Abstract
The precise structural and functional characteristics of input circuits targeting histaminergic neurons remain poorly understood. Here, using a rabies virus retrograde tracing system combined with fluorescence micro-optical sectioning tomography, we construct a 3D monosynaptic long-range input atlas of male mouse histaminergic neurons. We identify that the hypothalamus, thalamus, pallidum, and hippocampus constitute major input sources, exhibiting diverse spatial distribution patterns and neuronal type ratios. Notably, a specific layer distribution pattern and co-projection structures of upstream cortical neurons are well reconstructed at single-cell resolution. As histaminergic system is classically involved in sleep-wake regulation, we demonstrate that the lateral septum (predominantly supplying inhibitory inputs) and the paraventricular nucleus of the thalamus (predominantly supplying excitatory inputs) establish monosynaptic connections, exhibiting distinct functional dynamics and regulatory roles in rapid-eye-movement sleep. Collectively, our study provides a precise long-range input map of mouse histaminergic neurons at mesoscopic scale, laying a solid foundation for future systematic study of histaminergic neural circuits.
Similar content being viewed by others
Introduction
The neuronal histaminergic system plays crucial roles in multiple physiological functions, including sleep-wake regulation1, energy and endocrine homeostasis2, cognition and memory3,4, and motor functions5. Dysregulation or lesions of this system contribute to the pathogenesis of numerous neurological disorders, including insomnia and narcolepsy6,7, Alzheimer’s disease8, Parkinson’s disease9, multiple sclerosis10, depression11, and schizophrenia12,13. The potential interactions between histaminergic upstream and downstream neural circuits represent a key aspect for understanding the complicated functions of the central histaminergic network. Furthermore, several functionally characterized histaminergic circuits have demonstrated therapeutic potential for CNS disorders14,15,16,17, highlighting the importance of systemic investigations of the histaminergic network. Histaminergic neurons, localized exclusively in the tuberomammillary nucleus (TMN), project highly heterogeneous axonal fibers to nearly all major brain regions. These neurons regulate diverse physiological functions through distinct downstream circuits, which have been well reconstructed at mesoscopic resolution in our previous study18. However, the mechanism by which upstream input circuits regulate the histaminergic system remains another critical research question with profound physiological significance.
A previous study has mapped whole-brain upstream inputs to the TMN using cholera toxin subunit B (CTB)-based retrograde tracing. This approach identified 70 forebrain and brainstem neuronal groups projecting to the TMN, with particularly dense projections originating from the infralimbic cortex, lateral septum (LS), and preoptic region19. However, owing to the non-selective labeling properties of CTB and neuronal heterogeneity within the TMN20, this study could not achieve systematic, specific labeling of monosynaptic upstream inputs to histaminergic neurons. In recent decades, the development of genetically modified rabies virus (RV) systems has enabled monosynaptic retrograde tracing of defined neuronal populations21,22. Although Saito et al. employed RV-based retrograde tracing to investigate monosynaptic inputs to histaminergic neurons, their study only provided partial data on the distribution of these upstream cells in limited brain regions23. A comprehensive three-dimensional mapping of histaminergic upstream inputs across the whole-brain remains lacking. Recent advances in fluorescence micro-optical sectioning tomography (fMOST) technology have revolutionized precise circuit mapping studies for specific brain areas or neuronal types18,24,25,26,27. This cutting-edge methodology has significantly accelerated progress in precise functional circuit analysis across multiple neuroscience research domains. On the other hand, functional studies on the upstream circuits regulating histaminergic neurons remain relatively scarce28. Given the heterogeneous distribution of various neurotransmitter receptors on histaminergic neurons29, it is of critical importance—yet still lacks direct experimental evidence—to elucidate how distinct types of upstream input circuits directly modulate histaminergic neuronal activity and their potential functional consequences, including canonical histaminergic functions like sleep-wake regulation.
In this study, we develop a comprehensive and precise whole-brain atlas of monosynaptic long-range inputs to histaminergic neurons by integrating a cutting-edge fMOST system with HDC-CreERT2 mice and a retrograde RV tracing system. Importantly, we further select the LS and paraventricular nucleus of the thalamus (PVT) as representative inhibitory and excitatory upstream brain regions, to investigate their differential functional dynamics and causal roles in the sleep-wake cycle. These results provide a foundation for future functional studies of neural circuits projecting to the histaminergic nervous system.
Results
Whole-brain distribution of monosynaptic long-range inputs to histaminergic neurons
To genetically label the monosynaptic long-range input neurons projecting to histaminergic neurons, we first injected a mixture of rAAV-EF1α-DIO-mCherry-F2A-TVA-WPRE-hGH polyA and rAAV-EF1α-DIO-oRVG-WPRE-hGH polyA (TVA:RVG mixing ratio = 1:2) into the TMN of HDC-CreERT2 mice (Fig. 1a). These two helper viruses enabled TVA and RVG protein expression in Cre-positive neurons, where TVA mediated RV-ENVA-ΔG-EGFP entry into the cell body, and RVG facilitated monosynaptic retrograde tracing by RV-ENVA-ΔG-EGFP25. Tamoxifen (100 mg/kg, i.p.) was administered for 5 consecutive days starting one day after viral injection to induce Cre recombinase activity (all CreERT2 mice received tamoxifen administration18). Three weeks later, RV-ENVA-ΔG-EGFP was injected into the same region (Fig. 1a). Cells co-labeled with TVA-mcherry and RV-EGFP were defined as starter cells (Fig. 1b, d). RV-labeled upstream neurons were distributed across multiple brain regions at whole-brain scale, exhibiting distinct distribution patterns (Fig. 1c). When RV retrograde tracing system was injected unilaterally into the TMN (Supplementary Fig. 1a), RV-labeled upstream neurons were predominantly located in the ipsilateral hemisphere (Supplementary Fig. 1b).
a Experimental scheme and timeline for labeling long-range monosynaptic inputs to histaminergic neurons in mouse brain. rAAV-EF1α-DIO-mCherry-F2A-TVA-WPRE-hGH polyA, rAAV-EF1α-DIO-oRVG-WPRE-hGH polyA, and RV-ENVA-ΔG-EGFP was injected into TMN of HDC-CreERT2 adult mice. b Representative images of TVA-mcherry and RV-eGFP expression on TMN. This experiment was repeated independently with similar results for three times. c Representative images of coronal sections with several distance from bregma for RV-eGFP expression of mouse brains, with viruses injected into TMN bilaterally. d Drawings of the distribution of starter cells from several coronal sections containing TMN of sample mice. n = 3. e Main steps of data generation, acquisition, and processing in fMOST imaging. Raw data were analyzed by NeuroGPS system and manual correction. f Fluorescent-labeled information visualization of registered RV-eGFP-expression neurons of mouse brain (Top: horizontal and full view; Bottom: coronal and sagittal views). g Fluorescent-labeled neuron proportions of main upstream brain regions of histaminergic neurons from 5 mice. See the details and list of abbreviations in Supplementary Tables 1 and 2. All data are presented as mean ± SEM.
To verify the specificity and reliability of RV monosynaptic retrograde tracing system, we performed a series of control experiments. First, to assess viral expression specificity, we injected rAAV-EF1α-DIO-mCherry-F2A-TVA-WPRE-hGH polyA bilaterally into the TMN of HDC-CreERT2 mice (Supplementary Fig. 1c). Immunohistochemical staining for HDC revealed that the majority of TVA-labeled neurons were co-labeled with HDC (totally 87.27 ± 1.298 %, n = 3), indicating the convincing specificity of Cre-dependent expression manner of rAAV-EF1α-DIO-mCherry-F2A-TVA-WPRE-hGH polyA (Supplementary Fig. 1d, e). Furthermore, the expression specificity of starter cells (totally 86.50 ± 0.4513%, n = 3) was verified by immunohistochemistry in tested mice expressing the complete RV system (Supplementary Fig. 1a, f–h). To further evaluate potential leakage of RV expression, we conducted additional control experiments. Injection of RV-ENVA-ΔG-EGFP alone into the TMN of HDC-CreERT2 mice (Supplementary Fig. 1i) resulted in no labeled neurons throughout the brain (Supplementary Fig. 1f). When co-injecting rAAV-EF1α-DIO-mCherry-F2A-TVA-WPRE-hGH polyA and RV-ENVA-ΔG-EGFP into the TMN of HDC-CreERT2 mice without RVG component (Supplementary Fig. 1k), no labeled neurons were detected outside the TMN (Supplementary Fig. 1l). In wild-type (WT) mice receiving all three components (Supplementary Fig. 1m), only sparse labeling was observed in limited brain regions including medial prefrontal cortex (mPFC), anterior hypothalamus (AHY), and posterior amygdala nucleus (PA) (Supplementary Fig. 1n). This labeling was negligible compared to the extensive labeling observed in HDC-CreERT2 mice (Fig. 1c). These results convincingly demonstrate the specificity and reliability of the monosynaptic retrograde RV tracing methodology in HDC-CreERT2 mice.
To establish a precise and comprehensive whole-brain atlas of monosynaptic long-range inputs to histaminergic neurons, we employed high-resolution whole-brain sectioning, imaging, and reconstruction through fMOST system (Fig. 1e). Briefly, mouse brain samples were sectioned coronally at 1 μm thickness and automatically imaged with a resolution of 0.32 × 0.32 × 1 μm3. The raw data, comprising more than 10,000 coronal slices per brain, were registered to the Allen Reference Atlas Common Coordinate Framework (CCF v3.0)30 for spatial standardization (through careful delineation of major brain structures, manual verification, and automated computation and registration), consistent with our previous methodology18,25. Standardized whole-brain 3D reconstructions of monosynaptic long-range inputs to histaminergic neurons were visualized through coronal, sagittal, horizontal, and full-view projections (Fig. 1f), with representative coronal sections displayed at multiple rostrocaudal levels (Supplementary Fig. 2a). Quantitative distribution analysis of input neurons across all major brain regions was performed using NeuroGPS system followed by manual correction. To avoid potential interference from injection areas, we excluded brain regions containing histaminergic neuron somata (Supplementary Table 3), as implemented in our previous work18. In general, we found that the upstream cells targeting histaminergic neurons showed three key distribution patterns: (1) medial preference over lateral regions, (2) ventral preference over dorsal areas, and (3) concentration along part of the anterior-posterior axis (0 mm ~ −2.5 mm from bregma) (Supplementary Fig. 2b–d). At the regional level, hypothalamic nuclei contained the most upstream cells even after excluding the injection site (Fig. 1g), whereas the cerebellum (CB) showed the lowest density (Fig. 1g).
Quantitative analysis revealed the anterior hypothalamic nucleus (AHN, 8.20 ± 1.37%, n = 5) as the predominant source of upstream inputs, accounting for a significantly greater proportion than any other brain region (Fig. 1g and Supplementary Fig. 3b). Substantial inputs were also observed in the striatum, pallidum, hippocampal formation, and cortical subplate, with key nuclei including: the medial amygdalar nucleus (MEA, 3.83 ± 0.27%, n = 5), lateral septal nucleus, rostral part (LSr, 2.44 ± 0.08%, n = 5), and lateral septal nucleus, ventral part (LSv, 2.25 ± 0.36%, n = 5) in the striatum, the bed nuclei of the stria terminalis (BST, 4.74 ± 0.53%, n = 5) in the pallidum, field CA1 (CA1, 3.09 ± 0.44%, n = 5) in the hippocampal formation, and the posterior amygdalar nucleus (PA, 3.08 ± 0.28%, n = 5) in the cortical subplate (Fig. 1g and Supplementary Fig. 3c–f, h). Although the midbrain and thalamus only contained moderate input densities overall, specific nuclei, including the periaqueductal gray (PAG, 2.59 ± 0.41%, n = 5) and paraventricular nucleus of the thalamus (PVT, 2.00 ± 0.35%, n = 5) exhibited a relatively large number of upstream cells (Fig. 1g and Supplementary Fig. 3b, h). In contrast, fewer upstream cells were distributed in olfactory areas (OLF), hindbrain (HB), and the cortex. Spatially, upstream cells were diffusely distributed in OLF and HB (Fig. 1g and Supplementary Fig. 3a, g), whereas cortical inputs displayed a layer V-predominant distribution pattern (Fig. 1g and Supplementary Fig. 3d, f), which warrants further analysis. 3D visualization of major input brain regions (Supplementary Fig. 4) revealed several notable spatial preferences: (1) LS and CA1 showed ventral-dominant distributions (Supplementary Fig. 4a, i); (2) PVT inputs were exclusively localized to its anterior region (Supplementary Fig. 4g). Complete distributions across all sample mice are provided in Supplementary Tables 1 and 2, with major input areas summarized in Fig. 1g and Supplementary Fig. 3a–h.
Building upon our previous findings regarding the projection heterogeneity of histaminergic neurons18, we sought to examine whether subpopulations projecting to different downstream areas also exhibit differential input patterns. Firstly, we injected a 1:2 mixture of rAAV-EF1α-DIO-mCherry-F2A-TVA-WPRE-hGH polyA and rAAV-EF1α-DIO-oRVG-WPRE-hGH polyA into the bilateral TMN of HDC-CreERT2 mice. After three weeks, RV-ENVA-ΔG-EGFP was injected into either the MS or SC (Supplementary Fig. 7f). Notably, both MS- and SC-projecting histaminergic neuron subpopulations received dense inputs from the mPFC, LS, PVT, MEA, PA, and vCA1 (Supplementary Fig. 7g–l). However, comparative analysis revealed significant differences in input density (ratio of input cells/starter cells) from several upstream regions—including the mPFC, LS, PA, and vCA1—between MS- and SC-projection-defined subpopulations and the general TMN histaminergic neurons (Supplementary Fig. 7m–r). These findings support the hypothesis that histaminergic neurons with distinct efferent projections differentially integrate afferent inputs from upstream brain regions.
Specific layer distribution and co-projection characteristics of cortical inputs to histaminergic neurons
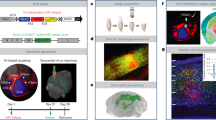
The cortex is well-established to play crucial roles in emotional and sensory processes31,32. Our analysis of fMOST raw data (Fig. 2c) and the quantified whole-brain input atlas (Fig. 1g) revealed a predominant layer V distribution of cortical inputs to histaminergic neurons, suggesting potential functional specialization. To characterize detailed cortical input organization, we first performed 3D visualization of all cortical input neurons (Fig. 2a). Notably, input neurons were widely distributed across most cortical regions, albeit with varying densities (Fig. 2b). Quantitative analysis demonstrated that most of cortical inputs originated from layer V (61.77 ± 5.54%, n = 5), followed by layer VIa (23.19 ± 4.35%, n = 5), while other cortical layers contained sparse inputs (Fig. 2d). At the regional level, the layer V preference was conserved across most cortical areas, with the notable exception of the infralimbic area (ILA), a subregion of mPFC that exhibited greater input density in layer VIa than in layer V (Fig. 2e).
a Fluorescent-labeled information visualization of registered RV-eGFP-expression cortical neurons of mouse brain (Top: coronal and sagittal views; Bottom: horizontal view and coronal sketch map). b Fluorescent-labeled neuron proportions of all cortical brain regions of histaminergic neurons. n = 5 (derived from different mice). c Representative image of the specific cell body layer distribution manner of one of the tested mice. d, e Fluorescent-labeled neuron proportions of different layers in all cortical brain regions (d) and each cortical brain region (e) of histaminergic neurons. n = 5 (derived from different mice). f Total 150 well reconstructed co-projection structure of upstream cortical neurons in different hemispheres (Top: horizontal view of cortical neurons in left and right hemisphere; Bottom: coronal views of cortical neurons in left and right hemisphere). g, h Co-projection proportion analyze (g) and simplified diagram (h) of well reconstructed neurons in different cortical brain regions. See the list of abbreviations in Supplementary Tables 1 and 2. All data are presented as mean ± SEM.
To further elucidate the co-projection patterns of different cortical input neurons and their potential regulatory mechanisms at the whole-brain circuitry level, we performed single-neuron morphological reconstruction using the fMOST raw data. Following semiautomatic tracing and manual verification, 150 well reconstructed neurons were obtained from various cortical areas. These neurons were then registered to CCF v3.0 for 3D visualization and projection target analysis (Fig. 2f–h and Supplementary Fig. 5). At the whole-cortex level, our analysis revealed that except for hypothalamus and cortex, the majority of input neurons co-projected to multiple subcortical regions, primarily the striatum, midbrain, pallidum, and thalamus (Fig. 2h). Notably, all reconstructed neurons innervated exclusively the ipsilateral hemisphere (Fig. 2f). Region-specific projection patterns were identified in certain cortical areas. For example, the entorhinal area was the sole cortical region exhibiting co-projections to HPF. Moreover, mPFC, orbital area (ORB), and agranular insular area (AI) displayed selective connectivity with OLF. Given the distinct somatic distribution patterns between two mPFC subregions, the ILA and PL, we further compared their co-projection patterns. Strikingly, ILA neurons preferentially co-projected to anterior regions (e.g., OLF and STR), whereas PL neurons preferentially co-projected to posterior areas (e.g., PAL, TH, and MB) (Supplementary Fig. 5b, c), suggesting distinct functional circuitry for these subregions.
Distinct organization of inhibitory and excitatory inputs to histaminergic neurons
Histaminergic neurons are known to widely express GABAA receptors and NMDA/AMPA receptors, mediating substantial inhibitory and excitatory inputs that modulate histaminergic signaling33,34,35. However, the precise distribution of GABAergic and glutamatergic neurons among most upstream brain areas remains poorly characterized, impeding functional studies of input circuits to histaminergic neurons. To address this, we performed RNAscope in situ hybridization on coronal slices containing major upstream brain regions of RV-labeled mouse brains (Fig. 3a). We selected GAD1 as the molecular marker for GABAergic neurons. For glutamatergic neurons, since vglut2 is widely expressed in subcortical brain areas but not in the cortex and hippocampus, while CaMKIIα serves as an appropriate marker for cortical and hippocampal areas36,37, we accordingly used GAD1-C1 and vglut2-C2/CaMKIIα-C3 probes. To compensate for the potential RV-EGFP quenching during the in situ hybridization process, we performed GFP immunohistochemical staining before confocal imaging. Quantitative analysis revealed that RV-EGFP labeled neurons in the LS and medial amygdalar nucleus (MEA) were predominantly GAD1-positive (LS: 98.82 ± 0.80%, MEA: 97.73 ± 2.27%, n = 4), with minimal vglut2/CaMKIIα signal (LS: 0.61 ± 0.36%, MEA: 0%, n = 4), indicating predominantly inhibitory inputs from these regions (Fig. 3b, i, j). BST also showed most RV-EGFP labeled neurons co-labeled with GAD1 (89.17 ± 1.78%, n = 4), with only a small fraction co-labeled with vglut2 (7.45 ± 1.29%, n = 4, Fig. 3f, j). In contrast, the PVT, CA1, and PA/BMAp contained primarily vglut2/CaMKIIα-positive inputs (PVT: 98.77 ± 0.95%, CA1: 97.09 ± 0.66%, PA/BMAp: 74.38 ± 2.92%, n = 4), with negligible GAD1 co-expression (PVT: 0.23 ± 0.23%, CA1: 0%, PA/BMAp: 2.1 ± 0.81%, n = 4), indicating basically excitatory inputs from them (Fig. 3d, g, h, j). On the other hand, PAG and anterior hypothalamic areas (AHY) exhibited mixed inputs, with substantial GAD1-positive (PAG: 70.14 ± 4.68%, AHY: 61.29 ± 4.82%, n = 4) and vglut2/CaMKIIα-positive populations (PAG: 26.56 ± 4.82%, AHY: 22.63 ± 1.43%, n = 4) (Fig. 3c, e, j). Given the heterogeneous composition of PAG and AHY input neurons, we further reconstructed the distribution of GABAergic and glutamatergic somata in coronal section. Spatial analysis revealed that GABAergic input neurons were widely distributed throughout the AHY, while glutamatergic input neurons were concentrated near the third ventricle (V3) and scattered across other areas. In PAG, GABAergic input neurons preferred ventral and lateral areas, whereas glutamatergic neurons were uniformly distributed (Supplementary Fig. 6a, b). These findings systematically characterize the major inhibitory and excitatory components of inputs to histaminergic neurons, establishing an important foundation for precise functional circuit studies in the future.
a Experimental scheme and timeline for viruses injection and RNAscope in situ hybridization. b–i Representative images of in situ hybridization labeled GABAergic (GAD1) or glutamatergic (Vglut2 or CaMKIIα) markers co-located with RV-eGFP labeled neurons in LS (b), AHY (c), PVT (d), PAG (e), BST (f), CA1 (g), PA/BMAp (h), and MEA (i). j Quantification of eGFP-labeled neurons in major upstream brain regions that are GABAergic (GAD1) or glutamatergic (Vglut2 or CaMKIIα). n = 4. All data are presented as mean ± SEM. All experiments in Fig. 4b–i were repeated independently with similar results for four times. AHY Anterior hypothalamus, BMA Basomedial amygdalar nucleus, BST Bed nuclei of the stria terminalis, CA1 field CA1, LS Lateral septal nucleus, MEA Medial amygdalar nucleus, PA Posterior amygdalar nucleus, PAG Periaqueductal gray, PVT Paraventricular nucleus of the thalamus.
Functional characterization of input circuits to histaminergic neurons remains limited compared to advances in histaminergic output circuit research, and functional analysis may in turn, validate the significance of established structural connections. Given the well-established role of the histaminergic system in sleep-wake regulation, we sought to preliminary validate the biological significance of differential modulation by excitatory and inhibitory inputs on histaminergic neurons. Accordingly, we selected two representative upstream brain regions, the LS (predominantly inhibitory neurons) and PVT (predominantly excitatory neurons), for the following study. These regions were chosen based on two key observations: (1) their substantial input neuron populations to histaminergic neurons (this study), and (2) the dense histaminergic fiber innervation revealed in our previous work18, suggesting their potential importance in both afferent and efferent histaminergic circuits. To further confirm monosynaptic connectivity between LS/PVT and histaminergic neurons, we employed a Cre-dependent WGA monosynaptic anterograde tracing system38. For LS GABAergic neurons, a 1:1 mixture of rAAV-VGAT1-CRE-EGFP-WPRE-hGH polyA and rAAV-CAG-DIO-mWGA-mCherry was injected into the bilateral LS of HDC; AI47 mice (Fig. 4a). Three weeks post-injection, WGA-mCherry/GFP co-labeled neurons in the TMN confirmed the monosynaptic LS→histaminergic neuron connection (Fig. 4b). Similarly, PVT glutamatergic neuron tracing was performed by injecting a 1:1 mixture of rAAV-VGLUT2-CRE-WPRE-hGH polyA and rAAV-CAG-DIO-mWGA-mCherry into the PVT of HDC; AI47 mice (Fig. 4c), and WGA-mCherry/GFP co-labeled neurons were also found in the TMN (Fig. 4d). On the other hand, considering different upstream brain regions may also exhibit input variability to histaminergic neurons, we performed WGA-based anterograde tracing from the PVT and LS in the same mouse. We injected a 1:1 mixture of rAAV-VGLUT2-CRE-WPRE-hGH polyA and rAAV-CAG-DIO-mWGA-mCherry into the PVT, and a 1:1 mixture of rAAV-VGAT1-CRE-EGFP-WPRE-hGH polyA and rAAV-CAG-DIO-WGA-EGFP into the bilateral LS of WT mice simultaneously (Supplementary Fig. 7a–c). Quantitative analysis revealed that while a substantial subset of histaminergic neurons received convergent inputs from both LS (38.54 ± 4.132%, n = 3) and PVT (41.54 ± 4.435%, n = 3), the majority exhibited selective innervation from either region (Supplementary Fig. 7d, e), demonstrating significant input variability of histaminergic neurons from different brain regions.
a Experimental scheme for labeling monosynaptic downstream neurons in TMN from long-range projection of LS GABAergic neurons. rAAV-VGAT1-CRE-EGFP-WPRE-hGH polyA mixed with rAAV-CAG-DIO-mWGA-mCherry was injected into the bilateral LS of HDC; AI47 mouse brain. b Representative images of mWGA-mCherry (monosynaptic input from LS) labeled neurons co-located with GFP (n = 3). c Experimental scheme for labeling monosynaptic downstream neurons in TMN from long-range projection of PVT glutamatergic neurons. rAAV-VGLUT2-CRE-WPRE-hGH polyA mixed with rAAV-CAG-DIO-mWGA-mCherry was injected into the PVT of HDC; AI47 mouse brain. d Representative images of mWGA-mCherry (monosynaptic input from PVT) labeled neurons co-located with GFP (n = 3). e Experimental scheme for viruses injection and electrophysiological test of histaminergic neurons. rAAV-VGAT1-CRE-EGFP-WPRE-hGH polyA mixed with AAV2/9-hSyn-FLEX-ChrimsonR-tdTomato-WPRE-pA was injected into LS of HDC; AI47 mouse brain. 5 ms, 1 Hz pulse of 635 nm red light was performed to evoke IPSCs on histaminergic neurons. f, g Representative images of clamped neuron (f) and all images of pulse laser evoked IPSCs (g), which still present after TTX and 4-AP administration but blocked by GABAA receptor antagonist bicuculline (n = 3). Traces coming from each cell are shown as same color. h Experimental scheme for viruses injection and electrophysiological test of histaminergic neurons. rAAV-VGLUT2-CRE-WPRE-hGH mixed with AAV2/9-hSyn-FLEX-ChrimsonR-tdTomato-WPRE-pA was injected into PVT of HDC; AI47 mouse brain. 5 ms, 1 Hz pulse of 635 nm red light was performed to evoke EPSCs on histaminergic neurons. i, j Representative images of clamped neuron (i) and all images of pulse laser evoked EPSCs (j), which still present after TTX and 4-AP administration but blocked by NMDA receptor antagonist APV and AMPA receptor antagonist CNQX (n = 3). Traces coming from each cell are shown as same color. APV 2-amino-5-phosphonovaleric, CNQX 6-cyano-7-nitroquinoxaline-2,3-dione, LS Lateral septal nucleus, PVT Paraventricular nucleus of the thalamus, TMN Tuberomammillary nucleus, TTX Tetrodotoxin, 4-AP: 4-aminopyridine.
To functionally characterize the monosynaptic connections between LS/PVT and histaminergic neurons, we performed whole-cell patch-clamp recordings in acute brain slices containing histaminergic neurons receiving red light-evoked opsin ChrimsonR-expressing inputs from LS/PVT39. For LS GABAergic inputs, we injected a 1:1 mixture of rAAV-VGAT1-CRE-EGFP-WPRE-hGH polyA mixed and AAV2/9-hSyn-FLEX-ChrimsonR-tdTomato-WPRE-pA into the bilateral LS of HDC; AI47 mice (Fig. 4e). Upon 5 ms red light stimulation, robust inhibitory post synaptic currents (IPSCs) were recorded in the GFP-labeled histaminergic neurons immediately (Fig. 4f, g). Notably, these IPSCs persisted in the presence of TTX and 4-AP but were abolished by the GABAA receptor antagonist bicuculline, confirming direct monosynaptic GABAergic transmission from the LS to histaminergic neurons (Fig. 4g). Analogously, for PVT glutamatergic inputs, we injected the same opsin-expressing virus mixture into the PVT of HDC; AI47 mice (Fig. 4h). Upon 5 ms red light stimulation, robust excitatory post synaptic currents (EPSCs) were recorded in the GFP-labeled histaminergic neurons immediately (Fig. 4i, j). Notably, these EPSCs persisted in the presence of TTX and 4-AP but were abolished by the NMDA receptor antagonist APV and AMPA antagonist CNQX, verifying direct monosynaptic glutamatergic input from the PVT to histaminergic neurons (Fig. 4j). Collectively, these results provide both structural and functional evidence for direct LS/PVT→histaminergic neuron connectivity.
The balanced roles of LS- and PVT-TMN circuits in sleep-wake regulation
Both LS and PVT exhibit high densities of histaminergic nerve fibers and substantial populations of input neurons projecting to histaminergic neurons, suggesting their importance in modulating core histaminergic functions. Given the well-established role of the histaminergic system in sleep-wake regulation1, and the reported involvement of PVT glutamatergic and LS GABAergic neurons in this process40,41, we sought to reconcile the apparent paradox that these functionally opposing nuclei appear to exert similar effects on sleep-wake transitions. To specifically examine histaminergic-projecting neurons in LS and PVT, we employed a Cre-dependent monosynaptic retrograde GCaMP system42,43. Briefly, we first injected a 1:2 mixture of rAAV-EF1α-DIO-mCherry-F2A-TVA and rAAV-EF1α-DIO-N2cG into the bilateral TMN of HDC-CreERT2 mice, followed by CVS-EnvA-ΔG-GCaMP6s injection three weeks later, with simultaneous optic fiber implantation in the LS or PVT (Fig. 5a, e). GCaMP6s-labeled neurons were found in the TMN and upstream brain regions (LS and PVT under optic fiber, Fig. 5b, c, f, g). Simultaneous EEG/EMG and fiber photometry recordings revealed that both LS and PVT input neurons exhibited decreased calcium signals during wake-to-non-rapid eye movement (NREM) sleep transitions (Fig. 5d, h, i, m), and increased calcium signals of input neurons during sleep (NREM or rapid eye movement (REM))-to-wake transitions (Fig. 5d, h, j, l, n, p). Notably, we observed divergent responses during NREM-to-REM transitions: PVT neurons exhibited decreased activity, whereas LS neurons showed no such reduction (Fig. 5d, h, k, o and Supplementary Fig. 8). These results demonstrate that histaminergic-projecting neurons in both LS and PVT coordinately modulate sleep-wake transitions, consistent with previous reports40,41, while revealing previously uncharacterized state-specific differences in NREM-to-REM sleep transitions.
a Experimental scheme for labeling and recording histaminergic neuron-projecting LS neurons. rAAV-EF1α-DIO-mCherry-F2A-TVA mixed with rAAV-EF1α-DIO-N2cG, and CVS-EnvA-ΔG-GCaMP6s were injected into the TMN of HDC-CreERT2 mice. Optical fiber was implanted into mouse brains above the LS for calcium signal recording. b, c Representative images of virus expression in TMN (b) and LS (c). d Representative EEG, EMG, and LS fiber photometry of tested mice. The signals of 5 trails are illustrated in the heatmap (from 2 mice); color scale indicates ΔF/F and warmer colors indicate higher fluorescence signal. Data are presented as mean values ± SEM. e Experimental scheme for labeling and recording histaminergic neuron-projecting PVT neurons. rAAV-EF1α-DIO-mCherry-F2A-TVA mixed with rAAV-EF1α-DIO-N2cG, and CVS-EnvA-ΔG-GCaMP6s were injected into the TMN of HDC-CreERT2 mice. Optical fiber was implanted into mouse brains above the PVT for calcium signal recording. f, g Representative images of virus expression in TMN (f) and PVT (g). h Representative EEG, EMG, and PVT fiber photometry of tested mice. The signals of 5 trails are illustrated in the heatmap (from 2 mice); color scale indicates ΔF/F and warmer colors indicate higher fluorescence signal. Data are presented as mean values ± SEM. i–l Analysis of area under the curve before and after state transitions of wake to NREM sleep (i), NREM sleep to wake (j), NREM sleep to REM sleep (k), and REM sleep to wake (l) in the calcium signal recording of LS. n = 5 (derived from different mice). Statistical analyses were conducted using two-tailed Student’s t-tests without adjustment for multiple comparisons. Significance levels were presented as *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, or ****p ≤ 0.0001. The exact p-values were as follows: 0.0008 (i), 0.0008 ( j), 0.0020 (k), and 0.0025 (l). m–p Analysis of area under the curve before and after state transitions of wake to NREM sleep (m), NREM sleep to wake (n), NREM sleep to REM sleep (o), and REM sleep to wake (p) in the calcium signal recording of PVT. n = 5 (derived from different mice). Statistical analyses were conducted using two-tailed Student’s t-tests without adjustment for multiple comparisons. Significance levels were presented as *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, or ****p ≤ 0.0001. The exact p-values were as follows: 0.0460 (m), 0.0004 (n), 0.0007 (o), and 0.0191 (p). LS Lateral septal nucleus, NREM non-rapid eye movement, PVT Paraventricular nucleus of the thalamus, REM rapid eye movement, TMN Tuberomammillary nucleus.
The differential functional dynamics during NREM to REM sleep transitions suggest differential regulatory roles of these input circuits. To assess their effects, we further employed chemogenetic activation approaches on LS/PVT input circuits during mouse sleep-wake cycle. Specifically, we injected a 1:1 mixture of rAAV-VGAT1-CRE-EGFP-WPRE-hGH polyA and rAAV-hSyn-hM3D(Gq)-EGFP-WPRE-hGH polyA into the bilateral LS, or a 1:1 mixture of rAAV-VGLUT2-CRE-WPRE-hGH polyA and rAAV-hSyn-hM3D(Gq)-EGFP-WPRE-hGH polyA into the PVT of WT mouse brain, with rAAV-CAG-DIO-mCherry-WPRE-hGH polyA as control (Fig. 6a–d). Following three weeks of expression, we implanted a bilateral cannula into the TMN for CNO administration delivery along with electrodes for EEG and EMG recording. After 7 days, we performed EEG/EMG recording during sleep-wake cycle under chemo-genetic manipulation, and found that consistent with previous reported regulatory roles of whole LS/PVT neurons, both LS-hM3Dq (54.22 ± 1.504%, n = 4) and PVT-hM3Dq (56.63 ± 0.9636%, n = 4) groups showed significantly increased wakefulness compared to the control group (48.31 ± 1.558%, n = 4)40,41 (Fig. 6e–i). However, REM sleep parameters revealed striking differences (Fig. 6e–g, i): LS-TMN activation enhanced REM duration (whole-day: 6.705 ± 0.695% vs control 2.650 ± 0.049%; sleep-period: 14.73 ± 1.657% vs control 5.150 ± 0.242%; n = 4), whereas PVT-TMN activation suppressed it (whole-day: 1.063 ± 0.259%; sleep-period: 2.420 ± 0.541%, n = 4) (Fig. 6i, j). Moreover, NREM-to-REM sleep transition frequency also differed markedly (LS: 371.0 ± 20.08; PVT: 82.50 ± 20.04; control: 177.3 ± 10.26; n = 4) (Fig. 6k). To further explore the sleep/wake situations of tested mice during the daytime and nighttime, we performed circadian analysis: (1) mice spend more time awake during the nighttime than daytime in both the mCherry, LS-hM3Dq, and PVT-hM3Dq group (Fig. 6l, p), (2) LS-hM3Dq mice showed elevated REM (day: 7.063 ± 1.138%; night: 6.333 ± 0.458%; n = 4) relative to controls (day: 3.483 ± 0.396%; night: 1.820 ± 0.392%; n = 4), while PVT-hM3Dq mice exhibited reduced REM (day: 1.268 ± 0.429%; night: 0.860 ± 0.141%; n = 4) (Fig. 6l–s). These findings reveal fundamental differences in how LS and PVT input circuits regulate sleep architecture, highlighting the complexity of histaminergic network regulation. Our whole-brain atlas thus provides a critical foundation for decoding these circuit-level mechanisms in the future.
a Experimental scheme for labeling input neurons in the LS. rAAV-VGAT1-CRE-EGFP-WPRE-hGH polyA mixed with AAV2/9-hSyn-DIO-hM3D(Gq)-mCherry-WPRE-pA or rAAV-CAG-DIO-mCherry-WPRE-hGH polyA were injected into the bilateral LS of WT mice. Bilateral cannula was implanted into mouse brains above the TMN for CNO infusion. b Representative images of hM3Dq expression in LS (left) and TMN (right). c Experimental scheme for labeling input neurons in the PVT. rAAV-VGLUT2-CRE-WPRE-hGH polyA mixed with AAV2/9-hSyn-DIO-hM3D(Gq)-mCherry-WPRE-pA or rAAV-CAG-DIO-mCherry-WPRE-hGH polyA were injected into the PVT of WT mice. Bilateral cannula was implanted into mouse brains above the TMN for CNO infusion. d Representative images of hM3Dq expression in PVT (left) and TMN (right). e–g Representative continuous EMG, EEG, wake, NREM sleep, and REM sleep scoring data recorded from group of mcherry (e), LS-hM3Dq (f), and PVT-hM3Dq (g) for 24 h (n = 4 for each group). h, i Ratio quantification of wake, NREM sleep, and REM sleep from different groups in 24 h (n = 4 for each group). The exact p-values were as follows: 0.0342 (mCherry-wake vs LS-wake), 0.0039 (mCherry-wake vs PVT-wake), 0.0060 (mCherry-NREM vs LS-NREM), 0.0091 (mCherry-NREM vs PVT-NREM), 0.0011 (mCherry-REM vs LS-REM), and 0.0009 (mCherry-REM vs PVT-REM). j Ratio quantification of NREM and REM sleep from different groups in sleep time. (n = 4 for each group). The exact p-values were as follows: 0.0012 (mCherry-REM vs LS-REM), and 0.0037 (mCherry-REM vs PVT-REM). k Transition number of NREM-REM from different groups in 24 h (n = 4 for each group). The exact p-values were as follows: 0.0011 (mCherry vs LS), 0.0009 (mCherry vs PVT), and <0.0001 (LS vs PVT). l, m Ratio quantification of wake, NREM sleep, and REM sleep from different groups during 12 h (light) of the daytime. The exact p-values were as follows: 0.1455 (mCherry-wake vs LS-wake), 0.0240 (mCherry-wake vs PVT-wake), 0.0476 (mCherry-NREM vs LS-NREM), 0.0384 (mCherry-NREM vs PVT-NREM), 0.0249 (mCherry-REM vs LS-REM), and 0.0090 (mCherry-REM vs PVT-REM). n Ratio quantification of NREM and REM sleep from different groups in daytime sleep. (n = 4 for each group). The exact p-values were as follows: 0.0149 (mCherry-REM vs LS-REM), and 0.0162 (mCherry-REM vs PVT-REM). o Transition number of NREM-REM from different groups during 12 h (light) of the daytime (n = 4 for each group). The exact p-values were as follows: 0.0096 (mCherry vs LS), 0.0298 (mCherry vs PVT), and 0.0006 (LS vs PVT). p, q Ratio quantification of wake, NREM sleep, and REM sleep from different groups during 12 h (dark) of the nighttime. The exact p-values were as follows: 0.0422 (mCherry-NREM vs LS-NREM), 0.3022 (mCherry-NREM vs PVT-NREM), 0.0003 (mCherry-REM vs LS-REM), and 0.0608 (mCherry-REM vs PVT-REM). r Ratio quantification of NREM and REM sleep from different groups in nighttime sleep. (n = 4 for each group). The exact p-values were as follows: <0.0001 (mCherry-REM vs LS-REM), and 0.0398 (mCherry-REM vs PVT-REM). s Transition number of NREM-REM from different groups during 12 h (light) of the nighttime (n = 4 for each group). The exact p-values were as follows: 0.0031 (mCherry vs LS), and 0.0004 (LS vs PVT). All data are presented as mean ± SEM. Statistical analyses were conducted using two-tailed Student’s t-tests without adjustment for multiple comparisons. Significance levels were presented as *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, or ****p ≤ 0.0001. LS Lateral septal nucleus, NREM non-rapid eye movement, PVT Paraventricular nucleus of the thalamus, REM rapid eye movement, TMN Tuberomammillary nucleus.
Discussion
In the present study, we construct a quantitative 3D whole-brain atlas of monosynaptic long-range inputs to mouse histaminergic neurons by combining RV retrograde tracing with fMOST reconstruction. Our analysis reveals that input cells are predominantly localized in hypothalamic nuclei, with additional clusters in specific brain regions of the thalamus, pallidum, striatum, and hippocampal formation, exhibiting distinct distribution patterns. Notably, we identify a layer-specific organization of cortical input neurons and characterize diverse co-projection patterns through single-neuron reconstruction at whole-brain scale. To identify the neuron types of major upstream brain regions, we perform RNAscope in situ hybridization to map GABAergic (GAD1+) and glutamatergic (vglut2+/CaMKIIα+) inputs among RV-labeled neurons. Furthermore, through integrate anterograde tracing, whole-cell patch clamp recordings, and fiber photometry, we demonstrate direct structural and functional connectivity between LS/PVT and histaminergic neurons, uncovering differential modulatory roles of LS- and PVT-TMN circuits in sleep-wake regulation. Our findings may provide a comprehensive foundation for future functional research of the input circuits targeting the histaminergic nervous system.
Previous studies have attempted to map whole-brain inputs to histaminergic neurons using either non-specific CTB tracing19 or Cre-dependent TVA-RV system23. While our findings in hypothalamic nuclei are largely consistent with prior RV-based work, we observed notable discrepancies in other regions. For example, whereas Saito et al. reported sparse inputs from amygdalar regions23, we identified substantial projections from the MEA, PA, and BMA (Fig. 1f). Moreover, we detected dense inputs from the PVT (Fig. 1f), particularly its anterior division (Supplementary Fig. 4g), contrasting with previous reports of minimal PVA (paraventricular thalamic nucleus, anterior part) inputs23. Notably divergent findings also emerged in hippocampal and septal regions. Whereas Saito et al. reported sparse inputs from all parts of the HPF, especially in CA1/CA3/DG (almost 0%)23, we observed substantial CA1 projections (Fig. 1f). In addition, although anatomically adjacent, our quantification revealed strikingly fewer MS versus LS inputs (Fig. 1f), contrasting with the comparable numbers reported previously23. However, a much earlier work using anterograde neuroanatomical tracing reported LS-dominant pattern44, which is somewhat coincide with our conclusion. The above discrepancies across multiple key input regions could significantly impact whole-brain mapping conclusions. Potential contributing factors include efficiency of TVA-RV tracing, integrity of slice imaging, and accuracy of cell counting and brain region segmentation in the fMOST approach. Moreover, through reviewing the distribution of starter cells in sample mice, we found more comprehensive starter cell labeling (Fig. 1d), particularly in dorsal TMN regions that appeared underrepresented in prior work23. However, there was also potential deficiency of our tracing approach. First, the potential inclusion of inputs targeting local histaminergic axonal varicosities, in addition to somatic inputs, should be noted. Detailed examination of TVA and RV expression in injection sites revealed predominant somatic localization, with limited axonal expression restricted to proximal regions (Fig. 1d), potentially introducing minor contamination of non-somatic inputs. On the other hand, we observed GFP+/mCherry− neurons within TMN (Fig. 1b), representing local microcircuit elements. HDC immunostaining confirmed these were predominantly non-histaminergic (Supplementary Fig. 1a, f–h), suggesting the existence of intrinsic microcircuits modulating histaminergic neurons within the TMN. These local networks may significantly contribute to histaminergic regulation and merit future investigation. Notwithstanding these considerations, our RV/fMOST methodology remains the high-level standard for long-range input mapping, as evidenced by its widespread adoption in circuit neuroscience. Besides, we observed substantial variability in input data from certain brain regions, including the CP, SNr, SCH, and PVa. We speculated this variability is due to differences between mice. Our findings confirmed the existence of such input variability, supporting the possibility that inter-mouse differences contribute to the observed regional variations. Corroborating this interpretation, immunohistochemical analysis of histidine decarboxylase (HDC) revealed that starter cells accounted for 26.79 ± 2.075% (n = 3) of total histaminergic neurons (Supplementary Fig. 1f–h). The variations in starter cell distribution between individual mice may underly the order-of-magnitude differences in input strength observed from certain upstream regions and more advanced retrograde viral tools capable of more comprehensive starter cell labeling may help resolve this issue.
The cortex is widely regarded as the supreme regulatory center for diverse physiological functions in the mammalian brain45,46. However, the structural and functional organization of cortical inputs to histaminergic neurons remains poorly characterized. Our study reveals the layer V and VIa distribution manner of most cortical input neurons, which consistent with the primary long-range output layer patterns of the cortex47,48,49. In addition, the mPFC, the dominant cortical input source, exhibits subregion-specific laminar distributions and co-projection patterns between its subregions ILA and PL (Fig. 2e and Supplementary Fig. 5b–e), suggesting functional heterogeneity that warrants further investigation. At whole cortical area scale, single-neuron reconstructions reveal preferential co-projection to the striatum, pallidum, thalamus, and midbrain areas (Fig. 2f), implying functional coupling between the histaminergic system and these regions mediated by cortical inputs. Surprisingly, cortical inputs exclusively innervate the ipsilateral hemisphere (Fig. 2f), contrasting with bilateral projections observed in mPFC-targeting cortical neurons using similar approaches25. Given the well-established laterality of cortical sensory/motor processing50,51, this striking ipsilateral preference may reflect previously unrecognized functional specialization in histaminergic regulation. Therefore, future studies on cortical input circuits may prioritize the specific co-projection manner and apparent differences between ILA and PL input neurons.
The central histaminergic neural network plays crucial roles in regulating diverse physiological functions, including sleep-wake regulation, sensory and motor function, cognition, learning and memory, and feeding and energy balance, with significant implications for CNS disorders52,53. Our whole-brain mapping reveals distinct input patterns that may reflect functional specialization across upstream regions. Hypothalamus, particularly the anterior hypothalamic area, accounts for the most input neurons of the whole mouse brain (Fig. 1f). This aligns with its established role in circadian regulation54,55,56. More importantly, a dense histaminergic nerve fiber distribution was also widely found in the anterior hypothalamic area in our previous work18. Notably, the reciprocal VLPO-TMN connectivity28 suggests a potential hypothalamic-histaminergic circuit modulating circadian behaviors. In addition, the BST, implicated in feeding behavior, anxiety, fear response, and learning memory57,58, exhibits both substantial inputs and dense histaminergic innervation, indicating bidirectional modulation. Besides, there are also major upstream brain regions that have only few or moderate histaminergic fiber density. For example, the amygdala complex plays important roles in fear and anxiety behaviors59,60, which were also reported to closely related to central histaminergic functions14,61. Multiple amygdalar nuclei (MEA, PA, BMAp) provide significant inputs despite limited histaminergic fibers. Conversely, the central amygdala (CEA) shows minimal inputs with substantial histaminergic fibers (0.33%), revealing region-specific connectivity patterns18. Given the reported histaminergic projection regulating activity of BLA-CEA microcircuit through presynaptic H3Rs62, and intrinsic amygdalar microcircuitry63, these observations suggest complex histaminergic-amygdalar interactions in fear/anxiety regulation. For hippocampus, CA1, especially ventral part (Supplementary Fig. 4i), emerges as a major input source, exhibited sparse histaminergic fibers (0.02%)18. CA1 is widely believed to play important role in learning and memory, which seems to be different from other major part of HPF, such as CA364,65. The vCA1 glutamatergic projection (Fig. 3g, j) may coordinate CA3 and histaminergic activity, potentially integrating memory processes66,67. While functional studies of histaminergic inputs remain limited, our anatomical framework provides a foundation for deciphering these complex circuit operations.
“Structure determines function” is a well-established concept in neurobiology. In our previous work on mapping histaminergic neural projections, we employed specific histamine sensor HA1m to preliminarily validate the relationship between histaminergic fiber density and histamine release capacity, further revealing its potential biological significance through behavioral studies of aversion responses18. In the current study, we similarly aim to enhance the biological significance of the histaminergic input map through functional investigations. Accordingly, we have selected two representative input circuits and focused on their connection with the classical physiological function of histaminergic systems—sleep-wake regulation—for further exploration. Our selection of LS and PVT for in-depth investigation was based on several key considerations. First, LS input cells were predominantly GABAergic, while PVT inputs were overwhelmingly glutamatergic (Fig. 3b, d, j). Second, we observed striking spatial correspondence between input neuron distributions and histaminergic fiber densities across LS/PVT subregions, suggesting functional reciprocity in these circuits. Specifically, LS inputs were concentrated in LSr (2.438%) and LSv (2.253%) rather than LSc (0.177%), mirroring histaminergic innervation patterns (LSr: 0.51%, LSv: 0.42%, LSc: 0.14%)18. For PVT, although CCF v3.0 does not distinguish anterior/posterior subdivisions, the structural and functional differences between them were widely recognized68,69. Our 3D reconstructions revealed preferential input localization to the anterior PVT (Supplementary Fig. 4g), paralleling histaminergic fiber distributions18. These observations, combined with established roles of LS/PVT circuits in sleep-wake regulation, prompted functional characterization. Calcium imaging revealed synchronous activity during sleep-wake transitions, but divergent patterns during NREM-to-REM sleep transitions (Fig. 5d, h). However, we also noted that the initial rising trend of neuronal activity in the LS during this transition was not pronounced. Therefore, we reanalyzed all calcium signal trends using an 8-s time window and found that LS neurons exhibited only a slight, statistically nonsignificant increase during NREM-to-REM sleep transitions (Supplementary Fig. 8c), while other experimental conditions still showed significant differences (Supplementary Fig. 8a, b, d–h). Consequently, we described this change as “no such reduction” rather than “increase”. Nevertheless, it remains undeniable that the calcium activity trends of LS and PVT neurons exhibit distinct patterns during NREM-to-REM sleep transitions. Given the importance of REM sleep in neuroplasticity70, and limited understanding of histaminergic regulatory roles on REM sleep, we further assessed circuit-specific manipulations. Notably, while both circuits similarly modulated sleep-wake ratios, they exerted opposing effects on REM sleep (Fig. 6e–i), consistent with their calcium dynamics. Moreover, the major effects of LS and PVT input circuits acting on the transition from NREM to REM sleep, which together indicated the potential critical role of LS and PVT input circuits targeting histaminergic neurons on sleep-wake regulation, especially on REM sleep. However, given that major inhibition inputs (e.g., VLPO) to histaminergic neurons tend to favor more NERM28,71,72, it seems difficult to understand why LS GABAergic inhibitory inputs leads to more wakefulness rather than NREM. This discrepancy may reflect histaminergic subpopulation specificity73, wherein LS-targeted neurons mediate distinct aspects of sleep regulation. This hypothesis merits future investigation, as our findings provide a framework for dissecting histaminergic control of REM sleep.
In general, we construct a comprehensive and precise whole-brain atlas of long-range monosynaptic inputs to mouse histaminergic neurons in this study. This atlas provides not only refined quantitative information on whole-brain input neurons, but also reveals their spatial distribution patterns and neuronal subtypes in major upstream regions, establishing a foundational resource for investigating functional input circuits to the histaminergic system. While histaminergic neurons demonstrate projection-specific integration properties, the fundamental organizational principles governing these selective connectivity patterns remain to be fully elucidated and require future functional studies.
Methods
Animals
HDC-CreERT2 and HDC;AI47 mice used in this study were generated by same approaches as previously descripted18. Wild-type mice (weight 22–26 × g) were purchased from SLAC Laboratory Animal Center (Shanghai, China). All mice used in this study belong to C57BL/6J strain. Mice were housed in the vivarium with a 12-h light/dark cycle: 22 ± 1 °C and 55 ± 5% humidity with food and water given ad libitum. Male mice aged 8 to 14 weeks were used. Animal experiments were performed in accordance with the guidelines of the Animal Advisory Committee of Zhejiang Chinese Medical University and the US NIH Guidelines for the Care and Use of Laboratory Animals. All procedures were approved by the Animal Advisory Committee of Zhejiang Chinese Medical University (No. 20230424-06).
Viruses
rAAV-EF1α-DIO-mCherry-F2A-TVA-WPRE-hGH polyA (viral titers: 5.33 × 1012 particles/mL), rAAV-EF1α-DIO-oRVG-WPRE-hGH polyA (viral titers: 5.27 × 1012 particles/mL), RV-ENVA-ΔG-EGFP (viral titers: 2.0 × 108 IFU/mL), AAV2/9-hSyn-DIO-hM3D(Gq)-mCherry-WPRE-pA (viral titers: 2.0 × 1012 particles/mL), and rAAV-CAG-DIO-mCherry-WPRE-hGH polyA (viral titers: 5.09 × 1012 particles/mL) were purchased from BrainVTA Co., Ltd (Wuhan, China); rAAV-VGAT1-CRE-EGFP-WPRE-hGH polyA (viral titers: 5.22 × 1012 particles/mL), rAAV-VGLUT2-CRE-WPRE-hGH polyA (viral titers: 5.40 × 1012 particles/mL), and AAV2/9-hSyn-FLEX-ChrimsonR-tdTomato-WPRE-pA (viral titers: 3.86 × 1012 particles/mL) were purchased from Taitool Bioscience Co., Ltd (Shanghai, China); rAAV-CAG-DIO-mWGA-mCherry (viral titers: 5.10 × 1012 particles/mL), rAAV-EF1α-DIO-mCherry-F2A-TVA (viral titers: 2.50 × 1012 particles/mL), rAAV-EF1α-DIO-N2cG (viral titers: 5.16 × 1012 particles/mL), and CVS-EnvA-ΔG-GCaMP6s (viral titers: 2.0 × 108 IFU/mL) were purchased from Braincase Co., Ltd (Shenzhen, China). All viral vectors were aliquoted and stored at −80 °C until use.
Stereotactic injections and surgery
Mice were anesthetized with 1% sodium pentobarbital (50 mg/kg, i.p., Sigma-Aldrich) and headfixed in a stereotaxic apparatus (RWD Life Science, Shenzhen, China). During the total operation, the body temperature of the anesthetized mice was kept constant at 37 °C using a heating pad. If the mice had a pain reflex response from a paw pinch, an additional 10% of the initial dose of sodium pentobarbital was given to guarantee a painless state. An incision was made in mice’s heads to expose the skull surface. After scraping the pericranium away, burr holes were stereo tactically made onto the skulls. For monosynaptic retrograde tracing of histaminergic neurons, 300 nL of rAAV-EF1α-DIO-mCherry-F2A-TVA-WPRE-hGH polyA mixed with rAAV-EF1α-DIO-oRVG-WPRE-hGH polyA (1:2, a mixed ratio used in same RV system tracing experiments25) was injected into each side of the TMN (bregma 2.0 mm, lateral 0.8 mm, depth 5.4 mm from skull surface) of HDC-CreERT2 adult mice. One day after virus injection, mice received tamoxifen injection (100 mg/kg/d, i.p.) for 5 consecutive days. Three weeks later, 300 nL of RV-ENVA-ΔG-EGFP was injected into the same site of HDC-CreERT2 mice, and RV was allowed to express for 7 days. For in vitro electrophysiological recording, 300 nL of rAAV-VGAT1-CRE-EGFP-WPRE-hGH polyA mixed with AAV2/9-hSyn-FLEX-ChrimsonR-tdTomato-WPRE-pA (1:1) was injected into each side of the LS (bregma −0.9 mm, lateral 0.4 mm, depth 3.5 mm from skull surface), or rAAV-VGLUT2-CRE-WPRE-hGH polyA mixed with AAV2/9-hSyn-FLEX-ChrimsonR-tdTomato-WPRE-pA (1:1) was injected into PVT (bregma 0.6 mm, lateral 0.0 mm, depth 3.5 mm from skull surface) of HDC;AI47 mice. One day after virus injection, mice received tamoxifen injection (100 mg/kg/d, i.p.) for 5 consecutive days. Four weeks later, in vitro electrophysiological recording was performed. For monosynaptic anterograde tracing, 300 nL of rAAV-VGAT1-CRE-EGFP-WPRE-hGH polyA mixed with rAAV-CAG-DIO-mWGA-mCherry (1:1) was injected into each side of the LS, or rAAV-VGLUT2-CRE-WPRE-hGH polyA mixed with rAAV-CAG-DIO-mWGA-mCherry (1:1) was injected into PVT of HDC;AI47 mice. One day after virus injection, mice received tamoxifen injection (100 mg/kg/d, i.p.) for 5 consecutive days. And WGA was allowed to express for three weeks. For calcium signal recording, 300 nL of rAAV-EF1α-DIO-mCherry-F2A-TVA mixed with rAAV-EF1α-DIO-N2cG (1:2) was injected into each side of the TMN (bregma 2.0 mm, lateral 0.8 mm, depth 5.4 mm from skull surface) of HDC-CreERT2 mice. Three weeks later, 300 nL of CVS-EnvA-ΔG-GCaMP6s was injected into the same site, and optic fiber was implanted into the LS (bregma −0.9 mm, lateral 0.4 mm, depth 3.3 mm from skull surface) or PVT (bregma 0.6 mm, lateral 0.0 mm, depth 3.3 mm from skull surface) at the same time. Meanwhile, two gold-plated EEG electrodes were inserted through the skull onto the dura mater for EEG recording, and two lengths of Teflon-insulated stainless-steel wire were inserted in the neck muscle for EMG recording. 7 days later, calcium signal recording was performed. For chemo-genetic manipulation, 300 nL of AAV2/9-hSyn-DIO-hM3D(Gq)-mCherry-WPRE-pA or rAAV-CAG-DIO-mCherry-WPRE-hGH polyA were injected into the PVT or bilateral LS of WT mouse brain. After three weeks of virus expression, bilateral cannula (62058, RWD Life Science, Shenzhen, China) was implanted into the bilateral TMN (bregma 2.0 mm, lateral 0.8 mm, depth 4.8 mm from skull surface). After 7 days recovery, CNO (3 μM, 200 nL per side) was administrated into the bilateral TMN through cannula, and EEG/EMG recording was performed 30 min later for 24 h.
Fiber photometry and EEG/EMG recording
The fiber photometry system (Nanjing Thinkertech) used a 488 nm diode laser (OBIS 488LS; Coherent), reflected by a dichroic mirror (MD498, Thorlabs) and coupled into a 0.23-mm 0.37 NA optical fiber using a 10× objective lens (Olympus) and fiber launch (Thorlabs). The laser intensity at the interface between the fiber tip and the animal ranged from 0.01 to 0.03 mW, to minimize bleaching. The GCaMP fluorescence was bandpass filtered (MF525-39, Thorlabs) and collected by a photomultiplier tube (R3896, Hamamatsu). An amplifier (C7319, Hamamatsu) was used to convert the photomultiplier tube current output to voltage signals, which were then further filtered through a low-pass filter (40 Hz cut-off). The analog voltage signals were digitized at 500 Hz and recorded. Two EEG and one EMG channels were recorded for each mouse by turn-key recording system (Pinnacle Technology Inc.). For LS/PVT input neuron calcium dynamic research, mice were allowed to recover for 7 days after RV-GCaMP system injection and surgery. To facilitate environmental adaptation before testing, mice were carefully placed inside the recording room with their home cage for 30 min. When recording start, the tested mouse was linked to optic fiber with fiber photometry system and recording electrode with turn-key recording system. The recording time is from 8 A.M. to 8 P.M. for each mouse. Photometry data were exported as MATLAB Mat files for further analysis. We segmented the data based on individual laser stimulation trials and derived the values of fluorescence change (ΔF/F) by calculating (F–F0)/F0. ΔF/F values are presented via heatmaps or average plots. The sampling rate was set up to 400 Hz. EMG was filtered by low-pass 100 Hz. EEG frequency was filtered by low-pass 40 Hz. EEG and EMG data were analyzed by Sirenia Sleep Pro (Version 1.8.0). EEG power was analyzed using FFT (Fast Fourier transform of the autocorrelation function) power spectra. Wake, NREM, and REM states were semi-automatically classified by Sirenia Sleep Pro for each 10 s epoch. Wake: desynchronized EEG with high EMG activity; NREM: synchronized EEG with high power at 0.5–4 Hz (defined as δ wave) and low EMG activity; REM: desynchronized EEG with low power at 0.5–4 Hz and high power at 6–9 Hz (defined as θ wave), and low EMG activity. Then the state of each epoch was manually checked.
RNAscope in situ hybridization and immunohistochemical staining
Mice were sacrificed and perfused with saline and 4% paraformaldehyde (PFA) in PBS (pH 7.4). The harvested brains were quickly dissected and further fixed in 4% PFA for 24 h before consecutive dehydration in 10%, 20%, and 30% sucrose. 14 μm thickness coronal brain slices containing major upstream brain regions were collected by freezing microtome (Thermo NX70). RNAscope Multiplex Fluorescent Reagent Kit v2 (Advanced Cell Diagnostics, 323110) was used for checking GAD1 (Advanced Cell Diagnostics, 400951), VGLUT2 (Advanced Cell Diagnostics, 319171), and CaMKIIα (Advanced Cell Diagnostics, 445231) expression. For further GFP immunohistochemical staining, the slices were then incubated with antibody against GFP (Abcam, ab13970, 1:100) and further photographed by Olympus FV3000 confocal microscope. For the labeling specificity analysis of TVA-mCherry and starter cells in Supplementary Fig. 1, anti-HDC (America Research Products, 03-16045, dilution 1:400) was used, followed by Goat anti-Rabbit IgG H&L (Alexa Fluor® 647) (Abcam, ab150079, dilution 1:800) as secondary antibody. The images were quantified by FIJI (ImageJ-win64) in a blind manner.
Electrophysiology
After four weeks of virus expression, the mice were quickly perfused with ice-cold artificial cerebrospinal fluid (ACSF, in mM: 120 NaCl, 11 Dextrose, 2.5 KCl, 1.28 MgSO4, 3.3 CaCl2, 1 NaH2PO4, and 14.3 NaHCO3). The brain was quickly transferred to a vibratome (VT1000 mol/L/E, Leica) to prepare coronal slices at 300-µm thickness and immersed in ice-cold ACSF constantly bubbled with 95 % O2 and 5 % CO2 for 1 min. Coronal brain slices containing TMN were recovered in a chamber filled with ACSF saturated with 95 % O2 and 5 % CO2 at 37 °C for 30 min and incubated at 25 °C for 1 h before recording. Then the slices were transferred into a recording chamber at 25 °C for patch clamp recording. Signals were amplified and recorded by HEKA EPC10 amplifier (HEKA Instruments, Germany). To record light-evoked synaptic currents, a low divalent ion ACSF (in mM: 125 NaCl, 3.5 KCl, 1.25 NaH2PO4, 0.5 MgCl2, 26 NaHCO3, 25 Dextrose, and 1 CaCl2) was used. Using pipettes filled with cesium-based internal fluid (in mM: 100 CsCH3SO3, 20 KCl, 10 HEPES, 4 Mg-ATP, 0.3 Tris-GTP, 7 Tris2-Phosphocreatine, and 3 QX-314), EPSCs were recorded at a holding potential of −60 mV and IPSCs were recorded at a holding potential of +10 mV. And naive EPSCs/IPSCs were recorded for about 20 s, then 1 Hz, 10 ms, 635 nm red light was performed for 10 s to optogenetic ally activate nerve fibers from PVT/LS. If light-evoked EPSCs/IPSCs were recorded, TTX (1 μM) and 4-AP (100 μM) were performed for avoiding potential muti-synaptic effect. Then, bicuculline (20 μM) or APV (100 μM) and CNQX (40 μM) were performed for blocking GABA-mediated IPSCs or glutamate-mediated EPSCs. Individual events were counted and analyzed with MiniAnalysis software (version 6.0.3).
fMOST tissue preparation
Mice were anesthetized and perfused with 0.01 M PBS and PBS containing 4% PFA, respectively. Brains were post-fixed in 4% PFA for 24 h. Fixed brains were transferred into 0.01 M PBS and incubated at 4 °C overnight, then dehydrated in graded ethanol solutions. Individual brains were then impregnated with Glycol Methacrylate (GMA, Ted Pella Inc.) and embedded in a vacuum oven.
Whole-brain imaging
The whole-brain imaging procedure is based on a Brain-wide Position System (BPS)74, with simultaneous propidium iodide (PI) staining to label the cytoarchitectonic landmarks and a brainwide fluorescence micro-optical sectioning tomography (fMOST) via structured illumination. For whole-brain imaging, each sample was immobilized in a water bath on a 3D nano-precision translation stage. The brain samples were then sectioned coronally at an interval of 1 μm in an antero-posterior (AP) direction to achieve the axial scan, counterstained in PI, and then underwent fluorescence image acquisition via mosaic scan. The combination of mosaic x-y-scan and axial z-scan produced a continuous dataset at a voxel resolution of 0.32 × 0.32 × 1 μm3, enabling 3D reconstruction. For individual mouse brains, ~10,000 coronal slices were sectioned with an original data size of 2 TB.
Image pre-processing
The raw data acquired by the BPS necessitated mosaic stitching and illumination correction image pre-processing. Briefly, the mosaics of each coronal section were stitched to obtain an entire section based on accurate spatial orientation and adjacent overlaps. Lateral illumination correction was performed section by section. Image pre-processing results were implemented in C++ and optimized in parallel using the Intel MPI Library (v.3.2.2.006, Intel). The whole data sets were executed on a computing server (72 cores, 2 GHz per core) within 6 h.
Cell counting and registration
For GFP-labeled neuron counting, the GFP-labeled dataset of whole mouse brain was sampled at 1 × 1 × 1 μm3 using the Cell Counter ImageJ plug-in and then automatically identified by NeuroGPS, which saved the data in an SWC format. These data were then manually corrected in Amira. We used the BrainsMapi for brain structure registration; the dataset of reconstructed brain was rotated and resampled at a resolution of 10 × 10 × 10 μm3 for registering in the template dataset of Allen CCF v3.0. Then, we manually segmented several brain regions as landmarks by Amira software. Finally, we registered these results by ANTs tools75 and loaded the outline of the mouse brain and the results into Amira simultaneously to generate figures.
Single neural structure tracing
We used GTree software76 with TDat plugin (Thermo Fisher Amira)77 through human–computer interaction to trace and reconstruct the single neural structure. The reconstruction of each neuron usually starts from the cell body; we identified a soma as the seed point and then navigated through the image data blocks to connect the signal node to the seed point until all the branches are traversed. To improve the tracing efficiency, we chose semiautomatic tracing at sparsely distributed long-distance fibers. Briefly, automatic reconstruction calculation is performed first, and then, a skilled annotator manually checks the results of the automatic calculation. If potential errors (when fluorescent signal is too dense or too weak to be automatically traced) are found, manual correction is carried out immediately. To minimize subjective errors, when an annotator finds it difficult to judge, it is recorded as a special event. Then three skilled annotators work independently on the reconstruction of each neuron. Finally, a neuroanatomy expert is responsible for checking all special events and acceptance of the results. All reconstructed results were saved as SWC files. MATLAB (2017a, MathWorks) was used to calculate the morphological parameters of the reconstructed neurons. Due to the exceptionally high density of local fibers in RV-labeled brains, we were unable to reliably distinguish traced axons from endogenous fibers within the TMN, precluding complete reconstruction of cortical axon terminals in this region.
Statistics
All statistical graphs were generated using Graphpad Prism v.6.01. All data are presented as means ± s.e.m. Number of experimental replicates (n) is indicated in the figure legends and refers to the number of experimental subjects independently treated in each experimental condition. A statistical significance threshold was set at 0.05, and significance levels are presented as *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, or ****p ≤ 0.0001 in all figures.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data analyzed in this research are included in this published paper and its Supplementary Information/Source data file. Source data are deposited in the Figshare under accession code https://doi.org/10.6084/m9.figshare.28900511. Source data are provided with this paper.
Code availability
We used codes in reconstruction of long-range inputs to histaminergic neurons. The NeuroGPS-Tree is open source and available as https://static-content.springer.com/esm/art%3A10.1038%2Fnmeth.3662/MediaObjects/41592_2016_BFnmeth3662_MOESM213_ESM.zip. A user guide can be found in the Supplementary information. We used codes in analyses of fiber photometry data. Through correspondence with the company, we confirmed that these codes are proprietary and commercially licensed. Consequently, in compliance with the company’s licensing agreements, it cannot be approved by the Open Source Initiative or deposited in open-source repositories.
References
Yoshikawa, T., Nakamura, T. & Yanai, K. Histaminergic neurons in the tuberomammillary nucleus as a control centre for wakefulness. Br. J. Pharmacol. https://doi.org/10.1111/bph.15220 (2020).
Xu, L., Lin, W., Zheng, Y., Wang, Y. & Chen, Z. The diverse network of brain histamine in feeding: dissect its functions in a circuit-specific way. Curr. Neuropharmacol. https://doi.org/10.2174/1570159X21666221117153755 (2022).
de Almeida, M. A. & Izquierdo, I. Memory facilitation by histamine. Arch. Int. Pharmacodyn. Ther. 283, 193–198 (1986).
Arumuham, A. et al. The histamine system and cognitive function: an in vivo H3 receptor PET imaging study in healthy volunteers and patients with schizophrenia. J. Psychopharmacol. 37, 1011–1022 (2023).
Inoue, I. et al. Impaired locomotor activity and exploratory behavior in mice lacking histamine H1 receptors. Proc. Natl. Acad. Sci. USA 93, 13316–13320 (1996).
Krystal, A. D., Richelson, E. & Roth, T. Review of the histamine system and the clinical effects of H1 antagonists: basis for a new model for understanding the effects of insomnia medications. Sleep. Med. Rev. 17, 263–272 (2013).
Nishino, S. et al. Decreased CSF histamine in narcolepsy with and without low CSF hypocretin-1 in comparison to healthy controls. Sleep 32, 175–180 (2009).
Zlomuzica, A. et al. Neuronal histamine and cognitive symptoms in Alzheimer’s disease. Neuropharmacology 106, 135–145 (2016).
Rinne, J. O. et al. Increased brain histamine levels in Parkinson’s disease but not in multiple system atrophy. J. Neurochem. 81, 954–960 (2002).
Tuomisto, L., Kilpelainen, H. & Riekkinen, P. Histamine and histamine-N-methyltransferase in the CSF of patients with multiple sclerosis. Agents Actions 13, 255–257 (1983).
Kano, M. et al. Decreased histamine H1 receptor binding in the brain of depressed patients. Eur. J. Neurosci. 20, 803–810 (2004).
Cheng, L. et al. Histamine H1 receptor deletion in cholinergic neurons induces sensorimotor gating ability deficit and social impairments in mice. Nat. Commun. 12, 1142 (2021).
Ma, Q. et al. Histamine H2 receptor deficit in glutamatergic neurons contributes to the pathogenesis of schizophrenia. Proc. Natl. Acad. Sci. USA 120, e2207003120 (2023).
Zhang, X. Y. et al. Targeting presynaptic H3 heteroreceptor in nucleus accumbens to improve anxiety and obsessive-compulsive-like behaviors. Proc. Natl. Acad. Sci. USA 117, 32155–32164 (2020).
Xu, L. et al. An H2R-dependent medial septum histaminergic circuit mediates feeding behavior. Curr. Biol. https://doi.org/10.1016/j.cub.2022.03.010 (2022).
Zheng, Y. et al. Postsynaptic histamine H3 receptors in ventral basal forebrain cholinergic neurons modulate contextual fear memory. Cell Rep. 42, 113073 (2023).
Xu, H.-T. et al. Histaminergic innervation of the ventral anterior thalamic nucleus alleviates motor deficits in a 6-OHDA-induced rat model of Parkinson’s disease. Neurosci. Bull. https://doi.org/10.1007/s12264-024-01320-0 (2024).
Lin, W. et al. Whole-brain mapping of histaminergic projections in mouse brain. Proc. Natl. Acad. Sci. USA 120, e2216231120 (2023).
Ericson, H., Blomqvist, A. & Kohler, C. Origin of neuronal inputs to the region of the tuberomammillary nucleus of the rat brain. J. Comp. Neurol. 311, 45–64 (1991).
Fujita, A. et al. Hypothalamic tuberomammillary nucleus neurons: electrophysiological diversity and essential role in arousal stability. J. Neurosci. 37, 9574–9592 (2017).
Wickersham, I. R. et al. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron 53, 639–647 (2007).
Wall, N. R., Wickersham, I. R., Cetin, A., De La Parra, M. & Callaway, E. M. Monosynaptic circuit tracing in vivo through Cre-dependent targeting and complementation of modified rabies virus. Proc. Natl. Acad. Sci. USA 107, 21848–21853 (2010).
Saito, Y. C. et al. Monoamines inhibit GABAergic neurons in ventrolateral preoptic area that make direct synaptic connections to hypothalamic arousal neurons. J. Neurosci. 38, 6366–6378 (2018).
Li, X. et al. Generation of a whole-brain atlas for the cholinergic system and mesoscopic projectome analysis of basal forebrain cholinergic neurons. Proc. Natl. Acad. Sci. USA 115, 415–420 (2018).
Sun, Q. et al. A whole-brain map of long-range inputs to GABAergic interneurons in the mouse medial prefrontal cortex. Nat. Neurosci. 22, 1357–1370 (2019).
Munoz-Castaneda, R. et al. Cellular anatomy of the mouse primary motor cortex. Nature 598, 159–166 (2021).
Gao, L. et al. Single-neuron projectome of mouse prefrontal cortex. Nat. Neurosci. https://doi.org/10.1038/s41593-022-01041-5 (2022).
Cheng, J. et al. The interaction between the ventrolateral preoptic nucleus and the tuberomammillary nucleus in regulating the sleep-wakefulness cycle. Front. Neurosci. 14, 615854 (2020).
Blandina, P., Munari, L., Provensi, G. & Passani, M. B. Histamine neurons in the tuberomamillary nucleus: a whole center or distinct subpopulations?. Front. Syst. Neurosci. 6, 33 (2012).
Wang, Q. et al. The allen mouse brain common coordinate framework: a 3D reference atlas. Cell 181, https://doi.org/10.1016/j.cell.2020.04.007 (2020).
Mesulam, M. M. From sensation to cognition. Brain 121, 1013–1052 (1998).
Etkin, A., Egner, T. & Kalisch, R. Emotional processing in anterior cingulate and medial prefrontal cortex. Trends Cogn. Sci. 15, 85–93 (2011).
Yang, Q. Z. & Hatton, G. I. Electrophysiology of excitatory and inhibitory afferents to rat histaminergic tuberomammillary nucleus neurons from hypothalamic and forebrain sites. Brain Res. 773, 162–172 (1997).
Faucard, R. et al. N-methyl-D-aspartate receptor antagonists enhance histamine neuron activity in rodent brain. J. Neurochem. 98, 1487–1496 (2006).
Yin, D. et al. Glutamate activates the histaminergic tuberomammillary nucleus and increases wakefulness in rats. Neuroscience 413, 86–98 (2019).
Park, S.-G. et al. Medial preoptic circuit induces hunting-like actions to target objects and prey. Nat. Neurosci. 21, 364–372 (2018).
Fremeau, R. T., Voglmaier, S., Seal, R. P. & Edwards, R. H. VGLUTs define subsets of excitatory neurons and suggest novel roles for glutamate. Trends Neurosci. 27, 98–103 (2004).
Tsai, N. Y. et al. Trans-Seq maps a selective mammalian retinotectal synapse instructed by Nephronectin. Nat. Neurosci. 25, 659–674 (2022).
Klapoetke, N. C. et al. Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014).
Wang, D. et al. GABAergic neurons in the dorsal-intermediate lateral septum regulate sleep-wakefulness and anesthesia in mice. Anesthesiology 135, 463–481 (2021).
Ren, S. et al. The paraventricular thalamus is a critical thalamic area for wakefulness. Science 362, 429–434 (2018).
Bouin, A. et al. New rabies viral resources for multi-scale neural circuit mapping. Mol. Psychiatry https://doi.org/10.1038/s41380-024-02451-6 (2024).
Chen, A.-X. et al. Specific hypothalamic neurons required for sensing conspecific male cues relevant to inter-male aggression. Neuron 108, https://doi.org/10.1016/j.neuron.2020.08.025 (2020).
Wouterlood, F. G., Gaykema, R. P., Steinbusch, H. W., Watanabe, T. & Wada, H. The connections between the septum-diagonal band complex and histaminergic neurons in the posterior hypothalamus of the rat. Anterograde tracing with Phaseolus vulgaris-leucoagglutinin combined with immunocytochemistry of histidine decarboxylase. Neuroscience 26, 827–845 (1988).
Buzsáki, G. & Draguhn, A. Neuronal oscillations in cortical networks. Science 304, 1926–1929 (2004).
Jin, H., Fishman, Z. H., Ye, M., Wang, L. & Zuker, C. S. Top-down control of sweet and bitter taste in the mammalian brain. Cell 184, https://doi.org/10.1016/j.cell.2020.12.014 (2021).
Harris, K. D. & Shepherd, G. M. G. The neocortical circuit: themes and variations. Nat. Neurosci. 18, 170–181 (2015).
Economo, M. N. et al. Distinct descending motor cortex pathways and their roles in movement. Nature 563, 79–84 (2018).
Naka, A. & Adesnik, H. Inhibitory circuits in cortical layer 5. Front. Neural Circuits 10, 35 (2016).
Li, A., Yetkin, F. Z., Cox, R. & Haughton, V. M. Ipsilateral hemisphere activation during motor and sensory tasks. AJNR Am. J. Neuroradiol. 17, 651–655 (1996).
Cramer, S. C., Finklestein, S. P., Schaechter, J. D., Bush, G. & Rosen, B. R. Activation of distinct motor cortex regions during ipsilateral and contralateral finger movements. J. Neurophysiol. 81, 383–387 (1999).
Panula, P. & Nuutinen, S. The histaminergic network in the brain: basic organization and role in disease. Nat. Rev. Neurosci. 14, 472–487 (2013).
Shan, L., Bao, A. M. & Swaab, D. F. The human histaminergic system in neuropsychiatric disorders. Trends Neurosci. 38, 167–177 (2015).
Mohawk, J. A., Green, C. B. & Takahashi, J. S. Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 35, 445–462 (2012).
Boes, A. D. et al. Connectivity of sleep- and wake-promoting regions of the human hypothalamus observed during resting wakefulness. Sleep 41, https://doi.org/10.1093/sleep/zsy108 (2018).
Chokroverty, S. Overview of sleep & sleep disorders. Indian J. Med. Res. 131, 126–140 (2010).
Gungor, N. Z. & Paré, D. Functional heterogeneity in the bed nucleus of the stria terminalis. J. Neurosci. 36, 8038–8049 (2016).
Kim, S.-R. & Kim, S.-Y. Functional dissection of glutamatergic and GABAergic neurons in the bed nucleus of the stria terminalis. Mol. Cells 44, 63–67 (2021).
Sah, P., Faber, E. S. L., Lopez De Armentia, M. & Power, J. The amygdaloid complex: anatomy and physiology. Physiol. Rev. 83, 803–834 (2003).
Janak, P. H. & Tye, K. M. From circuits to behaviour in the amygdala. Nature 517, 284–292 (2015).
Cheng, L. et al. Histamine H1 receptors in dentate gyrus-projecting cholinergic neurons of the medial septum suppress contextual fear retrieval in mice. Nat. Commun. 15, 5805 (2024).
Zhang, B.-B. et al. Suppression of excitatory synaptic transmission in the centrolateral amygdala via presynaptic histamine H3 heteroreceptors. J. Physiol. https://doi.org/10.1113/JP286392 (2024).
Duvarci, S. & Pare, D. Amygdala microcircuits controlling learned fear. Neuron 82, 966–980 (2014).
Dong, C., Madar, A. D. & Sheffield, M. E. J. Distinct place cell dynamics in CA1 and CA3 encode experience in new environments. Nat. Commun. 12, 2977 (2021).
Schumacher, A. et al. Ventral hippocampal CA1 and CA3 differentially mediate learned approach-avoidance conflict processing. Curr. Biol. 28, https://doi.org/10.1016/j.cub.2018.03.012 (2018).
Watkins de Jong, L., Nejad, M. M., Yoon, E., Cheng, S. & Diba, K. Optogenetics reveals paradoxical network stabilizations in hippocampal CA1 and CA3. Curr. Biol. 33, https://doi.org/10.1016/j.cub.2023.03.032 (2023).
Guzowski, J. F., Knierim, J. J. & Moser, E. I. Ensemble dynamics of hippocampal regions CA3 and CA1. Neuron 44, 581–584 (2004).
Curtis, G. R., Oakes, K. & Barson, J. R. Expression and distribution of neuropeptide-expressing cells throughout the rodent paraventricular nucleus of the thalamus. Front. Behav. Neurosci. 14, 634163 (2020).
Shima, Y. et al. Distinctiveness and continuity in transcriptome and connectivity in the anterior-posterior axis of the paraventricular nucleus of the thalamus. Cell Rep. 42, 113309 (2023).
Peever, J. & Fuller, P. M. The biology of REM sleep. Curr. Biol. 27, R1237–R1248 (2017).
Sherin, J. E., Elmquist, J. K., Torrealba, F. & Saper, C. B. Innervation of histaminergic tuberomammillary neurons by GABAergic and galaninergic neurons in the ventrolateral preoptic nucleus of the rat. J. Neurosci. 18, 4705–4721 (1998).
Steininger, T. L., Gong, H., McGinty, D. & Szymusiak, R. Subregional organization of preoptic area/anterior hypothalamic projections to arousal-related monoaminergic cell groups. J. Comp. Neurol. 429, 638–653 (2001).
Mickelsen, L. E. et al. Cellular taxonomy and spatial organization of the murine ventral posterior hypothalamus. Elife 9, https://doi.org/10.7554/eLife.58901 (2020).
Gong, H. et al. High-throughput dual-colour precision imaging for brain-wide connectome with cytoarchitectonic landmarks at the cellular level. Nat. Commun. 7, 12142 (2016).
Avants, B. B., Epstein, C. L., Grossman, M. & Gee, J. C. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 12, 26–41 (2008).
Zhou, H. et al. GTree: an open-source tool for dense reconstruction of brain-wide neuronal population. Neuroinformatics 19, 305–317 (2021).
Li, Y. et al. TDat: an efficient platform for processing petabyte-scale whole-brain volumetric images. Front. Neural Circuits 11, 51 (2017).
Acknowledgements
This project was supported by grants from National Key R&D Program of China (2021ZD0202803, received by Z.C.), STI2030-Major Projects (Grant No. 2021ZD0201002, received by Q.L.), the National Natural Science Foundation of China (82300818, received by W.L., T2122015, received by Q.L., U21A20418, received by Z.C., and U23A20533, received by Y.W.), Natural Science Foundation of Zhejiang Province (LD25H090002, received by Y.Z.), and China Postdoctoral Science Foundation (2023M733191, received by W.L.).
Author information
Authors and Affiliations
Contributions
Q.L., and Z.C. designed research; W.L., X.Z., X.Y., M.L., and X.Q. performed experiments; W.L., Z.F., Q.X., S.A., C.D., and A.L. analyzed data; L.C., L.X., Y.Z., W.H., and P.P. contributed to discussion, and W.L., Y.W., and Z.C. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lin, W., Zhu, X., Yu, X. et al. A whole-brain male mouse atlas of long-range inputs to histaminergic neurons. Nat Commun 16, 8092 (2025). https://doi.org/10.1038/s41467-025-63394-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-63394-2