Abstract
Since multiple and unpredicted influenza viruses cause seasonal epidemics and even high-risk pandemics, developing a universal influenza vaccine is essential to provide broad protection against various influenza subtypes. Combined with the mRNA lipid nanoparticle-encapsulated (mRNA-LNP) vaccine platform and chimeric immunogen strategy, we developed a novel cocktail mRNA vaccine encoding chimeric HAs (cH5/1-BV, cH7/3) and intact M2 (termed Fluaxe), which confers broad protection against major circulating IAVs and IBVs, as well as highly pathogenic avian influenza. Two-dose intramuscular immunization of Fluaxe in mice elicited cross-reactive neutralizing antibodies, T cell responses, and long-lived immunity, resulting in robust protection against multiple lethal influenza virus infections and severe acute lung injuries. In particular, intramuscular administration stimulated systemic immunity together with a prominent lung tropism of memory cells. Moreover, Fluaxe immunization inhibited the inflammatory response induced by influenza infection. In summary, we conclude that Fluaxe can elicit broad cross-protection against numerous influenza subtypes.
Similar content being viewed by others
Introduction
Influenza causes at least three to five million severe respiratory diseases and 290,000–650,000 deaths yearly in the flu season, as reported by World Health Organization (WHO)1. Approved influenza vaccines, including live attenuated (LAIV), inactivated (IIV), and recombinant vaccines, have been used for over 70 years. Unfortunately, the virus escaped neutralization through antigenic drift and antigenic shift, resulting in the emergence of new subvariants2. Although surveillance programs and modeling studies are gradually improved, replication and transmission among various host species further increase mutation rates and frequent reassortment of the influenza genome, which interferes with the accurate prediction of influenza subtypes of the next high-risk pandemic3,4. Although seasonal quadrivalent influenza vaccines are commonly administered to the public and iterated yearly to match circulating influenza variants, 10–60% of vaccine effectiveness could hardly control seasonal epidemics, let alone potential pandemics5. In addition, the H5 and H7 subtypes have been causing almost uninterrupted outbreaks in wild and domestic birds, and cases in humans with a mortality rate close to 50%, posing a significant threat to public health. Therefore, the National Institute of Allergy and Infectious Diseases recommends the necessity and urgency of the development of a universal influenza vaccine (UIV)6. Several vaccine technology platforms in combination with different vaccine designs have been explored for developing UIVs, including live attenuated, inactivated, and recombinant vaccines, with no UIV product approved for commercial use.
One direction for UIV development is inducing cross-reactive humoral-cellular immune responses by optimizing vaccine immunogens. Conserved domains of influenza hemagglutinin (HA), neuraminidase (NA), matrix (M), polymerase basic protein (PB), and nucleoprotein (NP) have been tested for UIV7,8,9. HA is activated upon cleavage by host proteases into disulfide-linked HA1 and HA2 subunits. The concept of chimeric HAs (cHAs) has been proposed, which consists of the globular HA head (part of HA1) and stem (part of HA1 and full length of HA2) domains derived from different virus subtypes. Such HA-based UIVs not only provide full or partial protection against homologous and heterologous viral challenges but also simplify vaccine formula10,11. The HA head domain contains some major neutralizing epitopes. Moreover, conserved headless HA antigens exhibit broad cross-protection against influenza subvariants12,13. Serum anti-stem domain IgGs have been found stable for at least 6 months after immunization14. As the immunodominance of the HA head limited antibody response against the stem domain from the same virus strain, heterologous cHAs could effectively redirect the humoral response from the head to the stem domain. Besides, a chimeric fusion strategy of heterologous viral proteins has been applied to recombinant vaccines15, such as M2e (extracellular domain of matrix 2)-H3 stalk and trimeric HA stem, conferring robust immunogenicity16,17. Thus, combining heterogeneous HAs by flexible linkers can potentially enhance cross-protection against more influenza subtypes. In addition, due to the high conservation of the M2 proteins between human and avian influenza virus. The importance of the M2 cytoplasmic tail binding with M1 in viral production has been reported18. Modest immunogenicity of an extracellular domain of M2 (M2e)-based vaccine and low abundance of M2 on virions limited the potential of M2 truncation as a substitute for existing influenza vaccines, yet little attention has been paid to the full-length M2 protein19. Therefore, the combination of cHA and intact M2 as vaccine immunogens can be an attractive strategy for UIV development.
mRNA vaccines have been very successful during the COVID-19 pandemic. Effective expression of transmembrane proteins, native post-translational modifications, accurate protein folding and complex assembly, and appropriate cellular trafficking of viral proteins in host cells promote optimal immunogenicity of mRNA-based vaccines, superior to recombinant HA proteins produced from prokaryotic systems20. Likewise, a trial on COVID-19 vaccines has confirmed that an mRNA vaccine stimulated higher neutralizing antibody titers than an inactivated vaccine21. So far, the tandem of conserved antigens (HA2, M2e, NP) has been verified to protect mice from some IAV infections, but with different degrees of morbidity and weight loss22. A quadrivalent mRNA vaccine consisting of the HA stalk domain, NA, M2, and NP was reported to stimulate both humoral and cellular immune responses against corresponding IAV and IBV influenza lineages. Still, relatively low levels of neutralizing antibody and cross-protection limited its efficacy23,24. Whereas mRNA vaccines based on single full-length HAs boosted excellent humoral immunity in neutralizing specific influenza subtypes25,26,27, additional HAs have been supplemented into vaccine formula to expand their antiviral spectrum, such as mRNA-1010 by Moderna28. It reportedly comprises 20 individual mRNAs encoding different influenza subtype HAs29, protecting against corresponding subtypes across influenza A virus (IAV) and influenza B virus (IBV)30. However, homogenous strain-specific HA hardly elicited broad-spectrum protection.
In this study, we tested the strategy of combining cHAs and intact M2 using mRNA technology for developing UIV. We constructed a mRNA vaccine encoding two novel chimeric HAs, cH5/1-BV and cH7/3, as well as the intact M2, to protect at least six influenza subtypes, including two circulating IAV group 1 (H1N1 and H5N1), two IAV group 2 (H3N2 and H7N9), and two IBV lineages (Victoria and Yamagata). Besides the major circulating influenza viral antigens of quadrivalent IIVs and LAIVs, we introduced HAs from two highly pathogenic avian influenza (HPAI), namely H5N1 and H7N9, into our mRNA-based vaccine to further combat the severe hazard of major poultry epizootics to human health, on which the WHO recently put great emphasis31,32. This novel cocktail mRNA vaccine induces robust humoral and cellular responses against a range of influenza viruses and protects mice from lethal viral challenges.
Results
Immunogen design and vaccine quality assessment
Based on the IAV HA sequences of current human and avian influenza viral strains circulating around the world, we constructed two heterogenic head-stem cHAs, namely cH5/1 and cH7/3, composed of the head domains from avian influenza H5N1 (A/Vietnam/1203/2004(H5N1)) and H7N9 (A/mallard/South Korea/20X-20/2021) formed by a disulfide bond (Cys58-Cys290 [H5 numbering], Cys60-Cys286 [H7 numbering]) and stem domains from H1N1 (A/Wisconsin/588/2019(H1N1)) and H3N2 (A/District Of Columbia/01/2021(H3N2)), respectively. An intact HA of IBV Victoria lineage (B/Washington/02/2019) was fused with cH5/1 by a flexible linker (triple GGGGS) to generate cH5/1-BV. Besides, the addition of highly conserved full-length M2 protein (A/Wisconsin/588/2019(H1N1) aims to cross-protect against more influenza subtypes. Nucleoside-modified mRNAs encoding the aforementioned chimeric HAs and complete M2 were synthesized by in vitro transcription (IVT) and encapsulated into LNPs, resulting in the candidate vaccine—Fluaxe (Fig. 1A).
A mRNA constructs of Fluaxe expressing the two chimeric HA and M2. Created in BioRender. Li (2025) https://BioRender.com/bithl81. B Monomeric structures of HA for four subtypes of IAV (H1N1, H5N1, H3N2, and H7N9) and IBV/Victoria. The disulfide bonds presented at the junction between the head and stem domains in all four IAV HA proteins are highlighted. For IBV/Victoria HA A56 and G302 residues are situated at the two equivalent structural positions instead of cysteines. C The predicted three-dimensional structure of Fluaxe chimeric sequences. The disulfide bond connecting the original head and stem fragments persists in the chimera. All structural figures were prepared using the PyMOL Molecular Graphics System, Version 3.0 (Schrödinger, LLC).
To understand the chimeric HA structure, we employed AlphaFold2 (AF2) to predict the three-dimensional structure of our custom-designed chimeric polypeptides. Although the primary protein sequences of monomeric HAs differed among H1N1, H3N2, H5N1, H7N9, and IBV/Victoria, their folded structures appeared remarkably similar. We noted a disulfide bond at the junction between the head and stem domains in all four IAV HA proteins. As for IBV/Victoria HA, A56 and G302 residues, rather than two cysteines, are situated at equivalent structural positions. Interestingly, the overall structure of IBV HA still adopts the same folding pattern as observed in the four IAV HAs (Fig. 1B). Moreover, the HA stem sequences among IAVs exhibit higher similarity than the head domains. Therefore, we selected major circulating H1N1 and H3N2 HA stems as the chimeric HA stem fragment to gain broad cross-protection against IAVs. For the two IBV lineages, both the HA head and stem regions share very strong similarities between their primary sequences but differ significantly from those of IAVs. Thus, we introduced only one IBV lineage HA into the chimeric structure (Supplementary Fig. 1A, B). As shown in Fig. 1C, cH5/1-BV folds together to create a back-to-back, axe-like structure, with the two fragments connected by a flexible linker. For cH5/1 and cH7/3, despite their heads and stems originating from different strains, the disulfide bond connecting the original head and stem fragments persists in the chimera. The structure of IBV/Victoria HA appears identical to its wild-type monomer structure, and the formation of the chimera does not change its folding modeling.
After IVT, the three mRNAs migrated at specific lengths of ~3788 nt for cH5/1-BV, ~2003 nt for cH7/3, and ~593 nt for M2 by microfluidic capillary electrophoresis, with >90% fragment integrity (Supplementary Fig. 2A). To confirm the size and subcellular distribution of polypeptides translated from Fluaxe mRNAs, western blotting analysis indicated strong expression of cH5/1-BV, cH7/3, and M2 in HEK293T cells with molecular weights of about 150 kDa (Cleaved fragments of ~75 kDa and ~100 kDa), ~75 kDa, and ~20 kDa, respectively Supplementary Fig. 2B). Moreover, there was a consistent skewing of cHAs and M2 toward cytoplasmic and membrane-associated distributions in MDCK cells by immunofluorescence, which probably facilitated antigen processing and presentation (Supplementary Fig. 2C).
Fluaxe broadly elicited cross-reactive antibodies
As HA is a major immunogen that provoke humoral response, we primarily monitored HA-specific antibody levels on Days 7, 14, 21, and 28 in mice intramuscularly vaccinated with two doses of Fluaxe (Fig. 2A). As expected, serum IgGs specific to H1N1, H3N2, H5N1, H7N9, and two IBV lineages HAs were simultaneously stimulated by the initial vaccination. The following booster immunization dramatically elevated the IgGs to a higher level measured by their respective geometric mean titers (GMTs) on Day 28 (Fig. 2B). The ratios of IgG2a/IgG1 and (IgG2a + IgG2c)/IgG1 for all six viruses of vaccine group were consistently higher than those from the LNP only control group without mRNA payloads (Supplementary Fig. 3A, B), suggesting a Th1-polarized immune response which is beneficial for viral clearance. Interestingly, although the antigen sequence of IBV/Yamagata was not included in the Fluaxe design, IBV/Yamagata HA-specific antibodies were detected. To characterize any cross-reactivity elicited by vaccination, a cell-based ELISA assay was developed to evaluate antigen-specific IgGs against different influenza strains after mice were vaccinated with mRNA-LNP expressing individual or combined antigens, respectively. By comparing the IgG GMTs of each vaccine groups against wild type H1N1 (A/Puerto Rico/8/34 (PR8)), H3N2 (A/Aichi/2/1968 (X31)), IBV/Victoria (B/Brisbane/60/2008), and IBV/Yamagata (B/Phuket/3073/2013) viruses, we found that antibodies induced by the cH5/1-BV immunogen poorly cross-reacted with H3N2. On the other hand, the cH7/3 immunogen failed to stimulate any significant antibody reactivity toward H1N1 (Supplementary Fig. 4A, B). Additionally, cH5/1-BV immunogen successfully stimulated broad antibody against cross-activity toward both IBV lineages (Supplementary Fig. 4C, D). The M2-elicited antibodies exhibited strong reactivity against IAV-infected cells but minimal reactivity toward IBV-infected cells (Supplementary Fig. 4). Together, our data suggested broad-spectrum antiviral characteristics of Fluaxe.
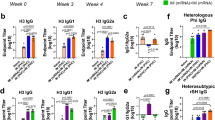
A A schematic diagram of mice immunization and prime-boost vaccination was executed on Days 0 and 14. Female BALB/c mice were injected via i.m. with Fluaxe (5 μg for each mRNA) or empty LNP per mouse (n = 5) and boosted with an equivalent dose. Sera were collected on Days 7, 14, 21, and 28, respectively, for antibody analysis. Lungs and spleens were harvested on Day 28 to analyze cellular immunity. Splenocytes were collected on Day 104 for memory T or B cell detection. Created in BioRender. Li (2025) https://BioRender.com/bithl81. B Specific IgG antibodies against H1N1, H3N2, H5N1, H7N9, IBV/Victoria, and IBV/Yamagata were measured by ELISA. Data were shown as Mean ± SEM. Significance was determined by two-way ANOVA with Bonferroni’s test for multiple comparisons (**p < 0.01, ***p < 0.001). Neutralizing antibodies of sera collected on Day 28 against H1N1, H3N2, IBV/Victoria, and IBV/Yamagata influenza viruses were detected by C MN and D HAI assays, respectively. E Neutralizing responses against H5- and H7-containing viruses were measured by HAI assay. Each symbol represents one mouse, and sera from five mice were assessed. Multiple t-tests with correction for multiple comparisons by the Holm-Šídák method were used to calculate p values relative to the control LNP group (n.s. not significant; *p < 0.05, **p < 0.01, ***p < 0.001). F Pseudotype neutralizing assays were performed using influenza pseudovirus bearing multiple group 1 and group 2 HAs (HA1-16). Detection of 50% pseudovirus neutralization titer (PNT50) of sera from immunized mice. The sera from five mice per group were pooled and tested in triplicate. Significance was calculated using two-tailed unpaired t-tests.
Both microneutralization (MN) and hemagglutination inhibition (HAI) assays were performed with two IAVs and two IBVs to further assess vaccine-elicited neutralizing antibodies. Generally, Fluaxe-vaccinated mouse sera exhibited substantial neutralizing effects in both the MN assay and the HAI assay. We detected the highest neutralizing antibody titers against IBV/Victoria in both experiments, which promoted significant cross-protection against the other IBV lineage B/Yamagata. Neutralizing titers of vaccination groups for IAVs increased about 9.6-fold for H1N1 and H3N2 (MN) and 2.4-fold for H1N1, 3.1-fold for H3N2 (HAI) compared with LNP only control group (Fig. 2C, D). Moreover, corresponding antibodies elicited by conserved immunogens of H1N1 or H3N2 HA stem further counteracted infection of three other H1N1 or H3N2 strains (Supplementary Fig. 5), demonstrating significant humoral immunity against H1N1 and H3N2 subtypes by Fluaxe vaccination. The H5 or H7 HA head domains encoded by Fluaxe also elicited vigorous neutralization activity against HPAI compared with the LNP group (4.4-fold for H5 and 4-fold for H7, respectively) measured by an inactivated virus-based HAI assay (Fig. 2E).
Apart from the in vitro neutralizing tests above, we evaluated protection by Fluaxe immunization in animals. Sera from vaccinated mice were incubated with a lethal dose (10 lethal dose 50 (10LD50)) of influenza virus, followed by trachea inoculation of the mixture into naive mice. Near-complete (80%–100%) protections against H1N1 (A/Puerto Rico/8/34), H3N2 (A/Aichi/2/1968), IBV/Victoria (B/Brisbane/60/2008), and IBV/Yamagata (B/Phuket/3073/2013) were observed, at 1:40, 1:40, 1:160, and 1:40 dilutions of Fluaxe-immunized mice sera, respectively, ensuring the efficacy of Fluaxe in stimulating neutralizing antibodies against various influenza viruses (Supplementary Fig. 6). We also performed a pseudovirus-based neutralization assay to evaluate the Fluaxe-mediated cross-group protection against IAV H1-H16, but excluding two bat influenza viruses (H17 and H18). Besides of H1, H3, H5, and H7, Fluaxe-immunization also counteracted infections of pseudoviruses bearing numerous Group 1 IAV HAs, including H2, H6, H8, H11, H13, and H16, as well as a pseudovirus bearing a Group 2 HA, namely H15 (Fig. 2F). Taken together, Fluaxe strongly elicited cross-protecting neutralizing antibodies against several IAVs and IBVs.
Fluaxe strongly elicited immunogen-specific T cell immune responses
The chimeric HA strategy, in addition to the intact M2, has the potential to provide not only neutralizing antibodies but also broadly protective cellular immunity. We evaluated the T cell response in spleens and lungs from Fluaxe-vaccinated mice with overlapping peptide pools of cH5/1-BV, cH7/3, and M2 stimulation on Day 28 (Fig. 2A). Intramuscular administration of Fluaxe significantly increased the percentage of CD3+ T cells in the lungs and bronchoalveolar lavage fluid (BALF) than that of LNP-only control group. However, the splenic CD3+ T cells remained similar to that of placebo-treated animals (Fig. 3A). Furthermore, the ratio of CD8+/CD4+ within the CD3+ population significantly increased in lung, BALF, and spleen, indicating a skewing of CD8+ T cell response by Fluaxe-vaccination (Fig. 3B). To investigate immunogen-specific T cell phenotype further, we measured the percentages of interferon-γ (IFN-γ), interleukin-2 (IL-2), and tumor necrosis factor-α (TNF-α) in CD4+ and CD8+ T cells by flow cytometry (FACS) or enzyme-linked immunospot (ELISpot), respectively. The average frequencies of cytokine-producing CD4+ and CD8+ in spleen dramatically increased by 6.7-fold and 4.8-fold for IFN-γ, 19-fold and 13.4-fold for IL-2, 4-fold and 6.9-fold for TNF-α compared with LNP group, respectively (Fig. 3C), strongly correlating with ELISpot results (Supplementary Fig. 7). Moreover, Fluaxe immunization elevated IFN-γ, IL-2, and TNF-α positive CD4+ and CD8+ T cells with significant fold change in the lungs, though less than those in the spleens (Fig. 3D). CD8+/CD4+ ratios within the CD3+cytokine+ population in the spleen tended to have an approximate average of 1 (IFN-γ: 1.584, IL-2: 1.726, TNF-α: 1.114, respectively), suggesting a balanced CD4+ and CD8+ T cell responses. In contrast, the more skewed ratios in the lungs (IFN-γ: 2.256, IL-2: 2.646, TNF-α: 1.392) strongly suggested specific CD8+ T cell responses (Fig. 3E).
Lungs, spleens, and BALFs of vaccinated mice were harvested and homogenized on Day 28 post-immunization. A CD3+ T cell ratio and B CD8+/CD4+ within CD3+ population were determined, respectively. Lymphocytes were stimulated with a peptide cocktail containing cH5/1-BV, cH7/3, and M2 peptide pools. CD4+ or CD8+ cells from C splenocytes or D lungs were stained for intracellular IFN-γ, IL-2, and TNF-α followed by FACS analysis. Percentages of cytokines+ cells within CD8+ and CD4+ T cell populations were shown. E Ratios of CD8+ /CD4+ T cells within the CD3+cytokines+ population were exhibited. The red dotted line represents a ratio of 1. Each dot or square in the bar graphs represents one individual (n = 5). Error bars indicate SEM. p values were determined using multiple t-tests with correction for multiple comparisons by the Holm-Šídák’s method (n.s. not significant; *p < 0.05, **p < 0.01, ***p < 0.001).
To evaluate possible broad-spectrum T cell responses, we assessed the single immunogen-specific CD4+ and CD8+ T cells in splenocytes after full-length HAs of H1N1, H3N2, H5N1, H7N9, B/Victoria, B/Yamagata, and intact M2 peptide cocktails stimulation, respectively (Supplementary Fig. 8A, B). Fluaxe significantly elicited broad T cell responses against all vaccine-associated influenza viruses. Interestingly, a clear skewing towards CD8+ T cell response was induced by H1N1, H7N9, B/Victoria, and B/Yamagata HA peptide pools compared to their CD4+ T cell counterparts. In contrast, H3N2, H5N1, M2 peptide cocktails tended to stimulate more robust CD4+ T cell response (Supplementary Fig. 8C), suggesting a balanced and broad distribution between CD4+ and CD8+ T cell responses by Fluaxe immunization.
Fluaxe induced long-term immunity against influenza viruses
Since transition and maintenance of influenza virus-specific memory cells is a critical factor for UIV evaluation, we examined both memory T and B cells in mice at 3 months post Fluaxe vaccination (Fig. 2A). The populations of Immunogen-specific CD4+ central memory T cells (Tcm) marked by CD44+CD62L+IFN-γ+ in splenocytes of Fluaxe-immunized mice resembled those of CD8+ Tcm cells (Fig. 4A and Supplementary Fig. 9A). Furthermore, CD4+ and CD8+ effector memory T cells (Tem) (CD44+CD62L-IFN-γ+) dramatically elevated following Fluaxe vaccination, respectively (Fig. 4B and Supplementary Fig. 9A). Tissue-resident memory (Trm) CD8+ and CD4+ T cells with CD69- and CD103-high expression also increased by peptide cocktail stimulation, suggesting heterosubtypic protection against influenza virus (Fig. 4C and Supplementary Fig. 9A). As CXCR3 mediates localization of circulating memory T cells to the respiratory tract in defending against influenza virus infection33, we corroborated that Fluaxe effectively elicited CD8+ and CD4+ circulating memory T cells with high CXCR3 expression defined by CD44+CD62L−CD69−CXCR3+ (Fig. 4D and Supplementary Fig. 9A), suggesting a robust lung tropism of memory T cells.
Spleens of prime-boost vaccinated mice were collected and homogenized on Day 104, lymphocytes were stimulated with a peptide cocktail containing cH5/1-BV, cH7/3, and M2 peptide pools. Memory cells were stained for intracellular IFN-γ. A Frequency of antigen-specific CD4+ and CD8+ Tcm (CD44+CD62L+IFN-γ+) in splenocytes. B Frequency of antigen-specific CD4+ and CD8+ Tem (CD44+CD62L−IFN-γ+) in splenocytes. C Frequency of antigen-specific CD4+ and CD8+ Trm (CD44+CD62L−CD69+CD103+IFN-γ+) in splenocytes. D Frequency of antigen-specific CD4+ and CD8+ CXCR3+Tm (CD44+CD62L−CD69−CXCR3+IFN-γ+) in splenocytes. E Frequency of H1N1 HA, H3N2 HA, H5N1 HA, H7N9 HA, IBV/Victoria HA, and IBV/Yamagata HA-specific MBC in splenocytes. Five mice per group, data are presented as Mean ± SEM. p values were determined using multiple t-tests followed by Holm-Šídák’s multiple comparisons.
To understand the formation of HA-specific memory B cells (MBC) by Fluaxe immunization, we used fluorescence-labeled HA proteins from H1N1, H3N2, H5N, H7N9, B/Victoria, and B/Yamagata subtypes to mark class-switched memory B cells classified by CD19+IgD−CD38+, respectively. Six influenza viral HA-specific MBCs were generally produced in spleens from vaccine-administered mice, suggesting that the chimeric strategy can induce multiple memory B responses against various influenza subtypes (Fig. 4E and Supplementary Fig. 9B). The strong formation of broad memory T and B cells suggested long-term immune activity of Fluaxe.
Fluaxe vaccination protects mice from a broad panel of influenza viruses
To further evaluate the potential protection provided by a prime-boost administration of Fluaxe, mice were challenged with current circulating influenza sublineages consisting of H1N1, H3N2, B/Victoria HA, and B/Yamagata. 10LD50 of A/PR/8/1934 (H1N1) virus, A/Aichi/2/1968 (H3N2), B/Brisbane/60/2008 (IBV/Victoria), or B/Phuket/3073/2013 (IBV/Yamagata) viruses were used for infection, followed by monitoring for weight loss and survival for 14 days (Fig. 5A). All vaccinated mice were protected from H1N1, IBV/Victoria, and IBV/Yamagata infection with minor morbidity and weight loss (Fig. 5B, F, H), compared with the control mice. The protection against H3N2 and was weaker than other viruses, although mice received the Fluaxe all survived, some morbidity and weight loss were observed, possibly due to high and fast pathogenicity of the ancient mouse-adapted H3N2 strain (Fig. 5D). Lungs were removed for viral load analysis on the Day 5 when the virus undergoes massive replication in mice (Fig. 5A). Fluaxe-immunization caused significantly decrease of viral loads by an average 13.6-fold for H1N1 (Fig. 5C), 5.7-fold for H3N2 (Fig. 5E), 180.9-fold for IBV/Victoria (Fig. 5G), and 3.4-fold for IBV/Yamagata (Fig. 5I). The viral load dynamics of the four influenza subtypes in Fluaxe-immunized mouse lungs showed a strong correlation with the levels of vaccine-induced neutralizing antibodies (Fig. 2C, D). Overall, Fluaxe immunization displayed strong and broad protections against IAV and IBV infections.
A Schematic diagram of mice immunization and influenza virus challenge. Created in BioRender. Li (2025) https://BioRender.com/bithl81. Prime-boost vaccination was executed on Days 0 and 14. Four influenza subtypes (10LD50) were used to challenge mice via intratracheal administration. Survival rate, weight loss, viral RNA in lung, and pathologic level were measured during 14 days post infection. The survival rate and weight loss of Fluaxe vaccinated mice were monitored post-challenge with B A/PR/8/1934 (H1N1) virus, D A/Aichi/2/1968 (H3N2) virus, F B/Brisbane/60/2008 (IBV/Victoria) virus, and H B/Phuket/3073/2013 (IBV/Yamagata) virus, respectively. Weight loss data are exhibited as Mean ± SD. The dotted lines in weight loss figures indicate the maximum body weight loss (25%) for the experiment. Kaplan–Meier survival curves of mice are shown, and significance testing was done by log-rank test (**p < 0.01). Viral RNA (vRNA) load in lungs of Fluaxe vaccinated mice were monitored post-challenge with C A/PR/8/1934 (H1N1) virus, E A/Aichi/2/1968 (H3N2) virus, G B/Brisbane/60/2008 (IBV/Victoria) virus, and I B/Phuket/3073/2013 (IBV/Yamagata) virus on Day 5 post-infection. Data of viral RNA are shown as Mean ± SEM (n = 5 per group) and significance was calculated using two-tailed unpaired t-tests (***p < 0.001), data are normalized to the LNP group, with the control value arbitrarily set to 1.
Next, we compared Fluaxe with the traditional quadrivalent inactivated influenza vaccine (QIIV). Under the same conditions of immunization dosage and schedule, Fluaxe administration induced higher levels of IgG against H3N2 and IBV/Victoria compared to QIIV, while the antibody levels against H1N1 and IBV/Yamagata were comparable (Supplementary Fig. 10A). Consistent with the IgG level, neutralizing titers of Fluaxe-vaccination group for H3N2 and IBV/Victoria increased about 2.3-fold and 2.3-fold compared with QIIV-vaccination group (Supplementary Fig. 10B). Additionally, all Fluaxe-vaccinated mice were protected from IAVs and IBVs with less morbidity and weight loss compared with the LNP group. Although QIIV could fully protect mice from death caused by IBVs infection, protection against H1N1 and H3N2 were obviously weaker than Fluaxe (Supplementary Fig. 10C–F). Collectively, Fluaxe immunization exhibited improved immunogenicity and protection.
Although preliminary experiments suggested that M2 could induce a certain level of humoral and cellular immune responses, these responses were relatively weak compared to those elicited by the chimeric HA antigen. Therefore, we further investigated the role of M2 in immune protection. A half dose of Fluaxe (Fluaxe-2) and a half dose of Fluaxe without M2 (Fluaxe-1) were used to vaccinate the mice. Although the antibody levels induced by Fluaxe-1 and Fluaxe-2 were generally similar (Supplementary Fig. 10A, B), the superior immune protection of Fluaxe-2 against H1N1 (survival rate: 80%) and H3N2 (survival rate: 40%) were observed compared to Fluaxe-1 (survival rate: 40% for H1N1; 0% for H3N2) (Supplementary Fig. 10C, D). This suggests that the addition of M2 provides higher efficacy of protection against IAV infection.
Fluaxe vaccination protects mice from lung injuries during influenza infection
As influenza infection causes acute lung injury by triggering complicated host responses, we further explored protection by Fluaxe against influenza-induced pathologic reactions. As all mouse lungs infected with different influenza subtypes present with the same visible signs of pathology, we selected the lungs of PR8-infected mice for display. Compared with normally pinkish mouse lung, lungs from virus-infected mice appear reddened due to inflammation and hemorrhage. Besides, an increased opacity and a swollen were also observed. Fluaxe-immunization obviously relieved such appearance changes of lung tissue compared to LNP group (Fig. 6A). Histologic examination of the lungs from the Fluaxe-vaccinated group revealed mild immune cell infiltration and various degrees of thickened alveolar septa. In contrast, mice inoculated with empty LNP control developed severe lesions within the lungs with higher pathological scores (Supplementary Fig. 11B), characterized by moderate infiltration of inflammatory lymphocytes, thickened alveolar septa accompanied by very severe vascular thrombus, mild epithelial exfoliation and pulmonary edema (Fig. 6B and Supplementary Fig. 11A). Moreover, compared with placebo mice, PR8 infection caused 2.9–6.7 folds decrease for several cell junction proteins in the lung, such as E-cadherin and ZO-1. In contrast, Fluaxe immunization effectively retained these proteins at a normal level regardless of PR8 infection, indicating a robust protection of lung barrier integrity34 (Fig. 6C and Supplementary Fig. 11C). Overall, our results strongly suggested that Fluaxe protected animals from respiratory failure and death caused by influenza infection.
A Images of lungs dissected from mice (Fluaxe and LNP groups) challenged with A/PR/8/1934 (H1N1) on 7 dpi. B H&E staining of PR8-infected lung tissue pathology. Tissue lesions were indicated by narrows as the following: inflammatory cell infiltration predominantly composed of granulocytes (black arrow); congestion of capillaries and venous vessels (yellow arrow); epithelial exfoliation (brown arrow); perivascular edema (purple arrow); loose connective tissue accompanied by mild hemorrhage (green arrow); lymphocyte infiltration (red arrow). Representative images from 5 mice are shown. Scale bar = 100 μm. C Representative images of immunofluorescence staining of (upper panel, green) gap junction protein E-cadherin and (lower panel, green) tight junction protein ZO-1 expressed in lung tissues from 5 mice challenged with PR8. DNA was stained by DAPI in blue. Scale bars = 100 μm. Inflammation-associated cells in lungs were detected on Day 5 post-PR8 infection by FACS. D Frequencies of alveolar macrophages (CD64+, CD11c+, Siglec F+) and interstitial peribronchial macrophages (CD11b+, CD11c+, CD64+, Siglec F−, Ly6C−) in lungs after mock (PBS) or PR8 infection. Inflammatory macrophages were stained for intracellular NLRP3. E Frequencies classical monocytes (CD11b+, Ly6C+, CD64−) of monocytes-derived cells (CD11b+, Ly6C+, CD64+) and in lungs after mock or PR8 infection. Inflammatory monocytes were stained for intracellular NLRP3. n = 5, data were presented as Mean ± SEM. Statistics were calculated using multiple t-tests with Holm-Šídák’s multiple comparisons. F Influenza virus-induced cytokine expression in the BALFs. BALF cytokine concentrations were determined by a 23-plex assay kit. n = 3. Mean values were shown. p values were calculated by comparing the LNP group and Fluaxe-vaccinated group animals that all were infected with PR8. Significance was calculated using two-tailed unpaired t-tests.
Acute lung injury (ALI) is tightly associated with host inflammatory response to influenza viral infection instead of viral load35. Two main types of inflammatory cells, macrophages and monocytes, in lung and BALF were measured for Fluaxe efficacy in eliminating respiratory inflammation induced by PR8 infection (Fig. 5A). On Day 5 post infection, mice of the LNP group infected with PR8 successively developed symptoms including febrile shaking, coat ruffling, and lethargy. Furthermore, alveolar macrophages (CD64+, CD11c+, Siglec F+) and monocytes-derived cells (CD11b+, Ly6C+, CD64+) in lungs respectively increased by 4.3-fold and 12.1-fold compared with PBS-treated animals, whereas such elevation was strongly suppressed by Fluaxe vaccination. In addition, viral infection caused a reduction of interstitial peribronchial macrophages (CD11b+, CD11c+, CD64+, Siglec F−, Ly6C−) and classical monocytes (CD11b+, Ly6C+, CD64−) in lungs of LNP-vaccinated mice, whereas Fluaxe vaccination retained the interstitial peribronchial macrophages at a normal level and further down-regulated classical monocytes (Fig. 6D, E and Supplementary Fig. 12C). These inflammatory cells in BALFs were significantly recruited by IAV infections, with a slight increase in Fluaxe-vaccinated mice. In the non-infected groups, besides other inflammatory cells, more alveolar macrophages were recruited in the BALFs of the Fluaxe group, probably reflecting some regulation of innate immunity by Fluaxe immunization (Supplementary Fig. 12A, B). Because activation of NLRP3 inflammasome is associated with influenza-induced inflammatory diseases, we performed intracellular NLRP3 staining in macrophages and monocytes. Fluaxe immunization strongly suppressed NLRP3 compared with the LNP group during PR8 infection, exhibiting a significant potential in regulating inflammation elicited by influenza infection (Fig. 6D, E).
To elucidate the effect of Fluaxe-immunization on severe inflammation, we profiled a panel of cytokines in BALFs. During viral infection, a dynamic balance of pro- and anti-inflammatory cytokines is critical for host protection. Compared with blank control, Fluaxe immunization rarely led to the secretion of inflammatory cytokines in BALFs without viral infection. PR8 infection stimulated a broad range of inflammatory cytokines, whereas BALFs from Fluaxe-vaccinated animals exhibited complex patterns on pro- and anti-inflammatory cytokines to guard against viral infection and control inflammation. Specifically, during PR8 infection, Fluaxe induced higher levels of Th1 (IFN-γ, IL-2, TNF-α) and Th2 (IL-4, IL-5, IL-9, IL-10) cytokines compared with the LNP only group, which helped to activate and enhance cellular and humoral immunity to eradicate the virus. Of note, IL-10 is an anti-inflammatory cytokine with roles in preventing inflammatory and autoimmune pathologies36. In addition, Fluaxe vaccination suppressed pro-inflammatory cytokines and chemokines such as IL-1α, MCP-1, MIP-1α (1β), and G-CSF, leading to decreased recruitment of macrophages and monocytes with excessive NLRP3 inflammasome in the lungs of Fluaxe-vaccinated mice (Fig. 6F). Together, Fluaxe immunization not only protects host animals from viral infection, but also alleviates pathological symptoms induced by excessive inflammation.
Discussion
The successful development of UIV will significantly reduce the global public health burden exerted by seasonal influenza infections. In this study, we constructed an mRNA vaccine Fluaxe, encoding two novel chimeric HAs and the intact M2, protecting against at least six major influenza subtypes from circulating IAV group 1, IAV group 2, and two IBV lineages. Our vaccine design of including only the B/Victoria HA sequence in Fluaxe takes advantage of the high degree of similarity shared between the two IBV HA proteins, as supported by strong cross-reactivity against B/Yamagata by Fluaxe vaccination. Inclusion of complete M2 further enhanced and expanded the protection power provided by HA immunogens. This novel cocktail mRNA vaccine stimulates robust humoral and cellular responses against a wide range of influenza viruses as shown by our in vitro and in vivo data, including strong protection of mice from lethal viral challenges.
Fluaxe promoted broad and effective neutralization activity against influenza viruses. In our chimeric design, heterogenous head-stem fusion constructs of HAs, namely cH5/1-BV and cH7/3, not only retained the strong immunogenicity of HAs but also encoded five major influenza antigens within two mRNA sequences. Compared with relatively low levels of neutralizing activities induced by some influenza vaccines containing cHA or HA stem region10,14, broadly neutralizing antibodies (bnAbs) elicited by two doses of Fluaxe were verified by both in vitro and in vivo neutralization assays. Several properties of Fluaxe contributed to bnAb generation. First, the M2 sequence is more conserved among influenza subtypes, with the potential to elicit bnAbs and cross-protection37. Indeed, our data demonstrated that M2-elicited antibodies are highly cross-reactive to heterologous influenza subtypes, and M2 potentiated the cross-protection of Fluaxe against IAVs (Supplementary Figs. 4 and 10C, D). Second, the two IBV lineages shared a very high degree of similarity in their HA sequences, thus immunogenicity. Third, proper translation, folding and assembly of chimeric HA proteins in host’s native cellular environment may enhance heterosubtypic protective immunity38,39. Consistently, Fluaxe vaccination resulted in a robust cross-protective response against IAV group 1 (H2, H6, H8, H11, H13, H16), IAV group 2 (H15), and IBV/Yamagata, supporting the tandem arrangement of divergent HA proteins as an effective chimeric strategy for stimulating broad cross-protection and simplifying the vaccine formula.
Fluaxe exhibited excellent protection against influenza viral challenges. It has been reported that protection from influenza not only depends on robust adaptive immunity but also is mediated by inhibiting inflammatory responses. Despite that both vaccine administration and viral infection are inflammation stimuli, Fluaxe vaccination effectively decreased hyper-inflammation and ultimately the degree of ALI induced by viral infection. Multifunctional innate immune cells and inflammatory cytokines could be activated or induced by two doses of Pfizer-BioNTech BNT162b2 mRNA vaccine to guard patients from COVID-19 infection and over-activated inflammation40. Influenza virus infection recruited alveolar macrophages and monocytes with excessive NLRP3 inflammasome activation in the lung. Fluaxe administration significantly improved such inflammatory responses and maintained the levels of these cells close to normal. Opposite to alveolar macrophages, interstitial macrophages, which are immune homeostasis regulatory cells41, turned out to decrease during influenza infection. Interestingly, Fluaxe significantly prevented such reduction (Fig. 6D). Both a single-cycle influenza virus vaccine and an influenza-vectored COVID-19 vaccine have been reported to significantly suppress both total pro- and anti-inflammatory cytokines during viral infection42,43. In contrast, Fluaxe immunization selectively upregulated Th-associated cytokines and downregulated pro-inflammatory cytokines, which might be attributed to the robust stimulation of T cell responses and homeostasis maintenance by the mRNA vaccine. This more balanced regulation of inflammatory cytokines induced by Fluaxe not only accelerated viral clearance but also protected the host from excessive inflammation.
Moreover, robust memory B and T cells elicited by Fluaxe supported durable protection. Despite the HA head domain is more likely to trigger specific antibody production than the stem domain, chimeric HA stem domains (H1N1 and H3N2) of Fluaxe resulted in more specific MBCs than the head domains from H5N1 and H7N9, suggesting a crucial role of the conserved stem region in the long-term immunogenicity of Fluaxe. Although emerging influenza subtypes may effectively evade antibody neutralization, it is more difficult to escape from T cell immunity because viral proteins have multiple conserved T cell epitopes44. On one hand, memory CD4+ T cells contribute to recalling memory T follicular helper cells and enhancing generation of neutralizing antibodies45; on the other hand, CD8+ Trm cells co-expressing CD103 and CD69 provide heterosubtypic protection against influenza virus46. Multifunctional CD4+ and CD8+ Tm cells were successfully boosted by Fluaxe vaccination (Fig. 4A–C). Moreover, CXCR3hi Tm cells are inclined to a respiratory tract localization (Fig. 4D), which can promote their protective capacity47.
T cell Immunity In the lung is essential for clearing virus-infected cells and protecting the host from severe symptoms due to influenza infection. Compared to traditional influenza vaccines, intramuscular administration of mRNA-LNP vaccines induced potent pulmonary T cell responses46. Prime-boost vaccination with Fluaxe led to broadly reactive cellular responses and a significant CD8+ T cell bias in the lung, compared to a more balanced CD8+/CD4+ ratio in the T cell population in the spleen (Fig. 3E). As conserved influenza viral proteins such as NP, M1 or PB1 have often been used to stimulate broad cellular responses in previous studies48,49,50, we reported here that mRNA-based Fluaxe immunization also elicited HA- and M2-specific T cell immunity against heterosubtypic influenza viruses. Interestingly, multi-targeting HA overlapping peptide pools stimulation caused inconsistent T cell skewing. It might be attributed to complex antigen recognition and processing mechanisms of chimeric HA proteins.
LNPs have been reported to possess adjuvant activity, which can induce robust T follicular helper cell and humoral responses51,52. In line with this, the LNPs used in Fluaxe were also shown to enhance class-switched B cell levels classified by CD19+CD38+IgD−IgG+, but negligibly impact the ratio of CD8+/CD4+ within the CD3+ T cells in spleen (Supplementary Fig. 13A, B). In addition, LNPs promote the increase in alveolar macrophages but not interstitial peribronchial macrophages, which is consistent with Fluaxe-immunization, suggesting its potential involvement in regulating pulmonary inflammatory responses (Fig. 6D and Supplementary Fig. 13C). Meanwhile, either empty LNPs or Fluaxe administration rarely induce aberrant secretion of cytokines in BALFs without viral infection (Fig. 6F and Supplementary Fig. 13D). Although comprehensive investigations are yet to be conducted, current data suggest that LNPs function as potent adjuvants to boost host immune responses.
Taken together, our mRNA-based Fluaxe vaccine featuring chimeric HAs and intact M immunogens strongly activated comprehensive host immunity against multiple influenza viruses and protected mice from severe lung injury induced by viral infection, warrant further investigations as a potential UIV.
Methods
Ethics statement
Mice protocols were performed following the recommendations in the Guidelines for the Care and Use of Laboratory Animals at the Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences (SYXK(JING)2022-0023). All experimental operations were executed under anesthesia conditions and abide by relevant ethical regulations. Female BALB/c mice were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd (SCXK(JING)2021-0006), and executed and housed in the Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences, Beijing, China.
Cells and viruses
HEK293T (ATCC) and MDCK (ATCC) were maintained in the Dulbecco’s modified Eagle’s medium (DMEM; Gibico) supplemented with 10% fetal bovine serum (FBS; Gibico) and 1× penicillin-streptomycin (Macgene, CC004). A/Puerto Rico/8/34 (H1N1), A/California /07/2009 (H1N1), A/Hong Kong/4801/2014 (H3N2), B/Brisbane/60/2008 (B/Victoria), and B/Phuket/3073/ 2013 (B/Yamagata) were kindly provided by the Institute of Medical Biology, Chinese Academy of Medical Sciences. A/Aichi/2/1968 (X31) (H3N2), A/Du Jun (Gui Zhou)/1230/2013 (H3N2), A/Yu Zhou (Chong Qing)/1230/2013 (H3N2) were provided by the Chinese National Influenza Center, CNIC. A/WSN/1933 was kept in our own lab.
Vaccines
mRNA sequences of the designed Fluaxe immunogens were individually optimized with a sequence optimization algorithm developed and owned by Rinuagene, Inc. All mRNAs of Fluaxe were produced by IVT and encapsuled into LNPs, then adjusted to desired concentrations, passed through a 0.22 µm filter, and stored at −20 °C until use. All formulations were tested for particle size, distribution, RNA concentration, purity, and encapsulation efficiency.
Mouse studies
Six-week-old female specific-pathogen-free BALB/c mice (n = 5) were immunized intramuscularly with Fluaxe containing a total 15 μg mRNA (5 μg for every component) in a volume of 100 μL per hind limb or with 100 μL LNP as a negative control and boosted with an equivalent dose on Day 14 post initial immunization. The mice were euthanized by CO2 asphyxiation, and the tissues were removed from each animal immediately after euthanasia.
For antibody response evaluation, blood of all groups of BALB/c mice was collected via retro-orbital bleeding on Days 7, 14, 21, and 28 after initial immunization, followed by an enzyme-linked immunosorbent assay (ELISA) or cell-based ELISA to detect the influenza virus-specific IgG. In addition, sera collected on Day 28 were used for neutralizing antibody analysis.
For T cell response analysis, mice were euthanized by CO2 asphyxiation on Day 28, and spleen, lung, and BALF were harvested for cellular immunity and long-lived immunity analysis by ELISpot assay or flow cytometry as described below.
To evaluate the protective efficacy against influenza, vaccinated mice were anesthetized with anesthetic tribromoethanol (200 mg/kg) for 5 min prior to receiving a viral challenge. The mice were positioned on a 45° tilting intubation stand to visualize the trachea, and a 1.22 mm endotracheal tube was used to deliver 50 μL of one of the following virus strains: A/Puerto Rico/8/34 (H1N1), A/Aichi/2/1968 (X31) (H3N2), B/Brisbane/60/2008 (B/Victoria), or B/Phuket/3073/2013 (B/Yamagata), each containing 10LD50. This procedure was performed on Day 28 post-vaccination. The mice were monitored daily for body weight loss over a period of 2 weeks, and those reaching a 25% reduction in their initial body weight were humanely euthanized. Lung tissue samples were collected from each group for pathological analysis during this procedure. For viral load analysis, lung tissue samples were harvested from each group on Day 5 post-challenge and influenza virus RNA were quantified by RT-qPCR.
For inflammatory response analysis, mice challenged with A/Puerto Rico/8/34 (H1N1) were euthanized by CO2 asphyxiation on Day 5 post-challenge. Lungs and BALFs were collected and applied for the detection of inflammatory cells and cytokines.
Tissue processing
Serum collection
Blood samples were drawn from the retro-orbital sinus of the mice and collected into 1.5 mL microtubes without anticoagulants. The samples were incubated at 37 °C for 1 h and then centrifuged at 900 × g for 30 min. The separated serum was then stored at −20 °C until further use.
Spleen and lung
Spleen and lung tissues were cut into ~5 mm3 cubes and homogenized using a 70 μm sieve (BD Falcon) using the plunger of a 5 mL sterile injection syringe to remove larger debris. The resulting single-cell suspension was collected in RPMI-1640 complete culture medium (Gibco) supplemented with 10% FBS and 1×penicillin-streptomycin. The cells were then washed twice with PBS and treated with red blood cell lysis buffer at room temperature for 3 min to remove remaining red blood cells. The cells were subsequently resuspended in RPMI-1640 complete culture medium containing 10% FBS and 1×penicillin-streptomycin for use in ELISpot assays and flow cytometry.
BALF
The trachea of euthanized mouse was exposed and inserted with a puncture needle, and the lungs were lavaged three times with 500 μL PBS. Cells derived from BALFs were collected by centrifugation for 10 min at 500 × g and resuspended in RPMI-1640 (Gibco) complete culture medium containing 10% FBS and 1×penicillin-streptomycin for flow cytometry.
ELISA
The serum IgG antibody was measured using an ELISA. Briefly, ELISA plates were coated with 2 μg/well recombinant HA proteins (Recombinant H1N1 HA (40787-V08H), H3N2 HA (40789-V08H, H5N1 HA (40022-V08H), H7N9 HA (40325-V08H), IBV (B/Washington/02/2019) HA (40722-V08H), IBV (B/Phuket/3037/2013) HA (40722-V08H)) from homologous wild-type viruses (Sino Biological) at 4 °C overnight. ELISA plates were blocked with 2% bovine serum albumin (BSA; Sigma) in PBS-0.05% Tween 20 (PBST) for 1 h at 37 °C, and then dry at room temperature. A proper dilution (initial dilution 1:100) of serum samples was added to wells (100 μL/well) and incubated for 1 h at 37 °C. After washing five times with PBST, HRP-conjugated goat anti-mouse IgG antibody (Proteintech) diluted at 1:2500 in PBST was added to the plate and incubated for another 1 h at 37 °C. Substrate tetramethylbenzidine (TMB; Tiangen Biotech) was added to the plate after washing and incubated for 30 min. The reaction was stopped with the addition of 0.5 M H2SO4. Plates were read using a spectrophotometer (EnSpire) at a wavelength of 450 nm. The cut-off value was defined as 2.1×OD value of blank control. Within the appropriate OD value range, the formula (OD value of the sample well/cut-off value × dilution) can be used to calculate the endpoint antibody titer. ELISA kits for mouse IFN-γ (PI507), IL-2 (PI575), and TNF-α (PT513) were purchased from Beyotime. All procedures were executed strictly according to the manufacturer’s instructions provided with the kits. Cytokine concentrations were determined by plotting sample optical densities against the standard curve.
Cell-based ELISA were performed by using 96-well plates seeded with 2 × 104 cells/mL MDCK cells which respectively infected with A/Puerto Rico/8/34 (H1N1), A/Aichi/2/1968 (X31) (H3N2), B/Brisbane/60/2008 (B/Victoria), and B/Phuket/3073/2013 (B/Yamagata) at a multiplicity of infection (MOI) of 1 for 24 h at 37 °C with 5% CO2. Then the virus-infected cells were fixed with 4% paraformaldehyde at 4 °C for 24 h and permeabilized with 0.1% Triton X-100 for 8 min at RT. The cells were washed three times with PBS. The remainder of the cell-based ELISA was performed as described above.
Hemagglutination inhibition (HAI) assay
Sera samples from mice were tested using an HAI in V-bottom 96-well microtiter plates with 1% guinea pig red blood cells. In brief, sera were incubated for 24 h at 37 °C with receptor-destroying enzyme (RDE) (1 volume sera: 3 volume RDE) and then heated to inactivation at 56 °C for 30 min. Sera were diluted serially starting at 1:8 in PBS and then added to the each well with an equal volume of four agglutinating doses of virus (H1N1, H3N2, IBV/Victoria, IBV/Yamagata) at room temperature for 1 h. Commercial HAI antigens (HVRIWK) containing H5-Re13 or H7-Re4 inactivated viruses were used to detect the HAI titers against H5- and H7-containing viruses. RDE-treated sera were mixed with an equal volume of four agglutinating doses of H5-Re13 or H7-Re4 inactivated virus. Finally, an equal volume of 1% (v/v) guinea pig red blood cells was added to each well. The HAI titers were read after 45 min at RT.
Microneutralization (MN) assay
MDCK cells were subcultured into 96-well plates at a concentration of 2 × 104 cells for 24 h. Two-fold dilutions of RDE-treated sera starting at 1:10 were incubated with an equal volume of 100× median tissue culture infectious dose (TCID50) of influenza virus for 1 h at 37 °C in new 96-well plates. Then the sera-virus mixture was transferred to MDCK cells for another 4 h. Then the sera-virus mixture was replaced by new cell culture medium. The cells were observed for the presence of cytopathic effect after 3–5 days at 37 °C. Neutralization titer was defined as the reciprocal of the highest diluted serum that completely neutralizes virus infection of MDCK cells.
Pseudovirus-based neutralization assay
Pseudovirus-based neutralization activity of immunized mouse serum was measured by using single-round influenza pseudoviruses (H1–H16) generated by reverse genetics. pHW2000 eight-plasmid system, which displaces HA coding sequence with the Gaussia luciferase sequence and 16 HAs expressing plasmid were kept in our lab. In brief, 2 × 104 cells/well MDCK cells were seeded in a 96-well plate and cultured for approximately 24 h. 2-fold serially diluted sera from immunized mice commencing with an initial dilution of 1:10 were incubated with an equal volume of 100TCID50 influenza pseudotyped virus for 1 h at 37 °C. Then the mixture was added to pre-seeded MDCK cells for 4 h incubation. Then the sera-virus mixture was replaced by new cell culture medium and kept incubating for another 48 h. The Gaussia luciferase activity in supernatants was measured by using the Luciferase Assay System (Promega) with Centro LB960 Microplate Luminometer (Berthold Technologies). The PNT50 was defined as the dilution of the serum at which the relative luminescence units were reduced by 50% compared with those derived from cells infected with influenza pseudoviruses in the absence of serum. The PNT50 was interpolated from the neutralization curves determined by 4-parameter nonlinear regression, i.e., the log (inhibitor) vs. normalized response (Variable slope), using GraphPad Prism 8.0. Each sample was tested three times.
Passive protection of sera
Sera from Fluaxe-vaccinated mice were collected and diluted with PBS at 1:20, 1:40, 1:80 and 1:160. Then diluted sera were respectively incubated with 10LD50 A/Puerto Rico/8/34 (H1N1), A/Aichi/2/1968 (X31) (H3N2), B/Brisbane/60/2008 (B/Victoria), and B/Phuket/ 3073/2013 (B/Yamagata) for 1 h at 37 °C. Naïve mice were intratracheally injected with 50 μL sera-virus mixture, and the survival rate was recorded for 14 days post infection.
Flow cytometry
The mice spleen, lung, or BALF samples were harvested from sacrificed mice for the preparation of a single-cell suspension. HA-specific overlapping peptide pools (2.5 µg/mL, SBS Genetech) and Brefeldin A (10 µg/mL, Solarbio) were added into the cell suspension. After 24 h stimulation, cells were washed with PBS and incubated with the surface-staining antibodies at a ratio of 1:100 (T cells: anti-CD3 APC, anti-CD4 APC/Cyanine7, anti-CD8a FITC, anti-CD44 PerCP, anti-CD62L Alexa Fluor 700, anti-CD69 PE/Cyanine7, anti-CD103 BV510, anti-CXCR3 BV421; B cells: anti-CD19 PE, anti-CD38 APC/Cyanine7, anti-IgD PE/Cyanine7, anti-IgG FITC, anti-Streptavidin PE/Cyanine5, anti-Streptavidin BV421; Inflammatory cells: anti-CD64 FITC, anti-Ly-6C APC, anti-CD11c PerCP, anti-CD11b APC/Cyanine7, anti-Siglec-F PE/Cyanine7; All antibodies purchased from Biolegend) for 1 h on ice. After washing twice with PBS, all cells were fixed and permeabilized using Fixation/Permeabilization kit (Invitrogen). Intracellular cytokines or inflammatory factors were respectively stained with anti-IFN-γ PE, anti-IL-2 PE, anti-TNF-α PE (Biolegend), or anti-NLRP3 (Abcam) conjugated with anti-rabbit Alexa Fluor 555 (Invitrogen) and detected by flow Cytometry (Cytoflex, Beckman Coulter).
For HA-specific memory B cell detection, Influenza HA proteins (SinoBiological) were biotinylated by EZ-Link™ Sulfo-NHS-LC-Biotinylation Kit (Thermo Fisher) and quantified by Pierce BCA Assay (Thermo Fisher) according to the manufacturer’s instructions. Biotinylated proteins were incubated with anti-Streptavidin PE/Cyanine5 at a mass ratio of 6.25:1(100 ng HA with 16 ng SA) of for 1 h at 4 °C. Streptavidin-BV421 was not multimerized with biotinylated HA proteins and was used as a decoy probe to rule out cells that non-specifically bind streptavidin. A total of 200 ng HA-PE/Cyanine5 were used to label HA-specific memory B cells.
Enzyme-linked immunospot (ELISpot) assay
The mouse spleen lymphocytes were collected, and antigen-specific cytokine-producing T cells were visualized by using IFN-γ, IL-2, or TNF-α ELISpot assay kits (Mabtech) as described in the instruction manual. In brief, 100,000 mouse spleen lymphocyte cells were added to cytokine coated 96-well plates and cocultured with HA-specific overlapping peptide pools (2.5 µg/mL, SBS Genetech) for 24 h at 37 °C, while cells without peptide served as negative control. Then incubated with biotinylated anti-mouse IFN-γ or IL-2 or TNF-α detection antibody for 1 h at room temperature. Finally, the alkaline phosphatase-labelled streptavidin (ALP-SA) was added until the spots appeared. Count spots using a CTL ImmunoSpot S6 Universal Analyzer and calculate the number of spot-forming cells (SFC) per million cells.
Western blotting
Vaccine antigen expression was detected by Western blot analysis. A proper number of HEK293T cells were plated in a 6-well plate and 1 μg vaccines of Influenza virus were added after 24 h. After incubation for 48 h, cells were collected, washed with PBS (PH 7.4), and lysed in loading buffer. Proper amounts of lysate samples were electrophoresed on 10% SDS-polyacrylamide gels and then transferred to 0.45 μm polyvinylidene difluoride (PVDF) membranes (Millipore). The membranes were blocked with 5% skim milk, and probed with primary antibody that specifically recognizes cH5/1-BV (SinoBiological, 11062-T62), H7/3 HA (SinoBiological, 11082-MM04) or M2 (ab56086) of Influenza virus. After being washed with PBST (PBS plus 0.1% Tween 20), the blots were incubated with HRP-conjugated antibodies (1:5000), which obtained from Beyotime Biotechnology for 1 h. Blots were developed using Western Lighting chemiluminescence reagent (Millipore) and the images were acquired on ChemiDocTM MP Imaging System (BIO-RAD).
H&E staining
Lung fragments from influenza virus-infected mice were fixed in 4% paraformaldehyde for more than 2 days, and then embedded in paraffin and sectioned to 5 μm slices. The fixed lung sections were subjected to hematoxylin and eosin (H&E) staining. Images were captured by 3DHISTECH (Pannoramic MIDI) scanner. Pulmonary pathological scores were measured according to 4-grade scoring system.
Immunofluorescence staining
For cell sample preparation, a proper number of MDCK cells were plated in 20 mm glass bottom dishes and 1 μg vaccines of Influenza virus were added for incubation 48 h, and fixed with 4% (w/v) paraformaldehyde for 20 min at room temperature. For mouse tissue sample preparation, the paraffin slices were rehydrated and incubated in an antigen retrieval solution. For immunofluorescence staining, the cells and tissue samples were permeabilized with 0.2% Triton X-100 and blocked with 2% BSA and 5% donkey normal serum for 1 h, respectively. All samples were stained with the primary antibodies. After washing with PBS, the cells or tissue sections were incubated with the appropriate secondary antibody (Thermo Fisher) at room temperature for 1 h. Primary antibodies include anti-influenza A virus HA for H5/1-BV (SinoBiological, 11062-T62), anti-influenza A virus HA for H7/3 (SinoBiological, 11082-MM04), anti-M2 (Abcam, ab56086), anti-ZO-1 (Abcam, ab221547), and anti-E-Cadherin (CST, 3195T). Nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI). Images were acquired with a ×100 Oil objective on Olympus FV3000 and 3DHISTECH (Pannoramic MIDI) scanner, respectively.
RNA isolation and RT-qPCR
To detect the influenza virus vRNA levels in lung tissues of infected mice, total RNA was extracted using TRIzol reagent (Sigma) according to the manufacturer’s protocol. Real-time quantitative PCR was carried out with the One Step PrimeScript™ RT-PCR Kit (Takara, RR064). Influenza virus genomic RNA quantification was performed by targeting the NP gene with specific primers: H1N1 forward primer: 5’-CCACAAGAGGGGTCCAGATT-3’; H1N1 reverse primer: 5’-GCACTGAGAATGTAGGCTGC-3’; H3N2 forward primer: 5’- GCAGTCAAAGGAATCGGGAC-3’; H3N2 reverse primer: 5’-TCCACCATTGCTCTTTGTGC-3’; IBV/Victoria forward primer: 5’-AATGGGAGCAGCTCTGATGT-3’; IBV/Victoria reverse primer: 5’-TCTGCATCACGTCCCTCAAT-3’; IBV/Yamgara forward primer: 5’-GGTTGGC ACGGATACACATC-3’ IBV/Yamgara reverse primer: 5’-GAGTTCATCCATGGCACCAC-3’. Mouse glyceraldehyde-3-phosphate dehydrogenase (mGAPDH) RNA were amplified with primer pair: 5’- GGGTCCCAGCTTAGGTTCAT-3’ and 5’- CATTCTCGGCCTTGACTGTG -3’, the results served as an internal control to normalize influenza virus RNA. The reverse transcription condition was 1 cycle at 42 °C for 5 min. The PCR conditions were 40 cycles at 95 °C for 5 s, and 60 °C for 34 s. Formula 2−ΔΔCt was used to calculate the relative RNA levels of influenza subtypes in lungs, which represents the fold changes in the expression of the virus RNA in the Fluaxe group relative to the LNP group.
Measurement of BALF cytokines and chemokines
BALF samples were collected and stored at −80 °C. The Concentration of cytokines and chemokines was determined by the Bio-Plex Pro Mouse Cytokine 23-plex Assay kit (BIO-RAD, M60009RDPD) and detected on a Luminex (Luminex, X-200) according to the manufacturer’s instructions. Data were analyzed using Milliplex Analyst software.
Statistical analysis
All data were analyzed and processed using GraphPad Prism 8 software. Quantification of immunofluorescent images was performed by using Image-Pro 10 software. All grouped data are reported as means ± Standard Error of the Mean (SEM) or Standard deviation (SD) and statistical differences between groups were analyzed using two-way ANOVA with multiple comparisons test, log-rank test, unpaired t-tests, or multiple t-tests. The levels of significance were indicated as * (p < 0.05); ** (p < 0.01); *** (p < 0.001); and n.s. denotes not significant (p ≥ 0.05).
Data availability
The vaccine sequence data are not publicly available due to intellectual property restrictions, but may be obtained from the corresponding author upon reasonable request. Any additional information required to use the data reported in this study is also available from the corresponding author upon request.
References
Iuliano, A. D. et al. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet 391, 1285–1300 (2018).
McLean, H. Q. & Belongia, E. A. Influenza vaccine effectiveness: new insights and challenges.Cold Spring Harb. Perspect. Med 11, a038315 (2021).
Neher, R. A., Bedford, T., Daniels, R. S., Russell, C. A. & Shraiman, B. I. Prediction, dynamics, and visualization of antigenic phenotypes of seasonal influenza viruses. Proc. Natl Acad. Sci. USA 113, E1701–E1709 (2016).
Luksza, M. & Lässig, M. A predictive fitness model for influenza. Nature 507, 57–61 (2014).
Uyeki, T. M., Hui, D. S., Zambon, M., Wentworth, D. E. & Monto, A. S. Influenza. Lancet 400, 693–706 (2022).
Erbelding, E. J. et al. A universal influenza vaccine: the strategic plan for the National Institute of Allergy and Infectious Diseases. J. Infect. Dis. 218, 347–354 (2018).
Paules, C. I., Marston, H. D., Eisinger, R. W., Baltimore, D. & Fauci, A. S. The pathway to a universal influenza vaccine. Immunity 47, 599–603 (2017).
Skarlupka, A. L., Bebin-Blackwell, A. G., Sumner, S. F. & Ross, T. M. Universal influenza virus neuraminidase vaccine elicits protective immune responses against human seasonal and pre-pandemic strains. J. Virol. 95, e0075921 (2021).
Lo, C. Y. et al. Universal influenza vaccine based on conserved antigens provides long-term durability of immune responses and durable broad protection against diverse challenge virus strains in mice. Vaccine 39, 4628–4640 (2021).
Margine, I. et al. Hemagglutinin stalk-based universal vaccine constructs protect against group 2 influenza A viruses. J. Virol. 87, 10435–10446 (2013).
Steel, J. et al. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. mBio 1, https://doi.org/10.1128/mbio.00018-10 (2010).
McCraw, D. M. et al. Designed nanoparticles elicit cross-reactive antibody responses to conserved influenza virus hemagglutinin stem epitopes. PLoS Pathog. 19, e1011514 (2023).
Moin, S. M. et al. Co-immunization with hemagglutinin stem immunogens elicits cross-group neutralizing antibodies and broad protection against influenza A viruses. Immunity 55, 2405–2418.e7 (2022).
Nachbagauer, R. et al. A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nat. Med. 27, 106–114 (2021).
Mallajosyula, V. V. et al. Influenza hemagglutinin stem-fragment immunogen elicits broadly neutralizing antibodies and confers heterologous protection. Proc. Natl Acad. Sci. USA 111, E2514–E2523 (2014).
Subbiah, J. et al. A chimeric thermostable M2e and H3 stalk-based universal influenza A virus vaccine. NPJ Vaccines 7, 68 (2022).
Impagliazzo, A. et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 349, 1301–1306 (2015).
McCown, M. F. & Pekosz, A. Distinct domains of the influenza a virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production. J. Virol. 80, 8178–8189 (2006).
Neirynck, S. et al. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat. Med. 5, 1157–1163 (1999).
Barbier, A. J., Jiang, A. Y., Zhang, P., Wooster, R. & Anderson, D. G. The clinical progress of mRNA vaccines and immunotherapies. Nat. Biotechnol. 40, 840–854 (2022).
Leung, N. H. L. et al. Comparative antibody and cell-mediated immune responses, reactogenicity, and efficacy of homologous and heterologous boosting with CoronaVac and BNT162b2 (Cobovax): an open-label, randomised trial. Lancet Microbe 4, e670–e682 (2023).
Xiong, F. et al. An mRNA-based broad-spectrum vaccine candidate confers cross-protection against heterosubtypic influenza A viruses. Emerg. Microbes Infect. 12, 2256422 (2023).
Freyn, A. W. et al. A multi-targeting, nucleoside-modified mRNA influenza virus vaccine provides broad protection in mice. Mol. Ther. 28, 1569–1584 (2020).
Pardi, N. et al. Development of a pentavalent broadly protective nucleoside-modified mRNA vaccine against influenza B viruses. Nat. Commun. 13, 4677 (2022).
Furey, C. et al. Development of a nucleoside-modified mRNA vaccine against clade 2.3.4.4b H5 highly pathogenic avian influenza virus. Nat. Commun. 15, 4350 (2024).
Ma, N. et al. Development of an mRNA vaccine against a panel of heterologous H1N1 seasonal influenza viruses using a consensus hemagglutinin sequence. Emerg. Microbes Infect. 12, 2202278 (2023).
Xu, S., Zhang, B., Yao, J. & Ruan, W. A new H9 influenza virus mRNA vaccine elicits robust protective immunity against infection. Vaccine 41, 2905–2913 (2023).
Lee, I. T. et al. Safety and immunogenicity of a phase 1/2 randomized clinical trial of a quadrivalent, mRNA-based seasonal influenza vaccine (mRNA-1010) in healthy adults: interim analysis. Nat. Commun. 14, 3631 (2023).
Arevalo, C. P. et al. A multivalent nucleoside-modified mRNA vaccine against all known influenza virus subtypes. Science 378, 899–904 (2022).
Krammer, F. et al. Influenza. Nat. Rev. Dis. Prim. 4, 3 (2018).
Taubenberger, J. K. & Kash, J. C. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 7, 440–451 (2010).
Shi, J., Zeng, X., Cui, P., Yan, C. & Chen, H. Alarming situation of emerging H5 and H7 avian influenza and effective control strategies. Emerg. Microbes Infect. 12, 2155072 (2023).
Slütter, B., Pewe, L. L., Kaech, S. M. & Harty, J. T. Lung airway-surveilling CXCR3(hi) memory CD8(+) T cells are critical for protection against influenza A virus. Immunity 39, 939–948 (2013).
Schmit, T. et al. Interferon-γ promotes monocyte-mediated lung injury during influenza infection. Cell Rep. 38, 110456 (2022).
Coates, B. M. et al. Inflammatory monocytes drive influenza A virus-mediated lung injury in juvenile mice. J. Immunol. 200, 2391–2404 (2018).
Saraiva, M. & O’Garra, A. The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 10, 170–181 (2010).
Kim, M. C. et al. Virus-like particles containing multiple M2 extracellular domains confer improved cross-protection against various subtypes of influenza virus. Mol. Ther. 21, 485–492 (2013).
Krammer, F., Pica, N., Hai, R., Margine, I. & Palese, P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J. Virol. 87, 6542–6550 (2013).
Schlake, T., Thess, A., Thran, M. & Jordan, I. mRNA as novel technology for passive immunotherapy. Cell Mol. Life Sci. 76, 301–328 (2019).
Li, C. et al. Mechanisms of innate and adaptive immunity to the Pfizer-BioNTech BNT162b2 vaccine. Nat. Immunol. 23, 543–555 (2022).
Schyns, J. et al. Non-classical tissue monocytes and two functionally distinct populations of interstitial macrophages populate the mouse lung. Nat. Commun. 10, 3964 (2019).
Zhang, L. et al. Intranasal influenza-vectored COVID-19 vaccine restrains the SARS-CoV-2 inflammatory response in hamsters. Nat. Commun. 14, 4117 (2023).
Zheng, M. Z. M. et al. Single-cycle influenza virus vaccine generates lung CD8(+) Trm that cross-react against viral variants and subvert virus escape mutants. Sci. Adv. 9, eadg3469 (2023).
Noh, J. Y., Jeong, H. W., Kim, J. H. & Shin, E. C. T cell-oriented strategies for controlling the COVID-19 pandemic. Nat. Rev. Immunol. 21, 687–688 (2021).
Sircy, L. M. et al. Generation of antigen-specific memory CD4 T cells by heterologous immunization enhances the magnitude of the germinal center response upon influenza infection. PLoS Pathog. 20, e1011639 (2024).
Zens, K. D., Chen, J. K. & Farber, D. L. Vaccine-generated lung tissue-resident memory T cells provide heterosubtypic protection to influenza infection. JCI Insight 1, e85832 (2016).
Jiang, X. et al. Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature 483, 227–231 (2012).
Grant, E. J. et al. Broad CD8(+) T cell cross-recognition of distinct influenza A strains in humans. Nat. Commun. 9, 5427 (2018).
Tu, W. et al. Cytotoxic T lymphocytes established by seasonal human influenza cross-react against 2009 pandemic H1N1 influenza virus. J. Virol. 84, 6527–6535 (2010).
van de Ven, K. et al. A universal influenza mRNA vaccine candidate boosts T cell responses and reduces zoonotic influenza virus disease in ferrets. Sci. Adv. 8, eadc9937 (2022).
Alameh, M. G. et al. Lipid nanoparticles enhance the efficacy of mRNA and protein subunit vaccines by inducing robust T follicular helper cell and humoral responses. Immunity 55, 1136–1138 (2022).
Verbeke, R., Hogan, M. J., Loré, K. & Pardi, N. Innate immune mechanisms of mRNA vaccines. Immunity 55, 1993–2005 (2022).
Acknowledgements
We thank the National Microbial Resource Center (No. NMRC-2020-3) and the CAMS Collection Center of Pathogenic Microorganisms (CAMS-CCPM-A) for providing valuable reagents. This work was supported by the CAMS Innovation Fund for Medical Sciences (Grant Nos. 2021-I2M-1-043, 2021-I2M-1-038, and 2022-I2M-2-002) and the Beijing Natural Science Foundation (Grant No. 7242097).
Author information
Authors and Affiliations
Contributions
S.C. and Y.D. conceived the project. W.Z. and D.Y. designed and supervised the study. D.Y. drafted the original paper, and S.C., C.H., W.Z. and Y.S. revised the paper. D.Y., Q.L., Y.Z., N.L., Q.Z. and S.G. performed the experiments and analyzed the data. Q.L. performed the molecular modeling. N.A., K.L., L.H., H.C., Y.W., C.C., H.S., J.W., X.L., L.B., D.W., Y.S. and G.L. contributed specific experiments and data analysis. All authors read and approved the contents of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
S.C., Y.D. and D.Y. are co-inventors on pending patent applications related to the IAV mRNA vaccine. N.L., Q.Z., Y.D., C.H., L.H., C.C., Y.W., H.C. and W.Z. are employees of RinuaGene Biotechnology Co., Ltd. All other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yi, D., Liu, Q., Guo, S. et al. Chimeric hemagglutinin and M2 mRNA vaccine for broad influenza subtype protection. npj Vaccines 10, 113 (2025). https://doi.org/10.1038/s41541-025-01178-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41541-025-01178-x