Abstract
Regulating the occurrence and progression of tumor cells at the epigenetic level is a new insight of clear cell renal cell carcinoma (ccRCC). Chromatin accessibility is an important pathway of epigenetic regulation, which may explain the mystery of tumor occurrence. Assay for transposase-accessible chromatin sequencing (ATAC-seq) provides insight into the epigenetic regulatory features of ccRCC, especially at the single-cell level. In this study, we performed scATAC-seq of 3 ccRCC samples and captured a total of 18,703 high-quality cell nuclei and 104,818 unique peaks. Our protocol for nuclear extraction was reliable and stable, which can be used to deal with fresh and frozen single-cell suspensions. We presented basic methods for scATAC-seq data analysis, such as cell clustering, gene activity scoring, cell subtype specific peaks, transcription factors, motif and motif footprinting analysis. Taken together, our data indicated the valuable epigenetic features of ccRCC, which will provide more references for the study of ccRCC.
Similar content being viewed by others
Background & Summary
Kidney cancer is one of the most common malignancies of the urinary system, second only to prostate cancer and bladder cancer, with 431,288 new cases worldwide in 20201. Clear cell renal cell carcinoma (ccRCC) is the most common type of histology, accounting for about 75% of cases2. The genetic characteristics of ccRCC are associated with potentially high levels of tumour heterogeneity and genetic susceptibility3,4. The previous TCGA studies5 have found a wide range of genetic alterations in ccRCC, including VHL, PBRM1, and SETD2 mutations, which reveals the comprehensive features of ccRCC. In addition, the epigenetic regulation of tumour cells is a new mode to drive tumour occurrence and progression6. The organization of accessible chromatin across the genome plays an essential role in establishing and maintaining cellular identity, reflecting a network of epigenetic regulation through which enhancers, promoters, insulators and chromatin-binding factors cooperatively regulate gene expression7. In particular, alterations in chromatin accessibility have been implicated in driving cancer initiation, progression and metastasis8.
In the past, there have been many DNA sequencing-based methods for mapping nucleosomes and chromatin accessibility, such as ATAC-seq9, DNase-seq10 and MPE-seq11. In recent years, with the development of single-cell sequencing technology, it has become possible to perform single-cell assay for transposase-accessible chromatin sequencing (scATAC-seq)12, especially the high-throughput single-cell analysis13. Although the characteristics of chromatin accessibility on ccRCC have been reported in a previous study14, the large number of cells (a total of 61,693 nuclei and 190,916 unique peaks) provided by the study is difficult to reutilize and reanalyze, which depends on high speed calculator. Another study was limited to chromatin accessibility features of immune cells, while did not focus on tumor cells15.
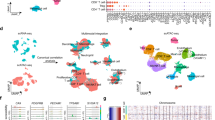
To address this problem, we provide a high-quality chromatin accessibility data for ccRCC at single-cell level. Performing high-throughput scATAC-seq on 3 human ccRCC samples (RCC30, RCC61 and RCC76), we obtained a total of 18,703 high quality nuclei and 104,818 unique peaks, including coding, non-coding regions, promoters and enhancers (Fig. 1). After quality control (QC) and downstream analysis, our data may reveal comprehensive epigenetic characteristics of ccRCC, including tumour cells, endothelial cells (EC), cancer-associated fibroblasts (CAF) and immune cells. In addition, we demonstrate a complete analysis process for scATAC-seq data, and makes the application more feasible and convenient. Taken together, the scATAC-seq data of ccRCC can provide valuable information and new strategies for study and treatment of ccRCC in the future.
Methods
Human ccRCC samples
These three participants (RCC30, RCC61 and RCC76) were diagnosed with renal tumor before surgery and underwent laparoscopic radical nephrectomy in The First Affiliated Hospital of Guangxi Medical University (Table 1). After detailed communications with the three patients, they voluntarily donated the tumor tissues. The postoperative pathological results of all samples indicated ccRCC. This study was approved by the Institution Review Board (IRB) from the First Affiliated Hospital of Guangxi Medical University (No. Expedited trial 2018003), which agreed to conduct the study and share the data. And all participants signed informed consent forms and agreed to share the data.
Single nuclei preparation
Here, we need to perform this in two steps. Firstly, we should prepare ccRCC single-cell suspension. By means of multi-point sampling, we can obtain representative tumour tissues, and totally select 1 cm3 tissues. Washing by DPBS (WISENT, 311-425-CL) twice, the tissue specimens were digested by a solution (1 mg/mL collagenase I (Gibco, 5401020001) and 1 mg/mL DNaseI (Roche, 10104159001) in HBSS) for 30 min at 37 °C. We terminated the digestion by DMEM (WISENT, 319-006-CL) with 10% FBS (Gibco, 10099141). 70 μm cell strainer (Falcon) can be used for filtering out large tissue fragments, which did not fully digeste into a single-cell state by the above digestive process. Then, we removed red blood cells by RBC lysis buffer (10X diluted to 1X; BioLegend, 420301) for 5 min on ice and filtered by 40 μm cell strainer (Falcon) again. At last, we obtained single-cell suspension that can be counted by trypan blue (Gibco, 15250-061) staining. In this study, RCC61 and RCC76 were isolated from single-cell nuclei, immediately. And RCC30 was frozen in liquid nitrogen for a period of time and then isolated from single-cell nuclei.
The second step for isolating single-cell nuclei can refer to our previous study14. Briefly, we need to prepare the lysis buffer (10 mM Tris-HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2, 0.1% Tween-20, 0.1% Nonidet P40 Substitute, 0.01% digitonin and 1% BSA) and wash buffer (10 mM Tris-HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2, 0.1% Tween-20 and 1% BSA). The lysis buffer (100 µl) was incubated with single-cells on ice for 4 different time gradients (3 min, 3.5 min, 4 min and 4.5 min), because cells were sensitive to the duration of lysis buffer. Then, we added the wash buffer (1 ml) for terminating the lysis process. The samples were centrifuged at 500 g for 5 min at 4 °C and removed the supernatant. Single-cell nuclei were resuspended by PBS and examined the quality of each gradients by microscopy. Finally, we selected the optimal samples that were resuspended in chilled 1x Nuclei Buffer (10x Genomics, 2000153) at approximately 5,000–7,000 nuclei/μL.
DNA library construction and preliminary sequencing results
The number of target nuclei captured in each sample was 7,000. The DNA library construction for scATAC can be refer to 10X Genomics ‘Chromium Single Cell ATAC Reagent Kits User Guide’ (https://support.10xgenomics.com/single-cell-atac/library-prep/doc/technical-note-chromium-next-gem-single-cell-atac-v11-reagent-workflow-and-software-updates). Then, DNA libraries were sequenced by Novaseq6000 (Illumina, San Diego, CA). The sequencing parameters can be set to the 2 × 50 paired-end. Preliminary sequencing files (.bcl) were converted to FASTQ files by CellRanger ATAC (version 1.2.0, https://support.10xgenomics.com/single-cell-atac/software/pipelines/latest/what-is-cell-ranger-atac). After running cellranger-atac mkfastq options, the read1, barcode, read2, and sample index were associated with R1, R2, R3, I1, respectively (Table 1). Then FASTQ files were compared with the human genome reference sequence GRCh37 by the cellranger-atac count function. Finally, the summary results were generated in a file, which we can indicate an overview of the scATAC-seq data (Table 2).
Secondly analysis for scATAC-seq data
Here, we demonstrated the use of Seurat16,17 (version 4.0.0) and Signac18 (version 1.0.0) R packages for data downstream analysis. Before quality control (QC), we had to understand these important parameters, such as transcriptional start site (TSS) enrichment score, nucleosome signal (NS), number of fragments in peaks, fraction of fragments in peaks (pct reads in peaks) and ratio reads in genomic blacklist regions (Fig. 2a–c, Table 2). According to the parameters (peak region fragments > 1000 & peak region fragments < 20000 & pct reads in peaks > 15 & blacklist ratio < 0.05 & nucleosome signal < 4 & TSS enrichment > 1) reported in the previous study14, we filtered out low-quality nuclei and finally obtained 18,703 high-quality nuclei (Fig. 2a–d, Table 2).
Data integration and quality control (QC) of the three ccRCC samples. (a,b) Performing transcriptional start site (TSS) enrichment score and the nucleosome signal (NS) of the data. (c) We showed the quality parameters of each cell nucleus by scatterplot. (d) A quality control strategy is applied to the data, with the blue dots representing cells and the cells in the red box being high-quality cells for downstream analysis. (e) We showed the correlation between depth and reduced dimension components from 1 to 50 by latent semantic indexing (LSI) analysis.
After QC, we performed term frequency-inverse document frequency (TF-IDF) normalization and obtained a total of 104,818 unique peaks. We identified gene annotations by GRanges function, referencing ‘hg19’ from University of California Santa Cruz (UCSC19). Then, we run singular value decomposition (SVD) on the TD-IDF matrix by using the peaks in each cell. After completing the above two steps, we can perform latent semantic indexing (LSI) analysis20. We calculated the correlation between depth and reduced dimension components from 1 to 50 and selected 35 as the parameter for downstream analysis, which the correlation is close to zero (Fig. 2e). Cell nuclei can be unbiased clustered by FindClusters function with a resolution of 0.5.
Calculation of gene activity score and differential peaks
The gene activity score is a method of quantifying the activity of each gene in the genome by assessing the accessibility of chromatin associated with a gene. Gene activity matrix can be created by extracting gene coordinates and including the 2 kb upstream region. This step can be performed with the GeneActivity function. To understand the chromatin state between each cell cluster, calculating differential peaks is a valuable method. Here, we applied the FindAllMarkers function to obtain differential peaks between each cell cluster (Table S1).
Motif and transcription factor (TF) footprinting analysis
Based on the chromVAR21 R package that was integrated into Signac, we performed the AddMotifs function to add the DNA sequence motif information for motif analyses. We could calculate the motif activity score in each cell and identify differential activity scores between cell types by FindAllMarkers function. Then we enriched the most significant motifs in each cell subtype (Table S2). After normalization by z-scores, differential activity scores between cell types referred to “avg_diff”. We selected the most significant motifs (top 1 or 2) in each cell subtype for secondary analysis. In addition, we visualized the above motifs and identified the corresponding TFs by the MotifPlot function. After confirming the cell type-specific TFs, we can gather all the required data and stores it in an assay by the Footprint function. Finally, we presented the footprint analysis of the above transcription factors by the PlotFootprint function.
Definition of each cell type
Compared with single-cell RNA sequencing, the results of scATAC-seq are more challenging for cell definition. As previously reported in the literature13, we recommend the integration of three dimensions for cell definition: (1) based on the gene activity matrix that extract gene coordinates and extend them to include the 2 kb upstream region, we can assess the chromatin accessibility associated with the marker genes; (2) calculating the differential peaks of each cell subtype, which can be matched to specific locations in the gene sequence (Table S1); (3) TF analysis that cell-type specific TFs were discovered and then combined with the reported literature.
Data Records
All the processed scATAC-seq data can be access in NCBI GEO database. The GEO accession number is GSE27127322. The data was obtained after preliminary analysis by Cellranger software. The raw data (.fastq files) have been deposited in the NCBI Sequence Read Archive (SRA) and the project accession number is PRJNA113084223. After secondary analysis by Signac, we saved the object file (.rds), Tables S1 and S2, which can be access in figshare24
Technical Validation
Here, we presented three high-quality scATAC-seq data from human ccRCC samples, including appropriate quality control parameters, such as fraction of fragments overlapping TSS > 30%, fraction of transposition events in peaks in cell barcodes > 40% and fraction of fragments overlapping any targeted region > 60% (Table 2). By applying Signac18 to unbiased clustering of the single-cell nuclei, a total of 18,703 high quality cells and 20 cell subtypes can be identified (Fig. 3a). According to cell annotation (Methods), we classified these cells into tumour cells, endothelial cells, T cells, macrophage, CAF, NK cells and B cells (Fig. 3a). Given that two of the three samples (RCC61 and RCC76) were fresh nuclei and one (RCC30) was frozen nuclei, we compared the cell subpopulations of the three samples. We found that in addition to the tumour cell subtypes, other cell subtypes were contributed by cells from almost each sample (Fig. 3b). This result was similar to previous scATAC-seq studies13,14. The samples included both male and female. For example, RCC30 and RCC61 obtained from male, while RCC76 obtained from female (Table 1). In addition, given that the samples we used for scATAC-seq included both fresh samples (RCC61 and RCC76) and frozen stored sample (RCC30), we randomly detected 3 regions of the chromatin accessibility, and found a high degree of similarity between the three samples (Fig. 3c–e), which further indicated the reliability of the scATAC-seq data.
Three ccRCC samples were merged by Signac. (a) A total of 18,703 nuclei were unbiased clustering and classified into 20 different cell subtypes, which were projected by UMAP. (b) Spatial distribution of three different ccRCC samples on UMAP. (c–e) The chromatin accessibility of three different samples on chr 3, chr 9 and chr 17 were randomly selected to demonstrate.
We detected the gene activity scores of major cell subtypes, of which these marker genes were consistent with previous studies, such as CA9 in tumour cells25 in tumour cells, VWF in endothelial cells26, RGS5 in CAF27, PTPRC in immune cells28, MSR1 in macrophage29, IL7R in T cells30, KLRD1 in NK cells31 and SDC1 in B cells13 (Fig. 4a). Meanwhile, this data can reveal the universal regions of chromatin accessibility in all cells (Fig. 4b,c). Interestingly, we discovered the specific peaks of each cell cluster and the chromatin location of these regions (Fig. 5a and Table S1). Here, we showed the specific peaks in tumour cell subtypes (cluster 1,2,3,14), which located on CTB-164N12.1, ATRNL1, KRT14, and RP11-118K6.3, respectively (Fig. 5–e).
scATAC-seq revealed the epigenetic regulatory features of ccRCC. (a) Cell type-specific gene activity scores, the colour gradient indicated the level of the score, with yellow representing high and purple representing low. (b,c) scATAC-seq identified the universal regions of chromatin accessibility.
Finally, based on the Signac18 and chromVAR21 R packages, we presented a method to discover cell type-specific transcription factors (TFs) and motifs that included variable 200 TFs and motifs (Fig. 6a). In addition, we enriched some of the most significant TFs and motifs, which have been verified by previous studies13,14,15, such as HNF1B/HNF1A in tumor cells, SOX8/SOX9 in endothelial cells, EBF2/EBF3 in CAF, SPIB/SPIC in macrophage, EOMES/TBR1 in NK cells and ETS1/FLI1 in T cells (Fig. 6b). Subsequently, we can perform footprint analysis for the above motifs in each cell cluster (Fig. 7). Collectively, our data provided high-quality epigenetic information on ccRCC and more references for future treatment and diagnosis.
Code availability
The R code used in the analysis of the scATAC-seq data is available on figshare24
References
Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: a cancer journal for clinicians 71, 209–249, https://doi.org/10.3322/caac.21660 (2021).
Linehan, W. M. & Ricketts, C. J. The Cancer Genome Atlas of renal cell carcinoma: findings and clinical implications. Nature reviews. Urology 16, 539–552, https://doi.org/10.1038/s41585-019-0211-5 (2019).
Linehan, W. M., Srinivasan, R. & Schmidt, L. S. The genetic basis of kidney cancer: a metabolic disease. Nature reviews. Urology 7, 277–285, https://doi.org/10.1038/nrurol.2010.47 (2010).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England journal of medicine 366, 883–892, https://doi.org/10.1056/NEJMoa1113205 (2012).
Network, T. C. G. A. R. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49, https://doi.org/10.1038/nature12222 (2013).
Feinberg, A. P. & Levchenko, A. Epigenetics as a mediator of plasticity in cancer. Science (New York, N.Y.) 379, eaaw3835, https://doi.org/10.1126/science.aaw3835 (2023).
Klemm, S. L., Shipony, Z. & Greenleaf, W. J. Chromatin accessibility and the regulatory epigenome. Nature reviews. Genetics 20, 207–220, https://doi.org/10.1038/s41576-018-0089-8 (2019).
Terekhanova, N. V. et al. Epigenetic regulation during cancer transitions across 11 tumour types. Nature 623, 432–441, https://doi.org/10.1038/s41586-023-06682-5 (2023).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods 10, 1213–1218, https://doi.org/10.1038/nmeth.2688 (2013).
Boyle, A. P. et al. High-resolution mapping and characterization of open chromatin across the genome. Cell 132, 311–322, https://doi.org/10.1016/j.cell.2007.12.014 (2008).
Ishii, H., Kadonaga, J. T. & Ren, B. MPE-seq, a new method for the genome-wide analysis of chromatin structure. Proceedings of the National Academy of Sciences of the United States of America 112, E3457–3465, https://doi.org/10.1073/pnas.1424804112 (2015).
Buenrostro, J. D. et al. Integrated Single-Cell Analysis Maps the Continuous Regulatory Landscape of Human Hematopoietic Differentiation. Cell 173, 1535–1548.e1516, https://doi.org/10.1016/j.cell.2018.03.074 (2018).
Satpathy, A. T. et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nature biotechnology 37, 925–936, https://doi.org/10.1038/s41587-019-0206-z (2019).
Yu, Z. et al. Integrative Single-Cell Analysis Reveals Transcriptional and Epigenetic Regulatory Features of Clear Cell Renal Cell Carcinoma. Cancer research 83, 700–719, https://doi.org/10.1158/0008-5472.can-22-2224 (2023).
Kourtis, N. et al. A single-cell map of dynamic chromatin landscapes of immune cells in renal cell carcinoma. Nature cancer 3, 885–898, https://doi.org/10.1038/s43018-022-00391-0 (2022).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nature biotechnology 33, 495–502, https://doi.org/10.1038/nbt.3192 (2015).
Stuart, T. et al. Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e1821, https://doi.org/10.1016/j.cell.2019.05.031 (2019).
Stuart, T., Srivastava, A., Madad, S., Lareau, C. A. & Satija, R. Single-cell chromatin state analysis with Signac. Nature methods 18, 1333–1341, https://doi.org/10.1038/s41592-021-01282-5 (2021).
Raney, B. J. et al. The UCSC Genome Browser database: 2024 update. Nucleic acids research 52, D1082–d1088, https://doi.org/10.1093/nar/gkad987 (2024).
Cusanovich, D. A. et al. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science (New York, N.Y.) 348, 910–914, https://doi.org/10.1126/science.aab1601 (2015).
Schep, A. N., Wu, B., Buenrostro, J. D. & Greenleaf, W. J. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nature methods 14, 975–978, https://doi.org/10.1038/nmeth.4401 (2017).
NCBI GEO https://identifiers.org/geo/GSE271273 (2024).
NCBI Sequence Read Archive https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA1130842&o=acc_s%3Aa (2024).
Zhenyuan, Y. Single-cell assay for transposase-accessible chromatin sequencing of human clear cell renal cell carcinoma. figshare https://doi.org/10.6084/m9.figshare.26131405 (2024).
Young, M. D. et al. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science (New York, N.Y.) 361, 594–599, https://doi.org/10.1126/science.aat1699 (2018).
Brehm, M. A. Von Willebrand factor processing. Hamostaseologie 37, 59–72, https://doi.org/10.5482/hamo-16-06-0018 (2017).
Wu, X. et al. Integrated analysis of single-cell RNA-seq and bulk RNA-seq unravels the heterogeneity of cancer-associated fibroblasts in TNBC. Aging 15, 12674–12697, https://doi.org/10.18632/aging.205205 (2023).
Al Barashdi, M. A., Ali, A., McMullin, M. F. & Mills, K. Protein tyrosine phosphatase receptor type C (PTPRC or CD45). Journal of clinical pathology 74, 548–552, https://doi.org/10.1136/jclinpath-2020-206927 (2021).
Chevrier, S. et al. An Immune Atlas of Clear Cell Renal Cell Carcinoma. Cell 169, 736–749.e718, https://doi.org/10.1016/j.cell.2017.04.016 (2017).
Bi, K. et al. Tumor and immune reprogramming during immunotherapy in advanced renal cell carcinoma. Cancer cell 39, 649–661.e645, https://doi.org/10.1016/j.ccell.2021.02.015 (2021).
Su, C. et al. Single-Cell RNA Sequencing in Multiple Pathologic Types of Renal Cell Carcinoma Revealed Novel Potential Tumor-Specific Markers. Frontiers in oncology 11, 719564, https://doi.org/10.3389/fonc.2021.719564 (2021).
Acknowledgements
Thanks to Dr. Yufang Lv for her help in this study, who is now at Southern Medical University. This work was supported by grants from the Natural Science Foundation of Guangxi Province (2024GXNSFBA010055), the China Postdoctoral Science Foundation (2024MD763944), the Youth Science Foundation of Guangxi Medical University (GXMUYSF202409), the Natural Science Foundation of Guangxi Province (2018GXNSFAA138192) and the Medical Excellence Award Funded by the Creative Research Development Grant from the First Affiliated Hospital of Guangxi Medical University, and Innovation Project of Guangxi Graduate Education (YCBZ2023091).
Author information
Authors and Affiliations
Contributions
W. Lu. and Y. Liu. performed the scATAC-seq. J. Li. provided professional data analysis. S. Huang., Z. Wen. and C. Su., dissected and supervised ccRCC tissues. Z. Lu. and Z. Mo. provided comments and revisions to this project. Z. Yu conceived and supervised the project, analysed the data, created the figures, and wrote the paper. All the authors read the manuscript and made comments.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lu, W., Liu, Y., Li, J. et al. Single-cell assay for transposase-accessible chromatin sequencing of human clear cell renal cell carcinoma. Sci Data 12, 334 (2025). https://doi.org/10.1038/s41597-025-04666-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-04666-w