Abstract
Acetyl-CoA synthetases (Acs) have emerged as drug targets for the treatment of cancer, metabolic diseases as well as fungal and parasitic infections. Although a variety of small molecule Acs inhibitors have been discovered, the systematic optimization of these molecules has been slowed by a lack of structural information regarding their mechanism of inhibition. Through a chemical genetic-based, synthetic lethal screen of the human fungal pathogen Cryptococcus neoformans, we identified an isoxazole-based Acs inhibitor with antifungal activity and high selectivity for the C. neoformans Acs1 relative to human ACSS2 as well as to other fungal Acs enzymes. Xray crystallography of the isoxazole-CnAcs1 complex revealed that the isoxazole occupies both the acetyl- and CoA-binding sites of CnAcs1. Biochemically, the isoxazoles display uncompetitive inhibition kinetics that are similar to antimalarial Acs inhibitors also proposed to target the CoA binding site. Consequently, these data provide structural and mechanistic insights into the remarkable selectivity of CoA pocket-targeting Acs inhibitors. As such, targeting fungal and parasitic Acs enzymes for the development of novel anti-infectives can be achieved with high selectivity and, thereby, low host toxicity.
Similar content being viewed by others
Introduction
Each year, fungal infections affect millions of people across the globe and cause diseases ranging from superficial skin dermatoses to life-threatening diseases of the bloodstream and deep organs such as the liver, kidney, and brain1. The majority of people at-risk for invasive fungal infections have altered immune function due to other diseases such as HIV/AIDs or because they are receiving immunosuppressive drugs used to treat cancer, inflammatory diseases, or manage organ transplantation2. People with fully functional immune systems are also at risk for life-altering fungal infections such as fungal keratitis, a common cause of infection-related vision loss, or chronic aspergillosis-associated asthma1,2. Human fungal infections are caused by an evolutionarily divergent set of pathogens with a broad range of pathobiological characteristics and features. Together, these characteristics of human fungal diseases make them challenging to treat.
Currently, only three classes of antifungal drugs are available as primary therapies for life-threatening fungal infections3: (1) polyenes such as amphotericin B; (2) azoles, including fluconazole and voriconazole; and (3) echinocandin 1,3-β-glucan synthase inhibitors exemplified by micafungin. Other drugs, such as 5-flucytosine or the novel triterpenoid 1,3-β-glucan synthase inhibitor ibrexafungerp, are used as adjuvant agents (5-flucytosine, ref. 4) or are currently limited to the treatment of mucosal infections (ibrexafungerp, ref. 5). The set of mechanistically distinct classes of antifungal drugs is quite limited compared to that available for treating bacterial infections. For example, there are more distinct classes of antibiotics with clinically useful activity against methicillin-resistant Staphylococcus aureus (MRSA) than the total number of antifungal drug classes. As resistance to frontline antifungal drugs continues to rise in species such as Candida auris and Aspergillus fumigatus6, the urgency of developing chemically and mechanistically novel antifungal drugs increases.
The unmet clinical need for new antifungal drugs has not gone unnoticed, and, encouragingly, two mechanistically and structurally novel candidates are currently in Phase II/III clinical trials. Specifically, fosmanogepix is a broad-spectrum agent that targets GPI anchor biosynthesis, critical for fungal cell wall biosynthesis7, while olorofim has activity against difficult-to-treat molds and selectively inhibits fungal dihydroorotate dehydrogenase8. However, even if these agents advance to clinical use, the threat of resistance remains acute, with only four to five drugs available to treat the broad variety of human fungal pathogens. Indeed, agricultural fungicides based on dihydroorotate dehydrogenase inhibition are already proceeding to use and could generate olorofim-resistant A. fumigatus before olorofim finishes Phase III clinical trials9. Therefore, the discovery and development of new antifungal drug candidates remains an important endeavor for medical mycology.
Acetyl-CoA synthetase (Acs) is an Acyl-CoA/Non-ribosomal peptide synthetase/Luciferase (ANL)-family, adenylating enzyme10 that converts acetate to the key metabolic molecule, acetyl-CoA (AcCoA). In mammalian cells, the majority of AcCoA is generated from glucose via citrate from the tricarboxylic acid (TCA) cycle11. TCA-generated citrate is exported from the mitochondria and converted to AcCoA by ATP-citrate lyase (Acl) in the cytosol and the nucleus12. Overall, acetate-derived AcCoA represents ~10% of the total AcCoA pool in human cells under a normal state of homeostasis, while AcCoA derived from glucose/Acl makes up the majority of the cellular pool11. In contrast, the metabolic origin of AcCoA is reversed in multiple cancer types, and acetate-derived AcCoA makes up the majority of the pool13,14. As such, the primary human Acs, ACSS2, has emerged as a cancer chemotherapy target, and one ACSS2 inhibitor has progressed to early-stage clinical trials14. Importantly, ACSS2 is not essential in mammals13 because Acl maintains the AcCoA pool under normal homeostasis; therefore, ACCS2 inhibitors have a reduced likelihood of causing toxicity in humans.
Acs has also generated interest as a target for the development of anti-infective agents, including antifungal15,16 and anti-malarial agents17,18,19. Previously, we found that the pyrazole AR-12 inhibits fungal Acs15, has broad-spectrum antifungal activity, and is efficacious in combination with fluconazole in a mouse model of disseminated cryptococcosis16. These data provided validation of ACS as a potential antifungal drug target. Unfortunately, the pharmacology of AR-12 is not suitable for further pre-clinical development. Therefore, new chemical classes of antifungal Acs inhibitors are needed to further explore this promising target.
The rationale for Acs as an antifungal drug target is based on the following considerations. First, medically important yeasts such as Candida albicans and non-albicans Candida spp. lack Acl enzymes and, therefore, are dependent on Acs for the generation of critical pools of AcCoA20. Multiple genetic analyses indicate that Acs enzymes are essential in C. albicans and C. glabrata21,22, and the lack of Acl in other Candida spp. such as C. auris strongly support the conclusion that ACS are required for viability in those species as well.
Second, genetic studies in Cryptococcus, the species of yeast that is the second most common cause of human disease after Candida species, support Acs inhibition as a therapeutic strategy. Hu et al. have shown that deletion of CnACS1, the only Acs expressed in C. neoformans23, reduces virulence in a mouse model of cryptococcosis24. This is a particularly important observation because C. neoformans expresses Acl. Both ACL1 and ACS1 are, therefore, necessary to generate sufficient AcCoA to maintain fitness during mammalian infection. Our group has also found that loss of either ACS1 or ACL1 significantly reduces the ability of C. neoformans to replicate in macrophages, a key niche during infection23.
Third, we have shown that the acs1∆ mutant is hypersensitive to fluconazole in vitro and that deletion of ACS1 increases the efficacy of fluconazole during mammalian infection23. Cryptococcosis is treated with combinations of amphotericin B, fluconazole, and/or 5-flucytosine4; therefore, Acs1 inhibitors could also be used in combination with fluconazole to both reduce the emergence of resistance and improve efficacy. Taken together, these previously reported studies and data support the development of Acs as an antifungal drug target.
In C. neoformans, the acl1∆ mutant has reduced growth on glucose-containing, nutrient-rich medium, while deletion of ACS1 causes reduced growth in nutrient-poor medium containing non-glucose carbon sources24. Consequently, ACS1 expression is increased in tissue culture-based in vitro medium designed to mimic the relatively nutrient-poor infection environment23, suggesting that the reduced fitness of the acs1∆ in vivo is due to its increased importance to AcCoA production under those nutrient conditions. Consistent with the in vitro observations, ACS1 expression is increased in vivo as well23. We have been unable to generate an acs1∆ acl1∆ double mutant under any in vitro conditions, indicating that ACS1 and ACL1 both make important contributions to AcCoA homeostasis23.
We took advantage of the synthetic lethal relationship between C. neoformans ACS1 and ACL1 to bias a cell-based, phenotypic screen toward potential CnAcs1 inhibitors by screening an acl1∆ mutant against a library of small molecules (Fig. 1A). Because of the highly conserved nature of most fungal ACS enzymes with respect to the residues within the substrate binding sites and the correspondingly similar overall structures of previously reported Acs-inhibitor complexes, we expected that Acs inhibitors identified from this cell-based screen in C. neoformans would also inhibit other fungal Acs enzymes. To our surprise, we identified a structurally novel isoxazole (1) Acs inhibitor that is highly specific for CnAcs1 (Fig. 1B, C). X-ray crystallography and biochemical analysis indicate that 1 is an uncompetitive inhibitor that interacts primarily with the CoA substrate binding pocket. Interestingly, additional biochemical and mutational studies suggest that a recently discovered anti-malarial Acs inhibitor (MMV084978, ref. 17) has a similar mechanism of inhibition. Both 1 and MMV084978 are selective for fungal/Plasmodium falciparum Acs relative to the human enzyme ACSS2. Accordingly, these studies not only further validate Acs as an antifungal drug target in C. neoformans but also highlight a surprisingly high level of target specificity that Acs inhibitors show despite the apparent structural and sequence conservation among Acs enzymes.
A The number of compounds evaluated in the primary screen, hit validation, and counter screening steps of the screening campaign are shown. B The structure of the hit isoxazole 1 is shown with the functionally distinct regions of the molecule highlighted. C The IC50 of isoxazole 1 towards CnAcs1 is indicated; the curve is representative of two independent experiments. The mean IC50 calculated from the two independent replicates performed in technical triplicate is shown with the standard deviation. R2 value for goodness-of-fit for the two experiments used to compute the IC50 were 0.996 and 0.995. Source data are provided as a Source data file.
Results
Chemical-genetic, synthetic lethal screen in C. neoformans identifies a CnAcs1 inhibitor
As introduced above, we took advantage of the synthetic lethality of the acs1∆ acl1∆ mutant in C. neoformans23 to design a whole-cell, high-throughput screen to identify potential inhibitors of CnAcs1 (Fig. 1A). Specifically, we employed a two-stage, screen/counter-screen strategy. First, the acl1∆ mutant was screened against a compound library for molecules that inhibit growth in standard yeast peptone-2% dextrose (YPD) medium. The synthetic lethal relationship between ACS1 and ACL1 was expected to sensitize the acl1∆ mutant to CnAcs1 inhibitors. Second, we counter-screened hits against WT cells in the same medium. The acs1∆ mutant has no growth phenotype in YPD and, therefore, on-target CnAcs1 inhibitors should have reduced antifungal activity against strains expressing both ACS1 and ACL1. Third, we determined the antifungal activity of hits that advanced through the first two steps against WT strains in medium containing acetate as the sole carbon source. Under these conditions, CnAcs1 is essential for growth23,24. We expected that this screening funnel would provide a phenotypic, cell-based assay favoring on-target Acs1 inhibitors; we also hypothesized that the highly conserved nature of fungal Acs25 enzymes would allow our strategy to identify broad-spectrum ACS inhibitors.
We optimized growth conditions for the acl1∆ mutant in 384-well format at 30 °C in YPD for 48 h using cell density (OD600) as the readout for fungal growth. Fluconazole was used as a positive control to validate the robustness of the assay for the detection of antifungal activity. Full plate assays with alternating rows of DMSO (1%) and fluconazole (64 µg/mL) generated a Z’ score of 0.65, which is compatible with high-throughput screening (Fig. S1). A library of 55,264 drug-like molecules (DIVERSet) purchased from ChemBridge was screened and yielded 175 primary hit molecules (0.3% hit rate) that reduced the growth of the acl1∆ mutant (Z ≥ 3) relative to the mean of each plate (Fig. 1A). This set of hits was re-tested under the screening conditions to identify molecules that inhibited growth by 50% in the primary screen and in the validation screen (Fig. 1A).
Independent samples of the 40 molecules that met these criteria were re-purchased and counter-screened against WT (strain H99) and the acl1∆ mutant under screening conditions. Five molecules showed decreased antifungal activity against the WT strain compared to the acl1∆ mutant. Finally, the antifungal activity of these five molecules against the WT strain was determined in minimal medium with 2% acetate as the carbon source. Of these five, isoxazole 1 was dramatically more active in acetate medium with a minimum inhibitory concentration (MIC) of 2 µg/mL compared to >64 µg/mL in rich, YPD medium (Fig. 2A). To further confirm the structure of isoxazole 1 and the reproducibility of its activity, it was re-synthesized using the route described in Fig. S2 and its activity confirmed using growth assays with WT, the acl1∆ mutant, and acetate-containing media. Finally, isoxazole 1 inhibited CnAcs1 enzyme with an IC50 of 8.6 ± 4.4 µM using our previously reported assay of CnAcs1 enzyme activity (Fig. 1C, ref. 25). These data strongly support the conclusion that isoxazole 1 inhibits CnAcs1.
A The minimum inhibitory concentration (MIC) of 1 against C. neoformans reference strain H99 and acl1∆ mutant strains in YPD, YNB + 2%acetate, and RPMI-MOPS buffer at 37 °C. The values were identical for three independent experiments performed in technical duplicate. B MIC values against C. albicans reference strain SC5314 and C. glabrata CBS138. Fractional inhibitor concentrations against H99 for fluconazole (C) and rapamycin (D). Growth in wells is indicated by tan fill, while empty wells indicate no growth. The wells with red fill indicate the fractional inhibitory concentration (FIC). E HepG2 cells were exposed to the indicated concentrations of isoxazole 1 for 24 h. The release of LDH into the medium was determined as described in “Methods” and normalized to detergent-induced lysis. Data are means of two independent experiments performed in technical triplicate with error bars indicating standard deviation. F Structures of isoxazole 1 and 2. G In vitro stability of isoxazole 1 in human and mouse liver microsomes. The times indicate t1/2 in minutes. H In vitro stability of isoxazole 2 in the presence and absence of pan-cytochrome inhibitor 1-amino-benzotriazole (ABT). Source data are provided as a Source data file.
Isoxazoles 1 and 2 have anticryptococcal activity in host-like media and are synergistic with fluconazole in vitro
CnAcs1 is required for in vitro growth in nutrient-poor conditions and has reduced fitness in animal models of infection24. We, therefore, reasoned that under host-like, in vitro conditions, isoxazole 1 may show antifungal activity toward C. neoformans. The standard clinical microbiology (Clinical and Laboratory Standards Institute (CLSI)) antifungal susceptibility testing conditions (RPMI medium buffered with 0.165 M MOPS, ref. 26) are a tissue culture medium with much lower concentrations of glucose and gluconeogenic amino acids compared to nutrient-rich YPD medium routinely used in the laboratory and in our primary and secondary screening assays27. Under these more host-like conditions, 1 inhibited the growth of H99 with a MIC of 32 µg/mL (Fig. 2A). The increased activity of 1 in RPMI relative to rich YPD medium is likely due to this difference in nutrient availability. Supporting that hypothesis, we previously showed that host-like in vitro conditions induce CnAcs1 expression23, suggesting that the Acs1-derived pool of AcCoA may become more important under these conditions. Furthermore, the acs1∆ mutant has reduced competitive fitness relative to H99 in RPMI medium (Fig. S3).
Surprisingly, 1 has no antifungal activity against C. albicans or C. glabrata, two Candida spp. which lack Acl enzymes and, therefore, are susceptible to other Acs inhibitors (Fig. 2B, refs. 15, 16). The primary sequences of Acs enzymes are highly conserved across multiple pathogenic fungi, including C. neoformans, C. albicans, and C. glabrata (Fig. S4). Additionally, crystallography of the C. neoformans and C. albicans proteins also shows that the secondary and tertiary protein structures of the enzymes are very similar to one another as well as to Acs enzymes from other species25. As the data presented below indicate, isoxazole 1 inhibits CnAcs1 activity but, unexpectedly, has little to no activity against other fungal Acs or the human enzyme ACSS2.
Fluconazole combined with 5-flucytosine is emerging as an important approach to the treatment of cryptococcosis because it avoids the toxicities associated with amphotericin B28. However, 5-flucytosine is not readily available in resource-limited regions with high rates of cryptococcosis and is also toxic to bone marrow. Therefore, the identification of new drugs that improve the activity of fluconazole is of interest4,28. We previously reported23 that the acs1∆ mutant shows increased susceptibility to fluconazole in vitro as well as in a mouse model of cryptococcosis. Therefore, if the antifungal activity of isoxazole 1 was due to inhibition of CnAcs1, we would expect it to show synergy with fluconazole. Indeed, isoxazole 1 is synergistic with fluconazole in vitro, with a Fractional Inhibitory Concentration Index (FICI) of 0.5 (Fig. 2C). In contrast, 1 has no interaction with either amphotericin B (FICI = 2) or 5-flucytosine (FICI = 2), the other two antifungal drugs used to treat cryptococcal meningitis28. In mammalian systems, Acs1 activity is regulated by the Target-of-Rapamycin pathway29. Consistent with these previous findings, 1 is also synergistic with rapamycin against C. neoformans (FICI = 0.5, Fig. 2D).
Based on our previously reported genetic data indicating that genetic disruption of Acs1 increases fluconazole efficacy and synergy of isoxazole 1 with fluconazole23, we further investigated the in vitro toxicity and in vitro pharmacodynamic properties of 1 as a prelude to potential efficacy studies in a mouse model of infection. As shown in Fig. 2E, 1 shows very little toxicity up to the limit of its solubility against Hep2G cells using an LDH assay. Inspection of 1 reveals a potential metabolic liability in the form of an oxidation-prone, electron-rich benzofuran ring. Therefore, we synthesized isoxazole 2, in which the benzofuran was removed and a fluorine-substituent was placed at the para-position of the aryl ring; isoxazole 2 maintains an inductively electronegative substituent (F) at the para-position of the aryl ring (Fig. 2F). The antifungal activity of 2 (MIC 32 µg/mL, RPMI) was similar to isoxazole 1.
Consistent with our concerns, isoxazole 1 was rapidly metabolized by murine microsomes (t1/2 7 min) (Fig. 2G). Unfortunately, the t1/2 of the isoxazole 2 was only slightly longer (t1/2 12 min, Fig. 2H). This metabolism was blocked by the addition of the pan-cytochrome P450 inhibitor 1-amino-benzotriazole (t1/2 > 120 min, ref. 30), confirming the susceptibility of 2 to cytochrome-mediated metabolism (Fig. 2H). Both 1 and 2 showed longer t1/2 in human microsomes (t1/2 83 min and 84 min, respectively) (Fig. 2H). Mass spectrometry of the metabolites of 1 also confirmed that the benzofuran ring was the primary site of oxidation. As predicted, replacement of the benzofuran with a fluoro-substituent reduced oxidation of the aryl ring, but 2 underwent oxidative de-methylation of the tertiary amide moiety (Table S1). Consequently, additional medicinal chemistry-based optimization of the structure will be needed to improve its pharmacodynamic and pharmacokinetic properties before efficacy studies of this scaffold in mouse models of infection can be undertaken.
Mutations in the SAGA (Spt-Ada-Gcn5-Acetyltransferase) complex component SGF29 cause resistance to isoxazole 1
Our chemical-genetic observations strongly support the conclusion that isoxazoles 1 and 2 inhibit CnAcs1 as the primary mechanism of their anticryptococcal activity. To identify other potential targets or pathways that may mediate resistance, we generated isoxazole 1-resistant mutants using an in vitro microevolution approach (Fig. 3A). Three parental strains were used. In addition to the H99 reference strain, we used the acl1∆ mutant because it is hyper-susceptible to isoxazole 1. The MIC of isoxazole 1 against H99 (32 µg/mL) is near the limit of solubility (64 µg/mL) and might limit our ability to apply sufficient selective pressure. The MIC of isoxazole 1 toward the acl1∆ mutant is 8-fold lower (4 µg/mL, Fig. 2A) and, therefore, this strain extends the range of concentrations under which selection would occur. Finally, we also used a double mutant lacking both ACL1 and KBC1; Kbc1 is an aceto-acetyl-CoA synthetase that also contributes to the AcCoA pool in C. neoformans23.
A Schematic of resistant mutant identification through iterative passaging into higher concentrations of isoxazole 1 using three parental strains: H99, the acl1∆ mutant, and the kbc1∆ mutant. B Diagram showing domains of the Sgf29 protein and the truncation mutant isolated from the passaging experiment. The green color indicates the Tudor domain of SGF29. The cross-hatches denote the regions of the proteins that are not translated in the truncation mutants. C Competitive growth assay between mNEON-tagged H99 and the sgf29∆ mutant at the indicated concentrations of isoxazole 1. The ratio of the two strains was determined by flow cytometry with data represented as mean and standard deviation of three independent replicates. The difference between the sgf29∆ mutant and H99 was statistically significant at 16 µg/mL (p = 0.0465 two-sided, unpaired Student’s t-test). Source data are provided as a Source data file.
The stains were incubated in microtiter plates containing RPMI/MOPS medium and isoxazole 1 at an initial concentration of ½ MIC for each strain. After 72 h, surviving cells were diluted 1:1000 and transferred to plates with 2-fold higher concentrations of 1 compared to the previous incubation. This transfer was repeated to a maximum concentration of 64 µg/mL, which is the limit of solubility for 1 in these conditions. Approximately 90% of lineages went extinct before the 64 µg/mL concentration was reached. Randomly selected surviving clones from each of the three lineages were passaged on YPD without isoxazole 1 and re-tested at 64 µg/mL of 1 to confirm stable resistance. Four isolates encompassing the three parental lineages were initially screened by Sanger sequencing of the ACS1 gene; however, no ACS1 mutations were identified in these isolates.
To identify mutations that may contribute to the resistance phenotype, we performed whole-genome sequencing of the parental and isoxazole 1-resistant isolates as described in “Methods.” The sequences were mapped to the H99 reference strain, and SNPs in each parental strain relative to the reference were initially identified. Consistent with the Sanger sequencing results, no SNPs were identified in ACS1. One strain derived from H99 and one derived from the acl1∆ kbc1∆ mutant contained insertion mutations leading to frameshift truncations (Fig. 3B) of the highly conserved SAGA complex component SGF29 (CNAG_06392). Based on extensive studies in the model yeast S. cerevisiae, Sgf29 recognizes methylated histones via its Tudor domains and recruits the SAGA complex, where it mediates acetylation of the histones and transcriptional activation31, and this function has been confirmed in C. neoformans32. Additionally, loss of Sgf29 does not affect the overall structure of the complex or its other functions. In the other two lineages, missense mutations within SGF29 were also present. No other mutations were identified in coding regions that were shared by all four isolates. Five additional proteins had non-synonymous SNPs, but none were clear loss-of-function mutations; although these may contribute to resistance, we focused additional experiments on SGF29 because the truncation mutations were present in multiple isolates (Table S2).
If the structural loss of function (truncation) mutations in SGF29 were responsible for the isoxazole 1-resistant phenotype of these strains, then a strain with a deletion mutation of SGF29 would also be resistant. Consistent with that hypothesis, the sfg29∆ mutant is also resistant to isoxazole 1 relative to its H99 parental strain (MIC > 64 µg/mL). We also compared the competitive fitness of the sgf29∆ deletion mutant to its H99 parental strain using a flow-cytometry-based assay33. In that assay, a 1:1 mixture of mNEON-labeled H99 and the unlabeled sgf29∆ mutant was incubated with DMSO solvent or isoxazole 1 (32 µg/mL) in RPMI/MOPS medium overnight at 37 °C. No difference in competitive fitness was noted in the absence of 1, but the sfg29∆ mutant was significantly more fit than H99 in the presence of 1 (Fig. 3C). These data suggest that loss of SAGA acetyltransferase activity improves the fitness of C. neoformans when Acs activity is compromised.
Interestingly, our prior work showed that serial passage of the Acs inhibitor AR-12 in S. cerevisiae led to a resistant isolate with a mutation in TRA1, another component of the SAGA complex16. Furthermore, Summers et al. found that the histone acetyltransferase inhibitor garcinol antagonizes the activity of the Plasmodium falciparum Acs inhibitor MMV01972117. Together, these data indicate that reduced histone acetyltransferase activity reduces dependence on Acs activity in multiple eukaryotes. This consistency further supports the conclusion that the antifungal activity of isoxazole 1 is due in large part to its effect on Acs1 and AcCoA homeostasis.
Isoxazole 1 and 2 are highly selective inhibitors of CnAcs1
The isoxazole CnAcs1 inhibitors are structurally distinct from other fungal, human, and Plasmodium falciparum Acs inhibitors reported in the literature11,17,18,34,35. Therefore, we were interested in determining their spectrum of activity as Acs inhibitors. In prior work25, we characterized the enzymology and structural basis of substrate selectivity for CnAcs1 as well as multiple other fungal Acs enzymes. As part of those studies25, His-tagged Acs enzymes from C. neoformans, C. albicans, Aspergillus fumigatus, and Coccidioides immitis were expressed in E. coli and purified by immobilized metal affinity chromatography (IMAC, Fig. S5). Acs activity was then measured in a coupled kinetic assay that detects pyrophosphate product release as previously reported25. We used a 6-point 5-fold concentration series of 1 to assess initial IC50 values for each enzyme. As discussed above, the structures and sequences of the fungal ACS enzymes are highly similar and conserved, respectively. Consistent with the selective antifungal activity of isoxazole 1 toward C. neoformans, it only inhibited CnAcs1 (IC50 = 8.6 ± 4.4 µM) (Figs. 1 and 4, Table 1). This observation further supports the conclusion that inhibition of CnAcs1 makes a significant contribution to the antifungal activity of isoxazoles 1 and 2.
To further explore the selectivity of isoxazole 1, we expressed and purified the human ACSS2 (Fig. S5). Consistent with the selectivity observed amongst fungal Acs enzymes, 1 did not inhibit ACSS2 up to the limit of its solubility (Fig. 4B). Interestingly, we previously reported that VY-3-248, an isoxazoline inhibitor of ACSS2, has no activity toward CnAcs1 (Table 1, ref. 23). A second class of ACSS2 inhibitor, MTB-9655 has become commercially available and is currently in clinical trials for cancer therapy (Clinicaltrials.gov/study/NCT04990739). Again, MTB-9655 has no activity against CnAcs1 (Fig. 4C, Table 1). Next, we tested MMV084978 and MMV019721, two molecules that inhibit PfAcAS and have anti-malarial activity17. MMV084978 inhibited CnAcs1 with an IC50 of 2.8 µM, which is ~8-fold higher than the IC50 reported for PfAcAS (370 nM) inhibition (Fig. 4D, ref. 17) but similar to isoxazole 1. Like isoxazole 1, MMV084978 did not inhibit CaAcs2 (Table 1). MMV019721, on the other hand, did not inhibit CnAcs1 below its solubility limit (Fig. 4E) but has sub-micromolar activity against PfAcAS (Table 1, ref. 17). Because the potency of MMV084978 toward CnAcs1 is similar to isoxazole 1 (Table 1), we determined its antifungal activity toward C. neoformans and found it has an MIC of 64 µg/ML while MMV019721 has no activity. As such, chemically distinct inhibitors of CnAcs1 show consistent albeit modest in vitro antifungal activity against C. neoformans, while molecules with no CnAcs1 activity also have no antifungal activity. These observations also further support the conclusion that structurally distinct Acs inhibitors can show selectivity toward enzymes from different species.
A The activity of isoxazole 1 against purified Saccharomyces cerevisiae Acs1 (ScAcs1), Candida albicans Acs2 (CaAcs2), Aspergillus fumigatus Acs1 (AfAcs1), Coccidioides immitis Acs1 (CiAcs1), and Cryptococcus neoformans Acs1 (CnAcs1). The inhibition curves are representative of three independent experiments showing similar results. B Isoxazole 1 has minimal activity toward human ACSS2. C Single-dose experiment assessing the activity of the human ACSS2 inhibitor MTB-9655 against CnAcs1 at the maximum soluble concentration. Bar indicates the mean of two independent experiments, with the value of the individual replicate shown by dots. D Anti-malarial, PfAcAS inhibitor MMV084978 inhibits CnAcs1 with IC50 = 2.8 µM (R2 = 0.99 for goodness-of-fit by non-linear regression analysis). The IC50 toward PfAcAS is indicated by the arrow. E PfAcAS inhibitor MMV019721 does not inhibit CnAcs1 at the maximum soluble concentration; bar indicates mean with value of individual replicates shown by dots. F Isoxazole 1 minimally inhibits the C. neoformans aceto-acetyl-CoA synthetase CnKbc1 at maximum soluble concentration. Source data are provided as a Source data file.
Acs enzymes are part of the general family of mechanistically similar Acyl-CoA synthetases that includes enzymes with different carboxylic acid specificities10. Consequently, we next asked if isoxazole 1 was selective for Acs when compared to a different acyl-CoA synthetase. C. neoformans Kbc1 converts aceto-acetate, a substrate structurally very similar to acetate, to aceto-acetyl-CoA but has almost no activity in the conversion of acetate to AcCoA23. As shown in Fig. 4F, 1 did not inhibit the aceto-acetyl-CoA synthetase CnKbc1. Taken together, these data indicate that isoxazole 1 displays high selectivity for CnAcs1, the enzyme upon which the chemical-genetic screen was designed. This selectivity is observed despite the mechanistic, sequence, and structural similarities of other fungal Acs enzymes and the human acetyl-CoA synthetase, ACSS225.
Isoxazole 1 is an uncompetitive inhibitor of CnAcs1
Acs is a multi-substrate enzyme that catalyzes the two-step conversion of acetate to AcCoA10,23. In the first step of the reaction, ATP condenses with acetate to generate the reactive acetyl-adenylate intermediate with release of pyrophosphate; in the second step, CoASH reacts with the acetyl-adenylate intermediate to generate the final product AcCoA and release AMP (Fig. 5A). The biochemical mechanism of this two-step reaction has been well-characterized and is classified as a bi-uni-uni-bi ping pong reaction, indicating an ordered binding of substrates25. During this two-step biochemical reaction, the ACS enzyme undergoes a set of conformational changes as outlined in Fig. 5A10. The apo enzyme binds ATP and acetate, which leads to the AD (adenylation) conformation. Next, the acyl-AMP bound form of the enzyme to the TE or thioester conformation to allow CoASH to enter the active site.
A Schematic of the two-step reaction catalyzed by Acs and the conformational changes that occur during the reaction. APO indicates an enzyme without substrate or product bound. AD indicates conformation associated with the adenylation reaction that generates the Ac-AMP intermediate. TE indicates the conformation associated with the thio-esterification reaction of Ac-AMP with CoA to yield AcCoA. CTD indicates the C-terminal domain of the protein that undergoes rearrangement through the course of the reaction. B, C Determination of isoxazole 1 Ki values for ATP and CoA substrates and goodness-of-fit (R2) values for an uncompetitive model of inhibition using non-linear regression analysis. The heat scheme shows the color-concentration correlations for the reaction plots. D, E Lineweaver-Burke plots for isoxazole 1 with ATP and CoA. F, G Plot of Km v isoxazole 1 concentration for ATP and CoA. The R2 values for the goodness-of-fit of the non-linear regression were 0.92 and 0.91 for the ATP (F) and CoA (G), respectively. Source data are provided as a Source data file.
The majority of previously characterized Acs inhibitors, such as alkyl-AMP esters, are ATP competitive15,23. Consistent with these kinetic data, our previously reported structural analysis of co-crystals of fungal Acs enzymes and alkyl-AMP esters has shown that alkyl-AMP esters occupy the putative adenine pocket25. The PfAcAS inhibitor MMV084978, on the other hand, is an uncompetitive inhibitor with respect to ATP and CoA, while MMV019721 is competitive with ATP and uncompetitive with CoA17. To characterize the kinetic mechanism for CnAcs1 inhibition by isoxazole 1, we examined the dependence of CnAcs1 inhibition by isoxazole 1 on the three substrates, as shown in Figs. 5B, C and S6. These data were fit to competitive, uncompetitive, and mixed models using non-linear regression analysis (GraphPad Prism). The best fits were obtained with uncompetitive models of inhibition for all three substrates, although acetate had the poorest fit. Lineweaver-Burke plots of the data confirmed the uncompetitive inhibition mechanism with respect to ATP and CoA (Fig. 5D, E) as demonstrated by parallel lines for the substrate dependence of velocity at different inhibitor concentrations; the acetate data did not, however, fit well with this model using this method of analysis (Fig. S6).
Uncompetitive inhibition indicates that the inhibitor binds to the enzyme-substrate complex rather than directly competing with the binding of a substrate36,37,38. This mode of inhibition is characterized by an increase in substrate affinity (decrease in Km) as the inhibitor concentration increases; Fig. 5F–G demonstrates that this phenomenon is observed for isoxazole 1 with respect to both ATP and CoA. The kinetic mechanism of the PfAcA inhibitor MMV084978 is also uncompetitive relative to CoA. Because MMV084978 also has activity against CnAcs1, we asked if it has the same mechanism of CnAcs1. Indeed, MMV084978 has a Ki of 4.1 µM with CnAcs1, and the data are best fit to the uncompetitive model (R2 = 0.9841). Furthermore, Lineweaver-Burke plots also showed the parallel relationship between velocity plots at different drug concentrations expected for uncompetitive inhibition (Fig. S7). These data suggest that isoxazole 1 and MMV084978 have similar kinetic mechanisms of CnAcs1 inhibition.
Structural analysis of isoxazole 1-bound to CnAcs1
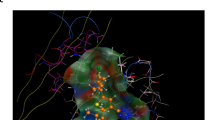
To better understand the mechanism by which isoxazole 1 inhibits CnAcs1, we obtained a co-crystal of its complex with CnAcs1 (PDB: 9CD8, Table S3). The overall structure of the enzyme is similar to previous structures obtained for CnAcs1 (e.g., the CnAcs1 complex with the ethyl-AMP ester; PDB 5K85, ref. 25). The CnAcs1-isoxazole 1 co-crystal structure showed an overall RMSD of 1.42 Å between Ca atoms (523 residues). Consistent with previous fungal Acs structures25,39, the CnAcs1-isoxazole 1 co-structure is a trimer (Fig. 6A). This trimeric structure was also observed using single-molecule solution-based mass photometry (Fig. 6B), suggesting that the trimeric conformation is unlikely to be an artifact of the crystallization process.
A Overall structure of the trimeric CnAcs1-isoxazole 1 complex with the C-terminal domains (CTD) shown in ribbon format. The blue, red, and tan regions of the trimer denote separate monomers comprising the overall trimer. B Mass photometry showing that CnAcs1 is most consistent with a trimer in solution at low concentrations and that the addition of supra-inhibitory of isoxazole 1 does not change the apparent size of the CnAcs1 protein complex. C Schematic comparing “open-TE” (tan) conformation of the CTD observed in the CnAcs1-isoxazole 1 complex to the CTD conformations in the uninhibited APO (blue), AD (green), and TE (orange) forms of the protein. D The region of the protein bound by isoxazole 1 (green) and its position within that pocket. E Overlay of isoxazole 1 (grey) with the bound pose of CoA (turquoise) and an ethyl-AMP inhibitor (yellow) in a previously reported structure of CnAcs1 (ref. 23), showing that 1 interacts with both the CoA pocket and the acetyl portion of the Ac-AMP/alkyl-AMP binding pocket. F The W334 residue functions to open the CoA tunnel by rotating upon CoA binding. The position of W334 CnAcs1-isoxazole 1 (grey) complex overlaps nearly perfectly with that observed in CnAcs1 structures with CoA bound. G Overall of the two CTD conformations in the isoxazole 1-CnAcs1 crystal structure. CTD1 conformation is shown in red, and CTD2 is shown in orange. H Overlap of the positions of isoxazole 1 in the two CTD conformations present in the unit cell of the isoxazole 1-CnAcs1 complex.
An important structural and mechanistic characteristic of ANL-family adenylating enzymes is that the C-terminal domain (CTD) undergoes large conformational changes between the two biochemical steps of the overall conversion of acetate to AcCoA10,25. In the Acs enzyme without bound substrates (Apo form), the CTD adopts a unique conformation (Fig. 6C). The first reaction is the adenylation step in which ATP reacts with acetate to generate an acetyl-AMP intermediate with the release of pyrophosphate. This step of the reaction leads to the AD conformation. Prior to the reaction between the acetyl-AMP and CoA, the acetyl-AMP-bound enzyme undergoes a conformational change in which the CTD rotates and leads to the TE conformation. In previous work, we obtained crystal structures of CnAcs1 in each of these conformations with either substrates, intermediates, or inhibitors bound25. The CTD in these structures is frequently observed in multiple conformations within the asymmetric units.
In the CnAcs1-isoxazole 1 structure, the CTD is observed in two conformations (the CTD is disordered in one chain of the asymmetric unit). First, the CTD adopts the Apo conformation indicative of an enzyme without bound substrates, while the second conformation is structurally distinct from the canonical conformations observed previously (Fig. 6A, C). This new conformation is most closely related to the TE conformation; the transition between the AD and TE conformations occurs through a hinge region upon which the CTD rotates. In the CnAcs1-isoxazole 1 structure, the CTD is rotated much further relative to the hinge region than in other structures, as shown in Fig. 6C. In the TE conformation, the CTD forms the interior of the CoA binding pocket. The “over-rotation” caused by isoxazole 1 binding prevents the formation of an intact, CoA-binding tunnel and opens the region to solvent; we refer to this new CTD conformation as the TE-open. Isoxazole 1-bound CnAcs1 enzyme, therefore, does not recapitulate the secondary and tertiary structure CnAsc1 conformations associated with the binding of substrates and substrate mimics. These observations are consistent with the fact that 1 is not competitive with Acs substrates and displays an uncompetitive mode of inhibition.
Isoxazole 1 binds in the region that accommodates CoA, with the biphenyl moiety placed adjacent to the CoA pantothenate chain binding site (Fig. 6D, E). The amide and isoxazole substituents of 1 extend from the CoA binding pocket into the region occupied by the acetyl group of the acetyl-AMP intermediate or the propyl group of propyl-AMP ester inhibitors (Fig. 6E). The size of the CoA tunnel nearest to the acetyl-AMP binding pocket is modulated by a key tryptophan residue (W334, ref. 23). In the absence of CoA, W334 rotates to hydrogen bond with substrates or intermediates within the ATP or acetyl-AMP pocket25. Upon CoA binding, W334 rotates to open up the CoA tunnel, presumably to allow binding of CoA and position it for the TE reaction. In the CnAcs1-isoxazole 1 co-structure, W334 adopts an orientation similar to that of the CoA-bound enzyme (Fig. 6F, ref. 25).
Like many Acs enzymes, CnAcs1 is exquisitely specific for acetate with little to no activity with carboxylic acids containing larger alkyl groups such as propionate and butyrate23,25. Based on structural and genetic data studies of Acs enzymes from multiple species, the indole ring of a highly conserved tryptophan (W439 in CnAcs1) limits the size of the acetyl-AMP pocket and is a key determinant of substrate specificity and inhibitor binding25. The cyclopropyl group of isoxazole 1 is positioned within the acetyl-AMP pocket in a manner similar to that observed in our previously reported structures of CnAcs1 bound to linear chain25, alkyl-AMP ester bi-substrate inhibitors.
Recently, we used an expanded series of alkyl-AMP ester bi-substrate inhibitors to probe the steric properties of alkyl groups that could be accommodated by the tryptophan wall in CnAcs137. The most potent alkyl-AMP-based inhibitor of CnAcs1 is ethyl-AMP (8 µM), while increasing the length of the alkyl chain to propyl and butyl decreases potency 3- and >50 fold, respectively; importantly, these trends parallel alkyl carboxylate substrate specificity23,40. Interestingly, the IC50 of the cyclopropyl-AMP ester (9 µM) is essentially identical to that of the ethyl-AMP inhibitor, indicating that the pocket is able to accommodate an alkyl group of this size and conformation. Furthermore, we obtained an X-ray crystal structure of CnAcs1 bound to the cyclopropyl-AMP ester and observed that the cyclopropyl moiety of the inhibitor and the W439 residues overlap almost exactly (PDB: 8G0T, Table S3, Fig. S8A; Fig. S8B shows the density map for the bound cyclopropyl-AMP ligand). Thus, the cyclopropyl-isoxazole portion of 1 is positioned in the portion of the enzyme that interacts with the methyl group of the acetyl-AMP during the reaction.
Integrating this structural information with the biochemical mechanism of inhibition leads us to propose the following model for isoxazole 1 inhibition. First, the enzyme proceeds through the adenylation step and conformational changes to the TE conf (Fig. 5A). Second, the CoA binding pocket and tunnel are generated by this conformational change. Third, the kinetic data would suggest that once CoA binds to CnAcs1, it may interact with isoxazole 1. This sequence of events is consistent with the uncompetitive mode of inhibition for both ATP and CoA. The structure shows that 1 partially overlaps the CoA binding site and disrupts the CoA tunnel needed for completion of the thio-esterification reaction. Once isoxazole 1 is bound, its interaction with CnAcs1 appears to be independent of the CTD conformation because the X-ray structure shows it bound to both an open AD-like conformation and the closed TE-like conformation. Figure 6G shows an overlay of the two CTD conformations present with isoxazole 1 bound, while Fig. 6H shows that the position of isoxazole 1 does not vary substantially between the two CTD conformers of the co-crystal structure. In this way, the ATP and CoA substrates are needed to generate the conformation to which isoxazole 1 can bind, but neither substrate directly competes with its binding. Isoxazole 1 binds both the open and closed conformation, leading to the release of substrate/AMP ester intermediate and a dead-end enzyme-inhibitor complex.
Molecular dynamics and structure-activity relationships identify molecular determinants of isoxazole 1 inhibition of CnAcs1
To further characterize the interactions that contribute to the binding of isoxazole 1 to CnAcs1, we performed molecular dynamics (MD) simulations using Schrödinger software as described in the “Methods” and Table S4. The interaction fraction data for residues that interacted with ligand 1 for >90% of the 1000-ns time-scale simulations are shown in Fig. 7A and summarized in the scheme shown in Fig. 7B. The majority of the interactions predicted to contribute to the binding of 1 to CnAcs1 are hydrophobic, which is consistent with inspection of the X-ray structure of the enzyme-inhibitor complex. Indeed, W334, which plays a key role in the modulation of the CoA binding pocket and is discussed above, is predicted to have a strong interaction. Similarly, W439 is predicted to interact with the isoxazole 1. W439 limits the size of the acetyl-AMP binding pocket and is a key determinant of substrate and inhibitor selectivity for AMP ester-class inhibitors25,40. As also discussed above, our structural data suggest that W439 is likely to interact with the cyclopropyl moiety of 1. The exception to this pattern of hydrophobic interactions is a predicted direct H-bond between T336 and the amide carbonyl of isoxazole 1. In the crystal structure, the distance between the threonine OH and the carbonyl of 1 is 3.7 Å, which is consistent with an H-bond due to an electrostatic rather than covalent interaction. The amide carbonyl of 1 and T336 also appears to participate in H2O-bridges involving D331. Finally, the MD modeling indicates that hydrophobic interactions between A382 and A330 may contribute to binding.
A Histogram of molecular dynamics interactions of isoxazole 1 with CnAcs1. B Schematic indicating residues predicted to contribute to the binding of isoxazole 1 to CnAcs1. The majority of residues predicted to contribute to binding participate in hydrophobic interactions (green). A key H-bonding interaction between T336 and the amide carbonyl of isoxazole 1 was identified in 81% of simulations. For panels A&B, charged interactions are shown in orange; hydrophobic in green; polar in blue; water in grey, and solvent exposed in light grey. C SAR of the isoxazole scaffold. IC50 data for isoxazoles 2–13 indicate that the amide carbonyl (isoxazole 3/4), isoxazole cyclopropyl moiety (isoxazoles 5–7), and an inductively electronegative substituent at the para-position of the aryl ring (isoxazoles 2, 9, and 11) are key drivers of CnAcs1 potency. Source data are provided as a Source data file, and the molecular dynamics files have been deposited at FigShare (see data availability statement for link).
The X-ray structure of 1-CnAcs1 and the MD simulations suggest that the amide carbonyl, the cyclopropyl, and the N-methyl substituent of the amide may be important for interactions between isoxazoles and CnAcs1. To test these predictions, we synthesized a set of molecules derived from isoxazole 2, the more metabolically stable and chemically accessible derivative of 1. First, we tested the necessity of the amide carbonyl group for inhibition by synthesizing a derivative of 2, which has a methylene group bridging the biphenyl with the nitrogen (3). Consistent with our hypothesis, 3 had no activity against CnAcs1 (Fig. 7C). We also synthesized the corresponding sulfonamide derivative 4, but it too was unable to inhibit CnAcs1. These data strongly support the hypothesis that the amide carbonyl group is important for CnAcs1 inhibition by the isoxazoles.
To test the importance of the cyclopropyl group, we first synthesized the corresponding isopropyl derivative (5). This is a relatively small steric change, but it eliminated inhibition entirely. The cyclopropyl group is positioned near W439 in CnAcs1 X-ray structures bound to both isoxazole 1 and the alkyl-AMP ester series of inhibitors25. However, the IC50 of the isopropyl-AMP ester toward CnAcs1 was equivalent to that of the cyclopropyl-AMP ester. The methyl-AMP ester was completely inactive against CnAcs1. The methyl isoxazole 6 was much less potent than 2 but did show 50% inhibition at 100 µM, while the unsubstituted isoxazole 7 was completely inactive. These data indicate that the cyclopropyl group is likely to interact with W439, but that this interaction is distinct from the manner in which the alkyl groups of alkyl-AMP ester inhibitors interact.
To test the potential interaction between A330/382 and the methyl group of the amide predicted by MD simulations, we synthesized a derivative of the benzofuran-substituted isoxazole lacking the amide N-methyl group (8) and found it had no activity against CnAcs1. This lack of activity may be due to loss of hydrophobic interactions, but it is also possible that the methyl group affects the conformation of the amide and leads to disruption of the H-bond between the carbonyl and T336.
The two active isoxazoles 1 and 2 have inductively electronegative substituents (O-alkyl and F) at the para position of the distal phenyl ring. Consistent with this pattern, the OMe derivative 9 had activity that was similar to 1 and more potent than 2, while placement of the OMe group at the ortho position 10 eliminated activity. Substitution of the F in isoxazole 2 with Cl in isoxazole 11 at the ortho position also increased potency relative to 2, while the unsubstituted derivative 12 had no activity, further supporting the importance of an inductively electronegative substituent at this position. The pyridine derivative 13 was synthesized to generate an unsubstituted electron-deficient ring system and was inactive. The OMe (9), Cl (11), and furan (14) substituted isoxazoles contain sterically larger para-substitutions than the F-substituted isoxazole (2) or pyridyl (13) containing isoxazoles and have increased potency. Thus, it seems that increased steric bulk as well as electronegativity may contribute to increased potency at this position.
Finally, we tested the antifungal activity of 13 to determine if molecules with no ability to inhibit CnAcs1 activity would also have no antifungal activity; consistent with that hypothesis, 13 had no antifungal activity against H99 at the limit of solubility. The structure-activity relationship studies identified three key regions of the isoxazoles that contribute to CnAcs1 activity: (1) the amide carbonyl and N-methyl group; (2) the cyclopropyl group; and (3) inductively electronegative substituents on the para position of the distal phenyl group. In addition, these data further support the conclusion that CnAcs1 inhibition can explain a good deal of the antifungal activity of this series of compounds.
Mutations in the CoA binding pocket of CaAcs2 designed to match CnAcs1 increase susceptibility to isoxazole 1 and MMV084978
The CnAcs1 residues predicted by MD to have interactions with isoxazole 1 were largely conserved among fungal Acs sequences. Therefore, this computational analysis did not provide immediate insights into the selectivity of 1 for CnAcs1 over the other fungal Acs enzymes. To identify amino acids that may contribute to this selectivity, we generated a homology model of CaAcs2 bound to isoxazole 1 based on the CnAcs1-isoxazole 1 structure, identified five residues within the binding pocket that were discordant between the two enzymes (Fig. 8A), and generated a mutant of CaAcs2 (CaCnAcs1) that matched the CnAcs1 binding pocket (Fig. 8B). Figure 8A, B shows a comparison of the CaAcs2 wild type and CnAcs1-like mutant. The CaCnAcs2 enzyme was expressed in E. coli and purified following previously described methods. The Km of CaCnAcs1 for ATP (82 ± 25 µM) and CoA (367 ± 129 µM) were similar to previously reported25 values for CaAcs2 (ATP: 83 ± 14 µM; CoA 471 ± 45 µM) and CnAcs1 (ATP: 59 ± 10 µM; CoA 880 ± 170). Indeed, the CaCnAcs2 enzyme was inhibited by the ethyl-AMP ester, which is competitive with ATP, with an IC50 nearly identical to CaAcs2 (19 ± 7 µM v 11 ± 5 µM, ref. 25).
A CoA binding site of CaAcs2 with isoxazole 1 was modeled based on structural alignment with the CnAcs1-isoxazole 1 complex. B The highlighted residues in CaAcs2 were mutated to the corresponding CnAcs1 residues to create CaCnAcs2. C Comparison of CaAcs2 and CaCnAcs2 inhibition by isoxazole 1 (R2 for CaCnAcs2 fit: 0.74). D CnM445QAcs1 mutant is inhibited by isoxazole 1 with similar potency; R2 = 0.93 for CnAcs1 and R2 = 0.82 for CnM445QAcs1. E The potency of MMV084978 is increased in CaCnAcs2 relative to CaAcs2 (R2 = 0.94). Error bars indicate the standard deviation of two independent replicates. F Overlap of 1 (grey) and MMV084978 (magenta) docked into CnAcs1 with binding affinity data (kcal/mol). Source data are provided as a Source data file.
The CaAcs2 enzyme is not inhibited by isoxazole 1 at 100 µM (2-fold below solubility limit). Interestingly, CaCnAcs2 is inhibited with an IC50 of 50 ± 23 with maximum inhibition of ~50% (Fig. 8C), indicating that the discordant residues within the binding pocket region of isoxazole 1 in CaAcs2 reduce its susceptibility relative to CnAcs1. Of the five residues that were mutated in CaCnAcs2, M445 is unique to CnAcs1 compared to all other fungal Acs enzymes, as well as the human ACSS2. We, therefore, mutated M445 to the conserved Q446 in the CnAcs1 enzyme. However, CnM445QAcs1 was inhibited with an IC50 similar to the wild-type enzyme (Fig. 8D). MMV084978 is also selective for CnAcs1 relative to CaAcs2. Therefore, we tested the activity of MMV84978 against CaCnAcs2 to determine if the mutations would increase its susceptibility in the manner observed for isoxazole 1 (Fig. 8E). Indeed, MMV084978 inhibited CaCnAcs2 with an IC50 of 88 ± 21 µM and a maximum inhibition of 93%. These data strongly suggest that MMV084978 and isoxazole 1 interact with CnAcs1 in a similar manner.
To further explore the potential effects of the mutations on selectivity, we took a modeling approach. The binding pose of isoxazole 1 was inserted into the crystal structure of CaAcs2 bound to an alkyl-AMP ester (PDB 8W0B) via structural alignment with isoxazole 1-bound CnAcs1. The same mutations made in the CaCnAcs2 enzyme were then recreated in the isoxazole 1-bound model of CaAcs2. Evaluation of binding energies showed a substantial improvement for CaCnAcs2 (−8.4 kcal/mol) vs CaAcs2 (−7.3 kcal/mol), recapitulating the improved activity against CaCnAcs2 in vitro. To better probe how binding site residues impact activity against CnAcs1, the same five non-conserved residues were mutated to make a CnAcs1 enzyme that matched the CaAcs2 binding site (CncaAcs1). When evaluating the binding energy of isoxazole 1 using this method, we found a much smaller effect (−8.3 kcal/mol for CnAcs1 vs −8.1 kcal/mol for CnCaAcs1), but one that still showed reduced preference for the CaAcs2 binding site residues. We also used this approach to model the binding of MMV084978 into the CnAcs1 CoA pocket and found a preferred orientation of the carbonyl (alpha to the phenyl) in close agreement with the crystal position of isoxazole 1 (Fig. 8F). This suggests an important role of T336 in stabilizing the bound state of MMV084978, as our data indicate for the isoxazole series. Similar to isoxazole 1, the estimated binding energy was reduced against the pocket for CnCaAcs1, though only a small effect was observed (−6.0 kcal/mol for CnAcs1 and −5.8 kcal/mol for CnCaAcs1). While limited in scope, this investigation supports that interaction with the conserved T336 residue is a key component of inhibition at the CoA binding site and is a shared feature among structurally distinct uncompetitive inhibitors of CnAcs1.
Discussion
Acs has emerged as a drug target for the treatment of a variety of human diseases and infections, including cancer, fatty liver disease, malaria, and mycoses13,14,15,16,17,18. One of the reasons that Acs is an attractive target for the treatment of human disease is that under normal physiological conditions, ACSS2 and its substrate, acetate, are minor contributors to the overall pool of AcCoA in human or mammalian cells13,14. Instead, glucose is the major carbon source from which AcCoA is derived, through the TCA cycle and the enzyme ATP-citrate lyase; accordingly, deletion of ACSS2 is possible in mice, while deletion of ATP-citrate lyase is not. However, ACSS2 has been shown to be involved in the generation of AcCoA in specific physiological circumstances in mammals39,41, indicating that loss of its function may have more subtle effects on cellular function. Consequently, in the context of targeting Acs as an approach to treating infections, the selective inhibition of microbial, parasitic, or fungal Acs enzymes relative to host ACSS2 would be likely to reduce potential adverse effects of those drugs.
With these considerations in mind, the first general conclusion from this work is that highly selective inhibition of Acs enzymes is chemically and biochemically feasible. To date, three other Acs inhibitors identified by phenotypic or target-based high-throughput screening have been reported11,17,34. Above, we described the identification of isoxazole 1 through a chemical-genetic, phenotypic screen using C. neoformans. Somewhat surprisingly, isoxazole 1 has no activity against other fungal Acs enzymes and is inactive toward the human enzyme ACSS2 (Table 1). The isoxazoline ACSS2 inhibitor VY-3-249 was identified by an in vitro screen directly against the ACSS2 enzyme11 and also has no activity against any of the fungal Acs enzymes examined to date25. Interestingly, a more potent analog, VY-3-135, is selective for ACSS2 over the related human enzymes ACSS334. Similarly, an ACSS2 inhibitor currently in clinical trials for a cancer indication, MTB-9655 (Clinicaltrials.gov/study/NCT04990739), also has no activity against CnAcs1 (Fig. 4C).
Acs has emerged as a particularly attractive target for the development of anti-malarial drugs. Two structurally distinct anti-malarial Acs inhibitors (MMV0894978 and MMV019721) were identified in a phenotypic screen for growth inhibition17. While MMV019721 does not significantly inhibit CnAcs1, MMV0894978 inhibits PfAcAs, CnAcs1, and ACSS2, but with IC50 values that are 8- and 50-fold higher toward CnAcs1 and ACSS2, respectively. Importantly, however, the potency of MMV089478 toward CnAcs1 (2.8 µM) is similar to that of isoxazole 1 (8.8 µM), and both have antifungal activity against C. neoformans. Furthermore, neither MMV089478 nor isoxazole 1 inhibited CaAcs2 significantly. These data suggested that MMV089478 and the isoxazole series of inhibitors share mechanistic characteristics. Indeed, both isoxazole 1 and MMV089478 are uncompetitive inhibitors of CnAcs1 and PfAcAs, respectively. We also found that MMV089478 is an uncompetitive inhibitor of CnAcs1. Mutations in the region of the CoA binding pocket of PfAcAs, based on homology models of the enzyme, confer resistance to MMV089478, and the inhibitor is uncompetitive relative to CoA. The PfAcAs mutations associated with MMV089478 resistance map to the region where isoxazole 1 binds to CnAcs1. Together, these data indicate that two structurally distinct Acs inhibitors show similar species selectivity, kinetic mechanism, and potentially similar interactions with the enzyme target.
The similarity of Acs binding sites for isoxazole 1 and MMV089478 is further supported by the fact that mutations in the CoA/isoxazole 1 binding region of CaAcs2 enzyme designed to match those found in CnAcs1 led to a CaCnAcs2 mutant enzyme that was inhibited by both compounds when the WT CaAcs2 was not. One of the sites that is mutated in CaCnAcs2 (CaAcs2: G335/CnAcs1: A330) aligns with PfAcAs A597 which is the site where one of the resistance mutations arose (A597V, ref. 17). It appears that variation in amino acid residues in, and around, the CoA pocket contribute to the selectivity of inhibitors that target this region of the enzyme. In the case of the fungal enzymes, the sequence variation in the CoA region that we probed does not entirely account for the level of observed selectivity. Additional factors must contribute to the observed selectivity.
One possibility is related to the uncompetitive mechanism of inhibition, which indicates that the inhibitor has significant interactions with a substrate-enzyme complex36. The tertiary complex that this mechanism of inhibition suggests is likely to have structural features that are distinct from the dead-end, CnAcs1-isoxazole 1 complex for which we have structural data. As such, the CnAcs1/AMP-Ac/CoA complex may bind 1 and/or MMV089478 through additional interactions not evident in the final CnAcs1-isoxazole 1 complex. Although our structural data do not explain the entire mechanism of selectivity of isoxazole 1, they do provide some of the first insights into how non-nucleoside-based Acs inhibitors interact with the enzyme.
The anticryptococcal activity of isoxazoles 1 and 2, as well as MMV089478, is in contrast to the non-essentiality of CnAcs1 under standard laboratory growth conditions when ACL1 is present to generate AcCoA23,24. Although the in vitro antifungal activity of these Acs inhibitors is modest, it is observed only in nutrient-limited medium designed to mimic the host environment27. Indeed, the acs1∆ mutant has previously been shown to have a growth defect under other in vitro conditions with low glucose or where non-glucose carbon sources are dominant24. In the same medium used to determine MIC values, the acs1∆ mutant has reduced fitness by competition assay (Fig. S3). Chemical inhibition leads to a rapid loss of protein function, whereas genetic deletion allows the cell to activate cellular mechanisms to compensate for that loss. In other systems, therefore, differences in the nature of phenotypes resulting from chemical inhibition and genetic deletion have been reported42. Therefore, we propose that chemical inhibition of Acs activity in the setting of relatively poor nutrient conditions leads to growth arrest because the cell is unable to rapidly compensate for the reduction in AcCoA.
The alternative explanation for the antifungal activity is that isoxazoles 1 and 2 have off-target activity against essential enzymes. Four lines of evidence argue against this explanation. First, three different Acs inhibitors have been reported to have antifungal activity: the pyrazole AR-12, the isoxazoles, and MMV08947815,16. The structural diversity of these inhibitors reduces the likelihood that they are inhibiting the function of similar off-target proteins. Second, resistance mutations in the SAGA histone acetylation complex have been observed for AR-12 and isoxazole 1, and inhibition of histone acetylation increases resistance to MMV08947816,17. These data indicate a common mechanism of resistance between the three chemically distinct Acs inhibitors, suggesting they are targeting similar proteins as part of their mechanism of antifungal activity. Third, close structural analogs of the isoxazoles that do not inhibit CnAcs1 do not have antifungal activity (Fig. 7C). Fourth, the isoxazole inhibitors show reduced activity in nutrient-rich conditions and increased activity when acetate is the sole carbon source, which is the pattern expected based on the phenotypes of the acs1∆ mutant. These observations cannot completely rule out that inhibition of another target may contribute to the antifungal activity of the isoxazoles. However, they strongly support the conclusion that inhibition of CnAcs1 makes a significant contribution to the antifungal activity of the isoxazoles.
As such, the isoxazole CnAcs1 inhibitors also provide proof-of-principle for inhibition of fungal Acs as an antifungal strategy even in species that can generate AcCoA through ATP-citrate lyases. Prior results from our lab have shown that the role of CnAcs1 in carbon metabolism is even more pronounced during infection23 than under in vitro conditions mimicking the host environment, supporting the possibility that the efficacy of CnAcs1 inhibitors is likely to be increased in vivo relative to in vitro. In addition, the AcCoA pool is critical for the synthesis of ergosterol, the key sterol of fungal plasma membranes and a target for two of the three antifungal drugs currently used to treat patients. Consistent with this role, the combination of Acs inhibitors with fluconazole is synergistic in vitro (Fig. 2C) and C. neoformans acs1∆ mutant is hypersensitive to fluconazole in vivo23. As a single therapy, fluconazole is inferior to combination therapy for the treatment of cryptococcal meningitis43. Therefore, there has been strong interest in developing new approaches to fluconazole-based combination therapy28, and Acs inhibitors appear to be strong candidates in this regard.
In summary, our discovery of the isoxazole Acs inhibitors and the structural and biochemical characterization of their mechanism of inhibition provides novel insights that should be useful to the design of Acs inhibitors for both infectious disease indications as well as the treatment of human cancers and metabolic diseases.
Methods
Strains, media, and growth conditions
Lab strains, clinical isolates, and genetically modified strains of C. neoformans or C. albicans were maintained in glycerol stocks that were stored at −80 °C and recovered on yeast peptone dextrose (YPD, 1% w/v yeast extract, 2% w/v peptone, 2% w/v dextrose, and 2% w/v agar). Yeast strains were re-struck from frozen stocks after 14 days. Overnight cultures grown into log phase, shaking at 200 rpm in liquid YPD, were used for each experiment. In cases where experimental procedures did not utilize YPD media, the appropriate media recipes are described in the associated methods sections. C. neoformans is of the H99-stud lineage, and the standard lab strain of C. albicans used in these studies is SC5314. The C. neoformans acl1Δ deletion mutant was generated and validated in previous work23. The C. neoformans sgf29Δ deletion mutant was generated as part of the Madhani knockout collection and obtained from the Fungal Genetics Stock Center. Artwork in the figures was generated using publicly available Inkscape software (https://inkscape.org).
Compound library
We obtained and screened 55,264 compounds from the ChemBridge DiverSET library designed by ChemBridge. The library was initially provided in 10 mM DMSO stocks and then diluted to 1.25 mM in 50% DMSO for screening purposes. All compounds were stored at −80 °C in 384-well plates with sealing foil. Each plate was thawed completely in a desiccating chamber before each use and minimally maintained at room temperature for the duration of assay assembly. For the initial hit validation screen, primary hits were manually picked from the original compound library, while secondary screening assays were performed with compounds that were re-ordered directly from ChemBridge.
Primary, validation, and secondary screens
Compounds were screened against the ACS-dependent C. neoformans acl1Δ deletion mutant in a high-throughput-based growth assay. On Day 0, the acl1Δ mutant was inoculated into 50 mL of YPD, where a 1:10 dilution and a 1:25 dilution culture was also created and grown shaking at 200 rpm at 30 °C to early log phase overnight. The culture was adjusted to a density of 5.33 × 105 cells/mL, and 15 µL was added to a 384-well plate containing 10 µl of YPD and 0.5 µl of compound (Nimbus, MicroLab). The final concentrations in each well were 8000 cells, 25 µM compound, and 1% DMSO in YPD. The cultures were incubated at 30 °C for 48 h before the optical density (OD600) for each plate was determined using a plate reader (SpectraMax i3X, Molecular Devices). Hits were verified using cherry-picked samples under identical conditions as the primary screen. Validated hits were re-ordered and counter-screened in a secondary assay where final concentrations of assay components were the same but scaled to 100 µl volume in a 96-well plate format and read as before.
Chemical synthesis
The synthetic methods and molecule characterization data are provided in the Supplementary Methods section.
Antifungal susceptibility assays
MICs were performed using a slightly modified CLSI micro-broth dilution method26. Overnight cultures were washed in sterile phosphate-buffered saline (PBS) brought up into either YPD, YNB-acetate, or RPMI supplemented with 165 mM MOPS pH 7.0, such that 1 × 103 cells would be delivered into each well with a final volume of 200 µl in a 96-well plate. Each tested drug was added such that the final DMSO concentration did not exceed 1 % and the maximum drug concentration tested was at least four-fold higher than the reported MIC or the limit of solubility. Plates were incubated at 37 °C for 72 h for C. neoformans and 24 h for C. albicans, unless otherwise stated. Each assay was performed in a minimum of technical duplicates with a minimum of two independent experimental replicates. Fractional inhibitory concentration assays were performed under similar conditions to MICs but set up with a standard checkerboard dilution for each paired16.
In vitro toxicity assay
HepG2 (ATCC, HB-8065) cells were maintained in Dulbecco’s modified Eagle medium (Gibco, Cat #11965–092) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Cells were cultured at 37 °C in a humidified atmosphere with 5% CO2. For experiments, cells were seeded into 96-well plates at a density of 1.25 × 10⁴ cells per well and incubated overnight under the same culture conditions. The following day, the medium was replaced with fresh medium containing a two-fold dilution series of the test drug, with an equal concentration of DMSO in all wells. After 24 h of incubation, the supernatant was collected to quantify lactate dehydrogenase (LDH) release using the CyQuant LDH assay kit (Invitrogen, Cat #C20300), following the manufacturer’s protocol. LDH levels were normalized to the maximum lysis control, achieved by treating cells with Triton X-100.
In vitro characterization of microsomal stability
Male ICR/CD-1 mouse microsome fractions (Cat M1000; Lot 2210246) and Human pooled gender microsomes (Cat X00807, Lot 2110263) were purchased from Xenotech/BioIVT (Baltimore, MD). Microsome protein (0.5 mg/mL) was placed in a glass screw cap tube; a 2 mM DMSO stock of each compound was spiked into a 50 mM Tris, pH 7.5 solution, and this was added to the microsome solution on ice. The final concentration of the compound after the addition of all reagents was 2 µM. An NADPH-regenerating system (1.7 mg/ml NADP, 7.8 mg/ml glucose-6-phosphate, 6 U/ml glucose-6-phosphate dehydrogenase in 2% w/v NaHCO3/10 mm MgCl2) was added for analysis of Phase I metabolism after heating both the regenerating solution and the sample tubes to 37 °C for 5 min in a 37 °C shaking water bath. Prior to adding compounds and the NADPH-Regenerating System, a subset of microsomes was preincubated with 500 µM final 1-ABT (MedChemExpress, Cat HY-103389; Lot 247772) for 30 min at room temperature. The incubation was continued in singlet and at varying time points after addition of phase I cofactors (0, 10, 30, 60, 120 min), the reactions were quenched with 0.5 mL (1:1) of methanol containing 0.2% formic acid, 4 mM ammonium acetate (Isoxazole 2 only), and 100 ng/mL N-benzyl-benzamide internal standard (0.1%FA, 2 mM NH4 acetate, and 50 ng/mL final concentration). Time 0 samples were stopped with the methanol solution while still on ice prior to the addition of the NADPH-regenerating system and compound, which were subsequently added. Samples were vortexed for 30 s, incubated at RT for 10 min, and spun for 5 min at 812 × g in a table-top centrifuge at RT. Supernatants were then transferred to an Eppendorf tube and spun in a 4 °C microfuge for 5 min at 16,100 × g. The resulting supernatant was analyzed by LC-MS/MS using an AB Sciex (Framingham, MA) 3200 QTRAP® mass spectrometer running Analyst 1.7.2 coupled to a Shimadzu (Columbia, MD) Prominence liquid chromatography system. Compounds were detected in positive MRM (multiple reaction monitoring) mode by following the precursor to fragment ion transition 375.1/223.0 (Isoxazole 1) and 351.098/199.2 (isoxazole 2). The internal standard (IS) N-benzylbenzamide (Sigma) was followed by the transition 212.1/91.1. An Agilent Poroshell EC-C18 column (2.7 micron packing, 50 × 3.0 mm size) was used for chromatography of all three compounds, but buffers and gradient conditions varied slightly. For isoxazole 1, Buffer A: dH20 + 0.1% formic acid, Buffer B: methanol + 0.1% formic acid. A flow rate of 0.7 ml/min was used with the following gradient conditions: 0–0.5 min 3% B, 0.5–3.0 min gradient to 100% B, 3.0–3.5 min 100% B, 3.5–3.6 min gradient to 3% B, 3.6–4.5 3% B. For isoxazole 2, Buffer A: dH20 + 0.1% formic acid + 2 mM NH4 acetate, Buffer B: methanol + 0.1% formic acid + 2 mM NH4 acetate. A flow rate of 0.7 ml/min was used with the following gradient conditions: 0–1.5 min 3% B, 1.5–2.0 min gradient to 100% B, 2.0–3.5 min 100% B, 3.5–3.6 min gradient to 3% B, 3.6–4.5 3% B. The method described in McNaney et al.44 was used with modification for the determination of metabolic stability half-life by substrate depletion. A “% remaining” value was used to assess the metabolic stability of a compound over time. The LC-MS/MS peak area of the incubated sample at each time point was divided by the LC-MS/MS peak area of the time 0 (T0) sample and multiplied by 100. The natural Log (ln) of the % remaining of the compound was then plotted versus time (in min), and a linear regression curve was plotted going through the y-intercept at ln(100). If the metabolism of a compound failed to show linear kinetics at a later time point, those time points were excluded. The half-life (T½) was calculated as T½ = 0.693/slope. The metabolism of 7-ethoxycoumarin was used to monitor microsome performance.
Characterization of in vitro metabolites of isoxazole 2
Isoxazole 2 was incubated with mouse and human microsomes as described above, except the concentration was increased to 10 µM to facilitate detection of metabolites, and the final supernatant was filtered through a 0.2-micron PVDF syringe filter prior to analysis by LC-TOF/MS (Sciex 6600 QTOF running Analyst TF 1.8.1). The same Agilent Poroshell EC-C18 column was utilized with slightly different chromatography but the same solvents (flow rate 0.8 ml/min; 0–0.5 min 3% B, 0.5–5.0 min gradient to 90% B, 5.0–7.0 min 90% B, 7.0–9.0 3% B). Samples were acquired using positive ESI, using the TOF-IDA-MS/MS mode. After sample acquisition, the data were transferred to a processing computer and loaded into MetabolitePilot 2.0.4 for metabolite characterization. Individual samples were screened for metabolites using a generic, predefined list of 80 phase 1 and 2 biotransformations, as well as using software-predicted compound-specific cleavages. Peaks were filtered to include those with (1) confirmation score above 45%, (2) charge state of +1, and (3) mass accuracy within 12 ppm of calculated formula. Confirmation score is an algorithmically derived weighted score assigned to peaks that indicates the likelihood that the peaks are a metabolite. All resulting peaks were visually inspected to confirm that no background peaks were included. After inspection, the remaining peaks were compared across all time point samples to identify those consistently present. Potential metabolites were reported if they appeared in three or more time point samples, or if they appeared in a single sample but were part of a documented multi-transformation pathway. Peak areas were summed (across time points and across discrete peaks of the same transformation type), and metabolites comprising greater than 5% of the total peak were investigated.
Whole-genome sequencing of resistant mutants
Isoxazole-resistant strains and their parents were grown overnight in 25 mL YPD. Pelleted cultures were transferred to a 2 mL screw cap tube with PBS, pelleted, and supernatant removed. Samples were lyophilized overnight, and 0.5 mm diameter glass beads (BioSpec Products) were added the following day. Freeze-dried material was subject to bead beating for 1 min or until cell pellets were broken into a fine powder, followed by the addition of 1 mL of CTAB extraction buffer [100 mM Tris-HCl, pH 7.5, 700 mM NaCl, 10 mM EDTA, 1% cetyltrimethylammonium bromide (CTAB), 1% β-mercaptoethanol (14 M)]. Samples were incubated at 65 °C for 30 min followed by the addition of 1 mL chloroform and gentle mixing. Samples were centrifuged for 10 min at 5000 × g, where the aqueous phase was transferred to a clean tube for a second round of chloroform extraction. The final aqueous phase was transferred to a clean tube and followed by standard ethanol precipitation. Final samples were resuspended in nuclease-free water and sent to SeqCoast for further analysis. Received samples were prepared for whole-genome sequencing using an Illumina DNA Prep tagmentation kit and unique dual indexes. Sequencing was performed on the Illumina NextSeq2000 platform using a 300-cycle flow cell kit to produce 2 × 150 bp paired reads. 1–2% PhiX control was spiked into the run to support optimal base calling. Read demultiplexing, read trimming, and run analytics were performed using DRAGEN v3.10.12, an on-board analysis software on the NextSeq2000. We include FastQC metrics as a best practice and for examination in the case of unexpected outputs. Reads were aligned to the C. neoformans H99 genome (FungiDB version 52) using bowtie2, followed by variant calling via samtools and vcftools with a minimum read depth = 10 and quality PHRED score = 37. Variants identified in each parental strain were filtered out of each respective isoxazole 1-resistant strain.
Flow cytometry competition assays
Following a previously published protocol33, overnight cultures of the H99 reference strain, H99-mNeonGreen expressing strain, and the sgf29∆ deletion strain were grown in standard YPD media. The sgf29∆ deletion strain and H99-mNeonGreen were washed with PBS, resuspended in RPMI-MOPS pH 7.0, and mixed 1:1 such that 1 × 104 cells/mL per strain were delivered with a final volume of 100 μl per well in a flat-bottom 96-well plate. A dilution series of isoxazole 1 was delivered such that the DMSO concentration remained constant and did not exceed 1.25%. Co-cultures were grown overnight at 37 °C for 24 h in ambient air before being analyzed on an Attune NxT Flow Cytometer with CytKick autosampler and Attune Cytometric software.
Recombinant enzyme expression and purification
Briefly, expression plasmids for the ACS enzymes (reported in ref. 25) were transformed into the Escherichia coli strain BL21 with appropriate antibiotic selection. Resistant colonies were used to start an overnight culture in standard LB broth with continued antibiotic selection, shaking at 200 rpm and 37 °C. The following morning, LB broth supplemented with 50 mM glucose and fresh antibiotic was inoculated with 1:1000 dilution of each respective overnight culture and allowed to grow shaking at 200 rpm and 37 °C until mid-log phase (OD600 0.5–0.8), then induced with 1 mM isopropyl-β-D thiogalactopyranoside (IPTG) for 2 h. Pelleted cells were lysed and protein was purified via IMAC (ref. 25). All proteins were dialyzed and stored at −80 °C in elution buffer.
Design and expression Ca CnAcs2 mutant
To select residues in the 1 binding pocket for mutagenesis, 1-bound CnAcs1 (PDB: 9CD8) was aligned to CaAcs2 (PDB: 8W0B, UniProt: Q8NJN3) using Pymol to identify non-conserved residues. Five mutations were chosen (A334M, G335A, F372Y, V386T, and Q445M), providing a final construct for expression in E. coli as described above. The construct was gene-synthesized and cloned into the expression vector at IDT (Coralville, IA). The protein sequence of the expressed enzyme is below with the N-terminal histidine tag:
MHHHHHHHHENLYFQGPTEQTHNVVHEANGVKLRETPKEFFERQPNKGHIHDVNQYKQMYEQSIKDPQGFFGPLAKELLSWDHDFHTVKSGTLKNGDAAWFLGGELNASYNCVDRHAFANPDKPALICEADDEKDSHILTYGDLLREVSKVAGVLQSWGIKKGDTVAVYLPMNAQAIIAMLAIARLGAAHSVIFAGFSAGSIKDRVNDASCKALITCDEGKRGGRTTNIKKLCDEALVDCPTVEKVLVYKRTNNPEIHLTEGRDYYWDVETAKFPGYLPPVSVNSEDPLFLLYTSGSTGTPKGVVHSTAGYLLGAALSTKYIFDIHPEDILFTMADVGWITGHTYALYGPLLLGVPTIIFEGTPAYPDYGRYWQIVEKHKATHFYTAPTALRLLRKAGEQEIAKYDLSSLRTLGSVGEPISPDIWEWYNEFVGKNQCHISDTYWMTESGSHLIAPLAGVVPNKPGSASYPFFGIDAALIDPVTGVEIEGNDAEGVLAIKDHWPSMARTVYKNHTKYMDTYMNPYPGYYFTGDGAARDHDGYYWIRGRVDDVVNVSGHRLSTAEIEAALIEDKKVSEAAVVGIHDDITGQAVIAYVALKEGNSDEDSEGLRKELVLQVRKTIGPFAAPKSVIIVQDLPKTRSGKIMRRILRKVSSNEADQLGDISTLSNPQSVEGIISAFGAQFGKK.
Enzymological characterization of Acs inhibitors
ACS activity was detected using the reported25 absorption-based coupled kinetic assay as modified from the EnzChek Pyrophosphate Assay Kit (Thermo). Substrate and coupling reagents were either prepared fresh or thawed from small aliquots stored at −80 °C for a maximum of two freeze-thaw cycles. All tested drug concentrations were diluted from stock such that DMSO concentrations did not exceed 5%. All reagents, including the compound and minus the start reagent, were mixed and aliquoted at room temperature, followed by a 15-min incubation at 37 °C. The start reagent, either acetate or aceto-acetate in the case of CnKbc1, was then added, and the reaction was followed continuously at 37 °C in a SpectraMax i3X Multi-Mode plate reader (Molecular Devices) at absorbance 360 nm. Single-point inhibition studies were performed with 50 µM compound, and percent inhibition was normalized to a DMSO control. Dose response curves were generated using a minimum 10-point two-fold drug dilution series using concentrations indicated for each compound tested, along with a DMSO control. Inhibition constants (Ki) were determined by varying one substrate while holding the other substrate pairs in excess across a dilution series of the inhibitor. Substrates were supplied in excess for the respective Ki determination, such that ATP = 2.5 mM, CoA = 1 mM, and acetate = 0.5 mM. The 50% inhibitory concentration (IC50) and Ki values were calculated using the non-linear regression analysis software, Prism (GraphPad). All inhibition studies were performed with a minimum of two experimental duplicates and always with a positive control inhibitor, the competitive inhibitor ethyl-AMP at 50 µM.
For compounds with high background in the kinetic assay, acetyl-CoA synthetase activity was measured using an endpoint FeCl3/hydroxamic acid assay15. For the IC50 determination, enzyme reactions contained sodium acetate (0.5 mM), CoA (1 mM), magnesium chloride (4 mM), ATP (1 mM), DMSO (5%), or inhibitor in DMSO (5%). The buffer for the reaction was potassium phosphate (125 mM) and contained freshly prepared hydroxylamine (200 mM, pH 7), reduced glutathione (10 mM), and potassium fluoride (50 mM). The reactions were carried out at 37 °C for 50 min and quenched by the addition of an aqueous solution of FeCl3 (370 mM) and trichloroacetic acid (3.3%). Absorption at 540 nm of quenched reaction solutions was determined with a SpectraMax i3X, and IC50 values were calculated using non-linear regression analysis with Prism software (GraphPad).
Crystallization conditions and procedures
Purified CnAcs1 was concentrated to 10 mg/mL in 20 mM NaCl, 20 mM Tris, pH 8.5, 1 mM TCEP for crystallization screening. All crystallization experiments were set up using an NT8 drop-setting robot (Formulatrix Inc.) and UVXPO MRC (Molecular Dimensions) sitting drop vapor diffusion plates at 18 °C. One hundred nanoliters of protein and 100 nL crystallization solution were dispensed and equilibrated against 50 µL of the latter. Isoxazole 1-CnAcs1: The complex with isoxazole 1 was prepared by adding the inhibitor, from a 100 mM stock in DMSO, to an aliquot of the protein to a final concentration of 2 mM and incubating for 30 min on ice. Crystals were obtained in 1–2 days from the Index HT screen (Hampton Research) condition F2 (0.2 M Ammonium sulfate, 0.1 M HEPES pH 7.5, 25% (w/v) PEG 3,350). Samples were transferred to a fresh drop composed of 80% crystallization solution and 20% (v/v) PEG 200 and stored in liquid nitrogen. Cyclopropyl-AMP-CnAcs1: 12.5% 8 K, 200 mM NaCl, 100 mM K/Na phosphate pH 6.2. 1 mM ligand was added to the protein prior to crystallization. Samples were transferred to 25% (v/v) PEG 200 + 75% crystallant for cryoprotection. X-ray diffraction data were collected at the National Synchrotron Light Source II (NSLS-II) beamline 19-ID (NYX).
Structure solution and refinement
Intensities were integrated using XDS45 via Autoproc46, and the Laue class analysis and data scaling were performed with Aimless47. Structure solution was conducted by molecular replacement with Phaser48 using a previously determined structure of CnAcs1 (PDB 8EPS) as the search model. Structure refinement and manual model building were conducted with Phenix49 and Coot50, respectively. Torsion angle non-crystallographic symmetry restraints were used during refinement. Additionally, TLS parameters were used in the later stages of refinement to model anisotropic atomic displacement parameters. Structure validation was conducted with Phenix51, and figures were prepared using the CCP4MG package52.
Molecular dynamics calculations
The docked ligand-receptor complex was subjected to MD simulation using the Desmond module of Schrodinger software (Schrodinger, LLC, New York, NY, 2024-1) with OPLS 4 force field53. First, the molecular system of the ligand-receptor complex was built with water molecules in a cubic box via the simple point charge method, which was later followed by ion neutralization by the addition of sodium, to balance the net charge of the solvated system. MD simulation was performed for 1000 ns using the Isothermal-isobaric (NPT) ensemble class, where temperature and pressure were reserved at 300 K and 1.01325 bar pressure employing Nose-Hoover temperature coupling and isotropic scaling. Equilibration of the systems and MD simulations was carried out using the default protocol provided in Desmond53. These simulations produced key results, including RMSD, RMSF, and ligand-protein interaction profiles. MD simulations were run 3 times using the parameters summarized in Supplementary Table 4. The output files for the three simulations have been deposited at FigShare (https://figshare.com/account/items/29896229/).
Molecular docking calculations
Molecular docking of 1 and MMV084978 into Acs enzymes was conducted using the Molecular Operating Environment (www.chemcomp.com, version 2020.01; Chemical Computing Group). The crystal structures of CnAcs1 (PDB: 9CD8) and CaAcs2 (PDB: 8W0B) were used as starting points for in silico mutagenesis. For CnAcs1, point mutations were made in Coot50 to match CaAcs2 as follows: M329A, A330G, Y367F, T384V, and M440Q. Side chains were manually reoriented to best match the starting crystal structure. This structure, now denoted as CncaAcs1, was protonated to pH=7 using ProPKa, and energy minimized in the presence of 1 within the Molecular Operating Environment using the Amber10:EHT force field. The binding energy of 1 was determined via the GBVI/WSA dG function without variable ligand placement to keep the ligand conformation as close to the crystal structure as possible. For MMV0894978, binding energy was determined following ligand placement (defined by selecting 1 as the binding site) and refinement using the London dG scoring function, followed by rescoring using the GBVI/WSA dG function. The top-scoring orientation was used for further structural analysis. The same protocol was applied in the creation of CaCnAcs2, where mutations were introduced as follows: A334M, G335A, F372Y, V386T, and Q445M. 1 was modeled into the binding site in COOT via structural alignment with 9CD8 (Chain A), and the structure was subsequently energy minimized prior to docking.
Statistical methods
Distinct samples were measured. Statistical analysis and curve fitting were performed using Prism (GraphPad) using linear regression analysis, parametric analysis, and a two-sided, unpaired Student t-test.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data are provided in the figures, tables, and Supplementary Information sections of the manuscript. The source data files for the data are also provided in the Source data File. Whole-genome sequencing data have been deposited at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under accession number PRJNA1190938. Coordinates and structure factors for CnAcs1 inhibitor complexes have been deposited to the Worldwide Protein Databank (wwPDB) with the accession codes 9CD8 (isoxazole 1) and 8G0T (cyclopropyl-AMP). The molecular dynamics simulations have been deposited at figshare.com (https://doi.org/10.6084/m9.figshare.29896229.v1). Source data are provided with this paper.
References
Denning, D. W. Global incidence and mortality of severe fungal disease. Lancet Infect. Dis. 24, e428–e438 (2024).
Vallabhaneni, S., Mody, R. K., Walker, T. & Chiller, T. The global burden of fungal diseases. Infect. Dis. Clin. N. Am. 30, 1–11 (2016).
Perfect, J. R. The antifungal pipeline: a reality check. Nat. Rev. Drug Discov. 16, 603–616 (2017).
Shiri, T. et al. Addition of flucytosine to fluconazole for the treatment of cryptococcal meningitis in Africa: a multi-country cost-effectiveness analysis. Clin. Infect. Dis. 70, 26–29 (2020).
Masone, M. C. Ibrexafungerp to treat vulvovaginal candidiasis. Nat. Rev. Urol. 18, 638 (2021).
Lockhart, S. R., Chowdhary, A. & Gold, J. A. W. The rapid emergence of antifungal-resistant human-pathogenic fungi. Nat. Rev. Microbiol 21, 818–832 (2023).
Vazquez, J. A. et al. Clinical efficacy and safety of a novel antifungal, fosmanogepix, in patients with candidemia caused by Candida auris: results from a Phase 2 trial. Antimicrob. Agents Chemother. 67, e0141922 (2023).
Wiederhold, N. P. Review of the novel investigational antifungal olorofim. J. Fungi 6, 122 (2020).
van Rhijn, N. et al. Aspergillus fumigatus strains that evolve resistance to the agrochemical fungicide ipflufenoquin are also resistant to olorofim. Nat. Microbiol. 9, 29–34 (2024).
Gulick, A. M. Conformational dynamics in the acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem. Biol. 4, 811–827 (2009).
Comerford, S. A. et al. Acetate dependence of tumors. Cell 159, 1591–1602 (2014).
Orsó, E. & Burkhardt, R. ATP-citrate lyase: a driver of metabolism and histone acetylation. Curr. Opin. Lipidol. 31, 362–363 (2020).
Huang, Z. et al. ACSS2 promotes systemic fat storage and utilization through selective regulation of genes in lipid metabolism. Proc. Natl. Acad. Sci. USA 115, e9499–e9506 (2018).
Schug, Z. T., Vande Voorde, J. & Gottlieb, E. The metabolic fate of acetate in cancer. Nat. Rev. Cancer 16, 708–717 (2016).
Koselny, K. et al. Antitumor/antifungal celecoxib derivative AR-12 is a non-nucleoside inhibitor of the ANL-family adenylating enzyme acetyl-CoA synthetase. ACS Infect. Dis. 2, 268–280 (2016).
Koselny, K. et al. The celecoxib derivative AR-12 has broad spectrum antifungal activity in vitro and improves the activity of fluconazole in a murine model of cryptococcosis. Antimicrob. Agents Chemother. 60, 7115–7127 (2016).
Summers, R. L. et al. Chemogenomics identifies acetyl-coenzyme A synthetase as a target for malaria treatment and prevention. Cell Chem. Biol. 29, 191–201.e8 (2022).
de Vries, L. E. et al. Preclinical characterization and target validation of the antimalarial MMV69318. Nat. Commun. 13, 2158 (2022).
Abt, E. R. et al. Metabolic modifier screen reveals secondary target of the protein kinase inhibitors within nucleotide metabolism. Cell Chem. Biol. 27, 197–205 (2020).
Carman, A. J., Vylkova, S. & Lorenz, M. C. Role of acetyl coenzyme A synthesis and breakdown in alternative carbon source utilization in Candida albicans. Eukaryot. Cell 7, 1733–1741 (2008).
Segal, E. S. et al. Gene essentiality analyzed by in vivo transposon mutagenesis and machine learning in a stable haploid isolate of Candida albicans. mBio 9, e02048–18 (2018).
Gale, A. N. et al. Identification of essential genes and fluconazole susceptibility genes in Candida glabrata. G3 10, 3859–3870 (2020).
Alden, K. M. et al. Genetic interaction analysis reveals that Cryptococcus neoformans utilizes multiple acetyl-CoA generating pathways during infection. mBio 13, e0127922 (2022).
Hu, G., Cheng, P. Y., Sham, A., Perfect, J. R. & Kronstad, J. W. Metabolic adaptation in Cryptococcus neoformans during early murine pulmonary infection. Mol. Microbiol. 69, 1456–1475 (2008).
Jezewski, A. J. et al. Structural characterization of the reaction and substrate specificity mechanism of pathogenic fungal acetyl-CoA synthetases. ACS Chem. Biol. 16, 1587–1599 (2021).
Wiederhold, N. P. Antifungal susceptibility testing of yeast and filamentous fungi by CLSI broth microdilution testing. Methods Mol. Biol. 2658, 3–16 (2023).
Reuwsaat, J. C. V. et al. The transcription factor Pdr802 regulates titan cell formation and pathogenicity of Cryptococcus neoformans. mBio 12, e03457-20 (2021).
Iyer, K. R., Revie, N. M., Fu, C., Robbins, N. & Cowen, L. E. Treatment strategies for cryptococcal infection: challenges, advances, and future outlook. Nat. Rev. Microbiol. 19, 454–466 (2021).
Martinez Calejman, C. et al. mTORC2-AKT signaling to ATP-citrate lyase drives brown adipogenesis and de novo lipogenesis. Nat. Commun. 11, 575 (2020).
Shaw, K. J. et al. In vitro and in vivo evaluation of APX001A/APX001 and other Gwt1 inhibitors against Cryptococcus. Antimicrob. Agents Chemother. 62, e00523-18 (2018).
Bian, C. et al. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J. 30, 2829–2842 (2011).
Arras, S. D. M. et al. Convergent microevolution of Cryptococcus neoformans hypervirulence in the laboratory and the clinic. Sci. Rep. 7, 17918 (2017).
Ristow, L. C. et al. Cryptococcus neoformans adapts to the host environment through TOR-mediated remodeling of phospholipid asymmetry. Nat. Commun. 14, 6587 (2023).
Miller, K. D. et al. Targeting ACSS2 with a transition-state mimetic inhibits triple-negative breast cancer growth. Cancer Res. 81, 1252–1264 (2021).
Sabnis, R. W. Novel substituted tetrazoles as ACSS2 inhibitors for treating cancer. ACS Med Chem. Lett. 12, 1894–1895 (2021).
Copeland, R. A. in Evaluation of Enzyme Inhibitors in Drug Discovery 2nd edn, Ch. 3a (Wiley, 2013).
Pesaresi, A. Mixed and non-competitive enzyme inhibition: underlying mechanisms and mechanistic irrelevance of the formal two-site model. J. Enzym. Inhib. Med. Chem. 38, 2245168 (2023).
Jogl, G. & Tong, L. Crystal structure of yeast acetyl-coenzyme A synthetase in complex with AMP. Biochemistry 43, 1425–1431 (2004).
Miller, K. D. et al. Acetate acts as a metabolic immunomodulator by bolstering T-cell effector function and potentiating antitumor immunity if breast cancer. Nat. Cancer 4, 1491–1507 (2023).
Jezewski, A. J. et al. A single Leishmania adenylate-forming enzyme of the ANL superfamily generates both acetyl- and acetoacetyl-CoA. J. Biol. Chem. 300, 107879 (2024).
Lin, Y. et al. ACSS2-dependent histone acetylation improves cognition in mouse models of Alzheimer’s disease. Mol. Neurodegener. 18, 47 (2023).
Knight, Z. A. & Shokat, K. M. Chemical genetics: where genetics and pharmacology meet. Cell 128, 425–430 (2007).
Hope, W. et al. Fluconazole monotherapy is a suboptimal option for initial treatment of cryptococcal meningitis because of emergence of resistance. mBio 10, e02575-19 (2019).
McNaney, C. A. et al. An automated liquid chromatography-mass spectrometry process to determine metabolic stability half-life and intrinsic clearance of drug candidates by substrate depletion. Assay. Drug Dev. Technol. 6, 121–129 (2008).
Kabsch, W. Automatic indexing of rotation diffraction patterns. J. Appl. Crystallogr. 21, 67–72 (1988).
Vonrhein, C. et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. Sect. D Biol. Crystallogr. 67, 293–302 (2011).
Evans, P. R. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. Sect. D Biol. Crystallogr. 67, 282–292 (2011).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 486–502 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Potterton, L. et al. Developments in the CCP4 molecular-graphics project. Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 (2004).
Bowers, K. J. et al. Scalable algorithms for molecular dynamics simulations on commodity clusters. Proceedings of the ACM/IEEE Conference on Supercomputing (SC06) 11–17 (Tampa, Florida, 2006).
Acknowledgements
This work was supported by NIH grants 5R01AI161973 (D.J.K.) and T32AI007511 (A.J.J. and J.P.) This project has been funded in whole or in part by Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health under Contract No. 75N93022C00036. This research used resources at the NYX beamline 19-ID, supported by the New York Structural Biology Center, at the National Synchrotron Light Source II, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under Contract No. DE-SC0012704. The NYX detector instrumentation was supported by grant S10OD030394 through the Office of the Director of the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The purchase of the NMR spectrometer used to obtain results included in this publication was supported by the National Science Foundation under the MRI award CHE-2117776 (T.J.H). The HRMS was supported by Northern Illinois University Molecular Analysis Core Facility (RRID:SCR 024586), which was established as a partnership with Shimadzu Scientific Instruments. Purchase of the Bruker Maxis Plus QTOF was made possible by the National Science Foundation under Grant No. 1726931.
Author information
Authors and Affiliations
Contributions
Data collection: A.J.J., K.M.A., J.P., D.G.D., C.L.L., M.E.H., A.J.F., J.C.F., L.L., K.P.B. Data analysis: A.J.J., K.M.A., J.P., D.G.D., C.L.L., M.E.H., A.J.F., J.C.F., L.L., K.P.B., N.S.W., B.L.S., S.L., T.J.H., D.J.K. Conceptualization: A.J.J., K.M.A., S.L., T.J.H., D.J.K. Writing: A.J.J., J.P., D.G.D., N.S.W., S.L., T.J.H., D.J.K. Supervision: N.S.W., B.L.S., S.L., T.J.H., D.J.K. Resources: A.J.J., J.P., B.L.S., S.L., T.J.H., D.J.K.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Iwao Ojima and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jezewski, A.J., Alden, K.M., Propp, J. et al. Discovery and mechanism of a highly selective, antifungal acetyl-CoA synthetase inhibitor. Nat Commun 16, 9118 (2025). https://doi.org/10.1038/s41467-025-64183-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-64183-7