Abstract
Up to an estimated 10% of women experience miscarriage in their lifetimes. Embryonic aneuploidy is a leading cause for miscarriage, infertility and congenital defects. Here we identify variants of ELL3, a gene encoding a transcription elongation factor, in couples who experienced consecutive early miscarriages due to embryonic aneuploidy. Maternal ELL3 knockout leads to mouse oocyte aneuploidy, subfertility and miscellaneous embryonic defects. Mechanistically, we find that ELL3 localizes to the spindle during meiosis, and that ELL3 depletion in both mouse and human oocytes increases the incidence of meiotic spindle abnormality. ELL3 coordinates with TPX2 to ensure the proper function of the microtubule motor KIF11. Live imaging analysis shows that ELL3 is paramount for promoting spindle assembly and driving chromosome movement. Together, our findings implicate maternal ELL3 deficiency in causing oocyte aneuploidy and early miscarriage.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Raw and processed RNA-seq data generated for this study are available on GEO (GSE268181). Source data are provided with this paper.
References
Quenby, S. et al. Miscarriage matters: the epidemiological, physical, psychological, and economic costs of early pregnancy loss. Lancet 397, 1658–1667 (2021).
Essers, R. et al. Prevalence of chromosomal alterations in first-trimester spontaneous pregnancy loss. Nat. Med. 9, 3233–3242 (2023).
Charalambous, C., Webster, A. & Schuh, M. Aneuploidy in mammalian oocytes and the impact of maternal ageing. Nat. Rev. Mol. Cell Biol. 24, 27–44 (2023).
So, C. et al. Mechanism of spindle pole organization and instability in human oocytes. Science 375, eabj3944 (2022).
Thomas, C., Cavazza, T. & Schuh, M. Aneuploidy in human eggs: contributions of the meiotic spindle. Biochem. Soc. Trans. 49, 107–118 (2021).
Holubcová, Z., Blayney, M., Elder, K. & Schuh, M. Human oocytes. Error-prone chromosome-mediated spindle assembly favors chromosome segregation defects in human oocytes. Science 348, 1143–1147 (2015).
Goodson, H. V. & Jonasson, E. M. Microtubules and microtubule-associated proteins. Cold Spring Harb. Perspect. Biol. 10, a022608 (2018).
Mogessie, B., Scheffler, K. & Schuh, M. Assembly and positioning of the oocyte meiotic spindle. Annu. Rev. Cell Dev. Biol. 34, 381–403 (2018).
Kyogoku, H. & Kitajima, T. S. Large cytoplasm is linked to the error-prone nature of oocytes. Dev. Cell 41, 287–298.e4 (2017).
Sang, Q., Ray, P. F. & Wang, L. Understanding the genetics of human infertility. Science 380, 158–163 (2023).
Van Der Kelen, A. et al. A systematic review and evidence assessment of monogenic gene-disease relationships in human female infertility and differences in sex development. Hum. Reprod. Update 29, 218–232 (2023).
Feng, R. et al. Mutations in TUBB8 and human oocyte meiotic arrest. N. Engl. J. Med. 374, 223–232 (2016).
Wu, T. et al. The mechanism of acentrosomal spindle assembly in human oocytes. Science 378, eabq7361 (2022).
Chen, B. et al. Novel mutations and structural deletions inTUBB8: expanding mutational and phenotypic spectrum of patients with arrest in oocyte maturation, fertilization or early embryonic development. Hum. Reprod. 32, 457–464 (2017).
Zhao, L. et al. Biallelic mutations in CDC20 cause female infertility characterized by abnormalities in oocyte maturation and early embryonic development. Protein Cell 11, 921–927 (2020).
Tyc, K. M. et al. Exome sequencing links CEP120 mutation to maternally derived aneuploid conception risk. Hum. Reprod. 35, 2134–2148 (2020).
Ottolini, C. S. et al. Genome-wide maps of recombination and chromosome segregation in human oocytes and embryos show selection for maternal recombination rates. Nat. Genet. 47, 727–735 (2015).
Nagaoka, S. I., Hassold, T. J. & Hunt, P. A. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 13, 493–504 (2012).
Mihajlović, A. I. & FitzHarris, G. Segregating chromosomes in the mammalian oocyte. Curr. Biol. 28, R895–R907 (2018).
Gruhn, J. R. et al. Chromosome errors in human eggs shape natural fertility over reproductive life span. Science 365, 1466–1469 (2019).
Miller, T., Williams, K., Johnstone, R. W. & Shilatifard, A. Identification, cloning, expression, and biochemical characterization of the testis-specific RNA polymerase II elongation factor ELL3. J. Biol. Chem. 275, 32052–32056 (2000).
Shilatifard, A., Lane, W. S., Jackson, K. W., Conaway, R. C. & Conaway, J. W. An RNA polymerase II elongation factor encoded by the human ELL gene. Science 271, 1873–1876 (1996).
Lin, C. et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell 37, 429–437 (2010).
Lin, C., Garruss, A. S., Luo, Z., Guo, F. & Shilatifard, A. The RNA Pol II elongation factor Ell3 marks enhancers in ES cells and primes future gene activation. Cell 152, 144–156 (2013).
Meng, S. et al. Young LINE-1 transposon 5′ UTRs marked by elongation factor ELL3 function as enhancers to regulate naive pluripotency in embryonic stem cells. Nat. Cell Biol. 25, 1319–1331 (2023).
Vallot, A. et al. Tension-induced error correction and not kinetochore attachment status activates the SAC in an aurora-B/C-dependent manner in oocytes. Curr. Biol. 28, 130–139 e3 (2018).
Madsen, B. E. & Browning, S. R. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 5, e1000384 (2009).
Yang, X. et al. Rare variants in FANCA induce premature ovarian insufficiency. Hum. Genet. 138, 1227–1236 (2019).
Olbrich, H. et al. Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left-right asymmetry. Nat. Genet. 30, 143–144 (2002).
Huang, C. J. et al. Maternal SENP7 programs meiosis architecture and embryo survival in mouse. Biochim. Biophys. Acta, Mol. Cell. Res. 1864, 1195–1206 (2017).
Clift, D., So, C., McEwan, W. A., James, L. C. & Schuh, M. Acute and rapid degradation of endogenous proteins by Trim-Away. Nat. Protoc. 13, 2149–2175 (2018).
Homer, H., Gui, L. & Carroll, J. A spindle assembly checkpoint protein functions in prophase I arrest and prometaphase progression. Science 326, 991–994 (2009).
Touati, S. A. et al. Mouse oocytes depend on BubR1 for proper chromosome segregation but not for prophase i arrest. Nat. Commun. 6, 6946 (2015).
Lan, Z. J., Xu, X. & Cooney, A. J. Differential oocyte-specific expression of Cre recombinase activity in GDF-9-iCre, Zp3cre, and Msx2Cre transgenic mice. Biol. Reprod. 71, 1469–1474 (2004).
Potter, H. Beyond trisomy 21: phenotypic variability in people with Down syndrome explained by further chromosome mis-segregation and mosaic aneuploidy. J. Syndr. Chromosom. Abnorm. 2, 109 (2016).
Beach, R. R. et al. Aneuploidy causes non-genetic individuality. Cell 169, 229–242.e21 (2017).
Roper, R. J. & Reeves, R. H. Understanding the basis for Down syndrome phenotypes. PLoS Genet. 2, e50 (2006).
Kapitein, L. C. et al. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 435, 114–118 (2005).
Hirokawa, N., Noda, Y., Tanaka, Y. & Niwa, S. Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10, 682–696 (2009).
Ma, N., Titus, J., Gable, A., Ross, J. L. & Wadsworth, P. TPX2 regulates the localization and activity of Eg5 in the mammalian mitotic spindle. J. Cell Biol. 195, 87–98 (2011).
Balchand, S. K., Mann, B. J., Titus, J., Ross, J. L. & Wadsworth, P. TPX2 inhibits Eg5 by interactions with both motor and microtubule. J. Biol. Chem. 290, 17367–17379 (2015).
Gable, A. et al. Dynamic reorganization of Eg5 in the mammalian spindle throughout mitosis requires dynein and TPX2. Mol. Biol. Cell 23, 1254–1266 (2012).
Helmke, K. J. & Heald, R. TPX2 levels modulate meiotic spindle size and architecture in Xenopus egg extracts. J. Cell Biol. 206, 385–393 (2014).
Eibes, S. et al. Nek9 phosphorylation defines a new role for TPX2 in Eg5-dependent centrosome separation before nuclear envelope breakdown. Curr. Biol. 28, 121–129.e4 (2018).
Wadsworth, P. Tpx2. Curr. Biol. 25, R1156–R1158 (2015).
Alfaro-Aco, R., Thawani, A. & Petry, S. Structural analysis of the role of TPX2 in branching microtubule nucleation. J. Cell Biol. 216, 983–997 (2017).
Zhang, R., Roostalu, J., Surrey, T. & Nogales, E. Structural insight into TPX2-stimulated microtubule assembly. eLife 6, e30959 (2017).
Kitajima, T. S., Ohsugi, M. & Ellenberg, J. Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in mammalian oocytes. Cell 146, 568–581 (2011).
Mayer, T. U. et al. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science 286, 971–974 (1999).
Kapoor, T. M., Mayer, T. U., Coughlin, M. L. & Mitchison, T. J. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J. Cell Biol. 150, 975–988 (2000).
Mailhes, J. B., Mastromatteo, C. & Fuseler, J. W. Transient exposure to the Eg5 kinesin inhibitor monastrol leads to syntelic orientation of chromosomes and aneuploidy in mouse oocytes. Mutat. Res. 559, 153–167 (2004).
Clift, D. & Schuh, M. A three-step MTOC fragmentation mechanism facilitates bipolar spindle assembly in mouse oocytes. Nat. Commun. 6, 7217 (2015).
Ma, N. et al. Poleward transport of TPX2 in the mammalian mitotic spindle requires dynein, Eg5, and microtubule flux. Mol. Biol. Cell 21, 979–988 (2010).
Brunet, S. et al. Meiotic regulation of TPX2 protein levels governs cell cycle progression in mouse oocytes. PLoS ONE 3, e3338 (2008).
Luo, Z., Lin, C. & Shilatifard, A. The super elongation complex (SEC) family in transcriptional control. Nat. Rev. Mol. Cell Biol. 13, 543–547 (2012).
Smith, E. R. et al. The little elongation complex regulates small nuclear RNA transcription. Mol. Cell 44, 954–965 (2011).
Hu, D. et al. The little elongation complex functions at initiation and elongation phases of snRNA gene transcription. Mol. Cell 51, 493–505 (2013).
Lin, C. et al. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC). Genes Dev. 25, 1486–1498 (2011).
Vale, R. D. & Milligan, R. A. The way things move: looking under the hood of molecular motor proteins. Science 288, 88–95 (2000).
Simmons, R. Molecular motors: single-molecule mechanics. Curr. Biol. 6, 392–394 (1996).
Arpağ, G., Shastry, S., Hancock, W. O. & Tüzel, E. Transport by populations of fast and slow kinesins uncovers novel family-dependent motor characteristics important for in vivo function. Biophys. J. 107, 1896–1904 (2014).
Paschal, B. M. et al. Isolated flagellar outer arm dynein translocates brain microtubules in vitro. Nature 330, 672–674 (1987).
Lee, S. et al. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am. J. Hum. Genet. 91, 224–237 (2012).
Xue, Z. et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500, 593–597 (2013).
Zhou, C. et al. The cohesin release factor Wapl interacts with Bub3 to govern SAC activity in female meiosis I. Sci. Adv. 6, eaax3969 (2020).
Courtois, A., Yoshida, S., Takenouchi, O., Asai, K. & Kitajima, T. S. Stable kinetochore–microtubule attachments restrict MTOC position and spindle elongation in oocytes. EMBO Rep. 22, e51400 (2021).
Paim, L. M. G. & FitzHarris, G. Tetraploidy causes chromosomal instability in acentriolar mouse embryos. Nat. Commun. 10, 4834 (2019).
Pedersen, T. & Peters, H. Proposal for a classification of oocytes and follicles in the mouse ovary. J. Reprod. Fertil. 17, 555–557 (1968).
Fuchs, M. et al. A role for the chaperone complex BAG3-HSPB8 in actin dynamics, spindle orientation and proper chromosome segregation during mitosis. PLoS Genet. 11, e1005582 (2015).
Guo, C. et al. ENL initiates multivalent phase separation of the super elongation complex (SEC) in controlling rapid transcriptional activation. Sci. Adv. 6, eaay4858 (2020).
Webb, M. R. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc. Natl Acad. Sci. USA 89, 4884–4887 (1992).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Strnad, P. et al. Inverted light-sheet microscope for imaging mouse pre-implantation development. Nat. Methods 13, 139–142 (2016).
Peng, H., Bria, A., Zhou, Z., Iannello, G. & Long, F. Extensible visualization and analysis for multidimensional images using Vaa3D. Nat. Protoc. 9, 193–208 (2014).
Acknowledgements
We thank all Lin and Luo laboratory members for critical discussion of the paper. We thank J. Tang (TimWinter company, China) for the technical support in living imaging. We thank X. Liu and J. Yue (Southeast University, China) for technical support. We also thank H. Fan (Zhejiang University, China) for providing H2B-mCherry and α-TUBULIN-eGFP vectors. Studies in this paper were supported by funds provided by National Natural Science Foundation of China (grant nos. 32350004 and 32030017 to C.L. and 32470627 to Z.L.), National Key R&D Program of China (grant no. 2018YFA0800100 to C.L.) and the Shenzhen Science and Technology Program (grant nos. JCYJ20220530160417038 to C.L. and JCYJ20220530160416037 to Z.L.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the paper.
Author information
Authors and Affiliations
Contributions
C.L. and Z.L. designed the research and supervised the project. S.Z. performed most of the experiments and analyzed the data. P.X., S.Z., J.Z., K.F. and K.G. analyzed the live imaging data. Y.Y., H.S. and J.L. performed the whole-exome sequencing data analysis. Z.X., Y.W., M.H., F.Q., H.H., H.F. and P.H. provided the whole-exome sequencing, Sanger sequencing and STR data. C.Z., H.Z. and K.W. performed ELL3 KD and Trim-Away assays in human GV oocytes. Y.Z and B.X. performed the microinjection and IVF. S.Z., Y.L., B.M. and J.X. collected the live imaging data. J.H. performed RNA-seq analysis. H.S. performed mouse ovary hematoxylin and eosin staining. J.L. and L.H. performed ELL3 TRIM-Away experiments in mouse oocytes. Y.S., L.D., D.L. and B.Z. provided light-sheet microscope, collected the live imaging data, and provided genetic counseling. Z.C., H.C., X.Y., W.X. and Y.S. provided the resources and discussed the paper. Z.L., C.L. and S.Z. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Kirstin Matthews and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: Dimitris Typas, in collaboration with the Nature Structural & Molecular Biology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Functional validation of the ELL3 variants.
a, Mini-gene splicing analysis of the ELL3 c.133-43 G > A, ELL3 c.366 A > G, and ELL3 c.967 A > G variants. b, Sanger sequencing showing the transcripts from the ELL3 c.133-43 G > A variant contained intron 1 (top left panel) and the transcripts from the ELL3 c.366 A > G variant contained intron 4 (top right panel), with both variations ultimately causing premature translational termination of ELL3. One type of the transcripts from the ELL3 c.967 A > G variant contained intron 8, leading to premature translational termination of ELL3 (bottom left panel), and another type of the transcripts from this variant skipped exon 10 (bottom right panel). c, Western analysis showing the protein levels of the ELL3 c.−32C > A and ELL3 c.*124 T > C variants were lower than WT control, respectively. Cycloheximide (CHX) chase assays showing the ELL3 D150H protein had a shorter half-life. d, The ELL3 c.649 C > T and ELL3 c.753 G > C variations caused a single mutation in the conserved amino acids. e, Representative images of RFP-hELL3 and RFP-hELL3 R217C localization in HeLa cells. Red, RFP; green, microtubules; blue, DNA. Scale bar, 10 μm.
Extended Data Fig. 2 Sanger sequencing and STR profiles of the 8 affected families.
a, Sanger sequencing showing the ELL3 vairations in the POC, father and mother from eight miscarriage families. Arrows indicating the positions of the variations. b, Capillary electrophoregrams illustrating the variability of STR markers across the 8 families. In the electrophoregrams, the bolded capitalized numbers/letters positioned above the peaks denoting the STR locus. X-axis representing the base pair length of the PCR products, whereas the y-axis corresponding to fluorescence intensity of these PCR products. Additionally, Ht signifying the peak height value, which aids in computing the peak height ratio for two alleles.
Extended Data Fig. 3 ELL3 deficieny leads to spindle abnormality in human oocytes.
a, Western blot showing the specificity of the ELL3 polyclonal antibody against the N-terminal region of ELL3 (pAb) using WT, ELL3 KO and ELL3 KO HCT 116 cells with GFP-ELL3 expression. Red asterisks pointing to the distinct bands corresponding to ELL3 or GFP-ELL3. Throughout this study, unless otherwise explicitly stated, this ELL3 pAb was used for IF, Western blot and IP experiments. b, Schematic diagram of ELL3 TRIM-Away assay in human oocytes. c, Western blot showing the specificity of the ELL3 monoclonal antibody (mAb) using WT, ELL3 KO and ELL3 KO HCT 116 cells with GFP-ELL3 expression. Red asterisks pointing to the distinct bands corresponding to ELL3 or GFP-ELL3. The ELL3 mAb was used in TRIM-Away experiments. d, Representative images showing ELL3 TRIM-Away efficiency, assayed by the validated ELL3-pAb, and spindles and chromosomes in control and ELL3 TRIM-Away human oocytes. Green, microtubules; blue, DNA. Scale bar, 5 μm. e, Spindle abnormality rate in control (n = 27) and ELL3 TRIM-Away (n = 29) human oocytes. f, Immunostaining showing ELL3 KD efficiency in human oocytes. Scale bar, 5 μm. g, PB1 extrusion rate in control (n = 24) and ELL3 KD (n = 27) human oocytes. h, Representative images showing chromosome spreading of control, ELL3, ELL3 D150H, ELL3 R217C, ELL3 E251D, ELL3 T323A, and ELL3 T323 overexpressing mouse MII oocytes. The number x/y marked on the lower right corner of each picture indicating that x oocytes are aneuploid among the y examined oocytes. i, PB1 extrusion rate in control (n = 54), ELL3 (n = 56), ELL3 D150H (n = 84), ELL3 R217C (n = 72), ELL3 E251D (n = 70), ELL3 T323A (n = 74), and ELL3 T323 (n = 73) overexpressing mouse oocytes. j, Representative images showing spindle morphologies in control, ELL3 WT, ELL3 R217C and ELL3 E251D overexpressing mouse oocytes. Scale bar, 10 μm. Data are presented as mean ± s.e.m. (g,i). Data are from three independent experiments (e,g,i). N indicated the cumulative number of independent oocytes and independent experiments are denoted by dots (g,i). Statistical analysis was performed using a two-tailed, unpaired Student’s t-test (g,i) or a two-sided, fisher’s exact test (e). ns, not significant.
Extended Data Fig. 4 ELL3 is localized at the meiotic spindle.
a, Representative images of ELL3-RFP localization in mouse oocytes at GV, GVBD, MI, and MII stages. Red, RFP; green, microtubules; blue, DNA. Scale bar, 20 μm. b, Western blot showing the specificity of the ELL3 polyclonal antibody against the C-terminal region of ELL3 (pAb-CT) using WT, ELL3 KO and ELL3 KO HCT 116 cells with GFP-ELL3 expression. Red asterisks pointing to the distinct bands corresponding to ELL3 or GFP-ELL3. c, Mouse oocytes were immunostained with the validated ELL3 pAb-CT. Green, ELL3; blue, DNA. Scale bar, 20 μm. d, Immunostaining showing Ell3 KD efficiency in mouse oocytes. Scale bar, 20 μm. e, ELL3 intensity in control (n = 6) and Ell3 KD (n = 6) mouse oocytes. f, Representative images of meiotic resumption in control and Ell3 KD mouse oocytes at GVBD stage. Scale bar, 100 μm. g, GVBD rate of control (n = 92) and Ell3 KD (n = 145) mouse oocytes. Data are presented as mean ± s.e.m. (e,g). Data are from more than three independent experiments (e,g). N indicated the cumulative number of independent oocytes and independent experiments are denoted by dots (g). Statistical analysis was performed using a two-tailed, unpaired Student’s t-test (e,g). ns, not significant.
Extended Data Fig. 5 Ell3 KD affects spindle assembly.
a, 3 h after GVBD, control and Ell3 KD mouse oocytes were immunostained with anti-BubR1 and anti-centromere (CREST) antibodies. Scale bar, 10 μm. b, BubR1 intensity in the oocytes depicted in a, for both the control (n = 24) and Ell3 KD (n = 23) groups. c, Control and Ell3 KD mouse oocytes at the MetI stage were immunostained with anti-BubR1 and CREST antibodies. Scale bar, 10 μm. d, BubR1 positive percentage of the oocytes depicted in c, for both the control (n = 19) and Ell3 KD (n = 23) groups. e, Representative images showing spindles and chromosomes in control, Ell3 KD and human ELL3-rescued mouse oocytes. Green, microtubules; blue, DNA. Scale bar, 10 μm. f, MI spindle abnormality rate in control (n = 131), Ell3 KD (n = 150) and human ELL3-rescued (n = 33) mouse oocytes. g, Schematic diagram of ELL3 TRIM-Away assay in mouse oocytes. h, Representative images showing ELL3 TRIM-Away efficiency, assayed by the validated ELL3-pAb, and and spindles and chromosomes in control and ELL3 TRIM-Away mouse oocytes. Scale bar, 20 μm. i, Percentage of abnormal MI spindles in control (n = 24) and ELL3 TRIM-Away (n = 31) mouse oocytes. j, Percentage of different types of spindle defects in ELL3 TRIM-Away (n = 31) mouse oocytes. Type I-short; type II-aMTOC clustering defects; type III-pole defocus; type IV-elongated; type V-chaos. k, Representative images of MII spindle morphologies in control and Ell3 KD mouse oocytes. Green, microtubules; blue, DNA. Scale bar, 20 μm. l, Percentage of abnormal MII spindles in control (n = 52) and Ell3 KD (n = 40) mouse oocytes. m, Representative images of ELL3-RFP localization at MI stage after cold-treatment. Red, RFP; green, microtubules; blue, DNA. Scale bar, 20 μm. Data are presented as mean ± s.e.m. (b,d,f,l). Data are from three independent experiments (b,d,f,i,l). N indicated the cumulative number of independent oocytes and independent experiments are denoted by dots (d,f,l). Statistical analysis was performed using a two-tailed, unpaired Student’s t-test (b,d,f,l) or a two-sided, fisher’s exact test (i). ns, not significant.
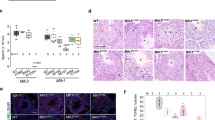
Extended Data Fig. 6 ELL3 deficiency in mouse oocytes.
a, Diagram depicting Ell3 oocyte-specific knockout strategy. Ell3fl/fl mice were crossed with mice expressing CRE recombinase under the control of the Gdf9 promoter (Gdf9-Cre) to generate oocyte-specific Ell3 KO mice. b, Genotyping analysis of Ell3 GcKO mice with PCR. c, Representative images of ovaries from the 4-week Ell3fl/fl and Ell3 GcKO mice. Scale bar, 1 mm. d, HE stainning of ovarian sections from the 4-week and 8-week Ell3fl/fl and Ell3 GcKO mice. Scale bar, 100 μm. e, Number of follicles at different developmental stages per ovary from the 4-week and 8-week Ell3fl/fl and Ell3 GcKO mice. Pmf, primordial follicle; PF, primary follicle; SF, secondary follicle; PAF, pre-antral follicle; AF, antral follicle. f, Representative images showing the meiotic spindles in Ell3fl/fl and the heterzygous Ell3fl/+;Gdf9-Cre oocytes immunostained with anti-ELL3 and anti-α-TUBULIN antibodies. Scale bar, 10 μm. The efficiency of GDF9-CRE driven Ell3 KO varied, with some Ell3fl/+;Gdf9-Cre oocytes exhibiting no reduction in ELL3 protein levels compared to Ell3fl/fl controls, while others demonstrating a noticeable decrease. g, ELL3 intensity in Ell3fl/+ (n = 23), Ell3fl/+;Gdf9-Cre (low KO efficiency, n = 17), and Ell3fl/+;Gdf9-Cre (strong KO efficiency, n = 10) oocytes. h, Percentage of normal and abnormal spindles in Ell3fl/+ (n = 23), Ell3fl/+;Gdf9-Cre (low KO efficiency, n = 17), and Ell3fl/+;Gdf9-Cre (strong KO efficiency, n = 10) oocytes. i, Numbers of the super-ovulated oocytes per mouse from Ell3fl/fl (n = 7) and Ell3 GcKO (n = 4) mice, respectively. Data are presented as mean ± s.e.m. (e,g,i). Data are from three (e,g,h) or more than three (i) independent experiments. Statistical analysis was performed using a two-tailed, unpaired Student’s t-test (e,g,i). ns, not significant.
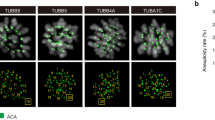
Extended Data Fig. 7 ELL3 deficiency in mouse oocytes causes aneuploidy.
a, Representative images showing chromosome spread for Ell3fl/fl♀× WT♂ and Ell3 GcKO♀× WT♂ blastocyst cells. Scale bar, 20 μm. b, Percentage of aneuploid cells in Ell3fl/fl♀× WT♂ (n = 32 cells) and Ell3 GcKO♀× WT♂ (n = 41 cells) blastocyst cells. Number of chromosomes per cell were assessed by chromosome spread. Ell3fl/fl♀× WT♂ aneuploid cells were 5 and Ell3 GcKO♀× WT♂aneuploid cells were 20. c, Representative images of blastocysts from Ell3fl/fl♀× WT♂ and Ell3 GcKO♀× WT♂ groups. Scale bar, 100 μm. d, Blastocyst rate of Ell3fl/fl♀× WT♂ (n = 47 cells) and Ell3 GcKO♀× WT♂ (n = 55 cells). e, Implantation sites marked by Chicago Sky Blue 6B in Ell3fl/fl and Ell3 GcKO mice. Scale bar, 5 mm. f, E10.0 embryos recovered from a single litter of control and Ell3 GcKO mice. Scale bar, 1 mm. Data are presented as mean ± s.e.m. (d). Data are from three independent experiments (b,d). N indicated the cumulative number of independent oocytes and independent experiments are denoted by dots (d). Statistical analysis was performed using a two-tailed, unpaired Student’s t-test (d) or a two-sided, fisher’s exact test (b). ns, not significant.
Extended Data Fig. 8 Validation of the interplay among ELL3, KIF11, and TPX2.
a, MA plot showing differentially expressed genes after Ell3 KD in mouse oocytes. Significantly up-regulated genes (n = 290) are shown in red; significantly down-regulated genes (n = 457) are shown in blue. b, Representative images of ELL3 localization in HeLa cells during cell cycle. Red, ELL3; green, microtubules; blue, DNA. Scale bar, 10 μm. c, Representative images of ELL3 localization in HEK-293 cells during cell cycle. HEK-293: Magenta, ELL3; green, microtubules; grey, DNA. Scale bar, 10 μm. d, Immunoprecipitation assay showing the binding between ELL3 and KIF11 was not largely affected by the R217C or E251D mutation. e, Kif11 KD efficiency in mouse oocytes with Kif11 siRNAs compared to control, shown by western blot. f, SDS-PAGE followed by silver staining showing the purified KIF11 and TPX2. Arrows pointing to the target protein bands. g, SDS-PAGE followed by coomassie staining showing the purified recombinant ELL3-mCherry. Arrow pointing to the target protein band. Asterisk pointing to the degraded band. h, Western blot indicating that the band shown in e can be recognized by specific ELL3 antibody. Arrow pointing to the target protein band. Asterisk pointing to the degraded band. i, Tpx2 KD efficiency in mouse oocytes (n = 150) with Tpx2 siRNA compared to control (n = 150), shown by western blot. j, Representative images showing localization of TPX2 and TUBULIN at the spindle in control and Ell3 KD mouse oocytes. Scale bar, 20 μm. k, TPX2/TUBULIN intensity ratio in control (n = 11) and Ell3 KD (n = 15) mouse oocytes. Box centres indicate median, boundaries represent 25th and 75th percentiles, and bars represent maximum and minimum values. Data are from three independent experiments (k). Statistical analysis was performed using a two-tailed, unpaired Student’s t-test (k). ns, not significant.
Extended Data Fig. 9 ELL3 promotes spindle assembly and chromosome movement.
a, Length of the spindle in oocytes used for live imaging assays (n = 17) and fixed IF assays (n = 31) was determined. b, Time-lapse 3D light-sheet microscopy imaging of control or Ell3 KD mouse oocytes expressing α-TUBULIN-eGFP and H2B-mCherry. Time, h:min from GVBD. Z projection, 1 μm. Scale bar, 20 μm. c, Quantitative analyses of spindle assembly stages across the indicated time points from data sets shown in a. d, Length of the spindle in control (n = 6) and Ell3 KD (n = 5) oocytes was determined from 3D reconstructed images. e, Paths of all single chromosome from GVBD to anaphase onset in control and Ell3 KD mouse oocytes. f, Distance to the equator plane across MI. Each colored curve matches one chromosome. g, Speeds of chromosome movement in three different Ell3 KD mouse oocytes. Data are presented as mean ± s.d. (a,d) or mean with shade represents 95 confidence interval (g). Data are from three independent experiments (a,c,d). Statistical analysis was performed using a two-tailed, unpaired Student’s t-test (a). ns, not significant.
Extended Data Fig. 10 ELL3 and TPX2 safeguard proper spindle assembly through balancing KIF11.
a, Time-lapse 3D light-sheet microscopy imaging of control and Kif11 KD mouse oocytes expressing H2B-mCherry. Scale bar, 20 μm. b, Left: Path analysis after Kif11 KD, showing the distance to the equator plane across MI. Each colored curve matches one chromosome. Right: Speed of chromosome movement in Kif11 KD mouse oocyte. c, Time-lapse 3D light-sheet microscopy imaging of DMSO and monastrol treated mouse oocytes expressing H2B-mCherry. Scale bar, 20 μm. d, Left: Path analysis of the monastrol treated mouse oocyte, showing the distance to the equator plane across MI. Each colored curve matches one chromosome. Right: Speed of chromosome movement in monastrol treated oocyte. e, Western blot showing TPX2 expression in control (n = 150), Tpx2 KD (n = 150) and Tpx2 KD + Tpx2ΔKID (n = 150) mouse oocytes. f, Representative images of 3D reconstructed spindle in control, Tpx2 KD + Tpx2ΔKID and Tpx2 KD + Tpx2ΔKID + Ell3 KD mouse oocytes at different stages. Oocytes at about 2 h after GVBD (early), about 4 h after GVBD (middle) and about 6 h after GVBD (late) are shown. Scale bar, 20 μm. g, Distance to the equator plane across MI. Each colored curve matches one chromosome. Data are presented as mean with shade represents 95 confidence interval (b,d).
Supplementary information
Supplementary Tables
Supplementary Tables 1–5.
Supplementary Video 1
WT oocyte.
Supplementary Video 2
Control oocyte.
Supplementary Video 3
Ell3 KD oocyte.
Supplementary Video 4
TPX2ΔKID oocyte.
Supplementary Video 5
TPX2ΔKID+Ell3 KD oocyte.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Unprocessed western blots.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Unprocessed western blots.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhu, S., Xie, P., Yang, Y. et al. Maternal ELL3 loss-of-function leads to oocyte aneuploidy and early miscarriage. Nat Struct Mol Biol 32, 381–392 (2025). https://doi.org/10.1038/s41594-024-01471-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41594-024-01471-8
This article is cited by
-
ELL3 regulates spindle assembly to prevent maternally inherited aneuploidy and infertility
Nature Structural & Molecular Biology (2025)