Abstract
The environment influences how an organism’s genotype determines its phenotype and how this phenotype affects its fitness. Here, to better understand this dual role of environment in the production and selection of phenotypic variation, we determined genotype–phenotype–fitness relationships for mutant strains of Saccharomyces cerevisiae in four environments. Specifically, we measured how promoter mutations of the metabolic gene TDH3 modified expression level and affected growth for four different carbon sources. In each environment, we observed a clear relationship between TDH3 expression level and fitness, but this relationship differed among environments. Mutations with similar effects on expression in different environments often had different effects on fitness and vice versa. Such environment-specific relationships between phenotype and fitness can shape the evolution of phenotypic plasticity. We also found that mutations disrupting binding sites for transcription factors had more variable effects on expression among environments than those disrupting the TATA box, which is part of the core promoter. However, mutations with the most environmentally variable effects on fitness were located in the TATA box, because of both the lack of plasticity of TATA box mutations and environment-specific fitness functions. This observation suggests that mutations affecting different molecular mechanisms contribute unequally to regulatory sequence evolution in changing environments.
Similar content being viewed by others
Main
An organism’s environment affects how its genotype determines its phenotype during the short-term process of development and how selection acts on that phenotype during the longer-term process of evolution (Fig. 1a). Often, a single genotype can produce different phenotypes in different environments; this is known as phenotypic plasticity1,2. Phenotypic plasticity is itself a variable and evolvable trait, and the same environmental change can induce different phenotypic changes among genetically distinct organisms through genotype-by-environment interactions1,3,4,5,6,7,8. The differences in reproductive success conferred by diverse phenotypes—known as relative fitness—can also differ among environments, because some trait values may be better suited to one environment than another9,10,11,12,13,14,15,16,17. This relationship between phenotypes and fitness, which can be described as a two-dimensional fitness function when considering variation in one particular trait18,19, determines which individuals are most likely to reproduce and influence evolution (Fig. 1a, dashed arrow).
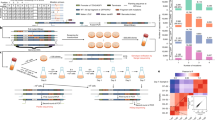
a, Schematic showing that genotype and environment both influence phenotype. Phenotype and environment determine an organism’s relative fitness. The grey dotted line connecting fitness to genotype reflects reproduction and potential evolutionary changes in allele frequencies from one generation to the next. b, Allelic variation for a gene’s promoter (arrowheads indicate transcription start site) is shown under genotype, leading to variation in the gene’s expression (curvy lines represent differences in RNA abundance in budding yeast cells), shown under phenotype, and variation in the abundance of different yeast genotypes due to differences in expression, shown under fitness. Hypothetical differences in RNA abundance and relative fitness between two environments (A and B) are also shown. c,d, A case where the alternate allele has no expression plasticity but environment-specific fitness effects (c) and a case where expression plasticity of the alternate allele results in the same fitness in both environments (d). Considering the wild-type and alternate alleles together, c shows a genotype-by-environment interaction for fitness but not expression (Extended Data Fig. 1a,b), and d shows a genotype-by-environment interaction for expression but not fitness (Extended Data Fig. 1c,e). Solid lines show the relationship between expression of a focal gene and fitness in two environments (green, environment A; purple, environment B). Black dot indicates expression and fitness of a ‘wild-type’ promoter allele, with both values defined as 1. Green and purple dots indicate expression and fitness of the same alternative promoter allele in environment A (green) and environment B (purple), with expression and fitness defined relative to the wild-type allele. The green and purple dotted arrows highlight the effects of genetic differences between the wild-type and alternative promoter alleles on gene expression and fitness in each environment. e, Abbreviated metabolic pathway illustrating how the carbon sources used in this study (glucose, galactose, glycerol and ethanol) relate to the glycolysis/gluconeogenesis pathway. TDH3p catalyses the interconversion of glyceraldehyde-3-phosphate (GAP) to 1,3-biphosphoglycerate (1,3BPG), as shown by the double-headed black arrow. Grey arrows highlight other metabolic steps; multiple grey arrows indicate reactions catalysed by two or more enzymes. Fit., fitness; Exp., expression; G6P, glucose 6-phosphate.
The importance of the dual role of the environment in the production and selection of phenotypic variation is widely recognized1,20. For example, empirical studies have demonstrated that phenotypic plasticity can be adaptive9,11,14,16,17,21,22, such as when dorsal head spikes that increase fitness develop in Daphnia water fleas in response to predator-associated chemicals9,17. However, phenotypic plasticity can also be maladaptive12,23,24,25; for example, mice adapted to low altitude conditions have a physiological response to low oxygen conditions that also causes overproduction of red blood cells and potential pulmonary hypertension in high altitude conditions25. Theoretical studies have articulated evolutionary scenarios in which plasticity may be selectively maintained or removed26,27 and described how genotype-by-environment interactions may allow accumulation of genetic variants that only affect phenotype under certain conditions28,29. A major remaining challenge, however, is to determine how the molecular mechanisms that generally underlie phenotypic plasticity shape—and are shaped by—evolutionary processes.
The plasticity of organismal phenotypes is often enabled by the plasticity of gene expression: changes in activation and repression of genes enable organisms to produce different phenotypes from the same genotype in response to environmental stimuli. In fact, investigations of how bacteria could transition from glucose to lactose metabolism without altering their DNA led Jacob and Monod to discover the foundational logic of molecular transcriptional regulation using the lac operon30. Over the past two decades, empirical studies—initially with microarrays in model organisms such as Saccharomyces cerevisiae31,32 and Caenorhabditis elegans33,34 and subsequently with RNA sequencing in a much greater diversity of taxa12,16,22,24,35—have found expression plasticity to be pervasive. Studies of S. cerevisiae showed that genetic variation, whether among multiple diverse strains32 or in recombinant genotypes produced by crossing two strains36, often affects gene expression plasticity. These findings further indicated that genotype-by-environment interactions, which are necessary for plasticity to evolve, are common within a species37,38. As plasticity in gene expression is expected to underlie plasticity in higher-order traits (including fitness), genetic changes in sequences regulating gene expression are likely to have a key role in the evolution of phenotypic plasticity12,14,15,16,22,24,39. Consequently, it is necessary to identify specific genetic changes in regulatory sequences that alter gene expression among environments and determine the fitness effects of these changes in multiple environments to understand how phenotypic variation emerges from the interplay of an organism’s internal molecular processes and external environment8. Such work can reveal, for example, whether mutations disrupting molecular processes of certain types may disproportionately produce environment-dependent phenotypes and are thus likely to be conditionally beneficial or deleterious. Further, such an approach should also enable comparison of environmental effects of mutations on gene expression and fitness; this is important, because the environment can potentially change both the phenotypes produced and the nature of selection acting on those phenotypes.
Experiments showing how specific genetic changes affect a focal trait in different environments and how the fitness of the resulting phenotypes differs among those environments are now possible, particularly in microbial systems40,41. The baker’s yeast, S. cerevisiae, provides a uniquely powerful eukaryotic system for measuring the environment-specific effects of genetic variants on gene expression and fitness for several reasons40,42,43. First, genetic changes that affect expression of a gene can be easily generated via site-directed mutagenesis of the promoter sequence located upstream of the gene’s coding sequence. Second, the quantitative effects of promoter mutations on expression of the focal gene can be measured with high precision and throughput using fluorescent reporters or RNA sequencing. Third, the impact of mutations on fitness in the unicellular eukaryote S. cerevisiae can be directly and precisely measured by comparing the relative growth rate of strains differing only by the mutations of interest. Finally, the environment of yeast cells can be easily controlled and changed in the laboratory.
By characterizing the expression and fitness of a set of yeast strains that differ only by mutations in the promoter of a focal gene under multiple environmental conditions (Fig. 1b), we can tease apart how different environments affect different mutations’ impact on expression level and on relative fitness. We can then interpret the environment-specific relationship between expression level and fitness using empirically determined fitness functions for the different environments. For example, if the fitness function (that is, the relationship between the focal gene’s expression and fitness) differs between two environments, a mutation could have the same effect on expression in both environments but different effects on fitness (Fig. 1c and Extended Data Fig. 1a,b), different effects on expression in the two environments but the same effect on fitness (Fig. 1d and Extended Data Fig. 1c,d) or some other combination of effects on expression and fitness.
Here, we used TDH3 in S. cerevisiae as a focal gene to examine the effects of promoter mutations on gene expression and fitness in four environments, using media containing glucose, galactose, glycerol or ethanol as a carbon source. Glucose and galactose are fermentable carbon sources that can be metabolized by S. cerevisiae under aerobic and anaerobic conditions via glycolysis and alcoholic fermentation, with galactose first requiring degradation via the Leloir pathway. Glycerol and ethanol are non-fermentable carbon sources that can only be metabolized by S. cerevisiae cells under aerobic conditions. The TDH3 gene encodes a GAPDH protein that catalyses the interconversion of glyceraldehyde-3-phosphate and 1,3-biphosphoglycerate, and this biological role makes the gene’s activity important for the metabolism of different carbon sources via glycolysis and gluconeogenesis (Fig. 1e). Past studies have demonstrated that some mutations in this promoter have effects on expression levels that vary among different carbon environments44, and studies in rich glucose media have shown that variation in TDH3 promoter activity affects organismal fitness45,46, but it has remained unknown how the relationship between expression level and fitness changes with the environment, and, in turn, how the effects of mutations on expression and fitness vary among environments.
To address this knowledge gap, we used 51 strains of S. cerevisiae carrying different alleles of the TDH3 promoter to empirically characterize the concurrent effects of the environment on expression level and fitness. We first used these data to construct environment-specific fitness functions relating TDH3 expression levels to relative growth rate, our proxy for fitness. Using these fitness functions, we then assessed the relationship between environment-dependent effects on expression and relative fitness for promoter variants. We found that both effects of mutations on gene expression and relationships between expression levels and fitness (fitness functions) varied among environments, with the plasticity of gene expression being beneficial in some contexts and detrimental in others. We also considered whether plasticity differed for mutations that affect transcription through different molecular mechanisms by comparing the effects of mutations in specific transcription factor binding sites and mutations in the TATA box on which the core transcriptional preinitiation complex assembles. We found that mutations in specific transcription factor binding sites generally had more variable effects on expression among environments than mutations in the TATA box; however, the mutations with the most variable effects on fitness among environments occurred in the TATA box. Together, these data show how the environment jointly affects the production and selection of phenotypic variation, revealing different environment-dependent effects of mutations that regulate gene expression through distinct molecular mechanisms and suggesting a mechanistic explanation for propensities in regulatory sequence evolution as organisms navigate life in different environments.
Results
Environments affect TDH3 expression and effect of TDH3 deletion on growth
Different environments pose different demands for growth on cells. S. cerevisiae can grow on a wide range of carbon sources, but it does so at different rates. To determine how the environments used in this study affected the growth of S. cerevisiae, we measured the growth rate of a haploid reference strain using batch cultures in four types of rich medium, each containing a different fermentable (glucose or galactose) or non-fermentable (glycerol or ethanol) carbon source. Significant differences in growth rate were observed among the environments (analysis of variance (ANOVA), F = 12252, d.f. = 3, P < 5.5 × 10−15). Adaptation to glucose is a hallmark of S. cerevisiae, and, as expected, this strain had the highest growth rate on glucose and grew more rapidly on fermentable than non-fermentable carbon sources (Fig. 2a).
a, Average numbers of cell divisions per hour (growth rate) for a wild-type strain of S. cerevisiae grown in media containing glucose, galactose, glycerol or ethanol as a carbon source. Growth rate was calculated from optical density measurements of batch cultures over time. As indicated, glucose and galactose are fermentable carbon sources; glycerol and ethanol are non-fermentable carbon sources. b, To measure activity of PTDH3 in different environments, a PTDH3-YFP reporter gene was integrated into the HO locus, cells were grown in each environment (media containing glucose, galactose, glycerol or ethanol as a carbon source) and YFP fluorescence was measured using flow cytometry. c, Violin plots showing arbitrary fluorescence units (AFU) for YFP (corrected for cell size) driven by the TDH3 promoter in the four different environments. In each case, the horizontal line shows the median expression in each environment, and the points show individual replicates. d, To quantify fitness of a wild-type reference strain or a TDH3 deletion strain (ΔTDH3) relative to a reference competitor genotype (Ref.), we marked the ∆TDH3 strain with YFP and the Ref. strain with GFP and then grew them competitively by mixing cells from the two genotypes. We tracked the genotypes’ relative abundance over 20–25 generations of log-phase growth (maintained by diluting the culture with new medium every 12 h) using flow cytometry every ~6–8 generations. e, Violin plots showing fitness of an unmutated reference strain (grey circles) and TDH3 deletion (red triangles) relative to the GFP-positive competitor strain in the different environments. For each environment, points show the relative fitness of individual replicates (four to six replicates per condition and genotype), and horizontal lines show the median relative fitness for each genotype. Ref., reference; gen., generation. Schematic in b created using BioRender.com.
To determine how these different environments affected expression of the TDH3 gene and its contribution to the rate of cell division, we measured the activity of the unmutated TDH3 promoter allele in the same genetic background in each environment and then determined how the loss of TDH3 affected the rate of cell division. Activity of the TDH3 promoter was quantified by using flow cytometry to measure the fluorescence of individual cells with the reference allele of the TDH3 promoter (PTDH3) driving expression of a yellow fluorescent protein (YFP) at the HO locus in each of the four environments (Fig. 2b). We found that the activity of the TDH3 promoter varied among environments (ANOVA, F = 2.8 × 1029, d.f. = 3, P < 2.2 × 10−16), with the highest activity in glucose and the lowest activity in galactose (Fig. 2c). We then examined the contribution of TDH3 to population growth in each environment by using flow cytometry to compare the relative growth rates of strains with and without a functional copy of the TDH3 gene. The reference and deletion strains were marked with green fluorescent protein (GFP) and YFP reporter genes, respectively (Fig. 2d). We found that the contribution of TDH3 to growth rate varied significantly among environments (Fig. 2e; ANOVA, F = 105, d.f. = 3, P < 2.1 × 10−12).
Comparison of the expression and fitness data suggested a complex relationship between the expression level of TDH3 in an environment and the importance of TDH3 for growth in that environment (Fig. 2c,e). For example, TDH3 had the highest expression and largest fitness impact upon deletion in glucose but the lowest expression level and the second largest fitness impact upon deletion in galactose. These observations underscore that a gene’s relative expression level in a given environment is not predictive of its contribution to fitness; a gene’s optimal expression level can vary among environments. This extreme case scenario—comparing reference and deletion strains—also demonstrates that even when a mutation has identical effects on expression in different environments (a complete loss of expression in this case), it can have different effects on relative fitness.
Environments alter the impact of expression on fitness
To better understand the relationship between TDH3 expression and its effects on growth in different environments, we identified 47 alleles of the TDH3 promoter that showed a wide range of effects on PTDH3 activity in glucose44,45,46 and measured their effects on expression of the YFP reporter gene inserted at the HO locus in each of the three other environments. We then used each of these mutant alleles to drive expression of the native TDH3 gene and measured its effects on relative growth rate in all four environments via competitive growth assays, as described above. For all TDH3 promoter alleles, point mutations were located in RAP1 or GCR1 transcription factor binding sites and/or in the TATA box (Extended Data Fig. 2a). The promoter alleles examined included 12 alleles with one mutation, 11 alleles with two mutations and 11 alleles with more than two mutations relative to the reference sequence (Extended Data Fig. 2b). The remaining 13 mutant genotypes included a duplication of the PTDH3-YFP reporter gene (when measuring expression) or of the full TDH3 gene (when measuring fitness) with or without promoter mutations in both copies of the gene (Extended Data Fig. 3).
Focusing first on PTDH3 activity, we compared YFP expression driven by each of the mutant promoter alleles with expression driven by the unmutated reference allele in the same environment. We found that the overall range of expression levels observed in this set of 47 PTDH3 alleles was similar in all four environments: 0% to 207% in glucose, 0% to 185% in galactose, 1% to 186% in glycerol and 1% to 182% in ethanol (Supplementary Data 1). Alleles carrying one promoter mutation had smaller effects on expression than alleles carrying multiple mutations, and alleles with duplications of the TDH3 promoter often had expression levels higher than the unmutated reference allele (Fig. 3a). Collectively, this allelic panel provided us with the tools necessary to examine the effects of environment across a wide range of gene expression levels.
a, Mean YFP expression with 95% confidence intervals from 4–8 replicates of each genotype in each environment for the 47 mutant PTDH3 alleles driving expression of YFP in glucose (blue), galactose (green), glycerol (purple) and ethanol (orange). Mutant strains are grouped by whether they had one mutation (1 mut.), two mutations (2 mut.), more than two mutations (>2 mut.) or two copies of PTDH3 with mutations (2 copies + mut.). YFP expression of each strain was normalized by the expression level of the strain carrying an unmutated PTDH3 grown in glucose. b, Pairwise comparisons for expression effects of mutant PTDH3 alleles in glucose, galactose, glycerol and ethanol. The mean expression levels (±s.d.) of the 47 PTDH3-YFP mutant genotypes relative to the reference strain were estimated from 4–8 replicates. Panels I and II show comparisons of genotypes between the two fermentable and two non-fermentable environments, respectively. Panels III–VI show comparisons between fermentable and non-fermentable environments. Relative expression levels of each mutant genotype in each environment were estimated from four to eight replicates. Genotypes with significantly different effects between the environments compared are shown in red. Significance was determined by two-sided t-tests, with P < 0.008 following Bonferroni correction for the six comparisons between pairs of environments. Pearson correlation coefficients (r) are also shown in each panel. c, Mean relative activity of PTDH3 (normalized to expression of the reference in glucose) is plotted against the relative fitness of the strain carrying the same PTDH3 allele at the native locus, with fitness normalized to the fitness of the unmutated reference strain in each environment. A LOESS regression line inferred from these points is shown for each environment (span = 0.75). d, The fitness functions shown in c are overlaid to show similarities and differences between environments. The coloured dots represent relative expression and fitness of the unmutated PTDH3 reference allele in glucose (blue), galactose (green), glycerol (purple) and ethanol (orange). The inset shows a higher magnification of the region of relative expression to better show expression plasticity of the reference allele. The lighter shaded areas in each environment indicate the inferred 95% confidence intervals in c and d.
To determine whether mutations in the TDH3 promoter altered its expression plasticity among environments (that is, showed genotype-by-environment interactions), we compared the differences in expression between each pair of environments for each promoter allele. Despite strong overall correlations between the effects of these PTDH3 alleles among environments (Fig. 3b; Pearson’s r ranging from 0.969 to 0.996 in each pair of environments), we found evidence of statistically significant genotype-by-environment interactions in many cases: 45 of the 47 genotypes (Supplementary Data 1) had effects that were significantly different between at least two environments based on t-tests with a Bonferroni correction for n = 6 (P < 0.0083), which is the number of pairwise comparisons for each genotype. These significant differences were generally small in magnitude, however (Fig. 3b). Overall, mutations tended to have more similar effects on expression in the two fermentable or two non-fermentable carbon sources than between fermentable and non-fermentable carbon sources (Fig. 3b and Supplementary Table 1), consistent with a prior study of 235 PTDH3 alleles carrying single point mutations and polymorphisms analysed in glucose, galactose and glycerol44.
To determine whether and how the relationship between TDH3 expression and fitness varies among environments, we plotted the effects of each mutant allele on gene expression. We then fit a line to these data using weighted local regression (LOESS), which makes no assumptions about the shape of the fitness function (Fig. 3c). We observed that the fitness functions seemed to be nonlinear and different among environments. Indeed, for each environment, a nonlinear, modified quadratic model fit the data better than a linear model (Extended Data Fig. 4a), and the quadratic model fit the data better with environment-specific parameters than without (Extended Data Fig. 4b). We expected that the fitness functions for TDH3 expression would be most similar for cells grown on the same type of carbon source (fermentable or non-fermentable), because they would be using TDH3 in more similar metabolic processes. For example, glucose and galactose are both hexose sugars that have the same chemical formula, and their metabolism shares nine of the ten steps of glycolysis. However, we found that this was not the case (Fig. 3c and Extended Data Fig. 4b); for example, the fitness function in glucose was more similar to the fitness function in glycerol than galactose (Fig. 3d).
Given that the optimal TDH3 expression level varied among environments (Fig. 3c and Extended Data Fig. 4b), and that the environment also affected expression driven by the reference PTDH3 allele (Fig. 2a), we next asked whether this plasticity increased the fitness of the reference allele. Between glucose and galactose, we found that the unmutated reference PTDH3 allele exhibited plasticity that resulted in 24% lower expression in galactose than glucose, but fitness remained near the optimum in both environments because of differences in the shape of the fitness functions (Fig. 3d and Extended Data Fig. 4b). This observation suggests that this plasticity is either neutral or beneficial. By contrast, the unmutated reference allele exhibited 7% lower expression in ethanol than in glucose, even though higher expression levels were associated with higher fitness in this environment. The fitness function observed in ethanol was distinct from the fitness functions in the other three environments in that it did not reach a fitness peak or plateau within the range of TDH3 expression levels sampled (Fig. 3c and Extended Fig. 4b). This unique relationship between TDH3 expression and fitness is consistent with S. cerevisiae using ethanol differently from the other three carbon sources (Fig. 1e): glucose, galactose and glycerol are precursors of GAP (the substrate that TDH3p, encoded by TDH3, converts into 1,3-biphosphoglycerate in the second part of the glycolysis metabolic pathway), whereas ethanol is not a precursor of GAP, and the role of TDH3p in ethanol metabolism probably involves gluconeogenesis.
a, Schematic of the S. cerevisiae TDH3 promoter showing locations of binding sites for the transcription factors RAP1 (yellow) and GCR1 (blue and red) as well as the location of the TATA box (grey) and transcription start site (arrow) for TDH3. b, Variance in effects of mutations on gene expression in the four environments tested (normalized to the reference allele in each environment) for PTDH3 alleles with a mutation only in either a RAP1 or GCR1 binding site (TFBS) or with a mutation(s) only in the TATA box (TATA). In each box plot, the centre line indicates the median effect size for the group, the bounds of the box indicate the interquartile range, and upper and lower whiskers extend from the bounds of the box to the largest and smallest values within 1.5× the interquartile range of the data. c, The same information is shown as in b but for variance in the effects of the promoter alleles on fitness rather than expression in the four environments. d, Mean relative effects of each of the 14 PTDH3 mutant alleles shown in b and c on gene expression and fitness in each of the four environments. Error bars indicate the 95% confidence interval for each genotype in each environment, calculated from four to eight replicates for gene expression and four replicates for fitness. In all panels, the colour of each point indicates whether the strain carried any mutations in the RAP1 binding site (yellow), GCR1 binding site (blue or red) or TATA box (grey).
Environmental specificity of molecular mechanisms
In any environment, the fitness function determines whether a mutation’s effect is visible to selection; selection is agnostic to the molecular mechanisms through which that mutation affects the phenotype. However, mutations affecting some molecular mechanisms may be more likely to be exposed to or hidden from selection because they have environment-specific effects. For example, promoters regulate gene expression by binding to context-specific transcription factors as well as core transcription factors that are part of the basal transcriptional machinery required every time a gene is transcribed. Mutations that alter these different types of binding sites are expected to differ in their environmental sensitivity to effects on gene expression and fitness, which could preferentially maintain some promoter mutations in populations when environments are changing. For example, mutations in binding sites for context-specific transcription factors that only function in one environment might have no effect on expression in other environments and thus be invisible to selection when the organism is in these other environments. By contrast, mutations that alter regulatory sequences used in all environments might always be subject to selection.
The S. cerevisiae TDH3 promoter contains a binding site for the RAP1 transcription factor, two binding sites for the GCR1 transcription factor and a TATA box sequence upon which the basal transcriptional machinery assembles (Fig. 4a). As prior work suggests that the RAP1 and GCR1 binding sites have different roles in regulation of TDH3 expression in fermentable and non-fermentable environments44,47, we hypothesized that mutations affecting the RAP1 and GCR1 binding sites would be more prone to environment-specific effects than those affecting the TATA box, which functions more similarly in all environments. To test this hypothesis, we examined the seven mutant genotypes in our datasets with a single copy of PTDH3 that carried a single point mutation in one of the RAP1 or GCR1 binding sites and compared their effects on gene expression and fitness with the seven mutant genotypes in our dataset with one or more mutation(s) in the TATA box (Extended Data Fig. 2b, highlighted with asterisk). Environmental variability in expression and fitness was estimated for each of the 14 mutant genotypes by calculating the variance of the relative PTDH3-YFP expression or relative fitness measures for each mutant allele among the four environments.
We observed the hypothesized pattern for the effects of mutations on gene expression but not fitness. The measures of expression variability between the set of alleles with mutations in either a RAP1 or GCR1 binding site tended to have effects on gene expression that were more variable than the set of alleles with a mutation or mutations in the TATA box (Fig. 4b; directional Mann–Whitney U-test, W = 38, P = 0.048; permutation test, N = 10,000, P = 0.038), with mutations that had the most variable effects on expression among environments located in the RAP1 binding site and the adjacent GCR1 binding site (GCR1a in Fig. 4a). Despite the more environmentally variable effects of mutations in the RAP1 and GCR1 binding sites on gene expression (Fig. 4b), the three mutant alleles with the most environmentally variable effects on fitness all carried a mutation or mutations located in the TATA box (Fig. 4c). Overall, the average variance of the seven TATA box mutants for fitness was higher than the average variance of the seven RAP1 or GCR1 binding site mutants (Fig. 4c), although this difference was not statistically significant (directional Mann–Whitney U-test, W = 17, P = 0.19; permutation test, N = 10,000, P = 0.108). Looking at the expression and fitness of each of these mutants individually in the four environments showed that the effects of these mutations on expression were generally largest in glucose and galactose, and the effects of TATA box mutations on fitness were often largest in ethanol (Fig. 4d), which has a steep fitness function (Fig. 3d). The differences in effects on expression and fitness we observed for the two classes of mutations are explained by their effects on plasticity and the different shapes of the fitness functions among environments.
Discussion
By measuring the effects of promoter mutations on both TDH3 expression and fitness in four different carbon environments, this study provides a rare empirical demonstration of the dual role of the environment in the production and selection of phenotypic variation. The plasticity in promoter activity, environment-dependent effects of mutations and differences in the shapes of fitness functions among environments that we observed are all expected to influence the evolutionary fate of genetic variants in the changing environments that yeast cells naturally experience. For example, when S. cerevisiae grows on a fermentable carbon source such as glucose, the non-fermentable carbon source ethanol accumulates, and, when glucose is exhausted, the ethanol produced is metabolized via respiration. Further, metabolic demands can vary significantly even between two fermentable carbon sources such as glucose and galactose that are metabolized through largely overlapping steps. In this case, a major selective constraint in galactose metabolism may be avoidance of the accumulation of a toxic intermediate, galactose-1-phosphate, which can happen if the relative activities of different genes in this pathway become mismatched48.
Our observations show that the same gene performing the same biochemical role can have different fitness functions under different conditions, and the differences in fitness functions we observed among environments with different carbon sources could favour promoter alleles that exhibit plasticity, conferring greater fitness in multiple environments. Such a pattern, in fact, could be predicted by metabolic control analysis, which posits that different relationships between gene activity and metabolic flux can emerge from the different structures of metabolic pathways49. Indeed, a prior study comparing the effects of mutations and polymorphisms on PTDH3 activity suggested that natural selection has favoured a particular degree of TDH3 expression plasticity between glucose and galactose44, although the differences in fitness functions reported here for these two environments suggest that greater plasticity (in the same direction) could be even more beneficial (Fig. 3d).
When considering how mutations in the TDH3 promoter affected expression and fitness, we found that mutations in the RAP1 and GCR1 binding sites caused greater variability in gene expression among the four environments tested. However, these mutations were no more variable in their effects on fitness than mutations in the TATA box. In fact, the three strains with the most variable effects on fitness had a mutation or mutations in the TATA box. This observation suggests that genetic variation in the RAP1 and GCR1 transcription factor binding sites is less likely to be deleterious when environments are changing than genetic variation in the TATA box; this could explain why genetic variants causing differences in gene expression between strains of S. cerevisiae are often found in transcription factor binding sites43 and show genotype-by-environment interactions41. For example, genetic variation in the RPN4 binding site of the ERG4 promoter and the GAL4 binding site of the PGM1 promoter have both been shown to have adaptive, environment-specific effects in S. cerevisiae and related species41,50,51,52. Arguments that condition-specific effects of cis-regulatory mutations in transcription factor binding sites are less deleterious than mutations affecting a gene’s expression constitutively have been made most often for tissue-specific enhancers found in metazoans53,54,55,56,57, but the same logic applies to environment-specific regulatory sequences in yeast promoters.
The different shapes of fitness functions we observed for TDH3 expression among environments also suggest a way for populations of S. cerevisiae to accumulate genetic variation that is inconsequential for fitness in one environment but affects fitness in another. Such genetic variation is often referred to as ‘cryptic’ because its effects are only seen in some environments58. For example, our data suggest that mutations with small to moderate effects on TDH3 expression (for example, up to a 50% decrease in expression relative to the wild type) could accumulate neutrally in a population growing on galactose as a carbon source but would then be selected against if the population shifted to growing on glucose, glycerol or ethanol (Fig. 3d). If the effects of these mutations were plastic, however, such that they caused different changes in TDH3 expression in different environments, they could be neutral in multiple environments, as we observed for the reference allele in glucose, galactose and glycerol (Fig. 3d). With limited data available showing the joint effects of the environment on the production of phenotypic variation and the shape of fitness functions, it remains to be seen how widespread these phenomena might be.
Understanding how the environment influences phenotypes and fitness is a shared goal of evolutionary and molecular genetics, yet studies documenting phenotypic plasticity and genotype-by-environment interactions are too rarely integrated with studies investigating environmental impacts on cellular processes. By considering the regulatory and fitness effects of individual mutations on TDH3 expression in the context of what is known about the molecular mechanism controlling TDH3 promoter activity, this study provides a bridge connecting these fields and suggests that historical and ongoing changes in environments might result in selection for phenotypic plasticity that favours particular types of evolutionary changes in regulatory sequences. This type of functional synthesis between molecular and evolutionary biology is critical for addressing fundamental questions in both fields59.
Methods
Yeast strains
All S. cerevisiae strains used in this work to measure the effects of PTDH3 mutations on gene expression and fitness (Supplementary Data 1 and Extended Data Figs. 2 and 3) were constructed as described by Duveau et al.46. Briefly, production of these haploid strains involved crossing strains BY4724, BY4722, BY4730 and BY4742, which are all derived from S288C45,46,60. The S288C alleles of RME1, TAO3, CAT5 and MIP1 were replaced with alleles (RME1 ins-308A, TAO3 Q1493, CAT5 M91 and MIP1 T661) that increase sporulation efficiency and decrease petite frequency relative to the alleles of the S288c laboratory strain61,62. The TDH3 promoter includes the 678 base pairs upstream of the TDH3 coding sequence in S288c with an A-293-G variant that has negligible effect on expression and fitness46. In strains used to quantify expression, the various TDH3 promoter alleles were first fused upstream of the coding sequence for a Venus YFP, a CYC1 terminator (TCYC1) and an independently transcribed KanMX4 resistance cassette. This whole construct was recombined into the HO locus. The HO gene encodes an enzyme involved in mating type switching that is inactive in laboratory strains of S. cerevisiae derived from S288c owing to missense mutations63, enabling maintenance of stable haploid strains. Site-specific integration was verified by Sanger sequencing (forward primer: 5′-CTCATCAGTAAGACCCGTTG; reverse primer: 5′-GATGAATGGGTCGTTCAAAG). In strains that had two copies of the PTDH3-YFP-TCYC1 construct, a URA3 marker was used to separate each copy (for example, PTDH3-YFP–TCYC1-URA3–PTDH3-YFP-TCYC1) to reduce the probability of cross-talk between the gene copies. The endogenous TDH3 promoter at the native locus was unaltered in these strains.
To measure the effects of these promoter alleles on fitness, a parallel set of strains was constructed in which the native PTDH3 promoter was replaced by each of the mutant PTDH3 alleles. Prior work46 has shown that the effects of mutant PTDH3 alleles on reporter gene expression at the HO locus are strongly correlated (R2 = 0.99) with the effects of the same alleles on TDH3 expression at the native TDH3 locus (as measured using a TDH3::PTDH3–TDH3–YFP gene encoding a fusion protein). These strains were constructed in two steps. First, the native copy of the TDH3 gene was replaced by a URA3–KanMX4 cassette to create an intermediate strain (TDH3::URA3–KanMX4). Then, the different mutant promoter alleles fused to the TDH3 protein were recombined into this locus to restore a functional copy of the TDH3 protein under the control of various promoters (for example, TDH3::PTDH3mut-TDH3). In strains with multiple copies of the TDH3 gene at the native locus, an independently transcribed URA3 resistance marker was used to separate the copies (for example, PTDH3-TDH3–URA3–PTDH3-TDH3). Site-specific integration and DNA sequences of the different alleles were verified by Sanger sequencing of the amplicon covering this region spanning from 849 bp upstream of the ATG stop codon to 395 bp downstream of the TAA stop codon (forward primer: 5′-CTCATCAGTAAGACCCGTTG, reverse primer: 5′-AGATGATACTGACTGCACGC).
Environments tested
To compare the growth of S. cerevisiae strains on four different carbon sources, cells were cultured in four types of medium that were identical except for the carbon source. Each environment contained 10 g l−1 of yeast extract and 20 g l−1 of peptone along with either 20 g l−1 of glucose (YPD), 20 g l−1 of galactose (YPGal), 30 ml l−1 of 99% glycerol (YPG) or 50 ml l−1 of 99% ethanol (YPE).
Measuring growth rate in four environments
To determine the growth rate of S. cerevisiae (strain YPW1160, which was also used as the competitor strain in the fitness assays described below), we measured its doubling time when grown in YPD (glucose), YPGal (galactose), YPG (glycerol) and YPE (ethanol). Three replicate cultures of YPW1160 were started in parallel in 5 ml of YPD, YPGal, YPG or YPE and incubated for 36 h at 30 °C with dilution to 5 × 105 cells ml−1 every 12 h. After the last dilution, cell density was quantified every 60 min for 10 h and then after another 800 min by measuring optical density at 660 nm. Doubling time was calculated as the inverse of the slope of the linear regression of log(cell density)/log(2) on time during log phase, when the relationship between log(cell density) and time is linear. The average doubling time was found to be 80 min in YPD, 108 min in YPGal, 165 min in YPG and 233 min in YPE. These data were converted to the doublings per hour shown in Fig. 2a by dividing each of these values by 60.
Measuring effects of P TDH3 alleles on gene expression level
Fluorescence attributable to expression of the PTDH3-YFP reporter gene was measured as a proxy for PTDH3 transcriptional activity using flow cytometry as previously described60. Briefly, all strains were revived from −80 °C glycerol stocks on YPG plates (10 g yeast extract, 20 g peptone, 30 ml glycerol, 20 g agar per litre) and, after 2 days of growth, arrayed in 96-well plates containing 0.5 ml YPD per well. In addition to the different samples, a reference strain YPW1002 was inoculated in 24 positions and used to correct for plate and position effects on fluorescence. Strain YPW978, which does not contain the YFP reporter construct60, was inoculated in one well per plate and served to correct for autofluorescence. Cells were maintained in suspension by fitting the culture plates on a rotator. After 22 h of growth in YPD, samples were diluted to one of four different environments that differed only by carbon source. Expression assays in glucose and galactose were performed in parallel but on a different day from the experiments in glycerol and ethanol. Samples were acclimated to each environment by dilutions to fresh medium every 12 h for 36 h. Before each dilution, cell density was measured for all samples using a Sunrise absorbance reader (Tecan), and one dilution factor was calculated for each 96-well plate so that the average cell density would reach 5 × 106 cells ml−1 after 12 h of growth. This procedure ensured that all samples were maintained in constant log phase after the first few hours of growth, because no sample reached a density above 107 cells ml−1, while limiting the strength of genetic drift because the smallest number of cells transferred during dilution was ~10,000. After 36 h of growth, samples were diluted to 2.5 × 106 cells ml−1 in phosphate-buffered saline (PBS), and fluorescence was acquired for 20,000 events per well on a BD Accuri C6 flow cytometer coupled to a HyperCyt autosampler (IntelliCyt Corp.). A 488-nm laser was used for excitation, and a 530/30 optical filter was used for acquisition of the YFP signal.
Flow cytometry data were analysed using R packages flowCore and flowClust as described in ref. 46. Briefly, single cells were separated from all events based on the height and the area of the forward scatter signal. Then, the intensity of the fluorescence signal was normalized by cell size in several steps46, and the YFP signal was adjusted for autofluorescence based on the signal from strain YPW978, which lacked YFP. The median expression level for each replicate of each genotype was calculated (six replicates per sample, 24–30 replicates of control strains without promoter mutations), and expression of each genotype was estimated as the mean of the median values. The plasticity of each mutant promoter variant was determined by dividing the YFP expression level of each mutant genotype in each environment by the YFP expression level of the unmutated reference strain (YPW1002 for strains with one YFP copy; YPW2675 for strains with two YFP copies) in that environment. Scripts used for this analysis are included in Supplementary File 1 in an R markdown file titled ‘ExpressionFitnessAnalysis.Rmd’.
Fitness assays
We used head-to-head competition assays between strains expressing different levels of TDH3p protein and a common reference to measure relative growth rate during log phase and used this as our proxy for fitness as described in ref. 46. Briefly, all strains with promoter mutations at the native TDH3 locus were engineered to also have an unmutated PTDH3-YFP reporter gene at the HO locus, allowing them to be recognized by their yellow fluorescence. A common competitor strain (YPW1160) with no mutations at the native TDH3 locus was engineered with a PTDH3-GFP allele at the HO locus that was recognized by its green fluorescence. The 47 YFP-marked mutant strains carrying different variants of the TDH3 promoter at the native locus (Supplementary Data 1) were arrayed on four 96-well plates, with two replicates of each strain on each plate. In parallel, the common competitor YPW1160 expressing GFP was also arrayed on four 96-well plates. After 24 h of growth in 0.5 ml of YPD at 30 °C, 0.1 ml of each culture was mixed with 23 μl of 80% glycerol in 96-well plates and stored at −80 °C. Of each of the eight culture plates, four plates were frozen to be used for the competition assays performed in the four different environments (YPD, YPGal, YPG and YPE). Competition assays were performed consecutively in the four environments over 3 weeks, starting with ethanol, then glucose, glycerol and galactose. The same batches of media were used for the expression and competition experiments, and cells were grown in 96-well plates at 30 °C and maintained in suspension on a rotor for all assays. A similar protocol was used for all environments, except for differences in the timing of dilutions mentioned below to adjust for variations in doubling time.
For each competitive growth experiment, YFP and GFP strains were thawed on eight separate OmniTrays filled with YPG agar medium. After 48 h of incubation at 30 °C, samples were transferred in 0.5 ml of YPD and grown for 24 h to saturation. Then, cell densities were measured using a Sunrise absorbance reader (Tecan). At this point, equal volumes of YFP and GFP cell cultures were mixed in 0.5 ml of either YPD, YPGal, YPG or YPE medium in four 96-well plates. The dilution factor was calculated for each plate based on the doubling time of the GFP strain so that the average cell density would reach ~5 × 106 cells ml−1 after 12 h of growth. This procedure of cell density measurement and dilution followed by 12 h of growth was repeated three times and constituted the acclimation phase of the experiment, during which the relative frequency of YFP and GFP strains was not recorded. After these first 36 h of growth, competitive growth was carried on for three additional cycles of dilution and growth, during which the ratio of YFP and GFP cells was recorded at four time points (before each cycle of growth and at the end of the last cycle). The duration of these three cycles of competitive growth depended on the environment: samples were diluted every 10 h in YPD, every 12 h in YPGal and every 24 h in YPG and YPE. Cell density was measured for all samples before each dilution. The relative frequency of YFP and GFP cells was quantified at each time point by flow cytometry. To this end, samples from incubated plates (not from freshly diluted cultures) were diluted in 0.3 ml of phosphate-buffered saline to a final density of 1.5 × 106 cells ml−1 in four 96-well plates and placed on ice to stop growth. These flow cytometry plates were prepared either immediately after samples had been diluted with fresh medium for growth (first three time points) or at the end of the last cycle of growth (last time point). Approximately 75,000 events were recorded for each sample on a BD Accuri C6 flow cytometer, using a 488-nm laser for excitation and two different optical filters (510/10 and 585/40) to acquire fluorescence. These filters allowed separation of the GFP and YFP signals. Therefore, the relative frequency of YFP and GFP cells was measured at four time points during the competition assays.
The number of cells expressing either YFP or GFP was counted for each sample using custom R scripts, as described by Duveau et al.46. Briefly, for each sample, we determined the number of cell generations that occurred during the three dilution cycles, using the median number of generations for all samples grown on the same 96-well plate as a robust estimator of the number of generations for all samples on that plate. The numbers of generations over the entire experiment were about 22 in YPD (glucose), 20 in YPGal (galactose), 26 in YPG (glycerol) and 13 in YPE (ethanol). The fitness of the YFP strain relative to the GFP competitor was calculated as the exponential of the slope of the ratio of YFP-positive over GFP-positive cells regressed on the number of generations across the four time points. For each PTDH3 variant, replicates for which fitness departed from the median fitness across all eight replicates by more than four times the median absolute deviation were considered to be outliers and excluded from further analysis. For each sample, the fitness relative to the GFP strain was then divided by the mean fitness for all replicates of the reference strain YPW1189 (for single-copy PTDH3 variants) or YPW2682 (for double-copy PTDH3 variants). We then calculated the mean relative fitness and standard deviation over the eight replicates of each variant. This measure of fitness expressed relative to a strain with the reference PTDH3 allele was used in all subsequent analyses.
Additional data analysis
All data analysis was done using custom R code included in Supplementary File 1, alongside the source data. ANOVA was used to test for significant effects of environment on doubling time, promoter expression and impact of a TDH3 deletion. We tested whether individual alleles had different effects on YFP expression level between all six pairs of environments using t-tests with Bonferroni correction (n = 6) for multiple tests. We also tested for statistically significant differences in Pearson correlation coefficients between some pairs of environments using Steiger’s Z-test. Fitness functions were initially examined using LOESS curves. To test for nonlinearity and significant differences among environment-specific fitness functions, we fit the data relating expression level and relative fitness to a linear model (lm(Fitness ~ Expression, data = data)) and a quadratic model lm(Fitness ~ poly(Expression, 2, raw = TRUE), data = data), and we chose the best-fit model based on the chi-squared statistic comparing likelihoods, as implemented in the anova() function in R. We tested whether the relationship between TDH3 expression and relative fitness differed significantly among environments by comparing the fit of quadratic models with lm(Fitness ~ Environment * poly(Expression, 2, raw = TRUE), data = data) and without lm(Fitness ~ poly(Expression, 2, raw = TRUE), data = data) environment terms using the Akaike information criterion. We compared the effects of mutations in the RAP1 and GCR1 binding sites with the effects of mutations in the TATA box using the non-parametric Mann–Whitney U-test, as well as permutation tests performed by shuffling the classification of each mutant relative to its measurements. The data and necessary code to reproduce each of these analyses and the plots displayed in the main text are included in Supplementary Data 1 and Supplementary File 1.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data necessary to reproduce the results are included in Supplementary Data 1. The raw flow cytometry.fcs files have been deposited to flowrepository.org (repository IDs FR-FCM-Z8EK and FR-FCM-Z8EQ).
Code availability
The R code necessary to reproduce the analyses and figures is provided separately as Supplementary File 1.
References
Scheiner, S. M. Genetics and evolution of phenotypic plasticity. Annu. Rev. Ecol. Syst. 24, 35–68 (1993).
Bradshaw, A. D. in Advances in Genetics Vol. 13 (eds Caspari, E. W. & Thoday, J. M.) 115–155 (Academic, 1965).
West-Eberhard, M. J. Developmental Plasticity and Evolution (Oxford Univ. Press, 2003).
Ghalambor, C. K., McKay, J. K., Carroll, S. P. & Reznick, D. N. Adaptive versus non-adaptive phenotypic plasticity and the potential for contemporary adaptation in new environments. Funct. Ecol. 21, 394–407 (2007).
Simpson, S. J., Sword, G. A. & Lo, N. Polyphenism in insects. Curr. Biol. 21, R738–R749 (2011).
Josephs, E. B. Determining the evolutionary forces shaping G × E. N. Phytol. 219, 31–36 (2018).
Paaby, A. B. & Testa, N. D. in Evolutionary Developmental Biology (eds Nuno de la Rosa, L. & Müller, G.) 1–14 (Springer International Publishing, 2018).
Goldstein, I. & Ehrenreich, I. M. in Phenotypic Plasticity & Evolution 1st edn (ed. Pfennig, D. W.) Ch. 4 (CRC, 2021).
Agrawal, A. A., Laforsch, C. & Tollrian, R. Transgenerational induction of defences in animals and plants. Nature 401, 60–63 (1999).
Reznick, D., Iv Butler, M. J. & Rodd, H. Life-history evolution in guppies. VII. The comparative ecology of high- and low-predation environments. Am. Nat. 157, 126–140 (2001).
Bashey, F. Cross-generational environmental effects and the evolution of offspring size in the Trinidadian guppy Poecilia reticulata. Evolution 60, 348–361 (2006).
Campbell-Staton, S. C., Velotta, J. P. & Winchell, K. M. Selection on adaptive and maladaptive gene expression plasticity during thermal adaptation to urban heat islands. Nat. Commun. 12, 6195 (2021).
Storz, J. F. & Scott, G. R. Phenotypic plasticity, genetic assimilation, and genetic compensation in hypoxia adaptation of high-altitude vertebrates. Comp. Biochem. Physiol. A 253, 110865 (2021).
Taylor, M. A. et al. Large-effect flowering time mutations reveal conditionally adaptive paths through fitness landscapes in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 116, 17890–17899 (2019).
Li, W. et al. A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell 170, 114–126.e15 (2017).
Kenkel, C. D. & Matz, M. V. Gene expression plasticity as a mechanism of coral adaptation to a variable environment. Nat. Ecol. Evol. 1, 14 (2016).
Becker, D. et al. Adaptive phenotypic plasticity is under stabilizing selection in Daphnia. Nat. Ecol. Evol. 6, 1449–1457 (2022).
Kimura, M. & Crow, J. F. Effect of overall phenotypic selection on genetic change at individual loci. Proc. Natl Acad. Sci. USA 75, 6168–6171 (1978).
Orr, H. A. Fitness and its role in evolutionary genetics. Nat. Rev. Genet. 10, 531–539 (2009).
Lewontin, R. C. The triple helix: gene, organism and environment. Nat. Med. 6, 1206 (2000).
Williams, D. G., Mack, R. N. & Black, R. A. Ecophysiology of introduced Pennisetum setaceum on Hawaii: the role of phenotypic plasticity. Ecology 76, 1569–1580 (1995).
Josephs, E. B., Van Etten, M. L., Harkess, A., Platts, A. & Baucom, R. S. Adaptive and maladaptive expression plasticity underlying herbicide resistance in an agricultural weed. Evol. Lett. 5, 432–440 (2021).
Velotta, J. P., Ivy, C. M., Wolf, C. J., Scott, G. R. & Cheviron, Z. A. Maladaptive phenotypic plasticity in cardiac muscle growth is suppressed in high-altitude deer mice. Evolution 72, 2712–2727 (2018).
Ghalambor, C. K. et al. Non-adaptive plasticity potentiates rapid adaptive evolution of gene expression in nature. Nature 525, 372–375 (2015).
Storz, J. F., Runck, A. M., Moriyama, H., Weber, R. E. & Fago, A. Genetic differences in hemoglobin function between highland and lowland deer mice. J. Exp. Biol. 213, 2565–2574 (2010).
Price, T. D., Qvarnström, A. & Irwin, D. E. The role of phenotypic plasticity in driving genetic evolution. Proc. Biol. Sci. 270, 1433–1440 (2003).
Lande, R. Adaptation to an extraordinary environment by evolution of phenotypic plasticity and genetic assimilation. J. Evol. Biol. 22, 1435–1446 (2009).
Paaby, A. B. & Rockman, M. V. Cryptic genetic variation: evolution’s hidden substrate. Nat. Rev. Genet. 15, 247–258 (2014).
Hermisson, J. & Wagner, G. P. The population genetic theory of hidden variation and genetic robustness. Genetics 168, 2271–2284 (2004).
Jacob, F. & Monod, J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3, 318–356 (1961).
Fay, J. C., McCullough, H. L., Sniegowski, P. D. & Eisen, M. B. Population genetic variation in gene expression is associated with phenotypic variation in Saccharomyces cerevisiae. Genome Biol. 5, R26 (2004).
Landry, C. R., Oh, J., Hartl, D. L. & Cavalieri, D. Genome-wide scan reveals that genetic variation for transcriptional plasticity in yeast is biased towards multi-copy and dispensable genes. Gene 366, 343–351 (2006).
Grishkevich, V. et al. A genomic bias for genotype–environment interactions in C. elegans. Mol. Syst. Biol. 8, 587 (2012).
Li, Y. et al. Mapping determinants of gene expression plasticity by genetical genomics in C. elegans. PLoS Genet. 2, e222 (2006).
Zhou, S., Campbell, T. G., Stone, E. A., Mackay, T. F. C. & Anholt, R. R. H. Phenotypic plasticity of the Drosophila transcriptome. PLoS Genet. 8, e1002593 (2012).
Smith, E. N. & Kruglyak, L. Gene–environment interaction in yeast gene expression. PLoS Biol. 6, e83 (2008).
Lehner, B. Conflict between noise and plasticity in yeast. PLoS Genet. 6, e1001185 (2010).
Chapal, M., Mintzer, S., Brodsky, S., Carmi, M. & Barkai, N. Resolving noise–control conflict by gene duplication. PLoS Biol. 17, e3000289 (2019).
Promislow, D. A regulatory network analysis of phenotypic plasticity in yeast. Am. Nat. 165, 515–523 (2005).
Keren, L. et al. Massively parallel interrogation of the effects of gene expression levels on fitness. Cell 166, 1282–1294.e18 (2016).
Chen, S.-A. A., Kern, A. F., Ang, R. M. L., Xie, Y. & Fraser, H. B. Gene-by-environment interactions are pervasive among natural genetic variants. Cell Genom. 3, 100273 (2023).
Rest, J. S. et al. Nonlinear fitness consequences of variation in expression level of a eukaryotic gene. Mol. Biol. Evol. 30, 448–456 (2013).
Sharon, E. et al. Functional genetic variants revealed by massively parallel precise genome editing. Cell 175, 544–557.e16 (2018).
Duveau, F., Yuan, D. C., Metzger, B. P. H., Hodgins-Davis, A. & Wittkopp, P. J. Effects of mutation and selection on plasticity of a promoter activity in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 114, E11218–E11227 (2017).
Duveau, F., Toubiana, W. & Wittkopp, P. J. Fitness effects of cis-regulatory variants in the Saccharomyces cerevisiae TDH3 promoter. Mol. Biol. Evol. 34, 2908–2912 (2017).
Duveau, F. et al. Fitness effects of altering gene expression noise in Saccharomyces cerevisiae. eLife 7, e37272 (2018).
Kuroda, S., Otaka, S. & Fujisawa, Y. Fermentable and nonfermentable carbon sources sustain constitutive levels of expression of yeast triosephosphate dehydrogenase 3 gene from distinct promoter elements. J. Biol. Chem. 269, 6153–6162 (1994).
Xu, H. et al. Synchronization of stochastic expressions drives the clustering of functionally related genes. Sci. Adv. 5, eaax6525 (2019).
Kacser, H. & Burns, J. A. The molecular basis of dominance. Genetics 97, 639–666 (1981).
Kuang, M. C. et al. Repeated cis-regulatory tuning of a metabolic bottleneck gene during evolution. Mol. Biol. Evol. 35, 1968–1981 (2018).
Boocock, J., Sadhu, M. J., Durvasula, A., Bloom, J. S. & Kruglyak, L. Ancient balancing selection maintains incompatible versions of the galactose pathway in yeast. Science 371, 415–419 (2021).
Siddiq, M. A. & Wittkopp, P. J. Mechanisms of regulatory evolution in yeast. Curr. Opin. Genet. Dev. 77, 101998 (2022).
Wray, G. A. et al. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 20, 1377–1419 (2003).
Wray, G. A. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 8, 206–216 (2007).
Wittkopp, P. J. & Kalay, G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13, 59–69 (2011).
Carroll, S. B. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134, 25–36 (2008).
Stern, D. L. & Orgogozo, V. The loci of evolution: how predictable is genetic evolution? Evolution 62, 2155–2177 (2008).
Gibson, G. & Reed, L. K. Cryptic genetic variation. Curr. Biol. 18, R989–R990 (2008).
Dean, A. M. & Thornton, J. W. Mechanistic approaches to the study of evolution: the functional synthesis. Nat. Rev. Genet. 8, 675–688 (2007).
Metzger, B. P. H. et al. Contrasting frequencies and effects of cis- and trans-regulatory mutations affecting gene expression. Mol. Biol. Evol. 33, 1131–1146 (2016).
Deutschbauer, A. M. et al. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics 169, 1915–1925 (2005).
Dimitrov, L. N., Brem, R. B., Kruglyak, L. & Gottschling, D. E. Polymorphisms in multiple genes contribute to the spontaneous mitochondrial genome instability of Saccharomyces cerevisiae S288C strains. Genetics 183, 365–383 (2009).
Meiron, H., Nahon, E. & Raveh, D. Identification of the heterothallic mutation in HO-endonuclease of S. cerevisiae using HO/ho chimeric genes. Curr. Genet. 28, 367–373 (1995).
Acknowledgements
We thank L. Kim for help with determining single-cell division rates, A. Hodgins-Davis and B. P. H. Metzger for helpful discussions, and all members of the Wittkopp laboratory for helpful comments on the paper. This work was supported by a European Molecular Biology Organization postdoctoral fellowship (EMBO ALTF 1114–2012) to F.D., National Institutes of Health National Research Service Award (5F32CA261115) and Michigan Pioneer Fellows to M.A.S., and grants from the National Science Foundation (MCB-1021398) and National Institutes of Health (R01GM108826 and 5R35GM118073) to P.J.W.
Author information
Authors and Affiliations
Contributions
F.D., M.A.S. and P.J.W. designed the study. F.D. collected the data. F.D. and M.A.S. analysed the data and produced figures. M.A.S., F.D. and P.J.W. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Ecology & Evolution thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Examples of genotype-by-environment interactions for gene expression and fitness.
(a, b) Relative gene expression (a) or relative fitness (b) is shown for the scenario depicted in Fig. 1c, with values for the wild-type allele shown in red and values for the alternate allele shown in blue. (c, d) Relative gene expression (c) or relative fitness (d) is shown for the scenario depicted in Fig. 1d, with values for the wild-type allele shown in blue and values for the alternate allele shown in red. In each of the four plots (a-d), lines that are not parallel indicate a genotype-by-environment interaction.
Extended Data Fig. 2 Mutations in strains carrying a single mutant copy of the TDH3 promoter.
(a) Schematic shows the reference sequence and mutations tested in the RAP1 binding site (yellow), two GCR1 binding sites (blue, red), and the TATA box (gray) of the TDH3 promoter. Coordinates shown are the number of bases upstream of the start codon of the downstream YFP or TDH3 protein. (b) The mutation(s) present in each of the 34 mutant PTDH3 alleles assayed with a single copy of the PTDH3-YFP reporter gene are shown. In each case, the first strain listed (YPW####) carries the mutant TDH3 promoter allele cloned upstream of a YFP reporter protein at the HO locus and was used to measure the effects of the mutation(s) on gene expression whereas the second strain listed carries the mutant PTDH3 allele at the native TDH3 locus and was used to measure the effects of the mutation(s) on fitness (Supplementary Table 1). Asterisks show alleles that were used to compare environment-dependent effects of disrupting TATA box or the TFBS.
Extended Data Fig. 3 Mutations in strains carrying two mutant copies of the TDH3 promoter.
The schematic at the top of the figure shows the arrangement of two copies of the TDH3 promoter driving expression of either a YFP reporter protein at the HO locus or the native TDH3 protein at the native locus, with each gene pair separated by a copy of the URA3 gene. The specific mutations carried in each of the 13 pairs of mutant strains carrying two copies of the PTDH3 allele are then shown with the name (YPW####) of the strain carrying the reporter genes used to measure effects on gene expression listed before the name of the strain with the duplication of TDH3 used to measure fitness (Supplementary Table 1). For each pair of strains, the mutated elements are underlined and the sequences of these elements are shown for each copy of the TDH3 promoter with the mutated sites shown in red. The color of the underline corresponds to the feature mutated: RAP1 binding site = yellow, GCR1a binding site = blue, GCR1b binding site = red, and TATA box = gray.
Extended Data Fig. 4 Growth curves relating expression levels and relative fitness among environments.
Figure 3c uses a weighted linear least squares regression (LOESS curve) to describe the relationship between TDH3 expression and fitness in each environment. This method fits a line to the same data without assuming a particular shape of the relationship, but it does not enable statistical comparisons of this relationship among environments. To enable such comparisons, we fit both linear and quadratic models to the data in each environment, determined which model fit better in each case, and then tested for significant differences in fit among environments. (a) Fit of linear (blue, dotted) and quadratic (red, solid) models to the data in each environment, with predicted 95% confidence intervals. Datapoints show mean relative expression and fitness for each TDH3 promoter allele. In each case, the quadratic model better described the relationship between expression and relative fitness, though the improvement in fit was only marginally significant in ethanol (ANOVA, Glucose: F = 156.62, P < 2.2 *10−16; Galactose: F = 168.65, P = < 2.2 *10−16; Glycerol: F = 126.46, P = 6.34*10−15; Ethanol: F = 4.18, P = 0.047). (b) Comparison of inferred models for the four environment-specific datasets. Datapoints show mean relative expression and fitness for each TDH3 promoter allele in each environment. To test whether the quadratic fitness functions varied among environments, we compared a quadratic model in which parameters were estimated separately for each of the four environments to a quadratic model in which the fitness function was estimated jointly for all environments. We found that the more complex model (that is, fitting a distinct quadratic model to data from each environment) provided a significantly better fit to the data (AIC: -1378.53 vs -1289.27; ANOVA F = 14.82, P < 2.2 *10−16), suggesting that different environments have distinct, non-linear fitness functions. The estimated optimal expression level (scaled to the expression level of the unmutated reference) ± the standard deviation from the inferred quadratic models in each of the four environments were 1.27 ± 0.04 for glucose (blue), 0.92 ± 0.03 for galactose (green), 1.33 ± 0.05 for glycerol (pink), and 2.44 ± 0.8 for ethanol (red).
Supplementary information
Supplementary Information
Supplementary Table 1.
Supplementary Data 1
Excel file with spreadsheets containing all the expression and fitness data for control and transgenic strains.
Supplementary File 1
R markdown file with all the code necessary to generate the analyses and figures provided in the main text using Supplementary Data 1 as input.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Siddiq, M.A., Duveau, F. & Wittkopp, P.J. Plasticity and environment-specific relationships between gene expression and fitness in Saccharomyces cerevisiae. Nat Ecol Evol 8, 2184–2194 (2024). https://doi.org/10.1038/s41559-024-02582-7
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41559-024-02582-7
This article is cited by
-
How genotype-by-environment interactions on fitness emerge
Nature Ecology & Evolution (2024)