least 70% of the copy number alteration in the same direction and with a length greater than 2Mbp were reported. The tumor suppressor (TSG) and oncogene (OG) of CNA score was then calculated using the equation reported previously56. The equation of CNA score was shown in Supplementary Table 4.
The LASSO implementation process is as follows: By constructing a penalty function, a more refined model was obtained, so that it compressed some coefficients and sets some coefficients to zero. It compressed the variables with large parameter estimates to 0, while the variables with small parameter estimates were compressed to 0, so as to achieve the effect of feature dimensionality reduction. This process was implemented using the LassoCV() function of the ‘sklearn’ package in Python. The LASSO inputs for NF, fragment, and end motif features were derived exclusively from the training cohort samples. For the NF input, we used the expression values of each gene per sample after filtering for a p-value less than 0.01. The fragment input was defined as the ratio of short to long fragments within each genomic region per sample, also filtered by a p-value threshold of <0.01. For the motif input, we calculated the proportion of various end motifs in each sample after filtering for a p-value less than 0.05. Features with non-zero coefficients in the ‘lasso.coef’ output were retained as the final selected features after dimensionality reduction.
The SVM method (support vector machine) was implemented for individual genomic feature-based model construction, based on three parameters: (1) C: Penalty coefficient; (2) Kernel function; and (3) Gamma. The input was the sample data of the training cohort, and the features of each dimension were filtered by LASSO dimensionality reduction. The GridSearchCV() function of the ‘sklearn’ package in python was used to find the optimal combination of three parameters in the training cohort, and the determination process of the optimal parameter combination used the 10X cross-validation method, that is, the training cohort samples were divided into 10 equal parts, of which 9 were used for parameter training fitting and the remaining 1 was used to verify the performance. Each dimension was trained separately to determine the optimal combination of parameters. The identified optimal parameters were applied directly to independent validation cohort samples. Finally, the predicted value of each sample in each of the three dimensions was the output.
Construction of PCM score
We integrated the fragment model, motif model, NF model, and CNA score using logistic regression method to construct a combined method—PCM scoring system (Fig. 2). The PCM score includes three components. The first component is the CNA score, which was previously introduced in the text. The second component is a logistic regression formula which allows the biomarkers, as a group, to be used to discriminate between pancreatic cancer and non-pancreatic cancer cases. Generally, the Wilcoxon rank-sum test was used to compare two datasets, pancreatic cancer vs non-pancreatic cancer. LASSO was applied to feature selection in the Training cohort. Features used for model construction was shown in Supplementary Tables 5–7. Data normalization was done using Z-score on Python. Support vector machine (SVM) was implemented for individual genomic feature-based model construction, based on three parameters: (1) C: Penalty coefficient; (2) Kernel function; and (3) Gamma. For the Training cohort, 10-fold cross-validation was employed to figure out the best combination of the parameters. The cutoff value was set at the point with the best diagnostic accuracy in testing cohort. To obtain the best diagnostic model, logistic regression model was generated using the results of the three individual models as input features. The Logistic Score was calculated as below.
The third component is individual genomic feature score (Single Score), calculated with the below formula.
Finally, CNA Score, Logistic Score and Single Score are subjected to a multivariate linear equation which generated the final PCM Score. The optimal cutoff of PCM Score was 0.75, determined by Youdens’ index. PCM Score ≥ 0.75 was regarded as positive, otherwise negative.
The detailed calculation equation of PCM score was shown in Supplementary Table 4.
The equation of PCM combined with CA19-9 model was: PCM score + log10(CA19-9), the unit of CA19-9 was U/ml.
Construction of PCP score
We constructed PCP scoring system using fragment, motif, and NF features of cfDNA. Pancreatic cancer patients with follow-up data were included in the analysis. Samples were separated to two groups, with recurrence or death within 1 year were classified as high-risk, while those without recurrence or death were classified as low-risk. Filtering features with significant p-values (p-value < 0.01), then further feature selection with LASSO. The samples in the Training cohort were comparable to those used in the PCM score model. Due to the absence of prognostic information in some samples, the remaining samples were grouped into the Combined cohort. The selected features used for model construction are listed in Supplementary Tables 8–10. Each of the three indicators was modeled independently using SVM, and the final integration was achieved through logistic regression.
Statistical analysis
Wilcoxon rank-sum test was applied to compare two groups of continuous variables and Fisher’s exact test was applied to categorical variables. P value was calculated using Python software (version 2.7.14), and p < 0.05 was considered as statistically significant. Area Under Curve (AUC) was applied to evaluate model performance. ROC curves were generated by using ‘pROC’ package (v1.16.2) in R software (v.3.6.3), ‘datatable’ (v1.14.2) was used to process the data and ‘pwr’(v1.3.0) was used to process the power analysis in R software (v.3.6.3). Survival curves were generated according to the Kaplan–Meier method and compared using the log-rank test. LASSO and SVM algorithms were performed with ‘sklearn’ in Python software (version 2.7.14).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw sequencing data have been deposited in Genome Sequence Archive in National Genomics Data Center (BioProject ID: PRJCA037852), under accession number HRA011014, accessible at https://ngdc.cncb.ac.cn/gsa-human. Source data are provided with this paper. The minimum data set has been uploaded to Zenodo with https://doi.org/10.5281/zenodo.15123508. Source data are provided with this paper.
Code availability
The codes are available on GitHub (https://github.com/JimmyWu2024/PAAD_pipline). The repository has been linked to Zenodo, with https://doi.org/10.5281/zenodo.15100396.
References
Chen, W. et al. Cancer statistics in China, 2015. CA Cancer J. Clin. 66, 115–132 (2016).
Tempero, M. A. et al. Pancreatic adenocarcinoma, Version 2.2017, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Canc Netw. 15, 1028–1061 (2017).
Kleeff, J. et al. Pancreatic cancer. Nat. Rev. Dis. Prim. 2, 16022 (2016).
Wang, X. et al. Antigen/HLA-agnostic strategies for characterizing tumor-responsive T cell receptors in PDAC patients via single-cell sequencing and autologous organoid application. Cancer Lett. 588, https://doi.org/10.1016/j.canlet.2024.216741 (2024).
Ma, J. et al. Early elevations of RAS protein level and activity are critical for the development of PDAC in the context of inflammation. Cancer Lett. 586 https://doi.org/10.1016/j.canlet.2024.216694 (2024).
Gromisch, C. et al. Pancreatic adenocarcinoma: unconventional approaches for an unconventional disease. Cancer Res. 80, 3179–3192 (2020).
Ermiah, E. et al. Prognostic value of serum CEA and CA19-9 levels in pancreatic ductal adenocarcinoma. Mol. Clin. Oncol. 17, 126 (2022).
Swords, D. S., Firpo, M. A., Scaife, C. L. & Mulvihill, S. J. Biomarkers in pancreatic adenocarcinoma: current perspectives. Onco Targets Ther. 9, 7459–7467 (2016).
Xu, C. et al. A circulating panel of circRNA biomarkers for the noninvasive and early detection of pancreatic ductal adenocarcinoma. Gastroenterology 166, 178–190.e116 (2024).
Holly, E. A., Chaliha, I., Bracci, P. M. & Gautam, M. Signs and symptoms of pancreatic cancer: a population-based case-control study in the San Francisco Bay area. Clin. Gastroenterol. Hepatol. 2, 510–517 (2004).
Oberstein, P. E. & Olive, K. P. Pancreatic cancer: why is it so hard to treat? Ther. Adv. Gastroenterol. 6, 321–337 (2013).
Fan, H. C., Blumenfeld, Y. J., Chitkara, U., Hudgins, L. & Quake, S. R. Analysis of the size distributions of fetal and maternal cell-free DNA by paired-end sequencing. Clin. Chem. 56, 1279–1286 (2010).
Snyder, M. W., Kircher, M., Hill, A. J., Daza, R. M. & Shendure, J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 164, 57–68 (2016).
Sun, K. et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc. Natl. Acad. Sci. USA 112, E5503–E5512 (2015).
Shapiro, B., Chakrabarty, M., Cohn, E. M. & Leon, S. A. Determination of circulating DNA levels in patients with benign or malignant gastrointestinal disease. Cancer 51, 2116–2120 (1983).
Mattox, A. K. et al. The origin of highly elevated cell-free DNA in healthy individuals and patients with pancreatic, colorectal, lung, or ovarian cancer. Cancer Discov. 13, 2166–2179 (2023).
Zhang, X. et al. Ultrasensitive and affordable assay for early detection of primary liver cancer using plasma cell-free DNA fragmentomics. Hepatology 76, 317–329 (2022).
Xu, R. H. et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat. Mater. 16, 1155–1161 (2017).
Cai, J. et al. Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma. Gut 68, 2195–2205 (2019).
Alunni-Fabbroni, M. et al. Circulating cell-free DNA combined to magnetic resonance imaging for early detection of HCC in patients with liver cirrhosis. Cancers (Basel) 13, https://doi.org/10.3390/cancers13030521 (2021).
Jamal-Hanjani, M. et al. Detection of ubiquitous and heterogeneous mutations in cell-free DNA from patients with early-stage non-small-cell lung cancer. Ann. Oncol. 27, 862–867 (2016).
Wang, S. et al. Multi-dimensional cell-free DNA fragmentomic assay for detection of early-stage lung cancer. Am. J. Respir. Crit. Care Med. https://doi.org/10.1164/rccm.202109-2019OC (2022).
Wu, X. & Tanaka, H. Aberrant reduction of telomere repetitive sequences in plasma cell-free DNA for early breast cancer detection. Oncotarget 6, 29795–29807 (2015).
Christensen, E. et al. Early detection of metastatic relapse and monitoring of therapeutic efficacy by ultra-deep sequencing of plasma cell-free DNA in patients with urothelial bladder carcinoma. J. Clin. Oncol. 37, 1547–1557 (2019).
Wan, N. et al. Machine learning enables detection of early-stage colorectal cancer by whole-genome sequencing of plasma cell-free DNA. BMC Cancer 19, 832 (2019).
Vandenberghe, P. et al. Non-invasive detection of genomic imbalances in Hodgkin/Reed-Sternberg cells in early and advanced stage Hodgkin’s lymphoma by sequencing of circulating cell-free DNA: a technical proof-of-principle study. Lancet Haematol. 2, e55–e65 (2015).
Ying, L. et al. Methylation-based cell-free DNA signature for early detection of pancreatic cancer. Pancreas 50, 1267–1273 (2021).
Jaworski, J. J., Morgan, R. D. & Sivakumar, S. Circulating cell-free tumour DNA for early detection of pancreatic cancer. Cancers (Basel) 12, https://doi.org/10.3390/cancers12123704 (2020).
Guler, G. D. et al. Detection of early stage pancreatic cancer using 5-hydroxymethylcytosine signatures in circulating cell free DNA. Nat. Commun. 11, 5270 (2020).
Risberg, B. et al. Effects of collection and processing procedures on plasma circulating cell-free DNA from cancer patients. J. Mol. Diagn. 20, 883–892 (2018).
Warton, K. et al. Evaluation of Streck BCT and PAXgene stabilised blood collection tubes for cell-free circulating DNA studies in plasma. Mol. Diagn. Ther. 21, 563–570 (2017).
Medina Diaz, I. et al. Performance of Streck cfDNA blood collection tubes for liquid biopsy testing. PLoS One 11, e0166354 (2016).
Cristiano, S. et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 570, 385–389 (2019).
Douville, C. et al. Assessing aneuploidy with repetitive element sequencing. Proc. Natl. Acad. Sci. USA 117, 4858–4863 (2020).
Cohen, J. D. et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc. Natl. Acad. Sci. USA 114, 10202–10207 (2017).
Jiang, P. et al. Plasma DNA end-motif profiling as a fragmentomic marker in cancer, pregnancy, and transplantation. Cancer Discov. 10, 664–673 (2020).
Lapin, M. et al. Fragment size and level of cell-free DNA provide prognostic information in patients with advanced pancreatic cancer. J. Transl. Med. 16, 300 (2018).
Yamamoto, Y. et al. Clinical significance of the mutational landscape and fragmentation of circulating tumor DNA in renal cell carcinoma. Cancer Sci. 110, 617–628 (2019).
Singhi, A. D., Koay, E. J., Chari, S. T. & Maitra, A. Early detection of pancreatic cancer: opportunities and challenges. Gastroenterology 156, 2024–2040 (2019).
Ballehaninna, U. K. & Chamberlain, R. S. The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: An evidence based appraisal. J. Gastrointest. Oncol. 3, 105–119 (2012).
Bie, F. et al. Multimodal analysis of cell-free DNA whole-methylome sequencing for cancer detection and localization. Nat. Commun. 14, 6042 (2023).
Ju, J. et al. Cell-free DNA end characteristics enable accurate and sensitive cancer diagnosis. Cell Rep. Methods 4, 100877 (2024).
Douville, C. et al. Machine learning to detect the SINEs of cancer. Sci. Transl. Med. 16, eadi3883 (2024).
An, Y. et al. DNA methylation analysis explores the molecular basis of plasma cell-free DNA fragmentation. Nat. Commun. 14, 287 (2023).
Liu, M. C. et al. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 31, 745–759 (2020).
Klein, E. A. et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann. Oncol. 32, 1167–1177 (2021).
Ben-Ami, R. et al. Protein biomarkers and alternatively methylated cell-free DNA detect early stage pancreatic cancer. Gut 73, 639–648 (2024).
Zill, O. A. et al. Cell-free DNA next-generation sequencing in pancreatobiliary carcinomas. Cancer Discov. 5, 1040–1048 (2015).
Martini, V., Timme-Bronsert, S., Fichtner-Feigl, S., Hoeppner, J. & Kulemann, B. Circulating tumor cells in pancreatic cancer: current perspectives. Cancers (Basel) 11, https://doi.org/10.3390/cancers11111659 (2019).
Bettegowda, C. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6, 224ra224 https://doi.org/10.1126/scitranslmed.3007094 (2014).
Yu, S. et al. Plasma extracellular vesicle long RNA profiling identifies a diagnostic signature for the detection of pancreatic ductal adenocarcinoma. Gut 69, 540–550 (2020).
Nakamura, K. et al. An exosome-based transcriptomic signature for noninvasive, early detection of patients with pancreatic ductal adenocarcinoma: a multicenter cohort study. Gastroenterology 163, 1252–1266.e1252 (2022).
Mouliere, F. et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 10, https://doi.org/10.1126/scitranslmed.aat4921 (2018).
Jiang, P. et al. Preferred end coordinates and somatic variants as signatures of circulating tumor DNA associated with hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 115, E10925–E10933 (2018).
Ulz, P. et al. Inferring expressed genes by whole-genome sequencing of plasma DNA. Nat. Genet 48, 1273–1278 (2016).
Davoli, T. et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155, 948–962 (2013).
Acknowledgements
This work was supported by the National Key R&D Program of China (2023YFC2507500 to W.X., 2022YFC3400903 to L.C.), the National Natural Science Foundation of China (U21A20376 to L.C., 82172712 to G.J., 81972913 to G.J., 82425038 to L.C., and 82421005 to L.C., 82588201 to Q.Z.), Postdoctoral Foundation of China 2023M734296 to T.W.; We thank the support of Shanghai Key Laboratory of Hepato-biliary Tumor Biology and Military Key Laboratory on Signal Transduction, and the Key Laboratory of Signaling Regulation and Targeting Therapy of Liver Cancer (SMMU), Ministry of Education. This study was also supported by Innovation Program of Shanghai Municipal Education Commission, Naval Medical University (2023MS020 to X.X.); Changhai Hospital of Naval Medical University (2023PY018 to X.X.). This study was also supported by Innovation Program of Shanghai Municipal Education Commission.
Author information
Authors and Affiliations
Contributions
J.W., X.X., Q.Z., P.L., and T.W. contributed equally to this work. J.W. performed experiments and wrote the manuscript, Q.Z. and T.W. performed experiments and analyzed the data, X.X. and P.L. performed experiments and contributed to the sample collection. S.G., L.D., S.S., J.H., and X.W. contributed to the sample collection. D.X., F.S., Y.W., L.Z., and J.B. analyzed the data, L.W., W.L., H.W., G.J., and L.C. designed the study and review the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wu, J., Xu, X., Zhang, Q. et al. Cell-free DNA testing for the detection and prognosis prediction of pancreatic cancer. Nat Commun 16, 6645 (2025). https://doi.org/10.1038/s41467-025-61890-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-61890-z