Abstract
Fibroblasts are non-haematopoietic structural cells that define the architecture of organs, support the homeostasis of tissue-resident cells and have key roles in fibrosis, cancer, autoimmunity and wound healing1. Recent studies have described fibroblast heterogeneity within individual tissues1. However, the field lacks a characterization of fibroblasts at single-cell resolution across tissues in healthy and diseased organs. Here we constructed fibroblast atlases by integrating single-cell transcriptomic data from about 230,000 fibroblasts across 17 tissues, 50 datasets, 11 disease states and 2 species. Mouse fibroblast atlases and a DptIRESCreERT2 knock-in mouse identified two universal fibroblast transcriptional subtypes across tissues. Our analysis suggests that these cells can serve as a reservoir that can yield specialized fibroblasts across a broad range of steady-state tissues and activated fibroblasts in disease. Comparison to an atlas of human fibroblasts from perturbed states showed that fibroblast transcriptional states are conserved between mice and humans, including universal fibroblasts and activated phenotypes associated with pathogenicity in human cancer, fibrosis, arthritis and inflammation. In summary, a cross-species and pan-tissue approach to transcriptomics at single-cell resolution has identified key organizing principles of the fibroblast lineage in health and disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Raw and processed RNA-seq, ATAC–seq and mouse healthy skin scRNA-seq datasets are available from the ArrayExpress repository under the accession numbers E-MTAB-10324, E-MTAB-10316 and E-MTAB-10315, respectively. Supplementary Table 3 lists the studies used to generate the Cd45− maps and fibroblast atlases. These integrated scRNA-seq objects used for analysis are provided in an online resource that can be accessed at https://fibroXplorer.com. Human pancreatic cancer single-cell data are available in the EGA database under accession EGAD00001005365. Source data are provided with this paper.
Code availability
No new algorithms were developed for this manuscript. All code generated for analysis is available from the authors upon request.
References
Koliaraki, V., Prados, A., Armaka, M. & Kollias, G. The mesenchymal context in inflammation, immunity and cancer. Nat. Immunol. 21, 974–982 (2020).
Hinz, B. & Lagares, D. Evasion of apoptosis by myofibroblasts: a hallmark of fibrotic diseases. Nat. Rev. Rheumatol. 16, 11–31 (2020).
Buechler, M. B. & Turley, S. J. A short field guide to fibroblast function in immunity. Semin. Immunol. 35, 48–58 (2018).
Gautier, E. L. et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13, 1118–1128 (2012).
Gosselin, D. et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159, 1327–1340 (2014).
Lavin, Y. et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014).
Croft, A. P. et al. Distinct fibroblast subsets drive inflammation and damage in arthritis. Nature 570, 246–251 (2019).
Wei, K. et al. Notch signalling drives synovial fibroblast identity and arthritis pathology. Nature 582, 259–264 (2020).
Zhang, F. et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. 20, 928–942 (2019).
Dominguez, C. X. et al. Single-cell RNA sequencing reveals stromal evolution into LRRC15+ myofibroblasts as a determinant of patient response to cancer immunotherapy. Cancer Discov. 10, 232–253 (2020).
Elyada, E. et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov. 9, 1102–1123 (2019).
Lambrechts, D. et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 24, 1277–1289 (2018).
Öhlund, D. et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 214, 579–596 (2017).
Rinkevich, Y. et al. Identification and isolation of a dermal lineage with intrinsic fibrogenic potential. Science 348, aaa2151 (2015).
Buechler, M. B. et al. A stromal niche defined by expression of the transcription factor WT1 mediates programming and homeostasis of cavity-resident macrophages. Immunity 51, 119–130.e5 (2019).
Krausgruber, T. et al. Structural cells are key regulators of organ-specific immune responses. Nature 583, 296–302 (2020).
Bellomo, A. et al. Reticular fibroblasts expressing the transcription factor WT1 define a stromal niche that maintains and replenishes splenic red pulp macrophages. Immunity 53, 127–142.e7 (2020).
Baryawno, N. et al. A cellular taxonomy of the bone marrow stroma in homeostasis and leukemia. Cell 177, 1915–1932.e16 (2019).
McCarthy, N. et al. Distinct mesenchymal cell populations generate the essential intestinal BMP signaling gradient. Cell Stem Cell 26, 391–402.e5 (2020).
Wirka, R. C. et al. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat. Med. 25, 1280–1289 (2019).
Tsukui, T. et al. Collagen-producing lung cell atlas identifies multiple subsets with distinct localization and relevance to fibrosis. Nat. Commun. 11, 1920 (2020).
Dahlgren, M. W. & Molofsky, A. B. Adventitial cuffs: regional hubs for tissue immunity. Trends Immunol. 40, 877–887 (2019).
Sitnik, K. M. et al. Context-dependent development of lymphoid stroma from adult CD34+ adventitial progenitors. Cell Rep. 14, 2375–2388 (2016).
Merrick, D. et al. Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science 364, eaav2501 (2019).
Zhou, X. et al. Circuit design features of a stable two-cell system. Cell 172, 744–757.e17 (2018).
Chai, Q. et al. Maturation of lymph node fibroblastic reticular cells from myofibroblastic precursors is critical for antiviral immunity. Immunity 38, 1013–1024 (2013).
Kapoor et al.
Maurer, C. et al. Experimental microdissection enables functional harmonisation of pancreatic cancer subtypes. Gut 68, 1034–1043 (2019).
Habermann, A. C. et al. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci. Adv. 6, eaba1972 (2020).
Emblom-Callahan, M. C. et al. Genomic phenotype of non-cultured pulmonary fibroblasts in idiopathic pulmonary fibrosis. Genomics 96, 134–145 (2010).
Kinchen, J. et al. Structural remodeling of the human colonic mesenchyme in inflammatory bowel disease. Cell 175, 372–386.e17 (2018).
Cremasco, V. et al. B cell homeostasis and follicle confines are governed by fibroblastic reticular cells. Nat. Immunol. 15, 973–981 (2014).
Kuehn, M. R., Bradley, A., Robertson, E. J. & Evans, M. J. A potential animal model for Lesch-Nyhan syndrome through introduction of HPRT mutations into mice. Nature 326, 295–298 (1987).
Smithies, O., Gregg, R. G., Boggs, S. S., Koralewski, M. A. & Kucherlapati, R. S. Insertion of DNA sequences into the human chromosomal β-globin locus by homologous recombination. Nature 317, 230–234 (1985).
Thomas, K. R., Folger, K. R. & Capecchi, M. R. High frequency targeting of genes to specific sites in the mammalian genome. Cell 44, 419–428 (1986).
Newman, R. J., Roose-Girma, M. & Warming, S. Efficient conditional knockout targeting vector construction using co-selection BAC recombineering (CoSBR). Nucleic Acids Res. 43, e124 (2015).
Gertsenstein, M. et al. Efficient generation of germ line transmitting chimeras from C57BL/6N ES cells by aggregation with outbred host embryos. PLoS ONE 5, e11260 (2010).
Hughes, E. D. & Saunders, T. L. Advanced Protocols for Animal Transgenesis, An ISTT Manual 291–325 (Springer, 2011).
Zhang, L. et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell 181, 442–459.e29 (2020).
Painter, M. W. et al. Transcriptomes of the B and T lineages compared by multiplatform microarray profiling. J. Immunol. 186, 3047–3057 (2011).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Johnson, W. E., Li, C. & Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007).
Korotkevich, G., Sukhov, V. & Sergushichev, A. Fast gene set enrichment analysis. Preprint at https://doi.org/10.1101/060012 (2021).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Corces, M. R. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017).
Wu, T. D. & Nacu, S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26, 873–881 (2010).
Zhang, Y. et al. Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137 (2008).
Li, Q., Brown, J. B., Huang, H. & Bickel, P. J. Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 5, 1752–1779 (2011).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Ho, D. E., Imai, K., King, G. & Stuart, E. A. MatchIt: nonparametric preprocessing for parametric causal inference. J. Stat. Softw. 42, 1–28 (2011).
McLeay, R. C. & Bailey, T. L. Motif enrichment analysis: a unified framework and an evaluation on ChIP data. BMC Bioinformatics 11, 165 (2010).
Wang, S. et al. Target analysis by integration of transcriptome and ChIP–seq data with BETA. Nat. Protoc. 8, 2502–2515 (2013).
Soliman, H. et al. Pathogenic potential of Hic1-expressing cardiac stromal progenitors. Cell Stem Cell 26, 205–220.e8 (2020).
Dobie, R. et al. Single-cell transcriptomics uncovers zonation of function in the mesenchyme during liver fibrosis. Cell Rep. 29, 1832–1847.e8 (2019).
Xiong, X. et al. Landscape of intercellular crosstalk in healthy and NASH liver revealed by single-cell secretome gene analysis. Mol. Cell 75, 644–660.e5 (2019).
Dahlgren, M. W. et al. Adventitial stromal cells define group 2 innate lymphoid cell tissue niches. Immunity 50, 707–722.e6 (2019).
Peyser, R. et al. Defining the activated fibroblast population in lung fibrosis using single-cell sequencing. Am. J. Respir. Cell Mol. Biol. 61, 74–85 (2019).
Xie, T. et al. Single-cell deconvolution of fibroblast heterogeneity in mouse pulmonary fibrosis. Cell Rep. 22, 3625–3640 (2018).
Koga, S. et al. Peripheral PDGFRα+ gp38+ mesenchymal cells support the differentiation of fetal liver–derived ILC2. J. Exp. Med. 278, jem.20172310-23 (2018).
Dell’Orso, S. et al. Single cell analysis of adult mouse skeletal muscle stem cells in homeostatic and regenerative conditions. Development 146, dev181743 (2019).
Scott, R. W., Arostegui, M., Schweitzer, R., Rossi, F. M. V. & Underhill, T. M. Hic1 defines quiescent mesenchymal progenitor subpopulations with distinct functions and fates in skeletal muscle regeneration. Cell Stem Cell 25, 797–813.e9 (2019).
Buechler, M. B. & Turley, S. J. Neutrophils follow stromal omens to limit peritoneal inflammation. Immunity 52, 578–580 (2020).
Davidson, S. et al. Single-cell RNA sequencing reveals a dynamic stromal niche that supports tumor growth. Cell Rep. 31, 107628 (2020).
Cheng, H.-W. et al. Origin and differentiation trajectories of fibroblastic reticular cells in the splenic white pulp. Nat. Commun. 10, 1739 (2019).
Burl, R. B. et al. Deconstructing adipogenesis induced by β3-adrenergic receptor activation with single-cell expression profiling. Cell Metab. 28, 300–309.e4 (2018).
Schwalie, P. C. et al. A stromal cell population that inhibits adipogenesis in mammalian fat depots. Nature 559, 103–108 (2018).
Micheli, A. J. D. et al. Single-cell transcriptomics identify extensive heterogeneity in the cellular composition of mouse achilles tendons. Preprint at https://doi.org/10.1101/801266 (2020).
Hepler, C. et al. Identification of functionally distinct fibro-inflammatory and adipogenic stromal subpopulations in visceral adipose tissue of adult mice. eLife 7, 771 (2018).
Boyd, D. F. et al. Exuberant fibroblast activity compromises lung function via ADAMTS4. Nature 587, 466–471 (2020).
Perez-Shibayama, C. et al. Type I interferon signaling in fibroblastic reticular cells prevents exhaustive activation of antiviral CD8+ T cells. Sci. Immunol. 5, eabb7066 (2020).
Guerrero-Juarez, C. F. et al. Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds. Nat. Commun. 10, 650 (2019).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
van den Brink, S. C. et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods 14, 935–936 (2017).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Gao, X., Hu, D., Gogol, M. & Li, H. ClusterMap: compare multiple single cell RNA-seq datasets across different experimental conditions. Bioinformatics 35, 3038–3045 (2019).
Schubert, M. et al. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun. 9, 20 (2018).
Street, K. et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018).
Bharat, A. et al. Lung transplantation for patients with severe COVID-19. Sci. Transl. Med. 12, eabe4282 (2020).
Maynard, A. et al. Therapy-induced evolution of human lung cancer revealed by single-cell RNA sequencing. Cell 182, 1232–1251.e22 (2020).
Tran, H. T. N. et al. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 21, 12 (2020).
Goldman, M. J. et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 38, 675–678 (2020).
Acknowledgements
We thank members of the Turley laboratory and individuals in the bioinformatics department for discussions; C. Dominguez for experimental assistance; S. Lianoglou, Y. Senbabaoglu and N. Lounsbury for bioinformatics assistance; facility staff at Genentech for vivarium maintenance and core facility assistance; and Turley laboratory members and A. Berkley for their careful review of this manuscript. This work was supported by Genentech.
Author information
Authors and Affiliations
Contributions
Conceptualization: M.B.B., R.N.P., S.M., R.B. and S.J.T. Methodology: M.B.B., R.N.P., S.M., R.B. and S.J.T. Software, formal analysis and data curation: R.N.P., S.M., M.B.B. and A.K.C. Investigation: M.B.B., R.N.P., A.T.K., C.C., A.W.W., Y.A.Y., L.T., R.C. and A.K.C. Writing: M.B.B., R.N.P., S.M., R.B. and S.J.T. Visualization: M.B.B., R.N.P., A.T.K., C.C., S.M. Supervision: M.R.-G., Z.M., J.R.A., S.M., R.B. and S.J.T.
Corresponding authors
Ethics declarations
Competing interests
All authors except for R.B. are employees of Genentech. R.B. is an employee of Freenome.
Additional information
Peer review information Nature thanks Christopher Buckley, Neil Henderson and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Mouse fibroblast bulk RNA-seq and ATAC–seq.
a, Diagram of tissues isolated for bulk RNA-seq and ATAC–seq (adipose tissue (brown, subcutaneous, epididymal), bone, liver, lung, lymph node, mesentery, omentum and pancreas) and experimental scheme. b, Correlation plot of bulk RNA sequencing samples based on top 1,000 most differentially expressed genes. c, Fraction of ATAC–seq reads identified as PCR duplicates, mitochondrial DNA, or unique non-duplicate non-mitochondrial DNA based on genomic mapping. d, Fragment lengths of ATAC–seq samples. e, Number of called peaks across ATAC–seq samples. Atlas describes the universe of all peaks. f, Distribution of ATAC–seq peaks across genomic regions. g, Aggregate signal around the TSS. SLN_2 samples shown, representative of all other samples. h, Heatmap of pairwise correlation coefficients of ATAC–seq samples. Ing, subcutaneous adipose; Epi, epididymal adipose; Panc, pancreas; Om, omentum; SLN, lymph node.
Extended Data Fig. 2 Bulk gene expression and open chromatin.
a, Heat map depicting enriched genes per tissue by bulk RNA-seq. Adjusted P ≤ 0.05, log2(fold change) ≥ 2 (DESeq2, two-sided). Row z-scored. Top, bar plot depicting number of signature genes per tissue. b, Heat map depicting regions of open chromatin per tissue by bulk ATAC–seq. Adjusted P ≤ 0.01, log2(fold change) ≥ 2 (DESeq2, two-sided). Row z-scored. Top, bar plot depicting number of open chromatin regions per tissue. c, Correlation (top) and BETA analysis (bottom) of bulk RNA-seq and ATAC–seq samples. d, BETA analysis of SLN evaluating enriched gene expression compared to enriched SLN OCRs. These data are representative of the rest of the dataset. e, Number of transcription factor binding motifs in signature OCRs per tissue. f, Statistical inference of transcription factor motif enrichment in fibroblasts. Bar colours denote RNA expression of transcription factor. Subcut., subcutaneous adipose; LN, lymph node.
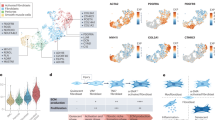
Extended Data Fig. 3 Steady-state fibroblast and Cd45− atlases.
a, UMAP embedding of 180,922 single cells in steady-state Cd45− atlas. Fourteen clusters identified through graph-based clustering are indicated by colour. b, Relative abundance of each tissue in steady-state Cd45− UMAP clusters. The sizes of bubbles indicate the contributions of cells from each tissue to a cluster, and grey bubbles indicate lack of contribution (number of cells <0.02% of tissue) of that tissue to the corresponding cluster. Graph to be read column-wise. c, Fibroblast- and other lineage-associated genes (in grey) in steady-state Cd45− atlas. The sizes of circles denote the percentages of cells from each cluster, and colour encodes the average expression across all cells within a cluster. The colour scale shows the expression level based on row z-score. d, Average bulk tissue-specific fibroblast gene signature scores across tissues represented in the steady-state atlas. Mean-centred values shown. e, Expression of cluster hallmark genes in steady-state fibroblast atlas. The sizes of circles denote the percentages of cells from each cluster, and colour encodes the average expression across all cells within a cluster. The colour scale shows the expression level based on row z-score. f, Expression of fibroblast- and other lineage-associated genes (in grey). Circle sizes denote percentages of cells from each cluster, and colour encodes average expression across all cells within cluster. Colour scale shows expression level (z-score by row). g, Relative abundance of each tissue in steady-state fibroblast UMAP clusters. The sizes of bubbles indicate the contributions of cells from each tissue to a cluster, and grey bubbles indicate lack of contribution (number of cells <0.02% of tissue) of that tissue to the corresponding cluster. Graph to be read column-wise. h, Expression of pathway-responsive genes in perturbed-state atlas clusters as assessed by PROGEN(y) analysis (z-scored per row). i, Volcano plot comparing genes enriched in Pi16+ cluster (left; blue) to Col15a1+ cluster (right; red). Select genes are labelled. Dots labelled in red or blue were determined as having a log2(fold change) of greater than 1 or less than −1 and an adjusted P value less than or equal to 0.05 (Bonferroni correction). j, Expression of Ly6a and Cd34 in steady-state clusters. Wilcoxon’s rank sum test P < 0.05.
Extended Data Fig. 4 Bulk sequencing validates single-cell atlas.
a–p, UMAPs highlighting distribution of cells from individual tissues in the steady-state fibroblast atlas. q, PCA of FACS-sorted bulk RNA-seq of fibroblasts and mesothelial cells, calculated for the 1,000 genes with the highest interquartile range. Circles represent fibroblasts and triangles are mesothelial cells. Each colour denotes a different tissue. r, Heatmap depicting top 20 Pi16+ (fibroblast) and Col15a1+ genes from steady-state fibroblast atlas in bulk RNA-seq data. Rows are z-scored. s, Gene enrichment analysis of top genes (log(fold change) > 0.5) from Pi16+ cluster and Col15a1+ cluster in loadings of PC1, which discriminates between mesothelial cells and fibroblasts (test: FGSEA, Benjamini–Hochberg correction). t, ATAC–seq traces of select genes from Pi16+ and Col15a1+ clusters genes, Ptprc (encoding CD45) and Hprt at ±2 kb of the TSS.
Extended Data Fig. 5 Supplementary FACS gating.
a, Representative FACS gating of live EpCAM−CD45−CD31−PDPN+PDGFRα+ fibroblasts in Figs. 2, 3, Extended Data Figs. 8, 9. b, c, Representative FACS gating on LY6C+SCA1+ (red), LY6C+SCA1− (blue) and LY6C−SCA1− (green) PDGFRα+ cells (as gated in Extended Data Fig. 7a) in lymph node of Ccl19YFP mice (b) and lymph node of Grem1creERT2Rosa26LSLYFP mice (c). Grem1creERT2Rosa26LSLYFP mice received tamoxifen via intraperitoneal injection for five consecutive days and were analysed on days 14–16 after first injection. d, Representative FACS gating on LY6C+SCA1+ (red), LY6C+SCA1− (blue) and LY6C−SCA1− (green) PDGFRα+ cells (as gated in Extended Data Fig. 7a) in tissues from DptIRESCreERT2Rosa26LSLYFP mice. Mice received tamoxifen chow for 14 days and were analysed on days 14–16. n = representative of 3 or more biologically independent experiments.
Extended Data Fig. 6 DptIRESCreERT2 marks universal fibroblasts.
a, Expression of Ly6c and Ly6a in the mouse steady-state fibroblast atlas. b, Representative gating for LY6C+SCA1+, LY6C−SCA1+ and SCA1− fibroblasts across indicated tissues. c, Quantification of LY6C+SCA1+, LY6C−SCA1+ and SCA1− fibroblasts across indicated tissues. Frequency (top), cell number per mg of tissue (bottom). d, Abundance of Pi16, Ccl19, Npnt, and Dpt transcripts in LY6C+SCA1+, LY6C−SCA1+ and SCA1− fibroblasts from lymph node (black) and lung (red). e, Expression of Pi16, Ccl19, Npnt and Dpt in mouse steady-state fibroblast atlas. f, RNAscope for Dpt (blue) and Pi16 (red) on lung (left) and small intestine (right). Scale bar, 50 μm. g, Schematic of generation of DptIRESCreERT2Rosa26LSLYFP mouse. YFP expression is achieved in Dpt-expressing cells following cre-mediated excision of a loxP-flanked transcriptional stop sequence. h, Representative gating for YFP (Dpt) in fibroblasts (live, EpCAM−CD45−CD31−PDPN+PDGFRα+) across indicated tissues. Red, shaded histogram is DptIRESCreERT2ki/kiRosa26LSLYFPwt/loxP and black is DptIRESCreERT2wt/wtRosa26LSLYFPwt/loxP. i, Representative gating for YFP (Dpt) in CD45+ cells (live, EpCAM−CD45+), EpCAM+ cells (live, EpCAM+), and endothelial cells (live, EpCAM−CD45−CD31+) from the pancreas. Red line is DptIRESCreERT2ki/kiRosa26LSLYFPwt/loxP and grey, shaded histogram is DptIRESCreERT2wt/wtRosa26LSLYFPwt/loxP. j, Quantification of YFP+ cells in DptIRESCreERT2ki/kiRosa26LSLYFPwt/loxP animals in indicated cell types across tissues. Percentage YFP+ was determined by subtracting fluorescence in DptIRESCreERT2ki/kiRosa26LSLYFPwt/loxP animals from DptIRESCreERT2wt/wtRosa26LSLYFPwt/loxP animals. k, Abundance of Pi16, Ccl19, Npnt, and Dpt transcripts in FACS-sorted YFP+ and YFP− cells from DptIRESCreERT2ki/kiRosa26LSLYFPwt/loxP and DptIRESCreERT2wt/kiRosa26LSLYFPwt/loxP animals 14–16 days after tamoxifen chow, in lymph node (top) and lung (bottom). l, Abundance of Csf1 transcripts in FACS-sorted YFP+ and YFP− cells from DptIRESCreERT2ki/kiRosa26LSLYFPwt/loxP and DptIRESCreERT2wt/kiRosa26LSLYFPwt/loxP animals 14–16 days after tamoxifen chow in lymph node and lung. n = 3 (c, d, k, l) or representative of 3 or more biologically independent experiments (b, f, h, i). Dot represents each mouse with bar at mean (c, j), plots show minimum and maximum (whiskers) and median (centre line). (d, k, l). h–l, Mice received tamoxifen chow for 14 days and were analysed on days 14–16.
Extended Data Fig. 7 Perturbed-state fibroblast and Cd45− atlases.
a, UMAP embedding of 182,609 cells in perturbed-state Cd45− atlas. Twelve clusters identified through graph-based clustering are indicated by colour. b, Relative abundance of each tissue in perturbed-state Cd45− UMAP clusters. The sizes of bubbles indicate the contributions of cells from each tissue to a cluster, and grey bubbles indicate lack of contribution (number of cells <0.02% of tissue) of that tissue to the corresponding cluster. Graph to be read column-wise. c, Fibroblast- and other lineage-associated genes (in grey) in Cd45− perturbed-state map. The sizes of circles denote the percentages of cells from each cluster, and colour encodes the average expression across all cells within a cluster. The colour scale shows the expression level based on row z-score. d, Expression of cluster hallmark genes in perturbed-state fibroblast atlas. The sizes of circles denote the percentages of cells from each cluster, and colour encodes the average expression across all cells within a cluster. The colour scale shows the expression level based on row z-score. e, Fibroblast- and other lineage-associated genes (in grey) in mouse perturbed-state fibroblast atlas. Circle sizes denote percentages of cells from each cluster; colour encodes average expression across all cells within cluster. Colour scale shows expression level (z-score by row). f, Relative abundance of each tissue in perturbed-state fibroblast UMAP clusters. The sizes of bubbles indicate the contributions of cells from each tissue to a cluster, and grey bubbles indicate lack of contribution (number of cells <0.02% of tissue) of that tissue to the corresponding cluster. Graph to be read column-wise. Clusters with analogues in steady state are depicted in grey, irrespective of the tissue contribution. g, Dendrogram representation of the hierarchical clustering of steady-state (blue) and perturbed-state (red) cluster marker genes. Similarity scores are indicated at the bottom of matching clusters from steady-state and perturbed-state tissues.
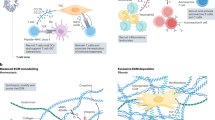
Extended Data Fig. 8 Validation of perturbed-state atlas.
a–o, UMAP representations of the distribution of fibroblasts across tissues and perturbations. p–u, Heat maps showing average relative gene expression in Pi16+, Col15a1+, Adamdec1+, Cxcl5+, and Lrrc15+ clusters (z-scored per row) in the following categories. p, Cytokines and chemokines. q, Wnt-associated genes. r, ECM-associated genes. s, Collagens and laminins. t, Matrix metalloproteases and cathepsins. u, Receptors and surface molecules. v, Expression of pathway-responsive genes in perturbed-state atlas clusters as assessed by PROGEN(y) analysis (z-scored per row). w, RNAscope for Dpt (blue) and Grem1 (red) in non-lesional colon (top) and lesional colon (bottom) on day 7 after induction of DSS colitis. Data are representative of three experiments. Scale bars, 50 μm (top) and 250 μm (bottom). x, Ly6a and Cd34 expression in perturbed-state clusters. Wilcoxon’s rank sum test, P < 0.05. y, Pseudotime(s) visualized using principal curves representing trajectories of fibroblast differentiation across perturbed-state fibroblast object. Blue lines show trajectory to activated clusters, grey lines show trajectory to clusters with a steady-state analogue. Pi16+ cluster set as root. z, Representative FACS strategy for subcutaneous tumour experiments. a′, Representative flow cytometry plots showing frequency of YFP+ cells in LRRC15+ fibroblasts from KPR3070 subcutaneous tumour at day 21 post-inoculation in DptIRESCreERT2wt/wtRosa26LSLYFPwt/loxP animals. b′, Quantification of FACS data (Fig. 3d, Extended Data Fig. 8z, a′). Data are from b′ or representative of 2 (z–a′) or 3 (w) experiments. Each dot represents one mouse (b′). n = 2 (b′) or representative of 2 (z–a′) or 3 (w) biologically independent experiments.
Extended Data Fig. 9 Human universal fibroblasts.
a, Top, UMAP embeddings of human pancreatic cancer tumour and normal adjacent tissue (n = 21,626 cells). Bottom, percentage of cells in each cluster coming from tumour or NAT. b, UMAP as in a, coloured by expression of indicated genes. c, Relative average expression of top 10 marker genes (sorted by log(fold change)) for each cluster in the pancreatic cancer single-cell dataset. Two representative genes highlighted per cluster. DEGs across clusters. d, Top, expression level of indicated marker genes (colour, y-axis) across 100 pseudo-bulk samples (x-axis) generated from human pancreatic cancer scRNA-seq data. The known percentage of cells from cluster 8 in each pseudo-bulk is shown by the dotted blue line. Bottom, boxplots representing the distributions of pairwise correlation coefficients of the top 20 marker genes for cluster 8 in pseudo-bulk samples containing (left) and not containing cells from cluster 8 (right). e, Boxplots summarizing DPT expression distributions across tissues from the GTEx portal. Tissues with mean above horizontal black line were included in correlation analysis (f). n = 7,851 biologically independent samples. f, Co-expression as in d, results from the gene-by-gene correlation matrices are summarized as boxplots for each individual tissue from GTEx. n = 5,957 biologically independent samples. g, Gene-by-gene correlation matrix of pairwise correlations in 205 normal pancreas bulk RNA-seq samples from GTEx. Blue indicates Pi16+ cluster signature gene, red indicates Col15a1+ signature gene. h, Human universal fibroblast score projected onto human pancreatic cancer samples. i, Human universal fibroblast score projected onto human subcutaneous adipose. d–f, Box and whisker plots show minimum and maximum (whiskers), interquartile range (box) and median (centre line).
Extended Data Fig. 10 Perturbed-state human fibroblasts.
a, Volcano plot visualizing log(fold change) (x-axis) and adjusted P value (y-axis) comparing fibroblasts from cluster 3 in Extended Data Fig. 9a compared to fibroblasts from cluster 8 (Wilcoxon’s rank sum test). b, Scatterplot visualizing the scores for a NAT (cluster 3) fibroblast expression gene set (y-axis) compared to scores for a CAF (cluster 8) expression gene set (x-axis) in 122 bulk RNA-seq samples of microdissected PDAC tissue. Each dot represents a sample, the regression line is given in blue. Error band, 95% confidence interval. c–f, UMAP representation of cells from the mouse perturbed-state atlas, each cell coloured by their score for gene sets corresponding to the following. c, Lrrc15+ myofibroblasts from human pancreatic cancer (cluster 3 CAF, ‘MyCAF’11), rheumatoid arthritis (‘Human RA F2’7), lung disease (‘Myofibroblast’29), IPF30 and ulcerative colitis (‘S2’31). d, Universal fibroblasts from human pancreatic cancer (cluster 8 NAT), lung disease (‘PLIN2+ fibroblast’29) and ulcerative colitis (‘S1’31). e, Inflammatory fibroblasts from pancreatic cancer (‘iCAF’11) and rheumatoid arthritis (‘Human RA F5’7). f, UMAP of IL-1 CAF cells derived from ref. 10, projected onto perturbed-state fibroblast atlas. g, Expression of human universal fibroblast module across human perturbed-state atlas clusters. h, Abundance of LRRC15 and CTHRC1 expression in human perturbed-state atlas. i, Heat maps showing average relative gene expression in human perturbed-state atlas (z-scored by row).
Supplementary information
Supplementary Table 1
Tissue-specific fibroblast and mesothelial cell genes by bulk RNAseq (related to Extended Data Fig. 2).
Supplementary Table 2
Tissue-specific OCRs and TFs by bulk ATACseq (related to Extended Data Fig. 2).
Supplementary Table 3
Tissue contributions to single cell RNAseq objects used in analyses of steady state and perturbed state murine tissues (related to Figs 1 and 3).
Supplementary Table 4
DEGs for steady-state mouse fibroblast atlas (related to Fig. 1).
Supplementary Table 5
DEGs for mouse perturbed-state fibroblast atlas (related to Fig. 3).
Supplementary Table 6
Patient information (related to Fig. 4 and Extended Data Figs 11 and 12).
Supplementary Table 7
DEGs for human pancreatic cancer samples (related to Fig. 4 and Extended Data Figs 11 and 12).
Supplementary Table 8
Human gene sets projected onto mouse perturbed-state fibroblast atlas (related to Extended Data Fig. 12).
Supplementary Table 9
DEGs for human perturbed-state atlas (related to Fig. 4).
Rights and permissions
About this article
Cite this article
Buechler, M.B., Pradhan, R.N., Krishnamurty, A.T. et al. Cross-tissue organization of the fibroblast lineage. Nature 593, 575–579 (2021). https://doi.org/10.1038/s41586-021-03549-5
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41586-021-03549-5
This article is cited by
-
Molecular profiling of skin cells identifies distinct cellular signatures in radiation-induced skin injury across various stages in the murine dataset
Experimental Hematology & Oncology (2025)
-
The type I collagen paradox in PDAC progression: microenvironmental protector turned tumor accomplice
Journal of Translational Medicine (2025)
-
Acidosis-induced p38-kinase activation triggers an IL-6-mediated crosstalk of renal proximal tubule cells with fibroblasts leading to their inflammatory response
Cell Communication and Signaling (2025)
-
Radiation-induced lung injury: from mechanism to prognosis and drug therapy
Radiation Oncology (2025)
-
Single-cell transcriptomic analysis reveals prognosis-related stromal signatures that potentiate stratification of patients with extrahepatic cholangiocarcinoma
BMC Gastroenterology (2025)