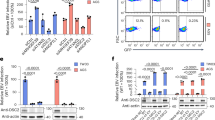
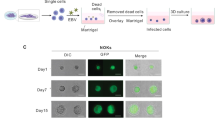
Abstract
Epstein–Barr virus (EBV) infects B and epithelial cells, causing various lymphomas and epithelial malignancies. Although cell-free infection of epithelial cells is inefficient, direct B–epithelial cell contact infection is highly efficient and probably the dominant route. To identify mechanisms of contact-mediated infection, we implemented a genome-wide CRISPR screen and uncovered desmocollin 2 (DSC2) as an EBV epithelial receptor and DSC3 as a co-factor for infection. DSC2 and DSC3 double knockout significantly inhibited both cell-free and cell–cell contact EBV infection of normal oral keratinocytes, while their overexpression permitted infection in receptor-negative cells. Antibodies to DSC2 blocked infection across normal oral keratinocytes, primary oral keratinocytes, and head and neck epithelial organoids. Combining DSC2 and DSC3 antibodies efficiently blocked cell–cell contact infection. Mechanistically, DSC2 interacted with the EBV gH/gL glycoprotein and facilitated epithelial fusion. Notably, EphA2 overexpression failed to restore infection in DSC2/3-deficient cells, indicating its dependence on DSC2/3. Our findings establish DSC2 as a principal EBV entry receptor and target for vaccine and therapeutic development.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The raw sequencing data of the CRISPR screen were deposited in the NCBI Sequence Read Archive (SRA BioProject ID: PRJNA1304944). Primers and synthetic sgRNA DNA oligos used in this study can be found in Supplementary Information. Source data are provided with this Article.
References
Gewurz, B. E., Longnecker, R. & Cohen, J. I. Epstein-Barr virus. in Fields Virology 7th edn (eds Knipe, D. & Howley, P.) 324–389 (Wolters Kluwer, 2021.
Epstein, M. A., Achong, B. G. & Barr, Y. M. Virus particles in cultured lymphoblasts from Burkitt’s lymphoma. Lancet 1, 702–703 (1964).
Cohen, J. I., Fauci, A. S., Varmus, H. & Nabel, G. J. Epstein–Barr virus: an important vaccine target for cancer prevention. Sci. Transl. Med. 3, 107fs107 (2011).
Wong, Y., Meehan, M. T., Burrows, S. R., Doolan, D. L. & Miles, J. J. Estimating the global burden of Epstein–Barr virus-related cancers. J. Cancer Res. Clin. Oncol. 148, 31–46 (2022).
Henle, G., Henle, W. & Diehl, V. Relation of Burkitt’s tumor-associated herpes-type virus to infectious mononucleosis. Proc. Natl Acad. Sci. USA 59, 94–101 (1968).
Hjalgrim, H. et al. Risk of Hodgkin’s disease and other cancers after infectious mononucleosis. J. Natl Cancer Inst. 92, 1522–1528 (2000).
Soldan, S. S. & Lieberman, P. M. Epstein–Barr virus and multiple sclerosis. Nat. Rev. Microbiol. 21, 51–64 (2023).
Bjornevik, K., Munz, C., Cohen, J. I. & Ascherio, A. Epstein–Barr virus as a leading cause of multiple sclerosis: mechanisms and implications. Nat. Rev. Neurol. 19, 160–171 (2023).
Bjornevik, K. et al. Longitudinal analysis reveals high prevalence of Epstein–Barr virus associated with multiple sclerosis. Science 375, 296–301 (2022).
Lanz, T. V. et al. Clonally expanded B cells in multiple sclerosis bind EBV EBNA1 and GlialCAM. Nature 603, 321–327 (2022).
Serafini, B. et al. Dysregulated Epstein–Barr virus infection in the multiple sclerosis brain. J. Exp. Med. 204, 2899–2912 (2007).
Goldacre, R. Risk of multiple sclerosis in individuals with infectious mononucleosis: a national population-based cohort study using hospital records in England, 2003–2023. Mult. Scler. 30, 489–495 (2024).
Harley, J. B. et al. Transcription factors operate across disease loci, with EBNA2 implicated in autoimmunity. Nat. Genet. 50, 699–707 (2018).
Mohl, B. S., Chen, J. & Longnecker, R. Gammaherpesvirus entry and fusion: a tale how two human pathogenic viruses enter their host cells. Adv. Virus Res. 104, 313–343 (2019).
Connolly, S. A., Jardetzky, T. S. & Longnecker, R. The structural basis of herpesvirus entry. Nat. Rev. Microbiol. 19, 110–121 (2021).
Hutt-Fletcher, L. M. Epstein–Barr virus entry. J. Virol. 81, 7825–7832 (2007).
Li, Q. et al. Epstein–Barr virus uses HLA class II as a cofactor for infection of B lymphocytes. J. Virol. 71, 4657–4662 (1997).
Wang, X., Kenyon, W. J., Li, Q., Mullberg, J. & Hutt-Fletcher, L. M. Epstein–Barr virus uses different complexes of glycoproteins gH and gL to infect B lymphocytes and epithelial cells. J. Virol. 72, 5552–5558 (1998).
Xiong, D. et al. Nonmuscle myosin heavy chain IIA mediates Epstein–Barr virus infection of nasopharyngeal epithelial cells. Proc. Natl Acad. Sci. USA 112, 11036–11041 (2015).
Wang, H. B. et al. Neuropilin 1 is an entry factor that promotes EBV infection of nasopharyngeal epithelial cells. Nat. Commun. 6, 6240 (2015).
Zhang, H. et al. Ephrin receptor A2 is an epithelial cell receptor for Epstein–Barr virus entry. Nat. Microbiol. 3, 1–8 (2018).
Chen, J. et al. Ephrin receptor A2 is a functional entry receptor for Epstein–Barr virus. Nat. Microbiol. 3, 172–180 (2018).
Imai, S., Nishikawa, J. & Takada, K. Cell-to-cell contact as an efficient mode of Epstein–Barr virus infection of diverse human epithelial cells. J. Virol. 72, 4371–4378 (1998).
Shannon-Lowe, C. D., Neuhierl, B., Baldwin, G., Rickinson, A. B. & Delecluse, H. J. Resting B cells as a transfer vehicle for Epstein–Barr virus infection of epithelial cells. Proc. Natl Acad. Sci. USA 103, 7065–7070 (2006).
Nanbo, A., Kachi, K., Yoshiyama, H. & Ohba, Y. Epstein–Barr virus exploits host endocytic machinery for cell-to-cell viral transmission rather than a virological synapse. J. Gen. Virol. 97, 2989–3006 (2016).
Nanbo, A., Terada, H., Kachi, K., Takada, K. & Matsuda, T. Roles of cell signaling pathways in cell-to-cell contact-mediated Epstein–Barr virus transmission. J. Virol. 86, 9285–9296 (2012).
Sokal, E. M. et al. Recombinant gp350 vaccine for infectious mononucleosis: a phase 2, randomized, double-blind, placebo-controlled trial to evaluate the safety, immunogenicity, and efficacy of an Epstein–Barr virus vaccine in healthy young adults. J. Infect. Dis. 196, 1749–1753 (2007).
Moutschen, M. et al. Phase I/II studies to evaluate safety and immunogenicity of a recombinant gp350 Epstein–Barr virus vaccine in healthy adults. Vaccine 25, 4697–4705 (2007).
Kanekiyo, M. et al. Rational design of an Epstein–Barr virus vaccine targeting the receptor-binding site. Cell 162, 1090–1100 (2015).
Bu, W. et al. Epstein–Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells. Immunity 57, 559–573.e6 (2024).
Wei, C. J. et al. A bivalent Epstein–Barr virus vaccine induces neutralizing antibodies that block infection and confer immunity in humanized mice. Sci. Transl. Med. 14, eabf3685 (2022).
Sun, C. et al. A gB nanoparticle vaccine elicits a protective neutralizing antibody response against EBV. Cell Host Microbe 31, 1882–1897.e10 (2023).
Li, W. et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014).
Sorem, J. & Longnecker, R. Cleavage of Epstein–Barr virus glycoprotein B is required for full function in cell–cell fusion with both epithelial and B cells. J. Gen. Virol. 90, 591–595 (2009).
Chen, W. H. et al. Epstein–Barr virus gH/gL has multiple sites of vulnerability for virus neutralization and fusion inhibition. Immunity 55, 2135–2148.e6 (2022).
Mohl, B. S., Chen, J., Park, S. J., Jardetzky, T. S. & Longnecker, R. Epstein–Barr virus fusion with epithelial cells triggered by gB is restricted by a gL glycosylation site. J. Virol. 91, e01255-17 (2017).
Matsuura, H., Kirschner, A. N., Longnecker, R. & Jardetzky, T. S. Crystal structure of the Epstein–Barr virus (EBV) glycoprotein H/glycoprotein L (gH/gL) complex. Proc. Natl Acad. Sci. USA 107, 22641–22646 (2010).
Program, C. Z. I. C. S. et al. CZ CELLxGENE Discover: a single-cell data platform for scalable exploration, analysis and modeling of aggregated data. Nucleic Acids Res. 53, D886–D900 (2025).
Uhlen, M. et al. A pathology atlas of the human cancer transcriptome. Science 357, eaan2507 (2017).
Wang, H. et al. Desmoglein 2 is a receptor for adenovirus serotypes 3, 7, 11 and 14. Nat. Med. 17, 96–104 (2011).
Su, C. et al. Molecular basis of EphA2 recognition by gHgL from gammaherpesviruses. Nat. Commun. 11, 5964 (2020).
Zhu, Q. Y. et al. A potent and protective human neutralizing antibody targeting a novel vulnerable site of Epstein–Barr virus. Nat. Commun. 12, 6624 (2021).
Wallaschek, N. et al. Ephrin receptor A2, the epithelial receptor for Epstein–Barr virus entry, is not available for efficient infection in human gastric organoids. PLoS Pathog. 17, e1009210 (2021).
Guo, R. et al. MYC controls the Epstein–Barr virus lytic switch. Mol. Cell 78, 653–669.e8 (2020).
Wang, C. et al. TAF family proteins and MEF2C are essential for Epstein–Barr virus super-enhancer activity. J. Virol. 93, e00513–e00519 (2019).
Kalla, M., Gobel, C. & Hammerschmidt, W. The lytic phase of Epstein–Barr virus requires a viral genome with 5-methylcytosine residues in CpG sites. J. Virol. 86, 447–458 (2012).
Zhang, S., Tan, H.C. & Ooi, E.E. Visualizing dengue virus through Alexa Fluor labeling. J. Vis. Exp. 53, e3168 (2011).
Liu, X. & Mondal, A. M. Conditional cell reprogramming for modeling host–virus interactions and human viral diseases. J. Med. Virol. 92, 2440–2452 (2020).
Timofeeva, O. A. et al. Conditionally reprogrammed normal and primary tumor prostate epithelial cells: a novel patient-derived cell model for studies of human prostate cancer. Oncotarget 8, 22741–22758 (2017).
Goldman, R. & Pollack, S. Electric fields and proliferation in a chronic wound model. Bioelectromagnetics 17, 450–457 (1996).
Bajic, G. et al. Influenza antigen engineering focuses immune responses to a subdominant but broadly protective viral epitope. Cell Host Microbe 25, 827–835.e6 (2019).
Clark, S. A. et al. SARS-CoV-2 evolution in an immunocompromised host reveals shared neutralization escape mechanisms. Cell 184, 2605–2617.e18 (2021).
Guo, W., Wang, M. & Chen, L. A co-expression vector for baculovirus-mediated protein expression in mammalian cells. Biochem. Biophys. Res. Commun. 594, 69–73 (2022).
Scheich, C., Kummel, D., Soumailakakis, D., Heinemann, U. & Bussow, K. Vectors for co-expression of an unrestricted number of proteins. Nucleic Acids Res. 35, e43 (2007).
Acknowledgements
We thank G. Sai Wah Tsao (University of Hong Kong) for the NP69 cells. We thank K. Munger (Tufts University) for the NOK cell lines. We thank J. M. Middeldorp (VU University Medical Center, Netherlands) for the OT 6-2 antibody. We thank E. Johannsen (University of Wisconsin) for EBV glycoprotein gL, gH and gB from pDONR221 and J. Doench (Broad Institute) for the pLX-TRC313 vector. We thank L. Swanback for technical assistance and B. Sahoo (Case Western Reserve University, School of Medicine) for helping with the MST experiment. This study was funded by NIAID AI123420 (B.Z.), DP2AI171139 (S.J.), NIGMS R35GM155298 (M.T.), NCI CA269043 (B.G.), NIAID AI137337 (B.G.), NCI CA228700 (B.G.), R33CA258016 (Xuefeng Liu), R01CA276474 (Xuefeng Liu) and U01CA278927 (Xuefeng Liu) from the National Institutes of Health, start-up funding from The Ohio State University Comprehensive Cancer Center (Xuefeng Liu) and a Burroughs Wellcome Career Award in Medical Sciences (B.G.). J.I.C. and W.B. are supported by the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases. B.Z. is also supported by the Fund to Sustain Research Excellence from Brigham Research Institute. We thank M. Kanekiyo and M. Gordon Joyce (NIAID) for help with the EBV gHgL monoclonal antibodies.
Author information
Authors and Affiliations
Contributions
B.Z., S.J. and H.W. conceived the study, oversaw the project and wrote the paper. H.W. conducted the experiments and analysed and interpreted the data. Z.M. performed the MST experiment. Q.G. conducted ALI experiments and performed data analysis. Z.M., Q.G., W.L., Y.N., C.W., Z.L., Z.M., J.L, W.B. and K.R.S.Z. conducted experiments and performed data analysis. Xiang Liu and M.T. performed bioinformatics and statistical analyses. Y.Y.Y. analysed immunohistochemistry data from the Human Protein Atlas and scRNA-sequencing data from the CellXGene database. Xuefeng Liu, B.G., M.T., J.I.C. and X.D. provided helpful suggestions for the study. All authors reviewed and approved the final paper.
Corresponding authors
Ethics declarations
Competing interests
S.J. is a co-founder of Elucidate Bio Inc. and has received research support from Roche and Novartis, both unrelated to this work. All other authors declare that they have no competing financial interests.
Peer review
Peer review information
Nature Microbiology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 High efficiency EBV cell-cell contact infection and genome-wide CRISPR screen, Related to Fig. 1.
a, NOK cells were co-cultured with cell-free EGFP EBV or anti-human IgG induced AKATA cells harboring EGFP EBV. Infection efficiencies were evaluated using fluorescence microscope. b, NOK cells were co-cultured with cell-free EGFP EBV or anti-human IgG induced EGFP EBV AKATA cells. Infection efficiencies were evaluated using FACS. c, The infection efficiencies of cell-free (n = 6) and cell-cell (n = 28) contact infection was determined by FACS. Each dot represents the efficiency of one experiment. d-e, anti-human IgG induced EGFP EBV AKATA cells were co-cultured with NOK directly or separated by 0.4 um filter in transwells. Infection efficiencies were determined by FACS, n = 6. f, Graphic description of genome-wide CRISPR screen for EBV entry receptors. g, Lentiviruses expressing control sgRNA or other sgRNAs from the screen were used to knock out genes in NOK cells stably expressing CAS9. Cells were co-cultured with EGFP EBV AKATA cells. The virus infection efficiencies were determined by FACS, n = 3. h, Same as g, except that the viral infection efficiencies were determined by florescence microscope. All FACS gating strategies use no EBV infection cells as negative control for EBV infection as shown in dot plot (b, d). The representative images (a, h), dot plot (b, and d) and EBV-infected cell percentage (e, g) were shown. Data represent as mean +/- S.D. from three (g) or six (e) biological replicates; P-values were derived from two-sided t-test without adjustments. Experiments were performed twice (d, e) or thrice (g, h) with similar results. (f) created using BioRender. Scale bars: 150 μm or 275 μm. Panel f created with BioRender.com.
Extended Data Fig. 2 DSC2 and DSC3 double knockout blocks EBV infection of NOK cellsRelated to Fig. 1.
a, Representative images of NOK cells with or double knockout infected by EBV through contact infection. The efficiencies were determined by fluorescence microscope. b, DSC2/3 expression for a. c, d, NOK cells with DSC2/3 double knockout were contact infected. The efficiencies were determined by FACS, n = 3. e. PE-conjugated control (mIgG-PE) or EphA2 antibody (EphA2-PE) stained NOK cells with double knockout were analyzed by FACS. f, Multiple sgRNAs depleted DSC2/3 in NOK cells. efficiently, determined by immunoblot. g, Multiple DSC2/3 sgRNAs inhibited contact infections, determined by FACS, n = 3. h, Multiple DSC2/3 sgRNAs reduced cell-free infections, determined by FACS, n = 3. i, DSC2/3 was completely depleted in NOK single clone cells (#23 NOK-DSC2/3- KO cells) shown by immunoblot. Puromycin and zerocin-resistance control sgRNA (sgCTL-PZ) transduced NOK cells were used as control. j, The cell – cell contact infection was completely abolished in DSC2/3 KO NOK cells. EBV infection efficiency was determined by flow cytometry, n = 3. k, CAS9-resistant DSC2/3 cDNAs were expressed in double knockout NOK cells as shown inimmunoblot. l, DSC2/3 cDNAs rescued cell - cell contact infection, determined by FACS, n = 3. Expression of DSC2/3 was determined by immunoblot (b, f, i and k), GAPDH served as an internal control. The representative images (a), histogram plot (c) and the percentage (d, g, h, i and l) of EBV infected cells were presented. Data are presented as mean values +/- S.D. from three biological replicates; (d, g,h, j, and l); P-values were derived from two-sided paired t-test without adjustments, n = 3. Experiments were performed twice (e, f, g, h, k, and l) and thrice (a, b, c, d, i and j) with similar results. Scale bars: 150 μm.
Extended Data Fig. 3 Expression of DSC2, DSC3, and EphA2 in epithelial cell lines and the primary Head and Neck Epithelial cells (HNE004). Related to Fig. 1.
a, Expression of DSC2, DSC3 and EphA2 in indicated cell lines by western blotting. b, c. EBV cell-cell contact infection efficiencies of indicated cell lines were determined by FACS, n = 3. d. Western blotting showing the expression of DSC2, DSC3, and EphA2 in NOK, HNE004, NP69, and Daudi cells. Data are presented as mean values +/- S.D. three biological replicates, (c) P-values were derived from two-sided t-test without adjustments. Experiments were performed thrice with similar results. GAPDH served as an internal control (a, and d).
Extended Data Fig. 4 DSC2 and DSC3 double knock out in AGS and 293 T cells blocks EBV infection, Related to Fig. 1.
a, Expression of DSC2 and DSC3 in AGS cells following double CRISPR knockout. b, Cell-cell contact infection of AGS cells with control or DSC2/3 double knock out determined by fluorescence microscope. c. Cell-cell contact infection of AGS cells with sgRNA control (sgCTL-PZ) or DSC2/3 double knock out determined by FACS, n = 3. d. Cell-free EBV infection of AGS cells with sgRNA control or DSC2/3 double knock out, determined by fluorescence microscope. e, Cell-free EBV infection of AGS cells with control or DSC2/3 double knock out determined by FACS, n = 3. f. AGS cells with DSC2 and DSC3 double knock out were stained with PE conjugated control (mIgG-PE) or EphA2 antibody (EphA2-PE) and analyzed by FACS. g, AGS cells expressing mCherry (AGS-mCherry) and with control (sgCTL-PZ) or DSC2/3 double knock out were cocultures with the GFP EBV AKATA cells for two hours. Unattached AKATA cells were washed off. The colocalization of AKATA (Green) and AGS (Red) cells was visualized using florescence microscope. h. Quantitation of AKATA cells attached to control and DSC2/3 double knock out AGS cells, n = 5. i, Expression of DSC2 and DSC3 in 293 T cells following single or double CRISPR knock out. j-k, 293 T cells with DSC2, DSC3, or doble knock out were infected by cell-cell contact (j-k), and cell-free (l-m) virus. The infection efficiencies were determined by fluorescence microscope (j,l) and FACS (k,m). Data are presented as mean values +/- S.D. three (c, e, k and m) or five (h) biological replicates. P-values were derived from two-sided t-test without adjustments. Experiments were performed twice (g, h, i, j, k, and m) or thrice (a, b, c, d, e, and f) with similar results. GAPDH served as an internal control (a, and i). Scale bars: 150 μm.
Extended Data Fig. 5 DSC2 and DSC3 expression in YCCEL1 and NP69 cells enables efficient EBV infection, Related to Fig. 1.
a, b, YCCEL1 cells expressing DSC2 or DSC3 were infected with EBV by cell-cell contact infection. The infection efficiencies were determined by fluorescence microscope (a) or FACS (b). c, d, NP69 cells expressing DSC2 or DSC3 were infected with EBV by cell-cell contact infection. The infection efficiencies were determined by fluorescence microscope (c) or FACS (d). Experiments were performed thrice with similar results.
Extended Data Fig. 6 DSC2 and DSC3 antibodies are specific for their targets, Related to Fig. 2.
a, DSC2 or DSC3 was expressed in NOK cells depleted for both DSC2/3. Sheep-anti DSC2 antibody only blocked cell-free EBV infection in DSC2 expression cells and had no effect on DSC3 expression cells. Sheep IgG as a control. b, Sheep-anti-DSC3 antibody did not block EBV infection of NOK cells expressing DSC2 only. c, Only sheep-anti-DSC3 antibody can block EBV infection of DSC3 expressing NOK cells. d, DSC2 antibody had no effect on EBV infection of Daudi cells. e, DSC2 antibody also reduced EBV cell-cell contact infection, but DSC3 antibody had no effect on EBV infection. f. The mouse monoclonal antibody #1H8G1 did not block EBV infection of NOK cells. Data are presented as mean values +/- S.D. three biological replicates, P-values were derived from two-sided t-test without adjustments, n = 3. Experiments were performed twice with similar results.
Extended Data Fig. 7 EBV gHgL interacted with DSC2 and DSC3, Related to Fig. 3.
a, Human monoclonal antibodies against gH/gL (769C2 and 769C5) or mouse monoclonal antibodies against gp350 (72A1) can immune precipitate gH-V5/gL or gp350-V5, respectively, but not immune precipitate EGFP-V5 in 293 T. 293 T cells were transiently transfected with V5-tagged EBV-gHgL and EBV-gp350 plasmids, EGFP-V5 as control. b, Anti gH/gL antibody 769C2 and gp350 antibody OT6-2 recognized gH/gL and gp350 protein in western blot, respectively. c, Anti gH/gL human antibodies co-immune precipitated gH/gL with DSC2 in NOK cells depleted for EphA2 and expressing DSC2-mCherry were co-cultured with anti-human IgG induced EGFP EBV AKATA cells. d. The direct interaction of purified His tagged EBV-gH/gL and Fc-tagged DSC2 extracellular domain (EC1-5) were shown by ELISA, but not with His tagged gp350, n = 6. e, The binding affinity of purified EBV-gH/gL and DSC2 were analysed by ELISA. EC50 as the binding affinity constant value. OD450max indicates maximum absorbance. Data are presented as mean values +/- S.D. (d, e) six biological replicates in two independent experiments, (c) P-values were derived from two-sided t-test without adjustments. Experiments were performed twice with similar results.
Extended Data Fig. 8 The DSC2 intracellular domain is dispensable for EBV infection, Related to Fig. 4.
a, DSC2 expression was detected in NP69 cells with DSC2 wild type (WT) or mutant (deleted intracellular domain, ΔIC) by western blot. b, Deletion of DSC2 intracellular domain (ΔIC) didn’t affect EBV infection. NP69 cells with DSC2 wild type (WT) or mutant (deleted intracellular domain, ΔIC) were infected with EBV through cell-cell contact or cell-free EBV. EBV infection efficiency was determined by fluorescence microscope. c, d, EBV gHgL interacted with DSC2 through its 1st and 2nd cadherin-like repeats in NOK and NP69 cells. FLAG tagged gH21-678gL were expressed with mCherry tagged wt or mutant (with deletion of EC1-2) DSC2 in NOK (c) and NP69 (d) cells. The anti-FLAG antibody M2 was used to immunoprecipitate EBV gHgL and the associated complex. WT or mutant DSC2 (ΔEC1-2) was detected by mCherry antibody in western blots. e, Expression of WT or mutant DSC2 in YCCEL1 and NP69 cells determined by western blotting. GAPDH served as an internal control. Represented images from two independent experiments. Scale bars: 150 μm.
Extended Data Fig. 9 Double knockout or over-expression of DSC2/3 did not affect EBV binding and cell growth, Related to Fig. 4.
a, Daudi cells with control or CD21 knock out were incubated with ALEXA Fluor 647 labelled EBV at 4 °C for an hour. The EBV bindings were determined by FACS following wash. b, NOK cells with control or DSC2/3 double knock out were incubated with ALEXA Fluor 647 labelled EBV at 4 °C for an hour, The EBV bindings were determined by FACS, following wash. c. DSC2/3 double knockout or EPHA2 knock out did not affect EBV binding. NOK cells with sgRNA control (sgCTL-PZ or sgCTL-P), EphA2 or DSC2/3 double knockout were incubated with EBV at 4 °C for three hours. After thoroughly wash to remove unbound virus, The EBV bindings were determined by qPCR, following cell wash, n = 3. d, DSC2/3 double knock out did not affect NOK cell growth, n = 4. e, DSC2 or DSC3 overexpression did not affect EBV binding. Data are presented as mean values +/- S.D. three (c) and four (d) biological replicates. (c, d) P-values were derived from two-sided t-test without adjustments; Experiments were performed twice (c, d) and thrice (a, b, and e) with similar results.
Extended Data Fig. 10 Overexpression of DSC2 mediated EBV epithelial entry is independent on EphA2, but EphA2 mediating EBV infection depended on DSC2/3. Related to Fig. 5.
a, NOK cells overexpressing mCherry or DSC2-mCherry, along with control or EPHA2 CRISPR knockout, were infected with EBV through cell-cell contact or cell free EBV. The infection efficiency was determined by fluorescence microscope. b, EphA2-mcherry was expressed in control NOK cells or NOK single clone cells with DSC2/3 double knockout. Expression of DSC2, DSC3 or EphA2-mCherry was assessed by western blotting. c, Expression of mCherry, DSC2-mCherry or EphA2-mCherry in YCCEL1 cells, was analyzed by western blotting with mCherry antibody. GAPDH served as an internal control. d, Cartoon schematic summarizing the role of DSC2/3 as receptors for EBV epithelial infection via the cell-free and cell-cell routes of infection, created with BioRender.com. Experiments were performed twice (a, b) or thrice (c) with similar results. Scale bars, 150 μm. Panel d created with BioRender.com.
Supplementary information
Supplementary Information
Supplementary Fig. 1 and Tables 1 and 2.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 7
Unprocessed western blots.
Source Data Extended Data Fig. 8
Unprocessed western blots.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Unprocessed western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, H., Mou, Z., Yeo, Y.Y. et al. Epstein–Barr virus exploits desmocollin 2 as the principal epithelial cell entry receptor. Nat Microbiol (2025). https://doi.org/10.1038/s41564-025-02126-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41564-025-02126-0
This article is cited by
-
Identified receptors shed light on Epstein–Barr virus infection
Nature Microbiology (2025)