Abstract
Chloroplasts are crucial players in immunity and photosynthesis. However, how chloroplasts arrange the transition between photosynthesis and immunity and how pathogens manipulate this transition remains elusive. Here we report an effector SsCm1 from Sclerotinia sclerotiorum, one of devastating phytopathogenic fungi, inhibits chloroplast immunity and resistance to pathogens, and alleviates photoinhibition in immune state. This is accomplished through stabilizing the conserved chloroplast protein MORF2, which is degraded during immunization and is a suppressor of photoinhibition, cell death, and chloroplast immunity. Overexpression of SsCm1 or MORF2 in plants represses basic immunity and resistance to pathogens, whereas deletion of SsCm1 reduces S. sclerotiorum virulence. Notably, SsCm1 possesses no chorismate mutase activity, which is different from the previously reported Cm effectors. This work reveals a strategy to fine-tune growth-defense balance in chloroplasts by manipulating MORF2, and a pathogenic strategy to subvert the process and promote infection via enzymatically nonfunctional effector stabilizing MORF2.
Similar content being viewed by others
Introduction
Plants perceive threats from pathogens through complex and ingenious mechanisms: the pattern-recognition receptors (PRRs) located on the plasma membrane detect pathogen-associated molecular patterns (PAMPs) and activate basic immunity (PAMP-triggered immunity, PTI); intracellular nucleotide-binding leucine-rich repeat receptors recognize pathogen effectors to activate more robust effector-triggered immunity (ETI)1. Both PTI and ETI are involved in overlapping downstream immune responses, including activation of mitogen-activated protein kinases (MAPKs), calcium-mediated signaling, reactive oxygen species (ROS) burst, transcriptional reprograming, callose deposition, and biosynthesis of salicylic acid (SA)2,3. Chloroplasts play vital roles in these immune responses and are crucial participants in plant-pathogen interactions4,5,6,7.
Chloroplasts are emerging as hubs in integrating environmental stimuli and determinants of biotic or abiotic stress responses7,8,9. The current views ascribe fundamental function to chloroplasts in orchestrating defense responses: upon plasma membrane sensing biological threats, signals are quickly relayed to chloroplasts through the plasma membrane/chloroplast dual-located proteins or calcium signal5,7. Then, chloroplasts produce calcium signatures, ROS, the by-products of tetrapyrrole pathway and communicate with the nucleus via retrograde signaling (RS)7,10,11,12,13. This signaling results in the expression of defense-related genes, such as genes involved in SA biosynthesis and encoding WRKY transcription factors14. When the threat persists or is fatal, MPK3/MPK6 actively inhibits photosynthesis to promote ROS accumulation in chloroplasts15. These processes represent the transformation from growth to defense, including immune transcriptional reprograming, SA biosynthesis, localized cell death, and resistance to pathogens16,17,18,19. Note that these changes come at the expense of the photosynthetic function of chloroplasts, including global down-regulation of photosynthetic genes, photoinhibition, and damage of photosystem II (PSII), which are essential for chloroplast immunity20,21,22,23,24. Therefore, chloroplasts require precise control to rapidly convert to the immune state upon pathogen infection while maintaining the photosynthetic state under normal conditions. How chloroplasts switch between these two opposite states and which components are responsible for this switch are unclear.
Consistent with the prominent role of chloroplasts in immunity and the arms race between pathogens and hosts, this organelle is a prime target for pathogen manipulation4,5,6. Convergently evolved effectors of pathogens from different kingdoms target chloroplasts and impair SA-dependent immunity to promote pathogenicity5. Virulent Pseudomonas syringae secretes multiple effectors that cooperate to reprogram the expression of photosynthesis-associated nuclear genes (PhANGs) and target the chloroplast to prevent the chloroplastic ROS burst by impairing PSI6. The effector Pst_12806 reduces the levels of photosynthesis and chloroplast-derived ROS to facilitate pathogen survival by targeting the chloroplastic cytochrome b6-f complex25. Similarly, Sclerotinia sclerotiorum also employs virulence factors, including oxalate and SsITL to increase chloroplast non-photochemical quenching (NPQ) and perturb immune function of the chloroplastic calcium-sensing receptor, respectively, to suppress host defense26,27,28. However, it is unclear how pathogens can manipulate the fine-tuned process of chloroplast switching from photosynthesis to immunity to promote infection.
As one of the polyphagous and devastating phytopathogens, S. sclerotiorum causes Sclerotinia stem rot of soybean and rapeseed29. A two-phase infection model based on cytological and genetic investigations has been widely accepted30. For successful infection and colonization, S. sclerotiorum needs to secrete oxalic acid (OA) and effectors to counteract plant immunity and ensure pathogen success31,32,33,34. Although the S. sclerotiorum genome encodes dozens (>70) of putative effectors, there are few studies on the molecular mechanism of their interaction with the host26,35,36,37,38. Here, we identified a chorismate mutase (Cm) effector SsCm1 by screening virulence-related secreted proteins (SsVSPs). Unlike the known Cm effectors39, SsCm1 shows no enzymatic activity and cannot interact with plant Cm. Functional genetic analyses and heterologous expression in plants confirmed that SsCm1, as an essential virulence factor, can inhibit basic immunity of plants and promote infection. Research on the target of effectors in hosts can serve as an important tool in identifying components of plant immune systems. We defined the conserved chloroplast RNA editing factor MORF2 as the functional target of SsCm1. Genetic analyses showed that MORF2 negatively regulates basic immunity and resistance to pathogens. Furthermore, biochemical evidences demonstrated that MORF2 is degraded during immunity stimulation, while SsCm1 stabilizes MORF2. We further showed that overexpression of SsCm1 or MORF2 in plants perturbed production of ROS, photoinhibition, and cell death. Overall, our results reveal that plants fine-tune the switch of chloroplast between photosynthesis and immunity by manipulating MORF2, and S. sclerotiorum secretes an enzymatically nonfunctional effector to promote infection via stabilizing MORF2.
Results
SsCm1 is an important effector secreted by S. sclerotiorum
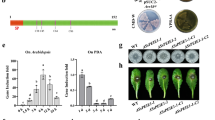
A number of putative secreted proteins are encoded by S. sclerotiorum, and many of them are up-regulated during infection. Here, we defined them as SsVSPs35,40,41. However, the functions of only a few SsVSPs have been characterized. By transiently expressing full-length SsVSPsFL or signal peptide (SP)-removed SsVSPsΔSP in Nicotiana benthamiana, we found that SsVSP31ΔSP promoted S. sclerotiorum infection (Fig. 1a). Due to the presence of a Cm domain (Supplementary Fig. 1a), SsVSP31 was renamed SsCm1. The expression of SsCm1 was continuously up-regulated in the early stage of infection in multiple hosts (Fig. 1b). The yeast invertase secretion assay confirmed that the SP of SsCm1 was functional and the specific detection of SsCm1-GFP in the culture supernatant confirmed that SsCm1 was secreted by S. sclerotiorum (Fig. 1c, d).
a Expression of SsVSP31ΔSP in N. benthamiana enhances infection of S. sclerotiorum. Leaf regions transiently expressing SsVSPs-FLAG or GFP-FLAG are inoculated with mycelium plugs of UF-1. The lesion areas were calculated using ImageJ at 24 hpi. Centre lines show the medians; box limits indicate the 25th–75th percentiles; whiskers extend to 1.5 × the interquartile range from the 25th–75th percentiles; the top lines represent maxima and the bottom lines represent minima; all individual data points (n = 36, n represents the total number of measured lesions with 3 biological replicates) are overlaid. FL full length, ΔSP deletion of the signal peptide. ns = no significance, ****p < 0.0001 (two-way ANOVA with Sidak’s multiple comparisons test, compared to GFP). b Expression profile of SsCm1 during UF-1 infection of multiple hosts. The Sstublin gene is used as an internal control, and the expression level of SsCm1 at 0 hpi is normalized to 1 (mean ± SD; n = 3 biological replicates). c Functional validation of the SP of SsCm1. The pSUC2 empty vector strain and the strain containing the SP of effector Avr1b are used as negative and positive controls, respectively. d Secretion of SsCm1. The strains carrying GFP or SsCm1-GFP are inoculated into PDB in the presence of N. benthamiana leaf discs. Total proteins are extracted from mycelia or freeze-dried concentrated culture supernatant at 24 hpi, and immunoblotted with anti-GFP antibody. CBB staining shows protein loading. e SsCm1 is an essential virulence factor of S. sclerotiorum. Disease symptoms on different hosts caused by UF-1, ΔSsCm1-17/19, and ΔSsCm1-17-C32. Source data are provided as a Source Data file.
To explore the function of SsCm1 in S. sclerotiorum, two knockout mutants (ΔSsCm1-17 and ΔSsCm1-19) were obtained in the background of UF-1, and SsCm1-GFP was introduced into the ΔSsCm1-17 as a genetically complemented strain (ΔSsCm1-17-C32) (Supplementary Fig. 1). The pathogenicity of the ΔSsCm1 mutants on different hosts was significantly reduced compared with the UF-1 and the complemented strain, whereas no significant variations among these strains were detected in colony morphology, hyphal growth rate, compound appressorium formation, sclerotium development, OA production, and content of cell wall hydrolase (Fig. 1e and Supplementary Figs. 2 and 3). Similarly, deleting SsCm1 in the oxalate minus mutant background (Δoah)42 further reduced virulence (Supplementary Fig. 4). These data demonstrate that SsCm1 is secreted by S. sclerotiorum and plays an important role in the virulence of S. sclerotiorum.
SsCm1 lowers plant basic immunity and resistance to pathogens
To investigate the function of SsCm1 in plants, we first evaluated the effects of SsCm1 on several immune responses of N. benthamiana. The results showed that the transient expression of SsCm1 inhibited callose deposition and the expression of pathogenesis-related genes (NbPR1a and NbPR2) induced by PAMPs, including Flg22, chitin, and Sclerotinia culture filtrate elicitor (SCFE), while no significant variation was detected in the ROS burst induced by PAMPs (Supplementary Fig. 5). Together with SsCm1 promoting the virulence of S. sclerotiorum on N. benthamiana (Fig. 1a, e), suggesting that SsCm1 inhibits the basic immunity and resistance to S. sclerotiorum in N. benthamiana.
Next, stable 35S:SsCm1 overexpressing lines of Arabidopsis were created. Compared with wild-type Arabidopsis thaliana (Col-0) and GFP-expressing control lines, plants overexpressing SsCm1 showed significantly enhanced their sensitivity to UF-1, Botrytis cinerea, and the Δoah mutant. All plants showed normal growth and morphology (Fig. 2a, b). Similarly, SsCm1-expressing transgenic lines were more susceptible to the hemi-biotroph pathogen Pseudomonas syringae pv. tomato strain DC3000 (DC3000) (Fig. 2c, d). This demonstrates that SsCm1 inhibits resistance to a broad range of Arabidopsis pathogens.
a, b SsCm1 enhances fungal infection in Arabidopsis. Morphology of Col-0, 35S:GFP-1/2, and 35S:SsCm1-1/2 lines grown for 3 weeks and disease symptoms of plants challenged with UF-1, Δoah, or B. cinerea (a). Centre lines show the medians; box limits indicate the 25th–75th percentiles; whiskers extend to 1.5×the interquartile range from the 25th–75th percentiles; the top lines represent maxima and the bottom lines represent minima; all points (n = 36, n represents the total number of measured lesions with 3 biological replicates) are overlaid (b). c, d SsCm1 enhances DC3000 infection in Arabidopsis. Leaves of Col-0 and 35S:SsCm1-1/2 were syringe-infiltrated with DC3000 (OD600 = 0.0005). Disease symptoms (c) and bacterial growth quantification (d) at 3 dpi. *p < 0.05 (one-way ANOVA with multiple comparisons test; mean ± SD; n = 4 biological replicates). e, f SsCm1 inhibits callose deposition induced by PAMPs in Arabidopsis. Leaves were infiltrated with water, 1 µM flg22, 200 µg/mL chitin, or 50 µg/mL SCFE, and aniline blue staining was performed at 24 hpi. SsCm1 inhibits stomatal closure (g) and ethylene production (h) induced by PAMPs in Arabidopsis. Leaves were treated with water or PAMPs for 6 h, stomatal aperture was calculated and ethylene emission was measured. i–n SsCm1 inhibits the expression of defense-related genes induced by PAMPs in Arabidopsis. Leaves were treated with water or PAMPs for 12 h and the expression of the immune marker genes was normalized to the levels of AtActin2 transcript. Data are presented as fold induction relative to the expression before treatment, which is set to baseline of 1. o SsCm1 decreases SA biosynthesis induced by PAMPs in Arabidopsis. Leaves were treated with water or PAMPs for 12 h and total SA content was quantified. For b, f–o, data were analyzed by two-way ANOVA with Sidak’s multiple comparisons test, and data points with different letters indicate significant differences of p < 0.05. For f–o, data represent the mean ± SD (n = 3 biological replicates). Source data are provided as a Source Data file.
Similarly, we also evaluated the effect of SsCm1-expression on the basic immunity processes in Arabidopsis. The results showed that compared with Col-0, 35S:SsCm1 overexpressing lines exhibited significantly decreased levels of callose deposition, stomatal closing, ethylene production, and the expression of immune marker genes, such as Flg22-induced receptor-like kinase 1 (FRK1), PR1, PR4, and phytoalexin deficient 3 (PAD3), following treatment with PAMPs (Fig. 2e–l). Furthermore, the expression of genes related to SA biosynthesis (ICS1 and SARD1) and the total SA content induced by PAMPs were also repressed in transgenic lines (Fig. 2m–o). Together, SsCm1 inhibits the basic immunity and resistance to pathogens in plants.
Enzymatically nonfunctional SsCm1 conservatively targets chloroplast RNA editing factors MORF2s
SsCm1 inhibits SA biosynthesis reminiscent of Cmu1 of Ustilago maydis which inhibits SA biosynthesis by interacting with the host Cm39. Here, we showed that although there was weak interaction between SsCm1 and three Cms of soybean based on yeast two-hybrid (Y2H) experiments, these interactions were not reproduced in split-luciferase, bimolecular fluorescence complementarity (BiFC), and co-immunoprecipitation (Co-IP) assays in planta, and SsCm1 could form a self-dimer (Supplementary Fig. 6). Importantly, the heterologous expression of SsCm1 into yeast aro7 mutant could not restore the ability of yeast to synthesize tyrosine and phenylalanine (Fig. 3a), and the purified SsCm1ΔSP showed lower Cm activity than that of UmCmu1 (Fig. 3b and Supplementary Fig. 7a). SsCm1 mixed with GmCms could not improve the enzyme activities in the reaction solutions (Fig. 3c and Supplementary Fig. 7a). Accordingly, the protein sequence of SsCm1 lacks a number of conserved amino acids reported in Cm and the amino acids key to Cm activity (Supplementary Fig. 7b)39,43. Protein structure comparison showed that SsCm1 was obviously different from other Cms in three-dimensional structure, and molecular docking simulation revealed that SsCm1 showed less affinity with chorismate (binding energy = −2.1 kcal/mol) than ScAro7 (−5.7 kcal/mol) (Supplementary Fig. 7c, d). Another gene (SsCm2) of S. sclerotiorum encoded Cm, which was predicted to contain no SP, and the ability of SsCm2 to complement Scaro7 mutant and its enzymatic activity in vitro was equivalent to that of UmCmu1 (Fig. 3a, b and Supplementary Fig. 7). These results suggest that SsCm1 might acted in a different way than UmCmu1 and did not have functional chorismate mutase activity.
a SsCm2 but not SsCm1 can complement the genetic defect of yeast aro7. SD-Leu synthetic medium (SD) without leucine, SD-Leu/Phe/Tyr SD without leucine, tyrosine, and phenylalanine. The pGADT7 was used as a negative control, and pGADT7-ScAro7 was used as a positive control. b SsCm2 but not SsCm1 has chorismate mutase activity in vitro. Recombinant SsCm1ΔSP-HA, SsCm2-HA, GST (negative control), and UmCmu1ΔSP-HA (positive control) were used to perform chorismate mutase activity assays. The absorbance of the phenylpyruvate was measured in alkaline solution at 320 nm at the indicated time points (min). c SsCm1 has no effect on the chorismate mutase activity of GmCms in vitro. The equal amount of recombinant GmCms-HA was incubated with GST or SsCm1ΔSP-HA, respectively, and the chorismate mutase activity assays were performed according to the methods in (b). For b, c, data represent the mean ± SD (n = 3 biological replicates), source data are provided as a Source Data file.
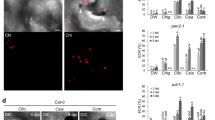
To identify potential plant targets of SsCm1, we used SsCm1 as a bait to screen a soybean cDNA library and identified 11 SsCm1-interacting protein candidates (Fig. 4a). Interaction with Multiple Organellar RNA Editing Factor 2 (GmMORF2a) was detected with the highest frequency and selected for further studies. The interaction between full-length GmMORF2a and SsCm1 was confirmed by Y2H (Fig. 4b). Subcellular localization experiments showed that SsCm1, when expressed alone, was localized to the cytoplasm, nucleus, and a few chloroplasts, but when SsCm1 and GmMORF2a were co-expressed, SsCm1 was co-localized with GmMORF2a within chloroplasts (Fig. 4c). Subsequent BiFC and Co-IP assays confirmed that SsCm1 interacted with GmMORF2a in chloroplasts (Fig. 4d, e).
a Plant proteins interacting with SsCm1 identified by a Y2H screen. The table includes the gene ID and description of the identified proteins, and their frequency of occurrence in the screening. b Detection of the interaction between SsCm1 and the full-length GmMORF2a protein by Y2H. SD-L/W SD without leucine and tryptophan, SD-L/W/H/A SD without leucine, tryptophan, histidine, and adenine. The pGBKT7-53 with pGADT7-T is used as a positive control, and pGBKT7-Lam with pGADT7-T is used as a negative control. Each yeast colony represents at least five similar results. c Subcellular localization and co-localization of SsCm1 and GmMORF2a. SsCm1-RFP and GmMORF2a-GFP were expressed into N. benthamiana, respectively, or co-expressed in N. benthamiana. d BiFC assays based on SsCm1 and GmMORF2a in N. benthamiana. SsCm1-cYFP and GmMORF2a-nYFP as well as the negative controls (SsCm1-cYFP and nYFP; cYFP and GmMORF2a-nYFP) were co-expressed into N. benthamiana, respectively. e Confirmation of the interaction between SsCm1 and GmMORF2a by Co-IP assays. SsCm1-FLAG and GmMORF2a-GFP as well as the negative controls (SsCm1-FLAG and GFP) were co-expressed into N. benthamiana, respectively. The protein samples were immunoprecipitated with Anti-GFP Affinity Beads, and immunoblotted with anti-GFP antibody and anti-FLAG antibody. Blots were stained with CBB. The protein band positions of GmMORF2a-GFP are marked with asterisks. f–h Confirmation of the interaction between SsCm1 and MORF2s in planta through split-luciferase and Co-IP. For f, g, the cLuc-SsCm1 and SsVSP25-nLuc combination was used as a negative control. For h, the SsCm1-FLAG and GFP combination was used as a negative control. The protein band positions of GmMORF2a-GFP and SsCm1-FLAG are marked with asterisks and #, respectively. For c, d, Chl, Chlorophyll b autofluorescence. The solid triangles represent the typical chloroplast co-localization. Scale bar, 25 µm. Uncropped blots are provided in Supplementary Fig. 18.
MORF2s are highly conserved chloroplast proteins encoded by nuclear genome44. There are two copies of MORF2s in soybean (GmMORF2a/b), three copies in N. benthamiana (NbMORF2a/b/c) and only one AtMORF2 in A. thaliana. Here, we confirmed that these MORF2s were localized in chloroplasts (Supplementary Fig. 8). Furthermore, the interaction between SsCm1 and MORF2s were confirmed in planta through split-luciferase and Co-IP assays (Fig. 4f–h). Therefore, SsCm1 conservatively targets MORF2s. To explore whether SsCm1 can also target the host MORF2s when S. sclerotiorum naturally infects, we inoculated the strains expressing SsCm1-GFP on wild-type N. benthamiana, expressing GmMORF2a, and silencing NbMORF2s, respectively. The results showed that SsCm1 was secreted by S. sclerotiorum into plant cells around the infection sites and targeted to chloroplasts in a manner that depends on the content of host MORF2s (Supplementary Fig. 9), suggesting that SsCm1 can target chloroplasts facilitated by host MORF2s.
MORF2s negatively regulates basic immunity and resistance to pathogens
To investigate the role of MORF2 in plant immunity, we silenced all three NbMORF2s genes in N. benthamiana by virus-induced gene silencing (VIGS). The results showed that specific silencing of NbMORF2s resulted in a mild mosaic phenotype (Supplementary Fig. 10a, b). Importantly, silencing NbMORF2s significantly enhanced the resistance of N. benthamiana to UF-1 and B. cinerea, whereas the transient expression of AtMORF2, GmMORF2a, NbMORF2a, and NbMORF2b reduced the resistance (Supplementary Fig. 10c–g). These findings suggest that MORF2s negatively regulates resistance to pathogens in N. benthamiana.
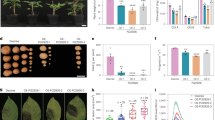
Subsequently, stable 35S:GmMORF2a Arabidopsis overexpressing lines were created. Compared with Col-0, the GmMORF2a-overexpressing lines exhibited normal growth and morphology. In contrast, the knockout mutants of AtMORF2 were either unable to grow in the soil or able to grow slowly with small rosette leaves, consistent with the report that AtMORF2 regulates chloroplast development44 (Fig. 5a). Importantly, the GmMORF2a-overexpressing lines were significantly more susceptible to UF-1, B. cinerea, Δoah mutant, and DC3000, whereas the morf2 mutants were significantly more resistant (Fig. 5a–d). Moreover, compared with Col-0, the GmMORF2a-overexpressing lines showed significantly lower levels in callose deposition, stomatal closing, ethylene production, accumulation of SA, and expression of basic immune marker genes and SA biosynthesis genes induced by PAMPs. Correspondingly, the morf2 mutants exhibited the characteristics of enhancing these immune responses, even in the absence of PAMP induction, suggesting a weak constitutive defense response (Fig. 5e–o). Therefore, our results demonstrate that MORF2 is negatively associated with basic immunity and resistance to pathogens in Arabidopsis.
a, b MORF2s attenuate resistance to fungi in Arabidopsis. Morphology Col-0, morf2-1/2, and 35S:GmMORF2a-1/2 lines at 3 weeks of growth and disease symptoms of plants challenged with UF-1, Δoah, or B. cinerea (a). Centre lines show the medians; box limits indicate the 25th–75th percentiles; whiskers extend to 1.5×the interquartile range from the 25th–75th percentiles; the top lines represent maxima and the bottom lines represent minima; all points (n = 36, n represents the total number of measured lesions with 3 biological replicates) are overlaid (b). c, d MORF2s attenuate resistance to DC3000 in Arabidopsis. Leaves were syringe-infiltrated with DC3000 (OD600 = 0.0005). Disease symptoms (c) and bacterial growth quantification (d) at 3 dpi. *p < 0.05 and **p < 0.01 (one-way ANOVA with multiple comparisons test, compared to Col-0; mean ± SD; n = 4 biological replicates). e, f MORF2s attenuate callose deposition induced by PAMPs in Arabidopsis. Leaves were infiltrated with water, 1 µM flg22, 200 µg/mL chitin, or 50 µg/mL SCFE, and aniline blue staining was performed at 24 hpi. MORF2s attenuate stomatal closure (g) and ethylene production (h) induced by PAMPs in Arabidopsis. Leaves were treated with water or PAMPs for 6 h, stomatal aperture was calculated and ethylene emission was measured. i–n MORF2s attenuate the expression of defense-related genes induced by PAMPs in Arabidopsis. Leaves were treated with water or PAMPs for 12 h and the expression of the immune marker genes was normalized to the levels of AtActin2 transcript. Data are presented as fold induction relative to the expression before treatment, which is set to baseline of 1. o MORF2s attenuate SA biosynthesis induced by PAMPs in Arabidopsis. Leaves were treated with water or PAMPs for 12 h and total SA content was quantified. For b, f–o, data were analyzed by two-way ANOVA with Sidak’s multiple comparisons test, and data points with different letters indicate significant differences of p < 0.05. For f–o, data represent the mean ± SD (n = 3 biological replicates). Source data are provided as a Source Data file.
Given the negative effects of MORF2s on plant immunity in N. benthamiana and A. thaliana, we also generated GmMORF2a and GmMORF2b loss-of-function mutants using CRISPR/Cas9 technology at two different targets. DNA sequencing identified two putative null mutants, Gmmorf2a/b-1 and Gmmorf2a/b-2, which harbored non multiple of 3-bp deletions in the first exon of GmMORF2a and GmMORF2b, causing frameshifts and premature termination of protein translation (Supplementary Fig. 11a). Morphological analysis showed that the Gmmorf2a/b mutants displayed dwarfish plants and spontaneous lesion mimic phenotypes on leaves (Supplementary Fig. 11b). Spontaneous lesion mimic mutants are usually accompanied by spontaneous cell death without pathogen infection and enhanced disease resistance45. Consistent with this, the Gmmorf2a/b mutants significantly enhanced resistance to UF-1 (Supplementary Fig. 11c, d). The results suggest that MORF2s also negatively regulates resistance to S. sclerotiorum in soybean.
To investigate the functional relevance of SsCm1 and MORF2, NbMORF2s-silenced N. benthamiana, Arabidopsis morf2 mutants, and soybean Gmmorf2a/b mutants were used in inoculation assays. In NbMORF2s-silenced plants, the ΔSsCm1-17 mutant no longer exhibited weakened virulence compared with the UF-1, and the transient expression of SsCm1 caused no enhanced sensitivity of N. benthamiana to UF-1 (Supplementary Fig. 12a–d). Similarly, the ΔSsCm1-17 mutant exhibited no reduced virulence compared with the UF-1 in the Arabidopsis morf2 mutants or soybean Gmmorf2a/b mutants (Supplementary Figs. 11c, d and 12e, f). These results indicate that SsCm1 depends on MORF2s to promote virulence in plants. Taken together, SsCm1 promotes infection by targeting the chloroplast development related protein MORF2s, which functions as a negative regulatory factor for basic immunity and resistance to pathogens.
SsCm1 stabilizes MORF2s from degradation during immune activation
AtMORF2 was down-regulated at transcriptional level during avirulent effector AvrRps4-activated ETI and MPK3/MPK6 were constitutively activated2,15. Reminiscent of the negative regulatory role of MORF2s in immunity, we surmise that MORF2s function is actively abolished during immune activation. In support of this hypothesis, the transcript of AtMORF2 was significantly down-regulated in response to PAMPs and pathogens infection (Supplementary Fig. 13a, b). Similarly, the transcript of NbMORF2s was significantly decreased following treatment with SCFE and UF-1 infection. Additionally, the transcript of GmMORF2a was also decreased after inoculation with UF-1 in soybean (Supplementary Fig. 13c, d). These results indicate that MORF2s are down-regulated at the transcriptional level during immune activation.
Interestingly, the co-expression of GmMORF2a-GFP and SsCm1-RFP showed stronger GFP fluorescence signal than that of GmMORF2a-GFP expressed alone (Fig. 6a, b). To more directly examine the effect of SsCm1 on MORF2s stability, we compared the accumulation of six MORF2s-GFP proteins in N. benthamiana co-expressed with SsCm1 or SsVSP25, another S. sclerotiorum effector that also targets chloroplasts but not interacting with GmMORF2a-GFP. The results showed that the six MORF2s-GFP proteins accumulated to much higher levels in the presence of SsCm1 than that of the co-expression with SsVSP25, while GmPETE, another SsCm1-interacting protein, accumulated at the similar level when co-expressed with SsCm1 or SsVSP25 (Fig. 6c). Furthermore, the cell-free degradation assays also confirmed that MORF2s would be degraded and SsCm1 could alleviate the degradation (Fig. 6d). There results suggest that MORF2s are unstable, while SsCm1 can specifically stabilize the MORF2s to promote their accumulation.
a, b SsCm1-RFP expression enhances the fluorescence signal of GmMORF2a-GFP in N. benthamiana. c SsCm1 stabilizes MORF2s to promote protein accumulation. MORF2s-GFP or GmPETE-GFP co-expressed with SsCm1-FLAG or SsVSP25-FLAG in N. benthamiana. d Cell-free assay shows His-SsCm1 inhibits MORF2s degradation. His-SsCm1 or His-SsVSP28 was purified and co-incubated with total protein extracted from N. benthamiana expressing GmMORF2a-GFP or AtMORF2-GFP for indicated times. e SsCm1 stabilizes GmMORF2a from degradation during immune activation. Leaves of N. benthamiana expressing GmMORF2a-GFP (Control) or co-expressed with SsCm1-FLAG were treated 36 h after agroinfiltration. The treatments include infiltration with H2O, 1 µM flg22, 200 µg/mL chitin, or 50 µg/mL SCFE for 30 min, and inoculation with DC3000 (OD600 = 0.0005), 2 × 106/mL spore suspension of B. cinerea, or mycelia of ΔSsCm1-17 for 3 h. f MORF2s were rapidly degraded after SCFE or Flg22 treatments. SsCm1 secreted by S. sclerotiorum inhibits endogenous AtMORF2 (g) and GmMORF2s (h) degradation. Leaves of Col-0 or soybean WT were treated indicated treatment and time. The treatments include infiltrated with H2O, SCFE of UF-1 or ΔSsCm1-17, and inoculation with mycelia of UF-1 or ΔSsCm1-17. i SsCm1 inhibits MORF2s degradation in chloroplasts. Chloroplast proteins from Col-0 or soybean WT were incubated with His-SsCm1, His-SsVSP28, SCFE of UF-1 or ΔSsCm1-17 for 2 h, respectively. Chloroplast proteins were probed with α-Rubisco large subunit (α-RbcL) and served as loading controls. j, k SsCm1 secreted by S. sclerotiorum inhibits GmMORF2a-GFP degradation in chloroplasts of N. benthamiana. For a, j, the solid triangles represent the typical chloroplast co-localization. Scale bar, 25 µm. For b, k, data were analyzed by two-tailed Student’s t test (*p < 0.05 and ****p < 0.0001). Error bars represent SD (n = 4 biological replicates). For c–h, blots stained with CBB served as loading controls. For c–i, numbers below the blot indicate relative abundances of the proteins quantified using ImageJ, the relative abundances when co-expressed with SsVSP25 (c), before incubation (d), H2O treatment (e, f), before treatments (g, h), and incubated with His-SsCm1 (i) are set to baseline of 1, respectively. Source data are provided as a Source Data file.
Previous study showed that AtMORF2 protein was reduced by about 90% in senescent leaves46. To explore the stability of the GmMORF2a protein during immune activation, we treated N. benthamiana leaves transiently expressing GmMORF2a-GFP with the three types of PAMPs (Flg22, Chitin, and SCFE) and three different pathogens (B. cinerea, ΔSsCm1-17 mutant, and DC3000). The results showed that, compared to the control treatment, the GmMORF2a-GFP protein degraded rapidly after PAMPs or pathogens treatments, particularly during the ΔSsCm1-17 treatment. Importantly, co-expression of SsCm1 significantly weakened the degradation of the GmMORF2a-GFP induced by PAMPs or pathogens treatments (Fig. 6e). Likewise, the other five MORF2s proteins were rapidly degraded after SCFE treatment, and NbMORF2a and AtMORF2 were also degraded following induction by Flg22, whereas the GmPETE protein level was not affected by SCFE treatment (Fig. 6f). These results indicate that MORF2s are rapidly degraded during immune activation, and SsCm1 can inhibit this degradation.
To investigate the stability of endogenous MORF2 proteins during immune activation, we synthesized the specific antibody of MORF2 and confirmed its applicability (Supplementary Fig. 14). Short-term treatment of A. thaliana Col-0 with SCFE of S. sclerotiorum UF-1 caused a slight decrease in the amount of endogenous MORF2 protein, while treatment with SCFE of the ΔSsCm1-17 mutant caused a more significant decrease. Moreover, UF-1 inoculation also resulted in less degradation of endogenous MORF2 protein than that of the ΔSsCm1-17 mutant inoculation (Fig. 6g). Likewise, the detection of endogenous MORF2s protein in soybean showed similar results (Fig. 6h). To confirm that SsCm1 secreted by S. sclerotiorum could reduce the degradation of MORF2s in chloroplasts, we incubated the extracted chloroplastic proteins with SCFE from UF-1 and the ΔSsCm1-17 mutant, respectively. The results of western blot revealed that compared with SCFE of the ΔSsCm1-17 mutant, SCFE of UF-1 could retain higher MORF2s protein level in chloroplasts, indicating that SsCm1 secreted by UF-1 could also inhibit the degradation of MORF2s in chloroplasts, similar to purified SsCm1, although the inhibition effect was not as strong as that of the purified SsCm1 (Fig. 6i). The fluorescence results also exhibited that inoculation with S. sclerotiorum could significantly reduce the content of GmMORF2a-GFP in chloroplasts, while SsCm1 secreted by UF-1 could significantly inhibit this reduction (Fig. 6j, k). These results suggest that endogenous MORF2s in plants can also be degraded after immune stimulation, and the physiological level of SsCm1 secreted by S. sclerotiorum in the early stage of infection can inhibit the degradation of MORF2s in chloroplasts to some extent.
SsCm1 and MORF2s inhibit the accumulation of ROS and cell death
When evaluating the effects of SsCm1 and GmMORF2a on various immune outputs, we found that the overexpression of SsCm1 and GmMORF2a not only widely inhibited basic immunity, but also inhibited N. benthamiana cell death triggered by different cell death inducing factors, including the constitutively active variant of AtMKK5 (AtMKK5DD, abbreviated as DD)47, the necrosis-inducing effector of S. sclerotiorum (SsNE2)48, and the Phytophthora elicitor (INF1)49, whereas SsCm1 and GmMORF2a could not induce cell death by themselves (Supplementary Fig. 15a). We observed that overexpression of GmMORF2a could significantly alleviate the level of cell death, while overexpression of SsCm1 slightly weakened cell death, whereas no significant difference in the score of hypersensitive response (HR) compared with GFP control (Supplementary Fig. 15b–d). Similarly, overexpression of NbMORF2a and AtMORF2 significantly alleviated cell death. Considering that SsCm1 targets MORF2s in chloroplasts, we generated a chimera cTP-SsCm1 (chloroplast targeting peptide of NbMORF2b added to the N-terminal of SsCm1-GFP) (Supplementary Fig. 16). Overexpression of cTP-SsCm1 significantly inhibited cell death, comparable to the overexpression of MORF2s. Importantly, after silencing NbMORF2s, cTP-SsCm1 lost its ability to inhibit cell death (Supplementary Fig. 15a–d). These results suggest that MORF2s is a general suppressor of cell death, and SsCm1 inhibits cell death by targeting MORF2s in the chloroplast. Furthermore, 3,3′-diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) staining of leaves prior to visible cell death also confirmed that overexpression of MORF2s or SsCm1 inhibited the accumulation of ROS before triggering cell death. It is worth noting that SsCm1-GFP can effectively inhibit the accumulation of ROS, suggesting that SsCm1 in planta can reduce the accumulation of ROS and delay the subsequent cell death (Supplementary Fig. 15e, f).
To evaluate the effects of SsCm1 and MORF2s on cell death in A. thaliana, dexamethasone-induced expression of DD in Arabidopsis transgenic plants leading to constitutive activation of MAPK3/650 were used to hybridize with morf2 mutant lines, GmMORF2a-overexpressing lines, and SsCm1-overexpressing lines, respectively. We found that overexpression of GmMORF2a or SsCm1 in DD background (DD/GmMORF2a or DD/SsCm1) inhibited cell death and ion leakage after dexamethasone treatment, and inhibited ROS accumulation prior to visible cell death (Fig. 7a–c). Moreover, up-regulated expression of defense genes PR1, PR2, and PAD4 caused by constitutive activation of MAPK3/6 was also weakened in DD/GmMORF2a and DD/SsCm1 plants, but was enhanced in DD/morf2-1 plants (Fig. 7d). These results suggest that overexpression of GmMORF2a or SsCm1 inhibits ROS accumulation and cell death induced by MAPK3/6 constitutive activation.
Overexpression of GmMORF2a or SsCm1 in DD (DEX:AtMKK5DD) inhibits cell death (a, upper), ROS accumulation (a, b), ion leakage (c), and up-regulation of defense genes (d) after dexamethasone (DEX) treatment. For cell death and ion leakages, 15 mΜ DEX was infiltrated into one half of a leaf and kept in light for 36 h. For ROS accumulation, DEX was infiltrated into one entire leaf and kept in light for 18 h, and cellular H2O2 and superoxide were visualized by DAB and NBT staining, respectively (a, middle and bottom), the fluorescence intensities were measured by microplate reader after vacuum-infiltrated with CellROX for 5 min (b). For quantify transcription levels, DEX was infiltrated into the entire leaves, and kept in light for 8 h. Overexpression of GmMORF2a or SsCm1 inhibits cell death (e) and ion leakages (f) induced by Pst-AvrRpt2. Pst-AvrRpt2 (OD600 = 0.02) was infiltrated into one half of a leaf, kept in light for 18 h. g Overexpression of GmMORF2a or SsCm1 inhibits ROS accumulation induced by pathogen. Pst-AvrRpt2 (OD600 = 0.02) or MgCl2 was infiltrated into leaves, and mycelia of UF-1 or PDB was inoculated onto the leaves, kept in light for 6 h and the fluorescence intensities of CellROX were measured. Effects of deleting SsCm1 on the ion leakage (h) and ROS accumulation (i) of infected areas of leaves during S. sclerotiorum infection. The PDB or pure hyphae of Δoah or Δoah/ΔSsCm1-9 was inoculated onto leaves, and the area of inoculation site after designated time was collected for measurement. Data (mean ± SD) were analyzed by two-way (b, d, g–i) or one-way ANOVA (c, f) with multiple comparisons test, data points with different letters indicate significant differences of p < 0.05 (b, d, g), or with ns = no significance, *p < 0.05, **p < 0.01, and ****p < 0.0001 (compared to DD or Col-0 for c, f; comparison between Δoah and Δoah/ΔSsCm1-9 for h, i). N = 4 biological replicates (b, g, i) or 3 biological replicates (c, d, f, h). Source data are provided as a Source Data file.
Chloroplast ROS accumulation and cell death is necessary for ETI and robust plant immunity15. Therefore, we evaluated the levels of ROS and cell death during infiltration with DC3000 carrying AvrRpt2 (Pst-AvrRpt2), a DC3000 strain carrying the avirulence effector (AvrRpt2) recognized by RPS2. Here, cell death, ion leakage, and ROS accumulation induced by high titer Pst-AvrRpt2 inoculation were weakened in both GmMORF2a-overexpressing lines and SsCm1-overexpressing lines (Fig. 7e–g). Similarly, overexpression of GmMORF2a or SsCm1 also alleviated ROS accumulation upon UF-1 treatment (Fig. 7g). These results suggest that MORF2 and SsCm1 negatively regulate the accumulation of ROS and cell death during pathogens infection.
We further investigated the role of SsCm1 in inhibiting plant cell death and ROS accumulation during the early infection of S. sclerotiorum. The inoculation experiments were carried out with weakly virulent strains Δoah and Δoah/ΔSsCm1-9 to avoid the strong pathogenicity of the UF-1. By measuring the ion leakage in the inoculation areas, it was found that strain Δoah caused significantly less ion leakage than strain Δoah/ΔSsCm1-9 (Fig. 7h). Consistently, in the early stage of infection, strain Δoah also caused less ROS accumulation than strain Δoah/ΔSsCm1-9 (Fig. 7i). These findings confirmed that S. sclerotiorum effectively suppressed host cell death and ROS accumulation through secreting SsCm1 in the early infection.
SsCm1 and MORF2s alleviate photosynthetic inhibition
MORF2s and SsCm1 inhibit the accumulation of ROS and cell death caused by MAPK3/6 constitutive activation, and MORF2s, as chloroplast protein, participates in chloroplast development and the maturation of RNA encoding photosynthetic elements44, which reminds us that MORF2s and SsCm1 may play roles in maintaining the efficient operation of photosynthetic apparatus to alleviate photosynthetic inhibition, which is necessary for ROS accumulation and cell death15. In support of this hypothesis, upon MPK3/MPK6 activation, the decrease of maximal PSII activity parameter Fv/Fm is significantly weakened in DD/GmMORF2a and DD/SsCm1 plants, and is further reduced in DD/morf2-1 plants (Fig. 8a). Additionally, the dramatic inhibition of photosynthesis related genes LHCB1.2, PETE, and PsbA after MPK3/MPK6 activation was also significantly alleviated in DD/GmMORF2a and DD/SsCm1 plants (Fig. 8b). Consistent with the finding that overexpression of GmMORF2a or SsCm1 inhibited the accumulation of ROS and cell death induced by Pst-AvrRpt2 inoculation, overexpression of GmMORF2a or SsCm1 also alleviated the decrease of Fv/Fm and the down-regulation of photosynthesis-related genes induced by Pst-AvrRpt2 inoculation, which is necessary for HR cell death in plant immunity15 (Fig. 8c–e). Importantly, inoculation with strain Δoah of S. sclerotiorum showed a slower rate of Fv/Fm decline than inoculation with strain Δoah/ΔSsCm1-9 (Fig. 8f), suggesting that physiological level of SsCm1 secreted by S. sclerotiorum could delay the photosynthetic inhibition caused by S. sclerotiorum infection. Similarly, SCFE treatment, as well as UF-1 and DC3000 inoculation also induced the decrease of Fv/Fm and the down-regulation of photosynthesis-related genes27, while overexpression of GmMORF2a or SsCm1 significantly alleviated these decreases, which were enhanced in morf2 mutants (Fig. 8g–j and Supplementary Fig. 17). These results suggest that GmMORF2a and SsCm1 alleviate immunization induced photosynthetic inhibition, which may lead to the inhibition of ROS accumulation and cell death.
Overexpression of GmMORF2a or SsCm1 in DD (DEX:AtMKK5DD) alleviates the decrease of Fv/Fm (a) and photosynthesis-related gene expression (b) after DEX treatment. 15 μM DEX was infiltrated into leaves and kept in light for 18 h to measure Fv/Fm and for 8 h to quantify gene transcript levels, EtOH was used as the solution control. c Overexpression of SsCm1 or GmMORF2a alleviates the decrease of Fv/Fm induced by Pst-AvrRpt2. Pst-AvrRpt2 (OD600 = 0.2) was infiltrated into leaves and Fv/Fm was measured in indicated periods of time. Overexpression of SsCm1 (d) or GmMORF2a (e) alleviates the decrease of photosynthesis-related genes induced by Pst-AvrRpt2. Pst-AvrRpt2 (OD600 = 0.2) or MgCl2 was infiltrated into leaves and gene transcript levels were quantified after 4 h. f Effects of deleting SsCm1 on the Fv/Fm of infected areas of leaves during S. sclerotiorum infection. The PDB or pure hyphae of Δoah or Δoah/ΔSsCm1-9 was inoculated onto leaves, and the area of inoculation site after designated time was collected for measuring the Fv/Fm. Overexpression of SsCm1 (g, i) or GmMORF2a (h, j) alleviates the decrease of Fv/Fm (g, h) and photosynthesis-related gene expression (i, j) induced by SCFE and UF-1. 50 µg/mL SCFE or PDB was infiltrated into leaves for 24 h to measure Fv/Fm and for 12 h to quantify gene transcript levels. Mycelia of UF-1 was inoculated onto the leaves for 6 h to measure Fv/Fm and for 4 h to quantify gene transcript levels. Data (mean ± SD) were analyzed by two-way (b, d–j) or one-way ANOVA (a) with multiple comparisons test, data points with different letters indicate significant differences of p < 0.05 (b, d, e, g–j), or with ns = no significance, **p < 0.01, and ****p < 0.0001 (compared to DD for a; comparison between Δoah and Δoah/ΔSsCm1-9 for f). N = 4 biological replicates (a, c, f–h) or 3 biological replicates (b, d, e, h–j). Source data are provided as a Source Data file.
Discussion
To cope with inevitable abiotic stresses and numerous invaders, plants have evolved mechanisms that sacrifice growth by consuming energy to build defensive barriers, adapt to environmental change, and eliminate invaders51,52. Chloroplast, as an integrator that perceives the environment, needs to dynamically switch the photosynthetic apparatus on and off to achieve a proper growth-defense tradeoff9. Firstly, chloroplasts actively down-regulate53 and reduce the processing of RNA encoding genes in photosynthesis-related apparatus54, control the routes of chloroplast protein import55, phosphorylate chloroplast proteins to inhibit electron transport rate56,57, reduce cyclic electron flow (CEF), reduce NPQ, and degrade PSII15,58, and finally inhibit photosynthesis to reduce energy obtained by other cellular activities and pathogens and prioritize energy investments. Secondly, when the threats are lethal, a robust defense, but not growth, is of the highest priority. Chloroplasts are also the main source of ROS. They actively disrupt the stoichiometry of photosynthetic proteins, degrade PSII, and change the redox state of the plastoquinone pool, resulting in the excessive production and accumulation of ROS, which eventually triggers a cell death that resembles the effector-triggered HR that limits the growth of pathogens17,23,24,59,60,61. Thirdly, 1O2 and tetrapyrrole are used as RS molecules to drive transcriptional reprogramming, which increases SA biosynthesis and nuclear-encoded defense gene expression, resulting in extensive basic immune output and ROS burst6,7,17,23,24,60. However, there are still confused issues about how chloroplasts can quickly and accurately achieve this transition from growth to defense and which components are responsible for this transition. Here, we found that plants fine-tune chloroplasts transitioning from photosynthetic state to immune state by reducing the level of MORF2s. Overexpression of MORF2 alleviates the immunization-induced photosynthetic inhibition and down-regulation of photosynthesis related genes and inhibits the accumulation of ROS and the subsequent cell death. In the meantime, MORF2 overexpression inhibits the output of various basic immunity and SA biosynthesis, leading to a weakened resistance to pathogens, while the corresponding loss of MORF2 shows the opposite phenomenon. In addition, the rapid degradation of MORF2s proteins and down-regulation of MORF2s expression during immune activation emphasizes that plants can spontaneously turn on the immune activation of chloroplasts. Importantly, S. sclerotiorum has evolved a strategy to subvert this process by secreting effector SsCm1 to stabilize MORF2s, effectively maintaining chloroplasts in a growth state and thereby facilitating infection.
AtMORF2 was originally characterized as an RNA editing factor of chloroplasts44, and the mitochondrial NbMORF8 was implicated as a negative regulator of immunity through the regulation of ROS accumulation and SA signaling62. OVEREXPRESSOR OF CATIONIC PEROXIDASE3 negatively regulates immunity and fungal resistance through controlling chloroplast NADH dehydrogenase (ndh) transcript editing54. Here, we find that MORF2s also negatively regulate immunity, possibly by mediating RNA editing of certain chloroplast proteins. Moreover, AtMORF2 may also be implicated in RS. AtMORF2 acts as chaperone that directly promotes the accumulation of several tetrapyrrole biosynthetic enzymes and participates in tetrapyrrole biosynthesis (TBS)46,63,64. This is reminiscent of PROGRAMMED CELL DEATH8, which negatively regulates RS, ROS accumulation and cell death by interacting with TBS enzymes65. RS promotes the expression of nuclear-encoded defense genes and SA synthesis12. AtMORF2 also directly interacts with GUN1 to participate in RS66 and several genome uncoupled (gun) mutants are susceptible to pathogens6. Consistent with the involvement of MORF2 in RS66, our results showed that overexpression of GmMORF2a or SsCm1 alleviates the down-regulation of immunization-induced RS marker gene LHCB1.2 (Fig. 8b, d, e, i, j). Therefore, we speculate that MORF2s inhibited immunization-induced RS by affecting TBS or directly interacting with GUN1, which could lead to the alleviation of photosynthetic inhibition, ROS accumulation, SA biosynthesis, basic immune output, and cell death, while SsCm1 inhibited RS by stabilizing MORF2s. Further research is needed to explore how MORF2s inhibits RS and how RS affects immunity.
Consistent with MORF2s functioning as negative regulators of plant immunity, the expression of MORF2s is suppressed during immune activation, which is similar to OCP3 and chloroplast RNA editor CRR2154,67, emphasizing that plants actively down-regulate immune negative regulators as a defense strategy. Similarly, stresses also regulate the degradation and input of chloroplast proteins through ubiquitination, protease, autophagy, senescence-associated vacuoles, and chloroplast vesiculation55,56,68,69,70,71,72,73, and the instability of AtMORF2 protein has been reported46,64. Here, we found that MORF2s were degraded within 30 min following immune activation, suggesting that plants directly activate unknown degradation systems to degrade MORF2s after sensing pathogen signals. Consistently, the pathogen-triggered rapid editing inhibition in ndh transcripts leads to NDH destabilization and subsequent inhibition of CEF54. This suggests that the rapid degradation of chloroplast editing factors may be ubiquitous, as such, the degradation of MORF2s may promote the inhibition of extensive RNA editing, thereby transitioning chloroplasts from the photosynthetic state to the immune state. These findings are consistent with the recent discovery, showing that MORF8 forms condensates under heat stress and recruits RNA editor elements, such as MORF2, to inhibit RNA editing and reduce photosynthesis74, implicating a common phenomenon, i.e., the reduction of the function of MORFs, such as the degradation of MORF2 and the formation of condensates of MORF8, decreases the level of photosynthesis under stress. To further reveal the key factors responsible for MORF2s degradation and the mechanism of immune signal-induced MORF2s degradation will provide new insights for understanding how chloroplasts balance photosynthesis and defense.
Correspondingly, S. sclerotiorum evolved effector SsCm1 to promote infection by directly targeting and stabilizing MORF2s to prevent chloroplasts from entering immune state. This is reflected in the phenotypic similarity between overexpressed SsCm1 and overexpressed GmMORF2a, which both alleviated immune-induced photoinhibition and inhibited ROS accumulation, cell death, basic immunity and pathogen resistance. In addition, the physiological level of SsCm1 secreted by S. sclerotiorum could also alleviate photoinhibition and inhibit ROS accumulation and cell death during infection. Crucially, the promotion of infection by SsCm1 depends entirely on MORF2s, and the virulence function of SsCm1 is lost after silencing or losing MORF2s, which further emphasizes that MORF2s are the physiological targets of SsCm1. Notably, based on the analysis of conserved domains, SsCm1 has always been regarded as the homologs of U. maydis effector Cmu134,35,40, a classical effector that relies on the activity of Cm and the interaction with ZmCm2, and in turn mediates metabolic priming, especially lowering the available substrate for SA biosynthesis39. However, we demonstrated in yeast complementation experiments and in vitro enzyme activity experiments that SsCm1 could not restore the growth of yeast aro7 mutant like Cmu1, nor did it have enzyme activity, indicating that SsCm1 is an enzymatically nonfunctional effector and has adopted different strategies to suppress plant immunity. This is different from the reported Cm effectors, which all have enzymatic activity39,75,76. A careful sequence alignment of SsCm1 and other Cms showed that SsCm1 has obvious differentiation in several reported essential amino acids for enzyme activity, such as A93 and V149 of SsCm1 are K and T respectively in the Cms with enzyme function, and also showed differences in many conserved amino acids39,43. The AlphaFold prediction also showed that the three-dimensional structure of SsCm1 is obviously different from the Cms with enzyme function, suggesting that the structural variation caused by the differentiation of key amino acids in enzyme activity may be the reason why SsCm1 loses its enzyme function. Notably, all known enzymatic functional Cm effectors are derived from biotrophic organisms39,75,76, which is different from SsCm1 from S. sclerotiorum, a necrotrophic or hemi-necrotrophic pathogens. Interestingly, the hypothesized Cm effectors are rare in necrotrophs and saprophytes, and is only identified in S. sclerotiorum39. We speculate that there are two possibilities for this difference in distribution of Cm effectors: (1) Metabolic priming by Cm effectors may lead to adverse reactions that are not conducive to necrotrophs, such as the accumulation of some harmful metabolites39,77 or the lack of preferred metabolites; (2) There is no urgent need for Cm effectors to inhibit certain specified plant defense responses in the pathogenic process of necrotrophs78, so the evolutionary selection pressure does not force necrotrophs to evolve enzymatic functional Cm effectors. Further detailed and comprehensive analysis of the evolution of Cm effectors will probably reveal the mystery. We noticed that SsCm1 does not contain the recognizable cTP, but it can be partially localized in chloroplasts, and artificially expressing SsCm1 in chloroplasts can more effectively inhibit elicitor-induced ROS accumulation and cell death, indicating that SsCm1 mainly functions in chloroplasts. This raises a question, how does SsCm1 target chloroplasts? Considering SsCm1 significantly increased chloroplast localization when co-expressed with MORF2, we speculate that SsCm1 may be migrated to chloroplast through interaction with MORF2. In addition, we cannot rule out the possibility that SsCm1 may enter chloroplasts through non-classical input mechanism or interaction with other chloroplast proteins6, and the explicit mechanism underlying the transportion of SsCm1 into chloroplasts needs further study.
Overall, we propose that when plants sense PAMPs or pathogens, they trigger unknown degradation systems to rapidly degrade MORF2s, which damages the RNA editing process of chloroplast proteins and TBS pathway, leading to photosystem damage, photosynthesis inhibition, ROS accumulation, triggering of RS, and transition of chloroplast from growth state to immune state. The RS is transmitted to the nucleus to promote the up-regulation of defense genes, such as SA biosynthesis genes, and down-regulation of PhANGs, ultimately leading to the synthesis of SA and the output of basic immunity, further strengthening the active inhibition of photosynthesis and the accumulation of ROS, and finally causing cell death and disease resistance (Fig. 9a). Correspondingly, the secreted enzymatically nonfunctional effector SsCm1 of S. sclerotiorum directly targets and stabilizes MORF2s, thus maintaining the photosynthetic state of chloroplasts, which reduces resistance of plants to pathogens (Fig. 9b).
a When plants are infected by S. sclerotiorum Sscm1 mutants, membrane receptors sense PAMPs or pathogens, and quickly triggers unknown degradation systems to rapidly degrade MORF2s, leading to the damage of photosystems, the inhibition of photosynthesis, the accumulation of ROS, the triggering of RS, and the transformation of chloroplast from the growth state to the immune state. RS is transmitted to the nucleus to promote the up-regulation of defense genes, such as SA biosynthesis genes, and down-regulation of photosynthesis genes, which leads to the synthesis of SA and the output of basic immunity, and further strengthens the active inhibition of photosynthesis and the accumulation of ROS, and finally causes cell death and disease resistance. b Correspondingly, the secretion of S. sclerotiorum enzymatically nonfunctional effector SsCm1 directly targets and stabilizes MORF2s, thus maintaining the photosynthetic state and subverting the immune output of chloroplasts, ultimately reducing resistance to pathogens. The thickness of the lines represents the intensity of the events. PhANGs photosynthesis-associated nuclear genes.
Methods
Plant materials and growth conditions
Nicotiana benthamiana wild-type and gene silencing lines, Phaseolus vulgaris, Solanum lycopersicum, and soybean wild-type (Glycine max cv. Williams 82) and the Gmmorf2a/b mutants were maintained at 22 °C in a growth chamber with a 16 h day/8 h night cycle (100–150 μmol photons m−2 s−1).
All Arabidopsis plants used in this study were in Col-0 background. Arabidopsis mutant seeds of morf2-1 (SALK_094930) and morf2-1 (SALK_149307) were obtained from the Nottingham Arabidopsis Stock Centre. The dexamethasone-inducible promoter-driven AtMKK5DD transgenic plants (DD) were reported previously50. Soil-grown Arabidopsis plants were grown in a growth chamber at 22 °C and approximately 70% relative humidity with a 10 h day/14 h night cycle (100–150 μmol photons m−2 s−1). For experiments related to cell death and photosynthesis, the treated Arabidopsis plants were kept for the times indicated in the corresponding experiments under continuous light (100 μmol photons m−2 s−1).
Pathogen strains and inoculation assays
The wild-type strain UF-1 and its derivative strains of S. sclerotiorum were initially grown on Potato Dextrose Agar (PDA) plates cultured at 25 °C. The oxalic acid minus mutant (Δoah 3-6-1) of S. sclerotiorum was reported previously42. Mycelium plugs or pure fresh mycelia were collected from fresh mycelia growing on PDA plates or cultured in Potato Dextrose Broth (PDB) for 2 d, inoculated onto plant leaves, and maintained at high humidity.
The B. cinerea strain B05.10 was used in this study. Mycelium plugs (0.5 cm in diameter) or 2 × 106/mL spore suspension with 0.01% Tween 20 were used to inoculate plant leaves and maintained at high humidity.
Pseudomonas syringae pv. tomato strain DC3000 containing an empty vector (DC3000) or AvrRpt2 (Pst-AvrRpt2) was cultured overnight at 28 °C in LB medium containing 25 μg/mL rifampicin or 25 μg/mL rifampicin with 50 μg/mL kanamycin. For disease evaluation caused by DC3000, the DC3000 strain was resuspended in 10 mM MgCl2 at OD600 = 0.0005. The bacterial suspensions were then infiltrated into 4-to-5-week-old Arabidopsis leaves. The numbers of bacterial cells were determined by colony-forming units per cm2 (CFU/cm2) of leaf tissue at 3 d post-infiltration (dpi) as previously described50. For the experiments related to photosynthesis, Arabidopsis leaves were infiltrated with DC3000 (OD600 = 0.2) or Pst-AvrRpt2 (OD600 = 0.2), and for the experiments related to cell death, Arabidopsis leaves were infiltrated with Pst-AvrRpt2 (OD600 = 0.02).
Transient expression, protein extraction, and western blots analysis
Agrobacterium-mediated transient expression in N. benthamiana and protein extraction were performed as previously described50. Before infiltration, the bacterial suspension carrying different constructs was adjusted to a final OD600 = 0.6. For experiments that required co-infiltration, Agrobacterium suspensions carrying different constructs were mixed at a 1:1 ratio before infiltration. Samples were collected at 36 h post inoculation (hpi) or 48 hpi for analysis based on experimental requirements. For the expression of cell death inducing factors, Agrobacterium expressing AtMKK5DD or INF1 was infiltrated with OD600 = 0.2, and Agrobacterium expressing SsNE2 was infiltrated with OD600 = 0.6.
Plant tissues were frozen and ground in liquid nitrogen and then extracted according to instructions of the Plant Protein Extraction Kit (CW0885, CWBIO) containing 1% (v/v) plant protease inhibitor cocktail (P9599, Sigma). For protein extraction of S. sclerotiorum, 0.2 g of fresh hyphae was used according to the MinuteTM Total Protein Extraction Kit for Microbes with Thick Cell Walls (YT-015, Invent). The supernatant of protein extracts was heated at 70 °C for 10 min in SDS loading buffer (2% (w/v) SDS, 10% (w/v) glycerol, 0.01% (w/v) bromophenol blue and 0.125 M Tris-HCl, 0.3 M DTT, pH 6.8) and loaded on SDS-PAGE acrylamide gels for western blotting. All immunoblots were analyzed using appropriate antibodies as indicated in the figures. Blots were stained with Coomassie Brilliant Blue (CBB) to verify equal loading. Uncropped blots are provided in Supplementary Fig. 18.
RNA isolation and quantitative RT-PCR
Samples for gene expression analysis were frozen and ground in liquid nitrogen and then total RNA purification, reverse transcription, and qRT-PCR were completed using one-step kit (TransGen Biotech). The S. sclerotiorum β-tubulin gene Sstublin, G. max actin gene GmActin, A. thaliana actin gene AtActin2, and N. benthamiana NbEF1α gene were used to normalize transcript levels. The relative expression levels were calculated with three technical replicates using the 2−ΔΔCt method. All primers used for qRT-PCR were listed in Supplementary Data 1.
Secretion assays and production of SCFE
The predicted N-terminal 20-amino acid SP sequences of SsCm1 were fused in frame to the invertase gene in the pSUC2 vector by gene synthesis in GenScript Biotech. The construct containing the SP of effector Avr1b were used as positive controls as previously described79. Recombinant constructs were transformed into yeast and SP secretion confirmation was performed using the Signal peptide secretion yeast detection kit (DZSL1561, Coolaber).
For the detection of SsCm1 secretion by S. sclerotiorum, the strains carrying GFP or SsCm1-GFP were inoculated into PDB in the presence of N. benthamiana leaf discs for 24 h. Culture medium was harvested by filtration through a 75-µm nylon mesh and the supernatant was concentrated by freeze-drying. Total proteins were extracted from mycelia or supernatant, and immunoblotted with rabbit polyclonal anti-GFP antibody (D110008, 1:5000; Sangon). To obtain SCFE, the UF-1 or the ΔSsCm1-17 mutant strain was inoculated into PDB in the presence of 10 µM 1,16-Hexadecanediol (S68032, Yuanye) for 24 h. Culture medium was harvested by filtration through a 75-µm nylon mesh and freeze-drying the concentrated supernatant. The dried material was resuspended in H2O (0.5 g dry weight/mL) and centrifuged twice for 20 min at 10,000 × g and 4 °C to remove insoluble particles. SCFE activity was identified by measurement of ethylene production in A. thaliana leaves following treatment with the SCFE solution as previously described80.
Genetic manipulation and developmental phenotype evaluation in S. sclerotiorum
Gene deletion mutants of the SsCm1 gene in isolate UF-1 and the Δoah strain of S. sclerotiorum were generated using a homologous recombination strategy using the protoplast transformation method81. Transformants were purified by hygromycin selection and were verified by PCR. Uncropped gels are provided in Supplementary Fig. 18. To genetically complement the SsCm1 knockout mutants, a SsCm1-GFP fusion complementation vector based on pYF11 was generated and was transformed into the ΔSsCm1-17 mutant. Transformants were purified using G418 selection and were verified by RT-PCR.
The evaluation of developmental phenotype of S. sclerotiorum was performed as previously described81. The strains were inoculated onto PDA plates for 2 d or 14 d at 25 °C and the dry weight of sclerotia was measured. Glass slides and onion epidermal tissue were used to induce the formation of infection cushions in vitro and in vivo, respectively. The strains cultured on PDA with 0.005% (w/v) bromophenol blue for 36 h were used to evaluate the production of OA. The enzymatic activities of cutinase and cellulase were measured by mycelium according to the previously described82 and the manufacturer’s instructions of the Cellulase test kit (A138, Nanjing Jiancheng), respectively.
Plasmid construction and generation of transgenic and gene silencing plants
To screen SsVSPs, the full-length coding region (SsVSPsFL) or the sequence with the predicted SP removed and artificial start codon added at the N-terminal (SsVSPsΔSP) was cloned into the pCHF1301-3×FLAG vector. Unless otherwise specified, the SsCm1ΔSP was used in this study. To generate the SsCm1-GFP and MORF2s-GFP constructs, the corresponding coding sequences were cloned into pCHF3301-GFP driven by the CaMV 35S promoter. All recombinant constructs were transformed into Agrobacterium GV3101 and then 35S:SsCm1 or 35S:GmMORF2a was transformed in Arabidopsis Col-0 through the floral dipping method. Transgenic Arabidopsis plants were selected using Basta (10 µg/mL). All experiments using transgenic Arabidopsis plants were performed using two independent T4 homozygous lines. The DD/morf2-1, DD/GmMORF2a, and DD/SsCm1 plants were created by crossing the homozygous plants, and homozygous lines were selected and confirmed by PCR.
VIGS assays were performed as previously described50. Briefly, independent cultures of Agrobacterium carrying pTRV1 or pTRV2-based constructs were resuspended in 10 mM MgCl2, 10 mM MES (pH 5.6) and 150 µM acetosyringone at OD600 = 0.8, and incubated for 2 h at 28 °C in the dark. Cultures were mixed at a 1:1 ratio and this suspension was used to infiltrate the stem and underside of cotyledons of 2-week-old N. benthamiana seedlings.
To generate the CRISPR/Cas9-engineered Gmmorf2a/b mutant, gRNAs were designed using the CRISPR-P website (http://crispr.hzau.edu.cn). Two 20-bp DNA fragments coding for the gRNA were inserted into the pCas9-MDC-sgRNA plasmid. Plasmids were individually introduced into Agrobacterium strain EHA105 via electroporation and then transformed into WT soybean (Williams 82) using the cotyledon-node method83. The mutant lines were identified by DNA sequencing. All oligonucleotide primers were listed in Supplementary Data 1.
PAMP treatments and determinations of basic immunity
Except for the luminol-based ROS burst assays, leaves were infiltrated with water (Mock), 1 µM flg22 (abs45152926, Absin), 200 µg/mL chitin (C9752, Sigma), or 50 µg/mL SCFE, and assayed for basic immunity or protein extractions at the designated times were indicated in the corresponding figures. The luminol-based ROS burst assays were performed as previously described50. Briefly, N. benthamiana leaf discs transiently expressing the SsCm1-FLAG or GFP-FLAG (as control) were placed in white 96-well plates (655074, Greiner) with 200 µL water overnight. Then, water was replaced by a solution containing individual PAMPs (100 nM flg22, 50 µg/mL chitin, or 2 µg/mL SCFE), 100 µM luminol (123072, Sigma), and 20 µg/mL HRP (HY-125859, MCE), and luminescence was measured in a microplate reader (PerkinElmer) for 60 or 90 min. For data analysis, both relative luminescence units (RLU) produced every minute and total RLU were plotted.
Callose quantification was performed as previously described5. The leaves of Arabidopsis or N. benthamiana were infiltrated with PAMPs or water using a needleless syringe and covered for 24 h. Chlorophyll was removed by incubating in ethanol (90% v/v). The destained leaves were vacuum-infiltrated with 0.01% aniline blue (S19056, Yuanye) and incubated for 1 h. Then, aniline blue was removed and callose deposits were visualized under UV illumination (excitation, 365 nm; emission, 460 nm) and quantified using ImageJ.
Stomatal aperture measurements were performed as previously described7. Arabidopsis leaves were detached and floated in the incubation buffer (10 mM MES, 10 mM KCl, 50 μM CaCl2) for 2 h at 22 °C with 100 μmol photons m−2 s−1 light. Then, leaves were treated with PAMPs or water and incubated for an additional 6 h. After the incubation, epidermal strips of the leaves were observed under microscopy. The stomatal aperture was calculated as the ratio of the pore width/guard cell length.
Ethylene and SA content were determined as previously described7,80. Leaves were treated with water or PAMPs for 6 h and ethylene emission was measured by gas chromatography. Leaves were treated with water (Mock) or PAMPs for 12 h and total SA content was quantified by high-performance liquid chromatography.
For the determination of defense-related genes expression, leaves of Arabidopsis plants or N. benthamiana were infiltrated with PAMPs or water for 12 h and tissues were snap-frozen. The expression of genes was normalized to the levels of AtActin2 or NbEF1α transcripts and presented as fold induction compared with the expression before treatment which was set as the baseline of 1.
Y2H screening and confirmation
The Y2H assay was used to screen a soybean cDNA library to identify potential targets of SsCm1 and to confirm the specific interaction. The coding sequence of SsCm1 (without the SP, SsCm1ΔSP) was cloned to the bait vector pGBKT7 and transformed into the Y2H Gold strain. The bait strain was mated with the Y187 yeast strain (harboring a soybean Mate & Plate Libraries), and the matings were selected on the synthetic medium (SD) without leucine, tryptophan, histidine, and adenine (SD-L/W/H/A) agar plates. The inserts in the prey vector were confirmed using yeast plasmid sequencing. The potential interactors were subjected to BLAST (http://www.ncbi.nlm.nih.gov/) analyses for identification and confirmation of the correct orientation of the insert sequences. The coding sequence of full-length potential interactors was cloned into the prey vector pGADT7 and co-transformed with pGBKT7-SsCm1ΔSP into Y2HGold yeast cells. All transformations were plated on SD-L/W, SD-L/W/H + X-α-Gal (630463, Clontech), and SD-L/W/H/A + X-α-Gal agar plates. For the quantitative evaluation of the results of Y2H, the transformed single colony with the same size on SD-L/W medium was transferred to the 10 mL liquid SD-L/W/H/A medium, and the OD600 values were determined after shaking culture for 24 h. The pGBKT7-53 with pGADT7-T was used as a positive control, and pGBKT7-Lam with pGADT7-T was used as a negative control.
Yeast complementation and determinations of chorismate mutase activity
The assays of gene functional complementation of ScARO7 in yeast were performed as previously described with modifications39. Briefly, yeast strain Y054679 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YPR060c::kanMX4, Aro7, Euroscarf) was transformed with the corresponding pGADT7 derivatives using standard protocols (Clontech) and tested for growth on SD without leucine, phenylalanine, and tryptophan.
Chorismate mutase activity assays were performed as previously described84. SsCm1ΔSP-HA, SsCm2-HA, GmCms-HA, and UmCmu1ΔSP-HA (positive control) were cloned into pGEX-6P-1 vector, respectively, and the proteins were obtained in Escherichia coli BL21 and purified by glutathione affinity purification (C600327, Sangon). Then, the GST moiety was removed by PreScission protease (C610303, Sangon) and chorismite mutase activity assays were performed. After acidic conversion of prephenate to phenylpyruvate, the reaction was basidified and the extinction at 320 nm was measured. The increase in extinction was plotted against time (in min) to visualize the formation of phenylpyruvate, and purified GST protein was used as a negative control.
Subcellular localization and BiFC assays
For subcellular localization, N. benthamiana leaves expressing GFP- or RFP-fused proteins at 2 dpi were imaged with a Leica confocal microscope (Leica Microsystems) using LAS-X software with the preset settings for GFP (excitation, 488 nm; emission, 500–550 nm), RFP (excitation, 554 nm; emission, 580–630 nm), and chlorophyll autofluorescence (excitation, 488 nm; emission, 630–670 nm). The laser intensity 5% and gain 10% were used to observe the chlorophyll autofluorescence, except that the SsCm1-GFP strains were inoculated onto N. benthamiana. For evaluating fluorescence intensity, gain and laser intensity were set uniformly and quantified using ImageJ.
BiFC assays were performed in N. benthamiana leaves. Agrobacterium mixtures carrying the appropriate BiFC constructs were infiltrated into N. benthamiana leaves. Samples were imaged with a Leica confocal microscope (Leica Microsystems) using LAS-X software with the preset settings for YFP (excitation, 514 nm; emission, 525–575 nm) at 2 dpi.
Co-IP and split-luciferase assays
Co-IP assays were performed as previously described50. Briefly, 1 g of N. benthamiana leaf tissue was collected 2 d post infiltration with Agrobacterium and frozen in liquid nitrogen. Total proteins were extracted with protein extraction buffer (100 mM Tris-HCl pH 8, 150 mM NaCl, 10% glycerol, 5 mM EDTA, 10 mM DTT, 2 mM PMSF, 10 mM NaF, 10 mM Na2MoO4, 2 mM NaVO3, 1%(v/v) NP-40, 1%(v/v) plant protease inhibitor cocktail). Immunoprecipitation was performed with 40 µL of anti-GFP affinity beads 4FF (SA070005, Smart-Lifesciences) incubated for 1 h at 4 °C. The beads were washed 4 times with 1 mL cold wash buffer (100 mM Tris-HCl pH 8, 150 mM NaCl, 10% glycerol, 2 mM DTT, 10 mM NaF, 10 mM Na2MoO4, 2 mM NaVO3, 0.5%(v/v) NP-40, 1%(v/v) plant protease inhibitor cocktail). Finally, the washed beads were resuspended in 100 µL SDS loading buffer and incubated for 10 min at 70 °C. The immunoprecipitated proteins and input proteins were separated on SDS-PAGE gels for western blotting with rabbit polyclonal anti-GFP (D110008, 1:5000; Sangon), rabbit polyclonal anti-FLAG (D110005, 1:2000; Sangon), and goat anti-rabbit IgG-HRP (A9169, 1:5000; Sigma) antibody. Chemiluminescence was imaged by Chemiluminescence Image System (Tanon) and blots were stained with CBB to verify equal loading.
Split-luciferase assays were performed as previously described50. Briefly, Agrobacterium strains containing the desired plasmids were infiltrated into N. benthamiana leaves. CCD imaging was performed at 2 dpi. Leaves were infiltrated with 0.5 mM luciferin (HY-12591B, MCE) and kept in the dark for 5 min before CCD imaging. The images were taken with Chemiluminescence Image System (Tanon).
Protein stability assays
For the protein stability assays in the nonimmunity state, MORF2s-GFP or GmPETE-GFP co-expressed with SsCm1-FLAG or SsVSP25-FLAG in N. benthamiana were used. Total proteins were extracted 48 h after Agrobacterium-infiltration. For the protein stability assays during immune activation, N. benthamiana leaves were expressed with MORF2s-GFP alone (Control) or co-expressed with SsCm1-FLAG, and the leaves were treated 36 h after agroinfiltration. The treatments included infiltration with H2O, 1 µM flg22, 200 µg/mL chitin, and 50 µg/mL SCFE, or inoculation with DC3000 (OD600 = 0.0005), a 2*106/mL spore suspension of B. cinerea, and mycelia of the ΔSsCm1-17 mutant. Total proteins were extracted 30 min after treatments were applied.
For cell free degradation assay, His-SsCm1 or His-SsVSP28 expressed in E. coli BL21 were purified and co-incubated with the total protein extracted from N. benthamiana leaves. After incubation for the indicated times, western blotting was performed.
For evaluate the stability of endogenous MORF2 proteins in plants. Firstly, the anti-MORF2 polyclonal antibodies were custom-developed by QiWei YiCheng Technology. Briefly, the synthesized AtMORF2159-174aa peptide (LPDSYVDPENKDYGAE-C) was conjugated to keyhole limpet hemocyanin carrier to immunize rabbits. The rabbit polyclonal antiserum was purified by affinity chromatography using AtMORF2159-174aa peptide, and the eluate was used as anti-MORF2 antibody. The applicability of antibody was evaluated by detecting MORF2 protein of Col-0, the morf2 lines, and 35S:GmMORF2a-GFP lines (Supplementary Fig. 14). The A. thaliana or soybean were treated according to the indicated treatment measures and times, and then the total protein was extracted.
For evaluate the stability of MORF2 protein in chloroplasts. Density gradient centrifugation was used to extract chloroplasts of Arabidopsis or soybean according to the manufacturer’s instructions of the Chloroplast Extraction Kit (PTE010, Coolaber). Subsequently, chloroplast proteins were extracted and incubated with about 50 µg of purified protein or SCFE for 2 h at 25 °C, then western blotting was performed. Chloroplast protein loading was indicated by detecting the protein content of rubisco large subunit (52.7 kDa, RbcL) without actin.
All protein samples were separated on SDS-PAGE gels for western blotting with mouse monoclonal anti-GFP (D191040, 1:5000; Sangon), rabbit polyclonal anti-FLAG (D110005, 1:2000; Sangon), mouse monoclonal anti-His (CW0286M, 1:1000; CWBIO), rabbit polyclonal anti-MORF2 (1:1000, this work), rabbit anti-RbcL (AS03037, 1:10000; Agrisera), rabbit polyclonal anti-actin (AS132640, 1:5000; Agrisera), goat anti-mouse IgG-HRP (D110087, 1:5000; Sangon), or goat anti-rabbit IgG-HRP (A9169, 1:5000; Sigma) antibodies. Chemiluminescence was imaged by Chemiluminescence Image System (Tanon) and blots were stained with CBB to verify equal loading. Protein abundance was quantified using ImageJ.
ROS staining, fluorescence quantification of CellROX, and ion leakage assays
For ROS staining of N. benthamiana, leaf discs from the area infiltrated with the designated Agrobacterium strains were collected 36 h after agroinfiltration. These leaf discs were vacuum-infiltrated with 1 mg/mL DAB (pH 5.5; D8001, Sigma) or 1 mg/mL NBT (A610379, Sangon) and incubated for 3 h at 28 °C. Chlorophyll was removed by boiling in ethanol (95% v/v) for 10 min. H2O2 production was visualized as a reddish-brown coloration based on DAB staining and superoxide was visualized as a dark blue-colored formazan deposit based on NBT staining. ROS staining in Arabidopsis was performed as previously described15. Briefly, 15 μM dexamethasone (D4902, Sigma) or 0.5 mM SA (S7401, Sigma) was infiltrated into leaves and kept in continuous light (100 μmol photons m−2 s−1) for 18 h or 24 h, respectively, and cellular H2O2 and superoxide were visualized by DAB and NBT staining, respectively.
Ion leakage assays were performed as previously described12. Briefly, 15 μM dexamethasone, Pst-AvrRpt2 (OD600 = 0.02), or 0.5 mM SA was infiltrated into leaves, which were kept in light for 36 h, 18 h or 36 h, respectively. Leaf discs were punched out and transferred to a tube containing 10 mL water. After 6 h of incubation at room temperature, conductivity of the solution (C1) was measured with conductivity meter, then the conductivity (C2) was measured after boiling for 10 min to completely kill the cells. The ion leakage ratio = (C2 − C1)/C2*100%. For measuring the ion leakage of leaves after inoculation of S. sclerotiorum, pure hyphae were inoculated onto leaves, and then the area of inoculation site after designated inoculation time was collected for measurement.
For CellROX fluorescence quantification, the leaves were vacuum-infiltrated with 10 μM CellROX™ Deep Red (C10422, Invitrogen) for 30 min after different treatments at specific time points designed for the experiment. Leaf discs were placed in 96-well plates and fluorescence intensity was measured with a microplate reader (Excitation/Emission = 640/665 nm). For measuring the CellROX fluorescence of leaves after inoculation of S. sclerotiorum, pure hyphae were inoculated onto leaves, and then the area of inoculation site after designated inoculation time was collected for measurement.
Determination of Fv/Fm
The maximum photochemical efficiency of PSII (Fv/Fm) was determined with a FluorCam system (FC800-C/1010GFP, Photon Systems Instruments) containing a CCD camera and an irradiation system according to the instrument manufacturer’s instructions after different treatments for the specifically designated time points.
Quantification and statistical analysis
Statistical analyses were performed with the Prism 8.0 software (GraphPad). Details about the statistical methods and number of samples (n) were described in the relevant figure legends.
Accession numbers
Accession numbers and sequence information of the genes investigated in this study can be found in the S. sclerotiorum genome database (https://www.ncbi.nlm.nih.gov/assembly/GCF_000146945.2) under the following accession numbers: SsVSP30 (Sscle12g088370), SsCm1 (SsVSP31, Sscle12g088370), SsVSP32 (Sscle06g055310), Sstublin (Sscle02g015170), SsVSP25 (Sscle04g038020), SsNE2 (Sscle03g024000), SsCm2 (Sscle10g078330), and SsVSP28 (Sscle01g006330); in the N. benthamiana genome database (https://solgenomics.net/organism/Nicotiana_benthamiana/genome) under the following accession numbers: NbMORF2a (Niben261Chr13g0626014.1), NbMORF2b (Niben101Scf07015g00008.1), NbMORF2c (Niben101Scf07015g00010.1), and NbMORF9 (Niben101Scf00176g00005.1); in the G. max genome database (https://phytozome-next.jgi.doe.gov/info/Gmax_Wm82_a4_v1) under the following accession numbers: GmMORF2a (Glyma.20G206400), GmMORF2b (Glyma.10G183800), GmActin (Glyma.18G290800), GmPETE (Glyma.06G020400), GmCm1 (Glyma.13G059400), GmCm2 (Glyma.17G222700), and GmCm3 (Glyma.01G054500); and in the Arabidopsis TAIR database (https://www.arabidopsis.org) under the following accession numbers: ACT2 (AT3G18780), AtMORF2 (AT2G33430), FRK1 (AT2G19190), PR1 (AT2G14610), PR4 (AT3G04720), PAD3 (AT3G26830), ICS1 (AT1G74710), SARD1 (AT1G73805), MKK5 (AT3G21220), LHCB1.2 (AT1G29910), PsbA (ATCG00020), PETE (AT1G20340), PR2 (AT3G57260), and PAD4 (AT3G52430).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The authors declare that the data supporting the findings of this study are available within the article, Supplementary Information, Supplementary Data, and Source Data. Uncropped gels and blots are provided in Supplementary Fig. 18. Source data are provided with this paper.
References
Jones, J. D., Vance, R. E. & Dangl, J. L. Intracellular innate immune surveillance devices in plants and animals. Science 354, https://doi.org/10.1126/science.aaf6395 (2016).
Ngou, B. P. M., Ahn, H. K., Ding, P. & Jones, J. D. G. Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110–115 (2021).
Peng, Y., van Wersch, R. & Zhang, Y. Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Mol. Plant Microbe Interact. 31, 403–409 (2018).
Yang, F., Xiao, K., Pan, H. & Liu, J. Chloroplast: the emerging battlefield in plant-microbe interactions. Front. Plant Sci. 12, 637853 (2021).
Medina-Puche, L. et al. A defense pathway linking plasma membrane and chloroplasts and co-opted by pathogens. Cell 182, 1109–1124.e1125 (2020).
de Torres Zabala, M. et al. Chloroplasts play a central role in plant defence and are targeted by pathogen effectors. Nat. Plants 1, 15074 (2015).
Nomura, H. et al. Chloroplast-mediated activation of plant immune signalling in Arabidopsis. Nat. Commun. 3, 926 (2012).
Zhu, J. K. Abiotic stress signaling and responses in plants. Cell 167, 313–324 (2016).
de Souza, A., Wang, J. Z. & Dehesh, K. Retrograde signals: integrators of interorganellar communication and orchestrators of plant development. Annu. Rev. Plant Biol. 68, 85–108 (2017).
Chan, K. X., Phua, S. Y., Crisp, P., McQuinn, R. & Pogson, B. J. Learning the languages of the chloroplast: retrograde signaling and beyond. Annu. Rev. Plant Biol. 67, 25–53 (2016).
Guo, H. et al. Plastid-nucleus communication involves calcium-modulated MAPK signalling. Nat. Commun. 7, 12173 (2016).
Lee, K. P. et al. Hierarchical regulatory module GENOMES UNCOUPLED1-GOLDEN2-LIKE1/2-WRKY18/40 modulates salicylic acid signaling. Plant Physiol. 192, 3120–3133 (2023).
Laloi, C. et al. Cross-talk between singlet oxygen- and hydrogen peroxide-dependent signaling of stress responses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104, 672–677 (2007).
Serrano, I., Audran, C. & Rivas, S. Chloroplasts at work during plant innate immunity. J. Exp. Bot. 67, 3845–3854 (2016).
Su, J. et al. Active photosynthetic inhibition mediated by MPK3/MPK6 is critical to effector-triggered immunity. PLoS Biol. 16, e2004122 (2018).
Grant, M. R. & Jones, J. D. Hormone (dis)harmony moulds plant health and disease. Science 324, 750–752 (2009).
Mühlenbock, P. et al. Chloroplast signaling and LESION SIMULATING DISEASE1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell 20, 2339–2356 (2008).
Zurbriggen, M. D. et al. Chloroplast-generated reactive oxygen species play a major role in localized cell death during the non-host interaction between tobacco and Xanthomonas campestris pv. vesicatoria. Plant J. 60, 962–973 (2009).
Trotta, A., Rahikainen, M., Konert, G., Finazzi, G. & Kangasjärvi, S. Signalling crosstalk in light stress and immune reactions in plants. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 369, 20130235 (2014).
Ballaré, C. L. Light regulation of plant defense. Annu. Rev. Plant Biol. 65, 335–363 (2014).
Göhre, V., Jones, A. M., Sklenář, J., Robatzek, S. & Weber, A. P. Molecular crosstalk between PAMP-triggered immunity and photosynthesis. Mol. Plant Microbe Interact. 25, 1083–1092 (2012).
Bilgin, D. D. et al. Biotic stress globally downregulates photosynthesis genes. Plant Cell Environ. 33, 1597–1613 (2010).
Lv, R. et al. Uncoupled expression of nuclear and plastid photosynthesis-associated genes contributes to cell death in a lesion mimic mutant. Plant Cell 31, 210–230 (2019).
Duan, J. et al. Impaired PSII proteostasis promotes retrograde signaling via salicylic acid. Plant Physiol. 180, 2182–2197 (2019).
Xu, Q. et al. An effector protein of the wheat stripe rust fungus targets chloroplasts and suppresses chloroplast function. Nat. Commun. 10, 5571 (2019).
Tang, L. et al. An effector of a necrotrophic fungal pathogen targets the calcium-sensing receptor in chloroplasts to inhibit host resistance. Mol. Plant Pathol. 21, 686–701 (2020).
Zhou, J., Zeng, L., Liu, J. & Xing, D. Manipulation of the xanthophyll cycle increases plant susceptibility to Sclerotinia sclerotiorum. PLoS Pathog. 11, e1004878 (2015).
Yang, C., Zhang, Z., Gao, H., Liu, M. & Fan, X. Mechanisms by which the infection of Sclerotinia sclerotiorum (Lib.) de Bary affects the photosynthetic performance in tobacco leaves. BMC Plant Biol. 14, 240 (2014).
Bolton, M. D., Thomma, B. P. & Nelson, B. D. Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 7, 1–16 (2006).
Liang, X. & Rollins, J. A. Mechanisms of broad host range necrotrophic pathogenesis in Sclerotinia sclerotiorum. Phytopathology 108, 1128–1140 (2018).
Williams, B., Kabbage, M., Kim, H. J., Britt, R. & Dickman, M. B. Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog. 7, e1002107 (2011).
Cessna, S. G., Sears, V. E., Dickman, M. B. & Low, P. S. Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. Plant Cell 12, 2191–2200 (2000).
Kabbage, M., Williams, B. & Dickman, M. B. Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLoS Pathog. 9, e1003287 (2013).
Xu, L., Li, G., Jiang, D. & Chen, W. Sclerotinia sclerotiorum: an evaluation of virulence theories. Annu. Rev. Phytopathol. 56, 311–338 (2018).
Derbyshire, M. et al. The complete genome sequence of the phytopathogenic fungus Sclerotinia sclerotiorum reveals insights into the genome architecture of broad host range pathogens. Genome Biol. Evol. 9, 593–618 (2017).
Lyu, X. et al. A small secreted virulence-related protein is essential for the necrotrophic interactions of Sclerotinia sclerotiorum with its host plants. PLoS Pathog. 12, e1005435 (2016).
Yang, G. et al. A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. N. Phytol. 217, 739–755 (2018).
Wei, W. et al. A fungal extracellular effector inactivates plant polygalacturonase-inhibiting protein. Nat. Commun. 13, 2213 (2022).
Djamei, A. et al. Metabolic priming by a secreted fungal effector. Nature 478, 395–398 (2011).
Westrick, N. M. et al. Gene regulation of Sclerotinia sclerotiorum during infection of Glycine max: on the road to pathogenesis. BMC Genomics 20, 157 (2019).
Guyon, K., Balagué, C., Roby, D. & Raffaele, S. Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen Sclerotinia sclerotiorum. BMC Genomics 15, 336 (2014).
Li, J. et al. Introduction of large sequence inserts by CRISPR-Cas9 to create pathogenicity mutants in the multinucleate filamentous pathogen Sclerotinia sclerotiorum. mBio 9, https://doi.org/10.1128/mBio.00567-18 (2018).
Schnappauf, G., Lipscomb, W. N. & Braus, G. H. Separation of inhibition and activation of the allosteric yeast chorismate mutase. Proc. Natl. Acad. Sci. USA 95, 2868–2873 (1998).
Takenaka, M. et al. Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc. Natl. Acad. Sci. USA 109, 5104–5109 (2012).
Lorrain, S., Vailleau, F., Balagué, C. & Roby, D. Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants?. Trends Plant Sci. 8, 263–271 (2003).
Yuan, J. et al. Two chloroplast-localized MORF proteins act as chaperones to maintain tetrapyrrole biosynthesis. N. Phytol. 235, 1868–1883 (2022).
Ren, D., Yang, H. & Zhang, S. Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J. Biol. Chem. 277, 559–565 (2002).
Seifbarghi, S. et al. Receptor-like kinases BAK1 and SOBIR1 are required for necrotizing activity of a novel group of Sclerotinia sclerotiorum necrosis-inducing effectors. Front. Plant Sci. 11, 1021 (2020).
Kamoun, S., van West, P., Vleeshouwers, V. G., de Groot, K. E. & Govers, F. Resistance of Nicotiana benthamiana to Phytophthora infestans is mediated by the recognition of the elicitor protein INF1. Plant Cell 10, 1413–1426 (1998).
Yu, G. et al. A bacterial effector protein prevents MAPK-mediated phosphorylation of SGT1 to suppress plant immunity. PLoS Pathog. 16, e1008933 (2020).
Todesco, M. et al. Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature 465, 632–636 (2010).
Huot, B., Yao, J., Montgomery, B. L. & He, S. Y. Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant 7, 1267–1287 (2014).
Zhang, Y., Tian, L. & Lu, C. Chloroplast gene expression: Recent advances and perspectives. Plant Commun. 4, 100611 (2023).
García-Andrade, J., Ramírez, V., López, A. & Vera, P. Mediated plastid RNA editing in plant immunity. PLoS Pathog. 9, e1003713 (2013).
Ling, Q. & Jarvis, P. Regulation of chloroplast protein import by the ubiquitin E3 ligase SP1 is important for stress tolerance in plants. Curr. Biol. 25, 2527–2534 (2015).
Wang, S. et al. YR36/WKS1-mediated phosphorylation of PsbO, an extrinsic member of photosystem II, inhibits photosynthesis and confers stripe rust resistance in Wheat. Mol. Plant 12, 1639–1650 (2019).
Liu, M. et al. Phosphorylation-guarded light-harvesting complex II contributes to broad-spectrum blast resistance in rice. Proc. Natl. Acad. Sci. USA 116, 17572–17577 (2019).
Wang, L. et al. Singlet oxygen- and EXECUTER1-mediated signaling is initiated in grana margins and depends on the protease FtsH2. Proc. Natl. Acad. Sci. USA 113, E3792–E3800 (2016).
Kim, C. et al. Chloroplasts of Arabidopsis are the source and a primary target of a plant-specific programmed cell death signaling pathway. Plant Cell 24, 3026–3039 (2012).
Li, M. & Kim, C. Chloroplast ROS and stress signaling. Plant Commun. 3, 100264 (2022).
Triantaphylidès, C. & Havaux, M. Singlet oxygen in plants: production, detoxification and signaling. Trends Plant Sci. 14, 219–228 (2009).
Yang, Y. et al. Cytidine-to-Uridine RNA editing factor NbMORF8 negatively regulates plant immunity to Phytophthora pathogens. Plant Physiol. 184, 2182–2198 (2020).
Yapa, M. M., Doroodian, P., Gao, Z., Yu, P. & Hua, Z. MORF2-mediated plastidial retrograde signaling is involved in stress response and skotomorphogenesis beyond RNA editing. Front. Plant Sci. 14, 1146922 (2023).
Zhang, F. et al. Tetrapyrrole biosynthetic enzyme protoporphyrinogen IX oxidase 1 is required for plastid RNA editing. Proc. Natl. Acad. Sci. USA 111, 2023–2028 (2014).
Geng, R. et al. PROGRAMMED CELL DEATH8 interacts with tetrapyrrole biosynthesis enzymes and ClpC1 to maintain homeostasis of tetrapyrrole metabolites in Arabidopsis. N. Phytol. 238, 2545–2560 (2023).
Zhao, X., Huang, J. & Chory, J. GUN1 interacts with MORF2 to regulate plastid RNA editing during retrograde signaling. Proc. Natl. Acad. Sci. USA 116, 10162–10167 (2019).
Coego, A. et al. An Arabidopsis homeodomain transcription factor, OVEREXPRESSOR OF CATIONIC PEROXIDASE 3, mediates resistance to infection by necrotrophic pathogens. Plant Cell 17, 2123–2137 (2005).
Gao, L. L., Hong, Z. H., Wang, Y. & Wu, G. Z. Chloroplast proteostasis: a story of birth, life, and death. Plant Commun. 4, 100424 (2023).
Woodson, J. D. Chloroplast stress signals: regulation of cellular degradation and chloroplast turnover. Curr. Opin. Plant Biol. 52, 30–37 (2019).
Wang, S. & Blumwald, E. Stress-induced chloroplast degradation in Arabidopsis is regulated via a process independent of autophagy and senescence-associated vacuoles. Plant Cell 26, 4875–4888 (2014).
Mohd Ali, S. et al. Multiple ubiquitin E3 ligase genes antagonistically regulate chloroplast-associated protein degradation. Curr. Biol. 33, 1138–1146.e1135 (2023).
Ling, Q., Huang, W., Baldwin, A. & Jarvis, P. Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science 338, 655–659 (2012).
Woodson, J. D. et al. Ubiquitin facilitates a quality-control pathway that removes damaged chloroplasts. Science 350, 450–454 (2015).
Wu, J. et al. Solid-like condensation of MORF8 inhibits RNA editing under heat stress in Arabidopsis. Nat. Commun. 16, 2789 (2025).
Bauters, L. et al. Chorismate mutase and isochorismatase, two potential effectors of the migratory nematode Hirschmanniella oryzae, increase host susceptibility by manipulating secondary metabolite content of rice. Mol. Plant Pathol. 21, 1634–1646 (2020).
He, Q. et al. A novel chorismate mutase from Erysiphe quercicola performs dual functions of synthesizing amino acids and inhibiting plant salicylic acid synthesis. Microbiol. Res. 242, 126599 (2021).
Wen, Z. et al. Integrating GWAS and gene expression data for functional characterization of resistance to white mould in soyabean. Plant Biotechnol. J. 16, 1825–1835 (2018).
Mengiste, T. Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 50, 267–294 (2012).
Oh, S. K. et al. In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell 21, 2928–2947 (2009).
Zhang, W. et al. Arabidopsis receptor-like protein 30 and receptor-like kinase suppressor of BIR1- 1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell 25, 4227–4241 (2013).
Xiao, K. et al. The Snf5-Hsf1 transcription module synergistically regulates stress responses and pathogenicity by maintaining ROS homeostasis in Sclerotinia sclerotiorum. N. Phytol. 241, 1794–1812 (2024).
Sooksai, T. et al. Production of cutinase from Fusarium falciforme and its application for hydrophilicity improvement of polyethylene terephthalate fabric. 3 Biotech 9, 389 (2019).
Lyu, X. et al. GmCRY1s modulate gibberellin metabolism to regulate soybean shade avoidance in response to reduced blue light. Mol. Plant 14, 298–314 (2021).
Gilchrist, D. G. & Connelly, J. A. Chorismate mutase from mung bean and sorghum. Methods Enzymol. 142, 450–463 (1987).
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (32172505 to J.L., 32272484 to H.P., 323B2055 to K.X.), the National Key Research and Development Program of China (2019YFE0114200 to H.P.), and the Natural Science Foundation of Jilin Province (20230101156JC to J.L.). We thank Professor Fengjie Sun of Georgia Gwinnett College, for help with editing the paper.
Author information
Authors and Affiliations
Contributions
H.P. and J.L. directed and supervised the research. H.P., J.L., and K.X. designed the research. K.X. and F.Y. conducted most of the experiments, analyzed the data, and wrote the original draft. W.C., J.G., and A.L. assisted with the qRT-PCR experiments and the acquisition of transgenic materials, A.L. and X.X. generated the CRISPR/Cas9-engineered Gmmorf2a/b mutants, and X.S. performed the luminol-based ROS burst assays and the subcellular co-location assays. F.W., Y.Z., and X.Z. assisted in writing. J.A.R., J.L., X.W., and H.P. revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xiao, K., Yang, F., Cui, W. et al. A fungal effector promotes infection via stabilizing a negative regulatory factor of chloroplast immunity. Nat Commun 16, 6970 (2025). https://doi.org/10.1038/s41467-025-62326-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-62326-4