Abstract
Large language models can lighten the workload of clinicians and patients, yet their responses often include fabricated evidence, outdated knowledge, and insufficient medical specificity. We introduce a general retrieval-augmented question-answering framework that continuously gathers up-to-date, high-quality medical knowledge and generates evidence-traceable responses. Here we show that this approach significantly improves the evidence validity, medical expertise, and timeliness of large language model outputs, thereby enhancing their overall quality and credibility. Evaluation against 15,530 objective questions, together with two physician-curated clinical test sets covering evidence-based medical practice and medical order explanation, confirms the improvements. In blinded trials, resident physicians indicate meaningful assistance in 87.00% of evidence-based medical scenarios, and lay users find it helpful in 90.09% of medical order explanations. These findings demonstrate a practical route to trustworthy, general-purpose language assistants for clinical applications.
Similar content being viewed by others
Introduction
Large language models (LLMs) hold significant promise for enhancing healthcare by assisting clinical decision-making, streamlining medical research, and improving patient care efficiency1,2,3,4,5. However, their integration into clinical practice remains hindered by several crucial limitations. First, LLMs frequently fabricate medical evidence content or sources6,7, which can lead to erroneous clinical decisions. Secondly, their responses frequently lack sufficient medical expertise and accuracy8, often failing to meet the needs of complex clinical scenarios. Third, LLMs rely on static training data; they fail to incorporate the latest medical guidelines and evidence9. These issues collectively undermine the quality and credibility of LLM-generated medical responses, particularly limiting their applicability in clinical scenarios that demand rigorous accuracy and verifiable evidence sources, such as evidence-based medicine10 (EBM) and medical order explanation (MOE).
Specifically, for EBM practice, while LLMs can rapidly generate evidence summaries through natural language interaction, studies show that over 60% of the evidence summaries generated by LLMs contain fabricated or erroneous associations11. Similarly, for the MOE task, the lack of reliable evidence in LLM-generated explanations, coupled with patients’ limited ability to discern medical information, ultimately leading them to seek clarification from their physicians, exacerbating clinical burdens. Therefore, enhancing the quality and credibility of existing LLM-generated medical responses, particularly ensuring the reliability of evidence content and sources, is crucial to overcoming these challenges and fully realizing their potential value in clinical settings.
Currently, several studies have attempted to mitigate these challenges. For instance, Med-PaLM4 enhances medical expertise through fine-tuning, while MEDAgents12 and MDAgents13 improve reasoning capabilities via multi-agent collaboration. However, these methods still face issues such as evidence fabrication and insufficient timeliness. Retrieval-augmented generation14 methods, including MedRAG15, Almanac16, and Clinfo.ai17, mitigate information timeliness by retrieving real-time data14,18. Still, traditional RAG approaches frequently retrieve ineffective information, struggle with complex clinical queries, and have not adequately resolved credibility issues due to evidence fabrication. Although several studies19,20,21 have increasingly recognized the importance of LLMs’ credibility in the medical field, existing research remains at the descriptive level, lacking actionable solutions. Overall, there is currently an absence of effective methods to enhance the quality and credibility of LLMs in clinical applications, which severely limits their clinical value.
To fill this gap, we propose a general medical Q&A framework named LINS, which effectively enhances the quality and credibility of existing LLMs’ medical responses. This advancement is driven by our newly introduced Multi-Agent Iterative Retrieval Augmented Generation (MAIRAG) and Keyword Extraction Degradation (KED) algorithms, which enable multi-agent collaboration to perform more granular problem decomposition and effective information filtering. By retrieving and integrating the latest effective medical information into its generative pipeline, coupled with an evidence-traceable output format, LINS significantly improves the evidence validity, medical expertise, and timeliness of LLM-generated responses. Consequently, LINS completes a comprehensive query-response-validation interactive loop, significantly enhancing the quality, credibility, and clinical applicability of responses from LLMs. To further validate the performance of LINS, we conducted a series of large-scale experiments, including general medical capability assessments and real-world clinical scenario testing.
The general medical capability evaluation of LINS consists of two components: a large-scale objective multiple-choice assessment and human evaluations. In the objective assessment, we not only covered six publicly available datasets but also developed Multi-MedCQA, a novel dataset curated by 50 attending physicians based on real-world clinical experience and rigorously cross-validated. This dataset addresses a significant challenge in medical evaluation—data leakage risks from publicly shared datasets, which can lead to overfitting in LLMs and inflated performance results. In the human evaluation component, we engaged both physicians and lay users to assess the responses generated by LINS and LLM to commonly searched consumer questions in medicine, evaluating quality across nine dimensions. The results demonstrate that LINS not only excels in the multiple-choice assessment, achieving SOTA performance, but also shows superior medical expertise and alignment with scientific consensus in human evaluations, significantly enhancing the quality of medical responses from LLMs.
Furthermore, the application of LINS in real-world clinical scenarios encompasses two major cases: assisting physicians in EBM practice and helping patients with MOE. In collaboration with four top hospitals in China, we engaged physicians to create the AEBMP and MOEQA datasets, which are based on medical records and clinical experiences. A randomized blind-controlled experiment was conducted, involving 113 physicians and 1207 complex clinical questions with medical records, totaling around 32,300 evaluations (each evaluation of a specific dimension of a question by an assessor counted as one evaluation). The experimental results demonstrated that LINS not only significantly improved the response quality of LLMs but also achieved a greater than 40% increase in credibility across various tasks (P < 0.001). This achievement is primarily attributed to LINS’s ability to provide fully authentic and traceable evidence, in contrast to the often fictitious evidence generated by LLMs.
Results
Overview of LINS
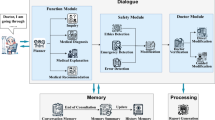
The LINS framework constructed in this study is primarily composed of a database module, a retrieval module, a multi-agent module, and a generator module (Fig. 1a and Methods). Upon receiving a user query, the retrieval module employs the KED algorithm (Fig. 1a and Methods) to search for relevant information within the database module. Subsequently, the multi-agent module utilizes the MAIRAG algorithm (Fig. 1b and Methods) to filter out effective information (information directly helpful for answering the question). Finally, the generator module produces a response in an evidence-traceable format, as indicated by the red box in Fig. 1a.
a Overview of the LINS framework. b Illustration of the KED and MAIRAG algorithms. c Clinical applications of LINS are demonstrated through assisting physicians in evidence-based medical practice and helping patients with medical order explanation. d Seven multiple-choice objective evaluation datasets and three human evaluation datasets, where blue indicates that the dataset is a public dataset, and pink indicates that the dataset is a physician-curated and internet-isolated dataset.
Specifically, the database module of LINS encompasses both local and online databases, offering a rich and continuously updated source of information. The retrieval module, comprising a retriever, a keyword extraction agent, and the KED algorithm (Fig. 1a, b, and Methods), is tasked with sifting through the vast and intricate database to pinpoint information relevant to the query at hand. The multi-agent module, equipped with four core agents and the MAIRAG algorithm (Fig. 1a, b, and Methods), serves two pivotal functions: (1) it filters out the truly effective information from the retrieved information, and (2) it breaks down complex questions that are challenging to retrieve directly into a series of easier sub-questions, enabling iterative retrieval and response. Finally, the generator module is responsible for synthesizing the gathered effective information, integrating it with the LLM’s inherent knowledge to produce a response in an evidence-traceable format, thereby further elevating the credibility of the responses. The construction and configuration of each module are detailed in the Methods section.
Notably, every module within the LINS framework is optional, allowing users to select models and configurations that best suit their specific requirements. Furthermore, we have developed an easy-to-use end-to-end python package (https://github.com/WangSheng21s/LINS) to streamline implementation and facilitate user accessibility.
Overall, LINS, as a general medical Q&A framework, demonstrates significant potential for diverse applications in the medical domain. In the subsequent sections, we will showcase its application in the medical field through examples such as LINS assisting physicians in evidence-based medical practice and helping patients with medical order explanation (Fig. 1c).
Novel expert-curated clinical datasets for evaluation
Currently, many publicly available multiple-choice evaluation datasets already exist (Fig. 1d), offering valuable resources for researchers. While these public datasets provide a consistent benchmark for comparing different methods, they also carry the risk of data leakage during model training. This issue can lead to inflated performance metrics, ultimately undermining the clinical validity and reliability of the results. To address this, we invited 50 physicians from four top hospitals in China to develop the Multi-MedCQA dataset. It consists of nine specialty-specific sub-datasets with 4685 multiple-choice questions, all manually created and reviewed by attending physicians based on real clinical experience. These sub-datasets cover areas including thoracic surgery, cardiology, hematology, respiratory medicine, urology, general surgery, nephrology, ophthalmology, and gastroenterology. Each question underwent multiple rounds of physician review and refinement, guaranteeing clinical relevance and accuracy. By addressing data leakage issues prevalent in public datasets, it establishes a robust benchmark for evaluating medical LLMs. The Multi-MedCQA dataset will be released without answers, with a testing interface to prevent data leakage, playing a crucial role in future medical LLM assessments. We describe the detailed construction process of Multi-MedCQA in the Methods section.
In real-world clinical settings, questions from physicians or patients are typically open-ended rather than multiple-choice. While publicly available medical Q&A datasets exist, there is a notable gap in datasets that assess LLMs’ abilities in two critical clinical scenarios: evidence-based medical (EBM) practice and medical order explanation (MOE). To address this, we developed two novel datasets—the AEBMP dataset and the MOEQA dataset—designed to evaluate LINS’s capabilities in assisting physicians in EBM practice and helping patients with MOE, respectively. These datasets feature open-ended Q&A questions paired with medical records, providing a more authentic assessment of LINS’s performance in real-world scenarios from both physician and patient perspectives. The AEBMP dataset includes clinical questions crafted by attending physicians based on real clinical experiences, while the MOEQA dataset consists of questions posed by lay users from a patient’s perspective. Together, these datasets effectively evaluate the real-world clinical applicability of LINS and LLMs, offering significant testing value. We provide a detailed description of the construction process for the AEBMP and MOEQA datasets in the Methods section.
Objective evaluations for General Medical Competency
To comprehensively evaluate the effectiveness of LINS in general medical capability, we first compared the performance of LINS-LLMs and LLMs across objective evaluation datasets using eight different LLMs as base models (Supplementary Fig. 1 and Methods). The eight LLMs included GPT-4o-mini, GPT-4o, o1-mini, o1-preview22, Llama3.1-70b23, Qwen2.5-72b24, Gemini-1.5-flash, and Gemini-1.5-pro25. The objective evaluation datasets comprised 9 human-created novel sub-datasets from Multi-MedCQA and 6 publicly available datasets (MedMCQA26, PubMedQA*27, MedQA-M, MedQA-U, MedQA-T28, and Geneturing-disease29).
First, we evaluated the performance of eight LLMs across 15 objective datasets. The results showed that o1-preview achieved the best performance on 11 out of the 15 objective datasets (Supplementary Fig. 1 and Supplementary Table 1), with an average rank score (calculated as the average ranking across all test datasets) of 1.33, securing the top position (Supplementary Table 2). These findings highlight the broader applicability and effectiveness of the o1-preview model in understanding and reasoning across various medical domains.
Next, we integrated the eight LLMs into the LINS framework to assess whether LINS could enhance their general medical competency. We observed that the performance of almost each LLM improved across the 15 objective datasets, with 114 out of 120 results showed improvement (Supplementary Fig. 1 and Supplementary Table 1). Notably, LINS-o1-preview achieved an average rank score of 1.4 (Supplementary Table 2), maintaining the top position. It consistently improved the performance of o1-preview across all datasets (Fig. 2a, b, Supplementary Table 1). On the six publicly available datasets, LINS-o1-preview achieved accuracies of 83.17%, 77.00%, 92.31%, 94.58%, 90.14%, and 86.54%, respectively, compared to the original o1-preview scores of 81.52%, 57.20%, 88.36%, 94.49%, 89.53%, and 65.45% (Fig. 2a). Similarly, LINS-o1-preview outperformed o1-preview on all nine novel human-created datasets (Fig. 2b). Comparative results for other LINS-LLMs are presented in Supplementary Table 1. Overall, these findings demonstrate that LINS consistently enhances the general medical competency of diverse LLMs, irrespective of their baseline capabilities.
To control for variables, all tests are conducted without using prompting techniques (e.g., few-shot, chain-of-thought reasoning). a Comparison between LINS-o1-preview and o1-preview on six public objective evaluation datasets. b Comparison between LINS-o1-preview and o1-preview on nine manually created objective evaluation datasets. c Performance comparison between LINS and other RAG frameworks across 15 objective evaluation datasets, with each dataset color-coded by performance ranking. d Comparison between LINS and previously reported scores on six public datasets. The previously reported scores is up to December 1, 2024. LINS reports the best score achieved using eight different base LLMs on each dataset. e Performance of LINS using eight base LLMs compared to the LLMs alone and LLMs with contextual information. The LLM score represents the direct score of the LLM on the PubMedQA* dataset, while the LINS score represents the performance of each LLM integrated with LINS on PubMedQA*. The LLM with contexts score indicates the performance of the LLM using contexts from the PubMedQA dataset as a reference when answering questions in PubMedQA*. The shaded areas in the bar chart indicate performance drops. f Human evaluation results by five clinicians comparing LINS-o1-preview and o1-preview on 300 real clinical questions randomly selected from the HealthSearchQA dataset. Statistical significance was assessed using the Kruskal-Wallis test (two-sided). No adjustments were made for multiple comparisons (p = 1.92e-130). g Human evaluation results by five lay users comparing LINS-o1-preview and o1-preview on 300 real clinical questions randomly selected from the HealthSearchQA dataset. Statistical significance was assessed using the Kruskal-Wallis test (two-sided). No adjustments were made for multiple comparisons (p = 2.07e-25). Source data are provided as a Source Data file.
Additionally, we compared LINS with two publicly available medical RAG frameworks, MedRAG15 and Clinfo17, across the aforementioned 15 datasets (Fig. 2c). To control variables, we selected GPT-4o-mini as the base model due to its high cost efficiency and the consistency observed across LLMs (consistency see Supplementary Fig. 2 and Supplementary Note). The results reveal that (Fig. 2c) Clinfo-GPT-4o-mini achieved better results than GPT-4o-mini on 6 datasets but underperformed on the remaining 9 datasets. MedRAG-GPT-4o-mini showed better overall performance compared to Clinfo. However, LINS-GPT-4o-mini performed the best, not only improving GPT-4o-mini across all datasets but also surpassing both Clinfo-GPT-4o-mini and MedRAG-GPT-4o-mini on 15 datasets, demonstrating its remarkable superiority.
Furthermore, we compared LINS with previously reported scores on 6 publicly available objective evaluation datasets. The results demonstrate that LINS outperformed the previously reported scores on all 6 datasets, achieving accuracies of 83.17%, 82.20%, 92.31%, 94.58%, 90.14%, and 86.66%, respectively (Fig. 2d). Details of the sources of previously reported scores, the LLMs, and the databases used for each dataset are provided in Supplementary Table 3.
We further evaluated the capability of the retrieval module in LINS. The PubMedQA* dataset is derived from the PubMedQA27 dataset by removing its contexts (see Methods), making the contexts included in the PubMedQA dataset the gold standard for retrieval in PubMedQA*. So we evaluated the effectiveness of the LINS retrieval module by comparing the performance of LLMs with contexts and LINS (Fig. 2e). First, we compared the performance of LLMs and LLMs with contexts on the PubMedQA* dataset. As expected, the performance of LLMs with contexts was consistently better than that of LLMs (Fig. 2e). Next, we assessed the performance of LINS compared to LLMs with contexts. Remarkably, LINS not only significantly improved the performance of all eight LLMs on the PubMedQA* dataset but also outperformed LLMs with contexts for six of the LLMs (Fig. 2e). This further demonstrates that the information retrieved by LINS is of higher quality than the gold-standard contexts, underscoring the superior capability of the LINS retrieval module.
In summary, LINS has demonstrated its effectiveness across both publicly available and novel human-created objective evaluation datasets, using eight different LLMs as base models, thereby showcasing its general medical capability. Additionally, considering the superior performance of LINS-o1-preview and the consistency observed across LLMs (Supplementary Table 2, Supplementary Fig. 2, and Supplementary Note), we selected o1-preview as the base model for subsequent human evaluations and application scenarios.
Human evaluations for general medical competency
To evaluate the general medical capability of LINS in common medical Q&A, we randomly selected 300 commonly searched consumer questions from the HealthSearchQA4 dataset for human evaluation. Subsequently, we conducted a randomized blind-controlled assessment of the responses generated by LINS-o1-preview and o1-preview (see Methods). The evaluation dimensions were categorized into three main aspects: utility, accuracy, and comprehensiveness. The utility aspect included helpfulness and harmfulness; the accuracy aspect includes scientific consensus, medical expertise, content relevance, and content completeness; and the comprehensiveness aspect includes answer diversity, empirical support, and bias (Supplementary Fig. 3). To ensure professional rigor, physicians conducted evaluations across nine dimensions, whereas patients focused solely on the utility aspect.
The evaluation results from 5 physicians (Figs. 2f and 5) lay users (Fig. 2g and Supplementary Table 4) reveal that LINS-o1-preview was deemed helpful in answering 94.40% and 99.00% of questions by physicians and patients, respectively, compared to 85.67% and 95.53% for o1-preview. These findings highlight the challenges in addressing complex clinical questions, while LINS-o1-preview demonstrates substantial potential in providing relevant assistance in most cases. Furthermore, physicians and lay users reported that LINS-o1-preview provided potentially misleading answers in only 1.27% and 0.20% of cases, respectively, outperforming o1-preview, which exhibited misleading responses in 2.27% and 1.13% of cases (Fig. 2f, g). In terms of accuracy, physicians noted that 99.40% of LINS-o1-preview’s answers adhered to scientific consensus, 99.47% demonstrated medical expertise, 99.67% aligned with the posed questions, and 94.87% were considered comprehensive (Fig. 2f). By comparison, o1-preview achieved scores of 97.80%, 90.27%, 96.80%, and 55.07% across these same metrics (Fig. 2f). Finally, we evaluated the responses based on their comprehensiveness, specifically whether they addressed questions from multiple perspectives, incorporated evidence support, and avoided bias. The two dimensions pose greater challenges for the model. The results showed that LINS-o1-preview scored 90.87%, 57.33%, and 0.40% in these aspects, respectively, while o1-preview achieved scores of 55.07%, 40.07%, and 1.73% (Fig. 2f).
Comparing the results of o1-preview and LINS-o1-preview, we found that LINS-o1-preview not only improved scientific consensus and medical expertise in the answers, enhanced the accuracy of objective question assessments, but also significantly enhanced answer diversity and comprehensiveness, providing more empirical support and effective assistance to users. Harmfulness and bias in the answers were also effectively suppressed, although content relevance slightly declined. A potential explanation for this observation is that the KED and MAIRAG algorithm retrieved numerous specialized and effective pieces of knowledge, providing LINS with valuable references, thus making its answers more professional, comprehensive, and reliable. Naturally, handling a vast array of complex and specialized medical knowledge may present significant challenges to the intrinsic capabilities of the LLM.
Overall, LINS demonstrates exceptional performance in the accuracy and professionalism of its responses to medical questions. Its answers align with scientific consensus, exhibit medical expertise, contain minimal bias, and include very few instances of misleading information, resulting in superior response quality. This provides strong support for its potential application in real-world medical scenarios.
LINS can assist physicians in evidence-based medicine practice
With the exponential growth of scientific literature and emerging evidence sources, evidence-based medicine (EBM)10 has become increasingly complex30. While LLMs can generate responses with evidence through prompt engineering, they suffer from limitations such as outdated knowledge and evidence hallucinations, often leading to inaccurate or irrelevant citations. In contrast, LINS retrieves high-quality evidence from authoritative databases with proper sourcing, making it a more reliable tool for EBM. We next evaluate the potential of LINS and LLMs in supporting physicians in EBM practice.
Overall, our evaluation was conducted in two phases. In the first phase, we invited 100 resident physicians to assess whether LINS-o1-preview could effectively assist their EBM practice. In the second phase, five attending physicians conducted a more granular evaluation, examining LINS’s ability to enhance the quality and credibility of o1-preview’s medical responses in EBM scenarios. The assessment was based on the AEBMP dataset, which we developed in collaboration with physicians to reflect real-world EBM practice. This dataset consists of two subdatasets: AEBMP_stage1 includes 1000 medical records and 1000 clinical questions for phase one, while AEBMP_stage2 contains 75 medical records and 100 clinical questions for phase two (Supplementary Fig.4 and Methods). Additionally, we developed HERD (Hierarchical Evidence Retrieval Database) for LINS, integrating an evidence-based local retrieval database, the PubMed online retrieval database, and the Bing online retrieval database to provide access to high-quality and comprehensive evidence (see Methods and Fig. 3b). To facilitate evaluation, we also built a dedicated website for assessing LINS’s utility in evidence-based medical practice (Supplementary Fig. 5). The detailed workflow of LINS in assisting physicians is comprehensively described in the Methods section and visually represented in Fig. 3a.
a Illustration of LINS assisting physicians in evidence-based medical practice. The PICO question represents a clinical question reformulated according to the PICO principle to make it searchable and answerable. b The Hierarchical Retrieval Database is specifically designed to support LINS in assisting evidence-based medical practice. c One hundred resident physicians were invited to conduct 3000 EBP practice evaluations. Firstly, they assessed whether LINS-o1-preview and o1-preview could assist them in evidence-based medical practice and listed the reasons if they could not. d Secondly, they evaluated which of LINS-o1-preview and o1-preview generated better evidence-based recommendations. The integers in the figure represent the number of questions. e Evaluation of resident physicians’ perceptions regarding the assistance of LINS-o1-preview in evidence-based medical practice. f Multidimensional evaluation results by five attending physicians assessing the quality of final answers obtained by resident physicians with assistance from LINS-o1-preview and o1-preview. g Assessment by five clinical physicians on whether the evidence citations in the final answers, facilitated by LINS-o1-preview and o1-preview, enhance the credibility of the responses. Statistical significance was assessed using the Kruskal-Wallis test (two-sided). No adjustments were made for multiple comparisons (p = 7.57e-7). Source data are provided as a Source Data file.
In the first phase, evaluations from 100 resident physicians showed that LINS-o1-preview effectively supported EBM practice in 87.00% of questions (Fig. 3c). Moreover, it significantly reduced workload, improved efficiency, and reduced stress in 85.03%, 85.10%, and 80.47% of cases, respectively (Fig. 3e). These findings underscore the potential of LINS in evidence-based clinical settings. Compared with o1-preview, LINS-o1-preview exhibited a 53.97% improvement in assisting EBM practice (Fig. 3c), and physicians considered its evidence-based recommendations superior in 78.40% cases (Fig. 3d). Further analysis indicated that, across 3000 cases (with 1000 questions repeated thrice), o1-preview provided invalid evidence in 1478 responses and partially irrelevant information in 233 responses, whereas the corresponding numbers for LINS-o1-preview were 61 and 69, respectively (Fig. 3c). Detailed evaluation procedures and methods are described in Methods.
In the second phase of the experiment, we invited five attending physicians to conduct an in-depth evaluation of the quality of the final answers generated by LINS-o1-preview and o1-preview in assisting resident physicians. The assessment was performed across six detailed dimensions (Supplementary Fig. 6): scientific consensus, content relevance, harmlessness, no bias, evidence validity, and credibility. The results showed that the overall quality of answers generated with LINS-o1-preview assistance was superior to that generated with o1-preview assistance. Scores for scientific consensus, content consistency, harmlessness, and no bias were 97.44%, 97.18%, 97.44%, and 95.64% for LINS-o1-preview, compared to 93.59%, 96.41%, 95.13%, and 89.23% for o1-preview (Fig. 3f). Notably, evidence validity in final answers generated with LINS-o1-preview assistance reached 86.41%, representing a 39.49% improvement over o1-preview (Fig. 3f). Moreover, in credibility (more credible: no change: less credible), LINS-o1-preview (329: 59: 2) significantly outperformed o1-preview (171: 207: 12) (Fig. 3g). Analysis of the evaluation results revealed a strong correlation between the credibility of the responses and the validity of the evidence within them, with a Pearson correlation coefficient of 0.75.
To provide a clearer illustration of the differences in workflow and quality between LINS and LLMs for assisting physicians in EBM practice, we visually showcased a comparative case based on a medical record and a PICO question (Transform clinical questions into searchable and answerable questions based on the PICO principle) in Supplementary Fig. 7. This example highlights the quality differences between LINS and LLMs in aiding physicians’ EBM. Notably, LINS-o1-preview retrieved a 2024 study recently published in Nature Medicine31 and provided a scientifically grounded response with valid evidence. In contrast, the o1-preview LLM (trained on data up to October 2023) generated a response citing three pieces of evidence, one of which was fabricated, and another irrelevant. Such issues can potentially undermine users’ expectations regarding the quality and credibility of evidence-based recommendations.
Overall, resident physicians exhibited significant enthusiasm when utilizing LINS-o1-preview to support EBM practice. Compared to o1-preview, LINS-o1-preview demonstrated greater effectiveness in assisting physicians in generating higher-quality evidence-based answers. While o1-preview performed well in terms of scientific consensus and content relevance, its overall performance fell short of LINS-o1-preview, particularly in the critical areas of evidence validity and credibility.
LINS can help patients with medical order explanation
Medical orders are essential for guiding patient care, but patients often struggle to understand them and cannot always consult doctors for clarification. While LLMs can generate explanations, they typically lack credible evidence or sources, making it hard for patients to trust or verify the information. In contrast, LINS offers reliable, evidence-backed explanations that are accessible at any time, helping patients better understand and follow medical orders. We next evaluate the potential of LINS and LLMs in helping patients with medical order explanations (MOE).
For the MOE, the workflow of LINS comprised three key processes, including medical terminology extraction, medical terminology explanation, and clinical Q&A. (Fig. 4a, with details provided in the Methods section). First, LINS assists in extracting medical terms from medical records that may require explanation. For each term, LINS retrieves relevant evidence and provides explanations in a traceable evidence format. Patients can continue to ask questions freely, and LINS will retrieve related evidence, offering reasonable and traceable responses tailored to the patient’s medical records. To comprehensively validate the effectiveness of LINS in MOE scenarios, 5 lay users and 5 physicians were invited to conduct a randomized, blind-controlled evaluation with MOEQA Datasets (see Methods). Details of the evaluation process are provided in Supplementary Fig. 8 and Methods, while the evaluation dimensions are outlined in Supplementary Fig. 9.
a Illustration of LINS helping patients with medical order interpretation and clinical Q&A. b Evaluation results from five lay users on whether the extraction of medical terms met their expectations. c Assessment by five lay users of the answer provided by LINS-o1-preview and o1-preview in medical term explanation tasks. d Assessment by five lay users of the answer provided by LINS-o1-preview and o1-preview in clinical Q&A tasks. e Credibility evaluation results from five lay users for LINS-o1-preview and o1-preview in both medical term explanation and clinical Q&A tasks. f Evaluation by five clinical physicians of the correctness of medical term interpretation by LINS-o1-preview and o1-preview. g Evaluation by five clinical physicians of the quality of clinical Q&A answers provided by LINS-o1-preview and o1-preview. Statistical significance was assessed using the Kruskal-Wallis test (two-sided). No adjustments were made for multiple comparisons. For physicians, p = 2.93e-40; for lay users, p = 4.01e-19. Source data are provided as a Source Data file.
From the patient evaluation results, an average of 87.72% of the 116 medical terms extracted by the Medical Terminology Extraction Agent (MTEA) from medical orders aligned with patient expectations (Fig. 4b), demonstrating that the majority of terms extracted were accurate and met patient needs. For medical terminology explanation, all five lay users reported that 99.14% of LINS-o1-preview’s explanations were helpful, while o1-preview achieved a helpfulness score of 87.93% (Fig. 4c). Moreover, 94.66% of the evidence provided by LINS-o1-preview was valid, in stark contrast to 25.00% for o1-preview (Fig. 4c). In the credibility scoring (more reliable: no change: less reliable), patients rated LINS-o1-preview at 471: 105: 4, significantly outperforming o1-preview, which received a score of 94: 478: 8 (Fig. 4e left). Similarly, for the clinical Q&A task, LINS-o1-preview achieved scores of 90.09% for helpfulness, 71.21% for evidence validity, and a credibility rating of 313: 220: 2. In contrast, o1-preview scored 87.86% for helpfulness, 14.02% for evidence validity, and a credibility rating of 62: 459: 14 (Fig. 4d, e right). These findings suggest that clinical Q&A for medical order are more challenging than terminology explanation. Nonetheless, LINS-o1-preview outperformed o1-preview in both tasks, with a particularly significant advantage in credibility. By analyzing the correlation coefficients between evidence validity and credibility in both the term explanation task and the clinical Q&A task, we found that the Pearson correlation coefficients were 0.78 and 0.83, respectively. This result further supports our finding that evidence validity plays a critical role in the credibility of LLM-generated medical responses.
We also invited five attending physicians to provide a more granular evaluation of LINS. According to their assessments, LINS-o1-preview achieved an accuracy rate of 98.45% in explaining the 116 medical terms, compared to 85.00% for o1-preview (Fig. 4f). In the clinical Q&A task, LINS-o1-preview scored 97.57% for scientific consensus, 99.25% for content relevance, 0.75% for harmfulness, and 5.23% for bias. In comparison, o1-preview achieved scores of 97.57%, 99.44%, 1.31%, and 6.17%, respectively (Fig. 4g). The observed bias in both LINS-o1-preview and o1-preview may partly result from their attempts to provide personalized answers tailored to the patient’s specific circumstances during the clinical Q&A process.
To provide a clearer illustration of the entire workflow, we present an example showcasing the processes of LINS and the LLM in MOE evaluation (Supplementary Fig. 10). Due to space limitations, we display results for two terminology explanations and one clinical Q&A. From this example, it can be observed that LINS-o1-preview delivers high-quality answers and valid evidence for each term and clinical question, whereas o1-preview includes some fabricated or irrelevant evidence, diminishing the overall quality of its responses.
In summary, compared to LLMs, LINS exhibits superior performance in medical terminology explanation and clinical Q&A, providing more accurate and expert responses backed by reliable medical evidence. This not only effectively addresses the issue of evidence hallucination in LLMs but also significantly enhances the credibility of its answers. This advantage helps overcome the trust barriers patients often encounter with LLMs, markedly increasing the clinical utility of LINS and underscoring its potential and practicality in assisting patients with complex medical information.
Evaluation system for evidence-traceable text
Evidence-traceable text, as shown in Fig. 5a, helps users locate sources of evidence and reduces hallucinations generated by the LLMs32. However, the quality assessment of evidence-traceable formatted text currently relies heavily on human evaluation, and there is a lack of a comprehensive and robust automated evaluation system for assessing such evidence-traceable formatted text. Human evaluation, as a scarce resource, is not always readily available, which hinders the widespread adoption of evidence-traceable formatted text. To address this, we propose Link-Eval. As illustrated in Fig. 5b, Link-Eval primarily evaluates two aspects: citations and statements. For citations, we introduce three evaluation metrics: citation set precision, citation precision, and citation recall, to assess the correctness and comprehensiveness of the citations. For statements, we propose statement correctness and statement fluency as evaluation metrics to assess the correctness and fluency of the statements. The specific calculation methods for these metrics are detailed in Fig. 5c–e and Methods.
a Example of the citation-based generated text format used by LINS. b Evaluation dimensions of Link-Eval. c Process for calculating the correct citation set, correct citations, and valid citations. d Example calculation of citation precision and citation recall scores. e Process for calculating statement correctness and statement fluency. f Test accuracy of the NLI models, clinical physician, and lay user on the MedNLI-mini dataset. g Cramér’s V correlation scores between the NLI models, physician, lay user, and gold label on the MedNLI-mini dataset. h Evaluation results of LINS using Link-Eval on PubMedQA* and HealthSearchQA. Source data are provided as a Source Data file.
Link-Eval integrates an NLI model, a fluency scorer, and a correlation scorer to assess quality across the aforementioned dimensions. The fluency scorer is adapted from the previously published UNIEVAL model33. The correlation scorer has undergone rigorous statistical validation (Supplementary Fig. 22). As a key component of the automated evaluation system, the NLI model is utilized to assess semantic relationships between sentence pairs, categorizing them as entailment, contradiction, or neutrality. To ensure the reliability of both the NLI model and Link-Eval, we first evaluated the performance and correlation of various NLI models with human assessments. We selected five NLI models (GPT-4o-mini, GPT-4o22, Qwen-2.5-72b24, T5-11b, and T5-3b34), and tested their performance on the Med-NLI-mini dataset, a subset of 415 randomly sampled questions from the publicly available Med-NLI medical inference dataset35. Additionally, one physicians and one lay users were invited for human evaluation. The results revealed that GPT-4o achieved the highest accuracy at 88.92% (Fig. 5f), followed by Qwen-2.5-72b (86.02%), GPT-4o-mini (80.48%), T5-11b (75.18%), and T5-3b (73.73%). In comparison, the accuracy rates for the physician and the lay users were 81.69% and 73.98%, respectively (Fig. 5f). These findings demonstrate that the Med-NLI-mini dataset presents a challenging task, as even experienced physicians are susceptible to errors. Notably, GPT-4o and Qwen-2.5-72b outperformed the physician, showcasing exceptional natural language reasoning capabilities. To further analyze the correlation between different methods, we introduced Cramer’s V coefficient36. The results (Fig. 5g) showed that GPT-4o exhibited the highest correlation with the standard answers (0.83) and a correlation of 0.7 with the physician, while the correlation between the physician and the standard answers was 0.74. This indicates that GPT-4o not only excels at capturing semantic relationships in complex medical contexts but also closely aligns with the reasoning patterns of professional physicians. In conclusion, GPT-4o demonstrated strong performance in complex medical NLI tasks, surpassing even human physicians. Consequently, GPT-4o has been selected as the preferred NLI engine in the Link-Eval automated evaluation system for subsequent experiments and tests.
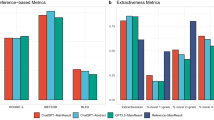
To test the effectiveness of Link-Eval on evidence-traceable text, we conducted Q&A using LINS on the PubMedQA* and 100 randomly sampled questions from the HealthSearchQA dataset and evaluated the results using Link-Eval. The evaluation results are shown in Fig. 5h. LINS performed exceptionally well in evidence-traceable text, achieving citation set precision scores of 96.44% and 92.49% on the PubMedQA* and HealthSearchQA datasets, respectively (Fig. 5h). Citation precision, compared to citation set precision, is a more challenging metric that more finely assesses the correctness of each citation within the citation set, judging the LLMs’ ability to analyze and organize retrieved passages from a more granular perspective. LINS achieved citation precision scores of 76.61% and 74.77% on the two datasets, respectively (Fig. 5h). Additionally, the citation recall score evaluates the comprehensiveness of the citations, with LINS achieving scores of 72.80% and 65.34% on the respective datasets, indicating that most effective retrieval passages were correctly cited (Fig. 5h). It is important to note that citation precision and citation recall scores are interdependent; striving for higher citation precision can lead to lower citation recall and vice versa. This demonstrates that Link-Eval effectively balances the accuracy and comprehensiveness of citations, avoiding an excessive focus on high-quality references that could result in the omission of valuable information.
In evaluating the statements, statement fluency assesses the fluency of the text, checking if the statements follows grammatical rules, has natural sentence structures, and uses appropriate words. Fluent text should read smoothly without causing stiffness or confusion for the reader. LINS achieved scores of 83.27% and 88.43% on the two datasets (Fig. 5h), respectively, indicating that its generated statements is fluent, logically coherent, and easy to read. Statement correctness evaluates the accuracy of the model, mainly to determine if there are any contradictory descriptions between statements. LINS achieved statement correctness scores of 98.07% and 98.76% on the two datasets, respectively, demonstrating its reliability in generating accurate statements (Fig. 5h).
In summary, Link-Eval serves as a complementary solution when professional human evaluation resources are limited, enabling efficient and objective automated assessment of evidence-traceable text quality while ensuring reproducible evaluation outcomes. Additionally, Link-Eval provides a standardized evaluation framework that fosters the advancement of evidence-traceable text research and applications.
Discussion
LLMs hold significant promise in enhancing clinical practice, medical research, and patient care. However, their integration into real-world clinical settings remains challenging due to issues such as fabricated evidence, lack of timeliness, and insufficient medical specificity. These limitations compromise the quality and credibility of LLM-generated medical responses, posing significant barriers to their widespread adoption and effective deployment in clinical setting.
To address the above limitations of LLMs in medical applications, we propose the LINS framework with the novel MAIRAG and KED algorithms. By dynamically retrieving and integrating effective medical information directly into the generative workflow, coupled with an evidence-traceable output format, LINS substantially improves the evidence validity, medical expertise, and timeliness of LLM-generated responses. Consequently, LINS completes a comprehensive query-response-validation interactive loop, significantly enhancing the quality, credibility, and clinical applicability of LLM-generated responses.
In our study, we conducted large-scale objective benchmarks and human evaluations to demonstrate LINS’ general medical capabilities. Additionally, we validated its clinical value in two real-world application scenarios: evidence-based medical practice and medical order interpretation. The results of our experiments demonstrate that LINS significantly improves the quality and credibility of medical responses generated by LLMs. For physicians, LINS offers a powerful tool for rapidly retrieving and summarizing evidence, thereby reducing the time and effort required for traditional evidence retrieval methods. For patients, LINS provides clear, personalized explanations of medical orders, which can improve their understanding and adherence to treatment plans. These advancements have the potential to alleviate clinical burdens and improve patient outcomes, making LINS a valuable asset in modern healthcare. Moreover, the creation of novel datasets such as Multi-MedCQA, AEBMP, and MOEQA addresses a critical need for high-quality, physician-curated data in the evaluation of LLMs. These datasets not only mitigate the risks of data leakage and overfitting but also provide a more accurate reflection of real-world clinical scenarios. By making these datasets publicly available, we hope to contribute to the ongoing development and evaluation of LLMs in the medical field, fostering further innovation and collaboration.
As we have mentioned, LLMs frequently fabricate medical evidence content or sources6,7. We define this phenomenon as “evidence hallucination”, a specific subset of AI hallucinations37. The root cause of these hallucinations lies in the generative nature of LLMs: the models predict the next word based on statistical correlations rather than aiming for factual accuracy38. This issue is particularly concerning in the medical field. If clinicians were to trust evidence fabricated by LLMs, they might unknowingly be led to incorrect diagnoses or treatment pathways, potentially causing severe consequences for patient health38. Over time, this could erode trust in LLMs within medical settings, leading to diminished confidence among both doctors and patients. To address these challenges, LINS innovatively integrates a retrieval strategy with an evidence-traceable mechanism, combining LLMs with external knowledge sources and verification mechanisms to ensure the traceability and reliability of generated answers. Specifically, LINS employs a dual-mechanism approach: First, LINS utilizes the MAIRAG and KED algorithms to retrieve relevant and reliable evidence, providing LLMs with real-time access to authoritative external information. This RAG14 strategy overcomes the limitations of traditional LLMs that rely solely on training, enabling the model to generate responses based on the latest and most credible sources. Second, LINS implements an evidence traceability mechanism that establishes a verifiable mapping between generated answers and their original sources. The mechanism not only annotates the sources of information in its responses but also maintains an internal evidence chain, ensuring that all cited literature, data, and other evidence are authentic and verifiable. All in all, through our framework, LINS teaches LLMs to consult sources and cite evidence rather than making assumptions, effectively mitigating evidence hallucination and enhancing the quality and credibility of LLM-generated medical responses. However, even with these technological safeguards, the use of medical AI requires caution: professionals must always verify AI-generated recommendations. Looking ahead, as such frameworks are refined and more widely deployed, we can expect to fully realize the potential of LLMs in healthcare while ensuring patient safety. This will allow us to benefit from the efficiency gains offered by LLMs without compromising scientific rigor or best practices.
In the medical field, information timeliness is of paramount importance, as medical knowledge and treatment protocols are constantly evolving. New research findings, clinical trial results, and treatment guidelines are published at a rapid pace39. However, LLMs are typically trained on static datasets40, which means they cannot reflect the latest medical advancements or real-time clinical information. Frameworks like LINS, which are based on retrieval-augmented generation14, address this limitation by enabling real-time retrieval of online information to supplement the knowledge referenced during LLM responses. Specifically, LINS, as a medical framework combining information retrieval and text generation, operates on the core principle of retrieving relevant information fragments from external knowledge bases as contextual input before the LLM generates its response. This framework ensures that LLMs are no longer confined to outdated knowledge from their training data but can instead leverage the latest research, clinical guidelines, and drug information to provide accurate and up-to-date answers.
In the medical domain, the importance of privacy protection cannot be overstated41,42,43. The LINS demonstrates strong capabilities in mitigating potential privacy risks. While some commercial APIs, such as the OpenAI API, are compliant with HIPAA (Health Insurance Portability and Accountability Act) standards, users with higher privacy requirements often demand that no sensitive data be uploaded or exposed to the internet under any circumstances. To address this, LINS offers flexible configuration options, enabling users to choose solutions tailored to their specific needs. For datasets without sensitive information, users can leverage commercial LLMs or online retrieval to enhance system functionality. However, for highly confidential data, LINS supports localized processing, recommending local deployment of LLMs and a retrieval database. This ensures that all sensitive data remains under the user’s control during processing and analysis, minimizing the risk of data leakage while maintaining robust functionality.
However, this study has several limitations. Firstly, although the human evaluation was conducted by a large and highly professional team of physicians, the current participants may not fully represent the diverse standards of medical practice across different regions and countries. Future work should aim to expand the size and diversity of the evaluation team to obtain more comprehensive and representative evaluation feedback. Secondly, although LINS’s local database retrieval enhancement function has advantages such as no need for training, low usage threshold, strong scalability, and good privacy protection, the current method of constructing local databases is commonly used in formats like TXT, JSON, and PDF. In future work, we plan to develop specialized retrieval mechanisms and processing workflows for diverse data formats, including tables and knowledge graphs, making LINS a powerful AI system capable of handling various retrieval enhancement formats. Thirdly, while the MAIRAG algorithm delivers excellent performance, it incurs additional token costs. Using MedQA-U and MedQA-M datasets as examples (MedQA-U contains many logic-reasoning questions, and MedQA-M focuses on questions requiring medical knowledge), we evaluated the performance and token usage (prompt_tokens + completion_tokens) of four methods: Chat (pure model), original RAG18, MAIRAG, and CoT44 (Supplementary Table 5) with GPT-4o-mini. For knowledge required questions in the MedQA-M dataset, the CoT method demonstrated the lowest token usage but achieved only moderate performance. In contrast, MAIRAG provided the best results with token costs comparable to those of the RAG algorithm. For reasoning required questions in the MedQA-U dataset, the original RAG performed poorly, while MAIRAG excelled by decomposing and reasoning through questions, albeit at a higher token cost (1.565 times that of the original RAG). Despite the increased expenditure, MAIRAG’s superior response quality makes it acceptable for users prioritizing accuracy. It is important to note that CoT and RAG methods are not mutually exclusive and can be combined to further enhance model performance. MAIRAG and RAG improve response accuracy by integrating external knowledge retrieval, enriching the generative model’s background information. Meanwhile, CoT aids in step-by-step reasoning, enabling more sophisticated logical responses. By combining these approaches, systems can balance rich contextual knowledge with effective multi-step reasoning, achieving higher-quality answers in knowledge-intensive scenarios while optimizing performance and cost. This synergistic approach holds great promise for delivering superior solutions in future applications.
The development of LINS marks a significant advancement in the integration of LLMs into clinical practice. By seamlessly integrating with any LLM, LINS enhances its versatility, offering healthcare providers and researchers a powerful tool for improving clinical decision-making and patient care. With its robust real-time evidence retrieval, integration, and extension capabilities, LINS aims to deliver accurate, professional, and credible support for medical researchers, clinicians, and patients alike. Moreover, its flexible design and adaptability to evolving medical dynamics are poised to elevate global healthcare quality, fostering progress in research and clinical practices across diverse medical fields. Furthermore, LINS’s open-source strategy and the release of its datasets are expected to catalyze deeper AI integration and innovation in medicine, paving the way for transformative breakthroughs in the field.
Methods
This study complies with all relevant ethical regulations. The research protocol was reviewed and approved by the Ethics Committee of Beijing Anzhen Hospital, Capital Medical University (Approval ID: KS2024118). All data were processed in accordance with applicable privacy and data protection standards, and no identifiable personal information was collected or used. All participants provided informed consent and received a small monetary compensation for their participation. Participants were randomly assigned to experimental conditions and were blinded to the study design. No deception was used. As the studies were conducted online, there was no direct interaction between researchers and participants.
In all studies, gender was determined based on participants’ self-identification. We did not consider or pre-register any analyses disaggregated by sex or gender, as these variables were not theoretically relevant to our research questions.
The construction and configuration of each module in LINS
As we show in Fig. 1a, the LINS framework constructed is primarily composed of a database module, a retrieval module, a multi-agent module, and a generator module. Each module communicates through natural language interfaces, achieving a synergistic and cooperative effect. Specifically:
Database module
The input to the database module is a natural language question, and the output is the information from the database that is relevant to the query. LINS supports both connecting to online databases for real-time data retrieval and allowing users to connect to local, private databases to meet personalized search needs. In this study, we provided two online databases, PubMed and Bing, and also constructed four local databases—OMIM, OncoKB, Textbooks, and Guidelines—as examples. The PubMed and Bing online databases rely on calling their respective online API interfaces, while the four local databases save data by encoding natural language text into vector database formats.
Retrieval module
The retrieval module takes as input the user’s question along with the relevant information provided by the database module, and produces as output the top-k most pertinent pieces of information corresponding to the query. Since the relevant information output by the database module may consist of hundreds or thousands of entries, the retrieval module is responsible for filtering out the top-k most relevant pieces of information using a finer-grained semantic vector similarity encoding method and passing them to the multi-agent module in natural language format. Here, top-k is a configurable hyperparameter. The retrieval module includes a retriever and a keyword extraction agent45. The KEA, implemented using a LLM based on specific prompts (Supplementary Fig. 12), is tasked with extracting a specified number of keywords from the query in order of importance, thereby facilitating the keyword extraction degradation algorithm (The Keyword Extraction Degradation Algorithm is described below).
Multi-agent module
The multi-agent module employs a distributed agent architecture, where each agent is built upon LLM and specializes in task processing through pre-defined prompts (Supplementary Fig. 11). The multi-agent module consists of four agents—PRA, SKA, QDA, and PCA. They are all constructed based on LLM through the use of prompts. The PRA aids in filtering effective knowledge, preventing interfering knowledge from disrupting the generation of answers. The SKA assesses the model’s internal knowledge reserves, thoroughly mining its internal knowledge in the absence of effective external knowledge. The QDA is responsible for breaking down complex questions into more manageable subproblems, iteratively delving into the core of the issue. The PCA ensures that the generated answers align with retrieved authoritative knowledge, avoiding contradictions with known information.
Generator module
The generator module is constructed based on a base LLM with pre-defined prompts (Supplementary Fig. 18). It is primarily responsible for generating responses in specific formats and content based on the user’s query, the retrieved information, and the model’s internal knowledge. LINS allows users to integrate existing open-source or commercial API LLMs as generators according to their needs. In this study, we comprehensively evaluated eight LLMs (GPT-4o-mini, GPT-4o, o1-mini, o1-preview, Gemini-1.5-flash, Gemini-1.5-pro, Qwen2.5-72b, and Llama3.1-70b) and provided detailed evaluation results and model selection guidelines in the Supplementary Note.
Multi-agent iterative retrieval augmented generation algorithm
To overcome the limitations of the original RAG algorithm, which suffers from insufficient retrieval quality and depth, we propose the MAIRAG algorithm. This algorithm enhances the quality of retrieved information and decomposes complex problems into subproblems for iterative processing. It primarily involves four main agents: PRA, SKA, QDA, and PCA. Specifically, when relevant information is retrieved, the PRA first filters for effective information—information that directly aids in answering the question. If effective information is identified, it is passed to the generator to produce a response. If PRA determines no effective information is available, the SKA evaluates whether the generator’s intrinsic knowledge is sufficient to answer the question. If not, the QDA breaks the query into sub-questions, retrieves information for each, and synthesizes the results into a reference passage for generating the final answer. When effective information is available, the PCA ensures the generated response aligns with this information. If inconsistencies arise, we prioritize the effective information and regenerate the response to align with it. Importantly, the activation of agents in the LINS framework is configurable. If none are enabled, the MAIRAG algorithm defaults to the original RAG algorithm.
Keyword extraction degradation algorithm
To overcome the issue of traditional retrieval methods potentially failing to retrieve results when handling complex and lengthy sentences, we propose the KED algorithm. The KED algorithm, as shown in Fig. 1b, effectively distills key information while maintaining a sufficient retrieval scope to ensure important results are not missed. We have developed a keyword extraction agent and a corresponding degradation strategy. The keyword extraction agent first extracts keywords from the retrieval passage in order of importance, from high to low, and users can control the maximum number of keywords extracted. During retrieval, each keyword essentially acts as a restriction. In the context of long-text retrieval, when the number of keywords is too few, the results may lack precision. By gradually removing the least important keywords and re-initiating the retrieval, effective results are obtained. This approach not only optimizes the query process but also ensures the relevance and comprehensiveness of the retrieval results. It is particularly suited for the complex retrieval demands of biomedical literature databases within the LINS system.
Database
In this study, we utilized an extensive set of online databases, including Bing, and PubMed27, tailored specifically for molecular biomedical research. Additionally, customizable local databases were constructed to meet specific user needs. For this study, we developed four representative local databases—OncoKB46, OMIM47, Textbooks28, and Guidelines—encompassing knowledge across diseases, cells, molecules, and drugs within specialized molecular biomedical domains. Notably, LINS uses the PubMed online database by default for information retrieval. However, users have the flexibility to specify alternative databases, such as Bing or local sources like OncoKB, through simple parameter settings. If users wish to perform combined searches across multiple databases, they can utilize the Hierarchical Retrieval Database (HRD) structure described below. By enumerating the desired database names, LINS will sequentially search each specified database in the defined priority order until relevant information is retrieved. Furthermore, users can conveniently add, delete, or modify knowledge within their local databases in natural language form and subsequently update these databases with a single command through our provided code. To ensure ease of use, we have included detailed, user-friendly tutorials within our codebase.
Online database
The online databases primarily leverage search engine APIs, offering advantages such as real-time updates, rapid iteration, and high integration. LINS integrates Bing and PubMed as part of its database ecosystem, allowing users to select based on their specific needs. This decision reflects the unique strengths and coverage of each source. Bing, as a general-purpose search engine, provides diverse and extensive information resources suitable for cross-disciplinary or general queries. In contrast, PubMed, a specialized database for biomedical literature, offers precise and in-depth resources tailored to medical researchers and professionals. This multi-interface integration enables LINS to address medical queries comprehensively, balancing broad general knowledge with authoritative, specialized insights. By incorporating diverse databases, the system enhances robustness, ensuring reliable, enriched, and comprehensive answers to a wide range of user inquiries.
Local database
In this study, we developed four local databases—OMIM, OncoKB, Textbooks, and Guidelines—while also demonstrating how users can construct personalized local databases within the LINS system. The specific implementation details are as follows.
Local OMIM database construction
The OMIM47 dataset is an authoritative online database of human genetic diseases. It includes detailed information about genetic disorders, such as descriptions of the diseases, inheritance patterns, related genes, genetic variations, and references to relevant literature. OMIM is widely used in medical genetic research and clinical diagnosis, serving as a valuable resource for understanding human genetic diseases and conducting related gene research. The data format in the OMIM database is shown in Supplementary Fig. 15b. For each human genetic disease, OMIM contains information described in natural language, such as TEXT, Description, Clinical Features, Diagnosis, Clinical Management, Pathogenesis, Molecular Genetics, Population Genetics, Animal Model, and History. We collected these pieces of information and segmented them based on line breaks, then removed invalid information with character lengths less than 100. Finally, we obtained a total of 68,626 segments as knowledge stored in jsonl format to form the local OMIM database.
Local OncoKB database construction
OncoKB46 is a knowledge-driven database specifically designed for precision oncology. It integrates information about tumor molecular profiles, clinical evidence, drug sensitivity, resistance mechanisms, and patient treatment outcomes. OncoKB aims to provide in-depth insights into cancer genomics, molecular targeted therapy, and immunotherapy for doctors, researchers, and patients, supporting clinical decision-making and the development of personalized treatment plans. The database synthesizes a vast amount of scientific literature, clinical trial data, and cancer genomic data to provide evidence for cancer treatment and is continuously updated to reflect the latest scientific advancements and clinical practices. The data format in the OncoKB database is shown in Supplementary Fig. 15c. For each gene mutation and corresponding cancer, it integrates relevant descriptions, drugs, and drug sensitivity. However, OncoKB does not describe this information in natural language but rather in tabular form. Therefore, we need to set a fixed template to convert this information into natural language. Specifically, if the level for drug D with mutation A in gene G causing cancer C is R1 or R2 (resistant), we will convert it into the following knowledge: “A patient with C, if A occurs at G, can become resistant to D, which is detrimental to treatment.” If the level is 1, 2, 3, or 4, we will convert it into: “A patient with C, if A occurs at G, the recommended drug is D.” Along with other natural language descriptions on OncoKB, our constructed local OncoKB database contains a total of 8150 pieces of knowledge, covering 858 genes, 7729 alterations, 137 cancer types, and 138 drugs.
Local Textbooks database construction
The Textbooks local database comprises textual material from 18 widely used English medical textbooks curated from the MedQA28 collection. These textbooks encompass a broad range of medical knowledge and have been meticulously organized to enable the model to extract relevant evidence for reasoning and clinical decision-making. As shown in Supplementary Fig. 15d, the content from these 18 textbooks was segmented by line breaks, resulting in 231,581 paragraphs containing a total of 12,727,711 words. The data was stored in JSON format as the foundation of the Textbooks local database. To integrate it into the LINS system, we encoded the content into a vector database using the text embedding model Text-embedding-3-large, facilitating efficient retrieval and use within the system.
Local guidelines database construction
As illustrated in Supplementary Fig. 15e, we collected 414 evidence-based medical guidelines from the internet, encompassing four specialties: nephrology, cardiothoracic surgery, orthopedics, and psychiatry. The dataset includes both Chinese and English guidelines. Using the E2M48 tool, we converted PDF documents into text (TXT) files. Given the strong contextual coherence inherent to medical guidelines, we structured the local library at the document level rather than passage-based segmentation, as was done for textbooks. However, the length of individual guidelines often exceeds the maximum input limits of retrieval models, and excessively long content can compromise retrieval precision. To address this, we employed the following strategy: For each of the 414 guidelines, we used the GPT-4o model to generate concise summaries and constructed a guideline map that links each summary to its corresponding txt file. The local guidelines library thus consists of these txt files alongside the guideline map. To integrate this library into LINS, we encoded the summaries into a vector database using the Text-embedding-3-large model. When LINS matches a guideline summary based on semantic similarity within the local guidelines library, the guideline map enables precise retrieval of the full guideline content for downstream applications.
Hierarchical retrieval database
The Hierarchical Retrieval Database (HRD) is a tiered retrieval data structure within LINS that enables users to seamlessly combine local and online databases for evidence search. It supports both user-defined local database and online database, allowing flexible selection of which libraries to use and their priority order. By default, the retrieval prioritizes local database over online databases. If relevant evidence is found in a higher-priority library, it is returned directly; otherwise, the search proceeds to the next library in the hierarchy. Users can customize local libraries by adding TXT or PDF files as retrieval content.
Hierarchical evidence retrieval database
As shown in Fig. 3b, the HERD is a specialized HRD designed to support evidence-based medical practice with LINS. It integrates three retrieval databases: a locally constructed Guidelines database, the PubMed online database, and the Bing database. The priority of these libraries is ranked from highest to lowest as follows: Guidelines, PubMed, and Bing.
Construction of novel expert-curated datasets
In this study, we invited 50 attending physicians from four top-tier hospitals in China to participate in the manual construction, inspection, and review of the Multi-MedCQA, AEBMP, and MOEQA datasets. Among them, Multi-MedCQA is used to assess the general medical capabilities of LLMs, while AEBMP and MOEQA are used to evaluate LINS and LLM performance in two real clinical scenarios: evidence-based medical practice and medical order explanation, respectively.
Multi-MedCQA
The Multi-MedCQA dataset consists of nine specialty-specific sub-datasets: thoracic surgery, cardiology, hematology, respiratory medicine, urology, general surgery, nephrology, ophthalmology, and gastroenterology. These sub-datasets collectively comprise 4685 multiple-choice questions, with 198–956 questions per specialty. Each question includes four options, with only one correct answer. The detailed construction process of the Multi-MedCQA dataset is illustrated in Supplementary Fig. 14. We invited attending physicians from nine medical specialties to manually create multiple-choice questions reflecting general medical knowledge within their respective fields. The nine specialties included thoracic surgery, cardiology, hematology, respiratory medicine, urology, general surgery, nephrology, ophthalmology, gastroenterology, with each contributing a minimum of 500 questions. This effort resulted in an initial set of nine sub-datasets containing 500, 523, 500, 550, 500, 1000, 200, 500, and 500 questions, respectively. To ensure the dataset’s quality and relevance, senior specialists reviewed each sub-dataset. Questions with issues such as missing answers, incorrect answers, identical content found online, unclear phrasing, redundancy, or inconsistent formatting were excluded. After this rigorous curation process, the finalized sub-datasets contained 500, 510, 499, 535, 498, 956, 198, 489, and 500 questions, respectively. To broaden the dataset’s applicability, a medical English specialist translated all questions and options into English, ensuring accuracy and consistency. The Multi-MedCQA dataset is thus available in both Chinese and English, making it accessible to a wider audience. To prevent the dataset from being used for training LLMs, we will release only the questions and answer options in both languages while withholding the answer keys. Instead, an open-access platform for accuracy evaluation will be provided, allowing researchers to assess LLM performance on Multi-MedCQA without compromising its integrity as an independent benchmark.
AEBMP
The AEBMP dataset consists of two subsets: AEBMP_stage1 and AEBMP_stage2, designed to evaluate the performance of LINS and LLM in EBM practice. AEBMP_stage1 contains 1000 medical records and 1000 corresponding clinical questions. First, we extracted 1600 real medical records from the Chinese Medical Record Repository, a collection of high-quality real-world medical cases. Then, we used GPT-4o to extract the patient information from each record and submitted them to 9 physicians for review. Based on their clinical experience, the physicians formulated a relevant clinical question for each medical record. In total, we obtained 1273 cases and corresponding clinical questions, from which 1000 were randomly selected to form the AEBMP_stage1 dataset. The AEBMP_stage2 dataset includes 75 medical records and 100 clinical questions. We used GPT-4o to categorize 411 medical records into nephrology (230 cases) and cardiac surgery (181 cases) from MedQA (USMLE)28. These records were then reviewed by nine physicians, who selected cases suitable for evidence-based medical practice and formulated related clinical questions. After this process, 178 medical records and 213 clinical questions were obtained, from which 75 medical records and 100 related clinical questions were randomly selected to form the final AEBMP_stage2 dataset.
MOEQA
The MOEQA dataset evaluates LINS’s ability to help patients with medical order explanation. It includes 24 medical records and 107 clinical questions. LINS analyzes user-provided medical records to extract relevant medical terminology, offering detailed explanations before addressing user-specific clinical questions based on the context of the records.
The construction process of the MOEQA dataset is outlined in Supplementary Fig. 9. Unlike AEBMP, this dataset required records containing comprehensive patient information, including diagnoses and treatments. We sourced cases from the Chinese Medical Case Repository, a collection of high-quality real-world medical cases. Three physicians selected and reformatted 48 cases into a standardized structure of ‘Patient Information’ plus ‘Examination’ plus ‘Diagnosis’ plus ‘Treatment’. From these, 24 cases were randomly selected. To generate clinical questions, five lay users simulated patient scenarios, formulating one or two questions for each case. After consolidation and deduplication, the final MOEQA dataset included 24 medical records and 107 clinical questions.
Setting of objective multiple-choice question evaluation
In this study, we built a large-scale objective multiple-choice question evaluation to assess the general medical capabilities of LINS. To more intuitively evaluate the performance of LINS, we employed a zero-shot setting across all datasets used in this experiment. We tested eight LLMs—GPT-4o-mini, GPT-4o, o1-mini, o1-preview22, Llama3.1-70b23, Qwen2.5-72b24, Gemini-1.5-flash, and Gemini-1.5-pro25—and their performance after integration with LINS on the nine subsets of our constructed Multi-MedCQA dataset and six publicly available datasets: MedMCQA26, PubMedQA*27, MedQA-M, MedQA-U, MedQA-T28, and Geneturing-disease29. Among them, LINS retrieves from the PubMedQA database for the PubMedQA* dataset, from the Textbooks database for the MedQA-U dataset, and from the Bing database for all other datasets. A brief description of the six publicly available datasets used in this study is provided below.
MedMCQA
MedMCQA26 is a large-scale, multidisciplinary multiple-choice question dataset designed specifically for question-answering tasks in the medical domain. It comprises over 194,000 high-quality multiple-choice questions sourced from AIIMS and NEET PG entrance exams, spanning 2400 medical topics across 21 medical disciplines. Each sample includes a question, the correct answer, and alternative options, aiming to assess various reasoning abilities of models across diverse medical topics and disciplines. For our evaluation, we utilized the validation subset, MedMCQA-dev, consisting of 4183 multiple-choice questions.
PubMedQA
The PubMedQA27 dataset is extracted from authentic literature, ensuring the data’s accuracy and reliability. It encompasses various fields such as clinical medicine, biomedical science, and pharmacology. The data format is shown in Supplementary Fig. 13a. Each entry includes PMID, Question, Contexts, Long Answer, and Final Decision. The Contexts section typically comprises Objective, Methods, and Results. Each PMID uniquely corresponds to a document in the PubMedQA30 database. The Question is either the title of the PMID-associated article or a question derived from the title, with an average length of 13.1 words. The Objective, Method, Results, and Long Answer sections correspond to parts of the article abstract, averaging 54.94 words in length. The Final Decision is an answer represented by Yes, No, or Maybe. The entire PubMedQA test dataset consists of 500 test questions that require inferential conclusions.
PubMedQA*
To simulate real-world biomedical application scenarios, we constructed PubMedQA* by following the methodology outlined in MedRAG15. This involved removing the provided contextual information (Contexts) from 500 expert-annotated test samples in the original PubMedQA dataset, leaving only the Questions and Final Decisions (Yes/No/Maybe). In real-world applications, user queries often lack accompanying contextual references, making PubMedQA* a more realistic benchmark for evaluating the performance of large models in practical settings compared to the original PubMedQA dataset.
MedQA-M
The MedQA-M28 test set consists of 3426 multiple-choice questions, each with four options and a single correct answer. These questions are sourced from the Medical Licensing Examination in mainland China (MCMLE). The test set includes both simple knowledge-based questions and complex case-based scenarios, covering a wide range of medical disciplines. Answering these questions requires a comprehensive understanding of various diseases and their treatments.
MedQA-U
The MedQA-U28 test set comprises 1273 multiple-choice questions, each with four options and a single correct answer. These questions are derived from the United States Medical Licensing Examination (USMLE). The test set focuses on clinical scenarios, emphasizing the need for deep understanding of medical concepts and multi-step logical reasoning. Most questions describe patient symptoms and require identification of the most likely diagnosis, appropriate treatment, or underlying disease mechanism, testing both reasoning ability and medical knowledge.
MedQA-T
The MedQA-T28 test set includes 1413 multiple-choice questions, each with four options and a single correct answer. These questions originate from medical licensing examinations in Taiwan (TWMLE) and are based on the same corpus as the MedQA-U subset. The questions typically present real-world clinical scenarios with detailed patient histories, requiring the model to make precise judgments among numerous medical options.
Geneturing-disease
The Geneturing-Disease dataset is a subset of the Geneturing dataset29, focusing on disease-related data. It contains 50 questions with standard answers, specifically designed to evaluate the ability of LLMs to identify genes associated with specific diseases. Model performance is assessed based on the proportion of correctly identified standard genes, providing a benchmark for evaluating capabilities in this critical genomics task.
Setting of Human Evaluation of Commonly Searched Consumer Medical Q&A
Evaluation dataset
In this study, we used HealthSearchQA4 for commonly searched consumer medical Q&A evaluation. HealthSearchQA consists of 3173 commonly searched consumer questions. This dataset contains only questions without providing answers or reference information, aiming to advance research in medical question-answering systems, particularly in open-domain and real-world applications. To evaluate LINS’s question-answering capabilities in clinical scenarios, we randomly selected 300 questions from the HealthSearchQA dataset.
Evaluation dimensions
We have established nine evaluation dimensions across three primary aspects (Supplementary Fig. 3), including utility (Helpfulness and Harmfulness), accuracy (Scientific Consensus, Medical Expertise, Content Relevance, and Content Completeness), and comprehensiveness (Answer Diversity, Empirical Support, and Bias). Each dimension corresponds to a question and two options, with -1 indicating a negative aspect and 1 indicating a positive aspect.
Evaluation methodology
We employed a randomized, blind-controlled experimental design for evaluation. The process involved a survey distributed to each evaluator, containing 300 questions. Each question corresponded to answers generated by LINS-o1-preview and o1-preview. For every answer, evaluators assessed nine dimensions, resulting in a total of 5400 individual evaluations per participant. To ensure impartiality, we removed citation formatting from LINS-o1-preview responses, making it impossible to distinguish between the two models based on answer formatting alone. The survey presented questions sequentially, with the two answers to each question randomly shuffled and displayed consecutively. This approach minimized user bias and preconceived impressions, ensuring fairness and accuracy in the evaluation process. The continuous arrangement of responses allowed evaluators to compare the answers directly without interruption, facilitating a clearer identification of each model’s strengths and weaknesses when addressing the same question. Randomizing the order of responses further mitigated potential biases stemming from consistent answer positioning and exposed subtle differences between the models. This rigorous methodology enhanced the precision and reliability of the evaluation, ensuring a more comprehensive and nuanced assessment of the models’ performance.
Settings of LINS Assists Physicians In Evidence-Based Medicine Practice
Workflow of LINS assisting in evidence-based medicine practice
LINS demonstrates robust capabilities in evidence collection and synthesis, enabling the generation of application-ready, evidence-traceable text to support clinicians in evidence-based medical practice. As illustrated in Fig. 3a, after a physician formulates a specific clinical question based on patient information, LINS leverages the PICO Agent (prompt details in Supplementary Fig. 18) to transform the query into a PICO question (Transform clinical questions into searchable and answerable questions based on the PICO principle). Alternatively, physicians may directly input a well-structured PICO question, in which case the PICO Agent is bypassed. Using the PICO question, LINS searches the HERD database for relevant, high-quality evidence and integrates the retrieved information with patient-specific data to generate traceable, evidence-based recommendations. These outputs serve to assist clinicians in making informed decisions.
Workflow of human evaluation
To comprehensively validate the efficacy of LINS in assisting physicians with evidence-based medical practice, we constructed the AEBMP dataset for systematic evaluation (Supplementary Fig. 14). The experiment was divided into two parts: first, we used the AEBMP_stage1 subset to evaluate whether LINS can assist resident physicians in evidence-based medical practice; second, we used AEBMP_stage2 to assess the specific quality of the final answers generated by LINS and LLMs in assisting resident physicians.
For AEBMP_stage1, we first used LINS-o1-preview and o1-preview to generate evidence-based recommendations for 1000 clinical questions in the dataset (prompt details in Supplementary Fig. 18). We then developed a web-based interface (Supplementary Fig. 6) and invited 100 resident physicians to conduct the experiment using the platform. To enhance the quality of the experiment, we set up three repetitions for each question, with each resident physician evaluating 30 random, non-repeating clinical questions and their corresponding 60 answers. LINS-o1-preview and o1-preview’s responses were paired and displayed for each question. To prevent format-based bias, all answers were anonymized and presented in a randomized order. To ensure the quality of the test, we also set a quality control question for each tester, and users who answered the quality control question incorrectly would have their test results reset. For each answer, the resident physicians were required to assess whether the response could assist in evidence-based medical practice, save time, reduce stress, and improve efficiency. If they selected “cannot assist in evidence-based medical practice”, testers were asked to choose one reason from five options (“lack of correctness, lack of evidence validity, harmfulness, bias, or other” with a space for further elaboration) for subsequent analysis.
For AEBMP_stage2, we first used LINS-o1-preview and o1-preview to generate evidence-based recommendations for 75 clinical cases and 100 clinical questions in the dataset (prompt details in Supplementary Fig. 18) and invited three resident physicians to choose either the LINS-o1-preview or o1-preview-generated evidence-based recommendations (presented in the same format, anonymized, and randomized in order) as assistance for each question and modify the answers to generate the final evidence-based answers. For 78 questions, resident physicians used both LINS-o1-preview and o1-preview recommendations to assist in generating the final answers. We then invited five attending physicians to evaluate the 156 answers for these 78 questions across multiple dimensions (Supplementary Fig. 7). The evaluation was conducted as a randomized, blind-controlled study. Expert evaluators received a questionnaire with paired responses from LINS-o1-preview and o1-preview for each question. To prevent format-based bias, all answers were anonymized and randomized. Each question’s paired answers were presented sequentially to minimize preconceived notions and ensure impartiality and accuracy in the evaluation.
Settings of LINS Helps Patients With Medical Order Explanation
Workflow of LINS helping with medical order explanation
As illustrated in Fig. 4a, LINS helps patients with medical order explanation through three key processes: medical terminology extraction, medical terminology explanation, and clinical question answering. When a patient or their family member (hereafter referred to as “the patient”) inputs their medical records (comprising Patient Information and Medical Order sections) into LINS, the system initiates the process by leveraging the Medical Terminology Extraction Agent (MTEA, prompt details provided in Supplementary Fig. 19) to extract medical terms from the Medical Order section. For each extracted medical term, LINS retrieves valid evidence from Bing online database. Bing was selected due to its alignment with patients’ typical search behaviors and its extensive database, which can address a wide array of patient queries effectively. LINS integrates the retrieved evidence with patient information to generate evidence-backed, traceable explanations for medical terms, helping patients comprehend complex medical concepts.
Additionally, patients can pose follow-up clinical questions to LINS (e.g., Will this disease be inherited by my child?). LINS utilizes the Retrieval Question Agent (QEA, prompt details provided in Supplementary Fig. 20) to convert these clinical questions into precise, retrieval-optimized queries (e.g., Will rheumatic heart disease be inherited by my child?). Subsequently, LINS retrieves evidence from the Bing database, synthesizes this evidence with patient information, and generates citation-based, informative responses. This approach not only addresses patients’ concerns but also enhances their trust and engagement in the treatment process.
Workflow of human evaluation
To comprehensively evaluate the effectiveness of LINS in assisting patients with understanding medical instructions, we developed the MOEQA dataset for validation. The experimental design involved two groups, o1-preview and LINS-o1-preview, and included assessments by five attending physicians and five lay users acting as patient proxies (Supplementary Fig. 9).
Initially, we utilized the medical terminology extraction agent to extract medical terms from the medical order sections of each medical record. Since both groups employed identical methods for this step, they produced the same results, yielding a total of 116 medical terms. Subsequently, both groups generated explanations for these 116 medical terms and answered 107 clinical questions from the MOEQA dataset. Importantly, both groups’ outputs were required to include traceable evidence, enabling users to verify the sources and fostering greater trust in the responses. To achieve this, we designed prompts specifically tailored to ensure evidence traceability for both groups (details in Supplementary Fig. 21).
The evaluation was conducted using a randomized, blinded, controlled design. Survey questionnaires were distributed to all evaluators, pairing explanations or answers from LINS-o1-preview and o1-preview for each term or question. To prevent format-based bias, responses were anonymized and randomized in order. For each term or question, the paired responses were presented sequentially but in a randomized order, minimizing evaluators’ preconceptions and ensuring fairness and accuracy in the assessments.
The five attending physicians evaluated the quality of the responses based on the accuracy of term explanations and the scientific consistency, comprehensiveness, potential harm, and bias in the clinical question answers. The five lay users acting as patients assessed whether the term extractions met expectations, the helpfulness of the term explanations and clinical answers, and the effectiveness of the evidence provided. Additionally, they evaluated whether the inclusion of evidence citations enhanced their credibility in the explanations or answers. Detailed evaluation dimensions are outlined in Supplementary Fig. 10.
Construction of Link-Eval
Evaluate dimensions
As shown in Fig. 5b, Link-Eval primarily evaluates the content based on two main components: citations and statements. For citations, it proposes three evaluation metrics: citation set precision, citation precision, and citation recall, to assess the accuracy and completeness of the citations. For statements, it introduces two evaluation metrics: statement correctness and statement fluency, to evaluate the correctness and fluency of the statements. In the citation-based generative text data format shown in Fig. 5a, there are \({{\rm{topk}}}({{\rm{topk}}}=5)\) Refs (retrieved passages). The generator uses these Refs to create statements and annotates each statement with a set of citations, which can contain 0 to topk citations. To calculate citation set precision, citation precision, and citation recall, the total number of citations, correct citations, valid Refs, correct and valid citations, and correct citation sets must first be determined.
Natural language inference system
A natural language inference system is essential to determine the entailment relationship between two natural language texts. In this study, we utilized GPT-4o as the inference system. Alternative models, such as the more resource-efficient GPT-4o-mini or the open-source Qwen2.5-72b, could also be employed depending on computational constraints. GPT-4o demonstrated strong performance on the MedNLI35 benchmark, surpassing human physicians in accuracy (see Fig. 5f, g). By using the instruction “nli premise: {premise} hypothesis: {hypothesis}”, GPT-4o can determine the entailment relationship between the premise and the hypothesis. In the process of determining the correct citation set in Link-Eval, S serves as the hypothesis, and the concatenated text of C serves as the premise, with the output labels {0, 1, 2} representing {entailment, contradiction, neutral} respectively.
Link-Eval scores calculation process
The methods for calculating correct citation sets and valid Refs are illustrated in Fig. 5c. We define a citation set C for a statement S as a correct citation set if the concatenated text of all citations in C contains the meaning of S. Therefore, the formula for calculating citation set precision is as follows:
where RC represents the total number of correct citation sets, and NC represents the total number of citation sets.
After obtaining the correct citation set, we need to further calculate the correct citations (Fig. 5d). Correct citations are those that play an irreplaceable role within the correct citation set. Suppose C is the correct citation set for statement S. We enumerate all citations c in C, and by removing c from C to get C', if the concatenated text corresponding to C' can’t entails S, then c is deemed irreplaceable in C, and thus considered a correct citation. If C is not the correct citation set for S, we directly determine that all citations in C are not correct citations.
Valid Ref refers to a Ref that is helpful in answering the question. To determine a valid Ref, we need a Ref evaluator to judge whether a retrieved Ref is indeed valid. Currently, there is no technology that can 100% accurately determine whether a retrieved Ref R is valid for a question Q. However, we have found a positive correlation between the ranking score of Q and R in the retriever and the validity of R. Through statistical analysis, we established a threshold value p = 0.60. When the ranking score > p, R has a 99.9% probability of being a valid Ref. The probability distribution graph is shown in Supplementary Fig. 22.
The total number of citations refers to the sum of citations across all citation sets in the response \(M={\sum }_{i=1}^{{{\rm{e}}}}|{C}_{i}|\). The total number of correct citations refers to the sum of correct citations across all correct citation sets in the response \(N={\sum }_{i=1}^{{{\rm{e}}}}|{C}_{i}^{*}|\). The number of valid Refs refers to the number of valid Refs among the topk Refs \(V=|{R}_{i}|\). The number of correct and valid citations refers to the number of valid Refs that are correctly cited \(W={{{|R}}_{i}\cap }(\cup _{i=1}^{{{\rm{e}}}}({C}_{i}^{*}))|\). Here, e denotes the number of citation sets in the response, \(|{C}_{i}|\) denotes the number of citations in the ith citation set, and \(|{C}_{i}^{*}|\) denotes the number of correct citations in the ith citation set. Therefore, citation precision and citation recall are calculated as follows:
Citation precision and citation recall are mainly used to evaluate the accuracy and comprehensiveness of citation generation in citation-based generative text. In addition, we define statement fluency and statement correctness to assess the fluency and correctness of the generated text. As natural language generation technology advances, traditional evaluation methods—such as calculating the text or semantic overlap of human-written reference summaries in the test set49 or BERTScore50)—are no longer sufficient to accurately distinguish the performance of different models14,33,51,52. Therefore, we adopt a more detailed evaluation metric, statement fluency, to measure the output quality of natural knowledge generation systems. This metric is directly related to the readability and naturalness of the generated text, focusing on the coherence of grammar, syntax, and overall expression. It ensures that the text reads as naturally and smoothly as human writing, rather than appearing stiff or unnatural as machine-generated text might. Highly fluent text is more easily accepted and understood by human readers, effectively enhancing the efficiency and effectiveness of information transmission. Specifically, we choose to use UNIEVAL33 as the tool for calculating statement fluency because it not only has a high correlation with human evaluation but also demonstrates strong zero-shot learning capabilities. UNIEVAL restructures the evaluation task as a Boolean QA problem, allowing a single model to assess the generated text from multiple dimensions, including but not limited to coherence, consistency, and relevance.
Statement correctness is used to evaluate the correctness of the model (Fig. 5f), particularly for questions with standard answers (such as multiple-choice questions). The presence of a standard answer in the response does not necessarily mean the answer is entirely correct; there may be contradictions within the answer. This means that the set of statements {S} may contain contradictory viewpoints. Therefore, for any pair of statements Si and Sj (i ≠ j) in {S}, we use an NLI model to determine if a conflict relationship exists. If a conflict is detected, then statement correctness = 0. When there is no conflict between any two statements, we check if {S} satisfies the correct answer. If it does, statement correctness = 1; otherwise, it is 0.
Statistics and reproducibility
This study presents a general retrieval-augmented framework, LINS, designed to effectively enhance the quality and credibility of medical responses generated by large language models. No statistical methods were used to predetermine sample sizes, and no data were excluded from the analysis. In the human evaluation experiments, randomization was applied, and the researchers were blinded to group allocation and outcome assessment to reduce potential bias. Statistical analyses included human raters’ quality assessments of the outputs, followed by two-sided paired significance tests. Reproducibility was confirmed through independent validation by licensed medical professionals in China. All code and data supporting the findings are publicly available in our GitHub repository: https://github.com/WangSheng21s/LINS [https://github.com/WangSheng21s/LINS].
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data used in LINS are publicly accessible. The MedMCQA dataset is available at https://medmcqa.github.io/ [https://medmcqa.github.io/] and the PubMedQA dataset can be obtained from https://pubmedqa.github.io [https://pubmedqa.github.io]. The MedQA dataset is available at https://drive.google.com/file/d/1ImYUSLk9JbgHXOemfvyiDiirluZHPeQw/view [https://drive.google.com/file/d/1ImYUSLk9JbgHXOemfvyiDiirluZHPeQw/view] and the HealthSearchQA data can be accessed from https://www.nature.com/articles/s41586-023-06291-2 #Sec59 [https://www.nature.com/articles/s41586-023-06291-2#Sec59]. Additionally, datasets such as PubMedQA*, GeneMedQA and Multi-MedCQA datasets is available at http://lins.drai.cn/ [http://lins.drai.cn/] with a testing interface. Source data are provided with this paper.
Code availability
LINS supports users in flexibly selecting modules such as generators, retrievers, databases, and more, allowing them to build an appropriate local database based on their specific needs. The Qwen models can be found at https://huggingface.co/Qwen [https://huggingface.co/Qwen], the llama models can be found at https://huggingface.co/meta-llama [https://huggingface.co/meta-llama], and the GPT models are available at https://openai.com/api/ [https://openai.com/api/]. The Gemini models can be accessed at https://ai.google.dev/gemini-api [https://ai.google.dev/gemini-api]. The recall model for the retriever is available at https://huggingface.co/BAAI/bge-m3 [https://huggingface.co/BAAI/bge-m3], and the ranking model is at https://huggingface.co/BAAI/bge-reranker-v2-m3 [https://huggingface.co/BAAI/bge-reranker-v2-m3]. Source data are provided with this paper.
To facilitate user usage, we have provided detailed, end-to-end, easy-to-use code and tutorials at https://github.com/WangSheng21s/LINS. https://doi.org/10.5281/zenodo.16215408.
References
Lu, M. Y. et al. A visual-language foundation model for computational pathology. Nat. Med. 30, 863–874 (2024).
Pais, C. et al. Large language models for preventing medication direction errors in online pharmacies. Nature Medicine. 1-9 (2024).
Saab, K. et al. Capabilities of Gemini models in medicine. arXiv preprint arXiv:2404.18416 (2024).
Singhal, K. et al. Large language models encode clinical knowledge. Nature 620, 172–180 (2023).
Van Veen, D. et al. Adapted large language models can outperform medical experts in clinical text summarization. Nature Medicine. 1-9 (2024).
Giuffrè, M., You, K. & Shung, D. L. Evaluating ChatGPT in medical contexts: the imperative to guard against hallucinations and partial accuracies. Clin. Gastroenterol. Hepatol. 22, 1145–1146 (2024).
Farquhar, S., Kossen, J., Kuhn, L. & Gal, Y. Detecting hallucinations in large language models using semantic entropy. Nature 630, 625–630 (2024).
Ullah, E., Parwani, A., Baig, M. M. & Singh, R. Challenges and barriers of using large language models (LLM) such as ChatGPT for diagnostic medicine with a focus on digital pathology–a recent scoping review. Diagnostic Pathol. 19, 43 (2024).
Mousavi, S. M., Alghisi, S. & Riccardi, G. in Findings of the Association for Computational Linguistics: EMNLP. 8014-8029 (2024).
Sackett, D. L. in Seminars in Perinatology. 3-5 (Elsevier).
Gravel, J., D’Amours-Gravel, M. & Osmanlliu, E. Learning to fake it: limited responses and fabricated references provided by ChatGPT for medical questions. Mayo Clin. Proc.: Digital Health 1, 226–234 (2023).
Tang, X. et al. MedAgents: Large Language Models as Collaborators for Zero-shot Medical Reasoning. Workshop on Large Language Model (LLM) Agents (ICLR, 2024).
Kim, Y. et al. In The Thirty-eighth Annual Conference on Neural Information Processing Systems.
Fabbri, A. R. et al. Summeval: Re-evaluating summarization evaluation. Trans. Assoc. Computational Linguist. 9, 391–409 (2021).
Xiong, G., Jin, Q., Lu, Z., & Zhang, A. Benchmarking retrieval-augmented generation for medicine. In Findings of the Association for Computational Linguistics ACL 2024. 6233–6251 (2024).
Zakka, C. et al. Almanac—retrieval-augmented language models for clinical medicine. NEJM AI 1, AIoa2300068 (2024).
Lozano, A., Fleming, S. L., Chiang, C.-C. & Shah, N. in Pacific symposium on biocomputing. 8-23 (World Scientific) (2024).
Lewis, P. et al. Retrieval-augmented generation for knowledge-intensive nlp tasks. Adv. Neural Inf. Process. Syst. 33, 9459–9474 (2020).
Gautam, D. & Kellmeyer, P. Exploring the Credibility of Large Language Models for Mental Health Support: Protocol for a Scoping Review. JMIR Res. Protoc. 14, e62865 (2025).
Quttainah, M., Mishra, V., Madakam, S., Lurie, Y. & Mark, S. Cost, Usability, Credibility, Fairness, Accountability, Transparency, and Explainability Framework for Safe and Effective Large Language Models in Medical Education: Narrative Review and Qualitative Study. JMIR AI 3, e51834 (2024).
Liu, J. & He, J. The Decoy Dilemma in Online Medical Information Evaluation: A Comparative Study of Credibility Assessments by LLM and Human Judges. arXiv preprint arXiv:2411.15396 (2024).
Achiam, J. et al. Gpt-4 technical report. arXiv preprint arXiv:2303.08774 (2023).
Touvron, H. et al. Llama: Open and efficient foundation language models. arXiv preprint arXiv:2302.13971 (2023).
Bai, J. et al. (2023). Qwen technical report. arXiv preprint arXiv:2309.16609.
Team, G. et al. Gemini: a family of highly capable multimodal models. arXiv preprint arXiv:2312.11805 (2023).
Pal, A., Umapathi, L. K. & Sankarasubbu, M. in Conference on health, inference, and learning. 248-260 (PMLR).
Jin, Q., Dhingra, B., Liu, Z., Cohen,W.W. & Lu, X. PubMedQA: A Dataset for Biomedical Research Question Answering. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP). (Association for Computational Linguistics, 2019).
Jin, D. et al. What disease does this patient have? a large-scale open domain question answering dataset from medical exams. Appl. Sci. 11, 6421 (2021).
Hou, W. & Ji, Z. GeneTuring tests GPT models in genomics. BioRxiv (2023).
Sood, A. & Ghosh, A. Literature search using PubMed: an essential tool for practicing evidence-based medicine. J.-Assoc. Physicians India 54, 303 (2006).
Pagano, G. et al. Prasinezumab slows motor progression in rapidly progressing early-stage Parkinson’s disease. Nature medicine, 1-8 (2024).
Gao, T., Yen, H., Yu, J. & Chen, D. Enabling Large Language Models to Generate Text with Citations. Proceedings of the 2023 Conference on Empirical Methods in Natural Language Processing. (Association for Computational Linguistics, 2023).
Zhong M. et al. Towards a Unified Multi-Dimensional Evaluator for Text Generation. In Proceedings of the 2022 Conference on Empirical Methods in Natural Language Processing. 2023–2038 (2022).
Raffel, C. et al. Exploring the limits of transfer learning with a unified text-to-text transformer. J. Mach. Learn. Res. 21, 1–67 (2020).
Shivade, C. In Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing, Brussels, Belgium. Association for Computational Linguistics. 1586-1596.
Cramér, H. Mathematical methods of statistics. Vol. 26 (Princeton University Press, 1999).
Salvagno, M., Taccone, F. S. & Gerli, A. G.Artificial intelligence hallucinations. Crit. Care 27, 180 (2023).
Roustan, D. & Bastardot, F. The Clinicians’ Guide to Large Language Models: A General Perspective With a Focus on Hallucinations. Interact. J. Med. Res. 14, e59823 (2025).
Dorsey, E. R., Venuto, C., Venkataraman, V., Harris, D. A. & Kieburtz, K. Novel methods and technologies for 21st-century clinical trials: a review. JAMA Neurol. 72, 582–588 (2015).
Zhu, C. et al. Is your LLM outdated? Evaluating LMS at temporal generalization. arXiv preprint arXiv:2405.08460 (2024).
Murdoch, B. Privacy and artificial intelligence: challenges for protecting health information in a new era. BMC Med. Ethics 22, 1–5 (2021).
Price, W. N. & Cohen, I. G. Privacy in the age of medical big data. Nat. Med. 25, 37–43 (2019).
Williamson, S. M. & Prybutok, V. Balancing Privacy and Progress: A Review of Privacy Challenges, Systemic Oversight, and Patient Perceptions in AI-Driven Healthcare. Appl. Sci. 14, 675 (2024).
Wei, J. et al. Chain-of-thought prompting elicits reasoning in large language models. Adv. neural Inf. Process. Syst. 35, 24824–24837 (2022).
Antaki, F. et al. Capabilities of GPT-4 in ophthalmology: an analysis of model entropy and progress towards human-level medical question answering. Br J Ophthalmol. 108, 1371–1378 (2024).
Chakravarty, D. et al. OncoKB: a precision oncology knowledge base. JCO Precis. Oncol. 1, 1–16 (2017).
Amberger, J. S., Bocchini, C. A., Schiettecatte, F., Scott, A. F. & Hamosh, A. O. M. I. M. org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic acids Res. 43, D789–D798 (2015).
Liu, L., Chen, J., Brocanelli, M. & Shi, W. In Proceedings of the 4th ACM/IEEE Symposium on Edge Computing. 59–73.
Lin, C.-Y. In Text summarization branches out. 74–81.
Zhang, T., Kishore, V., Wu, F., Weinberger, K. Q. & Artzi, Y. Bertscore: Evaluating text generation with bert. arXiv preprint arXiv:1904.09675 (2019).
Chen, Y., Wang, R., Jiang, H., Shi, S. & Xu, R. Exploring the Use of Large Language Models for Reference-Free Text Quality Evaluation: An Empirical Study. IJCNLP (Findings) https://doi.org/10.18653/v1/2023.findings-ijcnlp.32 (2023).
Shen, L., Liu, L., Jiang, H. & Shi, S. On the Evaluation Metrics for Paraphrase Generation. In Proceedings of the 2022 Conference on Empirical Methods in Natural Language Processing. 3178–3190 (2022).
Acknowledgements
This work was supported by the National Key R&D Program of China [2022YFF1203303 D.B.]; National Natural Science Foundation of China [92474204 Y.Z., 32341019 Y.Z., 32070670 Y.W.]; Ningbo Top Medical and Health Research Program [2023030615 Y.Z., 2024020919 Y.W.]; Beijing Natural Science Foundation [L222007 D.B.]; Ningbo Science and Technology Innovation Yongjiang 2035 Project [2023Z226 Y.Z., 2024Z229 Y.Z.]; Major Project of Guangzhou National Laboratory [GZNL2023A03001 Y.Z.]; State Key Laboratory of Systems Medicine for Cancer [KF2422-93 Y.Z.]. The funding bodies had no role in the design of the study, data collection, analysis, interpretation, or in writing the manuscript.
Author information
Authors and Affiliations
Contributions
W. (Sheng Wang), F.Z., D.B., and Y.Z. designed the study. S.W. (Sheng Wang) conducted data collection, data analysis, algorithm development, experimental design, experimental validation, visualization, and manuscript writing. F.Z. provided expertise, assisted with experimental design, visualization, and manuscript writing. M.G., X.Z., and X.W. participated in discussions and helped with experimental design. Y.L. and H.L. participated in discussions and assisted with data analysis. Z.Y. (Zhiyuan Yuan) provided suggestions, contributed to manuscript writing and visualization. B.W., W.H., and T.W. participated in data collection, experimental design, and experimental validation. Z.Y. (Zhaohui Yang) and J.S. contributed to data analysis and experimental validation. S.W. (Shu Wang), Y.W., and L.Z. assisted with manuscript writing. M.X., J.L. (Jingjia Liu), J.L. (Jianjun Luo), and J.Z. participated in discussions and helped with manuscript writing. W.Z., H.Z., K.Z. and R.C. provided knowledge support, interpreted the findings, helped with manuscript writing, and supervised the study. D.B. was involved in data collection, data analysis, experimental design, and experimental validation. Y.Z. provided expertise, interpreted the study results, designed experiments and algorithms, assisted with manuscript writing, and supervised the study. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, S., Zhao, F., Bu, D. et al. LINS: A general medical Q&A framework for enhancing the quality and credibility of LLM-generated responses. Nat Commun 16, 9076 (2025). https://doi.org/10.1038/s41467-025-64142-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-64142-2