Abstract
Understanding neurobiology and developing effective interventions for cognitive dysfunction in psychotic disorders remain elusive. Insufficient knowledge about the biological heterogeneity of cognitive dysfunction hinders progress. We aimed to identify subgroups of patients with psychosis and distinct patterns of functional brain alterations related to cognition (cognitive biotypes). We analyzed B-SNIP consortium data (2 270 participants including participants with psychotic disorders, relatives, and controls, 55% females). We used reference-informed independent component analysis with the standardized and fully automated framework NeuroMark and the 100k multi-scale intrinsic connectivity networks (ICN) template to obtain subject-specific ICNs and whole-brain functional network connectivity (FNC). FNC features associated with cognitive performance were identified using multivariate joint analysis. K-means clustering identified patient subgroups based on these features. Two biotypes with different functional brain alteration patterns were identified. Relative to controls, biotype 1 exhibited hypoconnectivity in cerebellar-subcortical and somatomotor-visual networks and worse cognitive performance. Biotype 2 exhibited hyperconnectivity in somatomotor-subcortical networks, hypoconnectivity in somatomotor-high cognitive processing networks, and better-preserved cognitive performance. Demographic, clinical, cognitive, and FNC characteristics of biotypes were consistent in discovery and replication sets and in relatives. 76.56% of relatives were assigned to a psychosis biotype, of those, 70.12% were to the same biotype as their affected family members. These findings suggest two distinctive psychosis-related cognitive biotypes with differing functional brain patterns shared with their relatives. Instead of traditional diagnosis, patient stratification based on these biotypes may help optimize future research and identify biological targets for the treatment of cognitive dysfunction in psychosis.
Similar content being viewed by others
Introduction
Cognitive dysfunction is widely recognized as a transdiagnostic dimension of psychotic disorders and is consistently associated with poorer functional outcomes1. While effective pharmacological treatments for positive psychotic symptoms have been available for decades2, treatments targeting cognitive dysfunction have not achieved the same level of efficacy3,4. Cognitive interventions have been typically designed for participants based on their DSM diagnosis5. DSM diagnoses rely on patterns of psychotic or affective symptoms, which do not necessarily map onto biological mechanisms. Brain alterations can vary significantly among individuals with the same DSM diagnosis6 and overlap with those with other diagnoses and relatives7. Distinct subtypes with different aberrations in neural circuits linked to cognitive dysfunction may exist within the broad group of psychotic disorders and may require different treatments8. However, traditional case-control comparisons may have masked the biological heterogeneity underlying cognitive dysfunction within DSM diagnostic categories6, all of it hindering the development of targeted and more effective therapeutic strategies for cognitive dysfunction8,9,10.
Previous efforts have aimed to identify subgroups of patients with psychosis to improve treatment development. Given the high dimensionality of the human population overall, and of psychotic disorders specifically, there are numerous valid ways in which participants with psychosis can be subgrouped10. However, any subgrouping might not help understand the biology of cognitive dysfunction10. Some studies have focused on identifying subgroups using cognitive performance alone, without incorporating biological measures, which limits their ability to pinpoint different brain circuits involved in cognitive dysfunction across various subgroups11. Other efforts have included cognitive performance along with other laboratory or neurophysiology parameters12,13. These studies have demonstrated differences in brain measures between the identified Biotypes12,13. However, these Biotypes were primarily based on multiple biomarkers related to psychosis in general, rather than those specifically associated with cognitive dysfunction, limiting its ability to identify specific brain mechanisms underlying cognitive dysfunction that could be used in treatment development.
Previous studies have tried to identify subgroups based on whole-brain neuroimaging data14,15,16,17,18,19. Clustering based on this type of data, specifically on rsfMRI, can be challenging due to the high dimensionality of the brain, and the usually low signal-to-noise ratio19. Consequently, the number of features used in clustering must be reduced to those of interest for the research question. Additionally, clustering based on whole-brain neuroimaging data can be easily driven by variability unrelated to psychosis but to confounds (e.g. age, sex, sites, ancestry) that may mislead the clustering. To overcome these problems, previous studies—using mostly structural magnetic resonance imaging (sMRI)—, have used different approaches to characterize the heterogeneity relevant to the research question9,10. Some examples have been HYDRA, which identifies clusters of patients based on their differences compared to controls20,21,22 and was used in schizophrenia21 and first-episode psychosis20, or a combination of clinical and brain clustering solutions in early psychosis23. However, the emphasis has remained on identifying general biological subgroups rather than focusing on the specific biological characteristics related to cognitive dysfunction20,21,22,24,25,26. Approaches that do not use cognitive performance as the primary dimension to parse variability across features are more likely to produce unhelpful subgroups in disentangling the biological diversity of cognitive dysfunction and in identifying potential biological targets for treatment in subsequent steps10.
Other types of approaches have tried to identify subgroups of mental health patients using data fusion techniques to combine symptoms and neuroimaging data27,28. These approaches had replication problems due to small sample sizes, instability of the results, and weak brain-symptom associations27. Cognition and rsfMRI29 have been suggested to present a stronger relationship compared with other combinations like structural MRI-psychopathology or rsfMRI-psychopathology30. Therefore, this type of approach along with larger sample sizes and new fMRI approaches may be suitable for the identification of cognition-related fMRI features, on which to base the identification of subgroups.
Approaches based on Independent Component Analysis (ICA) divide the functional brain into temporally coherent patterns, known as intrinsic connectivity networks (ICNs), which may be spatially overlapping but functionally distinct31,32,33,34,35. Recent advancements have led to a standardized and fully automated framework, called Neuromark36 for identifying ICNs across participants and datasets.
These advancements include a canonical template with 105 multi-scale ICNs from data collected from over 100,000 participants37. This ICN template was obtained through ICA with different model orders33, which allows the ICNs to arise from different spatial scales (multi-scale ICNs). The temporal coupling between these multi-scale ICNs is called multi-scale functional network connectivity (FNC).
This template can be used along with reference-informed ICA38,39 to identify these multi-scale ICNs and their multi-scale FNC in datasets, potentially leading to more replicable and stronger brain-cognition associations, subsequently helping to identify cognition-related FNC features that can serve to identify FNC-based cognition-related subgroups.
We hypothesize that subgroups of patients or biotypes with psychotic disorders with different patterns of multi-scale FNC related to cognitive dysfunction and different cognitive profiles can be identified. We used a large dataset of participants with schizophrenia (SZ), schizoaffective disorder (SAD), and bipolar disorder with psychotic symptoms (BDP), their first-degree relatives, and controls. First, we identified FNC features related to cognition in psychosis, and second, we used these features to identify subgroups of patients with different patterns of FNC alterations associated with cognitive performance or cognitive biotypes. We validated the subgroups in a replication sample and first-degree relatives.
Methods and materials
Participants
We analyzed data from 2270 participants recruited by the Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP) Consortium 1 and 2. We included participants meeting the criteria for BDP, SAD, or SZ (N = 1179), their first-degree relatives (N = 465), and healthy controls (N = 626). We used all available data from participants with complete rsfMRI and Brief Assessment of Cognition in Schizophrenia (BACS) data (Table 1).
Clinical and cognitive assessments
Demographic data were collected for all participants. For details about clinical assessment see Supplementary Material. All participants underwent cognitive testing with the Brief Assessment of Cognition in Schizophrenia (BACS)40,41 that includes six cognitive constructs: 1) Verbal Memory/List Learning Task, 2) Working Memory/Digit Sequencing Task, 3) Motor Speed/Token Motor Task, 4) Verbal Fluency/Category Instances Task, Controlled Oral Word Association Test, 5) Attention and Speed of processing/Symbol Coding Task, and 6) Executive functions/Tower of London and a composite score41. We also incorporated the Weschler Memory Scale (WMS) Backward and Forwards subtests for the cognitive assessment42. For patients recruited in B-SNIP2, information regarding childhood learning difficulties was collected.
Estimating subject-specific multi-scale intrinsic connectivity networks (ICNs) and multi-scale functional network connectivity (FNC)
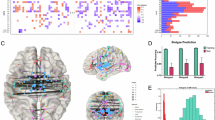
Imaging data acquisition and preprocessing details can be found in Supplementary Material. We used the GIFT software toolbox (http://trendscenter.org/software/gift)36 to perform multivariate-objective optimization ICA with reference (MOO-ICAR38,43) and generated subject-specific ICNs. As a reference, we used the Neuromark_fMRI_2.0 template (http://trendscenter.org/data), which includes highly replicated 105 ICNs across different spatial scales37 in over 100k individuals. Next, we computed subject-level static FNC by calculating pairwise Pearson correlations between the cleaned (see Supplementary material) time courses of ICNs. This process resulted in a 105 × 105 symmetric FNC matrix for each participant, representing the whole-brain functional connectome44,45 (Fig. 1A).
A We employed multivariate-objective optimization independent component analysis with reference (MOO-ICAR) to estimate 105 multi-scale intrinsic connectivity networks (ICNs) at the subject level. The reference for the 105 ICNs was derived from a large sample of over 100,000 participants. We computed subject-level static functional network connectivity (FNC) by calculating pairwise Pearson correlations between cleaned time courses of ICNs, resulting in a 105 × 105 symmetric FNC matrix for each participant. ICNs are grouped together based on their anatomical and functional properties. B Principal Component Analyses Plus Canonical Correlation (PCA-CCA) was fitted on the discovery set, including patients, relatives, and controls, to find FNC features associated with the Brief Assessment of Cognition in Schizophrenia and Wechsler Memory Scale Backward and Forwards Tests. The optimal number of principal components was estimated directly from the data using the Elbow criteria. Three canonical correlations remained statistically significant in the replication set and two remained statistically significant after adjusting for covariates. We conducted a pair-wise comparison between patients (N = 256) and controls (N = 126) for the remaining two canonical variates adjusting for covariates in the replication set. Similar comparisons including first degree relatives (N = 93) who make up the rest of the replication set (N = 475) can be found in Supplementary material. Patients and controls presented statistically significant differences in both cognitive canonical variates and in the first FNC canonical variate but did not in the second FNC canonical variate, therefore we did not include the second pair in subsequent analyses. C We selected FNC features with the highest correlation with the first canonical variate in patients in the discovery set. We selected FNC features with a loading >|0.1047| according to the elbow method (Cognitive FNC features in Psychosis or CFPs). Left: Correlation values of the 5460 functional network connectivity features and the first FNC canonical variate in 105 × 105 symmetric matrices in patients in the discovery set. Blue colors: negative correlation. Yellow-red colors: positive correlation. Lighter colors: correlation closer to 0. Middle: elbow plot of ranked absolute values of correlations with a vertical line at 0.1047, the value chosen as threshold. Right: FNC features with a correlation with absolute values lower than 0.1047 and therefore not included in k-means clustering are shown in white. The 1077 FNC features selected for k-means clustering are shown in color. D We conducted k-means clustering using patients from the discovery set to find subgroups of patients based on CFPs. Silhouette index solution for two clusters was statistically significant (p = 0.0005). E We assigned patients from the replication set to one of the clusters obtained in the discovery set based on the shortest Euclidean distance between each subject’s centroid and the clusters’ centroid for the 1077 CFPs. F We computed the centroid of the 1077 CFPs in the control group to simulate a control cluster. Like E, we assigned first-degree relatives to either one of the patients’ clusters or to the control cluster based on the shortest distance between the centroid of each subject and the centroids of the clusters.
Discovery and replication sets
We divided our sample into discovery and replication sets comprising about 80% and 20% of the sample. The proportion of patients, relatives, and controls was consistent in both sets. We regressed out the site-related and mean framewise displacement (see Supplementary materials for details).
Canonical FNC signatures of cognitive performance
First, we aimed to find FNC features highly associated with cognitive performance in psychosis that could serve to identify subgroups of patients. Secondly, we used these features to identify cognition-related neurobiology-based subgroups of patients or cognitive biotypes. For this purpose, we used a Principal Component Analysis Plus Canonical Correlation Analyses (PCA-CCA) model46,47,48,49,50. We performed PCA-CCA between cognitive variables (six BACS constructs and WMS Backward and Forwards tests), and the 5460 FNC variables on the discovery set, including patients, relatives, and controls (Fig. 1B). We excluded BACS composite score from the PCA-CCA model to avoid redundancy, as it combines individual BACS subtests. Combining the three groups allowed us to analyze between-group differences (see supplemental material for analyses involving first-degree relatives) and select the canonical variates with statistically significant differences between patients and controls, reducing the probability of merely capturing general population variability in the subsequent cluster analyses. Additionally, we achieved a larger sample size, boosting the statistical power and mitigating overfitting27,30.
We estimated the optimal number of principal components directly from the data. To tune the model, find an optimal number of principal components, and improve the robustness of our findings, we randomly split the discovery set into 80% train subset and 20% test subsets, estimated canonical pairs for a given number of principal components using 80% train subset, and estimated the correlation value for canonical pairs in the test subset (Fig. 1B). We selected the number of principal components with the highest average correlation value in the test subsets across 10,000 random splits (Supplementary material). We conducted additional approaches to ensure the robustness of the results (Supplementary material). We obtained the canonical variates in the replication set by applying the canonical weights from the discovery set in the replication data. Finally, we tested canonical correlations’ statistical significance in the hold-out replication set.
Influences of covariates on significant canonical pairs in the replication set
We analyzed how covariates (age, sex, race, socioeconomic status, and chlorpromazine equivalents) influenced the relationship between cognitive and FNC canonical variates in the replication set. We conducted linear models with canonical cognitive variates (CVCog) as dependent variables and canonical FNC variates (CVFNC) and covariates as independent variables. We aimed to identify FNC features associated with cognition that were less influenced by these covariates; therefore, we excluded canonical pairs without statistically significant associations when introducing covariates from subsequent analyses.
Determining cognition-related FNC features in participants with psychosis
To identify FNC features suitable for clustering, we focused on canonical variates that showed differences between patients and healthy controls. This approach aimed to reduce the risk of identifying subgroups that reflect general population variability rather than features specific to psychotic disorders. First, we compared canonical variates between patients and controls while controlling for the covariates. We selected canonical pairs with significant between-group differences in both cognitive and FNC canonical variates in the replication set (psychosis-related variates) for further analysis (Fig. 1B). Second, with the same purpose, we calculated Pearson correlations between these FNC canonical variates and the FNC features in the patient group in the discovery set. We ranked the correlation values and used the elbow method to establish a threshold. We selected the 1077 FNC features with a correlation higher than |0.1047| (Fig. 1C). For this work, we refer to these FNC features as Cognitive FNC Features in Psychosis or CFPs (Fig. 1C).
Cluster identification, significance, and stability
Clustering analysis procedures always identify clusters in a dataset, and indices for determining the optimal number of clusters always provide a result, however, these procedures do not indicate that the data genuinely exhibit a cluster structure beyond what would be expected by chance. To overcome this limitation, we used a statistical procedure (Supplementary material) to evaluate the presence of clusters in our data27,51. We obtained the optimal number of clusters with the Silhouette Index and evaluated the probability of observing this Index under the null hypothesis of no clusters (p-value)27,51,52.
We evaluated clustering stability with a bootstrapping resampling technique (n bootstraps=1000). We performed K-means clustering on each bootstrapped resample and we computed the Jaccard similarity average53,54.
Biotypes validation and characterization
In k-means clustering, each cluster is represented by its centroid. The centroid is the arithmetic mean position of all the observations in the cluster, representing the point in multi-dimensional space that minimizes the sum of squared distances to all other points in the cluster. New observations can be assigned to one of the established clusters based on the shortest Euclidean distance between the observation and cluster centroids55,56. We assigned patients from the replication set to clusters obtained in the discovery set using this method (Fig. 1E) to replicate the clusters.
First-degree relatives present intermediate cognitive performance between patients and controls7 (Table S1), therefore, we hypothesized that some relatives presented more similar CFP patterns to their affected family member than to controls, while other relatives may be more similar to controls than to patients. To evaluate this, we computed the centroid of the control group (healthy cluster or biotype) by calculating the average value of their CFPs. We assigned relatives to either one of the patients’ clusters or the healthy cluster based on their closest centroid (Fig. 1F). We calculated the proportion of relatives assigned to the same cluster as their affected family members. To validate the clusters, we compared clusters/biotypes of patients from discovery and replication sets and clusters/biotypes of relatives, for demographic, clinical, cognitive variables, and FNC features. We considered a result as replicated if it reached statistical significance in the discovery set in a two-tailed test after False Discovery Rate (FDR) correction and in a two-tailed test in the replication set. We use a consistent approach to evaluate the proportion of relatives that were assigned to the same DSM diagnosis as their family members (supplementary materials).
Results
Demographic, clinical, and cognitive characteristics of discovery and replication sets are shown in Table 1.
Canonical FNC signatures of cognitive performance and influence of covariates on statistically significant canonical pairs
Three canonical pairs presented statistically significant correlations in both the discovery (rDis1 = 0.49, pDis1 = 0.001; rDis2 = 0.34, pDis2 < 0.001; rDis3 = 0.33, pDis3 < 0.001, Fig. 2A) and replication set (rRep1 = 0.47, pRep1 = 0.001; rRep2 = 0.24, pRep2 < 0.001; rRep3 = 0.13, pRep3 = 0.002, Fig. 2B). Loadings are shown in Fig. 2C and D. Associations in the first and second pairs remained statistically significant after introducing the covariates, (βRep1 = 0.24, CIRep1: 0.16–0.33, pRep1 < 0.001; βRep2 = 0.12, CIRep2: 0.02–0.21, pRep2 = 0.015), but the third pair association did not (βRep3 = 0.03, CIRep3: −0.07–0.14, pRep3 = 0.500). Therefore, we did not include the third pair in subsequent analyses (supplementary material). We repeated these analyses excluding chlorpromazine equivalents and obtained similar results57.
A Scatterplots of the first (left), second (center), and third (right) canonical pairs in the discovery set. Canonical correlations: rDis1 = 0.49, pDis1 = 0.001; rDis2 = 0.34, pDis2 = 0.001; rDis3 = 0.33, pDis3 = 0.001. P-values computed with 1000 permutations. B Scatterplots of the first (left), second (center), and third (right) canonical pairs in the replication set. Canonical correlations: rRep1 = 0.47, pRep1 < 0.001; rRep2 = 0.24, pRep2 < 0.001; rRep3 = 0.13, pRep3 = 0.002. C Loadings (Pearson correlation between cognitive subtests and cognitive canonical variates) for Brief Assessment of Cognition in Schizophrenia (BACS, blue) subtests and Weschler Memory Scales (WMS, red) Backward and Forward for the first (left), second (center) and third (right) canonical pairs. D Loadings (Pearson correlation between Functional Network Connectivity (FNC) and FNC canonical variates) for the 5460 FNC and the first (left), second (center), and third (right) canonical pairs. Loadings are represented in 105 × 105 symmetric matrices. The 105 multiscale intrinsic connectivity networks are grouped as follows: Visual networks (VI), cerebellar networks (CB), temporal networks (TP), subcortical networks (SC), somatomotor networks (SM), and high cognitive processing networks (HCP). E–G Brain maps in MNI152 space, where nodes are the coordinates of peak activation points of the 105 intrinsic connectivity networks and edges are 10% FNC features with the highest loadings for each FNC canonical variate (E, first; F, second; G, third). H Violin plots of participants with psychosis and controls pairwise comparisons of the first (left) and second (right) canonical pairs in the replication set. Two-tailed t-tests obtained from linear models adjusting for covariates: First functional network canonical variate, d = 0.58, t(466) = 5.28, padj < 0.0001; First cognitive canonical variate, d = 0.53, t(466) = 5.05, padj < 0.0001; Second functional network connectivity canonical variate, d = 0.07, t(466) = 0.56, padj = 0.84; Second cognitive canonical variate, d = 0.35, t(466) = 3.05, padj = 0.007. t: t-statistic, t(degrees of freedom), d: difference. * statistically significant differences between groups. EMM: Estimated Marginal Mean. P-value: Tukey method for comparing a family of 3 estimates.
Determining cognition-related FNC features in participants with psychosis
We observed statistically significant differences between patients and controls in the first canonical FNC (controls–patients, d = 0.58, t = 5.28, padj < 0.0001), and cognitive variates (controls–patients, d = 0.53, t = 5.05, padj < 0.0001), and in the second cognitive canonical variate (controls–patients, d = 0.35, t = 3.05, padj = 0.007), but not in second FNC canonical variate (controls–patients, d = 0.07, t = 0.56, padj = 0.84) (Fig. 2H). As we aimed to identify canonical pairs that exhibited differences in both cognition and FNC between patients and controls, we included the first pair but not the second in subsequent analyses. We repeated these analyses excluding chlorpromazine equivalents and obtained similar results57. We selected FNC features with the highest correlation with the first FNC canonical variate in patients (Cognitive FNC Features in Psychosis, CFPs) for k-means clustering. Elbow’s method suggested 0.1047 as a threshold, comprising 1077 FNC features (Fig. 1C).
Clusters identification, significance, and stability. Validation and characterization of cognitive biotypes
The Silhouette index suggested two clusters as the optimal number and was statistically significant (p = 0.0005) against the null hypothesis of no clusters in our data (Fig. 1D). Bootstrapped Jaccard similarity values were 0.935 for Cluster 1 and 0.921 for Cluster 2, both significantly higher than the suggested threshold of 0.85 for highly stable clusters53,54. We repeated the analysis using hierarchical clustering (supplementary material). We calculated the intra-class correlation kappa coefficient to measure the agreement between the assignment of both methods and obtained a coefficient of 0.70 (indicative of substantial agreement: 0.61–0.8058) with a p < 0.001. Of those first-degree relatives with their family members included in analyses, 76.56% (N = 281/367) were assigned to one of the psychosis clusters. Of those, 70.12% (N = 197/281, χ2 = 7.03, p < 0.00001) were assigned to the same biotype as their family member (Fig. 1F). 23.4% (N = 86/367) of relatives were assigned to the control biotype (supplementary material). In the analyses using diagnostic-based clustering (SZ vs BD), only 54.85% of relatives assigned to psychosis clusters were assigned to the same diagnosis cluster as their family members (N = 96/175, χ2 = 1.22, p = 0.269).
Cognitive biotypes of patients in the discovery set
Functional network connectivity
Biotype 1 exhibited more extensive FNC alterations than biotype 2 when compared to controls. Relative to controls, biotype 1 displayed hypoconnectivity between visual-somatomotor, cerebellar-subcortical, high cognitive processing-cerebellar, somatomotor-subcortical, and high cognitive processing-temporal networks. Biotype 1 showed hyperconnectivity compared to controls in visual-subcortical, visual-cerebellar, somatomotor-subcortical, and high cognitive-somatomotor networks (Fig. 3A, B). Biotype 2 displayed hypoconnectivity relative to healthy controls in high cognitive processing-somatomotor, high cognitive processing-subcortical, and subcortical-cerebellar networks. Biotype 2 presented hyperconnectivity compared to controls in cerebellar-somatomotor, subcortical-somatomotor, and temporal-high cognitive processing networks (Fig. 3A and B, and Fig. S1A for between cluster comparison). comparisons were corrected with FDR for multiple comparisons.
Discovery set: biotype 1, N = 426; biotype 2, N = 497. Replication set: biotype 1, N = 110, biotype 2 N = 146. First-degree relatives: biotype 1, N = 153; biotype 2, N = 178, control biotype, N = 95. For each functional network connectivity (FNC) feature, we fit a linear model adjusting for sex, age, race, ethnicity, site, and head motion and conducted a two-tailed t-test to compare controls with each biotype of patients and relatives. Bottom-left triangle: t-statistic. Top-right triangle: p-values, those that reached statistical significance (p < 0.05) are shown with colors (–log10(p-value)×sign(t-statistic) scale); otherwise, they are shown in gray. The discovery set is corrected for multiple comparisons (False Discovery Rate). Yellow-Red colors: higher FNC in patients/relatives compared to controls. Blue colors: lower FNC in patients/relatives compared to controls. A Biotype 1 from discovery set. B Biotype 2 from discovery set. C Biotype 1 from replication set. D Biotype 2 from replication set. E First-degree relatives biotype 1. F First-degree relatives biotype 2. G First-degree relatives control biotype. Cognitive FNC features in psychosis (CFPs) included in k-means clustering are shown with a black line. Visual networks (VI), cerebellar networks (CB), temporal networks (TP), subcortical networks (SC), somatomotor networks (SM), and high cognitive processing networks (HCP). First-degree relatives with a diagnosis of a psychotic disorder were excluded from these analyses.
Demographic, cognitive, and clinical characteristics
Compared to biotype 2 and after FDR correction, biotype 1 presented a statistically significantly higher proportion of African Americans (VCramer=0.21, pFDR < 0.001), a lower proportion of Caucasians (VCramer=0.18, pFDR < 0.001), a higher proportion of SZ (VCramer=0.09, pFDR < 0.012), lower of BPD (VCramer=0.10, pFDR = 0.002), older age (dCohen=0.33, t = 4.93, pFDR < 0.001), lower socioeconomic status (dCohen=0.32, t = 4.80, pFDR < 0.001), more similarity to SZ than BPD (schizo-bipolar scale, SBS, dCohen = 0.27, t = 4.24, pFDR < 0.001) and lower score in the Global Assessment of Functioning (GAF, dCohen = −0.21, t = −3.14, pFDR = 0.004) and in Birchwood Social Functioning Scale (BSFS, dCohen = -0.25, t = −3.55, pFDR < 0.001) functioning. Biotype 1 presented significantly worse cognitive performance across all cognitive subtests (Fig. 4, see Table S2 for statistical details) and a higher proportion of patients with a history of childhood learning difficulties in all categories compared to biotype 2 (Fig. 5A and Table S2 for statistical details).
Discovery set: biotype 1, N = 426; biotype 2, N = 497. Replication set: biotype 1, N = 110, N = 146. First-degree relatives: biotype 1, N = 153; biotype 2, N = 178, control biotype, N = 95. WMS Weschler Memory Scale, BACS Brief Assessment of Cognition in Schizophrenia, YMRS Young Mania Rating Scale, BSFS Birchwood Social Functioning Scale, SES Socioeconomic Status (Hollingshead index), SBS Schizo-bipolar Scale, PANSS Positive and Negative Syndrome Scale for Schizophrenia, Chlorpromazine equivalentes: Average daily chlorpromazine dose; MADRS Montgomery-Asberg Depression Rating Scale, BACS Brief Assessment of Cognition in Schizophrenia, BSFS Birchwood Social Functioning Scale, SES Socioeconomic status, SBS Schizo-bipolar Scale, Chlorpromazine equivalents: Average daily chlorpromazine dose; MADRS Montgomery-Asberg Depression Rating Scale, GAF Global Assessment of Functioning Scale; Z-scores are shown. BACS z-scores were obtained from normative data stratified by age and sex. * Non-adjusted p-value < 0.05 from a two-tailed t-test; ** False discovery rate correction for multiple testing p-value < 0.05. Comparisons with three biotypes in first-degree relatives were adjusted with Tukey method for comparing a family of three estimates. WMS: Statistics were obtained from linear models that also accounted for the influence of age, sex, race, ethnicity, site, and socioeconomic status. BACS: Statistics were obtained from linear models that also accounted for the influence of race, ethnicity, site, and socioeconomic status (age and sex accounted for when computing z-scores). Statistical details are shown in Tables S2, S3, and S4. First-degree relatives with a diagnosis of a psychotic disorder were excluded from these analyses.
A Patients from discovery set: biotype 1, N = 426; biotype 2, N = 497. B Patients from replication set: biotype 1, N = 110, N = 146. C First-degree relatives: biotype 1, N = 153; biotype 2, N = 178, control biotype, N = 95. * Non-adjusted p-value < 0.05 from Pearson’s Chi-squared test; ** False discovery rate correction for multiple testing p-value < 0.05. DSM diagnosis in first-degree relatives refers to the diagnosis of the affected family member. Statistical details are shown in Tables S2, S3 and S4. First-degree relatives with a diagnosis of a psychotic disorder were excluded from these analyses.
Cognitive biotypes of patients in the replication set
A similar pattern of differences in the same direction was observed (Figs. 3C, D, 4 and 5B) but fewer differences in FNC, demographic, clinical, and cognitive characteristics reached statistical significance.
Demographic, cognitive, and clinical characteristics
Relative to biotype 2, biotype 1 presented a significantly higher proportion of African Americans (VCramer=0.15, p < 0.015), and older age (dCohen=0.52, t = 3.198, p < 0.002). Higher similarity to SCZ in the schizo-bipolar scale (SBS) was close to significance (dCohen=0.23, t = 1.815, p = 0.07) (Fig. 4 and Table S3). Regarding cognition, similar to the discovery set, biotype 1 displayed an average worse performance in all cognitive constructs and a higher proportion of patients with learning difficulties during childhood in all categories compared to biotype 2 (see Table S3 for statistical details). Differences in BACS composite (dCohen = −0.28, t = −2.10, p = 0.036), Tower of London (dCohen = −0.38, t = −2.87, p = 0.004), WMS backward (dCohen = −0.36, t = −2.62, p = 0.009), and forward (dCohen = −0.32, t = −2.364, p = 0.018) reached statistical significance. Symbol coding was close to significance (dCohen = −0.26, t = −1.94, p = 0.054) (Figs. 4, 5B and Table S3). Congruent patterns of differences also emerged in biotypes of first-degree relatives (Figs. 3E–G, 4, 5C, and Table S4) in FNC, demographic and clinical characteristics (see supplementary results). We repeated the cluster analysis using whole-brain FNC features and found no replicable differences in any of the nine cognitive measures (Supplementary material).
Cognitive biotypes of First-degree Relatives
367/465 relatives had their family members included in the analyses. 281/367 (76.57%) were assigned to psychosis clusters/biotype; of those, 197/281 (70.12%, χ2 = 7.0297, p < 0.00001) were assigned to the same cluster/biotype as their family member. 249/326 relatives of participants with SZ or BPP had their family members included in the analyses. 175/249 (70.28%) were assigned to BPP or SZ clusters; of those, 96/175 (54.85%, χ2 = 1.2215, p = 0.269) were assigned to the same cluster as their family members (Table S2).
Functional network connectivity
Compared to controls, biotype 2 also showed hypoconnectivity between visual-high cognitive performance networks and hyperconnectivity between visual-subcortical networks, a pattern not observed in the same biotype in patients. As expected, first-degree relatives assigned to the control biotype presented substantially fewer FNC differences compared with controls.
Demographic, cognitive, and clinical characteristics
After FDR correction and compared to biotype 2, biotype 1 exhibited statistically significant older age (t = 3.220, pFDR = 0.009), lower GAF (t = −3.467, pFDR = 0.006), and a higher proportion of African Americans (χ2 = 14.189, pFDR = 0.001). Compared to biotype 2 and after FDR correction, biotype 1 also presented a worse cognitive performance in the BACS composite (t = −0.345, pFDR = 0.006), verbal memory (t = −2.546, pFDR = 0.035), digit sequencing (t = −2.362, pFDR = 0.049), symbol coding (t = −3.104, pFDR = 0.009), and WMS backward (t = −3.120, pFDR = 0.009) and forward (t = −4.736, pFDR = 0.001). Compared to relatives in the control biotype, biotype 1 displayed worse performance in BACS composite (t = −2.879, pFDR = 0.016), verbal memory (t = −2.879 pFDR = 0.016), symbol coding (t = −2.44, pFDR = 0.042), digit sequencing (t = −3.119, pFDR = 0.009), and WMS forwards (t = −2.71, pFDR = 0.024) constructs after FDR correction. Biotype 2 (more cognitively preserved in patients) and the control biotype did not exhibit statistically significant differences in cognitive performance. This pattern mirrored the findings observed in patients.
Discussion
Our study aimed to identify neurobiology-based cognition-related biotypes in patients with psychotic disorders, characterized by distinct functional brain alterations related to cognition. Firstly, we identified a robust multivariate correlation between brain-wide FNC and cognitive performance across patients, relatives, and controls in a replication set. We found a higher correlation between the first canonical variates than previously reported30, potentially attributable to methodological improvements through our constrained ICA NeuroMark approach and the multi-scale ICNs template. Patients exhibited significantly lower scores in the first FNC and cognitive canonical variates which is in line with previous studies that suggested that disruptions in brain networks may be implicated in cognitive dysfunction in psychosis59,60.
Secondly, we identified two biotypes with distinct FNC characteristics linked to cognitive performance. Biotype 1 consistently exhibited poorer cognitive performance and a higher prevalence of childhood learning difficulties (Tables S2 and S3). BACS composite score, which provides a global measure of cognitive performance, and three other cognitive tests showed replicable differences between biotypes. We did not find replicable differences in five cognitive subtests, although BACS Symbol coding was close to significance (p = 0.054). However, except for BACS Tower of London whose effect size was reduced to -0.05 in the replication set, the remaining effect sizes were comparable to those observed in the discovery set, suggesting that the lack of significance were likely due to limited statistical power (see Supplementary material for power analyses). This is also supported by the similar patterns observed in demographic, clinical, cognitive, and FNC characteristics between biotypes in both the discovery and replication sets. Analogous patterns were observed in first-degree relatives: biotype 1 displayed inferior cognitive performance in six cognitive constructs compared to biotype 2 and in five compared to the healthy biotype (first-degree relatives more similar to controls). Biotypes in first-degree relatives also presented similar patterns in FNC alterations.
The two biotypes may represent distinct subgroups of patients, each characterized by divergent patterns of brain network alterations associated with cognitive dysfunction. These alterations appear to be distributed widely across the brain. Compared to biotype 2, biotype 1 exhibited increased FNC primarily between temporal-high cognitive processing and somatomotor-cerebellar networks and decreased FNC between somatomotor-high cognitive processing, some somatomotor-high cognitive processing networks, and temporal-cerebellar networks (Fig. S1). FNC between temporal-subcortical and temporal-somatomotor networks were largely spared (Figs. S1 and 3). Compared to healthy controls, biotype 1 was characterized by hypoconnectivity in cerebellar-subcortical and somatomotor-visual networks and worse cognitive performance. Biotype 2 was characterized by hyperconnectivity in somatomotor-subcortical networks and hypoconnectivity in somatomotor-high cognitive processing networks, and better preserved cognitive performance. This distinct pattern of hypo- and hyperconnectivity, with opposite directions and significant differences between biotypes, primarily involves the somatomotor-high cognitive processes, temporal-high cognitive processes, and subcortical-high cognitive processes networks (Figs. S1 and 3). These findings suggest that the biotypes may differ not only in the severity of alterations but also in the type of these alterations or potential compensatory mechanisms. Notably, not all FNC features that differed between biotypes or compared to controls were CFPs (Figs. 3 and S1).
Our results support that psychosis-free first-degree relatives presented similar biotypes as the participants with psychosis, suggesting that cognitive biotypes are not completely dependent on disease severity. Additionally, biotype 1 showed replicable differences in hospitalizations (but not in the ratio of hospitalizations to age), lifetime duration of psychotic symptoms, and the ratio of lifetime duration of psychosis to age (Tables S8 and S9). Cognitive impairment is associated with reduced treatment adherence, higher hospital admission rates, and longer hospital stays1, even if it is independent of positive and negative symptoms1, which could explain the observed differences without indicating that biotype 1 merely represents a more severe form of disease in terms of positive or negative symptoms.
To our knowledge, this is the first attempt to identify neurobiology-based psychosis subgroups associated with a specific symptom dimension in both patients and relatives. The FNC features that showed alterations compared to controls in each biotype, especially those with distinct patterns of dysconnectivity between biotypes (mainly somatomotor-high cognitive processing, temporal-high cognitive processing, and subcortical-high cognitive processing networks) and belonging to CFPs, may serve as the basis for identifying different altered brain circuits responsible for cognitive dysfunction in psychosis. These circuits could be targeted for therapeutic interventions. Subsequent investigations could explore the impact of treatments purportedly capable of modulating cognition in psychotic disorders1 on CFPs.
We consider our approach a potential advancement in unraveling the biological heterogeneity of cognitive dysfunction within psychosis, compared to previous efforts based on cognitive performance, other biomarkers or neuroimaging approaches11,12,20,21,22,26, more limited in identifying subgroups based on the specific brain networks related to cognitive dysfunction. One study13 used cognition and neurophysiological tests in the same sample and identified three Biotypes, where Biotypes 1 and 2 did not exhibit significant differences in general cognitive ability, though both differed from biotype 3 (Fig. S2)13. Two studies used structural brain metrics and three clusters in their respective discovery sets; one16 reported significant differences only between subgroups 1 and 2 on the N-back test, and another16 (with subsample from B-SNIP 1) only between biotypes 1 and 3 on the BACS composite and Digit Sequencing. A third study26 with structural brain metrics found no cognitive performance differences between the two identified subgroups. Similarly, we repeated the analysis using whole-brain FNC and found no replicable differences in any cognitive measures (supplementary material). Therefore, our approach appears to more effectively capture subgroups with distinct cognitive dysfunction and underlying brain mechanisms.
Our third key finding reveals that from 76.56% of relatives assigned to a psychosis biotype, 70.12% (χ2 = 7.0297, p < 0.00001) were assigned to the same biotype as their family member with psychosis, suggesting that relatives present similarities in the functional brain patterns related to cognition that delineate biotypes. Previous studies have suggested intermediate cognitive dysfunction in relatives in the same sample7 and altered network connectivity in relatives of participants with psychosis61. Intermediate neurobiology of biotypes may be present in psychosis-free first-degree relatives and contribute to their intermediate cognitive performance, even if not all relatives show clinically relevant cognitive dysfunction as those in biotype 2 and healthy biotype. Shared genetic and environmental backgrounds likely contribute to this phenomenon. Consistent differences in the prevalence of childhood learning difficulties in the discovery and replication set, suggest potential biotype-related distinctions in cognitive performance present, at least, since childhood. Notably, biotype 1 showed a higher representation of African Americans among patients and relatives, aligning with research on childhood adversity disparities in African Americans impacting structural brain differences62. This overrepresentation may reflect adverse environments for biotype 1, potentially influencing cognitive development, leading to learning difficulties since childhood and more pronounced FNC disruptions in adulthood.
The consistent observation of older age in biotype 1 might initially indicate a potential confounding factor. We found this unlikely given the modest age disparities (4-5 years), the age-adjusted analyses, and the disparities in childhood learning difficulties, all of which make it unlikely that age alone accounts for the differences between biotypes. Nevertheless, we conducted k-means clustering again after regressing the influence of age from FNC features and obtained similar results. (supplementary material).
Finally, the percentage of relatives in the same cluster is reduced to 54.85% (χ2 = 1.22, p = 0.269) when using SZ and BPD diagnoses instead of cognitive biotypes, suggesting that SZ and BPD groups are overlapping in CFPs. Neurobiologically driven stratification could be a better approach to foster treatment development for cognitive dysfunction, rather than approaches based on DSM diagnoses.
We did not explore solutions involving three or more clusters, as our analyses indicated that two clusters were the most appropriate. While we relied on Pearson correlation for computing static FNC it has been shown that new methodologies, such as nonlinear approaches63 or tangent space methods64, may offer advantages and improve sensitivity to differences between patients with schizophrenia and healthy controls63. Future studies could explore these methodological alternatives or combinations of them and compare their findings with the present study. Additional studies could evaluate the capacity of FNC-based or brain structure-based classification models to accurately distinguish between biotypes as has been done in previous work65.
Limitations
First, even though we have a relatively large sample size, it may have been insufficient for some analyses (see supplementary material for power analyses). Second, to avoid simply capturing general population variability in clustering, we selected 1) FNC canonical variates within pairs with differences between participants with psychosis and controls in both canonical variates and 2) FNC features with the highest correlation with this canonical variate in participants with psychosis. It still may be possible that identified biotypes correspond to general population variability and are not specific to psychotic disorders. Third, it remains possible that clusters may not conform the data and these subgroups represent degrees of severity of a single dimension. To address this possibility, 1) we conducted a statistical procedure to evaluate the presence of clusters in our data, with positive results; 2) we identified similar biotypes in first-degree relatives without psychosis, supporting that biotypes are at least partially independent of disease severity; 3) we found no differences between biotypes, even in the discovery set with greater statistical power, in key clinical markers of symptom severity such as PANSS, MADRS, YMRS, age of disease onset, suicidal tendencies, or antipsychotic dose (Tables S2, S3, S8 and S9), and 4) we observed distinct patterns of FNC alterations in biotypes that may suggest different brain mechanisms. Therefore, the identified biotypes may plausibly represent useful subgroups for advancing the understanding and treatment of cognitive dysfunction. However, potential overlap between disease severity and biotypes may exist, as poorer cognitive performance is linked to worse real-world outcomes1. This may partially explain the reported differences between biotypes in lifetime psychotic symptoms or psychosis duration relative to age. Although it is unlikely that biotypes were purely driven by disease severity given their presence in psychosis-free relatives, the extent to which these differences stem from cognitive performance or shared biological mechanisms remains unclear and warrants further investigation. Fourth, although we identified FNC features related to cognition, its translational utility for developing therapeutic targets is not straightforward. Fifth, we did not include cannabis or other substance use in the analyses. However, the biotypes showed no differences in cannabis use (Tables S8 and S9), reducing the likelihood of it acting as a confounding factor. However, the role of other substances or medical conditions known to impact cognition (i.e. metabolic disturbances)1,66,67 remains less clear. Sixth, about 60% of our sample were white/caucasian, limiting the generalization. Differences in the representation of races in biotypes may not extrapolate outside the US to countries with different social structures. Additionally, our sample included a higher proportion of women than other samples of psychosis, likely due to the inclusion of a representative sample of participants with affective psychosis, more frequent in women68.
Conclusions
We have identified and replicated, to a reasonable extent, two distinct cognitive biotypes in a large sample of patients with psychotic disorders. These biotypes exhibit disparities in cognitive dysfunction severity, demographics, and brain functional alterations with distinct patterns of hypo-hyperconnectivity. These biotypes may be partially present in first-degree relatives. Utilizing these biotypes as a stratification framework in future investigations focused on cognitive dysfunction may be promising for enhancing their success.
Data availability
The storage and management of the data that support the findings of this study are available from: B-SNIP1 (https://nda.nih.gov/edit_collection.html?id=2274) and B-SNIP2 (https://nda.nih.gov/edit_collection.html?id=2165). Access procedures are overseen by the National Institute of Mental Health (NIMH) through the National Data Archive (NDA), visit https://nda.nih.gov/.
Code availability
Preprocessing and data analysis were conducted using MATLAB 9.9.0.1857802 (R2020b) Update 7, alongside the Statistical Parametric Mapping toolbox (SPM 12), the FMRIB Software Library (FSL v6.0), the Group ICA of fMRI Toolbox (GIFT v4.0), RStudio (R v4.1.2), and Python 3.10.10. MATLAB R2020b can be downloaded from https://www.mathworks.com. The FSL v6.0 toolbox can be downloaded from https://fsl.fmrib.ox.ac.uk/fsl/fslwiki. The SPM 12 toolbox can be downloaded from https://www.fil.ion.ucl.ac.uk/spm/. GIFT v4.0 can be downloaded from https://trendscenter.org/software/gift/. R v4.1.2 can be downloaded from https://cran.r-project.org/. R packages used included ggplot2, stats, tidymodels, caret, tidyclust, cluster, emmeans, gtsummary, ggstatsplot, NbClust, fpc, MASS, hclust, and vcd. These packages can be installed directly from CRAN (https://cran.r-project.org/). Python 3.10.10 can be downloaded from https://www.python.org/downloads/release/python-31010/. The Netplotbrain library used to create brain maps can be downloaded from https://github.com/wiheto/netplotbrain. Additional MATLAB or R scripts used in this study are available from the corresponding authors upon reasonable request.
References
McCutcheon, R. A., Keefe, R. S. E. & McGuire, P. K. Cognitive impairment in schizophrenia: aetiology, pathophysiology, and treatment. Mol Psychiatry. https://doi.org/10.1038/s41380-023-01949-9 (2023).
Huhn, M. et al. Comparative efficacy and tolerability of 32 oral antipsychotics for the acute treatment of adults with multi-episode schizophrenia: a systematic review and network meta-analysis. Lancet 394, 939–951 (2019).
Nielsen, R. E. et al. Second-generation antipsychotic effect on cognition in patients with schizophrenia—a meta-analysis of randomized clinical trials. Acta Psychiatr. Scandinavica 131, 185–196 (2015).
Keefe, R. S. E. et al. Neurocognitive Effects of Antipsychotic Medications in Patients With Chronic Schizophrenia in the CATIE Trial. Arch. Gen. Psychiatry 64, 633–647 (2007).
Baldez, D. P. et al. The effect of antipsychotics on the cognitive performance of individuals with psychotic disorders: Network meta-analyses of randomized controlled trials. Neurosci. Biobehav. Rev. 126, 265–275 (2021).
Alnæs, D. et al. Brain Heterogeneity in Schizophrenia and Its Association With Polygenic Risk. JAMA Psychiatry 76, 739–748 (2019).
Hill, S. K. et al. Neuropsychological Impairments in Schizophrenia and Psychotic Bipolar Disorder: Findings from the Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP) Study. AJP 170, 1275–1284 (2013).
Zhang, W., Sweeney, J. A., Bishop, J. R., Gong, Q. & Lui, S. Biological subtyping of psychiatric syndromes as a pathway for advances in drug discovery and personalized medicine. Nat. Ment. Health 1, 88–99 (2023).
Feczko, E. & Fair, D. A. Methods and Challenges for Assessing Heterogeneity. Biol. Psychiatry 88, 9–17 (2020).
Feczko, E. et al. The Heterogeneity Problem: Approaches to Identify Psychiatric Subtypes. Trends Cogn. Sci. 23, 584–601 (2019).
Green, M. J., Girshkin, L., Kremerskothen, K., Watkeys, O. & Quidé, Y. A Systematic Review of Studies Reporting Data-Driven Cognitive Subtypes across the Psychosis Spectrum. Neuropsychol. Rev. 30, 446–460 (2020).
Clementz, B. A. et al. Identification of Distinct Psychosis Biotypes Using Brain-Based Biomarkers. Am. J. Psychiatry 173, 373–384 (2016).
Clementz, B. A. et al. Psychosis Biotypes: Replication and Validation from the B-SNIP Consortium. Schizophrenia Bull. 48, 56–68 (2022).
Planchuelo-Gómez, Á. et al. Identificacion of MRI-based psychosis subtypes: Replication and refinement. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 100, 109907 (2020).
Lubeiro, A. et al. Identification of two clusters within schizophrenia with different structural, functional and clinical characteristics. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 64, 79–86 (2016).
Pan, Y. et al. Morphological Profiling of Schizophrenia: Cluster Analysis of MRI-Based Cortical Thickness Data. Schizophrenia Bull. 46, 623–632 (2020).
Xiao, Y. et al. Subtyping Schizophrenia Patients Based on Patterns of Structural Brain Alterations. Schizophrenia Bull. 48, 241–250 (2022).
Sugihara, G. et al. Distinct Patterns of Cerebral Cortical Thinning in Schizophrenia: A Neuroimaging Data-Driven Approach. Schizophrenia Bull. 43, 900–906 (2017).
Yan, W. et al. Mapping relationships among schizophrenia, bipolar and schizoaffective disorders: A deep classification and clustering framework using fMRI time series. Schizophrenia Res. 245, 141–150 (2022).
Dwyer, D. B. et al. Psychosis brain subtypes validated in first-episode cohorts and related to illness remission: results from the PHENOM consortium. Mol. Psychiatry 28, 2008–2017 (2023).
Chand, G. B. et al. Two distinct neuroanatomical subtypes of schizophrenia revealed using machine learning. Brain 143, 1027–1038 (2020).
Chand, G. B. et al. Schizophrenia Imaging Signatures and Their Associations With Cognition, Psychopathology, and Genetics in the General Population. AJP 179, 650–660 (2022).
Dwyer, D. B. et al. Clinical, Brain, and Multilevel Clustering in Early Psychosis and Affective Stages. JAMA Psychiatry 79, 677–689 (2022).
Brucar, L. R., Feczko, E., Fair, D. A. & Zilverstand, A. Current Approaches in Computational Psychiatry for the Data-Driven Identification of Brain-Based Subtypes. Biol. Psychiatr. S0006322322018698 https://doi.org/10.1016/j.biopsych.2022.12.020 (2022).
Honnorat, N., Dong, A., Meisenzahl-Lechner, E., Koutsouleris, N. & Davatzikos, C. Neuroanatomical heterogeneity of schizophrenia revealed by semi-supervised machine learning methods. Schizophrenia Res. 214, 43–50 (2019).
Dwyer, D. B. et al. Brain Subtyping Enhances The Neuroanatomical Discrimination of Schizophrenia. Schizophrenia Bull. 44, 1060–1069 (2018).
Dinga, R. et al. Evaluating the evidence for biotypes of depression: Methodological replication and extension of. NeuroImage: Clin. 22, 101796 (2019).
Jacobs, G. R. et al. Integration of brain and behavior measures for identification of data-driven groups cutting across children with ASD, ADHD, or OCD. Neuropsychopharmacology 46, 643–653 (2021).
Voineskos, A. N. et al. Functional magnetic resonance imaging in schizophrenia: current evidence, methodological advances, limitations and future directions. World Psychiatry 23, 26–51 (2024).
Marek, S. et al. Reproducible brain-wide association studies require thousands of individuals. Nature 603, 654–660 (2022).
Calhoun, V. Data-driven approaches for identifying links between brain structure and function in health and disease. Dialogues Clin. Neurosci. 20, 87–99 (2018).
Calhoun, V. D. & Adali, T. Multisubject Independent Component Analysis of fMRI: A Decade of Intrinsic Networks, Default Mode, and Neurodiagnostic Discovery. IEEE Rev. Biomed. Eng. 5, 60–73 (2012).
Iraji, A. et al. Canonical and Replicable Multi-Scale Intrinsic Connectivity Networks in 100k+ Resting-State fMRI Datasets. https://doi.org/10.1101/2022.09.03.506487 (2022).
Iraji, A. et al. Moving beyond the ‘CAP’ of the Iceberg: Intrinsic connectivity networks in fMRI are continuously engaging and overlapping. NeuroImage 251, 119013 (2022).
Calhoun, V. D. & de Lacy, N. Ten Key Observations on the Analysis of Resting-state Functional MR Imaging Data Using Independent Component Analysis. Neuroimaging Clin. N. Am. 27, 561–579 (2017).
Du, Y. et al. NeuroMark: An automated and adaptive ICA based pipeline to identify reproducible fMRI markers of brain disorders. NeuroImage: Clin. 28, 102375 (2020).
Iraji, A. et al. Identifying canonical and replicable multi-scale intrinsic connectivity networks in 100k+ resting-state fMRI datasets. Hum. Brain Mapp. 44, 5729–5748 (2023).
Du, Y. & Fan, Y. Group information guided ICA for fMRI data analysis. NeuroImage 69, 157–197 (2013).
Lin, Q.-H., Liu, J., Zheng, Y.-R., Liang, H. & Calhoun, V. D. Semiblind spatial ICA of fMRI using spatial constraints. Hum. Brain Mapp. 31, 1076–1088 (2010).
Keefe, R. The Brief Assessment of Cognition in Schizophrenia: reliability, sensitivity, and comparison with a standard neurocognitive battery. Schizophrenia Res. 68, 283–297 (2004).
Keefe, R. et al. Norms and standardization of the Brief Assessment of Cognition in Schizophrenia (BACS). Schizophrenia Res. 102, 108–115 (2008).
Wechsler, D. Wechsler Adult Intell. Scale-Third Ed. https://doi.org/10.1037/t49755-000 (2019).
Duda, M. et al. Reliability and clinical utility of spatially constrained estimates of intrinsic functional networks from very short fMRI scans. Hum. Brain Mapp. 44, 2620–2635 (2023).
Jafri, M. J., Pearlson, G. D., Stevens, M. & Calhoun, V. D. A method for functional network connectivity among spatially independent resting-state components in schizophrenia. NeuroImage 39, 1666–1681 (2008).
Allen, E. A. et al. A baseline for the multivariate comparison of resting-state networks. Front. Syst. Neurosci. 5, 2 (2011).
Mihalik, A. et al. Canonical Correlation Analysis and Partial Least Squares for Identifying Brain–Behavior Associations: A Tutorial and a Comparative Study. Biol. Psychiatry.: Cogn. Neurosci. Neuroimaging 7, 1055–1067 (2022).
Mihalik, A., Adams, R. A. & Huys, Q. Canonical Correlation Analysis for Identifying Biotypes of Depression. Biol. Psychiatry.: Cogn. Neurosci. Neuroimaging 5, 478–480 (2020).
Uurtio, V. et al. A Tutorial on Canonical Correlation Methods. ACM Comput. Surv. 50, 1–33 (2018).
Wang, H.-T. et al. Finding the needle in a high-dimensional haystack: Canonical correlation analysis for neuroscientists. NeuroImage 216, 116745 (2020).
Zhuang, X., Yang, Z. & Cordes, D. A technical review of canonical correlation analysis for neuroscience applications. Hum. Brain Mapp. 41, 3807–3833 (2020).
Liu, Y., Hayes, D. N., Nobel, A. & Marron, J. S. Statistical Significance of Clustering for High-Dimension, Low-Sample Size Data. J. Am. Stat. Assoc. 103, 1281–1293 (2008).
Sæther, L. S. et al. Inflammation and cognition in severe mental illness: patterns of covariation and subgroups. Mol. Psychiatry. https://doi.org/10.1038/s41380-022-01924-w (2022).
Hennig, C. Cluster-wise assessment of cluster stability. Computational Stat. Data Anal. 52, 258–271 (2007).
Hennig, C. Dissolution point and isolation robustness: Robustness criteria for general cluster analysis methods. J. Multivar. Anal. 99, 1154–1176 (2008).
Hartigan, J. A. & Wong, M. A. Algorithm AS 136: A K-Means Clustering Algorithm. J. R. Stat. Soc. Ser. C. (Appl. Stat.) 28, 100–108 (1979).
Hvitfeldt, E. & Bodwin, K. tidyclust: A Common API to Clustering, (2023).
Miller, G. A. & Chapman, J. P. Misunderstanding analysis of covariance. J. Abnorm. Psychol. 110, 40–48 (2001).
McHugh, M. L. Interrater reliability: the kappa statistic. Biochem Med (Zagreb) 22, 276–282 (2012).
Sheffield, J. M. & Barch, D. M. Cognition and resting-state functional connectivity in schizophrenia. Neurosci. Biobehav. Rev. 61, 108–120 (2016).
Sheffield, J. M. et al. Transdiagnostic Associations Between Functional Brain Network Integrity and Cognition. JAMA Psychiatry 74, 605 (2017).
Meda, S. A. et al. Examining Functional Resting-State Connectivity in Psychosis and Its Subgroups in the Bipolar-Schizophrenia Network on Intermediate Phenotypes Cohort. Biol. Psychiatry.: Cogn. Neurosci. Neuroimaging 1, 488–497 (2016).
Dumornay, N. M., Lebois, L. A. M., Ressler, K. J. & Harnett, N. G. Racial Disparities in Adversity During Childhood and the False Appearance of Race-Related Differences in Brain Structure. AJP 180, 127–138 (2023).
Kinsey, S. et al. Networks extracted from nonlinear fMRI connectivity exhibit unique spatial variation and enhanced sensitivity to differences between individuals with schizophrenia and controls. Nat. Ment. Health 1–12. https://doi.org/10.1038/s44220-024-00341-y (2024).
Bijsterbosch, J. et al. Challenges and future directions for representations of functional brain organization. Nat. Neurosci. 23, 1484–1495 (2020).
Koen, J. D. et al. Supervised machine learning classification of psychosis biotypes based on brain structure: findings from the Bipolar-Schizophrenia network for intermediate phenotypes (B-SNIP). Sci. Rep. 13, 12980 (2023).
Takayanagi, Y., Cascella, N. G., Sawa, A. & Eaton, W. W. Diabetes is associated with lower global cognitive function in schizophrenia. Schizophrenia Res. 142, 183–187 (2012).
Bora, E., Akdede, B. B. & Alptekin, K. The relationship between cognitive impairment in schizophrenia and metabolic syndrome: a systematic review and meta-analysis. Psychological Med. 47, 1030–1040 (2017).
Riecher-Rössler, A., Butler, S. & Kulkarni, J. Sex and gender differences in schizophrenic psychoses—a critical review. Arch. Women’s Ment. Health 21, 627–648 (2018).
Acknowledgements
P.A.C. has received grant support from Programa Intramural de Impulso a la I + D + i 2023 (Instituto de Investigación Sanitaria Gregorio Marañón). C.D.C. has received grant support from Instituto de Salud Carlos III, Spanish Ministry of Science and Innovation (PI20/00721, PI23/00625, JR19/00024), and the European Commission (grant 101057182, project Youth-GEMs). V.C. has received grant support from the National Institutes of Health (R01MH123610). AI has received grant support from the National Institutes of Health (R01MH123610).
Author information
Authors and Affiliations
Contributions
P.A.C. and A.I. conceived and designed the study, with guidance from J.C. and V.C. R.B. was responsible for the processing and analysis of the neuroimaging data and contributed to the creation of the figures. P.A.C. and A.I. conducted the data analysis, with input from J.C., V.C., and C.D.C. J.C. and V.C. contributed to the interpretation of the findings, while C.D.C. also provided guidance on the clinical implications. P.A.C. drafted the manuscript and figures, and all authors critically reviewed and revised it for important intellectual content. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
P.A.C. has received travel support from Neuraxpharm, ROVI, and Casen Recordati. C.D.C. has received honoraria or travel support from Angelini, Viatris, and Janssen. J.C., R.B., V.C., and A.I. reported no biomedical financial interests or potential conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Andrés-Camazón, P., Diaz-Caneja, C.M., Ballem, R. et al. Neurobiology-based cognitive biotypes using multi-scale intrinsic connectivity networks in psychotic disorders. Schizophr 11, 45 (2025). https://doi.org/10.1038/s41537-025-00593-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41537-025-00593-2