Abstract
LRRK2-PD represents the most common form of autosomal dominant Parkinson’s disease. We identified the LRRK2 p.L1795F variant in three families and six additional unrelated cases using genetic data from over 50,000 individuals. Carriers with available genotyping data shared a common haplotype. The clinical presentation resembles other LRRK2-PD forms. Combined with published functional evidence showing strongly enhanced LRRK2 kinase activity, we provide evidence that LRRK2 p.L1795F is pathogenic.
Similar content being viewed by others
Pathogenic variants in the LRRK2 gene are among the most common causes of autosomal dominant Parkinson’s disease (PD)1,2 and are thought to act through a gain-of-function mechanism that increases kinase activity3. The LRRK2 p.L1795F variant (chr12:40322386:G:T, hg38, rs111910483) has been shown to significantly enhance kinase activity, supporting its pathogenic role4. It was previously identified in eight PD cases from 2007 to 20195,6,7, and most recently 20248 as well as suggested as a genetic risk factor with an odds ratio (OR) of 2.59. However, insufficient evidence of segregation precluded this variant from being considered “pathogenic”. Determining pathogenicity is crucial for diagnosis, genetic counseling, and even more for treatment, particularly now that LRRK2-specific clinical trials are underway10,11.
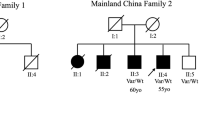
We screened a large cohort of PD cases and controls with short-read whole-genome sequencing (WGS) data, including 16,351 individuals from GP2 release 8 (DOI 10.5281/zenodo.13755496) and AMP-PD release 4 (for details see Methods and Supplementary Table 1) to identify recurrent rare coding variants of unknown significance co-segregating with PD in known PD genes (LRRK2, SNCA, VPS35, PINK1, PRKN, PARK7, and GBA1). We identified nine carriers of the LRRK2 p.L1795F variant (ENST00000298910.12:c.5385 G > T; chr12:40322386:G:T; Supplementary Figs. 1-6). Of these carriers, we identified two families based on kinship inference using genetic data (Fig. 1). The larger family (GP2-FAM-1) included four affected individuals showing the segregation of this variant with PD. The second family (AMP-FAM-1) consisted of three carriers, one clinically affected with PD and two asymptomatic carriers (at ages 55 and 76 years, respectively). The remaining two carriers were PD cases with a positive family history of PD, but no additional family members were available for genetic testing. Notably, rs111910483 is multiallelic, and we identified 7 additional carriers of the synonymous p.L1795L (ENST00000298910.12:c.5385 G > A; chr12:40322386:G:A) variant. However, this synonymous variant is very unlikely to be disease-causing and was therefore excluded from any further analyses. Additionally, we did not identify other recurrent variants in known PD genes with supporting segregation evidence.
Pedigree of Family GP2-FAM-1 (A), CANADA-FAM-1 (B), and AMP-FAM-1 (C) with the LRRK2 p.L1795F variant. The pedigrees were drawn based on reported family history and may be incomplete. The index cases are indicated with arrows. Affected individuals are indicated by black symbols: circles (female) and squares (male). Diamond is where sex is undefined. Unaffected individuals are indicated by open symbols. Unaffected variant carriers are indicated by open symbols with a dot in the middle. A diagonal line indicates deceased individuals. Red circle indicates individuals with genetic data available (WGS data for GP2-FAM-1 and AMP-FAM-1, single gene testing for CANADA-FAM-1). Heterozygous mutant (m) and wild-type (wt) genotypes are indicated with corresponding age at the sample collection (age) and age at motor symptom onset (if known; AAO). A The mother of GP2-FAM-1 index was reported to have eight additional siblings (#), several of whom are clinically affected with PD; however, no detailed family history is available for these relatives. B One maternal aunt (II-1) of the CANADA-FAM-1 index was reported to have had Alzheimer’s disease (##).
Next, we screened the genotyping data of 54,153 affected and unaffected individuals generated within GP2 (DOI: 10.5281/zenodo.10962119), where the LRRK2 p.L1795F variant was directly genotyped using the Neurobooster array. We identified three additional clinically affected variant carriers (Supplementary Fig. 7). We further screened the clinical exome data from 10,454 individuals from PDGENE which resulted in one additional variant carrier (Supplementary Fig. 8). Finally, querying the CENTOGENE proprietary Databank CentoMD®12, we identified another family with four individuals carrying the LRRK2 p.L1795F variant, three of whom were PD cases and one being an asymptomatic carrier. In total, we identified 17 individuals carrying this variant across all the datasets, including nine index cases with PD as well as five affected and three unaffected family members.
The demographic and clinical details of all identified variant carriers are displayed in Table 1. More than two-thirds were females (70.6%; n = 12/17). All affected and unaffected carriers had a positive family history of PD. Notably, among the six singleton cases, two reported only second-degree relatives with PD, while three reported a multi-incident family history of the disease. Ages of motor symptom onset (AAO) in affected individuals ranged from 36 to 66 years. The median AAO was 54.5 years (interquartile range 47-60 years). The asymptomatic carriers were 55, 76 and 76 years old, respectively, at the time of sample collection and clinical evaluation. Based on the available clinical data, the majority of affected individuals had classical PD with an asymmetric onset of symptoms and a good response to dopaminergic medication, and without obvious atypical signs suggestive of other diagnoses (missing data for up to 30%). Detailed data on non-motor symptoms and neuropsychiatric comorbidities were scarce. Cognition was reported to be unaffected in the majority of affected carriers with good scores in cognition tests (including Montreal Cognitive Assessment [MoCA] and Mini Mental State Examination [MMSE]); however, one clinically affected individual had significant cognitive impairment (MoCA score of 17 points) and one unaffected carrier also showed some cognitive deficits (MoCA score of 23 points). More detailed characteristics of the individuals from the three identified families are available in the Supplementary Material.
The p.L1795F (ENST00000298910.12:c.5385 G > T) variant is currently categorized as a variant of uncertain significance in ClinVar and shows conflicting evidence from various in-silico prediction tools and databases (Supplementary Table 2 and Supplementary Fig. 9). It is rare and confined to European populations in several investigated databases (including gnomAD v4.1, the Regeneron Genetics Center Million Exome Variant Browser13, and the UK Biobank14 500 K genomes). Similarly, all identified LRRK2 p.L1795F carriers in this study were of European ancestry, whereas the variant was absent in other ancestral populations (n = 15,316) within the GP2 genotyping cohort. In Europeans, it had an allele frequency of 0.00012 among PD cases (5 heterozygous carriers and 20,812 noncarriers) while being absent in controls (n = 9,032; Table 2). The logistic regression analysis using the European population of the GP2 genotyping cohort did not reveal a significant association between this variant and PD, likely due to insufficient controls available in the dataset given its rarity (P > 0.8, Supplementary Table 3). When comparing the distribution of carriers between PD cases from the combined genotyping and WGS dataset (6 heterozygous carriers and 23,270 noncarriers) and two non-Finnish European control populations: gnomAD v3.1.2 non-neuro (0 heterozygous carriers and 31,960 noncarriers) and gnomAD v4.1 (2 heterozygous carriers and 589,826 noncarriers), this variant was significantly associated with PD (P < 0.0056 using gnomAD v3.1.2 non-neuro, and P < 7.84e-08, OR = 76.04, 95% CI: 15.35–376.77 using gnomAD v4.1, two-tailed Fisher’s exact test). Given this variant was observed only in the European population, we searched for the overlapping IBD segments among the variant carriers using the genotyping data. The median length of an IBD segment over LRRK2 in these individuals was 7.05 cM (range: 2.1–96.3 cM, Fig. 2). All genotyped carriers shared a core haplotype of 2.825 Mbp at this locus (Supplementary Table 4), suggesting that the p.L1795F variant descended from a common founder.
Each line represents an IBD segment inferred between a unique pair of individuals. IBD segments are colored based on whether both individuals in a pair belong to the same family (GP2-FAM-1) or are considered unrelated (UR). FS indicates an IBD segment between full siblings, 2nd degree refers to a segment between a pair of second-degree relatives, and PO represents a segment between a parent and offspring. The vertical grey line marks the genomic position of the LRRK2 p.L1795F variant.
To our knowledge, we provide the largest number of LRRK2 p.L1795F variant carriers thus far, including 14 carriers clinically affected with PD and three asymptomatic carriers. The available data from the previously reported carriers5,6,7,8 do not align with our data, making an overlap of individuals between the studies unlikely. Including those reported in the literature, this brings the total to 22 clinically affected carriers of European ancestry. Still, the overall number of p.L1795F carriers is limited, and higher frequencies might be observed in specific European subpopulations. Our haplotype analysis indicating a common founder further supports this hypothesis, although we were only able to determine the geographical origin of one family of carriers in this study, which was of Ukrainian and Polish descent. Taken together with four recently published carriers of either Hungarian or Slovak origin, this likely indicates a Central-Eastern European origin8. Notably, we identified three asymptomatic p.L1795F carriers, who might still develop PD symptoms later in life. However, given the pedigree structure of these individuals, this may also reflect reduced penetrance - a common phenomenon in monogenic forms of PD, including other pathogenic LRRK2 variants.
Comparing the clinical phenotypes of p.L1795F carriers with those of other pathogenic LRRK2 variants, particularly p.G2019S15, revealed similarities among them and with idiopathic PD (iPD). While group differences in clinical phenotypes among LRRK2 variants may exist16, they do not enable meaningful genotype-phenotype correlations at an individual level. LRRK2-PD is clinically indistinguishable from iPD on an individual level. Most individuals with LRRK2-PD, including p.L1795F carriers, exhibit a classic PD phenotype with a good response to dopaminergic treatment. Atypical presentations have been described in single cases but are overall rare16. Notably, the p.L1795F variant is located in the COR-B domain, in close proximity to other pathogenic LRRK2 variants, namely p.Y1699C17 and p.F1700L18. Interestingly, for p.Y1699C carriers, a more heterogeneous phenotype has been reported, including atypical signs like amyotrophy, dementia and symptoms of behavioral disorders.17,19,20,21 However, this observation might be coincidental and biased by the small number of variant carriers. Atypical features, prominent non-motor features, or neuropsychiatric comorbidities haven’t been specifically reported for the majority of p.L1795F carriers, but the overall data is limited, making it difficult to draw meaningful conclusions. Overall, the p.L1795F phenotype aligns well with the general characteristics of LRRK2-PD and appears comparable to other LRRK2 variants with cautious interpretation given the limited number of identified carriers. The most significant differences between the genetic subtypes are their ancestral and geographical variability.
In conclusion, this is the first study providing evidence of the LRRK2 p.L1795F variant segregating with disease in multiplex families, missing from the previous reports5,6,7,8. Taken together with published functional data4, showing strongly enhanced LRRK2 kinase activity, our findings support the LRRK2 p.L1795F variant to be considered pathogenic. Large-scale studies can be helpful to identify novel rare causes of PD but also to re-evaluate previously identified variants by providing additional evidence of pathogenicity through an increased number of variant carriers and segregation. We therefore propose LRRK2 p.L1795F as a cause of PD, especially in the European population. Including this variant in the genetic screening of PD patients, particularly those of Central-Eastern European origin, may be beneficial for the variant carriers to be included in ongoing gene-specific clinical trials.
Methods
Ethics declaration
This study was conducted in accordance with the ethical standards of the institutional and national research committees. This study was approved by all ethics committees or institutional review boards of all sites participating in this study and providing samples and data, including the University of Cincinnati in Cincinnati (IRB#2017-5985), Ohio, USA, the Emory University School of Medicine in Atlanta, GA, USA, and the Michigan State University, MI, USA, and the University Health Network Research Ethics Board in Toronto, Canada. Informed consent for study participation was obtained from all participants.
Study design and participants
Our study workflow is highlighted in Fig. 3. Three sources of data were included in this study (Supplementary Table 1). First, we used the multi-ancestry whole-genome sequencing and genotyping data from the study participants recruited as part of GP222 (DOI 10.5281/zenodo.13755496) as previously described23,24. Individual-level demographic and clinical data were obtained from participating principal investigators and publicly available databases (e.g., for Coriell samples included in GP2). Second, we incorporated whole-genome sequencing data from AMP-PD. Participants in this initiative were recruited through multiple studies, including BioFIND, the Harvard Biomarkers Study (HBS), the Lewy Body Dementia Case-Control Cohort (LBD), the Parkinson’s Disease Biomarkers Program (PDBP), the Parkinson’s Progression Markers Initiative (PPMI), the LRRK2 Cohort Consortium (LCC), the Study of Isradipine as a Disease-Modifying Agent in Subjects with Early Parkinson Disease, Phase 3 (STEADY-PD3), and the Study of Urate Elevation in Parkinson’s Disease, Phase 3 (SURE-PD3). Clinical information and genetic samples from participants were obtained with appropriate written consent and local institutional and ethical approvals. Detailed information about these studies is available on the AMP-PD website (https://amp-pd.org) and the respective study websites. Third, we obtained the clinical exome sequencing data from PDGENE2, a large multi-center study in North America providing genetic testing and counseling to more than 15,000 participants.
Whole-genome sequencing (WGS) data
We included 9974 samples with the sequence alignment data available from BioFIND, HBS, LBD, PDBP, PPMI, STEADY-PD3, and SURE-PD3 cohorts through the AMP-PD release for joint genotyping with the GP2 cohort (Supplementary Table 5). Due to the unavailability of sequence alignment data from the LCC cohort, we used AMP-PD release 4 data to screen for potential pathogenic variants in this cohort.
Additionally, the DNA samples from 5,926 participants from the GP2 cohort (GP2 Data Release 8, DOI 10.5281/zenodo.13755496, Supplementary Table 5) were genome sequenced to an average of 30x coverage with 150 bp paired-end reads following Illumina’s TruSeq PCR-free library preparation protocol. We followed the same functional equivalence pipeline25 as AMP-PD to produce the sequence alignment against the GRCh38DH reference genome.
We used DeepVariant v.1.6.126 (https://github.com/google/deepvariant) to generate the single-sample variant calls for a total of 15,900 samples in GP2 and AMP-PD and performed joint-genotyping using GLnexus v1.4.3 (https://github.com/dnanexus-rnd/GLnexus) with the preset DeepVariant WGS configuration27. We set genotypes to be missing after variant quality control defined as genotype quality >=10, read depth >=10, and heterozygous allele balance between 0.2 and 0.8, and retained high-quality variants with a call rate > 0.95 after quality control. After the sample quality control following the quality metrics defined by AMP-PD28, we retained 15,752 samples (AMP-PD and GP2 combined) for the downstream analyses (Supplementary Table 5). Variant annotation was performed with Ensembl Variant Effect Predictor v111 (http://www.ensembl.org/info/docs/tools/vep/index.html, RRID:SCR_007931)29. We used KING v.2.3.0 (https://www.kingrelatedness.com, RRID:SCR_009251)30 to infer relatedness up to the second-degree relatives to confirm the known relationships and identify cryptic familial relationships. Genetic ancestry was determined using GenoTools v1.2.3 (https://github.com/GP2code/GenoTools) with the default settings31.
Genome-wide genotyping with the Neurobooster Array (GP2)
We screened the genotyping data published as part of GP2’s Data Release 732 (DOI: 10.5281/zenodo.10962119, Supplementary Table 6). Genotyping was performed by GP2 using the NeuroBooster Array (NBA; v.1.0, Illumina, San Diego, CA)33. Raw genotyping data underwent quality control and genetic ancestry prediction using GenoTools v1.2.3 with the default settings31. The LRRK2 p.L1795F variant was directly genotyped using NBA, and the quality of genotype calls was assessed by examining the signal intensity plots.
Clinical exome sequencing (PDGENEration)
We included 10,454 samples with clinical exome data available from PDGENE2 as part of GP2’s Data Release 8 (DOI 10.5281/zenodo.13755496)32. The sequence data processing followed the same pipeline of WGS data as mentioned above. We performed joint-genotyping using GLnexus v1.4.3 with the preset DeepVariant WES configuration and followed the same criteria for sample and variant quality control as for the WGS data.
Querying additional databases (CENTOGENE)
We queried the CENTOGENE proprietary Databank CentoMD®12 to identify potential additional variant carriers. CENTOGENE is a globally operating genetic diagnostic lab. Genetic data included in this manuscript was generated by exon-wise PCR amplification followed by Sanger sequencing.
Statistical analyses
To estimate the allele frequency of LRRK2 p.L1795F variant in multi-ancestral populations, we analyzed the GP2 genotyping data, the largest available dataset in this study. We excluded related individuals and samples from targeted recruitment, such as LRRK2 and GBA1 variant carriers within specific efforts of PPMI and LCC. Subsequently, we performed an association analysis of this variant with PD using the European population. We fitted the logistic regression model with PD status as binary outcome variable and the covariates as the genotype of LRRK2 p.L1795F variant, sex, age, family history, and the first six principal components to account for the population stratification. For cases, age at onset (AAO) or age at diagnosis was used, while for controls, age at sampling was used. Additionally, we merged GP2 genotyping data with the combined AMP-PD and GP2 WGS data, resulting in a cohort of 23,276 PD cases of European ancestry after excluding duplicated, related, and targeted recruitment samples as mentioned above. This allowed us to compare the carrier distribution between PD cases and non-Finnish European population from the Genome Aggregation Database (gnomAD v.3.1.2 non-neuro and v4.1, http://gnomad.broadinstitute.org/, RRID:SCR_014964) as external population controls using Fisher’s exact test. We excluded the PDGENE clinical exome data from this analysis as we could not estimate the genetic ancestry in the same manner as with the other datasets. The P value ≤ 0.05 was considered statistically significant for all the analyses.
To determine if carriers of the LRRK2 p.L1795F variant shared recent common ancestry, we phased the genotyping data from chromosome 12 in the European population using Beagle 5.4 (https://faculty.washington.edu/browning/beagle/beagle.html) with default settings34 and searched for identical-by-descent (IBD) segments with the length ≥2 cM shared across the carriers using hap-ibd v1.0.0 (https://github.com/browning-lab/hap-ibd) with default setting35.
Data availability
GP2 partnered with the online cloud computing platform Accelerating Medicines Partnership - Parkinson’s Disease (AMP PD; https://amp-pd.org) to share data generated by GP2. Qualified researchers are encouraged to apply for direct access to the data through AMP PD. The GP2 and AMP-PD datasets analysed during the current study are available through AMP-PD (https://amp-pd.org). Additional data analysed during this study (Centogene) are included in this published article.
Code availability
All scripts used for this study can be found in the public domain on GitHub (https://github.com/GP2code/EUR_LRRK2_pL1795F).
References
Westenberger, A. et al. Relevance of genetic testing in the gene-targeted trial era: the Rostock Parkinson’s disease study. Brain 147, 2652–2667 (2024).
Cook, L. et al. Parkinson’s disease variant detection and disclosure: PD GENEration, a North American study. Brain 147, 2668–2679 (2024).
Taylor, M. & Alessi, D. R. Advances in elucidating the function of leucine-rich repeat protein kinase-2 in normal cells and Parkinson’s disease. Curr. Opin. Cell Biol. 63, 102–113 (2020).
Kalogeropulou, A. F. et al. Impact of 100 LRRK2 variants linked to Parkinson’s disease on kinase activity and microtubule binding. Biochem. J 479, 1759–1783 (2022).
Nichols, W. C. et al. LRRK2 mutation analysis in Parkinson disease families with evidence of linkage to PARK8. Neurology 69, 1737–1744 (2007).
Benitez, B. A. et al. Resequencing analysis of five Mendelian genes and the top genes from genome-wide association studies in Parkinson’s Disease. Mol. Neurodegener. 11, 29 (2016).
Illés, A. et al. The role of genetic testing in the clinical practice and research of early-onset parkinsonian disorders in a hungarian cohort: increasing challenge in genetic counselling, improving chances in stratification for clinical trials. Front. Genet. 10, 1061 (2019).
Ostrozovicova, M. et al. p.L1795F LRRK2 variant is a common cause of Parkinson’s disease in Central Europe. Res. Sq. https://doi.org/10.21203/rs.3.rs-4378197/v1 (2024).
Pitz, V. et al. Analysis of rare Parkinson’s disease variants in millions of people. NPJ Parkinsons Dis 10, 11 (2024).
McFarthing, K. et al. Parkinson’s Disease Drug Therapies in the Clinical Trial Pipeline: 2022 Update. J. Parkinsons. Dis. 12, 1073–1082 (2022).
Kluss, J. H., Lewis, P. A. & Greggio, E. Leucine-rich repeat kinase 2 (LRRK2): an update on the potential therapeutic target for Parkinson’s disease. Expert Opin. Ther. Targets 26, 537–546 (2022).
Trujillano, D. et al. A comprehensive global genotype-phenotype database for rare diseases. Mol. Genet. Genomic Med. 5, 66–75 (2017).
Sun, K. Y. et al. A deep catalog of protein-coding variation in 985,830 individuals. bioRxiv https://doi.org/10.1101/2023.05.09.539329 (2023).
Sudlow, C. et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 12, e1001779 (2015).
MDSGene. Movement Disorder Society Genetic mutation database (MDSGene) https://www.mdsgene.org/.
Saunders-Pullman, R., Raymond, D. & Elango, S. LRRK2 Parkinson Disease. in GeneReviews® (eds. Adam, M. P. et al.) (University of Washington, Seattle, Seattle (WA), 2006).
Zimprich, A. et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 44, 601–607 (2004).
Borsche, M. et al. The New p.F1700L LRRK2 variant causes Parkinson’s disease by extensively increasing kinase activity. Mov. Disord. 38, 1105–1107 (2023).
Kim, J. S. et al. A Korean Parkinson’s disease family with the LRRK2 p.Tyr1699Cys mutation showing clinical heterogeneity. Mov. Disord. 27, 320–324 (2012).
Wszolek, Z. K. et al. German-Canadian family (family A) with parkinsonism, amyotrophy, and dementia - Longitudinal observations. Parkinsonism Relat. Disord. 3, 125–139 (1997).
Khan, N. L. et al. Mutations in the gene LRRK2 encoding dardarin (PARK8) cause familial Parkinson’s disease: clinical, pathological, olfactory and functional imaging and genetic data. Brain 128, 2786–2796 (2005).
Global Parkinson’s Genetics Program. GP2: The Global Parkinson’s Genetics Program. Mov. Disord. 36, 842–851 (2021).
Lange, L. M. et al. Elucidating causative gene variants in hereditary Parkinson’s disease in the Global Parkinson’s Genetics Program (GP2). NPJ Parkinsons Dis 9, 100 (2023).
Towns, C. et al. Defining the causes of sporadic Parkinson’s disease in the global Parkinson’s genetics program (GP2). NPJ Parkinsons Dis. 9, 131 (2023).
Regier, A. A. et al. Functional equivalence of genome sequencing analysis pipelines enables harmonized variant calling across human genetics projects. Nat. Commun. 9, 1–8 (2018).
Poplin, R. et al. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 36, 983–987 (2018).
Yun, T. et al. Accurate, scalable cohort variant calls using DeepVariant and GLnexus. Bioinformatics 36, 5582–5589 (2021).
Iwaki, H. et al. Accelerating medicines partnership: Parkinson’s disease. Genetic Resource. Mov. Disord. 36, 1795–1804 (2021).
McLaren, W. et al. The Ensembl Variant Effect Predictor. Genome Biol. 17, 122 (2016).
Manichaikul, A. et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010).
Vitale, D. et al. GenoTools: An Open-Source Python Package for Efficient Genotype Data Quality Control and Analysis. bioRxiv https://doi.org/10.1101/2024.03.26.586362 (2024).
Leonard, H. et al. Global Parkinson’s Genetics Program data release 7. Zenodo https://doi.org/10.5281/ZENODO.10962119 (2024).
Bandres-Ciga, S. et al. NeuroBooster Array: a genome-wide genotyping platform to study neurological disorders across diverse populations. medRxiv https://doi.org/10.1101/2023.11.06.23298176 (2023).
Browning, B. L., Tian, X., Zhou, Y. & Browning, S. R. Fast two-stage phasing of large-scale sequence data. Am. J. Hum. Genet. 108, 1880–1890 (2021).
Zhou, Y., Browning, S. R. & Browning, B. L. A fast and simple method for detecting identity-by-descent segments in large-scale data. Am. J. Hum. Genet. 106, 426–437 (2020).
Acknowledgements
Genotyping data and whole-genome sequencing data (DOI: 10.5281/zenodo.10962119, Release 7 and DOI 10.5281/zenodo.13755496, Release 8) used in the preparation of this article were obtained from Global Parkinson’s Genetics Program (GP2). GP2 is funded by the Aligning Science Against Parkinson’s (ASAP) Initiative and implemented by The Michael J. Fox Foundation for Parkinson’s Research (https://www.gp2.org). For a complete list of GP2 members see http://www.gp2.org. Whole-genome sequencing data used in the preparation of this article were obtained from the Accelerating Medicine Partnership® (AMP®) Parkinson’s Disease (AMP PD) Knowledge Platform. For up-to-date information on the study, visit https://www.amp-pd.org. The AMP® PD program is a public-private partnership managed by the Foundation for the National Institutes of Health and funded by the National Institute of Neurological Disorders and Stroke (NINDS) in partnership with the Aligning Science Across Parkinson’s (ASAP) initiative; Celgene Corporation, a subsidiary of Bristol-Myers Squibb Company; GlaxoSmithKline plc (GSK); The Michael J. Fox Foundation for Parkinson’s Research; Pfizer Inc.; AbbVie Inc.; Sanofi US Services Inc.; and Verily Life Sciences. ACCELERATING MEDICINES PARTNERSHIP and AMP are registered service marks of the U.S. Department of Health and Human Services. Clinical data used in preparation of this article were obtained from the MJFF-sponsored LRRK2 Cohort Consortium (LCC). For up-to-date information on the study, visit www.michaeljfox.org.lcc. The LRRK2 Cohort Consortium is coordinated and funded by The Michael J. Fox Foundation for Parkinson’s Research. The investigators within the LCC provided data, but did not participate in the analysis or writing of this report. The full list of LCC investigators can be found at www.michaeljfox.org/lccinvestigators. PD GENEration is a study funded by the Parkinson’s Foundation and supported by GP2, a program of Aligning Science Across Parkinson’s (ASAP). Variant queries in the UKBB were conducted under approved project 82590 (to Z.-H.F). Additional data used for this article was obtained from PD GENEration. PD GENEration is a study funded by the Parkinson’s Foundation and supported by GP2, a program of Aligning Science Across Parkinson’s (ASAP). This work was supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services, project ZIA AG000949.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Consortia
Contributions
L.M.L. and Z.-H.F. were responsible for the study conceptualization and execution. They analyzed and interpreted the generated genotyping, clinical-exome, and whole-genome sequencing data and wrote the first draft of the manuscript. L.M.L. analyzed and interpreted the clinical data. Z.-H.F. performed sequencing data processing. H.I., J.M., N.K., K.Le., D.V., H.L., M.A.N., and C.B. were involved in sample and genotyping data acquisition and access to raw data. K.Lo., N.E.M., A.B.S., C.K., and C.B. contributed to the genetic data analysis and interpretation. H.I., H.R.M., and C.K. contributed to clinical data collection and analysis. L.M., A.E., and H.C. contributed samples from affected individuals to GP2 that were identified to carry the LRRK2 variant and their respective demographic and clinical data included in this manuscript. S.A.F. and L.A.H. contributed clinical and genetic data generated as part of the PDGENEration study. S.F., N.A., and C.M. contributed clinical data for individuals included in this study. P.B. and C.B. contributed genetic data generated by CENTOGENE GmbH. All co-authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
L.M.L., N.A., K. Lo., and L.A.H. declare no competing interests. C.M. receives research funding from the Michael J Fox Foundation, the Parkinson’s Foundation (US) and holds the Catherine Manson Chair in Movement Disorders, funded by the Mayvon Foundation. D.V., H.L.L., H.I., K.Le., and M.A.N.’s participation in this project was part of a competitive contract awarded to DataTecnica LLC by the National Institutes of Health to support open science research. M.A.N. also currently serves on the scientific advisory board for Character Bio Inc plus is a scientific founder at Neuron23 Inc and owns stock. L.M. has received honoraria from the International Association of Parkinsonism and Related Disorders (IAPRD) Society for social media and web support, and personal compensation as a consultant/scientific advisory board member for Acadia. He has received a grant (collaborative research agreement) from the International Parkinson and Movement Disorders Society for the MDS-UTRS Validation Program (Role: PI), Non-Profit. A.J.E. has received grant support from the NIH and the Michael J Fox Foundation; personal compensation as a consultant/scientific advisory board member for Neuroderm, Amneal, Acadia, Avion Pharmaceuticals, Acorda, Kyowa Kirin, Supernus (formerly, USWorldMeds), NeuroDiagnostics, Inc (SYNAPS Dx), Intrance Medical Systems, Inc., Praxis Precision Medicines, and Herantis Pharma; and publishing royalties from Lippincott Williams & Wilkins, Cambridge University Press, and Springer. He co-founded REGAIN Therapeutics and is co-inventor of the patent “Compositions and methods for treatment and/or prophylaxis of proteinopathies. P.B. and C.B. are employees of CENTOGENE GmbH. S.A.F. received honoraria from Lundbeck, Biogen, Takeda, and Neurocrine and grants from Medtronics, Boston Scientific, Sun Pharmaceuticals Advanced Research Company, Aspen, Biohaven, Neurocrine, Voyager, Prilenia Therapeutics, CHDI Foundation, Michael J. Fox Foundation, NIH 1 P50 NS123103-01, NIH 1R01NS125294-01, and the Parkinson Foundation. Finally, he reports royalties from Demos, Blackwell Futura, Springer for textbooks, and Uptodate. N.E.M receives salary and research support from the NIH (1K08NS131581), the Parkinson’s Foundation and the Aligning Science Across Parkinson’s (ASAP) Global Parkinson’s Genetics Program (GP2). He serves as a member of the PDGENEration steering committee. H.R.M. is employed by UCL. In the last months, he reports paid consultancies from Roche, Aprinoia, AI Therapeutics and Amylyx; lecture fees/honoraria from BMJ, Kyowa Kirin, and the Movement Disorders Society; and research Grants from Parkinson’s UK, Cure Parkinson’s Trust, PSP Association, Medical Research Council, and the Michael J Fox Foundation. H.R.M. is also a co-applicant on a patent application related to C9ORF72 - Method for diagnosing a neurodegenerative disease (PCT/GB2012/052140). A.B.S. is an Associate Editor of Movement Disorders. He is an unpaid member of the Scientific Advisory Board of Cajal Neuroscience. C.K. has received grant support from The Michael J. Fox Foundation for Parkinson’s Research and the Aligning Science Across Parkinson’s Initiative. She serves as a medical advisor to Centogene, Retromer Therapeutics, and Takeda and received speakers’ honoraria from Bial and Desitin. Z-H.F. is supported by the Aligning Science Across Parkinson’s (ASAP) Global Parkinson’s Genetics Program (GP2) and receives GP2 salary support from The Michael J. Fox Foundation for Parkinson’s Research. K.Lo., S.F., and A.B.S. are Associate Editors of npj Parkinson’s disease. They were not involved in the journal’s review of or decisions related to this manuscript.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lange, L.M., Levine, K., Fox, S.H. et al. The LRRK2 p.L1795F variant causes Parkinson’s disease in the European population. npj Parkinsons Dis. 11, 58 (2025). https://doi.org/10.1038/s41531-025-00896-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41531-025-00896-2