Abstract
Inflammatory diseases are often chronic and recurrent, and current treatments do not typically remove underlying disease drivers1. T cells participate in a wide range of inflammatory diseases such as psoriasis2, Crohn’s disease3, oesophagitis4 and multiple sclerosis5,6, and clonally expanded antigen-specific T cells may contribute to disease chronicity and recurrence, in part by forming persistent pathogenic memory. Chronic rhinosinusitis and asthma are inflammatory airway diseases that often present as comorbidities7. Chronic rhinosinusitis affects more than 10% of the general population8. Among these patients, 20–25% would develop nasal polyps, which often require repeated surgical resections owing to a high incidence of recurrence9. Whereas abundant T cells infiltrate the nasal polyps tissue10,11, T cell subsets that drive the disease pathology and promote recurrence are not fully understood. By comparing T cell repertoires in nasal polyp tissues obtained from consecutive surgeries, here we report that persistent CD8+ T cell clones carrying effector memory-like features colonize the mucosal tissue during disease recurrence, and these cells characteristically express the tryptase Granzyme K (GZMK). We find that GZMK cleaves many complement components, including C2, C3, C4 and C5, that collectively contribute to the activation of the complement cascade. GZMK-expressing CD8+ T cells participate in organized tertiary lymphoid structures, and tissue GZMK levels predict the disease severity and comorbidities better than well-established biomarkers such as eosinophilia and tissue interleukin-5. Using a mouse asthma model, we further show that GZMK-expressing CD8+ T cells exacerbate the disease in a manner dependent on the proteolytic activity of GZMK and complements. Genetic ablation or pharmacological inhibition of GZMK after the disease onset markedly alleviates tissue pathology and restores lung function. Our work identifies a pathogenic CD8+ memory T cell subset that promotes tissue inflammation and recurrent airway diseases by the effector molecule GZMK and suggests GZMK as a potential therapeutic target.
Similar content being viewed by others
Main
To determine whether T cell memory is involved in the recurrence of nasal polyps (NPs), we first searched for shared T cell clones in samples obtained from repeated surgeries (Fig. 1a,b). T cell repertoire analyses were performed using paired frozen samples from six patients (NP1–6), with intervals between two surgeries ranging from 0.2 to 3.1 years. Nasal tissues from seven patients undergoing septoplasty were included as healthy controls (HC1–7) (Fig. 1b and Supplementary Table 1). Single transcripts of T cell receptors (TCRs) α and β were barcoded and sequenced, and the full-length variable regions were assembled and assigned to different clones (Fig. 1a and Supplementary Fig. 2a,b). As expected, T cell repertoires were distinct among individuals, with only 5 out of 1,585 TCRα and 1 out of 3,888 of TCRβ chains being shared between different patients (Supplementary Fig. 2c). On the contrary, we consistently identified common TCRα and TCRβ chains in NP tissues obtained from the two surgeries on the same patient (Fig. 1c and Supplementary Fig. 2d), suggesting that cells of the same clonal lineages recolonized the NP tissue during disease recurrence. Moreover, some of the persisting clones were among the most dominant ones in both surgical samples (Fig. 1c and Supplementary Fig. 2), suggesting local proliferation and expansion.
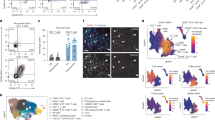
a, Workflow for analysis of surgically removed nasal tissues. RT, reverse transcription. b, Overview of surgical intervals for paired samples. See Extended Data Table 1 for more patient information. c, Persistent clones, identified by matched TCRβ sequences, found in paired surgical samples by bulk sequencing analyses. Each symbol represents a single clone. UMI, unique molecular identifier. d,e, UMAP visualization of 15 αβ T cell clusters (T0–T14) found in NP tissues (d) and their relative abundances in each individual (e). f,g, Persistent T cell clones in patients NP11–13. f, Clones with both TCRα and β chains identified in paired samples are superimposed on the UMAP of total T cells from the same patient. Only the top ten clones are shown. g, Abundance and cluster composition of individual persistent clones detected in the scRNA-seq analysis of the most recent NP resection (top) and bulk-seq analysis of the previous resection (bottom). Clones are ranked and numbered by their abundance in the scRNA-seq data.
To explore the possibility that clonal T cell populations might contribute to disease pathology and recurrence, we conducted single-cell messenger RNA sequencing (scRNA-seq) of total CD45+ cells from seven NP (NP7–13) and four HC surgical samples (HC6, 8, 9 and 10) (Fig. 1a,b and Supplementary Table 1). Three of the seven NP patients (NP11–NP13) underwent the second surgery to treat their NP relapse at the time of the study, and their previous NP biopsies from the first surgery were cryogenically (cryo)-preserved. By comparing TCR sequences from the two NP biopsies for each of these three patients, we can identify common clones of T cells (Fig. 1b). A total of 42,466 immune cells were included in the analysis and clustered according to their transcriptomes; subsets of lymphocytes, dendritic cells, monocytes and mast cells were identified by respective signature gene expression (Extended Data Fig. 1a,b). After full-length variable regions of αβ TCRs were assembled (Extended Data Fig. 1c), T cells bearing functional receptors could be clustered into diverse subsets in both HC and NP tissues (Fig. 1d,e, Extended Data Fig. 1d,e and Supplementary Table 2). As expected, the abundance of the T helper 2 cell (TH2) (T14) subset, a known disease driver, was significantly increased in patients with NP compared to the HC group8 (Extended Data Fig. 1f). In addition, one CD8+ subset (T1) and four CD4+ T cell subsets (T2, T7, T8 and T12) were also more abundant in NP than in HC tissues (Extended Data Fig. 1f).
We identified 24, 44 and 48 shared T cell clones in patients NP11–NP13, respectively. Most of those shared clones were mapped to T0 and T1, two CD8+ T cell subsets, instead of CD4+ T cells (Figs. 1f,g and 2a). Thus, NP recurrence seems to be accompanied by tissue colonization of CD8+ T cell clones already present in the previous disease episode. Given that the two surgeries were at least 7 months apart, those T cells are probably derived from disease-associated memory CD8+ T cells. For simplicity, we call these shared clones persistent clones.
a, Dot plots showing the expression of selected surface markers by different T cell clusters. Dot size and colour intensity indicate the percentage and the level of expression, respectively. b,c, Representative FACS plots (b) and summary statistics (c) of indicated CD8+ T cell subsets found in HC (n = 8), patients with NP (n = 20) and patients with svNP (n = 20). P values by two-sided Mann–Whitney tests. d, UMAP showing 14 peripheral blood CD8+ T cells clusters (B0–13). Cells isolated from NP11 and NP13 are shown, respectively. e, Clones commonly found in tissue and blood CD8+ T cells, projected onto total tissue T cells from the same individual. f, Abundance and cluster composition of clones found in the blood and paired surgical samples. Clones are numbered as in Fig. 1g. g, Heat maps showing Spearman’s correlation coefficient between T0, T1 and blood CD8+ T cell clusters.
A CD8+ T cell subset correlates with disease severity
We further profiled CD8+ T cell subsets containing those persistent clones. T0 cells showed a conventional resident memory T (TRM) cell phenotype, expressing genes essential for tissue residency, such as ITGAE, CD69, PRDM1, ZNF683 and RUNX3 (ref. 12). The T1 subset contained relatively few TRM cells but many expressing effector memory T cell (TEM) markers including KLRG1 and CD27 (Fig. 2a and Extended Data Fig. 1e). On the other hand, these cells did not highly express genes associated with effector functions, such as TBX21 and CX3CR1. T1 cells also expressed a relatively high level of EOMES, a transcription factor associated with long-term persistence12 (Extended Data Fig. 1e). These data indicate that persistent clones are mostly TRM- and TEM-like cells.
To validate whether these residence- and effector-associated surface markers of T0 and T1 could be used for distinguishing them by flow cytometry, we sort-purified different CD8+ T cell subsets from NP tissues (NP14) and performed scRNA-seq analyses (Extended Data Fig. 2a). When mapped onto the reference dataset (Fig. 1d), the sorted KLRC1+ITGAE+ TRM population mainly contained T0 cells, whereas KLRG1+CD27+ITGAE+ and KLRG1+CD27+ITGAE− cells were mainly mapped to the T1 cluster (Extended Data Fig. 2b,c).
Next, we compared relative abundance of T0-corresponding KLRC1+ITGAE+ TRM cells, T1-corresponding KLRG1+CD27+ITGAE+ TRM and T1-corresponding KLRG1+CD27+ITGAE− TEM-like cells in HC and NP tissues. We further stratified NP tissues by disease severity, with cases of recurrent NP or comorbid asthma defined as severe NP (svNP). We found the abundance of the KLRC1+ITGAE+ TRM subset significantly decreased in patients with NP, whereas the abundance of total T1-corresponding KLRG1+CD27+ cells increased. The resident T1 fraction, those ITGAE+KLRG1+CD27+ cells, progressively increased with disease severity, being more abundant in svNP than in NP (Fig. 2b–c).
In an attempt to predict potential antigens recognized by T1 cells, we integrated 37,483 annotated TCRs from public databases13,14 with 1,311 T1 TCRs identified in our own dataset, and performed network analyses using the GLIPH2 algorithm15. Our results identified 36 major TCR clusters, among which 27 involved TCRs with known antigens, including viral proteins, self-antigens and gliadin, a frequently identified allergen in humans16 (Supplementary Fig. 3).
Tissue T1 cells and the circulating compartment
Most T1 cells are ITGAE−. To test the relationship of T1 to T cells in circulation, we sorted peripheral blood CD8+ T cells from patients NP11 and NP13 to perform scRNA-seq and TCR analyses (Extended Data Fig. 2d). A total of 10,897 blood CD8+ T cells were analysed and clustered into 14 different subsets on the basis of transcriptomic features (B0–B13; Fig. 2d and Supplementary Table 3), revealing a general pattern of cell clustering very similar between the two patients. On the basis of TCR sequences, we found several clones in the blood matching clones populating tissue T1 cells in both patients (Fig. 2e and Supplementary Fig. 4a). Most such blood cells were in the B2 subset, with a few found in B4 and B6 as well (Supplementary Fig. 4b). Some of these clones shared between the blood and NP tissues were persistent clones already seen in NP tissues preserved from the first surgery (Fig. 2f). Among all blood CD8+ T cells, the B2 cluster was the one most similar to tissue T1 cells (Fig. 2g and Supplementary Fig. 4c), however, they lacked the expression of TRM signatures and expressed higher levels of genes associated with T cell stemness, such as TCF7, IL7R and SELL12 (Supplementary Fig. 4d). Therefore, T1 cells in NP tissues are probably derived from blood B2 cells, a model also supported by pseudotime analyses (Supplementary Fig. 5a,b).
Taken together, our data support the idea that persistent clones from a T cell memory reservoir can be continuously deployed into the tissue, potentially promoting disease pathology and recurrence.
Tissue KLRG1+CD27+ cells highly express GZMK
Next, we carefully searched for effector molecules that may be used by those T1 cells in the disease process. Type 2 cytokines (for example, IL-4, IL-5 and IL-13) promote eosinophil recruitment, tissue remodelling and B cell activation in the airway, representing the most well-established drivers of chronic rhinosinusitis and asthmatic tissue pathology17. However, T1 cells expressed almost no mRNA for type 2 cytokines (Fig. 3a). Granzymes are important effector molecules expressed by CD8+ T cells. Among all CD8+ T cells in NP tissues, T1 cells uniquely expressed a high level of Granzyme K (GZMK) transcripts (Fig. 3b). GZMK-expressing T cells have been found in ageing-related inflammation and various inflammatory diseases18,19,20. By intracellular staining, GZMK was highly expressed in both ITGAE+ and ITGAE− fractions of T1 cells but not in KLRC1+ITGAE+ T0 cells (Fig. 3c). GZMK were secreted by these cells on TCR stimulation (Extended Data Fig. 3). Besides T cells, GZMK was also expressed by a small subset of natural killer cells (Extended Data Fig. 1b,d and Fig. 3c) that were significantly increased in patients with svNP, although these cells were less abundant compared to GZMK-expressing T1 cells (Extended Data Fig. 2e,f and Fig. 3c). To explore the potential role of GZMK in NP pathogenesis, we analysed the nasal tissues from a cohort of 148 patients (21 HC, 62 NP and 65 svNP) with balanced age and sex representation (Supplementary Fig. 6a–d), and found that tissue but not serum GZMK protein levels were elevated in NP and furthermore in svNP (Fig. 3d and Supplementary Fig. 6e). Tissue GZMK levels represented a risk factor for svNP as predicted by a multiple linear regression model adjusted for sex, age and body mass index (Extended Data Table 2). In fact, tissue GZMK levels were a better predictive biomarker for svNP than tissue eosinophilia and IL-5 levels, two well-established risk factors for svNP21,22 (Fig. 3e). In addition, the levels of GZMK positively correlated with the abundance of tissue eosinophils in patients with svNP (Supplementary Fig. 2a). Taken together, these data support the possibility that T1 KLRG1+CD27+ CD8+ cells may exacerbate airway inflammation and promotes recurrent airway diseases by the effector enzyme GZMK.
a,b, Dot plots showing the expression of cytokines (a) and granzymes (b) in NP tissue T cell clusters. c, Representative FACS plots (left) and summary statistics (right) showing GZMK expression in different CD8+ T cell subsets and natural killer (NK) cells from NP tissues, in comparison to plasma cells (CD79a+CD38+) as the negative control. d, Nasal tissue GZMK levels in HC, patients with NP and patients with svNP measured by ELISA. Each circle indicates one patient and lines denote means. P values from two-sided Mann–Whitney tests. e, Receiver-operating characteristics (ROC) curves for predicting svNP on the basis of GZMK levels, IL-5 levels and number of eosinophils in NP tissues, respectively (n = 127). Areas under the curves are indicated and colour-coded. HPF, high-power magnification field.
GZMK cleaves C3 to release anaphylatoxin
To investigate whether and how GZMK might promote inflammation, we profiled its potential substrates in NP tissues. To this end, we expressed recombinant wild-type human GZMK and its inactive Ser214-to-Ala mutant (GZMK-S214A) in 293-F cells (Extended Data Fig. 4a). Their respective enzymatic activity and inactivity were confirmed in vitro using surrogate substrates (Extended Data Fig. 4b,c). Subsequently, we immunoprecipitated potential GZMK substrates by incubating GZMK-S214A-bound beads with NP tissue lysates obtained from three patients who underwent surgical NP removal. Through analysis of pulled-down proteins by liquid chromatography-mass spectrometry (Fig. 4a,b), we identified 161, 1,300 and 3,116 GZMK-associated proteins from those three patients (NP15–17), respectively (Fig. 4c and Supplementary Tables 1 and 4). Among those 56 proteins shared by all patients, known substrates of GZMK were present, including SET, HNRNPK and TUBB23 (Fig. 4c,d). We confirmed the cleavage of SET by performing an in vitro digestion assay using the wild-type GZMK (Extended Data Fig. 4d). Furthermore, we confirmed the cleavage of a newly identified substrate, DMBT1, which was shown to reduce inflammation in a mouse rhinitis model24 (Extended Data Fig. 4e).
a, Workflow for the GZMK pull-down assay. b, Representative Coomassie Blue-stained gel showing proteins pulled down by enzymatically inactive GZMK-S214A. Data are representative of three independent experiments using different samples from patients. c, Unique and overlapping GZMK-interacting proteins identified in patients NP15–17. d, Dot plots showing the normalized abundance and SEQUEST scores of the 56 common GZMK-interacting proteins found in all patients. See Methods for more details. e, Cleavage of serum-purified C3 by GZMK. A Coomassie Blue-stained gel (left) and the schematic graph showing the cleavage site (right), as determined by Edman sequencing, are shown. S, serine; N, asparagine; L, leucine. f, Immunoblots showing GZMK cleavage of serum-purified C3. Data are representative of at least three independent experiments (e and f). Source images for gels and blots are presented in Supplementary Fig. 1.
Complement C3 was among new substrates identified. Given its central position in the complement cascade and prominent role in promoting inflammation, we characterized its activation by GZMK in detail. Using serum-purified C3 as the testing substrate, we found that GZMK readily cleaved C3 (Fig. 4e,f), showing a C3-converting efficiency at 35% of the classical C3 convertase C4b2a (Extended Data Fig. 4f,g). By Edman sequencing and immunoblots, we confirmed that the cleavage site was at R748 in the α chain of C3, leading to the release of C3a (Fig. 4e,f), a potent inflammation mediator that has been implicated in inflammatory airway diseases25,26,27. We further verified that GZMK-converted C3a induced comparable levels of ERK phosphorylation in THP1 cells as did serum-purified C3a or C4b2a-converted C3a (Extended Data Fig. 4h–j). Combined, these data establish that GZMK can function as a C3-activating protease to generate active C3a capable of promoting inflammation.
Because several complement proteins were identified as potential substrates in the GZMK-S214A-interactome (Supplementary Table 4), we further tested whether GZMK could target several steps in the complement cascade. By using serum-purified complement proteins as substrates, we found that GZMK was able to cleave C2 into C2a and C2b (Extended Data Fig. 5a–c), and C4 into C4a and C4b (Extended Data Fig. 5d–f). Therefore, GZMK can also in principle promote the formation of the classical C3 convertase C4b2a. Furthermore, GZMK can cleave C5 (Extended Data Fig. 5g–i) in an unconventional way28 to enhance the activity of the membrane attack complex (Extended Data Fig. 5j). Therefore, GZMK is a pro-inflammatory effector molecule that can function by activating the complement cascade.
GZMK+ T cells associate with inflammatory aggregates
We next performed immunohistochemistry analysis of nasal tissues from HC and patients with NP. Whereas very few CD8+ T cells were found in HC, large numbers of CD8+ T cells, including those expressing GZMK, were found in NP tissues, particularly from patients suffering from more severe diseases; many of those cells formed aggregates around blood vessels, reminiscent of tertiary lymphoid structures (Extended Data Fig. 6a–c). In addition, C3 was more abundantly present in GZMK+ cell aggregate areas (Supplementary Fig. 7a,b), suggestive of in situ C3 conversion by GZMK. To characterize inflammatory contents in these aggregates of GZMK-expressing CD8+ T cells, we excised aggregate-containing tissues by laser-capture microdissection and performed bulk mRNA sequencing (Extended Data Fig. 6d–f). As expected, higher levels of GZMK and C3 mRNA were found in GZMK+ areas than in tissue areas that contained few CD8+ T cells (Extended Data Fig. 6g and Supplementary Fig. 7c,d). Areas rich in GZMK-expressing CD8+ T cells also contained higher levels of transcripts associated with TH2 cells, B cells and myeloid cells (Extended Data Fig. 6g and Supplementary Table 5), but not TH1, TH17 and regulatory T cells (Supplementary Fig. 7e). Cell aggregate regions also contained higher levels of chemokines and chemokine receptors important for inflammatory cell recruitment (for example, CXCR3, CXCR4, CCL4 and CCL5) and for organization of tertiary lymphoid structures (for example, CCR7, CXCR5 and their respective ligands)29; higher levels of inflammatory cytokines (for example, IL6, IL16, IL32 and IL27) were also seen in these aggregates (Extended Data Fig. 6g and Supplementary Table 5).
Overall, GZMK-expressing cells accounted for roughly 20% of total cells in these aggregates (Extended Data Fig. 6g–i), where TH2 cells are also abundantly present (Extended Data Fig. 6h,j), suggesting that such structures might promote the crosstalk between these two pathways to promote disease pathogenesis. Statistical analyses using CellPhoneDB30 did not predict strong interactions between GZMK+ CD8+ T cells and TH2 cells (Supplementary Fig. 8a). However, T1 cells highly expressed the chemokine CCL5 and CCL4 (Supplementary Fig. 8b), which might help draw type 2 immune cells expressing CCR1, CCR3 or CCR5. Consistent with this notion, our analyses predicted that GZMK+ CD8+ T cells interact extensively with pro-inflammatory myeloid cells, such as TGM2+ macrophages and ALOX15+ cDC2 (ref. 10) (Supplementary Fig. 8c–f), as these cells expressed matching pairs of chemokine–chemokine receptors (Supplementary Fig. 8g). In addition, the C3a locally produced by GZMK could serve as a chemoattractant to type 2 effector cells expressing C3AR, including eosinophils and mast cells (Supplementary Fig. 8h), which will in turn secrete cytokines and inflammatory mediators to promote tissue damage and type 2 immunity31.
Disease-promoting effects of T cell-derived GZMK
To directly test whether GZMK from CD8+ T cells promotes the airway inflammatory disease, we established a mouse asthma model using ovalbumin (OVA) as antigen. C57BL/6 mice were first immunized with OVA emulsified in 50% alum and lipopolysaccharide (LPS), and then challenged by inhalation of aerosolized OVA 1 week later. OVA inhalation was repeated twice to mimic disease recurrence, and fluorescence-activated cell sorting (FACS) analyses of bronchoalveolar lavage fluid (BALF) cells were performed 2 days after each challenge (Extended Data Fig. 7a,b). Diverse immune cell populations were recruited to the airway on OVA challenge, including eosinophils, neutrophils, CD4+ T cells, CD8+ T cells and dendritic cells (Extended Data Fig. 7c). Gzmk expression was largely restricted to CD8+ T cells but almost undetectable in CD4+ T cells, and was progressively increased with repeated OVA challenges (Extended Data Fig. 7d). By scRNA-seq analyses we confirmed Gzmk expression in a subset of BALF CD8+ T cells, and some of these Gzmk-expressing T cells also upregulated Itgae (Extended Data Fig. 7e,f), very similar to human NP tissue T1 cells.
Next, we took a loss-of-function approach to examine the development of airway inflammation in CD4-cre;Gzmkfl/fl mice, which lack Gzmk in T cells (Extended Data Fig. 8a). Compared to the CD4-cre;Gzmk+/+ control, CD4-cre;Gzmkfl/fl animals harboured significantly decreased numbers of eosinophils, neutrophils, CD4+ T cells, dendritic cells and macrophages in BALF (Extended Data Fig. 8b). Lung function of these latter mice was improved, showing better tolerance towards induced airway constriction and reduced goblet cell hyperplasia (Extended Data Fig. 8c–e). Therefore, T cell-derived GZMK promotes inflammation in the airway.
To more directly evaluate the role of CD8+ T cell-derived GZMK, we took two complimentary approaches. In the first set of experiments, we transferred Gzmk+/+ CD4+ T cells together with Gzmk+/+ or Gzmk−/− CD8+ T cells into Tcrb−/−Tcrd−/− recipient mice that lack T cells (Extended Data Fig. 8f). Following asthma induction, those mice reconstituted with Gzmk−/− CD8+ T cells had significantly lower numbers of eosinophils in the BALF (Extended Data Fig. 8g). In the second set of experiments, we adoptively transferred CD45.1+ OVA-specific OT-I cells overexpressing GZMK, its inactive GZMK-S213A mutant or GZMB into congenic CD45.2 hosts and acutely induced asthma with OVA (Extended Data Fig. 9a,c). After a single dose of aerosolized OVA challenge, more eosinophil infiltration into the airway was found in recipients of GZMK-overexpressing OT-I cells, whereas OT-I cells overexpressing the inactive GZMK-S213A or GZMB failed to make a difference (Extended Data Fig. 9b,d). The expansion or accumulation of OT-I T cells were not affected by the overexpression of GZMB or GZMK (Extended Data Fig. 9b,d), indicating that activation and memory formation of these cells were not grossly altered. When adoptively transferred into C3−/− mice, OT-I cells overexpressing GZMK could no longer promote eosinophil infiltration to the airway (Extended Data Fig. 9e,f), indicating that C3 is essential in the process, although immune deficiencies in these animals31 prevented us from investigation of more long-term effects. Taken together, our results indicate that CD8+ T cell-derived GZMK promotes eosinophil infiltration to the airway by activating C3.
Alleviation of airway inflammation by targeting GZMK
To examine whether targeting GZMK may provide therapeutic benefits, we bred the Gzmkfl/fl strain with the Rosa26-CreERT2 strain, which enables tamoxifen-induced Gzmk ablation. As shown in Fig. 5a, Rosa26-CreERT2;Gzmkfl/fl mice were treated with tamoxifen after the priming immunization with OVA but before the first inhalation challenge (group B) or after the first but before the second inhalation challenge (group C). Control Rosa26-creERT2;Gzmk+/+ animals were also treated with tamoxifen (group A). Without tamoxifen treatment, Rosa26-creERT2;Gzmkfl/fl and Rosa26-creERT2;Gzmk+/+ mice developed comparable airway inflammation (Supplementary Fig. 9). When Gzmk deletion was induced before any inhalation challenge, significantly fewer eosinophils, neutrophils, CD4+ T cells and dendritic cells were found in the BALF (Fig. 5b, comparing group B to A). When Gzmk deletion was induced after the first inhalation challenge but before a new episode of asthmatic attack, significantly fewer eosinophils and dendritic cells were found in BALF cells (Fig. 5b, comparing group C to A). To determine whether Gzmk deletion would lead to restoration of lung function, we measured forced expiration volumes in these three groups of mice on methacholine challenge. Compared to group A, both group B and group C mice had much improved lung functions (Fig. 5c) and reduced goblet cell hyperplasia (Fig. 5d,e).
a, Experimental setup. b, Numbers of indicated immune cells in the BALF. Each symbol indicates one mouse, and lines denote means. DC, dendritic cell. c, Lung function and airway hypersensitivity in response to increasing doses of methacholine, measured by the forced oscillation technique (see Methods for more details). Each symbol indicates one mouse and bars denote means. d,e, Lung histology. d, Representative AB-PAS staining of lung tissue sections. e, PAS+ areas in the airway (see Methods for more details). Each symbol indicates one mouse, and lines denote means. Data were pooled from three independent experiments, with at least two animals included in each group. P values by two-sided unpaired t-tests. Scale bars, 100 μm. i.p., intraperitoneal; i.h., inhalation; DPI, days postimmunization.
Furthermore, we also tested whether pharmacological inhibition of GZMK could alleviate mouse asthma. Following asthma induction, treatment of these mice every 2 days with a synthetic drug that potently inhibits both human and mouse GZMK activity, d-Phe-Pro-Arg-chloromethylketone (PPACK)32,33, but not the GZMB inhibitor carbobenzoxy-Ile-Glu-Thr-Asp-fluoromethylketone (Z-IETD-FMK)34 significantly decreased eosinophil infiltration to the airway (Extended Data Fig. 10a–c). PPACK also significantly reduced goblet cell hyperplasia and improved lung function in mouse asthma (Extended Data Fig. 10d–f). Taken together, our data support that GZMK produced by CD8+ T cells is an important driver of airway inflammation and disease progression and that GZMK removal or inhibition could be a useful therapeutic strategy.
Discussion
In this study, we identified a subset of tissue-engrafted, TEM-like CD8+ T cells that promote airway inflammation through its characteristic effector molecule GZMK. GZMK-expressing CD8+ T cells have been found in tumours35,36 and disease tissues of many autoimmune disorders such as rheumatoid arthritis and lupus nephritis18,19,36,37,38,39. However, functional importance of these cells in disease pathology has not been defined. By both loss- and gain-of-function approaches in mouse asthma models, our study demonstrates a pivotal role for GZMK-expressing CD8+ T cells in promoting airway inflammation.
CD8+ T cells are known to play disease-promoting or regulatory roles in inflammatory disorders, primarily by GZMB-mediated killing of target cells. For example, GZMB-expressing CD8+ T cells contribute to pathogenesis of type I diabetes by killing beta cells40,41,42. On the other hand, a subset of GZMB-expressing KIR+ CD8+ T cells suppress autoimmunity by eliminating autoreactive CD4+ T cells43. GZMK is not known as a major cytotoxic granzyme in CD8+ T cell-mediated killing. Our work defines GZMK as a new complement cascade-activating protease, a finding that is also supported by a recent preprint44. Previous research has identified the role of complement in T cell homeostatic survival45 and costimulation46. Yet our work shows a previously unappreciated T cell effector molecule in activating the complement system, which would help coordinate the activation of adaptive and innate immunity. In addition, GZMK is probably also used by other immune cells, such as innate lymphoid cells, as a general mechanism to promote complement activation and inflammatory responses.
GZMK not only cleaves C3 into C3a and C3b, but also targets both C2 and C4 to promote the generation of the classical C3 convertase C4b2a. In these ways, GZMK helps generate the pro-inflammatory C3a factor, a potent chemoattractant to C3AR-expressing immune cells31. In animal models, we show that GZMK contributes to early airway recruitment of eosinophils in a C3-dependent manner. Eosinophils are important pathogenic effectors that could subsequently promote inflammation, tissue damage and remodelling by the release of cytokines and cationic proteins47. In addition, C3a and C3b can signal in different immune cells, including subsets of myeloid cells and fibroblasts, to induce inflammatory responses48. By cleaving C2, C4 and C5, GZMK could also promote the formation of the terminal membrane attack complex, leading to further tissue damage and inflammation48. On the other hand, it is important to note that other GZMK substrates including DMBT1, a negative regulator for mouse rhinitis24, would probably also contribute to the overall disease-exacerbating effect of GZMK.
The separation between GZMB-expressing T0 and GZMK-expressing T1 cells in the NP tissue seems to mirror the bifurcation of type 1 and type 2 helper cells, which is based on expression of distinct cytokines. These two subsets have minimal clonal overlap but are connected to different blood CD8+ subsets in the patient cohort we analysed. We speculate a lineage choice between GZMB and GZMK effector pathways might be made early during the response. Recent studies suggested that molecular mimicry might underly aberrant T cell activation and autoimmunity in inflammatory diseases such as multiple sclerosis and ankylosing spondylitis49,50. T cells recognizing both foreign- and self-peptides would form immune memory as a result of infection, and become autoreactive later in life. T1 cells include those predicted to recognize allergens, viral and self-proteins by GLIPH2, offering a potential explanation for their clonal expansion in the NP tissue.
Our observation that the GZMK-expressing T cells tend to aggregate within tertiary lymphoid structures where TH2 cells are hints at the possibility that these two pathways might collaborate to promote the disease. Eosinophils and mast cells recruited by GZMK-generated C3a are sources of IL-4 and IL-13 (ref. 47), which promote type 2 immune cell differentiation. In addition, C3a and chemokines secreted by GZMK+ T cells might collectively contribute to the recruitment of pro-inflammatory myeloid cells, which in turn fuels the activation of type 2 effectors. On the other hand, we also found high GZMK levels in a subset of patients with svNP without tissue eosinophilia and type 2 signatures, including patient NP12 who we analysed in-depth, suggesting that GZMK might also promote airway inflammation independently in these patients. This notion is further supported by our findings that tissue GZMK better predicts disease severity than established type 2 signatures, and suggest that treatments targeting GZMK might provide additional benefits for patients with non-type 2 diseases.
The fact that GZMK-expressing CD8+ T cells can circulate, as documented here and elsewhere18,19, would explain why NP resection may not be curative and suggest that targeted deletion of these cells could reduce disease recurrence. In fact, by analysing a unique set of matching samples collected from patients with recurring diseases, our study documents the persistence and re-engraftment of GZMK-expressing CD8+ clones in disease tissues, providing strong evidence for adaptive immune memory in driving the recurrence of inflammatory diseases in human. Results from genetic ablation and pharmacological inhibition studies in animal models highlight the continuing operation of GZMK-dependent pathways in disease progression and provide a rationale for developing GZMK-targeted therapies in controlling inflammatory diseases.
Methods
Patient recruitment
A total of 206 participants undergoing nasal surgery were recruited between 2011 and 2024 from the Department of Rhinology and Allergy, Beijing TongRen Hospital (Extended Data Table 2). Among them, 172 were diagnosed with NP and 34 diagnosed with septal deviation (considered to be HCs in the current study). Patients who had immunodeficiency, pregnancy, chronic rhinosinusitis without NPs, allergic fungal sinusitis, cystic fibrosis or had taken oral corticosteroids 4 weeks before the surgery were excluded from the study. See Extended Data Table 1 for patient details. The study was approved by the Medical Ethics Committee of Beijing TongRen Hospital (TREC2009-27, TRECKY2019-027 and TREC2022-KY127). All participants provided written informed consent.
The diagnosis of NP was based on the European Position Paper on Rhinosinusitis and Nasal Polyps (EPOS) 2007 guidelines51. The comorbid asthma diagnosis was confirmed by pulmonologists according to the clinical presentation and results from the pulmonary function tests, following the Global Strategy for Asthma 2008 guidelines52. For research purposes, patients with NP were further grouped on the basis of their disease severity, with those placed in the svNP group having disease recurrence or comorbid asthma up to 1 year after surgery, or both.
Patient tissue processing
Polyps tissues from patients with NP and uncinate tissues from patients with septal deviation were collected during endoscopic sinus surgery. One part of the tissue was immediately digested into single-cell suspension for scRNA-seq and flow cytometry analyses. Another part of the tissue was either fixed in the 4% paraformaldehyde for histology or snap-frozen in liquid nitrogen for long-term storage at −80 °C for subsequent enzyme-linked immunosorbent assay (ELISA) and bulk sequencing analyses.
Preparation of single-cell suspension from human nasal tissues
Fresh human nasal tissues were first sliced into small pieces in supplemented Roswell Park Memorial Institute (RPMI) medium (Invitrogen) containing 2% FBS (Invitrogen), 2 mM l-Glutamine (Invitrogen), 50 IU ml−1 penicillin and 50 μg ml−1 streptomycin (Invitrogen). The nasal tissues were then collected by centrifugation and fragmented by the gentleMACS dissociator (Miltenyi Biotec) according to the manufacturer’s instructions. Next, nasal tissues were again collected by centrifugation and further digested with 2 mg ml−1 collagenase (Worthington) and 0.04 mg ml−1 DNases I (Roche) in supplemented RPMI medium for 40 min at 37 °C. The released cells were resuspended and filtered through a 70 μm cell strainer (BD Bioscience). Red blood cells were lysed by using Versalyse (Beckman Coulter).
PBMC isolation
Whole blood was collected during regular follow-up visits, and peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation using the Biosci human lymphocyte separation medium (Dakewe) following the manufacturer’s instructions.
T cell stimulation
For degranulation assays, lymphocytes were first enriched from human nasal cells by density centrifugation. Briefly, cells were resuspended in 40% Percoll (GE), layered on top of 70% Percoll and centrifuged at 1,500g for 15 min. Subsequently, the cells were stimulated by either plate-bound anti-CD3 (9 μg ml−1, clone OKT3) and anti-CD28 (9 μg ml−1, clone CD28.2) antibodies or PMA (50 ng ml−1) and ionomycin (1 μg ml−1) for 4 h in the presence of the FITC-anti-LAMP1 antibodies (clone H4A3, BioLegend), and monensin (5 μΜ, BD) was added after 1 h.
Mice
C57BL/6J (Jax 664), Rosa26-creERT2 (Jax 8463), CD45.1 (Jax 2014), OT-I (Jax 3831), Cd4-cre (Jax 22071) and Tcrb−/−Tcrd−/− (Jax 2122) mice were originally from the Jackson Laboratory. Gzmkfl/fl mice and Gzmk−/− mice (constructed on C57BL/6J background) were from GemPharmatech and Cyagen, respectively. All animals were maintained in specific pathogen-free facilities at Tsinghua University (THU), with filtered air, sterile pellet food, an acidified watering system and a 12-h light/12-h dark cycle. The temperature was kept at 22–26 °C and humidity at 40–70%. Experiments were performed according to the governmental and institutional guidelines for animal welfare and approved by the Institutional Animal Care and Use Committee at THU.
For each experiment, at least two animals were included in each group, and data were pooled from 2–4 independent experiments. Sex- and age-matched animals of the indicated genotypes were randomly assigned to different groups in each experiment. The mouse lung function assays were run by researchers blinded to group allocations. For other analyses that were based on subjective instrumental measurements, blinding was not performed.
Mouse airway inflammatory disease model construction
Mouse airway inflammatory disease models were built by two approaches. In the first approach, mice were first primed by intraperitoneal immunization with 50 μg of OVA (Sigma-Aldrich) emulsified in 50% alum (Invitrogen) with 1 μg of LPS (Sigma-Aldrich). The mice were then challenged by 5% aerosolized OVA (w/v) in a whole-body exposure chamber (Yiyankeji) for 30 min per day for three consecutive days starting from 1 week postimmunization. The process was repeated one or twice at a 7 day interval, and analyses were performed at the time indicated. For the induction of Gzmk deletion, the Rosa26-creERT2 Gzmk+/+ and Gzmkfl/fl mice were gavaged daily with 2 mg of tamoxifen (ApexBio) dissolved in 200 μl of sunflower seed oil (JSENB) for five consecutive days at the indicated time. In some experiments, Tcrb−/−Tcrd−/− mice were used for asthma induction. These mice were adoptively transferred with 6 × 106 splenic CD4+ T cells and 3 × 106 splenic CD8+ T cells from the Gzmk+/+ or Gzmk−/− mice, purified by magnetic beads (Miltenyi Biotec), 5 days before the intraperitoneal OVA immunization. PPACK (MedChemExpress) and Z-IETD-FMK (Selleck) was prepared in PBS containing 3% DMSO at a concentration of 312.5 μg ml−1 for the inhibition of GZMK and GZMB activity, respectively. Each mouse was injected intraperitoneally with 62.5 μg inhibitor every 2 days from 8 to 18 days postimmunization as indicated.
Alternatively, airway inflammation was in induced in animals adoptively transferred with OT-I T cells. Briefly, B6 mice were primed by intraperitoneal immunization of OVA as described above. Then 1 × 106 OT-I cells were transferred into the primed mice 6 days postimmunization, which were boosted with OVA intraperitoneally the following day. The mice were challenge with OVA inhalation for three consecutive days 1 week later, and the analyses were performed at the indicated time. For experiments involving C3−/− and matched control animals, 0.5 μg of LPS were used in the first OVA immunization and 2 × 106 OT-I cells were transferred.
Cell culture and retroviral transduction
Splenic CD8+ T cells were isolated from OT-I mice by CD8a Microbeads (Miltenyi), and expanded in vitro by plate-bound anti-CD3 and anti-CD28 antibodies (bioXcell) in complete RPMI medium (Invitrogen) supplemented with 10% FBS, 2 mM l-glutamine, 50 IU ml−1 penicillin, 50 μg ml−1 streptomycin and 10 ng ml−1 recombinant IL-2 (Peprotech). Subsequently, these OT-I cells were transduced with retrovirus encoding only green fluorescent protein (GFP), GFP with either Gzmk or GzmkS213A or GFP with Gzmb. GFP+ OT-I cells were sorted on a AriaIII sorter (BD) and cultured in vitro for two more days before being adoptively transferred into recipient mice.
Bronchoalveolar fluid collection
The mice were anaesthetized by intraperitoneal injection of 1.2% avertin (Sigma-Aldrich) at a dose of 300 mg kg−1. The tracheas were cannulated and lavaged three times, each time with 800 μl of cold PBS. The collected BALF was centrifuged at 300g for 3 min, and the cell pellets were subjected to FACS analyses.
Flow cytometry
Human cells isolated from nasal tissues and peripheral blood were washed once with PBS, blocked by the human TruStain blocking reagents (BioLegend) and stained with different surface antibodies in FACS buffer (PBS with 1% FBS and 5 mM EDTA) for 30 min on ice. For intracellular staining, the cells were fixed and permeabilized by the CytoFix/Perm kit (BD), and stained with antibodies against intracellular antigens for 30 min on ice. Staining reagents include zombie yellow, PerCP-Cy5.5-anti-CD45 (clone HI30), PE-anti-CD8 (clone HIT8a), BV711-anti-CD8 (clone HIT8a), APC-Cy7-anti-NCAM (clone HCD56), PE-cy7-anti-KLRG1 (clone SA231A2); PE-Cy5-anti-KLRC1 (clone S19004C), FITC-anti-ITGAE (clone Ber-ACT8), PE-Dazzle594-anti-ITGAE (clone Ber-ACT8), BV421-anti-CD27 (clone O323), PE-anti-GZMK (clone GM26E7) and APC-anti-GZMB (clone GB11) from BioLegend, APC-H7-anti-CD19 (clone SJ25C1) from BD Biosciences. For mouse models, BALF cells were first blocked with 10 μg ml−1 2.4G2 antibodies (BioXCell), and stained with different antibodies in FACS buffer on ice for 30 min. Staining reagents include AF700-anti-CD11c (clone HL3, BD), eFluor660-anti-CD3 (clone 17A2, eBioscience), APC-anti-I-Ab (clone AF6-120.1, BioLegend), FITC-anti-Ly6G (clone IA8, BD), PE-Cy7-anti-CD11b (clone M1/70, BD), PE-anti-Siglec-F (clone E50-2440, BD), APC-Cy7-anti-CD8 (clone 53-6.7, BioLegend), Percp-Cy5.5-anti-CD4 (clone RM4-5, BD) and zombie yellow (BioLegend). Data were acquired on the Aurora full-spectrum cytometer (Cytek). All FACS data were analysed with the FlowJo software (Treestar).
For single-cell sequencing analyses, 10,000-20,000 cells were sorted into 5 μl of PBS containing 1% FBS in microtubes using an Aria xsIII sorter. For human NP tissue cells, CD45+ cells were sorted as indicated in Extended Data Fig. 3a. Human peripheral CD8+ T cells were first enriched from total PBMCs, and sorted as shown in Extended Data Fig. 3d. Briefly, PBMCs were incubated with anti-human CD8a antibody (clone OKT8, Invitrogen) on ice for 30 min, and antibody-bound cells were isolated by using the streptavidin-coupled microbeads (Miltenyi) following the manufacturer’s instructions. For the analyses of mouse BALF CD8+ T cells, CD45+ cells were sorted with the exclusion of eosinophils and neutrophils as indicated in Extended Data Fig. 5b.
Immunohistochemistry analyses
Nasal tissues were fixed in 4% formaldehyde for 4 h, dehydrated in 30% sucrose at 4 °C overnight and frozen in the Tissue-Tek O.C.T. Compound (Sakura Finetek). Cryo-sections of 8 µm were prepared using a microtome (Leica). The sections were rehydrated in staining buffer (0.05% Tween 20 in 0.1 M Tris-HCl), and blocked with the immunofluorescence blocking buffer (CST) for 30 min. After staining with different antibodies in staining buffer at 4 °C overnight, the sections were incubated with 4′-6-diamidino-2-phenylindole dihydrochloride (DAPI, 1 μg ml−1, Sigma-Aldrich) for 10 min as indicated, and mounted with the fluoromount aqueous mounting medium (Sigma-Aldrich). Images of intact tissue sections were acquired using the Zeiss Axio Scan Z1 slide scanner (Zeiss); alternatively, zoom images of the GZMK-expressing T cell aggregates were acquired using the LSM 710 confocal microscope (Zeiss). Staining reagents include PE-anti-GZMK (clone GM26E7, BioLegend), APC-anti-CD8 (clone HIT8a, BD), FITC-anti-KLRG1 (clone 2F1/KLRG1, BioLegend), APC-anti-CD4 (clone OKT4, BioLegend), FITC-anti-PTGDR2 (clone BM16, BioLegend) and FITC-anti-complement C3b/iC3b (3E7/C3b, BioLegend).
Areas of GZMK-expressing T cell aggregation from whole tissue scanned images were quantified independently by investigators blinded to the patient information using ZEN (Zeiss). The numbers of different cell subsets were quantified from confocal images by Imaris (Bitplane) on the basis of expression of different marker genes.
Histologic evaluation of NP immune cell subsets
Fixed NP tissues were embedded in paraffin and stained with haematoxylin and eosin. The absolute numbers of eosinophils were quantified in three randomly selected high-power magnification fields (×400) by two pathologists.
Bulk TCR sequencing
Total RNAs were extracted from the nasal tissues by using the HiPure Total RNA Mini Kit (Magen). Reverse transcription of 10–500 ng input RNA was performed following the standard Smart-seq2 protocol53, with the following modifications: The polyT reverse transcription primer was replaced by the T30VN oligo (5′ TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTVN-3′) and the original template switch oligo (TSO) was replaced by a 26N unique molecular identifier (UMI)-containing TSO (5′-CTACACGACGCTCTTCCGATCTNNNNNNNNNNNNNNNNNNNNNNNNNNTTTCTTATATrGrGrG-3′, rG represents riboguanosines). TSO was removed by addition of 1 μl of ExoI (NEB) to each reverse transcription reaction and incubation at 37 °C for 15 min, which was subsequently inactivated by incubation at 85 °C for 5 min. TCR enrichment was done in two steps. The reverse transcription product was first mixed with 25 μl of 2× KAPA HiFi HotStart ReadyMix (KAPA Biosystems), 10 μM B/T enrich F1 primer (5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTC-3′; Sangon Biotech), 50 μM of each TCR outer primers (TCRα, 5′-TGAAGGCGTTTGCACATGCA-3′; TCRβ, 5′-TCAGGCAGTATCTGGAGTCATTGAG-3′; Sangon Biotech) to a total volume of 50 μl, and amplification was done with the following program (98 °C 45 s, 11 cycles of 98 °C 20 s, 67 °C 30 s, 72 °C 1 min; 72 °C 1 min). The PCR products was then purified by VAHTS DNA Clean Beads (Vazyme) at a 0.8 to 1 (beads to sample) ratio according to the manufacturer’s instructions. Afterwards, the second step of TCR enrichment was performed by mixing the purified products with 25 μl of 2× KAPA HiFi HotStart ReadyMix, 10 μM B/T enrich F2 primer (5′-AATGATACGGCGACCACCGA-3′; Sangon Biotec), 50 μM of each TCR inner primers (TCRα, 5′-AGTCTCTCAGCTGGTACACG-3′; TCRβ, 5′-TCTGATGGCTCAAACACAGC-3′; Sangon Biotec) to a total volume of 50 μl and amplified by the following program (98 °C 45 s, eight cycles of 98 °C 20 s, 67 °C 30 s, 72 °C 1 min; 72 °C 1 min). The products were put through a double-sided size selection by the VAHTS DNA Clean Beads, at 0.5 to 1 and 0.3 to 1 (bead to sample) ratios, respectively. Subsequently, 1 ng of the purified products was tagmented by Tn5 transposase (Vazyme) in 20 μl of TD reaction buffer (10 mM Tris-Cl pH 7.6, 5 mM MgCl2, 10% DMF) at 55 °C for 10 min. The reaction was mixed with 2 μl of 0.2% SDS and incubated at 70 °C for 20 min for transposase inactivation. Index PCR was performed by mixing the tagmentation product with 3 μl of 10 μM unique dual index primers and 25 μl of Q5 High-Fidelity 2× Master Mix (NEB), and amplified with the following program (72 °C 3 min, 98 °C 30 s, 12 cycles of 98 °C 15 s, 60 °C 30 s, 72 °C 3 min; 72 °C 5 min). The PCR products were then purified with VAHTS DNA Clean Beads at a ratio of 0.8 to 1 (bead to sample). The constructed libraries were sequenced on Novaseq 6000 System (Illumina) in a 2× 150 paired-end mode.
scRNA and scTCR sequencing
For human tissue CD45+ cells and tissue CD8+ T cell subsets, the scRNA and scTCR libraries were prepared using the Chromium Single Cell V(D)J Reagent Kits (10X Genomics). And the Chromium Next GEM Single Cell 5′ Kit v2 (10X Genomics) were used to construct libraries for human peripheral blood samples and mouse BALF cells. The library sequencing was performed on an NovaSeq 6000 System.
Laser-captured microdissection and bulk RNA sequencing analyses
RNA sequencing of the GZMK+ cell aggregated tissue areas was performed following the Geo-seq protocol54. Briefly, NP tissues were embedded in the Tissue-Tek O.C.T. compound, snap-frozen by liquid nitrogen and sectioned into 10 μm consecutive slices. The GZMK+ cell aggregated areas were first confirmed by immunohistochemistry staining with DAPI and PE-anti-GZMK antibodies, then an adjacent slice was attached to the PEN MembraneSlide (catalogue no. 11505151, Leica) and stained with crystal violet. The matched GZMK+ cell aggregated and control tissue areas were excised and added to 50 μl of guanidine thiocyanate, and incubated at 42 °C for 15 min. The RNA was then precipitated by mixing the samples with 150 μl of deionized H2O, 600 μl of anhydrous ethanol, 20 μl of 1.5 M sodium acetate (pH 6.5) and 1 μl of glycogen (20 mg ml−1). The mixtures were placed at −80 °C for 30 min and centrifuged at 12,000g for 30 min at 4 °C. After washed by 75% (v/v) ethanol, the precipitated RNAs were resuspended in H2O for library construction using the Smart-seq2 protocol53.
The sequencing data were mapped and quantified by Salmon55. Samples with mapping rates lower than 20% was excluded from the analyses. Differential expression analyses were performed by DESeq2 (ref. 56), and principal component analyses was done by using the ‘plotPCA’ function after variance-stabilizing transformation.
scRNA-seq analyses
CellRanger (v.3.0) was used to generate gene expression matrix for each cell, which was further processed by Seurat (v.3.0.2)57 for data combination, dimension reduction, clustering and gene differential expression analysis. For human tissue CD45+ cells, data from different samples were first pooled by the ‘merge’ function. Subsets containing αβ T cells and myeloid cells were identified, and batch effects were removed by the ‘IntegrateData’ function. For the analyses of human tissue CD8+ T cell subsets, data from different samples were first pooled by the ‘merge’ function, then the ‘FindTransferAnchors’, ‘TransferData’, ‘MapQuery’ and ‘IntegrateEmbeddings’ functions in Seurat v.4.3.0.1 were used for cluster prediction and projection of the query onto the reference uniform manifold approximation and projection (UMAP) structure on the basis of the total tissue αβ T cell dataset. For transcriptome association analyses in Fig. 2g, we first calculated the average gene expression in human tissue αβ T cell and blood CD8+ T cell clusters, and ranked the genes by their standard deviations across different clusters. Then the top 2,000 genes were used to form the average expression matrix, which was used to calculate the Spearman’s rank correlation coefficient. For the comparison between T1 and B2 cells described in Extended Data Figs. 5 and 6, we first pooled the T1 and blood CD8+ T cells from NP11 and NP13 by the ‘merge’ function in Seurat. The average UMI for each gene in the two subsets were then graphed and the Spearman correlation coefficient was calculated. For trajectory analyses, clones shared between tissue and blood were subsampled and used for UMAP dimensional reduction, and the pseudotime for these cells was calculated by using Monocle3 (ref. 58) (v.1.3.1). For cell–cell interaction analyses described in Supplementary Fig. 8, each indicated immune cell subset was downsampled to ≤100 cells for downstream analyses by using the ‘subset’ function in Seurat. The gene expression matrix was input into CellphoneDB30 (v.3) to calculate the receptor-ligand interactions. Network diagrams were graphed by Cytoscape59 (v.3.10.1) and bubble plots were graphed by gglot2 package.
For mouse BALF cells, we integrated data from two biological replicates with the ‘IntegrateData’ function and performed UMAP dimension reduction as well as clustering. Then the CD8+ T cells were put through another round of dimension reduction and clustering for further analyses.
TCR repertoire analyses
For the bulk TCR-seq dataset, adaptors were first removed by Cutadapt60, then the TCR sequences were assembled by TRUST4 (ref. 61), which identified the first N16 of N26 within the TSO oligos as UMI and resulted in filtered contig files containing high-quality assembled TCR contigs for each UMI. UMIs with more than one TCR contigs were excluded from the analyses. For the scTCR-seq datasets, CellRanger was used to assemble TCRs for single cells. TCR contigs were annotated using Igblast62 (v.1.16.0), and only productive TCR contigs with complete VDJ regions were used for downstream analysis. In addition, cells with many productive TCRα or TCRβ contigs were removed from the scTCR dataset. Common TCR clones between different datasets were identified by the shared CDR3 nucleic acid sequences and common V(D)J germline usage. In the case of scTCR-seq data, both α and β chains were considered. The circus plots were graphed by using the circlize package, and the rest of the graphs were visualized using ggplot2 and Seurat.
For TCR specificity prediction, 37,483 αβ TCR pairs with annotated cognate antigens were collected from VDJdb database13 and the Immune Epitope Database14 and integrated with 1,311 T1 TCRs from our own scTCR-seq dataset. The resulted TCR list was analysed using the GLIPH2 (ref. 15) web portal (http://50.255.35.37:8080). TCRs were clustered on the basis of both local and global similarity, and the clusters were filtered by the following criteria: Fisher_score ≤0.05 and vb_score ≤0.05. All clusters containing T1 cell TCRs were graphed using Cytoscape (v.3.10.1).
ELISA and Luminex assay
Nasal tissues were weighed and homogenized mechanically using the TissueLyser LT bead mill (Qiagen). RIPA buffer containing the complete protease inhibitor cocktail (Roche) was added to the homogenates (10 ml g−1). The lysates were frozen and thawed twice, centrifuged at 12,000g for 5 min at 4 °C and the supernatants were used for GZMK measurements by ELISA (Reddot Biotech) and IL-5 measurements by Luminex xMAP assay (Thermo Fisher) on a Bio-Plex 200 system (BioRad). All analyses were done following the manufacturer’s instructions.
Lung function assay
Airway responsiveness of mice was assessed using the FlexiVent system (SCIREQ) following the manufacturer’s instructions. Tracheotomy and endotracheal intubation were performed after anaesthetizing the mice with 80 mg kg−1 pentobarbital sodium (Sigma-Aldrich). Airways resistance and compliance were measured on sequential exposure to increasing doses of methacholine (Sigma-Aldrich) dissolved in sterile normal saline (0, 6, 12, 24 and 48 mg ml−1).
AB-PAS staining
The left mouse lungs were fixed in 4% formaldehyde for 4 h, dehydrated in 30% sucrose for 2 days at room temperature and frozen in Tissue-Tek O.C.T. Compound at −80 °C. The tissue blocks were cryo-sectioned into 10 μm slices, and Alcian Blue-Periodic acid Schiff (AB-PAS) staining was performed using the AB-PAS solution set (Servicebio) according to the manufacturer’s instructions. The panoramic images of a whole lung section were acquired for each animal using the Zeiss Axio Scan Z1 slide scanner, and zoom-in views were acquired by a Nikon eclipse Ts2R microscope (Nikon). PAS+ areas in each airway were quantified using ImageJ (NIH) with the same threshold and normalized to the perimeter of the airway. The mean value was calculated from the four airways with the highest normalized PAS+ area for each section, and assigned to the corresponding animal.
Quantitative PCR
RNA was extracted from sorted mouse cells by trizol and reverse transcribed into complementary DNA (cDNA) by 5× All-In-One MasterMix (abm). Quantitative PCR were performed with Blastaq 2× quantitative PCR Mix (abm) on the CFX Connect Real-Time System (BioRad). Primers used were listed as follows: Actb-F 5′-CCTAAGAGGAGGATGGTCGC, Actb-R 5′-CTCAAGTCAGTGTACAGGCCA; Gzmk-F 5′-TGTCCAACTGCTTCACCTGGG and Gzmk-R 5′-GCCACCAGAGTCACCCTTGCA. Gzmk level was normalized to the level Actb.
Expression and purification of recombinant human GZMK
Recombinant human GZMK were produced and purified as described previously63. Briefly, the cDNA for human GZMK was cloned into the pHL vector. PCR mutagenesis was performed to produce the enzymatic inactive GZMK-S214A mutant. 293-F cells (obtained from Thermo Fisher; validated by the provider; mycoplasma negative) were seeded in 293-TII medium (Sino Biological) at a concentration of 2 × 106 cells per ml one night before the transfection. Subsequently, the plasmids were mixed with linear polyethylenimine (molecular weight 40,000, Yeasen Biotechnology) at a ratio of 1:3, and transfected into 293-F cells. The supernatants were harvested after 3 days, and the granzymes containing a C-terminal His-tag were captured onto a nickel affinity column (Yeasen Biotechnology). The eluted proteins (containing an engineered enterokinase site at the N terminus) were digested by the enterokinase (Beyotime Biotechnology), and cation exchange chromatography was carried out using the Resource S column (Cytiva) on the AKTA Purifier 10 system (GE) to further purify the protein.
Protease activity assay of recombinant GZMK
The protease activity of GZMK was measured by a FRET-based assay (Extended Data Fig. 4b) using a synthetic fluorogenic peptide substrate, DABCYL-GDGRSIMTE-EDANS (Sangon Biotechnology). The reactions were performed in 20 mM HEPES, pH 7.0 at 37 °C with 0.5 µM protease and 7.5 µM peptide substrate. After 15 min, the fluorescence was monitored at 490 nm with an excitation wavelength of 340 nm using Varioskan Flash reader (Thermo Fisher) in a 96-well microplate. Trypsin (Promega) was used as a positive control, and lysozyme (Sigma-Aldrich) was used as a negative control.
GZMK pull-down assay
Purified GZMK-S214A was first biotinylated by reacting with the EZ-Link NHS-LC-Biotin (Thermo Fisher) according to the manufacturer’s instructions, and coupled to the M-280 Streptavidin Dynabeads (Thermo Fisher). To do that, 200 µl of Dynabeads were equilibrated in PBS and incubated with 20 µg biotinylated GZMK-S214A for 30 min at 4 °C. NP lysates were prepared as described except with a different lysis buffer (20 mM Tris, 150 mM NaCl, 1% Triton X-100) supplemented with protease and phosphatase inhibitors (Beyotime Biotechnology). Subsequently, 300 µl of the lysates from each sample were precleared by 200 µl of Dynabeads at 4 °C for 4 h, then the precleared lysates were incubated with the biotinylated GZMK-S214A-Dynabeads overnight at 4 °C. The Dynabeads were washed with the lysis buffer, and the associated proteins were eluted with 0.1 M Glycine (pH 2.5) and neutralized with 1 M Tris (pH 7.5).
Mass spectrometry and data analysis
Proteins pulled down by GZMK-S214A-Dynabeads or Dynabeads only were resolved by 4–20% SDS–PAGE and visualized by Coomassie brilliant blue staining. The protein bands were excised and subjected to in-gel digestion. Gel pieces were washed in 50 mM NH4HCO3 (pH 8) and destained in 50% 50 mM NH4HCO3 in 50% ACN. The proteins were then incubated with 10 mM DTT (Sigma-Aldrich) for 60 min at 60 °C followed by 20 mM IAA (Sigma-Aldrich) for 30 min in the dark at room temperature. Trypsin (Promega) was added at 1:50 ratio (w/w), and the mixture was incubated overnight at 37 °C. Finally, the samples were cleaned on a C18 cartridge (Waters Corporation) and ready for LC–MS analyses. LC–MS was carried out using an UltiMate 3000 UHPLC System (Thermo Fisher) connected to a Fusion Lumos Tribrid mass spectrometer (Thermo Fisher). Mass spectrometry data were analysed using the SEQUEST HT search engine against a UniProt Swiss-Prot database with Proteome Discoverer v.2.3 (Thermo Fisher). Among the identified GZMK-interacting proteins, 56 appeared in all samples analysed. These proteins were ranked by SEQUEST scores and the abundance ratio (samplepeptide counts/controlpeptide counts). The protein–protein interaction analysis of GZMK-interacting proteins was done with Cytoscape59 (v.3.9.1).
Edman sequencing
The phenylthiohydantoin amino acid was separated in the reversed-phase mode of high-performance liquid chromatography using the differences between the retention times of different amino acids, and the amount of UV (ultraviolet light) absorbance at specific wavelengths was detected. The samples were transferred to the polyvinyl difluoride membrane and five cycles were set. The amino acid sequences of each sample were determined from the chromatograms obtained in each cycle evaluation performed by comparing chromatograms with those in the previous and subsequent cycles and identifying the phenylthiohydantoin amino acids that had the greatest increase in abundance.
GZMK cleavage assay
The recombinant SET protein (Solarbio) and serum-purified complement C2, C3, C4 and C5 (Complement Technology) at a concentration of 120 μg ml−1 were incubated with 24 μg ml−1 recombinant GZMK in PBS at 37 °C for the indicated period of time. To test the cleavage of DMBT1, NP tissue lysates were incubated with increasing dose of recombinant GZMK at 37 °C for 1 h. The reactions were stopped by the addition of 6× SDS loading buffer (reducing, TransGen), and the samples were then boiled for 15 min and resolved by SDS–PAGE. Serum-purified C3a, C3b, C2a, C4b (Complement Technology) and recombinant GZMK were separately loaded as controls. Target proteins were either visualized by Coomassie brilliant blue staining or detected by immunoblotting with different antibodies. The antibodies used include anti-SET (clone EPR12973, Abcam), anti-GZMK (clone EPR24601-164, Abcam), anti-C2 (clone EPR17979, Abcam), anti-C3 (clone EPR19394, Abcam), anti-C3a/C3a-desArg (clone 2991, Hycultbiotech), anti-C4α (clone C-2, Santa Cruz), anti-C5a/C5a-desArg (clone C17/5, Abcam), anti-DMBT1 (clone G-4, Santa Cruz), anti-ACTB (clone C4, Santa Cruz), HRP-Goat-anti-Mouse IgG (H+L) (Beyotime Biotechnology) and HRP-Goat-anti-Rabbit IgG (H+L) (Beyotime Biotechnology). Immunoblots were developed by the Super ECL Detection Reagent (Yeasen Biotechnology).
For the comparison between GZMK and different C3 convertases, serum-purified human C3 (Complement Technology, 1.75 µM) were incubated with GZMK (1 µM) or other C3 convertases (C3bBb or C4b2a; 1 µM) for 20 min at 37 °C. C3bBb was formed by mixing C3b (Complement Technology, 2 µM), FB (Complement Technology, 1 µM) and FD (Complement Technology, 500 nM) in HBS-Mg buffer (20 mM HEPES, 140 mM NaCl, 5 mM MgCl2) for 2 min at 37 °C and the reaction was terminated by 5 mM EDTA. C4b2a was formed by incubation of C4b (Complement Technology, 1 µM), C1s (Complement Technology, 0.58 µM) and C2 (Complement Technology, 1 µM) in PBS with 0.5 mM CaCl2, 2 mM MgCI2 and 40 mM NaCl for 5 min at 37 °C, and the reaction was terminated by 2 mM EDTA. The amount of C3a shown on the Coomassie brilliant blue-stained gels were quantified by using ImageJ, and the data were normalized to the C4b2a group.
Purification of C3a and measurement of C3a activity
C3a converted by different C3 convertases were purified by size-exclusion chromatography (SEC). Specifically, the cleavage products were separated by an SEC column (Acclaim SEC-1000, 4.6 × 300 mm, Thermo Fisher) on a Vanquish HPLC system (Thermo Fisher) with the UV detector set to detect absorbance at 280 nm. PBS buffer (pH 7.4) was used as the mobile phase at a 0.25 ml min−1 flow rate for elution. SEC fractions were collected automatically and further concentrated by the Amicon ultra centrifugal unit (3 kDa molecular weight cutoff, Millipore). The purity of C3a was confirmed by SDS–PAGE (4–20%) and Coomassie brilliant blue staining.
THP1 cells (obtained from the National Infrastructure of Cell Line Resource, China; validated by the provider; mycoplasma negative) were maintained in RPMI-1640 medium (10% FBS, 1% penicillin and 1% streptomycin) and used to measure C3a activities. Briefly, the cells were serum-starved overnight, incubated with 5 nM C3a from the indicated sources at 37 °C for 5 min, and lysed with RIPA buffer containing protease inhibitors (Roche) and phosphatase inhibitors (Thermo Scientific). Immunoblotting was performed using anti-phospho-Erk1/2 (Cell Signaling Technology) and anti-Erk1/2 (clone 137F5, Cell Signaling Technology) antibodies. Data were quantified by ImageJ and normalized to the untreated control.
Haemolytic assay
Serum-purified human C5b6 (600 pM) was first incubated with recombinant GZMK or Trypsin (600 pM) at 37 °C for the indicated time. The reaction mix was then added to chicken erythrocytes (3.3 × 107 per ml, Sbjbio) suspended in the Veronal Buffered Saline (pH 7.4), and incubated at 37 °C for 5 min. Subsequently, serum-purified C7 was added (15 nM) to the erythrocytes and the mixture was incubated at 37 °C for 15 min. Finally, a mix of serum-purified C8 (10 nM) and C9 (25 nM) were add to the cells, and further incubated at 37 °C for 30 min. The remaining intact erythrocytes were removed by centrifugation at 1,000g 4 °C for 2 min, and haemolysis was determined by absorbance measurement at 405 nm.
Statistical analyses
Statistical analyses were performed by Prism v.9 (GraphPad). Mann–Whitney U-tests were used to calculate P values from the human data, and two-sided Student’s t-tests were used for mouse data analyses. Categorical variables were analysed using the Chi-square test. Receiver-operating characteristics analysis was also performed in R (v.4.3.3) with the pROC package for prediction of polyp recurrence or comorbid asthma. The multiple linear regression analyses were performed using the emmeans package in R (v.4.3.3).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw sequencing data have been deposited in the GSA (Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, http://gsa.big.ac.cn) under the accession number HRA005529 for the human data and CRA012671 for the mouse data. For GLIPH2 analyses, annotated TCR sequences were obtained from VDJdb (https://vdjdb.cdr3.net) and Immune Epitope Database (https://www.iedb.org).
Code availability
The original code for sequencing data analyses has been deposited at Zenodo (https://doi.org/10.5281/zenodo.14030622)64.
Change history
04 February 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41586-025-08712-w
References
McInnes, I. B. & Gravallese, E. M. Immune-mediated inflammatory disease therapeutics: past, present and future. Nat. Rev. Immunol. 21, 680–686 (2021).
Matos, T. R. et al. Clinically resolved psoriatic lesions contain psoriasis-specific IL-17-producing alphabeta T cell clones. J. Clin. Invest. 127, 4031–4041 (2017).
Camus, M. et al. Oligoclonal expansions of mucosal T cells in Crohn’s disease predominate in NKG2D-expressing CD4 T cells. Mucosal Immunol. 7, 325–334 (2014).
Morgan, D. M. et al. Clonally expanded, GPR15-expressing pathogenic effector TH2 cells are associated with eosinophilic esophagitis. Sci. Immunol. https://doi.org/10.1126/sciimmunol.abi5586 (2021).
Skulina, C. et al. Multiple sclerosis: brain-infiltrating CD8+ T cells persist as clonal expansions in the cerebrospinal fluid and blood. Proc. Natl Acad. Sci. USA 101, 2428–2433 (2004).
Babbe, H. et al. Clonal expansions of CD8(+) T cells dominate the T cell infiltrate in active multiple sclerosis lesions as shown by micromanipulation and single cell polymerase chain reaction. J. Exp. Med. 192, 393–404 (2000).
Jarvis, D. et al. Asthma in adults and its association with chronic rhinosinusitis: the GA2LEN survey in Europe. Allergy 67, 91–98 (2012).
Bachert, C. et al. Adult chronic rhinosinusitis. Nat. Rev. Dis. Primers 6, 86 (2020).
Schleimer, R. P. Immunopathogenesis of chronic rhinosinusitis and nasal polyposis. Annu. Rev. Pathol. 12, 331–357 (2017).
Wang, W. et al. Single-cell profiling identifies mechanisms of inflammatory heterogeneity in chronic rhinosinusitis. Nat. Immunol. 23, 1484–1494 (2022).
Ma, J. et al. Single-cell analysis pinpoints distinct populations of cytotoxic CD4(+) T cells and an IL-10(+)CD109(+) TH2 cell population in nasal polyps. Sci. Immunol. https://doi.org/10.1126/sciimmunol.abg6356 (2021).
Buggert, M., Price, D. A., Mackay, L. K. & Betts, M. R. Human circulating and tissue-resident memory CD8+ T cells. Nat. Immunol. 24, 1076–1086 (2023).
Bagaev, D. V. et al. VDJdb in 2019: database extension, new analysis infrastructure and a T-cell receptor motif compendium. Nucleic Acids Res. 48, D1057–D1062 (2020).
Vita, R. et al. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 47, D339–D343 (2019).
Huang, H., Wang, C., Rubelt, F., Scriba, T. J. & Davis, M. M. Analyzing the Mycobacterium tuberculosis immune response by T-cell receptor clustering with GLIPH2 and genome-wide antigen screening. Nat. Biotechnol. 38, 1194–1202 (2020).
Yang, F. et al. Positive rate of wheat allergens in the Chinese allergic population: a systematic review and meta-analysis. Sci. Rep. 13, 10579 (2023).
Gandhi, N. A. et al. Targeting key proximal drivers of type 2 inflammation in disease. Nat. Rev. Drug Discov. 15, 35–50 (2016).
Xu, T. et al. Single-cell profiling reveals pathogenic role and differentiation trajectory of Granzyme K+CD8+ T cells in primary Sjogren’s syndrome. JCI Insight https://doi.org/10.1172/jci.insight.167490 (2023).
Jonsson, A. H. et al. Granzyme K(+) CD8 T cells form a core population in inflamed human tissue. Sci. Transl. Med. 14, eabo0686 (2022).
Mogilenko, D. A. et al. Comprehensive profiling of an aging immune system reveals clonal GZMK(+) CD8(+) T cells as conserved hallmark of inflammaging. Immunity 54, 99–115 e112 (2021).
Bai, J. et al. Prognostic factors for polyp recurrence in chronic rhinosinusitis with nasal polyps. J. Allergy Clin. Immunol. 150, 352–361 e357 (2022).
Li, F. et al. Risk factors associated with comorbid asthma in patients with chronic rhinosinusitis with nasal polyps: a cross-sectional study. BMC Pulm. Med. 22, 338 (2022).
Bouwman, A. C., van Daalen, K. R., Crnko, S., Ten Broeke, T. & Bovenschen, N. Intracellular and extracellular roles of Granzyme K. Front. Immunol. 12, 677707 (2021).
Zhao, Y., Tao, Q., Wu, J. & Liu, H. DMBT1 has a protective effect on allergic rhinitis. Biomed. Pharmacother. 121, 109675 (2020).
Werner, U. et al. Linking complement C3 and B cells in nasal polyposis. J. Immunol. Res. 2020, 4832189 (2020).
Humbles, A. A. et al. A role for the C3a anaphylatoxin receptor in the effector phase of asthma. Nature 406, 998–1001 (2000).
Nakano, Y. et al. Elevated complement C3a in plasma from patients with severe acute asthma. J. Allergy Clin. Immunol. 112, 525–530 (2003).
Krisinger, M. J. et al. Thrombin generates previously unidentified C5 products that support the terminal complement activation pathway. Blood 120, 1717–1725 (2012).
Sato, Y., Silina, K., van den Broek, M., Hirahara, K. & Yanagita, M. The roles of tertiary lymphoid structures in chronic diseases. Nat. Rev. Nephrol. https://doi.org/10.1038/s41581-023-00706-z (2023).
Efremova, M., Vento-Tormo, M., Teichmann, S. A. & Vento-Tormo, R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 15, 1484–1506 (2020).
Zarantonello, A., Revel, M., Grunenwald, A. & Roumenina, L. T. C3-dependent effector functions of complement. Immunol. Rev. 313, 120–138 (2023).
Wilharm, E. et al. Generation of catalytically active Granzyme K from Escherichia coli inclusion bodies and identification of efficient Granzyme K inhibitors in human plasma. J. Biol. Chem. 274, 27331–27337 (1999).
Wilharm, E., Tschopp, J. & Jenne, D. E. Biological activities of Granzyme K are conserved in the mouse and account for residual Z‐Lys‐SBzl activity in Granzyme A‐deficient mice. FEBS Lett. 459, 139–142 (1999).
Thornberry, N. A. et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem. 272, 17907–17911 (1997).
Tiberti, S. et al. GZMK(high) CD8(+) T effector memory cells are associated with CD15(high) neutrophil abundance in non-metastatic colorectal tumors and predict poor clinical outcome. Nat. Commun. 13, 6752 (2022).
Wang, A. Z. et al. Glioblastoma-infiltrating CD8+ T cells are predominantly a clonally expanded GZMK+ effector population. Cancer Discov. https://doi.org/10.1158/2159-8290.CD-23-0913 (2024).
Koga, R. et al. Granzyme K- and amphiregulin-expressing cytotoxic T cells and activated extrafollicular B cells are potential drivers of IgG4-related disease. J. Allergy Clin. Immunol. 153, 1095–1112 (2024).
Arazi, A. et al. The immune cell landscape in kidneys of patients with lupus nephritis. Nat. Immunol. 20, 902–914 (2019).
Bottois, H. et al. KLRG1 and CD103 expressions define distinct intestinal tissue-resident memory CD8 T cell subsets modulated in Crohn’s disease. Front. Immunol. 11, 896 (2020).
Skowera, A. et al. CTLs are targeted to kill beta cells in patients with type 1 diabetes through recognition of a glucose-regulated preproinsulin epitope. J. Clin. Invest. 118, 3390–3402 (2008).
Pinkse, G. G. et al. Autoreactive CD8 T cells associated with beta cell destruction in type 1 diabetes. Proc. Natl Acad. Sci. USA 102, 18425–18430 (2005).
Gearty, S. V. et al. An autoimmune stem-like CD8 T cell population drives type 1 diabetes. Nature 602, 156–161 (2022).
Li, J. et al. KIR(+)CD8(+) T cells suppress pathogenic T cells and are active in autoimmune diseases and COVID-19. Science 376, eabi9591 (2022).
Donado, C. A. et al. Granzyme K drives a newly-intentified pathway of complement activation. Preprint at bioRxiv https://doi.org/10.1101/2024.05.22.595315 (2024).
Liszewski, M. K. et al. Intracellular complement activation sustains T cell homeostasis and mediates effector differentiation. Immunity 39, 1143–1157 (2013).
Arbore, G. et al. Complement receptor CD46 co-stimulates optimal human CD8(+) T cell effector function via fatty acid metabolism. Nat. Commun. 9, 4186 (2018).
Arnold, I. C. & Munitz, A. Spatial adaptation of eosinophils and their emerging roles in homeostasis, infection and disease. Nat. Rev. Immunol. https://doi.org/10.1038/s41577-024-01048-y (2024).
Mastellos, D. C., Hajishengallis, G. & Lambris, J. D. A guide to complement biology, pathology and therapeutic opportunity. Nat. Rev. Immunol. 24, 118–141 (2024).
Yang, X. et al. Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides. Nature 612, 771–777 (2022).
Wang, J. et al. HLA-DR15 molecules jointly shape an autoreactive T cell repertoire in multiple sclerosis. Cell 183, 1264–1281 e1220 (2020).
Fokkens, W., Lund, V., Mullol, J. European position paper on rhinosinusitis and nasal polyps 2007. Rhinol. Suppl. 20, 1–136 (2007).
Bateman, E. D. et al. Global strategy for asthma management and prevention: GINA executive summary. Eur. Respir. J. 31, 143–178 (2008).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Chen, J. et al. Spatial transcriptomic analysis of cryosectioned tissue samples with Geo-seq. Nat. Protoc. 12, 566–580 (2017).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 e1821 (2019).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J https://doi.org/10.14806/ej.17.1.200 (2011).
Song, L. et al. TRUST4: immune repertoire reconstruction from bulk and single-cell RNA-seq data. Nat. Methods 18, 627–630 (2021).
Ye, J., Ma, N., Madden, T. L. & Ostell, J. M. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 41, W34–W40 (2013).
Dotiwala, F. et al. A high yield and cost-efficient expression system of human granzymes in mammalian cells. J. Vis. Exp. https://doi.org/10.3791/52911 (2015).
Jizhou, L. et al. The original code for sequencing data analyses in the research paper ‘Granzyme K-expressing CD8 T cells promote recurrent airway inflammatory diseases’. Zenodo https://doi.org/10.5281/zenodo.14030622 (2024).
Acknowledgements
We thank Y. Li at the Tongren Hospital and B. Li at the China-Japan Friendship Hospital for assisting with the mouse lung function assays. We thank J. Xiao at Peking University for assisting with the haemolytic assay. This work was funded in part by the National Key R&D Program of China (Ministry of Science and Technology, grant no. 2022YFC2504100 to L.Z.), the National Science and Technology Major Project (National Health Commission of China, grant nos. 2023ZD0519900 to J.W. and 2023ZD0520300 to H.Q.), the Tsinghua University Initiative Scientific Research Program (Tsinghua University, H.Q.), the National Natural Science Foundation of China (grant nos. T2225005 and 21927802 to J.W., 32130037 to H.Q., 32422028 to X.L., and 81970851 and 82271140 to F.L.), SXMU-Tsinghua Collaborative Innovation Center for Frontier Medicine (H.Q.), the Program for Changjiang Scholars and Innovative Research Team (grant no. IRT13082 to L.Z.). H.Q. is a New Cornerstone Investigator.
Author information
Authors and Affiliations
Contributions
H.Q. and L.Z. initiated and codirected the collaboration. X.L. and H.Q. conceptualized and designed the study. S.D., X.W. and C.W. recruited patients and collected clinical samples under the guidance of L.Z. F.L., J.L. and Y.S. conducted experiments involving human samples under the supervision of H.Q. and X.L. J.L. conducted bioinformatic analyses under the supervision of J.W. and J.Y. W.M. conducted mouse experiments under the supervision of X.L. and H.Q. F.S. conducted substrate identification experiments under the supervision of J.W. and J.Y. L.Z. raised the most financial support to fund the study. X.L. and H.Q. interpreted the data and wrote the paper with inputs from all authors.
Corresponding authors
Ethics declarations
Competing interests
H.Q. is a cofounder of Emergent Biomed Solutions, Ltd.
Peer review
Peer review information
Nature thanks Claudia Kemper and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 The sc-RNAseq and Profiling of αβ T cells in nasal tissues.
a, UMAP visualization of total CD45+ cells in nasal tissues from all HCs and NP patients analyzed (related to Fig. 1b). b, Expression levels of CD3D, TRAC, TRDC, NKG7, PTGDR2, CD79A, HLA-DRA, CLEC4C, KIT and GZMK. c, Cells with fully assembled TCRα and β chains are superimposed on total CD45+ cells, visualized by UMAP. d, Dot plots showing expression of the indicated genes in innate lymphoid cells (ILCs)/T cells. The percentage and the level of expression are shown by the dot size and color intensity, respectively. e, Dot plots showing the expression of effector and memory T cell signature genes, based on which the αβ T cell subsets were annotated. The percentage and level of expression are indicated by dot size and colors, respectively. f, Statistics summarizing the relative abundance of different αβ T cell subsets in the nasal tissue from HC (n = 4) and NP (n = 7) patients. Each symbol indicates one patient, and lines denotes means. HC, healthy control; NP, nasal polyp. P values by Mann-Whitney tests.
Extended Data Fig. 2 Analyses of CD8 T cells and NK cells from NP tissue and peripheral blood.
a, Gating strategies for sorting CD8 T cell subsets from NP tissues. b-c, Validation of different NP tissue CD8 T cells subsets by scRNA-seq. b, Clustering of total sorted cells (left) or individual subsets (right). Data were matched to the reference dataset in Fig. 1d. c, Composition of each sorted subset. d, Sorting strategy for blood CD8 T cells. e, Gating strategies for NP tissue NK cells. f, Statistics summarizing the abundances of NK cells in the nasal tissue in HC (n = 8), NP (n = 20) and svNP (n = 20) patients. Each symbol indicates one patient, and lines denote means. P values by Mann-Whitney tests.
Extended Data Fig. 3 Secretion of GZMK by KLRG1+CD27+ T cells.
a-b, Representative FACS plots (a) and summary statistics (b) showing the surface expression of LAMP1 in the indicated cell subsets upon stimulation. c-d, Intracellular levels of GZMK in the LAMP1+ cells within each subset. Representative FACS plots (c) and summary statistics are shown (d). PMA, phorbol 12-myristate 13-acetate; MFI, mean fluorescence intensity. Data are representative of (a and c) or pooled from (b and d) three patients. P values by paired t-tests.
Extended Data Fig. 4 Characterization of GZMK substrates and Comparison between GZMK and other C3 convertases.
a, A representative Coomassie blue-stained gel showing the purified GZMK and GZMKS214A. b, Schematic of a fluorescence resonance energy transfer (FRET)-based protease assay for determining GZMK activity. c, Measurements of GZMK activity using the assay in (b). Lysozyme and Trypsin were included as negative and positive controls, respectively. Each symbol denotes a technical replicate (n = 3), and data are presented as mean±S.D. AU, arbitrary unit. d, Representative immunoblots showing cleavage of SET by GZMK. e, Representative immunoblots showing cleavage of tissue DMBT1 by different doses of GZMK, with β-actin as a sample processing control. Data are representative (a-e) of at least two independent experiments. f, Representative Coomassie Blue-stained gel showing the cleavage of serum-purified C3 by GZMK, C3bBb and C4b2a. g, Statistics summarizing the level of C3a converted by GZMK and C3bBb, as normalized to C4b2a. Each symbol denotes one experiment, and lines denote means. h, Representative HPLC trace (left) and commassie blue-stained gels (right) showing the isolation of C3a. Serum-purified C3 was incubated with recombinant human GZMK or C4b2a assembled from serum-purified components. mAU, milli-absorbance unit. i-j, Immunoblots (i) and summary statistics (j) showing the activation of ERK in THP1 cells by C3a from different sources. Data are representative (f, h and i) or pooled (g and j) from three independent experiments. Each symbol denotes one experiment, and lines denote means. P values by two-sided unpaired t tests. Source images for gels and blots are presented in Supplementary Fig. 1.
Extended Data Fig. 5 GZMK cleavage of additional complement components.
Representative Coomassie Blue-stained gels showing GZMK cleavage of serum-purified C2 (a), C4 (d) and C5 (g), as confirmed by Edman sequencing (b, e and h) and immunoblots (c, f and i). The incubation times are indicated. j, Hemolytic assays to measure the activity of serum-purified C5b6 with or without GZMK cleavage (See Methods for more details). Lysis of chicken erythrocytes were induced by the addition of C5b6 and an excess amount of serum-purified C7, C8 and C9. AU, arbitrary unit. Error bars indicate means± S.D. Each symbol denotes a technical replicate (n = 3), and lines denote means. Data are representative of at least two independent experiments. Source images for gels and blots are presented in Supplementary Fig. 1.
Extended Data Fig. 6 GZMK-expressing CD8 T cells localize to tertiary lymphoid structures.
a-c, IHC analyses of GZMK-expressing CD8 T cells in nasal tissues surgically removed from HCs or different groups of NP patients. a, Representative images of GZMK and CD8 staining. White squares indicate areas containing GZMK-expressing CD8 T cell aggregates; magnified display of the white-square areas provided as insets. Scale bar, 500 μm for the overall view and 50 μm for insets. b, Pie charts classifying individuals in each group into those who’s surgically removed tissues did or did not contain GZMK-expressing CD8 T cell aggregates. Numbers of individuals are indicated. c, Densities of GZMK-expressing CD8 T cell aggregate in NP and svNP patients, presented as fractional distributions. d-g, Expression profiling of tissue areas containing GZMK and cell aggregates by bulk mRNA sequencing analysis. d, Experimental setup. e, A brightfield image of one NP tissue section (Left, crystal violet-stained) and a fluorescent image of an adjacent section (Right, stained with DAPI and an anti-GZMK antibody) section, with pink and cyan circles indicating GZMK+ cell aggregates and control areas excised by laser-capture microdissection. Scale bar, 500 μm. f, Principal component analysis. Each symbol indicates one tissue area. g, Selected differentially expressed genes in the control and GZMK+ areas. h-j, IHC staining of indicated markers (h) and summary statistics of abundance of indicated cell types in control (n = 14) and GZMK+ areas (n = 22) are shown (i and j). Scale bar, 50 μm. Each symbol indicates one selected area, and lines denote means. Tissue sections from three (j) or four (i) patients were included in the analysis, and at least three control and GZMK+ areas were selected for each patient. P values by Mann-Whitney tests.
Extended Data Fig. 7 The murine asthma model.
a, Experimental setup. b, Gating strategies for different immune cell populations in BALF. c, Summary statistics showing the quantifications of BALF cell subsets. d, Expression of Gzmk in CD8 and CD4 T cells sorted from the lung tissue by quantitative RT-PCR. Data were normalized to the expression of Atcb. Data were pooled from two independent experiments, with at least 2 animals included in each group in a single experiment. P values by two-sided unpaired t tests. e-f, sc-RNA sequencing analyses of BALF CD8 T cells collected from C57BL/6 mice after the second OVA challenges as shown in a. e, UMAP visualization of 3 BALF CD8 T cell subsets. f, Violin plots showing the expression of Cd3e, Cd8a, Gzmk, Itgae. Data were combined from two biological replicates, each contains BALF cells pooled from 3 mice.
Extended Data Fig. 8 Functions of CD8 T cell-derived GZMK.
a-e, Induction of airway inflammation in Cd4-cre Gzmk+/+ and Gzmkfl/fl mice. a, Experimental design. b, Numbers of indicated immune cells. c, Lung function of the indicated mice as measured by airway hypersensitivity in response to an increasing dose of methacholine using the forced oscillation technique. Each symbol indicates one mouse, and bars denote means. d, Representative images of Alcian Blue-Periodic acid Schiff (AB-PAS) staining of lung tissue sections. Scale bar, 100 μm. e, Quantifications of PAS+ areas in the airway. f-g, CD8 T cell intrinsic role of GZMK. f, Experimental setup. g, Statistics summarizing the number of immune cells detected in BALF. i.p., intraperitoneal; i.h., inhalation; DPI, days post immunization. Each symbol indicates one mouse, and lines denote means. Data were pooled from two (d-e) or three (b, c and g) independent experiments, with at least two animals included in each group. P values by two-sided unpaired t tests.
Extended Data Fig. 9 CD8 T cell-derived GZMK promotes airway eosinophil recruitment through C3.
a-d, Induction of airway inflammation based on adoptive transfer of OT-I T cells over-expressing Gzmk, GzmkS213A (a and b) and Gzmb (c and d). a and c, Experimental setup. b and d, Numbers of indicated immune cell subsets in BALF. e-f, Asthma induction in C3-/- animals. d, Experimental schedule. e, Statistics summarizing the number of different immune cells in the BALF. Each symbol indicates one mouse, and lines denote means. Data were pooled from two (d and f) or four (b) independent experiments, with at least two animals included in each group. P values by two-sided unpaired t tests.
Extended Data Fig. 10 Effect of granzyme inhibitors.
a, Experimental schedule. PPACK or Z-IETD-FMK was administered intraperitoneally every two days during the indicated period of time. b and c, Summary statistics showing the number of different immune cells in the BALF of PPACK (b) or Z-IETD-FMK (c) treated mice. Each symbol indicates one mouse, and lines denote means. d, Airway hypersensitivity of the vehicle or PPACK treated mice measured using the forced oscillation technique. Each symbol indicates one mouse, and bars denote means. e, Representative AB-PAS staining of lung tissue sections of vehicle or PPACK treated mice. Scale bar, 100 μm. f, Statistics summarizing the PAS+ areas in the airway of indicated mice. Each symbol indicates one mouse, and lines denote means. Data were pooled from two (b, c, e and f) or four (d) independent experiments, with at least two animals included in each group. P values by two-sided unpaired t tests.
Supplementary information
Supplementary Information
This file contains Supplementary Figs. 1–9.
Supplementary Table 1
Detailed demographic and clinical information for patients analysed by bulk sequencing, single-cell sequencing, mass spectrometry and laser-capture microdissection.
Supplementary Table 2
Differentially expressed genes in T0 to T14. P values by two-sided Wilcoxon rank sum tests.
Supplementary Table 3
Differentially expressed genes in B0 to B13. P values by two-sided Wilcoxon rank sum tests.
Supplementary Table 4
A list of GZMK-interacting proteins.
Supplementary Table 5
Expression profiling of GZMK+ cell aggregates. P values by two-sided Wald tests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lan, F., Li, J., Miao, W. et al. GZMK-expressing CD8+ T cells promote recurrent airway inflammatory diseases. Nature 638, 490–498 (2025). https://doi.org/10.1038/s41586-024-08395-9
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-024-08395-9
This article is cited by
-
Spilling the T: T cells in tauopathy mechanisms, disease progression, and therapeutic horizons
Molecular Neurodegeneration Advances (2026)
-
ZNF683+ NK cells govern chemotherapy sensitivity in advanced HPSCC via reshaping immune microenvironment
Nature Communications (2026)
-
IL-4–STAT6 signaling delays protective CD8+ T cell bystander activation by antagonizing IL-18 sensing
Nature Immunology (2026)
-
A distinct monocyte transcriptional state links systemic immune dysregulation to pulmonary impairment in long COVID
Nature Immunology (2026)
-
Chronic rhinosinusitis with nasal polyps and asthma: unraveling shared inflammatory pathways and evolving therapeutic synergies
The Egyptian Journal of Bronchology (2025)