Abstract
A strong association between obesity and COVID-19 complications and a lack of prognostic factors that explain the unpredictable severity among these patients still exist despite the various vaccination programs. The expression of angiotensin converting enzyme 2 (ACE2), the main receptor for severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), is enhanced in obese individuals. The occurrence of frequent genetic single nucleotide polymorphisms (SNPs) in ACE2 is suggested to increase COVID-19 severity. Accordingly, we hypothesize that obesity-associated ACE2 polymorphisms increase the severity of COVID-19. In this study, we profiled eight frequently reported ACE2 SNPs in a cohort of lean and obese COVID-19 patients (n = 82). We highlight the significant association of rs2285666, rs2048683, rs879922, and rs4240157 with increased severity in obese COVID-19 patients as compared to lean counterparts. These co-morbid-associated SNPs tend to positively correlate, hence proposing possible functional cooperation to ACE2 regulation. In obese COVID-19 patients, rs2285666, rs879922, and rs4240157 are significantly associated with increased blood nitrogen urea and creatinine levels. In conclusion, we highlight the contribution of ACE2 SNPs in enhancing COVID-19 severity in obese individuals. The results from this study provide a basis for further investigations required to shed light on the underlying mechanisms of COVID-19 associated SNPs in COVID-19 obese patients.
Similar content being viewed by others
Obesity, a major epidemic and chronic disease, is a key contributing factor to adverse outcomes and hospitalization of patients with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus responsible for COVID-19. Up to 40% of the world’s population is categorized as overweight and obese, with increasing incidence by 20301,2. This is alarming, as obesity has been associated with severe infections. Moreover, obese individuals are characterized by their dysregulated immune system and impaired immune responses to vaccines. This could partially explain the increased risk of mortality in COVID-19 obese patients3,4. Recalling the 2009 influenza pandemic, obese individuals were at higher risk of death due to the viral infection. Obese individuals were twice likely to develop influenza despite vaccination5,6. Taken together, this has led to extensive clinical investigations regarding COVID-19. Recent studies have highlighted the effect of obesity as a unifying risk factor for increased hospitalization and the COVID-19 mortality rate. Analysis of COVID-19 individuals with high BMI and increased waist circumference displayed an increased risk of COVID-19-related mortality7,8,9. Similarly, the increased risk of hospitalization was directly linked to increased BMI in COVID-19 subjects9,10. In UAE, the presence of underlying co-morbidities and high BMI work synergistically to affect the clinical outcomes of COVID-1911. Despite this clinical association, there is still a lack of prognostic factors that explains the variability and unpredictable severity among COVID-19 obese patients.
SARS-CoV-2 entry into the cell is dependent on Angiotensin-Converting Enzyme 2 (ACE2) receptor. ACE2 is a type I transmembrane metallocarboxypeptidase with homology to ACE, an enzyme long known to be a key player in the renin-angiotensin system (RAS), and a target for the treatment of hypertension12,13. The secreted protein catalyses the cleavage of the C-terminal dipeptide of Angiotensin I to produce Angiotensin 1–9 and Angiotensin II to produce Angiotensin 1–714. It has been suggested that genetic variations, including single nucleotide polymorphisms (SNPs) in the ACE2 gene, may account for the differences in symptoms and severities seen in COVID-19 patients, leading to altered immune responses and greater viral susceptibility. For example, rs182366225 and rs2097723 are two polymorphisms that may increase the expression of ACE2 with a higher prevalence in the East Asian population. On the other hand, the rs142017934 SNP has also been associated with increased expression of ACE2; however, it is exclusive to Africans15. Residue changes in ACE2 may potentially affect its expression or its binding affinity to the virus and raise the vulnerability of individuals to SARS-CoV-2 infection16. Therefore, identifying ACE2 SNPs can shed light on the involvement of genetic variations in the epidemiological differences in COVID-19 susceptibility. Although earlier studies failed to show an association between ACE2 polymorphisms and susceptibility or severity of COVID-19 in the general population17, a more recent study was able to identify four SNPs in ACE2 which were associated with the severity of the disease. Among these, rs2106809 and rs2285666 correlated with an increased risk of hospitalization and severity of COVID-19. The presence of co-morbid conditions such as obesity increased the risk for ICU admission and death in these patients18.
We have previously shown that lung epithelial cells of obese patients express higher levels of ACE2, which may explain the increased susceptibility to infection and severe outcomes in obese patients19. Moreover, our group showed that obesity affects the metabolome of COVID-19 patients, where we proposed n6-acetyl-l-lysine and p-cresol as metabolic signatures having crucial roles in the poor prognosis of COVID-19 obese patients20. SARS-CoV-2 entry into the host cell depends on ACE2, thus we propose that genetic variations in ACE2 may account for the differences in symptoms and severities seen in COVID-19 patients. To our knowledge, no studies have focused on the ACE2 genetic variations in the COVID-19 obese population. We hypothesize that obesity is associated with genetic variants which may modulate the expression of ACE2 and, therefore be responsible for the increased susceptibility or severity of COVID-19. This study provides insight into the correlation of frequently reported ACE2 variants and increased severity, in association to obesity, in COVID-19 patients, which might further explain the complications seen in obese patients compared to their lean counterparts.
Results
Targeted Next-Generation Sequencing detected eight ACE2 polymorphisms in COVID-19 patients
Targeted Next-Generation Sequencing was performed in 82 COVID-19 patients for ACE2 variants screening using the Fluidigm Access Array. Our analysis revealed 8 ACE2 single nucleotide variants to be present among the studied population (Table 1). Interestingly, a splice variant represented as rs2285666 or as COSV53024795, at chromosome position X:g.15610348 (Fig. 1), is found to be the most frequent variant in our study cohort, being present in 78 out of the 82 COVID-19 patients (Fig. 1). Two out of the detected 8 variants, X:g.15,582,209 (rs35803318) and X:g.15,5824,429 (COSV53023851), are synonymous mutations located, respectively, at exons 18 and 17 (ENST00000427411.1; Table 1). rs35803318 was detected in only two patients that are obese and overweight, while COSV53023851 is detected in one obese patient (Fig. 1). One undetermined missense mutation (X:g.15603635) is found to be located in the coding region of ACE2 (Table 1) and is present in only one obese patient (Fig. 1). Also, our data revealed that out of the 8 assessed variants, only 4 show potential significance in the studied population (rs2285666, rs4240157, rs879922, rs2048683). Regardless of their BMI or clinical manifestations, these mutations are present in almost up to 69 COVID-19 patients; except the rs2074192 SNP is found to be present in 29 patients, as shown in Fig. 1.
Distribution of ACE2 variants among the COVID-19 lean and obese patients. rs2285666 (chrX:15,610,348, C > T) has the highest distribution among the studied cohort; present in 79 patients; rs2048683 is present in 68 patients; rs879922 is present in 64 patients and rs4240157 is present in 61 patients.
Potential association of rs2285666, rs4240157, rs879922, and rs2048683 with increased SARS-CoV-2 susceptibility and severity in obese COVID-19 patients
Next, we evaluated the possible involvement of the identified 8 genetic variants in the ACE2 gene in COVID-19 susceptibility by assessing the frequency distribution of each variant among the different BMI groups (Fig. 2), including the symptoms development status, referred as asymptomatic versus symptomatic; (Fig. 3). Our data showed a significantly higher distribution of rs2285666 (p = 0.05), rs2048683 (p = 0.0005) and rs4240157 (p = 0.0005) in lean COVID-19 patients (Fig. 2a,b,d), while the rs879922 SNP was almost potentially significant in COVID-19 obese and overweight (Ob/Ov) patients (p = 0.06; Fig. 2c). Furthermore, our data showed that the SNPs rs2285666, rs2048683, and rs879922 are significantly correlated with symptoms development of COVID-19 in Ob/Ov subjects (Fig. 3a–c), with p = 0.04, p = 0.0001, and p = 0.0001, respectively. Only rs4240157 is mainly correlated to the incidence of symptoms in COVID-19 lean subjects (p = 0.01; Fig. 3d). Consequently, these SNPs could be associated with symptomatic risk and severity of COVID-19 infection. On the other hand, our analysis displayed no significant correlation between rs2074192, rs35803318, COSv53023851, and the exonic variant chrX:15,603,635, among the studied cohort and their BMI (Table 2). Similarly, no significant correlation was detected between these SNPs and the symptom phenotype of COVID-19 (Table 2). Table 2 summarizes the significance of each SNP with BMI and their potential for COVID-19 symptom development.
ACE2 variants are correlated to the BMI of COVID-19 patients. (a) rs2285666 and (b) rs2048683 (c) rs879922 and (d) rs4240157 are the most significantly correlated to the body mass index (BMI) of COVID-19 lean and obese/overweight subjects. The variant (c) rs879922 is almost significantly expressed in the COVID-19 obese subjects (70%) more readily than in the COVID-19 lean subjects (54%). Contingency test using Chi-square analysis was applied for each variant.
COVID-19 obese subjects expressing ACE2 variants are more symptomatic than COVID-19 lean subjects. (a) rs2285666, (b) rs2048683, and (c) rs879922 are significantly correlated to the incidence of symptoms in COVID-19 obese/overweight subjects. The variant (d) rs4240157 is significantly correlated to the incidence of symptoms in COVID-19 lean subjects. A contingency test using Chi-square analysis was applied for each variant.
Given the potential significance of these findings, we then examined the frequency of the significantly correlated SNPs in our cohort by addressing the most extensive database for the COVID-19 associated variants, “A catalogue of associations between rare coding variants and COVID-19 outcomes” 21,22, using the online platform (https://rgc-covid19.regeneron.com/). Tables provided as supplementary (Tables S1, S2, and S3) show the list of studies with a significant association of each of the detected SNPs from our study in patients with COVID-19. Figure 4 shows the meta-analysis Odds ratio between different studies that included the alternative allele frequency (AAF) for these SNPs: rs2285666 (AAF: 021,162); rs2048683 (AAF: 0.63685); rs879922 (AAF: 0.62782). Interestingly, the African ancestry was one of the populations where the effect of rs2285666, rs2048683, and rs879922 are shown to direct towards a significant detrimental effect (Fig. 4a–c) when compared to other population groups. This implies that these SNPs might have a different impact based on different ethnicities.
Association between COVID-19 outcomes and the most significant variants: rs2285666, rs2048683, and rs879922. Results combined across cohorts using an inverse variance meta-analysis showing Odd ratios (Confidence interval; [95% CI) for (a) rs2285666 (n = 241 COVID-19-positive cases, n = 21,536 COVID-19-negative), (b) rs2048683 (n = 371 COVID-19-positive cases, n = 83,355 COVID-19-negative), and (c) rs879922(n = 20,177 COVID-19-positive cases, n = 653,646 COVID-19-negative). RR, individuals who have genotype reference/reference for all variants included in burden test; RA, individuals who have genotype reference/alternate for at least one variant; AA, individuals who have genotype alternate/alternate for at least one variant; AAF, alternative allele frequency.
ACE2 variants increase blood biomarkers in COVID-19 patients
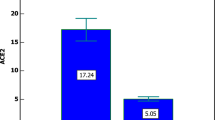
To better understand the relationship between the identified SNPs, the BMI, and the severity of symptoms due to COVID-19, a Pearson correlation analysis was performed between the occurrence of the identified SNPs and the number of patients with COVID-19. A graphical representation of the association is shown in Fig. 5. All identified SNPs were positively correlated, irrespective of the severity and BMI. For instance, rs2048683 is significantly correlated with rs879922 (p = 0.03) and with rs2285666 (p = 0.04), and rs4240157 is almost significantly correlated with rs879922 (p = 0.05), as shown in Fig. 5. These identified correlated SNPs are known to affect ACE2-SARS-CoV-2 binding affinity and COVID-19 severity23,24. Moreover, correlation analysis has been performed between the BMI of the studied cohort expressing these SNPs and their clinical information. Our analysis showed that BMI correlates significantly with blood urea nitrogen (BUN) and creatinine blood levels (Fig. 6), implying kidney complications or failure.
Positive correlation between ACE2 variants. Correlation plot showing the correlation between the different detected SNPs. The heatmap colors reflect rs2285666, rs2048683, rs4240157, and rs879922 are the most significantly correlated. Pearson correlation coefficients with blue for positive correlation and red for negative correlation. *p < 0.05; **p < 0.01.
A further analysis was performed to understand the exact correlation between rs2285666, rs4240157, rs879922, and rs2048683 and the clinical variability among the COVID-19 patients. Interestingly, our data showed that each of rs2285666, rs4240157, rs879922, and rs2048683 are significantly associated with increased levels of either of alanine aminotransferase (ALT), aspartate aminotransferase (AST), haemoglobin (Hb), absolute lymphocyte count (ALC), lactate dehydrogenase (LDH), BUN and creatinine, in addition to white blood cells (WBC) and platelets count (Fig. 7). COVID-19 patients expressing rs2285666 mainly display significant increase in ALT (p = 0.0315), AST(p = 0.0261), Hb (p = 0.02), ALC (p = 0.039), creatinine (p = 0.008), and BUN (p = 0.0027) blood levels (Fig. 7a). COVID-19 patients expressing rs879922 revealed significant increase in ALC (p = 0.048), creatinine (p = 0.0041), BUN (p = 0.0215), and LDH (p = 0.0026) blood levels, in addition to platelets count (p = 0.018; Fig. 7b). COVID-19 patients expressing rs2048683 and rs4240157 displayed increase in LDH (p = 0.0112) and creatinine (p = 0.005) blood levels, and in WBC count (p = 0.0444; Fig. 7c,d), respectively.
Significant association between rs228566, rs2048683, rs4240157 and rs879922 and elevated levels of blood biomarkers. Significant increase in the levels of ALT, AST, Hb, ALC, creatinine, and BUN clinical tests in COVID-19 patients expressing (a) rs2285666. In COVID-19 patients expressing (b) rs879922 revealed significant increase in the levels of ALC, creatinine, BUN, and LDH, in addition to platelets count. Increased levels in creatinine, LDH and WBC count in COVID-19 patients with (c) rs2048683 and (d) rs4240157. *p < 0.05; **p < 0.01. ALT alanine aminotransferase; AST aspartate aminotransferase; Hb hemoglobin; ALC absolute lymphocyte count; BUN blood urea nitrogen; LDH lactate dehydrogenase; WBC white blood cells.
This analysis was further stratified into assessing the correlation of these SNPs with BMI in the studied cohort. Interestingly, COVID-19 Ob/Ov patients expressing rs2285666, rs879922, and rs4240157 displayed significant increase in creatinine (p = 0.0394 and p = 0.0292) and BUN blood levels (p = 0.047 and p = 0.0023; Fig. 8a–c), respectively. This comes in parallel with our correlation analysis in Fig. 6, which further indicates the potential association of these SNPs with kidney health.
Significant association between ACE2 variants and blood biomarkers in COVID-19 obese patients. Creatinine and BUN levels are significantly increased in COVID-19 patients obese patients expressing (a) rs2285666, (b) rs879922, and (c) rs4240157 variants as compared to their lean counterparts. *p < 0.05; **p < 0.01.
Taken together, these data point out that the association of these ACE2 SNPs with COVID-19 severity is also in part related to obesity and possibly obesity-associated co-morbidities in these patients.
Discussion
Many studies have shown a positive association between obesity and COVID-19 severity. However, and to our knowledge, this is the first study to evaluate the contribution of obesity in SARS-CoV-2 infection susceptibility that may be associated with certain genomic variants within the ACE2 gene and thus might modulate its expression and function. We report the profiling and possible involvement of four out of eight detected ACE2 single nucleotide polymorphisms (SNPs; rs2285666, rs2048683, rs879922, and rs4240157) in offering a plausible biological explanation for the increased expression of ACE2 and health complications seen in obese COVID-19 subjects.
Polymorphisms of more than twenty obesity-related genes have been documented to correlate with metabolic dysregulation and low-grade inflammation in different diseases, including viral infections25,26. Moreover, it was suggested that these metabolically related polymorphisms might interfere with SARS-CoV-2 intrusions and antiviral responses27. Considering this association between gene polymorphisms, obesity and SARS-CoV-2 infection, it is important to decipher the effect of obesity on ACE2 polymorphisms, as being the primary entry site for SARS-CoV-2, with evidence of higher expression in obese individuals19,28. In this study, we have identified eight ACE2 genetic variations, among which rs2285666, rs2048683, rs879922, and rs4240157 are the most significantly present in the studied cohort that comprises lean and obese COVID-19 subjects. These SNPs were found to be commonly distributed in both lean and obese COVID-19 subjects. On the other hand, our analysis has shown that overweight and obese COVID-19 subjects who have these SNPs tend to be significantly more symptomatic as compared to lean COVID-19 subjects. Importantly, the rs879922 SNP was significantly correlated with both BMI and severity in COVID-19 obese and overweight subjects as compared to COVID-19 lean subjects. Among the detected ACE2 variants, the rs2285666 SNP is the most distributed polymorphism in the studied population. Several studies have highlighted a genetic association of ACE2 rs2285666 polymorphism with elevated ACE2 gene expression in different populations that involves Europeans, Asians, and Indians. The upregulated ACE2 expression is reported to be up to 50%, despite the striking difference in allele frequency in the different populations29,30,31,32 , hence suggesting a positive association between rs2285666 and increased susceptibility to COVID-19. Of note, rs2285666 is considered one of the variants that enhances the binding affinity of SARS-CoV-2 to ACE2, enhancing its infection, and thus impacting severe clinical outcomes related to SARS-CoV-2 infection23,33,34. On the other hand, a few other reports also suggested a positive correlation between the SNPs rs4240157, rs204683 and rs879922 with higher tissue expression of ACE2 and correlated it with increased hospitalisations of COVID-19 patients24. This aligns with our data, where our analysis showed that these SNPs, rs2285666, rs4240157, rs204683 and rs879922, positively correlate with each other, thus indicating possible functional cooperation and contribution to ACE2 regulation. In addition, our data showed a significant and positive association between the BMI of these patients and health complications, including kidney and liver malfunctions and associated inflammation due to increased WBC and ALC levels. Interestingly, clinical information of COVID-19 obese subjects expressing ACE2 rs2285666, rs879922, and rs4240157 variants reflected increased kidney complications due to the significant increase of creatinine and BUN blood levels. The rs2285666, rs2048683, rs879922, and rs4240157 variants are known to be associated with type 2 diabetes mellitus (T2DM) and obesity related co-morbidities such as cardiovascular disease and hypertension30,32,35. Overall, these data suggest the coordination of these SNPs in contributing to ACE2 expression that may possibly be a predisposing factor associated with the co-morbidities observed in COVID-19 patients, especially in obese subjects.
In summary, this study lays the foundation for future studies to bridge the gap between the polymorphisms and their possible involvement in the dysregulation of ACE2 in COVID-19. A limitation to our study includes the lack control samples (non-COVID-19) due to the difficulty in achieving patients’ samples that were not vaccinated, as vaccination process in the United Arab Emirates (UAE) has been initiated effectively by treated all people as emergency patients36,37. Moreover, additional information regarding demographic and pre-existing conditions such as smoking status for each patient were not provided. Despite the low number of non-vaccinated COVID-19 patients’ samples that we obtained, our data has highlighted the correlation of the frequently reported ACE2 SNPs (rs2285666, rs2048683, rs879922, and rs4240157) and symptoms development of COVID-19 patients. Unifying our understanding of obesity with severe health complications in COVID-19, we propose an underlying health complication that correlates ACE2 SNPs with poor COVID-19 outcomes (Fig. 9). We highlight the importance of ACE2 genetic polymorphisms, as it may be an effective method to enhance the efficacy of immunotherapeutic and conventional treatments in COVID-19 patients and specifically in obese COVID-19 patients.
Materials and methods
Patients and sample processing
Fasting blood samples were collected from a cohort of COVID-19 patients (n = 82) who visited Rashid Hospital (Dubai, UAE) between June and July 2020. Blood samples were then recruited from the COVID-19 Biobank for further analysis through a collaboration with the University of Sharjah (Sharjah, UAE). The studied population was categorized according to their body mass index (BMI) as follows; Patients with BMI ≥ 30 kg/m2, BMI = 25–29.9 kg/m2, and BMI < 25 kg/m2 are classified as obese (n = 28), overweight (n = 19), and lean (n = 35), respectively. Table 3 describes patients’ characteristics. Vaccinated COVID-19 samples have been excluded from the selection criteria. Patients’ COVID-19 clinical manifestations ranged from severe to moderate and mild symptoms and asymptomatic status.
Briefly, 8 ml of peripheral venous blood were collected in Ethylenediamine Tetra Acetic Acid (EDTA)- blood collection tubes. Within 24 h, collected blood samples were processed using density gradient media by diluting them with Phosphate Buffered Saline (PBS (1x); Sigma-Aldrich, USA) containing 2% human fetal bovine serum (FBS; Sigma-Aldrich, USA), in a 1:1 ratio, mixed thoroughly, and layered onto a HISTOPAQUE®-1077gradient media (Sigma-Aldrich, USA). Several centrifugations were performed at 400 × g for 30 min at room temperature to separate blood components. Blood clots were collected and stored at − 80 °C until DNA extraction.
All patients provided written informed consent. Ethical approval was obtained from the Dubai Scientific Research Ethics committee (DSREC-04/2020_19) of the Dubai Health Authority (DHA) in the United Arab Emirates (UAE).
DNA extraction
Genomic DNA was extracted using DNA Mini Kit -QIAMP (QIAGEN, Germany). Briefly, 20 µl of Proteinase K was added to a 200 µl of blood clot sample in an Eppendorf. Lysis buffer was then added to the mixture, where it was allowed for 10 min incubation at 70 °C. A volume of 200 µl of ethanol (96–100%) was then added to minimize DNA solubility, and the mixture was then transferred into a DNeasy Mini spin column, where several centrifugation steps were performed at room temperature; as per the manufacturer’s protocol. Eventually, DNA was eluted in 30 µl of Nuclease Free water (Invitrogen, UK). Using Nano-drop 8000 (Thermo-Scientific, USA), the quantity of extracted DNA from 200 μl of blood clot samples ranged from 24.5 ng/μl to 199.3 ng/μl, and the purity (OD260/OD280) range was 1.8–1.9; for all samples.
ACE2 primers designing and validation
Primers were designed to cover all exonic and some intronic regions of the ACE2 gene (Gene ID: 59,272, NM_001371415.1). The details of the ACE2 primers are listed in Supplementary Table (S4). The primers were then assessed using control DNA samples, and the expected amplicon sizes were confirmed using agarose gel electrophoresis.
Targeted next-generation sequencing (NGS)
The library for DNA sequencing using NGS was generated using the Fluidigm Access Array microfluidic chip as previously described38. The confirmed target-specific ACE2 primers were then tagged with Fluidigm specific tag sequences and used for targeted next-generation sequencing38. The prepared amplicon libraries were purified using AMPure XP beads (Beckman Coulter, USA) and quantified using a High Sensitivity DNA assay kit on BioAnalyzer (Agilent, USA). The libraries were further diluted to 1 pg for direct input into the emulsion polymerase chain reaction (PCR) with Ion SphereTM particles using the Ion Template OT2 kit (Ion OneTouch™ instrument). Enrichment of the clonal beads was carried out using the Ion OneTouch™ ES system following the manufacturer’s instructions (Thermo Fischer, USA). The pooled libraries were then sequenced using the Ion 520™ Chip on the Ion S5 XL Semiconductor sequencer, following the manufacturer’s instructions (Thermo Fisher).
Data processing and targeted genetic analysis
Reads were aligned to the reference Human genome (NCBI37/hg19) by burrows wheeler aligner (BWA) on default settings39. Unmapped reads were excluded from the analysis. Mapped reads were visualized using an integrative genomics viewer (IGV) with the relevant browser extensible data (BED) file for the ACE2 gene. Reads were analyzed using an in-house bioinformatics pipeline that incorporates samtools mpileup. Post-processing quality filters were applied to the output to improve specificity of downstream analysis. Resulting variants were annotated according to the Ensemble release 105 (Dec 2021). Exonic variants were further tested for pathogenicity using the in-silico tools PolyPhen240, SIFT41 , and Mutation Taster42.
Pan-ancestry exome-wide association analyses of COVID-19 outcomes
Exome-wide association analyses for the identified SNPs from our cohort were done using a publicly available database using the “A catalogue of associations between rare coding variants and COVID-19 outcomes”21,22, retrieved from the online platform (https://rgc-covid19.regeneron.com/). This catalogue was generated to identify rare variants (RVs, minor allele frequency [MAF]. < 1%) associated with COVID-19 susceptibility and severity, an exome-wide sequencing data was generated for 543,213 individuals from different studies (Geisinger Health System [GHS], Penn Medicine BioBank [PMBB] and UK Biobank [UKB]) and three ancestries (African, European and South Asian). The same phenotypes obtained from these studies were used to validate the association with common risk variants reported in our study, thus demonstrating that our phenotypes are calibrated with those used in other studies.
Statistical analysis
Microsoft Excel and GraphPad Prism software were used to perform statistical analysis. Results are expressed as individual data or as mean ± standard deviation. Regarding COVID-19 patients, and prior to analysis, data were tested for normality using Shapiro Wilk’s normality tests. Frequencies (number of mutated patients) and percentages were used as appropriate. Data were analyzed via the Chi-square test. An unpaired t-test with Welch’s correction was used to compare clinical data among the groups of COVID-19 patients. A correlation plot was generated using Pearson correlation test. Statistical significance was accepted at p < 0.05.
Ethical approval
The study was conducted in accordance with the Declaration of Helsinki, and ethical approval was obtained from Dubai Scientific Research Ethics committee (DSREC-04/2020_19) of Dubai Health Authority (DHA) in the United Arab Emirates (UAE).
Informed consent
Informed consent was obtained from all subjects involved in the study.
Data availability
All data are contained within the manuscript.
References
Landecho, M. F., Marin-Oto, M., Recalde-Zamacona, B., Bilbao, I. & Frühbeck, G. Obesity as an adipose tissue dysfunction disease and a risk factor for infections–Covid-19 as a case study. Eur. J. Intern. Med. 91, 3–9 (2021).
Kelly, T., Yang, W., Chen, C. S., Reynolds, K. & He, J. Global burden of obesity in 2005 and projections to 2030. Int. J. Obes. 32(9), 1431–1437 (2008).
Sanchis-Gomar, F., Lavie, C. J., Mehra, M. R., Henry, B. M. & Lippi, G. Obesity and outcomes in COVID-19: when an epidemic and pandemic collide. Mayo Clin. Proc. 95(7), 1445–1453 (2020).
Tagliabue, C., Principi, N., Giavoli, C. & Esposito, S. Obesity: Impact of infections and response to vaccines. Eur. J. Clin. Microbiol. Infect. Dis. 35(3), 325–331 (2016).
Louie, J. et al. California Pandemic (H1N1) Working Group. Clin. Infect. Dis. 52(53), 301–312. https://doi.org/10.1093/cid/ciq152 (2009).
Neidich, S. D. et al. Increased risk of influenza among vaccinated adults who are obese. Int. J. Obes. 41, 1324–1330 (2017).
Drucker, D. J. Diabetes, obesity, metabolism, and SARS-CoV-2 infection: The end of the beginning. Cell Metab. 33(3), 479–498 (2021).
Peters, S. A., MacMahon, S. & Woodward, M. Obesity as a risk factor for COVID-19 mortality in women and men in the UK Biobank: Comparisons with influenza/pneumonia and coronary heart disease. Diabetes Obes. Metab. 23(1), 258–262 (2021).
Recalde, M. et al. Body mass index and risk of COVID-19 diagnosis, hospitalisation, and death: A population-based multi-state cohort analysis including 2,524,926 people in Catalonia, Spain. J. Clin. Endocrinol. Metab. 106(112), e5030–e5042 (2021).
Hamer, M., Gale, C. R., Kivimäki, M. & Batty, G. D. Overweight, obesity, and risk of hospitalization for COVID-19: A community-based cohort study of adults in the United Kingdom. Proc. Natl. Acad. Sci. 117(35), 21011–21013 (2020).
Al Heialy, S. et al. Combination of obesity and co-morbidities leads to unfavorable outcomes in COVID-19 patients. Saudi J. Biol. Sci. 28(2), 1445–1450 (2021).
Li, W. et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454 (2003).
Shi, C.-S. et al. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J. Immunol. 193, 3080–3089 (2014).
Lu, R. et al. Caracterización genómica y epidemiología del nuevo coronavirus 2019: Implicaciones para los orígenes del virus y la unión al receptor. Lancet 395, 566–568 (2020).
Khayat, A. S. et al. ACE2 polymorphisms as potential players in COVID-19 outcome. PLoS ONE 15, e0243887 (2020).
Paniri, A., Hosseini, M. M., Moballegh-Eslam, M. & Akhavan-Niaki, H. Comprehensive in silico identification of impacts of ACE2 SNPs on COVID-19 susceptibility in different populations. Gene Rep. 22, 100979 (2021).
LoPresti, M., Beck, D. B., Duggal, P., Cummings, D. A. & Solomon, B. D. The role of host genetic factors in coronavirus susceptibility: Review of animal and systematic review of human literature. Am. J. Human Genet. 107(3), 381–402. https://doi.org/10.1016/j.ajhg.2020.08.007(2020) (2020).
Sabater Molina, M. et al. Polymorphisms in ACE, ACE2, AGTR1 genes and severity of COVID-19 disease. PLoS ONE 17, e0263140 (2022).
Al Heialy, S. et al. Regulation of angiotensin-converting enzyme 2 in obesity: Implications for COVID-19. J. Front. Physiol. 11, 555039 (2020).
Jalaleddine, N. et al. N6-acetyl-L-lysine and p-cresol as key metabolites in the pathogenesis of COVID-19 in obese patients. Front. Immunol. https://doi.org/10.3389/fimmu.2022.827603 (2022).
Kosmicki, J. et al. A catalog of associations between rare coding variants and COVID-19 outcomes. medRxiv, 2020.2010.2028.20221804 (2021).
Horowitz, J. et al. Common genetic variants identify targets for COVID-19 and individuals at high risk of severe disease. medRxiv, 2021.2006.2015.21258703 (2021).
Hashemi, S. M. A. et al. Human gene polymorphisms and their possible impact on the clinical outcome of SARS-CoV-2 infection. J Archiv. Virol. 166(168), 2089–2108 (2021).
Chen, J. et al. Individual variation of the SARS-CoV-2 receptor ACE2 gene expression and regulation. J. Aging Cell 19, e13168 (2020).
Razquin, C., Marti, A. & Martinez, J. A. Evidences on three relevant obesogenes: MC4R, FTO and PPARγ. Approaches for personalized nutrition. Mol. Nutr. Food Res. 55(1), 136–149 (2011).
Short, K. R., Kedzierska, K. & Van de Sandt, C. E. Back to the future: Lessons learned from the 1918 influenza pandemic. Front. Cell. Infect. Microbiol. 8, 343 (2018).
Yan, T., Xiao, R., Wang, N., Shang, R. & Lin, G. Obesity and severe coronavirus disease 2019: Molecular mechanisms, paths forward, and therapeutic opportunities. Theranostics 11(17), 8234 (2021).
Yan, T., Xiao, R. & Lin, G. Angiotensin-converting enzyme 2 in severe acute respiratory syndrome coronavirus and SARS-CoV-2: A double-edged sword?. FASEB J. 34(5), 6017–6026 (2020).
Srivastava, A. et al. Genetic association of ACE2 rs2285666 polymorphism with COVID-19 spatial distribution in India. J. Front. Genet. 11, 1163 (2020).
Wu, Y. H., Li, J. Y., Wang, C., Zhang, L. M. & Qiao, H. The ACE 2 G8790A polymorphism: Involvement in type 2 diabetes mellitus combined with cerebral stroke. J. Clin. Lab. Anal. 31(2), e22033 (2017).
Asselta, R., Paraboschi, E. M., Mantovani, A. & Duga, S. J. A. ACE2 and TMPRSS2 variants and expression as candidates to sex and country differences in COVID-19 severity in Italy. J. Aging 12, 10087 (2020).
Li, Y. Y. Lack of association of ACE2 G8790A gene mutation with essential hypertension in the Chinese population: A meta-analysis involving 5260 subjects. Front. Physiol. 3, 364. https://doi.org/10.3389/fphys.2012.00364 (2012).
Pouladi, N., Abdolahi, S. J. M. G. & Medicine, G. Investigating the ACE2 polymorphisms in COVID-19 susceptibility: An in silico analysis. J Mol. Genet. 9(6), e1672 (2021).
Alimoradi, N. et al. SNPs of ACE1 (rs4343) and ACE2 (rs2285666) genes are linked to SARS-CoV-2 infection but not with the severity of disease. Virol. J 19(11), 48 (2022).
Gómez, J. et al. Angiotensin-converting enzyme (ACE1, ACE2) gene variants are associated with COVID19 severity depending on the hypertension status. MedRxiv 762, 145102 (2020).
Suliman, D. M. et al. UAE efforts in promoting COVID-19 vaccination and building vaccine confidence. Vaccine 39, 6341–6345 (2021).
Abbas Zaher, W., Ahamed, F., Ganesan, S., Warren, K. & Koshy, A. COVID-19 crisis management: Lessons from the United Arab Emirates leaders. Front. Public Health 9, 724494 (2021).
Presneau, N. et al. Diagnostic value of H3F3A mutations in giant cell tumour of bone compared to osteoclast-rich mimics. J. Pathol. Clin. Res. 1, 113–123 (2015).
Thorvaldsdóttir, H., Robinson, J. T. & Mesirov, J. P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14(2), 178–192 (2013).
Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Ng, P. C. & Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31(13), 3812–3814 (2003).
Schwarz, J. M., Cooper, D. N., Schuelke, M. & Seelow, D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat. Methods 11(4), 361–362 (2014).
Acknowledgements
This work was supported in part by the Al Jalila Foundation, the MBRU internal grant (MBRU CMRG-2020-06), MBRU publication fund, and by the -Human Disease Biomarkers Discovery Research Group- study.
Funding
This project was funded by the Mohammed Bin Rashid University of Medicine and Health Sciences Internal Research Grant (MBRU-CM-RG2020-06), UAE. In addition to COVID-19 research grant (COV19-0307), University of Sharjah, UAE and Al Jalila Foundation Seed Grant (AJF202019) to R.H.; and by Prince Abdullah Ben Khalid Celiac Disease Research Chair, under the Vice Deanship of Research Chairs, King Saud University, Riyadh, Kingdom of Saudi Arabia.
Author information
Authors and Affiliations
Contributions
Conceptualization, S.A.H.; Methodology, N.J., S.A.H., A.S., M.H.; Sample collection, R.H.; Formal analysis, N.J., and A.B. Supervision of analysis, M.H., and R.A.H.; Resources, S.A.H.; Data curation, N.J. and S.A.H.; Writing—original draft preparation, N.J.; Writing—review and editing, N.J., S.A.H, A.B., M.H., A.S., R. H., R. A. H.; Supervision, S.A.H.; Funding Acquisition: Dubai Healthcare Authority (DHA). All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jalaleddine, N., Bouzid, A., Hachim, M. et al. ACE2 polymorphisms impact COVID-19 severity in obese patients. Sci Rep 12, 21491 (2022). https://doi.org/10.1038/s41598-022-26072-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-26072-7
This article is cited by
-
Development and characterization of a fully humanized ACE2 mouse model
BMC Biology (2025)
-
Severe COVID-19 disease is associated with genetic factors affecting plasma ACE2 receptor and CRP concentrations
Scientific Reports (2025)
-
Long non-coding RNAs in biomarking COVID-19: a machine learning-based approach
Virology Journal (2024)