Abstract
Survival strategy of bacteria is expanded by extrachromosomal elements (ECEs). However, their genetic diversity and functional roles for adaptability are largely unknown. Here, we discover a novel family of intracellular ECEs using 56 saliva samples by developing an efficient microbial DNA extraction method coupled with long-read metagenomics assembly. Even though this ECE family was not hitherto identified, our global prevalence analysis using 476 salivary metagenomic datasets elucidates that these ECEs reside in 74% of the population. These ECEs, which we named, “Inocles”, are giant plasmid-like circular genomic elements of 395 kb in length, including Streptococcus as a host bacterium. Inocles encode a series of genes that contribute to intracellular stress tolerance, such as oxidative stress and DNA damage, and cell wall biosynthesis and modification involved in the interactions with oral epithelial cells. Moreover, Inocles exhibit significant positive correlations with immune cells and proteins responding to microbial infection in peripheral blood. Intriguingly, we examine and find their marked reductions among 68 patients of head and neck cancers and colorectal cancers, suggesting its potential usage for a novel biomarker of gastrointestinal cancers. Our results suggest that Inocles potentially boost the adaptive capacity of host bacteria against various stressors in the oral environment.
Similar content being viewed by others
Introduction
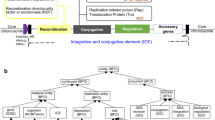
Bacteria residing in humans are exposed to various stressors, including nutrient competition, drugs for humans, antibiotics, and human immune responses. To survive these multiple stressors, bacteria expand their adaptive capacity by acquiring accessory genes corresponding to environmental stress tolerance. Mobile genetic elements (MGEs) are the major mechanism by which bacteria obtain accessory genes from other bacteria. Integrative and conjugative elements (ICEs) and transposons are intrachromosomal MGEs that transfer the accessory genes to host bacteria by integrating their sequences into the host bacterial chromosome1,2. In addition, bacteria obtain accessory genes via extrachromosomal elements (ECEs), which are genetic elements independent of chromosomes3. Microbiological research targeting pathogenic bacteria has revealed that ECEs, such as plasmids, confer environmental stress resistance on host bacteria, including antibiotic resistance2. This infers that ECEs contribute to expanding the adaptive capacity of human commensal bacteria.
Several advanced strategies for predicting ECEs from metagenomic sequences have revealed that human commensal bacteria harbor diverse ECEs, such as bacteriophages (phages)4,5,6,7,8,9, plasmids10, and viroid-like elements11. However, current metagenomic analysis based on a limited set of reference genomes may overlook ECEs that could not be classified using existing knowledge, and it is largely unknown the genetic diversity and functional roles of ECEs in human commensal bacteria.
Here, we identified the novel giant ECEs from the oral cavity of most individuals in the world, which are considered plasmid-like elements encoding multiple genes related to adaptation to intracellular and extracellular stresses. We have characterized this novel plasmid-like element, termed “Inocle,” focusing on its genetic and ecological significance. Moreover, we elucidated the significant associations of Inocles with human immune statuses and certain cancer types. This study demonstrates that human commensal bacteria adapt to changes in their habitat conditions and human physiological alterations, utilizing giant ECEs.
Results
Long-read metagenomic sequencing optimized for human saliva samples
Long-read sequencing improves metagenomic assembly for the reconstruction of ECEs with high completeness12,13,14,15,16; however, human saliva contains 85–95% human genomic DNA, making it difficult to obtain sufficient sequencing depth from microbes17. Therefore, we developed a simple method to extract high-molecular-weight DNA with reduced human genomic DNA from saliva samples. After collecting bacterial pellets by centrifugation, we removed extracellular DNA, which should be mostly human DNA17, by treating the bacterial pellet with nuclease under optimal conditions for nuclease activity. We refer to this method as the “preNuc”. After the treatment of preNuc, we extracted high-molecular-weight DNA using enzymatic bacterial cell lysis (Fig. 1A). Notably, preNuc reduced the average percentage of human DNA from 90% to 40% in short-read sequences prepared from fresh and frozen saliva samples (p = 0.01) (Fig. 1B and Supplementary Table S1). The preNuc treatment resulted in a two-fold increase in the total microbial bases compared to that in preNuc-untreated samples when using the PromethION sequencer of Oxford Nanopore Technologies (Fig. 1C) without shortened read length (Fig. 1D). Moreover, Spearman’s rho for bacterial composition averaged 0.83 ± 0.08 at the species level between preNuc-treated and -untreated saliva samples (Fig. 1E), indicating that our method easily reduces the human DNA in saliva samples and extracts high-molecular-weight DNA while minimally affecting bacterial composition.
A Schematic workflow for high-molecular DNA extraction from human saliva samples. Blue arrows indicate processes of the preNuc. B Effect of preNuc treatment on the removal of human DNA from saliva samples. Each plot shows the percentage of human reads in NovaSeq reads obtained using preNuc-treated and untreated saliva samples. *p < 0.05, N.S., not significant based on two-sided paired Student’s t test. C Effect of preNuc treatment on the sequence depth of microbial-derived long reads. Bar plots show the total bases of PromethION reads without low-quality and human reads per flow cell acquired from preNuc-treated and untreated samples. D Effect of preNuc treatment on the sequence length of long reads. Bar plots show the N50 length of PromethION reads without low-quality and human reads per flow cell of the preNuc-treated and -untreated samples. E Effect of preNuc treatment on bacterial composition. Scatter plots show the correlation of species-level bacterial composition between preNuc-treated and untreated saliva samples.
Subsequently, we obtained PromethION sequences from 46 Japanese saliva samples with an average of 3.2 million high-quality and non-human reads, including 23.4 Gb with an N50 length of 12.5 kb per sample (Supplementary Table S2). We performed De novo metagenomic assembly using metaFlye18 (Supplementary Table S3) and used contigs with a depth exceeding 10 as high-quality contigs in subsequent analyses because they had an average of 98.95% similarity with the corresponding short-read contigs assembled using NovaSeq sequences (Supplementary Fig. S1).
Discovery of unrecognized ECEs
We attempted to identify hitherto unrecognized ECEs to expand our knowledge of the genetic diversity of ECEs in the human oral microbiome. First, we classified 69,881 high-quality contigs, which are >3 kb in length and unaligned to the human genome, into known genetic elements (Supplementary Fig. S2A) and classified 85.8% potential bacterial chromosomal, 1.5% phage, 2.9% plasmid, and 9.8% unclassified contigs (Fig. 2A).
A The ratio of each microbial genetic element of long-read high-quality contigs. Among the 6852 unclassified contigs, 85 were further analyzed as strong candidates of unrecognized elements. B Abundance and prevalence of candidate contigs of unrecognized genetic elements. The scatter plot shows the average abundance (y-axis), prevalence (x-axis), and number of contigs in each operational contig cluster (plot size) obtained from 85 contigs. C An abundance of Cluster 0001 contigs among all contigs in each sample. The violin plot shows the distribution of the abundance of all contigs in each sample, which includes contigs of Cluster_0001. The red arrow indicates the abundance of Cluster 0001 contigs and their rank in all contig abundance in each sample. Box plots represent the inter-quartile range (IQR), and lines inside the box indicate the median. Whiskers show 1.5 IQR. D The contig length and (E) number of ORFs of 13 contigs of the Cluster 0001. Box plots represent the inter-quartile range (IQR), and lines inside the box indicate the median. Whiskers show 1.5 IQR. F Representative replichore structure of Cluster 0001 contig (Inocle_001). The green line indicates cumulative GC skew, and the gray plots indicate GC skew in each genomic position. The candidate replication origin and terminus are represented by vertical red and blue lines, respectively. G Representative genomic structure of Inocle: from inner to outside, GC skew, GC content, forward strand of ORFs, and reverse strand of ORFs. H Effect of preNuc treatment on the sequence depth of the Cluster 0001 contig. Bar plot shows the mean depth of the Cluster 0001 contig (Inocle_004) obtained from preNuc-treated and untreated saliva samples. I Comparison of the long-read contig and corresponding short-read contigs of the Cluster 0001 contig. The blue bar represents the contig constructed by the long-read assembly, and the red boxes indicate IS regions. The arrows represent fragmented positions in short-read assembly. The upper gray bar plot indicates the depth of the mapped long reads. Colored bars in the depth bar plots indicate single-nucleotide variants: Green = A, red = T, orange = G, and blue = C.
Among 6852 unclassified contigs, 85 were particularly considered unrecognized genetic elements with many unknown functional genes (Fig. 2A). Specifically, over 50% of the genes in these 85 contigs did not align with the UniRef90 database, which is a lower frequency than that of known microbial genetic elements (Supplementary Fig. S2B). For these contigs, we obtained 19 clusters (Cluster 0001–0019) without singletons by clustering them with contigs that shared over 80% of their genes within a cluster and >30% average amino acid identity (Fig. 2B). Quantification of cluster abundance through long-read mapping showed that Cluster 0001, comprising 13 contigs, exhibited the highest abundance and prevalence (Fig. 2B). Within this cluster, 8 of the 13 contigs ranked in the top 50% in terms of all contig abundance, and one of the Cluster_0001 contig (Sample ID: OO1116B8239) was among the 20 most highly abundant contigs, suggesting that Cluster 0001 presents an abundant genetic element in human saliva (Fig. 2C). None of the contigs of Cluster 0001 could be aligned to the plasmid or phage database with >95% identity and >85% coverage. These results indicate that Cluster 0001 comprises a potentially novel genetic element prevalent in the human oral microbiome.
The average contig size of Cluster 0001 was 352 kb (293–395 kb) (Fig. 2D), and the average number of open reading frames (ORFs) was 313 (Fig. 2E). The topology of the 13 contigs was identified as circular based on the assembly of the metaFlye18. Long-read mapping revealed an even sequence depth throughout the contigs, with no regions lacking mapped sequences, suggesting that these contigs are unlikely to be artificially misassembled (Supplementary Fig. S3). Notably, we observed no accumulation of alignment start and stop positions within the contigs, which is a characteristic feature of linear contigs with terminal direct repeats14, supporting the circular topology of these contigs (Supplementary Fig. S3). Moreover, we identified the candidate replication origin and terminus via GC skew analysis (Fig. 2F) and observed a switch in the orientation of ORFs at the replication origin and terminus (Fig. 2G and Supplementary Fig. S4), suggesting that this Cluster is a circular replicon.
We further examined the possible reasons why these abundant genetic elements have been overlooked in previous metagenomic studies. One of the reasons could be the increased sequencing depth. Specifically, the preNuc increased the long-read sequence coverage of Cluster 0001 in a given sample from 9.6 to 30-fold, which contributed to the reconstruction of a high-quality contig (Fig. 2H). Another reason is the challenge of assembling several regions using short reads. Although short-read assembly successfully reconstructed some parts of the contigs, these assemblies were fragmented, with 48% (48/102 points) of the breakpoints overlapping with repetitive insertion sequences (ISs) (Fig. 2I and Supplementary Fig. S5). We also noted that 17% (17/102 points) of the breakpoints overlapped with other hypervariable regions (Fig. 2I and Supplementary Fig. S5). We have designated this previously unrecognized genetic element as “Inocle” (Insertion sequence encoded; oral origin; circle genomic structure). Considering the lack of bacterial marker genes, Inocles are likely novel ECEs in the human oral microbiome.
Discovery of the Inocle marker gene expands the family members of Inocle
Upon discovering the Inocle, we first investigated how many sequences within its family remain undiscovered. To this end, we searched for any marker gene that could represent an efficient strategy for identifying Inocle contigs from other metagenomic contigs. To identify such a marker gene, we clustered all Inocle ORFs and identified a single gene that satisfied the following criteria: having >70% amino acid identity and > 50% alignment coverage across all initially identified Inocle contigs. We named this gene as “InoC” (Inocle conserved gene). InoC is a single-copy gene with an average amino acid length of 454 and a replication-relaxation domain (Pfam ID: PF13814) in the N-terminus domain (Fig. 3A). No genes similar to InoC were identified in the NCBI nucleotide database, indicating that InoC is an Inocle-specific gene.
A Workflow for identification of additional Inocle contigs. Discovery analysis using long-read contigs acquired from 46 Japanese participants identified 13 Inocle contigs and InoC genes. The representative predicted 3D structure of InoC is shown; the green region indicates the replication-relaxation domain in the InoC. To expand the number of Inocle contigs, circular contigs encoding an InoC were identified using high-quality long-read contigs obtained from 46 Japanese, 9 Indonesian, and 1 Thailand participants. B Phylogenetic tree of the 29 InoC amino acid sequences. The red, blue, purple, and yellow colors indicate Inocle-α, β, γ, and δ, respectively. C Comparative protein repertoires of 29 Inocles. The horizontal axis indicates protein families (PFs) of Inocles, and the vertical axis represents each Inocle. The heatmap shows the presence (gray) or absence (white) of each PF in each Inocle. The taxon of each Inocle corresponds with the color code used in (B). D Prevalence of Inocles across the seven countries. The size of each circle shows the prevalence of each Inocle taxon in each country. E The average abundance of each Inocle taxon in each body site.
Using InoC, we conducted an expansion analysis to identify additional Inocle family sequences from the high-quality circular contigs, which were initially obtained from the 46 Japanese saliva samples (Fig. 3A). To further examine the global prevalence of the Inocle family contigs, we also applied the same strategy to long-read contigs, which were assembled from saliva samples of nine Indonesian and one Thailand healthy participants (Supplementary Table S2 and S3). We identified 16 additional circular contigs encoding InoC (Fig. 3A), all containing candidates for replication origin and terminus (Supplementary Fig. S6). Finally, we identified 29 closed circular Inocle contigs, including 21 contigs from 17 Japanese participants, 7 contigs from four Indonesian participants, and 1 contig from a Thailand participant (Supplementary Table S4).
Discovery of four Inocle taxa and their genetical and ecological differences
The phylogenetic tree of the InoC genes of the 29 Inocle contigs revealed four distinct groups. We named these groups as Inocle-α, β, γ, and δ (Fig. 3B). The average amino acid identity of the InoC within the same group was 94.5%, and that among different groups was 64.1% (Supplementary Fig. S7A). Inocle-α had the largest contig size (368 kb on average), whereas Inocle-δ had the smallest (245 kb on average) (Supplementary Fig. S7B). The average nucleotide identity within each group was 88.9%, and we could not obtain sufficient alignment coverage ( >20%) between the groups (Supplementary Fig. S7C). The number of tRNAs differed in each group (Supplementary Fig. S7D). The GC content of Inocle-α, β, and γ averaged 32%, whereas that of Inocle-δ averaged 24% (Supplementary Fig. S7E). All Inocles had IS, with Inocle-α being particularly enriched with ISs (nine ISs/contig on average), suggesting that Inocles expand their genetic diversity by the mobile genetic element of ISs (Supplementary Fig. S7F). A total of 1619 protein families (PFs) were identified from across the Inocle contigs, having >50% amino acid identity (Supplementary Data 1). Among them, 1537 PFs (95%) were only observed in a specific group (Fig. 3C). The genetic differences between these four groups indicate that these four putative taxonomies of Inocles have different evolutionary histories.
To investigate the geographic distribution of each Inocle taxon, we downloaded salivary shotgun metagenomic sequences of healthy individuals from China (n = 47), the Philippines (n = 24), France (n = 16), the United States (n = 64), and Fiji (n = 237) from public databases (Supplementary Table S5). We also obtained short-read metagenomic sequences of saliva samples from 68 healthy Japanese and 20 healthy Indonesian participants (Supplementary Table S5). Inocle-positive samples were identified by mapping the short reads to the InoC genes, and we found that 74% of people are Inocle-positive on average (Fig. 3D). Indonesia exhibited a higher prevalence (90%) of Inocles than other countries, whereas Japan had the lowest prevalence (64%) (Fig. 3D). Across all countries, Inocle-α was the dominant group with 57% prevalence on average, although non-industrialized countries of Indonesia, the Philippines, and Fiji exhibited a higher prevalence of Inocle-β than that of other countries (Fig. 3D).
Next, we mapped the shotgun metagenomic sequences of various body sites collected by the Human Microbiome Project (HMP)19 to the InoC genes to clarify the localization of each Inocle taxon in the human body. We detected the Inocles from oral sites, but they were rare in feces, external nares, posterior fornix of the vagina, and retroauricular crease (Fig. 3E). Within the oral cavity, we revealed that the primary habitat of Inocle-α is at the tongue dorsum, that of Inocle-β and Inocle-δ is at the buccal mucosa, and that of Inocle-γ is at the gingiva (Fig. 3E), suggesting that each taxon has a distinct habitat within the oral cavity.
Inocles are giant plasmid-like elements of the Streptococcus
Bacterial cells and small extracellular particles such as phages can be separated by centrifugation followed by 0.45-μm filtration into centrifuged pellets and 0.45 μm filtered fraction, respectively20. We found that the number of Inocle-derived sequences was 16-fold higher in the bacterial pellet fraction than in the 0.45 μm filtered fraction on average after fractionating the tested saliva samples, suggesting Inocles are intracellular ECEs (Fig. 4A). Some intracellular ECEs have homologous genes with their host bacteria21. Therefore, to identify the host bacterium of Inocles, we performed a homology search using the Inocle genes against all bacterial genera in the UniRef90 database. The results revealed that an average of 53 ORFs/contig were homologous to Streptococcus genes (Fig. 4B), suggesting that Streptococcus is the candidate host bacteria of Inocles.
A Abundance of Inocles in the intracellular and extracellular particle fractions. Comparative abundance of 15 Inocles (Inocle_001, 002, 003, 004, 005, 006, 007, 008, 010, 017, 018, 019, 023, 024, and 028) in the bacterial pellets and 0.45 μm filtered fraction of saliva samples. B The distribution of the number of ORFs of 29 Inocles aligned to genus-defined genes in the UniRef90 database. Box plots represent the inter-quartile range (IQR), and lines inside the box indicate the median. Whiskers show 1.5 IQR. C Experimental workflow for the isolation of Streptococcus strains, including Inocle_004. The saliva sample, which contained Inocle_004, was streaked onto the MRS agar plate. The five colonies were subjected to Sanger sequencing of the colony PCR product, amplified using Inocle_004 specific primers. Three colonies were identified as positive for Inocle_004, as their PCR products perfectly matched with Inocle_004. Subsequently, three Inocle_004-positive colonies (colony-1, − 2, and − 3) were subjected to liquid culturing in the MRS medium. The absence of Inocle_004 in the colony-1 strain cultured in the liquid medium was verified using whole-genome sequencing. The absence of Inocle_004 in colony-2 and − 3 strains cultured in liquid media was confirmed via PCR using Inocle_004-specific primers. The bacterial taxonomic classification of colony-1 was obtained using GTDBtk based on whole genome sequences, while that of colony-2 and − 3 was obtained from the sequences of the 16S rRNA (V1-V2 region) PCR product. The red spectrum figure depicts the PCR product size (bp). N/A; not available. D Comparison of the tetranucleotide frequency between Inocles and chromosomes, plasmids, and phages of Streptococcus. The heatmap presents the tetranucleotide frequency, and the dendrogram was obtained based on the similarity of tetranucleotide frequency. E Comparison of the codon usage between Inocles and chromosomes, plasmids, and phages of Streptococcus. The heatmap depicts the relative synonymous codon usage, and the dendrogram was obtained based on the similarity of synonymous codon usage.
Given that Inocles are intracellular ECEs of oral bacteria, we investigated whether there are genomic datasets of orally isolated bacteria containing Inocle-derived sequences from the NCBI database. Our search across over 3000 oral bacterial isolates did not identify any samples with sequences mapped to InoC genes. Therefore, we attempted to isolate and culture bacterial strains containing one example of Inocle (Inocle_004). We cultured bacteria in saliva containing Inocle_004 on a De Man–Rogosa–Sharpe agar plate (Fig. 4C). Colony PCR using Inocle_004 specific primers for five colonies identified three colonies whose PCR product sequences perfectly matched the Inocle_004 contig (Fig. 4C and Supplementary Table S6). We then propagated one of the three strains (colony-1) in the MRS liquid medium to obtain sufficient genomic DNA for whole genome sequencing, and obtained short reads. Our sequences had more than 3 million reads but did not include Inocle_004-derived sequences, suggesting the loss of the Inocle_004 in host cells during culturing in the liquid medium. To confirm the stability of Inocle_004 during liquid culture in other isolates, we performed the PCR using Inocle_004-specific primers for liquid-cultured colony-2 and −3 strains. Expectedly, we were unable to obtain the PCR product (Fig. 4C), confirming that Inocle_004, which was detected on the agar plate immediately after isolation, was lost from host cells during culture in the liquid medium. This may explain why Inocles were not detected in the publicly available genomic sequences. The whole genome sequences (colony-1 strain) and 16S rRNA gene sequences (colony-2 and −3 strains) of the isolated strains revealed that all strains were Streptococcus salivarius (Fig. 4C and Supplementary Table S6). These results suggest that S. salivarius is a candidate host species for Inocle_004. However, it remains unclear which species serves as the host for the other Inocles. To estimate the copy number of Inocle_004 for the candidate host bacteria in the saliva, the saliva metagenomic reads were mapped to the S. salivarius genome of colony-1 and the Inocle_004 contig, and the estimated ratio of Inocle_004 to S. salivarius was 1:15.
We further compared the genomic signatures between Inocles and chromosomes, plasmids, and phages of Streptococcus. Although the tetranucleotide frequency of Inocles was relatively similar to plasmids compared to chromosomes and phages, Inocles formed a unique cluster in the dendrogram generated from the similarity of tetranucleotide frequency (Fig. 4D). We observed the same trend with the codon usage (Fig. 4E), indicating that compared to known plasmids, Inocles exhibit distinct nucleotide composition and codon usage. We identified the VirB4 and VirD4 in all Inocle contigs (Supplementary Data 1), which are required to transfer plasmid from donor to recipient cells via the type IV secretion system22. Hence, Inocles are considered novel giant plasmid-like elements of Streptococcus, possibly having the ability for horizontal transmission.
Gene annotation of the Inocles
To reveal the biological roles of Inocles in host bacteria, we conducted functional annotation of 1619 PFs encoded by the Inocle contigs (Supplementary Data 1). First, we considered the subcellular localization of these PFs and identified 644 (40%) and 620 (38%) PFs as cytoplasmic and unknown subcellularly localized proteins, respectively (Fig. 5A). Among these 644 cytoplasmic proteins, we annotated 21 functional genes using Prokka23. Among the 11 conserved cytoplasmic genes in all Inocle taxa, 6 were annotated as genes associated with DNA repair and recombination (ko03400) based on the Kyoto Encyclopedia of Genes and Genome (KEGG) BRITE database. All these genes contained DNA damage repair-related functions, such as those of exonuclease, DNA gyrase, and DNA polymerase III (Fig. 5B). We identified a RecD-like DNA helicase with unknown subcellular localization genes, which was also related to DNA damage repair-related genes (Fig. 5C). We found other stress response-related genes detected in multiple taxa, such as RpoS contributing stress responses against multiple stressors24 and NrdH conferring oxidative stress tolerance25. We confirmed the transcriptional activity for six of seven DNA damage repair genes, RpoS, and NrdH (Fig. 5B, C), based on the metatranscriptome sequences of human saliva26.
A The ratio of subcellular localization in 1619 Inocle protein families. B–D Functionally annotated genes of Inocles in cytoplasmic genes (B), unknown subcellular localization genes (C), and cell wall/membrane-related genes (D). The gray and white heatmap indicates the existence of each gene in each Inocle taxon. The functional categories of the KEGG BRITE are shown for each gene. Genes with confirmed transcriptional activity in saliva are shown in a red heatmap. E–H Representative 2D structure of the bifunctional transglycosylase (E), sortase A (F), LPXTG-like motif protein (G), and signal peptidase (H) on the membrane. I Gene functions of the homologous genes between Inocles and Streptococcus chromosomes. The bar plot indicates the number of homologous genes between Inocles and Streptococcus chromosomes in each function.
Next, we focused on 329 PFs (20%) of possible cell wall/membrane-related proteins (Fig. 5A). Among them, we identified eight functionally annotated genes, two of which were conserved across all Inocle taxa, while one was classified as peptidoglycan biosynthesis/degradation protein based on the KEGG BRITE database (Fig. 5D). This protein comprises a transmembrane domain at its N-terminus as well as a transglycosylase domain and a penicillin-binding protein transpeptidase domain in the outer membrane region (Fig. 5E). Therefore, it is a potential bifunctional transglycosylase that catalyzes the polymerization and cross-linking of the glycan chain for peptidoglycan biosynthesis27. The other conserved cell wall/membrane-related genes were annotated as sortase A (Fig. 5D), which comprises a transmembrane domain at its N-terminus, with the sortase domain located in the outer membrane region (Fig. 5F). This protein possibly recognizes the LPXTG motif in cell wall proteins and anchors them to peptidoglycan28. We identified six LPXTG-like motif PFs presumed to be recognized by sortase A. All Inocle taxa encoded at least one LPXTG-like motif PF (Supplementary Fig. S8A). These LPXTG-like motif PFs contain two transmembrane domains, one at the N-terminus and one at the C-terminus, spanning the outer membrane region (Fig. 5G and Supplementary Fig. S8B). We found that an LPXTG-like motif is in the outer membrane region of the C-terminus, with the signal peptide for membrane transition and the signal peptidase cleavage position located at the N-terminus (Fig. 5G and Supplementary Fig. S8B). We identified signal peptidases from Inocle-α, β, and γ with the Signal peptidase S26 domain (PF10502) (Supplementary Fig. S8C) and a lysine-active site for cleaving signal peptides (Fig. 5H and Supplementary Fig. S8D). We confirmed the transcriptional activity for these cell wall-related genes, suggesting their bioactivity in a native oral environment (Fig. 5D and Supplementary Fig. S8A, S8C).
Next, we investigated whether Inocles exhibits functional redundancy with the Streptococcus chromosome, which is commonly observed between plasmids and host bacterial chromosomes3. We identified an average of 20 ORFs (6%) per Inocle as homologous genes between Streptococcus chromosomes and Inocles (Supplementary Fig. S9). These genes included RecD-like DNA helicases, DNA polymerase III subunit alpha, bifunctional transglycosylases, and sortase A (Fig. 5I).
Inocle-α is correlated with human systemic immune statuses
We then investigated the associations of Inocles with human physiologies. We first examined whether Inocle-α is associated with the sex and age of the host human (healthy participants) and observed no significant associations (Supplementary Table S7 and Supplementary Fig. S10A, B). Owing to the low prevalence of Inocle-β, γ, and δ in our Japanese sample population, corresponding data could not be statistically analyzed (Fig. 3D).
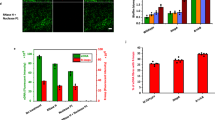
We further examined the associations using a multi-omics dataset of humans. We collected peripheral blood mononuclear cells (PBMCs) from the 29 individuals whose saliva was collected in this study and conducted the single-cell RNA sequencing (scRNA-seq) analysis (Supplementary Table S8). The cellular populations of PBMCs were annotated into 19 cell types based on Azimuth references29. The partial Spearman’s correlation, accounting for age and sex as confounders, revealed a significantly positive correlation of Inocle-α with intermediate (partial Spearman’s rho = 0.50 and p = 0.006), memory (partial Spearman’s rho = 0.42 and p = 0.02), and Naïve B cells (partial Spearman’s rho = 0.40 and p = 0.03) but not with plasma cells (Fig. 6A). We also observed a significantly negative correlation between Inocle-α and CD16 + (partial Spearman’s rho = − 0.60 and p = 0.0007) and CD14 + monocytes (partial Spearman’s rho = − 0.38 and p = 0.04) (Fig. 6A). These associations were not observed for Streptococcus (Fig. 6A).
A Correlations between Inocle-α and cell types in PBMC. The heatmap shows the partial Spearman’s rho, with age and sex considered as confounders between PBMC populations and the abundance of Inocle-α and Streptococcus. *p < 0.05, **p < 0.01, ***p < 0.001. B Plasma proteome pathways that were positively correlated with Inocle-α. The bar chart presents the results of Gene Ontology enrichment analysis using plasma proteins whose expression was significantly correlated with the abundance of the Inocle-α, based on partial Spearman’s rho with age and sex considered as confounders. C, D Comparison of the prevalence and abundance of Inocle-α (C) and Streptococcus (D) between healthy controls (HCs) (n = 53) and patients with head and neck cancer (HNC) (n = 45). The bar plots indicate the prevalence of Inocle-α and Streptococcus in the HC and HNC groups. The box plots indicate the abundance of Inocle-α and Streptococcus in the HC and HNC groups. The outliners are removed for visualization in box plots. *p < 0.05, ***p < 0.001, N.S., not significant. Statistical results were obtained using a two-sided Fisher’s exact test for prevalence. MaAsLin2 was performed for abundance, with the Benjamini–Hochberg method for multiple comparisons and accounting for confounders for age, sex, smoking status, and cancer. Box plots represent the inter-quartile range (IQR), and lines inside the box indicate the median. Whiskers show 1.5 IQR. E Comparison of the abundance of Inocle-α and Streptococcus between patients with different diseases and HCs in each study. The coefficient values and p-value calculated via MaAsLin2 are shown. MaAsLin2 was performed with the Benjamini–Hochberg method for multiple comparisons and accounting for confounders of age and sex. *p < 0.01, ***p < 0.001. Abbreviations; HSPC Hematopoietic stem and progenitor cell, MAIT Mucosal associated invariant T, CRC colorectal cancer, PDAC pancreatic ductal adenocarcinoma, RA rheumatoid arthritis.
To further scrutinize the associations between Inocle-α and human systemic immune systems, we analyzed the expression data of a total of 5420 plasma proteins using proximity extension assay technology (Olink) from 40 healthy Japanese participants (Supplementary Data 2). The partial Spearman’s correlation, accounting for age and sex as confounders, revealed 1187 and 62 proteins exhibiting significant positive and negative correlation with Inocle-α, respectively (Supplementary Data 3). GO enrichment analysis revealed enrichment in 43 pathways in the Inocle-α-positive group (Fig. 6B), including the B cell receptor signaling pathway, which probably corresponds to the positive correlation between Inocle-α and B cells (Fig. 6A). Among 15 particularly high-correlated pathways ( > 5 -log10 FDR), four were related to the response to bacterial and viral infections, including pathogenic Escherichia coli infection, Yersinia infection, hepatitis B, Salmonella infection, Kaposi sarcoma-associated herpesvirus infection, and Epithelial cell signaling in Helicobacter pylori infection (Fig. 6B). In addition, we noted elevated expression levels of proteins related to the induction of lipid and atherosclerosis, endocytosis, TNF signaling, NF-κB signaling, and apoptosis in the Inocle-α-positive group (Fig. 6B).
Inocle-α is negatively associated with patients of several cancer types
Notably, we found that Inocle-α was associated with several cancer-related pathways in the plasma proteome. These pathways included Proteoglycans in cancer, Chronic myeloid leukemia, Choline metabolism in cancer, Pancreatic cancer, and Bladder cancer (Fig. 6B). Therefore, we subsequently investigated the possible association between Inocle-α and cancers. We collected saliva samples from 45 patients with head and neck cancer (HNC), including 22 treatment-naïve patients, and obtained metagenomic short reads (Supplementary Table S8). We compared the abundance of Inocle-α among 53 healthy controls (HCs) and 45 patients with HNC while considering their ages, sexes, smoking status, and cancer treatment information as possible confounding factors (Supplementary Table S8). The prevalence of Inocle-α was significantly decreased in the HNC group (27%) compared to that in the HC group (60%) (p = 0.001), and correspondingly, the abundance of Inocle-α was decreased (p = 0.03) (Fig. 6C). However, we could not observe a significant reduction in the prevalence and abundance of Streptococcus in the HNC group compared to that in the HC group (Fig. 6D). These results suggest that host bacteria harboring Inocle-α are selectively decreased in the saliva of patients with HNC.
We further analyzed whether the associations of Inocles could be observed in other cancer types. To this end, we downloaded public salivary metagenomic short reads of patients with pancreatic ductal adenocarcinoma (PDAC)30 and those with colorectal cancer (CRC)31, as well as HCs from each study (Supplementary Table S9). As a reference for an inflammatory disease, the data of patients with rheumatoid arthritis (RA)32 were also obtained (Supplementary Table S9). Statistical analysis revealed that the abundance of Streptococcus was significantly decreased in PDAC, whereas that of Inocle-α was not altered (Fig. 6E). In contrast, Inocle-α was significantly decreased in the CRC group compared to the HC group (Fig. 6E), suggesting that the features of Inocle-α are specifically associated with disorders occurring in the gastrointestinal tract.
Discussion
Despite Inocles being globally distributed, they have not been previously identified nor included in reference genomes, partially due to their harboring ISs or hypervariable regions. The low score of the plasmid prediction tool for Inocles reflects the difficulty in detecting Inocles using current bioinformatics tools. These results demonstrate the importance of long-read metagenomics technology in a reference-independent strategy for identifying ECEs. The preNuc successfully reduced the amount of human DNA from saliva samples, contributing to high-throughput long-read metagenomic sequencing. The increase in the sequencing depth of Inocles following preNuc treatment suggests that this method effectively enriches intracellular ECEs. However, considering that the correlation between the bacterial composition of preNuc-treated and untreated samples is 0.83 (Spearman’s rhos), some bias is expected in the composition of ECEs due to preNuc treatment.
The biggest feature of Inocles is their large contig size, with a maximum size of 395 kb, possibly being one of the largest ECEs in the human commensal bacteria. We speculated that Inocles are plasmid-like elements based on nucleotide composition, suggesting that Inocles might be able to be categorized as megaplasmids33. Megaplasmids have been found in several human pathogens, and they carry multiple antimicrobial resistance genes to confer multidrug resistance and increase the infectious ability34,35. On the other hand, Inocles does not contain known antibacterial or virulent genes, suggesting that Inocles provides different benefits to the host bacteria than to the previously identified human-associated megaplasmids.
We identified ORFs of Inocles encode a variety of stress response proteins. For example, RpoS is a sigma factor that globally regulates bacterial transcription and induces stress responses, facilitating survival during environmental stresses35,36,37,38. Notably, we identified a series of genes related to the DNA damage repair pathway from Inocles, including DNA polymerase III, gyrase, RecD, RecJ, and DinG, which are activated by oxidative stress36,37,38,39. In addition, we identified the peroxidase cofactor (NrdH) in Inocle-α, β, and γ, which reduces intracellular reactive oxygen species (ROS) levels27. These results suggest that Inocles are a key factor in resisting harsh environments, including under oxidative stresses and DNA damage in the oral environment (Supplementary Fig. S11).
An additional feature of Inocle is that 20% of all genes are involved in cell wall/membrane-related functions. The discovery of a bifunctional transglycosylase27, signal peptidase, sortase A, and LPXTG-like motif protein suggests the following processes on the cytoplasmic membrane. First, the bifunctional transglycosylase of Inocles promotes peptidoglycan biosynthesis in host bacteria39. Following translation, LPXTG-like motif proteins are transported to the cytoplasmic membrane, where the signal peptidase cleaves the N-terminal signal peptide of LPXTG-like motif proteins40. Sortase A then cleaves the LPXTG-like motif and anchors the mature protein to peptidoglycans as a cell surface protein41. Several studies on biofilms on human epithelial cells have revealed that sortase A contributes to bacterial adhesion and evasion of the human immune system28,42,43. Although the functional roles of LPXTG-like motif proteins are uncharacterized, our findings suggest that cell wall-associated proteins of Inocles are involved in the colonization of Streptococcus to human epithelial cells.
Some plasmids exhibit functional redundancy with host chromosomes and contribute to increased genetic diversity and evolution, thus enhancing adaptive capacity3,33. Homologous genes between Inocles and Streptococcus chromosomes, including DNA damage repair, oxidative stress-related, and cell wall-related genes, likely contribute to enhanced adaptive capacity via functional redundancy. However, most of the genes of Inocles did not exhibit homology with the chromosomal genes of Streptococcus, indicating the functional independence of Inocles.
One of the highlights of this study is the elucidation of the association between Inocle-α and human physiology. We observed a clear positive correlation of the Inocle-α with B cells and signaling responses against infectious pathogens based on the scRNA-seq and plasma proteome dataset, respectively (Supplementary Fig. S11). Although this study does not provide mechanistic insights into the interactions between Inocles, host bacteria, and the human immune system, these results suggest that Inocle-α is involved in latent or tolerant immune responses. Another notable finding from our study is the disease-specific reduction in Inocle-α. Specifically, we observed a reduction in Inocle-α in HNC and CRC but not in PDAC and RA. One plausible explanation of these observations is that alterations in the immune conditions varying across cancer types44 change the necessity of Inocles for Streptococcus to adapt to immune responses. Another possibility is that Inocle-positive individuals are less likely to develop the corresponding cancers owing to their predisposed immune conditions being unfavorable for cancer development. In either case, Inocle has the potential to serve as a biomarker for a non-invasive test for those cancers (Supplementary Fig. S11). In the correlation analysis between Inocle-α and human physiological parameters, we were unable to consider several confounders, such as diet and lifestyle, which are known to impact the composition of the oral microbiome45. Therefore, further studies involving independent cohorts with a comprehensive collection of confounders are crucial to validate the associations between Inocle-α and human physiology.
The biological roles of Inocles remain largely unknown, as the functions of 95% of their genes remain elusive. Our experimental approach identified the host bacterial strain of one example of Inocle-α (Inocle_004). Some S. salivarius strains maintained Inocle_004 in colonies on agar plates, but all were lost in liquid culture. This observation underscores the difficulty of accurately identifying the host bacteria of Inocles. The candidate bacterial hosts of Inocles, except Inocle_004, remain unidentified. In addition, we cannot exclude the possibility for non-Streptococcus species to be hosts of Inocles. The establishment of culturing conditions that stably maintain Inocles in host bacterial cells could experimentally verify their functions and host bacterial range. Overall, despite only providing introductory findings, this study is anticipated to pave the way for further understanding the multi-faceted role or biological relevance of Inocles in modulating the adaptive capacity of oral bacteria and potentially impacting human health.
Methods
Human saliva sample collection
This study was approved by the Ethics Committee of the University of Tokyo, National Cancer Center Hospital East, and Sam Ratulangi University. Informed consent was obtained from all the participants. Four saliva samples were collected from four Japanese participants to develop a preNuc method. For long-read metagenomics, salivary samples were collected from 46 Japanese, 9 Indonesian, and 1 Thailand participants. To estimate the prevalence of Inocles, we collected salivary samples from 68 Japanese and 20 Indonesian participants. In total, 45 salivary samples were collected from patients with HNC. Freshly collected salivary samples were immediately frozen at − 80 °C and stored until use.
DNA extraction from saliva samples
In the preNuc method, a 1 mL saliva sample was suspended in 500 μL of phosphate-buffered saline (PBS) and centrifuged at 12,000 × g for 10 min at 4 °C. The supernatant was discarded, and the pellet was suspended using 500 μL of Tris/MgCl buffer (40 mM Tris/HCl, 10 mM Mg/Cl2). Next, DNase I (50 units) (NIPPON GENE, Tokyo, Japan, cat: 314-08071) and RNase I (5 μg/mL) (NIPPON GENE, Tokyo, Japan, cat: 312-01931) were added and incubated for 30 min at 37 °C followed by centrifugation at 12,000 × g for 10 min at 4 °C. The supernatant was discarded; subsequently, the pellet was suspended in 2 ml of PBS, centrifuged at 12,000 × g for 10 min at 4 °C. The supernatant was discarded, and the pellet was suspended in 800 μL of TE20 buffer (10 mM Tris, 20 mM EDTA). The mixture was mixed with 50 μL of Lysozyme solution (300 mg/mL) (Sigma-Aldrich, MA, USA, cat: L6876) and 20 μL of Achromopeptidase solution (100 units/μL) (FUJIFILM Wako Pure Chemical Corporation, Japan, cat: 015-09951), and then incubated for 2 h at 37 °C. The solution was incubated for 1 h at 55 °C with 10% SDS solution (final concentration 1%) and Proteinase K (0.5 mg) (Sigma-Aldrich, MA, USA, cat: 647-014-00-9). The lysate was treated with phenol/chloroform/isoamyl alcohol (NIPPON GENE, Tokyo, Japan, cat: 311-90151), and DNA was precipitated using isopropanol. DNA was further treated with RNase (NIPPON GENE, Tokyo, Japan, cat: 312-01931) and precipitated using polyethylene glycol solution. DNA was rinsed with 75% ethanol, dried, and dissolved in TE buffer (10 mM Tris-HCl, 1 mM EDTA)46.
For DNA extraction from small particle fractions in saliva, a 1 mL saliva sample was suspended in 1 ml of PBS, centrifuged at 5000 × g for 10 min at 4 °C. The supernatant was filtered using 0.45 μm PVDF pore membrane filters (Steriflip, Merck Millipore, Burlington, MA, USA). The filtrate was mixed with an equal volume of polyethylene glycol solution (20% PEG6000 in 2.5 M NaCl) and stored for 2 h at 4 °C. The solution was centrifuged at 20,000 × g for 45 min at 4 °C to collect small particles. After removing the supernatant, the pellets were suspended in 400 μL of TE20 buffer. The mixture was mixed with 50 μL of Lysozyme solution (300 mg/mL) (Sigma-Aldrich, MA, USA, cat: L6876) and 20 μL of Achromopeptidase solution (100 units/μL) (FUJIFILM Wako Pure Chemical Corporation, Japan, cat: 015-09951), and then incubated for 2 h at 37 °C. The solution was incubated for 1 h at 55 °C with 10% SDS solution (final concentration 1%) and Proteinase K (0.5 mg) (Sigma-Aldrich, MA, USA, cat: 647-014-00-9). The lysate was treated with phenol/chloroform/isoamyl alcohol (NIPPON GENE, Tokyo, Japan, cat: 311-90151), and DNA was precipitated using isopropanol. DNA was further treated with RNase (NIPPON GENE, Tokyo, Japan, cat: 312-01931) and precipitated using polyethylene glycol solution. DNA was rinsed with 75% ethanol, dried, and dissolved in TE buffer (10 mM Tris-HCl, 1 mM EDTA)46.
Long-read metagenomic sequencing and data processing
Library preparation was conducted using a Ligation Sequencing Kit (SQK-LSK114) following the protocol provided by the manufacturer (Oxford Nanopore Technologies, Oxford, UK). Sequencing was performed using PromethION (Oxford Nanopore Technologies) with R.10.4.1 flow cells. Base calling was performed using dorado-0.6.2, and long reads with an average Q score < 20 were removed using NanoFilt (v2.8.0)47. The quality-filtered long reads were mapped to the T2T genome48 using minimap2 (2.26-r1175)49, considering > 90% identity and > 85% alignment coverage, and mapped reads were removed.
Short-read metagenomic sequencing and data processing
The library was prepared using the TruSeq DNA Nano kit (Illumina, San Diego, CA, USA) or the TruSeq ChIP kit (Illumina), following the protocol provided by the manufacturers. Sequencing was performed using NovaSeq 6000 (Illumina, Inc., USA). Quality filtering of the NovaSeq reads was performed using fastp (v0.22.0)50 with the following options: n_base_limit 0 --trim_tail1 1 --cut_mean_quality 20 --qualified_quality_phred 20 --unqualified_percent_limit 50 --length_required 50 --detect_adapter_for_pe 2 --trim_poly_g --cut_tail --cut_tail_window_size 1 –dedup. Quality-filtered reads were mapped to the T2T genome using bowtie251 with default parameters, and mapped reads were removed.
Isolation of the Streptococcus strain, including Inocle
To isolate the Streptococcus strains with Inocle_004, a fresh saliva sample (sample ID: m_5) was plated on De Man–Rogosa–Sharpe (MRS) agar plate (Becton, Dickinson and Company, USA, cat: 288130) and incubated at 37 °C for one day. The presence of Inocle_004 was confirmed by Sanger sequencing of the colony PCR product of five colonies using Inocle_004 specific primer set (Inocle_004_forward 5’-AACGCCAGCTCTTCTGGATA-3’ and Inocle_004_reverse 5’-TGCTCGTGTAGGATCTGTCG-3’). The colony PCR was performed in 2 × KAPA HiFi DNA polymerase (Kapa Biosystems, Wilmington, MA, USA, cat: KK2602) and 10 μM primers under conditions of 3 min at 95 °C, 35 cycles of 98 °C for 20 sec, 60 °C for 30 sec, and 72 °C for 20 sec. Bacterial cells of five colonies were propagated in MRS liquid media at 37 °C for one day under aerobic conditions. Genomic DNA was extracted using the DNeasy PowerSoil Pro Kit (QIAGEN, Hilden, Germany, cat: 47014) from a cultured medium. PCR to check for the presence of Inocle_004 was performed on extracted DNA from liquid media, following the same procedure as the colony PCR. 16S rRNA PCR was performed using 10 μM of V1-V2 specific primers (27Fmod and 338 R)52 and 2 × KAPA HiFi DNA polymerase (Kapa Biosystems, Wilmington, MA, USA, cat: KK2602) under the following conditions: 3 min at 95 °C, 35 cycles of 98 °C for 20 s, 60 °C for 30 s, and 72 °C for 20 s. The Agilent 2100 Bioanalyzer (Agilent Technologies, USA) was used to determine the size of the PCR product. Sanger sequencing was used to sequence the PCR product of the 16S rRNA gene. The 16S rRNA PCR product sequences were then aligned with 16S rRNA sequences of the NCBI to identify the closest species. The whole-genome sequencing library of colony-5 was prepared using the NEBNext Ultra II DNA Library Prep Kit (New England Biolabs, USA, cat: #E7103). The genomic sequences were obtained using the AVITI sequencer (Element Biosciences, San Diego, CA, USA).
The genomic assembly of the AVITI sequences was performed using subsampled reads (500,000 reads) with the default parameters of SPAdes53. The bacterial taxonomy of the isolated strain was determined by GTDB-tk (v2.3.2)54. To calculate the Inocle-α/S. salivarius ratio in native saliva, metagenomic NovaSeq reads of the m_5 saliva sample (isolation source of Inocle_004) were mapped to the isolated S. salivarius and Inocle_004 contigs using bowtie2 (v2.4.1)51, and the RPKM was calculated from the RPB obtained using CoverM (v0.6.1) (--min-read-percent-identity 95 and --min-read-aligned-percent 85) normalized by the total number of reads. Subsequently, the Inocle-α/S. salivarius ratio was obtained by dividing the RPKM of Inocle-α by that of S. salivarius.
Long-read metagenomic assembly
Long-read assembly was performed using Flye (2.9.2-b1786)54 with a --meta and --read-error 0.03. The circularity and sequence depth of each contig were obtained from the metaFlye output file. Initially, two Inocle contigs (Inocle_008 and Inocle_018) were assembled as linear contigs. To validate the circularity of these two contigs, a reference-guided assembly was performed. To purify the long reads of two Inocle contigs assembled as linear contigs (Inocle_008 and Inocle_018), long reads were mapped to initially assembled contigs, considering > 95% identity and > 20% alignment coverage. Subsequently, mapped long reads were assembled using metaFlye, and circular contigs were obtained.
To verify the accuracy of long-read contigs, metagenomic short reads were obtained from two metagenomic DNA samples used to obtain the long reads. Short reads were assembled using MEGAHIT (v1.2.9)55. Short-read contigs were aligned to long-read contigs using minimap2 (2.26-r1175)49 with -cx asm20 parameter and obtained the similarity between short- and long-read contigs. All sequence statistics were obtained using Seqkit (v2.8.0)56.
Classification of long-read contigs into microbial genetic elements
Small contigs < 3 kb and T2T genome-aligned (> 80% identity and > 20% alignment coverage) contigs were removed from the > 10-depth long-read contigs. To identify the bacterial chromosomal contigs, rRNA marker gene sequences were predicted in long-read contigs using Barrnap (v0.9) (https://github.com/tseemann/barrnap) and Prokka (v 1.14.6)56. Other bacterial marker genes were predicted using fetchMG (v1.1)57, and contigs with bacterial marker genes were classified as bacterial chromosomal contigs. Binning analysis was performed using SemiBin2 (v2.0.2)58 with --environment human_oral --sequencing-type=long_read parameters, and contigs in a Bin, including bacterial chromosomal contigs, were classified as potential chromosomal contigs. Potential phage contigs were predicted in non-bacterial chromosomal contigs using VirSorter2 (v2.2.4)59 with > 0.9, and CheckV (v0.7.0)60 with contamination of < 10%. Potential plasmid contigs from non-phage contigs were predicted using PlasClass (v0.1.1)61 with a > 0.7 score. To identify eukaryotic viral and fungal contigs, non-plasmid contigs were aligned to RefSeq viral genomes (download date: 2023/07/06) and FugiDB (v64)62 using minimap2 (2.26-r1175)49 with parameter -cx asm20, > 70% identity, and > 50% alignment coverage. The remaining contigs were considered unclassified. The unclassified contigs were aligned to the plasmid63 and integrated phage genome database4,5,8,9,13,64,65.
ORFs of contigs were predicted using Prokka (v1.14.6)64 with default parameters. All ORFs were aligned to the UniRef9066 database using DIAMOND (v2.1.8)67 with --sensitive and an e-value < 1e-10 options. Unclassified contigs with > 50% ORFs unaligned with the UniRef90 database were classified as strong candidates for unrecognized genetic elements. Average amino acid identity (AAI)-based clustering was performed for contigs clustering49. All ORFs of the 85-strong candidate contigs of unrecognized genetic elements were subjected to “all vs. all aligned” by DIAMOND with --evalue 1e-5, --max-target-seqs 10000, --query-cover 50, and --subject-cover 50 options. All contig pairs satisfying a minimum of 80% shared ORFs, 30% average AAI, and > 10 shared ORFs were subjected to clustering analysis. Contig clustering was performed using MCL (v14-137) with -te 8 -I 2.0 –abc options. Contigs with > 1.5-fold smaller or larger than the average contig size in the same cluster were treated as different clusters. Long-read mapping was performed using minimap2 (2.26-r1175)66 with the map-ont option. The mapping results were visualized using the Integrative Genomics Viewer (IGV)68. To obtain the RPKM, first, we obtained the reads per base (RPB) using CoverM (v0.6.1) (https://github.com/wwood/CoverM) with --min-read-percent-identity 95 and --min-read-aligned-percent 85 options. Subsequently, the RPB was normalized to the total number of reads in each sample, and the RPKM was obtained.
Genomic analysis of Inocles
Based on cumulative GC skew, the replication origin and terminus were predicted using iRep (v1.10)69. Genome visualization was performed using Proksee70. The insertion sequences were predicted using ISEscan (v1.7.2.3)71, considering the default parameters. All high-quality circular contigs having ORFs aligned to at least one InoC gene using DIAMOND with < 1e-10 were identified as additional Inocle contigs. Average nucleotide identity was obtained using Pyani (v0.2.12)72 with the -m ANIb option. The PF of Inocles was obtained by ORF clustering using Roary (v3.13.0)73 with the -i 50 option. Based on the similarity of the PFs repertoire, a dendrogram was created using the ward.D2 method, considering the Euclidean distance calculated using the presence/absence information of the PFs. The short-read contigs assembled using MEGAHIT from the Inocle contig-reconstruction sample were aligned to Inocle contigs using minimap2 (2.26-r1175)64 with > 99% identity and 500 bp alignment length; fragmented positions in short-read contigs were determined based on aligned short-read contigs.
Host bacterial prediction of Inocles
To identify the ORFs of Inocles homologous with host bacterial ORFs, all Inocle ORFs were aligned to the UniRef9066 by DIAMOND (v2.1.8)67 with an e-value < 1e-10, and best-hit results were used for further analysis. The number of Inocle genes homologous to the host bacteria was calculated by summing the UniRef90 genera with the Inocle gene aligned.
Tetranucleotide frequency was obtained using CheckM (v1.1.3)74 from Inocles and chromosomes, plasmids, and phages of Streptococcus. The relative synonymous codon usage was obtained using the Codon Usage Generator (ver2.4) (http://bioinfo.ie.niigata-u.ac.jp/?Codon+Usage+Generator)75. All Streptococcus chromosomes, plasmids, and phages were obtained from the RefSeq database. The dendrograms based on the similarity of tetranucleotide frequency and codon usage were obtained using the ward.D2 method based on the Manhattan distance.
Short reads of the bacterial pellet and 0.45 μm-filtered fraction from the saliva sample were mapped to the Inocle contigs obtained from the same sample. The mapping was performed by bowtie2 (v2.4.1)74, and RPKM was calculated from the RPB obtained using CoverM (v0.6.1) (--min-read-percent-identity 95 and --min-read-aligned-percent 85) normalized to the total number of reads in each sample.
Functional gene annotation of Inocles
InoC was identified via ORF clustering using cd-hit (v4.8.1)76 with -c 0.7 -n 5 -aS 0.5. The 3D structure of InoC was obtained using ColabFold (v1.5.5)77. The phylogenetic tree of InoC was constructed via multiple alignments using MAFFT (v7.505)78 following FastTree (v2.1.11)79. The phylogenetic tree of InoC was constructed using the iToL80. Functional annotation of ORFs was conducted using Prokka (v1.14.6)79. Mobilization-related genes were predicted using ICEscreen (v1.2.0)81 with default parameters. Subcellular localization of ORFs was predicted using Psort (v3.0)82 with the --positive option. The KEGG orthology annotations for the ORFs were performed using the eggNOG mapper (v2.1.11)83 (--sensmode ultra-sensitive), and the KEGG BRITE hierarchy was generated based on the assigned K numbers84. The Pfam domains and active sites of the signal peptidase in the ORFs were predicted using InterProScan85. Transmembrane regions in the cell wall/membrane-related ORFs were predicted using TMHMM-2.086 with default parameters, and the protein 2D structure on the membrane was visualized using protter87. The LPXTG-like motif was predicted using CW-PRED88 with default parameters.
The homologous genes between Inocles and Streptococcus chromosomes were identified via a similarity search between the ORFs of Inocles and Streptococcus chromosomes using DIAMOND (v2.1.8)50 with the --sensitive and e-value < 1e-10 parameters applied.
Sequence analysis of deposited datasets
The deposited metagenomic sequences of HMP51 were downloaded from the NCBI SRA server. Low-quality reads were filtered using fastp51 with the following options --n_base_limit 0 --trim_tail1 1 --cut_mean_quality 20 --qualified_quality_phred 20 --unqualified_percent_limit 50 --length_required 50 --detect_adapter_for_pe 2 --trim_poly_g --cut_tail --cut_tail_window_size 1 –dedup. The quality-filtered reads were mapped to the T2T genome using bowtie251 with default parameters, and unmapped reads were extracted as filter-passed reads. Samples with > 4 million filter-passed reads were used for further analysis. Filter-passed short reads were mapped to non-redundant InoC genes (clustered by 100% identity) using bowtie2 (v2.4.1)51, and the RPKM was calculated from the RPB obtained using CoverM (v0.6.1) (--min-read-percent-identity 95 and --min-read-aligned-percent 85) normalized by the total number of reads in each sample. The RPKM of each Inocle taxon was obtained by summing all RPKM of the InoC gene for each Inocle taxon. To confirm the transcripts of Inocle ORFs, metatranscriptome short reads were mapped to nucleotide sequences of Inocle PFs using bowtie2 (v2.4.1)31, and the RPKM was calculated from the RPB obtained using CoverM (v0.6.1) (--min-read-percent-identity 95 and --min-read-aligned-percent 85) normalized by the total number of reads in each sample.
Genomic sequences (Illumina reads) from oral bacteria were obtained by searching the NCBI database with the keywords “oral,” “isolate,” and “bacteria,” while excluding metagenomic sequences. Short reads were mapped to the InoC genes using bowtie2 (v2.4.1)32.
To compare the abundance of Inocles, salivary metagenomic sequences from HCs and patients with PDAC86, RA88, or CRC31 in each cohort were collected from the NCBI SRA server; one sample was randomly selected from longitudinally sampled participants in the CRC study. Quality filtering and removal of human reads were performed using the same procedures as for the HMP study described above. Samples with > 4 million filter-passed reads were used for comparison analysis.
Single-cell RNA-seq analysis
Whole blood samples were centrifuged, and plasma was acquired from the supernatant for Olink analysis. After plasma was obtained, the remaining specimen was diluted with an equal volume of D-phosphate-buffered saline (PBS) (FUJIFILM, Japan, cat: 045-29795), loaded in SepMate-50 tubes (STEMCELL, Canada, cat: 15460) containing a Lymphoprep (Serumwerk Bernburg, Germany, cat: 1856), and centrifuged at 2000 × rpm for 10 minutes at room temperature. The top layer was poured into a new tube and washed twice with PBS at 1500 × rpm for 5 min at 4 °C. Peripheral PBMCs were obtained as precipitates and resuspended in Cell Banker2 (Nippon Zenyaku Kogyo, Japan, cat: CB031).
The peripheral blood mononuclear cells (PBMCs) were subjected to library preparation for the scRNA-seq using 10 × Genomics Chromium Next GEM Single Cell Multiome ATAC + Gene Expression according to the manufacturer’s user guide (CG000365 Rev A, CG000337 Rev A, 10 × Genomics). The scRNA-seq was performed using NovaSeq 6000 (Illumina). Sequencing datasets were processed using Cell Ranger ARC (Cellranger-arc-2.0.0, (default, intron-include mode, ref = GRCh38-2020-A-2.0.0)). Subsequently, NovaSeq reads were mapped to hg38 using STAR89 and obtained count data of human genes using Seurat (v4.1.1). Subsequently, count data was normalized and integrated to minimize batch effects using Harmony (v0.1.0). Cell clustering was performed using Seurat, and the clusters were manually annotated based on canonical markers described in the Azimuth reference90. GO enrichment was performed using ShinyGO 8.091.
Proteome analysis of human plasma samples
Blood samples and plasma fractions were collected for proteome analysis using Olink. Reactions with antibodies targeting 5420 proteins were conducted following the protocol provided by the manufacturer (Olink Proteomics, Uppsala, Sweden) without bridging different experimental reactions for all samples. Sequence libraries were prepared following the protocol provided by the manufacturer (Olink Explore HT), followed by sequencing using a NovaSeq 6000 (Illumina). Sequence counts were transformed into normalized protein expression (NPX) values and further analyzed.
Association analysis between Inocle-α, Streptococcus, blood cells, and plasma proteome
The abundance of Inocle-α was obtained as RPKM, and the abundance of Streptococcus was obtained using MetaPhlAn4 (v4.1.0)92. Correlation analysis was conducted using partial Spearman’s correlation coefficients, accounting for confounders such as age and sex, with the psych package in RStudio (v2023.12.0 + 369). The p-value of the partial Spearman’s correlation analysis was adjusted by the Benjamini-Hochberg method.
Statistical analysis
All statistical analyses were performed using RStudio (v2023.12.0 + 369). A two-tailed paired student’s t test was used to compare the preNuc-treated and -untreated groups. Fisher’s exact test was used to compare the prevalence of Inocle-α and Streptococcus between the HNC and HC groups. For the association analysis between the Inocle-α and human diseases, the statistical test was performed using MaAsLin292 with default parameters, except for the specific random effects. Data from HCs and patients with HNC were compared using MaAsLin2, with random effects of sex, age, smoking status, and cancer treatment status. Similarly, MaAsLin2, with random effects of sex and age, was performed to compare HCs and patients with PDAC, RA, and CRC in each cohort. Statistical results with p < 0.05 were considered significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The short-read sequences, long-read sequences, and closed circular Inocles contigs data generated in this study have been deposited in the DDBJ/NCBI database under accession code PRJDB18431 [https://ddbj.nig.ac.jp/search/entry/sra-study/DRP012192]. Also, closed circular Inocles contigs data have been deposited in the zenodo under DOI 10.5281/zenodo.16656823 [https://zenodo.org/records/16656823]. The sequences of InoC generated in this study are provided in the zendo under https://doi.org/10.5281/zenodo.13138926 [https://zenodo.org/records/13138926]. The single-cell RNA sequencing data generated in this study have been deposited in the National Bioscience Database Center under accession code JGAS000637 [https://ddbj.nig.ac.jp/resource/jga-study/JGAS000321]. The short-read sequencing data from saliva samples in previous studies were downloaded from the SRA database as follows. CRC cohort (Accession number: PRJNA289586 and PRJEB28422), RA cohort (Accession number: PRJEB6997), and PDAC cohort (Accession number: PRJNA832909).
Code availability
R scripts used for the analyses and figure generation of this paper are available at https://github.com/yuyanoah/Inocle-R-scripts.
References
Wozniak, R. A. F. & Waldor, M. K. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8, 552–563 (2010).
Ross, K. et al. TnCentral: A prokaryotic transposable element database and web portal for transposon analysis. MBio 12, e0206021 (2021).
Rodríguez-Beltrán, J., DelaFuente, J., León-Sampedro, R., MacLean, R. C. & San Millán, Á. Beyond horizontal gene transfer: the role of plasmids in bacterial evolution. Nat. Rev. Microbiol. 19, 347–359 (2021).
Nayfach, S. et al. Metagenomic compendium of 189,680 DNA viruses from the human gut microbiome. Nat. Microbiol. 6, 960–970 (2021).
Gregory, A. C. et al. The gut virome database reveals age-dependent patterns of virome diversity in the human gut. Cell Host Microbe 28, 724–740 (2020).
Soto-Perez, P. et al. CRISPR-Cas System of a prevalent human gut bacterium reveals hyper-targeting against phages in a human virome catalog. Cell Host Microbe 26, 325–335 (2019).
Paez-Espino, D. et al. IMG/VR v.2.0: an integrated data management and analysis system for cultivated and environmental viral genomes. Nucleic Acids Res. 47, D678–D686 (2018).
Camarillo-Guerrero, L. F., Almeida, A., Rangel-Pineros, G., Finn, R. D. & Lawley, T. D. Massive expansion of human gut bacteriophage diversity. Cell 184, 1098–1109 (2021).
Nishijima, S. et al. Extensive gut virome variation and its associations with host and environmental factors in a population-level cohort. Nat. Commun. 13, 1–14 (2022).
Zorea, A. et al. Plasmids in the human gut reveal neutral dispersal and recombination that is overpowered by inflammatory diseases. Nat. Commun. 15, 1–13 (2024).
Zheludev, I. N. et al. Viroid-like colonists of human microbiomes. Cell 187, 6521–6536 (2024).
Warwick-Dugdale, J. et al. Long-read viral metagenomics enables capture of abundant and microdiverse viral populations and their niche-defining genomic islands. PeerJ 7, e6800 (2019).
Yuya, K., Nishijima, S., Kumar, N., Hattori, M. & Suda, W. Long-read metagenomics of multiple displacement amplified DNA of low-biomass human gut phageomes by SACRA pre-processing chimeric reads. DNA Res. 28, dsab019 (2021).
Suzuki, Y. et al. Long-read metagenomic exploration of extrachromosomal mobile genetic elements in the human gut. Microbiome 7, 119 (2019).
Moss, E. L., Maghini, D. G. & Bhatt, A. S. Complete, closed bacterial genomes from microbiomes using nanopore sequencing. Nat. Biotechnol. 38, 701–707 (2020).
Antipov, D., Rayko, M., Kolmogorov, M. & Pevzner, P. A. viralFlye: assembling viruses and identifying their hosts from long-read metagenomics data. Genome Biol. 23, 57 (2022).
Marotz, C. A. et al. Improving saliva shotgun metagenomics by chemical host DNA depletion. Microbiome 6, 1–9 (2018).
Kolmogorov, M. et al. metaFlye: scalable long-read metagenome assembly using repeat graphs. Nat. Methods 17, 1103–1110 (2020).
Consortium, T. H. M. P. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Shkoporov, A. N. et al. Reproducible protocols for metagenomic analysis of human faecal phageomes. Microbiome 6, 68 (2018).
Al-Shayeb, B. et al. Borgs are giant genetic elements with potential to expand metabolic capacity. Nature 610, 731–736 (2022).
Álvarez-Rodríguez, I. et al. Type IV coupling proteins as potential targets to control the dissemination of antibiotic resistance. Front. Mol. Biosci. 7, 201 (2020).
Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Bouillet, S., Bauer, T. S. & Gottesman, S. RpoS and the bacterial general stress response. Microbiol. Mol. Biol. Rev. 88, e0015122 (2024).
Si, M.-R. et al. NrdH Redoxin enhances resistance to multiple oxidative stresses by acting as a peroxidase cofactor in Corynebacterium glutamicum. Appl. Environ. Microbiol. 80, 1750–1762 (2014).
Belstrøm, D. et al. Metagenomic and metatranscriptomic analysis of saliva reveals disease-associated microbiota in patients with periodontitis and dental caries. NPJ Biofilms Microbi. 3, 23 (2017).
Chen, X., Wong, C.-H. & Ma, C. Targeting the bacterial transglycosylase: Antibiotic development from a structural perspective. ACS Infect. Dis. 5, 1493–1504 (2019).
Susmitha, A., Bajaj, H. & Madhavan Nampoothiri, K. The divergent roles of sortase in the biology of Gram-positive bacteria. Cell Surf. 7, 100055 (2021).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Nagata, N. et al. Metagenomic identification of microbial signatures predicting pancreatic cancer from a multinational study. Gastroenterology 163, 222–238 (2022).
Schmidt, T. S. et al. Extensive transmission of microbes along the gastrointestinal tract. Elife 8, https://doi.org/10.7554/elife.42693 (2019).
Zhang, X. et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 21, 895–905 (2015).
Hall, J. P. J., Botelho, J., Cazares, A. & Baltrus, D. A. What makes a megaplasmid? Philos. Trans. R. Soc. Lond. B Biol. Sci. 377, 20200472 (2022).
Cazares, A. et al. A megaplasmid family driving dissemination of multidrug resistance in Pseudomonas. Nat. Commun. 11, 1370 (2020).
Alba, P. et al. Molecular epidemiology of Salmonella Infantis in Europe: insights into the success of the bacterial host and its parasitic pESI-like megaplasmid. Microb. Genom. 6, https://doi.org/10.1099/mgen.0.000365 (2020).
Hagensee, M. E., Bryan, S. K. & Moses, R. E. DNA polymerase III requirement for repair of DNA damage caused by methyl methanesulfonate and hydrogen peroxide. J. Bacteriol. 169, 4608–4613 (1987).
Mishra, S., Tewari, H., Chaudhary, R., S Misra, H. & Kota, S. Differential cellular localization of DNA gyrase and topoisomerase IB in response to DNA damage in Deinococcus radiodurans. Extremophiles 28, 7 (2023).
Ajiboye, T. O., Skiebe, E. & Wilharm, G. Contributions of RecA and RecBCD DNA repair pathways to the oxidative stress response and sensitivity of Acinetobacter baumannii to antibiotics. Int. J. Antimicrob. Agents 52, 629–636 (2018).
Voloshin, O. N., Vanevski, F., Khil, P. P. & Camerini-Otero, R. D. Characterization of the DNA damage-inducible helicase DinG from Escherichia coli. J. Biol. Chem. 278, 28284–28293 (2003).
Kaushik, S., He, H. & Dalbey, R. E. Bacterial signal peptides- navigating the journey of proteins. Front. Physiol. 13, 933153 (2022).
Hendrickx, A. P. A., Budzik, J. M., Oh, S.-Y. & Schneewind, O. Architects at the bacterial surface - sortases and the assembly of pili with isopeptide bonds. Nat. Rev. Microbiol. 9, 166–176 (2011).
Nobbs, A. H., Lamont, R. J. & Jenkinson, H. F. Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev. 73, 407–450 (2009). Table of Contents.
Spirig, T., Weiner, E. M. & Clubb, R. T. Sortase enzymes in Gram-positive bacteria. Mol. Microbiol. 82, 1044–1059 (2011).
Wang, S., He, Z., Wang, X., Li, H. & Liu, X.-S. Antigen presentation and tumor immunogenicity in cancer immunotherapy response prediction. Elife 8, https://doi.org/10.7554/eLife.49020 (2019).
Odendaal, M.-L. et al. Host and environmental factors shape upper airway microbiota and respiratory health across the human lifespan. Cell 187, 4571–4585 (2024).
Said, H. S. et al. Dysbiosis of salivary microbiota in inflammatory bowel disease and its association with oral immunological biomarkers. DNA Res. 21, 15–25 (2014).
De Coster, W. & Rademakers, R. NanoPack2: population-scale evaluation of long-read sequencing data. Bioinformatics 39, btad311 (2023).
Nurk, S. et al. The complete sequence of a human genome. Science 376, 44–53 (2022).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Kim, S.-W. et al. Robustness of gut microbiota of healthy adults in response to probiotic intervention revealed by high-throughput pyrosequencing. DNA Res. 20, 241–253 (2013).
Bankevich, A. et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Chaumeil, P.-A., Mussig, A. J., Hugenholtz, P. & Parks, D. H. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36, 1925–1927 (2019).
Li, D., Liu, C.-M., Luo, R., Sadakane, K. & Lam, T.-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015).
Shen, W., Le, S., Li, Y. & Hu, F. SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE 11, e0163962 (2016).
Sunagawa, S. et al. Metagenomic species profiling using universal phylogenetic marker genes. Nat. Methods 10, 1196–1199 (2013).
Pan, S., Zhao, X.-M. & Coelho, L. P. SemiBin2: self-supervised contrastive learning leads to better MAGs for short- and long-read sequencing. Bioinformatics 39, i21–i29 (2023).
Guo, J. et al. VirSorter2: a multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses. Microbiome 9, 37 (2021).
Nayfach, S. et al. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat. Biotechnol. 39, 578–585 (2021).
Pellow, D., Mizrahi, I. & Shamir, R. PlasClass improves plasmid sequence classification. PLoS Comput. Biol. 16, 1–9 (2020).
Stajich, J. E. et al. FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res. 40, D675–D681 (2012).
Galata, V., Fehlmann, T., Backes, C. & Keller, A. PLSDB: A resource of complete bacterial plasmids. Nucleic Acids Res. 47, D195–D202 (2019).
Li, S. et al. A catalog of 48,425 nonredundant viruses from oral metagenomes expands the horizon of the human oral virome. iScience 25, 104418 (2022).
Roux, S. et al. IMG/VR v3: An integrated ecological and evolutionary framework for interrogating genomes of uncultivated viruses. Nucleic Acids Res. 49, D764–D775 (2021).
Suzek, B. E., Huang, H., McGarvey, P., Mazumder, R. & Wu, C. H. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23, 1282–1288 (2007).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2014).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Brown, C. T., Olm, M. R., Thomas, B. C. & Banfield, J. F. Measurement of bacterial replication rates in microbial communities. Nat. Biotechnol. 34, 1256–1263 (2016).
Grant, J. R. et al. Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 51, W484–W492 (2023).
Xie, Z. & Tang, H. ISEScan: automated identification of insertion sequence elements in prokaryotic genomes. Bioinformatics 33, 3340–3347 (2017).
Pritchard, L., Glover, R. H., Humphris, S., Elphinstone, J. G. & Toth, I. K. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal. Methods 8, 12–24 (2015).
Page, A. J. et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693 (2015).
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015).
Suzuki, H., Brown, C. J., Forney, L. J. & Top, E. M. Comparison of correspondence analysis methods for synonymous codon usage in bacteria. DNA Res. 15, 357–365 (2008).
Li, W. & Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650 (2009).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019).
Lao, J. et al. ICEscreen: a tool to detect Firmicute ICEs and IMEs, isolated or enclosed in composite structures. NAR Genom. Bioinform. 4, lqac079 (2022).
Yu, N. Y. et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615 (2010).
Cantalapiedra, C. P., Hernández-Plaza, A., Letunic, I., Bork, P. & Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol 38, 5825–5829 (2021).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 (2016).
Jones, P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014).
Krogh, A., Larsson, B., Von Heijne, G. & Sonnhammer, E. L. L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305, 567–580 (2001).
Omasits, U., Ahrens, C. H., Müller, S. & Wollscheid, B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30, 884–886 (2014).
Fimereli, D. K. et al. in Artificial Intelligence: Theories and Applications 285–290 (Springer Berlin Heidelberg, 2012).
Dobin, A. et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Ge, S. X., Jung, D. & Yao, R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36, 2628–2629 (2019).
Blanco-Míguez, A. et al. Extending and improving metagenomic taxonomic profiling with uncharacterized species using MetaPhlAn 4. Nat. Biotechnol. 41, 1633–1644 (2023).
Mallick, H. et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput. Biol. 17, e1009442 (2021).
Acknowledgements
The authors thank M. Tsubaki, K. Imamura, and K. Abe for technical support in the experiments. The authors thank E. Ishikawa, R. Fujinaga, and S. Takashima for technical support in informatics. The authors thank Prof. Dr. dr. Nova H. Kapantow, DAN, M.Sc, Sp.GK for the saliva sample collection from Indonesian participants. The authors thank K. Matsuura for collecting saliva samples from patients with HNC. The authors thank S. Stamouli and D. Takewaki for the critical reading of this manuscript. The authors thank M. Iwanaga for technical support in the isolation of Streptococcus. This study was supported by grants from the Institute for Fermentation, Osaka (Y-2022-1-010), the Japan Agency for Medical Research and Development (AMED) (22fk0108538s0201), and the JSPS KAKENHI (24K18092) to Y.Kiguchi. This study was supported by a grant from the JSPS KAKENHI (22K15833) to T.E. This study was supported by grants from the Platform for Advanced Genome Science (22H04925) and JST Moonshot R&D-MILLENIA Program (JPMJMS2025) to Y.S.
Author information
Authors and Affiliations
Contributions
Y.Kiguchi and Y.S. conceived and supervised this study. Y.S. managed sequencing and bioinformatics platforms. N.H., Y.Kiguchi, and T.M. developed the preNuc method. Y.Kiguchi, N.H., and T.M. contributed to the metagenomic DNA extraction, sequencing, and analysis. L.R.R., J.S.B.T., Y.Kiguchi, and T.M. collected saliva samples from Indonesian participants. A.I. and T.M. isolated the Streptococcus strain with Inocle. T.E., S.K., T.F., N.T., M.T. and R.Y. collected the saliva samples from patients with HNC. Y.Kashima and A.K. obtained sequencing data for scRNA-seq and the Olink proteome from the blood samples. Y.Kashima analyzed the scRNA-seq dataset. Y.Kiguchi analyzed the Olink proteome dataset. Y.Kuze developed the bioinformatics environment. Y.Kiguchi, N.H., T.M., and Y.S. wrote the manuscript. All authors have read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests, except for N.H. The N.H. declares the financial interests as an employee of Medical & Biological Laboratories Co., Ltd.
Peer review
Peer review information
Nature Communications thanks Jonathon Baker, Wei-Hua Chen, and the other anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kiguchi, Y., Hamamoto, N., Kashima, Y. et al. Giant extrachromosomal element “Inocle” potentially expands the adaptive capacity of the human oral microbiome. Nat Commun 16, 7397 (2025). https://doi.org/10.1038/s41467-025-62406-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-62406-5