Abstract
Oncogenic PIK3CA mutations generate large clones in aging human esophagus. Here we investigate the behavior of Pik3ca mutant clones in the normal esophageal epithelium of transgenic mice. Expression of a heterozygous Pik3caH1047R mutation drives clonal expansion by tilting cell fate toward proliferation. CRISPR screening and inhibitor treatment of primary esophageal keratinocytes confirmed the PI3K–mTOR pathway increased mutant cell competitive fitness. The antidiabetic drug metformin reduced mutant cell advantage in vivo and in vitro. Conversely, metabolic conditions such as type 1 diabetes or diet-induced obesity enhanced the competitive fitness of Pik3caH1047R cells. Consistently, we found a higher density of PIK3CA gain-of-function mutations in the esophagus of individuals with high body mass index compared with those with normal weight. We conclude that the metabolic environment selectively influences the evolution of the normal epithelial mutational landscape. Clinically feasible interventions to even out signaling imbalances between wild-type and mutant cells may limit the expansion of oncogenic mutants in normal tissues.
Similar content being viewed by others
Main
The accumulation of somatic mutations is a hallmark of aging1,2,3,4,5,6,7,8,9. One of the most mutated tissues is the esophagus, which develops into a patchwork of mutant clones by middle age1. The most prevalent mutant genes are under strong positive selection, suggesting that they confer a proliferative advantage over wild-type cells10. However, little is known about the mechanisms driving mutant clone expansion. Understanding these processes may open opportunities for cancer prevention by limiting the number of transformable cells in normal tissues.
PIK3CA encodes the p110α catalytic subunit of phosphoinositide 3-kinase (PI3K) and is recurrently mutated in normal human esophagus1,4,8. PI3K is activated by insulin and other growth factors, and regulates multiple processes including cell proliferation, survival, growth and metabolism, mainly through the activation of Akt–mTOR signaling11,12. Gain-of-function PIK3CA mutations, such as PIK3CAH1047R, are recurrently found in tumors, including esophageal squamous cell carcinoma (ESCC), benign overgrowth syndromes and vascular malformations12,13,14,15.
Analysis of published data identified 57 missense PIK3CA mutant clones in 17 cm2 of histologically normal human esophageal epithelium1. Missense PIK3CA mutations had the second-highest average variant allele fraction (VAF) of 72 cancer-related genes analyzed after inactivating NOTCH1 mutations, indicating that they form large clones (Fig. 1a,b)16. PIK3CA mutant clones were significantly enriched for pathogenic and/or gain of function (Path/GoF) missense mutations (MMs) (Methods), with 65% (37/57) Path/GoF mutations observed, well above the ~2% expected under neutrality (P = 1.22 × 10−47, two-tailed binomial test)1. Clones with PIK3CA Path/GoF mutations were significantly larger compared with unknown/no effect PIK3CA mutations or synonymous mutations in all genes (Fig. 1c). In particular, PIK3CAH1047R mutations generated larger clones than other PIK3CA missense mutants and were the most prevalent of all Path/GoF mutations (19%, 7/37) (Fig. 1d,e). These findings argue that activating PIK3CA mutants, particularly PIK3CAH1047R, drive large clonal expansions in normal human esophagus.
a, Schematic representation of mutant clones in an average 1 cm2 of normal esophageal epithelium from a 48–51-year-old male donor from ref. 1. To generate the figure, a number of samples from the donor are randomly selected and the mutant clones detected are represented as circles and randomly distributed in space. b, The average VAF (top graph) and frequency (bottom graph) of missense mutations (MMs) detected more than once per gene, arranged from largest to smallest. PIK3CA highlighted in red. n = 844 samples from 9 donors. c, The distribution of PIK3CA MMs classified into pathogenic/gain of function (Path/GoF) or unknown/no effect (Unkn/NE) (Methods). VAF distribution of synonymous mutations in all genes is also shown. Medians (red) and quartiles (gray lines) are represented. Two-tailed Mann–Whitney test. n = 23, 26 and 603 mutant clones, respectively, from 9 donors. d, The frequency of MM codons in the p110α protein. Path/GoF mutations are shown in red. n = 41 mutant clones from 9 donors. e, A comparison of the VAF distribution of PIK3CAH1047R with other PIK3CA MMs classified as Path/GoF or Unkn/NE. Medians (red) and quartiles (gray lines) are represented. Two-tailed Mann–Whitney test. n = 8, 15 and 26 mutant clones, respectively, from 9 donors.
These observations led us to investigate how PIK3CAH1047R mutant progenitors colonize the normal esophagus. By combining lineage tracing and clustered regularly interspaced short palindromic repeats (CRISPR) screens, we demonstrate a role for the PI3K–mTOR and downstream pathways in determining the fitness advantage of mutant clones. We find that the antidiabetic drug metformin and metabolic conditions such as type 1 diabetes and diet-induced obesity or increased body mass index (BMI) modulate the expansion of Pik3ca mutant clones in the normal esophagus of mice and humans, respectively.
Results
Generation of inducible Pik3ca H1047R-YFP knock-in mice
Most PIK3CA mutant clones present in normal human esophagus exhibit activating PIK3CAH1047R mutations (Fig. 1d). To model these, we generated a conditional mouse strain, Pik3cafl-H1047R-T2A-YFP-NLS (Pik3caH1047R-YFP), that allows heterozygous expression of the Pik3caH1047R mutation from the endogenous locus (Supplementary Note). After recombination mediated by Cre recombinase, the wild-type exon 20 of Pik3ca is excised and replaced by the mutant exon 20 encoding Pik3caH1047R (Extended Data Fig. 1a). The mutant protein is co-expressed with a nuclear localized yellow fluorescent protein (YFP) reporter linked to the C-terminus of the Pik3caH1047R protein by a T2A self-cleaving peptide17. Pik3caH1047R cells and their progeny express YFP, allowing lineage tracing of mutant clones (Methods and Extended Data Fig. 1b). Following T2A cleavage, a peptide remains at the C-terminus of Pik3caH1047R protein. The C-terminally extended p110αH1047R protein still activated the PI3K–Akt pathway in vitro and in vivo (Extended Data Fig. 1c–f).
Pik3ca H1047R mutation drives esophageal tumorigenesis
Genomic alterations in the PI3K signaling axis, including activating mutations in PIK3CA, are frequent in esophageal cancer15. We therefore investigated whether heterozygous Pik3caH1047R mutations in normal mouse esophageal progenitors affected tumorigenesis. Pik3caH1047R-YFP/wt mice were crossed with the Cre recombinase line AhCreERT to generate Cre-Pik3caH1047R-YFP/wt mice. This strain allows induction of recombination in scattered progenitor cells in esophageal epithelium18. Aging experiments revealed that expression of Pik3caH1047R did not generate tumors, alter the gross appearance of the esophagus or impact mouse survival, as compared with Cre-YFP-induced controls (Extended Data Fig. 2a–c). These results are consistent with reports that heterozygous Pik3caH1047R expressed at physiological levels requires additional mutations to drive tumor formation19,20,21.
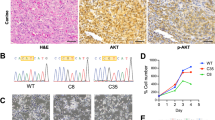
We then investigated Pik3caH1047R in two models of mutagen-induced esophageal tumorigenesis. The first consisted of administration of diethylnitrosamine (DEN), a carcinogen present in tobacco smoke22,23. Induction of Pik3caH1047R did not affect mouse survival following DEN treatment but increased the number of macroscopic tumors compared with controls (Fig. 2a–c and Extended Data Fig. 2d,e). In a second protocol, DEN administration was followed by treatment with the tumor promoter sorafenib (DEN + SOR)24. Pik3caH1047R induction did not affect tumor density but significantly increased tumor size compared with controls (Fig. 2d–f). ESCCs were only detected in Pik3caH1047R-induced mice (0.032 ESCCs per mouse from 35 DEN-treated and 0.231 ESCCs per mouse from 13 DEN–SOR-treated induced animals; no ESCCs were found in 33 and 7 DEN- and DEN–SOR-treated uninduced animals, respectively) (Extended Data Fig. 2f–i).
a, Protocol 1: Cre-Pik3caH1047R-YFP/wt mice were induced one or three times with Cre-inducing drugs (Methods) and treated with DEN for 8 weeks starting 4 weeks post-induction. Uninduced mice were used as controls. Tissues were collected before exceeding the permitted humane endpoint or at 1 year after DEN. b, Typical DEN-treated esophagus opened and flattened epithelial side up showing four tumors (yellow arrows). Scale bar, 2 mm. c, The number of macroscopic esophageal tumors in DEN-treated mice, uninduced or induced one or three times before DEN treatment. Two-tailed Mann–Whitney test versus uninduced (n = 33, 35 and 23 animals, respectively). The red lines indicate average values. d, Protocol 2. Cre-Pik3caH1047R-YFP/wt mice were treated with DEN for 8 weeks followed by Cre induction. A subgroup of animals was then treated with the tumor promoter sorafenib (SOR) for 6 weeks. Tissues were collected before exceeding the permitted humane endpoint (Methods) or 1 year post-DEN treatment. Control groups received all treatments but were uninduced. e,f, The number (e) and size (f) of macroscopic esophageal tumors. Two-tailed Mann–Whitney test (n = 33, 13, 7 and 13 mice, as they appear in the graph). The red lines indicate average values. g, The frequency of MMs for the indicated driver genes detected in human ESCCs from data collected from the TCGA and ICGC databases. Esophageal cancer (EC) driver genes were selected using the Intogen tool (https://www.intogen.org/search). Only driver genes with MM frequency >2% are shown.
These findings suggest a modest esophageal tumor-promoting role for Pik3caH1047R in mice, consistent with the observation of PIK3CA MMs in 10% of human ESCCs (Fig. 2g)15.
Pik3ca H1047R/wt mutant outcompete wild-type cells
Given the oncogenic potential of Pik3ca mutant clones, we investigated the cellular mechanism underlying their expansion by lineage tracing in Cre-Pik3caH1047R-YFP/wt mice. AhCreERTRosa26flYFP/wt (henceforth termed Cre-RYFP) mice, which express a neutral YFP reporter after induction, were used as controls25,26. At time points up to 6 months, the entire esophageal epithelium was imaged in three dimensions, and the number and location of cells in mutant and wild-type clones were recorded (Fig. 3a). Pik3caH1047R/wt mutant clones contained more cells than wild-type clones as early as 10 days post-induction, the difference increasing progressively over 6 months (Fig. 3b,c). We concluded that Pik3caH1047R/wt cells have a competitive advantage over their wild-type neighbors.
a, Protocol: Cre-RYFP and Cre-Pik3caH1047R-YFP/wt mice were induced, and wild-type and mutant clones were imaged at the time points shown. b, Total (basal + first suprabasal layer cells; left) and basal (right) cells per clone over time. Only clones with at least one basal cell were included. The dot indicates the average size of clones in each mouse. The lines and shaded areas represent the best-fitting model of clone size distributions and plausible intervals (Supplementary Note). Lines, mean ± s.e.m. Two-tailed unpaired t-test versus wild type (WT) (mice and clone numbers per time point indicated in a). c, Representative top-down confocal images of basal layer of wild-type (top) and Pik3caH1047R/wt mutant (bottom) clones at indicated time points. Clones, green; DAPI, blue. Scale bars, 20 μm. d, Heatmaps of clone size frequency; the number of basal and first suprabasal cells is shown for Cre-RYFP and Cre-Pik3caH1047R-YFP/wt animals. The black dots and dashed lines show the geometric median clone size. The graphs in the bottom row show differences between Cre-Pik3caH1047R-YFP/wt and Cre-RYFP animals for each time point. Two-tailed 2D Kolmogorov–Smirnov test. e,f, Confocal image (e) and quantifications (f) of EdU+ cells (red) in wild-type or Pik3caH1047R-/wt mutant areas (green) from the basal layer of Cre-Pik3caH1047R-YFP/wt esophagus, 3 months post-induction. EdU was injected 1 h before tissue collection. DAPI is blue. Scale bar, 20 μm. Each dot corresponds to an animal, two-tailed ratio paired t-test (52,898 cells from 3 animals, including 3,514 Pik3caH1047R/wt cells). The bars indicate mean ± s.d. g,h, The proportion of suprabasal cells (g) and clones with no basal cells (h) in wild-type (Cre-YFP) or Pik3caH1047R/wt mutant clones at the indicated time points post-induction, from data in d. Each dot corresponds to one animal; the lines connect mean values. Two tailed unpaired t-test. i, Schematic illustration of WT and Pik3caH1047R/wt cell behavior in esophageal epithelium. Model predictions for the proportions of cell division outcomes for each genotype. Pik3caH1047R/wt cells produce an excess of progenitor over differentiating cells per average cell division, driving clonal expansion even with the rate of mutant cell division being the same as WT cells.
To confirm that these results were not due to genetic differences between mutant and control mouse strains, we crossed Cre-Pik3caH1047R-YFP/wt mice with the Rosa26Confetti/wt strain27 (Extended Data Fig. 3a). This triple cross allows tracking of Pik3caH1047R/wt mutant (red fluorescent protein (RFP)+/YFP+) and wild-type clones (RFP+/YFP−) simultaneously (Methods and Extended Data Fig. 3b,c). Consistent with our previous results, Pik3caH1047R mutant clones expanded more rapidly than wild types within the same esophagus (Extended Data Fig. 3d,e).
As in humans, once the tissue had been colonized by mutant cells, it appeared normal with no change in the basal layer cell density and no gross tissue disruption (Extended Data Figs. 2c and 3f). Collectively, our results indicated that Pik3caH1047R/wt mutant progenitors have a competitive advantage over their wild-type counterparts.
Pik3ca H1047R mutation tilts cell fate toward proliferation
We next investigated the mechanisms of Pik3caH1047R mutant cell advantage over wild-type cells. One possibility is that mutant cells divide at a faster rate. To investigate this, we induced Cre-Pik3caH1047R-YFP/wt mice and aged them for 3 months. One hour before tissue collection, animals were injected with 5-ethynyl-2′-deoxyuridine (EdU), which labels S-phase cells. Mutant and wild-type cells within the same esophagus showed a similar proportion of EdU+ basal cells, arguing that the ratio of the length of S phase to total cell-cycle time, and the proportion of cycling basal cells, was not substantially altered by Pik3caH1047R expression (Fig. 3e,f). Another potential mechanism of growth advantage is by promoting apoptosis of neighboring cells28. However, there was negligible detectable apoptosis in wild-type cells, whether adjacent to or distant from mutant clones in induced Cre- Pik3caH1047R-YFP/wt animals (Extended Data Fig. 3g).
The advantage of Pik3caH1047R/wt cells may also be explained by altered progenitor cell fate16,18,29. The mouse esophagus consists of layers of keratinocytes, with progenitor cells residing in the deepest, basal cell layer. Differentiating cells exit the cell cycle and leave the basal layer, migrating toward the epithelial surface where they are shed25,26(Extended Data Fig. 3h). Each progenitor division generates either two progenitor daughters, two nondividing differentiating cells or one cell of each type (Extended Data Fig. 3i)25,26. In wild-type cells, the probabilities of these outcomes are balanced across the progenitor population so that, per average cell division, equal numbers of progenitor and differentiating cells are generated, maintaining tissue homeostasis (Extended Data Fig. 3i). Even when mutant and wild-type cell division rates are similar, mutant populations can still expand by producing more progenitor than differentiating daughter cells per average cell division18,26,29. We observed that Pik3caH1047R/wt clones contained a higher proportion of basal cells and fewer differentiated (suprabasal) cells compared with wild-type clones (Fig. 3d), suggesting that mutant progenitors generate a lower proportion of differentiating and more proliferating progeny than their wild-type equivalents. Mathematical modeling revealed that wild-type clones produced equal proportions of proliferating and differentiating cells (Supplementary Fig. 1a)25,26. However, Pik3caH1047R/wt clone dynamics could only be explained if the average division generated more proliferating than differentiating progeny (Supplementary Fig. 1b and Supplementary Note). This simple model fits both the observed basal cell and total (basal plus suprabasal) clone size distributions (Fig. 3b, Extended Data Fig. 3e and Supplementary Fig. 1c) and infers two further features of Pik3caH1047R/wt clones, a decreased proportion of suprabasal cells per clone (Supplementary Fig. 1d) and a reduction in the fraction of fully differentiated clones lacking any basal cells (Supplementary Fig. 1d and Supplementary Note). These predictions were both confirmed experimentally (Fig. 3g,h). We conclude that the increased fitness of Pik3caH1047R over wild-type cells is driven by a bias in mutant progenitor cell fate toward proliferation (Fig. 3i, Supplementary Note and Supplementary Video).
Increased PI3K pathway activity confers a fitness advantage
To further investigate mutant cell fitness we used epithelioid three-dimensional (3D) primary cultures30,31. We generated esophageal epithelioids RosaRYFP/RYFP (Pik3cawt/wt, henceforth referred to as WT-RYFP) and Pik3caH1047R-YFP/wt mice, and induced recombination by infecting these cultures with adenovirus encoding Cre recombinase (Extended Data Fig. 4a,b). Due to the much higher level of YFP expression in WT-RYFP compared with Pik3caH1047R/wt cells, the former can be identified by flow cytometry (Extended Data Fig. 4c).
We mixed WT-RYFP keratinocytes with either uninduced (Pik3cawt/wt) or induced (Pik3caH1047R/wt) cells and tracked the proportion of WT-RYFP cells over time (Fig. 4a and Extended Data Fig. 4c). We first confirmed that uninduced Pik3cawt/wt cells mixed with WT-RYFP cells competed neutrally (Fig. 4b, top and Fig. 4c,d). However, when induced Pik3caH1047R/wt and WT-RYFP cells were co-cultured, the Pik3caH1047R/wt cells almost completely took over the culture within 28 days (Fig. 4b, bottom and Fig. 4c). The suprabasal:basal cell ratio of induced Pik3caH1047R/wt cells was lower than for neighbor WT-RYFP cells (Fig. 4d), consistent with the reduced differentiation of mutant cells observed in vivo (Fig. 3g). These results indicate that Pik3caH1047R/wt mutant cells have an epithelial cell-autonomous competitive advantage over wild-type cells in vitro.
a, Protocol: Rosa26flYFP/flYFP (WT-RYFP) or Pik3caH1047R/wt (Pik3camut) cells were mixed with uninduced Pik3cawt/wt cells (Pik3cawt), from the same animal, cultured at confluence and analyzed at 14 and 28 days (Extended Data Fig. 4c). b, A typical confocal basal layer section of mixed culture. WT-RYFP cells, yellow; DAPI, blue. Scale bar, 20 μm. c, Proportion of WT-RYFP cells versus t = 0. d, The suprabasal:basal cell ratio at 14 days. +INS, treated with 5 μg ml−1 insulin. In c and d, n = 10–11 cultures from individual animals per condition. Two-tailed unpaired t-test. Mean ± s.d. e, Uninduced or induced cells were cultured overnight in starvation medium (STV) and then cultured for 1 h in STV, or STV plus LY294002 50 µM, or STV with insulin 5 µg ml−1, then lysed. Western blots for P-AKT(S473), P-AKT(T), AKT, P-GSK3β, GSK3β, P-S6, S6 and α-tubulin are shown, representative of three biological replicates. f, M, log ratio and A, mean average (MA) plots of RNA sequencing (RNA-seq) of induced Pik3caH1047R/wt and uninduced Pik3cawt/wt (WT) cultures comparing CTL and +INS treatments; red, differentially expressed transcripts (Wald test corrected for multiple testing, adjusted P < 0.05). g, Venn diagram of genes upregulated in Pik3caH1047R/wt cells also upregulated by insulin treatment of wild-type cells. h, A representative basal layer section of WT and Pik3caH1047R/wt mixed cultures after 28 days +INS or CTL. WT-RYFP cells, yellow; DAPI, blue. Scale bar, 20 μm. i, The proportion of WT-RYFP cells in mixed culture with Pik3caH1047R/wt cells, versus t = 0. +INS, treatment with 5 μg ml−1 insulin. Each dot represents a primary culture from an animal, lines connect means. n = 10–11 cultures from individual animals. Two-tailed paired t-test. j, The proportion of WT-RYFP cells mixed with Pik3caH1047R/wt cells, versus t = 0. Cells treated either in minimal medium or 0.5 µM LY294002. Each dot represents a primary culture from an animal; the lines connect means. n = 4–16 primary cultures from individual animals, per condition. Two-tailed paired t-test. k, Cell competition and PI3K activation. Increased PI3K pathway activity gives Pik3caH1047R/wt cells a competitive advantage over wild-type cells at physiological insulin levels. Leveling-up or leveling-down PI3K activity between Pik3caH1047R/wt mutant and wild-type cells with supraphysiological insulin (high INS) or PI3K inhibitor, respectively, reduces mutant competitive advantage.
We reasoned that the fitness advantage of Pik3caH1047R/wt mutants result from increased activation of the PI3K pathway compared with wild-type cells (Fig. 4e and Extended Data Fig. 1c–f). If so, increasing the PI3K activity in wild-type cells would reduce the signaling differences between the two genotypes and neutralize the mutant cell’s competitive advantage. Overactivation of PI3K signaling using supraphysiological doses of insulin (Extended Data Fig. 4d)32 abrogated the differences in differentiation and gene expression between wild-type and mutant cells (Fig. 4d–f). Indeed, 82% of the genes upregulated in Pik3caH1047R/wt mutant cells were also induced in wild-type cells upon insulin treatment (Fig. 4g). Importantly, this treatment significantly reduced the competitive advantage of Pik3caH1047R/wt mutant over Pik3cawt/wt wild-type cells in mixed cultures (Fig. 4h,i). Adding insulin within the physiological range, or another growth factor, epidermal growth factor (EGF), had no effect on mutant cell advantage (Extended Data Fig. 4e). The inhibitory effect of supraphysiological insulin on mutant cell advantage was reversible, as mutant cells were able to outcompete wild-type cells again when insulin was removed after 15 days of treatment (Extended Data Fig. 4f). Conversely, reduction in PI3K pathway activity in both wild-type and mutant cells by treating mixed cultures with the PI3K inhibitor LY294002 or the mTOR inhibitor rapamycin also reduced the mutant cell advantage (Fig. 4j and Extended Data Fig. 4g).
PI3K pathway modulates progenitor cell fitness
To confirm the role of the PI3K pathway in regulating cell competition genetically, we performed a targeted CRISPR–Cas9 competitive fitness screen in wild-type 3D mouse primary esophageal cultures (epithelioids) grown in minimal media. Cas9-expressing keratinocytes from Rosa26Cas9/wt mice were infected with a lentiviral library expressing 1,080 guide RNAs (gRNAs) including nontargeting gRNAs, gRNAs targeting essential genes and gRNAs targeting proteins related to the PI3K pathway (10 gRNAs per gene) (Fig. 5a and Supplementary Table 1). The gRNA representation at time 0 strongly correlated with that in the library (Extended Data Fig. 5a), indicating that representation was not affected by transduction or cell plating. The low infection rate ensured gene edited cells competed against wild-type neighbors over 3 weeks without passaging, after which the abundance of each gRNA was determined by DNA sequencing and compared with time 0 (Fig. 5a). As expected, there was no enrichment of nontargeting gRNAs and a strong depletion of the essential genes (Extended Data Fig. 5b,c). Targeting positive regulators of the PI3K pathway such as Igf1r, Irs2, Pik3ca, Pdpk1 and Akt1 reduced cell fitness, while the depletion of the pathway inhibitor Pten increased fitness (Fig. 5b and Extended Data Fig. 5c). Downstream of PI3K, depletion of mTOR pathway components such as Mtor, Rheb, Rptor or Rictor reduced cell fitness, while depleting mTOR pathway inhibitors such as Tsc1, Tsc2 and Tbc1d7 significantly enhanced competitiveness, confirming that the PI3K–mTOR pathway promotes esophageal progenitor cell fitness (Fig. 5b and Extended Data Fig. 5c).
a, Protocol: primary cultures from Pik3caH1047R/wt Rosa26Cas9/wt mice were induced (Pik3camut/wt) or uninduced (wild type, WT) and infected with a lentiviral gRNA library targeting PI3K–mTOR-related genes. The 0-week time point was assessed, and the remaining cells were cultured in minimal medium (CTL) or minimal medium with 5 μg ml−1 insulin (INS) for 3 weeks. The relative abundance of gRNA between the 3- and 0-week time points is expressed as log2(fold change) (LFC). The volcano plot shows the enrichment score and log2(fold change) of genes screened in WT cells in CTL medium. Significantly depleted or enriched gRNAs (false discovery rate (FDR) <0.1 and >10% fold change) are blue or orange, respectively, and unchanged gRNAs are gray. n = 2 biological replicates, 10 gRNA per gene. b, CRISPR screening results in WT (left) or Pik3caH1047R/wt (right) in CTL medium showing PI3K pathway and downstream genes including transcription factors (TF). Bold red: gRNAs that are significantly enriched or depleted (FDR <0.1 and >10% fold change difference). Pathway activation and inhibition is indicated by green and red arrows, respectively. The yellow boxes indicate the enzyme isoform most expressed in esophageal primary cells. c–e, Plots showing the correlation between average log2(fold change) of gRNA targeting indicated genes in the following cells and conditions: Pik3caH1047R/wt cells (y axis) versus WT cells (x axis) in control (CTL) condition (the panels indicate gene sets of PI3K pathway genes (top) and mTOR pathway genes (bottom)) (c); WT cells in CTL (x axis) versus INS (y axis) conditions (the panels indicate gene sets corresponding to PI3K pathway genes (left) and mTOR pathway genes (right)) (d); INS condition comparing Pik3caH1047R/wt cells (y axis) versus WT cells (x axis) (the panels indicate gene sets corresponding to PI3K pathway genes (left) and mTOR pathway genes (right)) (e). In c–e, the yellow and green areas of each graph indicate the higher absolute log2(fold change) in x-axis or y-axis condition, respectively. Linear regression (black) with slope and coefficient of determination R2. Identity line, orange. The error bars indicate the s.d. of n = 2–3 screen replicates.
The same CRISPR screen performed in Rosa26Cas9/wtPik3caH1047R/wt mutant cells in minimal medium yielded similar results to wild-type cells, with log2 fold changes of gRNAs targeting PI3K signaling after 3 weeks strongly correlated in both genotypes (Fig. 5b,c and Extended Data Fig. 5c). The log2(fold changes) of gRNAs targeting components of the mTOR pathway were less correlated (Fig. 5c), indicating that wild-type cells appear more dependent on the mTOR pathway for competitive fitness than mutant cells. Similar correlations were found in CRISPR–Cas9 screens performed in wild-type cells treated with supraphysiological doses of insulin that overactivated the PI3K pathway, confirming that the fitness of cells with higher PI3K activity is less dependent on the mTOR pathway. In agreement, differences between wild-type and mutant cells were reduced when screens were performed under insulin treatment (Fig. 5d,e). These results are consistent with Pik3caH1047R mutation promoting cell fitness by PI3K activation via mTOR-dependent and mTOR-independent pathways.
HIF1α and glycolysis contribute to mutant cell fitness
The PI3K–mTOR pathway modulates signaling and metabolism at transcriptional and posttranscriptional levels through multiple downstream effectors33. Although gRNAs targeting the Foxo transcription factors did not largely modify cell fitness, gRNAs targeting Atf4, Hif1α/Hif1β, Myc, Srebf1 or Srebf2 reduced cell fitness in both wild-type and mutant cells (Fig. 5b and Extended Data Fig. 5c), suggesting that PI3K might partially regulate cell fitness modulating gene expression through multiple transcription factors in parallel33,34. To identify the downstream pathways affected by the mutation, we compared the gene expression profiles of induced (Pik3caH1047R/wt) and uninduced (Pik3cawt/wt) primary cultures generated from the same mice. RNA sequencing revealed 301 upregulated and 195 downregulated transcripts (adjusted P value <0.05) in mutant cells (Extended Data Fig. 6a,b and Supplementary Table 2). As expected, gene set enrichment analysis showed that mutant cells have increased activation of the PI3K–mTOR pathway (Extended Data Fig. 6c,d). In addition, the expression of genes in both the HIF1 and MYC pathways33 was upregulated in mutant cells by gene set enrichment analysis (Extended Data Fig. 6e,f). Consistent with activation of HIF1 signaling, Hif1α/hypoxia signaling was one of the most enriched pathways in a Kyoto Encyclopedia of Genes and Genomes analysis of RNA sequencing data (Extended Data Fig. 6g). In total, 47% of the upregulated genes in mutant cells (transcripts with an adjusted P value <0.01) were known or predicted direct targets of the HIF1α transcription factor (Extended Data Fig. 6h). Messenger RNA of Hif1α and its canonical target gene Vegfa were upregulated in mutant cells or insulin-treated cells by RNA sequencing (Extended Data Fig. 6i).
A small increase in HIF1α protein levels was detected in mutant cells, which was dependent on PI3K activity (Extended Data Fig. 6j). Consistent with an upregulation in HIF1α signaling, we observed upregulation of glycolysis-related genes in mutant cells (Extended Data Fig. 6g,k,l). Such gene expression differences were abrogated by overactivating the PI3K pathway using supraphysiological insulin doses, which highly induce glycolysis gene expression in wild-type cells (Extended Data Fig. 6l,m). High-resolution respirometry demonstrated that such gene expression differences translated into metabolic differences, as mutant cells are significantly more glycolytic (lower OCR/ECAR ratio35) and differences are abrogated after insulin treatment (Extended Data Fig. 6n). In summary, we conclude that mutant cells show an upregulation of Hif1α signaling and a metabolic switch to aerobic glycolysis, as described in multiple PIK3CAH1047R mutant cell lines36.
To test if HIF1α pathway activation was one of the effectors of Pik3caH1047R/wt cell phenotype, we generated Pik3cawt/wt and Pik3caH1047R/wt primary cells stably expressing short hairpin RNA (shRNA) targeting Hif1a (shHif1a), or a nontargeting shRNA (shNT) (Extended Data Fig. 7a,b) and assessed their competitive fitness over WT-RYFP cells. Silencing Hif1a expression significantly reduced the advantage of Pik3caH1047R/wt mutant over wild-type cells (Extended Data Fig. 7c,d). These findings were corroborated pharmacologically, by treating mixed cultures of induced Pik3caH1047R/wt and WT-RYFP cells with the HIF1α inhibitor PX478, which reduced the advantage of mutant over wild-type cells (Extended Data Fig. 7e,f)37. We concluded that activation of HIF1 signaling partially contributes to the competitive advantage of Pik3caH1047R/wt mutant over wild-type cells, consistent with the results of the CRISPR screen.
We further investigated the metabolic changes in Pik3ca mutant cells (Extended Data Fig. 8a–k, Supplementary Fig. 2 and Supplementary Note). This led us to conclude that, as well as increased HIF1α signaling, glycolysis and lipogenesis also contribute to increased mutant cell fitness. Multiple metabolic changes are thus implicated in the mutant phenotype.
Organismal metabolism modulates Pik3ca H1047R mutant fitness
Collectively, our results argue that Pik3caH1047R/wt mutant cells have increased activation of the insulin–PI3K signaling pathway, which provides them with higher cell fitness. Therefore, conditions affecting the insulin–PI3K signaling pathway may alter the fitness of Pik3caH1047R mutant clones in vivo38. To test this hypothesis, we analyzed mutant clonal expansion in three different organismal metabolic scenarios. These were metformin treatment (a widely used antidiabetic agent that increases insulin sensitivity and mouse lifespan39,40), Ins2 mutant mice (Akita) (which model type I diabetes41,42) and a high-fat diet (HFD) model in mice (which alters PI3K signaling and promotes insulin resistance, increased body mass and hyperinsulinemia43,44,45).
Metformin treatment reduced mutant clone size and increased the proportion of differentiated mutant cells toward the levels observed in wild-type clones (Fig. 6a–c and Extended Data Fig. 9a). Metformin also significantly decreased the differences between wild-type and mutant basal cell clone size distributions (Extended Data Fig. 9b and Methods). Metformin treatment in vitro activates glycolysis in wild-type cells (Extended Data Fig. 9c) and reduced the expansion of Pik3caH1047R/wt cells in in vitro competition experiments (Extended Data Fig. 9d–f), suggesting that the in vivo effect of metformin could be partially explained by a direct reduction of the metabolic differences between wild-type and mutant esophageal cells. In summary, we conclude that metformin reduces the fitness advantage of Pik3ca mutant clones in mouse esophagus.
a, Protocol: Cre-RYFP control and Cre-Pik3caH1047R-YFP/wt mice were induced and treated with/without metformin (MET). Clones with at least one basal cell were analyzed at 28 days. b, Left and middle: heatmaps showing the frequency of basal and first suprabasal layer cells in clones; dots and dashed lines indicate geometric median clone size. Right: heatmaps showing differences between treatment and control in Cre-RYFP (top) or Cre-Pik3caH1047R-YFP/wt (bottom) animals. Two-tailed 2D Kolmogorov–Smirnov test. c, Average basal clone sizes for each strain and treatment. The bars are mean ± s.d. Two-tailed unpaired t-test. n = 431–917 clones from 5–10 animals per condition. d, Protocol: Cre-RYFP and Cre-Pik3caH1047R-YFP/wt mice in Akitawt (control) or AkitaHet (diabetic) backgrounds were induced after diabetes development in AkitaHet animals and tissues collected 28 days post-induction. e, Left and middle: heatmaps showing the frequency of basal and first suprabasal in clones from d. The dots and dashed lines indicate geometric median. Right: heatmaps showing the differences between each treatment and control in Cre-RYFP (top) or Cre-Pik3caH1047R-YFP/wt (bottom) mice. n = 518–589 clones from 4–8 animals per condition. Two-tailed 2D Kolmogorov–Smirnov test. f, Average basal clone sizes for each strain and treatment from d. The dots indicate the average clone size of a mouse. The bars indicate the s.d. (n = 4–8 mice). Two-tailed unpaired t-test. g, Protocol: Cre-RYFP and Cre-Pik3caH1047R-YFP/wt mice were induced, fed a chow or HFD, and tissues collected 28 days post-induction. h, Left and middle: heatmaps of frequency of basal and first suprabasal layer cells in clones from g. The dots and dashed lines indicate geometric median clone size. Right: heatmaps showing differences between treatment and control in Cre-RYFP (top) or Cre-Pik3caH1047R-YFP/wt (bottom) animals. n = 472–914 clones from 5–9 animals per condition. Two-tailed 2D Kolmogorov–Smirnov test. i, Average basal clone sizes for each strain and treatment from g. The dots indicate the average clone size of a mouse. The bars are s.d. (n = 5–9 mice). Two-tailed unpaired t-test. j, The effects of MET, Akita and HFD on mutant cell advantage. In a control situation, mutant clones show advantage that is reduced by MET treatment and increased in AkitaHet background or under HFD.
Next, we bred the Cre-RYFP and Cre-Pik3caH1047R-YFP/wt strains onto AkitaHet (diabetic) or Akitawt (nondiabetic) backgrounds. Diabetes development in AkitaHet mice was confirmed by measuring urinary glucose (Extended Data Fig. 9g). Clonal recombination was induced in both strains after the onset of diabetes in AkitaHet mice (Fig. 6d), and the number and size of clones were analyzed after 1 month. Type 1 diabetes did not modify the global clone size distribution or average size of wild-type clones. However, Pik3caH1047R/wt mutant clones were larger and the distribution of basal and suprabasal cells was altered in diabetic mice compared with nondiabetic littermates (Fig. 6e,f and Extended Data Fig. 9h,i). Differences in basal and suprabasal cell distributions between wild-type and mutant clones were further increased in AkitaHet mice (Extended Data Fig. 9h). We conclude that the fitness advantage of Pik3caH1047R mutant cells is increased in a diabetic background.
Finally, we examined the effects of HFD. Cre-RYFP and Cre-Pik3caH1047R-YFP/wt mice were induced and fed HFD or a control diet with matched ingredients, and the clone sizes were analyzed 1 month later (Fig. 6g). As previously reported, HFD significantly increased body weight and hyperinsulinemia, suggesting insulin resistance, compared with the control diet (Extended Data Fig. 9j,k)45,46,47. HFD caused small changes in the global clone size distributions of wild-type RYFP clones but did not alter average clone size or the proportion of suprabasal cells per clone. However, HFD substantially altered mutant global clone size distribution and average clone size, and decreased the proportion of suprabasal cells in mutant clones, in both males and females (Fig. 6h,i, Extended Data Fig. 9l,m, Supplementary Fig. 3 and Supplementary Tables 3 and 4). HFD further increased the differences between mutant and wild-type basal clone size distributions (Extended Data Fig. 9m and Methods).
Taken together, these results demonstrate that metabolic conditions affecting the insulin–PI3K axis modulate the selection of Pik3caH1047R mutant clones in the mouse esophagus (Fig. 6j). Further work will be needed to understand whether the modulation of mutant cell advantage in these conditions depends on the change in glycaemia, insulinemia or a combination of both.
PIK3CA mutant clone density is increased in overweight humans
These findings led us to explore the relationship between BMI and PIK3CA mutant clone size in human esophagus. We sequenced the PIK3CA gene in 698 samples of esophageal epithelium (covering a total of 13.96 cm2) from 10 individuals with no previous diagnosis of diabetes. We combined these data with published results shown in Fig. 1 (Supplementary Table 5)1. Donors were classified into nonoverweight (non-OW, BMI <25 kg m−2) and overweight–obese groups (OW–OB, BMI ≥25 kg m−2)48. The groups had an average age 58 and 62 years, respectively (Extended Data Fig. 10a). PIK3CA MMs were classified into Path/GoF or unknown/no evidence (Unkn/NE) (Methods). The distribution of PIK3CA mutations was similar between both groups, with PIK3CAH1047R being the most prevalent.
A multiple regression linear model was used to examine the mixed effects of mutation pathogenicity, donor age and bodyweight on the density of PIK3CA MM clones (Methods). The number of mutant clones significantly increased with donor age (analysis of variance (ANOVA) F-test, P = 0.0133). While the density of Unkn/NE PIK3CA mutations was not different between groups, Path/GoF mutations were significantly more frequent in OW–OB than in non-OW donors (Fig. 7a,b and Extended Data Fig. 10b). We estimate that, in the esophagus, Path/GoF PIK3CA mutant clones accumulate at a rate of 9.8 mutations dm−2 yr−1 in OW–OB donors (95% confidence interval (CI) 2.3–17.2), as opposed to 2.2 mutations dm−2 yr−1 in non-OW donors (95% CI −3.7 to 8.1). Consistently, the effect of pathogenic mutations on clonal density was significantly influenced by BMI (ANOVA F-test, P interaction 0.0217) (Fig. 7a and Extended Data Fig. 10b). These results suggest that Path/GoF PIK3CA MM are specifically selected in OW–OB individuals. As a result, these mutations cover a larger proportion of tissue than Unkn/NE mutations in OW–OB, while no differences are found in non-OW donors (Extended Data Fig. 10c). An increased density of pathogenic variants of the kinase domain was observed in the BMI ≥25 group (Extended Data Fig. 10d), consistent with the increased fitness of Pik3caH1047R clones in mice fed a HFD (Fig. 6g–i).
Human donors were classified into overweight, OW–OB (n = 10, BMI ≥25 kg m−2) and non-OW (n = 8, BMI <25 kg m−2). PIK3CA MMs were classified as Path/GoF and Unkn/NE (Methods) and analyzed separately in OW–OB and non-OW donors. a, The number of clones per centimeter squared carrying PIK3CA MM Path/GoF (top) or Unkn/NE (bottom) plotted against donor age. Each dot represents a donor. Simple linear regression for OW–OB and non-OW donors is shown. Tukey’s test. b, Representative patchwork plots. Each panel is a representation of PIK3CA mutant clones in an average 8 cm2 area of esophagus from non-OW (top) or OW-OB (bottom) donors. To generate each figure, a sample of biopsies from all donors >60 years of age was randomly selected from each weight category amounting to 8 cm2 of tissue. All PIK3CA mutant clones are represented as circles and randomly distributed in space.
We conclude that clones carrying pathogenic MMs in the kinase domain of PIK3CA, such as PIK3CAH1047R, have an additional competitive advantage in the esophagus of people with high BMI.
Discussion
Our results show that subtle activation of the PI3K pathway, caused by a heterozygous activating mutation in PIK3CA, is sufficient to drive clonal expansion in normal esophageal epithelium in humans and mice. The mechanism is a bias in mutant cell fate. We find genetic evidence for the PI3K–mTOR axis regulating progenitor fate. Pik3caH1047R/wt clones hijack this pathway driving clonal expansion. In vivo models of diabetes and diet-induced overweight increase mutant cell expansion, while the antidiabetic drug metformin decreases it (Fig. 8).
Top: overactivation of the PI3K–mTOR pathway increases the competitive fitness of Pik3caH1047R/wt mutant over wild-type cells, by reducing mutant cell differentiation, which drives clonal expansion. Middle: interventions that level out the differential activation of the PI3K–mTOR axis and cell fitness between wild-type and mutant cells neutralize the competitive advantage of the mutants. Bottom: on the contrary, metabolic conditions such as type 1 diabetes and overweight would further increase the cellular differences in the PI3K–mTOR axis between both cell types, resulting in a further increase in the competitive fitness of the mutant over wild-type cells.
The limitations of the study include the lack of direct evidence for signaling differences between Pik3caH1047R/wt clones and wild-type cells in vivo and showing that they are altered in the various metabolic states we studied. Changes induced by targeting oncogenic Pik3ca alleles to the endogenous allele in mice are difficult to detect with immunostaining, suggesting they are likely to be modest compared with those in Pik3ca mutant overexpression studies49,50,51. More work is also needed to characterize the relative contributions of the different metabolic changes induced by the mutant allele to cell fitness, for example, exploring the effects of multiple inhibitors and culture conditions on mutant fitness in vitro. Nevertheless, our results suggest that reducing insulin signaling in peripheral tissues via lower insulin levels or insulin sensitivity may enhance mutant fitness.
Our observations parallel studies in Drosophila where the fitness advantage gained by increased PI3K signaling can be modulated by insulin or metformin, suggesting a conserved mechanism across species52,53. However, the activity of the pathway is critical. For example, overexpression of Pik3caH1047R drives clonal expansion in the mouse intestine, while in the Drosophila wing disc PI3K overexpression has no effect54,55. In humans, heterozygous activating mutations in PIK3CA are positively selected in the skin56, indicating they provide competitive advantage; however, the biallelic expression of Pik3caH1047R/H1047R in mouse skin drives their elimination from the tissue possibly related to senescence induced by high-PI3K signaling57. These observations stress the importance of using mouse models that express heterozygous activating Pik3ca mutants from the endogenous promoter to model the corresponding mutants in human epithelia19,20.
We further demonstrate that overweight alters the selection of activating PIK3CA mutations in human esophagus. Indeed, our data argue that metabolic conditions alter the competitive selection of oncogenic mutant clones in normal tissues, which may impact cancer risk. Mutations in the insulin–PI3K pathway are common in breast and endometrial cancers, the incidence of which is positively correlated with BMI38,48. This correlation may be related to the expansion of PIK3CA mutations in overweight individuals before carcinogenesis. Metformin may suppress the expansion of this subset of oncogenic mutant cell clones in normal adult epithelia.
The concept of remodeling the mutational landscape of normal tissues described here sets the basis for studies on how metabolic conditions and treatments may affect mutant selection in human tissues and represents a new potential point of intervention in aging and cancer prevention58,59,60.
Methods
Ethical approval for animal experiments
All experiments were approved by the local ethical review committees at the Wellcome Sanger Institute and conducted according to Home Office project licenses PPL70/7543, P14FED054 and PF4639B40.
Ethical approval for human study
The study protocol was ethically reviewed and approved by the UK National Research Ethics Service Committee East of England, Cambridge South, Research Ethics Committee protocol reference 15/EE/0152 NRES. Esophageal tissue was obtained from deceased organ donors. Written Informed consent was obtained from the donor’s next of kin.
Mouse experiments
Animals were maintained on a C57/Bl6 genetic background. Mice were maintained in a specific-pathogen-free unit under a 12 h light and 12 h dark cycle with lights off at 19:30 and no twilight period. The ambient temperature was 21 ± 2 °C, and the humidity was 55 ± 10%. Mice were housed at three to five mice per cage (overall dimensions of caging: 365 mm × 207 mm × 140 mm (length × width × height), floor area, 530 cm2) in individually ventilated caging (Tecniplast, Sealsafe 1284L) receiving 60 air changes per hour. In addition to aspen bedding substrate and standard environmental enrichment of Nestlets, a cardboard tube/tunnel and wooden chew blocks were provided. Mice were given water and diet ad libitum. Animals were fed on standard chow unless otherwise specified. Experiments were carried out with male and female animals, and no sex specific differences were observed. Animals were randomly assigned to experimental groups. Pik3cafl-H1047R-T2A-EYFP-NLS/wt knock-in mice generation is described in the Supplementary Note. For lineage tracing experiments, the relevant floxed reporter lines were crossed onto the AhcreERT strain in which transcription from a transgenic CYP1A1 (arylhydrocarbon receptor, Ah) promoter is normally tightly repressed until is activated by β-napthoflavone61. In this model, tamoxifen promotes the nuclear translocation of CreERT protein to mediate recombination. For lineage tracing of control clones, the Rosa26flYFP/wt mice, which express YFP from the constitutively active Rosa26 locus, were used62. To assess the mutant and wild-type clone growth in the same mouse, Rosa26flConfetti animals were used27. Homozygous AhcreERTRosa26flConfetti animals were crossed onto Pik3cafl-H1047R-T2A-EYFP-NLS/fl-H1047R-T2A-EYFP-NLS to generate AhcreERTRosa26flConfetti-Pik3cafl-H1047R-T2A-EYFP-NLS/wt. Following induction, this strain yields cells expressing one of four colors of reporter from the Rosa26 locus (wild-type clones), just the YFP reporter of Pik3ca mutant cells (Pik3caH1047R/wt clones) or both (Pik3caH1047R/wt-Confetti Confetti clones) (Extended Data Fig. 2b). Because green fluorescent protein (GFP), YFP and cyan fluorescent protein (CFP) clones are recognized by the same anti-GFP antibody used to detect the EYFP co-expressed with the mutation, only RFP-expressing clones (wild-type or mutant) were analyzed. Where indicated, mice were treated with a well-tolerated dose of metformin (Sigma-Aldrich 317240, 2 mg ml−1) or sodium dichloroacetate (DCA; Sigma-Aldrich 347795, 0.5 mg ml−1) in drinking water containing 15% ‘Ribena’ to enhance palatability, for the duration of the experiment. In those cases, the results were compared with control mice treated with 15% ‘Ribena’ in water. For the analysis of Rosa26flYFP/wt wild-type or Pik3caH1047R/wt mutant clones in a hypoinsulinemic background, AhcreERT Rosa26flYFP/flYFP or AhcreERTPik3cafl-H1047R-T2A-EYFP-NLS/fl-H1047R-T2A-EYFP-NLS female mice were crossed with heterozygous C57BL/6-Ins2Akita/J males (AkitaHet) (The Jackson Laboratory, 003548) to produce AhcreERT- Rosa26flYFP/wt-Ins2Akita/wt and AhcreERT-Rosa26flYFP/wt-Ins2wt/wt or AhcreERT-Pik3cafl-H1047R-T2A-EYFP-NLS/wt-Ins2Akita/wt and AhcreERT-Pik3cafl-H1047R-T2A-EYFP-NLS/wt-Ins2wt/wt littermates. The development of diabetes phenotype was confirmed by analyzing the presence of glucose in urine at the start and end of the experiments (Extended Data Fig. 8g) using SURERS2/50 strips (SureScreen Diagnostics, UK) according to the manufacturer’s instructions. For the HFD experiments, mice were fed during the indicated times with irradiated rodent diet with 45 kcal% fat (D12451i, Researchdiets) or a chow diet with matched ingredients (D12450H, Researchdiets).
Lineage tracing
To induce low-frequency expression of EYFP in the mouse esophagus, 10–16-week-old transgenic mice were given a single intraperitoneal (i.p.) of 80 mg kg−1 β-naphthoflavone (MP Biomedicals 156738) and 1 mg tamoxifen (Sigma-Aldrich N3633) (AhcreERTRosa26flConfetti-Pik3cafl-H1047R-T2A-EYFP-NLS/wt) or 0.25 mg tamoxifen (other mouse strains), resulting in 2.9 ± 0.9% and 0.17 ± 0.04% (mean ± standard deviation (s.d.)) recombined epithelium by area, respectively, 10 days after induction. Following induction, between three and eight mice per time point were culled and the esophagus was collected. Time points analyzed included 10 days and 1, 3 and 6 months after induction. As expression from the endogenous Pik3ca locus is very low, immunostaining was necessary to detect EYFP-nuclear localization sequence (NLS) reporter expression (Extended Data Fig. 2b). Normalized, clone-size distributions were built for each experimental condition and time point from the observed relative frequencies fm,n of clones of a certain size, containing m basal and n suprabasal cells, resulting in two-dimensional (2D) histograms, (displayed as heatmaps using CloneSizeFreq_2Dheat package (https://github.com/gp10/CloneSizeFreq_2Dheat)). A 2D histogram of the residuals or differences observed between conditions in the relative frequencies of each particular clone size (that is, each cell on the grid) was generated when appropriate.
Chemically induced mutagenesis
To generate mutations in the esophageal epithelium, mice were treated with DEN (Sigma, catalog no. N0756) at 40 mg per 1,000 ml in sweetened drinking water for 24 h on 3 days a week (Monday, Wednesday and Friday) for 8 weeks. When indicated, mice were induced using one or three i.p. injections of 80 mg kg−1 β-naphthoflavone (MP Biomedicals 156738) and 1 mg tamoxifen (Sigma-Aldrich N3633), resulting in 2.9 ± 0.9% and 10.2 ± 2.8% (mean ± s.d.) recombined epithelium by area, respectively, 10 days after the last induction. After each dosage, mice received sweetened water until the next DEN treatment. Control mice received sweetened water as vehicle for the length of the treatment. After the 8 weeks, all mice were administered normal water.
Sorafenib (LC Laboratories S8599) treatment was performed as described24. Briefly, sorafenib was administered by i.p. injection at 10 mg ml−1 dissolved in cottonseed oil containing 5% dimethylsulfoxide. Control animals received a vehicle control (5% dimethylsulfoxide in cottonseed oil). Injections were given on alternate days for 6 weeks and starting after DEN treatment.
Animals were collected when the humane endpoint of the license was reached, that is, nontransient signs that deviate from normal health and/or reached a 20% weight loss over maximum weight.
Analysis of in vivo proliferation by EdU labeling
Three months after induction with one i.p. injection of 80 mg kg−1 β-naphthoflavone (MP Biomedicals 156738) and 1 mg tamoxifen (Sigma-Aldrich N3633), 10 μg of EdU in phospate-buffered saline (PBS) was administered by i.p. injection 1 h before culling. Tissues were collected and stained with EdU-Click-iT kit and immunofluorescence as explained below. EdU-positive basal cells were quantified from a minimum of ten z-stack images.
Human samples
A sample of mid-esophagus was removed, placed in University of Wisconsin organ preservation solution (Belzer UW Cold Storage Solution, Bridge to Life) and flash frozen in tissue freezing medium (Leica catalog no. 14020108926)1. The BMI of each donor was used to classify them into nonoverweight (BMI <25 kg m−2) or overweight–obese (BMI >25 kg m−2). None of the donors used in the study had a previous diagnosis of diabetes.
Human DNA sequencing
Data in Fig. 1 was from ref. 1. Figure 7 and Extended Data Fig. 9 human data combine the previously published donors with data from PIK3CA locus DNA sequencing from esophagus from ten additional transplant donors. Briefly, human esophagus epithelium was isolated and cut into a gridded array of 2 mm2 samples1. DNA was extracted and using QIAMP DNA microkit (Qiagen, 56304). DNA sequencing was performed as described1. For mutation calling, we used the ShearwaterML algorithm from the deepSNV package (v1.21.3, https://github.com/gerstung-lab/deepSNV)1,63. Instead of using a single-matched normal sample, the ShearwaterML algorithm uses a collection of deeply sequenced normal samples as a reference for variant calling that enables the identification of mutations at very low allele frequencies.
PIK3CA MM classified as pathogenic/gain of function (Path/GoF) were annotated either as pathogenic/likely pathogenic by the ClinVar mutation database (https://clinvarminer.genetics.utah.edu/variants-by-gene/PIK3CA) or as gain of function/predicted gain of function by the Clinical Knowledgebase (https://ckb.jax.org/gene/show?geneId=5290&tabType=GENE_VARIANTS). The remaining mutations, classified as unknown/no effect (Unkn/NE), did not appear in the datasets or were classified as uncertain significance/likely benign by Clinvar or unknown/no effect/no effect predicted by Clinical Knowledgebase. Pik3ca mutant clone density was calculated as the number of missense mutant clones classified as above and normalized by the total area of epithelium sequenced in each donor. All analyses were performed per donor and, therefore, are independent of the area of tissue sequenced in each donor.
Driver mutations in esophageal cancer
To detect the most frequent mutations found in esophageal cancer, we used the Integrative OncoGenomics pipeline (https://www.intogen.org/search) that collects and analyses somatic mutations in multiple datasets including The Cancer Genome Atlas Program (TCGA), to identify cancer driver genes using multiple driver discovery methods64. Then, we selected the frequency of MMs in the selected drivers in samples of ESCC from TCGA and International Cancer Genome Consortium (ICGC).
Primary epithelioid culture
After removing the muscle layer with fine forceps, esophageal explants were placed onto a transparent ThinCert insert (Greiner Bio-One) with the epithelium facing upward and the submucosa stretched over the insert membrane, and cultured in complete FAD medium (cFAD) (50:50) consisting of 4.5 g l−1 d-glucose, pyruvate and l-glutamine DMEM (Invitrogen 11971-025):DMEM/F12 (Invitrogen 31330-038), supplemented with 5 μg ml−1 insulin (Sigma-Aldrich I5500), 1.8 × 10−4 M adenine (Sigma-Aldrich A3159), 0.5 μg ml−1 hydrocortisone (Calbiochem 386698), 1 × 10−10 M cholera toxin (Sigma-Aldrich C8052), 10 ng ml−1 EGF (PeproTech EC Ltd 100-15), 5% fetal calf serum (PAA Laboratories A15-041), 5% penicillin–streptomycin (Sigma-Aldrich, P0781) and 5 μg ml−1 apo-transferrin (Sigma-Aldrich T2036). Explants were removed after 7 days once keratinocytes covered half of the membrane. Medium was changed every 3 days. Minimal FAD medium without cholera toxin, EGF, insulin and hydrocortisone was used from 2 weeks after establishing the culture.
Adenoviral infection
To establish either induced or uninduced mouse primary keratinocyte 3D cultures from Pik3cafl-H1047R-T2A-EYFP-NLS/wt mice and Rosa26flYFP/flYFP, cells were infected with either Cre-expressing adenovirus (Ad-CMV-iCre, Vectorbiolabs, #1045) or Null adenovirus (Ad-CMV-Null, Vectorbiolabs, #1300), respectively. Briefly, cells were incubated with adenovirus-containing medium supplemented with polybrene (Sigma-Aldrich, H9268) (4 μg ml−1) for 24 h at 37 °C, 5% CO2. Cells were washed, and fresh medium was added. Infection rates were >90%.
Generation of cultures stably expressing shRNA
MISSION lentiviral transduction particles expressing shRNA targeting Hif1a (Hif1a#1: TRCN0000232222) and lentiviral-negative control particles (SHC002V) were purchased from Sigma-Aldrich. Stably transduced cell lines were generated according to the manufacturer’s instructions. Briefly, cells were trypsinized and infected in suspension and seeded on a six-well insert. The following day, cells were infected. After 24 h, medium was changed. Cells were selected 7 days later by addition of 1 μg ml−1 puromycin (Sigma-Aldrich, P88338). Knockdown efficiency was determined by real-time PCR.
Cell competition assays
Fully induced Rosa26flYFP/flYFP cultures (WT-RYFP) were trypsinized and mixed, 1:1 for fluorescence-activated cell sorting analysis or 1:3 for microscopy, with Pik3cafl-H1047R-T2A-EYFP-NLS/wt either fully induced with Cre-expressing adenovirus (Pik3caH1047R/wt) or uninduced (Pik3cawt/wt) (infected with null adenovirus). After 1 week, when the cultures are fully confluent, cholera toxin, EGF, insulin and hydrocortisone were removed from the medium and the starting time point was collected and analyzed by flow cytometry on a BD LSRFortessa using Facs DIVA software. At the time points specified, cells were collected and analyzed by flow cytometry or fixed for microscopy. Where indicated, mixed cultures were treated with 2 ng ml−1 or 5 μg ml−1 insulin (Sigma-Aldrich I5500), 10 ng ml−1 EGF (PeproTech EC Ltd 100-15), 10 µM PX478 (Cambridge Bioscience 10005189), 0.5 μM LY294002 (Selleckchem S1105), 5 mM 2-deoxyglucose (Sigma-Aldrich D8375), 500 nM rapamycin (Sigma-Aldrich R0395), 6 µg ml−1 betulin (Merck B9757), 10 µM CB839 (Tocris 7591), 30 µM 5-tetradecyloxy-2-furoic acid (TOFA) (Santa Cruz sc-200653), 0.1 µM TVB2640 (Selleckchem S9714), 2.5 mM metformin (Sigma-Aldrich 317240) or 25 mM DCA (Sigma-Aldrich 347795).
Flow cytometry
Keratinocyte cultures were detached by incubation with 0.05% trypsin–EDTA for 20 min at 37 °C 5% CO2. Cells were pelleted for 5 min at 650g and resuspended in PBS to be immediately analyzed using a BD LSRFortessa. Were suprabasal and basal cells needed to be quantified, 2% PFA fixed cells were incubated in blocking solution (0.1% BSA and 0.5 mM EDTA in PBS) for 15 min and then with anti-ITGA6-647 antibody (1:125, BioLegend, UK 313610) in blocking solution for 45 min at room temperature. YFP fluorescence was collected using the 488 nm laser and the 530/30 bandpass filter, and ITGA6-647 fluorescence, to discriminate between basal and suprabasal cells, was collected using the 640 nm laser and the 670/14 bandpass filter. Data were analyzed using FlowJo software (version 10.5.3). Basal cells were defined as ITGA6-positive cells and suprabasal cells as ITGA6-negative cells.
Immunofluorescence and microscopy
For wholemount staining, the mouse esophagus was opened longitudinally, the muscle layer was removed and the epithelium was incubated for 1 h and 30 min in 20 mM EDTA–PBS at 37 °C. The epithelium was peeled from underlying tissue and fixed in 4% paraformaldehyde in PBS for 30 min. Wholemounts were blocked for 1 h in blocking buffer (0.5% bovine serum albumin, 0.25% fish skin gelatin, 1% Triton X-100 and 10% donkey serum) in PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA and 4 mM MgSO4·7H2O). Anti-GFP antibody (1:4,000, Life Technologies A10262) was incubated over 3–5 days using blocking buffer, followed by several washes over 24 h with 0.2% Tween-20 in PHEM buffer. Where indicated, an additional overnight incubation with anti-caspase-3 (1:500, Abcam ab2302) was performed. Finally, samples were incubated for 24 h with 1 μg ml−1 4′,6-diamidino-2-phenylindole (DAPI) and secondary antibody anti-chicken (1:2,000, Jackson ImmunoResearch 703-545-155) in blocking buffer. Clones were imaged on an SP8 Leica confocal microscope. Whole tissue or large tissue area images were obtained in most cases with the 20× objective with 1× digital zoom, optimal pinhole and line average, speed 600 Hz and a pixel size of 0.5678 µm per pixel. In YFP tissues at long time points, clones were manually detected in the microscope and individually imaged using 40× objective with 0.75× digital zoom, optimal pinhole and line average, speed 600 Hz and a pixel size of 0.3788 µm per pixel. The numbers of basal and suprabasal cells in each clone were counted manually. Representative images were produced by selecting a 120 × 120 µm area in the images obtained as previously stated. Images were processed using ImageJ software adjusting brightness and contrast and applying a Gaussian blur of 1. EdU incorporation was detected with Click-iT chemistry kit according to the manufacturer’s instructions (Invitrogen, 23227) using 555 Alexa Fluor azides. Confocal images for EdU–GFP staining were acquired on a Leica TCS SP8 confocal microscope (objective 20×; optimal pinhole and line average; speed 600 Hz; resolution 1,024 × 1,024, zoom ×2). Images were processed using ImageJ software adjusting brightness and contrast and applying a Gaussian blur of 1. For in vitro culture staining, inserts were fixed in 4% paraformaldehyde in PBS for 30 min, then blocked for 30 min in blocking buffer and incubated overnight with anti-GFP antibody (1:1,000, Life Technologies A10262) in blocking buffer followed by 4× 15 min washes with 0.2% Tween-20 in PHEM buffer. Finally, samples were incubated for 2 h with 1 μg ml−1 DAPI (Sigma-Aldrich D9542) and secondary antibody anti-chicken (1:500, Jackson ImmunoResearch 703-545-155) in blocking buffer. Afterward, inserts were washed 4× 15 min with 0.2% Tween-20 in PHEM buffer and mounted in Vectashield (Vector Laboratories H1000). Cultures were imaged on an SP8 Leica confocal microscope, obtained with the 40× objective with 0.75× digital zoom, optimal pinhole and line average, speed 600 Hz and a pixel size of 0.1893 µm per pixel. Then, 3D stacks were generated including all the cell layers of the culture, and where indicated, the basal layer plane was selected.
Histology
Part of the esophageal tumor was dissected, fixed in 10% formalin for at least 24 h and stored at 4 °C. Tissues were then embedded in paraffin and cut at 5 µm thickness. Sections were stained with hematoxylin and eosin and scanned. Some sections were immunostained. These were blocked for 1 h in blocking buffer (0.5% bovine serum albumin, 0.25% fish skin gelatin, 1% Triton X-100 and 10% donkey serum) in PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA and 4 mM MgSO4·7H2O). Next, sections were incubated for 1 h in blocking buffer with anti-GFP antibody (1:4,000, Life Technologies A10262) and anti-vimentin antibody (abcam ab92547 1:1,000). After three washes with 0.2% Tween-20 in PHEM buffer, sections were incubated for 1 h with 1 μg ml−1 DAPI and secondary antibodies anti-chicken (1:2,000, Jackson ImmunoResearch 703-545-155) and anti-rabbit (1:500, Thermo Fisher A32794).
Respirometry experiments
Fully recombined Pik3caH1047R/wt or uninduced (adenovirus-null infected) parallel cultures from the same mice were treated for at least 1 week in FAD medium supplemented with fetal calf serum, apo-transferrin and penicillin–streptomycin, with or without insulin. At the moment of the experiment, 5 mm circular sections of each culture were obtained using a biopsy punch (Stiefel BC-BI-1600) and transferred to XFe24 Cell Culture microplate wells with the basal layer facing up. Then, 675 μl of bicarbonate-free DMEM (Sigma-Aldrich, D5030) supplemented with 25 mM glucose, 1 mM pyruvate, 4 mM glutamine and 5 μg ml−1 apo-transferrin with or without 5 μg ml−1 insulin (Sigma-Aldrich I5500) was added to each sample. To eliminate residual carbonic acid from medium, cells were incubated for at least 30 min at 37 °C with atmospheric CO2 in a nonhumidified incubator. Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were assayed in a Seahorse XF-24 extracellular flux analyzer by three measurement cycles of 2 min mix, 2 min wait and 4 min measure. Then, the OCR/ECAR ratio was calculated using XF-24 software. Five technical replicates and four biological replicates were collected per condition.
Plasma and media biochemical analyses
Plasma samples or media samples collected after 3-day culture in the specified treatments were stored at −80 °C. Mouse insulin was measured simultaneously using a 2-plex Mouse Metabolic immunoassay kit (K15124C-3, MSD). The assay was performed according to the manufacturer’s instructions and using recombinant human insulin as calibrator. Lactate was analyzed using a commercial kit (DF16, Siemens Healthcare). All sample measurements were performed by the Medical Research Council Metabolic Diseases Unit Mouse Biochemistry Laboratory.
Quantification and statistical analysis
Unless otherwise specified, all data are expressed as mean values ± s.d. Differences between groups were assessed by two-tailed unpaired or paired t-test (specified in the figure legend) for normally distributed data or two-tailed Mann–Whitney U test for skewed data, using GraphPad Prism software V_8.3.1. For paired comparisons of clonal behavior across conditions, Peacock’s test was used, a 2D extension of a two-tailed Kolmogorov–Smirnov test (implemented as kstest_2s_2d function for MATLAB). The influence of metformin, DCA, HFD and Akita conditions on differences between Rosa26YFP/wt and Pik3caH1047R/wt basal clone sizes was assessed by multiple-regression linear modeling following a nonparametric two-factor design, of type: clone size ~ treatment (or genotype, in the case of Akita) × mutation. Given the skewness of the clone size distributions, we performed analysis of the variance of aligned-rank transformed (ART) data (contrast tests with ART (cran.r-project.org) ARTool (R) package vignette; updated 12 October 2021), reporting the P value of the interaction term (omnibus F-test). Exact P values for statistical tests are specified in the figures. No statistical method was used to predetermine sample size. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment.
The analysis of mutations and MM clone colonization in humans was done using a multiple-regression linear model with a three-factor design including possible interactions, of type: age × weight × mutation pathogenicity, using either the number of clones per centimeter squared or the summed VAF as the dependent variable. Age was modeled as a continuous factor, while BMI (below 25 or equal/above 25 kg m−2) and pathogenic status (Path/GoF versus Unkn/NE MM) as categorical factors. Results are reported following three-way ANOVA F-test, using Tukey’s test for post-hoc pairwise comparisons.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Mouse transcriptomic data can be viewed at https://www.ebi.ac.uk/ena using the following accession numbers: Animal 1 CTL; ERS2647403, Pik3cawt/wt Animal 2 CTL; ERS2647404, Pik3cawt/wt Animal 3 CTL; ERS2647405, Pik3cawt/wt Animal 4 CTL; ERS2647406, Pik3caH1047R/wt Animal 1 CTL; ERS2647407, Pik3caH1047R/wt Animal 2 CTL; ERS2647408, Pik3caH1047R/wt Animal 3 CTL; ERS2647409, Pik3caH1047R/wt Animal 4 CTL; ERS2647410, Pik3cawt/wt Animal 1 INS; ERS2515253, Pik3cawt/wt Animal 2 INS; ERS2515254, Pik3cawt/wt Animal 3 INS; ERS2515255, Pik3cawt/wt Animal 4 INS; ERS2515256, Pik3caH1047R/wt Animal 1 INS; ERS2515257, Pik3caH1047R/wt Animal 2 INS; ERS2515258, Pik3caH1047R/wt Animal 3 INS; ERS2515259, Pik3caH1047R/wt Animal 4 INS; ERS2515260. CTL refers to cultures incubated in minimal medium and insulin to cultures incubated with supraphysiological insulin dose. Human genomic sequencing data can be viewed at https://ega-archive.org, using the accession number EGAD00001008281. Source data are provided with this paper.
Code availability
Code used for modeling has been made publicly available and can be found via GitHub at https://github.com/gp10/DriverClonALTfate. Code used to generate 2D histograms of clone sizes, displayed as heatmaps, is available in the CloneSizeFreq_2Dheat package via GitHub at https://github.com/gp10/CloneSizeFreq_2Dheat.
References
Martincorena, I. et al. Somatic mutant clones colonize the human esophagus with age. Science 362, 911–917 (2018).
Martincorena, I. et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015).
Blokzijl, F. et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264 (2016).
Yokoyama, A. et al. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature 565, 312–317 (2019).
Lee-Six, H. et al. Population dynamics of normal human blood inferred from somatic mutations. Nature 561, 473–478 (2018).
Yoshida, K. et al. Tobacco smoking and somatic mutations in human bronchial epithelium. Nature 578, 266–272 (2020).
Moore, L. et al. The mutational landscape of normal human endometrial epithelium. Nature 580, 640–646 (2020).
Yizhak, K. et al. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science 364, eaaw0726 (2019).
Cagan, A. et al. Somatic mutation rates scale with lifespan across mammals. Nature 604, 517–524 (2022).
Fowler, J. C. & Jones, P. H. Somatic mutation: what shapes the mutational landscape of normal epithelia? Cancer Discov. 12, 1642–1655 (2022).
Fruman, D. A. et al. The PI3K pathway in human disease. Cell 170, 605–635 (2017).
Madsen, R. R., Vanhaesebroeck, B. & Semple, R. K. Cancer-associated PIK3CA mutations in overgrowth disorders. Trends Mol. Med 24, 856–870 (2018).
Castel, P. et al. Somatic PIK3CA mutations as a driver of sporadic venous malformations. Sci. Transl. Med 8, 332ra42 (2016).
Castillo, S. D. et al. Somatic activating mutations in Pik3ca cause sporadic venous malformations in mice and humans. Sci. Transl. Med. 8, 332ra43 (2016).
The Cancer Genome Atlas Research Network. Integrated genomic characterization of oesophageal carcinoma. Nature 541, 169–175 (2017).
Abby, E. et al. Notch1 mutations drive clonal expansion in normal esophageal epithelium but impair tumor growth. Nat. Genet 55, 232–245 (2023).
Trichas, G., Begbie, J. & Srinivas, S. Use of the viral 2A peptide for bicistronic expression in transgenic mice. BMC Biol. 6, 40 (2008).
Alcolea, M. P. et al. Differentiation imbalance in single oesophageal progenitor cells causes clonal immortalization and field change. Nat. Cell Biol. 16, 615–622 (2014).
Berenjeno, I. M. et al. Oncogenic PIK3CA induces centrosome amplification and tolerance to genome doubling. Nat. Commun. 8, 1773 (2017).
Kinross, K. M. et al. An activating Pik3ca mutation coupled with Pten loss is sufficient to initiate ovarian tumorigenesis in mice. J. Clin. Investig. 122, 553–557 (2012).
Mitchell, C. B. & Phillips, W. A. Mouse models for exploring the biological consequences and clinical significance of PIK3CA mutations. Biomolecules 9, 158 (2019).
Colom, B. et al. Spatial competition shapes the dynamic mutational landscape of normal esophageal epithelium. Nat. Genet. 52, 604–614 (2020).
Colom, B. et al. Mutant clones in normal epithelium outcompete and eliminate emerging tumours. Nature 598, 510–514 (2021).
Frede, J., Greulich, P., Nagy, T., Simons, B. D. & Jones, P. H. A single dividing cell population with imbalanced fate drives oesophageal tumour growth. Nat. Cell Biol. 18, 967–978 (2016).
Doupe, D. P. et al. A single progenitor population switches behavior to maintain and repair esophageal epithelium. Science 337, 1091–1093 (2012).
Piedrafita, G. et al. A single-progenitor model as the unifying paradigm of epidermal and esophageal epithelial maintenance in mice. Nat. Commun. 11, 1429 (2020).
Snippert, H. J. et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010).
de la Cova, C. et al. Supercompetitor status of Drosophila Myc cells requires p53 as a fitness sensor to reprogram metabolism and promote viability. Cell Metab. 19, 470–483 (2014).
Murai, K. et al. p53 mutation in normal esophagus promotes multiple stages of carcinogenesis but is constrained by clonal competition. Nat. Commun. 13, 6206 (2022).
Fernandez-Antoran, D. et al. Outcompeting p53-mutant cells in the normal esophagus by redox manipulation. Cell Stem Cell 25, 329–341 (2019).
Herms, A. et al. Self-sustaining long-term 3D epithelioid cultures reveal drivers of clonal expansion in esophageal epithelium. Nat. Genet. https://doi.org/10.1038/s41588-024-01875-8 (2024).
Boucher, J., Tseng, Y.-H. & Kahn, C. R. Insulin and insulin-like growth factor-1 receptors act as ligand-specific amplitude modulators of a common pathway regulating gene transcription. J. Biol. Chem. 285, 17235–17245 (2010).
Hoxhaj, G. & Manning, B. D. The PI3K–AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer 20, 74–88 (2020).
Sorge, S. et al. ATF4-induced Warburg metabolism drives over-proliferation in Drosophila. Cell Rep. 31, 107659 (2020).
Zhang, J. et al. Measuring energy metabolism in cultured cells, including human pluripotent stem cells and differentiated cells. Nat. Protoc. 7, 1068–1085 (2012).
Ilic, N. et al. PIK3CA mutant tumors depend on oxoglutarate dehydrogenase. Proc. Natl Acad. Sci. USA 114, E3434–E3443 (2017).
Welsh, S., Williams, R., Kirkpatrick, L., Paine-Murrieta, G. & Powis, G. Antitumor activity and pharmacodynamic properties of PX-478, an inhibitor of hypoxia-inducible factor-1α. Mol. Cancer Ther. 3, 233–244 (2004).
Hopkins, B. D., Goncalves, M. D. & Cantley, L. C. Insulin-PI3K signalling: an evolutionarily insulated metabolic driver of cancer. Nat. Rev. Endocrinol. 16, 276–283 (2020).
Martin-Montalvo, A. et al. Metformin improves healthspan and lifespan in mice. Nat. Commun. 4, 2192 (2013).
Wiernsperger, N. F. & Bailey, C. J. The antihyperglycaemic effect of metformin: therapeutic and cellular mechanisms. Drugs 58, 31–39 (1999).
Yoshioka, M., Kayo, T., Ikeda, T. & Koizumi, A. A novel locus, Mody4, distal to D7Mit189 on chromosome 7 determines early-onset NIDDM in nonobese C57BL/6 (Akita) mutant mice. Diabetes 46, 887–894 (1997).
Oyadomari, S. et al. Targeted disruption of the Chop gene delays endoplasmic reticulum stress-mediated diabetes. J. Clin. Invest. 109, 525–532 (2002).
Han, J.-W. et al. Impaired PI3K/Akt signal pathway and hepatocellular injury in high-fat fed rats. World J. Gastroenterol. 16, 6111–6118 (2010).
García-Prieto, C. F. et al. High-fat diet induces endothelial dysfunction through a down-regulation of the endothelial AMPK–PI3K–Akt–eNOS pathway. Mol. Nutr. Food Res. 59, 520–532 (2015).
Turner, N. et al. Distinct patterns of tissue-specific lipid accumulation during the induction of insulin resistance in mice by high-fat feeding. Diabetologia 56, 1638–1648 (2013).
Park, S.-Y. et al. Unraveling the temporal pattern of diet-induced insulin resistance in individual organs and cardiac dysfunction in C57BL/6 mice. Diabetes 54, 3530–3540 (2005).
Czech, M. P. Insulin action and resistance in obesity and type 2 diabetes. Nat. Med 23, 804–814 (2017).
Lauby-Secretan, B. et al. Body fatness and cancer—viewpoint of the IARC Working Group. N. Engl. J. Med. 375, 794–798 (2016).
Wilson, M. R. et al. ARID1A and PI3-kinase pathway mutations in the endometrium drive epithelial transdifferentiation and collective invasion. Nat. Commun. 10, 3554 (2019).
Deming, D. A. et al. PIK3CA and APC mutations are synergistic in the development of intestinal cancers. Oncogene 33, 2245–2254 (2014).
Adams, J. R. et al. Cooperation between Pik3ca and p53 mutations in mouse mammary tumor formation. Cancer Res. 71, 2706–2717 (2011).
Nowak, K., Seisenbacher, G., Hafen, E. & Stocker, H. Nutrient restriction enhances the proliferative potential of cells lacking the tumor suppressor PTEN in mitotic tissues. eLife 2, e00380 (2013).
Sanaki, Y., Nagata, R., Kizawa, D., Léopold, P. & Igaki, T. Hyperinsulinemia drives epithelial tumorigenesis by abrogating cell competition. Dev. Cell 53, 379–389.e5 (2020).
de la Cova, C., Abril, M., Bellosta, P., Gallant, P. & Johnston, L. A. Drosophila Myc regulates organ size by inducing cell competition. Cell 117, 107–116 (2004).
Yum, M. K. et al. Tracing oncogene-driven remodelling of the intestinal stem cell niche. Nature 594, 442–447 (2021).
Fowler, J. C. et al. Selection of oncogenic mutant clones in normal human skin varies with body site. Cancer Discov. 11, 340–361 (2021).
Ying, Z., Sandoval, M. & Beronja, S. Oncogenic activation of PI3K induces progenitor cell differentiation to suppress epidermal growth. Nat. Cell Biol. 20, 1256–1266 (2018).
Barzilai, N., Crandall, J. P., Kritchevsky, S. B. & Espeland, M. A. Metformin as a tool to target aging. Cell Metab. 23, 1060–1065 (2016).
Carstensen, B. et al. Cancer incidence in persons with type 1 diabetes: a five-country study of 9,000 cancers in type 1 diabetic individuals. Diabetologia 59, 980–988 (2016).
Bradley, M. C. et al. A cohort study of metformin and colorectal cancer risk among patients with diabetes mellitus. Cancer Epidemiol. Biomark. Prev. 27, 525–530 (2018).
Kemp, R. et al. Elimination of background recombination: somatic induction of Cre by combined transcriptional regulation and hormone binding affinity. Nucleic Acids Res. 32, e92 (2004).
Srinivas, S. et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4 (2001).
Gerstung, M., Papaemmanuil, E. & Campbell, P. J. Subclonal variant calling with multiple samples and prior knowledge. Bioinformatics 30, 1198–1204 (2014).
Martínez-Jiménez, F. et al. A compendium of mutational cancer driver genes. Nat. Rev. Cancer 20, 555–572 (2020).
Acknowledgements
We thank E. Choolun and T. Metcalf for their assistance in in vivo experiments, and Wellcome Sanger Institute RSF facilities for technical support. This work was supported by grants from the Wellcome Trust to the Wellcome Sanger Institute (098051, 296194 and 108413/A/15/D) and Cancer Research UK Programme Grants to P.H.J. (C609/A17257 and C609/A27326). A.H. benefited from the award of an EMBO long term fellowship (EMBO ALTF885-2015) and a Maria Zambrano Grant to attract international talent from Universitat de Barcelona and Ministerio de Universidades and cofunded with Next Generation EU funds. G.P. is supported by the Agencia Estatal de Investigación of Spain (grant no. PID2020-116163GA-I00). Work in the laboratory of B.V. is supported by Cancer Research UK (C23338/A25722) and the UK NIHR University College London Hospitals Biomedical Research Centre.
Author information
Authors and Affiliations
Contributions
A.H. and B.C. performed the experiments with support from E.A., I.C., A.P., D.F.-A. and C.B. A.H., A.K. and U.B. designed, performed and analyzed the CRISPR screen. K.M., B.V. and P.H.J. designed the mouse model and performed initial validations. C.K. and J.C.F. processed and analyzed the human sequencing data. G.P. analyzed clonal data and performed statistical analysis. M.W.J.H., S.H.O. and R.K.S. performed bioinformatics analysis. A.H., B.C. and P.H.J. wrote the manuscript with input from C.F. and B.V.
Corresponding author
Ethics declarations
Competing interests
B.C. is an employee of Altos Labs (Cambridge, UK). B.V. is a consultant for iOnctura (Geneva, Switzerland), Venthera (Palo Alto, CA, United States), Olema Pharmaceuticals (San Francisco, CA, United States) and Pharming (Leiden, The Netherlands). The other authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks the anonymous reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Generation of mice expressing a Pik3caH1047R mutation.
a, Schematic of the conditional targeted Pik3ca allele. Pik3ca exon 20 was flanked by loxP sites (triangles). The duplicate region of exon 20 encodes the H1047R mutation, a self-cleaving T2A peptide and an enhanced Yellow Fluorescent Protein (EYFP), followed by a nuclear localization signal (NLS). Wild-type p110α protein is expressed until Cre mediated recombination, after which the allele co-expresses p110αH1047R mutant protein and EYFP-NLS. Cre recombination was mediated by crossing the conditional mutant strain with AhCreERT mice which express the Cre recombinase upon induction with β-naphthoflavone (BNF) and tamoxifen (TAM). b, Typical top-down confocal image of esophageal epithelial whole-mount from an AhcreERTPik3caH1047R/wt mouse, 3 months post-induction. Optical section through basal cell layer is shown. Pik3caH1047R/wt clones, showing nuclear EYFP (green) indicated by white arrows. Nuclei are stained with DAPI (blue). Clones are delimited by dashed white lines. Scale bar, 20 μm. c, Immunoblots of protein expression of p110α, GFP and AKT (total and, phosphorylated Ser473 (top) or Thr308 (bottom)) in NIH3T3 cells transfected with empty vector, or vector carrying wild-type p110α, p110αH1047R or p110αH1047R -P2A-GFP. Cells were starved for 24 h and lysed. Images are representative of 3 separate experiments. d-f, Signaling in induced mutant cells. Protocol (d): primary esophageal keratinocytes from Pik3caH1047R/wt mice were infected either with null-adenovirus (uninduced Pik3cawt/wt controls) or Cre-adenovirus (induced Pik3caH1047R/wt). Cells were treated in medium containing either 0.1% serum and no added growth factors (Starved, STV), 20% serum and growth factors (FCS) or FCS and the PI3K inhibitor LY294002 (50 µM), lysed and the pAKT(Ser473)/Total-AKT (e) and p-PRAS40/Total-PRAS40 (f) analyzed by immune capillary electrophoresis. Two-tailed ratio paired t-test. n = 4 paired (Ad-Null and Ad-Cre infected) biological replicates (mice) per condition (paired samples are linked by lines). Each dot is a biological replicate culture, lines link uninduced and induced cultures derived from the same mouse.
Extended Data Fig. 2 Heterozygous Pik3caH1047R expression and mouse esophageal tumorigenesis.
a-c. Protocol (a) and survival (b) of AhCreERTRosa26flEYFP/wt (Cre-RYFP) and AhcreERTPik3caH1047R-YFP/wt (Cre-Pik3caH1047R-YFP/wt) mice induced and followed up to one-year post-induction. c Normal appearance of typical esophagi from a after longitudinal opening and flattening out. d-e, Carcinogenesis study. d. Protocol: Cre-Pik3caH1047R-YFP/wt mice were induced 1 or 3 times and treated with diethylnitrosamine (DEN) for 8 weeks starting 4 weeks post-induction. Uninduced mice were used as controls. Tissues were collected before exceeding the permitted humane endpoint or at 1 year after DEN. e. Survival curves of animals on protocol in d. Log-rank (Mantel-Cox) test. f-i, Pik3caH1047R-YFP/wt induced ESCC. Image in (f) shows unopened esophagus containing an ESCC (arrow) generated by DEN of an induced Cre-Pik3caH1047R-YFP/wt mouse. (g) Immunofluorescence of a section of the tumor in (f) showing DAPI (blue), Pik3caH1047R/wt (green) and Vimentin (red). Images in (h) and (i) depict Hematoxylin-Eosin staining of sections from the tumor in (f). Scale bars, 2 mm (f), 200 µm (g), 500 µm (h), 250 µm (i).
Extended Data Fig. 3 Mechanism of Pik3caH1047R-YFP/wt clonal expansion.
a, Multicolor lineage tracing in Pik3caH1047R-YFP/wtRosa26confetti/wtAhCreERT (Cre-Pik3caH1047R-YFP/wt-Confetti) animals. After recombination, one of four reporters is expressed. In some clones the Pik3ca locus is also recombined. b, Possible color combinations following immunostaining for GFP to detect Pik3caH1047R-YFP/wt. Only RFP+ in wild-type or Pik3caH1047R/wt clones can be distinguished. c, Top-down confocal image of Cre-Pik3caH1047R-YFP/wt -Confetti epithelium 84 days post-induction, showing RFP+ Pik3cawt/wt and YFP+/RFP+ Pik3caH1047R/wt clone. DAPI is blue. Scale bar, 20 μm. d, Heatmaps of number of basal and first suprabasal layer cells in RFP+ clones in Cre-Pik3caH1047R-YFP/wt-Confetti animals. Black dots and dashed lines show geometric median. Number of clones analyzed is shown, animal numbers are in brackets. Lower panels, differences between Pik3cawt/wt and Pik3caH1047R/wt clones. Two-tailed 2D Kolmogorov-Smirnov test. e, Average basal cells per clone from d, in clones with >1 basal cell. Dots indicate the average clone size per mouse. Black lines indicate mean±s.e.m. Lines represent the best fitting model shown in Fig. 3, shaded areas plausible intervals. f, Basal cell density in Pik3caH1047R/wt mutant clones and size-equivalent wild-type areas in esophagus of Cre-Pik3caH1047R-YFP/wt mice, 6 months post-induction. Red lines, mean ± S.D. Two tailed unpaired t-test (n = 63 areas per group, from 5 animals). g, Confocal images showing Caspase 3 (red), Pik3caH1047R/wt cells, green, and DAPI, blue staining in basal layer of a Cre-Pik3caH1047R-YFP/wt mouse, 1-month post-high dose induction (Methods). Images representative of 4 mice. Left panel, UV irradiated positive control sample. The middle and right panels are images from the same tissue showing apoptotic cell (red arrow) and Pik3caH1047R/wt clone (green arrow). Scale bars, 20 μm. Architecture h and dynamics i of mouse esophageal epithelium. h, The lowest basal layer contains proliferating progenitor cells and post-mitotic differentiating cells, which will migrate upwards through the suprabasal layers until they are shed. i, Progenitor divisions may generate two differentiating cells, two progenitor cells or one of each type. The probabilities of each outcome are balanced, producing equal numbers of progenitor and differentiating cells across the epithelium, maintaining tissue homeostasis. Tilting cell fate towards proliferation, produces excess mutant progenitors resulting in clone growth.
Extended Data Fig. 4 Mutant versus wild-type cell competition in culture.
a-b, Generation of induced WT-RYFP (a) and uninduced/induced Pik3caH1047R/wt (b) epithelioid cultures. Primary esophageal keratinocytes from uninduced Rosa26RYFP/RYFP (a) or uninduced Pik3caH1047R-YFP/wt animals (b) were incubated with Cre-expressing adenovirus (Ad-Cre) or null adenovirus (Ad-Null). Right panels, representative images of Ad-Cre or Ad-Null treated cultures stained for YFP (yellow) and DAPI (nuclei, blue). Scale bars, 20 μm. c, In vitro cell competition protocol. Pik3caH1047R/wt and Pik3cawt/wt keratinocytes from b, are mixed with WT-RYFP cells from a, and maintained at confluence. Proportion of WT-RYFP cells is quantified by flow cytometry, gating strategy is shown. The proportion of WT-RYFP cells after treatment was normalized to the initial WT-RYFP cell proportion. Scale bars, 20 μm. d, GSEA histograms of PI3K/Akt/mTOR (top) and mTOR signaling (bottom) Hallmark gene sets comparing RNA-seq data from control (CTL) and 5 μg/ml insulin (+INS) treated wild-type cells from the same animals. The nominal p-value, the normalized enrichment score (NES) and the false discovery rate (FDR) q-value are indicated. n = 4 independent replicates per condition from one animal each. e-g, Experimental scheme (top panel) and quantification by flow cytometry (bottom panel) of the proportion of WT-RYFP cells mixed with Pik3caH1047R/wt cells at the end of the experiment versus the start of each experiment. e, Cells were treated either in minimal FAD medium or treated with 10 ng/ml EGF, 2 ng/ml Insulin (physiological) or 5 μg/ml Insulin (supraphysiological) for one month. Each dot represents a biological replicate (n = 4-16 primary cultures from individual animals, per condition). f, Cells were cultured for 28 days in minimal medium (CTL), 28 days in minimal medium with 5 μg/ml insulin (INS), or 14 days in minimal medium with 5 μg/ml insulin followed by 14 days in minimal medium. Each dot represents a biological replicate (n = 4 primary cultures from individual animals, per condition). g, Cells were treated either in minimal FAD medium minus/plus Rapamycin 500 nM for 14 days. Each dot represents a biological replicate (n = 4-8 primary cultures from different animals, per condition). Red lines indicate mean values. Two-tailed paired t-test.
Extended Data Fig. 5 CRISPR screening of targets affecting mutant cell fitness.
a-b, CRISPR screening in uninduced (Pik3cawt/wt, WT) cells in minimal medium (CTL). a, Correlation between normalized read counts for the sequenced library and the average normalized read counts at Time 0. Orange line, linear regression between samples with the Pearson’s coefficient and two-tailed p-value of the correlation. b, Violin plots of distribution of average log2 fold change between Time 3 and Time 0 for each gRNA. n = 2 biological replicates, 10 gRNA per gene. c, Heatmaps of average log2 (fold change per gene of gRNA abundance after 3 weeks versus Time 0. Genes are grouped by pathway. Heatmaps show the enrichment of gRNAs targeting each gene at 3 weeks over 0 weeks in either minimal medium (CTL) or minimal medium supplemented with 5 μg/ml Insulin (INS), and either uninduced (WT) or induced (Pik3ca*/wt) cells from Rosa26Cas9/wt Pik3caH1047R/wt mice. Each column is a biological replicate. n = 2-3 biological replicates, 10 gRNA per gene.
Extended Data Fig. 6 Pik3caH1047R/wt cells activate Hif1α pathway and glycolysis.
a-b, RNA-seq analysis comparing Pik3cawt/wt (WT, uninduced) and Pik3caH1047R/wt mutant cultures from the same animal, in minimal medium. Heatmap shows altered transcripts (adjusted p < 0.05). n = 4 mice per group, paired induced-uninduced samples. Wald test corrected for multiple testing. c-f, Gene set enrichment analysis histograms of PI3K/Akt/mTOR (c), mTOR (d) Hypoxia (e) and myc targets (f) signaling Hallmark gene sets in WT and mutant cultures. Nominal p-value, normalized enrichment score (NES) and false discovery rate (FDR) q-value are indicated. g, KEGG pathway enrichment analysis for up-regulated genes in mutant vs WT (p-value adjusted for multiple hypotheses testing using the Benjamini-Hochberg method<0.01). Intensity indicates pathway enrichment. h, Proportion of HIF1α target genes significantly up-regulated (adjusted p-value (as in g)<0.01) in mutant vs WT. i, Transcripts per million (TPM) relative to WT CTL condition of Hif1a and Vegfa in Pik3cawt/wt and Pik3caH1047R/wt cells in control (CTL) versus cultures treated with 5 μg/ml insulin (INS). n = 4 biological replicates per condition. Red lines, median values. Wald test corrected for multiple testing. j, Uninduced (Pik3cawt/wt) and induced (Pik3caH1047R/wt) cells were cultured in starvation media (STV) +/− PI3K inhibitor LY294002 (0.5 µM), and HIF1α/α-Tubulin analyzed by immune capillary electrophoresis. HIF1α/α-Tubulin ratio versus the average of the WT STV shown. Two-tailed ratio paired t-test. n = 5 paired biological replicates per condition (lines link paired samples). k, GSEA histogram of Glycolysis Hallmark gene set comparing induced Pik3caH1047R/wt and WT cells in minimal medium. n = 4 biological replicates per condition. Statistics as in c-f. l, Heatmaps comparing WT and Pik3caH1047R/wt cultures in CTL and INS conditions. Two-tailed Wald test corrected for multiple testing comparing WT and Pik3caH1047R/wt CTL conditions. ***p < 0.001, n.s.= not significant. m, GSEA histograms of Glycolysis Hallmark set comparing control (CTL) and 5 μg/ml insulin (+INS) treated wild-type cells. n = 4 biological replicates per condition. Statistics as in c-f. n, Basal OCR to ECAR ratios of WT and Pik3caH1047R/wt cultures in CTL or INS conditions. Dots, average per animal (n = 4 mice). OCR/ECAR ratios are mean ± SD. Two-tailed paired t-test.
Extended Data Fig. 7 Fitness of Pik3caH1047R/wt cells depends on Hif1α.
a-d, Protocol (a): Pik3cawt/wt (wild-type) or Pik3caH1047R/wt mutant cells expressing shRNA against Hif1a (shHif1a) or control shRNA (shNT) were mixed with WT-RYFP and cultured for 28 days in minimal medium. b, Hif1α mRNA levels in shHif1a or shNT cells. Each dot is a culture from a different mouse (n = 4 mice). Red lines, median values. Two tailed paired t-test. c, Representative confocal images showing basal layer of an epithelioid culture generated as in a. n = 3 biological replicates. WT-RYFP cells, yellow, DAPI, blue. Scale bar, 20 μm. d, Proportion of WT-RYFP cells at 28 days versus day 0. Each dot is a culture from a different animal. n = 3-7 cultures per condition. Two-tailed paired t-test. e-f, In vitro cell competition of WT-RYFP mixed with Pik3caH1047R/wt cells or uninduced controls from the same mice. Cultures were then treated with HIF1α inhibitor PX-478 (10 µM) or vehicle for 28 days. (f). Each dot is mean of a culture from a different animal (n = 5-11). Bars are SD. Two tailed unpaired t-test.
Extended Data Fig. 8 Metabolic changes downstream of Pik3caH1047R/wt expression.
a-b, Proportion of WT-RYFP cells in mixed cultures with Pik3caH1047R/wt cells, after 15 days in minimal media normalized to baseline value. +/–Gluc indicates culture with/without glucose and 2-DG indicates culture with 5 mM 2-deoxyglucose. Each dot is a culture from a different animal, lines are mean values. n = 4 cultures per condition. c-e, In vitro cell competition assay. c, Protocol: Confluent mixed cultures of WT-RYFP and induced Pik3caH1047R/wt cells with/without DCA 25 mM for 28 days in minimal medium. d, Representative confocal images of basal cell layer of cultures from c, WT-RYFP cells, yellow, DAPI, blue. Scale bar, 20 μm. e, Flow cytometric analysis from mixed cultures in c. Each dot is a culture from a different mouse and lines connect mean values (n = 7-15 cultures). Two-tailed paired t-test. f, Protocol: Cre-RYFP control and Cre-Pik3caH1047R-YFP/wt mice were induced and treated with DCA. Clones with >1 basal cell sizes were analyzed at 28 days. Mice and clone numbers are shown. g, Heatmaps showing the differences between each treatment versus control in Cre-RYFP (upper panels) or Cre-Pik3caH1047R-YFP/wt (lower panels) mice from f. Two-tailed 2D Kolmogorov-Smirnov test. h-i, Average basal clone sizes h and proportion of first suprabasal layer cells i for each strain and treatment from f. Data includes clones with at least one basal cell. Bars are SD. Two-tailed unpaired t-test. j, Wild-type and Pik3caH1047R mutant basal cell clone size distributions from untreated or DCA-treated Cre-RYFP and Cre-Pik3caH1047R-YFP/wt mice, respectively, 28 days post- induction. n = 311-799 clones from 5-8 animals per condition. Dots indicate mean and lines standard deviation. Two-tailed Kolmogorov-Smirnov test and Contrast ART-C Post-hoc test of differences of differences between distributions. k. Proportion of WT-RYFP cells in mixed cultures with Pik3caH1047R/wt cells, at 15 days normalized to baseline value. Cells were culture in minimal media (CTL) or with Betulin (6 µg/ml), CB839 (10 µM), TOFA (30 µM) or TVB-2640 (0.1 µM). Each dot represents a culture from a different animal, lines correspond to mean values. n = 4 cultures from individual animals per condition. Two tailed paired t-test.
Extended Data Fig. 9 Metabolic conditions alter the Pik3caH1047R/wt fitness in vivo.
a-b, Cre-RYFP and Cre-Pik3caH1047R-YFP/wt mice were induced and treated with or without metformin (MET). First suprabasal layer cells per clone (a) and basal clone size distributions (b) analyzed at 28 days. Dots indicate mean and lines S.D. in b. Two-tailed unpaired t-test for suprabasal cells per clone. Two-tailed Kolmogorov-Smirnov test to compare wild-type and mutant basal clone distributions. Contrast ART-C Post-hoc test of differences of differences between distributions. n = 431-917 clones from 5-10 animals per condition. c, Lactate secretion in wild-type cells in minimal medium in control conditions (CTL) or with 2.5 mM MET. Bars indicate mean. Two-tailed paired t-test. d-f, In vitro competition assay. d, Protocol: Confluent cultures of WT-RYFP and Pik3caH1047R/wt cells in minimal medium with/without MET for 28 days. e, Representative confocal image of basal layer of culture from d. WT-RYFP cells, yellow, DAPI, blue. Scale bar, 20 μm. f, Flow cytometric analysis of cultures in d. Each dot represents a biological replicate, lines connect mean values (n = 3–12). Two-tailed paired t-test. g, Urine glucose levels in mice from Fig. 6d. Two-tailed Mann-Whitney test. n = 4 mice per group. Mean ± SD. h, Basal cells per clone in Fig. 6d. Dots and bars indicate mean and S.D. Two-tailed Kolmogorov-Smirnov and Contrast ART-C Post-hoc tests. i, Average proportion of suprabasal cells per clone for each strain and treatment from Fig. 6d, only the first suprabasal cell layer was counted. Each dot corresponds to one animal. Bars are mean ± SD. n = 431-917 clones from 5-10 animals per condition. Two-tailed unpaired t-test. j, Body weight measured at weeks 0 and 4 post diets. Each dot corresponds to one animal, lines link weights from same animal (n = 10-16 mice). Two-tailed paired t-test. k, Blood insulin levels at the end of experiment in j. Error bars are mean±s.e.m. Two-tailed unpaired t-test. l, Average proportion of suprabasal cells per clone for each strain and treatment from Fig. 6g, only first suprabasal cells were counted. Each dot corresponds to one animal. Bars are mean ± S.D. Two-tailed unpaired t-test. m, Distribution of basal cells per clone from Fig. 6g. Dots and bars indicate mean and S.D. Two-tailed Kolmogorov-Smirnov and Contrast ART-C Post-hoc tests.
Extended Data Fig. 10 Higher density of Pik3ca missense mutations in overweight humans.
a, Age distribution of all human donors, including donors from Fig. 1. Lines are mean±s.e.m. Two-tailed unpaired t-test. b-d, Human donors were classified into OW-OB (n = 10, BMI ≥ 25 kg m2) and non-OW (n = 8, BMI < 25 kg m2). PIK3CA missense mutations (MM) were classified as Path/GoF and Unkn/NE (see Methods) and analyzed separately in OW-OB and Non-OW groups. Each dot represents a donor. b, Number of PIK3CA missense mutant clones detected/cm2 in the specified groups. Bars are mean±s.e.m. Two-tailed unpaired t-test. c, Summed variant allele frequency (VAF) of all Path/GoF or Unkn/NE PIK3CA missense mutations per cm2 for each donor. Lines are mean±s.e.m. Two-tailed Mann-Whitney test. d, Number of clones/cm2 carrying Path/GoF PIK3CA missense mutations in each p110α domain. Bars are mean±s.e.m. Two-tailed Mann-Whitney test.
Supplementary information
Supplementary Information
Supplementary Note: Additional text, Methods and Figures.
Supplementary Video
2D simulation of the growth of wild-type and Pik3caH1047R/wt clones over 6 months starting from the same amount of induced cells and following the parameters described in Fig. 3i and Extended Data Fig. 3k,l.
Supplementary Tables 1–5
Supplementary Tables 1–5.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Source Data Scans of Western Blots
Source data: scans of uncropped western blots.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Herms, A., Colom, B., Piedrafita, G. et al. Organismal metabolism regulates the expansion of oncogenic PIK3CA mutant clones in normal esophagus. Nat Genet 56, 2144–2157 (2024). https://doi.org/10.1038/s41588-024-01891-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41588-024-01891-8
This article is cited by
-
A Mendelian randomization study of type 2 diabetes and cancer risk in East Asians
Cancer Cell International (2025)
-
Concurrent PIK3CA mutant promotes cachexia through inflammatory signaling in EGFR mutant lung cancer
Nature Communications (2025)
-
Host physiology shapes the mutational landscape of normal and carcinogenic tissue
Nature Genetics (2024)
-
Long-term 3D primary epithelioid cultures reveal genes that regulate esophageal cell fitness
Nature Genetics (2024)
-
Self-sustaining long-term 3D epithelioid cultures reveal drivers of clonal expansion in esophageal epithelium
Nature Genetics (2024)