Abstract
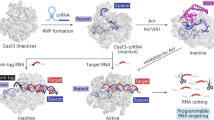
Cas13 is an RNA-guided RNA endonuclease derived from the type VI CRISPR–Cas system, which has been used in numerous RNA-targeting technologies, such as RNA knockdown, detection and editing. The catalytically inactive Prevotella sp. Cas13b (dPspCas13b) fused to the human adenosine deaminase acting on RNA 2 (ADAR2) deaminase domain can edit adenosine in target transcripts to inosine, in an RNA-editing technology called REPAIR (RNA editing for programmable A-to-I replacement), which has potential for gene therapy. Here we report the cryo-electron microscopy structures of the PspCas13b–guide RNA binary complex, the PspCas13b–guide RNA–target RNA ternary complex and the dPspCas13b–ADAR2–guide RNA–target RNA complex. These structures provide mechanistic insights into RNA cleavage and editing. We applied our structural insights to engineer a compact and efficient dPspCas13b–ADAR2 complex (REPAIR-mini). Overall, our findings advance the understanding of CRISPR–Cas13 effector nucleases and could enable the development of improved RNA-targeting technologies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The structural models were deposited to the PDB under the accession codes 8WF8 (binary complex), 8WF9 (state 1 ternary complex), 8WFA (state 2 ternary complex) and 8WFB (REPAIR complex). The raw images were deposited to the EM Data Bank under accession codes EMD-37486 (binary complex), EMD-37487 (state 1 ternary complex), EMD-37488 (state 2 ternary complex) and EMD-37489 (REPAIR complex). Source data are provided with this paper.
References
Hille, F. et al. The biology of CRISPR–Cas: backward and forward. Cell 172, 1239–1259 (2018).
Makarova, K. S. et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 18, 67–83 (2020).
Yang, H. & Patel, D. J. Structures, mechanisms and applications of RNA-centric CRISPR–Cas13. Nat. Chem. Biol. 20, 673–688 (2024).
Abudayyeh, O. O. et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573 (2016).
Shmakov, S. et al. Diversity and evolution of class 2 CRISPR–Cas systems. Nat. Rev. Microbiol. 15, 169–182 (2017).
Smargon, A. A. et al. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol. Cell 65, 618–630 (2017).
Konermann, S. et al. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 173, 665–676 (2018).
Yan, W. X. et al. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol. Cell 70, 327–339 (2018).
Xu, C. et al. Programmable RNA editing with compact CRISPR–Cas13 systems from uncultivated microbes. Nat. Methods 18, 499–506 (2021).
Kannan, S. et al. Compact RNA editors with small Cas13 proteins. Nat. Biotechnol. 40, 194–197 (2022).
Gootenberg, J. S. et al. Nucleic acid detection with CRISPR–Cas13a/C2c2. Science 356, 438–442 (2017).
Abudayyeh, O. O. et al. RNA targeting with CRISPR–Cas13. Nature 550, 280–284 (2017).
Cox, D. B. T. et al. RNA editing with CRISPR–Cas13. Science 358, 1019–1027 (2017).
Gootenberg, J. S. et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360, 439–444 (2018).
Ai, Y., Liang, D. & Wilusz, J. E. CRISPR/Cas13 effectors have differing extents of off-target effects that limit their utility in eukaryotic cells. Nucleic Acids Res. 50, e65 (2022).
Hu, W. et al. Single-base tiled screen unveils design principles of PspCas13b for potent and off-target-free RNA silencing. Nat. Struct. Mol. Biol. 31, 1702–1716 (2024).
Liu, L. et al. Two distant catalytic sites are responsible for C2c2 RNase activities. Cell 168, 121–134 (2017).
Liu, L. et al. The molecular architecture for RNA-guided RNA cleavage by Cas13a. Cell 170, 714–726 (2017).
Knott, G. J. et al. Guide-bound structures of an RNA-targeting A-cleaving CRISPR–Cas13a enzyme. Nat. Struct. Mol. Biol. 24, 825–833 (2017).
Zhang, C. et al. Structural basis for the RNA-guided ribonuclease activity of CRISPR–Cas13d. Cell 175, 212–223 (2018).
Zhang, B. et al. Structural insights into Cas13b-guided CRISPR RNA maturation and recognition. Cell Res. 28, 1198–1201 (2018).
Slaymaker, I. M. et al. High-resolution structure of Cas13b and biochemical characterization of RNA targeting and cleavage. Cell Rep. 26, 3741–3751 (2019).
Nakagawa, R. et al. Structure and engineering of the minimal type VI CRISPR–Cas13bt3. Mol. Cell 82, 3178–3192 (2022).
Deng, X. et al. Structural basis for the activation of a compact CRISPR–Cas13 nuclease. Nat. Commun. 14, 5845 (2023).
Matthews, M. M. et al. Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity. Nat. Struct. Mol. Biol. 23, 426–433 (2016).
Wu, Z., Yang, H. & Colosi, P. Effect of genome size on AAV vector packaging. Mol. Ther. 18, 80–86 (2010).
Bot, J. F., van der Oost, J. & Geijsen, N. The double life of CRISPR–Cas13. Curr. Opin. Biotechnol. 78, 102789 (2022).
Tong, H. et al. High-fidelity Cas13 variants for targeted RNA degradation with minimal collateral effects. Nat. Biotechnol. 41, 108–119 (2023).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A.cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Bepler, T. et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Tan, Y. Z. et al. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796 (2017).
Burnley, T., Palmer, C. M. & Winn, M. Recent developments in the CCP-EM software suite. Acta Crystallogr. D Struct. Biol. 73, 469–477 (2017).
Hoh, S. W., Burnley, T. & Cowtan, K. Current approaches for automated model building into cryo-EM maps using Buccaneer with CCP-EM. Acta Crystallogr. D Struct. Biol. 76, 531–541 (2020).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in PHENIX. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018).
Yamashita, K., Palmer, C. M., Burnley, T. & Murshudov, G. N. Cryo-EM single-particle structure refinement and map calculation using Servalcat. Acta Crystallogr. D Struct. Biol. 77, 1282–1291 (2021).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Joung, J. et al. Genome-scale CRISPR–Cas9 knockout and transcriptional activation screening. Nat. Protoc. 12, 828–863 (2017).
Acknowledgements
We acknowledge the staff scientists at the University of Tokyo’s cryo-EM facility, particularly Y. Sakamaki, for their assistance with cryo-EM data collection. J.I. is supported by Japan Society for the Promotion of Science (JSPS) KAKENHI grant number 23KJ0720. F.Z. is support by National Institutes of Health grants (1R01-HG009761, 1R01-MH110049 and 1DP1-HL141201), the Howard Hughes Medical Institute, Open Philanthropy, the G. Harold and Leila Y. Mathers Charitable Foundation, the Bill and Melinda Gates Foundation, the Edward Mallinckrodt, Jr. Foundation, the Poitras Center for Psychiatric Disorders Research at the Massachusetts Institute of Technology (MIT), the Hock E. Tan and K. Lisa Yang Center for Autism Research at MIT and the Yang-Tan Center for Molecular Therapeutics at the MIT McGovern Institute, as well as by L. Yang, the Phillips family and J. and P. Poitras. H.N. is supported by the Platform Project for Supporting Drug Discovery and Life Science Research from the Agency for Medical Research and Development under grant numbers JP21am0101115 (support number 2792) and JP19am0401005, JSPS KAKENHI grant numbers 21H05281 and 22H00403, the Takeda Medical Research Foundation, the Inamori Research Institute for Science and Japan Science and Technology Agency CREST grant number JPMJCR19H5.
Author information
Authors and Affiliations
Contributions
J.I. performed the sample preparation with assistance from K.K., S.O. and H.N. J.I., K.K. and H.N. performed the structural analysis with assistance from T.N. and K.Y. J.I., S.K., S.I., N.Y. and F.Z. performed the cellular experiments. J.I. and H.N. wrote the paper with help from all authors. H.N. supervised the research.
Corresponding author
Ethics declarations
Competing interests
F.Z. is a cofounder of Editas Medicine, Beam Therapeutics, Pairwise Plants, Arbor Biotechnologies and Sherlock Biosciences. The other authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Hong Li and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editors: Sara Osman and Dimitris Typas, in collaboration with the Nature Structural & Molecular Biology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Multiple sequence alignment of PspCas13b, PbuCas13b, and Cas13bt3.
The figure was prepared using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo) and ESPript3 (http://espript.ibcp.fr/ESPript/ESPript). Key residues are indicated by triangles.
Extended Data Fig. 2 Cryo-EM analysis of the PspCas13b–crRNA binary complex.
(a) Single-particle cryo-EM image processing workflow. (b) Representative micrograph at a magnification of ×105,000. (c) Representative 2D averaged class images. (d) FSC curves. The map-to-map FSC curve was calculated between the two independently refined half-maps after masking (blue line), and the overall resolution was determined by the gold standard FSC = 0.143 criterion. The map-to-model FSC curve was calculated between the refined atomic models and maps (red line). (e) Directional FSC plots calculated by the 3DFSC server. (f) Angular distribution of particles in the final reconstruction. (g and h) Cryo-EM density maps, colored according to the local resolution (g) and the structural domains (h).
Extended Data Fig. 3 Structural comparison of PspCas13b with PbuCas13b and Cas13bt3.
(a) HEPN active sites of the PspCas13b binary complex. The catalytic residues are shown as space-filling or stick models. (b and c) Ribbon (b) and surface (c) representations of the binary complexes of PspCas13b (left), PbuCas13b22 (PDB: 6DTD) (middle), and Cas13bt323 (PDB: 7VTI) (right). (d) Sequences and structures of the crRNAs of PspCas13b (left), PbuCas13b (PDB: 6DTD) (middle), and Cas13bt3 (PDB: 7VTI) (right).
Extended Data Fig. 4 crRNA recognition.
(a–d) Recognition of the crRNA (a), U4 and C5 (b), the possible seed sequence (c), and the 3ʹ nucleotide (d). Hydrogen bonds are depicted with green dashed lines. (e) SDS–PAGE analysis of the purified PspCas13b and its mutants. The proteins were visualized with CBB. All experiments were performed at least three times. (f) In vitro pre-crRNA processing experiments. The pre-crRNA was incubated with the purified WT PspCas13b or its mutants at 37 °C for 30 min, and then analyzed by denaturing urea–PAGE. The nucleic acids were visualized with SYBR Gold. All experiments were performed at least three times.
Extended Data Fig. 5 Schematics of RNA recognition.
Disordered regions are enclosed by dashed gray lines.
Extended Data Fig. 6 Cryo-EM analysis of the PspCas13b–crRNA–tgRNA ternary complex.
(a) Single-particle cryo-EM image processing workflow. (b) Representative micrograph at a magnification of ×105,000. (c) Representative 2D averaged class images. (d and e) Cryo-EM density maps of State 1 (d) and State 2 (e), colored according to the local resolution. (f and i) FSC curves of State 1 (f) and State 2 (i). The map-to-map FSC curves were calculated between the two independently refined half-maps after masking (blue line), and the overall resolutions were determined by the gold standard FSC = 0.143 criterion. The map-to-model FSC curves were calculated between the refined atomic models and maps (red line). (g and j) Directional FSC plots of State 1 (g) and State 2 (j), calculated by the 3DFSC server. (h and k) Angular distributions of the particles in the final reconstructions of State 1 (h) and State 2 (k). (l and m) Cryo-EM density maps of State 1 (l) and State 2 (m), colored according to the structural domains.
Extended Data Fig. 7 Structural comparison of the PspCas13b binary and ternary complexes.
(a) Surface representations of the PspCas13b binary complex (left) and ternary complexes in State 1 (middle) and State 2 (right). The active sites are indicated by dashed red lines. (b) Recognition of the guide–target duplex in the PspCas13b ternary complexes in States 1 (left) and State 2 (right). (c) Superimposition of the PspCas13b State 2 ternary complex (colored as in a) and the HEPN domains of the binary complex (colored blue), based on their HEPN1 domains. A potential steric clash between the State 2 ternary complex and the binary complex is indicated by a dashed orange circle. (d–f) Lid loop structures in the binary complex (d) and the ternary complexes in State 1 (e) and State 2 (f).
Extended Data Fig. 8 Cryo-EM analysis of the REPAIR complex.
(a) Domain structure of human ADAR2. dsRBD, dsRNA-binding domain. (b) Nucleotide sequence of the modified dsRNA used for the crystallographic analysis25. (c) Crystal structure of the ADAR2DD–dsRNA complex (PDB: 5ED1)25. Inositol hexaphosphate (IHP) is bound to the enzyme core and stabilizes the protein fold. The 8-azanebularine (8-AZ) is flipped out from the RNA duplex and recognized by the ADAR2DD active site. (d) Single-particle cryo-EM image processing workflow. (e) Representative micrograph at a magnification of ×105,000. (f) Representative 2D averaged class images. (g) FSC curves. The map-to-map FSC curve was calculated between the two independently refined half-maps after masking (blue line), and the overall resolution was determined by the gold standard FSC = 0.143 criterion. The map-to-model FSC was calculated between the refined atomic models and maps (red line). (h) Directional FSC plots calculated by the 3DFSC server. (i) Angular distribution of the particles in the final reconstruction. (j) Cryo-EM density maps, colored according to the local resolution. (k) Fitting of the ADAR2 domain to the cryo-EM density map. A close-up view of the density map around 8-AZ and IHP is shown on the right.
Extended Data Fig. 9 In vitro and in vivo RNA editing activities of the REPAIR variants.
(a) SDS–PAGE analysis of dPspCas13b–ADAR2DD and its variants. The proteins were visualized with CBB. All experiments were performed at least three times. (b) Western blot showing the expression levels of the WT dPspCas13b–ADAR2DD and the V2 variant in human cells. The two proteins were detected using an anti-FLAG antibody. The samples were normalized based on β-actin expression levels. All experiments were performed at least three times. (c) Design of the V4 dPspCas13b–ADAR2DD variant and crRNAs with varying guide lengths (GL) and mismatch distances (MD). (d) Preferences of the WT dPspCas13b–ADAR2DD and the V4 variant for editing positions. RNA editing activities were examined in human cells, using different crRNAs targeting the exogenous Cluc (W85X) transcript, the endogenous STAT1 (Y701C) transcript, or the endogenous CTNNB1 (T41A) transcript. Editing rates for A nucleotides in the target transcripts are displayed. Data are shown as mean ± S.D. (n = 4 biological replicates). The target A positions and the complementary guide regions are outlined in red and black, respectively. T, targeting guide; NT, nontargeting guide.
Supplementary information
Supplementary Information
Supplementary Fig. 1 and Tables 1 and 2.
Supplementary Video 1
Structural changes from the PspCas13b binary complex to the PspCas13b ternary complex.
Supplementary Video 2
Conformational flexibility of the guide–target duplex in the REPAIR complex.
Source data
Source Data Fig. 6 and Extended Data Fig. 9
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ishikawa, J., Kato, K., Kannan, S. et al. Structural insights into RNA-guided RNA editing by the Cas13b–ADAR2 complex. Nat Struct Mol Biol 32, 1567–1578 (2025). https://doi.org/10.1038/s41594-025-01529-1
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41594-025-01529-1