Abstract
The synaptonemal complex (SC) forms between homologous chromosomes during meiosis. In Arabidopsis thaliana, its central region (CR) is composed of the transverse filament protein ZYP1 and the central element proteins SCEP1 and SCEP2. Here we identify SCEP3 as a CR protein that is evolutionarily conserved across plant species. SCEP3 spatiotemporally overlaps with other CR proteins and localizes to the SC CR. The loss of SCEP3 prevents SC assembly, abolishes crossover (CO) assurance and interference, and eliminates sex-specific differences in CO rates (heterochiasmy) through increased CO in females. SCEP3 is required for a subset of COs in SC-deficient mutants, such as zyp1. Although SCEP3 physically interacts with ZYP1, it loads independently of other CR proteins. We propose that SCEP3 may associate with certain recombination intermediates, stabilizing them and/or recruiting additional factors, such as ZYP1, to a subset of these intermediates, thereby promoting and interlinking SC assembly and CO formation.
Similar content being viewed by others
Main
Meiotic homologous recombination assures genetic diversity in gametes1. The repair of programmed DNA double-strand breaks (DSBs) into interhomologue crossovers (COs) involves numerous proteins and several consecutive steps2,3. In most species, COs are divided into two classes. Class I COs, promoted by ZMM proteins (Zip1-4, Mer3 and Msh4/5)4,5 and MutL-γ (Mlh1/3)6, are interference-sensitive (one CO limits the probability of other COs nearby)7. Class II COs are insensitive to interference and form a minority in most species, including Arabidopsis thaliana, where their formation depends in part on MUS81 (refs. 8,9).
Meiotic recombination occurs during prophase I. Sister chromatids are initially organized into a linear loop–base array by a proteinaceous structure called the meiotic chromosome axis1. In Arabidopsis, the axis consists of ASY1 (refs. 10,11), ASY3 (ref. 12), ASY4 (ref. 13) and cohesion-associated proteins including REC8 (ref. 14). Initial repair of a large number of DSBs (~200 in Arabidopsis) leads to numerous meiotic recombination intermediates, including early interhomologue associations15. Upon installation of the transverse filament protein ZYP1, aligned chromosomes become physically connected at ~200 nm (ref. 16), leading to the formation of the tripartite synaptonemal complex (SC)1. In Arabidopsis, homologue alignment/pairing, albeit at a larger distance at ~300–400 nm, is found even in the absence of the SC16,17,18.
The SC structure, two lateral elements flanking a central region (CR), is conserved across species and regulates the number and distribution of COs1. Across species, the CR is composed of transverse filament proteins (such as Zip1 in Saccharomyces cerevisiae19, C(3)G in Drosophila melanogaster20, SYP-1/5/6 in Caenorhabditis elegans21,22,23, SYCP1 in Mus musculus24 and ZYP1 (ZYP1a/b) in A. thaliana16,17,25) and central element (CE) proteins (such as Ecm11 and Gmc2 in budding yeast26; Corona and Corolla in Drosophila27,28; SYP-2/3/4 and SKR-1/2 in worms29,30,31,32; SYCE1/2/3, TEX12 and SIX6OS1 in mice33,34,35,36,37; and SCEP1/2 in Arabidopsis18).
In most species, including flies, mice, worms and Sordaria, the absence of the SC impairs CO formation15. In plants, the SC is dispensable for CO formation in Arabidopsis and rice16,17,18,38 but probably required in barley39. Studies of Arabidopsis CR mutants suggest that the SC is critical for CO assurance, heterochiasmy and CO interference16,17,18,40. On the basis of Arabidopsis data, the coarsening of the ZMM protein HEI10 in the frame of the SC was proposed as the basis for CO interference40,41. However, the SC is not required for implementing CO interference in budding yeast or Sordaria1.
Interactions among CR proteins are linked to SC assembly—for example, in budding yeast, Ecm11–Gmc2 promotes Zip1 polymerization26, and in mice, SYCP1 tetramers remodelled by SYCE3 form a SYCP1–SYCE3 complex, and SYCE3 also interacts with TEX12–SYCE2 and SYCE1–SIX6OS1 (ref. 42). In budding yeast, SC assembly and CO formation are coupled via the interaction of Ecm11–Zip4 (in mouse via orthologues TEX12–TEX11)43. In worms, CO formation is regulated by SYP-4’s carboxy terminus, possibly through the recruitment of ZHP-3 (an orthologue of Arabidopsis HEI10) to the SC44. In Arabidopsis, SCEP1 and SCEP2 directly interact, while neither interacts with ZYP1 or ZIP4 (ref. 18), and HEI10 coarsening in the SC may underlie CO interference40,41. It is unclear whether further CE proteins exist in Arabidopsis that form a complex with ZYP1 and/or interconnect SC assembly and CO formation.
We previously identified proteins in proximity to ASY1 and ASY3 via proximity proteomics45. Here we functionally characterize the candidate ATC21, renamed SCEP3. SCEP3 is a CE protein that is conserved in plants and is probably the structural orthologue of SYP-4 in worms and SIX6OS1 in mammals31,36, suggesting conservation across kingdoms.
Results
SCEP3 is required for synapsis and obligate CO formation
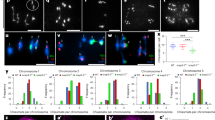
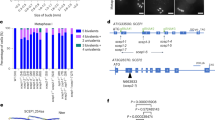
SCEP3 (AT4G18490) has 16 exons, encodes a protein of 803 amino acids (Fig. 1a) and is highly expressed in young flower buds46. According to structural prediction by AlphaFold2 (ref. 47), the last 70 amino acids at the C terminus form an α-helical domain, while the remaining 733 amino acids are disordered (Extended Data Fig. 1a). No obvious developmental differences were observed in any scep3 mutant (scep3-1, scep3-2, scep3-3 and scep3-1 scep3-2; Fig. 1a and Extended Data Fig. 1b) compared to the wild type (WT), except for slightly shorter siliques with seed gaps (Extended Data Fig. 1c), reduced seed fertility (Fig. 1b) and decreased pollen viability (Extended Data Fig. 1d). In the WT, chromosomes are fully synapsed at pachytene, five bivalents are invariably observed at metaphase I and chromosome segregation is balanced (Fig. 1c,d). In scep3, pachytene chromosomes appeared as pairs of parallel threads, indicating the absence of synapsis despite chromosome alignment/pairing (Fig. 1c), and ~50% of cells displayed one to three pairs of univalents (failure to form the obligate CO), leading to unequal chromosome segregation (Fig. 1c,d). Moreover, minimum chiasmata numbers (rod- and ring-shaped bivalents are scored as one and two chiasmata, respectively) in scep3 are reduced to ~70–75% of WT levels (Supplementary Table 1). Three-dimensional structural illumination microscopy (3D-SIM) inter-axis measurements confirmed that in scep3 chromosomes align/pair, albeit at a greater and more variable distance than in the WT (Fig. 1e). As in males, during scep3 female meiosis, achiasmatic and asynaptic yet aligned/paired chromosomes were found (Fig. 1e,f).
a, Gene model of SCEP3 (AT4G18490; confirmed by Sanger sequencing of flower bud complementary DNA), including exons (black boxes) and introns (black lines), and a schematic depiction of the SCEP3 protein. The locations of mutant alleles are indicated: scep3-1 (initially named atc21-1 (ref. 44)) and scep3-2 (transfer DNA insertions within exon 13 and intron 8, respectively) as well as scep3-3 and scep3-4 (CRISPR–Cas9-based mutagenesis); scep3-4 is in Ler-0, while all other alleles are in the Col-0 background. b, Seeds per silique in the WT (54.33 ± 5.38, n = 40), scep3-1 (37.98 ± 5.76, n = 40), scep3-2 (37.9 ± 4.75, n = 40), scep3-3 (38.05 ± 4.44, n = 40) and scep3-1 scep3-2 (36.33 ± 3.59, n = 40). No significant differences were found among the scep3 mutants (P = 0.30). However, all scep3 mutants produced significantly fewer seeds than the WT (P < 1 × 10−7). c, Male meiotic chromosome behaviour (scale bars, 10 μm; DNA counterstained with DAPI is shown in grey) in the WT and scep3 mutants. d, Frequency of cells with zero to three pairs of univalents, including the average bivalent number per cell (n is the number of cells analysed) in the WT and scep3 mutants. e, Left, 3D-SIM analysis of REC8 immunolocalization in male and female meiocytes of Col-0 and scep3-1. Along synapsed chromosomes, the two parallel lateral elements exhibit an average distance of 188 ± 21.6 nm (range, 148–243 nm; n = 26) in WT males and 187 ± 19.4 nm (range, 162–223 nm; n = 15) in WT females. In scep3-1, within regions of alignment, the average distance increased to 352 ± 87.5 nm (range, 189–592 nm; n = 37) in males and 311 ± 74.6 nm (range, 184–423 nm; n = 12) in females, showing greater variation. Single-slice images were used to measure the distances between two aligned axes. Scale bars, 2 μm. Right, quantification of aligned axis distances (the measurement points were randomly selected) in both Col-0 and scep3-1. ASY4-labelled lateral elements exhibit an average distance of 176 ± 13.4 nm (range, 149–197 nm; n = 28) in WT males. f, Female meiotic chromosome spreads of Col-0 and scep3-2 (DAPI-stained DNA is shown in grey). The experiments were repeated three times with similar results. Scale bars, 10 μm. Distinct plants from each mutant line (or the WT) were used for seed counting and chromosome spread analysis. Significance was evaluated using one-way ANOVA with post hoc Bonferroni multiple comparison. The data are presented as mean ± s.d. **P < 0.01. NS, not significant.
SCEP3 is found at the CR of the SC
To dissect the spatiotemporal localization of SCEP3, antibodies raised against its amino (SCEP3-N) or C terminus (SCEP3-C) were employed. Using SCEP3-N, SCEP3 is initially detected during early zygotene forming a limited number of foci (Fig. 2a). The foci numbers increase progressively, with initial SCEP3 stretches during zygotene and full polymerization at pachytene. During all stages, SCEP3 colocalizes with ZYP1 (Fig. 2a). SCEP3-C overlaps with SCEP3-N during zygotene/pachytene, albeit less continuously, leading to a patchier signal (Fig. 2b), as also revealed by both 3D-SIM and stimulated emission depletion (STED) microscopy of pachytene nuclei (Extended Data Fig. 2a). In any case, using SCEP3-C and SCEP3-N, SCEP3 also colocalizes with the CE proteins SCEP1 and SCEP2 at the SC (Fig. 2c). The absence of SCEP3-N and SCEP3-C signals in scep3 confirms the specificity of both antibodies (Extended Data Fig. 2b).
a–c, Immunolocalization in the WT of SCEP3-N, ZYP1-C and ASY1 during prophase I (a); SCEP3-C and SCEP3-N during zygotene and pachytene (b); and SCEP3-C with SCEP2 or SCEP3-N with SCEP1 at pachytene (c). DAPI-stained DNA is shown in grey. d,e, 3D-SIM of a pachytene nucleus in the WT immunolabelled with REC8, ZYP1-C and SCEP3-N (d) or ASY4, ZYP1-C and SCEP3-C (e). f,g, STED microscopy of a WT pachytene nucleus immunolabelled with ZYP1-C and SCEP3-N (f) or ZYP1-C and SCEP3-C (g). Scale bars, 10 μm in a–c, 2 μm in d–g. All experiments were repeated at least two times with similar results.
Using 3D-SIM, the following SC organization was detected at pachytene: REC8 and ASY4 each form two parallel lines separated by approximately 188 and 176 nm, respectively (Figs. 1e and 2d,e). Between these lines, ZYP1-C (its C termini localize towards the chromosome axis16) appears as either two distinct lines or a single bright line. Both SCEP3-N and SCEP3-C are centrally located within the single or two ZYP1-C lines (Fig. 2d,e). A similar Arabidopsis SC organization—that is, REC8 axes at a distance of 175–213 nm, two lines of ZYP1-C in between and between those centrally located CE proteins SCEP1/2—was found using STED microscopy16,18. To further investigate whether SCEP3 is indeed a CE protein within the SC CR, STED microscopy was performed. Similar to the two known CEs SCEP1/2 (ref. 18), both SCEP3-N and SCEP3-C were located centrally between the two ZYP1-C lines (Fig. 2f,g). Together, SCEP3’s localization to the SC CR supports its role as a CE protein. Hereafter, unless otherwise specified, SCEP3-N was employed.
SCEP3 is crucial for SC assembly and loads independently of the SC
In the WT, all CR proteins fully polymerize at pachytene, while ASY1 gets depleted from synapsed regions (Fig. 3a–d). In scep3-1 pachytene(-like) nuclei, no chromosome-associated ZYP1, SCEP1 or SCEP2 foci are detected (Fig. 3a–c), indicating the absence of SC assembly in scep3-1 and suggesting that chromosomal loading of ZYP1, SCEP1 and SCEP2 is SCEP3-dependent. The absence of synapsis is further reflected by the persistence of ASY1 at pachytene(-like) stages16,18 (Fig. 3a–c). In zyp1-2, scep1-1 and scep2-1 pachytene(-like) nuclei, axis-associated SCEP3 foci are found (Fig. 3d), indicating that SCEP3 loads independently of SC formation and of other CR proteins. Additionally, ZYP1 colocalizes with SCEP3 in scep1-1 and scep2-1 early pachytene(-like) nuclei (Fig. 3e), whereas neither SCEP1 nor SCEP2 is detected in zyp1-2 (Fig. 3f). This suggests that ZYP1 localization depends on SCEP3, but SCEP3 alone is not sufficient for SCEP1/SCEP2 localization. Together, while all CR proteins are required for SC assembly, SCEP3 loads independently of other CR components and of SC formation and recruits ZYP1, but SCEP3 is not sufficient for SCEP1/SCEP2 localization.
a–c, Immunolocalization in WT and scep3-1 pachytene nuclei of ZYP1-C and ASY1 (a), SCEP1 and ASY1 (b), and SCEP2 and ASY1 (c). d–f, Immunolocalization of SCEP3-N and ASY1 in the WT, zyp1-2, scep1-1 and scep2-1 (d); SCEP3-N and ZYP1-C in scep1-1 and scep2-1 (e); and SCEP1 or SCEP2 and ASY1 in zyp1-2 (f). DAPI-stained DNA is shown in grey. All experiments were repeated three times with similar results. Scale bars, 10 µm.
SCEP3 and its interaction with ZYP1 are conserved in plants
Given that SCEP3 and ZYP1 colocalize even in CE mutants, we investigated their relationship in mutants that form an SC despite impaired ZMM-dependent CO formation (msh5-2, hei10-2, zip4-2, mer3-1 and shoc1-1) or exhibit varying degrees of SC formation defects (spo11-2-3, mtopVIB-2, dmc1-2, asy3-1, rec8-1, pch2-1 and asy1-4). In all cases, SCEP3 and ZYP1 colocalization was found (Extended Data Fig. 3), either as short stretches or fully polymerized within partial or intact SCs, or as foci/protein aggregates in the absence of SC formation. We then tested whether this colocalization reflects a direct interaction. In yeast two-hybrid (Y2H) assays, a strong interaction of SCEP3–ZYP1 (both ZYP1a and ZYP1b) was found, mediated by the N terminus of ZYP1 (ZYP1b, amino acids 49–400) and the C terminus of SCEP3 (amino acids 734–803) (Fig. 4a,b). AlphaFold3 also predicts an interaction between the C-terminal α-helical domain of SCEP3 and the N-terminal region of ZYP1 (Fig. 4c).
a, Schematic depictions of ZYP1b (left) and SCEP3 (right), including fragments tested by Y2H assays. In ZYP1b, the central α-helical domain (orange) is flanked by two flexible regions (pink and green). SCEP3 consists of the C-terminal α-helical domain (blue) and the N-terminal disordered region (red). Green, grey and black lines indicate positive interaction, negative interaction and self-activation, respectively, in the Y2H experiments depicted below. b, Y2H interaction studies of ZYP1 and SCEP3. Both ZYP1a and ZYP1b interact with SCEP3 (full-length proteins). To determine the subregion within ZYP1 and SCEP3 responsible for their interaction, ZYP1 (ZYP1b was used) was divided into four fragments and SCEP3 into two. The N-terminal region of ZYP1 (amino acids 49–400) and the C-terminal region of SCEP3 (amino acids 734–803) were found responsible for the interaction between SCEP3 and ZYP1. Selection with minimal SD Base medium supplemented with DO Supplement −Leu/−Trp (SD/−LT (DDO)); DO Supplement −His/−Leu/−Trp (SD/−LT (TDO)); or DO Supplement −Ade/−His/−Leu/−Trp (SD/−LT (QDO)). c, AlphaFold3 predicts interaction between the C terminus of SCEP3 and full-length ZYP1b (~100–200 amino acids) in Arabidopsis. Predicted aligned error values are shown on the right; the interface predicted template modelling score is 0.48, and the predicted template modelling score is 0.24. Note that AlphaFold’s prediction of the global structure of ZYP1 is not consistent with cytological evidence that the N terminus and C terminus are apart, as previously described18.
Using PSI-BLAST, SCEP3 was found to be conserved across green plants, including many lower plants, but no homologues were identified outside Streptophyta (Extended Data Fig. 4). High sequence similarity was found at both the N- and C-terminal regions of SCEP3 (Supplementary Fig. 1). In most plants, homologues of all four CR components are present; however, in lower plants such as Taxus chinensis and Marchantia polymorpha, homologues of SCEP1/2 (but not SCEP3 and ZYP1) are absent (Extended Data Fig. 4). The close evolutionarily relationship between SCEP3 and ZYP1 may be linked to their direct interaction detected in A. thaliana. In Y2H assays, full-length barley SCEP3 and ZYP1 interact (Extended Data Fig. 5a), and AlphaFold3 predicts a conserved SCEP3–ZYP1 interaction across various plants, involving similar regions as in Arabidopsis (Extended Data Fig. 5b). Hence, a SCEP3–ZYP1 interface seems highly conserved across plants.
SCEP3 promotes a subset of COs upon impaired SC formation
Using the number of γH2AX foci as a proxy for the number of DSB sites48,49, we found no difference between scep3-2 and the WT (Extended Data Fig. 6a,b). Hence, the shortage of chiasmata in scep3 is probably not due to reduced DSB numbers.
Chiasma counts often underestimate CO numbers, as closely spaced COs cannot be microscopically resolved16,50. HEI10 foci undergo a dynamic reduction in number, accompanied by an increase in intensity and size, throughout prophase I. In late pachytene and diakinesis, bright HEI10 foci colocalize with CO-designated sites marked by MLH1 (ref. 51). We therefore quantified HEI10 foci as a proxy for class I COs (foci signal is absent in hei10-2; Extended Data Fig. 6c). Compared with other CR mutants, which had increased HEI10 foci numbers (13.18 ± 3.28 in zyp1, 13.94 ± 3.54 in scep1 and 14.59 ± 2.29 in scep2; Fig. 5a,b), scep3-1 displayed HEI10 foci counts (9.94 ± 2.43; P = 1, one-way analysis of variance (ANOVA) with post hoc Bonferroni multiple comparison) similar to those of the WT (10.34 ± 1.74) (Fig. 5a,b). Thus, in scep3, the number of class I COs appears unchanged. However, CO assurance is probably lost due to their random distribution across the genome; other CR mutants also exhibit CO assurance loss, even despite increased HEI10 foci numbers16,17,18. To address whether SCEP3 is required for the surplus HEI10-dependent COs found in zyp1, we generated scep3-1 zyp1-2 plants. In scep3-1 zyp1-2, the minimum chiasmata number was significantly reduced to 5.62 ± 1.91 compared with 7.69 ± 1.54 in zyp1-2 (P < 0.01, one-way ANOVA with post hoc Bonferroni multiple comparison) but was not significantly different from 5.93 ± 1.56 in scep3-1 (P = 0.71) (Extended Data Fig. 7 and Supplementary Table 1). Univalent frequency in scep3-1 zyp1-2 (58.7%) was similar to that in scep3-1 (55.3%) but higher than that in zyp1-2 (14.3%) (Fig. 5c). Additionally, HEI10 foci numbers in scep3-1 zyp1-2 (10.03 ± 2.80) were similar to those in scep3-1 (P = 1, one-way ANOVA with post hoc Bonferroni multiple comparison) but significantly lower than in zyp1-2 (13.18 ± 3.28; P < 0.01, one-way ANOVA with post hoc Bonferroni multiple comparison) (Fig. 5a,b). Hence, SCEP3 acts genetically upstream of ZYP1 in CO formation and is essential for the surplus HEI10-dependent COs observed in zyp1. In asy1 and asy3, with impaired SC formation and genetically upstream of ZYP1 (ref. 17), the depletion of SCEP3 further reduces bivalent/chiasma numbers (Fig. 5c, Extended Data Fig. 7 and Supplementary Table 1). SCEP3 is thus critical for a subset of COs that arise upon impaired SC formation.
a,b, Immunolocalization of ASY1 and HEI10 (a) and quantification of HEI10 foci number per diakinesis cell (b) in WT (10.34 ± 1.74, n = 41), scep3-1 (9.94 ± 2.43, n = 51), zyp1-2 (13.18 ± 2.94, n = 66), scep3-1 zyp1-2 (10.03 ± 2.43, n = 59), scep1-1 (13.94 ± 3.62, 18) and scep2-1 (14.59 ± 2.29, n = 17) male meiocytes, as well as WT (5.91 ± 1.86, n = 23) and scep3-1 (10.17 ± 3.31, n = 6) female meiocytes. Significant differences were found between scep3-1 and zyp1-2 (P < 1 × 10−7) as well as zyp1-2 and scep3-1 zyp1-2 (P < 1 × 10−7), but not between scep3-1 and the WT (P = 1) or scep3-1 and scep3-1 zyp1-2 (P = 1). DNA counterstained with DAPI is shown in merged and single-channel images in blue and grey, respectively. Scale bars, 10 μm. c, Frequency of cells with zero to five pairs of univalents including the average bivalent number per cell in a series of single or double mutants. Significant differences were found between zyp1-2 and scep3-1 zyp1-2 (P = 2.32 × 10−7), asy1-4 and scep3-2 asy1-4 (P < 1 × 10−7), scep3-2 and scep3-2 asy1-4 (P < 1 × 10−7), asy3-1 and scep3-2 asy3-1 (P = 3.26 × 10−6), scep3-2 and scep3-2 asy3-1 (P < 1 × 10−7), msh5-2 and scep3-1 msh5-2 (P = 0.016), scep3-1 and scep3-1 msh5-2 (P < 1 × 10−7), hei10-2 and scep3-2 hei10-2 (P = 4.56 × 10−7), scep3-2 and scep3-2 hei10-2 (P < 1 × 10−7), scep3-2 and scep3-2 mlh3-1 (P < 1 × 10−7), mlh3-1 and scep3-2 mlh3-1 (P < 1 × 10−7), and mus81-2 and scep3-1 mus81-2 (P < 1 × 10−7), but not between scep3-1 and scep3-1 zyp1-2 (P = 0.41) or scep3-1 and scep3-1 mus81-2 (P = 0.31). Distinct plants from each single or double mutant line (or the WT) were used for immunolocalization and chromosome spread analysis. Significance was assessed using one-way ANOVA with post hoc Bonferroni multiple comparison. The data are presented as mean ± s.d. *P < 0.05; **P < 0.01.
SCEP3 associates with HEI10 even independent of SC formation
Since SCEP3 loads and is required for the surplus HEI10-dependent COs in CR mutants (Figs. 3d and 5a,b), we asked whether SCEP3 colocalizes with HEI10. In the WT, during zygotene/early pachytene, discernible SCEP3 foci including those along SCEP3 stretches are closely associated with HEI10 foci, with 78% of HEI10 foci overlapping with SCEP3 foci and 72% of SCEP3 foci overlapping with HEI10 foci (Fig. 6a,b); however, as synapsis progresses, SCEP3 foci gradually transition into continuous linear signals decorated by bright HEI10 foci (Fig. 6c). In zyp1-2, scep1-1 and scep2-1, similar rates of association of HEI10 foci with SCEP3 foci (75–79%) and of SCEP3 foci with HEI10 foci (74–77%) are found in early pachytene(-like) nuclei (with ~100 HEI10 foci) (Fig. 6a,b). In late zyp1-2, scep1-1 and scep2-1 pachytene(-like) nuclei (with ~20 HEI10 foci), although the total numbers of both HEI10 and SCEP3 foci decrease, over 70% of prominent HEI10 foci remain associated with bright SCEP3 foci (Fig. 6b,c). By diplotene/diakinesis, bright HEI10 foci persist at reduced numbers, while bright SCEP3 foci are scarcely detectable (Figs. 5b and 6d). Together, these data suggest an association of SCEP3 and HEI10 throughout pachytene and an enrichment of SCEP3 at putative HEI10-marked CO sites in CR mutants. This association may be required for the surplus HEI10-dependent COs found in these mutants.
a, Immunolocalization of SCEP3-N and HEI10 in early pachytene nuclei of the WT, zyp1-2, scep1-1 and scep2-1. b, Quantification of SCEP3 and HEI10 foci numbers and their percentage of overlap in WT (HEI10, 83 ± 8.1; SCEP3, 94 ± 29.9; n = 7), zyp1-2 (HEI10, 88 ± 13.3; SCEP3, 95 ± 13.4; n = 10), scep1-1 (HEI10, 93 ± 24.2; SCEP3, 92 ± 23.9; n = 9) and scep2-1 (HEI10, 95 ± 16.7; SCEP3, 99 ± 22.1; n = 5) early pachytene (EP), as well as zyp1-2 (HEI10, 21 ± 3.4; SCEP3, 21 ± 0.9; n = 5), scep1-1 (HEI10, 20 ± 2.5; SCEP3, 20 ± 3.5; n = 3) and scep2-1 (HEI10, 24 ± 3.5; SCEP3, 21 ± 2.2; n = 4) late pachytene (LP). c,d, Same as a, but in late pachytene and diplotene nuclei, respectively. Distinct plants from each mutant line (or the WT) were used for immunolocalization analysis. DAPI-stained DNA is shown in grey. Scale bars, 10 µm. The data are presented as mean ± s.d.
In scep3-1, the number of HEI10 foci is similar to that observed in the WT and other CR mutants in early pachytene(-like) nuclei (Fig. 6a,b and Extended Data Fig. 8a–d), suggesting that early HEI10 localization is independent of all CR components. Additionally, ZYP1, SCEP1 and SCEP2 are absent in scep3-1 (Extended Data Fig. 8a–c). It is therefore tempting to speculate that SCEP3 may associate with a subset of HEI10-marked recombination intermediates, even independent of other CR proteins and SC formation. We noted that, similar to SCEP3 and ZYP1 (Extended Data Fig. 3), HEI10 was found in spo11-1-3 and mtopVIB-2 nuclei (Extended Data Fig. 8e). In these backgrounds, SCEP3 and HEI10 are found in association, but at lower numbers and frequencies than in the WT and CR mutants (Fig. 6a,b and Extended Data Fig. 8a–e). They even form aggregates or short stretches (Extended Data Fig. 8e), a pattern not observed in any CR mutant (Fig. 6a). This suggests that SCEP3 and HEI10 are expressed independently of meiotic DSB formation and have an intrinsic tendency to associate, even in the absence of meiotic recombination. Nonetheless, given that SCEP3 is required for the surplus HEI10-dependent COs in zyp1, colocalizes with HEI10 in both WT and SC-deficient cells, and is found at putative HEI10-marked CO sites during late pachytene in CR mutants, we propose that SCEP3 may associate with at least a subset of HEI10-marked recombination intermediates.
We next tested whether SCEP3’s axis localization or its spatiotemporal association with HEI10 might reflect a direct interaction with axis(-associated) or ZMM proteins. Interaction between SCEP3 and the axis proteins ASY1, ASY3 and REC8 was not found in Y2H45. In addition, we did not detect an interaction between SCEP3 and the other axis(-associated) proteins ASY4, COMET and PRD3, or between SCEP3 and the ZMM proteins ZIP4, HEI10, MER3 and PTD (Extended Data Fig. 8f). However, the absence of a Y2H interaction does not completely rule out the possibility of an interaction in planta.
SCEP3 is involved in both class I and class II CO formation
We further investigated the role of SCEP3 in CO formation. In mutants impaired in class I CO formation (msh5-2, hei10-2 and mlh3-1), SCEP3 depletion further reduces both chiasma and bivalent numbers (Fig. 5c, Extended Data Fig. 7 and Supplementary Table 1; reductions are significant in all cases except scep3-1 msh5-2 chiasmata). In addition, the depletion of SCEP3 in mus81-2 reduces both chiasma (not significantly) and bivalent (significantly) numbers compared with scep3-1 (Fig. 5c, Extended Data Fig. 7 and Supplementary Table 1). These data suggest that the majority of COs in scep3 are ZMM-dependent class I COs, while a small fraction rely on MUS81. Importantly, SCEP3 is required for some class II COs, at least in a zmm background, as well as for the surplus HEI10-dependent class I COs in the absence of the SC in CR mutants.
Heterochiasmy and CO interference are abolished in scep3
To dissect genome-wide male and female CO events, we isolated scep3-4 in Ler-0, with similar phenotypes as scep3 in Col-0 (Fig. 1 and Extended Data Fig. 9). By crossing scep3-2+/− with scep3-4+/−, we generated F1 hybrids of the WT (Col-0 × Ler-0) and scep3 (Col-0 × Ler-0). These hybrids were backcrossed with Col-0 as either the female or male parent, and the resulting four offspring groups (WT female/male and scep3 female/male) were sequenced.
In scep3, CO numbers significantly increased compared with the WT, by ~25% in males and ~105% in females (Fig. 7a). Heterochiasmy present in the WT vanished in scep3 (Fig. 7a). CO frequencies increased along chromosome arms, particularly towards chromosome ends, while decreasing in pericentromeric regions (Fig. 7b and Extended Data Fig. 10). In both sexes, CO interference present in the WT was abolished in scep3 (Fig. 7c,d). Despite comparable CO distributions (Fig. 7b and Extended Data Fig. 10), male and female scep3 CO levels were ~15% lower than those in zyp1 or scep1 (Fig. 7a)16,18. In scep3 males, this is consistent with the absence of increased HEI10 foci and the lower chiasma count than in zyp1 (Fig. 5b and Extended Data Fig. 7). In contrast, scep3 females exhibited a striking ~72% increase in HEI10 foci (10.17, n = 6) compared with the WT (5.91, n = 23) (Figs. 5b and 7e), largely explaining the female CO increase. Overall, CO numbers in scep3 offspring increased genome-wide, whereas heterochiasmy (primarily due to increased female CO levels) and CO interference vanished, similar to other CR mutants16,18.
a, Number of COs (the data are presented as mean ± s.e.m.) detected by sequencing of recombinant offspring of WT males (4.80 ± 0.17), WT females (3.00 ± 0.11), scep3-2 males (6.00 ± 0.24) and scep3-2 females (6.15 ± 0.23). The sample sizes used for analysis are indicated in parentheses. Significant differences were detected between WT males and females (P < 1 × 10−7), but not between scep3-2 males and females (P = 0.942) (nested ANOVA followed by Tukey’s honestly significant difference test, a two-sided test with adjustment for multiple comparisons). Compared with the WT, in scep3-2 CO numbers significantly increased in both males (P = 5.09 × 10−4) and females (P < 1 × 10−7). b, CO distribution along chromosome 2 in male and female WT, zyp1 (ref. 16), scep1-1 (ref. 18) and scep3-2. Centromere and pericentromere regions are indicated in grey and blue, respectively. CO data are presented within 1-Mb windows. Significant differences (based on χ2 tests) between the WT and scep3-2 (green dots), scep3-2 and zyp1 (red dots) and scep3-2 and scep1-1 (orange dots) are indicated. c, Distribution of inter-CO distances (only chromosomes with exactly two COs are included for analysis) in male and female WT and scep3-2. Calculated random distributions are shown in grey. The statistical significance is indicated in parentheses (nested ANOVA followed by Tukey’s honestly significant difference test, a two-sided test with adjustment for multiple comparisons). d, The coefficient of coincidence (CoC) was calculated for inter-interval distances ranging from 1 Mb to 15 Mb for each chromosome, and a LOESS curve was fitted (coloured lines). e, Immunolocalization of ASY1 and HEI10 in WT and scep3-1 female meiocytes. DAPI-stained DNA is shown in blue. Scale bars, 10 μm.
Discussion
Two plant CE proteins, SCEP1/2, were previously identified via transcriptomics18. We identified SCEP3 using TbID-based proteomics45. SCEP3 colocalizes with other CR components at the SC, is essential for SC assembly and interacts with the N terminus of ZYP1. In addition, both the N and C termini of SCEP3 are found at the centre of the SC. Taken together, these findings indicate that SCEP3 is a plant CE protein.
Spatiotemporal and functional overlap among CR proteins suggests potential interactions. SCEP1 and SCEP2 form a complex, yet neither interacts with ZYP1 (ref. 18). SCEP3 loads independently of SC formation and other CR proteins. In the absence of the SC, its localization is sufficient for the recruitment of ZYP1, but not for that of SCEP1 or SCEP2. SCEP3 also directly interacts with ZYP1, and this interaction appears to be conserved across plants. Thus, SCEP3 probably acts as a synapsis initiation factor, facilitating ZYP1 recruitment for synapsis initiation. Whether SCEP3 and ZYP1 load as a complex or SCEP3 recruits ZYP1 remains unclear. We prefer the latter possibility, as SCEP3 loads in zyp1. Which factor recruits SCEP3 remains unknown, as none of the tested axis or ZMM candidates directly interact with SCEP3 (at least in Y2H) or are critical for its localization. In budding yeast, Zip4 links recombination intermediates with SC assembly by recruiting and interacting with Ecm11 (ref. 43), and this mechanism seems conserved in mammals. In worms, SYP-4’s C terminus regulates CO formation, possibly through ZHP-3 (HEI10 orthologue) recruitment44. In Arabidopsis, the CE proteins SCEP1, SCEP2 (ref. 18) and SCEP3 do not directly interact with ZIP4 or HEI10 (at least in Y2H), nor is SC formation impaired in zip4 or hei10. SCEP1 and/or SCEP2 do not form a complex with SCEP3 and/or ZYP1, at least in the absence of the SC; only SCEP3 loads in zyp1, and only SCEP3–ZYP1 loads in scep1 or scep2 (ref. 18). Whether further proteins or modifications and/or the SC context are required for complex formation remains unclear. The Arabidopsis SC seems to be composed of at least two subdomains: SCEP3–ZYP1 and SCEP1–SCEP2. However, how they are functionally connected remains to be addressed.
In scep3, CO interference, CO assurance and heterochiasmy (primarily due to increased female COs) are lost, suggesting that these are the common phenotypes associated with SC depletion in CR mutants in Arabidopsis16,17,18. By contrast, in scep3, CO numbers (both male and female) are lower than in the other CR mutants. In scep3 females, the increase in CO numbers is largely ZMM-dependent (it results from additional HEI10 foci), while no additional HEI10 foci are found in scep3 males. In both scep3 sexes, HEI10 foci represent ~83% of the total COs in the offspring, suggesting that ~17% of COs are ZMM-independent class II COs. It is worth noting that the CO rates of scep3 offspring might be slightly overestimated, as meiotic cells with comparatively high CO numbers may preferentially form viable gametes in scep3. Although the majority of COs in scep3 are class I, SCEP3 is involved in the regulation of both class I and II CO formation. In males, SCEP3 is required for the surplus HEI10-dependent class I COs in zyp1, for a proportion of class II COs that arise ZMM-independently, and for some chiasmata in asy1 and asy3, which are impaired in SC formation. Despite no direct interaction in Y2H, SCEP3 associates with HEI10 in both WT and SC-deficient cells. Given its requirement for surplus HEI10-dependent COs in zyp1, we propose that the association of SCEP3 and HEI10 foci is probably required for the surplus HEI10-dependent COs found in CR mutants, although HEI10 and an association with SCEP3 are also found in cells deficient for meiotic DSB formation. SCEP3 and HEI10 foci also form exclusively—for example, SCEP3 may also localize at non-ZMM intermediates, as suggested by its requirement for some class II COs. We speculate that SCEP3’s association with certain recombination intermediates before synapsis, and independently of other CR proteins, may stabilize these intermediates and/or recruit additional factors, such as ZYP1, to a subset of them, thereby promoting synapsis initiation and the formation of designated class I and/or II COs.
In Arabidopsis, CO interference vanishes in CR mutants16,17,18 including scep3, suggesting that the SC per se is required for CO interference implementation. Our data are compatible with the HEI10 coarsening model in which the SC (the scaffold for HEI10 diffusion and condensation) is required for imposing CO interference40,41,52. Two non-exclusive scenarios may also contribute to increased CO numbers without signatures of CO interference in CR mutants—that is, longer axis persistence of ASY1 and/or lack of PCH2 (ref. 53) or absence of SC polymerization-mediated local downregulation of de novo DSBs54,55.
We identified a direct interaction, which seems conserved across plants, between SCEP3’s α-helical C terminus and ZYP1, suggesting that the C terminus of SCEP3 is probably responsible for recruiting ZYP1 to facilitate SC assembly. However, the functional importance of its conserved N terminus remains unclear. The N terminus of SCEP3 is largely disordered, a feature also found in other non-plant CE proteins, such as Ecm11 (yeast), SYP-4 (worms), Corolla (flies) and SIX6OS1 (mammals) (Supplementary Fig. 2). Recently, phenylalanine clusters in the C-terminal disordered region of SYP-4 (which has an α-helical domain at its N terminus) were found to be crucial for CO regulation44. SCEP3 also contains phenylalanine clusters at its N terminus, where phenylalanine makes up 12% of the conserved domain, compared with 2.5% in the whole protein, a situation reminiscent of SYP-4’s C terminus. These structural similarities between SCEP3 and SYP-4 strongly suggest that SCEP3 is probably the orthologue of SYP-4 and, by extrapolation, of SIX6OS1 (ref. 44). Whether the phenylalanine clusters in SCEP3 share a conserved function with those in SYP-4 is unclear.
We propose a dual role for SCEP3, one as a CR component of the SC, required for its assembly, and the other as a synapsis initiation factor, possibly associated with recombination intermediates. Together, these roles interlink SC and CO formation.
Methods
Plant materials and growth conditions
A. thaliana plants were grown under short-day conditions (8/16 h light/dark) for four weeks followed by long-day conditions (16/8 h light/dark) until maturity at constant 22 °C. Col-0 was used as the WT except where indicated. The transfer DNA insertion mutants, provided by the Nottingham Arabidopsis Stock Centre56, used in this study are scep3-1 (AT4G18490; SAILseq_210_G05), scep3-2 (AT4G18490; SALK_098044), spo11-1-3 (AT3G13170; SALK_146172)57, spo11-2-3 (AT1G63990; GABI_749C12)57, mtopVIB-2 (AT1G60460; GABI_314G09)58, msh5-2 (AT3G20475; SALK_026553)59, mus81-2 (AT4G30870; SALK_107515)9, asy1-4 (AT1G67370; SALK_046272)60, dmc1-2 (AT3G22880; SAIL_170_F08)61, asy3-1 (AT2G46980; SALK_143676)12, rec8-1 (AT5G05490; SALK_137095)62, pch2-1 (AT4G24710; SAIL_1187_C06)60, zip4-2 (AT5G48390; SALK_068052)63, hei10-2 (AT1G53490; SALK_014624)51, mer3-1 (AT3G27730; Salk_091560)64, shoc1-1 (AT5G52290; SALK_057589)65 and mlh3-1 (AT4G35520; SALK_015849)66. The mutants zyp1-2 (ref. 17), scep1-1 and scep2-1 (ref. 18) were described previously. The mutants scep3-3 and scep3-4 were isolated using CRISPR–Cas9 in this study. Details on the primers used for genotyping are provided in Supplementary Table 2.
Isolation of scep3-3 and scep3-4 using CRISPR–Cas9
Targeted mutagenesis in Arabidopsis via CRISPR–Cas9 was performed according to ref. 67. pMOD_A0503, pMOD_B2103, pMOD_C0000 and pTRANS_260d (Addgene nos 91013, 91061, 91081 and 91126) were used for assembling CRISPR constructs with Cas9 and guide RNAs both driven by the CmYLCV promoter. Two guide RNAs (#1: 5′-GAGCCAAAGCCAAAATCCATTGG-3′; #2: 5′-AACTAGACAAGTTCCCTCCAAGG-3′) addressing SCEP3 were Golden Gate assembled in pMOD_B2103. The final expression cassettes assembled in pTRANS_260d were transferred into ecotypes Col-0 and Ler-0 using Agrobacterium-mediated transformation via floral dip68. On the basis of Sanger sequencing of target sites, scep3-3 and scep3-4 were isolated in transgenic lines in the Col-0 and Ler-0 backgrounds, respectively. For primer details, see Supplementary Table 2.
Y2H assays
Full-length or truncated coding sequences of Arabidopsis SCEP3, ZYP1a, ZYP1b, C terminus of ZIP4 according to ref. 18, HEI10, MER3, PTD, COMET, ASY4 and PRD3 were PCR-amplified using Col-0 flower bud cDNA as the template and cloned into pGBKT7 and/or pGADT7 vectors (Takara) via Gibson Assembly (NEB). Cloning of barley SCEP3 and ZYP1 was done accordingly, but using cDNA prepared from barley anthers (cultivar Golden Promise). For primer details, see Supplementary Table 2.
Y2H assays were performed according to the manufacturer’s instructions (Takara). Bait and prey plasmids (empty vectors as controls) were co-transformed into the yeast strain Y2HGold (Takara, 630489) and grown at 30 °C for three to five days on plates with SD Base medium supplemented with DO Supplement −Leu/−Trp (DDO, 630417). Transformed clones underwent selection assays for five days on plates with Minimal SD Base medium supplemented with DO Supplement −His/−Leu/−Trp (TDO, 630419) or DO Supplement −Ade/−His/−Leu/−Trp (QDO, 630428).
AlphaFold protein structure modelling
The Arabidopsis SCEP3 protein structure (Extended Data Fig. 1a) was modelled using ColabFold v.1.5.3 (AlphaFold2 using MMseqs2)47. Modelling of protein–protein interactions (Fig. 4c and Extended Data Fig. 5b) and structures of SC proteins from different species (Supplementary Fig. 2) were performed with AlphaFold Server (AlphaFold3)69 and depicted with UCSF ChimeraX70.
Generation of polyclonal antibodies
Peptide synthesis and antibody production were performed by LifeTein LLC. The following peptides were selected the for respective Arabidopsis proteins and used for immunization: SCEP3-N (amino acids 69–87; C-GSSFKMDMPDFDFSSPAKK) in rat, SCEP3-C (amino acids 756–775; C-KKKHEEAKELLVRAVVDNNK) in rabbit, HEI10 (C-PKDEIWPARQNS, according to ref. 71) in rabbit and guinea pig, ZYP1-C (amino acids 833–851 in ZYP1b; C-SANIGDLFSEGSLNPYADD; peptides identical in ZYP1a/b) in rat and guinea pig, and ASY4 (C-AKLPDELDVDVSSDFKGI) and ASY1 (C-SKAGNTPISNKAQPAASRES, according to ref. 72) in rabbit and rat. All antibodies were affinity-purified against the synthetic peptide.
Cytological procedures
Pollen viability was assessed using Alexander’s stain73. Briefly, mature pollen grains were released from anthers in Alexander’s stain solution (MORPHISTO, 13441). After 30 min of staining at room temperature, pollen grains were analysed and counted (viable versus not viable) under a light microscope.
Male and female meiotic chromosome spread preparations and minimum chiasmata number counting were performed as described previously74. Briefly, young inflorescences were fixed in Carnoy’s solution (ethanol:acetic acid, 3:1, v/v) for at least 24 h at 4 °C, washed in citric buffer (0.01 M, pH 4.5) and dissected under a stereomicroscope. After dissected flower buds (males) and pistils (females) were digested in an enzyme solution (2% (w/v) cellulase R-10 (Duchefa Biochemie, 9012-54-8) and 1% (w/v) pectolyase Y-23 (Duchefa Biochemie, 9033-35-6), in citrate buffer) at 37 °C for ~70 min, the specimens were washed with 70% ethanol, placed on microscopic slides and macerated in a drop of water using fine forceps. Once the preparations had dried, 7 µl of 100% glacial acetic acid was added. The slides were placed in a humid chamber for 10 min, followed by air-drying and DAPI counterstaining.
Immunolocalization was performed as described previously74,75. Briefly, ~20 anthers (per slide) were dissected from young, fresh flower buds (0.35–0.55 mm). The anthers were then digested in 10 µl of an enzyme solution (0.1 g of cytohelicase (Sigma, 42613-29-6), 0.375 g of sucrose and 0.25 g of polyvinylpyrrolidone (Sigma, 9003-39-8) in 25 ml of ddH2O) on polysine-coated slides for 5 min at 37 °C, cut with a razor blade and squashed with a brass rod; another 10 µl of enzyme solution was then added. The mixture was then incubated at 37 °C for 7 min. After digestion, 20 µl of 1% lipsol (AZLON, 090844) was added and mixed with the cell suspension to spread the chromosomes. After incubation for 6 min at room temperature, 35 µl of 4% paraformaldehyde (Polysciences, 18814-10) was added to cover the cell area in a fume hood, after which the slides were incubated for at least 2 h in a humid chamber. The slides were then air-dried and treated with PBST (0.1% Triton X-100 in 1× PBS) for 5 min, followed by incubation with primary and secondary antibodies. The following primary antibodies and dilutions were used (conditions and concentrations were identical for the WT and mutants): anti-ASY1 (rabbit10; 1:2,000), anti-ASY1 (rabbit or rat, this study; 1:200), anti-ZYP1-C (guinea pig25; 1:2,000), anti-ZYP1-C (rat or guinea pig, this study; 1:200), anti-REC8 (rabbit76; 1:1,000), anti-ASY4 (rat, this study; 1:200), anti-γH2Ax (mouse, Sigma-Aldrich no. 05-636; 1:200), anti-SCEP3-N (rat, this study; 1:100), anti-SCEP3-C (rabbit, this study; 1:100), anti-HEI10 (guinea pig, this study; 1:200), anti-SCEP1 (rabbit18; 1:200) and anti-SCEP2 (rat18; 1:200). The following secondary antibodies were used (all diluted 1:500): anti-guinea-pig Cy5 (Abcam, ab102372), anti-guinea-pig Alexa 594 (Invitrogen, A11076), anti-guinea-pig Alexa 488 (Invitrogen, A11073), anti-rabbit Alexa 594 (Abcam, ab150076), anti-rabbit Alexa 488 (Abcam, ab150073), anti-rabbit Cy3 (Jackson ImmunoResearch, 111-165-003), anti-rat Alexa 488 (Jackson ImmunoResearch, 112-545-167) and anti-rat Alexa 594 (Abcam, ab150160).
Microscopy
Epifluorescence images were acquired using a Nikon Eclipse Ni-E microscope equipped with a Nikon DS-Qi2 camera and NIS-Elements-AR v.4.60 software (Nikon). The acquisition parameters were maintained for each antibody combination, independent of the material background (WT versus mutants), and chosen to be those that allowed capturing the most information while avoiding saturation to ensure accurate visualization and quantification. After image acquisition, deconvolution was performed using NIS-Elements-AR v.4.60 software with the method ‘Fast’ and the default parameters.
SCEP3–HEI10 colocalization analysis was performed using Imaris (Bitplane) v.10.1.0. Automatic spot detection with a size of 0.33 µm and background subtraction was applied for the red (HEI10) and the green (SCEP3) channels independently. Afterwards, a filter was chosen on the red channel to identify spots overlapping with spots in the green channel, allowing a maximum distance of 0.33 µm. The frequency of overlapping foci was afterwards obtained by calculating (number of overlapping foci × 100/total of red) and (number of overlapping foci × 100/total of green). To determine randomness of colocalization, the image was rotated 90°, spots were again detected for the green channel with the same parameters and these newly created spots were compared to the original red spots.
3D-SIM image stacks were acquired using an Elyra 7 microscope system and the software ZEN Black 3.0 v.16.0 (Carl Zeiss GmbH)77. Inter-axis distances were measured from centre to centre of each parallel axis in single-slice images (the positions were randomly picked).
Two-colour STED images were acquired with Abberior Expert Line with an Olympus UPlanSApo ×100/1.4 oil-immersion objective. Star-Orange and Star-Red were excited by 561-nm and 640-nm pulsed diode lasers, respectively, and the fluorescence signals were inhibited by a pulsed 775-nm depletion laser (total power, 3 W). For STED microscopy to perform at its full potential, alignment of the excitation and depletion beams was needed as described previously78. Briefly, the centres of the excitation and depletion beams were overlapped initially by scanning gold beads of 150 nm (BBI Solutions) in a reflection mode. Afterwards, TetraSpeck beads of four colours (TetraSpeckTM Microspheres; 100 nm; fluorescent blue, green, orange and dark red) were used to correct mismatches between the scattering mode and the fluorescence mode. To ensure correct and precise positioning of the same beads imaged by different laser lines, individual confocal and STED microscope channels were compared and adjusted respectively as the last step. During STED microscopy sample imaging, sequential scanning was applied to keep photobleaching of the samples at a minimum state, with first the Star-Red channel and then the Star-Orange channel. The images were acquired with a pixel size of 20 nm and a pixel dwell time of 10 µs, and a distance of 100 nm was applied between each slice for xyz stacks. The images were deconvolved with Huygens Professional v.24.10 (Scientific Volume Imaging, https://svi.nl/) using the classic maximum likelihood estimation algorithm with lateral drift stabilization and with 27 iterations (the detailed parameter selections were based on previously reported values16). Maximum intensity projections and contrast adjustments were applied to the deconvolved images using Fiji (open source)79. Additional image processing (adjusting brightness/contrast and merging of individual channels) was done with Fiji/ImageJ v.2.9.0 (NIH), Adobe Photoshop CS5 (Adobe) and ZEN v.3.1 (blue edition).
Genome-wide mapping of male and female COs in the WT and scep3
To generate male and female CO mapping populations, scep3-2+/− (Col-0) was crossed with scep3-4+/− (Ler-0), and F1 hybrids of the WT or biallelic for scep3 were crossed as a male or female with the WT Col-0. Total DNA samples were prepared from the four resulting backcross populations (WT male, 143 plants; WT female, 237 plants; scep3-2 scep3-4 male, 142 plants; scep3-2 scep3-4 female, 238 plants) using a ~150-mg leaf sample per individual plant and Econospin columns (96 well, Epoch Life Sciences) according to the manufacturer’s instructions adapted to plant samples. Whole-genome shotgun sequencing library preparation (Illumina DNA PCR-Free Library Prep, Tagmentation with standard DNA input amount) involved protocols from the manufacturer (Illumina). The library was quantified using quantitative PCR (KAPA Library Quant Kit; Roche Molecular Systems) and sequenced according to the manufacturer’s instructions using the NovaSeq 6000 device (Illumina; run type, SP flowcell with XP workflow and paired-end sequencing: 151 cycles (read 1) 10 cycles (index read 1), 10 cycles (index read 2) and 151 cycles (read 2)) at IPK Gatersleben.
Raw sequence reads were aligned to the A. thaliana Col TAIR10 reference genome80 using BWA-MEM81, converted to Binary/Alignment Map format, and sorted using SAMtools82. Variant calling was done using BCFtools83, filtering for a minimum mapping quality and minimum base quality of 30. The resulting variants matrix in Variant Call format was filtered using VCFtools84 for biallelic single nucleotide variants (SNVs), a minor allele frequency ranging between 0.2 and 0.3, a minimum read depth per site of 4, a maximum read depth per site of 100, a minimum mean read depth of 1 across all samples and a maximum mean read depth of 1.5. The resulting variant matrix contained 350,575 high-quality SNVs. Individuals with more than 75% missing data were removed (9 out of 760 samples). Genotype calls homozygous for the reference allele (Ler) were removed from further analysis (average of 8% per sample). Samples with SNV numbers below the 5% percentile or above the 95% percentile were removed from further analysis to avoid potential biases caused by extreme marker number deviations. To measure recombination events, SNV information was first aggregated in sliding windows of 20 consecutive SNVs with a step size of 1 by determining the modal SNV. Second, smoothed SNVs were further aggregated in non-overlapping windows of 1 Mb. For CO interference analysis, only chromosomes with exactly two COs were used. Observed inter-CO distances were compared against random inter-CO distances obtained via 500 permutations of the respective dataset (that is, male/female WT or male/female scep3). CO positions of zyp1 and scep1 were retrieved from refs. 16,18. The CoC was calculated in inter-interval distances from 1 Mb to 15 Mb for each chromosome (CoC = observed frequency of double COs / expected frequency of double COs). Statistical analysis of CO count and inter-CO distance was done in R v.4.4.1 via nested ANOVA (aov(CO count or inter-CO distance ~ sex/genotype)) followed by Tukey’s honestly significant difference test. Recombination landscapes were analysed via χ2 tests.
Material availability
Generated materials are available from the corresponding author upon reasonable request.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data supporting the findings of this research are presented in the main text, figures and supplementary information. The whole-genome resequencing raw data underlying Fig. 7 and Extended Data Fig. 10 have been deposited to the European Nucleotide Archive under accession number PRJEB81799 (http://www.ebi.ac.uk/ena/data/view/PRJEB81799). The gene/protein sequences and accession codes used in this study are available in the databases TAIR (https://www.arabidopsis.org/) and Ensembl Plants (http://plants.ensembl.org/index.html). The predicted protein structures are available in the AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk/). Source data are provided with this paper.
References
Zickler, D. & Kleckner, N. Meiosis: dances between homologs. Annu. Rev. Genet. 57, 1–63 (2023).
de Massy, B. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu. Rev. Genet. 47, 563–599 (2013).
Mercier, R., Mezard, C., Jenczewski, E., Macaisne, N. & Grelon, M. The molecular biology of meiosis in plants. Annu. Rev. Plant Biol. 66, 297–327 (2015).
Börner, G. V., Kleckner, N. & Hunter, N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117, 29–45 (2004).
Snowden, T., Acharya, S., Butz, C., Berardini, M. & Fishel, R. hMSH4–hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell 15, 437–451 (2004).
Cannavo, E. et al. Regulation of the MLH1–MLH3 endonuclease in meiosis. Nature 586, 618–622 (2020).
Zickler, D. & Kleckner, N. A few of our favorite things: pairing, the bouquet, crossover interference and evolution of meiosis. Semin. Cell Dev. Biol. 54, 135–148 (2016).
Berchowitz, L. E., Francis, K. E., Bey, A. L. & Copenhaver, G. P. The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet. 3, e132 (2007).
Higgins, J. D., Buckling, E. F., Franklin, F. C. H. & Jones, G. H. Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J. 54, 152–162 (2008).
Armstrong, S. J., Caryl, A. P., Jones, G. H. & Franklin, F. C. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 115, 3645–3655 (2002).
Sanchez-Moran, E., Santos, J. L., Jones, G. H. & Franklin, F. C. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 21, 2220–2233 (2007).
Ferdous, M. et al. Inter-homolog crossing-over and synapsis in Arabidopsis meiosis are dependent on the chromosome axis protein AtASY3. PLoS Genet. 8, e1002507 (2012).
Chambon, A. et al. Identification of ASYNAPTIC4, a component of the meiotic chromosome axis. Plant Physiol. 178, 233–246 (2018).
Cai, X., Dong, F., Edelmann, R. E. & Makaroff, C. A. The Arabidopsis SYN1 cohesin protein is required for sister chromatid arm cohesion and homologous chromosome pairing. J. Cell Sci. 116, 2999–3007 (2003).
Zickler, D. & Kleckner, N. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 7, a016626 (2015).
Capilla-Pérez, L. et al. The synaptonemal complex imposes crossover interference and heterochiasmy in Arabidopsis. Proc. Natl Acad. Sci. USA 118, e2023613118 (2021).
France, M. G. et al. ZYP1 is required for obligate cross-over formation and cross-over interference in Arabidopsis. Proc. Natl Acad. Sci. USA 118, e2021671118 (2021).
Vrielynck, N. et al. SCEP1 and SCEP2 are two new components of the synaptonemal complex central element. Nat. Plants 9, 2016–2030 (2023).
Sym, M., Engebrecht, J. & Roeder, G. S. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72, 365–378 (1993).
Page, S. L. & Hawley, R. S. c (3) G encodes a Drosophila synaptonemal complex protein. Genes Dev. 15, 3130–3143 (2001).
MacQueen, A. J., Colaiácovo, M. P., McDonald, K. & Villeneuve, A. M. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 16, 2428–2442 (2002).
Hurlock, M. E. et al. Identification of novel synaptonemal complex components in C. elegans. J. Cell Biol. 219, e201910043 (2020).
Zhang, Z. et al. Multivalent weak interactions between assembly units drive synaptonemal complex formation. J. Cell Biol. 219, e201910086 (2020).
de Vries, F. A. et al. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 19, 1376–1389 (2005).
Higgins, J. D., Sanchez-Moran, E., Armstrong, S. J., Jones, G. H. & Franklin, F. C. H. The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 19, 2488–2500 (2005).
Humphryes, N. et al. The Ecm11–Gmc2 complex promotes synaptonemal complex formation through assembly of transverse filaments in budding yeast. PLoS Genet. 9, e1003194 (2013).
Page, S. L. et al. Corona is required for higher-order assembly of transverse filaments into full-length synaptonemal complex in Drosophila oocytes. PLoS Genet. 4, e1000194 (2008).
Collins, K. A. et al. Corolla is a novel protein that contributes to the architecture of the synaptonemal complex of Drosophila. Genetics 198, 219–228 (2014).
Colaiácovo, M. P. et al. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell 5, 463–474 (2003).
Smolikov, S. et al. Synapsis-defective mutants reveal a correlation between chromosome conformation and the mode of double-strand break repair during Caenorhabditis elegans meiosis. Genetics 176, 2027–2033 (2007).
Smolikov, S., Schild-Prüfert, K. & Colaiacovo, M. P. A yeast two-hybrid screen for SYP-3 interactors identifies SYP-4, a component required for synaptonemal complex assembly and chiasma formation in Caenorhabditis elegans meiosis. PLoS Genet. 5, e1000669 (2009).
Blundon, J. M. et al. Skp1 proteins are structural components of the synaptonemal complex in C. elegans. Sci. Adv. 10, eadl4876 (2024).
Hamer, G. et al. Characterization of a novel meiosis-specific protein within the central element of the synaptonemal complex. J. Cell Sci. 119, 4025–4032 (2006).
Bolcun-Filas, E. et al. SYCE2 is required for synaptonemal complex assembly, double strand break repair, and homologous recombination. J. Cell Biol. 176, 741–747 (2007).
Schramm, S. et al. A novel mouse synaptonemal complex protein is essential for loading of central element proteins, recombination, and fertility. PLoS Genet. 7, e1002088 (2011).
Gómez-h, L. et al. C14ORF39/SIX6OS1 is a constituent of the synaptonemal complex and is essential for mouse fertility. Nat. Commun. 7, 13298 (2016).
Costa, Y. et al. Two novel proteins recruited by synaptonemal complex protein 1 (SYCP1) are at the centre of meiosis. J. Cell Sci. 118, 2755–2762 (2005).
Wang, M. et al. The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell 22, 417–430 (2010).
Barakate, A. et al. The synaptonemal complex protein ZYP1 is required for imposition of meiotic crossovers in barley. Plant Cell 26, 729–740 (2014).
Durand, S. et al. Joint control of meiotic crossover patterning by the synaptonemal complex and HEI10 dosage. Nat. Commun. 13, 5999 (2022).
Fozard, J. A., Morgan, C. & Howard, M. Coarsening dynamics can explain meiotic crossover patterning in both the presence and absence of the synaptonemal complex. eLife 12, e79408 (2023).
Crichton, J. H. et al. Structural maturation of SYCP1-mediated meiotic chromosome synapsis by SYCE3. Nat. Struct. Mol. Biol. 30, 188–199 (2023).
Pyatnitskaya, A., Andreani, J., Guérois, R., De Muyt, A. & Borde, V. The Zip4 protein directly couples meiotic crossover formation to synaptonemal complex assembly. Genes Dev. 36, 53–69 (2022).
Neves, A. R. R., Čavka, I., Rausch, T. & Köhler, S. Crossovers are regulated by a conserved and disordered synaptonemal complex domain. Nucleic Acids Res. 53, gkaf095 (2025).
Feng, C. et al. TurboID-based proteomic profiling of meiotic chromosome axes in Arabidopsis thaliana. Nat. Plants 9, 616–630 (2023).
Klepikova, A. V., Kasianov, A. S., Gerasimov, E. S., Logacheva, M. D. & Penin, A. A. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 88, 1058–1070 (2016).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Mah, L., El-Osta, A. & Karagiannis, T. γH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia 24, 679–686 (2010).
Su, H. et al. Arabidopsis RAD51, RAD51C and XRCC3 proteins form a complex and facilitate RAD51 localization on chromosomes for meiotic recombination. PLoS Genet. 13, e1006827 (2017).
Pochon, G. et al. The Arabidopsis Hop1 homolog ASY1 mediates cross-over assurance and interference. PNAS Nexus 2, pgac302 (2023).
Chelysheva, L. et al. The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet. 8, e1002799 (2012).
Morgan, C. et al. Diffusion-mediated HEI10 coarsening can explain meiotic crossover positioning in Arabidopsis. Nat. Commun. 12, 4674 (2021).
Yang, C. et al. ZYP1-mediated recruitment of PCH2 to the synaptonemal complex remodels the chromosome axis leading to crossover restriction. Nucleic Acids Res. 50, 12924–12937 (2022).
Mu, X., Murakami, H., Mohibullah, N. & Keeney, S. Chromosome-autonomous feedback down-regulates meiotic DNA break competence upon synaptonemal complex formation. Genes Dev. 34, 1605–1618 (2020).
Lee, M.-S. et al. The synaptonemal complex central region modulates crossover pathways and feedback control of meiotic double-strand break formation. Nucleic Acids Res. 49, 7537–7553 (2021).
Scholl, R. L., May, S. T. & Ware, D. H. Seed and molecular resources for Arabidopsis. Plant Physiol. 124, 1477–1480 (2000).
Hartung, F. et al. The catalytically active tyrosine residues of both SPO11-1 and SPO11-2 are required for meiotic double-strand break induction in Arabidopsis. Plant Cell 19, 3090–3099 (2007).
Vrielynck, N. et al. A DNA topoisomerase VI-like complex initiates meiotic recombination. Science 351, 939–943 (2016).
Higgins, J. D. et al. AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis. Plant J. 55, 28–39 (2008).
Lambing, C. et al. Arabidopsis PCH2 mediates meiotic chromosome remodeling and maturation of crossovers. PLoS Genet. 11, e1005372 (2015).
Pradillo, M. et al. Together yes, but not coupled: new insights into the roles of RAD51 and DMC1 in plant meiotic recombination. Plant J. 69, 921–933 (2012).
Watanabe, K. et al. The STRUCTURAL MAINTENANCE OF CHROMOSOMES 5/6 complex promotes sister chromatid alignment and homologous recombination after DNA damage in Arabidopsis thaliana. Plant Cell 21, 2688–2699 (2009).
Chelysheva, L. et al. Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet. 3, e83 (2007).
Chen, C., Zhang, W., Timofejeva, L., Gerardin, Y. & Ma, H. The Arabidopsis ROCK-N-ROLLERS gene encodes a homolog of the yeast ATP-dependent DNA helicase MER3 and is required for normal meiotic crossover formation. Plant J. 43, 321–334 (2005).
Macaisne, N. et al. SHOC1, an XPF endonuclease-related protein, is essential for the formation of class I meiotic crossovers. Curr. Biol. 18, 1432–1437 (2008).
Jackson, N. et al. Reduced meiotic crossovers and delayed prophase I progression in AtMLH3‐deficient Arabidopsis. EMBO J. 25, 1315–1323 (2006).
Čermák, T. et al. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 29, 1196–1217 (2017).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Meng, E. C. et al. UCSF ChimeraX: tools for structure building and analysis. Protein Sci. 32, e4792 (2023).
Huang, J., Li, X., Wang, C. & Wang, Y. Evaluation of crossover number, distribution, and interference using cytological assays in Arabidopsis. Curr. Protoc. 2, e599 (2022).
Fu, H. et al. Interfered chromosome pairing at high temperature promotes meiotic instability in autotetraploid Arabidopsis. Plant Physiol. 188, 1210–1228 (2022).
Alexander, M. Differential staining of aborted and nonaborted pollen. Stain Technol. 44, 117–122 (1969).
Armstrong, S. J., Sanchez-Moran, E., Chris, F. & Franklin, H. Cytological analysis of Arabidopsis thaliana meiotic chromosomes. Methods Mol. Biol. 558, 131–145 (2009).
Varas, J. & Pradillo, M. Immunolabeling protocols for studying meiosis in plant mutants defective for nuclear envelope components. Methods Mol. Biol. 1840, 237–247 (2018).
Cromer, L. et al. Centromeric cohesion is protected twice at meiosis, by SHUGOSHINs at anaphase I and by PATRONUS at interkinesis. Curr. Biol. 23, 2090–2099 (2013).
Weisshart, K., Fuchs, J. & Schubert, V. Structured illumination microscopy (SIM) and photoactivated localization microscopy (PALM) to analyze the abundance and distribution of RNA polymerase II molecules on flow-sorted Arabidopsis nuclei. Bio Protoc. 6, e1725–e1725 (2016).
Zhao, Z. et al. Super-resolution imaging of highly curved membrane structures in giant vesicles encapsulating molecular condensates. Adv. Mater. 34, 2106633 (2022).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Lamesch, P. et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210 (2012).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://doi.org/10.48550/arxiv.1303.3997 (2013).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. GigaScience 10, giab008 (2021).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Acknowledgements
We thank M. Grelon (IJPB, France) for providing REC8, SCEP1 and SCEP2 antibodies as well as scep1-1 and scep2-1 seeds, J. Higgins (University of Leicester, UK) and H. Puchta (JKIP and KIT, Germany) for sharing zyp1-2 seeds, and C. Franklin (University of Birmingham, UK) for sharing ASY1 and ZYP1 antibodies. We thank all lab members and colleagues at IPK for fruitful discussions, N. Bruhne for help with DNA sample collection, J. Pohl for help with whole-genome shotgun library preparation and sequencing, A. Fiebig for uploading of sequencing data and K. Weisshart (Carl Zeiss GmbH) for 3D-SIM image analysis. This work has received funding from the Deutsche Forschungsgemeinschaft (DFG, German Research foundation) within the framework of two projects (grant agreement nos 354617974 and 543670370.) to S.H. and from the IPK Gatersleben. S.H. acknowledges funding from the European Research Council under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 949618). B.W. is a holder of a China Scholarship Council fellowship (no. CSC202103250012). M.C. and F.H. acknowledge funding from the State of Sachsen-Anhalt (ZB I 180). C.E. and Z.Z. acknowledge funding from the DFG (Germany´s Excellence Strategy—EXC 2051—Project-ID 390713860; project no. 316213987—SFB 1278; instrument funding modular STED INST 1757/25-1 FUGG), the State of Thuringia (TMWWDG), the Free State of Thuringia (TAB; AdvancedSTED / FGZ: 2018 FGI 0022; Advanced Flu-Spec / 2020 FGZ: FGI 0031) and the BMBF, funding programme Photonics Research Germany (FKZ: 13N15713/13N15717), which is integrated into the Leibniz Center for Photonics in Infection Research (LPI). The LPI, initiated by Leibniz-IPHT, Leibniz-HKI, UKJ and FSU Jena, is part of the BMBF national road map for research infrastructures.
Funding
Open access funding provided by Leibniz-Institut für Pflanzengenetik und Kulturpflanzenforschung (IPK).
Author information
Authors and Affiliations
Contributions
C.F. conducted most of the research with assistance from J.L., B.W. and F.H. V.S. performed the 3D-SIM imaging. Z.Z., M.C. and C.E. performed the STED microscopy. A.S.C. and C.F. performed the AlphaFold predictions and interpretations. A.H. and S.D. performed whole-genome offspring resequencing and data analysis. N.F.-J. and M.C. contributed to the immunolocalization procedures and image analysis. S.H. acquired funding. C.F. and S.H. designed the experiments, analysed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Isolation of scep3-3 and phenotypic analysis of scep3 alleles.
a, SCEP3 full-length protein structure prediction by AlphaFold246. b, Isolation of scep3-3. Top: CRISPR/Cas9-mediated 22 bp deletion in exon 5 resulting in a predicted truncated protein of 201 AA; PAM sequence underlined and deleted bases as red dashes. Below: Alignment of SCEP3 WT and SCEP3-3 proteins. c, Seed abortion (indicated by asterisks) in the WT and scep3 alleles. d, Alexander staining of pollen grains (arrowheads indicate non-viable ones in blue; scale bar, 100 µm) and quantification of pollen viability in Col-0 (0.99 ± 0.01, n = 5), scep3-2 (0.92 ± 0.02, n = 5) and scep3-3 (0.93 ± 0.01, n = 3). Significance evaluated by one-way ANOVA with post-hoc Bonferroni multiple comparison (Col-0 and scep3-2, P = 1.26×10−4; Col-0 and scep3-3, P = 4.48×10−4; scep3-2 and scep3-3, P = 1). Data are presented as mean ± standard deviation (s.d.). **P < 0.01. N.S., not significant.
Extended Data Fig. 2 Both SCEP3-N and SCEP3-C antibodies localize to the SC and are specific for SCEP3 (signal absence in scep3).
Immunolocalization in pachytene nuclei of a, SCEP3-N and SCEP3-C in Col-0 (Images acquired by 3D-SIM or STED; Scale bar, 2 μm), and of b, SCEP3-N or SCEP3-C together with ASY1 in scep3-2. DNA counterstained with DAPI in gray. The experiments were repeated three times with similar results. Scale bar, 10 μm.
Extended Data Fig. 3 Co-localization of SCEP3 and ZYP1 in various meiotic mutants.
Immunolocalization of SCEP3-N, ZYP1-C and ASY1 or REC8 in spo11-2-3, mtopVIB-2, dmc1-2, asy3-1, rec8-1, pch2-1, asy1-4, msh5-2, hei10-2, zip4-2, mer3-1, and shoc1-1 pachytene(-like) nuclei. DNA stained with DAPI in gray. All experiments were repeated at least two times with similar results. Scale bar, 10 μm.
Extended Data Fig. 4 Evolutionary analysis of SCEP3 and other CR components.
The evolutionary tree is constructed based on SCEP3 homologous sequences (PSI blast results using Arabidopsis SCEP3 as seed sequence) from Streptophyta species. Accessions used: A. thaliana (NP_001328810.1), A. lyrata (XP_020875481.1), Capsella rubella (XP_023635313.1), Eutrema salsugineum (XP_024005618.1), Brassica rapa (XP_009136934.2), B. napus (XP_013737233.2), B. oleracea (XP_013594171.1), Gossypium hirsutum (XP_040935483.1), G. arboretum (KAK5792206.1), Theobroma cacao (XP_017978471.1), Citrus sinensis (XP_024950445.1), Phaseolus vulgaris (XP_007153704.1), Cajanus cajan (XP_029124714.1), Medicago truncatula (XP_039689560.1), Glycine max (XP_040873294.1), Ricinus communis (XP_048229528.1), Jatropha curcas (XP_020541047.2), Populus trichocarpa (KAI5570781.1), Cucumis sativus (XP_011658243.1), Solanum lycopersicum (XP_019067843.1), S. tuberosum (KAH0717859.1), Sorghum bicolor (XP_021312987.1), Zea mays (XP_008673176.1), Setaria italic (XP_004969993.2), Oryza sativa (KAF2952333.1), O. brachyantha (XP_040380573.1), Brachypodium distachyon (KQK09796.1), Ananas comosus (XP_020098929.1), H. vulgare (XP_044977082.1), Nicotiana tabacum (XP_016437386.1), Triticum aestivum (KAF7023752.1), Musa acuminate (XP_018686223.1), Amborella trichopoda (XP_020527858.1), Taxus chinensis (KAH9309445.1), Selaginella moellendorffii (XP_024528147.1), Nymphaea colorata (XP_049931681.1), Cryptomeria japonica (XP_059065081.1), Adiantum nelumboides (MCO5557227.1), Ceratopteris richardii (KAH7301291.1), Diphasiastrum complanatum (KAJ7553908.1), Marchantia polymorpha (PTQ45356.1), Chara braunii (GBG74560.1), Klebsormidium nitens (GAQ79536.1). Some Streptophyta and Chlorophyta species without SCEP3 homolog identified are also listed. The color-coded table includes SCEP3, ZYP1, SCEP1 and SCEP2 and shows the presence (green) or absence (purple) of respective homologs in different species.
Extended Data Fig. 5 Conservation of the SCEP3 and ZYP1 interaction in plants.
a, Y2H interaction of H. vulgare ZYP1 and SCEP3 (full-length proteins). Note, TDO (SD/-LTH) is a less and QDO (SD/-LTHA) a more stringent medium for selection. b, AlphaFold3 interaction prediction using the C-terminus (α-helical domain) of SCEP3 and full-length ZYP1 (ZEP1 is the rice ZYP1 homolog) from O. sativa, H. vulgare, G. max or B. napus. PAE values are shown on the right; ipTM and pTM values indicated in parentheses.
Extended Data Fig. 6 Similar γH2Ax foci numbers in scep3 when compared with the WT and absence of HEI10 immunofluorescence signals in hei10-2.
a, Immunolocalization of ASY1 and γH2Ax in WT and scep3-2 meiocytes. γH2Ax signal channel is depicted on the right (gray). Scale bar, 10 μm. b, Quantification of γH2Ax foci numbers in Col-0 (187 ± 51 nm, n = 31) and scep3-2 (177 ± 31 nm, n = 30) meiocytes (late leptotene/early zygotene). Data are presented as mean ± standard deviation (s.d.). No significant difference was found between Col-0 and scep3-2 (P = 0.36, two-sided Student’s t-test). N.S., not significant. c, Immunolocalization of HEI10 and ASY1 in hei10-2. The experiment was repeated two times with similar results.
Extended Data Fig. 7 SCEP3 is required for some chiasmata in SC-deficient or ZMM mutants.
Minimum chiasma number counts in single (scep3-1, scep3-2, zyp1-2, asy1-4, asy3-1, msh5-2, hei10-2, mlh3-1, mus81-2) and double (scep3-1zyp1-2, scep3-2asy1-4, scep3-2asy3-1, scep3-1msh5-2, scep3-2hei10-2, scep3-2mlh3-1, scep3-1mus81-2) mutants. Chromosome spreads were analysed for each mutant using flower buds from distinct plants. The number of cells (n) analysed is indicated at the top. Significance evaluated by one-way ANOVA with post-hoc Bonferroni multiple comparison (zyp1-2 and scep3-1zyp1-2, P < 1×10−7; zyp1-2 and scep3-1zyp1-2, P = 0.71; asy1-4 and scep3-2asy1-4, P = 1.96×10−7; scep3-2 and scep3-2asy1-4, P < 1×10−7; asy3-1 and scep3-2asy3-1, P = 6.39×10−4; scep3-2 and scep3-2asy3-1, P < 1×10−7; msh5-2 and scep3-1msh5-2, P = 0.069; scep3-1 and scep3-1msh5-2, P < 1×10−7; hei10-2 and scep3-2hei10-2, P = 4.72×10−3; scep3-2 and scep3-2hei10-2, P < 1×10−7; mlh3-1 and scep3-2mlh3-1, P < 1×10−7; scep3-2 and scep3-2mlh3-1, P < 1×10−7; mus81-2 and scep3-1mus81-2, P < 1×10−7; scep3-1 and scep3-1mus81-2, P = 1.51×10−3). Data (refer to Supplemental Table 1) are presented as mean ± standard deviation (s.d.). **P < 0.01. N.S., not significant.
Extended Data Fig. 8 Localization of CR proteins and HEI10 in scep3, spo11-1-3 and mtopVIB-2 as well as no direct interaction of SCEP3 with selected proteins in Y2H.
Immunolocalization of a, ZYP1-C and HEI10, b, SCEP1 and HEI10, and c, SCEP2 and HEI10 in early pachytene nuclei of WT and scep3-1. d, Quantification of SCEP3 and HEI10 foci numbers in scep3-1 (HEI10, 91 ± 8.1; n = 7), spo11-1-3 (HEI10, 17 ± 5.6; SCEP3, 18 ± 3.6; n = 15), and mtopVIB-2 (HEI10, 38 ± 9.7; SCEP3, 39 ± 8.1; n = 14) as well as their percentage of overlap in spo11-1-3 and mtopVIB-2. Data are presented as mean ± standard deviation (s.d.). e, Immunolocalization of SCEP3 and HEI10 in early pachytene nuclei of spo11-1-3 and mtopVIB-2. DAPI-stained DNA in gray. Scale bar, 10 µm. f, Interactions tested between SCEP3 and axis(-associated) (ASY4, COMET or PRD3) or ZMM proteins (ZIP4, HEI10, MER3 or PTD) in Y2H. TDO (SD/-LTH) is a less stringent and QDO (SD/-LTHA) is a more stringent medium used for selection. The experiments (a-c, e) were repeated at least three times with similar results.
Extended Data Fig. 9 Isolation of scep3-4 in Ler-0.
a, Details on the CRISPR/Cas9-mediated allele scep3-4: 2 bp deletion within exon five resulting in a predicted truncated protein of 182 AA. PAM sequence underlined and deleted bases as red dashes. Below: alignment of SCEP3 wildtype and truncated SCEP3-4 protein. b, Seed setting in siliques of Ler-0 and scep3-4. Seed abortion indicated by asterisks. c, Male meiotic chromosome spreads and d, quantification of univalent frequency in Ler-0 and scep3-4.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2.
Supplementary Data 1
Supplementary Tables 1 and 2.
Source data
Source Data Figs. 1, 2 and 5–7 and Extended Data Figs. 1 and 6–9
Statistical source data.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Feng, C., Lorenz, J., Dreissig, S. et al. The synaptonemal complex central element SCEP3 interlinks synapsis initiation and crossover formation in Arabidopsis thaliana. Nat. Plants 11, 1353–1366 (2025). https://doi.org/10.1038/s41477-025-02030-9
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41477-025-02030-9
This article is cited by
-
SCEP3 initiates synapsis and implements crossover interference in Arabidopsis
Nature Plants (2025)