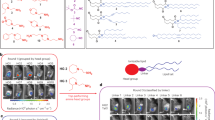
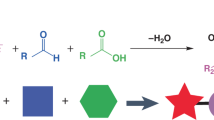
Abstract
The immunogenicity of lipid nanoparticles (LNPs) used for the delivery of nucleoside-modified messenger RNA limits the levels and durability of expression of the encoded protein. Here, by leveraging the Mannich reaction for ionizable lipid synthesis, and via the in vitro and in vivo screening of six combinatorial libraries of synthesized lipids, we report the identification of an antioxidant ionizable lipid, C-a16, exhibiting reduced immunogenicity. When incorporated into LNPs for mRNA delivery, C-a16 mitigated the generation of intracellular reactive oxygen species, thereby extending the duration of protein expression. In mice, and compared with commercial LNPs, LNPs incorporating C-a16 and co-delivering Cas9 mRNA and guide RNA for the editing of the transthyretin gene led to 2.8-fold higher editing efficiency; LNPs with C-a16 delivering fibroblast growth factor 21 mRNA increased the expression of the protein 3.6-fold; and when delivering mRNA encoding a tumour neoantigen or the spike protein of SARS-CoV-2, LNPs with C-a16 induced stronger antigen-specific immune responses. Our findings support the further testing of C-a16 as a promising ionizable lipid for mRNA delivery in therapeutic applications.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data that support the findings of this study are available within the paper and its Supplementary Information files. Source data are provided with this paper.
Change history
31 July 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41551-025-01486-6
References
Pardi, N., Hogan, M. J., Porter, F. W. & Weissman, D. mRNA vaccines—a new era in vaccinology. Nat. Rev. Drug Discov. 17, 261–279 (2018).
Karikó, K. et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 16, 1833–1840 (2008).
Karikó, K., Buckstein, M., Ni, H. & Weissman, D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 (2005).
Kulkarni, J. A., Witzigmann, D., Chen, S., Cullis, P. R. & van der Meel, R. Lipid nanoparticle technology for clinical translation of siRNA therapeutics. Acc. Chem. Res. 52, 2435–2444 (2019).
Cullis, P. R. & Hope, M. J. Lipid nanoparticle systems for enabling gene therapies. Mol. Ther. 25, 1467–1475 (2017).
Akinc, A. et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 14, 1084–1087 (2019).
Chen, S. et al. Nanotechnology-based mRNA vaccines. Nat. Rev. Methods Primers 3, 63 (2023).
Chaudhary, N., Weissman, D. & Whitehead, K. A. mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat. Rev. Drug Discov. 20, 817–838 (2021).
Igyártó, B. Z., Jacobsen, S. & Ndeupen, S. Future considerations for the mRNA-lipid nanoparticle vaccine platform. Curr. Opin. Virol. 48, 65–72 (2021).
Ndeupen, S. et al. The mRNA-LNP platform’s lipid nanoparticle component used in preclinical vaccine studies is highly inflammatory. iScience 24, 103479 (2021).
Moghimi, S. M. & Simberg, D. Pro-inflammatory concerns with lipid nanoparticles. Mol. Ther. 30, 2109–2110 (2022).
Minnaert, A.-K. et al. Strategies for controlling the innate immune activity of conventional and self-amplifying mRNA therapeutics: getting the message across. Adv. Drug Deliv. Rev. 176, 113900 (2021).
Siegwart, D. J. et al. Combinatorial synthesis of chemically diverse core-shell nanoparticles for intracellular delivery. Proc. Natl Acad. Sci. USA 108, 12996–13001 (2011).
Zhang, Y., Sun, C., Wang, C., Jankovic, K. E. & Dong, Y. Lipids and lipid derivatives for RNA delivery. Chem. Rev. 121, 12181–12277 (2021).
Eygeris, Y., Gupta, M., Kim, J. & Sahay, G. Chemistry of lipid nanoparticles for RNA delivery. Acc. Chem. Res. 55, 2–12 (2021).
Witten, J., Hu, Y., Langer, R. & Anderson, D. G. Recent advances in nanoparticulate RNA delivery systems. Proc. Natl Acad. Sci. USA 121, e2307798120 (2024).
Tramontini, M. Advances in the chemistry of Mannich bases. Synthesis 1973, 703–775 (1973).
Tramontini, M. & Angiolini, L. Further advances in the chemistry of Mannich bases. Tetrahedron 46, 1791–1837 (1990).
Tramontini, M., Angiolini, L. & Ghedini, N. Mannich bases in polymer chemistry. Polymer 29, 771–788 (1988).
Zhang, H. & Tsao, R. Dietary polyphenols, oxidative stress and antioxidant and anti-inflammatory effects. Curr. Opin. Food Sci. 8, 33–42 (2016).
Bucciantini, M., Leri, M., Nardiello, P., Casamenti, F. & Stefani, M. Olive polyphenols: antioxidant and anti-inflammatory properties. Antioxidants 10, 1044 (2021).
Yahfoufi, N., Alsadi, N., Jambi, M. & Matar, C. The immunomodulatory and anti-inflammatory role of polyphenols. Nutrients 10, 1618 (2018).
Grant, C. M. Regulation of translation by hydrogen peroxide. Antioxid. Redox Signal. 15, 191–203 (2011).
Devoldere, J., Dewitte, H., De Smedt, S. C. & Remaut, K. Evading innate immunity in nonviral mRNA delivery: don’t shoot the messenger. Drug Discov. Today 21, 11–25 (2016).
Linares-Fernández, S., Lacroix, C., Exposito, J.-Y. & Verrier, B. Tailoring mRNA vaccine to balance innate/adaptive immune response. Trends Mol. Med. 26, 311–323 (2020).
Forster Iii, J., Nandi, D. & Kulkarni, A. mRNA-carrying lipid nanoparticles that induce lysosomal rupture activate NLRP3 inflammasome and reduce mRNA transfection efficiency. Biomater. Sci. 10, 5566–5582 (2022).
Filomeni, G., De Zio, D. & Cecconi, F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 22, 377–388 (2015).
Dasuri, K., Zhang, L. & Keller, J. N. Oxidative stress, neurodegeneration, and the balance of protein degradation and protein synthesis. Free Radic. Biol. Med. 62, 170–185 (2013).
Cheng, Q. et al. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat. Nanotechnol. 15, 313–320 (2020).
Finn, J. D. et al. A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 22, 2227–2235 (2018).
Gillmore, J. D. et al. CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N. Engl. J. Med. 385, 493–502 (2021).
Fisher, F. M. & Maratos-Flier, E. Understanding the physiology of FGF21. Annu. Rev. Physiol. 78, 223–241 (2016).
Flippo, K. H. & Potthoff, M. J. Metabolic messengers: FGF21. Nat. Metab. 3, 309–317 (2021).
Geng, L., Lam, K. S. & Xu, A. The therapeutic potential of FGF21 in metabolic diseases: from bench to clinic. Nat. Rev. Endocrinol. 16, 654–667 (2020).
Jimenez, V. et al. FGF21 gene therapy as treatment for obesity and insulin resistance. EMBO Mol. Med. 10, e8791 (2018).
Gong, N. et al. Proton-driven transformable nanovaccine for cancer immunotherapy. Nat. Nanotechnol. 15, 1053–1064 (2020).
Buonaguro, L., Petrizzo, A., Tornesello, M. L. & Buonaguro, F. M. Translating tumor antigens into cancer vaccines. Clin. Vaccine Immunol. 18, 23–34 (2011).
Blass, E. & Ott, P. A. Advances in the development of personalized neoantigen-based therapeutic cancer vaccines. Nat. Rev. Clin. Oncol. 18, 215–229 (2021).
Ott, P. A. et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221 (2017).
Rojas, L. A. et al. Personalized RNA neoantigen vaccines stimulate T cells in pancreatic cancer. Nature 618, 144–150 (2023).
Hu, Z. et al. Personal neoantigen vaccines induce persistent memory T cell responses and epitope spreading in patients with melanoma. Nat. Med. 27, 515–525 (2021).
Sittplangkoon, C. et al. mRNA vaccine with unmodified uridine induces robust type I interferon-dependent anti-tumor immunity in a melanoma model. Front. Immunol. 13, 983000 (2022).
Berger, A. Th1 and Th2 responses: what are they? Br. Med. J. 321, 424 (2000).
Kaye, P. M., Curry, A. J. & Blackwell, J. M. Differential production of Th1- and Th2-derived cytokines does not determine the genetically controlled or vaccine-induced rate of cure in murine visceral leishmaniasis. J. Immunol. 146, 2763–2770 (1991).
Darrah, P. A. et al. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat. Med. 13, 843–850 (2007).
Precopio, M. L. et al. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8+ T cell responses. J. Exp. Med. 204, 1405–1416 (2007).
Giannotta, G. & Giannotta, N. mRNA COVID-19 vaccines and long-lived plasma cells: a complicated relationship. Vaccines 9, 1503 (2021).
Krienke, C. et al. A noninflammatory mRNA vaccine for treatment of experimental autoimmune encephalomyelitis. Science 371, 145–153 (2021).
Porto, E. M., Komor, A. C., Slaymaker, I. M. & Yeo, G. W. Base editing: advances and therapeutic opportunities. Nat. Rev. Drug Discov. 19, 839–859 (2020).
Acknowledgements
M.J.M. acknowledges support from a US National Institutes of Health (NIH) Director’s New Innovator Award (DP2 TR002776), a Burroughs Wellcome Fund Career Award at the Scientific Interface (CASI), a US National Science Foundation CAREER Award (CBET-2145491) and the American Cancer Society (number RSG-22-122-01-ET). E.L.H. acknowledges support from a US National Science Foundation Graduate Research Fellowship (DGE 1845298). C.G.F.-E. acknowledges support from a US National Science Foundation Graduate Research Fellowship (DGE 1845298), a GEM Fellowship and the NIH/National Cancer Institute Pre-doc to Post-doc Transition Award (F99 CA284294).
Author information
Authors and Affiliations
Contributions
N.G., D.K. and M.J.M. conceived of and designed the experiments. N.G., D.K., M.-G.A., R.E.-M., G.D., Q.S., X.H., L.X., J.X., R.P., Z.M., T.L. and C.G.F.-E. performed the experiments. N.G., D.K. and R.E.-M. analysed the data. D.W. and J.L. involved in results discussion. N.G., D.K., E.L.H., R.E.-M. and M.J.M. wrote and edited the paper. M.J.M. supervised the entire project. All authors discussed the results and commented on the paper.
Corresponding author
Ethics declarations
Competing interests
N.G. and M.J.M. have filed a patent application related to this study. The other authors declare no competing interests.
Peer review
Peer review information
Nature Biomedical Engineering thanks Bruno De Geest, Shuai Liu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Evaluation of LNP-induced innate immunity activation in vivo using complete blood counting assay and ELISA assay.
C-a16, C-a16-Q, DLin-MC3-DMA LNPs or PBS was intravenously injected, and a complete blood counting (CBC) assay was performed 24 hours post-LNP injection. a, The levels of white blood cells (WBC), b, neutrophils (Neu), c, lymphocytes (Lym), and d, monocytes (Mon), in the blood were measured (n = 4). ELISA assay was used to quantify the levels of IL-2 (e) and TNF-α (f) in the mouse serum (n = 4). Statistical differences were calculated using one-way ANOVA with Tukey’s post hoc test.
Extended Data Fig. 2 C-a16 LNPs induce antigen-specific immune responses.
a-d, Quantification of LNP distribution in immune cell populations, including DCs (a), macrophages (b), B cells (c), and T cells (d), assessed by flow cytometry. Data are presented as mean ± SD (n = 5), and statistical differences were determined by one-way ANOVA. e, Immunofluorescence images of lymph nodes in mice after intramuscular injection of DiR dye-labeled LNPs. Nuclei are represented in blue, CD11c+ DCs in green, and DiR-labeled LNPs in red. f, Evaluation of OVA-specific T cell generation following two doses of C-a16-mOVA vaccines. Seven days after the boost dose, blood T cells were stained with SIINFEKL-H2Kb tetramer-APC antibody. After that, SIINFEKL-H2Kb tetramer+ cells were detected by flow cytometry (f) and quantified (g). h and i, Investigation of vaccine-induced immune responses’ ability to eliminate SIINFEKL epitope+ target cells. A mixture of CFSElow SIINFEKL-loaded and CFSEhigh non-loaded splenocytes (in a 1:1 ratio) was intravenously injected into vaccinated mice. After 24 hours, splenocytes were collected and analyzed by flow cytometry (h), and target cell killing was quantified (i). Data in a-d, g and i are shown as mean ± SD (n = 5). Statistical differences were calculated using one-way ANOVA with Tukey’s post hoc test.
Extended Data Fig. 3 Body weight change and survival curves.
a, Body weight change after mice were treated with various LNP vaccines. b, Mice survival curves after treatment with various LNP vaccines encapsulating OVA mRNA. c, Survival curves of mice receiving various LNPs encapsulating Pbk-Actn4 mRNA. Data in a are shown as mean ± SD (n = 8).
Extended Data Fig. 4 T cell immune responses elicited by C-a16 LNP encapsulating mRNA encoding SARS-CoV-2 spike protein.
Mice received intramuscular immunization with various LNP vaccines on days 0 and 21 (0.25 mg/kg). On day 35, splenocytes were collected and stimulated with SARS-CoV-2 RBD peptide pools. a and b, T cells were assessed for Th2 (IL-4, IL-5) and c, Th17 (IL-17a) intracellular cytokine expression. d, Evaluation of the expression of the cytotoxic marker CD107α expression in CD8+ T cells. Data are shown as mean ± SD (n = 5). Statistical differences were calculated using one-way ANOVA with Tukey’s post hoc test.
Extended Data Fig. 5 Evaluation of polyfunctional CD4+ and CD8+ T cells.
Mice received intramuscular immunization with various LNP vaccines on days 0 and 21 (0.25 mg/kg). On day 35, splenocytes were collected and stimulated with SARS-CoV-2 RBD peptide pools. CD4+ (a-g) and CD8+ (h-n) polyfunctional T cell percentages (%) were assessed. Data are shown as mean ± SD (n = 5). Statistical differences were calculated using one-way ANOVA with Tukey’s post hoc test.
Extended Data Fig. 6 Investigation of humoral immune responses and memory B cells elicited by various LNPs.
a, Mice received intramuscular immunization with various LNP vaccines on days 0 and 21 (0.25 mg/kg). IgG2c to IgG1 ratio was determined. Data are presented as mean ± s.d. b, Enumeration of RBD-specific B cells per spleen. Splenocytes were stimulated with SARS-CoV-2 RBD peptide pools and stained with various antibodies before flow cytometry analysis. c, Percentage of RBD-specific B cells categorized by germinal center (GC) or memory phenotype. GC B cells were defined as CD38−GL7+, and memory B cells were defined as CD38+GL7−. Data are shown as mean ± SD (n = 5). Statistical differences were calculated using one-way ANOVA with Tukey’s post hoc test.
Supplementary information
Supplementary Information
Supplementary Figs. 1–37.
Source data
Source Data Figs. 1–5 and Extended Data Figs. 1–6
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gong, N., Kim, D., Alameh, MG. et al. Mannich reaction-based combinatorial libraries identify antioxidant ionizable lipids for mRNA delivery with reduced immunogenicity. Nat. Biomed. Eng 9, 2181–2195 (2025). https://doi.org/10.1038/s41551-025-01422-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41551-025-01422-8